Abstract
Extracellular vesicles (EVs) are essential intercellular communicators that are of increasing interest as diagnostic biomarkers. Exploring their biological functions and clinical values, however, remains challenging due to their small sizes and high heterogeneity. Herein, we report an ultrasensitive method that employs target-initiated construction of DNA nanostructure to detect single EVs with an input as low as 100 vesicles/μL. Taking advantage of both DNA nanostructure labeling and EV membrane staining, the method can also permit calibration-free analysis of the protein profiles among different EV samples, leading to clear EV differentiation by their cell of origin. Moreover, this method allows co-localization of dual protein markers on the same EV, and the increased number of EVs carrying dual tumor proteins present in human serum could differentiate cancer patients at the early developmental stage from healthy controls. Our results demonstrate the great potential of this single-EV visualization method in non-invasive monitoring the EV protein biomarkers for cancer diagnosis and treatment monitoring.
Keywords: Extracellular vesicle, DNA nanostructure, Fluorescence microscopy, Protein profiling, Cancer diagnosis
1. Introduction
Extracellular vesicles (EVs) are membrane-bound particles with diameters < 250 nm and secreted by nearly all types of cells (Théry et al., 2002; Gholizadeh et al., 2017). They carry diverse cargos like proteins, lipids, as well as nucleic acids, and transport them from cells to cells, acting as important mediators for cell-cell communication (Thompson et al., 2016; Koponen et al., 2020). EVs are stably present in biofluids, easily accessible with minimal invasion. Their biogenesis involves distinct intracellular regulatory processes that likely determines the close relationship between their molecular composition and the parental cell’s biological states; and their cargos have been reported to be highly relevant to pathological developments (Broekman et al., 2018; Kalluri and LeBleu, 2020). Thus, analyzing the molecular profiles of the circulating EVs may not only gain information about the remote tissues only reachable through surgery, but also reveal disease progression (Wang et al., 2016). These unique characteristics of EVs have attracted active research efforts to explore their potential as promising biomarkers in liquid biopsies (Kalluri, 2016; Oliveira-Rodríguez et al., 2017).
However, the EVs found in biofluids are highly heterogeneous, consisting of many sub-populations different in sizes, cargo composition, cell of origin, biogenesis pathways, and functions (Cocucci et al., 2019; Niel et al., 2018; Maas et al., 2017). Conventional EV analysis relies on bulk measurement methods, like enzyme-linked immunosorbent assays (ELISAs), Western blots (WB) and bead-based flow cytometry (FCM), that produce the ensemble average of the varying signals from a swarm of heterogeneous EVs (Shao et al., 2018). It is very likely for such bulk measurements to miss the unique molecular information carried by the few EVs derived from the diseased cells that are the most relevant to pathological development. Single EV phenotyping can well overcome the heterogeneity issue and differentiate EVs from different biological origins, as demonstrated by counting individual EVs under confocal fluorescence microscopy (CFM) with repeated dye bleaching and antibody binding cycles (Lee et al., 2018); or by the proximity barcoding assay that employs DNA ligation and barcoding, rolling circle amplification, and next-generation sequencing (Wu et al., 2019). While these techniques are powerful at EV differentiation and thus valuable in discovery of EV markers useful for disease diagnosis, they require lengthy and complicated sample treatments and sophisticated data analysis. Promising liquid biopsy tools often focus on detection of the specific EV sub-populations carrying a few markers among a heterogeneous mixture, which has also been achieved employing high-resolution FCM (Morales-Kastresana et al., 2019; Tian et al., 2018; Shen et al., 2018), super resolution microscopy (Nizamudeen et al., 2018), and total internal reflection fluorescent microscopy (TIRFM; He et al., 2019). Still, the small physical sizes of EVs and the limited amounts of cargo molecules enclosed represent great challenges for rapid, simple, and highly sensitive detection of single EVs.
Herein, we report an innovative method for detection of single EVs with low sample inputs that permits rapid and calibration-free comparison of surface protein expression among different EV samples. Our design takes advantage of a long ssDNA grown in situ on the EV surface by rolling circle amplification (RCA) upon recognition of the surface protein of single EV (Fig. 1). The ssDNA contains repeated regions or numerous labeled nucleotides for the binding of multiple fluorophores. With judicious design of the template sequence, the single-stranded RCA product can self-assemble into a compact structure, which is called DNA nanoflower (DNF), in the following text. Such a DNF not only enlarges the size of EV, but also amplify the signal from the surface protein, enabling easy visualization of individual EVs in the diffraction-limited CFM with an EV input EVs as low as ~ 100 vesicles/μL. Coupling with a membrane-staining dye, accurate comparison of the protein expression profiles in different EV samples can be achieved to differentiate the EVs by their cell-of-origin. By simply changing the primer sequence, dual protein markers can be simultaneously labeled by DNFs to realize duplex detection, enhancing the specificity in EV differentiation. Moreover, this method can directly detect EVs carrying tumor-specific proteins in biofluids, leading to clear recognition of cancer patients at early stages from healthy controls.
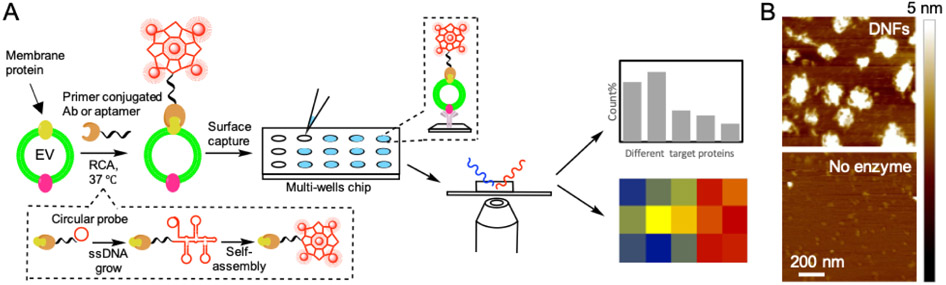
Figure 1.
(A) Schematic illustration of dual-color detection of single EV by dye staining and DNF tagging. (B) AFM images for RCA product (DNF) and the negative control produced without polymerase.
2. Materials and Methods
2.1. Chemicals and materials
The standard EVs were obtained from HansaBioMed Life Science Ltd (Tallinn, Estonia) (lyophilized EVs produced by COLO-1 cells) and System biosciences (lyophilized EVs produced by PC-3 cells and A549 Cells). Human breast cancer patient serum samples and healthy patient serum samples were provided by the NCI funded Cooperative Human Tissue Network (CHTN). Human cell lines of MCF-10A, MCF-7 and MDA-MB-231 were purchased from ATCC.
2.2. Cell culture and EV extraction
All cells were cultured at the recommended media containing 1% penicillin streptomycin. MCF-10A cells were cultured in the DMEM/F-12 media supplied with 5% horse serum, 0.1 μg/mL cholera toxin, 10 μg/mL insulin, 0.5 μg/mL hydrocortisone, and 20 ng/mL EGF. MCF-7 and MDA-MB-231 cells were cultured in DMEM media supplemented with 10% FBS. Patient-derived PDX265922 cancer cells (propagated in NSG mice and denoted as PDX) were cultured in Iscove’s Modified Dulbecco’s medium supplemented with 20% FBS. All cell lines were maintained at 37 °C in a humidified 5% CO2 incubator and routinely screened for Mycoplasa contamination. When the cells reached a confluency of 75%, the medium was replaced with the EV-depleted culture medium. After 24 hours incubation, the culture medium was collected and centrifuged at 500 g for 15 min and 15,000 g for 20 min to remove the cell debris. Next, the medium was ultra-centrifuged at 110,000 g for 70 min twice to pellet the EVs secreted by the cells. The EVs pellet was resuspended in 1 × PBS. Particle concentration in the EV solution was measured by NTA.
2.3. EV labeling and imaging by confocal fluorescence microscopy
EV labeling by the DNFs was carried out with one-pot incubation and sequential addition of the reagents for target recognition, RCA, and DNF/EV staining. Firstly, 2 μL of the EV sample in a microcentrifuge tube was incubated with 25 nM of the RCA initiator and 5 nM of the circular template (Supporting Methods) in 50 mM Tris-HCl, 10 mM MgCl2, 10 mM (NH4)2SO4 and 4 mM DTT for 30 min. Then, 80 μM of dNTPs, 20 μM of the biotinylated dATP and 2.5 U/ μL of phi29 DNA polymerase were added to grow the DNF at 37 °C for 0.5 hr. Next, 10 μg/mL of DiO dye and 50 nM of Alexa 633-conjugated streptavidin were added to stain the EV and DNFs, respectively. At last, the EV solution was added to the imaging surface modified with MFGE8 protein or anti-CD63 antibody, and washed 5 times 0.1% BSA, ready for imaging. For EV detection in serum samples, the only change was to first apply 10 μL of the serum sample on the imaging surface to capture the EVs for 1 hour at room temperature, before the one-pot reaction for DNF construction and EV staining. To improve sample throughput and reduce sample consumption, we fabricated a multi-well chip by attaching a PDMS layer to a glass cover slip (Supporting Methods), to simultaneously process up to 18 samples in one experiment, with each well containing up to 10 μL sample.
Fluorescence imaging was performed on a Zeiss 880 Inverted Confocal Microscope using an Argon laser at λex = 488 nm for fluorescence from DiO dye, and a HeNe laser at λex = 633 nm for fluorescence from the Alexa 633 dye. All CFM images were collected at a resolution of 512 × 512 pixels. The viewing area was 21 μm × 21 μm or 50 μm × 50 μm. For each sample, 10 independent images were acquired at 10 different locations from a single well. The image analysis was carried out using Image J. Each fluorescence particle that had intensity above the threshold (set as 3× of the standard deviation of background intensity), and a size of 2 – 10 pixels was counted computationally as one individual EV. The EV numbers counted in all acquired images were summed up to represent the expression level of the target protein in the EV sample.
3. Results and Discussion
3.1. EV labeling by DNA nanoflowers
We hypothesized that a relatively large DNA nanostructure formed in situ upon target recognition and carrying multiple fluorophores could enlarge the overall size of a single EV and enhance its visibility in a diffraction-limited confocal microscope (Fig. 1A). Such a DNA structure can be grown by RCA (Fig. 1A). RCA employs a polymerase capable of strand displacement to rapidly extend a primer (referred as “initiator” in the following text) that hybridizes to a circular template to a long ssDNA product at a constant temperature. The ssDNA product contains the repeated sequences complementary to the circular probe. Since each of the sequences can be bound with a labeling probe, the RCA product can be labeled by multiple signaling units, providing effective signal amplification for biomarker detection (Ali et al., 2014; Yao et al., 2013). More attractively, the circular template can be designed to adopt a T-shaped design (Table S1; Fig. S1A), so that the repeated sequences on the single-stranded RCA product can hybridize with each other (Fig. S1B), causing the self-assembly of the long ssDNA into a compact nanostructure, i.e. DNF (Hu et al., 2014).
Indeed, 1-hr RCA using a simple initiator (Initiator 1 in Table S1, with no target-binding sequence) and a T-shaped circular template (Template 1 in Table S1) produced long DNAs that could not migrate out of the agarose gel during electrophoresis (Fig. S1C). We also inspected the size and morphology of the RCA products by Atomic Force Microscopy (AFM) and Scanning Electron Microscopy (SEM). We can see from the AFM images that, the RCA products did display a globular structure with the average diameter around 100-150 nm, which were not observed if no polymerase was added to the reaction (Fig. 1B and Fig. S2A). Both the size and morphology of the DNF was also confirmed by SEM (Fig. S2B). In addition, we analyzed the hydrodynamic size of these RCA products by Nanoparticle Tracking Analysis (NTA). Particles larger than 150 nm were detected, the mean size of which gradually increased with the RCA time (Fig. S3A&C). In contrast, if a circular template without the self-hybridization regions was used for RCA, the RCA products did not show the reaction time-dependent increase in the mean size detected by NTA (Fig. S3B&C), because it could not fold into a compact, globular structure but rather form the flexible random coil structure.
We further optimized the RCA conditions by monitoring the product growth in a real-time PCR (rtPCR) instrument. We found that the rate of growth started to slow down after 1 hr (Fig. S4), at which time an obvious peak of the DNA particles larger than 150 nm detected by NTA (Fig. S3A). Although more large particles were produced with longer RCA times, 1 hr was chosen in our assay to keep the procedure short. The dNTP used for RCA contained the biotinylated dATP so that the RCA product can be labeled by the streptavidin conjugated with fluorescent probes afterwards. The optimal ratio of dATP/biotin-dATP was found to be 4:1 to gain a high number of labeling sites without compromising the reaction efficiency (Fig. S5). The reaction time of 1 hr yielded 10× increase in the fluorescence observed in the real-time PCR machine, which indicated the RCA product was probably 10× longer than the circular probe (84 nt). With a ratio of 4:1 of dATP : biotin-dATP used in RCA, the ~ 800 nt long RCA product could maximally contain 36 biotin-dATP for streptavidin labeling.
We hypothesized that EVs labeled by the DNF with comparable sizes (Fig. S2B, S3 & S6), and carrying tens of fluorophores through biotin-streptavidin interaction should be easily observable by the conventional CFM. Thus, we designed the RCA initiator that can recognize CD63 on the EV surface, a classic tetraspanin membrane protein with high abundance on a particular EV population, exosome (Jiang et al., 2017; Théry et al., 2002). This initiator contains the primer sequence complementary to a region on the circular template at its 3’ end for strand extension, and the anti-CD63 aptamer at the 5’ end for EV binding through CD63 recognition (Initiator 2 in Table S1). These two sequences were separated by a short spacer to ensure stable EV attachment. The initiator was incubated with the standard EVs derived from COLO-1 cells, and then the RCA reagents were added to the mixture for DNF construction. We confirmed that growing the DNF in situ on EV surface did not alter its compact structure of the DNF by SEM (Fig. S2B). At last, the DNF was labeled by the Alexa 488-conjugated streptavidin (SA-Alexa 488) before the EVs were captured onto the anti-CD9 modified coverslip for CFM (Fig. 1A). Indeed, many bright fluorescent particles were found by CFM, each occupying a region with a diameter of 5 – 9 pixels (200 – 400 nm) (Fig. S7, last image). In contrast, the EVs directly labeled with the Alexa 488-conjugated anti-CD63 Ab were not detectable (Fig. S7). In addition, labeling the EVs with the RCA product from a circular template without the self-assembly regions did not show countable fluorescent particles, other than very few but large particles (Fig. S7) that might be the result of the entanglement of multiple, long, freely straining single-stranded RCA products on the nearby EVs.
3.2. Calibration free detection of surface proteins on individual EVs
With the DNF allowing single EV visualization, the binding of the RCA initiator that recognizes a specific EV surface protein should be positively related to the target abundance on each EV. Therefore, the number of EVs illuminated by the DNFs in each EV sample could represent the sample’s target protein content. To prove this, we carried out the assay using the RCA initiator containing the anti-human epidermal growth factor receptor 2 (HER2; Gutierrez and Schiff, 2011) aptamer (Initiator 3 in Table S1) to grow the DNF, and capturing the EV sample to the imaging surface by anti-CD63 Ab (Fig. 2A, lower panel). The DNF was tagged by the Alexa 633-labeled streptavidin. We can see that the particle counts acquired in the red channel (detecting the DNF), PDNF, was indeed proportional to the EV concentration in the range of 100 – 5×104 particle/μl with a high linear regression coefficient (R2 = 0.982) (Fig. 2B; Fig. S8). Using the 3σ method, the limit of detection (LOD) with DNF labeling was 82 particle/μl, 100 times lower than that obtained by tagging single EVs with fluorescent antibodies (Min et al., 2020). This result confirms that we can “digitally” count the number of fluorescent particles for EV quantification with high sensitivity.
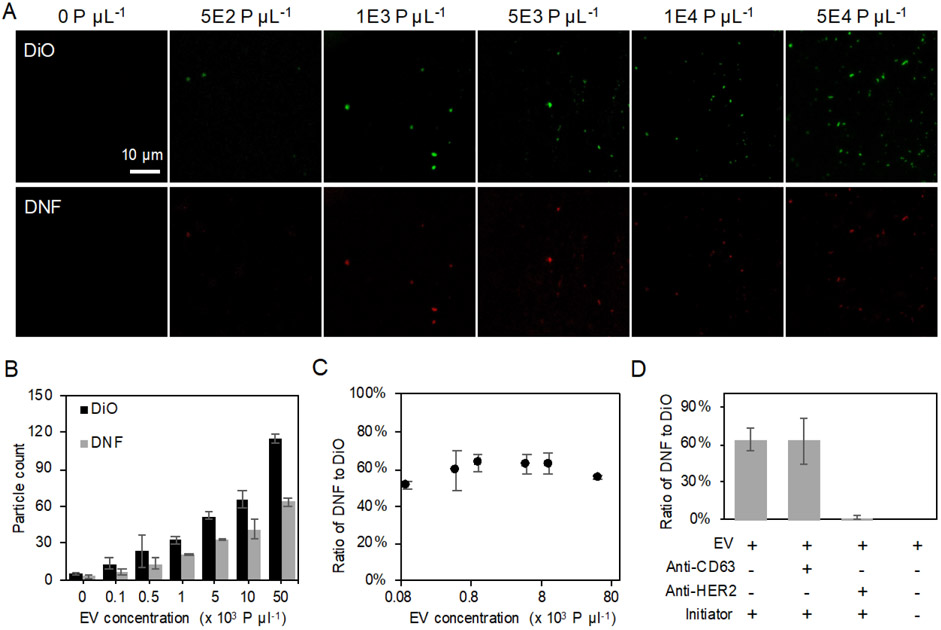
Figure 2.
(A) Representative CFM images of EV labeled with DiO and HER2-specific DNFs in EV-spiked solution with various EV concentration. All images share the same scale bar. (B) The EV particle count obtained from DiO- or DNF-staining increased with EV concentration. (C) Ratio of PDNFs/PDiO measured across different EV input concentration. (D) Percentage of EVs labeled with HER2-specific DNFs to DiO stained EVs detected in EV samples under different labeling conditions. All error bars represent the standard deviations from three repeated measurements.
However, EV capture efficiency on the imaging surface could vary and the input EV number could be different from sample to sample, preventing the usage of the absolute particle counts for accurate comparison of the EV protein expression profiles among samples. In conventional approaches like WB and ELISA, total EV counts and total protein amounts should be determined prior to protein expression analysis. To avoid these tedious sample characterization steps, we stained the EVs with the lipophilic fluorescent dye, DiO (Rodrigues et al., 2019), to directly count the total EVs captured on the viewing surface. The DiO-stained EVs were visible under the same imaging condition as that for the DNF-labeled EVs (Fig. 2A, upper panel; Fig. S7), and provided similar detection sensitivity as the DNF, probably because of the multiple dyes incorporated into the EV membrane (Fig. 2B; Fig. S8; R2 = 0.996). Since the number of DiO-stained EVs obtained in the green channel of the CFM (detecting the DiO dye), PDiO, should be proportional to all the EVs captured on the imaging surface, using PDiO as the internal standard to normalize PDNF, we can correct for variations in EV input or capture efficiency. Indeed, the ratio of PDNFs/PDiO remained constant and ~ 60% with different EV concentrations (Fig. 2C), meaning that ~ 60% of this EV population (standard EVs derived from PC-3 cells) carries HER2. This result proves that, using PDiO as the internal reference, we can compare the expression levels of the same protein marker in repeated measurements and across different EV samples, without the need of prior quantification of the total EV numbers or the total protein contents, nor to use any external calibration methods for quantification of the absolute target amount. Eliminating these steps could greatly save the time, effort, and sample consumption in evaluation of EV protein expression difference in samples. It is worth noting that our method does not rely on the particle fluorescence intensity for protein quantification, which could vary with the length of the RCA product and demands more time and efforts in data processing.
Another advantage of our assay is that, we only count the DNF-labeled particles co-localized with the DiO-stained ones, to eliminate the background DNF grown from the probes non-specifically adsorbed on the imaging surface, enhancing detection specificity. To verify the fluorescent particles detected by CFM were the EVs specifically labeled by DNF, we tested EV labeling by the HER2-targeting DNF: in the presence of 1) anti-HER2 Ab that should block the HER2 protein on EV surface and thus prevent the binding of the aptamer-based initiator probe needed for DNF construction; and 2) anti-CD63 Ab that does not compete with the initiator binding to HER2. We can see from Fig. 2D and S9 that, anti-HER2 Ab completely blocked the growth of DNF on the EVs, leaving no EVs stained by red fluorescence. On contrary, anti-CD63 Ab did not affect the DNF labeling. Additionally, if the RCA initiator contained only the template-hybridizing primer but not the anti-HER2 aptamer sequence (Initiator 1 in Table S1), no fluorescent particles was detected (Fig. S9). The same negative results were obtained in the controls that had no EV in the sample or no capturing Ab on the surface. These results demonstrate that, DNF labeling is specific to the EV target protein, and experiences little non-specific absorption to the detection surface. Since EV capture and DNF labeling target different surface proteins, our method can also avoid detection of the freely floating proteins in biospecimen, but report the co-localization of both targets on the same EV particle.
3.3. Analysis of protein expression profiles on EVs
The above works uses aptamers for target recognition, which can be conveniently incorporated into the sequence of the RCA initiator. To allow detection of more protein targets, we also conjugated the simple RCA initiator (Initiator 1 in Table S1) to the secondary Ab which can bind to the primary Ab used for recognition of specific surface proteins on EV surface (Schweitzer et al., 2020). To better preserve the secondary Ab’s affinity to the primary Ab, we employed the proximity-induced site-specific labeling strategy (Yu et al., 2018): the azide-modified primer was hybridized with an anti-IgG aptamer that recognizes the Fc region of IgG, so that crosslinking only occurred in the Fc region (Fig. S10). Using the primer-conjugated Ab, the PDNFs/PDiO of HER2 in the same EV sample was also found to be ~ 60% (Fig. S11), proving that both Ab and aptamer can be used in our design, and the ratio of PDNFs/PDiO is not affected by experimental conditions and consistent among different batches of measurements of the same EV population.
DNF can be used to label different target proteins by simply switching the aptamer sequence on the RCA initiator, or using the simple initiator-conjugated secondary Ab to bind to the primary Ab that recognizes different targets of interest. Thus, we attempted to profile the expression of different markers on EVs using our method. Since the standard EVs are derived from tumor cell lines, we chose to detect several well-known tumor proteins on the EV surface, including HER2, epidermal growth factor receptor (EGFR) (Voldborg et al., 1997); and CD44 (Chen et al., 2018). Exosomes are one of the EV sub-populations that have demonstrated to be highly relevant to cancer development (Becker et al., 2016; Rontogianni et al., 2019). Therefore, we also included two exosomal markers, CD63 and CD9. The 18-microwell chip was used to simultaneously process 18 samples in parallel, with each well detecting one EV protein in 10 μL of the dilute EV solution.
The standard EVs derived from COLO-1 were labeled by DNF and DiO, and captured by the Milk fat globule epidermal growth factor 8 (MFG-E8), which binds to the phosphatidylserine (PS) exposed on EVs (Ooishi et al., 2017). Individual EVs were counted in both the green and red channels, and the ratio of PDNS/PDiO for each of the protein target was measured. Higher PDNS/PDiO ratios (~ 80%) were found for CD63 and CD9 among 5 markers, agreeing with their roles as the general exosome markers (Fig. S12&S13). A high positive correlation (R2 = 0.91; Fig. S12) was detected between the PDNS/PDiO ratio and the chemiluminescence obtained by ELISA for these 5 markers, further confirming that the ratio of PDNS/PDiO is proportional to the expression level of the target protein in EVs. Our method only requires few μl of EVs sample and is much more sensitive than ELISA, avoiding EV enrichment prior to measurement that could take hours or even days (Greening et al., 2015).
We also tested the EVs derived from other cells, including PC-3, A549, and three breast-cancer (BC) related cells: MCF-7, MDA-MB-231, and the primary cells (PDX) taken from the tumor tissue of a female patient carrying triple-negative BC (Zhou et al., 2015; Yan et al., 2018; Fong et al., 2015). Strikingly, the protein expression profiles illustrated by the ratios of PDNS/PDiO were significantly different among all EV samples tested (Fig. 3A; Fig. S14 - S19). The expression levels of EGFR were the highest in the MDA-MB-231-derived EVs compared to those from other cells; and those from PDX cells had the highest PDNS/PDiO for HER2. The scores plot obtained from subjecting the profiles to Principal Component Analysis (PCA) also proved significant clustering of the EVs by the cell of origin (Fig. 3B); and even plotting the ratios of PDNFs/PDiO of HER2, EGFR and CD44 could lead to good clustering of the EVs originated from the same source (Fig. S13C). All of the lab-prepared EVs derived from the BC-related cell lines located closer to each other, but further apart from the standard EVs purchased from commercial sources with different cells of origin on the PCA plot. Although we could not exclude the contribution from the source of EVs to the protein expression difference observed, among the lab-prepared EVs, those originated from the two triple negative (Basel) cell lines – PDX and MDA-MB-231 – were clustered closer compared to their relative location to those from the MCF-7 cells (ER+/PR+/HER2−, Luminal A). This result strongly supports the feasibility of our method in EV differentiation based on the cell of origin.
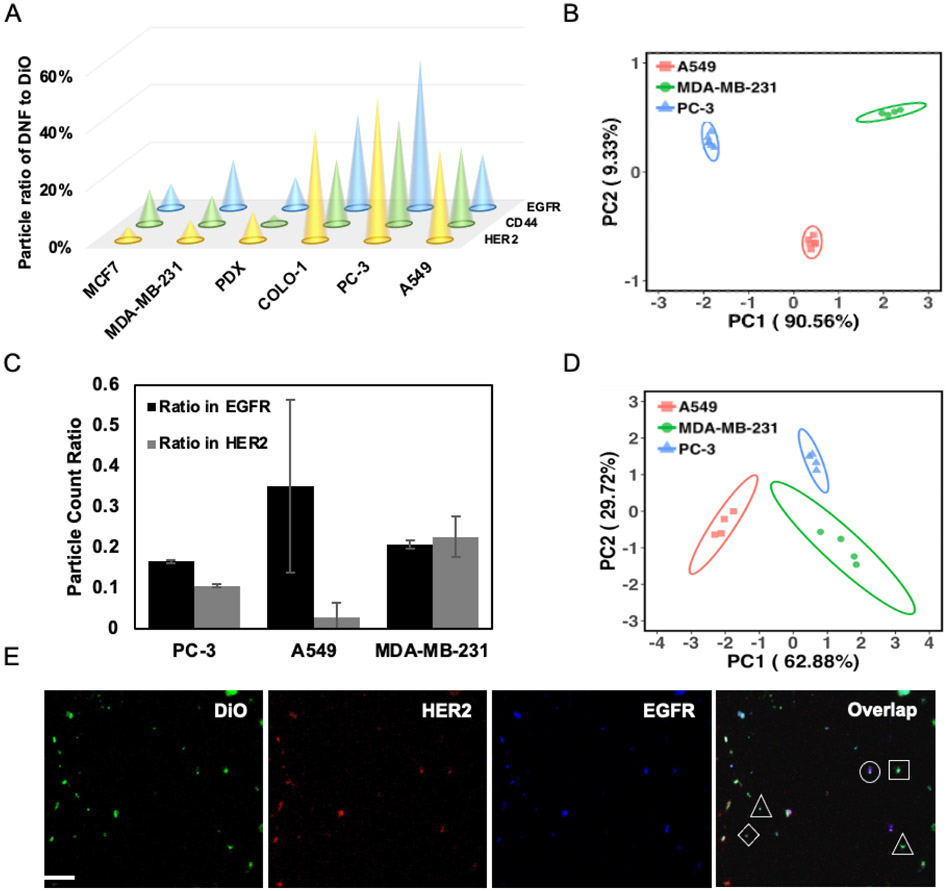
Figure 3.
(A) Comparison of the expression profiles, represented by the average particle ratios (n = 6), of 3 protein markers in the EVs derived from six cell lines. Average relative standard deviation of all measurements < 5%. (B) PCA scores plot to differentiate EVs derived from three cell lines based on the protein profiles shown in (A); ellipses indicate 95% confidence. (C) Ratio of EV carrying both markers detected in the EV population captured by anti-EGFR and anti-HER2 Ab, respectively, using the dual-marker colocalization assay. (D) PCA scores plot to differentiate EVs derived from three cell lines based on the protein profiles shown in (B); ellipses indicate 95% confidence. (E) Representative 3-color CFM images for the DiO-stained EV with dual-DNF labeling. Triangle – EV labeled with DiO; Square – EV labeled with both DiO and EGFR-specific DNFs; diamond – EV labeled with both DiO and HER2-specific DNFs; Circles – EV labeled with DiO, HER2-specific DNFs and EGFR-specific DNFs.
3.4. Co-localization of dual protein markers on the same EV
Co-localizing multiple biomarkers on the same EV can specifically detect the EV sub-groups carrying unique molecular signatures, as proved by the pioneering works (Lee et al., 2018; Wu et al., 2019). Different than these previous works that aim to discover the EV sub-groups with significantly different biological roles, our method focuses on sensitive detection of the EVs carrying dual protein targets in clinical samples for disease diagnosis. This can be simply done by targeting different proteins for EV capture and DNF construction, respectively. Using the anti-EGFR Ab for EV capture and the anti-HER2 Ab for DNF construction (labeled with the Alexa 633-conjugated streptavidin), we obtained the number of EVs carrying EGFR by counting the particles emitting green fluorescence (resulted from DiO staining), i.e. PEGFR, and those also having HER2 on the surface (emitting red fluorescence from Alexa-633), i.e. PEGFR/HER2. Switching the capture and labeling targets, we can then get the number of EVs carrying HER2 (PHER2) and the number of HER2+ EVs that also contain EGFR (PHER2/EGFR). The different expression situations of EGFR and HER2 in the EVs derived from PC-3, A549, and MDA-MB-231 cells were evaluated in Fig. 3C. Interestingly, we can see the proportion of EGFR+/HER2++ EVs among the EGFR+ or HER2+ EVs were comparable in the EVs derived from the MDA-MB-231 cells. In contrast, a significantly larger proportion of the EVs carrying both markers was detected in the EGFR+ EVs derived from the PC3 and A549 cells, compared to that in the HER2+ EVs. Moreover, dual-marker detection can lead to satisfactory EV differentiation (Fig. 3D), using the PEGFR, PEGFR/HER2, PHER2, and PHER2/EGFR found in the above capture/label schemes.
Protein co-localization can also be attained by simultaneously labeling two proteins with the DNF tagged by different fluorophores on the same EVs. Different than the biotin-streptavidin labeling strategy used above, fluorophore-conjugated short DNA probes were employed for dual-target labeling. These probes can complementarily bind to the repeated sequences on the DNFs. To demonstrate this, two DNFs were grown on the EV surface from two RCA initiators, one containing the anti-HER2 aptamer sequence (Initiator 5 in Table S1), and the other (Initiator 1 in Table S1) attached to the secondary Ab bound to the anti-EGFR primary Ab, with their corresponding templates (Template 1 & 2 in Table S1). The EVs were captured by anti-CD63 Ab; and the DNFs targeting HER2 and EGFR were tagged by Alexa 660 and Alexa 546, respectively, using Detection probe 1 and 2 (Table S1). The CFM images (Fig. 3E) revealed that, even among the EVs derived from the same cell of origin, i.e. PC3, there exist particles carrying different protein combinations: some particles emitted only green fluorescence (marked by a triangle), i.e. the CD63+/EGFR−/HER2− EVs; green plus red or yellow fluorescence (marked by a square or diamond), i.e. the CD63+/EGFR+/HER2− EVs or CD63+/EGFR−/HER2+ EVs; or all 3 fluorescence (marked by a circle), i.e. the CD63+/EGFR+/HER2+ EVs (Fig. S20). The ratio of PDNFs/PDiO for EGFR and HER2 is 69.6% and 64.6%, respectively, comparable to that obtained by single marker labeling (Fig. S21). This result hints that dual marker detection exhibits little space hindrance, probably owing to the compact structure of the DNF.
3.4. Detection of EVs carrying tumor markers in serum samples
Our method is simple (one-pot incubation) and sensitive (detecting down to 100 EV/μL) in detection of the specific EV sub-populations carrying the high potential markers, promising for disease diagnosis. To prove this, we detected the EVs in serum samples collected from healthy patients and breast cancer (BC) patients (Stage IA with the same molecular profiles of ER+/PR−/HER2+). The EVs were captured directly on the well surface without any sample pretreatment. Firstly, all three tumor markers of EGFR, HER2, and CD44 were assessed, with the ratios of PDNFs/PDiO obtained for each protein. We found that the PDNFs/PDiO ratios in the BC patients were all significantly higher than those in the healthy controls (Fig. S22, with p values < 0.01 by Student’s t test). The 3D plot of the particle ratios of these 3 markers also clearly separated the BC patients from the healthy control samples (Fig. 4A). In contrast, BC patient and healthy control differentiation cannot be attained by the total counts of particles stained by DiO (Fig. S23).
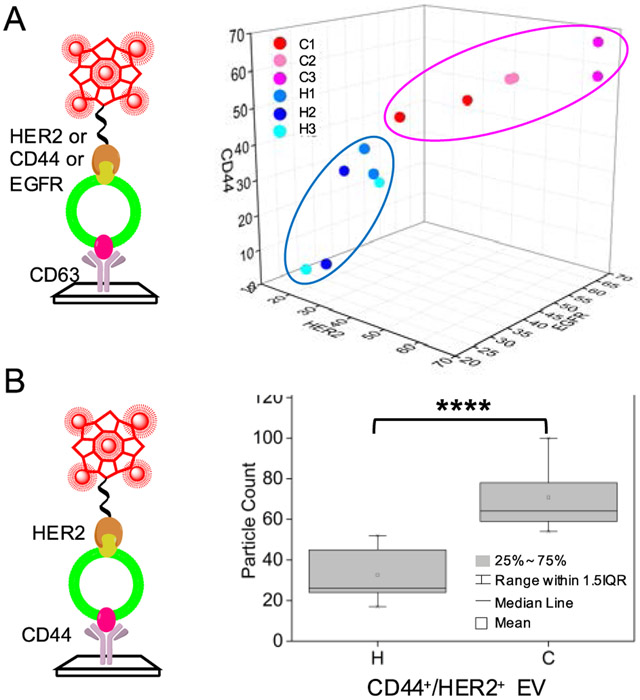
Figure 4.
(A) The plot of the particle ratios (PDNF/PDiO in %) of HER2, CD44, and EGFR detected in the serum samples collected from BC patients (C1, C2, & C3) and healthy controls (H1, H2, & H3) by the single-marker profiling assay. The ellipse shows arbitrary grouping of the cancer samples (red dots) and healthy controls (blue dots). (B) Box plot of the particle counts of the EV carrying both CD44 and HER2 collected from the same clinical samples as used in (A) by the dual-marker co-localization assay. ****p ≤ 0.0001, n = 3. IQR – interquartile range.
We also noticed that, the serum samples from BC patients and healthy controls in fact can be well separated by using the particle ratios of both HER2 and CD44 (Fig. S24A). Thus, we used the dual-marker co-localization assay to detect the EVs carrying both HER2 and CD44, by capturing the EVs via anti-CD44, and labeling HER2 by DNF. In this way, each sample (10 μL) was only measured one time to get the particle counts of the CD44+/HER2+ EVs, and the number of such particles was significantly higher in the BC patients than in the healthy controls (Fig. 4B, p ≤ 0.0001; Fig. S24B). These results prove that, co-localizing two markers on the same EV could enhance the detection throughput and exhibit more significant difference between BC patients and healthy controls than detection of single protein markers on the EVs.
4. Conclusion
To summarize, we have developed a technique to profile the surface protein contents on individual EVs. Different affinity probes, like aptamers, antibodies, and ligands, can be easily adopted to recognize diverse targets locating on the membrane of the EV. Construction of DNF on the surface of EVs not only enables single EV visualization by the diffraction-limited microscopes, but also greatly improves the sensitivity of EV detection. Evaluation of the surface protein expression profiles among different EV samples can be simply done with low sample consumption, and without the need of calibration curve construction, EV pre-concentration, and quantification of total EV counts or protein contents. Our method can also assess the co-localization of multiple markers on the same EV to improve EV differentiation, which can give higher specificity and sensitivity in EV detection. Overall, by enabling rapid and sensitive analysis of EV proteins on single EVs using a one-pot reaction on a multiwell chip and fluorescence microscopy, our technique could be a valuable tool for disease diagnosis and EV biogenesis and functional study that require rapid and high-throughput molecular analysis of the EVs in biofluids or secreted by stimulated cells.
Supplementary Material
Acknowledgements
This work was supported by the National Cancer Institute of the National Institutes of Health under Award Number R01CA188991, and by the University of California Cancer Research Coordinating Committee under the grant #CRR-19-585695 issued to W. Z. The work was also partially supported by R21AA025839. R. C. was supported by the UC-Hispanic Serving Institutions Doctoral Diversity Initiative (UC-HSI DDI) award. S. E. W. acknowledged the support from NCI through R01CA218140 and R01CA206911. The authors are also grateful to Prof. Meng Chen and Prof. Joseph Genereux for the usage of the ultracentrifuges in their laboratories for EV preparation.
References
- Théry C, Zitvogel L, Amigorena S, 2002. Nat. Rev. Immunol 2, 569–579. [DOI] [PubMed] [Google Scholar]
- Gholizadeh S, Darz MS, Zarghooni M, Sanati-Nezhad A, Ghavami S, Shafiee H, Akbari M, 2017. Biosens. Bioelectron 91, 588–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson AG, Gray E, Heman-Ackah SM, Mäger I, Talbot K, Andaloussi SE, Wood MJ, Turner MR, 2016. Nat. Rev. Neurol 12, 346–357. [DOI] [PubMed] [Google Scholar]
- Koponen A, Kerkelä E, Rojalin T, Lazaro-Ibanez E, Suutari T, Saari HO, Siljander P, Yliperttula M, Laitinen S, Viitala T, 2020. Biosens. Bioelectron 168, 112510. [DOI] [PubMed] [Google Scholar]
- Broekman ML, Maas SLN, Abels ER, Mempel TR, Krichevsky AM, Breakefield XO, 2018. Nat. Rev. Neurol 14, 482–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalluri R, LeBleu VS, 2020. Science 367, eaau6977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Khan A, Huang R, Ye S, Di K, Xiong T, Li. Z, 2020. Biosens. Bioelectron 154, 112056. [DOI] [PubMed] [Google Scholar]
- Kalluri R, 2016. J. Clin. Investig 126, 1208–1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveira-Rodríguez M, Serrano-Pertierra E, García AC, López-Martín S, Yañez-Mo M, Cernuda-Morollón E, Blanco-López MC, 2017. Biosens. Bioelectron 87, 38–45. [DOI] [PubMed] [Google Scholar]
- Cocucci E, Racchetti G, Meldolesi J, 2019. Trends Cell Biol. 19, 43–51. [DOI] [PubMed] [Google Scholar]
- Niel GV, D’Angelo G, Raposo G, 2018. Nat. Rev. Mol. Cell Biol 19, 213–228. [DOI] [PubMed] [Google Scholar]
- Maas SLN, Breakefield XO, Weaver AM, 2017. Trends Cell Biol. 27, 172–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao H, Im H, Castro CM, Breakefield X, Weissleder R, Lee H, 2018. Chem. Rev 118, 1917–1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K, Fraser K, Ghaddar B, Yang K, Kim E, Balaj L, Chiocca EA, Breakefield XO, Lee H, Weissleder R, 2018. ACS Nano 12, 494–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu D, Yan J, Shen X, Sun Y, Thulin M, Cai Y, Wik L, Shen Q, Oelrich J, Qian X, Dubois KL, Ronquist KG, Nilsson M, Landegren U, Kammali-Moghaddam M, 2019. Nat. Commun 20, 3854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morales-Kastresana A, Musich TA, Welsh JA, Telford W, Demberg T, Wood JCS, Bigos M, Ross CD, Kachynski A, Dean A, Felton EJ, Dyke JV, Tigges J, Toxavidis V, Parks DR, Overton WR, Kesarwala AH, Freeman GJ, Rosner A, Perfetto SP, Pasquet L, Terabe M, McKinnon K, Kapoor V, Trepel JB, Puri A, Kobayashi H, Yung B, Chen X, Guion P, Choyke P, Knox SJ, Ghiran I, Robert-Guroff M, Berzofsky JA, Jones JC, 2019. J. Extracell. Vesicles 8, 1597603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian Y, Ma L, Gong M, Su G, Zhu S, Zhang W, Wang S, Li Z, Chen C, Li L, Wu L, Yan X, 2018. ACS Nano 12, 671–680. [DOI] [PubMed] [Google Scholar]
- Shen W, Guo K, Adkins GB, Jiang Q, Liu Y, Sedano S, Duan Y, Yan W, Wang SE, Bergersen K, Worth D, Wilson EH, Zhong W, 2018. Angew. Chem 57, 15675–15680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nizamudeen Z, Markus R, Lodge R, Parmenter C, Platt M, Chakrabarti L, Sottile V, 2018. Biochim. Biophys. Acta Mol. Cell Res 1865, 1891–1900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He D, Ho S, Chan H, Wang H, Hai L, He X, Wang K, Li H, 2019. Anal. Chem 91, 2768 – 2775. [DOI] [PubMed] [Google Scholar]
- Ali MM, Li F, Zhang Z, Zhang K, Kang D, Ankrum JA, Le XC, Zhao W, 2014. Chem. Soc. Rev 43, 3324–3341. [DOI] [PubMed] [Google Scholar]
- Yao J, Flack K, Ding L, Zhong W, 2013. Analyst 138, 3121–3125. [DOI] [PubMed] [Google Scholar]
- Hu R, Zhang X, Zhao Z, Zhu G, Chen T, Fu T, Tan W, 2014. Angew. Chem 53, 5821–5826. [DOI] [PubMed] [Google Scholar]
- Jiang Y, Shi M, Liu Y, Wan S, Cui C, Zhang L, Tan W, 2017. Angew. Chem 56, 11916–11920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues M, Richards N, Ning B, Lyton CJ, Hu TY, 2019. Nano Lett. 19, 7623–7631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez C, Schiff R, 2011. Arch. Pathol. Lab. Med 135, 55–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min J, Son T, Hong J, Cheah PS, Wegemann A, Murlidharan K, Weissleder R, Lee H, Im H, 2020. Adv. Biosyst 4, 2000003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweitzer B, Wiltshire S, Lambert J, O’Malley S, Kukanskis K, Zhu Z, Kingsmore SF, Lizardi PM, Ward DC, 2000. Proc. Natl. Acad. Sci. U.S.A 97, 10113–10119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu C, Tang J, Loredo A, Chen Y, Jung SY, Jain A, Gordon A, Xiao H, 2018. Bioconjugate Chem. 29, 3522–3526. [DOI] [PubMed] [Google Scholar]
- Voldborg BR, Damstrup L, Spang-Thomsen M, Poulsen HS, 1997. Ann. Oncol 8, 1197–1206. [DOI] [PubMed] [Google Scholar]
- Chen C, Zhao S, Karnad A, Freeman JW, 2018. J. Hematol. Oncol. 11, 64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker A, Thakur BK, Weiss JM, Kim HS, Peinado H, Lyden D, 2016. Cancer Cell 30, 836–848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rontogianni S, Synadaki E, Li B, Liefaard MC, Lips EH, Wesseling J, Wu W, Altelaar M, 2019. Biol. Commun 2, 325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ooishi T, Nadano D, Matsuda T, Oshima K, 2017. Genes Cells 22, 885–899. [DOI] [PubMed] [Google Scholar]
- Greening DW, Xu R, Ji H, Tauro BJ, Simpson RJ, 2015. Methods Mol. Biol 1295, 179 – 209. [DOI] [PubMed] [Google Scholar]
- Zhou W, Fong MY, Min Y, Somlo G, Liu L, Palomares MR, Yu Y, Chow A, O’Conner STF, Chin AR, Yen Y, Wang Y, Marcusson EG, Chu P, Wu J, Wu X, Li AX, Li Z, Gao H, Ren X, Boldin MP, Lin PC, Wang SE, 2015. Cancer Cell 25, 501–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan W, Wu X, Zhou W, Fong MY, Cao M, Liu J, Liu X, Chen C, Fadare O, Pizzo P, Wu J, Liu L, Liu X, Chin AR, Ren X, Chen Y, Locasale JW, Wang SE, 2018. Nat. Cell Biol 20, 597–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fong MY, Zhou W, Liu L, Alontage AY, Chandra M, Ashby J, Chow A, O’Connor STF, Li S, Chin AR, Somlo G, Palomares M, Li Z, Tremblay JR, Tsuyada A, Sun G, Reid MA, Wu X, Piotr S, Ren X, Shi Y, Kong M, Zhong W, Chen Y, Wang S, 2015. Nat. Cell Biol 17, 183–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.