Summary
Mouse embryonic stem cells (ESCs) show cell-to-cell heterogeneity. A small number of two-cell-like cells (2CLCs) marked by endogenous retrovirus activation emerge spontaneously. The 2CLCs are unstable and they are prone to transiting back to the pluripotent state without extrinsic stimulus. To understand how this bidirectional transition takes place, we performed single-cell RNA sequencing on isolated 2CLCs that underwent 2C-like state exit and re-entry, and revealed a step-by-step transitional process between 2C-like and pluripotent states. Mechanistically, we found that cell cycle played an important role in mediating these transitions by regulating assembly of the nucleolus and peri-nucleolar heterochromatin to influence 2C gene Dux expression. Collectively, our findings provide a roadmap of the 2C-like state entry and exit in ESCs and also a causal role of the cell cycle in promoting these transitions.
Keywords: 2C-like cells, single-cell RNA-seq, nucleolus, peri-nucleolar heterochromatin
Highlights
-
•
The entry to and exit from the 2C-like state showed a step-by-step roadmap
-
•
Cell cycle participates in mediating dynamic transitions between ESCs and 2CLCs
-
•
G1/S phase arrest facilitates the Dux locus escape from heterochromatin
-
•
Nucleolus-heterochromatin remodeling is involved in 2C activation
In this article, Jin Zhang and colleagues show that cell-cycle variations distinguish 2CLCs, ESCs, and the intermediate states using single-cell RNA-seq. They found that cell-cycle-associated remodeling of nucleoli and peri-nucleolar heterochromatin was involved in the regulation of the 2C program. They provide novel advances in the fundamental field of 2C activation and stem cell fate decision.
Introduction
Mouse embryonic stem cells (mESCs) in culture exhibit cell-to-cell heterogeneity, with about 1% of them spontaneously emerging as two-cell-like cells (2CLCs) (Macfarlan et al., 2012; Zalzman et al., 2010). Unlike ESCs, 2CLCs display a signature of activated two-cell-specific genes and repeats such as Zscan4, Dux, and MERVL (Akiyama et al., 2015; De Iaco et al., 2017; Falco et al., 2007; Hendrickson et al., 2017; Whiddon et al., 2017). Previous studies on 2C-like state emergence from mESCs focused on mechanisms of overcoming genetic and epigenetic barriers. For example, expression of a pioneer transcription factor, Dux, can open up chromatin and activate endogenous retroviruses (ERVs) (De Iaco et al., 2017; Hendrickson et al., 2017; Yang et al., 2020). Depletion of repressive epigenetic factors such as Tet and Setdb1 can trigger this transition by releasing the epigenetic repression of 2C genes (Guallar et al., 2018; Wu et al., 2020). However, the 2C-like state is unstable and likely to transit back to the original pluripotent state, and relatively little is known about the mechanisms of the exit process (Macfarlan et al., 2012; Rodriguez-Terrones et al., 2018). A recent study shows that pluripotent genes are activated in two waves during the 2CLC-to-ESC transition with induced Dux overexpression, which is a system distinct from the spontaneous 2C-like state entry, and that Dux mRNA decay plays an important role in the 2C-like state exit (Fu et al., 2019; Xudong Fu and Zhang, 2020). However, the complete picture and molecular mechanisms involved in spontaneous 2C-like state entry and exit have yet to be fully elucidated.
The cell cycle is an ordered series of events involved in cell growth and division, during which dynamic genetic and epigenetic remodeling takes place in the nucleolus (Nagano et al., 2017; Padeken and Heun, 2014; Probst et al., 2009; Visintin and Amon, 2000; Zhang et al., 2019). It is tightly coupled to cell fate decisions such as differentiation and reprogramming, etc. (Gonzales et al., 2015; Gruenheit et al., 2018; Nemeth and Grummt, 2018). Former studies have the observation that 2C-like cells exhibit an extended G2/M phase (Atashpaz et al., 2020; Eckersley-Maslin et al., 2016). However, the detailed mechanistic link between 2C entry/exit and cell cycle is very poorly explored. Furthermore, nucleolus organization and heterochromatin assembly and disassembly exhibit different signatures during the cell cycle and are connected with cell fate decision (Boisvert et al., 2007; Borsos and Torres-Padilla, 2016; Guetg and Santoro, 2012; Müller and Almouzni, 2017).
In this study, we delineate the transcriptional dynamic shifts from the 2C-like to the pluripotent state through intermediate transitional states that exhibit different cell-cycle phase signatures, and 2CLCs show an enrichment of G1 and G2/M phases in comparison to S phase. By arresting mESCs at different cell-cycle phases, we demonstrate that blockage at G1/early S phase is sufficient for the activation of ERVs and the 2C transcriptional program. Mechanistically, we elucidate that cell-cycle-dependent nucleolus architecture and its associated peri-nucleolar heterochromatin remodeling are essential to orchestrate 2C-like state entry and exit.
Results
Reconstruction of the 2CLC-to-ESC transition intermediate states at single-cell resolution
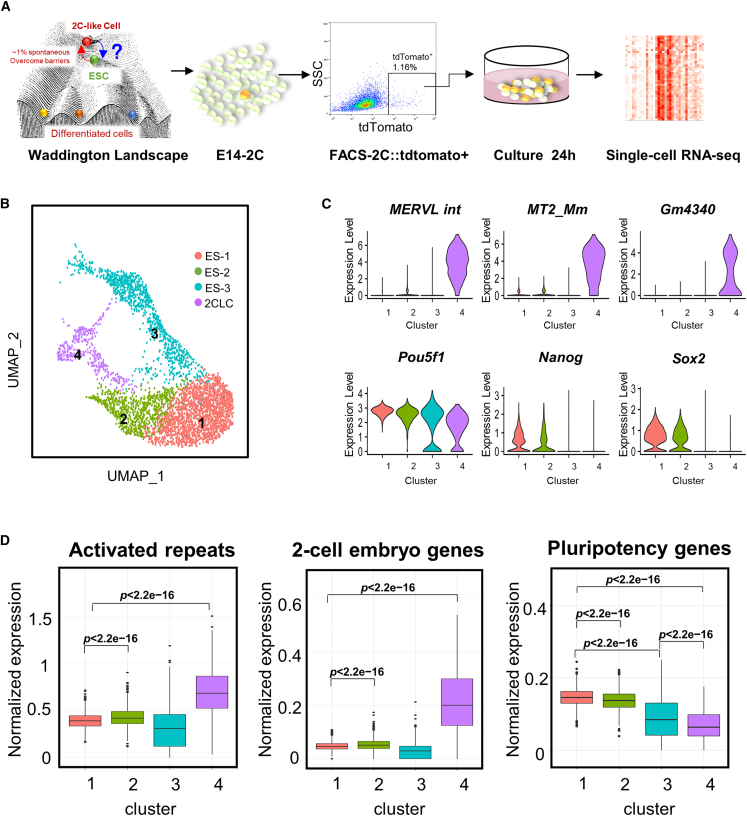
To characterize the roadmap underlying the emergence and exit of the 2C-like state, we used 2C::tdTomato to label the expression of MERVL and applied fluorescence-activated cell sorting (FACS) to sort the 2CLCs from mESCs, and subsequently released 2CLCs to exit the 2C-like state and to return to mESCs for 24 h. Then single-cell RNA sequencing (scRNA-seq) was performed to identify the dynamic cell states and cell fate determinants during the 2CLC-to-ESC transition process (Figure 1A).
Figure 1.
Single-cell RNA-seq uncovers transitional states between 2CLCs and ESCs in a sort-and-release system
(A) A schematic showing a sort-and-release system to dissect 2CLC-to-ESC transition dynamics in this study.
(B) UMAP plot of sorted 2C::TdTomato-positive cells by single-cell RNA-seq. Colors indicate different clusters.
(C) Violin plots showing the relative expression patterns of MERVL-int, MT2_mm, and Gm4340 (2C marker genes), as well as Pou5f1, Nanog, and Sox2 (pluripotency marker genes) of each cluster.
(D) Boxplots showing average expression of activated repeats (n = 301 repeats), two-cell embryo-specific genes (n = 75 genes), and pluripotency genes (n = 135 genes) of each cluster. The p values were calculated by two-tailed Wilcoxon test.
Uniform manifold approximation and projection (UMAP) showed the separated clusters of 3,839 sorted cells (Figure 1B). Clustering analysis indicated different ERV and pluripotency gene expression of the 2CLC and ESC subpopulations (Figure 1C). 2CLCs (cluster 4) displayed high MERVL int, MT2_mm, and Gm4340 long-terminal-repeat expression with low pluripotency, while ESCs separated into three subpopulations (clusters 1–3), and they all showed high expression of pluripotency marker genes such as Pou5f1, Nanog, and Sox2. Moreover, ES-3 (cluster 3) exhibited upregulation of MERVL int, MT2_mm, and Gm4340 (Figure 1C). We also demonstrated that the unsorted ESCs revealed less than 1% of 2CLCs with single-cell RNA-seq (Figure S1A and S1B), which confirmed this rare population in regular ESC culture. To distinguish whether the appearance of the three different ESC clusters was due to unique transitional states arising from sorted 2CLCs exiting the 2C-like state, or due to the cellular heterogeneity of unsorted ESC culture itself, we compared the UMAP clustering of the sorted-and-released 2CLCs and unsorted mESCs side by side (Figure S1A). We found that the sorted 2C-like state-exiting cells showed a significantly increased proportion of 2CLCs (cluster 4) and ES-3 (cluster 3), while the unsorted ESCs showed that nearly 80% of the cells were typical ESCs (Figure S1B).
To further dissect the transcriptional signatures of 2CLCs, ESCs, and their intermediate transitional states, we analyzed the expression pattern of 2C-specific elements, including activated repeats (Fadloun et al., 2013) and two-cell embryo genes (Falco et al., 2007; Hamatani et al., 2004) (Table S1). In addition to the obvious increase in two-cell-specific elements and decrease in pluripotency genes associated with the 2C-like state (cluster 4), we also detected that ES-3 showed significant downregulation of pluripotency genes (Table S1), while ES-2 (cluster 2) displayed significant upregulation of 2C-specific elements compared with ES-1 (cluster 1) (Figure 1D). We also observed that ES-2 and 2CLCs both showed significantly reduced levels of formative markers (Otx2, Dido1, and c-Myc) (Table S1, Figure S1C) (Smith, 2017), indicating that with pluripotency markers downregulated, ESCs could also be disposed toward a primed state other than the 2C state. Together, these data suggest that the sort-and-release system provides a model to study cellular dynamics after 2C exit and identifies transitional states during the 2CLC-ESC transition process.
Identification of a transcriptional roadmap during the 2CLC-to-ESC transition
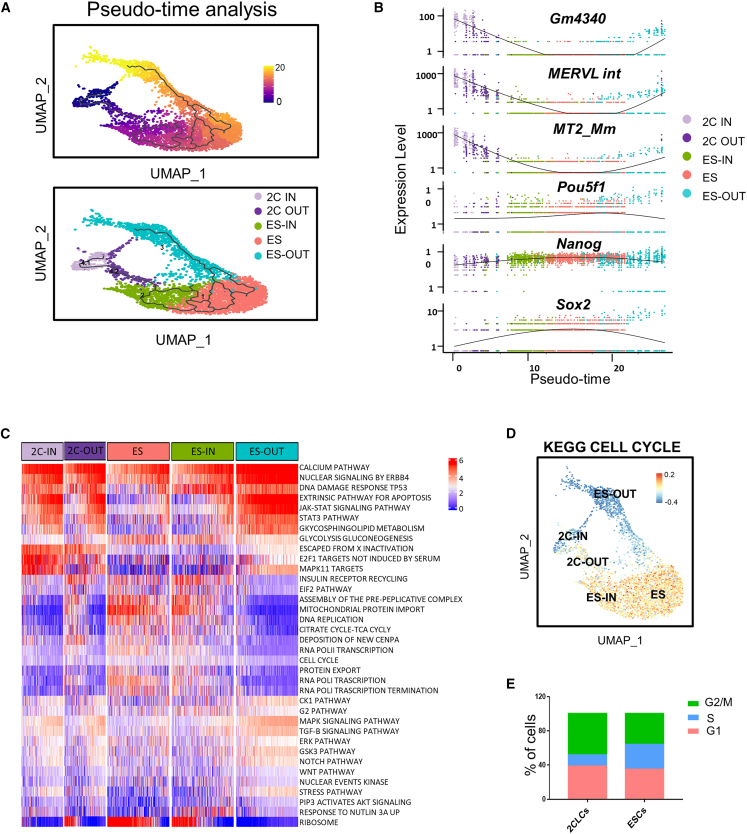
To clarify the transcriptional roadmap with the clusters identified above, we carried out pseudo-time trajectory analysis (Qiu et al., 2017; Trapnell et al., 2014) and further identified two subclusters in the 2CLCs. Cluster 4.1 at the beginning of the trajectory was named as 2C-IN cells, followed by 2C-OUT (cluster 4.2), ES-IN (cluster 2), ES (cluster 1), and ES-OUT (cluster 3) along the trajectory (Figure 2A).
Figure 2.
Identification of a transcriptional roadmap for 2C-like state exit and entry
(A) Pseudo-time analysis on UMAP of cells. Top: colors indicate trajectory order; each cell is colored according to its position in the pseudo-time. Bottom: colors indicate different clusters identified in Figure 1. The 2CLCs are further divided into two subclusters.
(B) Scatterplots showing the dynamics of expression of representative genes and repeats in each cell cluster along a pseudo-time axis.
(C) A gene set variation analysis (GSVA) heatmap showing the differentially enriched pathways in the 2C-IN, 2C-OUT, ES-IN, ES, and ES-OUT cell populations.
(D) Gene set variation analysis with KEGG cell-cycle pathway is shown on a UMAP plot. The intensity of colors indicates the enrichment score of the pathway.
(E) The cell-cycle distribution of 2CLCs and ESCs at the single-cell level.
To further delineate the relationship among these clusters, we used scatterplots to show the pseudo-time expression of MERVL int, MT2_mm, Gm4340, Pou5f1, Nanog, and Sox2, the representative 2C-specific and pluripotency genes, to display the 2CLC-to-ESC transition (Figure 2B). Gene expression patterns revealed that 2C and ERV gene expression first decreased from the 2C-IN to the 2C-OUT state, and then continued to decrease through the ES-IN and ES states, with a concomitant increase of pluripotent marker gene expression (Figure 2B). Importantly, the 2C genes and ERVs slightly increased with an obvious decrease in pluripotency genes in the ES-OUT stage compared with ES, consistent with previous research (Rodriguez-Terrones et al., 2018). This suggested that ES-OUT is an intermediate state of ESCs primed for 2C emergence.
To further characterize the global gene expression features of these clusters, the enriched pathways of differentially expressed genes (DEGs) were analyzed. We identified that the 2C-specific gene expression pattern in ES-OUT resembled more like 2CLCs, in terms of 2C gene expression (Figure S2A), which also indicated that ES-OUT is at an intermediate state of the ESC-to-2CLC transition, but closer to ESCs. In addition, 2C-IN showed higher expression of 2C-specific genes and lower expression of ribosome-related genes (Rpl10, Rpl27, and Rps12) compared with 2C-OUT (Figures 2C and S2A), indicating that 2C-OUT is also an intermediate state of the ESC-to-2CLC transition, and it may be primed for active translation to enter the fast-proliferating ES state.
Finally, we utilized gene set variation analysis (GSVA), which could provide greater sensitivity for finding gene expression changes of small magnitude that operate coordinately in specific sets of functionally related genes (Figures 2C and S2B) (Hänzelmann et al., 2013). ES and ES-IN showed enrichment in DNA replication, ribosomes, and formation of the ATP pathway compared with 2C-IN, 2C-OUT, and ES-OUT (Figures 2C and S2B), suggesting that the 2C-like and ES-OUT states are closer to a quiescent state, whereas ESCs are in a metabolically active state (Rodriguez-Terrones et al., 2020). In addition to the indicated gene expression differences and the connected pathway variations described above, our expression data also revealed a cell-cycle distinction in different clusters by GSVA (Figure 2D). We next analyzed the cell-cycle proportion at G1, S, and G2/M phase in each cluster with the single-cell RNA-seq data, and observed a significantly higher proportion of G2/M phase in 2CLCs compared with ESCs, as reported (Figure 2E) (Atashpaz et al., 2020; Eckersley-Maslin et al., 2016).
Cell-cycle variations distinguish 2CLC, ESCs, and the intermediate states
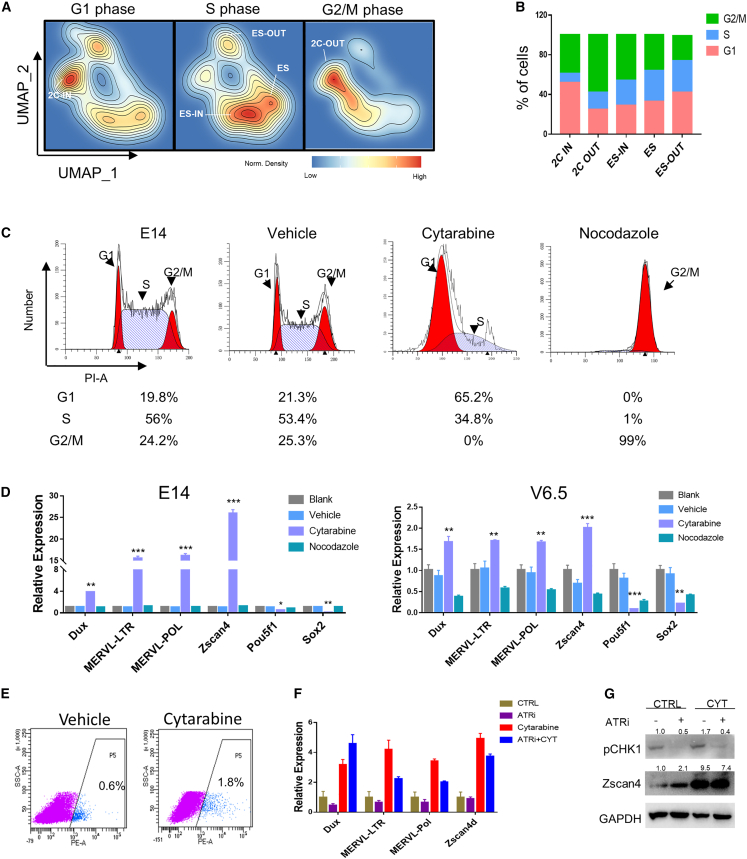
When we divided the 2CLCs into 2C-IN and 2C-OUT, we found that 2C-IN showed an enrichment of G1 phase, while 2C-OUT showed an enrichment of G2/M phase, and ES-IN, ES, and ES-OUT subclusters showed reduced G2/M phase by different degrees (Figures 3A and 3B). Moreover, the ES-OUT showed cell cycles similar to those of 2C-IN, with increased cell number in the G1-phase proportion, while the ES exhibited a large proportion in S phase and small proportion in G1 phase, which was in agreement with the fast self-renewal ability of ESCs (Figures 3A and 3B) (Liu et al., 2019). Re-analysis of single-cell RNA-seq data from a recent study on transitional 2CLCs to ESCs also showed an increased G1 phase in both 2C-like cells (D12C+) and transitional-state cells (D12C−) (Figures 3A and 3B) (Fu et al., 2019). We also sorted 2C+ and 2C− cells with a MERVL-2C: tdTomato reporter ESC line, and found that the 2C+ bulk population displayed a larger proportion of G2/M phase compared with ESCs or the 2C− population (Figure S3C), consistent with the single-cell RNA-seq results that the 2CLCs showed an enrichment of only G2/M phase, compared with ESCs (Figure S2E). Together, these data demonstrate that the enrichment of G1 and G2/M phases is a featured cell-cycle signature of the 2C-like state, whereas the enrichment of the DNA replication S phase is a signature of the ESC state.
Figure 3.
Cell-cycle variations distinguish 2CLCs, ESCs, and the intermediate states
(A) Normalized density plots showing the distribution of cells in different cell-cycle phases analyzed from the single-cell RNA-seq data.
(B) The cell-cycle distribution of 2CLCs, ESCs, and intermediate-state cells at the single-cell level.
(C) Flow cytometry analysis of cell-cycle distribution upon treatment with different cell-cycle inhibitors. The vehicle is a DMSO control. Cytarabine is a DNA replication inhibitor, and nocodazole is a G2/M inhibitor. n = 3 independent experiments.
(D) qRT-PCR analysis of 2C repeats and pluripotency gene expression with cell-cycle inhibitor-treated ESC lines E14 and V6.5. Data are presented as the mean ± SEM, n = 3 independent experiments.∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.005.
(E) Flow cytometry analysis of 2C::tdTomato-positive cells with cytarabine treatment.
(F) qRT-PCR for 2C-specific gene expression upon treatment with cytarabine and a replication stress inhibitor, ATRi. Data are presented as the mean ± SEM, n = 3 independent experiments.
(G) Immunoblotting showing expression of ZSCAN4 and p-CHK1 in ESCs with the indicated treatment. n = 2 independent experiments.
Cell-cycle regulation is tightly coupled with cell fate decision. An expanded G1 phase may enable the accumulation of factors needed for the dissolution of pluripotency and cell fate determination (Dalton, 2015; Gonzales et al., 2015; Lange and Calegari, 2010). G2/M phase is connected with nucleolar disassembly, involving dynamic changes in nucleolus architecture and functions, which plays an important role in early embryo development and stem cell fate decision (Boisvert et al., 2007; Leung et al., 2004). To understand whether cell-cycle regulation plays a causal role in the 2CLC and ESC state transition, cell-cycle inhibitors were used to examine the effect of cell-cycle arrest on 2C activation (Figure 3C). The G2/M phase inhibitor nocodazole (NOC) significantly arrested cells in the G2/M phase but showed barely any effect on the expression of ERV repeats and pluripotency genes, while treatment with the DNA synthesis inhibitor cytarabine (CYT) arrested mESCs at G1/early S phase and showed significant upregulation of 2C genes and downregulation of pluripotency genes in both E14 and V6.5 ESC lines (Figure 3D). In addition, we found that cell-cycle inhibitors caused elevated P53 expression (Figure S3D). And it has been reported that altered p53 pulses lead to the expression of a different set of downstream genes and also alter cell fate (Purvis et al., 2012). So, we tested cell-cycle inhibitors in Trp53 knockout mESCs, and the results also showed significantly increased ERV expression with CYT treatment, suggesting that the effect of G1/early S arrest-mediated ERV activation was not dependent on Trp53 (Figure S3E) (Reyes et al., 2018). In addition, flow cytometry analysis with the MERVL-2C: tdTomato mESCs also showed increased percentage of viable cells in 2CLCs upon CYT treatment excluded from the apoptosis effect (Figures 3E and S3F). Previous studies found that retinoblastoma (RB) protein and its phosphorylation played a regulatory role in controlling cell fate decision through mediation of the cell-cycle progress, and inactivation of RB triggers G2/M arrest (Conklin et al., 2012; Conklin and Sage, 2009; Li et al., 2018; Ruiz et al., 2011). We also found that the expression level of Rb1 (RB transcriptional corepressor 1) and Rbl2 (RB transcriptional corepressor-like 2) was enriched in the ES-IN and ES populations (Figure S3G), which implied that Rb1 and Rbl2 might involved in regulating of ES to 2CLC transition. Together, the above data demonstrate not only that the different states of 2CLCs identified by single-cell RNA-seq are associated with different cell-cycle phases, but also that arrest at G1/early S phase could drive 2C activation.
The G1/S arrest-induced 2C activation was not through the ATR pathway
To further confirm whether G1-phase cell-cycle arrest per se could activate ERVs, we synchronized cells to the G1 phase (treated by NOC and released for 3.5 h so that the cells reached the G1 phase), instead of treating the cells with CYT. Cells synchronized to the G1 phase also showed strong induction of the 2C gene Dux (Figure S3H). To further exclude non-cell-cycle-related side effects of CYT, as CYT also acts as a DNA polymerase inhibitor, which could probably trigger replication stress through the ATR pathway to activate ERVs (Atashpaz et al., 2020; Wang et al., 2018), we dissected whether the 2C activation by CYT was through the activated ATR pathway. We treated mESCs with CYT and ATR inhibitors (ATRi) and found that ATRi treatment could not abrogate the 2C activation caused by CYT (Figures 3F and 3G), even though western blotting analysis showed a clear reduction of phosphorylation of the replication stress marker CHK1 (Figure 3G). Overall, these data indicated that the 2C activation upon CYT treatment was at least not completely a result of ATR signaling.
Cell-cycle arrest leads to dynamic changes in nucleolar architecture
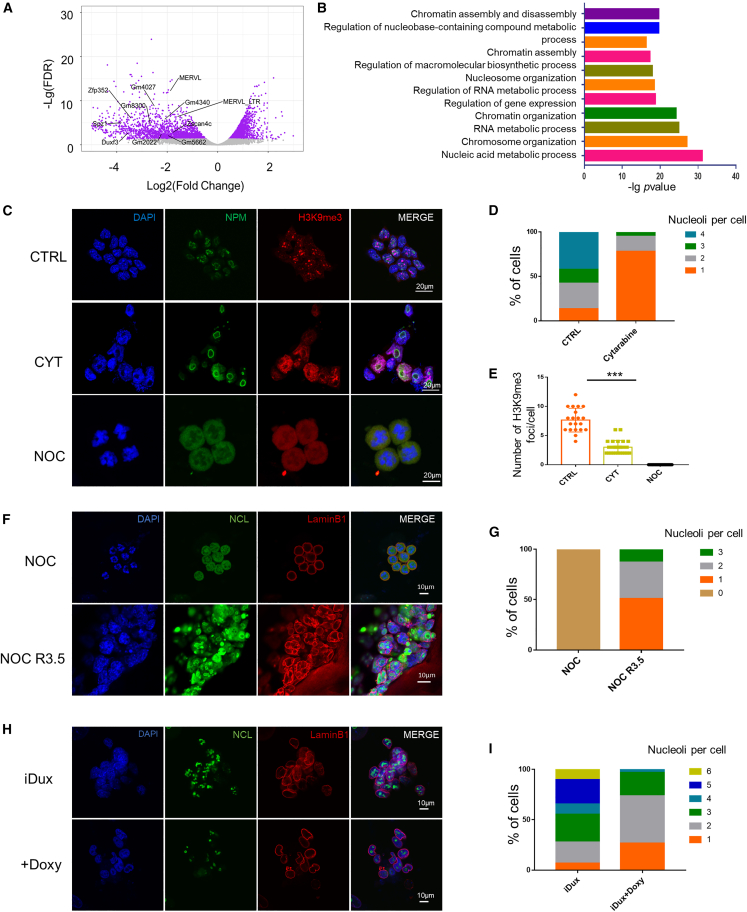
Having proved that G1/early S-phase arrest played an important role in the 2CLC and ESC state transition, we performed bulk RNA-seq with CYT treatment to dissect the potential pathways involved in regulating this transition. Analysis of DEGs (false discovery rate [FDR] <0.05 and log2 fold change >1) identified 1,570 upregulated genes and 1,279 downregulated genes upon CYT treatment (Table S2). We found that a significant fraction of CYT-induced DEGs overlapped with 2C-specific genes or elements, including MERVL, MT2_Mm(MERVL_LTR), Zscan4c, Duxf3, Gm4340, and Zfp352 (Figure 4A). Gene ontology (GO) analysis revealed that CYT treatment caused downregulation of genes in chromatin assembly and disassembly and nucleosome organization (Figure 4B), which is consistent with the fact that the nucleosome is assembled with DNA replication (Takami et al., 2007) and depletion of chromatin disassembly factors can trigger 2C activation (Ishiuchi et al., 2015). Previous studies show that the nucleolus exhibits different architecture during cell growth and division (Caragine et al., 2019; Sirri et al., 2008) and serves as a site for heterochromatin (Guetg and Santoro, 2012). To delineate nucleolus variations underlying different cell-cycle phases, we first sorted the 2C+ (2CLCs), 2C−/G1+, and 2C−/G1− cells using the Hoechst 33342 dye as a cell-cycle indicator from the MERVL-2C: tdTomato mESCs. We examined the nucleolar granular component (GC) marker protein nucleophosmin 1 (NPM1) with immunofluorescence and observed that NPM1 was enriched at the nucleolar periphery (Figure S4A). The 2C+ cells displayed reduced quantity of NPM1-labeled nucleolus with more fused and round morphology compared with the 2C−/G1− cells, and the 2C−/G1+ cells showed nucleolar characteristics similar to those of 2C+ cells (Figures S4A and S4B), which implied that the 2CLCs and ESCs displayed different nucleolar features.
Figure 4.
Cell-cycle arrest leads to dynamic changes in nucleolar architecture and nucleolus-associated heterochromatin
(A) An M-versus-A plot (MA) plot showing gene expression in ESCs treated with CYT in comparison to control. Key 2C-specific genes and elements are highlighted.
(B) A bar plot showing the −log10 (p value) of enrichment scores of the pathways enriched in differentially expressed genes upon CYT treatment. All data were derived from two independent replicates of RNA sequencing.
(C) Representative NPM1 and H3K9me3 immunofluorescence with CYT- and NOC-treated ESCs. CYT, cytarabine, NOC, nocodazole. Scale bars are indicated.
(D) The percentage of numbers of nucleoli per cell was quantified in CTRL and CYT treated conditions. The data represent averaged nucleoli number from four fields in two independent experiments.
(E) Numbers of H3K9me3 foci per cell in control, CYT and NOC treated conditions. ∗∗∗p < 0.005 by t test, n = 26 cells, shown is the mean ± SD. The data represent averaged H3K9me3 focus number from four fields in two independent experiments.
(F) Immunofluorescence of NCL and LaminB1 with nocodazole arrest at G2/M and release for 3.5 h (NOC R3.5) in ESCs synchronized at G1 phase.
(G) The percentages of the number of nucleoli per cell were quantified under NOC treatment and NOC treatment released for 3.5 h. The data represent averaged nucleoli number from four fields in two independent experiments.
(H) Immunofluorescence of NCL and LaminB1 in untreated (iDux) and doxycycline-induced Dux-overexpressing (+Doxy) ESCs.
(I) The percentages of the number of nucleoli per cell were quantified in iDux and Dux-overexpressing ESCs. The data represent averaged nucleoli number from four fields in two independent experiments.
To further investigate the architecture of the nucleolus and chromatin state during the cell-cycle process, we co-stained the nucleolar marker protein NPM1 and the heterochromatin marker histone H3 lysine 9 trimethylation (H3K9me3) with immunofluorescence to indicate whether the peri-nucleolar heterochromatin (Guetg and Santoro, 2012) is involved in 2C activation with cell-cycle inhibitor treatment. We found that mESCs treated with CYT showed reduced nucleoli counts per nucleus and rounder morphology like 2CLCs, while NOC-treated mESCs showed a disassembled nucleolus (Figure 4C and 4D). Moreover, the foci of H3K9me3 became diffused in the CYT-treated group (Figures 4C and 4E). Furthermore, we also detected dynamic morphological changes in the dense fibrillar component (DFC) with fibrillin immunofluorescence upon cell-cycle inhibitor treatment, namely, the DFC coalesced with CYT treatment and disappeared with NOC treatment (Figure S4C). To further confirm the nucleolar pattern of G1-phase cells, we stained nucleolin (NCL) in NOC-synchronized cells (treated by NOC and released for 3.5 h so that cells reached the G1 phase), and we found that the cells synchronized in G1 phase also showed reduced nucleoli number and round nucleolar shape, like 2C+ ESCs (Figures 4F and 4G). In conclusion, our study revealed that mESCs arrested at G1/early S phase exhibited a reduced and round nucleolar pattern, and diffused heterochromatin foci, while G2/M-phase mESCs showed a disassembled nucleolar pattern compared with untreated mESCs. It is reported that Dux is a major factor driving transcription in the 2C-like state by activating ERVL family repeats and ERVL-linked genes in mESCs (De Iaco et al., 2017; Hendrickson et al., 2017). Consequently, we carried out Dux overexpression and knockdown to see its effect on the cell cycle and nucleolus (Figures S4D and S4E). We found that more cells were at G1 and G2/M phase with Dux overexpression, and Dux knockdown showed reduced G1 phase (Figure S4F). And immunofluorescence showed that the nucleolus was also reduced and became round with Dux overexpression (Figures 4H and 4I), which suggested that the expression of Dux is connected with nucleolar structure. However, knockdown of Dux barely has an effect on the nucleolus and nuclear lamina (Figure S4H).
Cell-cycle arrest caused Dux release from the peri-nucleolar heterochromatin and 2C activation
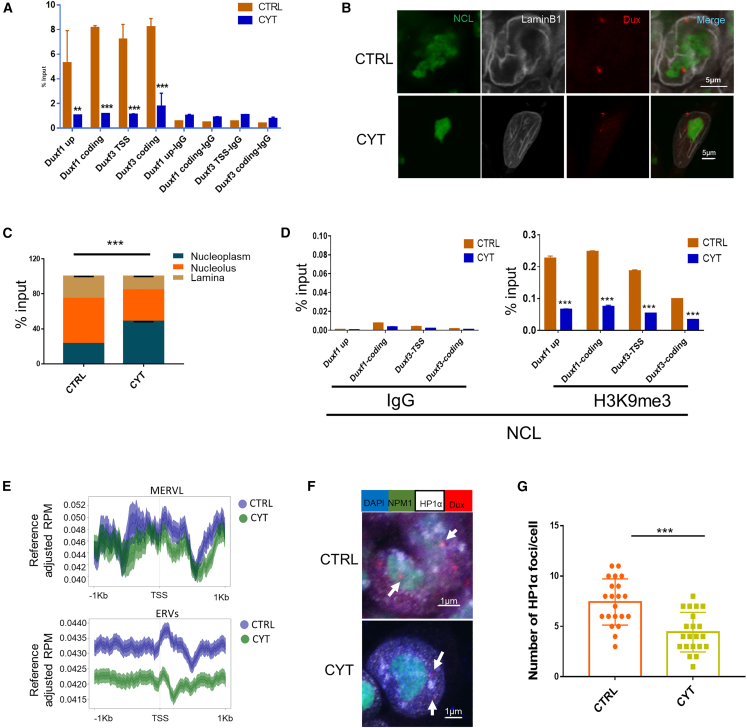
It has been reported that the nucleolar protein NCL can form a complex with TRIM28 and mediate the repression of the 2C gene Dux (Percharde et al., 2018). And we found that CYT-treated mESCs showed downregulated ribosome pathway genes and increased Zscan4 expression (Figures S5A–S5C), consistent with the abnormal nucleolar morphology shown above. Thus, we carried out chromatin immunoprecipitation (ChIP)-qPCR for NCL and found that NCL bound the Dux locus in mESCs and that the levels of NCL at the Dux locus were significantly reduced upon CYT treatment (Figure 5A). To further determine the interaction between Dux and heterochromatin regions, we examined the localization of the Dux locus using DNA fluorescence in situ hybridization (FISH) in combination with immunofluorescence of NCL and the nuclear lamina marker LaminB1. Nuclear lamina is a fibrous layer and often exhibits low transcriptional activity (Lanctôt et al., 2007). Similar to the ChIP results, the Dux locus tended to escape from the peri-nucleolar or laminar regions to the nucleoplasm with CYT treatment (Figures 5B and 5C), indicating that the Dux locus moved away from the transcriptionally repressive heterochromatin environment (Guetg and Santoro, 2012).
Figure 5.
G1/early S-phase arrest-mediated nucleolar architecture and peri-nucleolar heterochromatin remodeling modulates 2C gene activation
(A) ChIP assays showing NCL binding at the Dux locus, with or without CYT treatment. Data are shown as a percentage of input. ∗∗p < 0.01,∗∗∗p < 0.005 by one-way ANOVA, n = 3, error bars represent SE. Shown is a representative result from two independent experiments.
(B) Representative images of distinct localization patterns of the Dux locus stained with DNA FISH (red) under control and cytarabine-treated conditions, and co-stained by immunofluorescence for NCL (green) and LaminB1 (gray). Scale bars are indicated.
(C) Statistical analysis of the proportion of the Dux locus localization in lamina, nucleolus, and nucleoplasm. ∗∗∗p < 0.005 by t test, the data represent averaged Dux locus from six fields in three independent experiments, and error bars represent SE. Shown is a representative result from three independent experiments.
(D) ChIP-Re-ChIP qPCR assays for NCL binding and the H3K9me3 mark signal at the Dux locus, with and without CYT treatment. Data are shown as a percentage of input. ∗∗∗p < 0.005 by one-way ANOVA, error bars represent SE. Shown is a representative result from two independent experiments.
(E) Aggregated ChIP-Re-ChIP-seq signals of MERVL (top) and ERVs (bottom) with NCL as primary antibody and H3K9me3 as Re-ChIP secondary antibody; mark signals are in the range of ±1 kb around the transcription start site (TSS) in CTRL and CYT-treated ESCs. Data are shown as normalized to input. Shown is an average signal of two independent experiments.
(F) Representative images of distinct localization patterns of the Dux locus (red) under control versus CYT-treated conditions by immunofluorescence for NPM1 (green) and HP1α (white). Scale bars are indicated, the white arrow showing Dux locus.
(G) Quantification of the number of HP1α foci per cell in control and CYT-treated ESCs. ∗∗∗p < 0.005 by t test, error bars represent SE. The data represent averaged number of HP1α foci from four fields in two independent experiments.
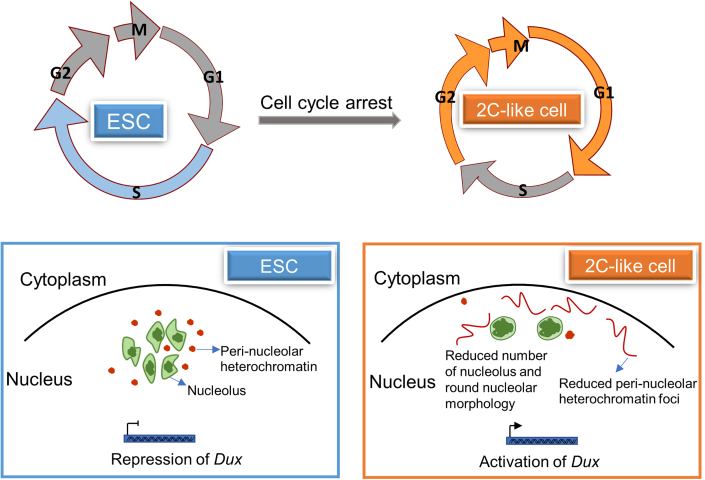
Next, we used H3K9me3 or heterochromatin protein 1 (HP1α) in combination with NCL to indicate peri-nucleolar heterochromatin. We performed ChIP-Re-ChIP analysis with NCL as a primary antibody and H3K9me3 as a secondary antibody, and it revealed that the levels of DNA pulled down by both NCL and H3K9me3 at the Dux locus were significantly reduced upon CYT treatment (Figure 5D). Furthermore, ChIP-Re-ChIP-seq results also revealed reduced NCL binding and H3K9me3 signals at the MERVL or ERV class gene promoters with CYT treatment (Figure 5E). Last, we performed Dux DNA FISH combined with NPM1 and HP1α immunofluorescence. DNA FISH showed that Dux signals became more separated from NPM1 and HP1α in the CYT-treated group, and the HP1α foci also became more scattered, like H3K9me3 foci, at the nucleolar periphery (Figures 5F and S5D), and the count of HP1α foci per cell was also reduced with CYT treatment (Figure 5G). Taken together, these data demonstrated that reduced peri-nucleolar heterochromatin around the Dux locus at the G1/early S phase might facilitate 2C gene expression (Figure 6).
Figure 6.
Models of cell-cycle-mediated nucleolar and heterochromatin regulation of 2C activation
Discussion
The 2C-like transcriptional program is spontaneously activated in less than 1% mESCs, and the process is transient and reversible. Previous studies characterized the transcriptional dynamics during ESC-to-2CLC transition and identified factors regulating this process (Fu et al., 2019; Rodriguez-Terrones et al., 2018). Recently, Xudong Fu et al. reported a roadmap for 2CLC exit to ESCs and found that mRNA degradation of key 2C genes contributes to the 2C-like state exit (Xudong Fu and Zhang, 2020). However, the mechanisms involved in entering and exiting the 2C-like state may not be fully unraveled. Here, we performed single-cell RNA-seq to dissect the transcriptional roadmap during the 2CLC-to-ESC transition. Our analysis identified different subpopulations of 2CLCs. We also identified intermediate states with different cell-cycle-phase signatures along the road of 2CLC entry and exit.
Cell-cycle states regulate pluripotency dissolution and lineage-specific differentiation (Gonzales et al., 2015; Pauklin and Vallier, 2013; Shin et al., 2015). Despite evidence showing that the cell-cycle phase varies from 2CLCs to ESCs (Atashpaz et al., 2020; Eckersley-Maslin et al., 2016), there is no direct evidence that cell-cycle phases regulate 2C-like state activation. Here, we demonstrated that the cell-cycle phases were not only associated with the 2C-like state, but also might drive the 2C emergence. Specifically, the 2CLCs with higher 2C-specific ERV expression (2C-IN) exhibited prolonged G1 phase, while 2CLCs with lower ERV expression (2C-OUT) exhibited prolonged G2/M phase. Particularly, the cell-cycle inhibitor CYT triggered cell-cycle arrest and 2C gene activation independent of the ATR-CHK1-pathway and the p53 pathway. Particularly, we have noted that the fluorescence ubiquitination cell-cycle indicator (FUCCI) system has been widely used in cell-cycle studies, as it enables real-time monitoring of different phases of the cell cycle in living cells (Chetty et al., 2013; Sakaue-Sawano et al., 2017; Shcherbina et al., 2019), which is a powerful tool for elaborating the duration of 2C emergence and exit at different cell-cycle phases in our next study.
The nucleolus has dynamic disassembly and reassembly in the cell cycle, and the nucleolar protein NCL has been reported to have an important role in mediating 2C gene activation (Hernandez-Verdun, 2011; Percharde et al., 2018). Previous studies showed that the transition from ESC to 2CLC undergoes drastic chromatin remodeling, such as discrete foci of heterochromatin (Akiyama et al., 2015) and decondensation of heterochromatin (Ishiuchi et al., 2015). And our study revealed that cell-cycle arrest at G1/early S phase showed reduced nucleolus and heterochromatin foci formation, especially, reduced peri-nucleolar heterochromatin around the Dux locus, which facilitated the Dux locus’s escape from the nucleoli or lamina to the nucleoplasm. Together, the results of our study revealed a cell-cycle-associated mechanism related to remodeling of the peri-nucleolar heterochromatin and Dux gene expression in 2CLC emergence. However, other cell-cycle-associated mechanisms promoting transition between 2CLC and ESC states cannot be excluded and merit further investigation.
Experimental procedures
mESC culture
ES-E14 cells were cultured on MEF feeder with knockout DMEM (Gibco, cat. no. 10829018) containing 15% FBS (VISTECH, cat. no.59216269), 1,000 U/mL mouse LIF (Peprotech, cat. no. 250-02), 0.1 mM non-essential amino acids (Gibco, cat. no. 11140), 0.1 mM β-mercaptoethanol (Sigma, cat. no. M6250), and 2 mM L-glutamine (Gibco, cat. no. A2916801). For culture of mESC lines, the medium was changed daily, and cells were routinely passaged every other day. The E14 cell line was kindly provided by the laboratory of George Daley. The MERVL-LTR-tdTomato reporter constructs were transfected into E14 cells by lipofection with Lipofectamine 2000 (Thermo Fisher, cat. no. 11668019). The next day, 150 μg/mL hygromycin (Gibco, cat. no. 10687010) was added to the medium for selection. Then, colonies containing tdTomato-positive cells were picked and expanded.
Fluorescence-activated cell sorting
Flow cytometry analysis was performed using the Beckman CytoFLEX LX, and cell sorting was performed on the Beckman MoFlo Astrios EQ. Data and images were analyzed and generated using FlowJo (v.10) software.
Single-cell RNA sequencing
Single-cell RNA-seq was performed with 10× platform as described previously (Zheng et al., 2017). FACS-sorted mESCs with the MERVL reporter that were cultured for 24 h were collected for single-cell RNA-seq.
Cell-cycle arrest
E14 cells were treated with 2 μM CYT (MedChemExpress [MCE], cat. no. 147-94-4) for 24 h, 1 μM NOC (MCE, cat. no. 31430-18-9) for 6 h.
Cell-cycle analysis
Cells were fixed with 70% ethanol at −20°C overnight. Next day, the fixed cells were centrifuged at 4°C, 2,000 rpm. Then they were washed once with PBS. The cells were stained with 10 μg/mL propidium iodide (PI) containing 100 μg/mL RNase A at 37°C for 30 min. Then cell-cycle analysis was performed on a BD Fortessa. Cell cycle was analyzed with modfit LT software.
RNA isolation and qPCR
Cellular RNA was collected using the FastPure Cell/Tissue Total RNA Isolation Kit (Vazyme, cat. no. RC101-01). cDNA was generated using the HiScript Q RT SuperMix for qPCR (Vazyme, cat. no. R122-01) and qRT-PCR was performed using the ChamQ Universal SYBR qPCR Master Mix (Vazyme, cat. no. Q711-02). Relative quantification was performed on a C1000 thermal cycler and CFX96 Real-Time System (Bio-Rad) and normalized with GAPDH. qPCR data were analyzed and visualized with Bio-Rad CFX manager and GraphPad Prism 7. The qPCR primers are provided in Table S3.
Immunofluorescence
ESCs were plated onto MEF-coated 24-well plate covered with slides. After being treated with cell-cycle inhibitors, the mESCs were fixed for 20 min in 4% paraformaldehyde (PFA), and blocked and permeabilized in IF blocking buffer (5% donkey serum, 2.5% BSA plus 0.3% Triton X-100 in PBS). The following antibodies and dilutions in IF buffer were used: Zscan4 (Sigma, AB4340; 1:400), NCL (Cell Signaling Technology [CST], 14,574; 1:1,000), NPM1 (Abcam, ab10530; 1:1,000), HP1α (CST, 2616; 1:200), H3K9me3 (CST, 13,969; 1:200). The next day, the slides were washed three times in PBST and incubated for 2 h at room temperature (RT) in the relevant fluorescently conjugated secondary antibodies (Abcam, ab150077 and ab150114). Slides were stained with DAPI and imaged on a Nikon A1R microscope.
Chromatin immunoprecipitation
Cells were cross-linked with 1% formaldehyde at room temperature for 10 min. Chromatin was prepared utilizing the reagents and protocol from the Pierce Magnetic ChIP Kit (Thermo, 26,157). For ChIP-Re-ChIP, immunoprecipitates (IPs) were eluted with 25 μL 10 mM DTT at 37°C for 30 min. Then they were diluted 20× with Re-ChIP buffer (1% Triton X-100, 2 mM EDTA, 150 mM NaCl, 20 mM Tris-HCl, and 1× cocktail) on ice. Then they were incubated with secondary antibody at 4°C overnight with mixing. Washes and IP elution were performed according to kit protocols. DNA was purified with thermo DNA purification columns and ChIP enrichments analyzed by qPCR. The qPCR primers are provided in Table S3. The ChIP-seq library was constructed with 100 ng purified DNA and input with a PCR-free DNA library prep kit (Vazyme, ND607).
DNA FISH
FISH samples were prepared using a protocol as previously described (Wang et al., 2016). Fixed cells were treated with freshly prepared 1 mg/mL sodium borohydride solution at room temperature for 10 min. They were washed and treated with 0.1 M HCl for 5 min at room temperature. Then they were digested with 100 μg/mL RNase A at 37°C for 60 min. Next, the cells were pre-hybridized at 80°C for 30 min in buffer containing 2× SSC, 50% formamide, and 0.1% v/v Tween 20. For hybridization, 20 μL hybridization buffer composed of 2× SSC, 40% formamide, 20% dextran sulfate, and 40–60 μM Dux probe was dropped onto a 24-well plate, covering the cells. The hybridization was processed at 80°C for 20 min and then incubated at 37°C for 18 h. The next day, the cells were washed twice with 2× SSC 0.1% v/v Tween 20 at 45°C for 15 min each and once with 2× SSC at room temperature for 15 min. The following immunofluorescence was proceeded as described before.
Data and code availability
All data that support the findings are available at GEO access (GSE171257, GSE171258). Custom code for the analysis can be found at https://github.com/chengarthur/CellCycleStem.
Author contributions
Y.Z., C.C., and L.C. contributed equally to this work. J.Z. conceived the project; Y.Z. and J.Z. designed the experiments; Y.Z. and L.C. performed the experiments; C.C. performed the bioinformatics analyses. All authors were involved in the interpretation of data. Y.Z. and J.Z. wrote the manuscript.
Conflict of interests
The authors declare that they have no competing interests.
Acknowledgments
We thank the Core Facilities of Zhejiang University School of Medicine, especially the Fluorescence Activated Cell Sorting (FACS) center for technical support. We thank Hongqing Liang (Zhejiang University), Yi Zhang (Harvard Medical School) and Wei Li (George Washington University) for discussion and support. J.Z. is supported by the National Key Research and Development Program of China (2018YFA0107100, 2018YFA0107103, 2018YFC1005002), the National Natural Science Foundation of China projects of China (31871453, 91857116), a Zhejiang Innovation Team grant (2019R01004), and the Zhejiang Natural Science Foundation projects of China (LR19C120001).
Published: October 7, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.stemcr.2021.09.003.
Supplemental information
References
- Akiyama T., Xin L., Oda M., Sharov A.A., Amano M., Piao Y., Cadet J.S., Dudekula D.B., Qian Y., Wang W., et al. Transient bursts of Zscan4 expression are accompanied by the rapid derepression of heterochromatin in mouse embryonic stem cells. DNA Res. 2015;22:307–318. doi: 10.1093/dnares/dsv013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atashpaz S., Samadi Shams S., Gonzalez J.M., Sebestyén E., Arghavanifard N., Gnocchi A., Albers E., Minardi S., Faga G., Soffientini P., et al. ATR expands embryonic stem cell fate potential in response to replication stress. eLife. 2020;12:e54756. doi: 10.7554/eLife.54756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boisvert F.M., van Koningsbruggen S., Navascués J., Lamond A.I. The multifunctional nucleolus. Nat. Rev. Mol. Cell Biol. 2007;8:574–585. doi: 10.1038/nrm2184. [DOI] [PubMed] [Google Scholar]
- Borsos M., Torres-Padilla M.-E. Building up the nucleus: nuclear organization in the establishment of totipotency and pluripotency during mammalian development. Genes Dev. 2016;30:611–621. doi: 10.1101/gad.273805.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caragine C.M., Haley S.C., Zidovska A. Nucleolar dynamics and interactions with nucleoplasm in living cells. Elife. 2019;26:e47533. doi: 10.7554/eLife.47533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chetty S., Pagliuca F.W., Honore C., Kweudjeu A., Rezania A., Melton D.A. A simple tool to improve pluripotent stem cell differentiation. Nat. Methods. 2013;10:553–556. doi: 10.1038/nmeth.2442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conklin J.F., Sage J. Keeping an eye on retinoblastoma control of human embryonic stem cells. J. Cell. Biochem. 2009;108:1023–1030. doi: 10.1002/jcb.22342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conklin J.F., Baker J., Sage J. The RB family is required for the self-renewal and survival of human embryonic stem cells. Nat. Commun. 2012;3:1244. doi: 10.1038/ncomms2254. [DOI] [PubMed] [Google Scholar]
- Dalton S. Linking the cell cycle to cell fate decisions. Trends Cell Biol. 2015;25:592–600. doi: 10.1016/j.tcb.2015.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckersley-Maslin M.A., Svensson V., Krueger C., Stubbs T.M., Giehr P., Krueger F., Miragaia R.J., Kyriakopoulos C., Berrens R.V., Milagre I., et al. MERVL/Zscan4 network activation results in transient genome-wide DNA demethylation of mESCs. Cell Rep. 2016;17:179–192. doi: 10.1016/j.celrep.2016.08.087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fadloun A., Le Gras S., Jost B., Ziegler-Birling C., Takahashi H., Gorab E., Carninci P., Torres-Padilla M.-E. Chromatin signatures and retrotransposon profiling in mouse embryos reveal regulation of LINE-1 by RNA. Nat. Struct. Mol. Biol. 2013;20:332–338. doi: 10.1038/nsmb.2495. [DOI] [PubMed] [Google Scholar]
- Falco G., Lee S.-L., Stanghellini I., Bassey U.C., Hamatani T., Ko M.S.H. Zscan4: a novel gene expressed exclusively in late 2-cell embryos and embryonic stem cells. Dev. Biol. 2007;307:539–550. doi: 10.1016/j.ydbio.2007.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu X., Wu X., Djekidel M.N. Myc and Dnmt1 impede the pluripotent to totipotent state transition in embryonic stem cells. Nat. Cell Biol. 2019;21:835–844. doi: 10.1038/s41556-019-0343-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzales K.A., Liang H., Lim Y.S., Chan Y.S., Yeo J.C., Tan C.P., Gao B., Le B., Tan Z.Y., Low K.Y., et al. Deterministic restriction on pluripotent state dissolution by cell-cycle pathways. Cell. 2015;162:564–579. doi: 10.1016/j.cell.2015.07.001. [DOI] [PubMed] [Google Scholar]
- Gruenheit N., Parkinson K., Brimson C.A., Kuwana S., Johnson E.J., Nagayama K., Llewellyn J., Salvidge W.M., Stewart B., Keller T., et al. Cell cycle heterogeneity can generate robust cell type proportioning. Dev. Cell. 2018;47:494–508 e494. doi: 10.1016/j.devcel.2018.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guallar D., Bi X., Pardavila J.A., Huang X., Saenz C., Shi X., Zhou H., Faiola F., Ding J., Haruehanroengra P., et al. RNA-dependent chromatin targeting of TET2 for endogenous retrovirus control in pluripotent stem cells. Nat. Genet. 2018;50:443–451. doi: 10.1038/s41588-018-0060-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guetg C., Santoro R. Formation of nuclear heterochromatin: the nucleolar point of view. Epigenetics. 2012;7:811–814. doi: 10.4161/epi.21072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamatani T., Carter M.G., Sharov A.A., Ko M.S.H. Dynamics of global gene expression changes during mouse preimplantation development. Dev. Cell. 2004;6:117–131. doi: 10.1016/s1534-5807(03)00373-3. [DOI] [PubMed] [Google Scholar]
- Hänzelmann S., Castelo R., Guinney J. GSVA: gene set variation analysis for microarray and RNA-Seq data. BMC Bioinformatics. 2013;14:7. doi: 10.1186/1471-2105-14-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrickson P.G., Doráis J.A., Grow E.J., Whiddon J.L., Lim J.-W., Wike C.L., Weaver B.D., Pflueger C., Emery B.R., Wilcox A.L., et al. Conserved roles of mouse DUX and human DUX4 in activating cleavage-stage genes and MERVL/HERVL retrotransposons. Nat. Genet. 2017;49:925–934. doi: 10.1038/ng.3844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez-Verdun D. Assembly and disassembly of the nucleolus during the cell cycle. Nucleus. 2011;2:189–194. doi: 10.4161/nucl.2.3.16246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Iaco A., Planet E., Coluccio A., Verp S., Duc J., Trono D. DUX-family transcription factors regulate zygotic genome activation in placental mammals. Nat. Genet. 2017;49:941–945. doi: 10.1038/ng.3858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishiuchi T., Enriquez-Gasca R., Mizutani E., Boskovic A., Ziegler-Birling C., Rodriguez-Terrones D., Wakayama T., Vaquerizas J.M., Torres-Padilla M.E. Early embryonic-like cells are induced by downregulating replication-dependent chromatin assembly. Nat. Struct. Mol. Biol. 2015;22:662–671. doi: 10.1038/nsmb.3066. [DOI] [PubMed] [Google Scholar]
- Lanctôt C., Cheutin T., Cremer M., Cavalli G., Cremer T. Dynamic genome architecture in the nuclear space: regulation of gene expression in three dimensions. Nat. Rev. Genet. 2007;8:104–115. doi: 10.1038/nrg2041. [DOI] [PubMed] [Google Scholar]
- Lange C., Calegari F. Cdks and cyclins link G1 length and differentiation of embryonic, neural and hematopoietic stem cells. Cell Cycle. 2010;9:1893–1900. doi: 10.4161/cc.9.10.11598. [DOI] [PubMed] [Google Scholar]
- Leung A.K., Gerlich D., Miller G., Lyon C., Lam Y.W., Lleres D., Daigle N., Zomerdijk J., Ellenberg J., Lamond A.I. Quantitative kinetic analysis of nucleolar breakdown and reassembly during mitosis in live human cells. J. Cell Biol. 2004;166:787–800. doi: 10.1083/jcb.200405013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Narayanan C., Bian J., Sambo D., Brickler T., Zhang W., Chetty S. A transient DMSO treatment increases the differentiation potential of human pluripotent stem cells through the Rb family. PLoS One. 2018;13:e0208110. doi: 10.1371/journal.pone.0208110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L., Michowski W., Kolodziejczyk A., Sicinski P. The cell cycle in stem cell proliferation, pluripotency and differentiation. Nat. Cell Biol. 2019;21:1060–1067. doi: 10.1038/s41556-019-0384-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macfarlan T.S., Gifford W.D., Driscoll S., Lettieri K., Rowe H.M., Bonanomi D., Firth A., Singer O., Trono D., Pfaff S.L. Embryonic stem cell potency fluctuates with endogenous retrovirus activity. Nature. 2012;487:57–63. doi: 10.1038/nature11244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller S., Almouzni G. Chromatin dynamics during the cell cycle at centromeres. Nat. Rev. Genet. 2017;18:192–208. doi: 10.1038/nrg.2016.157. [DOI] [PubMed] [Google Scholar]
- Nagano T., Lubling Y., Várnai C., Dudley C., Leung W., Baran Y., Mendelson Cohen N., Wingett S., Fraser P., Tanay A. Cell-cycle dynamics of chromosomal organization at single-cell resolution. Nature. 2017;547:61–67. doi: 10.1038/nature23001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemeth A., Grummt I. Dynamic regulation of nucleolar architecture. Curr. Opin. Cell Biol. 2018;52:105–111. doi: 10.1016/j.ceb.2018.02.013. [DOI] [PubMed] [Google Scholar]
- Padeken J., Heun P. Nucleolus and nuclear periphery: Velcro for heterochromatin. Curr. Opin. Cell Biol. 2014;28:54–60. doi: 10.1016/j.ceb.2014.03.001. [DOI] [PubMed] [Google Scholar]
- Pauklin S., Vallier L. The cell-cycle state of stem cells determines cell fate propensity. Cell. 2013;155:135–147. doi: 10.1016/j.cell.2013.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Percharde M., Lin C.J., Yin Y., Guan J., Peixoto G.A., Bulut-Karslioglu A., Biechele S., Huang B., Shen X., Ramalho-Santos M. A LINE1-nucleolin partnership regulates early development and ESC identity. Cell. 2018;174:391–405 e319. doi: 10.1016/j.cell.2018.05.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Probst A.V., Dunleavy E., Almouzni G. Epigenetic inheritance during the cell cycle. Nat. Rev. Mol. Cell Biol. 2009;10:192–206. doi: 10.1038/nrm2640. [DOI] [PubMed] [Google Scholar]
- Purvis J.E., Karhohs K.W., Mock C., Batchelor E., Loewer A., Lahav G. p53 dynamics control cell fate. Science. 2012;336:1440–1444. doi: 10.1126/science.1218351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu X., Mao Q., Tang Y., Wang L., Chawla R., Pliner H.A., Trapnell C. Reversed graph embedding resolves complex single-cell trajectories. Nat. Methods. 2017;14:979–982. doi: 10.1038/nmeth.4402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes J., Chen J.-Y., Stewart-Ornstein J., Karhohs K.W., Mock C.S., Lahav G. Fluctuations in p53 signaling allow escape from cell-cycle arrest. Mol. Cell. 2018;71:581–591.e585. doi: 10.1016/j.molcel.2018.06.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Terrones D., Gaume X., Ishiuchi T., Weiss A., Kopp A., Kruse K., Penning A., Vaquerizas J.M., Brino L., Torres-Padilla M.E. A molecular roadmap for the emergence of early-embryonic-like cells in culture. Nat. Genet. 2018;50:106–119. doi: 10.1038/s41588-017-0016-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Terrones D., Hartleben G., Gaume X., Eid A., Guthmann M., Iturbide A., Torres-Padilla M.-E. A distinct metabolic state arises during the emergence of 2-cell-like cells. Embo Rep. 2020;21:e48354. doi: 10.15252/embr.201948354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz S., Panopoulos A.D., Herrerías A., Bissig K.D., Lutz M., Berggren W.T., Verma I.M., Izpisua Belmonte J.C. A high proliferation rate is required for cell reprogramming and maintenance of human embryonic stem cell identity. Curr. Biol. 2011;21:45–52. doi: 10.1016/j.cub.2010.11.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakaue-Sawano A., Yo M., Komatsu N., Hiratsuka T., Kogure T., Hoshida T., Goshima N., Matsuda M., Miyoshi H., Miyawaki A. Genetically encoded tools for optical dissection of the mammalian cell cycle. Mol. Cell. 2017;68:626–640.e625. doi: 10.1016/j.molcel.2017.10.001. [DOI] [PubMed] [Google Scholar]
- Shcherbina A., Li J., Narayanan C., Greenleaf W., Kundaje A., Chetty S. Brief report: cell cycle dynamics of human pluripotent stem cells primed for differentiation. Stem Cells. 2019;37:1151–1157. doi: 10.1002/stem.3041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin J., Berg D.A., Zhu Y., Shin J.Y., Song J., Bonaguidi M.A., Enikolopov G., Nauen D.W., Christian K.M., Ming G.L., et al. Single-cell RNA-seq with waterfall reveals molecular cascades underlying adult neurogenesis. Cell Stem Cell. 2015;17:360–372. doi: 10.1016/j.stem.2015.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirri V., Urcuqui-Inchima S., Roussel P., Hernandez-Verdun D. Nucleolus: the fascinating nuclear body. Histochem. Cell Biol. 2008;129:13–31. doi: 10.1007/s00418-007-0359-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith A. Formative pluripotency: the executive phase in a developmental continuum. Development. 2017;144:365–373. doi: 10.1242/dev.142679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takami Y., Ono T., Fukagawa T., Shibahara K.-i., Nakayama T. Essential role of chromatin assembly factor-1-mediated rapid nucleosome assembly for DNA replication and cell division in vertebrate cells. Mol. Biol. Cell. 2007;18:129–141. doi: 10.1091/mbc.E06-05-0426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C., Cacchiarelli D., Grimsby J., Pokharel P., Li S., Morse M., Lennon N.J., Livak K.J., Mikkelsen T.S., Rinn J.L. The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat. Biotechnol. 2014;32:381–386. doi: 10.1038/nbt.2859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visintin R., Amon A. The nucleolus: the magician’s hat for cell cycle tricks. Curr. Opin. Cell Biol. 2000;12:372–377. doi: 10.1016/s0955-0674(00)00102-2. [DOI] [PubMed] [Google Scholar]
- Wang S., Su J.H., Beliveau B.J., Bintu B., Moffitt J.R., Wu C.T., Zhuang X. Spatial organization of chromatin domains and compartments in single chromosomes. Science. 2016;353:598–602. doi: 10.1126/science.aaf8084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Chen Z., Mishra A.K., Silva A., Ren W., Pan Z., Wang J.H. Chemotherapy-induced differential cell cycle arrest in B-cell lymphomas affects their sensitivity to Wee1 inhibition. Haematologica. 2018;103:466–476. doi: 10.3324/haematol.2017.175992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whiddon J.L., Langford A.T., Wong C.J., Zhong J.W., Tapscott S.J. Conservation and innovation in the DUX4-family gene network. Nat. Genet. 2017;49:935–940. doi: 10.1038/ng.3846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu K., Liu H., Wang Y., He J., Xu S., Chen Y., Kuang J., Liu J., Guo L., Li D., et al. SETDB1-Mediated cell fate transition between 2C-like and pluripotent states. Cell Rep. 2020;30:25–36 e26. doi: 10.1016/j.celrep.2019.12.010. [DOI] [PubMed] [Google Scholar]
- Xudong Fu M.N.D., Zhang Yi. A transcriptional roadmap for 2C-like–to–pluripotent state transition. Sci. Adv. 2020;6:22–29. doi: 10.1126/sciadv.aay5181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang F., Huang X., Zang R., Chen J., Fidalgo M., Sanchez-Priego C., Yang J., Caichen A., Ma F., Macfarlan T., et al. DUX-miR-344-ZMYM2-Mediated activation of MERVL LTRs Induces a totipotent 2C-like state. Cell Stem Cell. 2020;26:234–250.e237. doi: 10.1016/j.stem.2020.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zalzman M., Falco G., Sharova L.V., Nishiyama A., Thomas M., Lee S.L., Stagg C.A., Hoang H.G., Yang H.T., Indig F.E., et al. Zscan4 regulates telomere elongation and genomic stability in ES cells. Nature. 2010;464:858–863. doi: 10.1038/nature08882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Emerson D.J., Gilgenast T.G., Titus K.R., Lan Y., Huang P., Zhang D., Wang H., Keller C.A., Giardine B., et al. Chromatin structure dynamics during the mitosis-to-G1 phase transition. Nature. 2019;576:158–162. doi: 10.1038/s41586-019-1778-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng G.X., Terry J.M., Belgrader P., Ryvkin P., Bent Z.W., Wilson R., Ziraldo S.B., Wheeler T.D., McDermott G.P., Zhu J., et al. Massively parallel digital transcriptional profiling of single cells. Nat. Commun. 2017;8:14049. doi: 10.1038/ncomms14049. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data that support the findings are available at GEO access (GSE171257, GSE171258). Custom code for the analysis can be found at https://github.com/chengarthur/CellCycleStem.