Abstract
ISP-1 is a new type of immunosuppressant, the structure of which is homologous to that of sphingosine. In a previous study, ISP-1 was found to inhibit mammalian serine palmitoyltransferase, the primary enzyme involved in sphingolipid biosynthesis, and to reduce the intracellular pool of sphingolipids. ISP-1 induces the apoptosis of cytotoxic T cells, which is triggered by decreases in the intracellular levels of sphingolipids. In this study, the inhibition of yeast (Saccharomyces cerevisiae) proliferation by ISP-1 was observed. This ISP-1-induced growth inhibition was also triggered by decreases in the intracellular levels of sphingolipids. In addition, DNA duplication without cytokinesis was detected in ISP-1-treated yeast cells on flow cytometry analysis. We have cloned multicopy suppressor genes of yeast which overcome the lethal sphingolipid depletion induced by ISP-1. One of these genes, SLI2, is synonymous with YPK1, which encodes a serine/threonine kinase. Kinase-dead mutants of YPK1 did not show any resistance to ISP-1, leading us to predict that the kinase activity of the Ypk1 protein should be essential for this resistance to ISP-1. Ypk1 protein overexpression had no effect on sphingolipid biosynthesis by the yeast. Furthermore, both the phosphorylation and intracellular localization of the Ypk1 protein were regulated by the intracellular sphingolipid levels. These data suggest that the Ypk1 protein is a downstream kinase in the sphingolipid-mediated signaling pathway of yeast. The Ypk1 protein was reported to be a functional homologue of the mammalian protein kinase SGK, which is a downstream kinase of 3-phosphoinositide-dependent kinase 1 (PDK1). PDK1 phosphotidylinositol (PI) is regulated by PI-3,4,5-triphosphate and PI-3,4-bisphosphate through the pleckstrin homology (PH) domain. Overexpression of mammalian SGK also overcomes the sphingolipid depletion in yeast. Taking both the inability to produce PI-3,4,5-triphosphate and PI-3,4-bisphosphate and the lack of a PH domain in the yeast homologue of PDK1, the Pkh1 protein, into account, these findings further suggest that yeast may use sphingolipids instead of inositol phospholipids as lipid mediators.
Recent reports have shown that sphingolipids are involved in several biological functions, including adhesion, differentiation, growth, and apoptosis (3, 22, 23, 25, 26, 44, 58). Ceramide and sphingosine-1-phosphate, which are metabolic products of sphingolipids, are thought to be upregulated by several stimuli and to serve as second messengers in signal transduction pathways (24, 27, 50, 55). Sphingosine-1-phosphate is upregulated by the platelet-derived growth factor stimulus (51), and the increase in sphingosine-1-phosphate promotes the growth of 3T3 fibroblast cells (66). Recently, G-protein-coupled cell surface receptors for sphingosine-1-phosphate were found in several cell lines (4, 40, 68). Furthermore, the possible involvement of intracellular sphingosine-1-phosphate in several signal transduction pathways has been postulated (31, 59). In addition to sphingosine-1-phosphate, intracellular ceramide upregulation through the sphingomyelin cycle on stimulation by tumor necrosis factor alpha (56), Fas ligand (11, 20, 21), or UV or γ irradiation (24) causes apoptosis of Jurkat T cells and several other cell lines, although exogenously added ceramide does not exert all of the downstream effects of these stimulants (30, 57, 64).
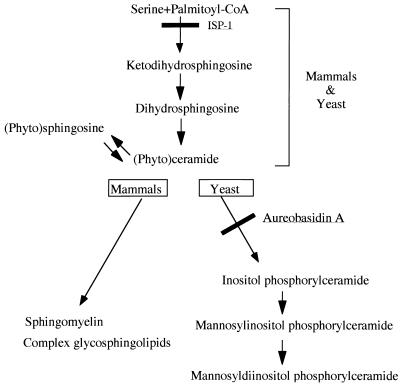
The yeast Saccharomyces cerevisiae synthesizes sphingolipids similarly to mammalian sphingolipids except that phytosphingosine rather than sphingosine is the predominant sphingoid base (Fig. 1), and it lacks cell surface receptors for sphingosine-1-phosphate (16). Therefore, yeast is a useful system for studying the intracellular sphingolipid-mediated signaling pathway. Indeed, yeast has a sphingosine kinase which produces sphingosine-1-phosphate from sphingosine, and the former is a crucial substance involved in the respiration of yeast cells (39). Ceramide has been shown to induce G1 arrest in yeast via a ceramide-activated protein phosphatase (49). A new phosphatase, which modulates stress responses through sphingolipid metabolites, has also been isolated from yeast (42). However, most of the downstream pathway of intracellular sphingolipid-mediated signaling has not been well clarified in yeast or mammals.
FIG. 1.
Major sphingolipid biosynthetic pathways in yeast and mammalian cells.
ISP-1 was isolated from Isaria sinclairii as a new type of potent immunosuppressant, the structure of which is homologous to that of sphingosine (17). In our previous studies, ISP-1 was found to inhibit mammalian serine palmitoyltransferase (SPT), which catalyzes the first step of sphingolipid biosynthesis. ISP-1 induced the apoptosis of mouse cytotoxic T cells (17, 45, 48) and rat Purkinje cells (18), which was triggered by reduction of the intercellular pools of sphingolipid metabolites. These results suggest that ISP-1 is a useful tool for the depletion of intracellular sphingolipids and for studying the sphingolipid-mediated signaling pathway in both mammals and yeast.
We report that ISP-1 prevents yeast proliferation due to the inhibition of sphingolipid biosynthesis and that a multicopy suppressor gene of ISP-1 encodes a downstream kinase of the sphingolipid-mediated signaling pathway in yeast.
MATERIALS AND METHODS
Strains, plasmids, and reagents.
S. cerevisiae KMY1005α (MATα leu2-3 ura3-52 his3-200 trp1-901 lys2-801 ADE2) and S. cerevisiae KMY1006a (MATa leu2-3 ura3-52 his3-200 trp1-901 lys2-801 ADE2) were used. A yeast genomic DNA library in YEp13 (an episomal, multicopy yeast vector containing the LEU2 selectable marker) was obtained from the American Type Culture Collection. ISP-1 was obtained as previously described (45) and stored in a methanol solution at −20°C.
Molecular cloning of ISP-1-resistant genes.
Plasmid pSLI, carrying the SLI gene, was selected by screening the yeast genomic DNA library in YEp13 on SD plates (0.67% Bacto Yeast Nitrogen Base without amino acids, 2% dextrose, 2% Bacto Agar) containing 1.25 μM ISP-1. Yeast cells were transformed by the lithium acetate method (33). pSLI was propagated in Escherichia coli DH5α. The 2.8-kb BstBI-BstBI fragment containing the wild-type YPK1 gene from pSLI was subcloned into YEp351 (an episomal, multicopy yeast vector containing the LEU2 selectable marker).
Yeast culture conditions.
The SD medium (SD agar plates) consisted of yeast nitrogen base, essential amino acids, and glucose. Colonies selected on agar plates were inoculated into SD medium and then incubated overnight at 30°C. Exponentially growing cultures were diluted to the desired concentrations, aliquoted into sterile culture tubes, and then treated with ISP-1 (1.25 μM) or methanol as a control. Proliferation was determined as the absorbance (optical density) at 600 nm (OD600) measured with a Shimadzu UV-160 spectrophotometer. For yeast viability measurement, a yeast culture was mixed with an equal volume of a methylene blue solution (0.2 mg/ml in 0.1 M phosphate buffer, pH 4.6). The mixture was left for 5 min at room temperature, and then the total cells and stained (dead) cells were counted under a microscope. Viability was expressed as the mean of the percentage of unstained cells.
Construction of plasmids.
Mutant alleles of the YPK1 gene were obtained as described by Deng and Nickoloff (12). A 1.2-kb PstI-SacI fragment of the wild-type YPK1 gene was subcloned into pBluescript II SK+. Two kinds of point mutants of 376-Lys were generated with a Transformer site-directed mutagenesis kit (Clontech Laboratories, Inc.). The mutant alleles of YPK1 gene constructs were generated by swapping the mutant region between the PstI and SacI sites with the corresponding region of wild-type YPK1. To construct the Ypk1-enhanced green fluorescent protein (EGFP) fusion protein, a YPK1 DNA fragment, in which the stop codon of YPK1 was replaced with a leucine codon and a HindIII site, was introduced after the leucine codon had been generated by long and accurate PCR using primers 5′-TATCATGGAAGCGCCTGTTG-3′ and 5′-GACTGCAGAATTCGAAGCTTCAATCTAATGCTTCTACCTTGCA-3′. The PvuII-HindIII fragment of the PCR product was subcloned into pBluescript KS+. The YPK1 DNA fragment with a disrupted stop codon was isolated by XhoI and HindIII digestion and then ligated into pEGFP-N1 (Clontech). The YPK1-EGFP fragment was then inserted into YEp351 using linkers containing NotI, BamHI, XhoI, and XbaI sites. Expression of the Ypk1-EGFP fusion protein did not change on treatment with ISP-1 or phytosphingosine (data not shown). An SGK construct was prepared by amplifying rat SGK cDNA with primers 5′-CCGGAATTCCGGATGACGGTGAAAACTGAGGC and 5′-CGGGATCCCGAAACCAAGCCCTAACAGGG. The PCR product was digested with BamHI and EcoRI and then subcloned into YEp351 with the ADH1 promoter. A PKH1 construct were prepared by amplifying the yeast genomic DNA library in YEp13 with primers 5′-AGCAGACACTATCTTGGTTTC and 5′-TGTACGTACGTACACTATCCTGAAATACATGCCCG. The PCR product was subcloned between the SacI and SphI sites of YEp352 (an episomal, multicopy yeast vector containing the URA3 selectable marker). All constructs were confirmed by automated DNA sequencing.
Preparation of antibodies specific to the Ypk1 protein.
The 0.9-kb EcoRI-EcoRI fragment corresponding to amino acids (aa) 1 to 258 of the Ypk1 protein was ligated in frame to the 3′ end of the glutathione S-transferase (GST) gene coding sequence in the cytoplasmic expression vector pGEX-4T-1 (Pharmacia Biotech). Ypk1 protein (aa 1 to 258)-GST binding protein was expressed in E. coli DH5α, followed by induction with isopropyl-β-thiogalactopyranoside. After sonication, the fusion protein was recovered in the insoluble fraction, purified on a glutathione-Sepharose 4B (Pharmacia Biotech) column, and then eluted from a sodium dodecyl sulfate gel. The antigen obtained was then injected into DA rats three times at 2-week intervals. The rats were killed under deep anesthesia. Following its collection, blood was allowed to clot at 4°C overnight and then centrifuged at 1,500 × g for 10 min at 4°C. The supernatant was then used as antibodies specific to the Ypk1 protein.
Immunoblotting.
Total protein extracts were prepared by growing cells to the mid-log phase, harvesting, resuspension in lysis buffer (50 mM Tris-HCl buffer [pH 8.0] containing 1 mM phenylmethylsulfonyl fluoride, 1 μg of leupeptin per ml, 1 μg of pepstatin per ml, 50 mM NaF, 1 mM Na3VO4, 1.5 mM MgCl2, 150 mM NaCl), and vortexing to lyse cells for 3 min with a half volume of 0.3- to 0.5-mm-diameter glass beads as described by Kohno et al. (38). Unbroken cells and debris were removed by centrifugation of the homogenates at 4,000 × g for 10 min. The resulting supernatant was separated on a sodium dodecyl sulfate-polyacrylamide gel. The Ypk1 protein was detected using anti-Ypk1 protein (aa 1 to 258) antibodies. Blots were examined using a diaminobenzidine kit (Vector Laboratories, Inc.) for the detection of overexpressed Ypk1 protein or a chemiluminescent reagent (Pierce) for the detection of endogenous Ypk1 protein. Protein concentrations were measured with a Bio-Rad protein assay kit.
SPT assay.
SPT assays were carried out by a modification of the method of Buede et al. (5). For each assay, the following components were used, in a final volume of 0.2 ml: 0.1 M HEPES (pH 8.3), 0.5 mM dithiothreitol (DTT), 2.5 mM EDTA (pH 7.4), 50 mM pyridoxal phosphate, 40 mM palmitoyl coenzyme A (palmitoyl-CoA), 5 mM l-serine, 2 μCi of l-[3H]serine, 0.01 to 100 nM ISP-1, and 0.5 mg of yeast total protein extract prepared with phosphate buffer (50 mM sodium phosphate [pH 7.0] containing 5 mM DTT and 1 mM phenylmethylsulfonyl fluoride). After incubation for 30 min at 30°C, 0.4 ml of 0.5 M NH4OH was added, and then the lipid products were extracted with chloroform. The chloroform layer was transferred to a scintillation vial and then dried down completely before the radioactivity was measured with a Beckman LS5000 liquid scintillation counter. The result for control without palmitoyl-CoA was subtracted from all the measurements to calculate the specific activity.
Synchronization of yeast.
For α-factor synchronization (67), strain KMY1006a was first grown in SD medium at 30°C to a density of 0.5 × 107 cells/ml. α-Factor (Sigma) was added to a final concentration of 0.5 U/ml. After incubation at 30°C for 2 h, more than 95% of the cells showed the shmoo terminal morphology, as judged microscopically. The cells were then spun down and washed three times with prewarmed SD medium. The cells were diluted to the desired concentration, aliquoted into sterile culture tubes, and then incubated at 30°C in the presence of ISP-1 or methanol as a control.
Flow cytometry.
Yeast samples were prepared for flow cytometric analysis by a modification of the method of Lew et al. (41). Cultures were fixed in 70% ethanol overnight at 4°C, followed by 4 h of digestion with RNase A. The cells were stained with propidium iodide. Samples were diluted and then sonicated for 10 s. Fluorescence was measured with a Becton Dickinson FACScan.
Alkaline phosphatase treatment of the Ypk1 protein.
Alkaline phosphatase treatment was carried out as described by Kohn et al. (37). Yeast cells transfected with YEp351 containing YPK1 were lysed with Tris buffer, and then total yeast protein extracts were prepared as described above. Immunoprecipitates were prepared by preabsorbing 5 μl of anti-Ypk1 protein antibodies to 20 μl of a 50:50 slurry of protein G-Sepharose. The beads were washed three times with 25 mM HEPES (pH 7.6) containing 0.1% bovine serum albumin, 10% glycerol, and 0.1% Triton X-100 and then twice with phosphatase buffer (10 mM Tris [pH 8], 1 mM MgCl2, 1 mM DTT). The beads were then incubated with shaking at 37°C for 1 h in phosphatase buffer with and without 20 U of calf intestinal alkaline phosphatase. When required, samples containing phosphatase inhibitors were supplemented with 10 mM NaF and 1 mM Na3VO4. The beads were then washed once with 25 mM HEPES (pH 7.6) containing 0.1% bovine serum albumin 10% glycerol, and 0.1% Triton X-100 and resuspended in the gel loading buffer.
Sphingolipid synthesis.
Two-milliliter cultures were grown to 2 × 106 cells/ml in SD medium at 30°C and then labeled with [3H]serine (20 μCi/ml) for 4 h. The cultures were chilled on ice after the addition of 0.5 ml of unlabeled stationary-phase cells and then subjected to centrifugation at 2,800 × g for 10 min at 4°C. The cells were washed two times with 5 ml of cold H2O and then treated with 5% trichloroacetic acid at 4°C for 20 min. Lipids were extracted twice with 1 ml of ethanol-water-diethyl ether-pyridine-NH4OH (15:15:5:1:0.018) at 60°C as described elsewhere (28). The [3H]serine-labeled extract was subjected to mild alkaline methanolysis with 0.1 N KOH. The lipid products were extracted with chloroform as described above for the SPT assay. The lipid products were dried under N2, resuspended in 0.04 ml of chloroform-methanol-H2O (16:16:5), applied to a silica gel thin-layer chromatography plate, and then resolved with CHCl3-methanol-4.2 N NH4OH (9:7:2). Radioactive bands were visualized with a BAS 3000 image analyzer.
Microscopy.
The Ypk1-EGFP fusion protein was observed by confocal microscopy (Olympus) without fixation. Samples were treated for 4 h with methanol (as a control), ISP-1, and phytosphingosine.
RESULTS
Inhibition of yeast cell growth by ISP-1 was prevented by the addition of phytosphingosine.
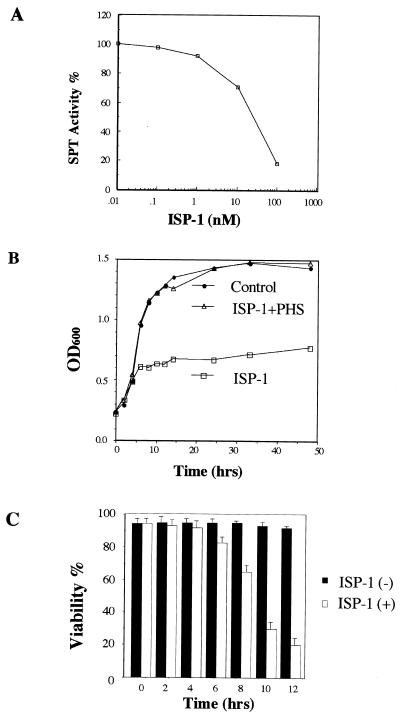
In our previous study, ISP-1 was found to inhibit the SPT activity of CTLL-2 cells (45). The similarity of the sphingosine biosynthesis pathways in mammalian and yeast cells prompted us to examine whether ISP-1 has any influence on SPT in yeast. As shown in Fig. 2A, the SPT activity of yeast was inhibited in a dose-dependent manner with nanomole concentrations.
FIG. 2.
Effects of ISP-1 on yeast cells. (A) In vitro SPT activity was inhibited by ISP-1 in a dose-dependent manner. The effect of ISP-1 on SPT was studied using a low-speed supernatant of sonicated yeast cells as the enzyme source. Samples were prepared and analyzed as described in Materials and Methods. (B) Growth inhibition of ISP-1 was abolished by the addition of phytosphingosine (PHS). KMY1005α cells were grown in medium containing 1.25 μM ISP-1 and 5 μM phytosphingosine. Proliferation was determined by measuring the OD600. (C) Effect of ISP-1 on yeast cell viability. Values represent the means ± standard deviations for three determinations.
The effect of ISP-1 on yeast growth was examined since the genes encoding SPT are essential for yeast cell growth (47, 53). When yeast cells were treated with ISP-1, cell growth was inhibited after about 5 h (Fig. 2B). The viability of yeast cells was markedly decreased after the growth inhibition (Fig. 2C). Because SPT is the primary enzyme involved in sphingolipid biosynthesis, ISP-1 also reduced the intracellular pools of sphingolipids in yeast (see Fig. 5B).
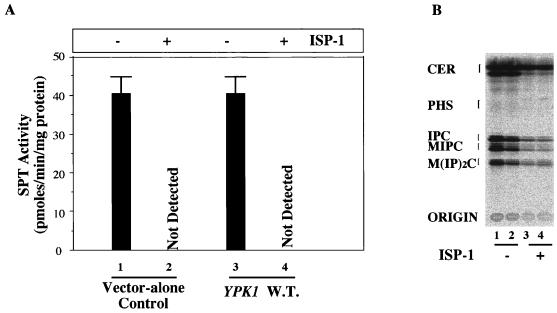
FIG. 5.
Effects of overexpression of the Ypk1 protein on de novo sphingolipid synthesis and SPT activity. (A) SPT activity was measured as described for Fig. 2, using a low-speed supernatant of the sonicated yeast as the enzyme source. The transformants in lines 1 and 3 were cultured for 4 h in the absence of ISP-1, and those in lines 2 and 4 were cultured for 4 h in the presence of 1.25 μM ISP-1 before sonication. W.T., wild type. (B) Ypk1 protein overexpression did not interfere with sphingolipid biosynthesis. The sphingolipids of YPK1 (lanes 2 and 4) and vector transformants (lanes 1 and 3) were labeled with [3H]serine for 4 h in the absence (lanes 1 and 2) or presence (lanes 3 and 4) of 1.25 μM ISP-1. Sphingolipids were analyzed as described in Materials and Methods. The positions of standard lipids are indicated by brackets. CER, ceramide; PHS, phytosphingosine; IPC, inositol phosphorylceramide; MIPC, mannose inositol phosphorylceramide; M(IP)2C, mannose diinositol diphosphoryl-ceramide.
To determine whether the sphingolipid depletion is a direct trigger of yeast cell death, the effect of exogenously added phytosphingosine, a metabolite of yeast sphingolipids, on ISP-1-induced yeast growth inhibition was studied. Phytosphingosine is easily incorporated into yeast cells and then converted to other sphingolipids (19). Yeast cells were grown in medium containing 1.25 μM ISP-1 and 5 μM phytosphingosine. The growth inhibition induced by ISP-1 was completely abolished (Fig. 2B). These results confirmed that ISP-1 induced yeast growth inhibition triggered by reductions in the intracellular level of sphingolipids due to the inhibition of SPT by ISP-1.
ISP-1 inhibits cytokinesis but not DNA replication.
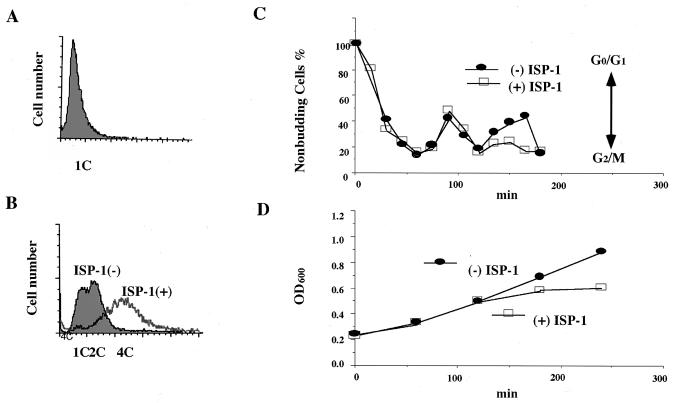
To further clarify the mechanism underlying the inhibition of yeast growth by ISP-1, we examined the effect of ISP-1 on α-factor-arrested yeast cells. KMY1006a, which has the same genotype as KMY1005α except that its mating type is the a type, was used in this experiment. Cells were synchronized at the G1 phase of the cell cycle with α-factor (Fig. 3A) and then released from the G1 block in the absence or presence of ISP-1. The percentage of unbudded cells was determined microscopically as a parameter of the cell cycle every 15 min (Fig. 3C), and cell numbers were determined by measuring the OD600 (Fig. 3D). The growth of synchronized cells was arrested primarily, with a late S or G2-like morphology, in the second cell cycle (Fig. 3C). However, the DNA content increased after the arrest, and the average DNA content finally reached that in the 4C state, at which ISP-1-treated cells died (Fig. 3B); this result was consistent with a mismatch between cell division and DNA synthesis of yeast due to the long-chain-base starvation (52). These findings suggest that ISP-1 inhibits cytokinesis but not DNA replication.
FIG. 3.
Effect of ISP-1 on the yeast cell cycle. The DNA contents of α-factor synchronized cells were determined by flow cytometry before (A) and after (B) 3 h of incubation with 1.25 μM ISP-1 or methanol as a control. During the incubation, the percentage of unbudded cells (C) and yeast growth (D) were determined every 15 min. Yeast cells were incubated with 1.25 μM ISP-1 or the methanol vehicle (control) for 4 h at 30°C.
Isolation of SLI genes.
The above findings prompted us to isolate multicopy suppressor genes for ISP-1-induced sphingolipid depletion as candidates for the downstream components of the sphingolipid-mediated signaling pathway in yeast. A yeast multicopy plasmid-based genomic DNA library was used to screen for ISP-1 resistance genes. Approximately 3,400 transformants were screened on plates containing 1.25 μM ISP-1. As previously described, 1.25 μM is the lowest concentration that can completely inhibit yeast cell growth. Twenty colonies showed ISP-1 resistance. Based on the results of DNA sequence analysis, the inserts of the plasmids isolated from the 20 positive yeast colonies were classified as four genes, referred to as SLI1, SLI2, SLI3, and SLI4 (SLI denotes sphingosine-like immunosuppressant resistance gene). SLI2 is a synonym of YPK1 (7). Below we focus on YPK1.
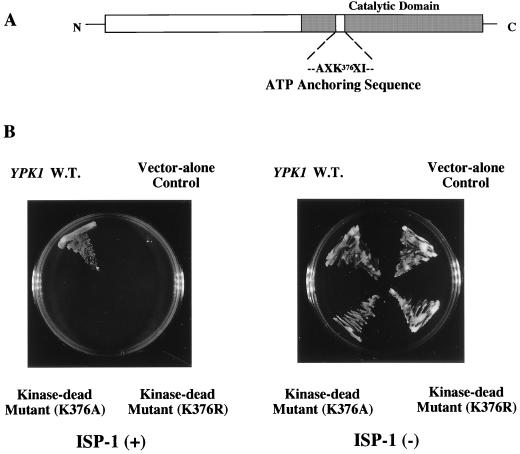
Previously, YPK1 was isolated by hybridization to bovine cyclic AMP-dependent protein kinase cDNA and mapped to the left arm of chromosome XI (43). The protein kinase catalytic domain is located in the carboxy-terminal portion of the Ypk1 protein (Fig. 4A).
FIG. 4.
Involvement of the protein kinase activity of the Ypk1 protein in the ISP-1-resistant phenotype. (A) Schematic representation of the primary structure of the Ypk1 protein. The protein kinase catalytic domain is located in the carboxy-terminal portion of the protein. Lys-376 of the catalytic domain is predicted to be directly involved in anchoring of the γ phosphate group of ATP. (B) Effect of protein kinase activity of the Ypk1 protein on ISP-1 resistance. Strain KMY1005α was transformed with plasmid YEp351 containing wild-type (W.T.) YPK1 or one of its point mutants (K376R and K376A). Transformants were plated on leucine-deficient SD plates containing 1.25 μM ISP-1 or methanol. Strain KMY1005α transformed with plasmid YEp351 was used as a control. The plates were incubated at 30°C for 2 days.
Kinase-dead mutants of the Ypk1 protein did not exhibit any resistance to ISP-1.
As shown in Fig. 4B, yeast cells overexpressing the Ypk1 protein showed considerable resistance to ISP-1. To determine whether the protein kinase activity is essential for the function of YPK1, we mutated Lys-376 in the ATP-binding domain of the Ypk1 protein to arginine or alanine. This lysine residue is predicted to be directly involved in the anchoring of the γ phosphate group of ATP (Fig. 4A) (35). In addition, substitution with any other amino acid, including arginine, would be expected to abolish the ability of the Ypk1 protein to transfer phosphate to its target amino acids. Indeed, the two mutants were found to be kinase dead when the kinase activity was measured using Crosstide (GRPRTSSFAEG in single-letter amino acid code) (1, 6), a peptide substrate (Y. Sun and Y. Kozutsumi, unpublished data). Mutation of these sites had no effect on either the expression levels of the Ypk1 protein or the growth phenotype of yeast in the absence of ISP-1 (data not shown). However, these kinase-dead mutants showed no resistance to ISP-1 (Fig. 4B). These results demonstrated that the kinase activity of the Ypk1 protein is indispensable for the resistance to ISP-1.
Overexpression of the Ypk1 protein has no effect on SPT activity or sphingolipid metabolism.
A possibility for the ISP-1 resistance of the Ypk1 protein may be that it exerts ISP-1 resistance activity through the upregulation of sphingolipid biosynthesis. To examine this possibility, the effects of Ypk1 protein overexpression on both in vitro SPT activity and the de novo synthesis of sphingolipids were studied. As shown in Fig. 5A, overexpression of the Ypk1 protein had no effect on the activity of SPT. For the de novo synthesis of sphingolipids, yeast cells were labeled with [3H]serine in the presence and absence of ISP-1. The lipid extracts were analyzed by thin-layer chromatography after mild alkaline methanolysis. As shown in Fig. 5B, ISP-1 decreased the de novo synthesis of sphingolipids in both the control and overexpressing YPK1 cells. Therefore, the ISP-1 resistance activity of the Ypk1 protein is not due to upregulation of sphingolipid biosynthesis. These observations suggested that the Ypk1 protein does not act upstream of sphingolipid biosynthesis in yeast.
Phosphorylation of the Ypk1 protein is regulated by sphingolipids.
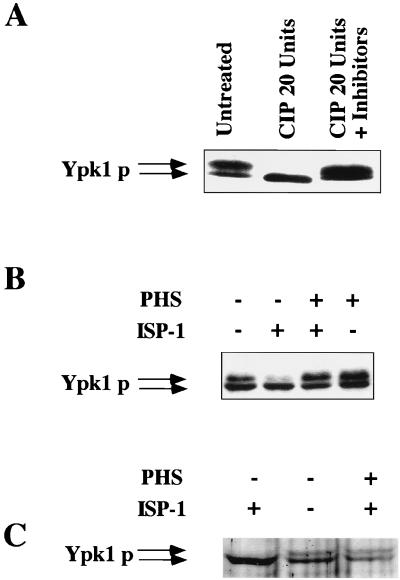
The Ypk1 protein overexpressed in yeast was detected with anti-Ypk1 protein antibodies as a doublet band. The upper band material was the phosphorylated form, since upon phosphatase treatment it was converted to the lower band material (Fig. 6A). Interestingly, the phosphorylated form of the Ypk1 protein was dominant when the intracellular sphingolipid concentration was increased by the addition of phytosphingosine. In contrast, the upper bands of the overexpressed and endogenous Ypk1 proteins decreased with ISP-1-induced sphingolipid depletion (Fig. 6B and C, respectively). Furthermore, the addition of phytosphingosine to ISP-1-treated cells suppressed the reduction of the upper band (Fig. 6B and C). These observations suggest that the phosphorylation of the Ypk1 protein is regulated by the intracellular sphingolipid concentration. Phosphorylation of the Ypk1 protein was not due to autophosphorylation, because the two kinase-dead mutant proteins were also phosphorylated, and this phosphorylation was regulated by the sphingolipid level (data not shown).
FIG. 6.
Phosphorylation of the Ypk1 protein. (A) Calf intestinal alkaline phosphatase treatment of the Ypk1 protein. The Ypk1 protein was immunoprecipitated from extracts of YPK1 transformants using anti-Ypk1 protein antibodies, and then the immobilized Ypk1 protein was treated with 20 U of calf intestinal alkaline phosphatase (CIP) at 37°C for 30 min in the absence or presence of phosphatase inhibitors (10 mM NaF and 1 mM Na3VO4). The phosphatase-treated Ypk1 protein was analyzed by immunoblotting using anti-Ypk1 protein antibodies. Phosphorylation of the overexpressed Ypk1 protein (B) and the endogenous Ypk1 protein (C) decreased following treatment with ISP-1 and increased following treatment with phytosphingosine (PHS). When the effects of ISP-1 and/or phytosphingosine on phosphorylation were monitored, the YPK1 transformants and nontransformant (for the detection of endogenous Ypk1 protein) were cultured for 1 h in the presence of 1.25 μM ISP-1 and/or 5 μM phytosphingosine, and then the Ypk1 protein was analyzed.
The intracellular localization of the Ypk1 protein is regulated by sphingolipids.
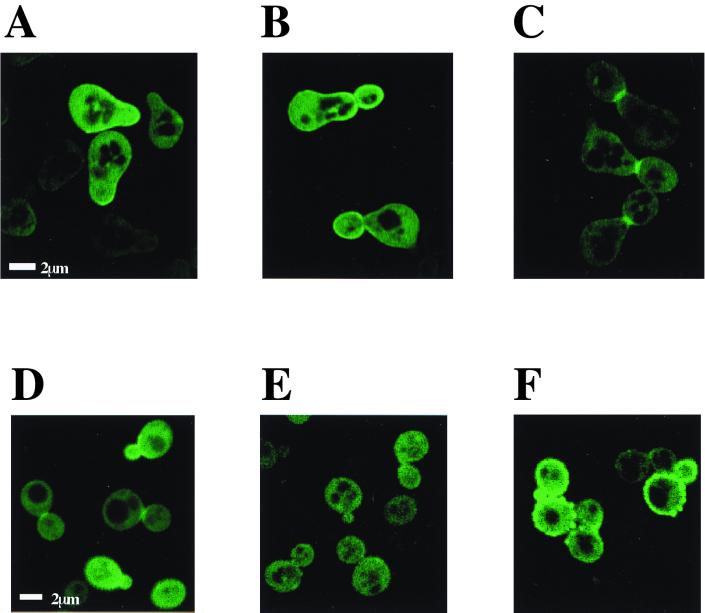
The intracellular localization of the Ypk1 protein was visualized by means of the fusion protein with EGFP. The ISP-1 resistance activity of the Ypk1-EGFP protein was weaker than that of the wild-type Ypk1 protein itself, but it still had significant activity (data not shown). During the yeast cell cycle, the Ypk1-EGFP fusion protein was distributed both on the plasma membrane and in the cytosol (Fig. 7). Greater accumulation was detected on the plasma membrane, such as in the budding area (Fig. 7A), a daughter cell and the neck region of the mother cell in the S phase (Fig. 7B), and the septum between a mother cell and a daughter cell in the G2 phase (Fig. 7C). On the other hand, such plasma membrane localization was almost absent in the case of ISP-1-induced sphingolipid depletion (Fig. 7E). The addition of phytosphingosine induced high-level accumulation throughout the plasma membrane (Fig. 7F). These findings suggest that the intracellular localization of the Ypk1 protein is also regulated by the sphingolipid level.
FIG. 7.
Intracellular localization of the Ypk1 protein. Ypk1-EGFP fusion protein-expressing yeast cells were synchronized with α-factor and then released. G1-phase (A), S-phase (B), and M-phase/cytokinesis (C) yeast cells were obtained. The Ypk1-EGFP fusion protein (A to C) was visualized. The Ypk1-EGFP fusion protein-expressing cells were incubated without synchronization and then treated with the methanol vehicle (D), 1.25 μM ISP-1 (E), or 5 μM phytosphingosine (F) for 4 h. The Ypk1-EGFP fusion protein was observed by confocal laser microscopy.
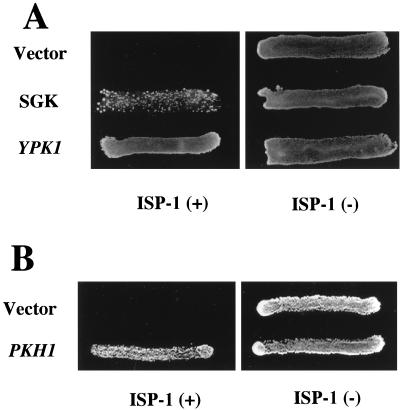
Overexpression of the SGK or Pkh1 protein conferred resistance to ISP-1.
YPK1 encodes a serine/threonine protein kinase which is homologous to mammalian serum- and glucocorticoid-inducible kinase SGK (6). SGK is a novel serine/threonine protein kinase transcriptionally regulated by serum and glucocorticoids in rat mammary tumor cells (65) and also regulated through alterations in cell volume in human hepatoma cells (63). As shown in Fig. 8A, overexpression of SGK also resulted in considerable resistance to ISP-1 under the control of the yeast ADH1 promoter. On the other hand, the upstream kinase of SGK in mammals is known to be 3-phosphoinositide-dependent kinase 1 (PDK1). The yeast homologue of PDK1 is the Pkh1 protein, which phosphorylates the Ypk1 protein (6). Indeed, overexpression of the Pkh1 protein overcame the ISP-1-dependent growth inhibition of yeast (Fig. 8B).
FIG. 8.
Overexpression of SGK or the Pkh1 protein overcame ISP-1-induced cell death. Yeast was transformed with plasmids containing the wild-type YPK1 and SGK gene (A) or PKH1 (B). Transformants were plated on leucine-deficient SD plates containing 1.25 μM ISP-1 or methanol. Strain KMY1005α transformed with plasmid YEp351 or YEp352 was used as a control. The plates were incubated at 30°C for 2 days.
DISCUSSION
In this report, we showed that ISP-1 inhibited SPT activity in yeast and reduced intracellular sphingolipid levels. This sphingolipid depletion consequently inhibited yeast growth.
A gene encoding the protein kinase, SLI2 (YPK1) was isolated as an ISP-1-resistant gene. Kinase-dead mutants of YPK1 did not show any resistance to ISP-1, leading us to predict that the kinase activity of the Ypk1 protein should be essential for its function as to the resistance to ISP-1. There are two possibilities that might explain how the Ypk1 protein shows resistance to ISP-1: (i) overexpression of the Ypk1 protein may upregulate intracellular sphingolipid biosynthesis; (ii) the Ypk1 protein may act downstream of the sphingolipid-mediated signaling pathway. The latter possibility was further supported by several results obtained in this study. Overexpression of the Ypk1 protein had no effect on SPT activity or sphingolipid synthesis, suggesting that the Ypk1 protein does not act upstream of sphingolipid biosynthesis in yeast. The phosphorylated form of the Ypk1 protein was decreased by ISP-1-induced sphingolipid depletion but was dominant when the intracellular sphingolipid concentration was increased by the addition of phytosphingosine, suggesting that the concentrations of sphingolipids controls Ypk1 protein phosphorylation. Finally, the overexpressed Ypk1-EGFP protein was found to be predominantly localized to the plasma membrane in a sphingolipid-dependent manner. The pattern and timing of the change of the protein localization are quite similar to those in the cases of Rho1, Cdc42p, and Spa2p (34), which function in bud growth and cytokinesis of yeasts. Overexpression of the Ypk1 protein overcame the ISP-1-induced G2 arrest in yeasts. Overall, the Ypk1 protein is thought to play important roles in yeast bud growth and cytokinesis. However, both a change in the subcellular localization and abrogation of G2 arrest were detected using the overexpressed protein, and therefore the endogenous Ypk1 protein's function should be examined more carefully.
Yeast cells contain both simple and complex sphingolipids. The simple sphingolipids are mainly phytosphingosine and phytoceramide, which are also present in mammals. On the other hand, the complex sphingolipids containing phosphoinositol, such as inositol phosphorylceramide, mannose inositol phosphorylceramide, and mannose diinositol diphosphorylceramide, are not detected in mammals (Fig. 1). Specific sphingolipids capable of regulating the phosphorylation and intracellular localization of the Ypk1 protein have not yet been found. Aureobasidin A is an inhibitor of yeast inositol phosphorylceramide synthase that catalyzes the transfer of inositol phosphate from phosphatidylinositol (PI) to ceramide to give inositol phosphorylceramide (Fig. 1) (15, 32, 46). Treatment of yeast with aureobasidin A results in a reduction of inositol-containing sphingolipids and growth inhibition. Overexpression of the Ypk1 protein did not cause any resistance to aureobasidin A-mediated growth inhibition (data not shown), suggesting that the sphingolipid involved in the regulation of the Ypk1 protein may not be an inositol-containing sphingolipid.
The detailed mechanism underlying the observation that the overexpressed Ypk1 protein overcomes cell death triggered by sphingolipid depletion, even under conditions where most of the overexpressed Ypk1 protein is dephosphorylated in the presence of ISP-1, is not known. One possibility is that the dephosphorylated Ypk1 protein may have slight kinase activity, and an excess amount of the dephosphorylated Ypk1 protein may be sufficient to activate the pathway. Alternatively, the dephosphorylated Ypk1 protein may be activated through an unknown pathway and then overcome the ISP-1-triggered lethal sphingolipid depletion.
Yeast contains a homologue of YPK1, called YPK2 (7). The YPK2 null mutant yeast does not show any specific phenotype, and the YPK1 null mutant shows slower cell growth. However, the YPK1 YPK2 double null mutation is completely lethal (7). Taking the yeasticidal activity of ISP-1 on sphingolipid depletion into account, both the Ypk1 and Ypk2 proteins may act in parallel downstream of the sphingolipid-mediated signaling pathway. The Ypk1 protein predominantly contributes to the pathway, and the Ypk2 protein may support its effects.
The two kinase-dead mutants of the Ypk1 protein were also phosphorylated, suggesting that the sphingolipid levels control the phosphorylation level of the Ypk1 protein through a putative sphingolipid-dependent kinase acting upstream of the Ypk1 protein. The Ypk1 protein is phosphorylated and activated by the Pkh1 protein, which is a homologue of mammalian PKD1 (6). PDK1 is thought to serve as a central signaling integrator for receptor-mediated activation of PI 3-kinase, because PDK1 phosphorylates numerous protein kinases including protein kinase B (PKB) (60), p70 S6 kinase (2, 54), PKA (8), PKCζ (13), and SGK, which is the mammalian homologue of the Ypk1 protein (36); this finding is consistent with data showing that the overexpressed SGK or Pkh1 protein exhibited ISP-1-resistant activity in yeast.
PDK1 has a pleckstrin homology domain which binds to PI-3,4,5-triphosphate or PI-3,4-bisphosphate (61). However, the yeast PDK1 homologue Pkh1 protein lacks a pleckstrin homology domain. Furthermore, yeast lacks the ability to generate PI-3,4,5 triphosphate or PI-3,4-bisphosphate (14, 29). These observations suggest that yeast uses sphingolipids instead of phosphoinositides as lipid activators. On the other hand, ISP-1 had various effects on mammalian cells, including the induction of apoptosis of mouse cytotoxic T cells (17, 45, 48) and rat Purkinje cells (18), resulting from the sphingolipid depletion caused by ISP-1. Therefore, sphingolipid signaling may occur in mammals similarly to in yeast, and SGK may be involved in this pathway similarly to the Ypk1 protein.
The ISP-1 used in this study for the depletion of sphingolipids in yeast is known to be an apoptosis-inducing immunosuppressant (17, 45, 48). FTY720, which is an analogue of ISP-1, can be tested in humans for clinical immunosuppressive regimens because it exhibits little apparent toxicity in vivo (9, 10, 62). The findings in this study will facilitate elucidation of the details of the mechanisms of action of these immunosuppressants.
REFERENCES
- 1.Alessi D R, Cohen P, Ashworth A, Cowley S, Leever S J, Marshall C J. Assay and expression of mitogen-activated protein-kinase, Map kinase, and Raf. Methods Enzymol. 1995;255:279–290. doi: 10.1016/s0076-6879(95)55031-3. [DOI] [PubMed] [Google Scholar]
- 2.Alessi D R, Kozlowski M T, Weng Q P, Morrice N, Avruch J. 3-Phosphoinositide-dependent protein kinase 1 (PDK1) phosphorylates and activates the p70 S6 kinase in vivo and in vitro. Curr Biol. 1998;8:69–81. doi: 10.1016/s0960-9822(98)70037-5. [DOI] [PubMed] [Google Scholar]
- 3.Ameisen J C. The origin of programmed cell death. Science. 1996;272:1278–1279. doi: 10.1126/science.272.5266.1278. [DOI] [PubMed] [Google Scholar]
- 4.An S, Bleu T, Huang W, Hallmark O G, Coughlin S R, Goetzl E J. Identification of cDNAs encoding two G protein-coupled receptors for lysosphingolipids. FEBS Lett. 1997;417:279–282. doi: 10.1016/s0014-5793(97)01301-x. [DOI] [PubMed] [Google Scholar]
- 5.Buede R, Rinker S C, Pinto W J, Lester R L, Dickson R C. Cloning and characterization of LCB1, a Saccharomyces gene required for biosynthesis of the long-chain base component of sphingolipids. J Bacteriol. 1991;173:4325–4332. doi: 10.1128/jb.173.14.4325-4332.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Casamayor A, Torrance P D, Kobayashi T, Thorner J, Alessi D R. Functional counterparts of mammalian protein kinases PDK1 and SGK in budding yeast. Curr Biol. 1999;9:186–197. doi: 10.1016/s0960-9822(99)80088-8. [DOI] [PubMed] [Google Scholar]
- 7.Chen P, Lee K S, Levin D E. A pair of putative protein kinase genes (YPK1 and YPK2) is required for cell growth in Saccharomyces cerevisiae. Mol Gen Genet. 1993;236:443–447. doi: 10.1007/BF00277146. [DOI] [PubMed] [Google Scholar]
- 8.Cheng X, Ma Y, Moore M, Hemmings B A, Taylor S S. Phosphorylation and activation of cAMP-dependent protein kinase by phosphoinositide-dependent protein kinase. Proc Natl Acad Sci USA. 1998;95:9849–9854. doi: 10.1073/pnas.95.17.9849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chiba K, Hoshino Y, Suzuki C, Masubuchi Y, Yanagawa Y, Ohtsuki M, Sasaki S, Fujita T. FTY720, a novel immunosuppressant possessing unique mechanisms. I. Prolongation of skin allograft survival and synergistic effect in combination with cyclosporine in rats. Transplant Proc. 1996;28:1056–1059. [PubMed] [Google Scholar]
- 10.Chiba K, Yanagawa Y, Masubuchi Y, Kataoka H, Kawaguchi T, Ohtsuki M, Hoshino Y. FTY720, a novel immunosuppressant, induces sequestration of circulating mature lymphocytes by acceleration of lymphocyte homing in rats. I. FTY720 selectively decreases the number of circulating mature lymphocytes by acceleration of lymphocyte homing. J Immunol. 1998;160:5037–5044. [PubMed] [Google Scholar]
- 11.Cifone M G, De-Maria R, Roncaioli P, Rippo M R, Azuma M, Lanier L L, Santoni A, Testi R. Apoptotic signaling through CD95 (Fas/Apo-1) activates an acidic sphingomyelinase. J Exp Med. 1994;180:1547–1552. doi: 10.1084/jem.180.4.1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Deng W P, Nickoloff J A. Site-directed mutagenesis of virtually any plasmid by eliminating a unique site. Anal Biochem. 1992;200:81–88. doi: 10.1016/0003-2697(92)90280-k. [DOI] [PubMed] [Google Scholar]
- 13.Dong L Q, Zhang R B, Langlais P, He H, Clark M, Zhu L, Liu F. Primary structure, tissue distribution, and expression of mouse phosphoinositide-dependent protein kinase-1, a protein kinase that phosphorylates and activates protein kinase Czeta. J Biol Chem. 1999;274:8117–8122. doi: 10.1074/jbc.274.12.8117. [DOI] [PubMed] [Google Scholar]
- 14.Dove S K, Cooke F T, Douglas M R, Sayers L G, Parker P J, Michell R H. Osmotic stress activates phosphatidylinositol-3,5-bisphosphate synthesis. Nature. 1997;390:187–192. doi: 10.1038/36613. [DOI] [PubMed] [Google Scholar]
- 15.Endo M, Takesako K, Kato I, Yamaguchi H. Fungicidal action of aureobasidin A, a cyclic depsipeptide antifungal antibiotic, against Saccharomyces cerevisiae. Antimicrob Agents Chemother. 1997;41:672–676. doi: 10.1128/aac.41.3.672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Erickson J R, Wu J J, Goddard J G, Tigyi G, Kawanishi K, Tomei L D, Kiefer M C. Edg-2/Vzg-1 couples to the yeast pheromone response pathway selectively in response to lysophosphatidic acid. J Biol Chem. 1998;273:1506–1510. doi: 10.1074/jbc.273.3.1506. [DOI] [PubMed] [Google Scholar]
- 17.Fujita T, Inoue K, Yamamoto S, Ikumoto T, Sasaki S, Toyama R, Chiba K, Hoshino Y, Okumoto T. Fungal metabolites. Part 11. A potent immunosuppressive activity found in Isaria sinclairii metabolite. J Antibiot (Tokyo) 1994;47:208–215. doi: 10.7164/antibiotics.47.208. [DOI] [PubMed] [Google Scholar]
- 18.Furuya S, Mitoma J, Makino A, Hirabayashi Y. Ceramide and its interconvertible metabolite, sphingosine, the function as indispensable lipid factors involved in the survival and dendritic differentiation of cerebellar Purkinje cells. J Neurochem. 1998;71:366–377. doi: 10.1046/j.1471-4159.1998.71010366.x. [DOI] [PubMed] [Google Scholar]
- 19.Gerald B, Lester R L. The isolation and characterization of a mutant strain of Saccharomyces cerevisiae that requires a long chain base for growth and for synthesis of phosphosphingolipids. J Biol Chem. 1983;258:10200–10203. [PubMed] [Google Scholar]
- 20.Gill B M, Nishikata H, Chan G, Delovitch T L, Ochi A. Fas antigen and sphingomyelin-ceramide turnover-mediated signaling: role in the life and death of T lymphocytes. Immunol Rev. 1994;142:113–125. doi: 10.1111/j.1600-065x.1994.tb00885.x. [DOI] [PubMed] [Google Scholar]
- 21.Gulbins E, Bissonnette R, Mahboubi A, Martin S, Nishioka W, Brunner T, Baier G, Baier-Bitterlich G, Byrd C, Lang F, Kolesnick R, Altman A, Green D. FAS-induced apoptosis is mediated via a ceramide-initiated RAS signaling pathway. Immunity. 1995;2:341–351. doi: 10.1016/1074-7613(95)90142-6. [DOI] [PubMed] [Google Scholar]
- 22.Hakomori S. Bifunctional role of glycosphingolipids. Modulators for transmembrane signaling and mediators for cellular interactions. J Biol Chem. 1990;265:18713–18716. [PubMed] [Google Scholar]
- 23.Hakomori S. Structure and function of sphingoglycolipids in transmembrane signaling and cell-cell interactions. Biochem Soc Trans. 1993;21:583–595. doi: 10.1042/bst0210583. [DOI] [PubMed] [Google Scholar]
- 24.Haimovitz-Friedman A, Kan C C, Ehleiter D, Persaud R S, McLoughlin M, Fuks Z, Kolesnick R N. Ionizing radiation acts on cellular membranes to generate ceramide and initiate apoptosis. J Exp Med. 1994;180:525–535. doi: 10.1084/jem.180.2.525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hannun Y A. Function of ceramide in coordinating cellular responsed to stress. Science. 1996;274:1855–1859. doi: 10.1126/science.274.5294.1855. [DOI] [PubMed] [Google Scholar]
- 26.Hannun Y A, Bell R M. Functions of sphingolipids and sphingolipid breakdown products in cellular regulation. Science. 1989;243:500–507. doi: 10.1126/science.2643164. [DOI] [PubMed] [Google Scholar]
- 27.Hannun Y A, Obeid L M. Ceramide: an intracellular signal for apoptosis. Trends Biochem Sci. 1995;20:73–77. doi: 10.1016/s0968-0004(00)88961-6. [DOI] [PubMed] [Google Scholar]
- 28.Hanson B A, Lester R L. The extraction of inositol-containing phospholipids and phosphatidylcholine from Saccharomyces cerevisiae and Neurospora crassa. J Lipid Res. 1981;21:309–315. [PubMed] [Google Scholar]
- 29.Hawkins P T, Stephens L R, Piggott J R. Analysis of inositol metabolites produced by Saccharomyces cerevisiae in response to glucose stimulation. Biol Chem. 1993;268:3374–3383. [PubMed] [Google Scholar]
- 30.Heu S C, Wu C C, Luh T Y, Chou C K, Han S H, Lai M Z. Apoptotic signal of Fas is not mediated by ceramide. Blood. 1998;91:2658–2663. [PubMed] [Google Scholar]
- 31.Hla T, Lee M J, Ancellin N, Liu C H, Thangada S, Thompson B D, Kluk M. Sphingosine-1-phosphate: extracellular mediator or intracellular second messenger? Biochem Pharmacol. 1999;58:201–207. doi: 10.1016/s0006-2952(99)00086-6. [DOI] [PubMed] [Google Scholar]
- 32.Ikai K, Takesako K, Shiomi K, Moriguchi M, Umeda Y, Yamamoto J, Kato I, Naganawa H. Structure of aureobasidin A. J Antibiot (Tokyo) 1991;44:925–933. doi: 10.7164/antibiotics.44.925. [DOI] [PubMed] [Google Scholar]
- 33.Ito H, Fukuda Y, Murata K, Kimura A. Transformation of intact yeast cells treated with alkali cations. J Bacteriol. 1983;153:163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kevin M, Michael S. Cell polarity and morphogenesis in budding yeast. Annu Rev Microbiol. 1998;52:687–744. doi: 10.1146/annurev.micro.52.1.687. [DOI] [PubMed] [Google Scholar]
- 35.Knighton D R, Pearson R B, Sowadski J M, Means A R, Ten E L F, Taylor S S, Kemp B E. Structural basis of the intrasteric regulation of myosin light chain kinases. Science. 1992;258:130–135. doi: 10.1126/science.1439761. [DOI] [PubMed] [Google Scholar]
- 36.Kobayashi T, Cohen P. Activation of serum- and glucocorticoid-regulated protein kinase by agonists that activate phosphatidylinositide 3-kinase is mediated by 3-phosphoinositide-dependent protein kinase-1 (PDK1) and PDK2. Biochem J. 1999;339:319–328. [PMC free article] [PubMed] [Google Scholar]
- 37.Kohn A D, Kovacina K S, Roth R A. Insulin stimulates the kinase activity of RAC-PK, a pleckstrin homology domain containing ser/thr kinase. EMBO J. 1995;14:4288–4295. doi: 10.1002/j.1460-2075.1995.tb00103.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kohno K, Normington K, Sambrook J, Gething M J, Mori K. The promoter region of the yeast KAR2 (BiP) gene contains a regulatory domain that responds to the presence of unfolded proteins in the endoplasmic reticulum. Mol Cell Biol. 1993;13:877–890. doi: 10.1128/mcb.13.2.877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lanterman M M, Saba J D. Characterization of sphingosine kinase (SK) activity in Saccharomyces cerevisiae and isolation of SK-deficient mutants. Biochem J. 1998;332:525–531. doi: 10.1042/bj3320525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lee M J, Van-Brocklyn J R, Thangada S, Liu C H, Hand A R, Menzeleev R, Spiegel S, Hla T. Sphingosine-1-phosphate as a ligand for the G protein-coupled receptor EDG-1. Science. 1998;279:1552–1555. doi: 10.1126/science.279.5356.1552. [DOI] [PubMed] [Google Scholar]
- 41.Lew D J, Marini N J, Reed S I. Different G1 cyclins control the timing of cell cycle commitment in mother and daughter cells of the budding yeast S. cerevisiae. Cell. 1992;69:317–327. doi: 10.1016/0092-8674(92)90412-6. [DOI] [PubMed] [Google Scholar]
- 42.Mandala S M, Thornton R, Tu Z, Kurtz M B, Nickels J, Broach J, Menzeleev R, Spiegel S. Sphingosine base 1-phosphate phosphatase: a key regulator of sphingolipid metabolism and the stress response. Proc Natl Acad Sci USA. 1998;95:150–155. doi: 10.1073/pnas.95.1.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Maurer R A. Isolation of a yeast protein kinase gene by screening with a mammalian protein kinase cDNA. DNA. 1988;7:469–474. doi: 10.1089/dna.1.1988.7.469. [DOI] [PubMed] [Google Scholar]
- 44.Merrill A H J, Stevens V L. Modulation of protein kinase C and diverse cell functions by sphingosine, a pharmacologically interesting compound linking sphingolipids and signal transduction. Biochim Biophys Acta. 1989;1010:131–139. doi: 10.1016/0167-4889(89)90152-3. [DOI] [PubMed] [Google Scholar]
- 45.Miyake Y, Kozutsumi Y, Nakamura S, Fujita T, Kawasaki T. Serine palmitoyltransferase is the primary target of a sphingosine-like immunosuppressant, ISP-1/myriocin. Biochem Biophys Res Commun. 1995;211:396–403. doi: 10.1006/bbrc.1995.1827. [DOI] [PubMed] [Google Scholar]
- 46.Nagiec M M, Nagiec E E, Baltisberger J A, Wells G B, Lester R L, Dickson R C. Sphingolipid synthesis as a target for antifungal drugs. Complementation of the inositol phosphorylceramide synthase defect in a mutant strain of Saccharomyces cerevisiae by the AUR1 gene. J Biol Chem. 1997;272:9809–9817. doi: 10.1074/jbc.272.15.9809. [DOI] [PubMed] [Google Scholar]
- 47.Nagiec M M, Baltisberger J A, Wells G B, Lester R L, Dickson R C. The LCB2 gene of Saccharomyces and the related LCB1 gene encode subunits of serine palmitoyltransferase, the initial enzyme in sphingolipid synthesis. Proc Natl Acad Sci USA. 1994;91:7899–7902. doi: 10.1073/pnas.91.17.7899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nakamura S, Kozutsumi Y, Sun Y, Miyake Y, Fujita T, Kawasaki T. Dual roles of sphingolipids in signaling of the escape from and onset of apoptosis in a mouse cytotoxic T-cell line, CTLL-2. J Biol Chem. 1996;271:1255–1257. doi: 10.1074/jbc.271.3.1255. [DOI] [PubMed] [Google Scholar]
- 49.Nickels J T, Broach J R. A ceramide-activated protein phosphatase mediates ceramide-induced G1 arrest of Saccharomyces cerevisiae. Genes Dev. 1996;10:382–394. doi: 10.1101/gad.10.4.382. [DOI] [PubMed] [Google Scholar]
- 50.Okazaki T, Bielawska A, Bell R M, Hannun Y A. Role of ceramide as a lipid mediator of 1 alpha,25-dihydroxyvitamin D3-induced HL-60 cell differentiation. J Biol Chem. 1990;265(26):15823–15831. [PubMed] [Google Scholar]
- 51.Olivera A, Spiegel S. Sphingosine-1-phosphate as a second messenger in cell proliferation induced by PDGF and FCS mitogens. Nature. 1993;365:557–560. doi: 10.1038/365557a0. [DOI] [PubMed] [Google Scholar]
- 52.Pinto W J, Srinivasan B, Shepherd S, Dickson R A, Lester R L. Sphingolipid long-chain-base auxotrophs of Saccharomyces cerevisiae: genetics, physiology, and a method for their selection. J Bacteriol. 1992;174:2565–2574. doi: 10.1128/jb.174.8.2565-2574.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pinto W J, Wells G W, Lester R L. Characterization of the enzymatic synthesis of sphingolipid long-chain bases in Saccharomyces cerevisiae: mutant strains exhibiting long-chain-base auxotrophy are deficient in serine palmitoyltransferase activity. J Bacteriol. 1992;174:2575–2581. doi: 10.1128/jb.174.8.2575-2581.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pullen N, Dennis P B, Andjelkovic M, Dufner A, Kozma S C, Hemmings B A, Thomas G. Phosphorylation and activation of p70s6k by PDK1. Science. 1998;279:707–710. doi: 10.1126/science.279.5351.707. [DOI] [PubMed] [Google Scholar]
- 55.Saba J D, Obeid L M, Hannun Y A. Ceramide: an intracellular mediator of apoptosis and growth suppression. Philos Trans R Soc Lond B. 1996;351:233–240. doi: 10.1098/rstb.1996.0021. [DOI] [PubMed] [Google Scholar]
- 56.Schutze S, Potthoff K, Machleidt T, Berkovic D, Wiegmann K, Kronke M. TNF activates NF-kappa B by phosphatidylcholine-specific phospholipase C-induced “acidic” sphingomyelin breakdown. Cell. 1992;71:765–776. doi: 10.1016/0092-8674(92)90553-o. [DOI] [PubMed] [Google Scholar]
- 57.Sillence D J, Allan D. Evidence against an early signaling role for ceramide in Fas-mediated apoptosis. Biochem J. 1997;324:29–32. doi: 10.1042/bj3240029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Spiegel S, Foster D, Kolesnick R. Signal transduction through lipid second messengers. Curr Opin Cell Biol. 1995;8:159–167. doi: 10.1016/s0955-0674(96)80061-5. [DOI] [PubMed] [Google Scholar]
- 59.Spiegel S. Sphingosine 1-phosphate: a prototype of a new class of second messengers. J Leukoc Biol. 1999;65:341–344. doi: 10.1002/jlb.65.3.341. [DOI] [PubMed] [Google Scholar]
- 60.Stephens L, Anderson K, Stokoe D, Erdjument-Bromage H, Painter G F, Holmes A B, Gaffney P R, Reese C B, McCormick F, Tempst P, Coadwell J, Hawkins P T. Protein kinase B kinases that mediate phosphatidylinositol 3,4,5-trisphosphate-dependent activation of protein kinase B. Science. 1998;279:710–714. doi: 10.1126/science.279.5351.710. [DOI] [PubMed] [Google Scholar]
- 61.Stokoe D, Stephens L R, Copeland T, Gaffney P R, Reese C B, Painter G F, Holmes A B, McCormick F, Hawkins P T. Dual role of phosphatidylinositol-3,4,5-trisphosphate in the activation of protein kinase B. Science. 1997;277:567–570. doi: 10.1126/science.277.5325.567. [DOI] [PubMed] [Google Scholar]
- 62.Troncoso P, Stepkowski S M, Wang M E, Qu X, Chueh S C, Clark J, Kahan B D. Prophylaxis of acute renal allograft rejection using FTY720 in combination with subtherapeutic doses of cyclosporine. Transplantation. 1997;67:145–151. doi: 10.1097/00007890-199901150-00024. [DOI] [PubMed] [Google Scholar]
- 63.Waldegger S, Barth P, Raber G, Lang F. Cloning and characterization of a putative human serine/threonine protein kinase transcriptionally modified during anisotonic and isotonic alterations of cell volume. Proc Natl Acad Sci USA. 1997;94:4440–4445. doi: 10.1073/pnas.94.9.4440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Watts J D, Gu M, Polverino A J, Patterson S D, Aebersold R. Fas-induced apoptosis of T cells occurs independently of ceramide generation. Proc Natl Acad Sci USA. 1997;94:7292–7296. doi: 10.1073/pnas.94.14.7292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Webster M K, Goya L, Ge Y, Maiyar A C, Firestone G L. Characterization of sgk, a novel member of the serine/threonine protein kinase gene family which is transcriptionally induced by glucocorticoids and serum. Mol Cell Biol. 1993;13:2031–2040. doi: 10.1128/mcb.13.4.2031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhang H, Desai N N, Olivera A, Seki T, Brooker G, Spiegel S. Sphingosine-1-phosphate, a novel lipid, involved in cellular proliferation. J Cell Biol. 1991;114:155–167. doi: 10.1083/jcb.114.1.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhou C, Jong A. CDC6 mRNA fluctuates periodically in the yeast cell cycle. J Biol Chem. 1990;265:19904–19909. [PubMed] [Google Scholar]
- 68.Zondag G C, Postma F R, Etten I V, Verlaan I, Moolenaar W H. Sphingosine 1-phosphate signaling through the G-protein-coupled receptor Edg-1. Biochem J. 1998;330:605–609. doi: 10.1042/bj3300605. [DOI] [PMC free article] [PubMed] [Google Scholar]