Summary
Proteins interacting with ADP-ribosyl groups are often involved in disease-related pathways or viral infections, making them attractive drug targets. We present a robust and accessible assay applicable to both hydrolyzing or non-hydrolyzing binders of mono- and poly-ADP-ribosyl groups. This technology relies on a C-terminal tag based on a Gi protein alpha subunit peptide (GAP), which allows for site-specific introduction of cysteine-linked mono- and poly-ADP-ribosyl groups or analogs. By fusing the GAP-tag and ADP-ribosyl binders to fluorescent proteins, we generate robust FRET partners and confirm the interaction with 22 known ADP-ribosyl binders. The applicability for high-throughput screening of inhibitors is demonstrated with the SARS-CoV-2 nsp3 macrodomain, for which we identify suramin as a moderate-affinity yet non-specific inhibitor. High-affinity ADP-ribosyl binders fused to nanoluciferase complement this technology, enabling simple blot-based detection of ADP-ribosylated proteins. All these tools can be produced in Escherichia coli and will help in ADP-ribosylation research and drug discovery.
Keywords: ADP-ribosylation, macrodomain, high-throughput screening, binding assay, protein labeling, inhibitors, post-translational modification, SARS-CoV-2
Graphical abstract
Highlights
-
•
The GAP-tag allows site-specific introduction of cysteine-ADP-ribosyl groups
-
•
Binding assays for 22 different ADP-ribosyl binding proteins
-
•
Nanoluciferase-fused ADP-ribosyl binders are used as blot detection reagents
-
•
Suramin was identified as a non-specific inhibitor for the macrodomain of SARS-CoV-2
Motivation
Specific and potent inhibitors against ADP-ribosyl binding proteins are scarcely available but would be highly useful in research and as potential therapeutics. Assay technologies to screen for inhibitors of these proteins are often limited because they work on only a subset of proteins, are not suited for high-throughput applications, or require expensive reagents. We developed several molecular tools that are easy to produce and aid in the setup of high-throughput screening assays for a wide range of hydrolyzing and non-hydrolyzing ADP-ribosyl binding proteins.
ADP-ribosyl binding proteins are integral actors in many cellular processes, but only a few inhibitors have been discovered thus far. Sowa et al. report a range of easy-to-produce in vitro tools for detection of ADP-ribosylation that also facilitate assay development, allowing for screening of inhibitors in a high-throughput setup.
Introduction
ADP-ribosylation is a post-translational modification involved in the regulation of many diverse processes in the cell. Despite its physiological importance, the intricate interplay of ADP-ribose transfer, detection, and removal and its connection to specific pathways is not well understood at the molecular level (Gupte et al., 2017; O’Sullivan et al., 2019; Palazzo and Ahel, 2018). ADP-ribosyltransferases (“writers”) such as PARP enzymes of the ARTD family catalyze the transfer of mono- or poly-ADP-ribosyl groups to target proteins. These ADP-ribosyl groups can then be recognized by ADP-ribosyl binding proteins (“readers”), which often serve to recruit further proteins. ADP-ribosyl groups may also be removed by hydrolyzing proteins (“erasers”), reversing the action of writers and thereby impacting respective signaling processes.
The human genome encodes many different readers and erasers of ADP-ribosyl groups. Macrodomains represent a large class of ADP-ribosyl binders known in humans. Many of them are encoded as part of other multidomain proteins such as ADP-ribosyltransferases (PARP9, PARP14, PARP15) or histones (macroH2A variants). Other macrodomains such as MDO1, MDO2, poly(ADP-ribose)glycohydrolase [PARG], or TARG1 possess hydrolytic activity and are integral actors in ADP-ribose signaling pathways (Feijs et al., 2013; Rack et al., 2016, 2020a). Viruses such as coronaviruses or togaviruses are known to harbor macrodomains that can remove ADP-ribose from proteins inside the host cell. These viral macrodomains are implied to weaken the host virus defense mechanism by interfering with the host ADP-ribosylation signaling machinery and have been shown to be necessary for virus replication and pathogenesis (Abraham et al., 2018; Fehr et al., 2018; Leung et al., 2018; McPherson et al., 2017). Viruses with these macrodomains include chikungunya virus, MERS-CoV (camel flu), and SARS-CoV-2 (COVID-19). In addition to macrodomains, ADP-ribosyl glycohydrolases of another human enzyme family, ARH1 and ARH3, have been reported to hydrolyze ADP-ribose linked especially to arginine and serine residues, respectively (Abplanalp et al., 2017; Rack et al., 2018, 2020a). Several other domain types exist that are primarily associated with binding of poly-ADP-ribosyl (PAR) groups. Examples of these are the PAR binding zinc finger domain of APLF (Ahel et al., 2008), WWE domain in multiple ADP-ribosyltransferases and E3 ubiquitin ligases (Aravind, 2001; Wang et al., 2012), and PAR binding phosphate pocket in the BRCT1 domain of XRCC1 (Li et al., 2013).
While development of inhibitors for ADP-ribosyltransferases has been investigated for multiple decades mainly in the context of cancer therapeutics, the development of inhibitors against binders or hydrolyzers of ADP-ribose has only gained momentum in recent years (Palazzo and Ahel, 2018). Inhibitors of ADP-ribosyl binding and hydrolyzing proteins would be valuable tools that could help to decipher the complex ADP-ribosyl signaling machinery inside the cell. These inhibitors might also display therapeutic potential as recently demonstrated with novel PARG inhibitors that can impair cancer cell survival (Houl et al., 2019). Furthermore, the inhibition of viral macrodomains to combat diseases caused by these viruses was suggested (Fehr et al., 2018; Rack et al., 2020b; Shimizu et al., 2020) and is currently being explored extensively for the SARS-CoV-2 nsp3 macrodomain (Brosey et al., 2021; Cantini et al., 2020; Michalska et al., 2020; Ni et al., 2021; Russo et al., 2021; Schuller et al., 2021; Virdi et al., 2020).
We sought to set up a system that enables easy development of binding assays suited for a wide variety of ADP-ribosyl binders and hydrolases alike, could further be used for inhibitor screening in a high-throughput setting, and would be simple and easily accessible. Many previous binding assays could only be applied to either ADP-ribosyl readers (Ekblad et al., 2018) or erasers (Haikarainen et al., 2018; Wazir et al., 2021) or are not suitable for high-throughput setups such as mass spectrometry and immunoblot-based methods (Haikarainen and Lehtiö, 2016; Hirsch et al., 2014). Many assays also rely on expensive reagents as used in AlphaScreen or time-resolved fluorescence resonance energy transfer (TR-FRET) technologies or use custom synthesized reagents. Voorneveld et al. (2021) developed a method to synthesize peptides containing mono-ADP-ribosyl (MAR) groups in a selected serine, threonine, or cysteine residue. These synthetic peptides provided excellent control on the sample homogeneity and were used to study ADP-ribose hydrolysis by different erasers. In another study, Schuller et al. (2017) used a synthetic ring-opened ADP-ribosyl group linked to a biotinylated peptide to generate a robust binding assay based on AlphaScreen technology. This modified ADP-ribosyl group was shown to be non-hydrolyzable, so that this assay technology could be successfully applied to readers and erasers. Even though these methods were shown to work robustly, the reagents used may not be easily accessible.
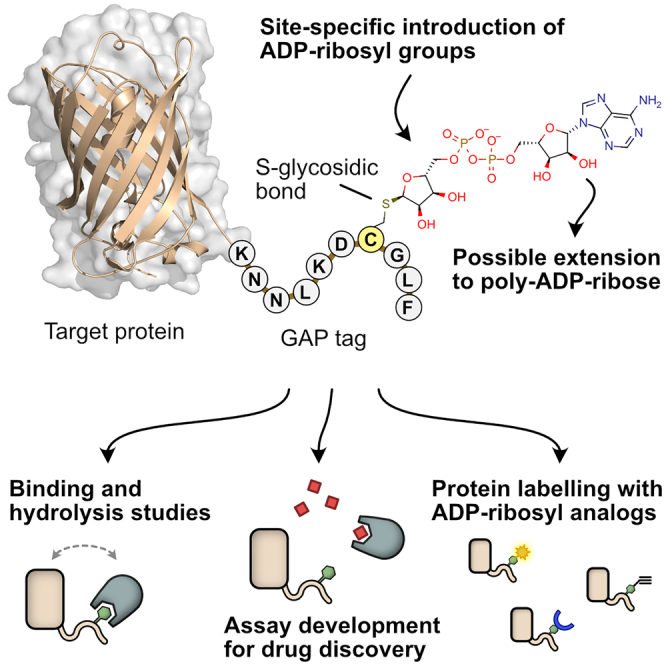
In proteins, many different residues can serve as acceptors of ADP-ribose. Residues such as serine, aspartate, or glutamate form an O-glycosidic bond and lysine, arginine, or asparagine form an N-glycosidic bond with ADP-ribose. Additionally, the linkage via an S-glycosidic bond can be formed with cysteine residues (Cohen and Chang, 2018). While many different ADP-ribosyl-hydrolases exist and can remove ADP-ribose from O- or N-glycosidic bonds, to date there is no human enzyme reported that is able to reverse the S-glycosidic linkage (Voorneveld et al., 2021). We reasoned that a naturally occurring S-glycosidically linked ADP-ribosyl group may serve as a non-hydrolyzable ADP-ribosyl binding probe and therefore could be used to measure the interaction to both hydrolyzing- and non-hydrolyzing ADP-ribosyl binders.
To generate S-glycosidically linked ADP-ribosyl groups in a controlled manner, we used the S1 subunit of pertussis toxin from the bacterium Bordetella pertussis, which is known to efficiently catalyze the transfer of a single ADP-ribosyl group to a specific C-terminal cysteine residue in the αi subunits of heterotrimeric G proteins (Gαi) (Ashok et al., 2020; Katada, 2012). We recombinantly produced proteins with a C-terminal 10-mer peptide of Gαi and still observed efficient modification. This allows, in theory, site-specific addition of ADP-ribose to any protein with an accessible C terminus. We used this system to generate a MARylated YFP protein with stable S-glycosidic bond able to bind ADP-ribosyl readers or erasers. We were able to further extend this system by using PARP enzymes that extend the single residue linked MAR to PAR, allowing us to probe the binding of both MAR and PAR binders. The in vitro system allows for simple and efficient setup of binding assays for ADP-ribosyl readers and erasers based on site-specific cysteine ADP-ribosylation of a Gαi-based tag we termed the Gi protein Alpha subunit Peptide (GAP). We further demonstrated the possibility to modify GAP-tagged proteins with chemically modified NAD+ analogs. To complement the binding assays, we developed a fast and simple detection method of mono- or poly-ADP-ribosylation on blots by fusing ADP-ribosyl binders to nanoluciferase (Nluc). These methods open ways for the development of various in vitro assay systems (Figure 1). To show the applicability for screening, we set up a binding assay for the macrodomain of SARS-CoV-2 non-structural protein 3 and identified the drug suramin as a moderate inhibitor.
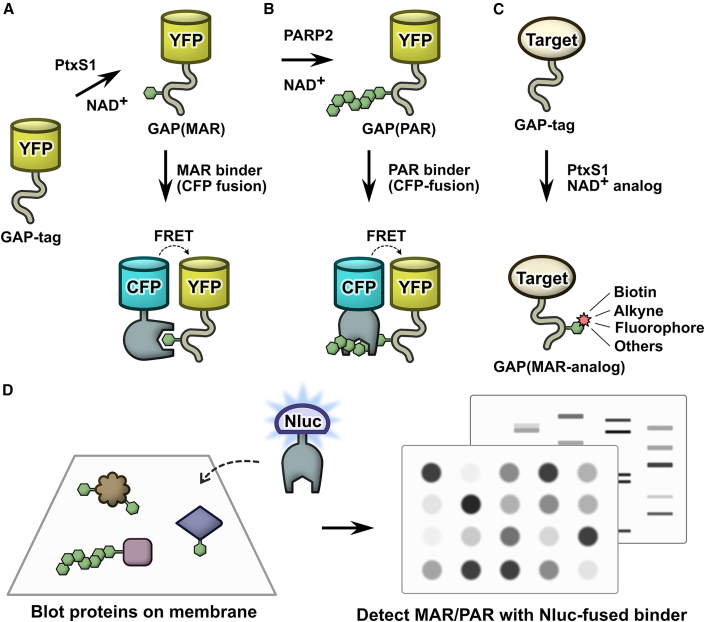
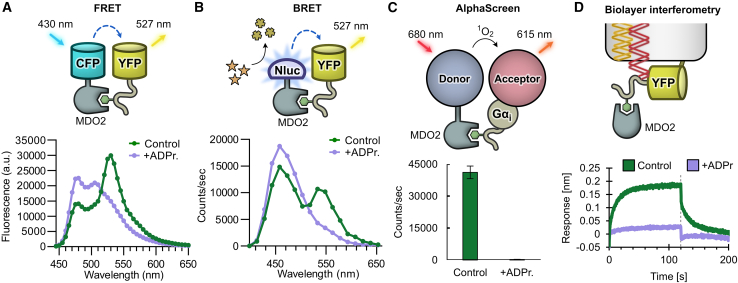
Figure 1.
A molecular toolbox for in vitro interaction studies and assay development of ADP-ribosyl binding proteins
(A) Site-specific ADP-ribosylation of a C-terminal Gαi-based 10-mer peptide (GAP-tag) by pertussis toxin subunit S1 (PtxS1) allows for generation of single S-glycosidically linked mono-ADP-ribosyl (MAR) groups.
(B) The MAR group of the GAP-tag can be extended to a poly-ADP-ribosyl (PAR) group by PARP2. This system can be used to measure binding of proteins interacting with mono- or poly-ADP-ribosyl groups by FRET or other binding technologies.
(C) The GAP-tag can be used for site-specific labeling with NAD+ analogs.
(D) High-affinity ADP-ribosyl binders fused to nanoluciferase (Nluc) can be used as luminescent probes for fast, sensitive, and selective detection of mono- and poly-ADP-ribosylated proteins in blot-based methods.
Results
Initial preparation of proteins for toolbox studies
The proteins used in this study were recombinantly produced in Escherichia coli. We reasoned that mono-ADP-ribosylation (MARylation) of Gαi by pertussis toxin would provide a good probe to test binding to ADP-ribosyl groups because the modification is well defined at a single residue and is linked to cysteine via an S-glycosidic linkage. We initially assumed this linkage to be stable against enzymatic hydrolysis by many of the erasers, which later during our work was also experimentally confirmed by others (Voorneveld et al., 2021). It was previously shown that the recombinantly produced pertussis toxin subunit S1 (PtxS1) could be used to ADP-ribosylate Gαi proteins in vitro (Ashok et al., 2020). We tested the ability of PtxS1 to modify unlabeled and YFP-fused full-length Gαi as well as a 10-residue C-terminal peptide of Gαi (GAP) when fused to YFP (Figure 2A). While ADP-ribosylation by PtxS1 was confirmed for these Gαi constructs, the lower signal for the GAP-tag indicates less efficient modification by PtxS1 compared with full-length constructs. Similar results were reported recently for synthetic Gαi peptides (Eskonen et al., 2020) and were further confirmed by NAD+-consumption assay (Figure S1). Despite this, we found that ADP-ribosylation activity by PtxS1 was sufficient to produce MARylated GAP-tagged YFP (Figure S1).
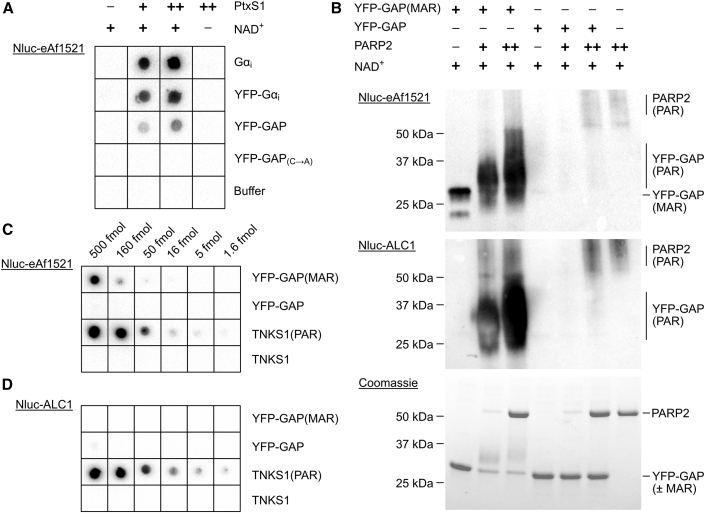
Figure 2.
Initial development and preparation of toolbox components
(A) Testing ADP-ribosylation by PtxS1 with different Gαi constructs. Unlabeled or YFP-fused full-length Gαi constructs and GAP-tagged YFP were tested as cysteine-ADP-ribose acceptors when treated with 50 nM (+) or 250 nM (++) PtxS1. As controls, buffer or YFP-GAP in which the acceptor cysteine was mutated to alanine were used. Reactions were blotted on a nitrocellulose membrane, and detection was done using Nluc-eAf1521.
(B) The mono-ADP-ribosyl (MAR) group in the GAP-tag can be extended to poly-ADP-ribose by PARP2. YFP-GAP or YFP-GAP(MAR) (10 μM) were mixed with 1 mM NAD and 400 nM (+) or 4 μM (++) PARP2 or buffer as control. The reactions were run on SDS-PAGE and visualized using Coomassie blue or by western blot and detection using Nluc-eAf1521 or Nluc-ALC1.
(C) Detection of MAR and PAR by Nluc-eAf1521.
(D) Selective detection of PAR by Nluc-ALC1. Dilution series of YFP-GAP(±MAR) or TNKS1(±PAR) were blotted on nitrocellulose membranes. YFP-GAP: 1 fmol = 31 pg; TNKS1: 1 fmol = 75 pg.
Next, we aimed to introduce a PAR chain to the GAP-tag in order to generate a probe that could be used to detect interaction of PAR binding proteins. We reasoned that the MAR group of the previously modified GAP-tag could serve as the starting point for elongation to poly-ADP-ribose by PARP enzymes. Detection of PAR chains by western blot shows that the MARylated GAP-tag, but not the unmodified GAP-tag, can be PARylated by PARP2 in the presence of NAD+ (Figures 2B and S2). A similar extension can be created by using TNKS1 instead of PARP2 (Figure S2).
As a tool to detect ADP-ribosylation, we adapted a method for blot-based detection by Nluc luminescence (Boute et al., 2016). Instead of fused antibodies, we produced the recently reported engineered ADP-ribosyl superbinder eAf1521 (Nowak et al., 2020) or high-affinity PAR binding macrodomain of ALC1 (Singh et al., 2017) as fusion proteins with Nluc. We found that these constructs are easy to produce in E. coli and work in a simple and fast protocol for sensitive detection of mono- and poly-ADP-ribosylated proteins (Figures 2C and 2D).
The GAP-tag for site-specific labeling of proteins using NAD+ analogs
While we were able to use the GAP-tag to site-specifically label proteins with ADP-ribose, we sought to demonstrate that this system could be extended with NAD+ analogs to introduce various chemical groups to the C terminus of proteins. Many NAD+ analogs already exist and are commercially available, such as biotinylated, fluorescent, and click-chemistry-ready NAD+ analogs (Depaix and Kowalska, 2019). We first tested the modification of GAP-tagged YFP with 6-biotin-17-NAD+ and confirmed that this NAD+ analog serves as substrate for PtxS1-based modification of the GAP-tag (Figure 3A). The site-specific modification was detected using dot blot with streptavidin-conjugated horseradish peroxidase (HRP). We further tested modification of the GAP-tag with 6-propargyladenine-NAD+. The NAD+ analog is accepted as a substrate by PtxS1 and, as the resulting ADP-ribosyl-group contains an alkyne moiety, it can be used in a copper(I)-catalyzed alkyne-azide cycloaddition reaction with azide-labeled Cy3 or Cy5 fluorophores to label the proteins (Figure 3B).
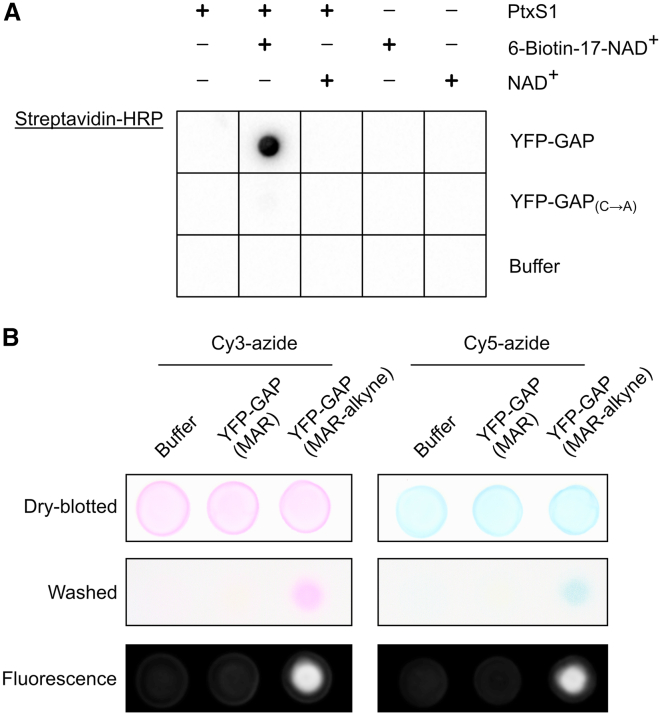
Figure 3.
The GAP-tag can be used to introduce site-specific modifications with NAD+ analogs
(A) Site-specific biotinylation of the GAP-tag. GAP-tagged YFP was mixed with NAD+ or 6-biotin-17-NAD+ in absence or presence of PtxS1. The reactions were blotted on a nitrocellulose membrane, and detection of biotin was done with streptavidin-HRP.
(B) YFP-GAP was MARylated with PtxS1 using NAD+ or 6-propargyladenine-NAD+ containing an alkyne group. The resulting proteins YFP-GAP(MAR) or YFP-GAP(MAR-alkyne) or buffer were mixed with Cy3-azide or Cy5-azide, and the copper(I)-catalyzed alkyne-azide cycloaddition (CuAAC) reaction was performed by addition of 5 mM sodium ascorbate, 300 μM CuSO4, and 600 μM L-histidine. The samples were incubated for 3 h at room temperature and blotted on nitrocellulose membranes, and visible-light images were taken. Unreacted Cy3-azide or Cy5-azide was removed by washing of the membranes in Tris-buffered saline/Tween-20. Visible-light and fluorescent images were taken.
Evaluation of binding with confirmed and potential ADP-ribosyl binders
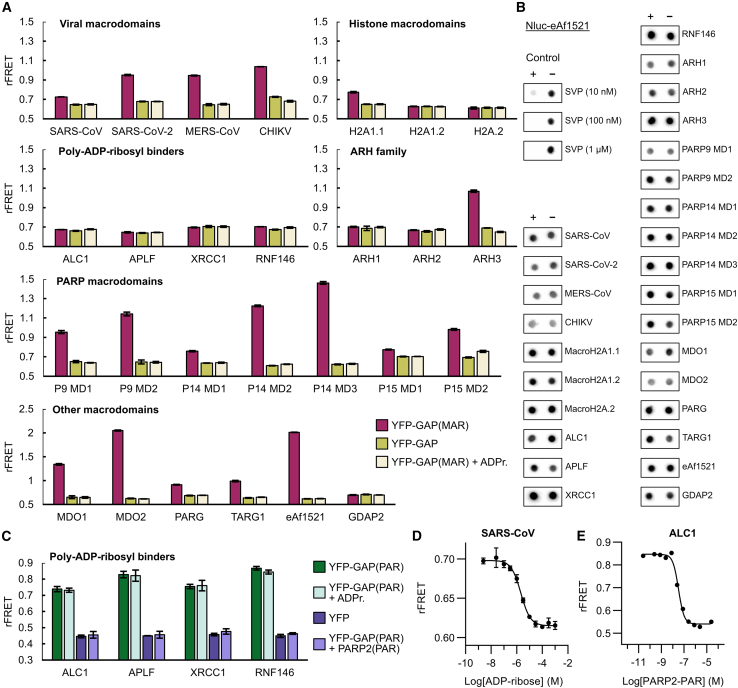
We selected a set of 27 different proteins to be tested for binding to the MARylated GAP-tag. We recombinantly produced these proteins in E. coli as fusions with CFP and tested ratiometric FRET signals (rFRET) upon binding to the MARylated YFP-GAP construct (Figure 4A). We used non-MARylated YFP-GAP construct as a control as well as MARylated YFP-GAP containing 200 μM ADP-ribose to compete with the interaction. A higher FRET signal correlates with a higher occupancy and, thus, binding affinity of the binding partners, but it is also affected by the distance and orientation of the fluorophores (Kashida et al., 2017). We found that the proteins previously reported to bind ADP-ribose showed a higher FRET signal compared with the controls, indicating binding to the MARylated GAP-tag. ARH1 was reported to bind ADP-ribose with a low affinity (Rack et al., 2018), which is likely the reason that we could not measure an FRET signal for this construct. While all three histone macrodomains macroH2A1.1, macroH2A1.2, and macroH2A2 have highly similar sequences, previous reports have concluded that only macroH2A1.1 has the ability to bind ADP-ribose (Kozlowski et al., 2018; Kustatscher et al., 2005). This is in agreement with our observations, wherein only macroH2A1.1 shows an elevated FRET signal over the controls. The GDAP2 protein has a macrodomain with similarities to MDO1 and MDO2; however, it was reported to be unable to bind ADP-ribose (Neuvonen and Ahola, 2009), which is consistent with our findings.
Figure 4.
Testing interactions of reported and potential readers and erasers with YFP-GAP
(A) Interactions of CFP-fused potential and confirmed ADP-ribosyl binders with MARylated YFP-GAP. CFP-fusion proteins (1 μM) were mixed with 5 μM YFP-GAP or with 5 μM YFP-GAP(MAR) in absence or presence of 200 μM ADP-ribose (ADPr.). The ratiometric FRET signals were measured. P9, P14, and P15 denote PARP9, PARP14, and PARP15.
(B) Test of ADP-ribosyl removal from GAP-tag. YFP-GAP(MAR) (10 μM) was prepared in absence (−) or presence (+) of 1 μM CFP-fused proteins or 0.01 μM–1 μM snake venom phosphodiesterase I (SVP). Samples were incubated for 24 h at room temperature and blotted on a nitrocellulose membrane. Detection was done with Nluc-eAf1521.
(C) Interactions of poly-ADP-ribosyl binders with PARylated YFP-GAP. CFP-fusion proteins (250 nM) were mixed with 500 nM YFP or 500 nM YFP-GAP(PAR) in absence or presence of 100 μM ADP-ribose or 2.5 μM automodified PARP2. The ratiometric FRET signals were measured.
(D) Representative dose-response curve of 1 μM CFP-SARS-CoV nsp3 and 5 μM YFP-GAP(MAR) upon competition with ADP-ribose. The control containing no ADP-ribose was set one logarithmic unit below the lowest concentration. Dose-response curves with ADP-ribose for all CFP-fusion proteins are shown in Figure S4.
(E) Representative dose-response curve of 250 nM CFP-ALC1 and 500 nM YFP-GAP(PAR) upon competition with PARylated PARP2. The control containing no PARP2(PAR) was set one logarithmic unit below the lowest concentration, while the control using YFP-GAP(MAR) instead of YFP-GAP(PAR) was set one logarithmic unit above the highest PARP2(PAR) concentration. Data shown for FRET assays are mean ± standard deviation with n = 4 replicates.
While cysteine-(ADP-ribosyl)hydrolase activity was detected in human erythrocytes and mitochondria (Herrero-Yraola et al., 2001; Tanuma and Endo, 1990), no specific enzymes in humans with this activity could be identified to date. To test whether any of the proteins show S-glycosylhydrolase activity, we mixed the aforementioned CFP-fused constructs with MARylated YFP-GAP protein and tested the hydrolysis of ADP-ribose using dot-blot analysis after a 24-h incubation period (Figure 4B). While the controls including snake venom phosphodiesterase I (SVP) showed loss of the signal through cleavage of the ADP-ribose diphosphate, none of the tested proteins were able to substantially hydrolyze the S-glycosidic bond under the conditions tested, which is in agreement with recently reported findings (Voorneveld et al., 2021). ARH family members also showed no hydrolysis upon addition of MgCl2 (Figure S3). These findings highlight the versatility of this system for measuring the ADP-ribosyl binding of proteins that can otherwise hydrolyze O- or N-glycosidic linkages such as MDO1, MDO2, TARG1, ARH3, or SARS-CoV-2 nsp3.
The PAR binders ALC1, APLF, XRCC1, and RNF146 WWE domain did not show binding to MARylated YFP-GAP (Figure 4A); however, they showed increased FRET signals when we used YFP-GAP PARylated by PARP2 (Figure 4C). For proteins binding to the MARylated GAP-tag, even comparatively low ratiometric FRET values showed good signals in dose-response experiments by competition with ADP-ribose (Figures 4D and S4). Similarly, a representative curve for ALC1 with PARylated YFP-GAP was recorded and shows loss of the FRET signal with higher concentrations of auto-PARylated PARP2 protein (Figure 4E).
We used FRET for testing the interaction of proteins with the ADP-ribosylated GAP-tag; however, we sought to demonstrate that binding to MARylated Gαi could also be measured with different binding technologies, for which we used MDO2 as an example protein. Similarly to the binding of CFP-fused MDO2 to MARylated YFP-GAP to measure FRET (Figures 5A and S5, dissociation constant KD = 654 nM), we used Nluc-fused MDO2 to generate a bioluminescence resonance energy transfer (BRET) signal upon interaction with MARylated YFP-GAP (Figure 5B). Conveniently, the same YFP variant can serve as acceptor in FRET and BRET applications. AlphaScreen protocols for ADP-ribosyl readers or erasers exist (Ekblad et al., 2018; Haikarainen et al., 2018; Schuller et al., 2017), and we could use our system adapted to AlphaScreen technology to directly probe binding to MARylated Gαi by MDO2 (Figure 5C). We further showed binding of MDO2 to MARylated YFP-GAP using biolayer interferometry (BLI) (Figure 5D), which in recent years has gained popularity as a method for the screening of small-molecule compounds (Kaminski et al., 2017; Overacker et al., 2021; Peltomaa et al., 2018). This method also allows for simple quantification of kinetic binding parameters such as the KD (Figure S5).
Figure 5.
Various assay technologies can be utilized to detect binding to the MARylated Gαi
(A) Measurement of interaction by FRET. Fluorescence emission spectra of CFP-MDO2 and YFP-GAP(MAR) in absence (control) or presence of 200 μM ADP-ribose (ADPr.).
(B) Measurement of interaction by BRET. Luminescence emission spectra of Nluc-MDO2 and YFP-GAP(MAR) in absence (control) or presence of 200 μM ADP-ribose.
(C) Measurement of interaction by AlphaScreen. Biotinylated MDO2 and His-tagged MARylated Gαi were mixed with streptavidin donor beads and chelate acceptor beads in absence (control) or presence of 10 μM ADP-ribose. The luminescence signal was detected upon excitation of donor beads. Data shown are mean ± standard deviation with n = 4 replicates.
(D) Measurement of interaction by biolayer interferometry. His-tagged YFP-GAP(MAR) was bound to the optical sensor surface, and the change of signal after association (0 s) or dissociation (120 s, dotted line) of unlabeled MDO2 protein was determined in absence or presence of 3.16 μM ADP-ribose.
Application example: Screening for inhibitors against the SARS-CoV-2 nsp3 macrodomain
In light of the current situation regarding the COVID-19 pandemic, we chose to demonstrate the applicability of this binding system for the screening of small-molecule inhibitors against the macrodomain of SARS-CoV-2 nsp3. Presently, efforts are being made by researchers worldwide to find inhibitors against this macrodomain (Brosey et al., 2021; Cantini et al., 2020; Michalska et al., 2020; Ni et al., 2021; Russo et al., 2021; Schuller et al., 2021; Virdi et al., 2020). The nsp3 macrodomain of coronaviruses was shown to be critical for viral replication (Fehr et al., 2015, 2016), and therefore small-molecule inhibitors might show promise as therapeutic agents to fight infections caused by SARS-CoV-2 and other viruses.
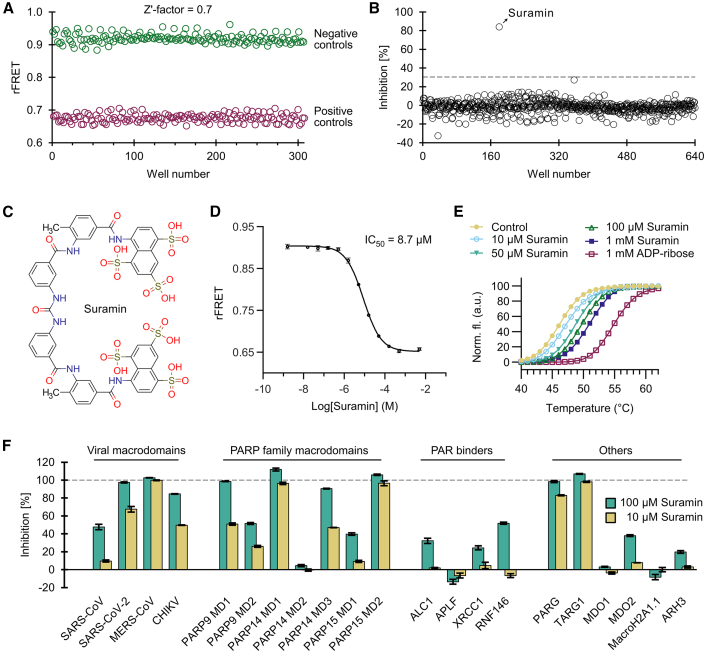
We assessed the quality of the FRET signals of the alternating positive and negative controls in a 384-well plate by mixing CFP-fused SARS-CoV-2 nsp3 macrodomain with MARylated YFP-GAP in the absence and presence of 200 μM ADP-ribose (Figure 6A). While the SARS-CoV-2 macrodomain was reported to have a relatively low binding affinity to ADP-ribose (KD = 17 μM, Alhammad et al., 2021), we still found that the FRET signal showed sufficient separation of positive and negative controls. The Z′ factor was calculated to be 0.7, indicating that this assay is well suitable for high-throughput screening (Zhang et al., 1999).
Figure 6.
Development of a screening assay for the SARS-CoV-2 nsp3 macrodomain
(A) Signal validation for a screening assay with CFP-SARS-CoV-2. SARS-CoV-2 (1 μM) was mixed with 5 μM YFP-GAP(MAR) in absence (negative control) or presence (positive control) of 200 μM ADP-ribose, and a Z′ factor of 0.7 was calculated.
(B) Screen of ENZO FDA-approved drug library comprising 640 compounds. Only the compound suramin showed inhibition above 30% and was taken to further validation.
(C) Structure of the hit compound suramin.
(D) Dose-response curve with suramin shows an IC50 of 8.7 μM for the SARS-CoV-2 nsp3 macrodomain in the FRET-based assay. The control containing no compound was set one logarithmic unit below the lowest concentration, while the control containing 200 μM ADP-ribose was set one logarithmic unit above the highest suramin concentration.
(E) Suramin shows stabilization of SARS-CoV-2 nsp3 macrodomain by DSF.
(F) Inhibition profile of suramin against human and viral ADP-ribosyl binders used in this study. The inhibition was calculated based on the ratiometric FRET signals of the CFP-fused binders mixed with YFP-GAP(MAR) or YFP-GAP(PAR).
Data shown for (D) and (F) are mean ± standard deviation with n = 4 replicates.
We screened against a Food and Drug Administration (FDA)-approved drug library comprising 640 small-molecule compounds at 20 μM compound concentration (Figure 6B). From the screening, only the compound suramin was regarded as a hit with 82% inhibition (Figure 6C). The IC50 value of this compound was determined to be 8.7 μM against the SARS-CoV-2 nsp3 macrodomain in the FRET-based assay (Figure 6D). To confirm binding of the compound to the SARS-CoV-2 nsp3 macrodomain, we performed differential scanning fluorimetry (DSF) analysis and showed stabilization of the protein by suramin in a concentration-dependent manner (Figure 6E). Suramin, like ADP-ribose, also reduced the hydrolysis activity of the macrodomain (Figure S6). Intriguingly, suramin is used as a broadband antiviral and antiparasitic drug and was recently reported to inhibit SARS-CoV-2 infection in cell-culture-based models (Salgado-Benvindo et al., 2020) and to bind with high affinity to the RNA polymerase of SARS-CoV-2 (Yin et al., 2021). After reviewing the literature associated with suramin, we found that it is reported to inhibit a plethora of protein targets such as DNA and RNA polymerases, sirtuins, ATPases, and G-protein-coupled receptors (Freissmuth et al., 1996; Torrente et al., 2014; Trapp et al., 2007; Wiedemar et al., 2020), indicating that it exhibits low target specificity. We next tested the inhibition of suramin against the viral and human ADP-ribose readers and erasers produced in this study using the FRET-based assay (Figure 6F). Not surprisingly, suramin showed strong inhibition even at 10 μM against many of the proteins tested, confirming the low target specificity of this compound. To our knowledge, inhibition of macrodomains by suramin was not previously reported. We show that suramin also inhibits the nsp3 macrodomain of CHIKV, and inhibition of CHIKV pathogenesis by suramin was shown in multiple studies (Albulescu et al., 2015, 2020; Henß et al., 2016; Ho et al., 2015; Kuo et al., 2016; Lu et al., 2017). While multiple mechanisms for this inhibitory activity against CHIKV were reported, it is tempting to speculate that the inhibition of the nsp3 macrodomain poses another yet overlooked mechanism.
Discussion
The high complexity of ADP-ribosyl-associated pathways makes the involved proteins notoriously difficult to study (Bonfiglio et al., 2020; Lüscher et al., 2018). While potent and specific inhibitors against many of the ADP-ribosyltransferases fundamentally helped to broaden our understanding of these enzymes (Durkacz et al., 1980; Huang et al., 2009; Kirby et al., 2018; Venkannagari et al., 2016; Wang et al., 2018), such inhibitors against ADP-ribosyl readers and erasers are still scarcely available or in early stages of development (Harrision et al., 2020; James et al., 2016; Liu et al., 2020; Palazzo and Ahel, 2018; Schuller et al., 2017). It was suggested that a possible bottleneck for the discovery of inhibitors arises from the lack of accessible high-throughput technologies (Schuller et al., 2017). Many of the assay systems work for only a subset of MAR or PAR readers or erasers, require expensive or custom-made reagents, or are not suited for high-throughput screening.
We have shown that the MARylated GAP-tag provides an easy-to-use and stable template for the development of binding assays for ADP-ribosyl binding proteins. This system can be adapted to different binding technologies such as FRET, BRET, AlphaScreen, or BLI as shown in this study, but is not limited to these and could be extended to work with technologies such as protein-fragment complementation assays or TR-FRET methods. The GAP-tag can also be used to site-specifically introduce chemical ADP-ribose analogs to the protein of interest, adding another technology to the chemical biology toolkit of protein labeling tags (Lotze et al., 2016). The ADP-ribosyl labeling technology ELTA might further be used to extend this functionality (Ando et al., 2019). We have further shown that the MAR group attached to the GAP-tag can be subsequently extended to poly-ADP-ribosyl chains by PARP2, which we used to detect interactions with PAR binding proteins. Similarly, Prokhorova et al. (2021) recently reported that a histone H3 peptide could be PARylated by PARP1 in vitro only after having been primed by a mono-ADP-ribose unit. The generation of single poly-ADP-ribose chains at the protein-fusible GAP-tag might also prove useful in other applications in the future. We tested in this study a total of 27 proteins that were either confirmed to bind to ADP-ribosyl groups or were shown not to bind them, despite being macrodomains. For 22 of these proteins, we confirmed binding to either MAR or PAR groups attached to the GAP-tag. The remaining five proteins not showing binding were either reported not to bind ADP-ribose or bind it only with very low affinity. The systems described in this work can be entirely recombinantly produced in E. coli with good yields for most of the proteins (>200 mg/L culture). While we have produced and tested a large set of proteins in this study, the production of only three proteins is required to set up a high-throughput assay for an ADP-ribosyl binder of interest (e.g., CFP-fusion protein, YFP-GAP, and PtxS1). In the frame of our work, we also made use of Nluc-fused high-affinity ADP-ribose binders eAf1521 and ALC1 as sensitive detection agents of MAR and PAR groups for blot-based studies, respectively. Gibson et al. (2017) described a similar system based on ADP-ribosyl binding proteins fused to the Fc region of rabbit immunoglobulin, which can subsequently be detected by commercial antibodies targeting the Fc region. Due to simple recombinant expression, we used Nluc and not the more established HRP as luminescent reporter. We found that our system, which does not require a secondary antibody, takes only little working time of about 1 h from blotting to imaging in practice. This method may require more optimization efforts for quantitative blots compared with the more established reporter HRP, but it works readily for semi-quantitative or qualitative detection as shown in this work. We used these reporters to confirm mono- or poly-ADP-ribosylation of the GAP-tag and other Gαi constructs and to test for potential hydrolysis of S-glycosidic bonds by the proteins used in this study, but it could also be used for studying ADP-ribosylation by PARP family members or might find applications in cell-based studies.
The presence of a hydrolytically active macrodomain in the nsp3 protein of SARS-CoV-2 has led to multiple efforts in finding inhibitors against this protein. Thus far, many different techniques have been utilized in the search for inhibitors. In a recent study by Schuller et al. (2021), screening for small molecules binding to the SARS-CoV-2 nsp3 macrodomain was performed using a combination of crystallographic and computational fragment screening. While this is a promising setup to generate high-affinity binders of this macrodomain, conditions to produce high-quality protein crystals are needed, which may hinder the transfer of this technology to other ADP-ribosyl binding proteins. Additionally, initial fragment hits often have relatively low binding affinity and need to be chemically linked or modified to yield more potent binders. Virdi et al. (2020) utilized a DSF-based approach to screen for inhibitors of SARS-CoV-2 nsp3 macrodomain. This technique may yield more strongly binding initial hits compared with fragments-based screening, although compounds are commonly found to interfere with the signal. DSF is typically performed with large amounts of protein in low-density 96-well plates and a time-consuming denaturation is required, limiting the throughput capabilities of this technique.
As an example of an application of the GAP-based binding assay, we screened a small-molecule library of existing drugs against the nsp3 macrodomain of SARS-CoV-2. We validated suramin as a hit compound, confirming that the assay system is suitable for screening of inhibitors. Furthermore, the high yield of the proteins produced and low complexity of the assay setup in a mix-and-measure manner highlight the applicability of this assay for high-throughput screening. We used our assay with the other ADP-ribosyl binding proteins and found that suramin showed significant inhibition against the majority of these proteins. We believe that this simple setup of profiling for potential inhibitors against many different ADP-ribosyl binders is a valuable application of this technology and may thus complement other screening technologies such as the ones already mentioned herein.
In summary, the tools presented in this study are very accessible and allow the setting up of robust and adaptable ADP-ribosyl binding assays, and will therefore aid in the investigation of ADP-ribosyl binders and the discovery of chemical probes targeting them.
Limitations
In the frame of this work, we developed the GAP-tag, which allows for a simple setup of binding assays with mono- or poly-ADP-ribosyl binders. Many of the MAR binding proteins have only weak micromolar binding affinity to ADP-ribose, which in turn requires low-micromolar concentrations of the proteins in the binding assays to be present, thereby limiting the sensitivity of the method. This could be circumvented to some extent by applying the GAP-tag to amplified luminescent techniques such as AlphaScreen (Figure 5C). Other assay systems that measure, for example, enzymatic activities may require only nanomolar or even subnanomolar amounts of protein. While the majority of the proteins used in this study can be recombinantly produced in substantial amounts sufficient for screening several tens of thousands of compounds, some of the proteins such as macrodomain 1 of PARP9 and PARP15 are expressed in quantities too small to be considered for such screening efforts (Table S2). We were, however, still able to use them for testing the inhibition profile of suramin. We could so far not come up with a strategy that would allow us to transfer this assay technology to cell-based systems, as many binders and ADP-ribosyl groups present in the cell would readily compete with the binding interactions of interest. The fluorescently labeled macrodomains described here, however, could be used in a fashion similar to that of the recently described ADP-ribosyl detection reagent MacroGreen (García-Saura et al., 2021). On a related note, the labeling of GAP-tagged proteins with NAD+ analogs is likely reserved for in vitro applications, as naturally occurring NAD+ in cells would compete with such analogs. However, labeling of proteins on the cell surface may be possible, which is a common application with current labeling techniques.
STAR★Methods
Key resource table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Bacterial and virus strains | ||
| E. coli BL21 (DE3) | New England BioLabs | C2527H |
| E. coli Rosetta2 (DE3) | Sigma-Aldrich | 71400-3 |
| E. coli NEB5α | New England BioLabs | C2987H |
| Chemicals, peptides, and recombinant proteins | ||
| NAD+ | Sigma-Aldrich | N0632 |
| 6-Biotin-17-NAD+ | Biolog | N 012 |
| 6-propargyladenine-NAD+ | Biolog | N 051 |
| Cy3-azide | Sigma-Aldrich | 777315 |
| Cy5-azide | Sigma-Aldrich | 777323 |
| Streptavidin-HRP | PerkinElmer | NEL750001EA |
| Phosphodiesterase I (Crotalus adamanteus venom) | Worthington Biochemical Corporation | LS003926 |
| TB Auto induction media | Formedium | AIMTB0210 |
| Nano-Glo luciferase substrate | Promega | N1110 |
| ADP-ribose | Sigma-Aldrich | A0752 |
| WesternBright ECL HRP substrate | Advansta | K-12045 |
| AlphaScreen Histidine (Nickel Chelate) Detection Kit | PerkinElmer | 6760619C |
| Recombinant DNA | ||
| Expression vectors used in this study are described in Table S1 | This work | N/A |
| pNIC28-Bsa4 | Addgene | 26103 |
| pNH-TrxT | Addgene | 26106 |
| pET Biotin His6 mCitrine LIC cloning vector (H6-mCitrine) | Addgene | 29724 |
| pET mCerulean LIC cloning vector (H6-mCerulean) | Addgene | 29726 |
| pUAS-NanoLuc | Addgene | 8769 |
| pNIC-CFP | Addgene | 173074 |
| pNH-Nluc | Addgene | 173075 |
| Software and algorithms | ||
| Prism (ver. 8.0.2) | GraphPad Software | https://www.graphpad.com/scientific-software/prism/ |
| ImageJ (ver. 1.53k) | NIH | https://imagej.nih.gov/ij/ |
| MicroArray profile plugin | OptiNav | https://www.optinav.info/MicroArray_Profile.htm |
| Other | ||
| OptiPlate-384, White Opaque microplates | PerkinElmer | 6007290 |
| AlphaPlate-384, Light gray microplates | PerkinElmer | 6005350 |
| Fisherbrand 384-Well ShallowWell black polypropylene microplates | Fisher Scientific | 13595450 |
| Infinite M1000 Pro multimode microplate reader with AlphaScreen module | Tecan | Discontinued |
| Spark multimode microplate reader | Tecan | https://lifesciences.tecan.com/multimode-plate-reader |
| Octet RED384 | ForteBio | https://www.fortebio.com/node/150 |
Resource availability
Lead contact
-
•
Further information and requests for resources and reagents should be directed to the lead contact, Lari Lehtiö (lari.lehtio@oulu.fi).
Materials availability
-
•
Expression constructs generated in this study that can be used to set up the technology can be requested from the lead contact. Most of the expression vectors generated in this study are available through Addgene.
Experimental model and subject details
While no experimental model organism was used in the study, we used commercially available E. coli strains for cloning procedures and recombinant protein production. E. coli NEB5α (New England BioLabs) was used for cloning procedures. E. coli BL21 (DE3) (New England BioLabs) and E. coli Rosetta2 (DE3) (Sigma-Aldrich) were used for the recombinant protein production.
Method details
Cloning
Constructs were cloned into expression vectors by SLIC (Jeong et al., 2012). E. coli NEB5α was used for cloning procedures. The expression vectors are based on pNIC28-Bsa4 (Addgene Plasmid #26103) or pNH-TrxT (Addgene Plasmid #26106). For pNIC-MBP, we inserted the sequence for E. coli maltose binding protein (UniProt ID: P0AEX9, residues 27-392) between His6-tag and TEV protease cleavage site of pNIC28-Bsa4 as previously described (Sowa et al., 2020). In the same manner for pNIC-YFP and pNIC-CFP, the sequences encoding mCitrine (Addgene Plasmid #29724) or mCerulean (Addgene Plasmid #29726) were inserted between His6-tag and TEV protease cleavage site of pNIC28-Bsa4. For pNH-Nluc, the sequence encoding Nluc (Addgene Plasmid #87696) was inserted into pNH-TrxT in place of the sequence encoding TrxT. Sequence boundaries of the cloned expression constructs are shown in Table S1.
Protein expression
Expression and purification procedures for full length Gαi (Ashok et al., 2020), MDO2 (Wazir et al., 2021), PARP2 constructs (Obaji et al., 2021), SRPK2 (Venkannagari et al., 2013) and YFP (Sowa et al., 2020) were performed as previously described. The following constructs were expressed in E. coli strain Rosetta2 (DE3): CFP-SARS-CoV, CFP-MacroH2A2, CFP-PARP9 (MD2), CFP-PARP14 (MD1), CFP-PARP14 (MD2), CFP-PARP14 (MD3), CFP-PARP15 (MD2), CFP-MDO2, CFP-PARG and CFP-GDAP2. All other constructs were expressed in E. coli strain BL21(DE3). The respective chemically competent cells were transformed with the plasmids encoding the expression constructs described in Table S1. 500 ml Terrific Broth (TB) autoinduction media including trace elements (Formedium, Hunstanton, Norfolk, England) were supplemented with 8 g/l glycerol and 50 μg/ml kanamycin and inoculated with 5 ml of overnight preculture. The media was additionally supplemented with 34 μg/ml chloramphenicol for Rosetta2 (DE3) cells. The flasks were incubated shaking at 37°C until an OD600 of about 1 was reached. The temperature was thereafter set to 15°C for constructs PtxS1, CFP-PARP14 (MD3) and CFP-PARP15 (MD1) or to 16°C for constructs CFP-SARS-CoV-2 and SARS-CoV-2 or to 18°C for all other constructs and incubation continued overnight for about 20–22 hours. The cells were collected by centrifugation at 4,200×g for 15–30 min at 4°C. The pellets were resuspended in respective lysis buffers (Table S2) and stored at −20°C until purification.
Protein purification
IMAC
All constructs were initially purified by immobilized metal affinity chromatography (IMAC). The cells were thawed, supplemented with 0.1 mM Pefablock SC (Roche) and 20 μg/ml DNase I (Roche) and lysed by sonication. The lysate was centrifuged (16,000×g, 4°C, 30 min), filtered and loaded onto an IMAC column equilibrated with lysis buffer and charged with Ni2+ or Zn2+. The column was washed with lysis buffer and wash buffer and the protein was eluted with elution buffer. Detailed information with the column, buffers and volumes used for each construct can be found in Table S2.
Reverse IMAC
After purification by IMAC, a reverse IMAC step was performed for PARP10 and SARS-CoV-2 nsp3 proteins. The His6- or His6-TrxT-tag was removed by digestion with TEV-protease (1:30 molar ratio) while the protein was dialyzed against 30 mM HEPES pH 7.5, 300 mM NaCl, 10% glycerol, 0.5 mM TCEP (4°C, 20 hours). The protein was thereafter loaded to a 5 ml HiTrap Chelating HP column charged with Ni2+. The column was washed with 3 column volumes of lysis buffer. The flowthrough and wash fractions were collected for further purification by size-exclusion chromatography.
MBP affinity chromatography
After purification by IMAC, a final MBP affinity purification step was performed for MBP-tagged TNKS1-SAM-Catalytic construct. The IMAC eluate was loaded onto a 5 ml MBPTrap HP column equilibrated with MBPTrap loading buffer (50 mM HEPES pH 7.5, 500 mM NaCl, 0.5 mM TCEP). The column was washed with 3 column volumes of the MBPTrap loading buffer and eluted with the same buffer containing 10 mM maltose. The protein was aliquoted and flash frozen in liquid nitrogen and stored at −70°C.
Size exclusion chromatography
With exception of Nluc-eAf1521 and MBP-TNKS1, all constructs were purified by a final size exclusion chromatography step. Size exclusion chromatography was carried out on a S75 16/600 size-exclusion chromatography column with the previously purified IMAC eluates or reverse IMAC fractions of the constructs. The buffer used was 20 mM HEPES pH 7.5, 300 mM NaCl, 0.5 mM TCEP for PtxS1, 20 mM HEPES pH 7.5, 250 mM NaCl, 1 mM TCEP, 5% glycerol for CFP-SARS-CoV, CFP-PARP9 (MD2) and CFP-PARP15 (MD2). For all other constructs, 30 mM HEPES pH 7.5, 300 mM NaCl, 10% Glycerol, 0.5 mM TCEP was used. Purified fractions were combined, aliquoted and flash frozen in liquid nitrogen and stored at −70°C.
Preparation of mono- and poly-ADP-ribosylated GAP-tagged proteins
YFP with C-terminal GAP-tag was purified by IMAC and dialyzed against 20 mM HEPES pH 7.5, 350 mM NaCl. YFP-GAP was diluted to 100 μM in 50 mM sodium phosphate buffer pH 7.0 and mixed with 1.5 μM PtxS1 and 150 μM NAD+. The reaction was incubated for an hour at room temperature. To ensure completeness of the reaction, NAD+ (150 μM) was added a second time to the reaction. Incubation was continued for 1 h at room temperature. The reaction mixture was loaded to an IMAC column to remove pertussis toxin, hydrolysis products and unreacted NAD+. IMAC was carried out as described in the purification procedures above. The buffer was exchanged to 20 mM HEPES pH 7.5, 150 mM NaCl, 0.5 mM TCEP and the MARylated YFP-GAP was subsequently concentrated to about 1 mM concentration using a Amicon Ultra-15 Centrifugal Filter Unit (MWCO: 10kDa). The protein was flash frozen in liquid nitrogen and stored at −70°C.
PARylated YFP-GAP for FRET experiments was prepared from MARylated YFP-GAP. MARylated YFP-GAP (10 μM) was incubated in the presence of 400 nM PARP2 (residues 90-583) and 1 mM NAD+ in a buffer solution containing 50 mM Tris pH 8.0 and 5 mM MgCl2 for 2 h at room temperature. The reacted sample was then purified using IMAC as described above to remove PARP2. The sample buffer was exchanged to 30 mM HEPES pH 7.5, 150 mM NaCl, 10% glycerol, 0.5 mM TCEP using an Amicon Ultra-15 Centrifugal Filter Unit (MWCO: 10kDa). The protein was aliquoted and flash frozen in liquid nitrogen and stored at −70°C.
For PARylated YFP-GAP production by TNKS1, MARylated YFP-GAP was incubated with 200 nM TNKS1 SAM-catalytic domain dimer and 1 or 10 mM NAD+ in a buffer solution containing 10 mM Bis-Tris-Propane pH 7.0 and 0.01% Triton X-100. The reaction has carried out for 16 h at room temperature.
Blot-based detection of mono- and poly-ADP-ribosylation
For dot blot experiments, we transferred 0.5 μl per spot of the sample solution to dry nitrocellulose membranes using Echo 650. All following steps were performed at room temperature. After drying of the spots, the membrane was blocked on a shaker for 10 min in 15 ml 5%(w/v) skimmed milk powder in TBS-T. The blocking solution was discarded, and the membrane was incubated on a shaker for 10 min with 15 ml of 0.1 μg/ml Nluc-eAf1521 or Nluc-ALC1 in 1%(w/v) skimmed milk powder in TBS-T. After discarding the Nluc solution, the membrane was rinsed with 15 ml TBS-T and incubated on a shaker with 15 ml TBS-T for 15 min. After a final rinsing with 15 ml TBS-T, the membrane was imaged using 500 μl of 1:1000 NanoGlo substrate (Promega, catalogue number: N1110) diluted in 10 mM sodium phosphate buffer pH 7.0.
For western blot, 10 μl samples were first run in SDS-PAGE (Mini-Protean TGX 4–20% gradient gel, BioRad). The proteins were then transferred to a nitrocellulose membrane (Trans-Blot Turbo, BioRad) using TransBlot semi dry system (BioRad). After transfer, the membranes were treated following the same procedure as described above for dot blot. Nluc-eAf1521 and Nluc-ALC1 were used at 0.1 μg/ml.
Testing Nluc-eAf1521 and Nluc-ALC1 sensitivity and selectivity
Dilution series of YFP-GAP, MARylated YFP-GAP, MBP-TNKS1 construct or auto-PARylated MBP-TNKS1 construct were blotted on a nitrocellulose membrane and detected with Nluc-eAf1521 or Nluc-ALC1 as described above.
To generate auto-PARylated TNKS1, purified TNKS1 construct (10 μM) was mixed with 1 mM NAD+ in 50 mM Bis-Tris-Propane pH 7.0, 0.01% Triton X-100, 0.5 mM TCEP. To generate a control containing TNKS1 without PAR, partial auto-PARylation that occurred during recombinant expression in E. coli was removed by mixing TNKS1 construct with 2 μM snake venom phosphodiesterase I from Crotalus adamanteus (Worthington Biochemical Corporation).
Modification test of YFP-GAP with 6-Biotin-17-NAD+
YFP-GAP or YFP-GAP(cysteine to alanine mutant) (10 μM) was mixed with 1 μM NAD+ or 6-Biotin-17-NAD+ (Biolog). Reactions were prepared in absence or presence of 0.5 μM PtxS1 and were incubated for 1 h at room temperature and then blotted to a dry nitrocellulose membrane (0.5 μl per spot of the sample solution to dry nitrocellulose membranes using Echo 650). The membrane was let dry and thereafter blocked on a shaker for 10 min in 15 ml blocking buffer (1% casein in TBS, BioRad). The blocking solution was discarded, and the membrane was incubated on a shaker for 1 hour with 15 ml of 1:5000 Streptavidin-HRP in blocking buffer. After discarding the Streptavidin-HRP solution, the membrane was rinsed with 15 ml TBS-T and incubated on a shaker with 15 ml TBS-T for 15 min. After a final rinsing with 15 ml TBS-T, the membrane was imaged using ECL solution (Advansta).
Modification test of YFP-GAP with 6-propargyladenine-NAD+ and addition Cy3 and Cy5 azides by CuAAC
YFP-GAP(6-propargyladenine-MAR) was prepared as described above for YFP-GAP using 6-propargyladenine-NAD+ instead of NAD+. To test the addition of Cy3 or Cy5 to YFP-GAP(6-propargyladenine-MAR) by CuAAC, reactions were prepared in 25 mM HEPES pH 7.5 by mixing 15 μM YFP-GAP(6-propargyladenine-MAR) or YFP-GAP(MAR) with 5 mM sodium ascorbate, 50 μM Cy3-azide or Cy5-azide and pre-mixed 300 μM CuSO4 and 600 μM L-histidine. Additionally, controls without protein were prepared. The reactions were let incubate for 3 hours at room temperature and afterwards blotted on a nitrocellulose membrane (5 μl per spot). The membrane was washed in 15 ml TBS-T for 30 min and imaged. Fluorescence imaging was done with an Azure 600 imaging system (Azure Biosystems) using Cy3 or Cy5 filter settings, respectively.
FRET measurement
The measurements were done as previously described (Sowa et al., 2020). Samples were prepared in 384-well black low-volume polypropylene plates (Fisherbrand). Briefly, the samples were excited at 410 nm and emission at 477 nm and 527 nm wavelengths were measured. The ratiometric FRET value (rFRET) was calculated by dividing the fluorescence intensity at 527 nm by the fluorescence intensity at 477 nm. The experiments with MARylated YFP-GAP were carried out in assay buffer (10 mM Bis-Tris-Propane pH 7.0, 3%(w/v) PEG20,000, 0.01%(v/v) Triton X-100 and 0.5 mM TCEP) in 10 μl volume per well. The assay buffer contained additionally 25 mM NaCl for RNF146, PARG and PARP15 macrodomain 2 and 150 mM NaCl for TARG1, XRCC1, ALC1 and PARP15 macrodomain 1. Measurements with PARylated YFP-GAP were done in 10 mM Tris pH 8.0, 150 mM NaCl, 0.01% Tween-20 in 20 μl, unless stated otherwise. In dose-response experiments where the decrease of the rFRET signal by addition of ADP-ribose or suramin was measured, a non-linear fit was used in GraphPad Prism 8 with the model “log(inhibitor) vs. response – Variable slope (four parameters)”. Fluorescence emission spectra were recorded using an excitation wavelength of 410 nm (20 nm bandwidth) with 1 µM CFP-MDO2 and 5 µM YFP-GAP(MAR).
BRET measurement
The reactions were performed in 384-well white OptiPlates (PerkinElmer). A reaction volume of 40 μl per well was used. Nluc-MDO2 (50 nM) was mixed with 1 μM MARylated YFP-GAP. The reaction was started by addition of 1:4000 NanoGlo substrate (Promega, catalogue number N1110). The reaction was incubated for 5 minutes and the emission spectra were measured using Tecan Spark multimode plate reader with luminescence readout and a settle time of 10 ms and integration time of 500 ms. The experiments were carried out in assay buffer (10 mM Bis–Tris-Propane pH 7.0, 3% (w/v) PEG20,000, 0.01%(v/v) Triton X-100 and 0.5 mM TCEP).
AlphaScreen
AlphaScreen technology was utilized to demonstrate the assay principle as described previously (Haikarainen et al., 2018). The reaction was performed in a 384-well flat-grey Alphaplate (PerkinElmer) in a total volume of 25 μl. The reaction consisted of 300 nM His-tagged Gαi(MAR) mixed with 300 nM biotinylated MDO2 in a buffer containing 25 mM HEPES pH 7.5, 100 mM NaCl and 0.1 mg/ml BSA. The plate was sealed and incubated for 80 min at room temperature with constant shaking at 300 rpm. Finally, 5 μg/ml nickel chelate acceptor and streptavidin donor beads were added to the plates followed by additional 3-h incubation. The plate contained blank wells (assay buffer and AlphaScreen beads only), control 1 (biotinylated MDO2, His-tagged Gαi) and control 2 (biotinylated MDO2, MARylated His-tagged Giα and 10 μM ADP-ribose). Luminescence was read using Tecan infinite M1000 Pro plate reader with AlphaScreen detection module.
Biolayer interferometry
Biolayer interferometry (BLI) assays were carried out in Octet Red system (ForteBio) in a buffer containing 10 mM Bis-Tris-Propane pH 7.0, 150 mM NaCl, 1% BSA, 0.02% Triton X-100 and at 30°C and shaking at 1500 rpm. 10 μg/ml YFP-GAP or MARylated YFP-GAP was loaded on Ni2+-NTA coated sensors, followed by a wash step in buffer. Association to MDO2 was measured by dipping the sensors in solution containing 0-2 μM MDO2 for 120 s, while for the dissociation step the sensors were dipped in buffer for 120 s.
For the ADP-ribose competition experiments, 10 μg/ml YFP-GAP or MARylated YFP-GAP were loaded onto Ni2+-NTA coated sensors. For association, sensors were dipped in 100 nM MDO2 mixed with a half-log dilution series of ADP-ribose (10 μM to 10 nM) for 120 s and then transferred to buffer for the dissociation step.
Cysteine-ADP-ribosyl hydrolysis assay
All samples were incubated for 24 hours at room temperature prior to blotting. CFP-fused proteins (1 μM) or SVP (0.01μM, 0.1 μM, 1 μM) were mixed with 10 μM MARylated YFP-GAP in 10 mM HEPES pH 7.5, 25 mM NaCl, 0.5 mM TCEP. After incubation, 0.5 μl per spot of the reaction mixtures were blotted next to 0.5 μl spots of 10 μM MARylated YFP-GAP. As control, non-MARylated YFP-GAP (0.5 μl, 10 μM) was blotted next to MARylated YFP-GAP (0.5 μl, 10 μM). Detection was done with Nluc-eAf1521.
Compound screening
For the screening, 40 nl of 10 mM compound stocks dissolved in DMSO from the FDA-approved drug library (Enzo Life Sciences) were transferred to 384-well black low-volume polypropylene plates (Fisherbrand). The sample mixture containing 1 μM CFP-SARS-CoV-2 nsp3 macrodomain and 5 μM MARylated YFP-GAP was prepared in assay buffer (10 mM Bis–Tris-Propane pH 7.0, 3%(w/v) PEG20,000, 0.01%(v/v) Triton X-100 and 0.5 mM TCEP) and 20 μl per well were dispensed using Mantis liquid dispenser (Formulatrix). The rFRET signal was determined after a 5-minute incubation time. The sample mixtures in presence or absence of 200 μM ADP-ribose were used as positive and negative controls, respectively.
Differential scanning fluorimetry
The SARS-CoV-2 nsp3 macrodomain without tags was diluted to 5 μM in 10 mM HEPES pH 7.5, 25 mM NaCl, 0.5 mM TCEP buffer and mixed with 5x SYPRO Orange. Samples were prepared with 10 μM, 50 μM, 100 μM or 1 mM of suramin. Samples in presence or absence of 1 mM ADP-ribose were used as controls. Samples were transferred to 96-well qPCR plates. Measurement was performed in a BioRad C1000 CFX96 thermal cycler. Data points for melting curves were recorded in 1 min intervals from 20–95°C, with the temperature increasing by 1°C/min. The analysis of the data was done in GraphPad Prism 8 using a nonlinear regression analysis (Boltzmann sigmoid equation) of normalized data.
Quantification and statistical analysis
Statistical details be found in the corresponding figure legends or STAR Methods sections. The mean was used with n representing the number of replicate samples measured. The standard deviation (SD) was determined from the measurements of replicate samples. An unpaired t-test was used for evaluation for statistical significance. The statistical significance is defined as ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001.
Acknowledgments
This work was funded by the Jane and Aatos Erkko and Sigrid Jusélius Foundations. The use of the facilities of the Biocenter Oulu Structural Biology core facility, member of Biocenter Finland, Instruct-ERIC Centre Finland, and FINStruct, as well as of Proteomics and Protein Analysis and Sequencing core facilities are gratefully acknowledged. We thank Dr. Yashwanth Ashok for cloning of the pNH-Nluc and Gαi vectors and Jere Hukkanen for assisting with ARH1-3 protein production.
Author contributions
L.L. conceived the research. S.T.S., A.G.-P., S.W., H.I.A., and M.M.M. cloned and produced the proteins used in the studies. S.T.S. and A.G.-P. developed the assay principles for MAR and PAR binders, respectively. S.T.S. developed the dot-blot detection method and carried out inhibitor screening. A.G.-P. carried out the BLI assay and S.W. the AlphaScreen assay. S.W. did KD measurements with the FRET assay. S.T.S., A.G.-P., and L.L. wrote the paper with contributions from all the authors.
Declaration of interests
S.T.S., A.G.P., and L.L. are inventors listed in a patent application related to the described methods, and these authors declare no additional interests. The remaining authors declare no competing interests.
Published: November 11, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.crmeth.2021.100121.
Supporting citations
The following references appear in the supplemental information: Schneider et al. (2012); Song et al. (2012); Thorsell et al. (2017).
Supplemental information
If only imidazole content is noted for the wash buffer or elution buffer, the other buffer components are identical to the respective lysis buffer used. We estimated the yield of protein after purification by IMAC per liter of culture. Wash volumes are shown in column volumes (CV).
Data and code availability
-
•
All data reported in this paper will be shared by the lead contact upon request.
-
•
This paper does not report original code.
-
•
Information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- Abplanalp J., Leutert M., Frugier E., Nowak K., Feurer R., Kato J., Kistemaker H.V.A., Filippov D.V., Moss J., Caflisch A., et al. Proteomic analyses identify ARH3 as a serine mono-ADP-ribosylhydrolase. Nat. Commun. 2017;8:2055. doi: 10.1038/s41467-017-02253-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abraham R., Hauer D., McPherson R.L., Utt A., Kirby I.T., Cohen M.S., Merits A., Leung A.K.L., Griffin D.E. ADP-ribosyl-binding and hydrolase activities of the alphavirus nsP3 macrodomain are critical for initiation of virus replication. Proc. Natl. Acad. Sci. U S A. 2018;115:E10457–E10466. doi: 10.1073/pnas.1812130115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahel I., Ahel D., Matsusaka T., Clark A.J., Pines J., Boulton S.J., West S.C. Poly(ADP-ribose)-binding zinc finger motifs in DNA repair/checkpoint proteins. Nature. 2008;451:81–85. doi: 10.1038/nature06420. [DOI] [PubMed] [Google Scholar]
- Albulescu I.C., van Hoolwerff M., Wolters L.A., Bottaro E., Nastruzzi C., Yang S.C., Tsay S.-C., Hwu J.R., Snijder E.J., van Hemert M.J. Suramin inhibits chikungunya virus replication through multiple mechanisms. Antivir. Res. 2015;121:39–46. doi: 10.1016/j.antiviral.2015.06.013. [DOI] [PubMed] [Google Scholar]
- Albulescu I.C., White-Scholten L., Tas A., Hoornweg T.E., Ferla S., Kovacikova K., Smit J.M., Brancale A., Snijder E.J., van Hemert M.J. Suramin inhibits chikungunya virus replication by interacting with virions and blocking the early steps of infection. Viruses. 2020;12:314. doi: 10.3390/v12030314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alhammad Y.M.O., Kashipathy M.M., Roy A., Gagné J.-P., McDonald P., Gao P., Nonfoux L., Battaile K.P., Johnson D.K., Holmstrom E.D., et al. The SARS-CoV-2 conserved macrodomain is a mono-ADP-ribosylhydrolase. J. Virol. 2021;95:e01969-20. doi: 10.1128/JVI.01969-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ando Y., Elkayam E., McPherson R.L., Dasovich M., Cheng S.-J., Voorneveld J., Filippov D.V., Ong S.-E., Joshua-Tor L., Leung A.K.L. ELTA: enzymatic labeling of terminal ADP-ribose. Mol. Cell. 2019;73:845–856.e5. doi: 10.1016/j.molcel.2018.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aravind L. The WWE domain: a common interaction module in protein ubiquitination and ADP ribosylation. Trends Biochem. Sci. 2001;26:273–275. doi: 10.1016/s0968-0004(01)01787-x. [DOI] [PubMed] [Google Scholar]
- Ashok Y., Miettinen M., Oliveira D.K.H.de, Tamirat M.Z., Näreoja K., Tiwari A., Hottiger M.O., Johnson M.S., Lehtiö L., Pulliainen A.T. Discovery of compounds inhibiting the ADP-ribosyltransferase activity of pertussis toxin. ACS Infect. Dis. 2020;6:588–602. doi: 10.1021/acsinfecdis.9b00412. [DOI] [PubMed] [Google Scholar]
- Bonfiglio J.J., Leidecker O., Dauben H., Longarini E.J., Colby T., San Segundo-Acosta P., Perez K.A., Matic I. An HPF1/PARP1-based chemical biology strategy for exploring ADP-ribosylation. Cell. 2020;183:1086–1102.e23. doi: 10.1016/j.cell.2020.09.055. [DOI] [PubMed] [Google Scholar]
- Boute N., Lowe P., Berger S., Malissard M., Robert A., Tesar M. NanoLuc luciferase—a multifunctional tool for high throughput antibody screening. Front. Pharmacol. 2016;7:27. doi: 10.3389/fphar.2016.00027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brosey C.A., Houl J.H., Katsonis P., Balapiti-Modarage L.P.F., Bommagani S., Arvai A., Moiani D., Bacolla A., Link T., Warden L.S., et al. Targeting SARS-CoV-2 Nsp3 macrodomain structure with insights from human poly(ADP-ribose) glycohydrolase (PARG) structures with inhibitors. Prog. Biophys. Mol. Biol. 2021;163:171–186. doi: 10.1016/j.pbiomolbio.2021.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantini F., Banci L., Altincekic N., Bains J.K., Dhamotharan K., Fuks C., Fürtig B., Gande S.L., Hargittay B., Hengesbach M., et al. 1H, 13C, and 15N backbone chemical shift assignments of the apo and the ADP-ribose bound forms of the macrodomain of SARS-CoV-2 non-structural protein 3b. Biomol. NMR Assign. 2020;14:339–346. doi: 10.1007/s12104-020-09973-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen M.S., Chang P. Insights into the biogenesis, function, and regulation of ADP-ribosylation. Nat. Chem. Biol. 2018;14:236–243. doi: 10.1038/nchembio.2568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Depaix A., Kowalska J. NAD analogs in aid of chemical biology and medicinal chemistry. Molecules. 2019;24:4187. doi: 10.3390/molecules24224187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durkacz B.W., Omidiji O., Gray D.A., Shall S. (ADP-ribose)n participates in DNA excision repair. Nature. 1980;283:593–596. doi: 10.1038/283593a0. [DOI] [PubMed] [Google Scholar]
- Ekblad T., Verheugd P., Lindgren A.E., Nyman T., Elofsson M., Schüler H. Identification of poly(ADP-ribose) polymerase macrodomain inhibitors using an AlphaScreen protocol. SLAS Discov. 2018;23:353–362. doi: 10.1177/2472555217750870. [DOI] [PubMed] [Google Scholar]
- Eskonen V., Tong-Ochoa N., Mattsson L., Miettinen M., Lastusaari M., Pulliainen A.T., Kopra K., Härmä H. Single-peptide TR-FRET detection platform for cysteine-specific post-translational modifications. Anal. Chem. 2020;92:13202–13210. doi: 10.1021/acs.analchem.0c02370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fehr A.R., Athmer J., Channappanavar R., Phillips J.M., Meyerholz D.K., Perlman S. The nsp3 macrodomain promotes virulence in mice with coronavirus-induced encephalitis. J. Virol. 2015;89:1523–1536. doi: 10.1128/JVI.02596-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fehr A.R., Channappanavar R., Jankevicius G., Fett C., Zhao J., Athmer J., Meyerholz D.K., Ahel I., Perlman S. The conserved coronavirus macrodomain promotes virulence and suppresses the innate immune response during severe acute respiratory syndrome coronavirus infection. MBio. 2016;7:e01721-16.. doi: 10.1128/mBio.01721-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fehr A.R., Jankevicius G., Ahel I., Perlman S. Viral macrodomains: unique mediators of viral replication and pathogenesis. Trends Microbiol. 2018;26:598–610. doi: 10.1016/j.tim.2017.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feijs K.L.H., Forst A.H., Verheugd P., Lüscher B. Macrodomain-containing proteins: regulating new intracellular functions of mono(ADP-ribosyl)ation. Nat. Rev. Mol. Cell Biol. 2013;14:443–451. doi: 10.1038/nrm3601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freissmuth M., Boehm S., Beindl W., Nickel P., Ijzerman A.P., Hohenegger M., Nanoff C. Suramin analogues as subtype-selective G protein inhibitors. Mol. Pharmacol. 1996;49:602–611. [PubMed] [Google Scholar]
- García-Saura A.G., Herzog L.K., Dantuma N.P., Schüler H. MacroGreen, a simple tool for detection of ADP-ribosylated proteins. Commun. Biol. 2021;4:919. doi: 10.1038/s42003-021-02439-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson B.A., Conrad L.B., Huang D., Kraus W.L. Generation and characterization of recombinant antibody-like ADP-ribose binding proteins. Biochemistry. 2017;56:6305–6316. doi: 10.1021/acs.biochem.7b00670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupte R., Liu Z., Kraus W.L. PARPs and ADP-ribosylation: recent advances linking molecular functions to biological outcomes. Genes Dev. 2017;31:101–126. doi: 10.1101/gad.291518.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haikarainen T., Lehtiö L. Proximal ADP-ribose hydrolysis in trypanosomatids is catalyzed by a macrodomain. Sci. Rep. 2016;6:24213. doi: 10.1038/srep24213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haikarainen T., Maksimainen M.M., Obaji E., Lehtiö L. Development of an inhibitor screening assay for mono-ADP-ribosyl hydrolyzing macrodomains using AlphaScreen technology. SLAS Discov. 2018;23:255–263. doi: 10.1177/2472555217737006. [DOI] [PubMed] [Google Scholar]
- Harrision D., Gravells P., Thompson R., Bryant H.E. Poly(ADP-Ribose) glycohydrolase (PARG) vs. Poly(ADP-Ribose) polymerase (PARP)—function in genome maintenance and relevance of inhibitors for anti-cancer therapy. Front. Mol. Biosci. 2020;7:191. doi: 10.3389/fmolb.2020.00191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henß L., Beck S., Weidner T., Biedenkopf N., Sliva K., Weber C., Becker S., Schnierle B.S. Suramin is a potent inhibitor of Chikungunya and Ebola virus cell entry. Virol. J. 2016;13:149. doi: 10.1186/s12985-016-0607-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrero-Yraola A., Bakhit S.M., Franke P., Weise C., Schweiger M., Jorcke D., Ziegler M. Regulation of glutamate dehydrogenase by reversible ADP-ribosylation in mitochondria. EMBO J. 2001;20:2404–2412. doi: 10.1093/emboj/20.10.2404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirsch B.M., Burgos E.S., Schramm V.L. Transition-state analysis of 2-O-acetyl-ADP-ribose hydrolysis by human macrodomain 1. ACS Chem. Biol. 2014;9:2255–2262. doi: 10.1021/cb500485w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho Y.-J., Wang Y.-M., Lu J., Wu T.-Y., Lin L.-I., Kuo S.-C., Lin C.-C. Suramin inhibits chikungunya virus entry and transmission. PLoS One. 2015;10:e0133511. doi: 10.1371/journal.pone.0133511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houl J.H., Ye Z., Brosey C.A., Balapiti-Modarage L.P.F., Namjoshi S., Bacolla A., Laverty D., Walker B.L., Pourfarjam Y., Warden L.S., et al. Selective small molecule PARG inhibitor causes replication fork stalling and cancer cell death. Nat. Commun. 2019;10:5654. doi: 10.1038/s41467-019-13508-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S.-M.A., Mishina Y.M., Liu S., Cheung A., Stegmeier F., Michaud G.A., Charlat O., Wiellette E., Zhang Y., Wiessner S., et al. Tankyrase inhibition stabilizes axin and antagonizes Wnt signalling. Nature. 2009;461:614–620. doi: 10.1038/nature08356. [DOI] [PubMed] [Google Scholar]
- James D.I., Smith K.M., Jordan A.M., Fairweather E.E., Griffiths L.A., Hamilton N.S., Hitchin J.R., Hutton C.P., Jones S., Kelly P., et al. First-in-class chemical probes against poly(ADP-ribose) glycohydrolase (PARG) inhibit DNA repair with differential pharmacology to olaparib. ACS Chem. Biol. 2016;11:3179–3190. doi: 10.1021/acschembio.6b00609. [DOI] [PubMed] [Google Scholar]
- Jeong J.-Y., Yim H.-S., Ryu J.-Y., Lee H.S., Lee J.-H., Seen D.-S., Kang S.G. One-step sequence- and ligation-independent cloning as a rapid and versatile cloning method for functional genomics studies. Appl. Environ. Microbiol. 2012;78:5440–5443. doi: 10.1128/AEM.00844-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaminski T., Gunnarsson A., Geschwindner S. Harnessing the versatility of optical biosensors for target-based small-molecule drug discovery. ACS Sens. 2017;2:10–15. doi: 10.1021/acssensors.6b00735. [DOI] [PubMed] [Google Scholar]
- Kashida H., Kurihara A., Kawai H., Asanuma H. Orientation-dependent FRET system reveals differences in structures and flexibilities of nicked and gapped DNA duplexes. Nucleic Acids Res. 2017;45:e105. doi: 10.1093/nar/gkx200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katada T. The inhibitory G protein G(i) identified as pertussis toxin-catalyzed ADP-ribosylation. Biol. Pharm. Bull. 2012;35:2103–2111. doi: 10.1248/bpb.b212024. [DOI] [PubMed] [Google Scholar]
- Kirby I.T., Kojic A., Arnold M.R., Thorsell A.-G., Karlberg T., Vermehren-Schmaedick A., Sreenivasan R., Schultz C., Schüler H., Cohen M.S. A potent and selective PARP11 inhibitor suggests coupling between cellular localization and catalytic activity. Cell Chem Biol. 2018;25:1547–1553.e12. doi: 10.1016/j.chembiol.2018.09.011. [DOI] [PubMed] [Google Scholar]
- Kozlowski M., Corujo D., Hothorn M., Guberovic I., Mandemaker I.K., Blessing C., Sporn J., Gutierrez-Triana A., Smith R., Portmann T., et al. MacroH2A histone variants limit chromatin plasticity through two distinct mechanisms. EMBO Rep. 2018;19:e44445. doi: 10.15252/embr.201744445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo S.-C., Wang Y.-M., Ho Y.-J., Chang T.-Y., Lai Z.-Z., Tsui P.-Y., Wu T.-Y., Lin C.-C. Suramin treatment reduces chikungunya pathogenesis in mice. Antivir. Res. 2016;134:89–96. doi: 10.1016/j.antiviral.2016.07.025. [DOI] [PubMed] [Google Scholar]
- Kustatscher G., Hothorn M., Pugieux C., Scheffzek K., Ladurner A.G. Splicing regulates NAD metabolite binding to histone macroH2A. Nat. Struct. Mol. Biol. 2005;12:624–625. doi: 10.1038/nsmb956. [DOI] [PubMed] [Google Scholar]
- Leung A.K.L., McPherson R.L., Griffin D.E. Macrodomain ADP-ribosylhydrolase and the pathogenesis of infectious diseases. PLoS Pathog. 2018;14:e1006864. doi: 10.1371/journal.ppat.1006864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M., Lu L.-Y., Yang C.-Y., Wang S., Yu X. The FHA and BRCT domains recognize ADP-ribosylation during DNA damage response. Genes Dev. 2013;27:1752–1768. doi: 10.1101/gad.226357.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., Xie R., Yu L.L., Chen S.-H., Yang X., Singh A.K., Li H., Wu C., Yu X. AI26 inhibits the ADP-ribosylhydrolase ARH3 and suppresses DNA damage repair. J. Biol. Chem. 2020;295:13838–13849. doi: 10.1074/jbc.RA120.012801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lotze J., Reinhardt U., Seitz O., Beck-Sickinger A.G. Peptide-tags for site-specific protein labelling in vitro and in vivo. Mol. Biosyst. 2016;12:1731–1745. doi: 10.1039/c6mb00023a. [DOI] [PubMed] [Google Scholar]
- Lu J.-W., Hsieh P.-S., Lin C.-C., Hu M.-K., Huang S.-M., Wang Y.-M., Liang C.-Y., Gong Z., Ho Y.-J. Synergistic effects of combination treatment using EGCG and suramin against the chikungunya virus. Biochem. Biophys. Res. Commun. 2017;491:595–602. doi: 10.1016/j.bbrc.2017.07.157. [DOI] [PubMed] [Google Scholar]
- Lüscher B., Bütepage M., Eckei L., Krieg S., Verheugd P., Shilton B.H. ADP-ribosylation, a multifaceted posttranslational modification involved in the control of cell physiology in health and disease. Chem. Rev. 2018;118:1092–1136. doi: 10.1021/acs.chemrev.7b00122. [DOI] [PubMed] [Google Scholar]
- McPherson R.L., Abraham R., Sreekumar E., Ong S.-E., Cheng S.-J., Baxter V.K., Kistemaker H.A.V., Filippov D.V., Griffin D.E., Leung A.K.L. ADP-ribosylhydrolase activity of Chikungunya virus macrodomain is critical for virus replication and virulence. Proc. Natl. Acad. Sci. U S A. 2017;114:1666–1671. doi: 10.1073/pnas.1621485114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michalska K., Kim Y., Jedrzejczak R., Maltseva N.I., Stols L., Endres M., Joachimiak A. Crystal structures of SARS-CoV-2 ADP-ribose phosphatase: from the apo form to ligand complexes. IUCrJ. 2020;7:814–824. doi: 10.1107/S2052252520009653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuvonen M., Ahola T. Differential activities of cellular and viral macro domain proteins in binding of ADP-ribose metabolites. J. Mol. Biol. 2009;385:212–225. doi: 10.1016/j.jmb.2008.10.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni X., Schröder M., Olieric V., Sharpe M.E., Hernandez-Olmos V., Proschak E., Merk D., Knapp S., Chaikuad A. Structural insights into plasticity and discovery of remdesivir metabolite GS-441524 binding in SARS-CoV-2 macrodomain. ACS Med. Chem. Lett. 2021;12:603–609. doi: 10.1021/acsmedchemlett.0c00684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowak K., Rosenthal F., Karlberg T., Bütepage M., Thorsell A.-G., Dreier B., Grossmann J., Sobek J., Imhof R., Lüscher B., et al. Engineering Af1521 improves ADP-ribose binding and identification of ADP-ribosylated proteins. Nat. Commun. 2020;11:5199. doi: 10.1038/s41467-020-18981-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obaji E., Maksimainen M.M., Galera-Prat A., Lehtiö L. Activation of PARP2/ARTD2 by DNA damage induces conformational changes relieving enzyme autoinhibition. Nat. Commun. 2021;12:3479. doi: 10.1038/s41467-021-23800-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Sullivan J., Tedim Ferreira M., Gagné J.-P., Sharma A.K., Hendzel M.J., Masson J.-Y., Poirier G.G. Emerging roles of eraser enzymes in the dynamic control of protein ADP-ribosylation. Nat. Commun. 2019;10:1182. doi: 10.1038/s41467-019-08859-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overacker R.D., Plitzko B., Loesgen S. Biolayer interferometry provides a robust method for detecting DNA binding small molecules in microbial extracts. Anal Bioanal. Chem. 2021;413:1159–1171. doi: 10.1007/s00216-020-03079-5. [DOI] [PubMed] [Google Scholar]
- Palazzo L., Ahel I. PARPs in genome stability and signal transduction: implications for cancer therapy. Biochem. Soc. Trans. 2018;46:1681–1695. doi: 10.1042/BST20180418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peltomaa R., Glahn-Martínez B., Benito-Peña E., Moreno-Bondi M.C. Optical biosensors for label-free detection of small molecules. Sensors. 2018;18:4126. doi: 10.3390/s18124126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prokhorova E., Agnew T., Wondisford A.R., Tellier M., Kaminski N., Beijer D., Holder J., Groslambert J., Suskiewicz M.J., Zhu K., et al. Unrestrained poly-ADP-ribosylation provides insights into chromatin regulation and human disease. Mol. Cell. 2021;81:2640–2655.e8. doi: 10.1016/j.molcel.2021.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rack J.G.M., Perina D., Ahel I. Macrodomains: structure, function, evolution, and catalytic activities. Annu. Rev. Biochem. 2016;85:431–454. doi: 10.1146/annurev-biochem-060815-014935. [DOI] [PubMed] [Google Scholar]
- Rack J.G.M., Ariza A., Drown B.S., Henfrey C., Bartlett E., Shirai T., Hergenrother P.J., Ahel I. (ADP-ribosyl)hydrolases: structural basis for differential substrate recognition and inhibition. Cell Chem. Biol. 2018;25:1533–1546.e12. doi: 10.1016/j.chembiol.2018.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rack J.G.M., Palazzo L., Ahel I. (ADP-ribosyl)hydrolases: structure, function, and biology. Genes Dev. 2020;34:263–284. doi: 10.1101/gad.334631.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rack J.G.M., Zorzini V., Zhu Z., Schuller M., Ahel D., Ahel I. Viral macrodomains: a structural and evolutionary assessment of the pharmacological potential. Open Biol. 2020;10:200237. doi: 10.1098/rsob.200237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russo L.C., Tomasin R., Matos I.A., Manucci A.C., Sowa S.T., Dale K., Caldecott K.W., Lehtiö L., Schechtman D., Meotti F.C., et al. The SARS-CoV-2 Nsp3 macrodomain reverses PARP9/DTX3L-dependent ADP-ribosylation induced by interferon signaling. J. Biol. Chem. 2021;297:101041. doi: 10.1016/j.jbc.2021.101041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salgado-Benvindo C., Thaler M., Tas A., Ogando N.S., Bredenbeek P.J., Ninaber D.K., Wang Y., Hiemstra P.S., Snijder E.J., van Hemert M.J. Suramin inhibits SARS-CoV-2 infection in cell culture by interfering with early steps of the replication cycle. Antimicrob. Agents Chemother. 2020;64:e00900-20. doi: 10.1128/AAC.00900-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider C.A., Rasband W.S., Eliceiri K.W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuller M., Riedel K., Gibbs-Seymour I., Uth K., Sieg C., Gehring A.P., Ahel I., Bracher F., Kessler B.M., Elkins J.M., et al. Discovery of a selective allosteric inhibitor targeting macrodomain 2 of polyadenosine-diphosphate-ribose polymerase 14. ACS Chem. Biol. 2017;12:2866–2874. doi: 10.1021/acschembio.7b00445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuller M., Correy G.J., Gahbauer S., Fearon D., Wu T., Díaz R.E., Young I.D., Carvalho Martins L., Smith D.H., Schulze-Gahmen U., et al. Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking. Sci. Adv. 2021;7:eabf8711. doi: 10.1101/2020.11.24.393405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu J.F., Martins D.O.S., McPhillie M.J., Roberts G.C., Zothner C., Merits A., Harris M., Jardim A.C.G. Is the ADP ribose site of the Chikungunya virus NSP3 Macro domain a target for antiviral approaches? Acta Trop. 2020;207:105490. doi: 10.1016/j.actatropica.2020.105490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh H.R., Nardozza A.P., Möller I.R., Knobloch G., Kistemaker H.A.V., Hassler M., Harrer N., Blessing C., Eustermann S., Kotthoff C., et al. A poly-ADP-ribose trigger releases the auto-inhibition of a chromatin remodeling oncogene. Mol. Cell. 2017;68:860–871.e7. doi: 10.1016/j.molcel.2017.11.019. [DOI] [PubMed] [Google Scholar]
- Song Y., Rodgers V.G.J., Schultz J.S., Liao J. Protein interaction affinity determination by quantitative FRET technology. Biotechnol. Bioeng. 2012;109:2875–2883. doi: 10.1002/bit.24564. [DOI] [PubMed] [Google Scholar]
- Sowa S.T., Vela-Rodríguez C., Galera-Prat A., Cázares-Olivera M., Prunskaite-Hyyryläinen R., Ignatev A., Lehtiö L. A FRET-based high-throughput screening platform for the discovery of chemical probes targeting the scaffolding functions of human tankyrases. Sci. Rep. 2020;10:12357. doi: 10.1038/s41598-020-69229-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanuma S., Endo H. Purification and characterization of an (ADP-ribose)n glycohydrolase from human erythrocytes. Eur. J. Biochem. 1990;191:57–63. doi: 10.1111/j.1432-1033.1990.tb19093.x. [DOI] [PubMed] [Google Scholar]
- Thorsell A.-G., Ekblad T., Karlberg T., Löw M., Pinto A.F., Trésaugues L., Moche M., Cohen M.S., Schüler H. Structural basis for potency and promiscuity in poly(ADP-ribose) polymerase (PARP) and tankyrase inhibitors. J. Med. Chem. 2017;60:1262–1271. doi: 10.1021/acs.jmedchem.6b00990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torrente M.P., Castellano L.M., Shorter J. Suramin inhibits Hsp104 ATPase and disaggregase activity. PLoS One. 2014;9:e110115. doi: 10.1371/journal.pone.0110115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapp J., Meier R., Hongwiset D., Kassack M.U., Sippl W., Jung M. Structure-activity studies on suramin analogues as inhibitors of NAD+-dependent histone deacetylases (sirtuins) ChemMedChem. 2007;2:1419–1431. doi: 10.1002/cmdc.200700003. [DOI] [PubMed] [Google Scholar]
- Venkannagari H., Fallarero A., Feijs K.L.H., Lüscher B., Lehtiö L. Activity-based assay for human mono-ADP-ribosyltransferases ARTD7/PARP15 and ARTD10/PARP10 aimed at screening and profiling inhibitors. Eur. J. Pharm. Sci. 2013;49:148–156. doi: 10.1016/j.ejps.2013.02.012. [DOI] [PubMed] [Google Scholar]
- Venkannagari H., Verheugd P., Koivunen J., Haikarainen T., Obaji E., Ashok Y., Narwal M., Pihlajaniemi T., Lüscher B., Lehtiö L. Small-molecule chemical probe rescues cells from mono-ADP-ribosyltransferase ARTD10/PARP10-induced apoptosis and sensitizes cancer cells to DNA damage. Cell Chem. Biol. 2016;23:1251–1260. doi: 10.1016/j.chembiol.2016.08.012. [DOI] [PubMed] [Google Scholar]
- Virdi R.S., Bavisotto R.V., Hopper N.C., Vuksanovic N., Melkonian T.R., Silvaggi N.R., Frick D.N. Discovery of drug-like ligands for the Mac1 domain of SARS-CoV-2 Nsp3. SLAS Discov. 2020;25:1162–1170. doi: 10.1177/2472555220960428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voorneveld J., Rack J.G.M., van Gijlswijk L., Meeuwenoord N.J., Liu Q., Overkleeft H.S., van der Marel G.A., Ahel I., Filippov D.V. Molecular tools for the study of ADP-ribosylation: a unified and versatile method to synthesise native mono-ADP-ribosylated peptides. Chemistry. 2021;27:10621–10627. doi: 10.1002/chem.202100337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Michaud G.A., Cheng Z., Zhang Y., Hinds T.R., Fan E., Cong F., Xu W. Recognition of the iso-ADP-ribose moiety in poly(ADP-ribose) by WWE domains suggests a general mechanism for poly(ADP-ribosyl)ation-dependent ubiquitination. Genes Dev. 2012;26:235–240. doi: 10.1101/gad.182618.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Grosskurth S.E., Cheung T., Petteruti P., Zhang J., Wang X., Wang W., Gharahdaghi F., Wu J., Su N., et al. Pharmacological inhibition of PARP6 triggers multipolar spindle formation and elicits therapeutic effects in breast cancer. Cancer Res. 2018;78:6691–6702. doi: 10.1158/0008-5472.CAN-18-1362. [DOI] [PubMed] [Google Scholar]
- Wazir S., Maksimainen M.M., Alanen H.I., Galera-Prat A., Lehtiö L. Activity-based screening assay for mono-ADP-ribosylhydrolases. SLAS Discov. 2021;26:67–76. doi: 10.1177/2472555220928911. [DOI] [PubMed] [Google Scholar]
- Wiedemar N., Hauser D.A., Mäser P. 100 years of suramin. Antimicrob. Agents Chemother. 2020;64:e01168-19. doi: 10.1128/AAC.01168-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin W., Luan X., Li Z., Zhou Z., Wang Q., Gao M., Wang X., Zhou F., Shi J., You E., et al. Structural basis for inhibition of the SARS-CoV-2 RNA polymerase by suramin. Nat. Struct. Mol. Biol. 2021;28:319–325. doi: 10.1038/s41594-021-00570-0. [DOI] [PubMed] [Google Scholar]
- Zhang J.-H., Chung T.D.Y., Oldenburg K.R. A simple statistical parameter for use in evaluation and validation of high throughput screening assays. J. Biomol. Screen. 1999;4:67–73. doi: 10.1177/108705719900400206. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
If only imidazole content is noted for the wash buffer or elution buffer, the other buffer components are identical to the respective lysis buffer used. We estimated the yield of protein after purification by IMAC per liter of culture. Wash volumes are shown in column volumes (CV).
Data Availability Statement
-
•
All data reported in this paper will be shared by the lead contact upon request.
-
•
This paper does not report original code.
-
•
Information required to reanalyze the data reported in this paper is available from the lead contact upon request.