Integrating somatic mutation analysis and gene expression profiling distinguishes pediatric AML subtypes with differential prognoses and clinical risks.
Abstract
Genomic characterization of pediatric patients with acute myeloid leukemia (AML) has led to the discovery of somatic mutations with prognostic implications. Although gene-expression profiling can differentiate subsets of pediatric AML, its clinical utility in risk stratification remains limited. Here, we evaluate gene expression, pathogenic somatic mutations, and outcome in a cohort of 435 pediatric patients with a spectrum of pediatric myeloid-related acute leukemias for biological subtype discovery. This analysis revealed 63 patients with varying immunophenotypes that span a T-lineage and myeloid continuum designated as acute myeloid/T-lymphoblastic leukemia (AMTL). Within AMTL, two patient subgroups distinguished by FLT3-ITD and PRC2 mutations have different outcomes, demonstrating the impact of mutational composition on survival. Across the cohort, variability in outcomes of patients within isomutational subsets is influenced by transcriptional identity and the presence of a stem cell–like gene-expression signature. Integration of gene expression and somatic mutations leads to improved risk stratification.
Significance:
Immunophenotype and somatic mutations play a significant role in treatment approach and risk stratification of acute leukemia. We conducted an integrated genomic analysis of pediatric myeloid malignancies and found that a combination of genetic and transcriptional readouts was superior to immunophenotype and genomic mutations in identifying biological subtypes and predicting outcomes.
This article is highlighted in the In This Issue feature, p. 549
Introduction
Acute myeloid leukemia (AML) comprises a heterogeneous group of malignancies that are linked by the presence of blasts displaying morphologic and immunophenotypic features of myeloid cell differentiation. These characteristics served as the initial approach to subdivide AML into distinct clinical entities (1). Morphology and immunophenotype, however, are limited in biological, prognostic, and therapeutic significance. The identification of cytogenetic alterations and molecular lesions has allowed newer classification schemes to be developed with the most recent widely used approach being the World Health Organization classification of AML (2). Although the latter classification scheme divides AML into many distinct clinical, morphologic, and/or molecular subtypes, from a clinical perspective most current therapeutic pediatric protocols stratify patients into favorable, intermediate, and poor prognostic groups (3). Therapy in these groups is based on the relative risk of relapse, with poor prognostic groups proceeding to allogeneic hematopoietic stem cell (HSC) transplantation in first remission when a suitable donor is available.
With the development of genome-wide gene-expression profiling, array-based comparative genomic hybridization methodologies, and next-generation sequencing technologies, the field has gained a greater understanding of the molecular features involved in the occurrence of pediatric myeloid malignancies. Several pathologic lesions have been found to have prognostic implications contributing to a continuous refinement of risk stratification over time in the context of modern therapy. We previously applied an integrated analysis to a large cohort of pediatric acute megakaryoblastic leukemia (AMKL) that underwent next-generation sequencing with the goal of identifying biologically and clinically relevant subtypes so that we could gain a greater understanding of the biology of the disease as well as inform clinical decision-making (4). In that study, using gene-expression profiling coupled with somatic variants and outcome data, we were able to identify distinct molecular subtypes with varying outcomes. These results led to a recommendation to limit high-risk designation to a subset of patients, which has already been instituted in the ongoing multi-institutional AML16 trial for newly diagnosed pediatric patients with AML (NCT03164057) and several other collaborative group protocols. Here we apply a similar approach to a cohort of 435 pediatric patients with a spectrum of myeloid-related malignancies to provide a comprehensive view of this clinical entity and propose a refined classification scheme with clinical utility. Using this approach, we identify a previously undescribed subtype that spans a T-lineage and myeloid continuum, as well as new prognostic mutational events within previously described subtypes. Further, we demonstrate that mutational events, transcriptional profile, and evidence of a primitive hematopoietic progenitor gene-expression signature all associate independently with outcome. The most significant association occurs when all three of these factors are combined, arguing in favor of subgroup classification by comprehensive molecular profiling to optimize risk stratification in pediatric AML.
Results
Genomic Landscape of Normal and Complex Karyotype Pediatric AML
The Children's Oncology Group (COG)–NCI TARGET AML initiative molecularly characterized 993 pediatric AML cases, including 197 specimens that underwent comprehensive whole-genome sequencing (WGS; ref. 5). Of these, 94 carried one of three oncogenic fusions known to be strong drivers of leukemogenesis: RUNX1–RUNX1T1, CBFB–MYH11, and KMT2A rearrangements (KMT2Ar). Among all other somatic alterations detected, only 10 occurred in more than 5% of subjects, all of which had been described previously. This suggested that low-frequency molecular subsets may exist that require larger cohorts to fully elucidate. To address this limitation, we selected 122 pediatric AML normal, noncomplex, and complex karyotype specimens from five cooperative study groups (SJCRH, DCOG, NOPHO, AIEOP, and BFM) that lacked RUNX1–RUNX1T1, CBFB–MYH11, and KMT2Ar by clinical testing for WGS and/or whole-exome sequencing (WES) and RNA sequencing (RNA-seq) to enrich for cases that carry low-frequency events (Supplementary Tables S1 and S2; Fig. 1A). Structural variations (SV), copy-number alterations (CNA), single-nucleotide variations (SNV), and indels were determined by our established pipelines, as well as an evaluation for regulatory rearrangements driving oncogene overexpression through enhancer hijacking (Supplementary Tables S3–S9 and Supplementary Figs. S1 and S2; ref. 6). When considering exonic SNV/indel, CNA, and SV calls, mutational burden ranged from 1 to 101 somatic events, including a case with TP53-associated chromothripsis that carried 89 lesions in total (Supplementary Table S9; Supplementary Figs. S1 and S3). In addition to known AML somatic mutations in genes such as CEBPA, GATA2, NPM1, WT1, FLT3, NRAS, KRAS, ETV6, RAD21, SMC1A, STAG1, STAG2, STAG3, SMC3, and rearrangements in NUP98 and KAT6A, we identified rare events in known oncogenic drivers. These include internal tandem duplications (ITD) in GATA2, RUNX1, and CEBPA, as well as the repositioning of a distal ZEB2 enhancer, MYC enhancer, or ETV6 enhancer to ectopically activate BCL11B, MECOM, and MNX1 loci, respectively (Supplementary Table S6; Supplementary Figs. S4 and S5). Interestingly, 15 AML cases (12.3%) carrying loss-of-function mutations in polycomb repressive complex 2 (PRC2) genes were found to resemble an early T-cell precursor acute lymphoblastic leukemia (ETP-ALL) gene-expression profile (GEP) by gene set enrichment analysis (GSEA; Supplementary Fig. S6). ETP-ALL exhibits aberrant expression of stem cell and myeloid markers and has been shown to have a GEP consistent with transformation of a stem cell progenitor (7, 8). Further, mixed phenotype acute leukemias (MPAL) with T and myeloid lineage characteristics have previously been suggested to be in this spectrum of immature leukemias (9). We therefore hypothesized that these PRC2-mutated AML cases represented the myeloid end of this continuum. To provide global transcriptional context to these ETP-like AMLs and evaluate a comprehensive cohort encompassing a range of pediatric myeloid malignancies, we integrated results from previously published AML (N = 169), MPAL (N = 80), AMKL (N = 45), and ETP-ALL (N = 19) data sets that had RNA-seq and either WES or WGS available for a total of 435 cases (Supplementary Table S10 and Fig. 1A; refs. 4,5,7,8,9).
Figure 1.
Transcriptional identities correlate with key oncogenic driver events and are agnostic of immunophenotype. A, Study design. 122 normal, noncomplex, and complex karyotype pediatric specimens were selected. Exclusion criteria for sequencing include FAB M3 (acute promyelocytic leukemia, APML), FAB M7 (AMKL), core binding factor (CBF) leukemia (RUNX1–RUNX1T1, CBFB–MYH11), and KMT2Ar cases. Cases underwent WGS, WES, and RNA-seq. Data were combined with four other pediatric data sets, including FAB M7, early T-cell precursor acute lymphoblastic leukemia, the TARGET AML data set, and pediatric mixed phenotype acute leukemia for a total of 435 cases (4,5,7,8,9). Ten additional KMT2Ar AML cases were sequenced to increase the cohort size. Transcriptional clusters as identified by t-SNE, somatic calls, and outcome correlates were utilized to identify biological subtypes as previously described (4). NGS, next-generation sequencing. B, RNA-seq of 435 cases of pediatric AML, AMKL, MPAL, and ETP were combined and batch corrected. t-SNE visualization utilizing the top 100 differentially expressed genes within each data set. Immunophenotype of cases as determined by flow cytometry at diagnosis is shown. AMTL, acute myeloid/T-lymphoblastic leukemia; AUL, acute undifferentiated leukemia; B/M, B-lymphoid and myeloid coexpression; MK, mixed karyotype; T/B, T-lymphoid and B-lymphoid coexpression; T/B/M, T-lymphoid, B-lymphoid, and myeloid coexpression; T/M, T-lymphoid and myeloid coexpression. C, Key oncogenic driver mutations as determined by next-generation sequencing. Ph-like, Philadelphia chromosome–like acute lymphoblastic leukemia; PTD, partial tandem duplication; Txn, transcription.
Figure 2.
Mutational composition of pediatric myeloid malignancies. Waterfall plot of major mutational events across the entire cohort. See Supplementary Table S12 for genomic details and Supplementary Table S13 for genes and fusion events included in each of the groupings on the y-axis. AMP, amplification; AUL, acute undifferentiated leukemia; DEL, deletion; IP, immunophenotype; LOH, loss of heterozygosity; Ph-like, Philadelphia chromosome–like acute lymphoblastic leukemia; PTD, partial tandem duplication; Txn, transcription.
Molecular Classifier of Pediatric Myeloid Malignancies Agnostic of Immunophenotype
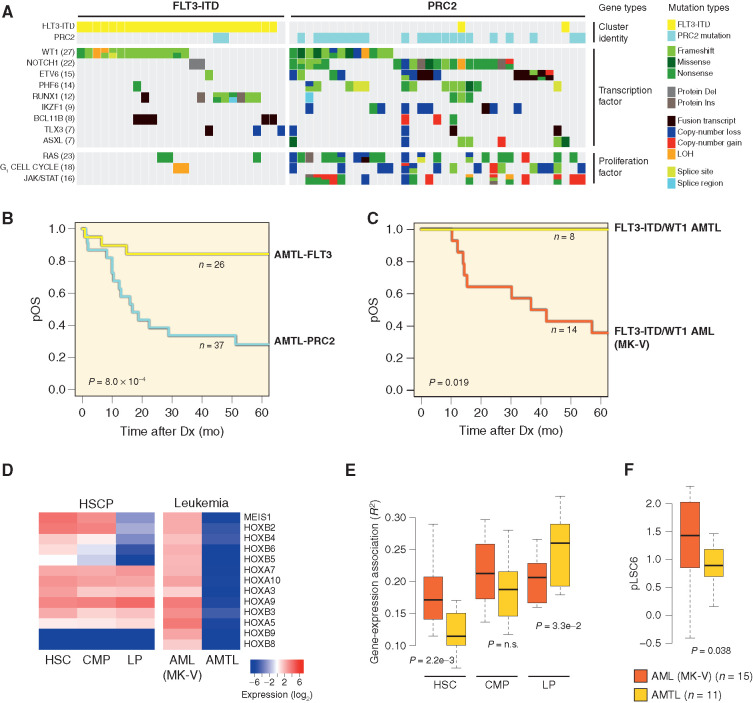
T-distributed Stochastic Neighbor Embedding (t-SNE) visualization using a 381-gene list derived from the top 100 most variably expressed transcripts within each of the five sequencing data sets revealed a clear molecular classifier, identifying groups that had consistent mutational compositions but were agnostic of immunophenotype (Figs. 1B and C and 2; Supplementary Tables S10–S13; Supplementary Fig. S7). A bootstrap hierarchical clustering procedure defined subgroups with an overall reproducibility of 97.4% and highly concordant with the t-SNE transcriptional subgroups (adjusted Rand index = 0.72; Supplementary Table S14), indicating the subgroups identified by t-SNE are statistically meaningful. This classifier allowed the distinction of 63 cases with an ETP-ALL GEP comprising a mixture of AML (N = 12/63, 19%), acute undifferentiated leukemia (AUL; N = 1/63, 1.6%), MPAL (N = 31/63, 49.2%), and ETP-ALL (N = 19/63, 30.2%) leukemias (bootstrap reproducibility = 93.6%; Fig. 1B). All but one MPAL case within this subgroup coexpressed T-lineage antigens in addition to either myeloid and/or B-lineage antigens (Fig. 1B; Supplementary Table S10). Expression of MPO and CD3E confirmed that the reported immunophenotypes of these cases were correct (Supplementary Fig. S8). A separate validation cohort of 399 pediatric AML cases with microarray data confirmed the presence of this entity with 23 cases identified (Supplementary Fig. S9; Supplementary Tables S15 and S16). A five-gene classifier consisting of CD3G, COCH, SLC35D2, SPTLC3, and TOR4A was able to predict these cases in both the discovery and validation cohorts (AUC 0.977 and 0.88, respectively). A molecularly distinct subtype of acute leukemia termed acute myeloid/T-lymphoblastic leukemia (AMTL), with shared myeloid and T-lineage features, has previously been proposed by Gutierrez and Kentsis (10). In support of this entity, they noted shared gene mutations in prior sequencing reports of T-lineage and AML studies, including WT1, PHF6, RUNX1, and BCL11B. Consistent with this, transcriptionally defined AMTL cases in our discovery cohort carried mutations in these genes and were found to fall into one of two subgroups: a group characterized by FLT3-ITD (N = 26/63, 41.3%) and a second group enriched for loss-of-function alterations in one of three core PRC2 complex genes, including EZH2, SUZ12, and EED, or a splicing factor mutation that leads to inclusion of a cryptic exon resulting in truncated EZH2 transcripts predicted to undergo nonsense mediated decay (N = 37/63, 58.7%; Fig. 3A; Supplementary Fig. S10; ref. 11). Both subsets were found to carry cooperating events in transcription factors (WT1, NOTCH1, ETV6, PHF6, RUNX1, IKZF1, BCL11B TLX3); unique to PRC2 cases were activating events in RAS (NRAS, KRAS, NF1) and JAK/STAT (JAK1, JAK3, IL7R, SH2B3) signaling cascades, as well as loss-of-function mutations in genes that play a role in G1 checkpoint arrest (RB1, CCDN3, CDKN1B, and CDKN2A/B; Fig. 3A). In particular, network analyses identified a strong association between transcription factors associated with T-lineage differentiation (NOTCH1, PHF6, BCL11B, TLX3, TAL1, and IKZF2), PRC2 loss-of-function mutations, and JAK/STAT pathway alterations, whereas FLT3-ITD cases were enriched for RUNX1 and WT1 transcription factors (Supplementary Fig. S11; ref. 12). A comparison of overall survival clearly demonstrated that outcomes of the isotranscriptional AMTL subset are influenced by the mutational spectrum. Irrespective of whether the patient received AML, ALL, or a hybrid treatment approach, FLT3-ITD–positive AMTL cases were associated with a favorable outcome, whereas those with PRC2 mutations had a poor prognosis (P = 8 × 10−4; Fig. 3B; Supplementary Table S10). Consistent with this, AMTL cases in our AML validation cohort for which mutational data were available (N = 16/23, 69.6%) were similarly composed of FLT3-ITD–positive (N = 8/16, 50%) and FLT3-ITD–negative cases (N = 8/16, 50%); a subset of the negative cases (N = 3/8) had copy-number data available that confirmed deletional events in PRC2 genes in all three cases and an association with poor outcomes (P = 0.01; Supplementary Fig. S12; Supplementary Table S16). PRC2 loss-of-function mutations were also present in a subset of core binding factor cases (N = 8/61, 13.1%; Supplementary Table S12). To determine if the presence of PRC2 mutations confers a poor prognosis in these patients as well, we evaluated outcomes in pediatric core binding factor AML cases from two previously published cohorts and found an inferior event-free survival in patients carrying both KIT activating mutations and PRC2 loss-of-function mutations (N = 5/142, 3.5%; P = 0.026; Supplementary Table S17; Supplementary Fig. S13; refs. 5, 13). In alignment with these data, prior studies have shown chemoresistance as a result of PRC2 loss in AML and T-lineage ALL models (14, 15).
Figure 3.
Genomic and transcriptional features of AMTL. Sixty-three cases spanning AML, AUL, MPAL, and ETP immunophenotypes shared a common transcriptional identity (see Fig. 1B). A, Mutational spectrum of AMTL cases. Del, deletion; Ins, insertion; LOH, loss of heterozygosity. B, Outcomes of patients with AMTL according to FLT3-ITD and PRC2 transcriptional identity. Dx, diagnosis; pOS, probability overall survival. C, Outcomes of FLT3-ITD/WT1 double-mutant cases based on AMTL and MK-V transcriptional identity (see Fig. 1). D, Expression of HOX locus genes in normal hematopoietic progenitor subsets and FLT3-ITD/WT1 cases from AMTL and MK-V transcriptional clusters. CMP, common myeloid progenitor; HSCP, HSC progenitor; LP, lymphoid-restricted progenitor. E, Enrichment of gene-expression signatures from HSC, CMP, and LP in FLT3-ITD/WT1 cases from AMTL and MK-V transcriptional clusters. n.s., not significant. F, pLSC6 score of FLT3-ITD/WT1 cases from AMTL and MK-V transcriptional clusters.
Outcomes of Isomutational Subsets Are Influenced by Transcriptional Identity
The favorable prognosis of AMTL cases carrying FLT3-ITD included those with cooperating WT1 mutations, several of which were classified as AML by immunophenotype (N = 10/26; 38.5% of FLT3-ITD AMTL cases carried WT1 mutations, two of which were AML). Historically, pediatric patients with AML with FLT3-ITD and a WT1 mutation have been reported to have a dismal prognosis (16). A significant number of these FLT3-ITD/WT1 double-mutant cases were also found to associate within a different transcriptional cluster, AML MK-V (N = 14/25, 56% in MK-V; N = 10/25, 40% in AMTL; N = 1/25, 4% in MK-IV; Fig. 1B and C). In contrast to AMTL, FLT3-ITD/WT1 double-mutant patients who fell into AML MK-V transcriptional cluster had an extremely poor outcome consistent with prior reports (Fig. 3C). Thus, the presence of these somatic events alone is insufficient to distinguish high-risk status. A comparison of differentially expressed genes between AMTL FLT3-ITD/WT1 and AML MK-V FLT3-ITD/WT1 identified significant upregulation of genes within the HOX locus in AML MK-V cases (Fig. 3D). Although the mutational spectrum is known to influence the transcriptional profile of leukemia, the cell that acquires the mutations (“cell of origin”) may also be reflected. To look at this further, we evaluated expression of the HOX locus in a normal hematopoietic progenitor data set and found elevated expression of the HOX genes upregulated in our AML MK-V FLT3-ITD/WT1 patients in both HSC and common myeloid progenitor (CMP) compartments compared with lymphoid progenitors (LP; Fig. 3D), suggesting that the differential HOX expression between the two subsets may reflect a stem cell–like state (17).
We, therefore, identified gene-expression signatures for the different hematopoietic subsets and looked for enrichment of those signatures in our two subsets of FLT3-ITD/WT1 patients to determine whether the correlation of stem cell–associated genes extended beyond the HOX locus. This analysis confirmed an enrichment in our AML MK-V cluster cases for HSC as well as CMP signatures in contrast to AMTL cases that have a greater enrichment for LP signatures (Fig. 3E). We hypothesize that this reflects a more primitive cell of transformation in AML MK-V FLT3-ITD/WT1 cases that retain a stem cell progenitor–like state contributing to chemotherapy resistance. To assess whether this phenomenon is restricted to FLT3-ITD/WT1 genotypes, we applied the same analysis to KMT2Ar cases that fell into AML MK-V and the 11q23-rearranged transcriptional cluster (Fig. 1C; Supplementary Fig. S9). Consistent with the inferior event-free survival (EFS) of AML MK-V KMT2Ar cases compared with those in the 11q23-rearranged transcriptional cluster in both discovery and validation cohorts, we found a more pronounced enrichment for HSC and CMP signatures in AML MK-V KMT2Ar, suggesting a more primitive stem cell–like state (Supplementary Figs. S14–S16).
Leukemia Stemness Is Unevenly Distributed across Myeloid Leukemias
Ng and colleagues previously developed a 17-gene transcriptional score related to stemness, derived from functionally defined leukemia stem cells of adult patients with AML, which was predictive of prognosis (LSC17; ref. 18). More recently, a six-gene LSC score has been developed with significant prognostic value in pediatric AML (pLSC6; ref. 19). To determine if the more primitive nature of AML MK-V FLT3-ITD/WT1 cases was reflected in this score, we compared pLSC6 in AML MK-V and AMTL FLT3-ITD/WT1 patients (Fig. 3F). Consistent with enrichment of more primitive hematopoietic progenitor gene-expression signatures, AML MK-V FLT3-ITD/WT1 patients had a higher pLSC6 score (P = 0.038). To evaluate this more comprehensively across the cohort, we determined the pLSC6 score in normal hematopoietic progenitor subsets to define thresholds of low (lineage-committed cells), intermediate (multipotent progenitors), and high (pluripotent progenitors; Fig. 4A and B) values. Imposing these thresholds on our cohort, we identified a subset of patients with intermediate and high scores, which was significantly associated with an inferior overall survival (N = 302/435, 69.4% low pLSC6; N = 119/435, 27.4% intermediate pLSC6; N = 14/435, 3.2% high pLSC6; P = 9.3 × 10−7 discovery cohort; and N = 262/399, 65.7% low pLSC6; N = 124/399, 31.1% intermediate pLSC6; N = 13/399, 3.2% high pLSC6; P = 2.1 × 10−6 validation cohort; Fig. 4C; Supplementary Fig. S17). Although several subsets had uniform pLSC6 scores, such as CBFA2T3–GLIS2-, RUNX1–RUNX1T1-, CBFB–MYH11-, and MNX1-rearranged cases, other subsets had variable scores demonstrating heterogeneity in leukemia “stemness” (e.g., KMT2Ar cases), highlighting pLSC6 as an independent variable in addition to mutational type and overall transcriptional signature (Fig. 4D; Supplementary Table S17; Supplementary Figs. S18–S20).
Figure 4.
Leukemia stemness is associated with overall survival. A, pLSC6 score was determined in normal hematopoietic progenitor subsets as previously described (17, 19). DC, dendritic cell; E, erythrocyte; G, granulocyte; GMP, granulocyte/macrophage progenitor; MEG, megakaryocyte; MEP, megakaryocyte/erythrocyte progenitor; NK, natural killer cell. B, pLSC6 scores from normal hematopoietic progenitors were used to define thresholds of low (lineage-committed cells), intermediate (multipotent progenitors), and high (pluripotent progenitors) values in our cohort. C, Imposing pLSC6 thresholds on our cohort found a subset of patients with intermediate and high scores that were significantly associated with overall survival (P = 9.3 × 10−7). Dx, diagnosis; pOS, probability overall survival. D, t-SNE visualization of the cohort with pLSC6 levels indicated. Thresholds of low, medium, and high as determined in A. B/M, B-lymphoid and myeloid coexpression; Ph-like, Philadelphia chromosome–like acute lymphoblastic leukemia; PTD, partial tandem duplication; Txn, transcription.
Transcriptional Identity, Mutations, and Stemness All Contribute to Outcome
To evaluate the relative contribution of each of the factors identified in our study to carry an association with survival, we utilized a Cox proportional hazards model to look at associations with overall survival. Transcriptional identity, oncogenic drivers, and leukemia stemness were all independently found to associate with outcome (Figs. 4C and 5A; Supplementary Tables S18 and S19; Supplementary Figs. S17, S19, and S20). The greatest association occurred when all three of these factors were combined (P = 1.06 × 10−12 discovery cohort and P = 1.19 × 10−7 validation cohort). The impact of individual factors on outcome associations was variable in our discovery cohort, with CBFA2T3–GLIS2, ETS family rearrangements (FUS–ERG, EWSR1–ERG, FUS–FEV, FUS–FLI1, MN1–FLI1, and EWSR1–FEV), and high pLSC6 score having the greatest negative association with outcome, whereas CEBPA mutations (mono- and biallelic) and low pLSC6 carried the greatest positive association with outcome (Supplementary Tables S20 and S21; Supplementary Fig. S21). Within biological subgroups identified in pediatric AML, certain factors carried greater weight than others (Table 1). Utilizing these rules for risk stratification, we compared outcomes in our discovery and validation cohorts for our proposed genomic classification (low, intermediate, and high risk) to those of the ongoing multi-institutional AML16 prospective clinical trial for newly diagnosed pediatric patients with AML (NCT03164057; Fig. 5B and C validation cohort; Supplementary Figs. S22–S25 discovery and combined cohorts; Supplementary Tables S16, S22, and S23). For a given risk classification, we defined and computed the risk classification utility (RCU), which considers estimate outcomes for each risk group (outcome discrimination index) and the proportion of patients designated as high or low risk given that intermediate risk designates a patient lacking definitive high-risk or low-risk characteristics, and thus represents a patient whose status is unknown (Supplementary Table S24). A bootstrap procedure was then used to quantify the statistical variability and significance of comparisons of the RCU with the two classification schemes (Supplementary Table S25). In both the discovery and validation cohorts as well as in a combined analysis, our proposed classification was found to have a statistically significant greater RCU for EFS than AML16 (P = 0.036 discovery cohort, P = 0.018 validation cohort, and P = 0.036 combined cohorts; Fig. 5C; Supplementary Fig. S25). In particular, the proposed classification was superior at identifying high-risk patients within the intermediate- and low-risk groups, resulting in a lower proportion of intermediate-risk patients who had an improved EFS, which brings the proposed stratification closer to the ideal state—one in which there are only two risk groups: Patients who have an event (high risk) and those who do not (low risk; Fig. 5B).
Figure 5.
Oncogenic driver events, transcriptional identity, and leukemia stemness all contribute to outcome in pediatric myeloid-related acute leukemias. A, Integrative Cox proportional hazards model to look at associations with overall survival in the discovery cohort (38). Each bar represents the −log10 P value of covariates and their association with survival. The covariates used in the model to calculate the P value are indicated below the graph with a check mark. Immunophenotype as a single covariate failed to reach statistical significance. B, Probability of EFS (pEFS) of an ongoing multi-institutional prospective pediatric AML trial (AML16) and the proposed classification scheme based on this article for the validation cohort. See Supplementary Figs. S22 and S23 for results of each independent cohort. C, Performance of the proposed genomic classification relative to that utilized in an ongoing prospective upfront pediatric AML study (NCT03164057) in terms of discrimination capability (left) and percentage of high-risk or low-risk classified patients (right) culminating in a risk classification utility score (top, right) for the validation cohort. See Supplementary Figs. S24 and S25 for results of each independent cohort. D, Working model. Mutational events in distinct hematopoietic progenitor subsets lead to transformation, and both components contribute to the transcriptional identity and leukemia stemness. Chemotherapy sensitivity and therefore outcomes are a composite of these factors.
Table 1.
Biological subtypes identified in pediatric AML cases
| Subtype | Immunophenotypes across the entire cohort (N)a | Proposed risk status based on overall survival (reference)b,c |
|---|---|---|
| AMTL | AML (12), MPAL (30), AUL (1), ETP (19) |
|
| CEBPA (mono-and biallelic) | AML (28), MPAL (3) |
|
| RUNX1–RUNX1T1 | AML (27) |
|
| CBFB–MYH11 | AML (34) |
|
| MNX1-r | AML (3), AMKLd, AUL (1) |
|
| ETS-r | AML (10), AMKL (1) MPAL (2) |
|
| CBFA2T3–GLIS2 | AML (2), AMKL (11) | |
| KMT2A-r | AML (56), AMKL (10), MPAL (9), AUL (2), ETP (1) | |
| GATA1 | AML (3), AMKL (6), MPAL (1) |
|
| HOX-r | AML (2), AMKL (13) |
|
| NUP98-r | AML (17), AMKL (6), ETP (3) | |
| NPM1 | AML (25) | |
| DEK–NUP214 | AML (5) |
|
| FLT3-ITDe | AML (28), MPAL (18), ETP (6) |
|
| AML other | AML (39), AMKL (6), MPAL (24) |
|
aNumbers in parentheses indicate the number of cases across the discovery cohort with indicated immunophenotype. Genomic subtypes not identified in AML cases are not included in this table.
bOutcomes approaching 80% overall survival or greater are designated as low risk and survival less than 40% are designated as high risk. Literature support of previously described subtypes and risk status is indicated in parentheses.
cMinimal residual disease is considered an independent risk factor, and residual levels of disease following induction chemotherapy warrant escalation of risk status.
dReported in the literature (52).
eFLT3-ITD cases that are not included in the other subtypes (see Supplementary Fig. S26).
Discussion
Gene expression, genomic classification, and leukemia stemness have all been shown to affect prognosis to varying extents in both adult and pediatric AML (5,18,19,20,21,22,23). However, few studies to date and none in pediatric AML integrate all three of these aspects to determine the relative contribution to outcomes. Through this comprehensive approach and by including pediatric acute leukemias with myeloid characteristics, we were able to identify a previously undescribed subtype, AMTL, which spans a T-lineage and myeloid continuum as well as new prognostic mutational events within previously described subtypes, such as PRC2 mutations in core binding factor leukemias. Recently, two groups have reported on acute leukemias with T-lineage markers such as cytoplasmic CD3 and/or CD2 that carry BCL11B enhancer hijacking events similar to several cases within the AMTL subgroup (24, 25). Unique to our study is the identification of AMTL cases that are devoid of T-lineage markers by flow cytometry and the distinction of the two subsets within AMTL that have differing outcomes. It has been shown through murine modeling that T-LP retain a broad lineage potential when transformed with oncogenes and specifically have the ability to differentiate into myeloid leukemia while retaining a lymphoid epigenetic memory, consistent with our findings (26). In this study by Riemke and colleagues, a cohort of adult patients with AML was found to resemble the murine T-LP–derived myeloid leukemias by gene expression. This population, however, had a negative association with ETP-ALL by GSEA, and the mutation profile of these patients was predominated by mutations not found in pediatric AMTL, including NPM1, IDH2, and DNMT3A. This difference may be a result of distinct oncogenic events that are acquired by a T-LP as opposed to a difference in the cell of origin (Fig. 5D).
The existence of patients with FLT3-ITD/WT1 in AMTL that had superior outcomes, in contrast with previously published results, led us to compare outcomes of these patients across transcriptional subsets. The inferior overall survival of FLT3-ITD/WT1 double-mutant patients was restricted to those within the MK-V cluster. Of note, the vast majority of patients within this study were treated prior to the implementation of FLT3 inhibitors (2/291 patients with AML in the discovery cohort for whom treatment details were known received an FLT3 inhibitor at diagnosis, both of whom had events and are deceased; Supplementary Table S10). Although we cannot determine whether FLT3 inhibition would improve outcomes of MK-V FLT3-ITD/WT1 patients in our study, results from COG AAML1031 suggest that this targeted treatment approach can improve outcome in FLT3-ITD/WT1 patients with the caveat that the transcriptional identity in this study is unknown (4). The absence of FLT3 inhibition in our cohort allowed us to identify isomutational groups where disease outcome clearly associates with transcriptional identity and isotranscriptional groups where outcome clearly associates with mutational status. This finding has broad implications on variant interpretation in the era of precision medicine, as the impact on prognosis is not limited to the presence or absence of a given mutation. Furthermore, the incorporation of stem cell–associated signatures also allowed us to distinguish patients who have the same genomic classification but differing outcomes (Table 1). The highest power and outcome associations occur when all three of these factors are combined, arguing in favor of comprehensive diagnostics to optimize risk stratification in pediatric AML. A multivariate analysis to evaluate the prognostic informativeness of WT1 and FLT3-ITD mutational events after considering transcriptional identity, key driver mutation, and pLSC6 score supports this conclusion: Neither EFS nor overall survival was significantly associated with the presence of FLT3-ITD, a WT1 alteration, or the combination of these two after adjustment for pLSC6 score as a numeric predictor, transcriptional identity as a stratification factor, or driver mutation as a stratification factor (Supplementary Fig. S26; Supplementary Table S26). Further, neither EFS nor OS was significantly associated with FLT3-ITD, WT1, or the presence of both in models that considered only these variables as predictors (Supplementary Table S26).
The benefit of risk-adapted indications for HSC transplantation in pediatric AML has recently been shown by the BFM study group, with significantly higher EFS and higher rates of HSC transplants through improvements in genetic risk stratification (27). In a disease entity where the chemotherapy approach has remained largely unchanged over time with a limited number of novel therapeutic agents on the horizon, risk stratification, refined allograft indications, and supportive care continue to be major factors that have led to the improvement in outcome over time (28). It is, therefore, imperative that risk stratification be optimized to the maximum extent to cure more pediatric patients with AML. The vast majority of pathogenic calls and transcriptional information necessary to use our integrated approach can be obtained from paired WES and RNA-seq, which has been increasingly adopted in the clinical setting, arguing in favor of the feasibility of this approach (29,30,31). Targeted capture panels that detect SNV/indels and copy-number changes in combination with fusion detection assays are less comprehensive but also able to detect the vast majority of oncogenic lesions described in this study. In pediatric AML, all patients enrolled on the St. Jude AML16 study are already receiving Clinical Laboratory Improvement Amendments–certified WGS, WES, and RNA-seq on diagnostic blasts. Although next-generation sequencing approaches are becoming increasingly standardized and prevalent in the field, bioinformatic analyses and interpretation of mutational impact within a case based on transcriptional identity and leukemia stemness will require additional expertise to implement. To enhance the clinical applicability of this study, we developed a panel of five genes whose expression can distinguish AMTL cases that can be combined with the previously developed six-gene pLSC6 classifier—key determinants in our risk stratification model. In combination with key mutational events, this allows one to follow a hierarchical decision-making tree to stratify a patient (Fig. 6).
Figure 6.
Hierarchical decision-making tree for proposed risk stratification. *, T-MPAL, MPAL with T-lineage markers. MPAL cases coexpressing B-lineage markers contained ZNF384, Ph+, Ph-like, and KMT2Ar oncogenes and should be treated with ALL-directed therapy unless they prove nonresponsive to this approach. **, FLT3-ITD cases that are not AMTL and lack high-risk and low-risk features such as NUP98r, monosomy 7, NPM1, and CEBPA. HR, high risk; IR, intermediate risk; LR, low risk.
The cell of origin of leukemia is defined as the normal hematopoietic cell from which the disease develops through the acquisition of mutations. A subset of cells termed “leukemia stem cells” are felt to propagate the disease over time, and studies have shown that similar to normal hematopoiesis, a hierarchical structure exists in leukemia, with the most primitive clone being identifiable through functional assays (32). Given the differentiation spectrum seen in leukemias, it can be a challenge to infer the cell of origin in bulk tumor populations. Despite this potential limitation, we found significant enrichment for more primitive progenitor cell signatures in patients with higher LSC6 scores. Our data are consistent with a model whereby a cell of origin acquires oncogenic driver mutations, and these two factors both contribute to the transcriptional identity of the leukemia and the stemness, all of which influence outcome (Fig. 5D).
In summary, comprehensive next-generation sequencing of pediatric AML can be utilized beyond pathogenic mutation calls to optimize risk stratification. Incorporation of transcriptional identity and leukemia stemness in clinical decision-making will further improve the identification of patients who may benefit from stem cell transplant in first remission and those who can be cured with chemotherapy alone.
Methods
Cohort
Specimens sequenced in this study were provided from multiple institutions and collaborative groups. All samples were obtained with patient- or parent/guardian-provided written informed consent under protocols approved by the Institutional Review Board at each institution. Studies were conducted in accordance with the International Ethical Guidelines for Biomedical Research Involving Human Subjects. Samples were de-identified prior to nucleic acid extraction and analysis. WGS, WES, RNA-seq and analysis for SVs, SNVs, indels, and CNA were performed as previously described (4, 7). TARGET AML, ETP, MPAL, and AMKL cohorts have been previously published and were obtained with permission from database of Genotypes and Phenotypes (dbGaP) and/or St. Jude Children's Research Hospital (4,5,7,8,9). Transcript expression levels for gene-expression analyses were estimated from RNA-seq data as fragments per kilobase of transcript per million mapped fragments (FPKM) as previously described (4). Data for samples sequenced in this study have been deposited to the St. Jude Cloud (www.stjude.cloud; ref. 33) and European Genome-phenome Archive (study ID EGAS00001004701).
RNA-seq Read Mapping, Gene-Expression Summary, and Batch Correction
RNA reads were mapped using our StrongARM pipeline, described previously (13). Paired-end reads from RNA-seq were aligned to the following four database files using Burrows–Wheeler alignment: (i) the human GRCh37-lite reference sequence, (ii) RefSeq, (iii) a sequence file representing all possible combinations of nonsequential pairs in RefSeq exons, and (iv) the AceView database flat file downloaded from UCSC, representing transcripts constructed from human expression sequence tags. Additionally, they were mapped to the human GRCh37-lite reference sequence using STAR. The mapping results from the databases (ii–iv) were aligned to human reference genome coordinates. The final BAM file was constructed by selecting the best of the five alignments.
Reads from aligned BAM files were assigned to genes and counted using HTSeq with the GENCODE human release 15-gene annotation (34). The gene count matrix was used to generate an FPKM gene-expression data matrix using gene length information. A gene was called as “expressed” in a given sample if it had an FPKM value ≥0.01 based on the distribution of FPKM gene-expression values, and genes not expressed in any sample were excluded from downstream analysis. The gene-expression data were further quantile normalized using the normalizeBetweenArrays function available from the Limma R package (35). The detected batch effect due to data source of St. Jude versus TARGET was corrected using the ComBat method available from the R package sva (36).
381-gene Classifier
For construction of the 381-gene classifier, the top 100 most variant genes from each of the five data sets (this article, ETP-ALL, MPAL, TARGET AML, and AMKL) were combined using log2-transformed FPKM values and median-adjusted deviation (4,5,7,8,9). This procedure effectively eliminated remaining batch effects (Supplementary Fig. S27). Visualization was performed using t-SNE using a perplexity value of 10 and 10,000 iterations (37). t-SNE coordinates from the run with the lowest final error (out of 10 runs) were selected for further analysis.
HSC Progenitor Gene-Expression Analysis
Single-cell HSC progenitor (HSCP) counts, SPRING plot coordinates, and population assignments were taken from Pellin and colleagues (17). For comparing HSCP and leukemia gene expression, single-cell counts per gene were summed up for each of the 11 different HSCP populations, normalized to the number of cells in each population, and log2 transformed. Resulting gene-expression values were scaled together with log2FPKM expression values of the 435 leukemias using the normalize between arrays function of Limma (method quantile). pLSC6 scores and Spearman correlation coefficients were calculated using these values. For some analyses, multilymphoid progenitors and pre-B/natural killer values were averaged to generate LP values. pLSC6-high, -medium, and -low cutoff values were based on HSCP population values, with the most primitive populations designated as high (populations 1, 2, 3, 7, 9, 10, and 11 from Pellin and colleagues), the more committed populations designated medium (populations 4, 5, 6, and 8), and values lower than these low. Exact cutoff values were calculated using linear extrapolation.
Statistical Analysis
All analyses were done in R. Survival and global test analyses were performed as previously described (4). Treatment details for patients are included in Supplementary Tables S1 and S10. The integrative statistical model was evaluated using the global test assuming interaction between the explanatory variables (38). Transcriptional identity and key oncogenic driver were defined as categorical and leukemia stemness (pLSC6) as a continuous variable, and assuming interaction between these three exploratory variables. Individual associations are shown in Supplementary Fig. S18, and main contributing covariates clarified in Supplementary Table S15 by using pLSC6 as a categorical variable (low vs. medium/high).
Validation Cohort
A pediatric AML microarray gene-expression cohort of 443 cases was constructed based on previously published data (19, 39, 40). AML M5 cases with t(15;17) were excluded from this cohort prior to assembly, because this subclass was absent from the discovery cohort and has excellent therapy options and disease outcome. Of these, 44 were also included in the discovery cohort and functioned as controls for the equivalence of the RNA-seq and microarray measured gene expression. Three hundred ninety-nine cases, which did not overlap, were used for gene-expression validation of results obtained in the discovery cohort. For 386 of these cases, disease outcome data were available (Supplementary Table S15) and were used for outcome validation analyses.
Key oncogenic driver determination was based on a combination of clinical testing and/or laboratory testing from the cohorts as previously published (see Supplementary Table S15, column K). Cases in which mutational status was unknown were removed from analyses as appropriate.
Transcriptional identity of the validation cohort cases was determined by coclustering of microarray mRNA expression values of overlapping classifier genes (n = 249) of single cases with the complete RNA-seq cohort using Spearman correlation distance-based t-SNE, exactly as done for the RNA-seq cohort clustering. For overlapping genes, probe sets with highest specificity and selectivity (https://genecards.weizmann.ac.il/geneannot/index.shtml) were used, omitting probe sets recognizing more than one gene. For robustness assessment of transcriptional identity calls, we made use of the stochastic initial seeding of the t-SNE algorithm by performing 10 clustering repeats. Cases with clustering inconsistency in more than 2 of the 10 runs (25/443 cases, 5.6%) were not assigned a transcriptional identity label. Transcriptional identity of 95% (41/43) of the microarray profiled cases also present in the RNA-seq cohort were identical. In 9 of 327 cases, the transcriptional identity calls were inconsistent with oncogenic driver determination (2.8%), similar to the discovery cohort.
Transcriptional identity was further confirmed by clustering of the validation cohort using a classifier derived from the microarray expression values only. For this, the batch effect of AML02 and Rotterdam cohort expression values was removed using the ComBat function of the sva R package (Supplementary Fig. S11A). Clustering visualization was done by t-SNE using a 350-gene set consisting of the highest variant probe sets by least median square (Supplementary Fig. S11B), where only probe sets recognizing single genes were used and sex-specific and hemoglobin genes were removed.
pLSC6 scores of the validation cohort were calculated as previously described using log2 intensity values of Affimetrix probe sets 209543_s_at (CD34), 220668_s_at (DNMT3B), 220377_at (FAM30A), 212070_at (GPR56), 203373_at (SOC2), and 206310_at (SPINK2; ref. 19). Of the 44 cases with both microarray and RNA-seq data, pLSC6 values were highly correlated (r = 0.82). pLSC6 categories of low, medium, and high were determined by matched RNA-seq expression value pLSC6 quantiles (0%, 66.21%, 96.78%, and 100%). Eighty-two percent of overlapping cases (36/44) were assigned the same pLSC6 category using this method.
In the validation cohort, association between transcriptional identity, oncogenic driver, pLSC6 score, and overall survival was modeled using a Cox regression implementation in the global test, accounting for interactions between the three variables (Supplementary Table S19). Two hundred ninety-three cases had overall survival data and could be assigned both a transcriptional identity and an oncogenic driver label. Sparse transcriptional identities (3 or fewer cases) were removed, leaving 8 transcriptional identity and 14 oncogenic driver covariates, whereas pLSC6 was used as a continuous variable. Main covariates contributing (cases >1) to the global association are reported (Supplementary Table S20), with pLSC6 categorized as medium/high versus low. Because pLSC6 was developed using the AML02 validation cohort, association with overall survival was independently assessed excluding the AML02 cases from the validation cohort (Supplementary Fig. S19).
AMTL Five-Gene Classifier
A five-gene classifier to identify the AMTL subtype was developed as follows. First, using the RNA-seq cohort, the expression of each gene was summarized by computing median expression for each transcriptional subgroup and using the Wilcoxon test to compare medians across each pair of subgroups. The genes for which AMTL had the greatest or least median expression were selected and then ranked by the maximum of the Wilcoxon test P values comparing AMTL to other subgroups. The top 14 genes in this list were then considered as candidate predictor variables for a logistic regression predicting the AMTL versus non-AMTL class using the bestglm procedure in R. The bestglm procedure defined the model as logit(Prob(AMTL)) = −0.78 + 1.01 × CD3G −0.85 × x COCH-1.20 SLC35D2 + 0.81 SPTLC3 – 0.93 TOR4A (Supplementary Table S27). The model classified AMTL with an AUC of 0.977. In 1,000 rounds of leave-out 10% cross-validation, this model building procedure (median calculation, pairwise Wilcoxon tests, bestglm) achieved an average AUC of 0.973, with a range of 0.952 to 0.983. See Supplementary Table S27 for AMTL logistic regression classifier model terms, estimates, confidence intervals, and P values. We then went on to validate this five-gene classifier in our validation cohort. The Affymetrix microarrays (U133 v2.0) included six probe sets that measured the expression of the five genes in the classifier (gene symbol, probe set IDs: COCH, 205229_s_at; CD3G, 206804_at; SLC35D2, 213082_s_at; SLC35D2, 213083_at; TOR4A, 219620_x_at; and SPTLC3, 220456_at).
A principal component analysis of the two probe sets measuring SLC35D2 gave similar coefficients for 213082_s_at (0.74) and 213083_at (0.67). Thus, for each subject, the expression of SLC35D2 was computed as the simple arithmetic average of the expression of these two probe sets. The other four genes were measured by one probe set each. For each subject, a score was computed as the dot product of the microarray expression of the five genes with the coefficients from the RNA-seq cohort's logistic regression model. This score classified the AMTL/non-AMTL in the independent microarray cohort (those without RNA-seq data) with an AUC of 0.88.
RCU
For a given risk classification, censored event-time endpoint (such as EFS or overall survival), and cohort outcome data set, we defined and computed the RCU as follows: We computed the proportion of patients assigned to low-, intermediate-, and high-risk groups and the Kaplan–Meier estimates of outcome for each risk group (Fig. 4B and C; Supplementary Figs. S24 and S25). Then, for each observed event time, we plotted the utility curve as Kaplan–Meier survival estimate of the low-risk group versus that of the high-risk group (Fig. 4C; Supplementary Fig. S26). An ideal utility curve is a flat line at y = 1; in this case, there is some time point at which the Kaplan–Meier estimate of high-risk patients is 0 and that of low-risk patients is 1. A utility curve along the line y = x could reasonably be obtained by completely random assignment of patients into low-risk or high-risk groups. The “outcome discrimination index” was defined and computed as twice the area above the line y = x and below the utility curve. The outcome discrimination index is 1 if the utility curve is ideal and 0 if the utility curve does not have any point above the line y = x that can be obtained by random risk classification assignments. We defined and computed the “meaningful classification proportion” as the proportion of patients designated as high or low risk because intermediate risk typically designates a patient lacking definitive high-risk or low-risk characteristics (Fig. 4C; Supplementary Fig. S27). Finally, the RCU was defined and computed as the product of the meaningful classification proportion and the outcome discrimination index (Fig. 4C; Supplementary Fig. S27). The RCU equals 1 if and only if all patients have a meaningful classification and the outcome discrimination is 1.
A bootstrap procedure was used to quantify the statistical variability and significance of comparisons of RCU of four risk classification schemes. The RCU of each risk classification scheme was computed for the discovery cohort, and 100,000 bootstraps of the discovery cohort, the validation cohort, and the combined cohort was determined (Supplementary Table S25).
Authors' Disclosures
S. Noort reports grants from KiKa during the conduct of the study. J.K. Lamba reports a patent for 62/944523 and a patent for 62/904552 pending. D. Reinhardt reports other support from Bristol Myers Squibb, Novartis, bluebird bio, and Janssen outside the submitted work. S. Pounds reports grants from American Lebanese Syrian Associated Charities (ALSAC) and NIH during the conduct of the study, as well as a patent for pLSC6 gene signature pending. C.M. Zwaan reports other support from Pfizer, Jazz, AbbVie, Takeda, Incyte, Novartis, and Bristol Myers Squibb outside the submitted work. No disclosures were reported by the other authors.
Authors' Contributions
M. Fornerod: Conceptualization, data curation, formal analysis, supervision, validation, investigation, visualization, methodology, writing–review and editing. J. Ma: Formal analysis, visualization, writing–review and editing. S. Noort: Data curation, investigation, writing–review and editing. Y. Liu: Formal analysis, writing–review and editing. M.P. Walsh: Formal analysis, writing–review and editing. L. Shi: Formal analysis, writing–review and editing. S. Nance: Investigation, writing–review and editing. Y. Liu: Formal analysis, writing–review and editing. Y. Wang: Formal analysis, writing–review and editing. G. Song: Formal analysis, writing–review and editing. T. Lamprecht: Investigation, writing–review and editing. J. Easton: Investigation, writing–review and editing. H.L. Mulder: Investigation, writing–review and editing. D. Yergeau: Formal analysis, writing–review and editing. J. Myers: Formal analysis, writing–review and editing. J.L. Kamens: Investigation, writing–review and editing. E.A. Obeng: Supervision, writing–review and editing. M. Pigazzi: Resources, writing–review and editing. M. Jarosova: Resources, writing–review and editing. C. Kelaidi: Resources, writing–review and editing. S. Polychronopoulou: Resources, writing–review and editing. J.K. Lamba: Resources, investigation, writing–review and editing. S.D. Baker: Resources, writing–review and editing. J.E. Rubnitz: Resources, writing–review and editing. D. Reinhardt: Resources, writing–review and editing. M.M. van den Heuvel-Eibrink: Resources, writing–review and editing. F. Locatelli: Resources, writing–review and editing. H. Hasle: Resources, writing–review and editing. J.M. Klco: Supervision, writing–review and editing. J.R. Downing: Resources, supervision, funding acquisition, writing–review and editing. J. Zhang: Resources, formal analysis, supervision, writing–review and editing. S. Pounds: Formal analysis, supervision, visualization, writing–review and editing. C.M. Zwaan: Conceptualization, resources, supervision, writing–review and editing. T.A. Gruber: Conceptualization, resources, data curation, formal analysis, supervision, funding acquisition, validation, investigation, visualization, methodology, writing–original draft, project administration, writing–review and editing.
Supplementary Material
Acknowledgments
The authors thank St. Jude Tissue Resources Laboratory, the Flow Cytometry and Cell Sorting Core, and the Hartwell Center for Biotechnology and Bioinformatics. This work was supported by grants from the American Cancer Society (T.A. Gruber; RSG-16-046-01), Hyundai Hope on Wheels (T.A. Gruber), Dutch Cancer Society KWF (M. Fornerod), KiKa Children Cancer-free Foundation (S. Noort), Fondazione AIRC (Associazione Italiana Ricerca sul Cancro) IG 20562 and CARIPARO grant 17/04 (M. Pigazzi), NIH (J.K. Lamba and S. Pounds; R01-CA132946), and American Lebanese Syrian Associated Charities (ALSAC) of St. Jude Children's Research Hospital.
Footnotes
Note: Supplementary data for this article are available at Blood Cancer Discovery Online (https://bloodcancerdiscov.aacrjournals.org/).
#M. Fornerod, C.M. Zwaan, and T.A. Gruber are co–senior authors of this article.
##J. Ma and S. Noort contributed equally to this article.
Blood Cancer Discov 2021;2:586–99
References
- 1.Bennett JM, Catovsky D, Daniel MT, Flandrin G, Galton DA, Gralnick HR, et al. Proposed revised criteria for the classification of acute myeloid leukemia. A report of the French-American-British Cooperative Group. Ann Intern Med 1985;103:620–5. [DOI] [PubMed] [Google Scholar]
- 2.Arber DA, Orazi A, Hasserjian R, Thiele J, Borowitz MJ, Le Beau MM, et al. The 2016 revision to the World Health Organization classification of myeloid neoplasms and acute leukemia. Blood 2016;127:2391–405. [DOI] [PubMed] [Google Scholar]
- 3.Zwaan CM, Kolb EA, Reinhardt D, Abrahamsson J, Adachi S, Aplenc R, et al. Collaborative efforts driving progress in pediatric acute myeloid leukemia. J Clin Oncol 2015;33:2949–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.de Rooij JD, Branstetter C, Ma J, Li Y, Walsh MP, Cheng J, et al. Pediatric non-Down syndrome acute megakaryoblastic leukemia is characterized by distinct genomic subsets with varying outcomes. Nat Genet 2017;49:451–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bolouri H, Farrar JE, Triche T, Jr, Ries RE, Lim EL, Alonzo TA, et al. The molecular landscape of pediatric acute myeloid leukemia reveals recurrent structural alterations and age-specific mutational interactions. Nat Med 2018;24:103–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu Y, Li C, Shen S, Chen X, Szlachta K, Edmonson MN, et al. Discovery of regulatory noncoding variants in individual cancer genomes by using cis-X. Nat Genet 2020;52:811–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang J, Ding L, Holmfeldt L, Wu G, Heatley SL, Payne-Turner D, et al. The genetic basis of early T-cell precursor acute lymphoblastic leukaemia. Nature 2012;481:157–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu Y, Easton J, Shao Y, Maciaszek J, Wang Z, Wilkinson MR, et al. The genomic landscape of pediatric and young adult T-lineage acute lymphoblastic leukemia. Nat Genet 2017;49:1211–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Alexander TB, Gu Z, Iacobucci I, Dickerson K, Choi JK, Xu B, et al. The genetic basis and cell of origin of mixed phenotype acute leukaemia. Nature 2018;562:373–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gutierrez A, Kentsis A.Acute myeloid/T-lymphoblastic leukaemia (AMTL): a distinct category of acute leukaemias with common pathogenesis in need of improved therapy. Br J Haematol 2018;180:919–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shiozawa Y, Malcovati L, Galli A, Sato-Otsubo A, Kataoka K, Sato Y, et al. Aberrant splicing and defective mRNA production induced by somatic spliceosome mutations in myelodysplasia. Nat Commun 2018;9:3649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 2003;13:2498–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Faber ZJ, Chen X, Gedman AL, Boggs K, Cheng J, Ma J, et al. The genomic landscape of core-binding factor acute myeloid leukemias. Nat Genet 2016;48:1551–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gollner S, Oellerich T, Agrawal-Singh S, Schenk T, Klein HU, Rohde C, et al. Loss of the histone methyltransferase EZH2 induces resistance to multiple drugs in acute myeloid leukemia. Nat Med 2017;23:69–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Aries IM, Bodaar K, Karim SA, Chonghaile TN, Hinze L, Burns MA, et al. PRC2 loss induces chemoresistance by repressing apoptosis in T cell acute lymphoblastic leukemia. J Exp Med 2018;215:3094–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hollink IH, van den Heuvel-Eibrink MM, Zimmermann M, Balgobind BV, Arentsen-Peters ST, Alders M, et al. Clinical relevance of Wilms tumor 1 gene mutations in childhood acute myeloid leukemia. Blood 2009;113:5951–60. [DOI] [PubMed] [Google Scholar]
- 17.Pellin D, Loperfido M, Baricordi C, Wolock SL, Montepeloso A, Weinberg OK, et al. A comprehensive single cell transcriptional landscape of human hematopoietic progenitors. Nat Commun 2019;10:2395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ng SW, Mitchell A, Kennedy JA, Chen WC, McLeod J, Ibrahimova N, et al. A 17-gene stemness score for rapid determination of risk in acute leukaemia. Nature 2016;540:433–7. [DOI] [PubMed] [Google Scholar]
- 19.Elsayed AH, Rafiee R, Cao X, Raimondi S, Downing JR, Ribeiro R, et al. A six-gene leukemic stem cell score identifies high risk pediatric acute myeloid leukemia. Leukemia 2020;34:735–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Valk PJ, Verhaak RG, Beijen MA, Erpelinck CA, Barjesteh van Waalwijk van Doorn-Khosrovani S, Boer JM, et al. Prognostically useful gene-expression profiles in acute myeloid leukemia. N Engl J Med 2004;350:1617–28. [DOI] [PubMed] [Google Scholar]
- 21.Papaemmanuil E, Gerstung M, Bullinger L, Gaidzik VI, Paschka P, Roberts ND, et al. Genomic classification and prognosis in acute myeloid leukemia. N Engl J Med 2016;374:2209–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bullinger L, Dohner K, Bair E, Frohling S, Schlenk RF, Tibshirani R, et al. Use of gene-expression profiling to identify prognostic subclasses in adult acute myeloid leukemia. N Engl J Med 2004;350:1605–16. [DOI] [PubMed] [Google Scholar]
- 23.Ross ME, Mahfouz R, Onciu M, Liu HC, Zhou X, Song G, et al. Gene expression profiling of pediatric acute myelogenous leukemia. Blood 2004;104:3679–87. [DOI] [PubMed] [Google Scholar]
- 24.Montefiori LE, Bendig S, Gu Z, Chen X, Polonen P, Ma X, et al. Enhancer hijacking drives oncogenic BCL11B expression in lineage ambiguous stem cell leukemia. Cancer Discov 2021. Jun 8 [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Di Giacomo D, La Starza R, Gorello P, Pellanera F, Kalender Atak Z, De Keersmaecker K, et al. 14q32 rearrangements deregulating BCL11B mark a distinct subgroup of T and myeloid immature acute leukemia. Blood 2021;138:773–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Riemke P, Czeh M, Fischer J, Walter C, Ghani S, Zepper M, et al. Myeloid leukemia with transdifferentiation plasticity developing from T-cell progenitors. EMBO J 2016;35:2399–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rasche M, Steidel E, Kondryn D, Von Neuhoff N, Sramkova L, Creutzig U, et al. Impact of a risk-adapted treatment approach in pediatric AML: a report of the AML-BFM registry 2012. Blood 2019;134 (Supplement_1):293. [Google Scholar]
- 28.Alexander TB, Wang L, Inaba H, Triplett BM, Pounds S, Ribeiro RC, et al. Decreased relapsed rate and treatment-related mortality contribute to improved outcomes for pediatric acute myeloid leukemia in successive clinical trials. Cancer 2017;123:3791–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rusch M, Nakitandwe J, Shurtleff S, Newman S, Zhang Z, Edmonson MN, et al. Clinical cancer genomic profiling by three-platform sequencing of whole genome, whole exome and transcriptome. Nat Commun 2018;9:3962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Van Allen EM, Robinson D, Morrissey C, Pritchard C, Imamovic A, Carter S, et al. A comparative assessment of clinical whole exome and transcriptome profiling across sequencing centers: implications for precision cancer medicine. Oncotarget 2016;7:52888–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Uzilov AV, Ding W, Fink MY, Antipin Y, Brohl AS, Davis C, et al. Development and clinical application of an integrative genomic approach to personalized cancer therapy. Genome Med 2016;8:62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bonnet D, Dick JE.Human acute myeloid leukemia is organized as a hierarchy that originates from a primitive hematopoietic cell. Nat Med 1997;3:730–7. [DOI] [PubMed] [Google Scholar]
- 33.McLeod C, Gout AM, Zhou X, Thrasher A, Rahbarinia D, Brady SW, et al. St. Jude Cloud—a pediatric cancer genomic data sharing ecosystem. Cancer Discov 2021;11:1082–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Anders S, Pyl PT, Huber W.HTSeq–a Python framework to work with high-throughput sequencing data. Bioinformatics 2015;31:166–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, et al. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 2015;43:e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Leek JT, Johnson WE, Parker HS, Fertig EJ, Jaffe AE, Zhang Y, Storey JD, Torres LC.sva: Surrogate Variable Analysis. R package version 3.40.0. 2021. Available from: https://bioconductor.org/packages/release/bioc/html/sva.html.
- 37.Van der Maaten LJP HG.Visualizing data using t-SNE. J Mach Learn Res 2008;9:2579–605. [Google Scholar]
- 38.Goeman JJ, van de Geer SA, de Kort F, van Houwelingen HC.A global test for groups of genes: testing association with a clinical outcome. Bioinformatics 2004;20:93–9. [DOI] [PubMed] [Google Scholar]
- 39.Balgobind BV, Van den Heuvel-Eibrink MM, De Menezes RX, Reinhardt D, Hollink IH, Arentsen-Peters ST,, et al. Evaluation of gene expression signatures predictive of cytogenetic and molecular subtypes of pediatric acute myeloid leukemia. Haematologica 2011;96:221–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Buelow DR, Pounds SB, Wang YD, Shi L, Li Y, Finkelstein D, et al. Uncovering the genomic landscape in newly diagnosed and relapsed pediatric cytogenetically normal FLT3-ITD AML. Clin Transl Sci 2019;12:641–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ho PA, Alonzo TA, Gerbing RB, Pollard J, Stirewalt DL, Hurwitz C, et al. Prevalence and prognostic implications of CEBPA mutations in pediatric acute myeloid leukemia (AML): a report from the Children's Oncology Group. Blood 2009;113:6558–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rubnitz JE, Lacayo NJ, Inaba H, Heym K, Ribeiro RC, Taub J, et al. Clofarabine can replace anthracyclines and etoposide in remission induction therapy for childhood acute myeloid leukemia: the AML08 multicenter, randomized phase III trial. J Clin Oncol 2019;37:2072–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tosi S, Kamel YM, Owoka T, Federico C, Truong TH, Saccone S.Paediatric acute myeloid leukaemia with the t(7;12)(q36;p13) rearrangement: a review of the biological and clinical management aspects. Biomark Res 2015;3:21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Noort S, Zimmermann M, Reinhardt D, Cuccuini W, Pigazzi M, Smith J, et al. Prognostic impact of t(16;21)(p11;q22) and t(16;21)(q24;q22) in pediatric AML: a retrospective study by the I-BFM study group. Blood 2018;132:1584–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gruber TA, Gedman AL, Zhang J, Koss CS, Marada S, Ta HQ, et al. An Inv(16)(p13.3q24.3)-encoded CBFA2T3-GLIS2 fusion protein defines an aggressive subtype of pediatric acute megakaryoblastic leukemia. Cancer Cell 2012;22:683–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hollink IHIM, van den Heuvel-Eibrink MM, Arentsen-Peters STCJM, Pratcorona M, Abbas S, Kuipers JE, et al. NUP98/NSD1 characterizes a novel poor prognostic group in acute myeloid leukemia with a distinct HOX gene expression pattern. Blood 2011;118:3645–56. [DOI] [PubMed] [Google Scholar]
- 47.de Rooij JD, Hollink IH, Arentsen-Peters ST, van Galen JF, Beverloo HB, Baruchel A, et al. NUP98/JARID1A is a novel recurrent abnormality in pediatric acute megakaryoblastic leukemia with a distinct HOX gene expression pattern. Leukemia 2013;27:2280–8. [DOI] [PubMed] [Google Scholar]
- 48.Bisio V, Zampini M, Tregnago C, Manara E, Salsi V, Di Meglio A, et al. NUP98-fusion transcripts characterize different biological entities within acute myeloid leukemia: a report from the AIEOP-AML group. Leukemia 2017;31:974–7. [DOI] [PubMed] [Google Scholar]
- 49.Hollink IH, Zwaan CM, Zimmermann M, Arentsen-Peters TC, Pieters R, Cloos J, et al. Favorable prognostic impact of NPM1 gene mutations in childhood acute myeloid leukemia, with emphasis on cytogenetically normal AML. Leukemia 2009;23:262–70. [DOI] [PubMed] [Google Scholar]
- 50.Brown P, McIntyre E, Rau R, Meshinchi S, Lacayo N, Dahl G, et al. The incidence and clinical significance of nucleophosmin mutations in childhood AML. Blood 2007;110:979–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sandahl JD, Coenen EA, Forestier E, Harbott J, Johansson B, Kerndrup G, et al. t(6;9)(p22;q34)/DEK-NUP214-rearranged pediatric myeloid leukemia: an international study of 62 patients. Haematologica 2014;99:865–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Taketani T, Taki T, Sako M, Ishii T, Yamaguchi S, Hayashi Y.MNX1-ETV6 fusion gene in an acute megakaryoblastic leukemia and expression of the MNX1 gene in leukemia and normal B cell lines. Cancer Genet Cytogenet 2008;186:115–9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.