Summary
Retinoic acid (RA) signaling plays an important role during heart development in establishing anteroposterior polarity, formation of inflow and outflow tract progenitors, and growth of the ventricular compact wall. RA is also utilized as a key ingredient in protocols designed for generating cardiac cell types from pluripotent stem cells (PSCs). This review discusses the role of RA in cardiogenesis, currently available protocols that employ RA for differentiation of various cardiovascular lineages, and plausible transcriptional mechanisms underlying this fate specification. These insights will inform further development of desired cardiac cell types from human PSCs and their application in preclinical and clinical research.
In this article, Devalla and colleagues outline the role of retinoic acid (RA) signaling in the heart and provide an overview of human pluripotent stem cell-based cardiac differentiation approaches that employ RA. They offer new insights into molecular mechanisms underpinning the function of RA signaling and discuss the crosstalk with wingless-related integration site (WNT) and bone morphogenetic protein (BMP) signaling in cardiac cell fate specification.
Introduction
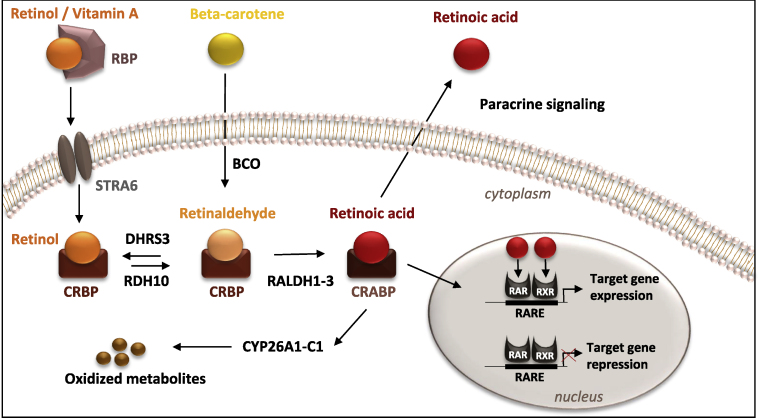
Retinoic acid (RA), the active derivative of vitamin A, has pleiotropic functions during vertebrate development, differentiation, and homeostasis. Vitamin A is obtained from diet, either as preformed vitamin A (retinol, retinyl esters) found in animal products or precursors such as carotenoids found in plant products. Retinol is bound by plasma retinol binding protein and transthyretin complexes for transport to target tissues, where it is taken up through STRA6 cell-surface receptors (Kanai et al., 1968; Kawaguchi et al., 2007). Once inside the cell, retinol forms a complex with intracellular retinol binding proteins (Napoli, 2016), and is processed by short-chain dehydrogenase/reductase, RDH10, for conversion to retinaldehyde (Sandell et al., 2007). Retinaldehyde can be reconverted back to retinol by DHRS3 for storage, which helps in maintaining RA homeostasis (Billings et al., 2013). Oxidation of retinaldehyde to RA is carried out by retinaldehyde dehydrogenases (RALDH1–3, also known as ALDH1A1–3) (Duester, 2008). Among RALDH enzymes, RALDH2 is the major source of RA during embryogenesis, and its expression was shown to closely correlate with active RA signaling (Niederreither et al., 1997; Moss et al., 1998). RA uptake is mediated by binding to cellular RA binding proteins, CRABP, which deliver RA to the nucleus for regulation of gene expression or to CYP26 enzymes (CYP26A1, CYP26B1, and CYP26C1) for degradation. Biosynthesis and metabolism of RA is briefly summarized in Figure 1 and extensively reviewed elsewhere (Kedishvili, 2016; Metzler and Sandell, 2016).
Figure 1.
Biosynthesis and metabolism of RA in vertebrates
Retinol obtained from diet is converted to retinaldehyde by retinal dehydrogenase enzymes. Retinaldehyde can also be synthesized from β-carotene. Oxidation of retinaldehyde to RA is carried out by RALDH enzymes. RA is a diffusible morphogen with autocrine as well as paracrine actions. RA is the ligand for heteromeric RAR and RXR receptors, which bind to RARE in promoters of target genes. In non-target tissues, RA is degraded by CYP26 enzymes.
Vitamin A is an essential nutrient required throughout life. Notably, RA, the physiologically active form of vitamin A, is critical for proper development of the embryo. The majority of our knowledge regarding the consequences of maternal vitamin A deficiency and the impact on fetal development came from studies in animal models. A wide spectrum of multi-organ abnormalities including those of the heart were observed in the progeny of vitamin A-deficient rats (Wilson et al., 1953; Wilson and Warkany, 1949). In humans, congenital heart defects have been reported in people carrying mutations in STRA6 and RALDH2, which results in RA deficiency (Pasutto et al., 2007; Pavan et al., 2009). Importantly, an excess of vitamin A during pregnancy has also been associated with heart defects among other malformations and was even found to be teratogenic (Lammer et al., 1985; Rothman et al., 1995). These findings suggest that a precise balance in RA availability underscores normal development.
The functions of RA in embryogenesis mainly stem from its role in gene regulation. RA acts as a ligand for members of the steroid hormone superfamily of transcription factors, the retinoic acid receptors (RARs) and the retinoic acid X receptors (RXRs). Both receptor subfamilies comprise three independent genes: RARα, RARβ, RARγ and RXRα, RXRβ, RXRγ. While RA is the sole ligand for RARs, RXRs can also be bound by other ligands such as polyunsaturated fatty acids (Dawson and Xia, 2012). RARs and RXRs form heterodimers and bind to RA-response element (RARE) in the regulatory regions (promoters, enhancers) of target genes and recruit co-activators or co-repressors to regulate gene expression in a ligand-dependent manner. In this review, we focus on the role of RA in heart development, particularly in the specification of various cardiac cell types and its application in differentiating these cell populations from human pluripotent stem cells (hPSCs).
Overview of RA signaling in cardiogenesis
Heart development begins at gastrulation, when cell populations in the primitive streak are patterned to form distinct cardiac cell types (Devine et al., 2014; Lescroart et al., 2014; Ivanovitch et al., 2021). Left ventricular progenitors are the first to emerge from distal primitive streak at mid-streak stage (between embryonic day 7 [E7.0] and E7.75), followed by right ventricular progenitors at late-streak stage (E7.25–E7.75) and outflow tract (OFT) progenitors at no-allantoic-bud to early-allantoic-bud stages (E7.25–E8.0). Atrial progenitors leave the primitive streak concomitant with OFT progenitors, albeit from a more proximal region (Ivanovitch et al., 2021). The cardiac progenitors migrate from the primitive streak to the anterior lateral plate mesoderm and form bilateral fields, termed the first heart field (FHF) and the second heart field (SHF). The FHF cells express Hand1 and Hcn4 among other markers. Recently, an additional multipotent progenitor field anterior to the FHF, also expressing Hand1, has been identified to contribute to myocardium of the left ventricle, atrioventricular canal, and the proepicardium (Tyser et al., 2021; Zhang et al., 2021). While the FHF differentiates and forms the linear heart tube, the SHF is maintained in a proliferative progenitor state (Prall et al., 2007). Subsequently, cells of the SHF are progressively added to the heart tube. The SHF dorsal of the forming heart tube is partitioned into an anterior component (aSHF) marked by Tbx1 and a posterior component (pSHF) marked by Tbx5 (Bruneau et al., 1999; De Bono et al., 2018). The aSHF progenitors of the right ventricle and outflow tract are contiguous with the arterial pole of the heart tube, and pSHF progenitors of the atria and sinus venosus are connected to the venous pole of the linear heart tube. The elongating heart undergoes rightward looping. Simultaneously, the proepicardium develops caudoventrally of the inflow tract, undergoes epithelial-to-mesenchymal transition (EMT), and forms the epicardium, the outer layer of the heart.
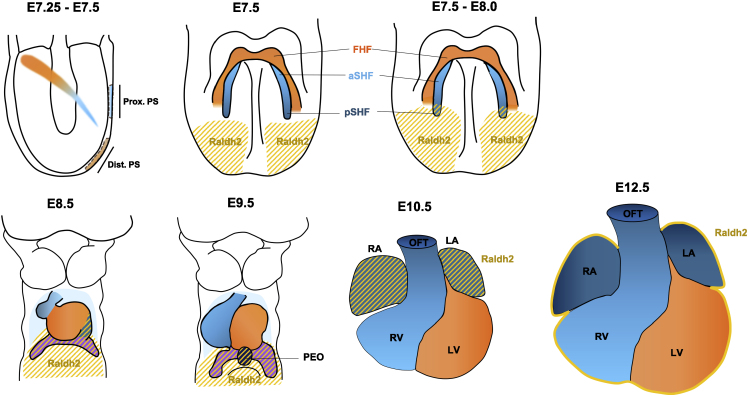
RA signaling has been implicated in multiple stages of heart development including formation of anterior-posterior boundaries of cardiac mesoderm, specification of cardiomyocyte subtypes, development of the epicardium, the outflow tract, and growth of the ventricular compact wall and coronary arteriogenesis. Early studies investigating the expression of Raldh2 (Figure 2), and the effects of its deletion provided insights into spatiotemporal requirements of RA in the developing heart. At primitive-streak to early-bud stages, cardiac progenitors do not express Raldh2 (Ivanovitch et al., 2021). At late-bud stages (E7.5–E8.0), Raldh2 is expressed in the posterior lateral plate mesoderm and expands anteriorly, encountering caudal cardiac precursors at early head fold stage (E7.5–E8.0) (Niederreither et al., 1997; Hochgreb et al., 2003). Subsequently, Raldh2 is expressed by pSHF progenitors themselves and remains confined to the prospective sinoatrial and atrial tissues until E9.5 (Moss et al., 1998; Hochgreb et al., 2003; Ivanovitch et al., 2021). At E9.5, the proepicardium is formed, which also shows robust expression of Raldh2 (Del Monte et al., 2011). At E10.5 and E11.5, Raldh2 expression persists in the atrial myocardium. At E12.5, Raldh2 expression is restricted to the epicardium of both the atria and the ventricles (Moss et al., 1998; Brade et al., 2011), and gradually decreases at late gestational stages.
Figure 2.
Expression of Raldh2 in the developing mouse heart
Cardiac progenitors migrate anterolaterally from the primitive streak. Raldh2 is expressed in the posterior part of the embryo at E7.5, which expands anteriorly, toward the cardiogenic fields. Thereafter, the pSHF progenitors express Raldh2 and the expression is found in the sinus venosus, atria, and the proepicardium at E9.5. Raldh2 expression is restricted to the atrial myocardium at E10.5–E11.5 and to the epicardium at E12.5.
The balance between RA synthesis by RALDH2 and degradation by CYP26 enzymes determines RA availability and distribution in the embryo. Interestingly, Raldh2 and Cyp26 expression domains are largely complementary (Niederreither and Dollé, 2008). In the heart, Cyp26a1 is expressed in the endocardium of the outflow tract, atria, and sinus venosus between E8.0 and E9.0 (MacLean et al., 2001). At E14.5, Cyp26a1 expression is restricted to atrioventricular valves, whereas Cyp26b1 is expressed in the endocardium of the outflow tract (Abu-Abed et al., 2002). Unlike the strict expression patterns of RALDH2 and CYP26 enzymes, RA receptors are expressed widely. In particular, Rarα and all three Rxr genes show ubiquitous expression in the developing heart. Rarb1 expression is restricted to the outflow tract mesenchyme, while Rarb2 is found throughout the developing myocardium. Rarγ is specifically expressed in the endocardial cushions and developing vessels at E12.5 (reviewed in Dollé, 2009). Insights on RA signaling obtained from experimental animal models greatly contributed to the understanding of organ development and have been successfully applied to in vitro differentiation protocols discussed below (Zhang et al., 2011; Devalla et al., 2015; Iyer et al., 2015; Guadix et al., 2017; Lee et al., 2017; Protze et al., 2017; Zhao et al., 2017).
RA in atrial development
The majority of the atrial compartments are formed by contributions from the pSHF progenitors. While the caudal pSHF gives rise to the sinus venosus and distal atria, cranial sections of the pSHF form the proximal portions of the atria closer to the atrioventricular junction (Domínguez et al., 2012). Left-right identity is conferred by Pitx2c expression in the left pSHF, which contributes to most of the left atrium (Galli et al., 2008). RA signaling is instrumental in maintaining the posterior limits of the SHF (Ryckebusch et al., 2008). Moreover, the boundary between aSHF and pSHF is facilitated by the expression of RA-responsive Hox genes as well as T-box transcription factors Tbx1 and Tbx5 (Bertrand et al., 2011; De Bono et al., 2018; Stefanovic et al., 2020). While Tbx1 expressed in aSHF antagonizes RA signaling, RA-induced Tbx5 in the pSHF is required to suppress the Tbx1-dependent aSHF program (De Bono et al., 2018). Abrogation of RA signaling in Raldh2 knockout mice results in hypoplastic atria and sinus venosus (Niederreither et al., 2001), while a more dramatic total lack of the sinus venosus was observed in vitamin A-deficient quail embryos (Kostetskii et al., 1999). Similarly, inhibition of RA signaling in Hamburger-Hamilton stage 4–7 chicken embryos using a pan RA receptor antagonist resulted in a smaller inflow tract compartment and enlarged ventricles (Hochgreb et al., 2003).
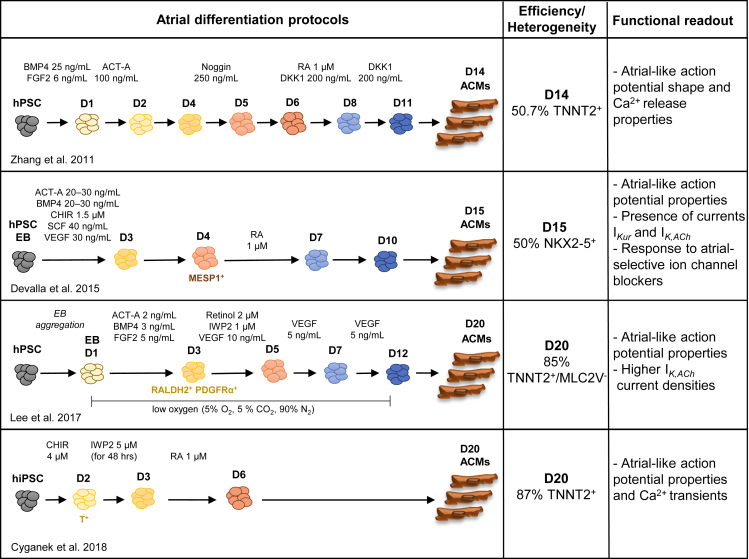
Consistent with in vivo studies, the role of RA in atrial differentiation has also been investigated and confirmed in several in vitro studies (Gassanov et al., 2008; Zhang et al., 2011; Devalla et al., 2015; Lee et al., 2017). The first report was based on experiments with transgenic mouse embryonic stem cells (mESCs), in which GFP expression was driven by the atrial-specific promoter of Nppa (Wobus et al., 1997; Gassanov et al., 2008). The authors examined the effect of varying concentrations of RA treatment for 5 days, during early or late stages of differentiation. They concluded that embryoid bodies treated with 100 nM and 1 nM RA from day 1–6 differentiated toward an atrial phenotype as assessed by an increase in Nppa-GFP+ areas. However, this increase was a moderate 6%–9% in the RA-treated group, and the changes in Myl7 (Mlc2a) and Myl2 (Mlc2v) mRNA expression were only significant in the 100 nM condition. Nonetheless, this study indicated a possible role for RA in atrial differentiation in vitro. Subsequently, a role for RA in directing atrial specification of human embryonic stem cells (hESCs) was described (Zhang et al., 2011). In this study, upon mesoderm formation, bone morphogenetic protein (BMP) signaling was inhibited using Noggin, followed by treatment with 1 μM RA and wingless-related integration site (WNT) inhibitor DKK1. The differentiation timeline suggested that RA treatment was performed at the cardiac progenitor stage in this approach; however, corroborating data are not available. In the study by Zhang et al., atrial versus ventricular phenotype of resulting cells was primarily assessed by a decrease in MLC2V and an increase in MLC2A, as well as ANP protein expression. Furthermore, action potential shape and calcium transient properties were used to differentiate atrial and ventricular cardiomyocytes. Studies performed in hPSCs are summarized in Figure 3.
Figure 3.
Summary of differentiation protocols for the generation of atrial cardiomyocytes from hPSCs
Further evidence for RA-driven atrial specification in hESCs was provided by our group (Devalla et al., 2015). In contrast to the study by Zhang et al. (2011), we found that atrial differentiation relies on the stimulation of RA signaling shortly after the peak expression of MESP1 at the cardiac mesoderm stage. In agreement with concentration-dependent effects of RA, we observed that treatment with a lower concentration of RA (1–10 nM) improved differentiation efficiency but had no effect on subtype specification. This is consistent with findings in mESCs that 1–10 nM RA accelerates cardiac differentiation (Wobus et al., 1997). However, treatment with a higher concentration of RA (1 μM) at late mesoderm stage (day 4) steered differentiation toward atrial fate. Moreover, we identified that atrial-specific genes NR2F1 (COUP-TFI) and NR2F2 (COUP-TFII) are induced in response to RA, as also observed in zebrafish mesoderm (Dohn et al., 2019). These transcription factors bind directly to regulatory DNA sequences of ion channel genes KCNA5 and KCNJ3, which confer unique electrophysiological properties to atrial cells. In addition, in vivo studies identified a role for Nr2f2 in maintaining atrial chamber identity and size (Wu et al., 2013; Duong et al., 2018).
RA-directed atrial fate specification in vitro has been shown to be orchestrated by MEIS2, which antagonizes ISL1, in order to induce the atrial transcription factor, NR2F1 (Quaranta et al., 2018). Mounting evidence suggests that the TALE family of genes such as MEIS1/2 and PBX1–3 are co-factors for HOX-mediated gene regulation (reviewed in Lescroart and Zaffran, 2018). Indeed, putative enhancer regions identified in pSHF cells revealed enrichment for HOX and TALE binding motifs (Stefanovic et al., 2020). While HOX and MEIS genes appear to be direct targets of RA itself (Mercader et al., 2000; Berenguer et al., 2020), studies have shown that they can also regulate Raldh2 expression and thereby RA levels through a positive feedback loop (Vitobello et al., 2011; López-Delgado et al., 2021). Interestingly, misexpression of Hoxb1 in the aSHF domain resulted in an upregulation of Tbx5 and Nr2f2 (Stefanovic et al., 2020), both of which are implicated in the differentiation of atrial and sinoatrial nodal cells in vivo and in vitro (Liberatore et al., 2000; Wu et al., 2013; Devalla et al., 2015; Protze et al., 2017). These findings suggest that co-operative binding of HOX and MEIS proteins to target genes is one of the earliest events directing specification toward atrial and sinoatrial lineages.
Current protocols yield 50%–90% TNNT2+ cells in atrial differentiations. Analyses of these cardiomyocytes by genetic marker expression, such as NR2F2 or by action potential morphology, indicate that a large majority of these cardiomyocytes have an atrial-like phenotype. Nonetheless, the expression of SHOX2 in cells obtained from atrial differentiations suggests the presence of a sinoatrial node-like cardiomyocyte (SANCM) side population in these cultures (Devalla et al., 2015; Cyganek et al., 2018). To improve efficiencies across various cell lines, it is important to better characterize the differentiation process. Besides optimizing timing and concentration of RA application, the induction of appropriate mesoderm has been proposed to regulate differentiation of hPSCs to atrial versus ventricular identities. The RALDH2-expressing mesodermal population has been shown to have a higher potential for atrial fate compared with CYP26A1 (enzyme that degrades RA)-expressing mesoderm, which is better suited for differentiation toward ventricular lineage (Lee et al., 2017). Even though exogenous RA induced >90% TNNT2+ cells from CYP26A1+ mesoderm, expression of atrial genes was lower than in RALDH2+ mesoderm treated with RA. Furthermore, RALDH2+ mesodermal cells treated with 2 μM retinol generated 85% TNNT2+ cells, suggesting that they efficiently produce RA required for differentiation toward the atrial lineage. Nevertheless, reduction in the expression of the ventricular gene MYL2 and increase in expression of the atrial gene KCNJ3 were more pronounced upon treatment with RA itself.
Due to applications in drug screening and disease modeling for atrial fibrillation, there has been a lot of interest in hPSC-derived atrial cells. To fully utilize the potential of these cells, further refinement of their phenotype is required. For example, hPSC-atrial cells exhibit relatively faster beating rates (about 1–1.2 Hz), comparable with hPSC-derived SANCMs, reflecting their immaturity (Devalla et al., 2015; Lee et al., 2017; Protze et al., 2017). Strategies to promote maturation of hPSC-atrial cells are thus important. Moreover, the right versus left atrial identity of these cells has not been evaluated. Right and left atrial cardiomyocytes display differences in gene expression as well as propensity for atrial fibrillation (Van Ouwerkerk et al., 2020). Directed approaches to generate right versus left atrial cells will open avenues for application of these cells in better understanding the etiology of atrial fibrillation.
RA in sinoatrial node development
The sinoatrial node (SAN) is referred to as the pacemaker of the heart, as it generates electrical impulses that set the rate and rhythm of cardiac contractions. At E7.5 in mice and 16- to 23-somite stage in zebrafish, a distinct progenitor population at the lateralmost edge of the pSHF downregulates Nkx2-5 and initiates Tbx18 expression (Mommersteeg et al., 2010; Ren et al., 2019). These progenitors also express Isl1 and are subsequently integrated into the differentiating sinus venosus myocardium at the inflow tract of the expanding heart tube (Christoffels et al., 2006; Mommersteeg et al., 2010). Following looping and remodeling of the linear heart tube, the SAN is established at the entrance of the superior caval vein in the right atrium and takes over as the primary pacemaker at E12.5 in mouse (Yi et al., 2012). The SAN is a structurally and functionally complex tissue, which can be broadly divided into three regions: the head region marked by both Tbx18 and Tbx3 and conspicuous by the absence of Nkx2–5; the tail region marked by the presence of Tbx3 and Nkx2–5 but devoid of Tbx18 (Wiese et al., 2009; Goodyer et al., 2019; Li et al., 2019); and a transition zone with a molecular and functional phenotype intermediate to that of SAN and the adjacent atrial tissue (Boyett et al., 2000; Goodyer et al., 2019; Li et al., 2019).
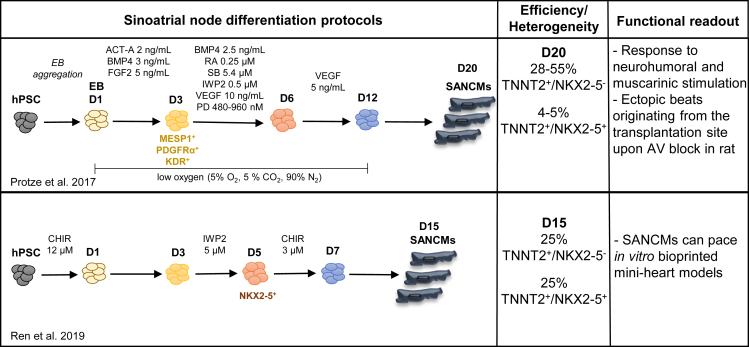
As discussed above, RA signaling is important for the development of atria and sinus venosus. In line with this, RA has also been implicated in the in vitro differentiation of hPSCs to SANCMs (Protze et al., 2017). Treatment of MESP1+ cardiac mesoderm with BMP4 and RA (hereafter BMP4 + RA) resulted in cardiomyocytes with SAN-like molecular and functional profile (Figure 4). The early requirement for BMP and RA signaling in the differentiation of inflow tract myocardium is evidenced by several studies. BMP ligands are involved in the recruitment of pSHF toward the myocardial lineage (Kruithof et al., 2006; Schlueter et al., 2006; van Wijk et al., 2009). Similarly, atrial and SAN progenitors in the pSHF are progressively exposed to RA. The Raldh2-expressing field is initially in close proximity to the pSHF progenitors followed by the expression of Raldh2 in the cardiac progenitor cells themselves (Hochgreb et al., 2003). While it is conceivable that the pSHF progenitors first receive RA via a paracrine action, RARE-lacZ data at these early stages is not available. Nevertheless, pharmacological inhibition of RA signaling in chicken embryos, using an RAR antagonist, revealed an early requirement for RA signaling in the pSHF, even before the onset of Raldh2 expression (Hochgreb et al., 2003).
Figure 4.
Summary of differentiation protocols for the generation of sinoatrial nodal cardiomyocytes from hPSCs
Although application of BMP4 alone at the cardiac mesoderm stage was sufficient to generate the majority of NKX2–5− SAN cardiomyocyte population, addition of RA was crucial to enhance the expression of SAN associated genes such as TBX5 (Protze et al., 2017). This is in accordance with in vivo findings that have demonstrated an association between RA and Tbx5 (Liberatore et al., 2000; De Bono et al., 2018). During embryonic development, Raldh2 is co-expressed with Tbx5 in a subset of pSHF progenitors (Hochgreb et al., 2003; Ryckebusch et al., 2008; De Bono et al., 2018), and Raldh2 knockouts display reduced Tbx5 expression (Niederreither et al., 2001; Sirbu et al., 2008). Furthermore, a recent study revealed that Tbx5 promotes pSHF identity through a positive feedback loop with Raldh2 (Rankin et al., 2021). Importantly, Tbx5 regulates the expression of a key pacemaker gene, Shox2 (Puskaric et al., 2010), which is in turn implicated in regulating other important SAN genes such as Bmp4 and Isl1 (Puskaric et al., 2010; Hoffmann et al., 2013). These findings suggest that RA promotes pacemaker identity by establishing an SAN gene-regulatory network via TBX5.
Interestingly, both in limb and lung development, where Tbx5 plays an important role, the transcription factor interacts with canonical WNT signaling. Whereas in limb development canonical WNT signaling is upstream of Tbx5, in lung development Tbx5 activates the expression of Wnt2 in cardiopulmonary progenitors (Ng et al., 2002; Nishimoto et al., 2015; Steimle et al., 2018; Rankin et al., 2021). WNT signaling has also been identified as a critical cue for pacemaker development (Bressan et al., 2013; Ren et al., 2019). An alternative approach described for the differentiation of hPSCs to SAN cardiomyocytes showed that the activation of canonical WNT signaling is sufficient to enforce pacemaker identity (Ren et al., 2019) (Figure 4). One of the key differences between the BMP4 + RA and the WNT protocols is the timing of signaling modulation. While BMP4 + RA is applied at the cardiac mesoderm stage (MESP1+), activation of canonical WNT signaling is performed at the cardiac progenitor stage (NKX2–5+). It is also noteworthy that BMP4 + RA treatment is performed in conjunction with inhibition of WNT signaling using small-molecule inhibitors such as IWP2. During early stages of cardiogenesis, a biphasic antagonistic role for WNT signaling has been identified (Naito et al., 2006; Ueno et al., 2007). These processes are simulated in cardiac differentiation protocols, where WNT signaling is first activated in hPSCs and subsequently inhibited upon cardiac mesoderm induction to promote cardiomyocyte formation. Following the inhibition of WNT signaling at cardiac mesoderm stage, which results in NKX2–5+ cardiac progenitors, canonical WNT signaling was activated to direct cells toward the SAN lineage.
Similar to the BMP4 + RA protocol for SANCM differentiation (Protze et al., 2017), the WNT protocol also resulted in cardiomyocytes expressing key SAN genes such as SHOX2, ISL1, and BMP4 (Ren et al., 2019). Bmp4, as well as Isl1, have been identified as downstream targets of canonical WNT signaling in cardiac progenitors (Klaus et al., 2007; Lin et al., 2007), which likely mediate the differentiation toward pacemaker cells along with Tbx5, as discussed above. Interestingly, Tbx5 and Bmp4 are also crucial for the specification of the proepicardium (Liu and Stainier, 2010). Previous studies have shown that the proepicardial cells and the myocardial cells of the inflow tract diversify from a common progenitor population (van Wijk et al., 2009; Mommersteeg et al., 2010). Comparably, the non-cardiomyocyte population obtained from both the BMP4 + RA and the WNT differentiation protocols is of epicardial identity (Ren et al., 2019; A.W., and H.D.D., unpublished observations). Further research is needed to evaluate the crosstalk between BMP, RA, and WNT signaling in SANCM and epicardial differentiation.
As the SAN is composed of several subpopulations, it remains to be evaluated whether both BMP4 + RA and WNT differentiation protocols generate the same cellular subtypes of the SAN. It has recently been demonstrated that a subpopulation of the SAN expressing Shox2 and Nkx2–5 (“SAN junction cells” of the transitional zone) plays a crucial role in atrial activation (Li et al., 2019). Both differentiation approaches generate around 55% of TNNT2+ cardiomyocytes (CMs). However, only about half of the TNNT2+ cells are NKX2–5− in the WNT-based protocol (Ren et al., 2019), a gene expression pattern found in SAN head cells in vivo (Wiese et al., 2009; Goodyer et al., 2019). Whether the remaining NKX2–5+ cardiomyocyte fraction corresponds to SAN transitional zone populations is not known (Ren et al., 2019). On the other hand, the BMP4 + RA-based protocol results in 55% NKX2–5− CMs and only 5% NKX2–5+ CMs (Protze et al., 2017), indicating preferential differentiation toward SAN head. The outcome of the two protocols might, however, be influenced not only by the different signaling modulators used but also by different culture conditions (two-dimensional [2D] versus three-dimensional [3D]).
All in all, a better understanding of the signaling pathways underlying SAN development including the association between BMP, RA, and WNT signaling would provide opportunities for improving differentiation efficiencies and obtain desired cellular fractions from hPSCs. Importantly, methods to differentiate cells to specific subpopulations of the SAN, such as the head, tail, and transition zone, will enable the creation of advanced cellular models relevant for disease modeling and drug screenings as well as informing the application of these cell types for therapeutic purposes.
RA in epicardial development
The epicardium develops from a transient structure called the proepicardium, located at the venous pole of the developing heart. The proepicardium is characterized by the ubiquitous expression of Tbx18, Tcf21, and Wt1 among other genes (Lupu et al., 2020). The precise origins of the proepicardium are poorly understood, and recent insights reveal that it receives contributions from multiple progenitor populations (Mommersteeg et al., 2010; Tyser et al., 2021; Zhang et al., 2021). It has been identified that a population anterior to the FHF has the potential to form myocardial components such as left ventricle, atrioventricular canal, and proepicardium (Tyser et al., 2021; Zhang et al., 2021). Similarly, tetramethylindocarbocyanine (DiI) labeling in chick and mouse embryos has shown that a Tbx18+ population lateral in the pSHF contributes to both the proepicardium and the sinus venosus myocardium (van Wijk et al., 2009; Mommersteeg et al., 2010). Although RA signaling in the pSHF is necessary for the differentiation of atrial and sinus venosus myocardium, its requirement for the formation of the proepicardium is not known. As depicted in Figure 2, Raldh2 expression begins in the proepicardium and is also found in the epicardium at E12.5. Loss of Raldh2 does not impair the formation of proepicardium itself. However, ventricular growth and formation of coronary vasculature were severely affected in Raldh2-deficient mice (Niederreither et al., 2001; Lin et al., 2010).
RA signaling in the epicardium is regulated by WT1 through direct activation of Raldh2 (Guadix et al., 2011). Raldh2 is expressed in a spatiotemporal pattern similar to that of Wt1, and structural defects in mice lacking either gene were found to be similar (Perez-Pomares et al., 2002; Guadix et al., 2011; von Gise et al., 2011). Raldh2 levels in Wt1 knockout mice were more severely reduced in ventricular epicardium compared with atrial epicardium (Guadix et al., 2011). Besides Wt1, another important proepicardial and epicardial gene, Tcf21, is also associated with RA signaling. Tcf21 is induced by RA treatment, which promotes fibroblast differentiation at the expense of smooth muscle cells (Acharya et al., 2012; Braitsch et al., 2012). Similarly, in Dhrs3−/− hearts with increased RA synthesis, an increase in fibroblasts was observed (Xiao et al., 2018). In vivo findings thus suggest that RA may not be required for proepicardial specification itself but that it has a critical role in the subsequent development of the epicardium and further differentiation toward fibroblasts.
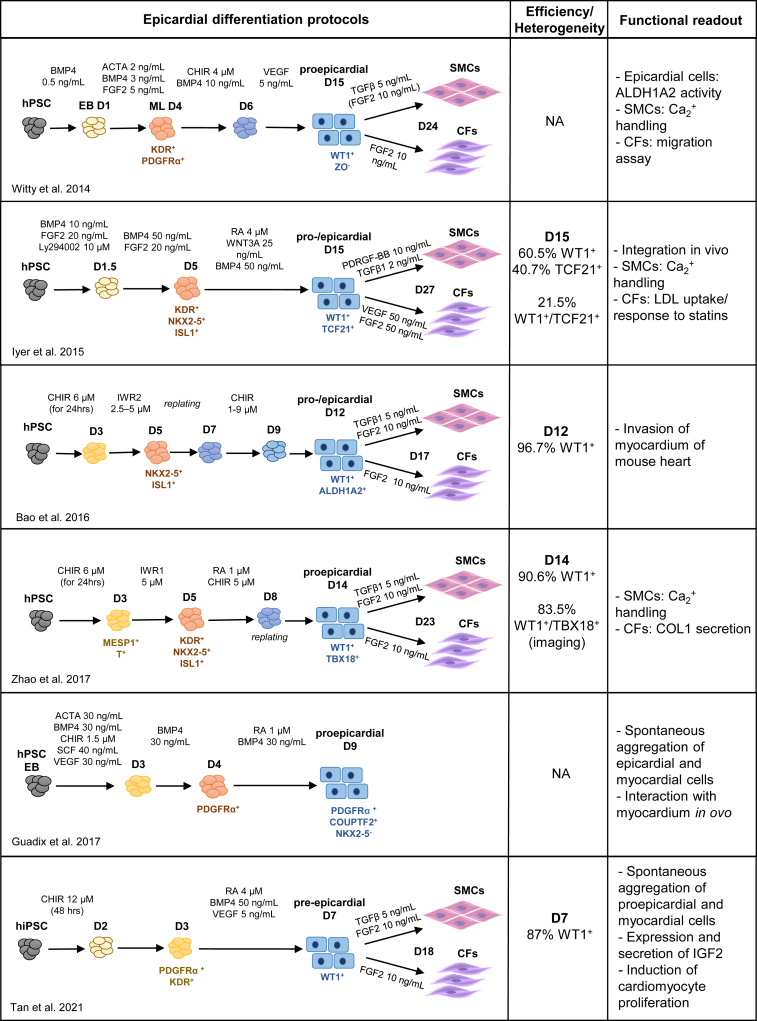
In contrast to in vivo findings, in vitro studies show that treatment with RA steers proepicardial differentiation of hPSCs (Iyer et al., 2015; Guadix et al., 2017; Zhao et al., 2017; Tan et al., 2021). Manipulation of BMP, WNT, RA signaling pathways, or a combination of these allowed the generation of epicardial-like cells from hPSCs (Figure 5) (Witty et al., 2014; Iyer et al., 2015; Bao et al., 2016; Guadix et al., 2017; Zhao et al., 2017). In a more recent protocol, BMP4 + RA + vascular endothelial growth factor (VEGF) was used to generate proepicardial cells; however, the added advantage of including VEGF as opposed to using BMP4 + RA alone was not investigated (Tan et al., 2021). Like RA, WNT signaling is also only implicated in later stages of epicardial development, epicardial EMT, and formation of coronary vessels in vivo (Zamora et al., 2007; von Gise et al., 2011). However, there is overwhelming evidence implicating Bmp4 in proepicardial development as well as epicardial migration (Schlueter et al., 2006; Ishii et al., 2010; Liu and Stainier, 2010). As discussed above, RA, BMP, and WNT pathways are also implicated in SAN differentiation, reflecting shared ontogeny with proepicardial cells (van Wijk et al., 2009; Mommersteeg et al., 2010) (Figure 6). Interestingly, the common link between RA, WNT, and BMP pathways appears to be TBX5. Besides its role in the pSHF, Tbx5 is also implicated in proepicardial development and epicardial migration (Liu and Stainier, 2010; Diman et al., 2014). Proepicardial cells generated from hPSCs in vitro using BMP4 + RA also express Tbx5 (Tan et al., 2021). Moreover, microarray analysis of Tbx5-induced genes identified Wt1 as a possible target (Plageman and Yutzey, 2006).
Figure 5.
Summary of differentiation protocols for the generation of (pro)epicardial cells from hPSCs
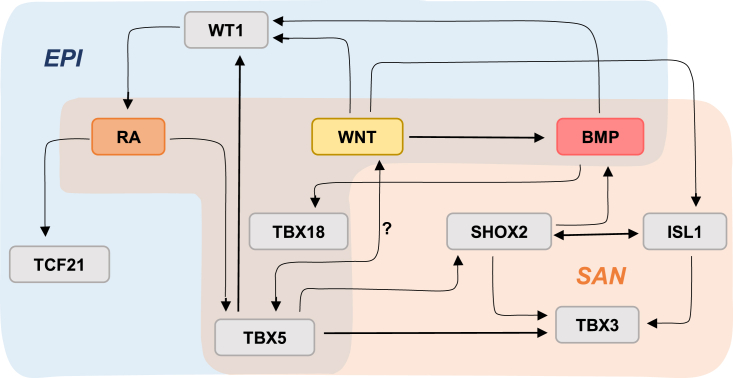
Figure 6.
Transcriptional networks involved in epicardial (EPI) and sinoatrial node (SAN) development
RA, WNT, and BMP signaling are implicated in EPI (blue) and SAN (orange) specification. Both TBX18 and TBX5 are expressed in EPI and SAN progenitors where they regulate a number of cell-type-specific target genes.
Timing of application and/or concentration of signaling modulators appears to delineate SAN versus epicardial differentiation. In about half of the epicardial differentiation protocols, signaling modulators are introduced at the cardiac progenitor stage marked by NKX2–5 and ISL1 expression (Iyer et al., 2015; Bao et al., 2016; Zhao et al., 2017). This is in accordance with in vivo findings that revealed the contribution of NKX2–5+/ISL1+ cardiac progenitors to the proepicardium (Zhou et al., 2008). In other protocols, an earlier population at the cardiac mesoderm stage marked by PDGFRα was steered toward the epicardial lineage (Witty et al., 2014; Guadix et al., 2017; Tan et al., 2021), implying that there are multiple differentiation routes toward epicardial cells. Whether these correspond to the different progenitor populations as shown in vivo is yet to be evaluated (Mommersteeg et al., 2010; Tyser et al., 2021; Zhang et al., 2021).
Activation either of canonical WNT signaling alone (Bao et al., 2016) or in combination with BMP4 (Witty et al., 2014) or RA (Zhao et al., 2017) or both BMP4 and RA (Iyer et al., 2015; Tan et al., 2021) was used in these methods (Figure 5). Inhibition of canonical WNT signaling resulted in a decrease in WT1, indicating that at least endogenous WNT signaling is required for epicardial differentiation (Witty et al., 2014; Iyer et al., 2015). The differences in the choice of signaling molecules and the timing of application likely explains the variations in efficiency and heterogeneity observed in the resulting epicardial populations (Figure 5). The percentage of WT1+ cells was greater (>90%) in protocols that used either WNT activation alone (Bao et al., 2016) or WNT + RA (Zhao et al., 2017). In the WNT + BMP4 + RA protocol (Iyer et al., 2015), only 20% of the end population was positive for both WT1 and TCF21. On the contrary, combined treatment of WNT + RA yielded 80% of cells positive for both WT1 and TBX18 (Zhao et al., 2017). Similarly, treatment with BMP4 + RA + VEGF yielded a high percentage of proepicardial cells that co-expressed WT1 and ZO1 (Tan et al., 2021). As shown in a recent study in the mouse heart, typical epicardial markers are co-expressed in the proepicardium and epicardium until E13.5, as opposed to the septum transversum, where a more heterogeneous expression of the same markers is found (Lupu et al., 2020). This finding suggests that better characterization is needed in order to determine the identity of WT1+ cells in vitro.
An important criterion for assessing the functionality of hPSC-derived epicardial cells is their ability to generate epicardial-derived cell populations (EPDCs). Epicardial-derived progenitor cells undergo EMT and give rise to EPDCs such as fibroblasts and smooth muscle cells (Quijada et al., 2020). These cell types are valuable for disease modeling and tissue engineering applications aimed at reconstructing the microenvironment of the heart. Epicardial cells generated from most protocols in vitro undergo EMT and further differentiate to smooth muscle cells and fibroblasts upon manipulation of transforming growth factor beta (TGFB) and fibroblast growth factor (FGF) pathways, respectively (Witty et al., 2014; Iyer et al., 2015; Zhao et al., 2017; Tan et al., 2021). Furthermore, epicardial cells and cardiac fibroblasts generated from such two-step protocols are being implemented in disease modeling, in maturation of hPSC CMs, and even in augmenting heart regeneration after myocardial infarction (Bargehr et al., 2019; Giacomelli et al., 2020; Tan et al., 2021). Collectively, data from the various differentiation approaches suggest that there are multiple strategies to obtain epicardial cells from hPSCs, which induce a transcriptional cascade involving WT1, RALDH2, and TBX18. However, additional insight into the heterogeneity in the resulting populations, as well as a uniform set of markers and parameters used for evaluation of epicardial and EPDC phenotypes, must still be established.
RA in ventricular specification and maturation
The FHF and the aSHF give rise to the left and right ventricles, respectively (Zaffran et al., 2004; Verzi et al., 2005). The majority of ventricular cardiomyocytes are derived from Mesp1+ cardiac progenitors (Saga et al., 1999). More recently, it has been found that derivatives of two temporally distinct Foxa2+ progenitor populations also contribute to 30%–50% of ventricular cardiomyocytes (Bardot et al., 2017; Ivanovitch et al., 2021). Whether these Foxa2+ cells constitute a subfraction of Mesp1+ progenitors or an altogether independent population remains to be determined. In the absence of RA signaling in mice, development of FHF-derived left ventricle proceeds normally (Ryckebusch et al., 2008). In zebrafish, however, RA-deficient embryos present significant expansion of a differentiating FHF population accompanied by a reduction in aSHF progenitors (Duong et al., 2021). In the SHF, RA maintains the border between aSHF and pSHF progenitors. Right ventricular progenitors located in the aSHF are demarcated by the expression of Tbx1. Tbx1 positively regulates Cyp26a1–c1 genes involved in RA degradation, thereby antagonizing RA (Roberts et al., 2006). Expectedly, the treatment of pregnant mice at E7.75–E8.25 with a pan-RAR antagonist did not affect the right ventricle of the examined embryos (De Bono et al., 2018). Moreover, exogenous RA resulted in a hypoplastic ventricle by impairing aSHF differentiation toward the ventricular lineage (Bardot et al., 2017; Gonzalez et al., 2021). Similarly, in zebrafish, RA-induced nr2f genes were responsible for restricting ventricular CM numbers while promoting pharyngeal muscle development (Duong et al., 2018). Altogether, these findings suggest that RA is not required and is even detrimental for ventricular specification. In contrast, strong evidence has underscored the requirement of RA in later stages of ventricular development and maturation.
At E9.5 of mouse development, the ventricular wall comprises a very thin layer of myocardium. Growth of the ventricles, which involves trabeculation and subsequent thickening of the compact layer, requires proliferative and other morphogenic signals that are initiated around E9.5–E10.5. A majority of the paracrine signals that drive ventricular compact wall expansion are thought to be derived from the epicardium (Sucov et al., 2009). Epicardial migration begins at E9.5 and entirely covers the outer surface of the heart by E10.5 (Viragh and Challice, 1981), coinciding with ventricular expansion. In Raldh2 knockout mouse embryos, abnormal looping results in a single ventricle, which lacks trabeculae (Niederreither et al., 1999). Ventricular hypoplasia was also observed in vitamin A-deficient rat progeny (Wilson and Warkany, 1949; Niederreither et al., 2001). Subsequent studies showed that the effect of RA on the ventricles is mediated by RXRα receptors in the epicardium (Merki et al., 2005). Moreover, ventricular defects observed in germline Rxrα knockout and Raldh2 knockout embryos resemble the ventricular phenotype caused by defective epicardium (Takahashi et al., 2014).
In the majority of hPSC differentiation studies, obtained cardiomyocytes are a heterogeneous mix of various cardiac subtypes, with ventricular cardiomyocytes being the predominant group (Blazeski et al., 2012; Cyganek et al., 2018; Friedman et al., 2018). It is worth noting that in the absence of RA or other posterior fate-inducing signals, the default differentiation path of MESP1+ cardiac mesoderm cells in vitro is toward the ventricular lineage in the majority of cell lines and existing differentiation protocols. The RA-degrading enzyme CYP26A1 is expressed at day 3 of differentiation, coinciding with peak MESP1 expression, and is more suited for differentiation toward ventricular cardiomyocytes (Lee et al., 2017; Calderon et al., 2021; H.D.D., unpublished observations). Furthermore, treatment with an RA antagonist during differentiation, presumably at the cardiac mesoderm stage (day 3) or cardiac progenitor stage (day 5), steered cells toward a ventricular fate (Zhang et al., 2011; Tan et al., 2021).
It is well known that PSC-derived cardiomyocytes exhibit immature structural, metabolic, and electrophysiological properties, corresponding to fetal rather than adult cardiomyocytes. The effect of RA on maturation of PSC-derived cardiomyocytes has not yet been assessed. Nevertheless, as the effect of RA is mediated by paracrine factors from the epicardium such as insulin-like growth factor (IGF) and FGF, approaches that recapitulate epicardial-myocardial interactions have been probed. Co-culture of hPSC-derived epicardial cells with cardiomyocytes in 2D cultures as well as 3D constructs enhanced contractility and calcium handling properties of the cardiomyocytes. It was shown that the proliferative effect of proepicardial cells on co-cultured ventricular cardiomyocytes was mediated at least in part by RA-dependent IGF2 signaling (Tan et al., 2021). IGF2 has also been utilized for the maturation of compact ventricular cells derived from hPSCs (Funakoshi et al., 2021). IGF2 was strongly decreased in both Raldh2−/− and Rxrα mutants and is implicated in ventricular wall proliferation (Brade et al., 2011; Li et al., 2011). Furthermore, several studies have shown that IGF1 in combination with thyroid hormone T3 and dexamethasone or neuregulin promotes maturation of hPSC-derived cardiomyocytes in 2D and 3D cultures (Birket et al., 2015; Rupert and Coulombe, 2017; Huang et al., 2020). A head-to-head comparison of the direct effects of secreted epicardial factors versus co-culture with epicardial cells or epicardial-derived fibroblasts on hPSC-derived cardiomyocytes is warranted for identification of the best strategies to promote maturation in vitro.
Concluding perspectives
Cardiac development is a complex process that relies on coordinated spatiotemporal interactions of signaling pathways, transcriptional regulators, and epigenetic modifiers. As presented in this review, RA signaling is indispensable in cardiogenesis for the development of atria and sinus venosus during early embryogenesis and for ventricular growth and expansion at later stages. In vitro, RA is utilized in the differentiation of multiple cardiac cell types from hPSCs (Figure 7). Addition of RA to atrial and sinoatrial differentiations from hPSCs mimics the earliest stages of heart development synonymous with the expression of Raldh2 in pSHF progenitors. Timing and concentration of signaling modulators are crucial for successful differentiation of the desired cardiac cell type. Whereas SANCM differentiation uses lower concentrations of BMP4 (2.5 ng/mL) and RA (0.25 μM), epicardial cell specification is induced by higher concentrations of BMP4 (10–50 ng/mL) and RA (1–4 μM). It is worth noting that the important difference in the SAN and epicardial differentiation protocols, which use canonical WNT signaling alone, is the timing of application (Bao et al., 2016; Ren et al., 2019). In SAN differentiation, WNT signaling is activated at day 5 upon the expression of NKX2–5. In epicardial differentiation, on the other hand, cells are replated and cultured for 48 h prior to WNT activation. For the differentiation of atrial cells from hPSCs, activation of RA signaling is the only known strategy thus far. The majority of the studies demonstrate that application of RA (0.5–1 μM) shortly after mesoderm induction directs hPSCs toward an atrial fate.
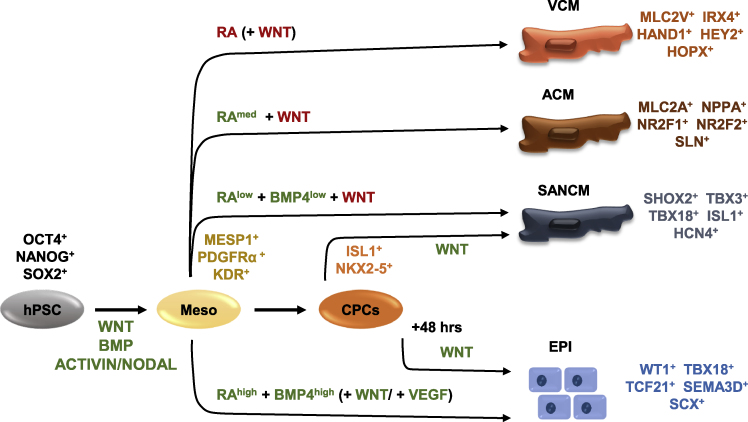
Figure 7.
Schematic overview of the differentiation of various cardiac cell types from hPSCs
Signaling pathways implicated in each step are indicated in green (activation) or red (inhibition). Genes that are typically used as markers for each cell type are also mentioned. The majority of the protocols rely on modulation of signaling pathways at the cardiac mesoderm stage, marked by MESP1, PDGFRα, or KDR. Inhibition of RA signaling in combination with inhibition of WNT or inhibition of WNT alone gives rise to VCM. Activation of RA signaling (using intermediate concentrations of 0.5–1 μM RA) in conjunction with inhibition of WNT signaling generates ACM. Activation of RA (using lower concentrations of 0.25 μM RA) along with activation of BMP signaling and inhibition of WNT signaling yields SANCM. SANCM can also be generated by activating WNT signaling at the cardiac progenitor stage, marked by ISL1 and NKX2–5. Replating ISL1+/NKX2–5+ cardiac progenitors and subsequent activation of WNT signaling results in (pro)epicardial cells. Alternatively, activation of RA signaling (using higher concentration of 1–4 μM RA) alongside activation of BMP and WNT/VEGF signaling also gives rise to (pro)epicardial cells. ACM, atrial-like cardiomyocytes; CPC, cardiovascular progenitor cells; EPI, proepicardial/epicardial cells; hPSCs, human pluripotent stem cells; KDR, kinase insert domain receptor; Meso, mesoderm; SANCM, sinoatrial nodal-like cardiomyocytes; VCM, ventricular-like cardiomyocytes.
Furthermore, aforementioned in vitro studies point to a concerted action of RA, BMP, and WNT pathways in the specification of SANCMs and epicardial cells. Interestingly, BMP and WNT signaling co-operatively induce Hox gene expression required for the specification of hematopoietic cells from ventral mesoderm (Lengerke et al., 2008). Similarly, WNT-dependent activation of Hox genes has been reported in the early embryo (Neijts et al., 2016). Whether a similar mechanism could explain the findings in the context of cardiac differentiations, in which BMP and/or WNT are used in lieu of RA, is yet to be determined. Moving forward, studies to better define the hierarchy and interaction between HOX, TALE family members such as MEIS1/2, NR2F transcription factors, and TBX5 will shed light on the molecular mechanisms underlying RA-driven cardiac cell specification. In particular, tissue-specific co-factors and targets of TBX5, which discern epicardial, atrial, or sinoatrial lineage identity, are to be investigated. Directed differentiation of hPSCs to cardiac cell types will serve as an excellent model in this context. Sampling of cells at various time points during differentiation and comprehensive analysis of the genetic and epigenetic changes will unravel cell-type-specific gene-regulatory networks. Such insights will in turn inform further improvement of in vitro differentiation protocols and even trigger the development of systems and synthetic biology approaches to precisely modulate cell fate in a dish.
Author contributions
Conceptualization, data curation, and writing, A.W. and H.D.D.; visualization, A.W.; review and editing, H.D.D., V.M.C., and G.J.J.B.; supervision, H.D.D.
Conflicts of interest
G.J.J.B. is a co-founder of Pacingcure.
Acknowledgments
This work is supported by funding from European Research Council starting grant 714866 and associated proof-of-concept grant 899422, Health Holland LentiPace II to G.J.J.B.; Netherlands Organization for Health Research and Development (ZonMW), ZonMW TOP 40-00812-98-17061 to V.M.C., ZonMW and the Dutch Heart Foundation MKMD grant 114021512, and Dutch Heart Foundation Dekker fellowship 2020T023 to H.D.D.
References
- Abu-Abed S., MacLean G., Fraulob V., Chambon P., Petkovich M., Dollé P. Differential expression of the retinoic acid-metabolizing enzymes CYP26A1 and CYP26B1 during murine organogenesis. Mech. Dev. 2002;110:173–177. doi: 10.1016/s0925-4773(01)00572-x. 10.1016/S0925-4773(01)00572-X [DOI] [PubMed] [Google Scholar]
- Acharya A., Baek S.T., Huang G., Eskiocak B., Goetsch S., Sung C.Y., Banfi S., Sauer M.F., Olsen G.S., Duffield J.S., et al. The bHLH transcription factor Tcf21 is required for lineagespecific EMT of cardiac fibroblast progenitors. Development (Cambridge) 2012;139:2139–2149. doi: 10.1242/dev.079970. 10.1242/dev.079970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao X., Lian X., Hacker T.A., Schmuck E.G., Qian T., Bhute V.J., Han T., Shi M., Drowley L., Plowright A.T., et al. Long-term self-renewing human epicardial cells generated from pluripotent stem cells under defined xeno-free conditions. Nat. Biomed. Eng. 2016;1 doi: 10.1038/s41551-016-0003. 10.1038/s41551-016-0003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bardot E., Calderon D., Santoriello F., Han S., Cheung K., Jadhav B., Burtscher I., Artap S., Jain R., Epstein J., et al. Foxa2 identifies a cardiac progenitor population with ventricular differentiation potential. Nat. Commun. 2017;8:1–15. doi: 10.1038/ncomms14428. 10.1038/ncomms14428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bargehr J., Ong L.P., Colzani M., Davaapil H., Hofsteen P., Bhandari S., Gambardella L., Le Novère N., Iyer D., Sampaziotis F., et al. Epicardial cells derived from human embryonic stem cells augment cardiomyocyte-driven heart regeneration. Nat. Biotechnol. 2019;37:895–906. doi: 10.1038/s41587-019-0197-9. 10.1038/s41587-019-0197-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berenguer M., Meyer K.F., Yin J., Duester G. Discovery of genes required for body axis and limb formation by global identification of retinoic acid-regulated epigenetic marks. PLoS Biol. 2020;18 doi: 10.1371/journal.pbio.3000719. 10.1371/journal.pbio.3000719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertrand N., Roux M., Ryckebüsch L., Niederreither K., Dollé P., Moon A., Capecchi M., Zaffran S. Hox genes define distinct progenitor sub-domains within the second heart field. Dev. Biol. 2011;353:266–274. doi: 10.1016/j.ydbio.2011.02.029. 10.1016/j.ydbio.2011.02.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Billings S.E., Pierzchalski K., Tjaden N.E.B., Pang X., Trainor P.A., Kane M.A., Moise A.R. The retinaldehyde reductase DHRS3 is essential for preventing the formation of excess retinoic acid during embryonic development. FASEB J. 2013;27:4877–4889. doi: 10.1096/fj.13-227967. 10.1096/fj.13-227967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birket M.J., Ribeiro M.C., Kosmidis G., Ward D., Leitoguinho A.R., van de Pol V., Dambrot C., Devalla H.D., Davis R.P., Mastroberardino P.G., et al. Contractile defect caused by mutation in MYBPC3 revealed under conditions optimized for human PSC-cardiomyocyte function. Cell Rep. 2015;13:733–745. doi: 10.1016/j.celrep.2015.09.025. 10.1016/j.celrep.2015.09.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blazeski A., Zhu R., Hunter D.W., Weinberg S.H., Zambidis E.T., Tung L. Cardiomyocytes derived from human induced pluripotent stem cells as models for normal and diseased cardiac electrophysiology and contractility. Prog. Biophys. Mol. Biol. 2012:166–177. doi: 10.1016/j.pbiomolbio.2012.07.013. 10.1016/j.pbiomolbio.2012.07.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Bono C., Thellier C., Bertrand N., Sturny R., Jullian E., Cortes C., Stefanovic S., Zaffran S., Théveniau-Ruissy M., Kelly R.G. T-box genes and retinoic acid signaling regulate the segregation of arterial and venous pole progenitor cells in the murine second heart field. Hum. Mol. Genet. 2018;27:3747–3760. doi: 10.1093/hmg/ddy266. 10.1093/hmg/ddy266 [DOI] [PubMed] [Google Scholar]
- Boyett M.R., Honjo H., Kodama I. The sinoatrial node, a heterogeneous pacemaker structure. Cardiovasc. Res. 2000:658–687. doi: 10.1016/s0008-6363(00)00135-8. 10.1016/S0008-6363(00)00135-8 [DOI] [PubMed] [Google Scholar]
- Brade T., Kumar S., Cunningham T.J., Chatzi C., Zhao X., Cavallero S., Li P., Sucov H.M., Ruiz-Lozano P., Duester G. Retinoic acid stimulates myocardial expansion by induction of hepatic erythropoietin which activates epicardial Igf2. Development. 2011;138:139–148. doi: 10.1242/dev.054239. 10.1242/dev.054239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braitsch C.M., Combs M.D., Quaggin S.E., Yutzey K.E. Pod1/Tcf21 is regulated by retinoic acid signaling and inhibits differentiation of epicardium-derived cells into smooth muscle in the developing heart. Dev. Biol. 2012;368:345–357. doi: 10.1016/j.ydbio.2012.06.002. 10.1016/j.ydbio.2012.06.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bressan M., Liu G., Mikawa T. Early mesodermal cues assign avian cardiac pacemaker fate potential in a tertiary heart field. Science. 2013;340:744–748. doi: 10.1126/science.1232877. 10.1126/science.1232877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruneau B.G., Logan M., Davis N., Levi T., Tabin C.J., Seidman J.G., Seidman C.E. Chamber-specific cardiac expression of Tbx5 and heart defects in Holt-Oram syndrome. Dev. Biol. 1999;211:100–108. doi: 10.1006/dbio.1999.9298. 10.1006/dbio.1999.9298 [DOI] [PubMed] [Google Scholar]
- Calderon D., Wickramasinghe N., Sarrafha L., Schaniel C., Chen S., Tomishima M., Dubois N.C. Identification and characterization of hPSC-derived FOXA2+ progenitor cells with ventricular cardiac differentiation potential. bioRxiv. 2021:2021. 10.1101/2021.07.18.452860 [Google Scholar]
- Christoffels V.M., Mommersteeg M.T.M., Trowe M.O., Prall O.W.J., De Gier-De Vries C., Soufan A.T., Bussen M., Schuster-Gossler K., Harvey R.P., Moorman A.F.M., et al. Formation of the venous pole of the heart from an Nkx2-5-negative precursor population requires Tbx18. Circ. Res. 2006;98:1555–1563. doi: 10.1161/01.RES.0000227571.84189.65. 10.1161/01.RES.0000227571.84189.65 [DOI] [PubMed] [Google Scholar]
- Cyganek L., Tiburcy M., Sekeres K., Gerstenberg K., Bohnenberger H., Lenz C., Henze S., Stauske M., Salinas G., Zimmermann W.H., et al. Deep phenotyping of human induced pluripotent stem cell-derived atrial and ventricular cardiomyocytes. JCI Insight. 2018;3 doi: 10.1172/jci.insight.99941. 10.1172/jci.insight.99941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawson M.I., Xia Z. The retinoid X receptors and their ligands. Biochim. Biophys. Acta - Mol. Cell Biol. Lipids. 2012:21–56. doi: 10.1016/j.bbalip.2011.09.014. 10.1016/j.bbalip.2011.09.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devalla H.D., Schwach V., Ford J.W., Milnes J.T., El-Haou S., Jackson C., Gkatzis K., Elliott D.A., Chuva de Sousa Lopes S.M., Mummery C.L., et al. ‘Atrial-like cardiomyocytes from human pluripotent stem cells are a robust preclinical model for assessing atrial-selective pharmacology. EMBO Mol. Med. 2015;7:394–410. doi: 10.15252/emmm.201404757. 10.15252/emmm.201404757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devine W.P., Wythe J.D., George M., Koshiba-Takeuchi K., Bruneau B.G. Early patterning and specification of cardiac progenitors in gastrulating mesoderm. eLife. 2014;3 doi: 10.7554/eLife.03848. 10.7554/eLife.03848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diman N.Y.S.G., Brooks G., Kruithof B.P.T., Elemento O., Seidman J.G., Seidman C.E., Basson C.T., Hatcher C.J. Tbx5 is required for avian and mammalian epicardial formation and coronary vasculogenesis. Circ. Res. 2014;115:834–844. doi: 10.1161/CIRCRESAHA.115.304379. 10.1161/CIRCRESAHA.115.304379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dohn T.E., Ravisankar P., Tirera F.T., Martin K.E., Gafranek J.T., Duong T.B., VanDyke T.L., Touvron M., Barske L.A., Crump J.G., et al. Nr2f-dependent allocation of ventricular cardiomyocyte and pharyngeal muscle progenitors. PLoS Genet. 2019;15:e1007962. doi: 10.1371/journal.pgen.1007962. 10.1371/journal.pgen.1007962 Edited by M.C. Mullins. Public Library of Science. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dollé P. Developmental expression of retinoic acid receptors (RARs) Nucl. Receptor Signal. 2009:6. doi: 10.1621/nrs.07006. 10.1621/nrs.07006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domínguez J.N., Meilhac S.M., Bland Y.S., Buckingham M.E., Brown N.A. ‘Asymmetric fate of the posterior part of the second heart field results in unexpected left/right contributions to both poles of the heart. Circ. Res. 2012;111:1323–1335. doi: 10.1161/CIRCRESAHA.112.271247. 10.1161/CIRCRESAHA.112.271247 [DOI] [PubMed] [Google Scholar]
- Duester G. Retinoic acid synthesis and signaling during early organogenesis. Cell. 2008:921–931. doi: 10.1016/j.cell.2008.09.002. 10.1016/j.cell.2008.09.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duong T.B., Ravisankar P., Song Y.C., Gafranek J.T., Rydeen A.B., Dohn T.E., Barske L.A., Crump J.G., Waxman J.S. Nr2f1a balances atrial chamber and atrioventricular canal size via BMP signaling-independent and -dependent mechanisms. Dev. Biol. 2018;434:7–14. doi: 10.1016/j.ydbio.2017.11.010. 10.1016/j.ydbio.2017.11.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duong T.B., Holowiecki A., Waxman J.S. Retinoic acid signaling restricts the size of the first heart field within the anterior lateral plate mesoderm. Dev. Biol. 2021;473:119–129. doi: 10.1016/j.ydbio.2021.02.005. 10.1016/j.ydbio.2021.02.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman C.E., Nguyen Q., Lukowski S.W., Tam P.P.L., Powell J.E., Palpant N.J. Single-cell transcriptomic analysis of cardiac differentiation from human PSCs reveals HOPX-dependent cardiomyocyte maturation. Cell Stem Cell. 2018;23:586–598. doi: 10.1016/j.stem.2018.09.009. 10.1016/j.stem.2018.09.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funakoshi S., Fernandes I., Mastikhina O., Wilkinson D., Tran T., Dhahri W., Mazine A., Yang D., Burnett B., Lee J., et al. Generation of mature compact ventricular cardiomyocytes from human pluripotent stem cells. Nat. Commun. 2021;12:1–23. doi: 10.1038/s41467-021-23329-z. 10.1038/s41467-021-23329-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galli D., Domínguez J.N., Zaffran S., Munk A., Brown N.A., Buckingham M.E. Atrial myocardium derives from the posterior region of the second heart field, which acquires left-right identity as Pitx2c is expressed. Development (Cambridge, England) 2008;135:1157–1167. doi: 10.1242/dev.014563. 10.1242/dev.014563 [DOI] [PubMed] [Google Scholar]
- Gassanov N., Er F., Zagidullin N., Jankowski M., Gutkowska J., Hoppe U.C. Retinoid acid-induced effects on atrial and pacemaker cell differentiation and expression of cardiac ion channels. Differentiation. 2008;76:971–980. doi: 10.1111/j.1432-0436.2008.00283.x. 10.1111/j.1432-0436.2008.00283.x [DOI] [PubMed] [Google Scholar]
- Giacomelli E., Meraviglia V., Campostrini G., Cochrane A., Cao X., van Helden R.W.J., Krotenberg Garcia A., Mircea M., Kostidis S., Davis R.P., et al. Human-iPSC-derived cardiac stromal cells enhance maturation in 3d cardiac microtissues and reveal non-cardiomyocyte contributions to heart disease. Cell Stem Cell. 2020;26:862–879.e11. doi: 10.1016/j.stem.2020.05.004. 10.1016/j.stem.2020.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Gise A., Zhou B., Honor L.B., Ma Q., Petryk A., Pu W.T. WT1 regulates epicardial epithelial to mesenchymal transition through β-catenin and retinoic acid signaling pathways. Dev. Biol. 2011;356:421–431. doi: 10.1016/j.ydbio.2011.05.668. 10.1016/j.ydbio.2011.05.668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez D.M., Schrode N., Ebrahim T., Beaumont K.G., Sebra R., Dubois N.C., Dubois -- N. Understanding mechanisms of chamber-specific differentiation through combination of lineage tracing and single cell transcriptomics. bioRxiv. 2021:2021. 10.1101/2021.07.15.452540 [Google Scholar]
- Goodyer W.R., Beyersdorf B.M., Paik D.T., Tian L., Li G., Buikema J.W., Chirikian O., Choi S., Venkatraman S., Adams E.L., et al. Transcriptomic profiling of the developing cardiac conduction system at single-cell resolution. Circ. Res. 2019;125:379–397. doi: 10.1161/CIRCRESAHA.118.314578. 10.1161/CIRCRESAHA.118.314578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guadix J.A., Ruiz-Villalba A., Lettice L., Velecela V., Muñoz-Chápuli R., Hastie N.D., Pérez-Pomares J.M., Martínez-Estrada O.M. Wt1 controls retinoic acid signalling in embryonic epicardium through transcriptional activation of Raldh2s. Development. 2011;138:1093–1097. doi: 10.1242/dev.044594. 10.1242/dev.044594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guadix J.A., Orlova V.V., Giacomelli E., Bellin M., Ribeiro M.C., Mummery C.L., Pérez-Pomares J.M., Passier R. Human pluripotent stem cell differentiation into functional epicardial progenitor cells. Stem Cell Rep. 2017;9:1754–1764. doi: 10.1016/j.stemcr.2017.10.023. 10.1016/j.stemcr.2017.10.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochgreb T., Linhares V.L., Menezes D.C., Sampaio A.C., Yan C.Y.I., Cardoso W.V., Rosenthal N., Xavier-Neto J. A caudorostral wave of RALDH2 conveys anteroposterior information to the cardiac field. Development. 2003;130:5363–5374. doi: 10.1242/dev.00750. 10.1242/dev.00750 [DOI] [PubMed] [Google Scholar]
- Hoffmann S., Berger I.M., Glaser A., Bacon C., Li L., Gretz N., Steinbeisser H., Rottbauer W., Just S., Rappold G. Islet1 is a direct transcriptional target of the homeodomain transcription factor Shox2 and rescues the Shox2-mediated bradycardia. Basic Res. Cardiol. 2013;108 doi: 10.1007/s00395-013-0339-z. 10.1007/s00395-013-0339-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C.Y., Peres Moreno Maia-Joca R., Ong C.S., Wilson I., DiSilvestre D., Tomaselli G.F., Reich D.H. Enhancement of human iPSC-derived cardiomyocyte maturation by chemical conditioning in a 3D environment. J. Mol. Cell Cardiol. 2020;138:1–11. doi: 10.1016/j.yjmcc.2019.10.001. 10.1016/j.yjmcc.2019.10.001 [DOI] [PubMed] [Google Scholar]
- Ishii Y., Garriock R.J., Navetta A.M., Coughlin L.E., Mikawa T. BMP signals promote proepicardial protrusion necessary for recruitment of coronary vessel and epicardial progenitors to the heart. Dev. Cell. 2010;19:307–316. doi: 10.1016/j.devcel.2010.07.017. 10.1016/j.devcel.2010.07.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanovitch K., Soro-Barrio P., Chakravarty P., Jones R.A., Bell D.M., Neda Mousavy Gharavy S., Stamataki D., Delile J., Smith J.C., Briscoe J. Ventricular, atrial, and outflow tract heart progenitors arise from spatially and molecularly distinct regions of the primitive streak. PLoS Biol. 2021;19:e3001200. doi: 10.1371/journal.pbio.3001200. 10.1371/journal.pbio.3001200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer D., Gambardella L., Bernard W.G., Serrano F., Mascetti V.L., Pedersen R.A., Talasila A., Sinha S. Robust derivation of epicardium and its differentiated smooth muscle cell progeny from human pluripotent stem cells. Development (Cambridge) 2015;142:1528–1541. doi: 10.1242/dev.119271. 10.1242/dev.119271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanai M., Raz A., Goodman D.S. Retinol-binding protein: the transport protein for vitamin A in human plasma. J. Clin. Invest. 1968;47:2025–2044. doi: 10.1172/JCI105889. 10.1172/JCI105889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawaguchi R., Yu J., Honda J., Hu J., Whitelegge J., Ping P., Wiita P., Bok D., Sun H. A membrane receptor for retinol binding protein mediates cellular uptake of vitamin A. Science. 2007;315:820–825. doi: 10.1126/science.1136244. 10.1126/science.1136244 [DOI] [PubMed] [Google Scholar]
- Kedishvili N.Y. Retinoic acid synthesis and degradation. Sub-Cellular Biochem. 2016;81:127–161. doi: 10.1007/978-94-024-0945-1_5. 10.1007/978-94-024-0945-1_5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klaus A., Saga Y., Taketo M.M., Tzahor E., Birchmeier W. Distinct roles of Wnt/β-catenin and Bmp signaling during early cardiogenesis. Proc. Natl. Acad. Sci. U S A. 2007;104:18531–18536. doi: 10.1073/pnas.0703113104. 10.1073/pnas.0703113104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kostetskii I., Jiang Y., Kostetskaia E., Yuan S., Evans T., Zile M. Retinoid signaling required for normal heart development regulates GATA- 4 in a pathway distinct from cardiomyocyte differentiation. Dev. Biol. 1999;206:206–218. doi: 10.1006/dbio.1998.9139. 10.1006/dbio.1998.9139 [DOI] [PubMed] [Google Scholar]
- Kruithof B.P.T., van Wijk B., Somi S., Kruithof-de Julio M., Pérez Pomares J.M., Weesie F., Wessels A., Moorman A.F.M., van den Hoff M.J.B. BMP and FGF regulate the differentiation of multipotential pericardial mesoderm into the myocardial or epicardial lineage. Dev. Biol. 2006;295:507–522. doi: 10.1016/j.ydbio.2006.03.033. [DOI] [PubMed] [Google Scholar]
- Lammer E.J., Chen D.T., Hoar R.M., Agnish N.D., Benke P.J., Braun J.T., Curry C.J., Fernhoff P.M., Grix A.W., Lott I.T., et al. Retinoic acid embryopathy. New Engl. J. Med. 1985;313:837–841. doi: 10.1056/NEJM198510033131401. 10.1056/NEJM198510033131401 [DOI] [PubMed] [Google Scholar]
- Lee J.H., Protze S.I., Laksman Z., Backx P.H., Keller G.M. Human pluripotent stem cell-derived atrial and ventricular cardiomyocytes develop from distinct mesoderm populations. Cell Stem Cell. 2017;21:179–194.e4. doi: 10.1016/j.stem.2017.07.003. 10.1016/j.stem.2017.07.003 [DOI] [PubMed] [Google Scholar]
- Lengerke C., Schmitt S., Bowman T.V., Jang I.H., Maouche-Chretien L., McKinney-Freeman S., Davidson A.J., Hammerschmidt M., Rentzsch F., Green J.B.A., et al. BMP and Wnt specify hematopoietic fate by activation of the Cdx-Hox pathway. Cell Stem Cell. 2008;2:72–82. doi: 10.1016/j.stem.2007.10.022. 10.1016/j.stem.2007.10.022 [DOI] [PubMed] [Google Scholar]
- Lescroart F., Chabab S., Lin X., Rulands S., Paulissen C., Rodolosse A., Auer H., Achouri Y., Dubois C., Bondue A., et al. Early lineage restriction in temporally distinct populations of Mesp1 progenitors during mammalian heart development. Nat. Cell Biol. 2014;16:829–840. doi: 10.1038/ncb3024. 10.1038/ncb3024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lescroart F., Zaffran S. Hox and tale transcription factors in heart development and disease. Int. J. Dev. Biol. 2018;62:837–846. doi: 10.1387/ijdb.180192sz. 10.1387/ijdb.180192sz [DOI] [PubMed] [Google Scholar]
- Li H., Li D., Wang Y., Huang Z., Xu J., Yang T., Wang L., Tang Q., Cai C.-L., Huang H., et al. Nkx2-5 defines a subpopulation of pacemaker cells and is essential for the physiological function of the sinoatrial node in mice. Development. 2019;146 doi: 10.1242/dev.178145. 10.1242/dev.178145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li P., Cavallero S., Gu Y., Chen T.H.P., Hughes J., Hassan A.B., Brüning J.C., Pashmforoush M., Sucov H.M. IGF signaling directs ventricular cardiomyocyte proliferation during embryonic heart development. Development. 2011;138:1795–1805. doi: 10.1242/dev.054338. 10.1242/dev.054338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liberatore C.M., Searcy-Schrick R.D., Yutzey K.E. Ventricular expression of tbx5 inhibits normal heart chamber development. Dev. Biol. 2000;223:169–180. doi: 10.1006/dbio.2000.9748. 10.1006/dbio.2000.9748 [DOI] [PubMed] [Google Scholar]
- Lin L., Cui L., Zhou W., Dufort D., Zhang X., Cai C.L., Bu L., Yang L., Martin J., Kemler R., et al. β-Catenin directly regulates Islet1 expression in cardiovascular progenitors and is required for multiple aspects of cardiogenesis. Proc. Natl. Acad. Sci. U S A. 2007;104:9313–9318. doi: 10.1073/pnas.0700923104. 10.1073/pnas.0700923104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin S.C., Dollé P., Ryckebüsch L., Noseda M., Zaffran S., Schneider M.D., Niederreithera K. Endogenous retinoic acid regulates cardiac progenitor differentiation. Proc. Natl. Acad. Sci. U S A. 2010;107:9234–9239. doi: 10.1073/pnas.0910430107. 10.1073/pnas.0910430107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Stainier D.Y.R. Tbx5 and Bmp signaling are essential for proepicardium specification in zebrafish. Circ. Res. 2010;106:1818–1828. doi: 10.1161/CIRCRESAHA.110.217950. 10.1161/CIRCRESAHA.110.217950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Delgado A.C., Delgado I., Cadenas V., Sánchez-Cabo F., Torres M. Axial skeleton anterior-posterior patterning is regulated through feedback regulation between Meis transcription factors and retinoic acid. Development (Cambridge) 2021;148 doi: 10.1242/dev.193813. 10.1242/dev.193813 [DOI] [PubMed] [Google Scholar]
- Lupu I.E., Redpath A.N., Smart N. Spatiotemporal analysis reveals overlap of key proepicardial markers in the developing murine heart. Stem Cell Rep. 2020;14:770–787. doi: 10.1016/j.stemcr.2020.04.002. 10.1016/j.stemcr.2020.04.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacLean G., Abu-Abed S., Dollé P., Tahayato A., Chambon P., Petkovich M. Cloning of a novel retinoic-acid metabolizing cytochrome P450, Cyp26B1, and comparative expression analysis with Cyp26A1 during early murine development. Mech. Development. 2001;107:195–201. doi: 10.1016/s0925-4773(01)00463-4. 10.1016/S0925-4773(01)00463-4 [DOI] [PubMed] [Google Scholar]
- Mercader N., Leonardo E., Piedra M.E., Martinez-A C., Ros M.A., Torres M. Opposing RA and FGF signals control proximodistal vertebrate limb development through regulation of Meis genes. Development. 2000;127:3961–3970. doi: 10.1242/dev.127.18.3961. 10.1242/dev.127.18.3961 [DOI] [PubMed] [Google Scholar]
- Merki E., Zamora M., Raya A., Kawakami Y., Wang J., Zhang X., Burch J., Kubalak S.W., Kaliman P., Belmonte J.C.I., et al. Epicardial retinoid X receptor α is required for myocardial growth and coronary artery formation. Proc. Natl. Acad. Sci. U S A. 2005;102:18455–18460. doi: 10.1073/pnas.0504343102. 10.1073/pnas.0504343102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metzler M.A., Sandell L.L. Enzymatic metabolism of vitamin A in developing vertebrate embryos. Nutrients. 2016 doi: 10.3390/nu8120812. 10.3390/nu8120812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mommersteeg M.T.M., Domínguez J.N., Wiese C., Norden J., De Gier-De Vries C., Burch J.B.E., Kispert A., Brown N.A., Moorman A.F.M., Christoffels V.M. The sinus venosus progenitors separate and diversify from the first and second heart fields early in development. Cardiovasc. Res. 2010;87:92–101. doi: 10.1093/cvr/cvq033. 10.1093/cvr/cvq033 [DOI] [PubMed] [Google Scholar]
- Del Monte G., Casanova J.C., Guadix J.A., MacGrogan D., Burch J.B.E., Pérez-Pomares J.M., De La Pompa J.L. Differential notch signaling in the epicardium is required for cardiac inflow development and coronary vessel morphogenesis. Circ. Res. 2011;108:824–836. doi: 10.1161/CIRCRESAHA.110.229062. 10.1161/CIRCRESAHA.110.229062 [DOI] [PubMed] [Google Scholar]
- Moss J.B., Xavier-Neto J., Shapiro M.D., Nayeem S.M., McCaffery P., Dräger U.C., Rosenthal N. Dynamic patterns of retinoic acid synthesis and response in the developing mammalian heart. Dev. Biol. 1998;199:55–71. doi: 10.1006/dbio.1998.8911. 10.1006/dbio.1998.8911 [DOI] [PubMed] [Google Scholar]
- Naito A.T., Shiojima I., Akazawa H., Hidaka K., Morisaki T., Kikuchi A., Komuro I. Developmental stage-specific biphasic roles of Wnt/β-catenin signaling in cardiomyogenesis and hematopoiesis. Proc. Natl. Acad. Sci. U S A. 2006;103:19812–19817. doi: 10.1073/pnas.0605768103. 10.1073/pnas.0605768103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Napoli J.L. Functions of intracellular retinoid binding-proteins. Sub Cell. Biochem. 2016;81:21–76. doi: 10.1007/978-94-024-0945-1_2. 10.1007/978-94-024-0945-1_2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neijts R., Amin S., Van Rooijen C., Tan S., Creyghton M.P., De Laat W., Deschamps J. Polarized regulatory landscape and Wnt responsiveness underlie Hox activation in embryos. Genes Dev. 2016;30:1937–1942. doi: 10.1101/gad.285767.116. 10.1101/gad.285767.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng J.K., Kawakami Y., Büscher D., Raya Á., Itoh T., Koth C.M., Rodríguez Esteban C., Rodríguez-León J., Garrity D.M., Fishman M.C., et al. The limb gene Tbx5 promotes limb initiation by interacting with Wnt2b and Fgf10. Development. 2002:5161–5170. doi: 10.1242/dev.129.22.5161. 10.1242/dev.129.22.5161 [DOI] [PubMed] [Google Scholar]
- Niederreither K., Mccaffery B’c P., Dr∼iger B’c U.C., Chambon P., Doll6 P. Restricted expression and retinoic acid-induced downregulation of the retinaldehyde dehydrogenase type 2 (RALDH-2) gene during mouse development. Mech. Development. 1997;62:67–78. doi: 10.1016/s0925-4773(96)00653-3. [DOI] [PubMed] [Google Scholar]
- Niederreither K., Subbarayan V., Dollé P., Chambon P. Embryonic retinoic acid synthesis is essential for early mouse post- implantation development. Nat. Genet. 1999;21:444–448. doi: 10.1038/7788. 10.1038/7788 [DOI] [PubMed] [Google Scholar]
- Niederreither K., Vermot J., Messaddeq N., Schuhbaur B., Chambon P., Dollé P. Embryonic retinoic acid synthesis is essential for heart morphogenesis in the mouse. Development (Cambridge, England) 2001;128:1019–1031. doi: 10.1242/dev.128.7.1019. http://www.ncbi.nlm.nih.gov/pubmed/11245568 [DOI] [PubMed] [Google Scholar]
- Niederreither K., Dollé P. Retinoic acid in development: towards an integrated view. Nat. Rev. Genet. 2008:541–553. doi: 10.1038/nrg2340. 10.1038/nrg2340 [DOI] [PubMed] [Google Scholar]
- Nishimoto S., Wilde S.M., Wood S., Logan M.P.O. RA acts in a coherent feed-forward mechanism with Tbx5 to control limb bud induction and initiation. Cell Rep. 2015;12:879–891. doi: 10.1016/j.celrep.2015.06.068. 10.1016/j.celrep.2015.06.068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Ouwerkerk A.F., Hall A.W., Kadow Z.A., Lazarevic S., Reyat J.S., Tucker N.R., Nadadur R.D., Bosada F.M., Bianchi V., Ellinor P.T., et al. Epigenetic and transcriptional networks underlying atrial fibrillation. Circ. Res. 2020:34–50. doi: 10.1161/CIRCRESAHA.120.316574. 10.1161/CIRCRESAHA.120.316574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasutto F., Sticht H., Hammersen G., Gillessen-Kaesbach G., FitzPatrick D.R., Nürnberg G., Brasch F., Schirmer-Zimmermann H., Tolmie J.L., Chitayat D., et al. Mutations in STRA6 cause a broad spectrum of malformations including anophthalmia, congenital heart defects, diaphragmatic hernia, alveolar capillary dysplasia, lung hypoplasia, and mental retardation. Am. J. Hum. Genet. 2007;80:550–560. doi: 10.1086/512203. 10.1086/512203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavan M., Ruiz V.F., Silva F.A., Sobreira T.J., Cravo R.M., Vasconcelos M., Marques L.P., Mesquita S.M.F., Krieger J.E., Lopes A.A.B., et al. ALDH1A2 (RALDH2) genetic variation in human congenital heart disease. BMC Med. Genet. 2009;10:113. doi: 10.1186/1471-2350-10-113. 10.1186/1471-2350-10-113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Pomares J.M., Carmona R., González-Iriarte M., Atencia G., Wessels A., Muñoz-Chapuli R. Origin of coronary endothelial cells from epicardial mesothelium in avian embryos. Int. J. Dev. Biol. 2002;46:1005–1013. 10.1387/ijdb.12533024 [PubMed] [Google Scholar]
- Plageman T.F., Yutzey K.E. Microarray analysis of Tbx5-induced genes expressed in the developing heart. Dev. Dyn. 2006;235:2868–2880. doi: 10.1002/dvdy.20923. 10.1002/dvdy.20923 [DOI] [PubMed] [Google Scholar]
- Prall O.W.J., Menon M.K., Solloway M.J., Watanabe Y., Zaffran S., Bajolle F., Biben C., McBride J.J., Robertson B.R., Chaulet H., et al. An Nkx2-5/Bmp2/Smad1 negative feedback loop controls heart progenitor specification and proliferation. Cell. 2007;128:947–959. doi: 10.1016/j.cell.2007.01.042. 10.1016/j.cell.2007.01.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Protze S.I., Liu J., Nussinovitch U., Ohana L., Backx P.H., Gepstein L., Keller G.M. Sinoatrial node cardiomyocytes derived from human pluripotent cells function as a biological pacemaker. Nat. Biotechnol. 2017;35:56–68. doi: 10.1038/nbt.3745. 10.1038/nbt.3745 [DOI] [PubMed] [Google Scholar]
- Puskaric S., Schmitteckert S., Mori A.D., Glaser A., Schneider K.U., Bruneau B.G., Blaschke R.J., Steinbeisser H., Rappold G. Shox2 mediates Tbx5 activity by regulating Bmp4 in the pacemaker region of the developing heart. Hum. Mol. Genet. 2010;19:4625–4633. doi: 10.1093/hmg/ddq393. 10.1093/hmg/ddq393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quaranta R., Fell J., Rühle F., Rao J., Piccini I., Araúzo-Bravo M.J., Verkerk A.O., Stoll M., Greber B. Revised roles of ISL1 in a hES cell-based model of human heart chamber specification. eLife. 2018;7 doi: 10.7554/eLife.31706. 10.7554/eLife.31706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quijada P., Trembley M.A., Small E.M. The role of the epicardium during heart development and repair. Circul. Res. 2020:377–394. doi: 10.1161/CIRCRESAHA.119.315857. 10.1161/CIRCRESAHA.119.315857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rankin S.A., Steimle J.D., Yang X.H., Rydeen A.B., Agarwal K., Chaturvedi P., Ikegami K., Herriges M.J., Moskowitz I.P., Zorn A.M. TBX5 drives Aldh1a2 expression to regulate a RA-Hedgehog-Wnt gene regulatory network coordinating cardiopulmonary development. bioRxiv. 2021:2021. doi: 10.7554/eLife.69288. 10.1101/2021.04.09.439219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren J., Han P., Ma X., Farah E.N., Bloomekatz J., Zeng X.X.I., Zhang R., Swim M.M., Witty A.D., Knight H.G., et al. Canonical Wnt5b signaling directs outlying Nkx2.5+ mesoderm into pacemaker cardiomyocytes. Dev. Cell. 2019;50:729–743.e5. doi: 10.1016/j.devcel.2019.07.014. 10.1016/j.devcel.2019.07.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts C., Ivins S., Cook A.C., Baldini A., Scambler P.J. Cyp26 genes a1, b1 and c1 are down-regulated in Tbx1 null mice and inhibition of Cyp26 enzyme function produces a phenocopy of DiGeorge Syndrome in the chick. Hum. Mol. Genet. 2006;15:3394–3410. doi: 10.1093/hmg/ddl416. 10.1093/hmg/ddl416 [DOI] [PubMed] [Google Scholar]
- Rothman K.J., Moore L.L., Singer M.R., Nguyen U.-S.D.T., Mannino S., Milunsky A. Teratogenicity of high vitamin A intake. N. Engl. J. Med. 1995;333:1369–1373. doi: 10.1056/NEJM199511233332101. 10.1056/NEJM199511233332101 [DOI] [PubMed] [Google Scholar]
- Rupert C.E., Coulombe K.L.K. IGF1 and NRG1 enhance proliferation, metabolic maturity, and the force-frequency response in hESC-derived engineered cardiac tissues. Stem Cells Int. 2017;2017 doi: 10.1155/2017/7648409. 10.1155/2017/7648409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryckebusch L., Wang Z., Bertrand N., Lin S.C., Chi X., Schwartz R., Zaffran S., Niederreither K. Retinoic acid deficiency alters second heart field formation. Proc. Natl. Acad. Sci. U S A. 2008;105:2913–2918. doi: 10.1073/pnas.0712344105. 10.1073/pnas.0712344105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saga Y., Miyagawa-Tomita S., Takagi A., Kitajima S., Miyazaki J.I., Inoue T. MesP1 is expressed in the heart precursor cells and required for the formation of a single heart tube. Development. 1999;126:3437–3447. doi: 10.1242/dev.126.15.3437. 10.1242/dev.126.15.3437 [DOI] [PubMed] [Google Scholar]
- Sandell L.L., Sanderson B.W., Moiseyev G., Johnson T., Mushegian A., Young K., Rey J.P., Ma J.X., Staehling-Hampton K., Trainor P.A. RDH10 is essential for synthesis of embryonic retinoic acid and is required for limb, craniofacial, and organ development. Genes Dev. 2007;21:1113–1124. doi: 10.1101/gad.1533407. 10.1101/gad.1533407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlueter J., Männer J., Brand T. BMP is an important regulator of proepicardial identity in the chick embryo. Dev. Biol. 2006;295:546–558. doi: 10.1016/j.ydbio.2006.03.036. 10.1016/j.ydbio.2006.03.036 [DOI] [PubMed] [Google Scholar]
- Sirbu I.O., Zhao X., Duester G. Retinoic acid controls heart anteroposterior patterning by down-regulating Isl1 through the Fgf8 pathway. Dev. Dyn. 2008;237:1627–1635. doi: 10.1002/dvdy.21570. 10.1002/dvdy.21570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefanovic S., Laforest B., Desvignes J.P., Lescroart F., Argiro L., Maurel-Zaffran C., Salgado D., Plaindoux E., De Bono C., Pazur K., et al. Hox-dependent coordination of mouse cardiac progenitor cell patterning and differentiation. eLife. 2020;9:1–32. doi: 10.7554/eLife.55124. 10.7554/ELIFE.55124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steimle J.D., Rankin S.A., Slagle C.E., Bekeny J., Rydeen A.B., Chan S.S.K., Kweon J., Yang X.H., Ikegami K., Nadadur R.D., et al. Evolutionarily conserved Tbx5-Wnt2/2b pathway orchestrates cardiopulmonary development. Proc. Natl. Acad. Sci. U S A. 2018;115:E10615–E10624. doi: 10.1073/pnas.1811624115. 10.1073/pnas.1811624115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sucov H.M., Gu Y., Thomas S., Li P., Pashmforoush M. Epicardial control of myocardial proliferation and morphogenesis. Pediatr. Cardiol. 2009:617–625. doi: 10.1007/s00246-009-9391-8. 10.1007/s00246-009-9391-8 [DOI] [PubMed] [Google Scholar]
- Takahashi M., Yamagishi T., Narematsu M., Kamimura T., Kai M., Nakajima Y. Epicardium is required for sarcomeric maturation and cardiomyocyte growth in the ventricular compact layer mediated by transforming growth factor β and fibroblast growth factor before the onset of coronary circulation. Congenit. Anom. 2014;54:162–171. doi: 10.1111/cga.12048. 10.1111/cga.12048 [DOI] [PubMed] [Google Scholar]
- Tan J.J., Guyette J.P., Miki K., Xiao L., Kaur G., Wu T., Zhu L., Hansen K.J., Ling K.-H., Milan D.J., et al. Human iPS-derived pre-epicardial cells direct cardiomyocyte aggregation expansion and organization in vitro. Nat. Commun. 2021;12:4997. doi: 10.1038/s41467-021-24921-z. 10.1038/s41467-021-24921-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyser R.C.V., Ibarra-Soria X., McDole K.A., Jayaram S., Godwin J., van den Brand T.A.H., Miranda A.M.A., Scialdone A., Keller P.J., Marioni J.C., et al. Characterization of a common progenitor pool of the epicardium and myocardium. Science. 2021:eabb2986. doi: 10.1126/science.abb2986. 10.1126/science.abb2986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueno S., Weidinger G., Osugi T., Kohn A.D., Golob J.L., Pabon L., Reinecke H., Moon R.T., Murry C.E. Biphasic role for Wnt/β-catenin signaling in cardiac specification in zebrafish and embryonic stem cells. Proc. Natl. Acad. Sci. U S A. 2007;104:9685–9690. doi: 10.1073/pnas.0702859104. 10.1073/pnas.0702859104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verzi M.P., McCulley D.J., De Val S., Dodou E., Black B.L. The right ventricle, outflow tract, and ventricular septum comprise a restricted expression domain within the secondary/anterior heart field. Dev. Biol. 2005;287:134–145. doi: 10.1016/j.ydbio.2005.08.041. 10.1016/j.ydbio.2005.08.041 [DOI] [PubMed] [Google Scholar]
- Viragh S., Challice C.E. The origin of the epicardium and the embryonic myocardial circulation in the mouse. Anat. Rec. 1981;201:157–168. doi: 10.1002/ar.1092010117. 10.1002/ar.1092010117 [DOI] [PubMed] [Google Scholar]
- Vitobello A., Ferretti E., Lampe X., Vilain N., Ducret S., Ori M., Spetz J.F., Selleri L., Rijli F.M. Hox and Pbx factors control retinoic acid synthesis during hindbrain segmentation. Dev. Cell. 2011;20:469–482. doi: 10.1016/j.devcel.2011.03.011. 10.1016/j.devcel.2011.03.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiese C., Grieskamp T., Airik R., Mommersteeg M.T.M., Gardiwal A., De Gier-De Vries C., Schuster-Gossler K., Moorman A.F.M., Kispert A., Christoffels V.M. Formation of the sinus node head and differentiation of sinus node myocardium are independently regulated by Tbx18 and Tbx3. Circ. Res. 2009;104:388–397. doi: 10.1161/CIRCRESAHA.108.187062. 10.1161/CIRCRESAHA.108.187062 [DOI] [PubMed] [Google Scholar]
- van Wijk B., van den Berg G., Abu-Issa R., Barnett P., van der Velden S., Schmidt M., Ruijter J.M., Kirby M.L., Moorman A.F.M., van den Hoff M.J.B. Epicardium and myocardium separate from a common precursor pool by crosstalk between bone morphogenetic protein- and fibroblast growth factor-signaling pathways. Circul. Res. 2009;105:431–441. doi: 10.1161/CIRCRESAHA.109.203083. 10.1161/CIRCRESAHA.109.203083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson J.G., Roth C.B., Warkany J. An analysis of the syndrome of malformations induced by maternal vitamin a deficiency. Effects of restoration of vitamin a at various times during gestation. Am. J. Anat. 1953;92:189–217. doi: 10.1002/aja.1000920202. 10.1002/aja.1000920202 [DOI] [PubMed] [Google Scholar]
- Wilson J.G., Warkany J. Aortic-arch and cardiac anomalies in the offspring of vitamin A. Am. J. Anat. 1949;85:113–155. doi: 10.1002/aja.1000850106. 10.1002/aja.1000850106 [DOI] [PubMed] [Google Scholar]
- Witty A.D., Mihic A., Tam R.Y., Fisher S.A., Mikryukov A., Shoichet M.S., Li R.K., Kattman S.J., Keller G. Generation of the epicardial lineage from human pluripotent stem cells. Nat. Biotechnol. 2014;32:1026–1035. doi: 10.1038/nbt.3002. 10.1038/nbt.3002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wobus A.M., Kaomei G., Shan J., Wellner M.C., Rohwedel J., Guanju J., Fleischmann B., Katus H.A., Hescheler J., Franz W.M. Retinoic acid accelerates embryonic stem cell-derived cardiac differentiation and enhances development of ventricular cardiomyocytes. J. Mol. Cell. Cardiol. 1997;29:1525–1539. doi: 10.1006/jmcc.1997.0433. 10.1006/jmcc.1997.0433 [DOI] [PubMed] [Google Scholar]
- Wu S., pin, Cheng C.M., Lanz R.B., Wang T., Respress J.L., Ather S., Chen W., Tsai S.J., Wehrens X.H.T., Tsai M.J., et al. Atrial identity is determined by a coup-tfii regulatory network. Dev. Cell. 2013;25:417–426. doi: 10.1016/j.devcel.2013.04.017. 10.1016/j.devcel.2013.04.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao Y., Hill M.C., Zhang M., Martin T.J., Morikawa Y., Wang S., Moise A.R., Wythe J.D., Martin J.F. Hippo signaling plays an essential role in cell state transitions during cardiac fibroblast development. Dev. Cell. 2018;45:153–169.e6. doi: 10.1016/j.devcel.2018.03.019. 10.1016/j.devcel.2018.03.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi T., Wong J., Feller E., Sink S., Taghli-Lamallem O., Wen J., Kim C., Fink M., Giles W., Soussou W., et al. Electrophysiological mapping of embryonic mouse hearts: mechanisms for developmental pacemaker switch and internodal conduction pathway. J. Cardiovasc. Electrophysiol. 2012;23:309–318. doi: 10.1111/j.1540-8167.2011.02191.x. 10.1111/j.1540-8167.2011.02191.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaffran S., Kelly R.G., Meilhac S.M., Buckingham M.E., Brown N.A. Right ventricular myocardium derives from the anterior heart field. Circ. Res. 2004;95:261–268. doi: 10.1161/01.RES.0000136815.73623.BE. 10.1161/01.RES.0000136815.73623.BE [DOI] [PubMed] [Google Scholar]
- Zamora M., Männer J., Ruiz-Lozano P. ‘Epicardium-derived progenitor cells require β-catenin for coronary artery formation. Proc. Natl. Acad. Sci. U S A. 2007;104:18109–18114. doi: 10.1073/pnas.0702415104. 10.1073/pnas.0702415104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q., Jiang J., Han P., Yuan Q., Zhang J., Zhang X., Xu Y., Cao H., Meng Q., Chen L., et al. Direct differentiation of atrial and ventricular myocytes from human embryonic stem cells by alternating retinoid signals. Cell Res. 2011;21:579–587. doi: 10.1038/cr.2010.163. 10.1038/cr.2010.163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q., Carlin D., Zhu F., Cattaneo P., Ideker T., Evans S.M., Bloomekatz J., Chi N.C. Unveiling complexity and multipotentiality of early heart fields. Circ. Res. 2021;129:474–487. doi: 10.1161/CIRCRESAHA.121.318943. 10.1161/CIRCRESAHA.121.318943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J., Cao H., Tian L., Huo W., Zhai K., Wang P., Ji G., Ma Y. Efficient differentiation of TBX18+/WT1+ epicardial-like cells from human pluripotent stem cells using small molecular compounds. Stem Cells Dev. 2017;26:528–540. doi: 10.1089/scd.2016.0208. 10.1089/scd.2016.0208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou B., Gise A., von, Ma Q., Rivera-Feliciano J., Pu W.T. Nkx2-5- and Isl1-expressing cardiac progenitors contribute to proepicardium. Biochem. Biophys. Res. Commun. 2008;375:450–453. doi: 10.1016/j.bbrc.2008.08.044. 10.1016/j.bbrc.2008.08.044 [DOI] [PMC free article] [PubMed] [Google Scholar]