Abstract
In the last decade, the focus of computational pathology research community has shifted from replicating the pathological examination for diagnosis done by pathologists to unlocking and discovering “sub-visual” prognostic image cues from the histopathological image. While we are getting more knowledge and experience in digital pathology, the emerging goal is to integrate other-omics or modalities that will contribute for building a better prognostic assay. In this paper, we provide a brief review of representative works that focus on integrating pathomics with radiomics and genomics for cancer prognosis. It includes: correlation of pathomics and genomics; fusion of pathomics and genomics; fusion of pathomics and radiomics. We also present challenges, potential opportunities, and avenues for future work.
Keywords: Radiomics, pathomics, genomics, prognosis, digital pathology
Introduction
Several recent works suggest that patterns discovered from high dimensional, multi-modal data could improve estimation of disease aggressiveness and patient outcomes (1-3) compared to monomodal data. Data across multiple scales and modalities including radiology images, histology images, genetic mutations, gene expression, etc. were used to create better companion diagnostic tools. Among these modalities, histological images are traditionally used for identifying and characterizing complex histopathological phenotypes, and the histological examination is generally considered as the “gold standard” for diagnosis of most solid tumors. With the advancements in high-speed high-resolution whole slide image scanning hardware, the histological tissue slides can be digitized and analyzed efficiently. Pathomics or quantitative histomorphometric analysis refers to the process of extraction and mining of computer derived measurements from digitized histopathology images. While the visual reading of routine histopathology slides of tumors by pathologists can help predict cancer behavior to a certain degree, sophisticated pathomics has the potential to “unlock” more revealing sub-visual attributes about tumors (4). Perhaps, even more importantly pathomics enables a detailed spatial interrogation of the entire tumor landscape and its most invasive elements from a standard hematoxylin and eosin (H&E) slide. The research community has developed approaches quantifying nuclear arrangement, texture, and orientation for disease presence, risk, aggressiveness, progression and survival. These include not only the nuclear architecture and graphical arrangement of a single histologic primitive, but also novel approaches that are focused on characterizing the spatial arrangement (5-7) of tumor infiltrated lymphocytes (TILs) and interplays between multiple different histological primitives simultaneously [e.g. interplay of lymphocytes and cancer cells (8-10)], thus potentially providing a comprehensive portrait of tumor’s morphologic heterogeneity.
On the other hand, radiological imaging, which typically involves non-invasive procedures, presents anatomic and functional characteristics at the macroscopic level. Imaging modalities [such as magnetic resonance imaging (MRI), ultrasound, computerized tomography (CT) and X-ray] are typically used in the initial stages for cancer detection, diagnosis and localization prior to biopsy of specific tissues for confirmatory tests. They are also used for treatment planning, delivery of therapy and monitoring. Radiomics refers to quantitative measurements of texture and shape attributes extracted using advanced image processing and computer vision techniques from imaging modalities. They quantify underlying sub-visual tissue heterogeneity that is not always apparent to a human reader. Since imaging is acquired at the macroscopic scale, radiomics allows for interrogating not only the disease regions of interest, but also surrounding structures such as the peri-tumoral region (11). Radiology images offer the opportunity to be used in conjunction with machine learning to build diagnostic, prognostic and predictive models (12-14).
Compared to imaging and pathology that quantify disease phenotypes, genomic analysis focuses on cellular activities measured at the molecular level. Bulk gene expression data have been used to understand molecular differences between disease phenotypes, socio-economic environments and response to therapies. The investigation on mutations, copy number changes, DNA methylation and gene expression that are correlated with tissue phenotypes enables discovery of new cancer genes and understanding of the underlying molecular mechanism and drivers of tumor morphology associated with diseases. A typical prognostic model using genomic data is OncotypeDX for breast cancer patients, in which a risk score of recurrence was generated by a linear combination of 21 genes expression (15).
In clinical setting, it may very often be likely that patient data comprising more than one of imaging, pathologic or genomic modalities are available in course of their diagnosis through treatment. Genomic data provide rich molecular resolution while imaging data provide spatial phenotype information of cancer in addition to pathology. Thus, multi-modal data offer a unique opportunity to comprehensively interrogate the cancer microenvironment thereby enabling a more accurate assessment of disease aggressiveness. The integration of imaging phenotypes and genotypes could help us 1) understand histological context of genetic data; 2) understand underlying biological basis/process of specific quantitative imaging features; 3) gain complimentary information for visualizing spatial and molecular context of cancer; 4) resolve confounding effects of tissue heterogeneity; 5) discover new diagnostic/prognostic signatures; and 6) build a holistic model/approach to understand the progression of different diseases.
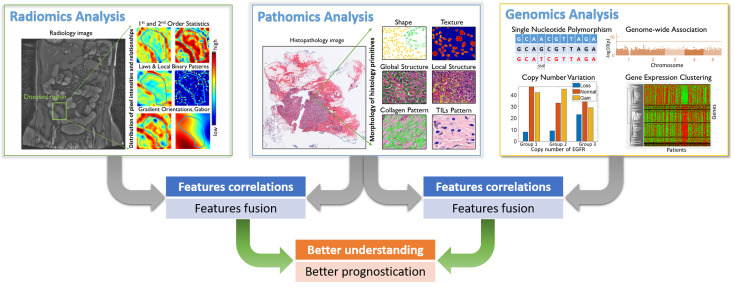
In this article, we provide a brief review of representative works that focus on integrating pathomics with radiomics and genomics for cancer diagnosis and prognosis. It includes: correlation of pathomics and genomics; fusion of pathomics and genomics; fusion of pathomics and radiomics. We also present challenges, potential opportunities and avenues for future work. An overview of the fusion of pathomics, radiomics and genomics analysis is shown in Figure 1 .
Figure 1.
An overview for the fusion of pathomics, radiomics and genomics analyses. In radiomics analysis, quantitative image features were derived from radiology images, which may include traditional hand-crafted features, e.g., 1st and 2nd order statistics, Laws & Local Binary Patterns, Gradient orientations and Gabor and features that learnt by deep learning model. In pathomics analysis, quantitative image features were derived from histopathological images, which may include hand-crafted features like nuclear shape, texture, global structure, local structure, stroma collagen pattern and TIL patterns and features that learnt by deep learning model. In genomics analysis, single nucleotide polymorphism (SNP), copy number variation (CNV), genome structure data and gene expression data [e.g., ribonucleic acid (RNA)-seq data] were analyzed. In the context of prognosis, features/signatures that associated with patient outcomes from different modalities can be associated and fused, in order to better understand the relationship of disease genotypes and genotypes and to create better prognostic tools.
Correlating pathomics and genomics
Correlating tumor morphology quantified by pathomics with large-scale genomic analyses is an emerging research topic in recent literatures, since the causal and inferential relationship between gene expression and pathomics is crucial in biomarker discovery. These association can be done via classical Pearson correlation (16), or advanced methods like sparse canonical correlation analysis (17,18) that can identify correlated sets of genes and histomorphometrics for more effective analysis. In 2013, Wang et al. (19) established an automated pipeline for correlating the histomorphometrics to gene expression data. In The Cancer Genomic Atlas (TCGA) triple negative breast cancer (TNBC) cohort, correlations between histomorphometrics and gene expression were first calculated. The histomorphometrics that can significantly separate high-risk and low-risk patients were then identified in a local TMA cohort. In other datasets, gene clusters with strong correlations to these histomorphometrics were validated as biomarkers. Similarly, Ash et al. (17) used image features learnt by convolutional auto-encoder and performed sparse canonical correlation analysis to identify sets of genes that correlate with histomorphometrics. Lu et al. (10) associated the cellular diversity features that derived from the non-small cell lung carcinomas with bulk gene data to investigate the underlying biological pathways of image features derived from the pathological image. Subramanian et al. (18) shows that integrative approaches combining tissue phenotypes from images with genomic analysis can resolve confounding effects of tissue heterogeneity and should be used to identify new drivers in other cancers. AbdulJabbar et al. (5) utilized the genomic data to validate the signature extracted from the histology image. Cooper et al. (20) illustrated how morphological features extracted from histology images can be integrated with clinical and genomic data in a study of glioblastomas (GBMs). More specifically, the tumor microenvironment and transcriptional classification of GBM were explored. In addition, the authors shown that molecular and clinical associations were revealed through quantitative nuclear morphometry. Barsoum et al. (21) provided a brief review on how to use morphological features extracted from histology image to correlate with clinical behavior, host immune response, and genomic information. They also discussed the combination of digital pathology and genetic studies and its correlation with tumor behavior. Table 1 gives an overview of research works relating to pathomics and genomics correlation.
Table 1. Overview of research works on correlating pathomics with genomics.
| References | Approach | Data used | Results | |
| CAE, convolutional autoencoder; BCa, breast cancer; H&E, hematoxylin and eosin; IHC, immunohistochemistry; WSI, whole-section image; NSCLC, non-small cell lung cancer; TCGA, The Cancer Genomic Atlas; CNN, convolutional neural networks; TMA, tissue microarrays; CCA, canonical correlation analysis. | ||||
| Ash et al.
(17) |
1) CAE was first applied to histology image to extracted features; 2) sparse canonical correlation analysis (CCA) was then applied to the image features and gene expression to find subsets of gene expression values that correlate to subsets of image features. | Three cohorts (BCa, lower grade glioma, and Genotype-Tissue Expression project) with histological images and bulk RNA-sequencing data from paired tissue samples. | 1) Gene sets associated with the structure of the extracellular matrix and cell wall infrastructure, implicating uncharacterized genes in extracellular processes; 2) found sets of genes associated with specific cell types; 3-image features that capture population variation in thyroid and in colon tissues associated with genetic variants. | |
| AbdulJabbar et al. (5) | 1) Train deep learning model to identify cancer cells, lymphocytes, stromal cells and an “other” cell class in H&E-stained images (validated by sequencing data, IHC, and pathologists); 2) define immune hot and cold regions based on lymphocytes percentage (validated by the RNA-seq classification). | WSI, RNA-seq from multiregion TRAcking Cancer Evolution through Therapy (Rx) (TRACERx, n=100); The Leicester Archival Thoracic Tumor Investigatory Cohort (LATTICe-A, n=970). | High geospatial immune variability between tumor regions; Tumors with more than one immune cold region had a higher risk of relapse in lung adenocarcinomas. | |
| Lu et al. (10) | Image features that captured cellular diversity in local region were correlated with bulk RNA expression data. | N=405 NSCLC histology image with bulk RNA expression data from TCGA | CellDiv features were found to be strongly associated with apoptotic signalling and cell differentiation pathways. | |
| Subramanian et al. (18) | Use CCA and sparse CCA to correlate gene expression and histological features describing nucleus shape, texture and intensity. | N=615 BCa samples from TCGA with histology images and gene expression data. | CCA found significant correlation of image features with expression of PAM50 genes. | |
| Martins et al. (16) | Stroma were segmented from H&E-stained images and quantified by a fraction score. The stroma score and gene expression were correlated using Pearson correlation. | Two independent cohorts of TMAs of ovarian cancer (n=521). | Stroma strongly biases estimate of PTEN expression | |
| Wang et al. (19) | Image features captured tumor morphology were correlated with gene expression data. The strong correlated image features and gene lists/clusters were test for prognostic ability in independent test cohorts. | TCGA Triple-Negative BCa (n=44) with image and gene data. Evaluating the image features in a local TMA cohort (n=143). | Forty-eight pairs of significantly correlated image features and gene clusters were identified; four image features were prognostic in a validation cohort; gene clusters correlated with these four image features were prognostic in public gene datasets. | |
Fusion of pathomics and genomics
Due to the intra-tumor heterogeneity, the expression level of certain genes may differ significantly in various regions within the same tumor. On the other hand, the diagnostic slide of tissue samples provides a global view of tumor morphology, and thus pathomic analysis could alleviate the sampling issues raised in genomic analysis. However, the pathomic features may not be able to correlate accurately with the clinical behavior of patients or difficult to provide a biological explanation for certain associations. Therefore, understanding the histological context of genomic data is essential for a full understanding of the clinical behavior of a tumor. Beyond the correlating of pathomics and genomics studies described in the last section, many researches attempted to combine these two to create better diagnostic companion tools.
A straight forward strategy to integrate pathomics and genomics signal is to perform the feature vectors concatenation (22-25). Shao et al. (26) introduced an ordinal multi-modal feature selection method that identified important features from each modality with the consideration of the intrinsic relationship between modalities. Chen et al. (27) proposed a sophisticated end-to-end integrated framework for fusing the learned deep features from histology image, at patch-level and cell graph-level, and learned genomic feature from genomic profile. A gating-based mechanism was first used to control the contribution of each modality, followed by the Kronecker product to model feature interactions across modalities. Table 2 summarizes the representative research works that combine pathomics and genomics for better prognosticating.
Table 2. Overview of research works on fusion of pathomics with genomics.
| References | Aims | Approach | Data used | Results | |
| CNN, convolutional neural networks; GCN, graph convolutional networks; H&E, hematoxylin and eosin; IHC, immunohistochemistry; CNV, copy number variant; CCRCC, clear cell renal cell carcinoma; CV, cross-validation; TCGA, The Cancer Genomic Atlas; WSI, whole-section image; HR, hazard ratio; 95% CI, 95% confidence interval; ROI, region of interest; HPF, high power field; DNN, deep neural network; SVM, support vector machine; BCa, breast cancer; ER, estrogen receptor. | |||||
| Chen
et al. (27) |
Constructing a prognostic models for glioma and CCRCC | Histologic image-based features extracted by CNN, and graph-based image features extracted by GCN, and genomic features learned by Feed Forward Network. All above mentioned data were integrated by a multimodal learning paradigm, which modeled on pairwise feature interactions across modalities by taking the Kronecker product of unimodal feature representations and gating attention mechanism, for prognostication. | Glioma: 1,505 H&E-stained images from 769 patient with 320 genomic features from CNV, mutation status and bulk RNA-Seq expression; 1,251 H&E-stained CCRCC images from 417 patients with 357 genomic features from CNV and RNA-Seq. | C-index=0.826 for Glioma; C-index=0.720 for CCRCC. Both models’ performance are higher than the corresponding unimodal models.
Results reported under CV scheme. |
|
| Shao
et al. (26) |
Proposing a framework combining pathological images and multi-modal genomic data for the prognosis of early-stage cancer patients. | 1) A generalized sparse canonical correlation analysis, named ordinal multi-modality feature selection (OMMFS) that captures the intrinsic relationship among multiple views, to identify important features from WSI and multi-modal data; 2) cox proportional hazard model was applied for prognosticating patients. | Kidney renal clear cell carcinoma, kidney renal papillary cell carcinoma, and lung squamous cell
carcinoma cohorts with WSI and multi-modal genomic data from TCGA. |
The identified image and multi-modal features were strongly correlated with patients survival outcome, thus enable effective stratification of patients. | |
| Cheerla
et al. (22) |
Constructing a deep learning based pancancer model for predicting survival of patients. | Auto encoder to extract four data modalities (gene expression, miRNA data, clinical data, and WSI) into a single feature vector for each patient, handling missing data through a resilient, multimodal dropout method. | Gene expression (n=10,198), miRNA data (n=10,125), clinical data (n=7,512), and WSI (n=10,914) from TGCA (20 different cancer types). | The pan-cancer prognostic model yielded a C-index of 0.78 overall. | |
| Cheng
et al. (23) |
Constructing a prognostic model for clear cell renal cell carcinoma | 1) Nuclear features (nucleus size, shape, texture, and distance to neighbors) were aggregated statistically into patient-level features; 2) gene co-expression network analysis (GCNA) to cluster genes into co-expressed modules (clusters of highly interconnected/correlated genes); 3) lasso-regularized Cox proportional hazards model was used to calculate the risk scores based on the feature from 1 and 2. | WSI, transcriptome, and somatic mutation. N=410 from TCGA. | 1) Patients with high percentage of stromal tissue are related to poor prognosis; 2) risk index is independent of known prognostic factors with HR (95% CI)=3.06 (2.10−4.45) P<0.005.
Note: Results reported under CV scheme. |
|
| Mobadersany
et al. (24) |
Predicting the overall survival of patients diagnosed with glioma | Hybrid architecture combing abstracted histologic image features from convolutional layers and genomic variables (IDH mutation status and 1p/19q codeletion) to fully connected layers. When predicting of a newly diagnosed patient, 9 HPFs were sampled from each ROI, and the median risk score was selected to represent that ROI. Second highest risk score among all ROIs of a WSI was used as the final risk score. | N=1,061 WSIs from 769 patients from TCGA. Genomic variables (IDH mutation status and 1p/19q codeletion). | Model achieved prognostic power with c index of 0.754 and correlate with molecular subtypes and histologic grade; the c-index boosted to 0.801 while integrating with genomic variables.
Note: Results reported under CV scheme. |
|
| Ren
et al. (25) |
Constructing a survival model for predicting the recurrent of prostate cancer patients with Gleason score 7 | 1) Pathway activities were quantified by pathway scores using RNA sequences; 2) image patches from WSI and pathway scores were integrated into DNN to extract “deep features”; 3) “deep features” and clinical prognostic factors were fed into a Cox model. | N=339 WSIs and RNA (Illumina HiSeq) sequencing data from TCGA. | Integrated model yielded C-index=0.74, and C-index=0.71 for histology image only. | |
| Yuan
et al. (7) |
Correlation between histology image features and genomic data; Prognosticating early-stage ER-BCa patients | Cancer cells, stroma cells and lymphocytes were detected from the histology image and the proportions of these cells are used as image features to correlate and combine with genomic data. | N=564 early-stage BCa patients with H&E-stained WSIs and genomic data. | A SVM predictor integrating gene expression and image features achieved 86%±3.0% cross-validation accuracy and improved stratification of the patient cohort. | |
Fusion of pathomics and radiomics
Radiomics involves high throughput extraction of computational features quantifying tissue heterogeneity at the macroscopic level using advanced image processing and computer vision techniques. Whereas pathomics provides quantitative information at the micro scale. Fusion of radiomics and pathomics provides an opportunity to combine tumor heterogeneity at the macro and micro scale, which may complement each other and result in a stronger integrated signature.
Some of the previous works have explored correlations between radiomics and pathomics to explain the morphological basis of signatures observed on imaging. For instance, studies conducted by Alvarez-Jimenez et al, Penzias et al., and Shiradkar et al. correlated pathomic features with radiomics and quantitative imaging features to establish the morphologic basis of imaging (28-30). While some other works combined radiomic and pathomics to build integrated models for disease characterization and classification (31-34). Vaidya et al. integrated radiomic and pathomic features to build an integrated radio-pathomic signature for cancer prognosis (31-34). Saltz et al. (35) introduced a suite of tools to support the fusion of radiomic and pathomic features and discussed how this toolset can help to investigate the correlations between image features, molecular data, and clinical outcome. Some of these works are summarized in Table 3 .
Table 3. Overview of research works on fusion of pathomics with radiomics.
| References | Aims | Approach | Data used | Results | |
| MRI, magnetic resonance imaging; H&E, hematoxylin and eosin; AUC, area under the curve; MRF, magnetic resonance fingerprinting; ADC, apparent diffusion coefficient; ROI, region of interest; CT, computed tomography; NPC, nasopharyngeal cancer; NSCLC, non-small cell lung cancer; nCRT, neoadjuvant chemoradiotherapy; SVM, support vector machine; GBM, glioblastoma; TCIA, The Cancer Imaging Archive; TCGA, The Cancer Genomic Atlas. | |||||
| Penzias
et al. (29) |
Identify morphologic basis of radiomic features for prostate cancer risk stratification | Radiomic features from T2W MRI that were associated with low- and high-risk prostate cancer were identified, pathomic features that were best correlated with these features were explored | A single institution cohort of 36 patient studies was used with T2W MRI, post-surgical H&E slides | Gabor features on T2W MRI performance (AUC=0.69) and gland lumen shape features (AUC=0.75) resulted in best classification performance | |
| Shiradkar
et al. (30) |
Establish the morphologic basis of MR fingerprinting values on the prostate. | Co-registration of whole mount pathology with MRI, MRF followed by correlation of tissue compartments with MR measurements within prostate cancer, prostatitis and normal prostate ROI | A set of 14 patient studies who underwent MRI, MRF scans followed by radical prostatectomy | Tissue compartments of epithelium, lumen and stroma were significantly correlated with T1, T2 MRF, ADC values (P<0.05) | |
| Alvarez-Jimenez
et al. (28) |
Association between radiomic and pathomic features that distinguish adenocarcinoma and squamous cell carcinoma | Pathomic features from digitized H&E slides of lung cancer; radiomic features from lung cancer CT scans; Cross scale associations were computed between radiomic and pathomic features to compare with individual feature classes | N=171 pathology studies, n=101 lung CT studies acquired from publicly available databases. | Cross-scale associated features resulted in better discrimination (AUC=0.78) of NSCLC subtypes compared to using individual feature classes | |
| Zhang
et al. (34) |
A prognostic nomogram integrating radiomics and pathology signature to prognosticate NPC | Radiomics from MRI images are combined with a pathomic signature obtained from a deep learning model along with clinical factors to build a multi-scale prognostic nomogram for nasopharyngeal cancer | N=220 NPC patients were divided into n=132 for training, n=88 for internal and external validation. | Multi-scale nomogram resulted in an improved predictor of survival (C-index 0.82 vs. 0.73) compared with clinical model and individual signatures. | |
| Vaidya
et al. (33) |
Integrating radiomic and pathomic signatures of NSCLC to predict cancer recurrence | Radiomic features from ROIs on lung CT were combined with pathomic eatures from H&E slides of resected tissue to build an integrated supervised machine learning classifier. | 50 NSCLC patients were used for training and 43 patients for external validation | The combined classifier resulted in higher AUC=0.78 compared to radiomic (AUC=0.74) and pathomic classifier (AUC=0.67) alone | |
| Braman
et al. (36) |
Deep learning prognostic model for gliomas integrating radiology, pathology, genomics and clinical data | Deep learning model where each modality embeddings are combined via attention gated tensor fusion. A multimodal orthogonalization loss is presented to maximize information from each modality so they are complementary. | 176 patients witn T1w and T2w-FLAIR sequences annotated by 7 radiologists, H& slides and DNA sequencing info | Presented model results in C-index of 0.788±0.067, significantly outperforming (P=0.023) the best performing unimodal (C-index of 0.718±0.064) | |
| Shao
et al. (32) |
Integrating radiological and pathological information on pre-treatment info to predict pathological response in rectal cancer | Computational features were derived from rectal pre-treatment MRI and digitized H&E slides, combined to create a radiopathomic signature (RPS) to predict treatment response | N=981 patients who received nCRT along with pretreatment MRI and biopsy whole slide images. | RPS resulted in AUC of 0.84−0.98 at each grade of pathological response with significantly higher performance compared to without integration. | |
| Rathore
et al. (31) |
Integrating radiomic and pathomic features for prognosis of GBM | Radoimic features from T1, T1-Gd, T2, T2-FLAIR, were combined with pathomic s of H&E slides to build a SVM classifier for differentiating long and short term survivors | N=107 GBM patients with MRI and pathology images obtained from TCIA and TCGA | AUC=0.74, 0.76 and 0.8 for radiomics, pathomics and combined model in predicting survival outcome | |
Fusion of pathomics, radiomics and genomics
Fusion of radiomics, pathomics, and genomics would further allow for integrating multiple scales of data. However, such data are not easy to obtain and there have been very few studies that have explored this aspect. Braman et al. (36) presented a strategy to intelligently fuse embeddings of radiomics, pathomics and genomics to derive an optimal complementary signature in order to predict outcome in glioblastoma patients. Vaidya et al. (37) looked at correlating lung CT derived radiomic features with pathomic and genomic signatures to provide a biological rational for radiomic signatures that were associated with better survival in non-small cell lung cancer patients.
Challenges and opportunities
The integration of quantitative measurements from multi-modality data for prognosis prediction remains a challenging task because of the high dimensionality and heterogeneity of the data.
Explainability & standardization
The understanding of how abstracted features from different modalities influencing the model’s inference remains another significant problem. The deep learning model was treated as a “black box” method since the learnt features and model decision making were difficult to explain. Researchers have tried to open this box by using activation maps (12,38) and providing visualization of learnt features (39). Compared to deep learning approaches, the hand-crafted features extracted from histology image and radiology image provide better explainability since the features were pre-defined, either in a domain agnostic (13,40) or domain inspired (8) way. In the multi-modality fusion study, the interpretation of the extracted feature becomes more difficult. A computational fusion method should not only consider the discriminative power of the extracted features in the task, but also need to consider the explainability of the extracted features. The fusion frameworks proposed by Shao et al. and Chen et al. (26,27) illustrated that they can visualize and understand the extracted features to some extent. Sharing the extracted features is still challenging since there lacks of standard on the naming, parameter setting of these features. Therefore, developing open-access software that provides transparent information on the computational process is required to evaluate clinical decision support systems. National Cancer Institute (NCI) launched National Interim Clinical Imaging Procedure (NICIP) Code Set to help facilitate the scientific collaboration in cancer research community, which could help researchers to reach a consensus on standardized methods/tools.
Generalizability
One barrier to translating the discovered digital “biomarkers” in pathology imaging-related studies into practice is the issue of generalizability. The discriminative features were mined from a limited number of samples, which easily led to the “overfitting” problem (41,42). That is, the discovered features and model perform well to differentiate patients with distinct outcome in the discovery or training cohort, however, fail in unseen validation cohorts. Therefore, besides the discovery cohort, independent validation cohorts are strongly recommended to further validate the robustness of the found biomarkers. One may claim that cross-validation may help to alleviate the overfitting issues, however, the result may still be biased toward the discovery cohort. The overfitting issue may be caused by the “batch effect”, e.g., artifacts associated with a specific scanner, thus a proper quality check should be performed first before the analysis of data (43,44). In addition, stain variation may hinder the pre-trained model to work well in unseen cohorts. Several approaches have been proposed to address the stain variation issues by using stain normalization (45,46) or training a robust model with a training set containing images with as much variation as possible, i.e., images scanned by different scanners and from different centers. We believe that the generalizability issue could be alleviated if there are more well-maintain benchmark datasets hosting pathology image, radiology image, and genomic data.
Source availability and customizability
Developing and implementing the multi-modal fusion model require access to matched pathology, radiology or genomic data. As we may have known that TCGA project (47) is a landmark cancer genomics program, which hosts over 2.5 petabytes genomic, epigenomic, transcriptomic, and proteomic data over 20,000 primary cancers of 33 cancer types. More importantly, it also includes the digital diagnostic FFPE histology tissue slides for most of the patients, along with clinical information. The Cancer Imaging Archive (TCIA) (48), on the other hand, providing radiology image and histology image, partially overlapped with patients in TCGA, for cross-modality studies. The TCGA-TCIA interface provides a valuable platform for scientists who would like to perform multi-omics investigations. It is common that we have missing data for a certain modality, either imaging or genomic data, or lack of data labeling. Therefore, the fusion approach should be robust enough to learn the representation of available data and is agnostic to data modality and availability. For better associations or integrated signatures between modalities, generating spatially co-registered data from different modalities is a promising approach. For instance, Bourne et al. (49) introduce an approach for aiding histological validation of MRI studies of human prostate, in which a 3D patient-specific mold was created that facilitates the co-registration of in vivo MRI and histology image.
In the last decade, the focus of computational pathology research community has shifted from replicating the pathological examination for diagnosis done by pathologists to unlocking and discovering “sub-visual” prognostic image cues from the histopathological image. While we are getting more knowledge and experience in digital pathology, and the emerging goal is to integrate other-omics or modalities that will contribute to building a better prognostic or predictive assay.
Conclusions
Correlations between pathomics and radiomics, genomics allowed for establishing domain specific biological understanding of cancer morphology. Integration of pathomics with radiomics, genomics resulted in improved comprehensive signatures that were better associated with cancer sub-types and prognosticating treatment outcome. While there is significant potential and promise in complementing pathomics with other-omics data, current studies have largely been limited to small and single institutional datasets. Efforts in making large-scale multi-modal datasets available to the research community will potentially allow for developing sophisticated fusion strategies furthering the potential of pathomics or quantitative histomorphometry.
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
Acknowledgements
This study is supported by the DoD Breast Cancer Research Program Breakthrough Level 1 Award W81XWH-19-1-0668, NIH-NCI R21 CA253108-01; DoD Prostate Cancer Research Program Idea Development Award W81XWH-18-1-0524; Key R&D Program of Guangdong Province, China (No. 2021B0101420006); National Science Fund for Distinguished Young Scholars, China (No. 81925023); and National Natural Science Foundation of China (No. 62002082, 62102103, 61906050, 81771912).
References
- 1.Bera K, Schalper KA, Rimm DL, et al Artificial intelligence in digital pathology — new tools for diagnosis and precision oncology. Nat Rev Clin Oncol. 2019;16:703–15. doi: 10.1038/s41571-019-0252-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Doherty GJ, Petruzzelli M, Beddowes E, et al Cancer treatment in the genomic era. Annu Rev Biochem. 2019;88:247–80. doi: 10.1146/annurev-biochem-062917-011840. [DOI] [PubMed] [Google Scholar]
- 3.Fournier L, Costaridou L, Bidaut L, et al Incorporating radiomics into clinical trials: expert consensus endorsed by the European Society of Radiology on considerations for data-driven compared to biologically driven quantitative biomarkers. Eur Radiol. 2021;31:6001–12. doi: 10.1007/s00330-020-07598-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bhargava R, Madabhushi A Emerging themes in image informatics and molecular analysis for digital pathology. Annu Rev Biomed Eng. 2016;18:387–412. doi: 10.1146/annurev-bioeng-112415-114722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.AbdulJabbar K, Raza SEA, Rosenthal R, et al Geospatial immune variability illuminates differential evolution of lung adenocarcinoma. Nat Med. 2020;26:1054–62. doi: 10.1038/s41591-020-0900-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Saltz J, Gupta R, Hou L, et al Spatial organization and molecular correlation of tumor-infiltrating lymphocytes using deep learning on pathology images. Cell Rep. 2018;23:181–93.e7. doi: 10.1016/j.celrep.2018.03.086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yuan Y, Failmezger H, Rueda OM, et al Quantitative image analysis of cellular heterogeneity in breast tumors complements genomic profiling. Sci Transl Med. 2012;4:157ra143. doi: 10.1126/scitranslmed.3004330. [DOI] [PubMed] [Google Scholar]
- 8.Corredor G, Wang X, Zhou Y, et al Spatial architecture and arrangement of tumor-infiltrating lymphocytes for predicting likelihood of recurrence in early-stage non-small cell lung cancer. Clin Cancer Res. 2019;25:1526–34. doi: 10.1158/1078-0432.CCR-18-2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lu C, Koyuncu C, Corredor G, et al Feature-driven local cell graph (FLocK): New computational pathology-based descriptors for prognosis of lung cancer and HPV status of oropharyngeal cancers. Med Image Anal. 2021;68:101903. doi: 10.1016/j.media.2020.101903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lu C, Bera K, Wang X, et al A prognostic model for overall survival of patients with early-stage non-small cell lung cancer: a multicentre, retrospective study. Lancet Digit Health. 2020;2:e594–e606. doi: 10.1016/s2589-7500(20)30225-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Beig N, Khorrami M, Alilou M, et al Perinodular and intranodular radiomic features on lung CT images distinguish adenocarcinomas from granulomas. Radiology. 2019;290:783–92. doi: 10.1148/radiol.2018180910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hiremath A, Bera K, Yuan L, et al. Integrated clinical and CT based artificial intelligence nomogram for predicting severity and need for ventilator support in COVID-19 patients: A multi-site study. IEEE J Biomed Health Inform 2021;PP. [Online ahead of print]
- 13.Khorrami M, Bera K, Leo P, et al Stable and discriminating radiomic predictor of recurrence in early stage non-small cell lung cancer: Multi-site study. Lung Cancer. 2020;142:90–7. doi: 10.1016/j.lungcan.2020.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Song B, Yang K, Garneau J, et al Radiomic features associated with HPV status on pretreatment computed tomography in oropharyngeal squamous cell carcinoma inform clinical prognosis. Front Oncol. 2021;11:744250. doi: 10.3389/fonc.2021.744250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sparano JA, Gray RJ, Makower DF, et al Adjuvant chemotherapy guided by a 21-gene expression assay in breast cancer. N Engl J Med. 2018;379:111–21. doi: 10.1056/NEJMoa1804710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Martins FC, Santiago Id, Trinh A, et al Combined image and genomic analysis of high-grade serous ovarian cancer reveals PTEN loss as a common driver event and prognostic classifier. Genome Biol. 2014;15:526. doi: 10.1186/s13059-014-0526-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ash JT, Darnell G, Munro D, et al Joint analysis of expression levels and histological images identifies genes associated with tissue morphology. Nat Commun. 2021;12:1609. doi: 10.1038/s41467-021-21727-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Subramanian V, Chidester B, Jian M, et al Correlating cellular features with gene expression using CCA. 2018 IEEE 15th International Symposium on Biomedical Imaging (ISBI 2018) 2018:805–8. doi: 10.1109/ISBI.2018.8363694. [DOI] [Google Scholar]
- 19.Wang C, Pécot T, Zynger DL, et al Identifying survival associated morphological features of triple negative breast cancer using multiple datasets. J Am Med Inform Assoc. 2013;20:680–7. doi: 10.1136/amiajnl-2012-001538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cooper LA, Kong J, Gutman DA, et al Novel genotype-phenotype associations in human cancers enabled by advanced molecular platforms and computational analysis of whole slide images. Lab Invest. 2015;95:366–76. doi: 10.1038/labinvest.2014.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Barsoum I, Tawedrous E, Faragalla H, et al Histo-genomics: digital pathology at the forefront of precision medicine. Diagnosis (Berl) 2019;6:203–12. doi: 10.1515/dx-2018-0064. [DOI] [PubMed] [Google Scholar]
- 22.Cheerla A, Gevaert O Deep learning with multimodal representation for pancancer prognosis prediction. Bioinformatics. 2019;35:i446–54. doi: 10.1093/bioinformatics/btz342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cheng J, Zhang J, Han Y, et al Integrative analysis of histopathological images and genomic data predicts clear cell renal cell carcinoma prognosis. Cancer Res. 2017;77:e91–100. doi: 10.1158/0008-5472.CAN-17-0313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mobadersany P, Yousefi S, Amgad M, et al Predicting cancer outcomes from histology and genomics using convolutional networks. Proc Natl Acad Sci U S A. 2018;115:E2970–9. doi: 10.1073/pnas.1717139115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ren J, Karagoz K, Gatza ML, et al Recurrence analysis on prostate cancer patients with Gleason score 7 using integrated histopathology whole-slide images and genomic data through deep neural networks. J Med Imaging (Bellingham) 2018;5:047501. doi: 10.1117/1.JMI.5.4.047501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shao W, Han Z, Cheng L, et al Integrative analysis of pathological images and multi-dimensional genomic data for early-stage cancer prognosis. IEEE Trans Med Imaging. 2020;39:99–110. doi: 10.1109/TMI.2019.2920608. [DOI] [PubMed] [Google Scholar]
- 27.Chen RJ, Lu MY, Wang J, et al. Pathomic fusion: An integrated framework for fusing histopathology and genomic features for cancer diagnosis and prognosis. IEEE Trans Med Imaging 2020;PP. [Online ahead of print]
- 28.Alvarez-Jimenez C, Sandino AA, Prasanna P, et al Identifying cross-scale associations between radiomic and pathomic signatures of non-small cell lung cancer subtypes: Preliminary results. Cancers (Basel) 2020;12:3663. doi: 10.3390/cancers12123663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Penzias G, Singanamalli A, Elliott R, et al Identifying the morphologic basis for radiomic features in distinguishing different Gleason grades of prostate cancer on MRI: Preliminary findings. PloS One. 2018;13:e0200730. doi: 10.1371/journal.pone.0200730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shiradkar R, Panda A, Leo P, et al T1 and T2 MR fingerprinting measurements of prostate cancer and prostatitis correlate with deep learning-derived estimates of epithelium, lumen, and stromal composition on corresponding whole mount histopathology. Eur Radiol. 2021;31:1336–46. doi: 10.1007/s00330-020-07214-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rathore S, Iftikhar M, Gurcan MN, et al Radiopathomics: Integration of radiographic and histologic characteristics for prognostication in glioblastoma. Neuro Oncol. 2019;21(Suppl 6):vi178–9. doi: 10.1093/neuonc/noz175.745. [DOI] [Google Scholar]
- 32.Shao L, Liu Z, Feng L, et al Multiparametric MRI and whole slide image-based pretreatment prediction of pathological response to neoadjuvant chemoradiotherapy in rectal cancer: A multicenter radiopathomic study. Ann Surg Oncol. 2020;27:4296–306. doi: 10.1245/s10434-020-08659-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vaidya P, Wang X, Bera K, et al RaPtomics: integrating radiomic and pathomic features for predicting recurrence in early stage lung cancer. Medical Imaging. 2018:105810M. doi: 10.1117/12.2296646. [DOI] [Google Scholar]
- 34.Zhang F, Zhong LZ, Zhao X, et al. A deep-learning-based prognostic nomogram integrating microscopic digital pathology and macroscopic magnetic resonance images in nasopharyngeal carcinoma: a multi-cohort study. Ther Adv Med Oncol 2020;12: 1758835920971416.
- 35.Saltz J, Almeida J, Gao Y, et al Towards generation, management, and exploration of combined radiomics and pathomics datasets for cancer research. AMIA Jt Summits Transl Sci Proc. 2017;2017:85–94. [PMC free article] [PubMed] [Google Scholar]
- 36.Braman N, Gordon JWH, Goossens ET, et al. Deep orthogonal fusion: Multimodal prognostic biomarker discovery integrating radiology, pathology, genomic, and clinical data. In: de Bruijne M, Cattin PC, Cotin S, et al. editors. Medical Image Computing and Computer Assisted Intervention - MICCAI 2021. Cham: Springer, 2021. P667-77.
- 37.Vaidya P, Bera K, Gupta A, et al CT derived radiomic score for predicting the added benefit of adjuvant chemotherapy following surgery in stage I, II resectable non-small cell lung cancer: a retrospective multi-cohort study for outcome prediction. Lancet Digit Health. 2020;2:e116–28. doi: 10.1016/s2589-7500(20)30002-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.He L, Long LR, Antani S, et al Histology image analysis for carcinoma detection and grading. Comput Methods Programs Biomed. 2012;107:538–56. doi: 10.1016/j.cmpb.2011.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Badea L, Stănescu E Identifying transcriptomic correlates of histology using deep learning. PLoS One. 2020;15:e0242858. doi: 10.1371/journal.pone.0242858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Whitney J, Corredor G, Janowczyk A, et al Quantitative nuclear histomorphometry predicts oncotype DX risk categories for early stage ER+ breast cancer. BMC Cancer. 2018;18:610. doi: 10.1186/s12885-018-4448-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Subramanian J, Simon R. Overfitting in prediction models — Is it a problem only in high dimensions? Contemp Clin Trials 2013;36:636-41.
- 42.Xie Z, He F, Fu S, et al Artificial neural variability for deep learning: On overfitting, noise memorization, and catastrophic forgetting. Neural Comput. 2021;33:2163–92. doi: 10.1162/neco_a_01403. [DOI] [PubMed] [Google Scholar]
- 43.Janowczyk A, Zuo R, Gilmore H, et al HistoQC: An open-source quality control tool for digital pathology slides. JCO Clin Cancer Inform. 2019;3:1–7. doi: 10.1200/CCI.18.00157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sadri AR, Janowczyk A, Zhou R, et al Technical note: MRQy - An open-source tool for quality control of MR imaging data. Med Phys. 2020;47:6029–38. doi: 10.1002/mp.14593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tellez D, Litjens G, Bándi P, et al Quantifying the effects of data augmentation and stain color normalization in convolutional neural networks for computational pathology. Med Image Anal. 2019;58:101544. doi: 10.1016/j.media.2019.101544. [DOI] [PubMed] [Google Scholar]
- 46.Zheng Y, Jiang Z, Zhang H, et al Stain standardization capsule for application — driven histopathological image normalization. IEEE J Biomed Health Inform. 2021;25:337–47. doi: 10.1109/JBHI.2020.2983206. [DOI] [PubMed] [Google Scholar]
- 47.Cancer Genome Atlas Research Network, Weinstein JN, Collisson EA, et al The cancer genome atlas pan-cancer analysis project. Nat Genet. 2013;45:1113–20. doi: 10.1038/ng.2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Prior F, Smith K, Sharma A, et al The public cancer radiology imaging collections of The Cancer Imaging Archive. Sci Data. 2017;4:170124. doi: 10.1038/sdata.2017.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bourne RM, Bailey C, Johnston EW, et al Apparatus for histological validation of in vivo and ex vivo magnetic resonance imaging of the human prostate . Front Oncol. 2017;7:47. doi: 10.3389/fonc.2017.00047. [DOI] [PMC free article] [PubMed] [Google Scholar]