Abstract
Erythrocytosis is a blood disorder characterised by an increased red blood cell mass. The most common causes of erythrocytosis are acquired and caused by diseases and conditions that are accompanied by hypoxaemia or overproduction of erythropoietin. More rarely, erythrocytosis has a known genetic background, such as for polycythaemia vera and familial erythrocytosis. The majority of cases of polycythaemia vera are associated with acquired variants in JAK2, while familial erythrocytosis is a group of congenital disorders. Familial erythrocytosis type 1 is associated with hypersensitivity to erythropoietin (variants in EPOR), types 2–5 with defects in oxygen-sensing pathways (variants in VHL, EGLN1, EPAS1, EPO), and types 6–8 with an increased affinity of haemoglobin for oxygen (variants in HBB, HBA1, HBA2, BPGM). Due to a heterogenic genetic background, the causes of disease are not fully discovered and in more than 70% of patients the condition remains labelled idiopathic.
The transfer of next-generation sequencing into clinical practice is becoming a reality enabling detection of various variants in a single rapid test. In this review, we describe the current research on erythrocytosis gene variants and the mechanisms associated with disease development, along with the currently used diagnostic tests.
Keywords: erythropoiesis, erythrocytosis, polycythaemia, genetic disease, genetic testing
INTRODUCTION
The process of erythropoiesis in the human body generates 2.4 million red blood cells (RBC) every second1,2. Haematopoietic stem cells differentiate from nucleated erythroid progenitors into mature non-nucleated erythrocytes2. Erythropoiesis is tightly controlled by erythropoietin (EPO) and signalling through its receptor, EPOR3. Defects in erythropoiesis can lead to erythrocytosis, a disorder that is characterised by elevated red cell mass (RCM)3–7.
Erythrocytosis is suspected when an abnormally high haematocrit, haemoglobin concentration, or RBC count are detected. These parameters depend on plasma volume, as reduced plasma volume leads to relative (false) erythrocytosis. Absolute (true) erythrocytosis is defined by a RCM >125% of predicted for age and sex. RCM measurement is rarely possible nowadays. It has been shown that a male/female with a haematocrit of ≥0.60/0.56 will always have absolute erythrocytosis7. According to the new 2016 World Health Organisation (WHO) criteria, erythrocytosis is diagnosed with lower thresholds for haemoglobin concentration and haematocrit (>165 g/L or >0.49 in males and >160 g/L or >0.48 in females)8. In some cases a haematocrit threshold of >0.52/>0.48 in males/females is still used9.
Erythrocytosis can be classified as congenital or acquired, and the origins of both can be primary (i.e., an intrinsic defect in RBC) or secondary (i.e., extrinsic to RBC)6,7,10,11. The most common causes of erythrocytosis are acquired, and they develop due to diseases and conditions that are accompanied by hypoxaemia or overproduction of EPO. Conditions that can lead to central hypoxic processes include smoking, sleep apnoea, carbon monoxide poisoning, and a high-altitude habitat6. Several heart, lung, and kidney diseases can also result in erythrocytosis6. Pathological EPO production (i.e., secondary acquired erythrocytosis) is observed in several cancers, such as cerebellar haemangioblastoma, meningioma, parathyroid carcinoma, and some other types of carcinoma3,6. Erythrocytosis following allogeneic haematopoietic stem cell transplant and renal transplant has also been reported7,12,13. Erythrocytosis is a common side effect of drug consumption, as with diuretics, recombinant human EPO, and testosterone replacement therapy14. Chronic diuretic use can lead to a decrease in the plasma volume and relative erythrocytosis. The use of androgens elevates endogenous EPO production and consequently haemoglobin concentration by a mechanism similar to that of recombinant EPO administration.
Less often, erythrocytosis has a genetic background, acquired in polycythaemia vera (PV), the most common myeloproliferative neoplasm, and congenital or familial erythrocytosis (ECYT), a rare genetic disorder. Acquired variants in the Janus tyrosine kinase 2 gene (JAK2) are indicative of PV15. Congenital ECYT has a heterogeneous genetic background and it has been divided into eight types, based on the genes affected (Table I). Primary erythrocytoses, PV and ECYT1, are the consequences of constant activation of the EPO-EPOR signalling pathway due to variants in the JAK2 or EPOR genes, with the levels of serum EPO usually being below normal3. Secondary congenital erythrocytoses (i.e., ECYT2-8) develop due to a mechanism extrinsic of the erythroid cells, and here the levels of serum EPO are usually normal or elevated3. The mechanism underlying ECYT2-5 is an altered hypoxia inducible factor (HIF)-EPO oxygen-sensing pathway which is due to inherited variants in the VHL, EGLN1, EPAS1, or EPO genes16. Finally, the mechanism that leads to ECYT6-8 is an increased affinity of haemoglobin for oxygen, due to variants in the HBB, HBA1, HBA2, and/or BPGM genes6.
Table I.
Genes associated with erythrocytosis
| Gene | Effect on protein | Type |
|---|---|---|
| JAK2 | Gain of function, impaired auto-inhibitory domain | PV |
| EPOR | Gain of function, increased activation of signalling cascade | ECYT1 |
| VHL | Loss of function, impaired degradation of HIF2α | ECYT2 |
| EGLN1 (PHD2) | Loss of function, impaired degradation of HIF2α | ECYT3 |
| EPAS1 (HIF2A) | Gain of function, increased stabilityin normoxia | ECYT4 |
| EPO | Gain of function, increased expression in normoxia | ECYT5 |
| HBB | Loss of function, increases Hb affinity for O2 | ECYT6 |
| HBA1 & HBA2 | Loss of function, increases Hb affinity for O2 | ECYT7 |
| BPGM | Loss of function, impaired synthesis of 2,3-BPG | ECYT8 |
PV: polycythaemia vera; ECYT: familial er ythroctyosis; HIF-2α: hypoxia-inducible factor 2-alpha; Hb: haemoglobin; O2: oxygen; 2,3-BPG: 2,3-bisphosphoglycerate.
The incidence of PV is 0.4 to 2.6 cases per 100,000 inhabitants, while the prevalence in Europe is undetermined17. The incidence of PV in the USA (more precisely, in Olmsted County, Minnesota) from 1935 to 1989 was estimated to be 1.9 cases per 100,000 inhabitants (males, 2.8 and females 1.3 per 100,000 inhabitants per year). The incidence in Japan is lower than in the USA or Europe. Both the incidence and prevalence of ECYT are low, but precise epidemiological data are unavailable because of the limited disease classification and incomplete genetic diagnosis.
Recently, knowledge about erythrocytosis has evolved relatively rapidly, as new genes involved in the development of ECYT have been described. Erythrocytosis detection tests have been updated regularly, including customised next-generation sequencing (NGS) gene panels18.
In this review, we initially describe the process of erythropoiesis and its regulation and review the gene variants associated with erythrocytosis. An up-to-date review of the HIF-EPO pathway was published in 2019 by Lappin and Lee19. In this paper, we focus on additional pathways that are associated with erythrocytosis and review the genetic testing in current clinical practice. A new overview on erythrocytosis will bring new insights into disease development and diagnostics.
Erythropoiesis and its regulation
Erythropoiesis is defined as the proliferation, differentiation, and maturation of haematopoietic stem cells into fully functional mature somatic RBC20–26 (Figure 1). This process is directed by the cell cytokines and the signalling pathways that are directly or indirectly involved in erythrocyte maturation and oxygen sensing27,28. The main, important cytokines are the granulocyte and macrophage colony-stimulating factors (i.e., GM-CSF, M-CSF), the interleukins (e.g., IL-3), and EPO, the concentrations of which are tightly regulated inside the bone marrow.
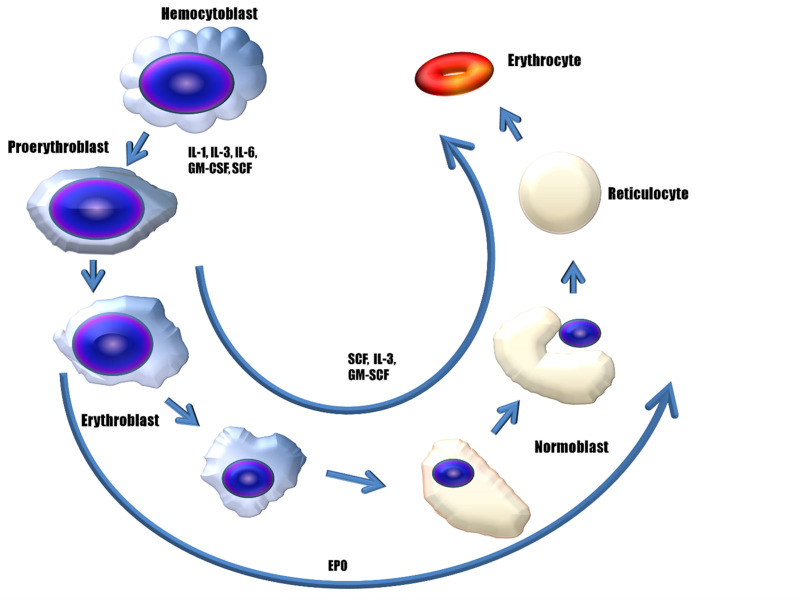
Figure 1. Role of erythropoietin and other cytokines in erythropoiesis.
Scheme of the maturation and differentiation of stem cells to fully developed erythrocytes. The cytokines interleukin (IL)-1, IL-3, IL-6, granulocyte-macrophage colony-stimulating factor (GM-CSF) and stem-cell factor (SCF) regulate the development of haematopoietic stem cells into proerythroblasts. Cytokines SCF, IL-3, GM-SCF, and erythropoietin (EPO) further evolve the proerythroblasts to erythrocytes. EPO influences the differentiation of cells expressing EPO receptor (EPOR) on the cell surface, up to the formation of reticulocytes. Erythroblasts evolve into normoblasts, which lose the nucleus and develop into reticulocytes, and later into erythrocytes.
Erythroid differentiation is divided into engagement and maturation phases. In the engagement phase, multipotent haematopoietic stem cells differentiate into unipotent erythroid progenitor proerythroblasts. In the maturation phase, these proerythroblasts further differentiate into erythroblasts (during a ribosome synthesis phase), normoblasts (during a haemoglobin accumulation phase), and reticulocytes (during the phase of ejection of the nucleus). Reticulocytes are released from the bone marrow into the bloodstream, where it takes them up to 2 days to develop into mature erythrocytes20,21,25.
Autophagy has an important role in the correct formation of erythrocytes, as it participates in the clearance of “unnecessary” cellular organelles, such as the nucleus, ribosomes, and mitochondria29. As erythrocytes do not have a nucleus, genes, and gene variants that might affect erythropoiesis can only be expressed up to the normoblast phase. Haemoglobin and other proteins remain active until programmed cell death kicks in, which is similar to apoptosis, and is known as eryptosis30.
MECHANISMS LEADING TO ERYTHROCYTOSIS
The hypoxia-inducible factor – erythropoietin pathway
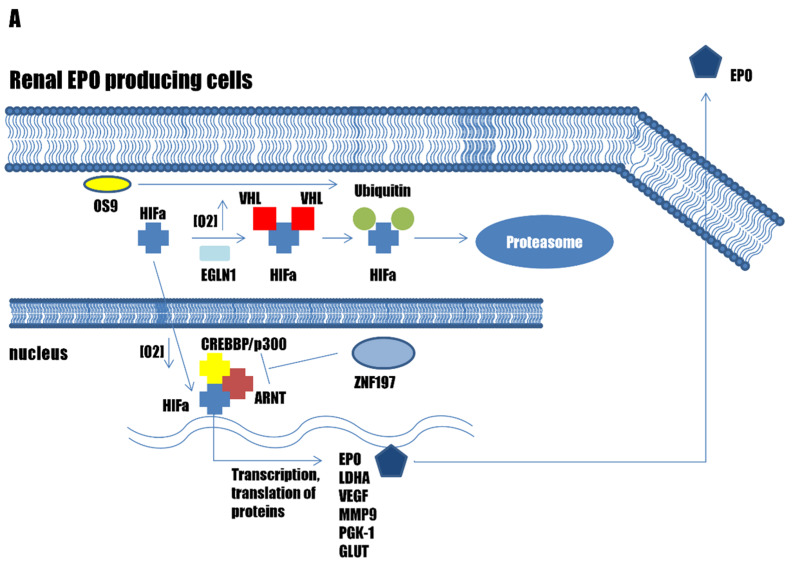
The prime regulator of erythropoiesis is EPO, which is a protein hormone that is produced mainly by the kidney19. The HIF transcription factor family regulates EPO expression. In humans there are three alpha subunit paralogues: HIF1A, EPAS1 (HIF2A), and HIF3A. All three HIFA paralogues form a transcription complex with HIF beta subunit, aryl hydrocarbon receptor nuclear translocator (ARNT, also known as HIF1B). ARNT is constitutively expressed, while HIFA are regulated through sensing of the oxygen concentrations in the cell cytoplasm31,32. The main regulator of EPO production in the kidneys is endothelial Per-Arnt-Sim (PAS)-domain-containing protein 1 (EPAS1, also known as HIF2A)33. Two additional proteins are crucial for this HIF–EPO pathway regulation: the Egl nine homolog 1 (EGLN1, also known as prolyl hydroxylase domain-containing protein 2; PHD2); and von Hippel-Lindau disease tumour suppressor (VHL)34–36 (Figure 2A).
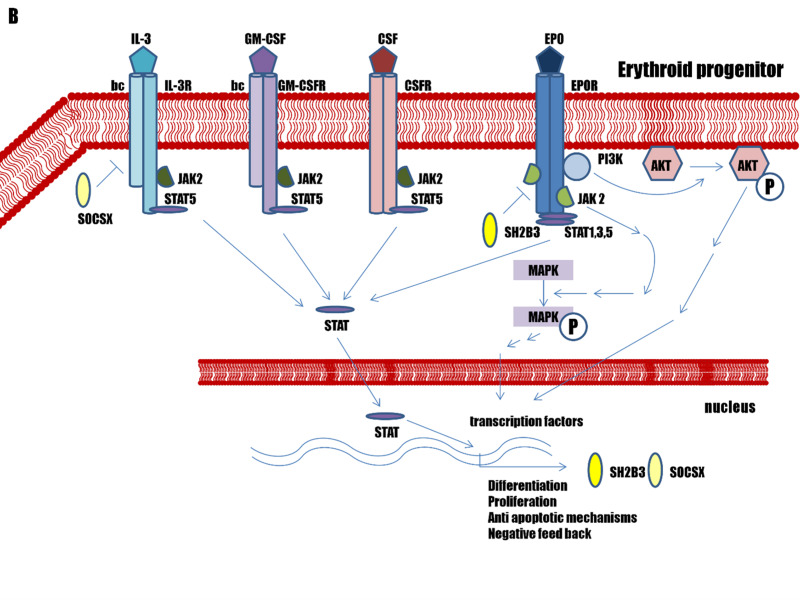
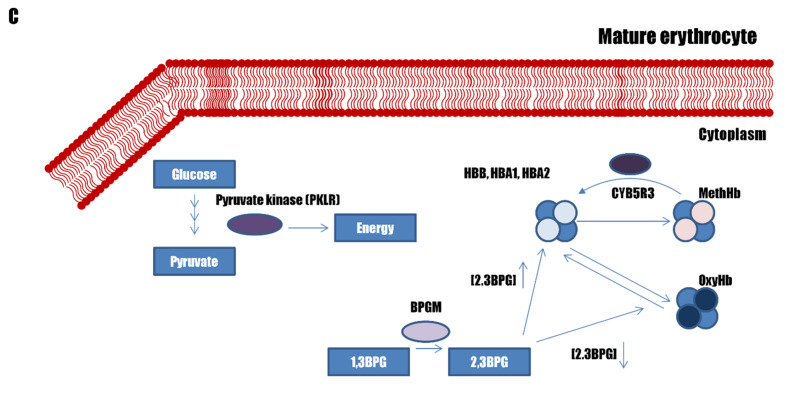
Figure 2.
Pathways associated with erythrocytosis
(A) The genes involved in the hypoxia-inducible factor (HIF) – erythropoietin (EPO) pathway in the kidneys. (B) Genes involved in the EPO – erythropoietin receptor (EPOR) signalling pathway in erythroid progenitors. (C) Genes affecting haemoglobin-oxygen affinity in erythrocytes.
Under normal oxygen conditions, HIFA is hydroxylated by EGLN1, with this hydroxylation being recognised by VHL, which leads to HIFA ubiquitination and degradation in the proteasome16,32,37. Under low oxygen concentrations, EGLN1 is not active, and HIFA is translocated into the nucleus, where it forms a transcription complex with ARNT and CREBBP/p300 HAT; this results in the transcription of numerous hypoxia-inducible genes, including EPO, which influence the growth of erythroid progenitor cells38. Once EPO is released into the bloodstream and transported to the bone marrow, it binds to EPOR on the surface of erythroblasts, thus leading to activation of the EPO–EPOR signalling pathway39.
The erythropoietin – erythropoietin receptor pathway
The EPO ligand can bind to two EPOR, which results in receptor homodimerisation and conformational change. The EPOR cytokine receptor does not have any kinase activity, but it is associated intracellularly with JAK2, which is crucial for receptor activation via phosphorylation. The conformational change in the EPOR dimer brings two JAK2 molecules into proximity, which leads to their autophosphorylation and transphosphorylation, as well as to phosphorylation of tyrosine residues in the EPOR cytoplasmic region. This results in activation of the downstream signalling molecules, which include the JAK2/STAT5, MAPK/ERK, PI3K/AKT, and protein kinase C pathways39. The JAK2 self-activation is prevented by the pseudokinase autoinhibitory domain on one of the JAK2 molecules, which blocks the kinase domain of the other JAK2. The EPO-EPOR signal is terminated via the EPOR intracellular inhibitory domain PTPN6/SOCS-3. The SH2B adapter protein 3 (SH2B3, also known as lymphocyte-specific adapter protein; LNK) works as a negative feedback mechanism for EPOR signal termination40–42 (Figure 2B).
Activation of the EPO-EPOR signalling pathway in erythroblasts results in the transcription of various genes that are involved in cell proliferation, differentiation, prevention of apoptosis, and iron regulation, leading to RBC maturation42.
Regulation of haemoglobin – oxygen affinity
The haemoglobin within mature RBC carries oxygen from the lungs to the peripheral tissues. Haemoglobin has a tetrahedral structure made up of four haem groups and four globin groups. Oxygen is bound to the iron atom within each haem group, enabling one haemoglobin to carry up to four oxygen atoms. In humans, several haemoglobin subunits exist: HBA1, HBA2, HBB, HBD, HBG1, and HBG2. The fully functional haemoglobin in adults, HbA, comprises two HBA and two HBB subunits. The HBA1 and HBA2 genes are paralogues, with an almost identical DNA sequence43. HBG1 and HBG2 are expressed only during foetal development, when two HBG together with two HBA subunits constitute foetal haemoglobin (HbF). HbF is normally replaced by adult haemoglobin (HbA) at birth. HBD integrates into only 3% of adult haemoglobin.
The enzyme bisphosphoglycerate mutase (BPGM) transforms 1,3-bisphosphoglycerate (BPG) into 2,3-BPG, which pushes the chemical equilibrium into the release of oxygen from the haemoglobin44 ( Figure 2C). A haemoglobin variant with a higher affinity for oxygen or lower BPGM activity can decrease the oxygen release from haemoglobin, which will lower the free oxygen in the blood, resulting in tissue hypoxia.
GENE VARIANTS ASSOCIATED WITH ERYTHROCYTOSIS
Erythrocytosis is associated with several gene variants that have been reported in patients with symptoms associated with erythrocytosis (i.e., increased haemoglobin concentration, haematocrit, RBC, RCM). The Online Mendelian Inheritance in Man (OMIM) database45 specifies that two genes are involved in somatic erythrocytosis or PV; JAK246 and SH2B338, and nine genes are involved in familial erythrocytoses ECYT1-8; EPOR47–49; VHL50–52; EGLN153–55; EPAS156,57; EPO58,59; HBB4,60; HBA143,60; HBA243,60; and BPGM44,60 ( Table I). Each of these genes is involved in a key regulatory mechanism of erythropoiesis including the HIF-EPO pathway in kidneys, the EPO-EPOR signalling pathway in the bone marrow, and the regulation of haemoglobin-oxygen affinity in RBC (Figure 2). Several other genes have been identified in patients with erythrocytosis, but their role in the development of erythrocytosis was not confirmed10.
The Leiden Open Variation Database (LOVD) is an open-source DNA variation database system that is designed to collect and display all variants of any specific gene61. Common variations are often non-pathogenic, while pathogenic variants are frequently present in populations at low proportions. Several hundred variants of several genes have been deposited in LOVD and have been associated with PV and ECYT. Although LOVD is regularly updated, not all identified variants are yet deposited in the database. In Table II we have listed 58 variants of the HBB, EPAS1, VHL, EGLN1, EPOR, EPO, and BPGM genes that have not yet been deposited in LOVD, which were extracted from the literature in the PubMed database.
Table II.
Genetic variants associated with congenital erythrocytosis
| Gene | Location | DNA change (published as) | Protein change | dbSNP ID | Number reported | Genotype | PMID (reference) | Remarks |
|---|---|---|---|---|---|---|---|---|
| EPOR | Exon 8 | NM_000121.4: c.1161_1186del | p.(Pro388Hisfs*3) | / | 1/1,192 | Heterozygous | 29790589 | |
| NM_000121.4: c.1166dup | p.(Gly390Trpfs*10) | / | 1/1,192 | Heterozygous | 29790589 | |||
| NM_000121.4: c.1202C>G | p.(Ser401*) | / | 1/1,192 | Heterozygous | 29790589 | |||
| NM_000121.4: c.1307T>A | p.(Leu436*) | / | 1/1,192 | Heterozygous | 29790589 | |||
| NM_000121.4: c.1310G>A | p.(Arg437His) | rs62638744 | 1/58 | 26010769 | ||||
| NM_000121.4: c.1362C>A | p.(Tyr454*) | / | 1/1,192 | Heterozygous | 29790589 | |||
| Other genetic variants are listed in Vo č anec et al . (2019) 59 | ||||||||
| VHL | Exon 1 | NM_000551.3: c.28G>T | p.(Glu10*) | / | 1/163 | Heterozygous | 24115288 (4) | |
| NM_000551.3: c.154G>T | p.(Glu52*) | rs373068386 | 1/125 | Heterozygous | 27651169 (10) | |||
| NM_000551.3: c.235C>T | p.(Arg79Cys) | rs200885420 | 1 | Compound heterozygous | 15642680 | Together with variant p.(Leu188Val) | ||
| NM_000551.3: c.241C>G | p.(Pro81Ala) | / | 1/163 | Compound heterozygous | 24115288 (4) | Together with variant p.(Gly144Arg) | ||
| NM_000551.3: c.290C>T | p.(Pro97Leu) | / | 1/1,192 | Heterozygous | 29790589 | |||
| NM_000551.3: c.311G>T | p.(Gly104Val) | rs869025630 | 1/43 | Heterozygous | 15642664 | |||
| Intron 1 (cryptic exon E1′) | NM_000551.3: c.340+574A>T | / | rs982745672 | 1 | Compound heterozygous | 29891534 (51) | Together with variant p.(Gln164His) | |
| NM_000551.3: c.340+694_711dup | p.(Trp159*) (impact on potential X1 protein) | / | 2 | Compound heterozygous | 29891534 (51) | Together with variant p.(Arg200Trp) or p.(Gly144Arg) | ||
| NM_000551.3: c.340+770T>C | p.(Ser179Pro) (impact on potential X1 protein) | rs1346312258 | 4 | Compound heterozygous | 29891534 (51) | Together with variant p.(Arg200Trp) or p.(Asp143Asp) | ||
| NM_000551.3: c.340+816A>C | p.(*194Serext*24) (impact on potential X1 protein) | rs1031288121 | 3 ( family members) | Homozygous / heterozygous | 29891534 (51) | |||
| Exon 2 | NM_000551.3: c.370A>G | p.(Thr124Ala) | / | 2 ( family members) | Compound heterozygous | 23772956 | Together with variant p.(Leu188Val) | |
| NM_000551.3: c.376G>T | p.(Asp126Tyr) | rs104893831 | 2 ( family members) | Heterozygous | 12393546 | |||
| NM_000551.3: c.413C>T | p.(Pro138Leu) | rs780178275 | 1 | Homozygous | 23538339 | |||
| NM_000551.3: c.429C>T | p.(Asp143Asp) | rs773556807 | 2 | Homozygous | 29891534 (51) | |||
| NM_000551.3: c.430G>A | p.(Gly144Arg) | rs869025650 | 2/163 | Compound heterozygous | 24115288 (4) | Together with variant p.(Arg200Trp) or p.(Pro81Ala) | ||
| 1/6 | Heterozygous | 15921386 | Stop gained variant | |||||
| Exon 3 | NM_000551.3: c.524A>G (523A>G) | p.(Tyr175Cys) | rs193922613 | 1/5 | Heterozygous | 15642680 | ||
| NM_000551.3: c.548C>T | p.(Ser183Leu) | rs5030823 | 1 | Heterozygous | 21454469 | |||
| NM_000551.3: c.562C>G | p.(Leu188Val) | rs5030824 | 2/7 | Compound heterozygous | 12844285 | Together with variant p.(Arg200Trp) | ||
| NM_000551.3: c.571C>G | p.(His191Asp) | rs28940301 | 1/7 | Homozygous | 12844285 | |||
| NM_000551.3: c.574C>T | p.(Pro192Ser) | rs28940300 | 1/7 | Compound heterozygous | 12844285 | Together with variant p.(Arg200Trp) | ||
| NM_000551.3: c.574C>A | p.(Pro192Thr) | / | 1/163 | Compound heterozygous | 24115288 (4) | Together with variant p.(Arg200Trp) | ||
| NM_000551.3: c.586A>G | p.(Lys196Glu) | rs281860296 | 1/70 | Homozygous | 23859443 | |||
| EGLN1 | Exon 1 | NM_022051.2: c.122A>G | p.(Tyr41Cys) | / | 2/1,192 | Heterozygous | 29790589 | |
| NM_022051.2: c.380G>C | p.(Cys127Ser) | rs12097901 | 8/163 | Heterozygous | 24115288 (4) | |||
| NM_022051.2: c.461C>A | p.(Ser154*) | rs1018129986 | 2/1,192 | Heterozygous | 29790589 | Stop gained variant | ||
| NM_022051.2: c.494delC | p.(Pro165Glnfs*9) | / | 1/1,192 | Heterozygous | 29790589 | |||
| NM_022051.2: c.494dupC | p.(Ser166Lysfs*81) | / | 1/1,192 | Heterozygous | 29790589 | |||
| NM_022051.2: c.610A>G (c.610G>A) | p.(Lys204Glu) | / | 1/163 | Heterozygous | 24115288 (4) | |||
| NM_022051.2: c.678dupG | p.(Arg227Alafs*20) | / | 3 (2 family members) | Heterozygous | 27774468 (55) | |||
| NM_022051.2: c.682G>T | p.(Ala228Ser) | / | 1/2 | Heterozygous | 25263965 | Comorbidity of phaeochromocytoma (PHEO) / paraganglioma (PGL) | ||
| NM_022051.2: c.715C>T | p.(Gln239*) | / | 1 | 27774468 (55) | ||||
| NM_022051.2: c.815T>C | p.(Leu272Pro) | / | 1/1,192 | Heterozygous | 29790589 | |||
| NM_022051.2: c.835_850del (c.835del14) | p.(Leu279Thrfs*43) | / | 1/38 | Heterozygous | 24482100 | |||
| NM_022051.2: c.836T>C | p.(Leu279Pro) | / | 1/125 | Heterozygous | 27651169 (10) | |||
| NM_022051.2: c.853G>C | p.(Gly285Arg) | rs1184568745 | 1/163 | Heterozygous | 24115288 (4) | |||
| NM_022051.2: c.867C>G | p.(Ser289Arg) | rs763045676 | 1/1,192 | Heterozygous | 29790589 | |||
| Exon 2 | NM_022051.2: c.896T>C | p.(Met299Thr) | / | 2/1,192 | Heterozygous | 29790589 | ||
| NM_022051.2: c.911C>T | p.(Pro304Leu) | rs1293106237 | 1 | 27774468 (55) | ||||
| NM_022051.2: c. 949_950delinsAG | p.(Pro317Arg) | / | 1/1,192 | Heterozygous | 29790589 | |||
| NM_022051.2: c.1000T>C | p.(Trp334Arg) | / | 5 (family members) | Heterozygous | 23859443 | Damaging effect on protein function (In silico predictions) | ||
| NM_022051.2: c.1001G>A | p.(Trp334*) | / | 1/32 | 27034858 | ||||
| Exon 3 | NM_022051.2: c.1012dup (c.1010dup) | p.(Val338Glyfs*18) | / | 1/163 | Heterozygous | 24115288 (4) | ||
| NM_022051.2: c.1030C>T | p.(Arg344*) | rs752961498 | 1/1,192 | Heterozygous | 29790589 | Stop gained variant | ||
| NM_022051.2: c.1096T>C | p.(Phe366Leu) | / | 2 (family members) | Heterozygous | 29881576 (53) | |||
| NM_022051.2: c.1111C>T | p.(Arg371Cys) | / | 1 | 27774468 (55) | ||||
| NM_022051.2: c.1132C>T | p.(Pro378Ser) | rs1437849917 | 1/1,192 | Heterozygous | 29790589 | |||
| Exon 4 | NM_022051.2: c.1153G>A | p.(Ala385Thr) | / | 2/1,192 | Heterozygous | 29790589 | ||
| NM_022051.2: c.1167G>T | p.(Trp389Cys) | / | 1/1,192 | Heterozygous | 29790589 | |||
| EPAS1 | Genetic variants are listed in Kristan et al . (2019) 38 | |||||||
| EPO | Genetic variants are listed in Vo č anec et al . (2019) 59 | |||||||
| HBB | Exon 2 | NM_000518.4:c.310T>C | p.(Phe104Leu) | / | 1 | Heterozygous | 28332377 (88) | |
| Exon 3 | NM_000518.4:c.412G>A | p.(Val138Met) | rs748704616 | 1/38 | Heterozygous | 24482100 | ||
| BPGM | Exon 3 | NM_199186.2:c.184C>T | p.(Arg62Trp) | rs1436218818 | 1/1,192 | Heterozygous | 29790589 | |
| NM_199186.2:c.344G>A | p.(Trp115*) | rs149329328 | 1/1,192 | Homozygous | 29790589 | Stop gained variant | ||
Revised genetic variants of the genes associated with congenital erythrocytosis ( ECYT) types 1-8, as an update of the Leiden Open Variation Database.
dbSNP ID: Single Nucleotide Polymorphism Database identifier; PMID: PubMed identifier.
JAK2 activation in polycythaemia vera
PV is a clonal haematopoietic stem-cell disorder that is characterised by increased total blood volume and proliferation of erythroid, granulocytic, and megakaryocytic elements of the bone marrow41. The somatic JAK2 V617F exon 14 variant is the cause of the disorder in 95% of affected patients, while 3% have variants in JAK2 exon 1241,62–64. These two exon site variants appear to have similar activating effects. The JAK2 V617F variant triggers JAK2 signalling, which leads to increased sensitivity to cytokines (including EPO) and cytokine-independent cell proliferation and survival.
The disease mechanism is based on inactivation of the JAK2 pseudokinase autoinhibitory domain, which is responsible for inhibition of JAK2 self-activation41. Two variants in exon 13 have been associated with erythrocytosis65,66, and heterozygous variants in JAK2 and EGLN1 with familial erythrocytosis67. Variants of the JAK2 gene may be present in several other diseases, such as essential thrombocythaemia and primary myelofibrosis, although the symptoms of affected patients can be very different41,68.
Specific mutations of the calreticulin (CALR) gene and thrombopoietin receptor (MPL) gene are very rare in PV. However, more than 50% of PV patients were found to have at least one mutation other than well-described driver mutations, with TET2 and ASXL1 being the most commonly involved genes41,68. SH2B3 variants, germline or somatic, are not frequently associated with erythrocytosis but may cause primary erythrocytosis similar to ECYT140,69,70.
Erythropoietin receptor activation in familial erythrocytosis type 1
Familial erythrocytosis type 1 (ECYT1) is an autosomal dominant genetic disorder that is associated with more than 28 germline EPOR variants59,71–78. Different frameshift variants in exon 8 have been identified in families in which several members show symptoms of the disease. The mechanism underlying the frameshift variants is a truncation of the EPOR intracellular C-terminal region, which is responsible for negative feedback regulation of the receptor79,80. These variants enable EPO-EPOR signal transduction even at low serum EPO levels, and they prevent feedback inhibition through the SH2B3 and PTPN6/SOCS-3 inhibitory domain47,50.
In 2018, Pasquier et al. reported an additional mechanism leading to erythrocytosis. An EPOR variant in exon 8 leads to the appearance of a new C-terminal tail that increases EPOR dimerisation, constitutive signalling, and hypersensitivity to EPO47.
Oxygen-sensing defects in familial erythrocytosis types 2-5
Familial erythrocytosis types 2-5 (ECYT2-5) are associated with genes that affect the HIF-EPO signalling pathway, including VHL (ECYT2), EGLN1 (ECYT3), EPAS1 (ECYT4), and EPO (ECYT5)32,72,73,81,82.
The disease mechanism underlying these variants is inactivation of the EGLN1 and VHL enzymes, EPAS1 stabilisation, or increased EPO transcription activation, all of which result in increased EPO production under normoxic conditions and, consequently, RBC overproduction. The promoter and enhancer regions surrounding the respective genes might also represent important variant sites51,58,59.
The VHL variants that cause ECYT2, an autosomal recessive disorder, lead to loss of function and can be homozygous, heterozygous, or compound heterozygous. Over 15 variants associated with ECYT2 are spread all over the coding regions of the gene. Recently, a variant in intron 1 which results in splicing alterations was linked with the disease51. Chuvash polycythaemia with variant VHL c.598C>T (p.Arg200Trp) is the most common form of ECYT255,83,84. Variants can also result in angiogenesis and other changes in metabolism51. VHL is a tumour suppressor gene, so its loss-of-function variants can also result in highly vascularised tumours in the central nervous system, retinal haemangioblastomas, pancreatic neuroendocrine tumours, phaeochromocytomas, and clear-cell renal carcinomas51.
ECYT3 is associated with more than 15 heterozygous loss-of-function EGLN1 variants near to or within the prolyl hydroxylase domain. It appears that specific variants in the MYND type zinc finger can also cause erythrocytosis54.
Most of the nine EPAS1 variants that cause ECYT4, an autosomal dominant disorder, are heterozygous variants of exon 12, which encode the oxygen-dependent degradation domain. These variants prevent the binding of EGLN1 hydroxylase to the EPAS1 protein and subsequent hydroxylation and binding of the VHL protein38,81.
EPO was added to the list of ECYT-causing genes most recently, which led to the definition of an autosomal dominant disorder ECYT5. It is known that two frameshift variants interrupt the transcription of the main EPO mRNA, which results in excess production of EPO through an alternative promoter in intron 158,59.
Haemoglobin-oxygen affinity defects in familial erythrocytosis types 6-8
Familial erythrocytosis types 6-8 (ECYT6-8) are linked to variants in genes that affect the affinity of haemoglobin for oxygen, including different haemoglobin subunits HBB (ECYT6), HBA1, and HBA2 (ECYT7) and the haemoglobin-modifying enzyme BPGM (ECYT8).
The autosomal dominant disorders ECYT6 and ECYT7 are caused by heterozygous variants in HBB and HBA that result in high oxygen affinity of haemoglobin85–87. The HBB gene variants (over 80 variants) are more frequent than those of HBA1 and HBA2 (over 24 variants)88. The three crucial regions that are responsible for haemoglobin stability are its β-chain C terminus, the α1β2 interface, and the 2,3-BPG) binding site85. HBB variants with high oxygen affinity affect the transition of the R-state (relaxed binding structure) to the T state (tight-binding structure). Variants in the 2,3-BPG binding site and the HBB haem pocket have also been reported. Substitutions are the major cause of defects in the function of HBA. A small proportion of survivable transferable variants has also been attributed to deletions and frameshift variants88.
The mechanism of erythrocytosis of the haemoglobin variants is based on lower concentrations of free oxygen, resulting in tissue hypoxia and increased EPO production. Haemoglobin variants can result in further pathologies, such as sickle cell anaemia, thalassaemia, and unstable haemolytic anaemias.
Rare compound heterozygous variants in BPGM reported to be associated with ECYT8 are the result of a BPGM deficiency due to mutations in proximity to active binding sites. Up to now, several variants have been found, although only four variants are known to cause ECYT844,89–91.
STEPWISE CLINICAL EXAMINATION AND GENETIC TESTING
Clinical examination
Normally a patient suspected of having erythrocytosis is referred to a haematologist by a general practitioner with a few old complete blood count results. A stepwise clinical and laboratory assessment follows4,5,92. It is a good decision to measure RCM at this point if it is possible to do this investigation. It is essential to take the patient’s history and conduct a thorough physical evaluation, focusing on symptoms, comorbidities, medications taken, habits, and family history. Diagnostic predictions are then followed by laboratory evaluation.
High haematocrit, haemoglobin concentration, and/or RCM for >2 months with an appropriate clinical picture and biochemistry confirm absolute erythrocytosis (first step). In the second step, acquired secondary erythrocytosis due to chronic diseases (heart, lung, kidney) and other conditions (smoking, sleep apnoea, therapy with steroids, or their misuse) should be excluded. The important third step is to determine whether the patient has PV by measuring serum EPO levels and screening for JAK2 variants. A single point JAK2 variant V617F indicative of PV can be identified by allele-specific polymerase chain reaction (PCR) analysis93. Cases without JAK2 V 617F should be screened for exon 12 variants by using the melting curve assay63. In the fourth step, genes related to ECYT are sequenced. When the EPO level is low, usually only the EPOR gene is sequenced; otherwise, the testing focuses on the HBB, HBA, BPGM, VHL, EPAS1, EGLN1, and EPO genes. If no known causative variant is found in these genes, extended NGS analysis should be performed6,60. A remaining group of patients with unexplained erythrocytosis is characterised as having idiopathic erythrocytosis.
Genetic testing
Different techniques are used for the detection of variants in the selected genes. Depending on the gene variant, the whole gene or specific regions of the gene can be sequenced. The most common analysis is PCR amplification of the targeted region, followed by Sanger sequencing94,95. Techniques such as comparative genomic hybridisation, quantitative PCR, and high resolution melting are also used41. However, NGS has gained ground here over the last decade, due to the possibility of analysing a large number of samples and genes in one test10,96,97. Nevertheless, Sanger sequencing is still used for validation of the NGS results or characterisation of additional members from the same ECYT family.
One of the most promising NGS approaches, besides whole-exome sequencing and whole-genome sequencing, is targeted gene sequencing with a NGS gene panel. The selection of the specific genes involved in the disease development increases the coverage and reduces the costs of targeted gene sequencing, compared to whole-genome/exome sequencing. Customised NGS panels are constantly evolving through the addition of new candidate genes to the currently known gene list (Table I). An important step in the process enabling extraction of rare genomic and potential erythrocytosis-causing variants is variant annotation. Since NGS technology is becoming more affordable, it is now possible to precisely screen more samples cost-effectively and detect multiple variants across different genes in less time.
Different erythrocytosis gene panels are now available for the detection of one or more types of erythrocytosis (see Table III). Along with the common genes involved, the NGS gene panels enable analysis of additional genes, depending on the specific needs and supplier’s options. Following clinical trials, several additional new gene panels that are awaiting approval can be used to sequence the JAK2, EPO, HBB, HBA1, and HBA2 genes (see ClinicalTrials.gov). The US Food and Drug Administration has approved a PV test that detects the JAK2 gene using PCR98. In the European Union, a panel for the detection of erythrocytoses has been approved which includes the EPOR, VHL, EGLN1, EPAS1, HBB, HBA1, HBA2, BPGM, PKLR, HIF1A, EGLN2, and EGLN3 genes99.
Table III.
List of diagnostic tests for erythrocytosis
| Company§ | Test* | Genes (including) | Technique |
|---|---|---|---|
| Ambry Genetics | VHL gene sequence and deletion/ duplication | VHL | NGS, MLPA |
| RenalNext | 19 genes (VHL) | NGS | |
| PGLNext | 12 genes (VHL) | NGS | |
| Baylor Miraca Genetics Laboratories | VHL comprehensive - sequence and deletion/duplication analysis | VHL | Sanger, MLPA |
| VHL deletion/ duplication analysis (different tests) | VHL | MLPA, Sanger | |
| Hereditary paraganglioma/pheochromocytoma panel | 9 genes (VHL) | NGS, MLPA | |
| Bioarray | Familial erythrocytosis | EPOR | NGS |
| BloodGenetics | NGS panel for congenital erythrocythosis or familiar polycythaemia | BPGM, EGLN1, EPAS1, EPOR, JAK2, SH2B3, VHL | NGS |
| CeGaT GmbH | Erythrocytes, anaemia panel | AMN, ANK1, C15orf41, CBLIF, CDAN1, COX4I2, CUBN, EGLN1, EPAS1, EPB42, EPOR, G6PD, HBA1, HBA2, HBB, HBD, HFE, KIF23, KLF1, LPIN2, RPL11, RPL35A, RPL5, RPS10, RPS17, RPS19, RPS24, RPS26, RPS7, SEC23B, SH2B3, SPTA1, SPTB | NGS/MPS |
| Single gene testing JAK2 | JAK2 | Sanger | |
| Single gene testing VHL | VHL | Sanger | |
| CEN4GEN Institute for Genomics and Molecular Diagnostics | Primary familial and congenital polycythaemia: gene sequencing | EPOR | NGS/MPS |
| Center for Human Genetics | Hereditary onco-endocrine tumours | 44 genes (EGLN1, EPAS1, VHL) | NGS/MPS |
| Centogene AG - the Rare Disease Company Germany | Erythrocytosis, familial type 1 | EPOR | qPCR, Sanger |
| Erythrocytosis, familial type 1 | SH2B3 | qPCR, Sanger | |
| Erythrocytosis, familial type 3 | EGLN1 | qPCR, Sanger | |
| Erythrocytosis, familial type 4 | EPAS1 | qPCR, Sanger | |
| CGC Genetics | Erythrocytosis, familial (sequence analysis of EPOR gene) | EPOR | Sanger |
| Erythrocytosis familial, 3 (sequence analysis of EGLN1 gene) | EGLN1 | Sanger | |
| Erythrocytosis familial, 4 | EPAS1 | Sanger | |
| Detection of V617F somatic mutation of JAK2 gene | JAK2 | RT-qPCR | |
| Pheochromocytoma and paraganglioma (NGS panel for 16 genes) | EGLN1, FH, GDNF, KIF1B, MAX, MEN1, NF1, PR-KAR1A, RET, SDHA, SDHAF2, SDHB, SDHC, SDHD, TMEM127, VHL | NGS/MPS | |
| Cincinnati Children’s Hospital Medical Center Laboratory of Genetics and Genomics | Erythrocytosis panel | BPGM, EGLN1, EPAS1, EPOR, HBA1, HBA2, HBB, JAK2, VHL | NGS |
| Thrombocytosis panel by NGS | CALR, JAK2, MPL, THPO | NGS | |
| EPOR sequencing | EPOR | Sanger | |
| EPAS1 sequencing | EPAS1 | Sanger | |
| Fulgent Genetics | Erythrocytosis NGS panel | ANK1, BPGM, CALR, EGLN1, EPAS1, EPB41, EPB42, EPOR, HBA1, HBA2, HBB, HIF1A, JAK2, KCNN4, PIEZO1, PKLR, RHAG, SH2B3, SLC4A1, SPTA1, SPTB, TET2, TET3, VHL | NGS |
| Clinical exome | 4,673 genes for 5156 conditions (VHL, EGLN1, EPAS1) | NGS | |
| Full comprehensive cancer panel | 127 genes for 329 conditions (VHL, EGLN1) | NGS/MPS | |
| Comprehensive thrombosis, platelet disorder and coagulation deficiency NGS panel | 82 genes for 146 conditions (EGLN1, EPAS1) | NGS | |
| Von Hippel-Lindau syndrome (VHL single gene test) | VHL | NGS/MPS | |
| Expanded polycystic kidney disease NGS panel | 34 genes (VHL) | NGS/MPS | |
| Comprehensive primary immunodeficiency NGS panel | 472 genes (SH2B3, JAK2) | NGS | |
| SPRTN single gene | SPRTN | NGS/MPS | |
| Paraganglioma-pheochromocytoma comprehensive panel | 11 genes (VHL) | NGS/MPS | |
| EPOR single gene | EPOR | NGS/MPS | |
| VHL single gene | VHL | NGS/MPS | |
| SH2B3 single gene | SH2B3 | NGS/MPS | |
| JAK2 single gene | JAK2 | NGS/MPS | |
| Myelofibrosis NGS panel | CALR, JAK2, MPL, SH2B3 | NGS/MPS | |
| EGLN1 single gene | EGLN1 | NGS/MPS | |
| GeneDx | VHL gene sequencing | VHL | Sanger |
| GENETAQ Molecular Genetics Centre and Diagnosis of Rare Diseases | Erythrocytosis, familial: EPOR gene sequence analysis | EPOR | Sanger |
| Erythrocytosis, familial: EGLN1 gene sequence analysis | EGLN1 | Sanger | |
| Erythrocytosis, familial: EPAS1 (HIF2A) gene sequence analysis | EPAS1 | NGS | |
| Instituto de Medicina Genomica | EPOR. Complete sequencing | EPOR | Sanger |
| Labor Dr. Wisplinghoff | Familial erythrocytosis, 1 | EPOR | Sanger |
| Laboratorio de Genetica Clinica SL | Familial erythrocytosis | EPOR | Sanger |
| Polycythaemia, secondary (autosomal dominant) | EGLN1, EPAS1 | Sanger | |
| Primary familial polycythaemia | EPOR | Sanger | |
| Mayo Clinic Genetic Testing Laboratories | 2,3-Bisphosphoglycerate mutase, full gene sequencing analysis | BPGM | Sanger |
| MedGene | Erythrocytosis, somatic | JAK2 | Sanger |
| Genome Diagnostics Lab. | VHL gene sequencing and deletion/ duplication analysis | VHL | NGS, MPS, MLPA |
| Custom gene sequencing panel | 31 genes (VHL) | NGS | |
| Praxis fuer Humangenetik Wien | Erythrocytosis, somatic | JAK2 | Sanger |
| PreventionGenetics | Primary familial and congenital polycythaemia (PFCP) via EPOR gene | EPOR | NGS, Sanger, MPS |
| Von Hippel-Lindau disease via VHL gene sequencing with CNV detection | VHL | NGS, Sanger | |
| Reference Laboratory Genetics | Familial erythrocytosis, panel massive sequencing (NGS) four genes | EGLN1, EPAS1, EPOR, VHL | NGS |
| Familial erythrocytosis, sequencing EPOR gene | EPOR | Sanger | |
| Familial erythrocytosis type 2, sequencing VHL gene | VHL | Sanger | |
| Familial erythrocytosis, sequencing EGLN1 gene | EGLN1 | Sanger | |
| Familial erythrocytosis type 4, sequencing exon 12 EPAS1 gene | EPAS1 | Sanger | |
| Familial erythrocytosis, sequencing EPAS1 gene | EPAS1 | Sanger | |
| Erythrocytosis due to bisphosphoglycerate mutase deficiency, sequencing BPGM | BPGM | Sanger |
See Appendix 1;
Tests names are protected under a trademark owned by a business company. Tests names may change, products can be removed from the market. The National Center for Biotechnology Information (NCBI) Genetic Testing Registry Site contains information about 92 genetic tests for the detection of erythrocytosis alone plus other groups of clinically important diseases. MLPA, multiplex ligation-dependent probe amplification; CGH, comparative genomic hybridisation; MPS, massive parallel sequencing; NGS, next-generation sequencing; qPCR, quantitative polymerase chain reaction; Sanger, sequencing by Sanger; RT-qPCR, quantitative reverse transcription-polymerase chain reaction; CNV: copy number variation.
The National Center for Biotechnology Information (NCBI) Genetic Testing Registry Site contains information about 92 genetic tests for erythrocytosis. These tests can detect one or more specific types of erythrocytosis in a group with other clinically significant diseases. The different companies diagnose erythrocytosis through the use of different sets of genes100.
Potential new candidate genes involved in erythrocytosis
With current NGS testing, over 70% of patients suspected of having familial erythrocytosis remain undiagnosed10,18,60. Therefore, several other genes must influence the onset of erythrocytosis. The most probable candidate genes are from pathways that alter EPO expression, regulate the EPO-EPOR signalling pathway, and influence erythrocyte production, function, and apoptosis. Their function may be direct or indirect, through their roles as enzymes, transporters, inhibitors, transcription factors, and other DNA and RNA interacting proteins. A detailed search of public databases (NCBI, GenCards, UniProt, Reactome, WikiPathways, etc.) could be performed in the future with the aim of identifying new candidate genes suitable for inclusion in extended targeted NGS.
CONCLUSIONS
The purpose of this review was to describe the current, rapidly evolving knowledge in the wide and diverse field of erythrocytosis. A group of patients with idiopathic erythrocytosis is of broad interest to clinicians, geneticists, and researchers. NGS techniques represent the preferred methods for the rapid discovery of new unknown variants involved in the development of erythrocytosis.
Further studies focusing on the discovery of new genes and variants that are responsible for erythrocytosis are urgently needed. In the future, several unknown causative genes are expected to be identified as indicative of different types of erythrocytosis. Major targets are the genes involved in three pathways: EPO-EPOR signalling, the HIF-EPO pathway, and haemoglobin–BPGM-regulated oxygen affinity.
ACKNOWLEDGEMENTS
All Authors contributed to this review. JG wrote the manuscript, performed a literature review, and developed the figures and tables. AK performed part of the literature review and prepared tables. TK drafted the manuscript and reviewed genetic variants. IPZ described the stepwise clinical examination and genetic testing. ND developed the concept for this article and edited the manuscript, tables and figures. All Authors reviewed and commented on the drafts and approved the final manuscript.
All Authors thank Iva Mencin, Nina Rupar, Ana Marentič, Ida Hramus, and Laura Albreht for their contributions to the review of the genetic variations involved in ECYT. The text was edited by Christopher Berrie. This study was supported by the Slovenian Research Agency (ARRS) through research project L3-9279, Young Researcher funding to AK and programmes P1-0390 and P4-0220.
APPENDIX 1.
| Company | Town | State |
|---|---|---|
| Ambry Genetics | Aliso Viejo | USA |
| Baylor Miraca Genetics Laboratories | Houston | USA |
| Bioarray | Elche | Spain |
| BloodGenetics | Llobregat | Spain |
| CeGaT GmbH | Tuebingen | Germany |
| CEN4GEN Institute for Genomics and Molecular Diagnostics | Edmonton | Canada |
| Center for Human Genetics | Brussels | Belgium |
| Centogene AG - the Rare Disease Company Germany | Rostock | Germany |
| CGC Genetics | Porto | Portugal |
| Cincinnati Children’s Hospital Medical Center Laboratory of Genetics and Genomics | Cincinnati | USA |
| Fulgent Genetics | Temple City | USA |
| GeneDx | Gaithersburg | USA |
| GENETAQ Molecular Genetics Centre and Diagnosis of Rare Diseases | Malaga | Spain |
| Instituto de Medicina Genomica | Paterna | Spain |
| Labor Dr. Wisplinghoff | Koeln | Germany |
| Laboratorio de Genetica Clinica SL | Madrid | Spain |
| Mayo Clinic Genetic Testing Laboratories | Rochester | USA |
| MedGene | Bratislava | Slovakia |
| Genome Diagnostics Lab | Toronto | Canada |
| Praxis fuer Humangenetik Wien | Wien | Austria |
| PreventionGenetics | Marshfield | USA |
| Reference Laboratory Genetics | Llobregat | Spain |
Footnotes
The Authors declare that they have no conflicts of interest.
REFERENCES
- 1.Orkin SH. Diversification of haematopoietic stem cells to specific lineages. Nat Rev Genet. 2000;1:57–64. doi: 10.1038/35049577. [DOI] [PubMed] [Google Scholar]
- 2.Zivot A, Lipton JM, Narla A, Blanc L. Erythropoiesis: insights into pathophysiology and treatments in 2017. Mol Med. 2018;24:11. doi: 10.1186/s10020-018-0011-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang JY, Hayashi Y, Yokota A, et al. Expansion of EPOR-negative macrophages besides erythroblasts by elevated EPOR signaling in erythrocytosis mouse models. Haematologica. 2018;103:40–50. doi: 10.3324/haematol.2017.172775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bento C, Percy MJ, Gardie B, et al. Genetic basis of congenital erythrocytosis: mutation update and online databases. Hum Mutat. 2014;35:15–26. doi: 10.1002/humu.22448. [DOI] [PubMed] [Google Scholar]
- 5.Debeljak N, Lazarevič J, Miskič D, et al. Characterization of erythrocytosis and a proposed diagnostic algorithm in Slovenia. Zdrav Vest. 2019;88:5–6. [Google Scholar]
- 6.Keohane C, McMullin MF, Harrison C. The diagnosis and management of erythrocytosis. BMJ Case Rep. 2013;347:f6667. doi: 10.1136/bmj.f6667. [DOI] [PubMed] [Google Scholar]
- 7.McMullin MF. Investigation and management of erythrocytosis. Curr Hematol Malig Rep. 2016;11:342–7. doi: 10.1007/s11899-016-0334-1. [DOI] [PubMed] [Google Scholar]
- 8.Arber DA, Orazi A, Hasserjian R, et al. The 2016 revision to the World Health Organization classification of myeloid neoplasms and acute leukemia. Blood. 2016;127:2391–405. doi: 10.1182/blood-2016-03-643544. [DOI] [PubMed] [Google Scholar]
- 9.Rumi E, McMullin MF, Harrison C, et al. Facing erythrocytosis: results of an international physician survey. Am J Hematol. 2019;94:E225–7. doi: 10.1002/ajh.25545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Camps C, Petousi N, Bento C, et al. Gene panel sequencing improves the diagnostic work-up of patients with idiopathic erythrocytosis and identifies new mutations. Haematologica. 2016;101:1306–18. doi: 10.3324/haematol.2016.144063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gordeuk VR, Stockton DW, Prchal JT. Congenital polycythemias/ erythrocytoses. Haematologica. 2005;90:109–16. [PubMed] [Google Scholar]
- 12.Chaudhry QUN, Ahmed P, Satti HS, et al. Post allogeneic hematopoietic cell transplant erythrocytosis in aplastic anemia: a review of 208 cases. Blood. 2017;130:5499. [Google Scholar]
- 13.Khanduja S, Takkar B, Khanduja N, Venkatesh P. Post-transplant erythrocytosis-related maculopathy: successful management of hyperviscosity with phlebotomy. Int Ophthalmol. 2018;38:2163–6. doi: 10.1007/s10792-017-0660-x. [DOI] [PubMed] [Google Scholar]
- 14.Motta G, Zavattaro M, Romeo F, et al. Risk of erythrocytosis during concomitant testosterone and SGLT2-inhibitor treatment: a warning from two clinical cases. J Clin Endocrinol Metab. 2018;104:819–22. doi: 10.1210/jc.2018-01702. [DOI] [PubMed] [Google Scholar]
- 15.Spivak JL. Polycythemia vera. Curr Treat Options Oncol. 2018;19:12. doi: 10.1007/s11864-018-0529-x. [DOI] [PubMed] [Google Scholar]
- 16.Semenza GL. Involvement of oxygen-sensing pathways in physiologic and pathologic erythropoiesis. Blood. 2009;114:2015–9. doi: 10.1182/blood-2009-05-189985. [DOI] [PubMed] [Google Scholar]
- 17.Moulard O, Mehta J, Fryzek J, et al. Epidemiology of myelofibrosis, essential thrombocythemia, and polycythemia vera in the European Union. Eur J Haematol. 2014;92:289–97. doi: 10.1111/ejh.12256. [DOI] [PubMed] [Google Scholar]
- 18.Girodon F, Airaud F, Garrec C, et al. Gene panel sequencing in idiopathic erythrocytosis. Haematologica. 2017;102:E30. doi: 10.3324/haematol.2016.158337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lappin TR, Lee FS. Update on mutations in the HIF: EPO pathway and their role in erythrocytosis. Blood Rev. 2019;37:100590. doi: 10.1016/j.blre.2019.100590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Broudy VC, Lin N, Brice M, et al. Erythropoietin receptor characteristics on primary human erythroid cells. Blood. 1991;77:2583–90. [PubMed] [Google Scholar]
- 21.Chida D, Miura O, Yoshimura A, Miyajima A. Role of cytokine signaling molecules in erythroid differentiation of mouse fetal liver hematopoietic cells: functional analysis of signaling molecules by retrovirus-mediated expression. Blood. 1999;93:1567–78. [PubMed] [Google Scholar]
- 22.Kolbus A, Blazquez-Domingo M, Carotta S, et al. Cooperative signaling between cytokine receptors and the glucocorticoid receptor in the expansion of erythroid progenitors: molecular analysis by expression profiling. Blood. 2003;102:3136–46. doi: 10.1182/blood-2003-03-0923. [DOI] [PubMed] [Google Scholar]
- 23.Robb L. Cytokine receptors and hematopoietic differentiation. Oncogene. 2007;26:6715–23. doi: 10.1038/sj.onc.1210756. [DOI] [PubMed] [Google Scholar]
- 24.Lapillonne H, Kobari L, Mazurier C, et al. Red blood cell generation from human induced pluripotent stem cells: perspectives for transfusion medicine. Haematologica. 2010;95:1651–9. doi: 10.3324/haematol.2010.023556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Franco RS. Measurement of red cell lifespan and aging. Transfus Med Hemother. 2012;39:302–7. doi: 10.1159/000342232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Klimmeck D, Hansson J, Raffel S, et al. Proteomic cornerstones of hematopoietic stem cell differentiation: distinct signatures of multipotent progenitors and myeloid committed cells. Mol Cell Proteomics. 2012;11:286–302. doi: 10.1074/mcp.M111.016790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rieger MA, Schroeder T. Hematopoiesis. Cold Spring Harb Perspect Biol. 2012;4:a008250. doi: 10.1101/cshperspect.a008250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sieweke MH, Allen JE. Beyond stem cells: self-renewal of differentiated macrophages. Science. 2013;342:1242974. doi: 10.1126/science.1242974. [DOI] [PubMed] [Google Scholar]
- 29.Grosso R, Fader CM, Colombo MI. Autophagy: a necessary event during erythropoiesis. Blood Rev. 2017;31:300–5. doi: 10.1016/j.blre.2017.04.001. [DOI] [PubMed] [Google Scholar]
- 30.Lang E, Lang F. Triggers, inhibitors, mechanisms, and significance of eryptosis: the suicidal erythrocyte death. Biomed Res Int. 2015;2015:513518. doi: 10.1155/2015/513518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang GL, Jiang BH, Rue EA, Semenza GL. Hypoxia-inducible factor-1 is a basic-helix-loop-helix-PAS heterodimer regulated by cellular O2 tension. Proc Natl Acad Sci U S A. 1995;92:5510–4. doi: 10.1073/pnas.92.12.5510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Franke K, Gassmann M, Wielockx B. Erythrocytosis: the HIF pathway in control. Blood. 2013;122:1122–8. doi: 10.1182/blood-2013-01-478065. [DOI] [PubMed] [Google Scholar]
- 33.Semenza GL, Wang GL. A nuclear factor induced by hypoxia via de novo protein-synthesis binds to the human erythropoietin gene enhancer at a site required for transcriptional activation. Mol Cell Biol. 1992;12:5447–54. doi: 10.1128/mcb.12.12.5447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jaakkola P, Mole DR, Tian YM, et al. Targeting of HIF-alpha to the von Hippel-Lindau ubiquitylation complex by O2-regulated prolyl hydroxylation. Science. 2001;292:468–72. doi: 10.1126/science.1059796. [DOI] [PubMed] [Google Scholar]
- 35.Ivan M, Kondo K, Yang HF, et al. HIF alpha targeted for VHL-mediated destruction by proline hydroxylation: implications for O2 sensing. Science. 2001;292:464–8. doi: 10.1126/science.1059817. [DOI] [PubMed] [Google Scholar]
- 36.Kaelin WG, Ratcliffe PJ, Semenza GL. Pathways for oxygen regulation and homeostasis. The 2016 Albert Lasker Basic Medical Research Award. JAMA. 2016;316:1252–3. doi: 10.1001/jama.2016.12386. [DOI] [PubMed] [Google Scholar]
- 37.Lee FS, Percy MJ. The HIF pathway and erythrocytosis. Annu Rev Pathol. 2011;6:165–92. doi: 10.1146/annurev-pathol-011110-130321. [DOI] [PubMed] [Google Scholar]
- 38.Kristan A, Debeljak N, Kunej T. Genetic variability of hypoxia-inducible factor alpha (HIFA) genes in familial erythrocytosis: analysis of the literature and genome databases. Eur J Haematol. 2019;103:287–99. doi: 10.1111/ejh.13304. [DOI] [PubMed] [Google Scholar]
- 39.Debeljak N, Solar P, Sytkowski AJ. Erythropoietin and cancer: the unintended consequences of anemia correction. Front Immunol. 2014;5:563. doi: 10.3389/fimmu.2014.00563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lasho TL, Pardanani A, Tefferi A. LNK mutations in JAK2 mutation-negative erythrocytosis. N Engl J Med. 2010;363:1189–90. doi: 10.1056/NEJMc1006966. [DOI] [PubMed] [Google Scholar]
- 41.Grinfeld J, Godfrey AL. After 10 years of JAK2V617F: disease biology and current management strategies in polycythaemia vera. Blood Rev. 2017;31:101–18. doi: 10.1016/j.blre.2016.11.001. [DOI] [PubMed] [Google Scholar]
- 42.Ginzburg YZ, Feola M, Zimran E, et al. Dysregulated iron metabolism in polycythemia vera: etiology and consequences. Leukemia. 2018;32:2105–16. doi: 10.1038/s41375-018-0207-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Moradkhani K, Prehu C, Old J, et al. Mutations in the paralogous human alpha-globin genes yielding identical hemoglobin variants. Ann Hematol. 2009;88:535–43. doi: 10.1007/s00277-008-0624-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Petousi N, Copley RR, Lappin TRJ, et al. Erythrocytosis associated with a novel missense mutation in the BPGM gene. Haematologica. 2014;99:E201–E4. doi: 10.3324/haematol.2014.109306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Amberger JS, Bocchini CA, Scott AF, Hamosh A. OMIM.org: leveraging knowledge across phenotype-gene relationships. Nucleic Acids Res. 2019;47:D1038–43. doi: 10.1093/nar/gky1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Levine RL, Wadleigh M, Cools J, et al. Activating mutation in the tyrosine kinase JAK2 in polycythemia vera, essential thrombocythemia, and myeloid metaplasia with myelofibrosis. Cancer Cell. 2005;7:387–97. doi: 10.1016/j.ccr.2005.03.023. [DOI] [PubMed] [Google Scholar]
- 47.Pasquier F, Marty C, Balligand T, et al. New pathogenic mechanisms induced by germline erythropoietin receptor mutations in primary erythrocytosis. Haematologica. 2018;103:575–86. doi: 10.3324/haematol.2017.176370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Peroni E, Bertozzi I, Gherlinzoni F, et al. Two novel missense mutations in EPOR gene causes erythrocytosis in two unrelated patients. Br J Haematol. 2018;180:908–11. doi: 10.1111/bjh.14486. [DOI] [PubMed] [Google Scholar]
- 49.Toriumi N, Kaneda M, Hatakeyama N, et al. A case of primary familial congenital polycythemia with a novel EPOR mutation: possible spontaneous remission/alleviation by menstrual bleeding. Int J Hematol. 2018;108:339–43. doi: 10.1007/s12185-018-2435-1. [DOI] [PubMed] [Google Scholar]
- 50.Bader HL, Hsu T. Systemic VHL gene functions and the VHL disease. FEBS Lett. 2012;586:1562–9. doi: 10.1016/j.febslet.2012.04.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lenglet M, Robriquet F, Schwarz K, et al. Identification of a new VHL exon and complex splicing alterations in familial erythrocytosis or von Hippel-Lindau disease. Blood. 2018;132:469–83. doi: 10.1182/blood-2018-03-838235. [DOI] [PubMed] [Google Scholar]
- 52.Sarangi S, Lanikova L, Kapralova K, et al. The homozygous VHLD126N missense mutation is associated with dramatically elevated erythropoietin levels, consequent polycythemia, and early onset severe pulmonary hypertension. Pediatr Blood Cancer. 2014;61:2104–6. doi: 10.1002/pbc.25056. [DOI] [PubMed] [Google Scholar]
- 53.Barradas J, Rodrigues CD, Ferreira G, et al. Congenital erythrocytosis - discover of a new mutation in the EGLN1 gene. Clin Case Rep. 2018;6:1109–11. doi: 10.1002/ccr3.1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sinnema M, Song DS, Guan W, et al. Loss-of-function zinc finger mutation in the EGLN1 gene associated with erythrocytosis. Blood. 2018;132:1455–8. doi: 10.1182/blood-2018-06-854711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gardie B, Percy M, Hoogewijs D, et al. The role of PHD2 mutations in the pathogenesis of erythrocytosis. Hypoxia (Auckl) 2014;2:71–90. doi: 10.2147/HP.S54455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tarade D, Robinson CM, Lee JE, Ohh M. HIF-2 alpha-pVHL complex reveals broad genotype-phenotype correlations in HIF-2 alpha-driven disease. Nat Commun. 2018;9:3359. doi: 10.1038/s41467-018-05554-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Liu QL, Tong DL, Liu GL, et al. HIF2A germline-mutation-induced polycythemia in a patient with VHL-associated renal-cell carcinoma. Cancer Biol Ther. 2017;18:944–7. doi: 10.1080/15384047.2017.1394553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zmajkovic J, Lundberg P, Nienhold R, et al. A gain-of-function mutation in EPO in familial erythrocytosis. N Engl J Med. 2018;378:924–30. doi: 10.1056/NEJMoa1709064. [DOI] [PubMed] [Google Scholar]
- 59.Vocanec D, Prijatelj T, Debeljak N, Kunej T. Genetic variants of erythropoietin (EPO) and EPO receptor genes in familial erythrocytosis. Int J Lab Hematol. 2019;41:162–7. doi: 10.1111/ijlh.12949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bento C. Genetic basis of congenital erythrocytosis. Int J Lab Hematol. 2018;40:62–7. doi: 10.1111/ijlh.12828. [DOI] [PubMed] [Google Scholar]
- 61.Fokkema IFAC, Taschner PEM, Schaafsma GCP, et al. LOVD v.2.0: the next generation in gene variant databases. Hum Mutat. 2011;32:557–63. doi: 10.1002/humu.21438. [DOI] [PubMed] [Google Scholar]
- 62.Scott LM, Tong W, Levine RL, et al. JAK2 exon 12 mutations in polycythemia vera and idiopathic erythrocytosis. N Engl J Med. 2007;356:459–68. doi: 10.1056/NEJMoa065202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Schnittger S, Bacher U, Haferlach C, et al. Detection of JAK2 exon 12 mutations in 15 patients with JAK2V617F negative polycythemia vera. Haematologica. 2009;94:414–8. doi: 10.3324/haematol.13223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Trifa AP, Banescu C, Bojan AS, et al. MECOM, HBS1L-MYB, THRB-RARB, JAK2, and TERT polymorphisms defining the genetic predisposition to myeloproliferative neoplasms: a study on 939 patients. Am J Hematol. 2017;93:100–6. doi: 10.1002/ajh.24946. [DOI] [PubMed] [Google Scholar]
- 65.Bahar B, Barton K, Kini AR. The role of the exon 13 G571S JAK2 mutation in myeloproliferative neoplasms. Leuk Res Rep. 2016;6:27–8. doi: 10.1016/j.lrr.2016.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Patel AB, Franzini A, Leroy E, et al. JAK2 ex13InDel drives oncogenic transformation and is associated with chronic eosinophilic leukemia and polycythemia vera. Blood. 2019;134:2388–98. doi: 10.1182/blood.2019001385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kapralova K, Horvathova M, Kucerova J, et al. JAK2 E846D germline mutation associated with erythrocytosis causes increased and prolonged Epo-induced activation of STAT5. Blood. 2014;124:4008. [Google Scholar]
- 68.Helbig G. Classical Philadelphia-negative myeloproliferative neoplasms: focus on mutations and JAK2 inhibitors. Med Oncol. 2018;35:119. doi: 10.1007/s12032-018-1187-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Maslah N, Cassinat B, Verger E, et al. The role of LNK/SH2B3 genetic alterations in myeloproliferative neoplasms and other hematological disorders. Leukemia. 2017;31:1661–70. doi: 10.1038/leu.2017.139. [DOI] [PubMed] [Google Scholar]
- 70.Spolverini A, Pieri L, Guglielmelli P, et al. Infrequent occurrence of mutations in the PH domain of LNK in patients with JAK2 mutation-negative ‘idiopathic’ erythrocytosis. Haematologica. 2013;98:E101–E2. doi: 10.3324/haematol.2013.090175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kralovics R, Sokol L, Prchal JT. Absence of polycythemia in a child with a unique erythropoietin receptor mutation in a family with autosomal dominant primary polycythemia. J Clin Invest. 1998;102:124–9. doi: 10.1172/JCI2886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Al-Sheikh M, Moradkhani K, Lopez M, et al. Disturbance in the HIF-1 alpha pathway associated with erythrocytosis: further evidences brought by frameshift and nonsense mutations in the prolyl hydroxylase domain protein 2 (PHD2) gene. Blood Cells Mol Dis. 2008;40:160–5. doi: 10.1016/j.bcmd.2007.07.017. [DOI] [PubMed] [Google Scholar]
- 73.Al-Sheikh M, Mazurier E, Gardie B, et al. A study of 36 unrelated cases with pure erythrocytosis revealed three new mutations in the erythropoietin receptor gene. Haematologica. 2008;93:1072–5. doi: 10.3324/haematol.12260. [DOI] [PubMed] [Google Scholar]
- 74.Arcasoy MO, Degar BA, Harris KW, Forget BG. Familial erythrocytosis associated with a short deletion in the erythropoietin receptor gene. Blood. 1997;89:4628–35. [PubMed] [Google Scholar]
- 75.Arcasoy MO, Karayal AF, Segal HM, et al. A novel mutation in the erythropoietin receptor gene is associated with familial erythrocytosis. Blood. 2002;99:3066–9. doi: 10.1182/blood.v99.8.3066. [DOI] [PubMed] [Google Scholar]
- 76.Kralovics R, Indrak K, Stopka T, et al. Two new EPO receptor mutations: truncated EPO receptors are most frequently associated with primary familial and congenital polycythemias. Blood. 1997;90:2057–61. [PubMed] [Google Scholar]
- 77.Le Couedic JP, Mitjavila MT, Villeval JL, et al. Missense mutation of the erythropoietin receptor is a rare event in human erythroid malignancies. Blood. 1996;87:1502–11. [PubMed] [Google Scholar]
- 78.Bento C, McMullin MF, Percy M, Cario H. Primary familial and congenital polycythemia. In: Adam MP, Ardinger HH, Pagon RA, et al., editors. GeneReviews. Seattle, WA: University of Washington; 1993–2020. [PubMed] [Google Scholar]
- 79.De La Chapelle A, Traskelin AL, Juvonen E. Truncated erythropoietin receptor causes dominantly inherited benign human erythrocytosis. Proc Natl Acad Sci U S A. 1993;90:4495–9. doi: 10.1073/pnas.90.10.4495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Furukawa T, Narita M, Sakaue M, et al. Primary familial polycythaemia associated with a novel point mutation in the erythropoietin receptor. Br J Haematol. 1997;99:222–7. doi: 10.1046/j.1365-2141.1997.3583172.x. [DOI] [PubMed] [Google Scholar]
- 81.Percy MJ, Beer PA, Campbell G, et al. Novel exon 12 mutations in the HIF2A gene associated with erythrocytosis. Blood. 2008;111:5400–2. doi: 10.1182/blood-2008-02-137703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Tan QL, Kerestes H, Percy MJ, et al. Erythrocytosis and pulmonary hypertension in a mouse model of human HIF2A gain of function mutation. J Biol Chem. 2013;288:17134–44. doi: 10.1074/jbc.M112.444059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Perrotta S, Nobili B, Ferraro M, et al. Von Hippel-Lindau-dependent polycythemia is endemic on the island of Ischia: identification of a novel cluster. Blood. 2006;107:514–9. doi: 10.1182/blood-2005-06-2422. [DOI] [PubMed] [Google Scholar]
- 84.Gardie B, Couve S, Ladroue C, et al. A comprehensive study of the VHL-R200W Chuvash polycythemia mutation reveals a gradual dysregulation of the hypoxia pathway in oncogenesis. Blood. 2014;124:4020. [Google Scholar]
- 85.Jorge SE, Bringas M, Petruk AA, et al. Understanding the molecular basis of the high oxygen affinity variant human hemoglobin Coimbra. Arch Biochem Biophys. 2018;637:73–8. doi: 10.1016/j.abb.2017.11.010. [DOI] [PubMed] [Google Scholar]
- 86.Brennan SO, Pullon B, Owen MC, Brittain T. Novel haemoglobin mutation (alpha 127Lys -> Glu) increases oxygen affinity and has a minor effect on haptoglobin binding. Clin Biochem. 2012;45:1587–90. doi: 10.1016/j.clinbiochem.2012.08.021. [DOI] [PubMed] [Google Scholar]
- 87.Wajcman H, Galacteros F. Hemoglobins with high oxygen affinity leading to erythrocytosis. New variants and new concepts. Hemoglobin. 2005;29:91–106. [PubMed] [Google Scholar]
- 88.Shin SY, Kim HY, Kim HJ, Kim HG. Hb Heathrow [beta 103(G5) Phe -> Leu], a first report in an Asian patient with erythrocytosis. Yonsei Med J. 2017;58:665–7. doi: 10.3349/ymj.2017.58.3.665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Lemarchandel V, Joulin V, Valentin C, et al. Compound heterozygosity in a complete erythrocyte bisphosphoglycerate mutase deficiency. Blood. 1992;80:2643–9. [PubMed] [Google Scholar]
- 90.Rosa R, Prehu MO, Beuzard Y, Rosa J. The first case of a complete deficiency of diphosphoglycerate mutase in human erythrocytes. J Clin Invest. 1978;62:907–15. doi: 10.1172/JCI109218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Hoyer JD, Allen SL, Beutler E, et al. Erythrocytosis due to bisphosphoglycerate mutase deficiency with concurrent glucose-6- phosphate dehydrogenase (G-6-PD) deficiency. Am J Hematol. 2004;75:205–8. doi: 10.1002/ajh.20014. [DOI] [PubMed] [Google Scholar]
- 92.McMullin MF, Harrison CN, Ali S, et al. A guideline for the diagnosis and management of polycythaemia vera. A British Society for Haematology guideline. Br J Haematol. 2019;184:176–91. doi: 10.1111/bjh.15648. [DOI] [PubMed] [Google Scholar]
- 93.Baxter EJ, Scott LM, Campbell PJ. Acquired mutation of the tyrosine kinase JAK2 in human myeloproliferative disorders. Lancet. 2005;365:1054–61. doi: 10.1016/S0140-6736(05)71142-9. [DOI] [PubMed] [Google Scholar]
- 94.Sanger F, Nicklen S, Coulson AR. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977;74:5463–7. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Smith LM, Sanders JZ, Kaiser RJ, et al. Fluorescence detection in automated DNA-sequence analysis. Nature. 1986;321:674–9. doi: 10.1038/321674a0. [DOI] [PubMed] [Google Scholar]
- 96.Basho RK, Eterovic AK, Meric-Bernstam F. Clinical applications and limitations of next-generation sequencing. Am J Hematol Oncol. 2015;11:17–22. [Google Scholar]
- 97.Hall N. Advanced sequencing technologies and their wider impact in microbiology. J Exp Biol. 2007;210:1518–25. doi: 10.1242/jeb.001370. [DOI] [PubMed] [Google Scholar]
- 98.Peyro-Saint-Paul H, Hermitte F. Detection of the JAK2V617F mutation with the Ipsogen MutaScreen kit: absence of JAK2V617F does not mean absence of myeloproliferative neoplasm. Blood. 2010;116:1994–5. doi: 10.1182/blood-2010-06-289819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Hussein K, Percy M, McMullin MF. Clinical utility gene card for: familial erythrocytosis. Eur J Hum Genet. 2012;20 doi: 10.1038/ejhg.2011.252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Rubinstein WS, Maglott DR, Lee JM, et al. The NIH genetic testing registry: a new, centralized database of genetic tests to enable access to comprehensive information and improve transparency. Nucleic Acids Res. 2013;41:D925–35. doi: 10.1093/nar/gks1173. [DOI] [PMC free article] [PubMed] [Google Scholar]