Abstract
We present an integrated analysis of urine and serum proteomics and clinical measurements in asymptomatic, mild/moderate, severe and convalescent cases of COVID-19. We identify the pattern of immune response during COVID-19 infection. The immune response is activated in asymptomatic infection, but is dysregulated in mild and severe COVID-19 patients. Our data suggest that the turning point depends on the function of myeloid cells and neutrophils. In addition, immune defects persist into the recovery stage, until 12 months after diagnosis. Moreover, disorders of cholesterol metabolism span the entire progression of the disease, starting from asymptomatic infection and lasting to recovery. Our data suggest that prolonged dysregulation of the immune response and cholesterol metabolism might be the pivotal causative agent of other potential sequelae. Our study provides a comprehensive understanding of COVID-19 immunopathogenesis, which is instructive for the development of early intervention strategies to ameliorate complex disease sequelae.
Keywords: COVID-19, Mass spectrometry, Proteomics, Immune response
1. Introduction
The outbreak of coronavirus disease 2019 (COVID-19) since late December 2019 has brought significant harm and major challenges to over 200 countries and regions world-wide. COVID-19 is caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), which infects the lungs, leading to fever, cough, and dyspnea, etc. [1]. Most patients present with mild disease symptoms but some go on to develop acute respiratory distress syndrome and multi-organ dysfunction, which may lead to death [2]. The whole COVID-19 pandemic has resulted in more than 145 million confirmed infections with more than 3 million deaths and has been documented as a public health emergency of international concern.
Since the outbreak of COVID-19, research has moved rapidly to develop diagnostic kits and test candidate vaccines [3]. Unfortunately, key questions about the pathogenic mechanisms underlying COVID-19 have not been satisfactorily answered. This information is crucial for the design of rational therapeutic strategies. Inflammation and immune activation are expected to accompany a viral infection, and these processes have been extensively studied in SARS-CoV-2 infection. It has been suggested that COVID-19 is characterized by lymphopenia and increased numbers of neutrophils [[4], [5], [6]]. A growing body of evidence indicates that most patients with severe COVID-19 have high levels of proinflammatory cytokines including interleukin-6 (IL-6) [[7], [8], [9]]. More detailed analysis using single-cell transcriptomics of peripheral-blood mononuclear cells (PBMCs) accompanied by targeted serum proteomics showed that severe COVID-19 is marked by dysregulation of blood myeloid cells, including accumulation of immature neutrophils with an immunosuppressive profile, and repressed monocyte activation, accompanied by release of massive amounts of calprotectin (S100A8/S100A9) [[10], [11], [12], [13], [14]]. Multiple untargeted quantitative proteomic studies of serum or urine samples showed that COVID-19 patients have significant immune dysregulation [[15], [16], [17], [18], [19], [20]]. However, current studies are mostly focused on the shift between mild/moderate and severe COVID-19, and therefore lack a panoramic view of the pathogenicity of the whole disease process.
An important feature of COVID-19 is the existence of asymptomatic carriers who are able to transmit the virus to others. These asymptomatic carriers make the diagnosis, control and prevention of COVID-19 even more complicated. Asymptomatic infections are reported to be more common in populations of young and middle-aged individuals with functional performance status and no underlying diseases [21]. However, the pathogenesis of asymptomatic infection is not clear. In addition, it was not until recently that COVID-19 survivors were reported to be troubled with fatigue or muscle weakness, sleep difficulties, and increased neurological and psychiatric morbidity in the 6 months after COVID-19 infection. More strikingly, patients discharged from hospital after COVID-19 had increased rates of multiorgan dysfunction [[22], [23], [24]]. How these sequelae are related to disease pathogenesis is completely unknown, which makes it impossible to design early and rational interventions to prevent long-term symptoms in survivors. Thus, urgent research is needed to establish the risk factors for sequelae.
It remains unclear to what extent immune responses are causative or exacerbated factors to the disease and could be used for accurate patient stratification, as in asymptomatic, mild/moderate, severe and convalescent cases. Alterations of proteins in human body fluids are well recognized as direct indicators of pathophysiological changes caused by diseases. In this study we profiled the protein level of urine samples from asymptomatic carriers and of serum sample from recovered patients for 6 or 12 months through the data-independent acquisition (DIA) proteomics approach. We integrated the previously acquired proteomics data from urine of mild and severe COVID-19 patients and from serum of recovered patients for less than 1 month to get a whole picture of the disease progress on molecular level. We found that distinct changes in the levels of proteins related to immune function marked the different clinical trajectories of the disease. We also identified prolonged changes in proteins related to cholesterol transport, myocardial and coagulation disorders, which provides hints to explain the fatigue or neurological outcomes in convalescing patients. More importantly, the differentially expressed proteins identified and validated among multiple groups could contribute to diagnosis or prognosis and might be potential therapeutic targets.
2. Results
2.1. Study design and patient cohorts
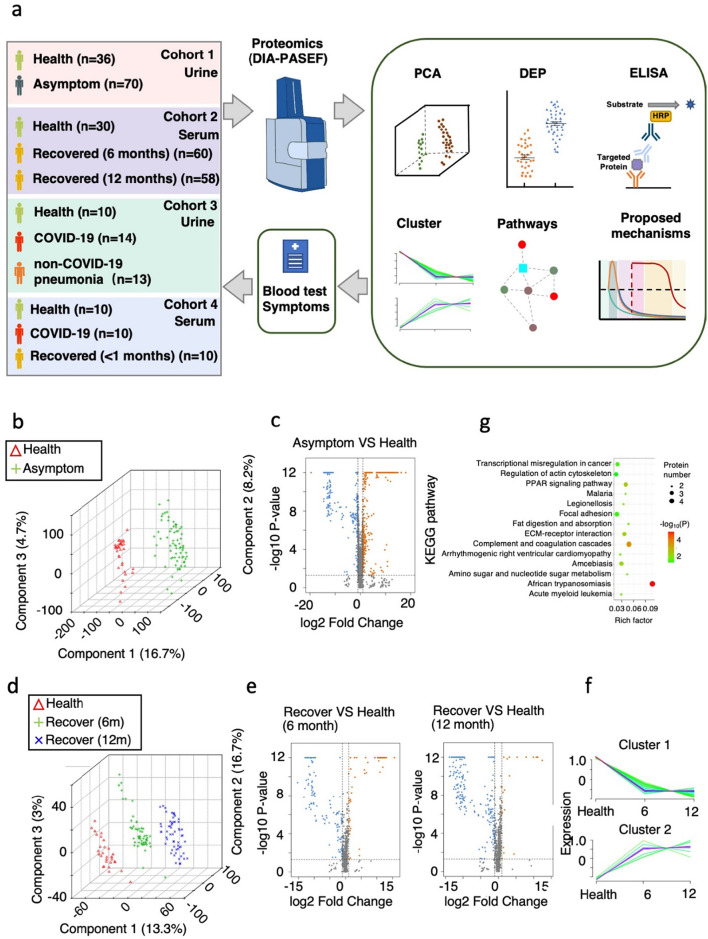
We aimed to obtain comprehensive understanding of the entire COVID-19 disease process from acute to convalescent phase including asymptomatic, mild/moderate, severe cases. We collected urine samples from 70 asymptomatic carriers and 36 healthy controls (cohort 1) (Table S1, Fig. 1a). Asymptomatic carriers refer to people with positive detection of SARS-CoV-2 nucleic acid by reverse transcriptase-polymerase chain reaction (RT-PCR), but with no typical clinical symptoms or signs, and no apparent abnormalities. We also collected serum samples from 30 healthy controls, 60 patients (7 severe) recovered from COVID-19 for 6 months and 58 (7 severe) patients recovered from COVID-19 for 12 months (cohort 2) (Table S1, Fig. 1a). For the recovered patients, serum samples were taken at 6 or 12 months after diagnosis. Serum samples were first subjected to depletion of high-abundance serum proteins. Total proteins from the low-abundance fraction of serum samples or from urine samples were extracted, denatured, and digested into peptides by trypsin for data-independent acquisition (DIA) mass spectrometry. The resulting quantitative proteomic data were subsequently analyzed for differentially expressed proteins (DEPs) and enriched pathways and further validated by ELISA (Fig. 1a).
Fig. 1.
Summary of COVID-19 patient samples and experimental design.
a. Experimental design for the quantitative proteomic analysis in this study. In cohort 1, a total of 106 urine samples were analyzed from 2 groups: healthy controls, n = 36; asymptomatic carriers, n = 70. In cohort 2, a total of 148 serum samples were taken from 3 groups: healthy controls, n = 30; convalescent patients (7 severe cases) recovered for 6 months, n = 60; convalescent patients (7 severe cases) recovered for 12 months, n = 58. The cohort 3 and 4 are data from other studies. All differentially expressed proteins (DEPs) selected for further analysis meet the criteria that fold change >2 or < 0.5, two-tailed t-test; p < 0.05).
b. Principal components analysis (PCA) showing the inter-group differences in cohort 1. Individuals in the healthy control group and the asymptomatic carrier group are indicated by coloured symbols in the figure.
c. Volcano plot of identified urine DEPs comparing asymptomatic carriers with the healthy control group in cohort 1 (two-tailed t-test; p < 0.05, fold change >2 or < 0.5).
d. Principal components analysis (PCA) showing the inter-group differences in cohort 2. Individuals in the healthy control group, and patients recovered for 6 or 12 months are indicated by coloured symbols in the figure.
e. Volcano plot of identified serum DEPs comparing patients recovered for 6 or 12 months with the healthy control group (cohort 2).
f. Hierarchical clustering shows 2 distinct groups differentiated according to similarity.
g. KEGG-based enrichment analysis of DEPs in the subclusters shown in f. KEGG terms were sorted by P value.
2.2. Quantitative proteomic profiling of urine and serum samples from asymptomatic carriers and convalescent patients
The raw data were firstly processed using a double boundary Bayes (DBB) imputation method and were standardized for further analysis. The data showed clear stratification of the different patient/control groups according to principal components analysis (PCA) (Fig. 1b, d). A total of 4930 and 1311 proteins were identified across all urine samples and across all serum samples, respectively. Volcano plot showed identified proteins that were significantly increased or decreased in asymptomatic carriers or recovered patients compared to healthy controls (Fig. 1c, e). The DEPs are mainly related to infection and metabolisms.
It is surprising that there are significantly more down-regulated proteins than up-regulated proteins in recovered patients, both at 6 months and 12 months (Fig. 1e). Subclustering analysis was performed on the significantly changed proteins to analyze their variation at healthy controls, and recovered patients for 6 or 12 months. Differentially expressed proteins (DEPs) were subjected to 2 unique subclusters, which showed long lasting changes (Fig. 1f). The enriched pathways of these proteins are related to infection and immune responses, in addition to complement and coagulation cascades and arrhythmogenic right ventricular cardiomyopathy (Fig. 1g). This long-lasting cardiomyopathy is in consistent with previous observation on COVID-19 patients recovered within 2 months [25].
We found that many proteins involved in viral latency, viral genome replication, viral entry into host cells and lysosome functions are increased in urine samples from asymptomatic carriers (Fig. S1a, b). This suggests an infection stress response as expected. In asymptomatic carriers, many proteins involved in tight junction organization and tight junction assembly are increased except for FZD5 (Fig. S1c). We previous observed that tight junction proteins are significantly decreased in urine samples of symptomatic COVID-19 patients [20]. This difference in tight junction protein levels between asymptomatic carriers and symptomatic COVID-19 patients might indicate a defense process at the very early stage of infection.
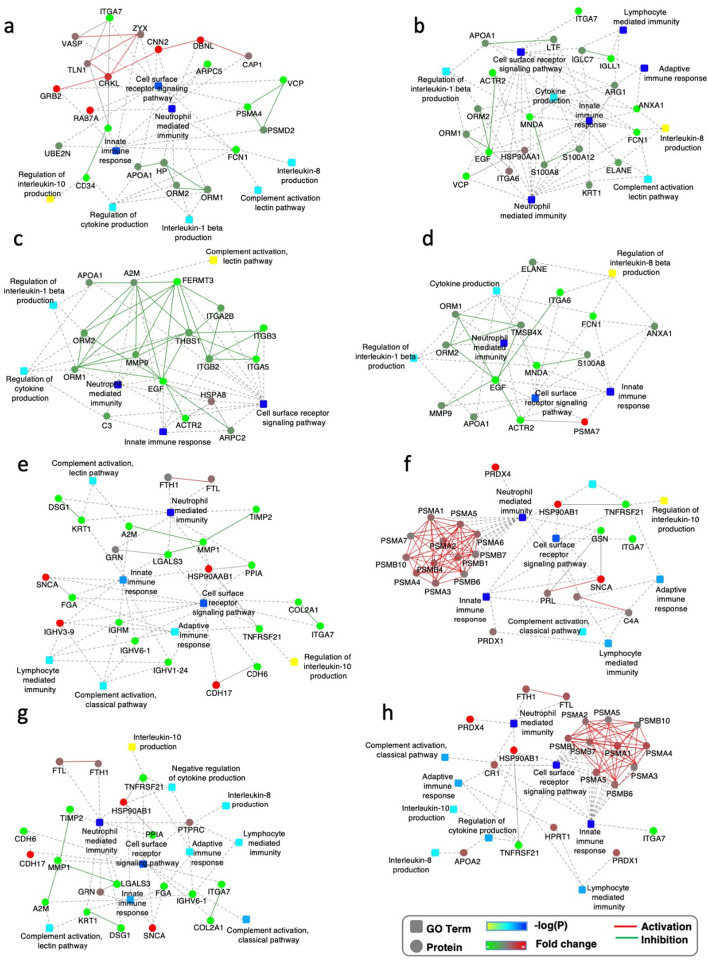
Most interestingly, we found that DEPs in asymptomatic carriers are highly enriched in immune response functions. Many proteins involved in the innate immune response, leukocyte-mediated immunity, neutrophil-mediated immunity and cytokine production were increased in urine samples from asymptomatic carriers (Fig. 2a). The individual scatter plots of differentially expressed proteins which are identified in all analyzed samples are shown in Supplementary Fig. 2.
Fig. 2.
Immune response transition from activation to suppression in asymptomatic carriers and symptomatic patients of COVID-19.
a. Interaction diagrams of the indicated pathways when urine proteins from asymptomatic carriers are compared to those from healthy controls (cohort 1). The significance of the pathways, represented by -log(P-Value) (Fisher's exact test), is shown by the color scale, with dark blue representing the highest significance. Color bar from orange to green represents the fold change of protein level from increasing to decreasing. Fold change indicates the protein level in asymptomatic carriers compared to healthy controls.
b. c. Fold change of urine DEPs related to the immune related pathways are shown comparing asymptomatic carriers to healthy controls (cohort 1) and comparing COVID-19 patients to healthy controls (cohort 3) (b); or comparing asymptomatic carriers to healthy controls (cohort 1) and comparing non-COVID patients to healthy controls (cohort 3) (c). X-axis is individual proteins represented by different bar colours. Only proteins of fold-change >2 or < 0.5, P < 0.05 (t-test) were selected.
d. Interaction diagrams of indicated pathways enriched by proteins with opposite expressing pattern as in asymptomatic carriers (left panel) and COVID-19 patients (right panel) for urine samples.
We calculated the fold change of the DEPs involved in the immune response in asymptomatic carriers, in COVID-19 patients and in non-COVID-19 pneumonia patients (Cohort 3 in Fig. 1a) to their respective health controls, the proteomics data of whom we reported before [20]. We compared the fold change of DEPs in asymptomatic carriers and in COVID-19 patients or fold change of DEPs in COVID-19 patients and non-COVID-19 pneumonia. Interestingly, the pattern in asymptomatic carriers is similar to non-COVID-19 pneumonia patients, but are sharply different to COVID-19 patients. Proteins that are elevated in urine from asymptomatic carriers are mostly decreased in COVID-19 patients (Fig. 2b, c). The biological functions of these proteins were related to neutrophil- and myeloid cell- mediated immunity and cellular response to cytokine stimulus (Fig. 2d). Specifically, calpain (CAPN1) is implicated in the expansion of the neutrophil plasma membrane that accompanies cell spreading and phagocytosis [26]. S100A12 (EN-RAGE) is mainly expressed and secreted by neutrophil granulocytes and is the ligand for the cell surface receptor RAGE (receptor for advanced glycation end products) and TLR4 (Toll-like receptor 4) on macrophages, endothelium, and lymphocytes [27] (Fig. S2). Thus, reduced neutrophil and myeloid cell functions might be the SARS-CoV-2-specific risk factors which determine the switch from asymptomatic infection to symptomatic COVID-19. This is partially consistent with other studies showing that altered neutrophil function may be a potential driver of COVID-19 and a predictor of poor outcome [[28], [29], [30], [31]]. Neutrophilia was reported in COVID-19 patients [7]. Although leukocytosis and neutrophilia are hallmarks of acute infection, in the case of COVID-19, we propose that neutrophilia is a sign of neutrophil anergy in SARS-CoV-2 infection.
2.3. Persistent immune suppression in convalescent patients
Reports of long-term consequences of COVID-19 in adult patients after discharge from hospital have raised serious concerns. In our study, more than 50% of discharged patients reported feeling unwell at 12 months (Supplementary Table 2). There is no molecular clue to how these sequelae occur, which makes diagnosis and intervention very difficult. Altered immune responses seem to play a vital role in the clinical trajectories of COVID-19. This prompted us to explore the mechanism, from the perspective of the immune response, underlying the sequelae in mild/moderate or severe COVID-19 patients after 6 or 12 months of recovery. To our surprise, immune pathways such as the innate immune response, the adaptive immune response, neutrophil-mediated immunity, lymphocyte-mediated immunity, regulation of cytokine production and complement activation are significantly suppressed in patients recovering from both mild/moderate and severe COVID-19, especially at the 12-month time point (Fig. 3a, b, c, d). It is worth mentioning that some proteins related to the cell surface receptor signalling pathway are up-regulated at 6 months, but this pathway is down-regulated at 12 months (Fig. 3a, c).
Fig. 3.
Persistent immune suppression in convalescent patients.
a, b. Interaction diagram of the indicated pathways when serum proteins from mild/moderate COVID-19 patients after 6 months recovery (a) or 12 months recovery (b) are compared to healthy controls (cohort 2).
c, d. Interaction diagram of the indicated pathways when serum proteins from severe COVID-19 patients after 6 months recovery (c) or 12 months recovery (d) are compared to healthy controls (cohort 2).
e, f. Interaction diagrams of the indicated pathways when serum proteins from mild/moderate patients (e) or severe patients (f) are compared to healthy controls (cohort 4).
g, h. Interaction diagrams of the indicated pathways when serum proteins from convalescent patients after about 10 days recovery from mild/moderate (g) or severe (h) COVID-19 are compared to healthy controls (cohort 4). The significance of the pathways represented by -log(P-Value) (Fisher's exact test) is shown by the color scale, with dark blue representing the highest significance. Color bar from orange to green represents the fold change of protein level from increasing to decreasing. Fold change indicates the protein levels in patients compared to healthy controls.
In another study by our group, we carried out quantitative proteomic analysis of serum samples from patients diagnosed with mild/moderate or severe COVID-19 at 2 distinct time points, shortly after diagnosis with COVID-19 and before discharge from hospital (cohort 4 in Fig. 1a) [32]. The immune response of mild/moderate patients is largely repressed at both time points (Fig. 3e, g). However, severe patients showed signs of an activated immune response (Fig. 3f, h). Up-regulated proteins include subunits of the proteasome system for protein degradation and antigen presentation. These results are consistent with previous proteomic studies on urine samples from mild/moderate and severe patients [20]. The increased level of α-Synuclein (SNCA) might also be informative. SNCA has been well studied for its neuropathological roles, and extracellular α-synuclein also triggers immune cell activation, proliferation, secretion of cytokines and other immune mediators, and phagocytosis [[33], [34], [35], [36]]. Interestingly, there is difference between the upregulated immune response proteins in severe patients and in asymptomatic carriers. It is still not clear, however, how these molecules determine the switch from asymptomatic infection to mild/moderate COVID-19 or from mild/moderate to severe status.
2.4. The persistent alteration cholesterol metabolism, blood coagulation and cardiomyopathy in convalescent patients
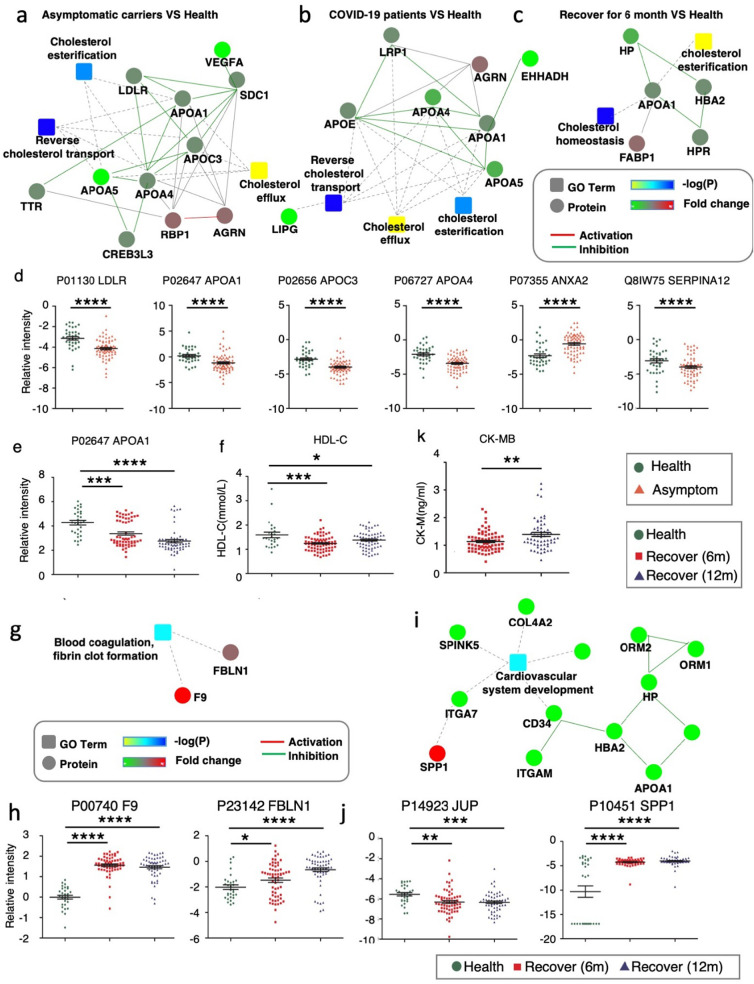
We and others have shown that cholesterol metabolism is directly related to the pathogenesis and severity of COVID-19, and an increased serum total cholesterol level seems to be specific for COVID-19 [32,37]. Interestingly, we found that cholesterol metabolism pathways all showed some degree of alteration with Apolipoprotein A-I (ApoA1) down regulated in asymptomatic patients, COVID-19 patients, and convalescent patients (Fig. 4a-e) [20,32]. ApoA1 is a major component of high-density lipoproteins (HDL), which are important for reverse cholesterol transport. The disturbance of serum HDL may suggest a defect in cholesterol reverse transport and cholesterol functions. We found that HDL-C levels were reduced at both 6 and 12 months of recovery (Fig. 4f), which further supports the notion that cholesterol transport is disrupted along the entire progression of COVID-19, including recovery. HDL also has major pleiotropic functions that could play a pivotal role in acute inflammatory conditions. Evidence suggests that HDLs are globally protective for the endothelial layer. One study reported that HDLs from COVID-19 patients were less protective for endothelial cells stimulated with TNFα than HDLs from healthy subjects [37]. Thus, we hypothesize that disrupted HDL involved cholesterol transport, which occur at the earliest stage of infection and persist throughout the entire period, might be an early inducing factor as well as a long-lasting adverse factor which determines disease outcome.
Fig. 4.
The immune system dysregulations are linked with cholesterol metabolism, blood coagulation and cardiomyopathy.
a. Interaction diagrams of the indicated pathways when urine proteins from asymptomatic carriers are compared to healthy controls (cohort 1).
b. Interaction diagrams of the indicated pathways when urine proteins from COVID-19 patients are compared to healthy controls (cohort 3).
c. Interaction diagrams of the indicated pathways when serum proteins from COVID-19 patients after recovery for 6 months are compared to healthy controls (cohort 2).
d, e. Scatter plot graphs showing DEPs from a-c. One-way ANOVA was used to analyze the data. Data are presented as mean ± SEM. * P-value <0.05, ** P-value <0.01, *** P-value <0.001, **** P-value <0.0001, t-test.
f. HDL-C levels in convalescent COVID-19 patients after recovery for 6 or 12 months (cohort 2). Data are presented as mean ± SEM. * P-value <0.05, ** P-value <0.01, *** P-value <0.001, **** P-value <0.0001, t-test.
g, i. Interaction diagrams of blood coagulation, fibrin clot formation (g) or cardiovascular system development (i) when serum proteins from convalescent COVID-19 patients after recovery for 6 months are compared to healthy controls (cohort 2).
h, j. Scatter plot graphs showing DEPs from g and i. One-way ANOVA was used to analyze the data. Data are presented as mean ± SEM. * P-value <0.05, ** P-value <0.01, *** P-value <0.001, **** P-value <0.0001, t-test.
k. Quantitative level of Creatine Kinase, MB Form in convalescent COVID-19 patients after 6 or 12 months of recovery.
It has been reported by multiple groups that blood coagulation is disturbed in COVID-19 patients [38]. We found that levels of the coagulation factors IX and FBLN1 were significantly increased at both 6 and 12 months of recovery (Fig. 4g, h). Blood coagulation and inflammation are universal responses to infection and there is close crosstalk between them. For instance, inflammatory cytokines and leukocyte elastase can down-regulate natural anticoagulant proteins that help to maintain endothelial-cell integrity and control clotting [39]. Thus, immune suppression might contribute to the tendency for clot formation during recovery.
Several studies have reported major cardiac complications in COVID-19 patients including COVID-19-related myocarditis [[40], [41], [42]]. Moreover, recovered patients also present impaired cardiac function independent of preexisting conditions, which indicates that COVID-19 results in long-term heart sequelae. We found DEPs enriched in cardiovascular system development at 6 and 12 months of recovery (Fig. 4i, j). Specifically, the level of junction plakoglobin (JUP) is significantly reduced. JUP is a binding partner for major proteins in desmosomes, which are junction structures important in cardiomyopathy. Evidence shows that impaired desmosome assembly reduces incorporation of JUP into the cell membrane in arrhythmogenic right ventricular cardiomyopathy (ARVC) [43]. The reduced level of JUP detected in serum might be a sign of disturbed JUP localization and junction structure disorders. OPN is an extracellular matrix protein that takes a role in increase of macrophage and T cells on inflammation areas [44]. Moreover, OPN increased the risk of atherosclerosis by leading vascular inflammation through increasing macrophage activation and release of other cytokines [45]. More evidence of cardiomyopathy comes from that the level of creatine kinase, MB form (CK-MB) increased significantly from 6 months to 12 months of convalescence (Fig. 4k) compared to healthy controls. This further raised awareness that ongoing investigation of the long-term cardiovascular consequences of COVID-19 is urgently needed. Among all identified DEPs of multiple functions, the Apolipoprotein A-I (APOA1), Osteopontin (SPP1), Haptoglobin (HP), Alpha-1-antitrypsin (SERPINA1), Neutrophil elastase (ELANE) level in recovered patients was validated by Elisa, which are proteins important in cholesterol transport, cardiovascular system and immune responses (Fig. S3).
There is evidence showing that immune responses are critical components of the host reaction to myocardial injury and subsequent remodeling [46]. The persistent immune dysregulation caused by SARS-CoV-2 infection prompt us to speculate that this might be an important causative agent of long-term myocardial disorders. Interestingly, we did not detect cardiovascular-related pathway disorders in asymptomatic infection (data not shown), possibly because the earliest infection has not yet exerted profound influence.
3. Discussion
Increasing evidence suggests that the heterogeneity of clinical manifestations and the complexity of host responses of COVID-19 require detailed analyses using high-resolution techniques and well- characterized clinical cohorts. Here, we profiled the serum proteome from 118 convalescent COVID-19 patients and the urine proteome from 70 asymptomatic COVID-19 patients using the DIA proteomics approach. This is the first MS-based body fluid proteomics study of asymptomatic infection and convalescent COVID-19 patients at 6- and 12- months after diagnosis. With a total of 106 urine samples and 148 serum samples, this is one of the largest untargeted proteomic studies on COVID-19.
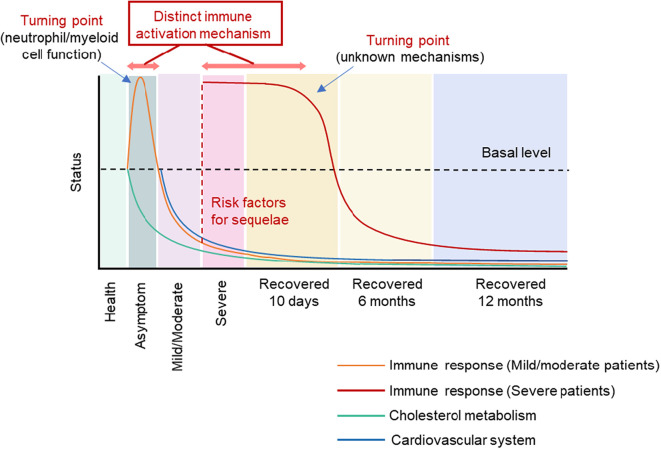
When we integrated the proteomics analysis of asymptomatic, mild/moderate, severe and convalescent cases, our main finding is that immune responses are activated in asymptomatic infection, suppressed in mild/moderate COVID-19 patients, and abnormally activated in severe COVID-19 patients (Fig. 5 ). The turning point might be defective function of myeloid cells and neutrophils, since proteins related to these functions were identified to switch from increasing to decreasing (Fig. 2d). Interestingly, the immune response activation in severe patients involves the proteasome system, which contrasts with the immune response activation seen in asymptomatic infection. Surprisingly, both mild/moderate and severe patients show prolonged immune suppression at 6 and 12 months of recovery. It is interesting to explore how the distinct immune patterns determine the disease trajectory. Tight junctions are partially activated in asymptomatic infection but are disrupted in COVID-19 patients, providing another line of evidence for disease progression. Cholesterol metabolism was disrupted throughout the entire progress from asymptomatic infection to recovery. Cardiovascular function and blood coagulation pathway was not significantly affected in asymptomatic infection, but was disrupted during mild/moderate/severe COVID-19 and recovery. It is worth mentioning that the results of blood tests (routine cell counts and biochemistry) appeared normal in these convalescent patients (Supplementary Table 2). Thus, the molecular alteration was more sensitive than blood tests for prediction of dysfunction. From the timeline of each observed disorders and the known inter-relationships of these processes, we speculate that immune responses and cholesterol metabolism might be the risk factors and pivotal causative agents of other molecular disorders, especially in the recovery stage, which are manifested as complex clinical symptoms. It remains to be explored why disorders of the immune system and cholesterol metabolism persist even when the virus is not detectable in recovering patients.
Fig. 5.
Schematic overview of the molecular pattern during multiple disease stages. Molecular pattern for immune response, cardiovascular system and cholesterol metabolism in asymptomatic infection, COVID-19 disease, and COVID-19 recovery for 10 days, 6 months and 12 months.
The limitations of this study lie in the possibility that changes in serum or urine protein levels partially but not completely represents the activation or suppression of the pathways that the proteins function in. More rigorous mechanistic studies in cellular or mouse models are required to trace the origin of these proteins, how they are released, and how they function in disease conditions. Single cell multi-omics studies for cell lineage tracing of samples from convalescent patients should be integrated with unbiased proteomics to dissect the relationship between disease pathogenicity and host cell response. Isolating neutrophils or monocytes from convalescent patients and testing their functions in vitro may also provide insight into specific immune mechanism. In addition, more personalized study requires molecular mapping of longitudinal samples from one patient across the disease progression.
In conclusion, our work gives a whole view of disease progression at the proteomic level and finds that distinct immune response patterns and altered cholesterol metabolism constitute the central hub for clinical trajectories of the disease and risk factors for long-term sequelae. Our study provides a valuable proteomics resource at multiple disease stages to comprehensively understand COVID-19-associated host responses. Our results shed light on the pathogenesis of COVID-19 and, more importantly, provide insight into the urgently needed mechanism of sequelae and early intervention strategies.
4. Materials and methods
4.1. Sample collection
Urine samples were collected based on the protocol for routine mid-stream urine in the morning. Serum samples were collected in pro-coagulation vacuum tubes using standard venepuncture protocols. Serum was extracted by centrifugation for 10 min at 3000 rpm. The urine and serum samples were inactivated at 65 °C for 1 h and subsequently stored at −80 °C before use. The study was approved by the Ethics Committee of the National Institution for Viral Disease Control and Prevention, China CDC (Ethical approval No.IVDC2020-021). Written informed consent was obtained from all participants.
4.2. Depletion of highly abundant proteins from serum samples
Abundant protein depletion was performed on the Agilent 1290 Infinity II liquid chromatography system coupled with the Multi Affinity Removal Column, Human-14. Protein concentration was measured using a BCA Protein Assay Kit (Thermo Scientific).
4.3. Serum and urine sample preparation for DIA analysis
Urine samples were processed as described previously (Tian et al., 2020). Firstly, urine proteins were precipitated by trichloroacetic (TCA) acid solution at 4 °C for 4 h, reduced by 20 mM (2-carboxyethyl) phosphine hydrochloride (TCEP) and alkylated with 40 mM iodoacetamide. The mixture was digested with 3 μg trypsin protease overnight. After desalting, 1/3 lysate was used to measure peptide concentration while the remaining 2/3 was dissolved in Milli-Q water with 0.1% formic acid (FA) for mass spectrometry analysis. Serum proteins were also precipitated by TCA solution, reduced by 20 mM TCEP and alkylated with 40 mM iodoacetamide. The mixture was digested with trypsin protease at a 1/100 (w/w) trypsin protease to protein ratio, at 37 °C overnight. The lysate was desalted using a Monospin C18 column (GL Science, Tokyo, Japan) and vacuum-centrifuged to dryness afterwards. The dried peptides were reconstituted before DIA analysis.
4.4. Sample preparation for spectral library generation
Peptides from each sample were mixed to obtain 100 μg purified mixture and then vacuum-centrifuged to dryness. The dried peptides were reconstituted in Milli-Q water with 2% acetonitrile (ACN) for fractionation.
4.5. High-pH reversed-phase fractionation
Peptides were fractionated at pH 10 on a chromatographic column (BEH C18, 300 Å, 1.7 μm, 1 mm × 150 mm) coupled to a Waters XevoTM ACQUITY UPLC (Waters, USA) as described previously (Tian et al., 2020). Approximately 100 μg purified peptides were separated into 62 fractions by collecting the fluid every 60 s. The fractions were mixed in pairs to give 31 samples and then vacuum-centrifuged to dryness. Dried peptides were dissolved in 10 μL Milli-Q water with 0.1% formic acid (FA) and then spiked with iRT peptides for chromatographic correction.
4.6. Liquid chromatography
Nanoflow reversed-phase chromatography was performed on a nanoElute liquid chromatography system (Bruker Daltonics). Peptides were separated with a non-linear gradient starting with 2% acetonitrile in 0.1% formic acid and increasing to 22% acetonitrile in 0.1% formic acid, then increasing to 37% within 8 min, then further increasing to 100% within 5 min, and finally maintained at 100% for 7 min before re-equilibration. A homemade column (25 cm × 75 μm, 1.5 μm C18-AQ particles) was used.
Mobile phases A and B were water and ACN with 0.1% formic acid respectively. Separation was performed at a flow rate of 300 nL/min.
4.7. Mass spectrometry
LC was coupled online to a timsTOF Pro (Bruker) via a CaptiveSpray nano-electrospray ion source. The fraction samples were analyzed in data-dependent mode, setting mass spectra in the range from m/z 100–1700 in positive electrospray mode for generation of the spectral library. The other analysis was in data-independent mode, consisting of an MS1 scan from m/z 400 to m/z 1200 and 64 MS2 windows in a diaPASEF acquisition scheme covering the mass range from m/z 400 to m/z 1200.
The ion mobility was scanned from 0.6 to 1.6 Vs/cm2. The collision energy was ramped linearly as a function of the mobility from 59 eV at 1/K0 = 1.6 Vs/cm2 to 20 eV at 1/K0 = 0.6 Vs/cm2.
4.8. Generation of spectral libraries and DIA data analysis
Spectral libraries were generated with Spectronaut version 14.2 (Biognosys) against a UNIPROT human database (only reviewed entries). All the parameters were default. DIA files were processed in a default mode, except that correction factor of XIC IM extraction window was set to 0.8.
4.9. Enzyme-linked immunosorbent assay (ELISA)
Human protein ELISA kits were used to quantify serum levels of endogenous protein according to manufacturers' instructions (R and D system). Briefly, serum samples were diluted according to the manufacturers' dilution guideline. Serum were diluted at 1: 20000 for detection of APOA1 and Haptoglobin, 1:10 or 1:100 for ELA2, 1:10 for Serpin A1and 1: 25 for OPN. Then, a total of 100 μL of fixed dilution plasma sample were added to the precoated plates, and the plates were incubated at 37 °C for 2 h. After washing, 100 μL Conjugate-specific antibody was added to each well, and the plates were incubated at 37 °C for 1 h. Followed by washing, 100 μL Substrate Solution was added and incubated at 37 °C for 1 h. Finally, the OD value at 450 nm were determined after addition of 50 μL Tetramethyl-benzidine (TMB) reagent and stop solution. The standard curve of each protein was generated by determination of OD values from serially dilutions of the standard samples with known protein concentrations provided by the manufacturers.
4.10. Statistical analysis
Omicsbean software was used for data analysis including data imputation, normalization, and principal component analysis (PCA). Fold-change of 2 and fold-change<0.5 and P-value (t-test) of 0.05 were used to filter differentially expression proteins. Mfuzz was used to detect different sub-clustering models of gene expression among groups. Network visualization was performed using the ggplot228 packages and Cytoscape v.3.5.129 implemented in the Omicsbean workbench. Ingenuity Pathway Analysis (IPA) was performed to explore the downstream effects in the dataset of significantly up- or down-regulated proteins.
Data availability
The experimental data that support the findings of this study have been deposited in iProX (integrated proteome resources) of ProteomeXchange with the accession code PXD026442.
Author contributions
Y.C., C.C.W., G.F.G. and W.J.L. conceived the project. J.Z., S.D., H.S., M.L., X.L., Y.G., B.Y., Y.Z., T.Y., J.Z. and Y.J. collected clinical samples and clinical test data. C.C.W. supervised the mass spectrometry proteomics experiments. N.Z., S.G., T.Z. and J.G. performed the experiments. Y.C., C.C.W., G.F.G. and W.J.L. analyzed the data and wrote the manuscript.
Declaration of Competing Interest
The authors declare no competing interests.
Acknowledgments
The authors thank Bruker Daltonics Inc. for their support in proteomic analysis. The authors are grateful to OmicsBean (Gene For Health Inc.) for their assistance in data analysis. The authors thank Xuezhao Feng (Xinjiang Medical University, Urumqi, China) for helping sample collection. This work is supported by the Big Science Strategy Plan 2020YFE0202200, the Ministry of Science and Technology of the People's Republic of China 2018YFA0507102 and National Natural Science Foundation of China 91754108 and to Y.C.; and the Fundamental Research Funds for the Central Universities (BMU2017YJ003, BMU2018XTZ002), Research Funds from the Health@InnoHK Program launched by Innovation Technology Commission of the Hong Kong Special Administrative Region, and the PKU-Baidu Fund 2019BD007. W.J.L. is supported by the Excellent Young Scientist Program of the National Natural Science Foundation of China (81822040).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bbapap.2021.140736.
Appendix A. Supplementary data
Supplementary material 1
Supplementary material 2
Supplementary material 3
References
- 1.Guan W.J., et al. vol. 382. 2020. Clinical Characteristics of Coronavirus Disease 2019 in China; pp. 1708–1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wu Z., McGoogan J.M. Characteristics of and important lessons from the coronavirus disease 2019 (COVID-19) outbreak in China: summary of a report of 72314 cases from the Chinese Center for Disease Control and Prevention. JAMA. 2020;323:1239–1242. doi: 10.1001/jama.2020.2648. [DOI] [PubMed] [Google Scholar]
- 3.Liu W.J., Wu G. Convincing the confidence to conquer COVID-19: from epidemiological intervention to laboratory investigation. Biosaf. Health. 2020;2:185–186. doi: 10.1016/j.bsheal.2020.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wu C., et al. Risk factors associated with acute respiratory distress syndrome and death in patients with coronavirus disease 2019 pneumonia in Wuhan, China. JAMA Intern. Med. 2020;180:934–943. doi: 10.1001/jamainternmed.2020.0994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kuri-Cervantes L., et al. Comprehensive mapping of immune perturbations associated with severe COVID-19. Sci. Immunol. 2020;5 doi: 10.1126/sciimmunol.abd7114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tan L., et al. Lymphopenia predicts disease severity of COVID-19: a descriptive and predictive study. Signal. Transduct. Target Ther. 2020;5:33. doi: 10.1038/s41392-020-0148-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Huang C., et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhu L., et al. Single-cell sequencing of peripheral mononuclear cells reveals distinct immune response landscapes of COVID-19 and influenza patients. Immunity. 2020;53 doi: 10.1016/j.immuni.2020.07.009. 685–696 e683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang P., et al. Downregulated miR-451a as a feature of the plasma cfRNA landscape reveals regulatory networks of IL-6/IL-6R-associated cytokine storms in COVID-19 patients. Cell. Mol. Immunol. 2021;18:1064–1066. doi: 10.1038/s41423-021-00652-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Arunachalam P.S., et al. Systems biological assessment of immunity to mild versus severe COVID-19 infection in humans. Science. 2020;369:1210–1220. doi: 10.1126/science.abc6261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Silvin A., et al. Elevated calprotectin and abnormal myeloid cell subsets discriminate severe from mild COVID-19. Cell. 2020;182 doi: 10.1016/j.cell.2020.08.002. 1401–1418 e1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schulte-Schrepping J., et al. Severe COVID-19 is marked by a dysregulated myeloid cell compartment. Cell. 2020;182 doi: 10.1016/j.cell.2020.08.001. 1419–1440 e1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Su Y., et al. Multi-omics resolves a sharp disease-state shift between mild and moderate COVID-19. Cell. 2020;183 doi: 10.1016/j.cell.2020.10.037. 1479–1495 e1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wilk A.J., et al. A single-cell atlas of the peripheral immune response in patients with severe COVID-19. Nat. Med. 2020;26:1070–1076. doi: 10.1038/s41591-020-0944-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Di B., et al. Identification and validation of predictive factors for progression to severe COVID-19 pneumonia by proteomics. Signal. Transduct. Target Ther. 2020;5:217. doi: 10.1038/s41392-020-00333-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hou X., et al. Serum protein profiling reveals a landscape of inflammation and immune signaling in early-stage COVID-19 infection. Mol. Cell. Proteomics. 2020;19:1749–1759. doi: 10.1074/mcp.RP120.002128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rieder M., et al. Serum protein profiling reveals a specific upregulation of the immunomodulatory protein Progranulin in coronavirus disease 2019. J. Infect. Dis. 2021;223:775–784. doi: 10.1093/infdis/jiaa741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shen B., et al. Proteomic and metabolomic characterization of COVID-19 patient sera. Cell. 2020;182 doi: 10.1016/j.cell.2020.05.032. 59–72 e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shu T., et al. Plasma proteomics identify biomarkers and pathogenesis of COVID-19. Immunity. 2020;53 doi: 10.1016/j.immuni.2020.10.008. 1108–1122 e1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tian W., et al. Immune suppression in the early stage of COVID-19 disease. Nat. Commun. 2020;11:5859. doi: 10.1038/s41467-020-19706-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gao Z., et al. A systematic review of asymptomatic infections with COVID-19. J. Microbiol. Immunol. Infect. 2021;54:12–16. doi: 10.1016/j.jmii.2020.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ayoubkhani D., et al. Post-covid syndrome in individuals admitted to hospital with covid-19: retrospective cohort study. BMJ. 2021;372 doi: 10.1136/bmj.n693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huang C., et al. 6-month consequences of COVID-19 in patients discharged from hospital: a cohort study. Lancet. 2021;397:220–232. doi: 10.1016/S0140-6736(20)32656-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Taquet M., Geddes J.R., Husain M., Luciano S., Harrison P.J. 6-month neurological and psychiatric outcomes in 236 379 survivors of COVID-19: a retrospective cohort study using electronic health records. Lancet Psychiatry. 2021;8:416–427. doi: 10.1016/S2215-0366(21)00084-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen Y., et al. Proteomic analysis identifies prolonged disturbances in pathways related to cholesterol metabolism and myocardium function in the COVID-19 recovery stage. J. Proteome Res. 2021 doi: 10.1021/acs.jproteome.1c00054. [DOI] [PubMed] [Google Scholar]
- 26.Campbell J.S., Hallett M.B. Active calpain in phagocytically competent human neutrophils: electroinjection of fluorogenic calpain substrate. Biochem. Biophys. Res. Commun. 2015;457:341–346. doi: 10.1016/j.bbrc.2014.12.113. [DOI] [PubMed] [Google Scholar]
- 27.Bagheri V. S100A12: friend or foe in pulmonary tuberculosis? Cytokine. 2017;92:80–82. doi: 10.1016/j.cyto.2017.01.009. [DOI] [PubMed] [Google Scholar]
- 28.Barnes B.J., et al. Targeting potential drivers of COVID-19: neutrophil extracellular traps. J. Exp. Med. 2020;217 doi: 10.1084/jem.20200652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Middleton E.A., et al. Neutrophil extracellular traps contribute to immunothrombosis in COVID-19 acute respiratory distress syndrome. Blood. 2020;136:1169–1179. doi: 10.1182/blood.2020007008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu Y., et al. Neutrophil-to-lymphocyte ratio as an independent risk factor for mortality in hospitalized patients with COVID-19. J. Inf. Secur. 2020;81:e6–e12. doi: 10.1016/j.jinf.2020.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Laforge M., et al. Tissue damage from neutrophil-induced oxidative stress in COVID-19. Nat. Rev. Immunol. 2020;20:515–516. doi: 10.1038/s41577-020-0407-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen Y., et al. Proteomic analysis identifies prolonged disturbances in pathways related to cholesterol metabolism and myocardium function in the COVID-19 recovery stage. J. Proteome Res. 2021;20:3463–3474. doi: 10.1021/acs.jproteome.1c00054. [DOI] [PubMed] [Google Scholar]
- 33.Kammerl I.E., Meiners S. Proteasome function shapes innate and adaptive immune responses. Am. J. Phys. Lung Cell. Mol. Phys. 2016;311:L328–L336. doi: 10.1152/ajplung.00156.2016. [DOI] [PubMed] [Google Scholar]
- 34.Jamaluddin M., et al. Role of peroxiredoxin 1 and peroxiredoxin 4 in protection of respiratory syncytial virus-induced cysteinyl oxidation of nuclear cytoskeletal proteins. J. Virol. 2010;84:9533–9545. doi: 10.1128/JVI.01005-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Grozdanov V., Danzer K.M. Intracellular alpha-Synuclein and immune cell function. Front. Cell Dev. Biol. 2020;8 doi: 10.3389/fcell.2020.562692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jayaprakash P., et al. Hsp90alpha and Hsp90beta together operate a hypoxia and nutrient paucity stress-response mechanism during wound healing. J. Cell Sci. 2015;128:1475–1480. doi: 10.1242/jcs.166363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Begue F., et al. Altered high-density lipoprotein composition and functions during severe COVID-19. Sci. Rep. 2021;11:2291. doi: 10.1038/s41598-021-81638-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Giannis D., Ziogas I.A., Gianni P. Coagulation disorders in coronavirus infected patients: COVID-19, SARS-CoV-1, MERS-CoV and lessons from the past. J. Clin. Virol. 2020;127 doi: 10.1016/j.jcv.2020.104362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Esmon C.T. Interactions between the innate immune and blood coagulation systems. Trends Immunol. 2004;25:536–542. doi: 10.1016/j.it.2004.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Puntmann V.O., et al. Outcomes of cardiovascular magnetic resonance imaging in patients recently recovered from coronavirus disease 2019 (COVID-19) JAMA Cardiol. 2020;5:1265–1273. doi: 10.1001/jamacardio.2020.3557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Siripanthong B., et al. Recognizing COVID-19-related myocarditis: the possible pathophysiology and proposed guideline for diagnosis and management. Heart Rhythm. 2020;17:1463–1471. doi: 10.1016/j.hrthm.2020.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kochi A.N., Tagliari A.P., Forleo G.B., Fassini G.M., Tondo C. Cardiac and arrhythmic complications in patients with COVID-19. J. Cardiovasc. Electrophysiol. 2020;31:1003–1008. doi: 10.1111/jce.14479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Marian A.J. On the diagnostic utility of junction plakoglobin in arrhythmogenic right ventricular cardiomyopathy. Cardiovasc. Pathol. 2013;22:309–311. doi: 10.1016/j.carpath.2013.05.002. [DOI] [PubMed] [Google Scholar]
- 44.Ashkar S., et al. Eta-1 (osteopontin): an early component of type-1 (cell-mediated) immunity. Science. 2000;287:860–864. doi: 10.1126/science.287.5454.860. [DOI] [PubMed] [Google Scholar]
- 45.Leali D., et al. Osteopontin (Eta-1) and fibroblast growth factor-2 cross-talk in angiogenesis. J. Immunol. 2003;171:1085–1093. doi: 10.4049/jimmunol.171.2.1085. [DOI] [PubMed] [Google Scholar]
- 46.Choo E.H., et al. Infarcted myocardium-primed dendritic cells improve remodeling and cardiac function after myocardial infarction by modulating the regulatory T cell and macrophage polarization. Circulation. 2017;135:1444–1457. doi: 10.1161/CIRCULATIONAHA.116.023106. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material 1
Supplementary material 2
Supplementary material 3
Data Availability Statement
The experimental data that support the findings of this study have been deposited in iProX (integrated proteome resources) of ProteomeXchange with the accession code PXD026442.