Abstract
The spike (S) protein is a leading vaccine candidate against SARS-CoV-2 infection. The S1 domain of S protein, which contains a critical receptor-binding domain (RBD) antigen, potentially induces protective immunoreactivities against SARS-CoV-2. In this study, we presented preclinical evaluations of a novel insect cell-derived SARS-CoV-2 recombinant S1 (rS1) protein as a potent COVID-19 vaccine candidate. The native antigenicity of rS1 was characterized by enzyme-linked immunosorbent assay with a neutralizing monoclonal antibody targeting the RBD antigen. To improve its immunogenicity, rS1-adjuvanted with fucoidan/trimethylchitosan nanoparticles (FUC-TMC NPs) and cytosine-phosphate-guanosine-oligodeoxynucleotides (CpG-ODNs) were investigated using a mouse model. The S1-specific immunoglobulin G (IgG) titers, FluoroSpot assay, pseudovirus- and prototype SARS-CoV-2-based neutralization assays were assessed. The results showed that the rS1/CpG/ FUC-TMC NPs (rS1/CpG/NPs) formulation induced a broad-spectrum IgG response with potent, long-lasting, and cross-protective neutralizing activity against the emerging SARS-CoV-2 variant of concern, along with a Th1-biased cellular response. Thus, the rS1/CpG/NPs formulation presents a promising vaccination approach against COVID-19.
Keywords: SARS-CoV-2 infection, Recombinant S1 protein, CpG adjuvant, Fucoidan-trimethylchitosan nanoparticles, Immunogenicity enhancement, Neutralizing antibody, Th1-biased cellular immune responses
1. Introduction
The ongoing COVID-19 pandemic caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is adversely affecting the world. Thus, the development of multiple safe, effective, and long-lasting vaccines is a major goal to end the SARS-CoV-2 pandemic. Accordingly, many preclinical and clinical trials are being conducted with various spike (S)-glycoprotein-based candidate vaccines (https://www.who.int/publications/m/item/draft-landscape-of-covid-19-candidate-vaccines). All the approved SARS-CoV-2 vaccines of Wuhan-Hu-1, including Pfizer, Moderna, AstraZeneca, and NVX-CoV2373, demonstrated 65–96% efficacy and reduced SARS-CoV-2-associated morbidity and mortality in phase 3 trials [1], [2]. S glycoprotein is a major surface protein of SARS-CoV-2, which is cleaved by furin into S1 and S2 domains, and it is responsible for receptor binding and membrane fusion [3]. S1 contains a RBD, which SARS-CoV-2 binds to its cellular receptor angiotensin-converting enzyme 2 (ACE2). The S2 domain is responsible for viral membrane fusion [4], [5]. Cryoelectron microscopy studies on the stabilized ectodomain of SARS-CoV-2 S protein in the prefusion conformation revealed an S trimer with three RBD domains for ACE2 binding [3]. However, the three RBDs on the S trimer are not cooperative with ACE-2 binding. Studies on human immunity against SARS-CoV-2 have shown that infected humans induce robust neutralizing antibody responses targeting the RBD [6], [7], [8] and cellular immune response [9] against the S protein. Reducing the size of the glycoprotein antigen to limit exposure to non-neutralizing epitopes could minimize the risk of undesired immunopathology. However, monomeric RBD has poor immunogenicity [10]. Sufficient amino acid sequences for maintaining native RBD structure and more T-cell epitopes might be necessary for better vaccine design. Therefore, S1 has been suggested as an ideal target for betacoronavirus vaccine development [11], [12], [13]. The SARS-CoV-2 S1 domain shares a lower sequence identity (<64%) among betacoronaviruses [14].
The insect cell expression system associated with post-translational modifications is an ideal eukaryotic expression system for the production of SARS-CoV-2 vaccines [15]. An insect-derived full-length SARS-CoV-2 S glycoprotein (Novavax) from the baculovirus/lepidopteran expression system (BEVS) induced immune responses that exceeded levels in COVID-19 convalescent serum in the Phase 1 and 2 trials [16], and a Phase 3 trial was launched [17]. Another insect (Drosophila cell)-derived SARS-CoV-2 S protein ectodomain formulated with CoVaccine HT™ adjuvant potently induced humoral and T helper 1 (Th1)-biased cellular immune responses in mice [18]. Furthermore, advances in nonreplicating viral-vectored transient gene expression (TGE) of insect cells could promote scalable production of the secreted recombinant protein. The baculovirus/mosquito (BacMos) system, which is a novel viral-vectored TGE of insect cells, has been used to express recombinant proteins from mosquito cell lines [19], [20].
An adjuvant is an immunological agent that improves the magnitude, quality, breadth, and/or longevity of specific immune responses against a coadministered antigen. Synthetic unmethylated CpG-ODNs are potent activators of innate and adaptive immune responses because CpG motifs are recognized by Toll-like receptor 9 (TLR9), which is found in antigen-presenting cells and B cells. CpG ODNs promote both antigen-specific antibodies and natural killer T-cell responses, and have significant potential as vaccine adjuvants for the treatment of infectious diseases and cancer [21]. However, CpG ODNs are anionic, and it is difficult to penetrate the cell membrane with a negative surface charge. Furthermore, natural CpG ODNs are easily degraded by nucleases under biological conditions; thus, their transitory biological activity and poor cellular uptake limit their therapeutic applications. Chemical modification of CpG ODNs is an alternative method for protecting them against degradation by nucleases. However, several serious side effects caused by coadministration of modified CpG ODNs is a major concern. Therefore, it is necessary to develop efficient delivery systems to enhance the immunostimulatory effect of CpG ODNs. The recent development in nanobiotechnology offers great promise for the development of effective nanocarriers for CpG ODN delivery. Various nanomaterials, such as liposomes, carbon nanotubes, gold nanoparticles (NPs), boron nitride nanospheres, and silica nanomaterials, have been employed as efficient nanocarriers for CpG ODN delivery [21]. Nanocarriers have contributed to improving the safety and efficacy of SARS-CoV-2 vaccines [22], [23].
Polysaccharide NPs from natural or synthetic biodegradable polysaccharides have been extensively used as vaccine adjuvants because of their biocompatibility and affordability. Positively or negatively surface-charged polysaccharide NPs can be easily prepared using a polyelectrolyte complexation (PEC) method with an anionic polyelectrolyte and a cationic polyelectrolyte through an environment-friendly process [24], [25]. Fucoidan (FUC) refers to a family of anionic fucose-containing sulfated polysaccharides extracted from marine brown algae [26]. FUC exhibits anticoagulant, antithrombotic, antiviral, antioxidant, anti-inflammatory, antitumor, anticomplementary, and immunomodulating properties. Previous experimental data also proved that FUC are agonists of innate immune receptors and strong inducers of cellular and humoral immune responses [27]. Cationic chitosan (CS) and its quaternary derivatives, such as trimethylchitosan (TMC) and N-(2-hydroxy-3-trimethylammonium) propyl-chitosan (HTCC), have various advantages as vaccine adjuvants that enhance both humoral and cellular immune responses after vaccination [28]. In addition, a CS-based adjuvant yielded better antigen-specific antibody titers and splenic CD4+ proliferation than an incomplete Freund's or an aluminum hydroxide adjuvant [29]. CS-based NPs have been employed as nanocarriers and/or adjuvants for delivering several antigens [30], [31], [32]. Our previous studies also proved that FUC/quaternary chitosan NP-adjuvanted anthrax vaccine strongly induces both cellular and humoral immunity in mice [33], [34]. The main objective of this study was to evaluate the in vitro and in vivo efficacies of FUC-TMC NPs as co-adjuvants and nanocarriers of CpG-adjuvanted SARS-CoV-2 S1. We investigated CpG/FUC-TMC NPs (CpG/NPs)-adjuvanted rS1 protein vaccine as a vaccine candidate in terms of S1-specific immunoglobulin G (IgG) titrations, FluoroSpot assay, and pseudovirus- and Prototype (WT)-SARS-CoV-2-based neutralization assays. Notably, rS1 protein along with CpG/NPs delivered intramuscularly induced broad and durable neutralizing antibodies as well as Th1-biased immune responses in mice.
2. Materials and methods
2.1. Materials
CS (viscosity: 3.6 mPa·s [5 g/L]; degree of deacetylation: 93.8%) was purchased from Wako Pure Chemical Industries, Ltd. (Osaka, Japan). Dimethyl sulfoxide (DMSO), absolute ethanol, acetone, N-methylpyrrolidinone, iodomethane, sodium chloride, sodium iodide, and sodium hydroxide were purchased from Merck (Darmstadt, Germany). FUC (from Fucus vesiculosus), carbonate-bicarbonate buffer, 5% skim milk, methylcellulose, and fluorescein 5(6)-isothiocyanate (FITC) were obtained from Sigma–Aldrich (St. Louis, MO, USA). ODN 1826 (CpG ODN, 5-tccatgacgttcctgacgtt-3, molecular weight: 6364 g/mol, bases were phosphorothioate, ammonium salt) was purchased from InvivoGen (CA, USA). Dulbecco's modified Eagle medium (DMEM), SFM-900, Grace's insect medium, fetal bovine serum (FBS), l-glutamine, antibiotic solution (penicillin and streptomycin), and phosphate-buffered saline (PBS; pH 7.4) were supplied by Gibco BRL (Thermo Fisher Scientific, Inc., Waltham, MA, USA). The peroxidase-labeled secondary antibody, 3,3′,5,5′-tetramethylbenzidine (TMB) substrate, and TMB stop solution were obtained from KPL (Gaithersburg, MD, USA). Ultrapure water was obtained using a Milli-Q system (Millipore, Billerica, MA, USA).
2.2. Cell lines
Spodoptera frugiperda IPLB-Sf21 (Sf21) cells were cultured in SFM-900 and Grace's insect medium (1:4) containing 10% FBS at 28 °C. C6/36 cells (Aedes albopictus clone) (ATCC® CRL-1660™) were grown in RPMI-1640 medium (HyClone) supplemented with 10% FBS, l-glutamine, and antibiotic–antimycotic at 28 °C in a 5% CO2 humidified incubator. HEK-293T-hACE2 cells (RNAi Core, Academia Sinica, Taiwan) were maintained in DMEM supplemented with 10% FBS, blasticidin (InvivoGen), l-glutamine, and penicillin–streptomycin at 37 °C in a 5% CO2 humidified incubator.
2.3. rS1 protein production and purification
A recombinant baculovirus contains the hr1pag1 promoter that drives a synthetic gene encoding a fusion protein comprising of mosquito cecropin B1 signal peptide (1st–24th amino acid residues, sequence ID: KJ439044.1) and an insect codon-optimized SARS-CoV-2 S1 (T22th-S680th amino acid residues of S protein, Sequence ID: MT459919.1) from pUC57-2019-nCoV-S(insect) (GenScript). Its C-terminally fused with enterokinase cut site-V5 epitope-8xHis. A total of 107 C6/36 cells were seeded into T-75 flasks (corning) and incubated for 16–18 hours (h). The cells were transduced with baculovirus at a Multiplicity of infection (MOI) of 20 in growth medium. After 20 h of incubation, cells were washed twice using PBS and refresh growth medium (RPMI-1640 containing 20% tryptose phosphate broth, l-glutamine, antibiotic, and HEPES). The culture medium was harvested and refreshed at 3, 5, and 7 dpt. The collected culture medium was stored at 4 °C until further use. The harvested culture medium was centrifuged at 6000 rpm for 20 minutes (min). The supernatant was filtrated (0.45 μm), concentrated (1/10) and then diafiltrated with PBS (original volume) using a KrosFlo® KR2i Tangential Flow Filtration (TFF) System with MidiKros Hollow Fiber Filter (10 K MPES 0.5 mm) (Spectrums). The resultant product was subjected to affinity purification using TALON metal affinity resin (TAKARA).
2.4. Western blot
To generate rabbit anti-SARS-CoV-2 RBD serum, a cDNA fragment of RBD from SARS-CoV-2 prototype was subcloned into pET32c plasmid. The expression, purification, and animal immunization of recombinant SARS-CoV-2 RBD were performed in accordance with methods described in a previous report [35]. Samples were resuspended in sample buffer, boiled, and loaded onto a sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) gel. Following separation, the proteins were transferred to a nitrocellulose membrane using semi-dry transfer (Hofer TE77). After blocking with 5% skim milk at 37 °C for 1 h, the membrane was incubated with rabbit anti-SARS-CoV-2 RBD serum at 4 °C overnight. The membrane was washed three times with PBS containing 0.05% Tween 20 (PBST) before incubating 1 h at room temperature (22–25 °C) with HRP-conjugated goat anti-mouse secondary antibody. After four times final washing with PBST, the membrane was incubated with ECL substrate and signals were captured using the Amersham Image 600 (GE Healthcare).
2.5. Preparation and characterization of FUC-TMC NPs
TMC was synthesized from chitosan via a two-step methylation procedure as described previously [25]. Positively charged FUC-TMC NPs were prepared using the PEC method, as previously reported [34]. Briefly, the FUC solution (4 mg/mL) was added dropwise into the TMC solution (4 mg/mL) at a mass ratio (FUC/TMC) of 0.6, and then stirred at 28 °C for 5 min. The particle size was characterized by photon correlation spectroscopy (Zetasizer Nano-ZS; Malvern Instruments, UK). All measurements were performed at a wavelength of 633 nm at 28 °C with a detection angle of 173°. The raw data were correlated to the mean hydrodynamic size by cumulant analysis (Z-average mean). The zeta potentials of all NPs were analyzed via laser Doppler velocimetry (Zetasizer Nano-ZS; Malvern Instruments, UK). NP morphology was examined using a Hitachi HT-7700 transmission electron microscope (Minato-ku, Tokyo, Japan). TEM samples were prepared as follows: one drop of FUC-TMC NP suspension (resuspended in water from the pellet after centrifugation) was deposited on a 200-mesh Formvar/carbon-coated copper grid, and the excess solution was removed by wicking it with filter paper to avoid particle aggregation. Samples were stained with 2% phosphotungstic acid and dried at 28 °C.
2.6. Native antigenicity assay with an indirect enzyme-linked immunosorbent assay (ELISA)
96-well plates were coated with purified rS1 or rS1 mixed with CpG ODN 1826 and FUC-TMC NPs with rS1 protein concentration of about 0.1 μg/well in carbonate-bicarbonate buffer overnight at 4 °C. Wells were blocked with 5% skim milk in PBST at 37 °C for 1 h. After blocking, 100 μL blocking buffer containing 0.4, 4, 10, 25, 50, or 100 ng of either anti-SARS-CoV-2 S RBD neutralizing monoclonal antibody (NT mAb) (GTX01556, GeneTex) or control antibody (6B6C, an anti-Flavivirus mAb) was added to each well, followed by two washes with PBST. After an hour of incubation at 37 °C, four washes with PBST were performed, which was followed by incubation with peroxidase-labeled secondary antibody for 1 h at room temperature. Further, five washes with PBST were performed, followed by the addition of 100 μL/well TMB substrate for 10 min incubation at room temperature in the dark. TMB stop solution (50 μL) was added to each well, and the absorbance was read at 450 nm within 15 min. Means of absorbance value calculated from 3 independent experiments in duplicate.
2.7. Animal studies
BALB/c mice were handled in strict accordance with regulations, as defined by the Council of Agriculture, Executive Yuan, Taiwan. The protocol was approved by the Institutional Animal Care and Use Committee of the Institute of Preventive Medicine, National Defense Medical Center (IACUC No. AN-109-20). 35 female mice 8 weeks old (n = 5 per group) were intramuscularly (IM) immunized with either 1 μg rS1 protein alone or 5 μg rS1 protein alone or with 10 μg CpG or 10 μg CpG plus 200 μg FUC-TMC NPs at 8, 11, and 14 weeks of age. The negative control group received PBS CpG and FUC-TMC NPs. Blood was collected from all mice at 10, 13, and 16 weeks of age for the antibody assay. Additionally, blood from three mice of each rS1/CpG/NPs group was collected at seven months of age for a lasting neutralizing antibody assay. Three weeks after the final vaccination, splenocytes from one mice per group (median response of total IgG in the group) were isolated to conduct FluoroSpot assay.
2.8. Evaluation of S1-specific IgG response using ELISA
The 96-well plates were coated with a commercial rS1 (HEK293-derived sheep Fc-Tagged rS1; The Native Antigen, UK) at a concentration of 1 μg/ml in carbonate-bicarbonate buffer overnight at 4 °C. Wells were blocked with 5% skim milk in PBST at 37 °C for 1 h. Further, two washes with PBST were performed, followed by the addition of 100 μL of 100 to 1.56 × 106 -fold diluted sera (5-fold serial dilution) to each well and incubation at 37 °C for 1 h. Four washes with PBST were performed, followed by incubation with either peroxidase labeled anti-mouse IgG (KPL), IgG1 (Zymed), IgG2a (Invitrogen) or IgG2b (Bethyl) antibodies for 1 h at room temperature. Furthermore, five washes with PBST were performed, followed by the addition of 100 μL TMB substrate/well for 10 min incubation at 28 °C in the dark. The TMB stop solution (50 μL) was added to stop the reaction. The optical density was measured at 450 nm within 15 min.
2.9. Pseudotyped neutralization test (NT)
WT-, D614G-, Alpha (B.1.1.7)-, Beta (501Y·V2)- and Delta (B.1.617.2)-S pseudoviruses (RNAi Core, Academia Sinica, Taiwan) were packaged from HEK293T cells using reporter plasmids (pCMVΔ8.91 and pLAS2w.FLuc.Ppuro). Sera were first heat-inactivated for 30 min at 56 °C and 2.5-fold serial dilution (either 1/16–1/3906 or 1/40–1/9766), incubated with 3 × 105 relative light unit (RLU) individual pseudovirus in growth medium (without blasticidin) at 37 °C for 1 h, along with virus-alone (positive control) and growth medium (negative control) wells. The mixtures were then added to HEK293T-ACE2 cells (2 × 104 cells/well in 96-well plates). After incubation for 16–20 h, the culture medium (without blasticidin) was refreshed (100 μL/well) and incubated for another 48 h at 37 °C. Cells were lysed with Bright-Glo reagent (Promega) for 3 min. The lysate was transferred to a white plate, and luminescence was measured using LB 960 Luminometer (Berthold). The ID50 (half maximal inhibitory dilution) of neutralizing antibody titer was calculated with GraphPad Prism as the reciprocal of the dilution: the sample showed that the RLUs were reduced by 50% as compared to pseudovirus-alone control wells.
2.10. Plaque reduction neutralization test (PRNT50)
Vero E6 cells were cultured in high-glucose DMEM (GeneDireX) supplemented with 10% (v/v) FBS in an incubator under a humidified atmosphere of 5% CO2 at 37 °C. The viral titer of the SARS-CoV-2 strain 3586 (TSGH_15 GISAID accession number EPI_ISL_436100) that propagated in the Vero E6 cells was determined via plaque assays in the Vero E6 cells in the BSL-3 laboratory. The Vero E6 cells (4 × 105 cells/well) were seeded into 12-well plates and incubated at 37 °C overnight. Pooled sera of mice and individual serum of SARS-CoV-2 patients inactivated at 56 °C for 30 min were first diluted (either 1/16–1/625 or 1/16–1/1562) in 2.5-fold DMEM medium. Serially diluted serum (250 μL) was incubated with an equal volume of medium containing approximately 100 plaque-forming unit of SARS-CoV-2. The mixtures were then added to Vero E6 cells and incubated at 37 °C for 1 h with occasional shaking. Cells were subsequently cultured in 4 ml DMEM containing 2% FBS and 1.55% (v/v) methylcellulose for 72 h. After culturing, the cells were fixed with 10% formaldehyde for 1 h at room temperature. Plaques were stained with 0.5% (w/v) crystal violet for at least 30 min at 28 °C and then counted. NT titers were defined as the reciprocal of the maximum dilution of serum that reduced the virus titer by 50%. Two independent experiments were performed with very similar results, and the data presented represent the results from one independent experiment.
2.11. FluoroSpot assay
Cellular immunity assay was performed using the Mouse IFN-γ/IL-4 FluoroSpotPLUS kit (Mabtech). Briefly, 5 × 105 splenocytes/well (in triplicate) in DMEM supplemented with 10% FBS and antibiotic–antimycotic was plated onto the precoated FluoroSpot plates. Further, the cells were incubated with either a peptide pool of SARS-CoV-2 S1 (Mabtech, 3629-1, Table S3) (5 μg/well), ConA (positive control, 2 μg/ml), or PBS (negative control) in RPMI 1640 medium at 37 °C for 48 h. Following incubation with monoclonal detection antibodies, secondary reagents conjugated to fluorophores, which is a fluorescence enhancer. Furthermore, positive cells were photographed and counted using an automated FluoroSpot reader equipped with filters for excitation 490 nm/emission 510 nm (LED490, FITC) for IFN-γ detection and excitation 550 nm/emission 570 nm (LED550, Cy3) for IL-4 detection.
2.12. Statistical analysis
Data were analyzed using the GraphPad Prism 6.01 software for statistically significant differences using a two-tailed, unpaired t-test. Statistical significance was set at p < 0.05. The p-values are indicated as follows: ns: not significant; *p < 0.05: significant; **p < 0.01: highly significant.
3. Results
3.1. Preparation and characterization of SARS-CoV-2 rS1 protein
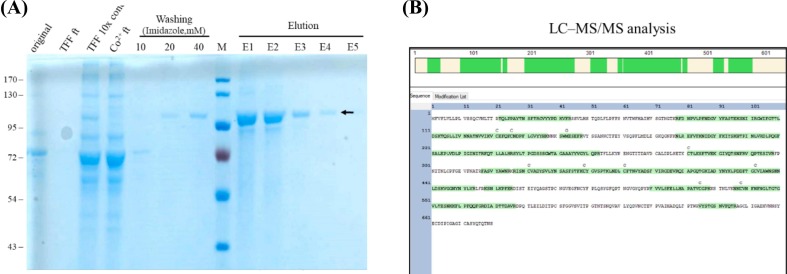
In this study, the BacMos system was employed to produce a recombinant SARS-CoV-2 S1 protein for vaccine development. After harvesting from the four-day batch serum-free culture supernatants, the collected culture medium was clarified and subsequently concentrated by TFF. Further, the crude material was purified using the TFF-Co2+ protocol. Western blot analysis (Fig. 1A) of the corresponding fractions confirmed that a protein of approximately 100 kDa molecular weight was present in the major band of the elution fraction using Coomassie blue staining (Fig. 1B). Furthermore, LC-MS/MS analysis revealed that this 100 kDa protein had a matching amino acid sequence of SARS-CoV-2 S1 protein with 59.41% coverage (Fig. 1C).
Fig. 1.
Purification and identification of rS1. (A) Western blot analysis to detect rS1 glycoprotein. Protein sizes (kDa) of markers are shown on the left. rS1 is indicated by the arrow. (B) Coomassie gel analysis of a representative purification run of rS1. Proteins were separated on SDS-PAGE and stained with SimplyBlue SafeStain. Original: starting material; TFF flow through (ft): TFF flow-through; TFF 10× con.: 10-fold TFF concentration; Co2+ ft.: flow-through from Co2+ column; Washing: 10, 20, and 40 mM imidazole wash; Elution: elution from HisTrap with 150 and 250 mM imidazole treatments; The respective molecular weights (kDa) of the proteins are indicated on the left. rS1 is indicated by the arrow. (C) Purified rS1 analyzed via mass spectrometry. The amino acid sequences of SARS-CoV-2 S glycoprotein are shown along with the matching peptides obtained from LC–MS/MS analysis (shown in green). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
3.2. Formulation of SARS-CoV-2 rS1
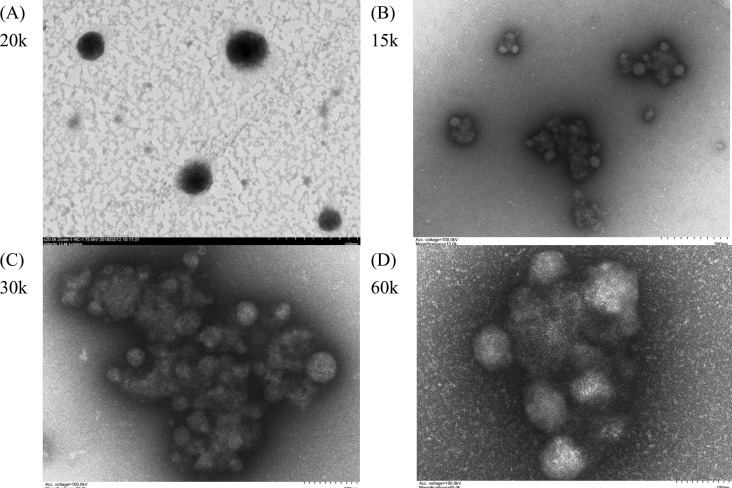
The formulation of SARS-CoV-2 rS1 used for immunization was developed by simply mixing the buffered solution of rS1, CpG, and FUC-TMC NPs. Nanosized FUC-TMC NPs with positive surface charges can be prepared via the PEC method, and the particle size and zeta potential of NPs depend on the mass ratio of anionic FUC to cationic TMC. rS1 and CpG are anionic; thus, the mass ratio of FUC/TMC must be carefully adjusted to avoid aggregation after mixing and to keep the surface positively charged. In this study, the mass ratio of FUC/TMC used to prepare NPs was 0.6, and the degree of quaternization of TMC was 52%. The particle sizes, polydispersity index (PDI), and zeta potentials of FUC-TMC NPs before and after mixing with rS1 and CpG are listed in Table S1. Fig. 2 shows the representative TEM images of FUC-TMC NPs and rS1/CpG/NPs. The TEM images of rS1/CpG/NPs (Fig. 2B–D) showed much smaller particle sizes than the original FUC-TMC NPs (Fig. 2A), and the particles loosely joined together to form agglomerates. Thus, the average particle size of rS1/CpG/NPs measured by Dynamic Light Scattering (DLS) was larger than that of the original FUC-TMC NPs (Table S1). This phenomenon may result from the anionic nature of rS1 and CpG. Furthermore, the addition of rS1 and CpG to FUC-TMC NPs changes the anion-to-cation ratio, and the particle rearranges to becomes smaller because PEC is a reversible process [33], [34]. Moreover, anionic rS1 and CpG can join more than one FUC-TMC NP via electrostatic interaction; thus, agglomerates were formed. A similar phenomenon was observed in our previous studies when polysaccharide NPs were mixed with anthrax vaccine adsorbed proteins [34].
Fig. 2.
Representative TEM images of (A) FUC-TMC NPs and (B–D) rS1/CpG/NPs of different magnification.
3.3. Formulation effect in antigenicity
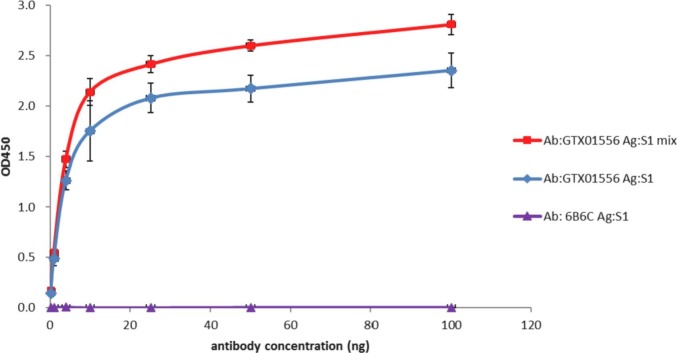
To determine if rS1 possesses a native RBD conformation before and after mixing with CpG/NPs, the purified rS1 alone or rS1/CpG/NPs mixture was subjected to an indirect ELISA with an anti-SARS-CoV-2 S RBD neutralizing mAb. The results (Fig. 3 ) show that this neutralizing mAb exhibits dose-dependent binding to rS1 alone and rS1/CpG/NPs. Binding of mAb to rS1 alone at a concentration of 25 ng mAb/well or higher approaches saturation. Notably, the binding avidity of this neutralizing mAb on rS1/CpG/NPs was significantly higher than that of rS1 alone. These results suggested that rS1 maintained a native RBD structure in the rS1/CpG/NPs formulation as a vaccine candidate for SARS-CoV-2.
Fig. 3.
rS1 possess a native RBD structure in rS1/CpG/NPs formulation. rS1 alone or rS1/CpG/NPs was coated onto a 96-well plate and incubated with a neutralizing monoclonal antibody (NT mAb) or control antibody (anti-Flavivirus mAb) at a range of antibody concentrations (0.1 to 100 ng). The absorbance value was measured at 450 nm.
3.4. Humoral response
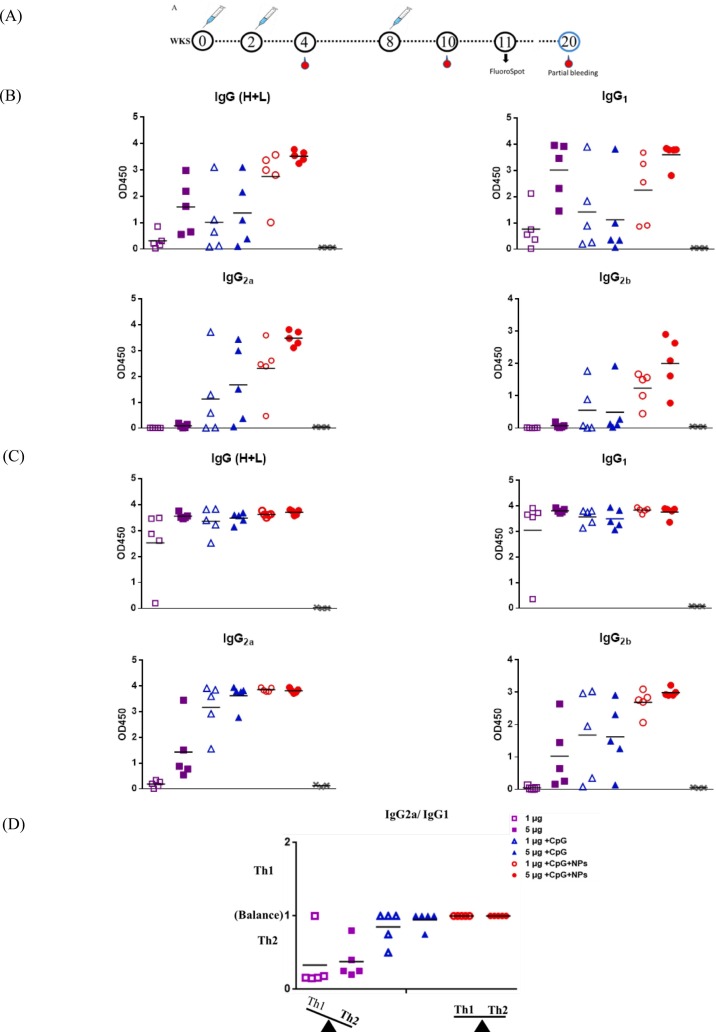
To evaluate the immunogenicity of rS1, BALB/c mice were immunized with rS1, rS1/CpG, or rS1/CpG/NPs at week 0 (prime), week 2 (second injection), and week 8 (third injection). ELISA assay results (Fig. 4 ) showed that mice immunized with rS1/CpG/NPs induced S1 protein-specific total IgG, IgG1, IgG2a, and IgG2b antibodies at significantly higher levels after two doses of inoculation and extremely high levels after three doses of inoculation (Fig. 4B). Notably, weak IgG2a and IgG2b responses were observed in the rS1 alone group. Conversely, dominant IgG1, IgG2a, and IgG2b responses were detected in the rS1/CpG/NPs groups. Interestingly, mice group of 5 μg rS1/CpG/NPs perform significant higher titers of total IgG, IgG2a and IgG2b subtypes (cellular immunity) after second boost. Stratified ratios of Th1 (IgG2a) vs. Th2 (IgG1) subclasses (Fig. 4C), and stratified results confirmed that a trend toward an IgG2a bias and a more balanced response for the 5 μg rS1/CpG/NPs formulation.
Fig. 4.
Immunization schedule and S-specific IgG responses. (A) Schedule presenting mice immunizations, bleeds, and FluoroSpot assay. (B) SARS-CoV-2 S-specific IgG responses after the second and third injections. (C) Stratified ratios of IgG2a (Th1) vs. IgG1 (Th2) subclasses.
To detect neutralizing antibody titers, an individual serum was measured by the WT S-pseudovirus-based neutralization assay (Fig. 5A) and compared with patient sera. After two doses of inoculation, the average ID50 titer (128 and 605) of rS1/CpG/NPs-immunized mice were significantly higher than the average ID50 titer (0 and 39) of rS1 alone. The average ID50 titer (605) of 5 μg rS1/CpG/NPs-immunized mice was higher than the average ID50 titer (340) of patient sera. After three doses of inoculation, the average ID50 titers (3805, 2045, 1840, 5528, and 10712) of all rS1-containing groups except the 1 μg rS1-alone group (ID50 = 30) were significantly higher than the average ID50 titer (250) of patient sera.
Fig. 5.
Neutralizing antibody responses. (A) NT with pseudotype virus. WT SARS-CoV-2 S protein pseudotyped virus applied to determine the individual ID50 titers of neutralization antibodies for comparison with ID50 titers of individual serum of SARS-CoV-2 patients. The statistical significance between the groups was analyzed using Student's t-test (*p < 0.05; **p < 0.01). (B) PRNT50 with WT SARS-CoV-2 virus. Pooled sera of mouse group and individual serum of COVID-19 patients were used to determine the PRNT50 titers with the prototype SARS-CoV-2 virus. (C) Cross reactivity of NT. Pooled sera of mouse group were subjected to NT assay with variant SARS-CoV-2 S proteins pseudotyped viruses. (D) Duration of NT induced by rS1/CpG/NPs. Sera of three mice immunized with rS1/CpG/NPs were harvested at 0.5 and 5 months after the third injection and subjected to NT assay with WT SARS-CoV-2 S protein pseudotyped virus.
3.5. Longevity and breadth of neutralizing antibody titers against emerging viral variants
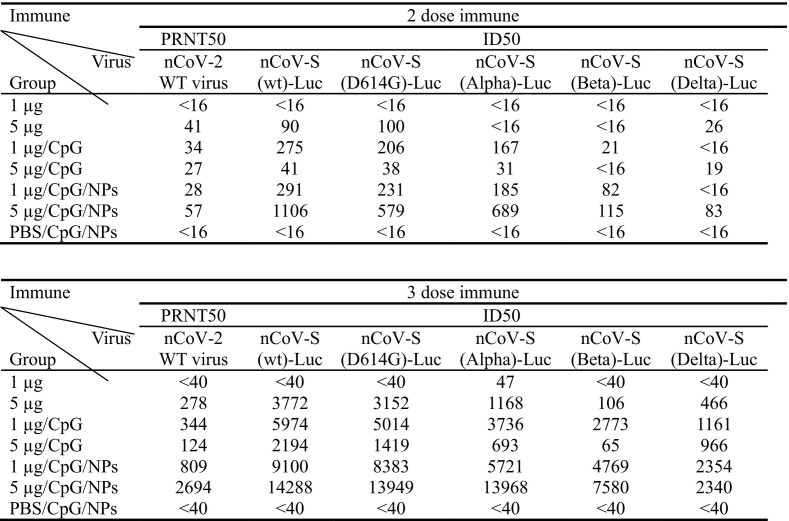
To confirm neutralizing antibody titers, pooled sera were subjected to WT SARS-CoV-2-based neutralization assay (Fig. 5B). Corresponding to the results of ID50 titers, the average PRNT50 titer (56) of 5 μg rS1/CpG/NPs-immunized mice were higher than the average PRNT50 titer (54) of patient sera after two doses of vaccination. The average PRNT50 titers (62, 277, 344, 124, 809, and 2693) of all rS-containing groups were significantly higher than the average PRNT50 titer (54) of patient sera after three doses of vaccination. To investigate if mouse sera cross-protected infections of clinical SARS-CoV-2 variants, neutralizing activities of pooled sera against D614G-, Alpha-, Beta-S or Delta-S pseudoviruses were measured (Fig. 5C). Inoculation of rS1/CpG/NPs induced substantial neutralizing titers against D614G, Alpha-, or Beta-S pseudoviruses. Inoculation of rS1/CpG/NPs induced neutralizing activities against D614G-, Alpha-, Beta-S or Delta-S pseudoviruses with comparable titers. As expected, titers (rS1/CpG/NPs) of neutralizing antibodies against WT S-pseudovirus were slightly higher than titers of neutralizing antibodies against D614G- and Alpha-S pseudoviruses, and significantly higher than Beta-S and Delta-S pseudoviruses. However, NT potency remained against these variants. Consistently, neutralization activities against authentic SARS-CoV-2 were lower than those against pseudovirus but they retained potency (median ID50 of 1501 and 3942 at weeks 6 and 12, respectively) (Fig. 5A and B).
The rS1/CpG/NPs vaccine, which induced a broadly neutralizing antibody response to four SARS-CoV-2 variants, could meet the needs of next-generation SARS-CoV-2 vaccines (Table 1 ). To investigate the duration of neutralizing titers induced by rS1/CpG/NPs, sera collected 0.5 and 5 months after the final boost were subjected to WT S-pseudovirus-based neutralization assay (Fig. 5D). The 1 μg dose of rS1/CpG/NPs remains an average ID50 titres of 3994 (range: 3396-5127) of 5 months as compared to an average ID50 titres of 7052 (range: 6050-7700) of 0.5 months, whereas 5 μg dose of rS1/CpG/NPs remains an average ID50 titres of 7373 (range: 5082-9131) of 5 months as compared to an average ID50 titres of 10,790 (range: 6010-16,483) of 0.5 months. This suggested that three doses of inoculation with rS1/CpG/NPs induced persistently high levels of neutralizing antibodies for at least 5 months in mice. Overall, these results suggest that the rS1/CpG/NPs formulation induces broadly durable NT activity against clinical SARS-CoV-2 variants.
Table 1.
Cross reactivity of SARS-CoV-2 neutralizing antibody responses. Pooled sera of mouse group (n = 5) were subjected to NT assay with either WT SARS-CoV-2 virus or variant SARS-CoV-2 S proteins pseudotyped viruses.
3.6. Cellular response
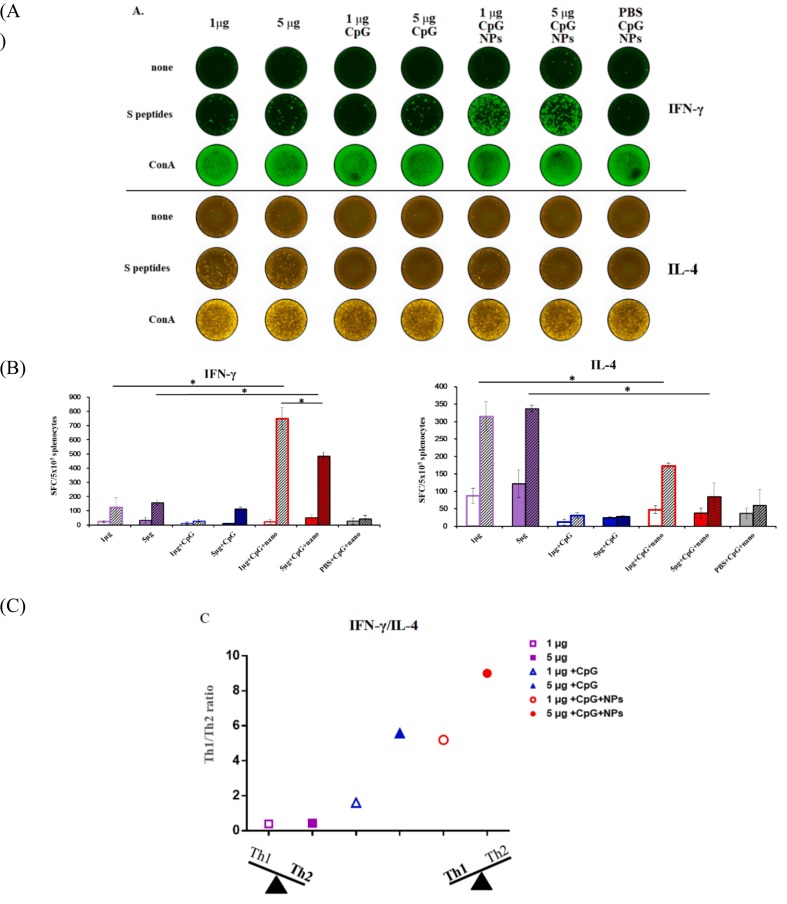
To confirm Th polarity, splenocytes (1 mouse per group) were subjected to FluoroSpot assay for detection of IFN-γ-and IL-4-secreting cells (Fig. 6A). We observed that splenocytes from mice immunized with rS1 alone presented a higher number of IL-4-secreting spots (315 and 338 spots/5 × 105 cells) than that of IFN-γ-secreting spots (125 and 158 spots/5 × 105 cells) upon peptide stimulation, thereby indicating the Th2-biased cellular responses in rS1 alone-immunized mice (Fig. 6B). Conversely, the densities of IFN-γ-secreting spots (750 and 486 spots/5 × 105 cells) induced by rS1/CpG/NPs were higher than those of IL-4-secreting spots (174 and 85 spots/5 × 105 cells), thereby indicating the Th1-biased cellular responses in rS1/CpG/NPs-immunized mice. The 1 μg rS1/CpG/NPs dose maintained higher numbers of IFN-γ and IL-4-secreting spots than the 5 μg rS1/CpG/NPs dose, thereby suggesting that 1 μg rS1 dose is sufficient for a robust Th1-focused T cell response in CpG/NPs formulation. A higher background signal of IL-4 (87 and 122 spots/5 × 105 cells, without stimulation) was obtained following immunization with rS1 alone. However, rS1 adjuvanted with either CpG/NPs (35 and 26 spots/5 × 105 cells) or CpG (10 and 22 spots/5 × 105 cells) decreased the background (without stimulation) signals of IL-4 in FluoroSpot assays. Notably, rS1 adjuvanted with CpG induced both lower IFN-γ (25 and 102 spots/5 × 105 cells) and IL-4 responses (31 and 28 spots/5 × 105 cells) under peptide stimulation. To further confirm the adjuvant effect on Th polarity, IFN-γ-and IL-4-secreting spots were stratified to analyze the Th1/Th2 ratio (Fig. 6C). Evidently, the rS1/CpG/NPs formulation skewed Th2-biased cellular responses of rS1 alone toward Th1-biased cellular responses in mice.
Fig. 6.
Cellular responses. Splenocytes from immunized mice (1 mouse/group) were collected and stimulated either without (none), with pooled peptides of S (S peptides) or with Con A followed by detection of IFN-γ or IL-4 cytokines in triplicate using FluoroSpot assay. (A) FluoroSpot wells show the IFN-γ or IL-4 released/5 × 105 cells. (B) Spot-forming cells (SFC) from (A) were quantitated and calculated. Empty and slash columns indicate none and S peptides stimulations, respectively. The data are presented as mean ± SEM, and the statistical significance between the groups under peptide stimulation was analyzed using Student's t-test (*p < 0.05). (C) Stratified SFC ratios of IFN-γ (Th1) vs. IL-4 (Th2).
4. Discussion
Different successful vaccines are required to overcome the global SARS-CoV-2 pandemic. Several vaccine strategies, such as using nucleic acid, viral vector, inactivated virus, and recombinant protein-based vaccines have been deployed, and some are under clinical use [36]. Regarding safety, subunit vaccines containing specific viral protein fragments eliminate the concerns of incomplete viral inactivation, virulence revertant, and pre-existing immunity against viral vectors. The S protein of SARS-CoV-2 is an important target for neutralizing antibodies. A S trimer induced high levels of neutralizing antibodies and Th1-biased cellular immune responses in rodents and protected nonhuman primates from the SARS-CoV-2 challenge [37]. Additionally, the neutralizing epitope-rich S1 protein has been proven as an alternative target for effective and safe betacoronavirus vaccines [11], [12], [13], [14]. However, subunit vaccines need to be combined with adjuvant(s) to improve immunogenicity and efficacy [38].
Control of immune systems via TLR9 by CpG ODNs has proven to be effective in preventing infectious diseases, cancers, and allergies. However, ODNs with only a phosphodiester backbone are easily degraded by deoxyribonuclease. Thus, CpG ODNs with a phosphorothioate backbone, such as ODN 1826, have been studied for the placement of phosphodiesters for clinical application; however, it has raised concerns regarding unfavorable side effects [39]. The advantages of using NPs as CpG ODN carriers include protection from DNase degradation, extension of retention time inside the body, improvement of cellular uptake efficiency, and delivery to target tissues. Furthermore, NPs allow the slow release of CpG ODNs over a long period of time and change localization inside the body, thereby reducing the frequency of vaccinations and the dose of vaccines. As described previously, CpG ODNs do not require encapsulation in NPs to be protected from DNase activity; adsorption to NPs is sufficient [40]. Thus, using NPs is a quick approach to develop an efficient delivery system for CpG ODNs. Generally, the method of attaching CpG ODNs by modifying negatively charged NPs with polycations is the most effective method [41]. However, polycations elicit nonspecific adsorption of negatively charged molecules and usually promote the formation of NP aggregates. Moreover, the disadvantages of using NPs include an inability to establish the safety of NPs or clarify the metabolic process [39]. Various studies have been conducted to avoid these shortcomings of polycations using PEG [42] and polyanions. Furthermore, it has been stated that ionizable cationic liposomes eliminate the disadvantages of polycations [43]. In this study, we used biodegradable FUC-TMC NPs as adjuvants and/or nanocarriers of rS1 and CpG ODN. Consequently, TMC and FUC exhibited some cytotoxicity and reduced viability of L929 cells, A549 cells, and JAWS II DCs. However, both positively and negatively charged FUC-TMC NPs were highly compatible with these cell lines and had low cytotoxicity (Table S2). NP formation via PEC reduced the cytotoxicity and immunotoxicity of the original composites, thereby rendering FUC-TMC NPs safer than TMC and FUC as drug carriers or vaccine adjuvants [33], [34]. This phenomenon is likely a result of materials with stronger cationic or anionic charges producing higher cytotoxicity in cells and both FUC-TMC NPs with milder surface charge being caused by the complexation of cationic and anionic groups, thereby resulting in less toxicity than the original composites. In addition, rS1 protein and CpG ODN were more easily adsorbed on the surface of positively charged FUC-TMC NPs through electrostatic attraction, and only formed some agglomerates (Fig. 2B–D). No aggregation was observed during formulation.
Here, the immunogenicity of rS1 alone or CpG-adjuvanted rS1 with or without the nanocarrier FUC-TMC NPs was investigated. Similar levels of IgG1/IgG2a and IgG1/IgG2b indicated that a balanced immune response was induced in the rS1/CpG/NPs groups (Fig. 4). CpG/NPs modulated the humoral response toward Th1-type relative to both groups of rS1 alone and rS1/CpG, as indicated by the significant higher levels of IgG2a and IgG2b. Variants of Concern (VOCs), such as B.1.1.7, 501Y·V2, and P.1 lineages, have emerged [44]. Some of these mutants within the S protein are resistant to the neutralizing activities of NTD- and RBD-specific mAbs, convalescent sera, RNA-based, and protein-based vaccines-elicited sera [45], [46], [47], [48], [49]. This raises a global concern about the efficacies of the current Wuhan-Hu-1-based vaccines, which highlights the necessity of a strategy to elicit a broadly neutralizing antibody response against SARS-CoV-2 variants. The Wuhan-Hu-1 rS1 protein, which contains major neutralizing epitopes, potentially induces cross-neutralizing antibodies against clinical variants. The results (Fig. 5A and Table 1) of the pseudovirus NT assay showed VOC D614G, B.1.1.7 (Alpha), and 501Y·V2 (Beta) that resulted in a 2-to-8-fold reduction and a slight but significant decrease compared to WT-S in neutralization by sera from S1/FUC-TMC NP-vaccinated mice. However, rS1/CpG/NPs-elicited mice sera retained potent cross-protective activity against the four clinical SARS-CoV-2 variants in the pseudovirus NT assay (Fig. 5A and C). In line with previous findings, vaccine-elicited sera maintain high NT effectivity against the Alpha lineage but less than 50% NT effectivity against the Beta lineage [46], [47], [50]. Antibody response established by infection or vaccination might be able to effectively neutralize SARS-CoV-2 B.1.1.7 (Alpha), but neutralizing titers against B.1.351 (Beta), B.1.617.2 (Delta) and P.1 (Gamma) suffered large reductions [51], [52], [53], [54]. The Beta lineage, which contains 10 amino acid substitutions on S glycoprotein (vs. prototype) and rapidly emerged in South Africa in late 2020, is resistant to convalescent (prototype infection) and vaccinated sera [49]. The D614G mutation pseudovirus slightly reduced the sensitivity of the virus to serum neutralizing antibodies in individual convalescent COVID-19 patients [55]. Notably, the pseudotyped NT response against Wuhan-Hu-1 SARS-CoV-2 was sustained for up to five months (Fig. 5D). This indicates that the rS1/CpG/NPs formulation could yield a broadly durable neutralizing repertoire and reduce the likelihood of the variant escaping host immunity. Overall, the rS1/CpG/NPs formulation exhibited the most potent neutralizing antibody and Th1 cellular immune responses. The median titer of ID50 in the group of 5 μg rS1/CpG/NPs after two doses of immunization was three times higher than the median titers of neutralizing antibodies in cohorts of six convalescent humans after recovery from SARS-CoV-2 infection (Fig. 5A). rS1/CpG/NPs-based vaccines can induce a robust Th1-prone cellular immune response (Figs. 4C and 6). Importantly, the Th1 response may reduce ADE and activate both arms of the immune system [56]. The results (Fig. 6) revealed that rS1 alone induces a predominantly Th2 type response in mice. In contrast, CpG/FUC-TMC NPs-based formulated with rS1 elicit more potent neutralizing antibodies while boosting Th1 responses, which may improve vaccine safety, efficacy, and durability. We also conducted immunogenicity assays of rS1/FUC-TMC NPs in mice (data not shown); however, the results showed lower titers of S-specific IgG and NT responses in rS1/FUC-TMC NP formulation. This indicates that CpG plus FUC-TMC NPs are required to maximize the immunogenicity of rS1 in mice. Immunization with SARS and MERS coronavirus vaccines leads to pulmonary immunopathology on virus challenge [57], [58], [59]. A similar phenomenon has also been reported in clinical trials with whole-inactivated virus vaccines against RSV and measles virus [60], [61]. In addition, the importance of Th1 cell responses has been highlighted in a recent study of asymptomatic and mild SARS-CoV-2 convalescent samples [62]. These results emphasize that vaccines capable of generating Th1-dominant responses are important for providing protection against SARS-CoV-2. T cells play an important role in SARS-CoV-2 infection. Along this line, rS1/CpG/NPs vaccine candidate induced a robust SARS-CoV-2 S-specific Th1 response, which involves a higher cytotoxicity of CD8+ T cells to eliminate virus-infected cells and induction of CD4+ T cells to facilitate humoral responses [9], [63].
Two doses of rS1/CpG/NPs formulation induced substantial neutralizing titers against the prototype and the D614G mutant of SARS-CoV-2 in mice. Furthermore, we confirmed the production of Th1-biased cellular responses in mice. This study is an important step toward the development of an efficacious vaccine for humans.
Immunization of two mammalian cell-derived rS1 can elicit high levels of neutralizing antibodies in various animals [64], [65]. In this study, we identify BacMos to be a potential clinically relevant platform for the production of rS1 protein as a vaccine immunogen. We formulated this rS1 with CpG/NPs, which induced a strong, broad, and durable neutralizing antibody response. Currently, animal experiments in hamster and human ACE2-transgenic mice are ongoing to evaluate if FUC-TMC NPs-adjuvanted SARS-CoV-2 S1 protein vaccine can protect against SARS-CoV-2 infection.
5. Conclusion
In this study, we verified that FUC-TMC NPs are good nanocarriers and adjuvant candidates for intramuscularly administered CpG ODN-adjuvanted SARS-CoV-2 rS1 protein vaccine. In vitro studies suggested that rS1 maintained correct folding in the formulation of rS1/CpG/NPs. Furthermore, FUC-TMC NPs are easily produced on a large scale via a simple mixing process of two oppositely charged polyelectrolytes; thus, they are more cost effective than chemically synthesized adjuvants, such as CpG ODNs. In the murine model, rS1 the CpG/NPs formulation induced a broad-spectrum IgG response, improved longevity and breadth of NT activity, and Th1-biased cellular responses. In summary, in this study, we suggest that rS1/CpG/NPs formulation is a promising COVID-19 vaccine candidate.
CRediT authorship contribution statement
Hui-Tsu Lin: Investigation, Formal analysis, Data curation. Cheng-Cheung Chen: Conceptualization, Methodology, Funding acquisition, Supervision, Resources. Der-Jiang Chiao: Investigation, Formal analysis, Data curation. Tein-Yao Chang: Methodology, Investigation, Formal analysis, Data curation, Resources. Xin-An Chen: Investigation, Formal analysis, Validation. Jenn-Jong Young: Conceptualization, Methodology, Writing – original draft, Writing – review & editing, Project administration, Funding acquisition. Szu-Cheng Kuo: Conceptualization, Methodology, Writing – original draft, Writing – review & editing, Project administration, Funding acquisition.
Acknowledgements
Financial support for this work was provided by the Ministry of Science and Technology (MOST 109-2327-B-016-002, MOST 109-2327-B-016-003 & MOST 110-2740-B-016-001), National Health Research Institutes (NHRI-109A1-MRCO-08202003 & NHRI-110A1-MRCO-08212101), Medical Affairs Bureau (MND-MAB-C13-111047-049 & MND-MAB-110-075), and National Defense Medical Center (IPM-110-G5 & IPM-110-G7) of the Republic of China.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ijbiomac.2021.11.020.
Appendix A. Supplementary data
Supplementary material
References
- 1.Polack F.P., Thomas S.J., Kitchin N., Absalon J., Gurtman A., Lockhart S., Perez J.L., Marc G.P., Moreira E.D., Zerbini C., Bailey R., Swanson K.A., Roychoudhury S., Koury K., Li P., Kalina W.V., Cooper D., Frenck R.W., Jr., Hammitt L.L., Türeci Ö., Nell H., Schaefer A., Ünal S., Tresnan D.B., Mather S., Dormitzer P.R., Şahin U., Jansen K.U., Gruber W.C. Safety and efficacy of the BNT162b2 mRNA Covid-19 vaccine. N. Engl. J. Med. 2020;383:2603–2615. doi: 10.1056/NEJMoa2034577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Voysey M., Clemens S.A.C., Madhi S.A., Weckx L.Y., Folegatti P.M., Aley P.K., Angus B., Baillie V.L., Barnabas S.L., Bhorat Q.E., Bibi S., Briner C., Cicconi P., Collins A.M., Colin-Jones R., Cutland C.L., Darton T.C., Dheda K., Duncan C.J.A., Emary K.R.W., Ewer K.J., Fairlie L., Faust S.N., Feng S., Ferreira D.M., Finn A., Goodman A.L., Green C.M., Green C.A., Heath P.T., Hill C., Hill H., Hirsch I., Hodgson S.H.C., Izu A., Jackson S., Jenkin D., Joe C.C.D., Kerridge S., Koen A., Kwatra G., Lazarus R., Lawrie A.M., Lelliott A., Libri V., Lillie P.J., Mallory R., Mendes A.V.A., Milan E.P., Minassian A.M., McGregor A., Morrison H., Mujadidi Y.F., Nana A., O’Reilly P.J., Padayachee S.D., Pittella A., Plested E., Pollock K.M., Ramasamy M.N., Rhead S., Schwarzbold A.V., Singh N., Smith A., Song R., Snape M.D., Sprinz E., Sutherland R.K., Tarrant R., Thomson E.C., Török M.E., Toshner M., Turner D.P.J., Villafana T.L., Watson M.E.E., Williams C.J., Douglas A.D., Hill A.V.S., Lambe T., Gilbert S.C., Pollard A.J. Safety and efficacy of the ChAdOx1 nCoV-19 vaccine (AZD1222) against SARS-CoV-2: an interim analysis of four randomised controlled trials in Brazil, South Africa, and the UK. Lancet. 2021;397:99–111. doi: 10.1016/S0140-6736(20)32661-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wrapp D., Wang N., Corbett K.S., Goldsmith J.A., Hsieh C.L., Abiona O., Graham B.S., McLellan J.S. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367:1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cai Y., Zhang J., Xiao T., Peng H., Sterling S.M., Rits-Volloch S., Chen B., Walsh R.M., Jr., Rawson S. Distinct conformational states of SARS-CoV-2 spike protein. Science. 2020;369:1586–1592. doi: 10.1126/science.abd4251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yao H., Song Y., Chen Y., Wu N., Xu J., Sun C., Zhang J., Weng T., Zhang Z., Wu Z., Cheng L., Shi D., Lu X., Lei J., Crispin M., Shi Y., Li L., Li S. Molecular architecture of the SARS-CoV-2 virus. Cell. 2020;183:730–738. doi: 10.1016/j.cell.2020.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zost S.J., Gilchuk P., Case J.B., Binshtein E., Chen R.E., Nkolola J.P., Schäfer A., Reidy J.X., Trivette A., Nargi R.S., Sutton R.E., Suryadevara N., Martinez D.R., Williamson L.E., Chen E.C., Jones T., Day S., Myers L., Hassan A.O., Kafai N.M., Winkler E.S., Fox J.M., Shrihari S., Mueller B.K., Meiler J., Chandrashekar A., Mercado N.B., Steinhardt J.J., Ren K., Loo Y.-M., Kallewaard N.L., McCune B.T., Keeler S.P., Holtzman M.J., Barouch D.H., Gralinski L.E., Baric R.S., Thackray L.B., Diamond M.S., Carnahan R.H., Crowe J.E., Jr. Potently neutralizing and protective human antibodies against SARS-CoV-2. Nature. 2020;584:443–449. doi: 10.1038/s41586-020-2548-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wec A.Z., Wrapp D., Herbert A.S., Maurer D.P., Haslwanter D., Sakharkar M., Jangra R.K., Dieterle M.E., Lilov A., Huang D., Tse L.V., Johnson N.V., Hsieh C.-L., Wang N., Nett J.H., Champney E., Burnina I., Brown M., Lin S., Sinclair M., Pudi S., Wirchnianski A.S., Laudermilch E., Florez C., Fels J.M., O'Brien C.M., Graham B.S., Nemazee D., Burton D.R., Voss J.E., Chandran K., Dye J.M., McLellan J.S., Walker L.M., Johnson C., Bortz R., III, Baric R.S. Broad neutralization of SARS-related viruses by human monoclonal antibodies. Science. 2020;369:731–736. doi: 10.1126/science.abc7424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Robbiani D.F., Gaebler C., Muecksch F., Lorenzi J.C.C., Wang Z., Cho A., Agudelo M., Barnes C.O., Gazumyan A., Finkin S., Hägglöf T., Oliveira T.Y., Viant C., Hurley A., Hoffmann H.-H., Millard K.G., Kost R.G., Cipolla M., Gordon K., Bianchini F., Chen S.T., Ramos V., Patel R., Dizon J., Shimeliovich I., Mendoza P., Hartweger H., Nogueira L., Pack M., Horowitz J., Schmidt F., Weisblum Y., Michailidis E., Ashbrook A.W., Waltari E., Pak J.E., Huey-Tubman K.E., Koranda N., Hoffman P.R., West A.P., Jr., Rice C.M., Hatziioannou T., Bjorkman P.J., Bieniasz P.D., Caskey M., Nussenzweig M.C. Convergent antibody responses to SARS-CoV-2 in convalescent individuals. Nature. 2020;584:437–442. doi: 10.1038/s41586-020-2456-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Grifoni A., Weiskopf D., Ramirez S.I., Mateus J., Dan J.M., Moderbacher C.R., Rawlings S.A., Sutherland A., Premkumar L., Jadi R.S., Marrama D., de Silva A.M., Frazier A., Carlin A.F., Greenbaum J.A., Peters B., Krammer F., Smith D.M., Crotty S., Sette A. Targets of T cell responses to SARS-CoV-2 coronavirus in humans with COVID-19 disease and unexposed individuals. Cell. 2020;181:1489–1501. doi: 10.1016/j.cell.2020.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Walls A.C., Fiala B., Schäfer A., Wrenn S., Pham M.N., Murphy M., Tse L.V., Shehata L., O’Connor M.A., Chen C., Navarro M.J., Miranda M.C., Pettie D., Ravichandran R., Kraft J.C., Ogohara C., Palser A., Chalk S., Lee E.-C., Guerriero K., Kepl E., Chow C.M., Sydeman C., Hodge E.A., Brown B., Fuller J.T., Dinnon K.H., III, Gralinski L.E., Leist S.R., Gully K.L., Lewis T.B., Guttman M., Chu H.Y., Lee K.K., Carter L., Pepper M., Sheahan T.P., Veesler D., King N.P., Kellam P., Fuller D.H., Baric R.S. Elicitation of potent neutralizing antibody responses by designed protein nanoparticle vaccines for SARS-CoV-2. Cell. 2020;183:1367–1382. doi: 10.1016/j.cell.2020.10.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang Y., Wang L., Cao H., Liu C. SARS-CoV-2 S1 is superior to the RBD as a COVID-19 subunit vaccine antigen. J. Med. Virol. 2021;93:892–898. doi: 10.1002/jmv.26320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang L., Shi W., Joyce M.G., Modjarrad K., Zhang Y., Leung K., et al. Evaluation of candidate vaccine approaches for MERS-CoV. Nat. Commun. 2015;6:7712. doi: 10.1038/ncomms8712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim E., Erdos G., Huang S., Kenniston T.W., Balmert S.C., Carey C.D., Raj V.S., Epperly M.W., Klimstra W.B., Haagmans B.L., Korkmaz E., Falo L.D., Jr., Gambotto A. Microneedle array delivered recombinant coronavirus vaccines: immunogenicity and rapid translational development. EBioMedicine. 2020;55 doi: 10.1016/j.ebiom.2020.102743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jaimes J.A., André N.M., Chappie J.S., Millet J.K., Whittaker G.R. Phylogenetic analysis and structural modeling of SARS-CoV-2 spike protein reveals an evolutionary distinct and proteolytically sensitive activation loop. J. Mol. Biol. 2020;432:3309–3325. doi: 10.1016/j.jmb.2020.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shi X., Jarvis D.L. Protein N-glycosylation in the baculovirus-insect cell system. Curr. Drug Targets. 2007;8:1116–1125. doi: 10.2174/138945007782151360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Keech C., Albert G., Cho I., Robertson A., Reed P., Neal S., Plested J.S., Zhu M., Cloney-Clark S., Zhou H., Smith G., Patel N., Frieman M.B., Haupt R.E., Logue J., McGrath M., Weston S., Piedra P.A., Desai C., Callahan K., Price-Abbott P., Formica N., Shinde V., Fries L., Lickliter J.D., Griffin P., Wilkinson B., Glenn G.M., Lewis M. Phase 1-2 trial of a SARS-CoV-2 recombinant spike protein nanoparticle vaccine. N. Engl. J. Med. 2020;383:2320–2332. doi: 10.1056/NEJMoa2026920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kyriakidis N.C., López-Cortés A., González E.V., Grimaldos A.B., Prado E.O. SARS-CoV-2 vaccines strategies: a comprehensive review of phase 3 candidates. NPJ Vaccines. 2021;6:28. doi: 10.1038/s41541-021-00292-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lai C.Y., To A., Wong T.A.S., Lieberman M.M., Clements D.E., Senda J.T., Ball A.H., Pessaint L., Andersen H., Donini O., Lehrer A.T. bioRxiv; 2021. Recombinant Protein Subunit SARS-CoV-2 Vaccines Formulated With CoVaccine HT Adjuvant Induce Broad, Th1 Biased, Humoral and Cellular Immune Responses in Mice. preprint. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Naik N.G., Lo Y.W., Wu T.Y., Lin C.C., Kuo S.C., Chao Y.C. Baculovirus as an efficient vector for gene delivery into mosquitoes. Sci. Rep. 2018;8:17778. doi: 10.1038/s41598-018-35463-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chang Y.H., Chiao D.J., Hsu Y.L., Lin C.C., Wu H.L., Shu P.Y., Chang S.-F., Chang J.-H., Kuo S.-C. Mosquito cell-derived japanese encephalitis virus-like particles induce specific humoral and cellular immune responses in mice. Viruses. 2020;12:336. doi: 10.3390/v12030336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang H., Gao X.-D. Nanodelivery systems for enhancing the immunostimulatory effect of CpG oligodeoxynucleotides. Mater. Sci. Eng., C. 2017;70:935–946. doi: 10.1016/j.msec.2016.03.045. [DOI] [PubMed] [Google Scholar]
- 22.Nel A.E., Miller J.F. Nano-enabled COVID-19 vaccines: meeting the challenges of durable antibody plus cellular immunity and immune escape. ACS Nano. 2021;15:5793–5818. doi: 10.1021/acsnano.1c01845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Machhi J., Shahjin F., Das S., Patel M., Abdelmoaty M.M., Cohen J.D., Singh P.A., Baldi A., Bajwa N., Kumar R., Vora L.K., Patel T.A., Oleynikov M.D., Soni D., Yeapuri P., Mukadam I., Chakraborty R., Saksena C.G., Herskovitz J., Hasan M., Oupicky D., Das S., Donnelly R.F., Hettie K.S., Chang L., Gendelman H.E., Kevadiya B.D. Nanocarrier vaccines for SARS-CoV-2. Adv. Drug Deliv. Rev. 2021;171:215–239. doi: 10.1016/j.addr.2021.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tsai T.N., Yen H.J., Chen C.C., Chen Y.C., Young Y.A., Cheng K.M., Young J.J., Hong P.D. Novel protein-loaded chondroitin sulfate-N-[(2-hydroxy-3-trimethylammonium) propyl]chitosan nanoparticles with reverse zeta potential: preparation, characterization, and ex vivo assessment. J. Mater. Chem. B. 2015;3:8729–8737. doi: 10.1039/C5TB01517K. [DOI] [PubMed] [Google Scholar]
- 25.Young J.J., Chen C.C., Chen Y.C., Cheng K.M., Yen H.J., Huang Y.C., Tsai T.N. Positively and negatively surface-charged chondroitin sulfate-trimethylchitosan nanoparticles as protein carriers. Carbohydr. Polym. 2016;137:532–540. doi: 10.1016/j.carbpol.2015.10.095. [DOI] [PubMed] [Google Scholar]
- 26.Li B., Lu F., Wei X., Zhao R. Fucoidan: structure and bioactivity. Molecules. 2008;13:1671–1695. doi: 10.3390/molecules13081671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kuznetsova T.A., Persiyanovaа E.V., Ermakovab S.P., Khotimchenkoc M.Y., Besednova N.N. The sulfated polysaccharides of brown algae and products of their enzymatic transformation as potential vaccine adjuvants. Nat. Prod. Commun. 2018;13:1083–1095. doi: 10.1177/1934578X1801300837. [DOI] [Google Scholar]
- 28.Wen Z.H., Xu Y.L., Zou X.T., Xu Z.R. Chitosan nanoparticles act as an adjuvant to promote both Th1 and Th2 immune responses induced by ovalbumin in mice. Mar. Drugs. 2011;9:1038–1055. doi: 10.3390/md9061038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zaharoff D.A., Rogers C.J., Hance K.W., Schlom J., Greiner J.W. Chitosan solution enhances both humoral and cell-mediated immune responses to subcutaneous vaccination. Vaccine. 2007;25:2085–2094. doi: 10.1016/j.vaccine.2006.11.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Malik A., Gupta M., Mani R., Gogoi H., Bhatnagar R. Trimethyl chitosan nanoparticles encapsulated protective antigen protects the mice against anthrax. Front. Immunol. 2018;9:562. doi: 10.3389/fimmu.2018.00562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Farhadian A., Dounighi N.M., Avadi M. Enteric trimethyl chitosan nanoparticles containing hepatitis B surface antigen for oral delivery. Hum. Vaccin. Immunother. 2015;11:2811–2818. doi: 10.1080/21645515.2015.1053663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Verheul R.J., Hagenaars N., van Es T., van Gaal E.V., de Jong P.H., Bruijns S., Mastrobattista E., Slutter B., Que I., Heldens J.G.M., van den Bosch J.F., Glansbeek H.L., Hennink W.E., Jiskoot W. A step-by-step approach to study the influence of N-acetylation on the adjuvanticity of N, N, N-trimethyl chitosan (TMC) in an intranasal nanoparticulate influenza virus vaccine. Eur. J. Pharm. Sci. 2012;45:467–474. doi: 10.1016/j.ejps.2011.10.001. [DOI] [PubMed] [Google Scholar]
- 33.Chuang C.C., Tsai M.H., Yen H.J., Shyu H.F., Cheng K.M., Chen X.A., et al. A fucoidan-quaternary chitosan nanoparticle adjuvant for anthrax vaccine as an alternative to CpG oligodeoxynucleotides. Carbohydr. Polym. 2020;229:1154032. doi: 10.1016/j.carbpol.2019.115403. [DOI] [PubMed] [Google Scholar]
- 34.Tsai M.H., Chuang C.C., Chen C.C., Yen H.J., Cheng K.M., Chen X.A., Chen C.C., Young J.J., Kau J.H. Nanoparticles assembled from fucoidan and trimethylchitosan as anthrax vaccine adjuvant: in vitro and in vivo efficacy in comparison to CpG. Carbohydr. Polym. 2020;236 doi: 10.1016/j.carbpol.2020.116041. [DOI] [PubMed] [Google Scholar]
- 35.Kuo S.C., Chen Y.J., Wang Y.M., Kuo M.D., Jinn T.R., Chen W.S., Chang Y.C., Tung K.L., Wu T.Y., Lo S.J. Cell-based analysis of chikungunya virus membrane fusion using baculovirus-expression vectors. J. Virol. Methods. 2011;175:206–215. doi: 10.1016/j.jviromet.2011.05.015. [DOI] [PubMed] [Google Scholar]
- 36.Jeyanathan M., Afkhami S., Smaill F., Miller M.S., Lichty B.D., Xing Z. Immunological considerations for COVID-19 vaccine strategies. Nat. Rev. Immunol. 2020;20:615–632. doi: 10.1038/s41577-020-00434-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liang J.G., Su D., Song T.Z., Zeng Y., Huang W., Wu J., Xu R., Luo P., Yang X., Zhang X., Luo S., Liang Y., Li X., Huang J., Wang Q., Huang X., Xu Q., Luo M., Huang A., Luo D., Zhao C., Yang F., Han J.-B., Zheng Y.-T., Liang P. bioRxiv; 2020. S-Trimer, A COVID-19 Subunit Vaccine Candidate, Induces Protective Immunity in Nonhuman Primates. preprint. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang N., Zheng B.J., Lu L., Zhou Y., Jiang S., Du L. Advancements in the development of subunit influenza vaccines. Microbes Infect. 2015;17:123–134. doi: 10.1016/j.micinf.2014.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hanagata N. Structure-dependent immunostimulatory effect of CpG oligodeoxynucleotides and their delivery system. Int. J. Nanomedicine. 2012;7:2181–2195. doi: 10.2147/IJN.S30197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhu Y., Meng W., Li X., Gao H., Hanagata N. Design of mesoporous silica/cytosine-phosphodiester-guanin oligodeoxynucleotide complexes to enhance delivery efficiency. J. Phys. Chem. C. 2011;115:447–452. doi: 10.1021/jp109535d. [DOI] [Google Scholar]
- 41.Nguyen D.N., Green J.J., Chan J.M., Langer R., Anderson D.G. Polymeric materials for gene delivery and DNA vaccination. Adv. Mater. 2009;21:847–867. doi: 10.1002/adma.200801478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pack D.W., Hoffman A.S., Pun S., Stayton P.S. Design and development of polymers for gene delivery. Nat. Rev. Drug Discov. 2005;4:581–593. doi: 10.1039/C5ME00015G. [DOI] [PubMed] [Google Scholar]
- 43.Wilson K.D., de Jong S.D., Tam Y.K. Lipid-based delivery of CpG oligonucleotides enhances immunotherapeutic efficacy. Adv. Drug Deliv. Rev. 2009;61:233–242. doi: 10.1016/j.addr.2008.12.014. [DOI] [PubMed] [Google Scholar]
- 44.Davies N., Abbott S., Barnard R.C., Jarvis C.I., Kucharski A.J., Munday J.D., Pearson C.A.B., Russel T.W., Tully D.C., Washburne A.D., Wenseleers T., Gimma A., Waites W., Wong K.L.M., van Zandvoort K., Silverman J.D., Diaz-Ordaz K., Keogh R.H., Eggo R.M., Funk S., Jit M., Atkins K.E., Edmunds W.J. Estimated transmissibility and severity of novel SARS-CoV-2 variant of concern 202012/01 in England. Science. 2020;372 doi: 10.1126/science.abg3055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang Z., Schmidt F., Weisblum Y., Muecksch F., Barnes C.O., Finkin S., Schaefer-Babajew D., Cipolla M., Gaebler C., Lieberman J.A., Oliveira T.Y., Yang Z., Abernathy M.E., Huey-Tubman K.E., Hurley A., Turroja M., West K.A., Gordon K., Millard K.G., Ramos V., Da Silva J., Xu J., Colbert R.A., Patel R., Dizon J., Unson-O'Brien C., Shimeliovich I., Gazumyan A., Caskey M., Bjorkman P.J., Casellas R., Hatziioannou T., Bieniasz P.D., Nussenzweig M.C. mRNA vaccine-elicited antibodies to SARS-CoV-2 and circulating variants. Nature. 2021;592:616–622. doi: 10.1038/s41586-021-03324-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Collier D.A., Marco A.De, Ferreira I., Meng B., Datir R., Walls A.C., Kemp S.A., Bassi J., Pinto D., Silacci-Fregni C., Bianchi S., Tortorici M.A., Bowen J., Culap K., Jaconi S., Cameroni E., Snell G., Pizzuto M.S., Pellanda A.F., Garzoni C., Riva A., Kingston N., Graves B., McCoy L.E., Smith K.G.C., Bradley J.R., Temperton N., Ceron-Gutierrez L., Barcenas-Morales G., Harvey W., Virgin H.W., Lanzavecchia A., Piccoli L., Doffinger R., Wills M., Elmer A.E., Veesler D., Corti D., Gupta R.K. Sensitivity of SARS-CoV-2 B.1.1.7 to mRNA vaccine-elicited antibodies. Nature. 2021;593:136–141. doi: 10.1038/s41586-021-03412-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Callaway E., Mallapaty S. Novavax offers first evidence that COVID vaccines protect people against variants. Nature. 2021;590:17. doi: 10.1038/d41586-021-00268-9. [DOI] [PubMed] [Google Scholar]
- 48.Wang P., Nair M.S., Liu L., Iketani S., Luo Y., Guo Y., Wang M., Yu J., Zhang B., Kwong P.D., Graham B.S., Mascola J.R., Chang J.Y., Yin M.T., Sobieszczyk M., Kyratsous C.A., Shapiro L., Sheng Z., Huang Y., Ho D.D. Antibody resistance of SARS-CoV-2 variants B.1.351 and B.1.1.7. Nature. 2021;593:130–135. doi: 10.1038/s41586-021-03398-2. [DOI] [PubMed] [Google Scholar]
- 49.Wibmer C.K., Ayres F., Hermanus T., Madzivhandila M., Kgagudi P., Oosthuysen B., Lambson B.E., de Oliveira T., Vermeulen M., van der Berg K., Rossouw T., Boswell M., Ueckermann V., Meiring S., von Gottberg A., Cohen C., Morris L., Bhiman J.N., Moore P.L. SARS-CoV-2 501Y.V2 escapes neutralization by South African COVID-19 donor plasma. Nat. Med. 2021;27:622–625. doi: 10.1038/s41591-021-01285-x. [DOI] [PubMed] [Google Scholar]
- 50.Muik A., Wallisch A.K., Sänger B., Swanson K.A., Mühl J., Chen W., Cai H., Maurus D., Sarkar R., Türeci Ö., Dormitzer P.R., Şahin U. Neutralization of SARS-CoV-2 lineage B.1.1.7 pseudovirus by BNT162b2 vaccine-elicited human sera. Science. 2021;371:1152–1153. doi: 10.1126/science.abg6105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang G.L., Wang Z.Y., Duan L.J., Meng Q.C., Jiang M.D., Cao J., Yao L., Zhu K.L., Cao W.C., Ma M.J. Susceptibility of circulating SARS-CoV-2 variants to neutralization, the. N. Engl. J. Med. 2021;384:2354–2356. doi: 10.1056/NEJMc2103022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Planas D., Veyer D., Baidaliuk A., Staropoli I., Guivel-Benhassine F., Rajah M.M., Planchais C., Porrot F., Robillard N., Puech J., Prot M., Gallais F., Gantner P., Velay A., Le Guen J., Kassis-Chikhani N., Edriss D., Belec L., Seve A., Courtellemont L., Péré H., Hocqueloux L., Fafi-Kremer S., Prazuck T., Mouquet H., Bruel T., Simon-Lorière E., Rey F.A., Schwartz O. Reduced sensitivity of SARS-CoV-2 variant Delta to antibody neutralization. Nature. 2021;596:276–280. doi: 10.1038/s41586-021-03777-9. [DOI] [PubMed] [Google Scholar]
- 53.Bernal J.Lopez, Andrews N., Gower C., Gallagher E., Simmons R., Thelwall S., Stowe J., Tessier E., Groves N., Dabrera G., Myers R., Campbell C.N.J., Amirthalingam G., Edmunds M., Zambon M., Brown K.E., Hopkins S., Chand M., Ramsay M. Effectiveness of Covid-19 vaccines against the B.1.617.2 (Delta) variant. N. Engl. J. Med. 2021;385:585–594. doi: 10.1056/NEJMoa2108891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Liu C., Ginn H.M., Dejnirattisai W., Supasa P., Wang B., Tuekprakhon A., Nutalai R., Zhou D., Mentzer A.J., Zhao Y., Duyvesteyn H.M.E., López-Camacho C., Slon-Campos J., Walter T.S., Skelly D., Johnson S.A., Ritter T.G., Mason C., Clemens S.A.Costa, Naveca F.Gomes, Nascimento V., Nascimento F., da Costa C.Fernandes, Resende P.C., Pauvolid-Correa A., Siqueira M.M., Dold C., Temperton N., Dong T., Pollard A.J., Knight J.C., Crook D., Lambe T., Clutterbuck E., Bibi S., Flaxman A., Bittaye M., Belij-Rammerstorfer S., Gilbert S.C., Malik T., Carroll M.W., Klenerman P., Barnes E., Dunachie S.J., Baillie V., Serafin N., Ditse Z., Silva K.Da, Paterson N.G., Williams M.A., Hall D.R., Madhi S., Nunes M.C., Goulder P., Fry E.E., Mongkolsapaya J., Ren J., Stuart D.I., Screaton G.R. Reduced neutralization of SARS-CoV-2 B.1.617 by vaccine and convalescent serum. Cell. 2021;184:4220–4236. doi: 10.1016/j.cell.2021.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hu J., He C.L., Gao Q.Z., Zhang G.J., Cao X.X., Long Q.X., Deng H.-J., Huang L.-Y., Chen J., Wang K., Tang N., Huang A.-L. bioRxiv; 2020. The D614G Mutation of SARS-CoV-2 Spike Protein Enhances Viral Infectivity and Decreases Neutralization Sensitivity to Individual Convalescent Sera. preprint. [DOI] [Google Scholar]
- 56.Gupta T., Gupta S.K. Potential adjuvants for the development of a SARS-CoV-2 vaccine based on experimental results from similar coronaviruses. Int. Immunopharmacol. 2020;86 doi: 10.1016/j.intimp.2020.106717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tseng C.T., Sbrana E., Iwata-Yoshikawa N., Newman P.C., Garron T., Atmar R.L., Peters C.J., Couch R.B. Immunization with SARS coronavirus vaccines leads to pulmonary immunopathology on challenge with the SARS virus. PLoS One. 2012;7 doi: 10.1371/journal.pone.0035421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bolles M., Deming D., Long K., Agnihothram S., Whitmore A., Ferris M., Funkhouser W., Gralinski L., Totura A., Heise M., Baric R.S. A double-inactivated severe acute respiratory syndrome coronavirus vaccine provides incomplete protection in mice and induces increased eosinophilic proinflammatory pulmonary response upon challenge. J. Virol. 2011;85:12201–12215. doi: 10.1128/jvi.06048-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Agrawal A.S., Tao X., Algaissi A., Garron T., Narayanan K., Peng B.H., Couch R.B., Tseng C.-T.K. Immunization with inactivated Middle East respiratory syndrome coronavirus vaccine leads to lung immunopathology on challenge with live virus. Hum. Vaccin. Immunother. 2016;12:2351–2356. doi: 10.1080/21645515.2016.1177688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kim H.W., Canchola J.G., Brandt C.D., Pyles G., Chanock R.M., Jensen K., Parrott R.H. Respiratory syncytial virus disease in infants despite prior administration of antigenic inactivated vaccine. Am. J. Epidemiol. 1969;89:422–434. doi: 10.1093/oxfordjournals.aje.a120955. [DOI] [PubMed] [Google Scholar]
- 61.Fulginiti V.A., Eller J.J., Downie A.W., Kempe C.H. Altered reactivity to measles virus. Atypical measles in children previously immunized with inactivated measles virus vaccines. JAMA. 1967;202:1075–1080. doi: 10.1001/jama.202.12.1075. [DOI] [PubMed] [Google Scholar]
- 62.Sekine T., Perez-Potti A., Rivera-Ballesteros O., Strålin K., Gorin J.-B., Olsson A., Llewellyn-Lacey S., Kamal H., Bogdanovic G., Muschiol S., Wullimann D.J., Kammann T., Emgård J., Parrot T., Folkesson E., Rooyackers O., Eriksson L.I., Henter J.-I., Sönnerborg A., Allander T., Albert J., Nielsen M., Klingström J., Gredmark-Russ S., Björkström N.K., Sandberg J.K., Price D.A., Ljunggren H.-G., Aleman S., Buggert M. Robust T cell immunity in convalescent individuals with asymptomatic or mild COVID-19. Cell. 2020;183:158–168. doi: 10.1016/j.cell.2020.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Le Bert N., Tan A.T., Kunasegaran K., Tham C.Y.L., Hafezi M., Chia A., Chng M.H.Y., Lin M., Tan N., Linster M., Chia W.N., Chen M.I.-C., Wang L.-F., Ooi E.E., Kalimuddin S., Tambyah P.A., Low J.G.-H., Tan Y.-J., Bertoletti A. SARS-CoV-2-specific T cell immunity in cases of COVID-19 and SARS, and uninfected controls. Nature. 2020;584:457–462. doi: 10.1038/s41586-020-2550-z. [DOI] [PubMed] [Google Scholar]
- 64.Ren W., Sun H., Gao G.F., Chen J., Sun S., Zhao R., Gao G., Hu Y., Zhao G., Chen Y., Jin X., Fang F., Chen J., Wang Q., Gong S., Gao W., Sun Y., Su J., He A., Cheng X., Li M., Xia C., Li M., Sun L. Recombinant SARS-CoV-2 spike S1-fc fusion protein induced high levels of neutralizing responses in nonhuman primates. Vaccine. 2020;38:5653–5658. doi: 10.1016/j.vaccine.2020.06.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Haun B.K., Lai C.Y., Williams C.A., Wong T.A.S., Lieberman M.M., Pessaint L., Andersen H., Lehrer A.T. CoVaccine HT™ adjuvant potentiates robust immune responses to recombinant SARS-CoV-2 spike S1 immunization. Front. Immunol. 2020;11 doi: 10.3389/fimmu.2020.599587. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material