Summary
We propose a new concept that human somatic cells can be converted to become male germline stem cells by the defined factors. Here, we demonstrated that the overexpression of DAZL, DAZ2, and BOULE could directly reprogram human Sertoli cells into cells with the characteristics of human spermatogonial stem cells (SSCs), as shown by their similar transcriptomes and proteomics with human SSCs. Significantly, human SSCs derived from human Sertoli cells colonized and proliferated in vivo, and they could differentiate into spermatocytes and haploid spermatids in vitro. Human Sertoli cell-derived SSCs excluded Y chromosome microdeletions and assumed normal chromosomes. Collectively, human somatic cells could be converted directly to human SSCs with the self-renewal and differentiation potentials and high safety. This study is of unusual significance, because it provides an effective approach for reprogramming human somatic cells into male germ cells and offers invaluable male gametes for treating male infertility.
Keywords: human Sertoli cells, reprogramming, DAZL/DAZ2/BOULE genes, male germline stem cells, self-renewal and differentiation
Highlights
-
•
Human Sertoli cells can be converted into SSCs by overexpressing DAZ family genes
-
•
Human Sertoli cell-derived cells have phenotypic features with SSCs in vivo
-
•
Human Sertoli cell-derived SSCs differentiate into haploid spermatids in vitro
-
•
Human Sertoli cell-derived SSCs have normal chromosomes and no gene microdeletion
In this article, Zuping He and colleagues show that human Sertoli cells can be converted directly into human spermatogonial stem cells that are able to self-renew in vivo and differentiate into spermatocytes and haploid spermatids in vitro via the overexpression of three DAZ family genes.
Introduction
Infertility has become one of the most serious diseases affecting human reproduction, as demonstrated by the fact that around 15% of couples worldwide are infertile (Handel et al., 2014). There are 50 million patients with infertility in China, and about half of them are attributed to male factors. Notably, azoospermia comprises 25% of male infertility (Wu et al., 2007). There has been a significant decline of the newborn population, by 7.82 million in 2020 compared with that in 2016 in China, around 50% of which may be due to male infertility. Spermatogenesis is a highly organized process by which spermatogonial stem cells (SSCs) self-renew and differentiate into spermatocytes and spermatids. Most infertile males have gametogenesis failure, e.g., azoospermia and oligospermia.
DAZ gene has four copies, namely DAZ1, DAZ2, DAZ3, and DAZ4, and it has been found in gonocytes, spermatogonia, and spermatocytes of adult testes. Different DAZ copy cluster deletions have different effect on spermatogenesis. DAZ1/DAZ2 deletions cause spermatogenic impairment, whereas DAZ3/DAZ4 knockout has no influence on fertility (Li et al., 2013). In humans, deletion of DAZ2 gene copies is associated with male infertility (Ghorbel et al., 2014), while DAZ2 protein is present in the tails of mature spermatids (Habermann et al., 1998), suggesting that the DAZ2 gene is involved in the late stage of normal spermatogenesis. There are two homologous genes of DAZ in the autosome, i.e., DAZL and BOULE. DAZ family proteins are found exclusively in germ cells, reflecting that they may be associated with gametogenesis. Deletion or mutation of their encoding genes usually causes severe azoospermia or oligospermia.
DAZ family proteins mediate the proliferation, differentiation, and maturation of germ cells by regulating the expression, transport, and location of targeting genes and proteins (Fu et al., 2015; Jenkins et al., 2011; Jiao et al., 2002; Maegawa et al., 2002; Maines and Wasserman, 1999; Venables et al., 2001). BOULE protein plays a pivotal role in germline specification and meiosis. It has been reported that the BOULE gene is essential for meiosis in male germ cells (Eberhart et al., 1996). BOULE protein is expressed in the meiotic stages from leptotene spermatocytes to late spermatocytes, and it is undetectable in the testes of men with meiotic arrest (Luetjens et al., 2004). The overexpression of human BOULE can restore fertility of flies with the Boule mutation (Xu et al., 2003). As such, the ectopic expression of BOULE makes it feasible for human Sertoli cell-derived SSCs to differentiate into spermatocytes. Depending on the species, the DAZL gene is present in primordial germ cells (PGCs) and/or pre-meiotic and meiotic germ cells of both sexes. Mice with a deficiency in DAZL are infertile (Saunders et al., 2003; Schrans-Stassen et al., 2001), and microdeletion of the DAZL gene results in male infertility in humans (Reijo et al., 1995), implicating that DAZL is crucial for normal spermatogenesis. DAZL plays a critical role in primordial germ cell formation, while DAZ and BOULE promote meiosis and haploid gamete formation. Germ cells can be derived from human embryonic stem cells (ESCs) by overexpressing DAZL, DAZ, and BOULE genes, and they can further enter and progress through meiosis, which results in haploid spermatids (Kee et al., 2009). Compared with human ESCs as the initial cells (Kee et al., 2009), this study using human Sertoli cells as starting cells has the following advantages: (1) it is more important to reprogram Sertoli cells (somatic cells) into male germline stem cells with an aim to gain novel knowledge about the gap between male germ cells and somatic cells; and (2) generation of spermatids from human Sertoli cells could offer valuable male gametes for Sertoli cell-only syndrome (SCOS). As such, we asked whether human Sertoli cells could be reprogrammed to generate male germ cells by overexpression of DAZL, DAZ, and BOULE genes. Human ESCs and the induced pluripotent stem cells (iPSCs) can be coaxed to differentiate into male germ cells and progress meiosis through the overexpression of DAZL and/or VASA (Medrano et al., 2012). In addition, PAX5-OCT4-PRDM1 proteins form the transcriptional network, which facilitates the germ cell differentiation of human PGCs (Fang et al., 2018). Very recently, oocyte-like cells have been generated from mouse PSCs by the enforced expression of eight transcription factors in vitro (Hamazaki et al., 2021). These studies illustrate that it is feasible to generate human male germ cells in vitro via the defined factors.
Generation of human germ cells, especially male gametes in vitro, has significant applications in reproductive medicine and cell biology. Various methods for obtaining male germ cells have been developed. PGCs can be generated from mouse ESCs and iPSCs via overexpressing Blimp1, Prdm14, and Tfap2c, and these cells differentiate into spermatids after transplantation to recipient mice (Hayashi et al., 2011; Nakaki et al., 2013). It is worth noting that PGCs derived from mouse ESCs complete the meiosis in vitro and give rise to spermatids (Zhou et al., 2016). Human PSCs can differentiate into PGCs and TFAP2A+ progenitors for initiating in vitro gametogenesis (Chen et al., 2019). Recently, it has been demonstrated that PRDM14 is essential for the generation of PGCs from human ESCs (Sybirna et al., 2020). Significantly, human ESCs and iPSCs have been demonstrated to differentiate into SSCs, spermatocytes, and spermatids (Easley et al., 2012; Kee et al., 2009; Sasaki et al., 2015; Zhao et al., 2018). We have demonstrated the generation of haploid spermatids from human SSCs of obstructive azoospermia (OA) and cryptorchid patients (Sun et al., 2018; Yang et al., 2014).
SCOS is the most severe disease in which only Sertoli cells but no male germ cells exist in the seminiferous tubules of patients. It is an unsolved and key issue to generate male germ cells from SCOS patients and other patients without spermatids. Lineage reprogramming in which the initiating cells are somatic cells might be a novel alternative strategy to obtain germ cells. Significantly, in January, 2018, Dr. M Azim Surani, Professor at the University of Cambridge, highlighted that it is essential to close the gap between soma and germ cells with an aim to generate germ cells from somatic cells, which could be a milestone for obtaining gametes in vitro. Here, we propose a novel concept that human somatic cells can be directly reprogrammed to male germline stem cells using the defined factors. As such, the starting cells do not need to be pluripotent or reprogrammed to become pluripotent, which provides a very useful approach for manual intervention in cell fates. For example, it has been shown to reprogram liver cells to pancreas progenitors (Cerda-Esteban et al., 2017), fibroblasts to Sertoli-like cells (Buganim et al., 2012), Sertoli cells to Leydig cells (Zhang et al., 2015), mouse fibroblasts to neural progenitors (Kim et al., 2011), human fibroblasts to hepatocytes (Zhu et al., 2014), and fibroblasts to cardiomyocytes (Ieda et al., 2010). These studies demonstrate the unlimited potentials of cellular plasticity. Notably, Sertoli cells have certain advantages over other types of cells, since they can be expanded in vitro to acquire a large number of cells for translational medicine and can be reprogrammed to other cell lineages (Sheng et al., 2012; Zhang et al., 2015). In this study, we have demonstrated, for the first time, that overexpression of three genes of the DAZ family, namely DAZL, DAZ2, and BOULE, can directly reprogram human Sertoli cells to cells with biochemical phenotypes, and self-renewal and differentiation capacities of human SSCs. This study thus offers invaluable male gametes for treating the infertility of azoospermia patients.
Results
Isolation and identification of human primary Sertoli cells
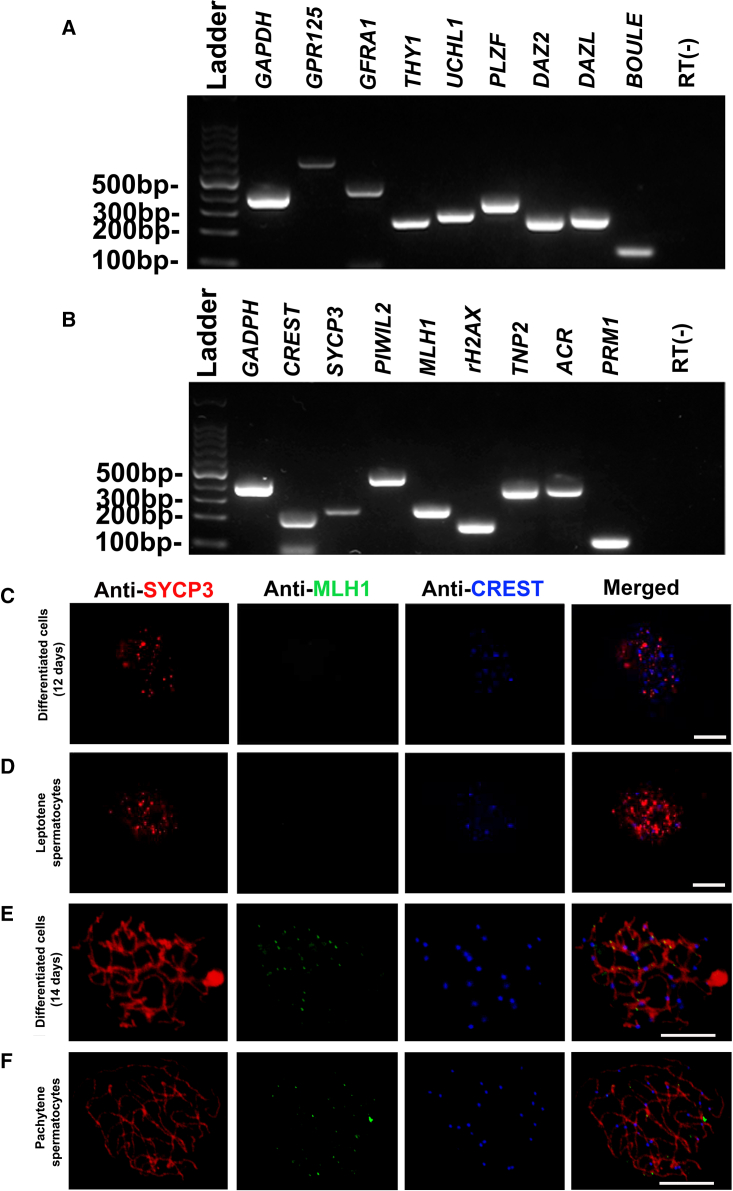
Human Sertoli cells were isolated from OA patients with normal spermatogenesis utilizing two-step enzymatic digestion followed by differential plating as described previously (He et al., 2010). Human seminiferous tubules were obtained from the testicular tissues by first using enzymatic digestion and then washing extensively with DMEM to remove potential contamination of peritubular myoid cells. A cell mixture containing Sertoli cells and male germ cells was isolated by a second enzymatic digestion. RT-PCR revealed that a number of genes, including GFRA1, GPR125, UCHL1, PLZF, THY1, RET, SYCP3, CREST, MLH1, rH2AX, ACR, TNP2, PRM1, PRM2, AR, SOX9, WT1, BMP4, SCF, and VASA were detected in the cell mixture containing human male germ cells and Sertoli cells (Figure S1A). After culturing the cell mixture with DMEM/F12 and 10% fetal bovine serum for 3 h, human Sertoli cells attached to the culture plates (Figure S2A), whereas male germ cells were suspended in the culture medium. RT-PCR showed that the transcripts of GFRA1, GPR125, UCHL1, PLZF, THY1, SYCP3, MLH1, TNP1, TNP2, PRM1, PRM2, and ACR were observed in the isolated human male germ cells (Figure S1B), whereas the mRNA of SCF, BMP4, WT1, and FSHR was undetectable in these cells (Figure S1B). The transcription of FSHR, GATA4, BMP4, SOX9, GDNF, WT1, SCF, and AR was seen in the isolated human Sertoli cells (Figure S1C) and, conversely, the mRNA of GPR125, PLZF, and THY1, markers for human SSCs, was undetected in these cells (Figure S1C), which excludes the contamination of human primary SSCs in isolated human Sertoli cells.
A number of hallmarks for Sertoli cells were employed to identify the initial human cells used for reprogramming. RT-PCR showed that the transcripts of SOX9, AR, BMP4, WT1, GATA4, and FSHR were present in isolated human Sertoli cells (Figure S2B), whereas the mRNA of VASA GFRA1, UCHL1, RET, PLZF, THY1, GPR125, SYCP3, MLH1, CREST, TNP2, ACR, PRM1, and PRM2 was undetectable in these cells (Figure S2B). Immunocytochemistry further revealed that over 96% of the isolated human cells were positive for WT1 (Figure S2C), GDNF (Figure S2D), SOX9 (Figure S2E), GATA4 (Figure S2F), and VIM (Figure S2G), but were negative for VASA (Figure S2I). Meanwhile, more than 95% of the isolated human cells expressed Ki-67 (Figure S2H), a hallmark for proliferating cells, reflecting that these cells have a high level of proliferation potential. After replacement of primary antibodies with mouse or rabbit normal IgGs, no immunostaining was seen in the isolated human cells (Figure S2J), thus demonstrating the specific immunostaining of the antibodies mentioned above. Fluorescence-activated cell sorting (FACS) showed that 97.3% of human Sertoli cells were negative for THY1 (Figure S2K, left panel), the SSC marker, thus excluding SSC contamination in human Sertoli cells. Considered together, these results implicated that the starting human cells are human Sertoli cells phenotypically without contamination of male germ cells.
Human Sertoli cells overexpress DAZL, DAZ2, and BOULE genes and proteins
We asked whether overexpression of DAZ family genes could reprogram human Sertoli cells to male germline stem cells. Plasmids overexpressing DAZL, DAZ2, and BOULE genes were sequenced, and the sizes of the three plasmids were 9,421, 10,210, and 9,385 bp, respectively (Figures S3A–3C; Table S1). The promoter for DAZL, DAZ2, and BOULE genes was EF1a, and the structures of three plasmids with blasticidin resistance were shown in Figures S3A–3C.
Plasmids overexpressing DAZL, DAZ2, and BOULE were packaged into gag/pol-293T cells with the packaging plasmids of Δ8.9 and VSVG, and the lentivirals were collected and transfected to human Sertoli cells by polybrene. The transfected cells were then selected by 4 μg/mL blasticidin as determined by preliminary experiments. As a control, p2k7-null vector was packaged into gag/pol-293T cells, and there were no positive cells after blasticidin selection in human Sertoli cells by introducing p2k7-null lentiviral (data not shown). After 2 weeks of selection, RT-PCR showed that the transcription of DAZ2, DAZL, and BOULE genes was present in human transfected cells (Figure S4A), whereas these genes were undetected in the primary Sertoli cells (Figure S4A). Immunocytochemistry demonstrated that human transfected cells were positive for DAZL (Figure S4B), DAZ2 (Figure S4C), and BOULE (Figure S4D) proteins. These data indicate that the transfected cells overexpress DAZ2, DAZL, and BOULE genes and their respective proteins.
Human Sertoli cells overexpressing DAZL, DAZ2, and BOULE genes are human SSCs in phenotype
To examine whether human Sertoli cells overexpressing DAZ2, DAZL, and BOULE genes were putative SSCs, we characterized these cells by phenotypic and morphological characteristics. The mRNA of genes for SSCs, spermatocytes, spermatids, and germ cells, including PLZF, GFRA1, GPR125, UCHL1, SYCP3, MLH1, CREST, ACR, TNP2, PRM1, PRM2, and VASA, was undetectable in human Sertoli cells without overexpression of DAZ family genes but with culture with conditioned medium (Figure S5A). Notably, DAZ2, DAZL, and BOULE transcripts were detected in the transfected cells (Figure S5B), whereas mRNA of SOX9, BMP4, SCF, GDNF, WT1, AR, and FSHR was undetectable in these cells (Figure S5B), which reflects the successful reprogramming of the fate of human Sertoli cells by overexpressing DAZ family genes. Morphologically, human Sertoli cells had a fibroblast-like appearance (Figures S5C and S2A), while the transfected cells assumed an oval shape (Figure S5D), which was similar to primary human SSCs in culture. The transcripts of GFRA1 (Figures 1A and S5B), GPR125 (Figures 1A and S5B), RET (Figures 1A and S5B), THY1 (Figures 1A and S5B), UCHL1 (Figures 1B and S5B), and PLZF (Figure S5B) were expressed in the transfected cells. In contrast, mRNA of PIWIL2 (Figure 1B), SYCP1 (Figure 1B), and SYCP3 (Figure 1B), markers for spermatocytes, was undetected in these cells, suggesting that these cells were not converted into spermatocytes. GFRA1, GPR125, THY1, and PLZF have been regarded as the markers of human SSCs (Easley et al., 2012; Fend-Guella et al., 2019; Hermann et al., 2018; Zhao et al., 2018). Immunocytochemistry demonstrated that the transfected cells stained positively for THY1 (Figure 1C), UCHL1 (Figure 1D), GPR125 (Figure 1E), GFRA1 (Figure 1F), and MAGEA4 (gift from Professor Giulio C. Spagnoli, University Hospital of Basel, Switzerland) (Figure 1G). We also evaluated the proliferation potential of the transfected cells using PCNA, a marker for cell proliferation. Immunocytochemistry illustrated that all transfected cells were positive for PCNA, indicating that these transfected cells have a high proliferation capacity (Figure 1H). After replacement of primary antibodies with rabbit or mouse normal IgGs, no immunostaining was seen in isolated human cells (Figures S3D and S3E), thus verifying specific immunostaining of the antibodies mentioned above. Flow cytometric analysis showed that 94.2% of the transfected cells expressed GPR125 (Figure 1I), indicating a high efficiency of the conversion of human Sertoli cells to human SSCs. Collectively, these results suggest that human Sertoli cells overexpressing DAZ2, DAZL, and BOULE genes are phenotypically human SSCs with high proliferation potential.
Figure 1.

Phenotypic characterization of human Sertoli cells overexpressing DAZL, DAZ2, and BOULE genes
(A and B) RT-PCR demonstrated the transcripts of GFRA1 (A), GPR125 (A), RET (A), THY1 (A), UCHL1 (B), PIWIL2 (B), SYCP1 (B), and SYCP3 (B) in human Sertoli cells overexpressing DAZL, DAZ2, and BOULE genes (the transfected cells). GAPDH served as a loading control of total RNA, and cDNAs from human testis tissues of OA patients were utilized as the positive controls. Replacement of RNA with water and PCR using GAPDH primers was used as a negative control. The data shown in (A and B) are from at least three independent experiments.
(C–H) Immunocytochemistry demonstrated that the presence of THY1 (C), UCHL1 (D), GPR125 (E), GFRA1 (F), MAGEA4 (G), and PCNA (H) proteins in human Sertoli cells overexpressing three DAZ family genes. Scale bars, 10 μm in (C–F) and right panel in (G); 20 μm left panel in (G) and (I).
(I–J) Flow cytometric analysis revealed the expression of GPR125 (I) and mouse or rabbit IgG (J) in human Sertoli cells overexpressing three DAZ family genes. Three independent experiments were performed.
Human Sertoli cell-derived SSCs colonize and proliferate in vivo in the recipient nude mice
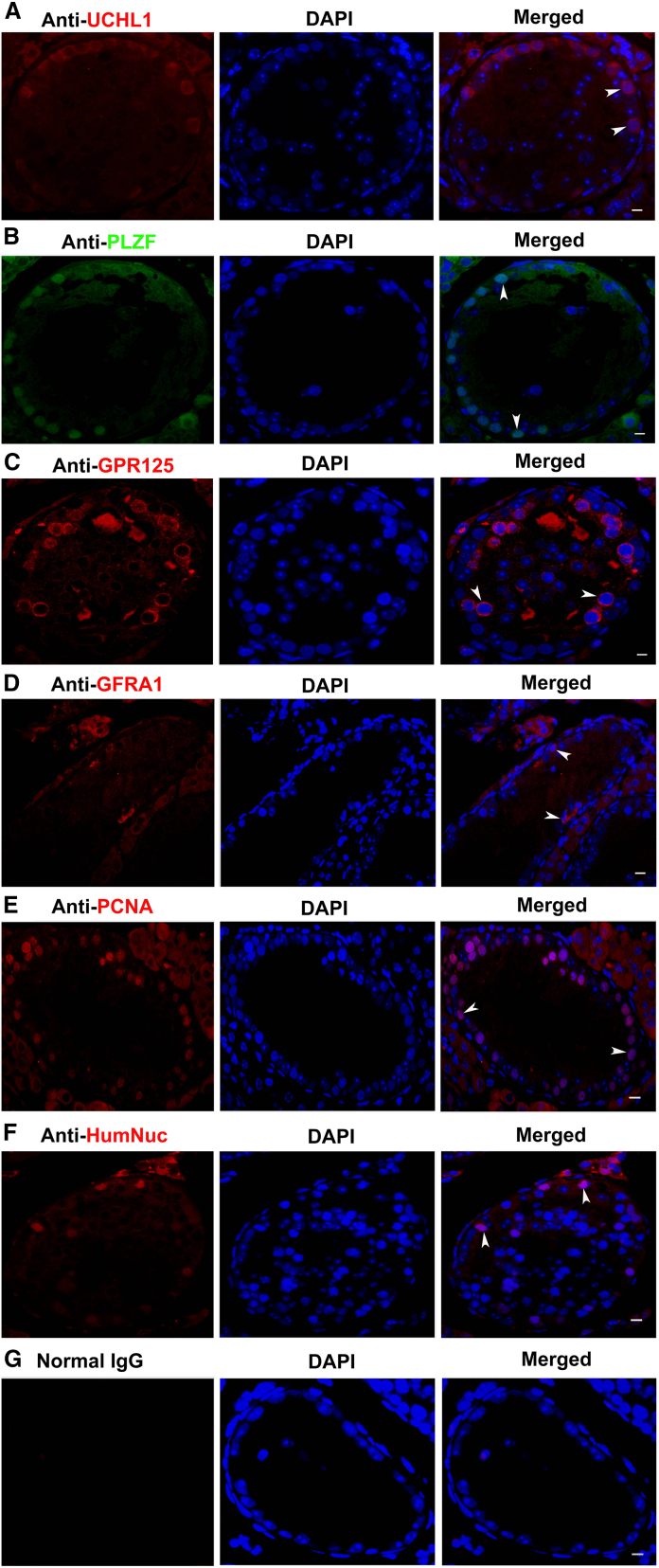
We further determined whether human Sertoli cell-derived SSCs could colonize and proliferate in vivo in busulfan-treated nude mice. Four weeks after treatment of busulfan, H&E staining showed that male germ cells were depleted, whereas only Sertoli cells remained in the seminiferous tubules of mice (Figure S6A). Around 90% of the testes of recipient nude mice were transplanted with human Sertoli cell-derived cells (Figure S6B). Two months after transplantation of human Sertoli cell-derived SSCs, immunohistochemistry revealed that UCHL1 (Figure 2A), PLZF (Figure 2B), GPR125 (Figure 2C), and GFRA1 (Figure 2D), hallmarks for human SSCs, were expressed in the cells (indicated by arrowheads) within seminiferous tubules of recipient nude mice. Meanwhile, PCNA was strongly expressed in the cells of seminiferous tubules of recipient nude mice (Figure 2E). Furthermore, we utilized an antibody against human nuclear antigen (HumNuc) that specifically identifies the cells of human source rather than recipient mice, and found that HumNuc was detected in the cells within the seminiferous tubules of the recipient mice (Figure 2F). After replacement of primary antibodies with normal rabbit IgG, no immunostaining was seen in the cells within the seminiferous tubules of recipient nude mice (Figure 2G). Double immunostaining was performed using antibodies against HumNuc and PLZF, and we observed that a number of cells within the seminiferous tubules of recipient nude mice with human Sertoli cell-derived SSC transplantation were coexpressing HumNuc and PLZF (Figures S6C and S7A). No staining of HumNuc or PLZF in the cells within the seminiferous tubules of recipient nude mice without cell transplantation was used as a negative control (Figure S7B), whereas the coexpression of HumNuc and PLZF in the testis of OA patients served as a positive control (Figure S7C). These results clearly indicate that human Sertoli cell-derived SSCs can survive, colonize, and proliferate in vivo in the recipient nude mice.
Figure 2.
Phenotypic characterization of human Sertoli cell-derived SSCs in vivo
(A–G) (A) Immunohistochemistry revealed the expression of UCHL1 (A), PLZF (B), GPR125 (C), GFRA1 (D), PCNA (E), human nuclear antigen (HumNuc) (F), and normal mouse or rabbit IgG (G) in the seminiferous tubules of recipient nude mice with transplantation of human Sertoli cell-derived SSCs. The typically positive cells for the antibodies are indicated by arrowheads. Scale bars, 10 μm in (A–G). Three independent experiments were performed.
Human Sertoli cell-derived SSCs have similar transcriptomes and proteomics with human SSCs
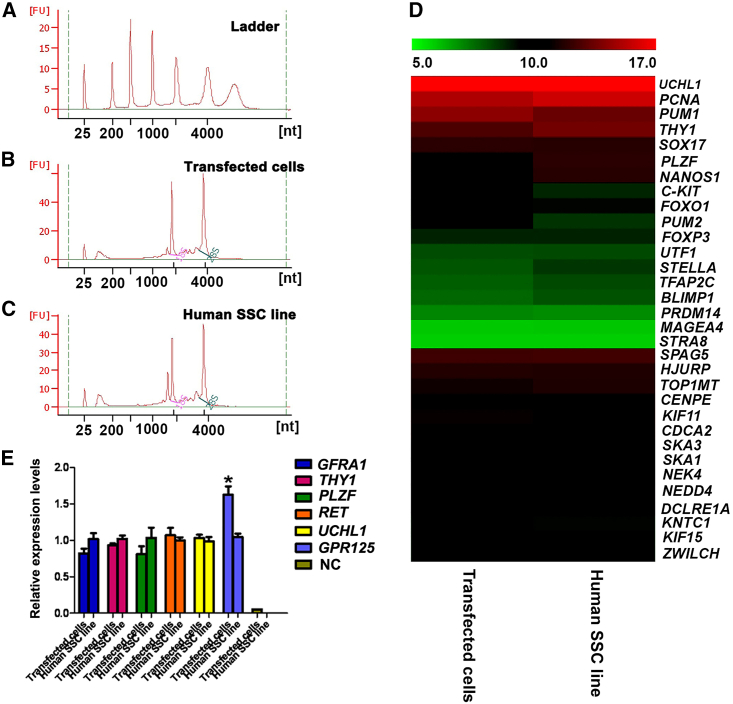
We compared the global gene expression profiles between human Sertoli cell-derived SSCs and the human SSC line that was established by our group in primary human SSCs expressing human SV40 large T antigen (Hou et al., 2015). An electropherogram indicated the high quality of total RNA isolated from human Sertoli cell-derived SSCs (transfected cells) (Figure 3B) and the human SSC line (Figure 3C). In total, 28,226 genes were detected by microarray analysis in both cell types, and 876 (3.1%) and 429 (1.5%) genes were upregulated and downregulated in human Sertoli cell-derived SSCs compared with the human SSC line, respectively. As such, 95.4% of global gene profiles in human Sertoli cell-derived SSCs were similar to that of the human SSC line. Microarray analysis revealed that there was no significant difference in the expression levels of UCHL1, GFRA1, THY1, PLZF, RET, and MAGEA4, markers for human SSCs and spermatogonia, between human Sertoli cell-derived SSCs and the human SSC line (Table S2). A number of genes that were related to reproduction and cell division were chosen for further hierarchical clustering analysis, which showed that there was no significant difference in the levels of UCHL1, PCNA, THY1, SOX17, PLZF, NANOS1, C-KIT, FOXP3, UTF1, and MAGEA4 between human Sertoli cell-derived SSCs and the human SSC line (Figure 3D). Real-time PCR was performed to verify the results of microarray analysis. We found that there was no significant difference in the expression levels of GFRA1, THY1, PLZF, RET, and UCHL1 between human Sertoli cell-derived cells and the human SSC line (Figure 3E), except that the expression level of GPR125 was higher in human Sertoli cell-derived SSCs than in the human SSC line (Figure 3E). Together, microarray analyses further demonstrate that human Sertoli cell-derived SSCs have similar transcriptomes to human SSCs.
Figure 3.
Global gene expression of human Sertoli cell-derived SSCs
(A–C) Electropherogram using the Agilent bioanalyzer indicated the concentrations and nucleotides (nt) of total RNA isolated from human Sertoli cell-derived SSCs (transfected cells) (B) and the human SSC line (C). The RNA ladders are shown in (A).
(D) Microarray clustering analysis of a number of genes related to reproduction and cell division in human Sertoli cell-derived SSCs (transfected cells) and the human SSC line. Two independent experiments were performed.
(E) Real-time PCR verified the expression levels of numerous genes, including GFRA1, THY1, PLZF, RET, UCHL1, and GPR125 between human Sertoli cell-derived SSCs (transfected cells) and the human SSC line. ∗Statistically significant differences (p < 0.05) between human Sertoli cell-derived SSCs (transfected cells) and the human SSC line. NC, negative control. Three independent experiments were performed.
Label-free quantitative proteomics was performed to compare the large scale of protein expression profiling between human Sertoli cell-derived SSCs and the human SSC line. The proteins of human Sertoli cell-derived SSCs and the human SSC line were separated and effectively ranged from 15 to 220 kDa without any degradation (Figure 4A). In total, 4,333 proteins were detected in both kinds of cell types, while 568 and 632 proteins were especially expressed in human Sertoli cell-derived SSCs and the human SSC line, respectively. In addition, 150 and 219 proteins were upregulated and downregulated, respectively, in human Sertoli cell-derived SSCs compared with the human SSC line (Figure 4B; Table S3). Therefore, 91.48% of proteins in human Sertoli cell-derived SSCs were similar to that in the human SSC line (Table S3). The Pearson correlation among human Sertoli cell-derived SSCs (transfected cells) and the human SSC line ranged from 0.88 to 0.95 (Figure 4C), reflecting that human Sertoli cell-derived SSCs assume similar proteomics with human SSCs. We then selected 48 proteins that were related to stem cell and reproduction for further hierarchical clustering analysis, and showed that there was no significant expression difference in these proteins between human Sertoli cell-derived SSCs and the human SSC line (Figure 4D). Western blots were performed to verify the results of label-free quantitative proteomics, and no statistically significant difference was seen in the levels of UCHL1 (Figure 4E) and THY1 (Figure 4F) proteins between human Sertoli cell-derived SSCs and the human SSC line, which was consistent with the results of label-free quantitative proteomics.
Figure 4.
Label-free quantitative proteomics of human Sertoli cell-derived SSCs
(A) The protein quality assessment of human Sertoli cell-derived SSCs (transfected cells) and the human SSC line.
(B) Volcano plot of global protein profiles in human Sertoli cell-derived SSCs (transfected cells) and the human SSC line. The upregulated and downregulated proteins are represented by red and green dots, respectively.
(C) Pearson correlation between human Sertoli cell-derived SSCs (transfected cells) and the human SSC line.
(D) Hierarchical clustering analysis of proteins related to stem cell and reproduction in human Sertoli cell-derived SSCs (transfected cells) and the human SSC line. Two independent experiments were performed.
(E and F) Western blots illustrate the relative expression levels of UCHL1 (E) and THY1 (F) in human Sertoli cell-derived SSCs (transfected cells) and the human SSC line after normalization to ACTB. The results shown in (E and F) are from three independent experiments.
Human Sertoli cell-derived SSCs assume normal chromosomes and exclude Y chromosome microdeletions
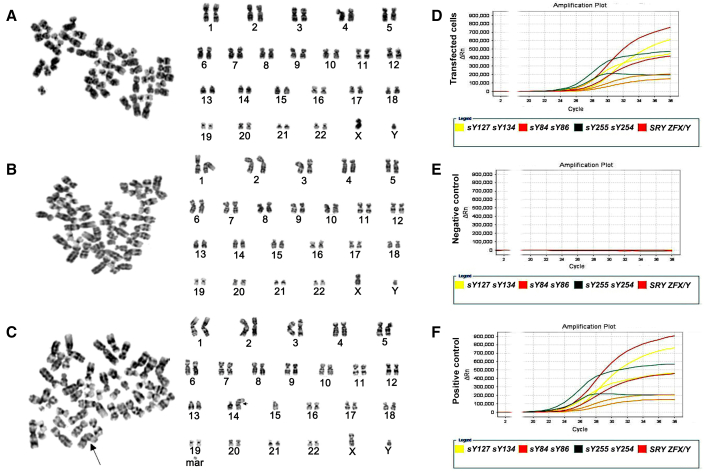
Transferring exogenous genes into cells may cause an abnormal karyotype, and thus we checked the chromosomes of human Sertoli cell-derived SSCs overexpressing DAZ family genes. Karyotype analysis revealed that 92.5% of the transfected cells had normal karyotype with 23 pairs of chromosomes (Figures 5A and 5B), while 7.5% of these cells showed an abnormal karyotype with an unbalanced translocation or chromosomal numerical aberrations (Figure 5C). Together, these data suggest that human Sertoli cell-derived SSCs assume almost normal chromosomes.
Figure 5.
Karyotype and Y chromosome microdeletion analyses of human Sertoli cell-derived SSCs
(A–C) Cytogenetic assay revealed a normal karyotype with 23 pairs of chromosomes (A and B) and abnormal karyotype unbalanced translocation or chromosomal numerical aberrations (C) in human Sertoli cell-derived SSCs.
(D–F) Multiplex real-time PCR displays the expression of all eight specific STS markers from AZFa, AZFb, and AZFc regions, including sY127, sY134, sY86, sY84, sY254, sY255, SRY, and ZFX/Y, in human Sertoli cell-derived SSCs (D). Water substituting for DNA was used as a negative control (E), and DNA from normal human blood served as a positive control (F). The results shown in (A–F) are from three independent experiments.
Multiplex PCR was used to evaluate whether human Sertoli cell-derived SSCs had Y chromosome microdeletions. As shown in Figure 5D, eight specific genes, including sY127, sY134, sY86, sY84, sY254, sY255, SRY, and ZFX/Y, were detected in human Sertoli cell-derived SSCs. DNA was substituted by water and used as a negative control (Figure 5E), while DNA from normal blood served as a positive control (Figure 5F). These data implicate that human Sertoli cell-derived SSCs exclude Y chromosome microdeletions.
Human Sertoli cell-derived SSCs can be induced to differentiate into spermatocytes and spermatids in vitro
We finally determined whether human Sertoli cell-derived SSCs could initiate and progress through meiosis in vitro. Before induction, RT-PCR revealed that human Sertoli cell-derived SSCs were expressing GPR125, GFRA1, THY1, UCHL1, RET, and PLZF, markers for human SSCs, as well as DAZ2, DAZL, and BOULE (Figure 6A), suggesting that the identity of these cells were human SSCs. After 12–14 days of induction, the transcripts of CREST, SYCP3, MLH1, PIWIL2, rH2AX, TNP2, ACR, and PRM1 were detected in these cells (Figure 6B). Triple immunostaining for detecting SYCP3, MLH1, and CREST was performed. Meiotic spreads revealed that SYCP3- and CREST-positive cells were observed in human Sertoli cell-derived SSCs after 12 days of induction (Figure 6C) and that a subset of cells had punctuate SYCP3 staining (Figure 6C), whereas MLH1 (DNA repair protein) was not detected in these cells (Figure 6C), implicating that these cells were likely leptotene spermatocytes of meiotic prophase I. Notably, we found that human Sertoli cell-derived SSCs after 14 days of induction coexpressed SYCP3, MLH1, and CREST (Figure 6E), indicating that these cells are probably pachytene spermatocytes. The expression of SYCP3, CREST, and MLH1 in leptotene spermatocytes (Figure 6D) and pachytene spermatocytes (Figure 6F) from OA patients served as positive controls.
Figure 6.
Phenotypic characterization of the differentiated cells from human Sertoli cell-derived SSCs after 12–14 days of induction
(A) RT-PCR showed the transcripts of GPR125, GFRA1, THY1, UCHL1, RET, PLZF, DAZ2, DAZL, and BOULE in human Sertoli cell-derived SSCs.
(B) RT-PCR demonstrated the mRNA of CREST, SYCP3, MLH1, PIWIL2, rH2AX, TNP2, ACR, and PRM1 in the differentiated cells obtained from human Sertoli cell-derived SSCs for 12–14 days of induction. GAPDH was employed as a loading control of total RNA, and RNA without RT (RT−) but with PCR of GAPDH primers utilized as a negative control. The results shown in (A and B) are from at least three independent experiments.
(C–F) Meiotic spreads show the coexpression of SCP3 (red fluorescence), MLH1 (green fluorescence), and CREST (blue fluorescence) in human Sertoli cell-derived SSCs after 12 days of induction (C) and 14 days of induction (E). The expression of SYCP3, CREST, and MLH1 in leptotene spermatocytes (D) and pachytene spermatocytes (F) from OA patients was used as the positive controls. Three independent experiments were performed in (C–F). Scale bars, 10 μm in (C–F).
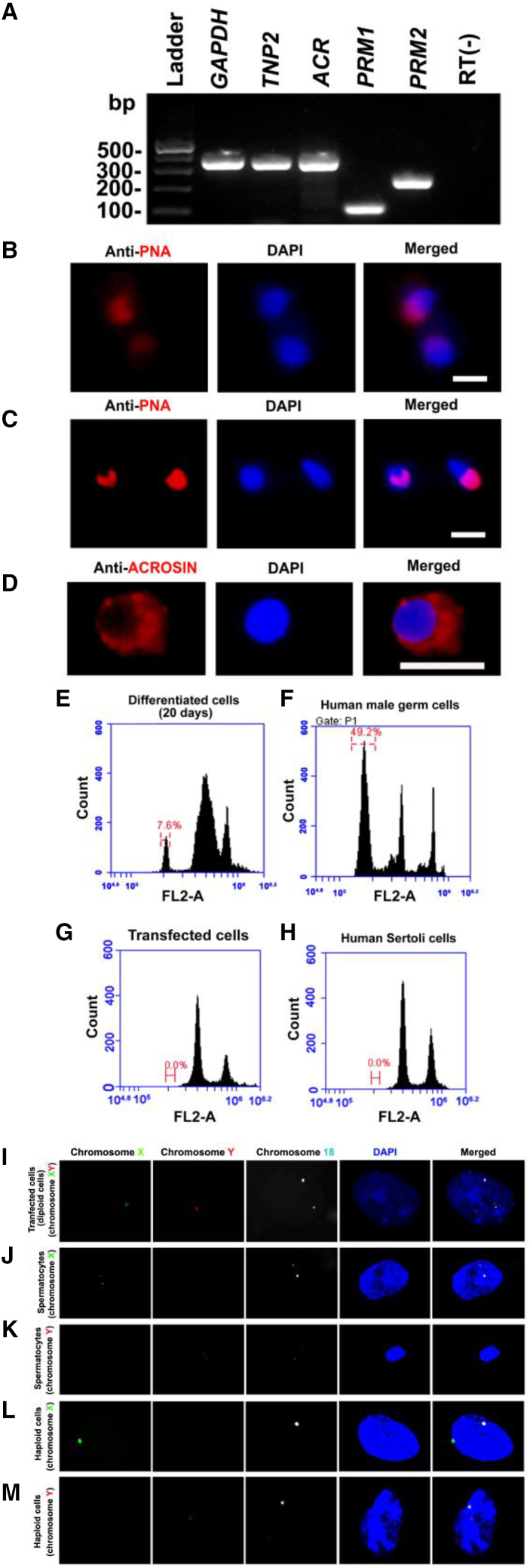
RT-PCR revealed that the transcripts of TNP2, ACR, PRM1, and PRM2, hallmarks for haploid spermatids, were detected in human Sertoli cell-derived SSCs after 15–20 days of induction (Figure 7A). Immunocytochemistry illustrated the expression of PNA (Figures 7B and 7C) at various stages and ACROSIN (Figure 7D) in differentiated cells. FACS of DNA contents showed that 7.6% of haploid (1N) cells could be obtained from human Sertoli cell-derived SSCs at day 20 of differentiation by induction (Figure 7E), whereas no haploid (1N) cells could be detected in the transfected cells (human Sertoli cell-derived SSCs) (Figure 7G) or untransfected cells (human Sertoli cells) (Figure 7H). The percentage of haploid (1N) cells in human male germ cells of OA patients was utilized as positive cells (Figure 7F). Fluorescence in situ hybridization (FISH) further demonstrated that haploid (1N) cells with chromosome X (Figure 7L) and chromosome Y (Figure 7M), as well as the secondary spermatocytes (4N) with chromosome X (Figure 7J) and chromosome Y (Figure 7K), were obtained from the human Sertoli cell-derived SSCs (2N) (Figure 7I) after differentiation, which illustrates the generation of haploid cells (1N) from human Sertoli cell-derived SSCs. Collectively, these data indicate that human SSCs derived from human Sertoli cells overexpressing of DAZ family genes can be induced to differentiate into spermatocytes and haploid spermatids.
Figure 7.
Phenotypic characterization and DNA contents assays of the differentiated cells from human Sertoli cell-derived SSCs after 15–20 days of induction
(A) RT-PCR displays the transcription of TNP2, ACR, PRM1, and PRM2 in human Sertoli cell-derived SSCs after 15–20 days of induction. GAPDH was used as a loading control of total RNA, and RNA without RT (RT−) but with PCR of GAPDH primers served as a negative control. The results shown in (A) are from at least three independent experiments.
(B–D) Immunocytochemistry revealed the expression of PNA (B and C) and ACROSIN (D) in the differentiated cells from human Sertoli cell-derived SSCs after 15–20 days of induction. Scale bars, 5 μm in (B–D).
(E–H) FACS analysis indicated the percentages of haploid cells (1N) in the differentiated cells from human Sertoli cell-derived SSCs at day 20 of induction (E), male germ cells of OA patients (F), the transfected cells (human Sertoli cell-derived SSCs) (G), and human Sertoli cells (H).
(I–M) FISH showed the ploidy of the secondary spermatocytes with chromosome X (J) and chromosome Y (K) as well as the haploid cells with chromosome X (L) and chromosome Y (M) derived from human Sertoli cell-derived SSCs (I). Three independent experiments were performed in (B–M).
Discussion
Germ cells have recently been generated in vitro from mouse and human ESCs as well as iPSCs (Easley et al., 2012; Kee et al., 2009; Zhou et al., 2016). In this study, we demonstrate that the overexpression of DAZL, DAZ2, and BOULE genes could effectively reprogram human Sertoli cells into human SSCs with self-renewal and differentiation potentials. To our knowledge, this is the first report highlighting the generation of phenotypic attributes of human germline stem cells directly from human somatic cells. Significantly, our human Sertoli cell-derived SSCs could be further coaxed to differentiate into spermatocytes and spermatids by a specific medium in vitro, which could offer an invaluable source of male gametes to treat male infertility in patients with azoospermia.
We identified human Sertoli cells overexpressing DAZL, DAZ2, and BOULE as putative human SSCs, since they possess the phenotypic characteristics of human primary SSCs both in vitro and in vivo. First of all, these cells shared a number of markers for human SSCs, including UCHL1, PLZF, GFRA1, RET, GPR125, and THY1, at the transcriptional and translational levels. Secondly, microarray analysis highlighted that the transfected cells have 95.4% similarity of global gene profiles with the human SSC line that possesses the features of human primary SSCs (Hou et al., 2015), implying that the transfected cells from Sertoli cells possess biochemical characteristics of human SSCs. Thirdly, SSC transplantation has been generally regarded as the gold standard assay for evaluating the function of SSCs in vivo (Brinster and Zimmermann, 1994). In this study, we transplanted the transfected cells into the testis of recipient nude mice. Notably, the transfected cells could survive for 2 months, colonize, and proliferate in the seminiferous tubules of recipient nude mouse testes, as demonstrated by the expression of SSC markers, e.g., GPR125, GFRA1, UCHL1, PLZF, HumNuc, and PCNA in the recipient mice, which was consistent with the typical attribute of primary human SSCs for xenotransplantation. Finally, human Sertoli cell-derived SSCs can be induced to initiate and progress meiosis to become spermatocytes and haploid spermatids, as demonstrated by RT-PCR, immunocytochemistry, meiotic spread assays, FACS analysis of DNA contents, and FISH of chromosomes.
To evaluate the safety of the process of transfection, we checked the karyotype and Y chromosome microdeletions of human Sertoli cell-derived SSCs. Chromosomal changes, e.g., chromosomal numbers and chromosomal banding pattern, may occur when transferring exogenous genes into cells. It is worth noting that 92.5% of human Sertoli cell-derived SSCs had normal chromosomal characteristics, which is in accordance with the previous studies showing that transferring exogenous genes into cells causes less change in chromosomes (Akimov et al., 2005; Balducci et al., 2014). Using multiplex PCR with a number of genes, we found that no Y chromosome microdeletion was seen in human Sertoli cell-derived SSCs. As such, human Sertoli cell-derived SSCs may have significant applications in both reproductive and regenerative medicine.
Mammalian spermatogenesis starts with the self-renewal and differentiation of SSCs, giving rise to certain kinds of spermatocytes and haploid spermatids (Bellve et al., 1977). To induce human Sertoli cell-derived SSCs to initiate meiosis and gametogenesis in vitro, we used the conditioned medium containing testosterone, BMP4, activin A, retinoic acid (RA), stem cell factor (SCF), EGF, BPE, and KSR, which are essential for spermatogenesis. BMP4 and activin A promote the self-renewal and proliferation of germ cells (Carlomagno et al., 2010; Mithraprabhu et al., 2010). RA has been shown to be sufficient for inducing spermatocytes from mouse SSCs and is required for the development of complete meiosis and generation of haploid cells from both mouse ESCs and human iPSCs (Eguizabal et al., 2011; Nayernia et al., 2006; Wang et al., 2016). We have reported that RA and SCF induce human SSCs to differentiate into haploid spermatids (Yang et al., 2014). BMP4 cooperates with the RA to stimulate the differentiation of mouse spermatogonia (Yang et al., 2016). In this study, we found expression of SYCP3, MLH1, and CREST in human Sertoli cell-derived SSCs after 14 days of induction, indicating the formation of pachytene spermatocytes. We next asked if human Sertoli cell-derived spermatocytes further develop into haploid spermatids, and thus we checked the expression of PNA, ACROSIN, and PRM2, which are markers of haploid spermatids. As expected, the expression of these three genes or proteins was detected in human Sertoli cell-derived SSCs after 15–20 days of induction. Notably, 7.6% of haploid cells, as assessed by FACS and FISH, can be obtained by our conditioned medium, which was comparable with the efficiency for our generation of haploid spermatids from human SSCs (Sun et al., 2018).
In summary, we have demonstrated, for the first time, the direct reprogramming human Sertoli cells into human germline stem cells, with phenotypic characteristics and high safety, by overexpressing three DAZ family genes. Significantly, human Sertoli cell-derived SSCs can colonize and renew in vivo and differentiate into spermatocytes and haploid spermatids in vitro. Our ability to directly reprogram human Sertoli cells and other somatic cells into male germline stem cells and haploid spermatids may provide invaluable male gametes for azoospermia patients.
Experimental procedures
Reprogramming of human Sertoli cells into human SSCs
The plasmids used in this study, including p2k7-DAZL, p2k7-DAZ2, p2k7-BOULE, and p2k7-null, were kindly provided by Dr. Kehkooi Kee, Professor at Tsinghua University School of Medicine, China. Lentivirus packaging plasmids including Δ8.9 and Vsvg were packaged, respectively, using two packaging plasmids and plasmids p2k7-DAZL, p2k7-DAZ2, p2k7-BOULE, or p2k7-null into gag/pol-293T cells with lipofectamine 3000 (Invitrogen) according to the manufacturer’s instructions. Expression titers were determined by infecting the 293T cells with series of dilutions. About 1.5 × 105 human Sertoli cells and 1.5 × 105 human skin fibroblast HFF-1 (ATTC no. SCRC-1041) were transfected with 1–10 μL of lentiviral overexpression vectors for DAZL, DAZ2, BOULE, and p2k7-null with 8 μg/mL polybrene (Sigma-Aldrich) in DMEM medium for 24 h in 5% CO2 incubator and recovered for another 24 h. Culture medium was changed every 2 days with new medium plus 4 μg/mL blasticidin for about 14 days for selection of cells. The positive cells after blasticidin selection were used for further identification and following studies.
Chromosomal karyotype analysis
Chromosomal karyotype analysis of the exponentially growing human Sertoli cell-derived SSCs at passage 8 was performed in terms of the method as described previously (Ma et al., 2013; Yang et al., 2014). In brief, the cells were treated with 5 μg/mL colcemid for 3 h and by 0.075 M KCl solution for 25 min at 37°C, and they were fixed twice in fixative (methanol:glacial acetic acid = 3:1) for 20 min. Cell suspension was cytospun onto the slides. Cell slides were stained with Giemsa solution and counted under a microscope. The chromosome status was analyzed with the recommendation of the International System for Human Cytogenetic Nomenclature.
Label-free quantitative proteomics
The transfected cells (human Sertoli cell-derived SSCs) and the human SSC line were lysed with RIPA (Santa Cruz) buffer for 30 min on ice. Cell lysates were cleared by centrifugation at 12,000 × g for 20 min at 4°C, and the concentrations of the proteins were determined using the Qubit fluorescent protein quantification kit (Invitrogen) according to the manufacturer’s instruction. Proteins were reduced by DTT (DL-dithiothreitol) to a final concentration of 10 mM at 56°C for 1 h, and they were diluted by 10 times with 250 mM iodoacetamide and kept in the dark for 1 h. Protein samples were digested with trypsin at 37°C for 12 h. The digested supernatant fractions were stored at −80°C until high performance liquid chromatography (HPLC)-tandem mass spectrometry analysis.
Label-free analysis was performed using an Agilent 1100 quaternary HPLC system, micrOTOF-Q II mass spectrometer (Bruker Scientific) coupled with QExactive mass spectrometer (Thermo Scientific, Bremen, Germany). Bioinformatics analysis was performed using Mascot2.2 and Profile Analysis software 2.0. For comparison of differences, the differentially expressed proteins were selected as fold changes ≥1.5 and p < 0.05.
Xenotransplantation of human Sertoli cell-derived SSCs
The xenotransplantation of human Sertoli cell-derived SSCs was performed according to the procedure as described previously (Hermann et al., 2012). In brief, 20 male nude mice 6–8 weeks old were used as recipients, and they were treated with 40 mg/kg body weight of busulfan (Sigma, St Louis, MO, USA) to remove endogenous male germ cells. Four weeks after busulfan treatment, human Sertoli cell-derived SSCs were harvested using 0.25% trypsin and resuspended in DMEM/F12 at a cell concentration of 1–3 × 107/mL. Filtered trypan blue was added to cell suspension and agitated before cell transplantation. Approximately 10 μL of cell suspension was transplanted into seminiferous tubules of one testis via rete testis, while the other testis without cell injection served as internal control. Eight weeks after cell transplantation, recipient mice were killed, and testes were collected for preparing the paraffin sections and immunohistochemistry.
FISH
FISH was conducted to further assess the ploidy of various kinds of cells, including human Sertoli cell-derived SSCs and the secondary spermatocytes as well as the haploid cells derived from human Sertoli cell-derived SSCs pursuant to the methods as we described previously (Sun et al., 2018).
Statistical analysis
All values were presented as mean ± SEM from at least three independent experiments. Statistical analyses were evaluated using Student’s t test, and p < 0.05 was considered statistically significant.
Author contributions
W.Z. and W.C. performed the experiments, wrote the manuscript, and helped with data analysis. Y.C., L.W., Q.Y., F.Z., Q.Q., and M.S. assisted with the experiments and data analysis. Z.L. assisted with the testicular tissue collection and experiments. Z.H. was responsible for the conception and design, supervision of all aspects of the laboratory experiments, data analysis, writing the manuscript, and final approval of the manuscript. All authors approved the manuscript.
Conflicts of interest
The authors declare no competing interests.
Acknowledgments
The authors thank Dr. Kehkooi Kee, Professor at Tsinghua University School of Medicine, China, for providing the plasmids overexpressing DAZL, DAZ2, and BOULE. This work was supported by grants from the National Nature Science Foundation of China (32170862, 31872845) and the Chinese Ministry of Science and Technology (2016YFC1000606), the High Level Talent Gathering Project in Hunan Province (2018RS3066), the Major Scientific and Technological Projects for Collaborative Prevention and Control of Birth Defect in Hunan Province (2019SK1012), and Key Grant of Research and Development in Hunan Province (2020DK2002).
Published: October 14, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.stemcr.2021.09.011.
Supplemental information
Data and code availability
Microarray data are available under NCBI GEO accession no: GSE184088.
References
- Akimov S.S., Ramezani A., Hawley T.S., Hawley R.G. Bypass of senescence, immortalization, and transformation of human hematopoietic progenitor cells. Stem Cells. 2005;23:1423–1433. doi: 10.1634/stemcells.2005-0390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balducci L., Blasi A., Saldarelli M., Soleti A., Pessina A., Bonomi A., Cocce V., Dossena M., Tosetti V., Ceserani V., et al. Immortalization of human adipose-derived stromal cells: production of cell lines with high growth rate, mesenchymal marker expression and capability to secrete high levels of angiogenic factors. Stem Cell Res. Ther. 2014;5:63. doi: 10.1186/scrt452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellve A.R., Cavicchia J.C., Millette C.F., O'Brien D.A., Bhatnagar Y.M., Dym M. Spermatogenic cells of the prepuberal mouse. Isolation and morphological characterization. J. Cell Biol. 1977;74:68–85. doi: 10.1083/jcb.74.1.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brinster R.L., Zimmermann J.W. Spermatogenesis following male germ-cell transplantation. Proc. Natl. Acad. Sci. U S A. 1994;91:11298–11302. doi: 10.1073/pnas.91.24.11298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buganim Y., Itskovich E., Hu Y.C., Cheng A.W., Ganz K., Sarkar S., Fu D., Welstead G.G., Page D.C., Jaenisch R. Direct reprogramming of fibroblasts into embryonic Sertoli-like cells by defined factors. Cell Stem Cell. 2012;11:373–386. doi: 10.1016/j.stem.2012.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlomagno G., van Bragt M.P., Korver C.M., Repping S., de Rooij D.G., van Pelt A.M. BMP4-induced differentiation of a rat spermatogonial stem cell line causes changes in its cell adhesion properties. Biol. Reprod. 2010;83:742–749. doi: 10.1095/biolreprod.110.085456. [DOI] [PubMed] [Google Scholar]
- Cerda-Esteban N., Naumann H., Ruzittu S., Mah N., Pongrac I.M., Cozzitorto C., Hommel A., Andrade-Navarro M.A., Bonifacio E., Spagnoli F.M. Stepwise reprogramming of liver cells to a pancreas progenitor state by the transcriptional regulator Tgif2. Nat. Commun. 2017;8:14127. doi: 10.1038/ncomms14127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D., Sun N., Hou L., Kim R., Faith J., Aslanyan M., Tao Y., Zheng Y., Fu J., Liu W., et al. Human primordial germ cells are specified from lineage-primed progenitors. Cell Rep. 2019;29:4568–4582.e4565. doi: 10.1016/j.celrep.2019.11.083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Easley C.A.t., Phillips B.T., McGuire M.M., Barringer J.M., Valli H., Hermann B.P., Simerly C.R., Rajkovic A., Miki T., Orwig K.E., Schatten G.P. Direct differentiation of human pluripotent stem cells into haploid spermatogenic cells. Cell Rep. 2012;2:440–446. doi: 10.1016/j.celrep.2012.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberhart C.G., Maines J.Z., Wasserman S.A. Meiotic cell cycle requirement for a fly homologue of human deleted in azoospermia. Nature. 1996;381:783–785. doi: 10.1038/381783a0. [DOI] [PubMed] [Google Scholar]
- Eguizabal C., Montserrat N., Vassena R., Barragan M., Garreta E., Garcia-Quevedo L., Vidal F., Giorgetti A., Veiga A., Izpisua Belmonte J.C. Complete meiosis from human induced pluripotent stem cells. Stem Cells. 2011;29:1186–1195. doi: 10.1002/stem.672. [DOI] [PubMed] [Google Scholar]
- Fang F., Angulo B., Xia N., Sukhwani M., Wang Z., Carey C.C., Mazurie A., Cui J., Wilkinson R., Wiedenheft B., et al. A PAX5-OCT4-PRDM1 developmental switch specifies human primordial germ cells. Nat. Cell Biol. 2018;20:655–665. doi: 10.1038/s41556-018-0094-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fend-Guella D.L., von Kopylow K., Spiess A.N., Schulze W., Salzbrunn A., Diederich S., El Hajj N., Haaf T., Zechner U., Linke M. The DNA methylation profile of human spermatogonia at single-cell- and single-allele-resolution refutes its role in spermatogonial stem cell function and germ cell differentiation. Mol. Hum. Reprod. 2019;25:283–294. doi: 10.1093/molehr/gaz017. [DOI] [PubMed] [Google Scholar]
- Fu X.F., Cheng S.F., Wang L.Q., Yin S., De Felici M., Shen W. DAZ family proteins, key players for germ cell development. Int. J. Biol. Sci. 2015;11:1226–1235. doi: 10.7150/ijbs.11536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghorbel M., Baklouti-Gargouri S., Keskes R., Chakroun N., Sellami A., Fakhfakh F., Ammar-Keskes L. Combined deletion of DAZ2 and DAZ4 copies of Y chromosome DAZ gene is associated with male infertility in Tunisian men. Gene. 2014;547:191–194. doi: 10.1016/j.gene.2014.05.061. [DOI] [PubMed] [Google Scholar]
- Habermann B., Mi H.F., Edelmann A., Bohring C., Backert I.T., Kiesewetter F., Aumuller G., Vogt P.H. DAZ (Deleted in AZoospermia) genes encode proteins located in human late spermatids and in sperm tails. Hum. Reprod. 1998;13:363–369. doi: 10.1093/humrep/13.2.363. [DOI] [PubMed] [Google Scholar]
- Hamazaki N., Kyogoku H., Araki H., Miura F., Horikawa C., Hamada N., Shimamoto S., Hikabe O., Nakashima K., Kitajima T.S., et al. Reconstitution of the oocyte transcriptional network with transcription factors. Nature. 2021;589:264–269. doi: 10.1038/s41586-020-3027-9. [DOI] [PubMed] [Google Scholar]
- Handel M.A., Eppig J.J., Schimenti J.C. Applying "gold standards" to in-vitro-derived germ cells. Cell. 2014;159:216. doi: 10.1016/j.cell.2014.09.018. [DOI] [PubMed] [Google Scholar]
- Hayashi K., Ohta H., Kurimoto K., Aramaki S., Saitou M. Reconstitution of the mouse germ cell specification pathway in culture by pluripotent stem cells. Cell. 2011;146:519–532. doi: 10.1016/j.cell.2011.06.052. [DOI] [PubMed] [Google Scholar]
- He Z., Kokkinaki M., Jiang J., Dobrinski I., Dym M. Isolation, characterization, and culture of human spermatogonia. Biol. Reprod. 2010;82:363–372. doi: 10.1095/biolreprod.109.078550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermann B.P., Cheng K., Singh A., Roa-De La Cruz L., Mutoji K.N., Chen I.C., Gildersleeve H., Lehle J.D., Mayo M., Westernstroer B., et al. The mammalian spermatogenesis single-cell transcriptome, from spermatogonial stem cells to spermatids. Cell Rep. 2018;25:1650–1667.e1658. doi: 10.1016/j.celrep.2018.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermann B.P., Sukhwani M., Winkler F., Pascarella J.N., Peters K.A., Sheng Y., Valli H., Rodriguez M., Ezzelarab M., Dargo G., et al. Spermatogonial stem cell transplantation into rhesus testes regenerates spermatogenesis producing functional sperm. Cell Stem Cell. 2012;11:715–726. doi: 10.1016/j.stem.2012.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou J., Niu M., Liu L., Zhu Z., Wang X., Sun M., Yuan Q., Yang S., Zeng W., Liu Y., et al. Establishment and characterization of human germline stem cell line with unlimited proliferation potentials and no tumor formation. Sci. Rep. 2015;5:16922. doi: 10.1038/srep16922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ieda M., Fu J.D., Delgado-Olguin P., Vedantham V., Hayashi Y., Bruneau B.G., Srivastava D. Direct reprogramming of fibroblasts into functional cardiomyocytes by defined factors. Cell. 2010;142:375–386. doi: 10.1016/j.cell.2010.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkins H.T., Malkova B., Edwards T.A. Kinked beta-strands mediate high-affinity recognition of mRNA targets by the germ-cell regulator DAZL. Proc. Natl. Acad. Sci. U. S. A. 2011;108:18266–18271. doi: 10.1073/pnas.1105211108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao X., Trifillis P., Kiledjian M. Identification of target messenger RNA substrates for the murine deleted in azoospermia-like RNA-binding protein. Biol. Reprod. 2002;66:475–485. doi: 10.1095/biolreprod66.2.475. [DOI] [PubMed] [Google Scholar]
- Kee K., Angeles V.T., Flores M., Nguyen H.N., Reijo Pera R.A. Human DAZL, DAZ and BOULE genes modulate primordial germ-cell and haploid gamete formation. Nature. 2009;462:222–225. doi: 10.1038/nature08562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J., Efe J.A., Zhu S., Talantova M., Yuan X., Wang S., Lipton S.A., Zhang K., Ding S. Direct reprogramming of mouse fibroblasts to neural progenitors. Proc. Natl. Acad. Sci. U. S. A. 2011;108:7838–7843. doi: 10.1073/pnas.1103113108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q., Qiao D., Song N.H., Ding Y., Wang Z.J., Yang J., Wang W., Yin C.J. Association of DAZ1/DAZ2 deletion with spermatogenic impairment and male infertility in the South Chinese population. World J. Urol. 2013;31:1403–1409. doi: 10.1007/s00345-013-1058-7. [DOI] [PubMed] [Google Scholar]
- Luetjens C.M., Xu E.Y., Rejo Pera R.A., Kamischke A., Nieschlag E., Gromoll J. Association of meiotic arrest with lack of BOULE protein expression in infertile men. J. Clin. Endocrinol. Metab. 2004;89:1926–1933. doi: 10.1210/jc.2003-031178. [DOI] [PubMed] [Google Scholar]
- Ma M., Yang S., Zhang Z., Li P., Gong Y., Liu L., Zhu Y., Tian R., Liu Y., Wang X., et al. Sertoli cells from non-obstructive azoospermia and obstructive azoospermia patients show distinct morphology, Raman spectrum and biochemical phenotype. Hum. Reprod. 2013;28:1863–1873. doi: 10.1093/humrep/det068. [DOI] [PubMed] [Google Scholar]
- Maegawa S., Yamashita M., Yasuda K., Inoue K. Zebrafish DAZ-like protein controls translation via the sequence ‘GUUC’. Genes Cell: Devoted Mol. Cell. Mech. 2002;7:971–984. doi: 10.1046/j.1365-2443.2002.00576.x. [DOI] [PubMed] [Google Scholar]
- Maines J.Z., Wasserman S.A. Post-transcriptional regulation of the meiotic Cdc25 protein twine by the Dazl orthologue Boule. Nat. Cell Biol. 1999;1:171–174. doi: 10.1038/11091. [DOI] [PubMed] [Google Scholar]
- Medrano J.V., Ramathal C., Nguyen H.N., Simon C., Reijo Pera R.A. Divergent RNA-binding proteins, DAZL and VASA, induce meiotic progression in human germ cells derived in vitro. Stem Cells. 2012;30:441–451. doi: 10.1002/stem.1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mithraprabhu S., Mendis S., Meachem S.J., Tubino L., Matzuk M.M., Brown C.W., Loveland K.L. Activin bioactivity affects germ cell differentiation in the postnatal mouse testis in vivo. Biol. Reprod. 2010;82:980–990. doi: 10.1095/biolreprod.109.079855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakaki F., Hayashi K., Ohta H., Kurimoto K., Yabuta Y., Saitou M. Induction of mouse germ-cell fate by transcription factors in vitro. Nature. 2013;501:222–226. doi: 10.1038/nature12417. [DOI] [PubMed] [Google Scholar]
- Nayernia K., Nolte J., Michelmann H.W., Lee J.H., Rathsack K., Drusenheimer N., Dev A., Wulf G., Ehrmann I.E., Elliott D.J., et al. In vitro-differentiated embryonic stem cells give rise to male gametes that can generate offspring mice. Dev. Cell. 2006;11:125–132. doi: 10.1016/j.devcel.2006.05.010. [DOI] [PubMed] [Google Scholar]
- Reijo R., Lee T.Y., Salo P., Alagappan R., Brown L.G., Rosenberg M., Rozen S., Jaffe T., Straus D., Hovatta O., et al. Diverse spermatogenic defects in humans caused by Y chromosome deletions encompassing a novel RNA-binding protein gene. Nat. Genet. 1995;10:383–393. doi: 10.1038/ng0895-383. [DOI] [PubMed] [Google Scholar]
- Sasaki K., Yokobayashi S., Nakamura T., Okamoto I., Yabuta Y., Kurimoto K., Ohta H., Moritoki Y., Iwatani C., Tsuchiya H., et al. Robust in vitro induction of human germ cell fate from pluripotent stem cells. Cell Stem Cell. 2015;17:178–194. doi: 10.1016/j.stem.2015.06.014. [DOI] [PubMed] [Google Scholar]
- Saunders P.T., Turner J.M., Ruggiu M., Taggart M., Burgoyne P.S., Elliott D., Cooke H.J. Absence of mDazl produces a final block on germ cell development at meiosis. Reproduction. 2003;126:589–597. doi: 10.1530/rep.0.1260589. [DOI] [PubMed] [Google Scholar]
- Schrans-Stassen B.H., Saunders P.T., Cooke H.J., de Rooij D.G. Nature of the spermatogenic arrest in Dazl –/– mice. Biol. Reprod. 2001;65:771–776. doi: 10.1095/biolreprod65.3.771. [DOI] [PubMed] [Google Scholar]
- Sheng C., Zheng Q., Wu J., Xu Z., Wang L., Li W., Zhang H., Zhao X.Y., Liu L., Wang Z., et al. Direct reprogramming of Sertoli cells into multipotent neural stem cells by defined factors. Cell Res. 2012;22:208–218. doi: 10.1038/cr.2011.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun M., Yuan Q., Niu M., Wang H., Wen L., Yao C., Hou J., Chen Z., Fu H., Zhou F., et al. Efficient generation of functional haploid spermatids from human germline stem cells by three-dimensional-induced system. Cell Death Differ. 2018;25:749–766. doi: 10.1038/s41418-017-0015-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sybirna A., Tang W.W.C., Pierson Smela M., Dietmann S., Gruhn W.H., Brosh R., Surani M.A. A critical role of PRDM14 in human primordial germ cell fate revealed by inducible degrons. Nat. Commun. 2020;11:1282. doi: 10.1038/s41467-020-15042-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venables J.P., Ruggiu M., Cooke H.J. The RNA-binding specificity of the mouse Dazl protein. Nucleic Acids Res. 2001;29:2479–2483. doi: 10.1093/nar/29.12.2479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S., Wang X., Ma L., Lin X., Zhang D., Li Z., Wu Y., Zheng C., Feng X., Liao S., et al. Retinoic acid is sufficient for the in vitro induction of mouse spermatocytes. Stem Cell Reports. 2016;7:80–94. doi: 10.1016/j.stemcr.2016.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu B., Lu N.X., Xia Y.K., Gu A.H., Lu C.C., Wang W., Song L., Wang S.L., Shen H.B., Wang X.R. A frequent Y chromosome b2/b3 subdeletion shows strong association with male infertility in Han-Chinese population. Hum. Reprod. 2007;22:1107–1113. doi: 10.1093/humrep/del499. [DOI] [PubMed] [Google Scholar]
- Xu E.Y., Lee D.F., Klebes A., Turek P.J., Kornberg T.B., Reijo Pera R.A. Human BOULE gene rescues meiotic defects in infertile flies. Hum. Mol. Genet. 2003;12:169–175. doi: 10.1093/hmg/ddg017. [DOI] [PubMed] [Google Scholar]
- Yang S., Ping P., Ma M., Li P., Tian R., Yang H., Liu Y., Gong Y., Zhang Z., Li Z., He Z. Generation of haploid spermatids with fertilization and development capacity from human spermatogonial stem cells of cryptorchid patients. Stem Cell Reports. 2014;3:663–675. doi: 10.1016/j.stemcr.2014.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y., Feng Y., Feng X., Liao S., Wang X., Gan H., Wang L., Lin X., Han C. BMP4 cooperates with retinoic acid to induce the expression of differentiation markers in cultured mouse spermatogonia. Stem Cell Int. 2016;2016:9536192. doi: 10.1155/2016/9536192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Chen M., Wen Q., Li Y., Wang Y., Wang Y., Qin Y., Cui X., Yang L., Huff V., Gao F. Reprogramming of Sertoli cells to fetal-like Leydig cells by Wt1 ablation. Proc. Natl. Acad. Sci. U. S. A. 2015;112:4003–4008. doi: 10.1073/pnas.1422371112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y., Ye S., Liang D., Wang P., Fu J., Ma Q., Kong R., Shi L., Gong X., Chen W., et al. In vitro modeling of human germ cell development using pluripotent stem cells. Stem Cell Reports. 2018;10:509–523. doi: 10.1016/j.stemcr.2018.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Q., Wang M., Yuan Y., Wang X., Fu R., Wan H., Xie M., Liu M., Guo X., Zheng Y., et al. Complete meiosis from embryonic stem cell-derived germ cells in vitro. Cell Stem Cell. 2016;18:330–340. doi: 10.1016/j.stem.2016.01.017. [DOI] [PubMed] [Google Scholar]
- Zhu S., Rezvani M., Harbell J., Mattis A.N., Wolfe A.R., Benet L.Z., Willenbring H., Ding S. Mouse liver repopulation with hepatocytes generated from human fibroblasts. Nature. 2014;508:93–97. doi: 10.1038/nature13020. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Microarray data are available under NCBI GEO accession no: GSE184088.