Abstract
Purpose
To determine the influence of different genotypes of Ala307Thr and Asn680Ser FSHr polymorphisms on controlled ovarian stimulation (COS) outcome and pregnancy.
Methods
This study collected blood and physiological and clinical parameters of 517 Caucasian patients (Statistical power ≥ 80%) that underwent COS treatment. Genotypes of Ala307Thr and Asn680Ser polymorphisms were determined using PCR amplification followed by Bsu36I and BsrI digestion, respectively.
Results
Ala307Ala and Ser680Ser genotypes associated to worse parameters of COS outcome (preovulatory follicles P = 0.05, in both), justifying their lower pregnancy rate than Non-Ala307Ala, P = 0.01 and Non-Ser680Ser, P = 0.004, respectively or together, (P = 0.003). Within the Non-Ala307Ala group, Thr307Thr genotype showed higher number of fertilized oocytes (P = 0.04) and embryos (P = 0.01) than Non-Thr307Thr, but no influence on pregnancy rate. Ala307Ala and Ser680Ser patients doubled probability of non-pregnancy than Non-Ala307Ala (odds ratio = 2.0) and Non-Ser680Ser (odds ratio = 2.11), respectively. Ala307Ala and Ser680Ser genotypes tend to appear together (P < 0.0001), which increases the probability of non-pregnancy.
Conclusions
Ala307Ala and Ser680Ser genotypes of 307 and 680 FSHr polymorphisms associate to worse COS outcome than its respective Non-Ala307Ala and Non-Ser680Ser. Within the Non-Ala307Ala genotypes, Thr307Thr, although shows higher Fertilized Oocytes and Embryos, do not influence on pregnancy rate. Ala307Ala and Ser680Ser genotypes double the probability of Non-Pregnancy than their respective Non-Ala307Ala and Non-Ser680Ser genotypes. Furthermore, the strong tendency of these genotypes to appear together worsens the probability of pregnancy in these patients.
Supplementary Information
The online version contains supplementary material available at 10.1007/s10815-021-02276-0.
Keywords: Assisted reproductive techniques;, Controlled ovarian stimulation (COS);, FSHr polymorphisms;, Ala307Thr polymorphism;, Asn680Ser polymorphism
Introduction
Assisted reproductive techniques (ART) have increased steadily over the years. From 1997 to 2015, the European Society of Human Reproduction and Embryology (ESHRE) has reported about 9 million ART treatments leading to the birth of 1.6 million children [1]. Only in 2015, 7.3% of all children born in Europe was achieved due to ART. In spite of these encouraging results, little is known about the different treatments, factors and genetic biomarkers that influence COS response resulting in improved outcomes.
Since the ovarian response is highly unpredictable, one of the main factors to obtain an optimal ovarian response is the choice of the correct COS protocol [2]. Different studies have attempted to design individualized strategies of COS as a solution to avoid the ovarian hyperstimulation syndrome (OHSS) [3–5] or, on the contrary, unexpected poor response that decreases the chance of obtaining an euploid embryo [6, 7], without obtaining conclusive results.
New specific biomarkers and pharmacogenetic strategies, such as improvement of the design, adjustment, and individualization of the COS protocol, are being explored to improve COS success [8]. One of the most studied factors has been the interaction between FSH and its receptor (FSHr) [9, 10]. FSH has been described as the essential hormone in reproduction, due to its role in promoting follicle growth and regulating ovarian function by binding to FSHr. It has been well-established that accurate FSH-FSHr interaction is essential to allow correct follicular development and oocyte maturation [11].
Functional FSHr is a homodimer of a polypeptide of 695 amino acids, expressed in ovarian granulose cells and anchored in their membrane [12–14]. This protein consists of three domains: extracellular, transmembrane, and intracellular. The intracellular domain is rich in Ser and Thr, potentially phosphorylatable zones, which appear to serve as anchors for the cytoplasmic tail of FSHr and, once FSH interacts with FSHr, to initiate signal transduction of the Estradiol synthesis pathway.
The monomer polypeptide belongs to the protein G group and is encoded by a gene localized in chromosome 2p21 in humans (NCBI database), consisting of 10 exons. Most of the extracellular domain is encoded by 1–9 exons ranging from 69–251 bp each. The last C-terminal part of the extracellular domain, the transmembrane domain, and the intracellular domain are encoded by the exon 10 (1234 bp) [10], in which five (out of approximately 2000) single nucleotide polymorphisms (SNPs) are localized in the FSHr [15]. Two of these SNPs of cDNA, localized in exon 10, have been repeatedly studied in relation to fertility problems in women: FSHr SNP (rs6165) (variant ID 2-49191041-C-T) characterized by a nucleotide change c.919G>A, resulting in amino acid change from Ala to Thr (p.A307T) and the other, FSHr SNP (rs6166) (Variant ID 2-49189921-C-T) characterized by nucleotide substitution A for G at position 2039 (c.2039A>G), determining an Asn to Ser change at position 680 of the amino acid chain (p.N680S) (Web Resources).
Several authors have reported that some genotypes of Ala307Thr and Asn680Ser polymorphisms of FSHr are associated to good or poor response to COS treatment [16–18]. Despite this, an association between the different genotypes of these polymorphisms and infertility has not been documented [14], nor statistical differences achieved in Basal FSH, Luteinizing Hormone (LH) and Estradiol between patients and controls [19], or in different physiological parameters as preovulatory follicles (PF), recruited oocytes (RO) or fertilized oocytes (FO) [20].
Some authors have reported correlation between Ser680Ser [21–25] or Ala307Ala [18] genotypes and poor COS outcome, while others correlated them with higher Basal FSH [26]. This last parameter is considered an indicator of diminished efficiency of FSHr, since patients with both genotypes, together or separately, induce a poor response to exogenous FSH during COS treatment [27].
Other interesting retrospective works, that enrolled patients of different ethnicities and studied COS ovarian response and ovarian reserve, exist; however, their results are inconclusive. This was in part due to standardization of a concrete strategy based on the type of ovarian reserve at the beginning of stimulation [19, 28, 29].
Considering previous investigations, this study focused on study two well-characterized FSHr polymorphisms, Ala307Thr and Asn680Ser [30–34] to determine the influence of their different genotypes on COS outcome and, overall, on pregnancy success in order to design, if necessary, particular COS strategies depending on those genotypes and the ovarian reserve of the patients.
Subjects and methods
The blood samples of this study have been collected over 10 years, only from the patients who signed the informed consent and their partners have normal seminogram or non-severe oligozoospermia or asthenozoospermia, until sample (n=517) achieved a statistical power ≥ 80%, and we got the financial grant to carry it out. This has determined that the study has been extended in time for a decade. Every patient was attended at the Human Assisted Reproduction Unit at the Hospital Universitario Príncipe de Asturias of Alcalá de Henares, Madrid (Spain).
All of patients were younger than 35 years (32.15 ± 2.5), had a personal history of infertility for durations longer than 12 months, normal karyotype, determined by G banding (n = 153) and chromosome painting techniques (n = 364) in external laboratories, in both members of the couple and Body Mass Index (BMI) (weight in kg/(height in m)2) < 30, which rules out spurious associations.
Subjects were fully informed of the aims of the work before signing informed consent forms. The study conforms to the code of Ethics of the World Medical Association (Declaration of Helsinki) and was approved (January 22nd, 2003) by the Committee of Clinical Trials of the Hospital Universitario Príncipe de Asturias of Alcalá de Henares, Madrid (Spain).
Patient treatment
Previously to initiate the COS treatment, when patients were diagnosed of tubal factor infertility problem, the corresponding treatment was applied to ensure sufficient tubal permeability. Patients underwent COS treatment according to the standard stimulation protocols of the medical reproductive unit [34–37]. Most of the patients were stimulated using antagonist protocol with 100–300 daily IU of recombinant FSH or human menopausal gonadotropin (hMG). The initial gonadotropin dose was chosen depending on other IVF cycles previous response, in case other stimulations had been done. Patients with history of poor response were stimulated with higher initial doses, comparing to those with normal COS response. Naïf patients were stimulated according to ovarian reserve biomarkers. Total dose of FSH (TD FSH) have depended on initial dose and length of treatment, 10–12 days in average. An antagonist of gonadotropin-releasing hormone (GnRH) was administered when two follicles reached 14 mm. Ovulation trigger was performed when two follicles almost reached 16 mm, as described in literature [34–37].
Initially, follicular growth and development were evaluated by transvaginal ultrasound on the fifth or sixth day at the beginning and on every other day thereafter, until the triggering day. Ovulation was induced by a single dose of 6500 IU of recombinant human chorionic gonadotropin (r-hCG). In a case of moderate or severe hyperstimulation syndrome risk, triggering was induced using an analogous GnRH bolus. Oocyte retrieval took place 34–36 h after triggering, as usually described in protocols [2, 34–37].
In regard the treatment of the semen samples, we used a concentration gradient (90%, 70%, 50%; Gradient 100 medium, Origio) followed by swim-up in the same medium, to select the best spermatozoa. In the case of non-severe oligo or asthenozoospermia, only swim-up was used. In any way, we make sure that sufficient number of well-formed and progressive spermatozoa were collected to perform IVF or ICSI techniques, reducing, in this way, the possible influence of the male factor of infertility.
Conventional in vitro fertilization (IVF) was performed in 259 women and intracytoplasmatic sperm injection (ICSI) in 258 women, according to standard stimulation protocols [38]. Embryos were scored according to the Spanish Association for the Study of the Biology of Reproduction (ASEBIR) guidelines and pregnancy was confirmed by a blood hCG analysis 14 days after embryo was transferred.
Poor/good responder definition
Patients were classified as poor (<4 oocytes) or good (≥4 oocytes) responders based on the number of oocytes picked up after COS treatment. These classifications were made according to the most accepted criterion proposed by different authors [39–42], although the concept of poor responders is still a major challenge in assisted reproduction.
DNA extraction
Total DNA was extracted from the blood samples of the patients. Extraction was performed according to standard procedures by using the method “phenol-chloroform” with some modifications [43]. Total DNA was recovered, air-dried to eliminate ethanol and finally resuspended in sterile water.
Genotyping Ala307Thr and Asn680Ser polymorphisms of the FSHr
The cDNA positions of the polymorphisms Ala307Thr and Asn680Ser in the FSHr are located at 919 (Variant ID 2-49191041-C-T) and 2039 (Variant ID 2-49189921-C-T), respectively. For determining Ala307Thr polymorphism genotypes, we used nested polymerase chain reaction (PCR). We first amplified a 409-bp fragment in exon 10 by PCR-1 using two specific primers: primer-1 forward 5´-TCTGAGCTTCATCCAATTTG-3´; and primer-1 reverse of a new design 5´-CAACTGATGCAATGAGCAG-3´ according to [28]. The PCR product was then amplified by nested PCR of 364 bp with primers proposed by Sudo et al. [44], primer-2 forward 5´-CAAATCTATTTTAAGGCAAGAAGTTGATTATATGCCTCAG-3´, which included a mismatch nucleotide (bold and underlined) to introduce a Bsu36I restriction site for restriction fragment length polymorphism (RFLP) analysis and the primer-2 reverse 5´-GTAGATTCCAATGCAGAGATCA-3´. The cycling conditions were as follows: initial denaturation at 95°C for 2 min, followed by 40 cycles of 95°C for 1 min, annealing at 58°C for 50 s then elongation at 72°C for 30 s, and a final elongation step at 72°C for 10 min in both, first and nested, PCR. The PCR fragment following Bsu36I digestion (37°C, overnight) and agarose (3.0%) gel electrophoresis with ethidium bromide staining reveals three different patterns. The presence of 328/36-bp fragments corresponded to Ala/Ala, 364/328/36 to Ala/Thr and 364 to Thr/Thr genotypes (Supplementary Table 1).
For determining Asn680Ser polymorphism genotypes, we amplified a region of 687 bp in exon 10 using the following primer pair designed by Primer 3 Plus program: forward 5´-CATGGATATTGACAGCCCTT-3´ and reverse 5´-GGAATTAATAGTTCCTGACC-3´. The cycling conditions were as follows: initial denaturation at 94°C for 5 min, followed by 35 cycles at 94°C for 90 s, annealing at 58°C for 1 min then at 72°C for 2 min, and a final elongation step at 72°C for 5 min. The PCR fragment following BsrI digestion (65°C, overnight) and agarose (3.0%) gel electrophoresis with ethidium bromide staining reveal three different patterns. The presence of 500/187-bp fragments corresponds to Ser/Ser; 687/500/187 to Asn/Ser; and 687 to Asn/Asn genotypes (Supplementary Table 1).
As previously mentioned, this is a study based on samples collected and analyzed over more than a decade. This fact has determined that all samples have been genotyped using the RFLP technique, regardless of the time of collection, to maintain uniformity. Unfortunately, at this moment, we do not have DNA to check, by sequencing, the genotyping performed by RFLP.
Statistical analysis
The statistical power and effect size of the study was determined with Epidat 3.1 (Web resources).
The allele frequencies, Inheritance Model analysis according to [45], the Permutation Test and Odds rate were carried out with the R package, version 2.0-2. Software: R Core Team (2020) (Web resources).
Results were expressed as mean ± standard deviation (SD). We checked normal distribution of parameters by the Kolmogorov-Smirnov (K-S) test. Since normality was observed (P > 0.05) in all of them, the Student’s t-test and χ2-independence test were applied. Statistical analysis was carried out with the statistical package StatView 5.0 for Macintosh (SAS, USA). A P value < 0.05 was considered statistically significant.
Results
The statistical power of the study
Considering that the two main objectives of the present work have been to determine the influence of the Ala307Thr and Asn680Ser FSHr polymorphisms on the COS response and on pregnancy success, we calculated the statistical power of the study between Ala307Thr polymorphism and both, Poor/Good responder (96%) and Pregnancy/Non-Pregnancy (94.8%); as well as between Asn680Ser polymorphism and both, Poor/Good responder (71.5%) and Pregnancy/Non-Pregnancy (94%), obtaining a mean statistical power of the sample ≥ 80%, as required.
Analysis of physiological parameters in pregnancy and non-pregnancy groups
Patients were divided into Pregnancy (n=193) and Non-Pregnancy (n=324) groups. The normal distribution of the recruited population was initially analyzed by clinical and biological characteristics of both groups and infertility etiology factors (Table 1). The Pregnancy group required a lower TD FSH (IU) (P = 0.02) in COS treatment and showed a higher level of Estradiol than the Non-Pregnancy group (P = 0.02) on the trigger day (Fig. 1). In addition, when we performed a Student’s t-test of Basal FSH, Preovulatory Follicles (PF), Recruited Oocytes (RO), Fertilized Oocytes (FO), and Embryos (E) in both groups, the Pregnancy group showed a statistically significant higher number of PF (P = 0.007), RO (P = 0.04), FO (P = 0.0003), and E (P <0.0001) than the Non-Pregnancy group, but Basal FSH showed non-significant difference (Fig. 2).
Table 1.
Characteristics of studied population
| All Patients | Pregnancy | Non-Pregnancy | |
|---|---|---|---|
| n = 517 | n = 193 | n = 324 | |
| a Characteristic of women presented as mean ± SD value | |||
| Age | 32.15 ± 2.5 | 32.1 ± 2.1 | 32.2 ± 2.6 |
| FSH basal (IU/mL) | 7.4 ± 2.1 | 7.5 ± 2.1 | 7.3 ± 2.2 |
| AFC* | 9.9 ± 5.6 | 10.2 ± 5.1 | 9.8 ± 5.8 |
| b Infertility etiology of couples | |||
| Male factor** | 37% | 39% | 37% |
| Unexplained | 12% | 11% | 14% |
| Tubal Factor | 18% | 17% | 18% |
| Endometriosis | 5% | 4% | 6% |
| Poor response | 28% | 28% | 26% |
*AFC antral follicle count. **Male factor [non-severe oligozoospermia (n = 85); non-severe asthenozoospermia (n = 106)
Fig. 1.

TD FSH and Estradiol in Pregnancy (n= 193) and Non-Pregnancy (n= 324) groups. Student’s t-test analysis: TD FSH (IU) [Pregnancy (1.684.5 ± 654.8) vs. Non-Pregnancy (1.832.1 ± 705.1); P = 0.02]; Estradiol (pg/mL) [Pregnancy (1.382.9 ± 789.1) vs. Non-Pregnancy (1.228.1 ± 728.9); P = 0.02]
Fig. 2.
Physiological parameters after COS treatment. Pregnancy (n=193), Non-Pregnancy (n=324). Student’s t-test analysis: Basal FSH (IU/mL) [Pregnancy (7.3 ± 1.99) vs. Non-Pregnancy (7.4 ± 2.31); P >0.05]; Preovulatory follicles (PF) [Pregnancy (10.7 ± 5.6) vs. Non-Pregnancy (9.4 ± 5.6); P = 0.007]; Recruited Oocytes (RO) [Pregnancy (8.7 ± 4.2) vs. Non-Pregnancy (7.9 ± 4.1); P = 0.04], Fertilized Oocytes (FO) [Pregnancy (5.8 ± 3.4) vs. Non-Pregnancy (4.8 ± 3.3); P = 0.0003] and Embryos (E) [Pregnancy (5.4 ± 3.2) vs. Non-Pregnancy (4.3 ± 2.9); P <0.0001]
Alleles and genotypes frequencies of 307 and 680 FSHr polymorphisms
The allele and genotype frequencies of these polymorphisms were calculated both, in whole population and in Pregnancy and Non-Pregnancy groups (Table 2). The results showed that, in whole population, these frequencies were similar as those described in other Caucasian populations [5, 46], which rules out any bias of our population. However, frequencies in Pregnancy and Non-Pregnancy groups differed between them in both polymorphisms. Interestingly, the probability of Non-Pregnancy in patients harboring Ala307Ala or Ser680Ser genotypes was twice than Non-Ala307Ala or Non- Ser680Ser, Odds Ratio (OR) = 2.0 and OR = 2.11, respectively.
Table 2.
Allele and frequencies of 307 and 680 FSHr polymorphisms.
| Total patients n = 517 | Pregnancy n = 193 | Non-Pregnancy n = 324 | |||
|---|---|---|---|---|---|
| FSHr gene Polymorphisms | Allele Amino acid Allele frequency | Genotype (Phenotype) | Distribution | Distribution Allele frequency | Distribution Allele frequency |
| Variant ID 2-49191041-C-T rs6165 c.919G>A Ala307Thr | G Ala q ≈ 0.45 | GG (Ala/Ala) | 85 (≈16%) | 21 (≈11%) q ≈ 0.40 | 64 (≈ 20%) q ≈ 0.48 |
| AG (Thr/Ala) | 298 (≈58%) | 113 (≈ 59%) | 185 (≈ 57%) | ||
| A Thr p ≈ 0.55 | AA (Thr/Thr) | 134 (≈26%) | 59 (≈ 31%) p ≈ 0.60 | 75 (≈ 23%) p ≈ 0.52 | |
| Variant ID 2-49189921-C-T rs6166 c.2039A>G Asn680Ser | A Asn p ≈ 0.62 | AA Asn/Asn | 182 (≈35%) | 67 (≈ 35%) q ≈ 0.64 | 115 (≈ 36%) q ≈ 0.61 |
| AG Asn/Ser | 275 (≈53%) | 112 (≈ 58%) | 163 (≈ 50%) | ||
| G Ser q ≈ 0.38 | GG Ser/Ser | 60 (≈12%) | 14 (≈ 7%) p ≈ 0.36 | 46 (≈ 14%) p ≈ 0.39 |
A and G correspond to Adenine and Guanine. Sample size n=517
Determination of the best inheritance model of 307 and 680 FSHr polymorphisms for parameters of COS outcome
After reviewing the bibliography about the polymorphisms under study, we considered that it was necessary to check the inheritance model (recessive, codominant or dominant genotype) that best fits each of parameters of COS outcome. For this purpose, we carried out the analysis proposed by Akaike [45], according to which the best inheritance model would correspond that shows the lowest value of the Akaike’s Information Criterion (AIC). This AIC is a measure of the relative quality of a statistical model, for a given set of data and, as such, provides a means for model selection, but cannot say anything about the quality of the model in an absolute sense.
Once we tested the inheritance models, the results showed that the most accurate varied in the different parameters, being the recessive model of polymorphism 307 (Ala307Ala vs Non-Ala307Ala) the most suitable for Basal FSH, PF and Pregnancy and the dominant one (Thr307Thr vs Non-Thr307Thr) for RO, FO and E. In a similar way, the recessive model of polymorphism 680 (Ser680Ser vs Non-Ser680Ser) was the best fits to Basal FSH, PF, RO and Pregnancy and the dominant one (Asn680Asn vs Non-Asn680Asn) to FO and E (Supplementary table 2).
To confirmed these results, we also performed the Permutation Test with Monte-Carlo simulation, an analysis of statistical significance for the study of differences between groups. The distribution of the mean studied was obtained by calculating the p-value of the mean difference for all possible rearrangements in no real groups with the same number of samples that real ones. If the shuffled data sets look like the real data (P > 0.05), there isn’t difference between real groups. Otherwise, if they look different from the real data (P < 0.05), we conclude that real groups are different. Since it involves calculating all possible situations, it is an exact test. The application of this analysis validated results obtained by inheritance model analysis (Supplementary table 3).
For Pregnancy vs Non-Pregnancy, the best model was Recessive in both 307 and 680 polymorphisms (Ala307Ala vs Non-Ala307Ala and Ser680Ser vs Non-Ser680Ser) (Supplementary table 2). The Permutation Test also confirmed results obtained by inheritance model analysis.
307 and 680 FSHr genotypes and parameters of COS outcome and pregnancy
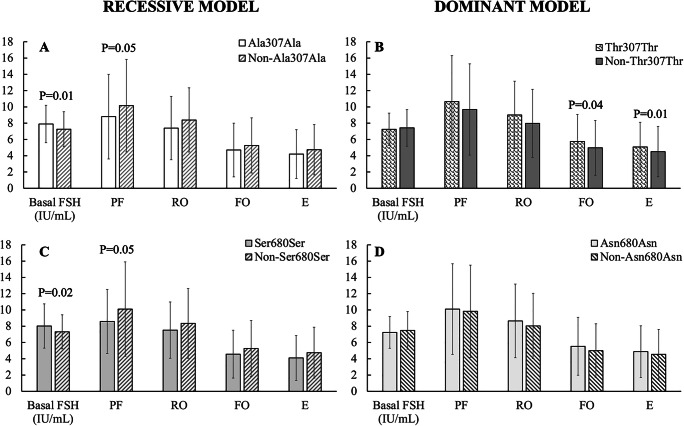
Since AIC did not provide a test of a model in the sense of testing a null hypothesis, we applied the Student’s t-test to different parameters of COS response in recessive/dominant model of 307 and 680 polymorphisms. In 307, we found statistically significant higher Basal FSH in Ala307Ala vs Non-Ala307Ala, (P = 0.01) and diminished PF (P = 0.05) (Fig. 3A), while dominant model (Thr307Thr vs Non-Thr307Thr) showed significant higher FO, (P = 0.04) and E, (P = 0.01) (Fig. 3B).
Fig. 3.
Analysis of inheritance models of 307 and 680 FSHr polymorphisms genotypes for Basal FSH (B FSH), Preovulatory Follicles (PF), Recruited Oocytes (RO), Fertilized Oocytes (FO) and Embryos (E) when Student’s t-test were applied. A: Recessive model in 307 [Ala/Ala (n= 85); Non-Ala/Ala (n= 432)]: statistically significant differences have been found in Basal FSH (IU/mL) [Ala/Ala (7.9 ± 2.3) vs. Non-Ala/Ala (7.3 ± 2.1); P = 0.01] and PF [Ala/Ala (8.8 ± 5.2) vs. Non-Ala/Ala (10.2 ± 5.7); P = 0.05]. However, non-significant differences have been found in RO [Ala/Ala (7.4 ± 3.9) vs. Non-Ala/Ala (8.4 ± 4.29) (P > 0.05)]; FO [Ala/Ala (4.7 ± 3.3) vs. Non-Ala/Ala (5.3 ± 3.4) (P > 0.05)]; nor in E [Ala/Ala (4.2 ± 3.0) vs. Non-Ala/Ala (4.7 ± 3.1) (P > 0.05)]. B: Dominant model in 307 [Thr/Thr (n= 134); Non-Thr/Thr (n= 383)] showed non statistically significant differences in Basal FSH (IU/mL) [Thr/Thr (7.2 ± 2.0) vs. Non-Thr/Thr (7.4 ± 2.2) (P > 0.05)]; PF [Thr/Thr (10.7 ± 5.6) vs. Non-Thr/Thr (9.7 ± 5.6) (P > 0.05)] and RO [Thr/Thr (9.0±4.1) vs. Non-Thr/Thr (9.0±4.2) (P > 0.05)]. However, FO [Thr/Thr (5.7±3.3) vs. Non-Thr/Thr (5.0±3.4) (P = 0.04)] and E [Thr/Thr (5.1±3.0) vs. Non-Thr/Thr (4.5±3.1) (P = 0.01)] showed statistically significant differences. C: Recessive model in 680 [Ser/Ser (n= 60) and Non-Ser/Ser (n= 457)]. Statistically significant differences have also been found in Basal FSH (IU/mL) [Ser/Ser (8.0 ± 2.7) vs. Non-Ser/Ser (7.3 ± 2.1); P = 0.02], PF [Ser/Ser (8.6 ±3.9) vs. Non-Ser/Ser (10.1 ± 5.8), P = 0.05]. Nevertheless, no statistically significant differences have been found in RO [Ser/Ser (7.5 ± 3.5) vs. Non-Ser/Ser (8.3 ± 4.3); P > 0.05]; FO [Ser/Ser (4.6 ± 2.9) vs. Non-Ser/Ser (5.3 ± 3.4); P > 0.05] nor in E [Ser/Ser (4.1 ± 3.1) vs. Non-Ser/Ser (4.7 ± 3.1); P > 0.05]. D: Dominant model in 680 [Asn/Asn (n= 182) and Non-Asn/Asn (n= 335)]. Basal FSH (IU/mL) [Asn/Asn (7.2 ± 1.9) vs. Non-Asn/Asn (7.5 ± 2.3); P > 0.05]; PF [Asn/Asn (10.1 ± 5.6) vs. Non-Asn/Asn (9.8 ± 5.7); P > 0.05]; RO [Asn/Asn (8.7 ± 4.5) vs. Non-Asn/Asn (8.0 ± 4.0); P > 0.05]; FO [Asn/Asn (5.5 ± 3.6) vs. Non-Asn/Asn (5.0 ± 3.3); P > 0.05]; E [Asn/Asn (4.9 ± 3.2) vs. Non-Asn/Asn (4.6 ± 3.0); P > 0.05], without statistically significant differences in all parameters
In regarding 680, in recessive model (Ser680Ser vs Non-Ser680Ser), we found statistically significant higher Basal FSH, (P = 0.02) and PF (P = 0.05) (Fig. 3C), while for dominant model we found that there was no statistically significant difference between genotypes in any parameter (Fig. 3D).
χ2-independence test from contingency tables of different variables
In order to check if Ala307Ala vs Non-Ala307Ala are or nor independent of Ser680Ser vs Non-Ser680Ser, the χ2-independence test from contingency tables was performed (χ2 = 183.081, P <0.0001, 1df), being Ala307Ala and Ser680Ser genotypes (contribution to χ2= 135.437) those that significantly appeared together in the same patient (Table 3).
Table 3.
Resume of χ2-independence tests of 307 and 680 FSHr genotypes and Poor/Good responder or Pregnancy/Non-pregnancy groups (see the “Results” section for details)
| Variables | DF | χ2 |
Contribution to χ2 |
P | Result |
|---|---|---|---|---|---|
|
Ala307Ala vs Non-Ala307Ala vs Ser680Ser vs Non-Ser680Ser |
1 | 183.081 | 135.437 | <0.0001 | Ala307Ala and Ser680Ser genotypes are not independent |
|
Ala307Ala vs Non-Ala307Ala vs Poor/Good responder |
1 | 5.601 | 2.338 | 0.01 | Ala307Ala associates to Poor responder |
|
Ser680Ser vs Non-Ser680Ser vs Poor/Good responder |
1 | 4.908 | 2.169 | 0.03 | Ser680Ser associates to Poor responder |
|
Ala307Ala vs Non-Ala307Ala vs Pregnancy/Non-Pregnancy |
1 | 6.63 | 3.493 | 0.01 | Ala307Ala is underrepresented in Pregnancy |
|
Ser680Ser vs Non-Ser680Ser vs Pregnancy/Non-Pregnancy |
1 | 8.193 | 4.581 | 0.004 | Ser680Ser is underrepresented in Pregnancy |
|
Ala307Ala and Ser680Ser association vs Pregnancy/Non-Pregnancy |
1 | 8.881 | 5.087 | 0.003 | Ala307Ala and Ser680Ser were underrepresented in Pregnancy |
We also analyzed the possible influence of Ala307Ala vs Non-Ala307Ala, on the one hand, and Ser680Ser vs Non-Ser680Ser, by the other, on Poor/Good responder, performing the χ2-independence test (307, χ2 = 5.601, P = 0.01, 1 df. 680, χ2 = 4.908 P = 0.03, 1 df). The analysis showed that COS response was not independent of 307 and 680 genotypes, since Ala307Ala (χ2 contribution = 2.338) and Ser680Ser (χ2 contribution = 2.169) genotypes were significantly increased in Poor responder (Table 3).
Finally, we applied the χ2-independence test to analyze the influence of Ala307Ala vs Non-Ala307Ala and Ser680Ser vs Non-Ser680Ser on Pregnancy/Non-Pregnancy. The results showed that Pregnancy was not independent of the genotypes of FSHr polymorphisms (307, χ2 = 6.63, P = 0.01, 1 df. 680, χ2 = 8.193, P = 0.004, 1 df) (Table 3). Upon examination of the contribution of the different genotypes to χ2, we found the Ala307Ala (χ2 contribution = 3.493) and Ser680Ser (χ2 contribution = 4.581) were underrepresented in the Pregnancy group and, therefore, responsible for the high χ2 value. In addition, when both polymorphisms were simultaneously analyzed, we found a χ2 = 8.881, P = 0.003, 1 df, with a diminished presence of the Ala307Ala-Ser680Ser in the Pregnancy group (χ2 contribution = 5.087) responsible for the high χ2 value (Table 3).
Discussion
In a first approximation, we checked if our population responded to COS treatment as other authors reported. The results confirmed that patients with pregnancy success required significantly lower TD FSH (P = 0.02) during treatment, showed statistically significant higher Estradiol (P = 0.002), before the triggering day (Fig. 1), and presented better fertility parameters of COS response (Fig. 2). On the other hand, we ruled out any bias of the sample due to the homogeneity of our population, since patients and their partners fulfill a series of conditions to be selected.
Since FSHr polymorphisms were first described in 1995 [9], different authors have proposed them as modifiers of FSH-FSHr binding [13, 47–51]; however, to our knowledge, non-conclusive results have been reported allowing the use of FSHr genotypes as predictive biomarkers for a successful ART outcome. FSHr polymorphisms are well known to show different genotypic frequencies depending on a patient’s ethnic background [29, 52], which suggests that ethnicity is a potential influencer on the outcomes of conventional COS treatment [27, 53]. Due to this speculation, we examined the FSHr genotypes and alleles frequencies of our patients (Table 2), confirming its similarity to those previously reported in Caucasian population [22, 34, 54].
In the present work, we analyzed the influence of 307 and 680 FSHr polymorphisms on different parameters of ovarian response in patients undergoing COS treatment in a large and homogeneous sample of 517 Caucasian patients (Statistical power of the study ≥80%), a higher population size than other published works, which reinforces the reliability of our results.
After determining the adequate inheritance models, we confirmed that patients harboring Ala307Ala genotype showed statistically significant higher Basal FSH (P = 0.01) and lower PF (P = 0.05) (Fig. 3A), both parameters previously related to poor COS response by other authors [16–18]. In addition, we found that Thr307Thr genotype showed a statistically significant higher FO and Embryos (Fig. 3B) than Non-Thr307Th.
Similarly, the most suitable 680 polymorphism inheritance model was checked for the same parameters (Fig. 3 C, D), finding that Ser680Ser showed statistically significant higher Basal FSH (P = 0.02) and a lower PF (P = 0.05) than Non- Ser680Ser, confirming previous reports [54]. In fact, this last study shows that increasing levels of FSH in patients seem to be ineffective in enhancing ovarian response, observing either a decrease in number of follicles or mature oocytes associated with these genotypes. Furthermore, these genotypes also associated to a lower ovarian reserve and Estradiol on the trigger day [27, 46, 55] which designates them as responsible for diminished ovarian response capacity [17, 18]. The same results have been reported in patients of similar age, applying comparable protocols of ovarian stimulation [28, 56, 57] and warranting proposal of Asn680Ser polymorphism as a biomarker in COS prognosis [57].
However, other publications do not find correlation between COS response and 307 and 680 FSHr polymorphisms [58] or between Asn680Ser polymorphism and female infertility [59]. In a small sample of patients (n = 105), Ser680Ser genotype do not associate with a poor response, but shows three times higher implantation and pregnancy rates than patients harboring Asn680Asn genotype [60], which completely contradicts our results. It has been also reported that infertile Brazilian women with heterozygous Ala307Thr receptors showed more mature oocytes and embryos, which contradicts our study and other previous findings [23]. We consider that these discrepancies could be explained by different ethnicities, sample size and/or applied COS protocols. Since multiple factors modify COS response, the maximum homogeneity of the samples is essential to compare different works and reach definitive conclusions.
The results of the present work also confirmed that both genotypes, Ala307Ala and Ser680Ser, have a statistically significant tendency to appear together (P<0.0001), suggesting a coevolution of these genotypes, as previously proposed [27]. These FSHr variants seem to alter the FSHr expression pattern on granulosa cells [59, 61], the Estradiol secretion and the follicle response [62, 63]. We propose that lower frequencies of Ala307Ala (16%) and Ser680Ser (12%) could suggest a progressive elimination from population due to the defective interaction of FSH-FSHr.
Our study also focused on investigating the influence of these polymorphisms (307/680 FSHr) on pregnancy success. The findings showed that not only Ser680Ser (P = 0.004), as reported [18, 46], but also Ala307Ala (P = 0.01) are genetic variants that negatively influence on getting pregnant. Other authors, however, have reported that the association of genotypes Ser680Ser of FSHr and Ser312Ser of LHCGr in patients have a 40% higher chance of achieving pregnancy [25], which apparently opposes precedent works. Despite the large sample size of this last study, we consider that the number of samples that harbored both genotypes was too small to draw definitive conclusions.
From a biochemical point of view, the substitution of one amino acid for another in both polymorphisms does not seem to be indifferent: Ala is a small non-polar amino acid with isoelectric point (pI) 6, while Thr is polar, although uncharged, larger and pI 5.87 and Ser, polar amino acid, uncharged, smaller volume and pI 5.68, while Asn, also polar and uncharged, greater volume, with two amino groups and pI 5.41. Taken together, these little changes suggest that Ala307Ala and Ser680Ser could determine, together or separately, a defective coupling of the FSHr monomers, FSH-FSHr binding, anchorage in the granulosa cell membrane and/or signal transduction to cytoplasma, justifying the negative influence on COS response we have found, by a diminish synthesis of Estradiol, as observed by [27], result not found in our study (data not showed). The lower concentration of this hormone in blood would induce, by negative feedback in the pituitary gland, higher basal FSH that associates to poor COS treatment response, as documented by our study and previous publications [22, 26, 64]. This interpretation seems to be confirmed by the low pregnancy rate of these genotypes, separately (Ala307Ala, P = 0.01 and Ser680Ser, P = 0.004) or together (P = 0.003) in our samples. The fact that Ala307Ala (OR = 2.0) and Ser680Ser (OR = 2.11) double probability of Non-Pregnancy that its respective Non-Ala307Ala and Ser680Ser reinforces this interpretation.
This study has limitations that we cannot ignore. Not checking by sequencing, due to lack of DNA, the genotyping carried out by RFLP is one of them. On the other hand, it must be accepted that the studied polymorphisms of FSHr are among many other factors that undoubtedly influence on COS outcome. Despite the fact that we selected patients and their partners with standard parameters of normality or almost normality, on the oocyte fertilization and in vitro embryo development, some other unknown factors, as genetic background of gametes, could be influencing. In a similar way, psychological, anatomical, and physiological factors, not analyzed in the present work neither in previous studies, could also influence on implantation and subsequent embryo development in uterus, and perhaps should be considered in future works. Finally, we cannot forget that the statistical significance found in different parameters of COS outcome and in pregnancy success, in this and other studies, does not always translate into clinical consequences in particular couples. For this reason, this study provides conclusions that, together with those of other researchers, will contribute to a better understanding of infertility and its causes, but in no case can we consider that they have a cause-effect relationship.
On the opposite, the robustness of the study is supported by the homogeneity and large sample size (517 patients) of the studied population. Our work not only confirms the negative influence of Ala307Ala and Ser680Ser variants on the number of preovulatory oocytes after COS treatment, but it also quantifies, as double, the probability of Non-Pregnancy of these genotypes comparing with Non-Ala307Ala and Non-Ser680Ser, respectively. In addition, the fact that, within the Non-Ala307Ala genotypes, Thr307Thr has showed higher fertilized oocytes and embryos that Non-Thr307Thr, although no influence on pregnancy success, is an interesting result that must be deeper analyzed in further works.
Although new studies must be carried out to confirm our findings, we consider that the results of the present work provide strong evidence of the effect that certain genotypes of both, 307 and 680, polymorphisms could have on the final result of ART. From our point of view, the routinely determination of 307/680 FSHr polymorphisms prior to ART could allow to design ad hoc clinical protocols and provide a more accurate prognosis of pregnancy, avoiding side effects and unnecessary delays in getting pregnant. However, we are aware that genotyping would complicate and make ART more expensive, so we consider that it could at least be performed in those patients who present an abnormal COS response.
Conclusions
Our study provides a comprehensive relationship between 307 and 680 FSHr polymorphisms genotypes and COS outcome and pregnancy success. Ala307Ala and Ser680Ser variants influence negatively on COS outcome. Both variants associate to higher Basal FSH and lower Estradiol and PF than Non-Ala307Ala and Non-Ser680Ser, respectively. Within the Non-Ala307Ala group, is Thr307Thr that shows higher Fertilized Oocytes and Embryos than Non-Thr307Thr, but non influence on pregnancy rate. Patients harboring Ala307Ala or Ser680Ser variants have a double probability of Non-Pregnancy than their respective Non-Ala307Ala and Non-Ser680Ser. Coherently, Ala307Ala and Ser680Ser together significantly increase Non-Pregnancy probability.
Supplementary information
(DOCX 17 kb)
(DOCX 14 kb)
(DOCX 17 kb)
Acknowledgements
The authors would like to express their gratitude to all patients and to their partners, who contributed genetic material for this study.
Author contribution
BM-O performed the laboratory work of the study and contributed to wrote the manuscript information. FC and LM collected data and provided subjects’ treatment. EGA contributed to wrote the manuscript information. ML-P, JM and ER-P Laboratory studies for the T307A and N680S polymorphisms. ML-P and FC designed the study. BM-O and CD-S performed statistical analysis of data. ML-P and CD-S supervised quality control of molecular biology studies and designed RFLP studies. CD-S contributed to study design and wrote the manuscript information.
Funding
BM-O was supported by a grant of the Foundation for Biomedical Research of the Hospital Universitario Príncipe de Asturias of Alcalá de Henares, Madrid (Spain). The study was supported by a grant from the Government of Aragon (Spain) (Applied Research Group B33 of 2009. Main Researcher Dr Julio Montoya). The funding sources had no involvement in study design, collection, analysis, interpretation of data, writing the report or decision to submit the article for publication.
Data availability
All data are available upon request.
Declarations
Ethics approval and consent to participate
The subjects were fully informed of the aims of the work before signing the informed consent form. The study conforms to the code of Ethics of the World Medical Association (Declaration of Helsinki) and was approved (January 22nd, 2003) by the Committee of Clinical Trials of the Hospital Universitario Príncipe de Asturias of Alcalá de Henares, Madrid (Spain).
Consent for publication
All authors agree in publishing the data contained in this article.
Conflict of interest
The authors declare no competing interests.
Web resources
NCBI Data Base: https://www.ncbi.nlm.nih.gov
Variant ID of polymorphisms FSHr: https://bibliome.ai/gene/FSHR
Statistical power of the study: http://scielo.isciii.es/scielo.php?script=sci_arttext&pid=S1135-57272004000200013&lng=es&nrm=iso>. ISSN 2173-9110.
R software: https://CRAN.R-project.org/package=SNPassoc. https://www.R-project.org/). Library SNPassoc: SNPs-Based Whole Genome Association Studies.
Footnotes
Manuel J. López-Pérez and Francisco de Castro are deceased.
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Wyns C, Bergh C, Calhaz-Jorge C, De Geyter C, Kupka MS, Mocanu E, et al. ART in Europe, 2015: results generated from European registries by ESHRE The European IVF-monitoring Consortium (EIM) for the European Society of Human Reproduction and Embryology (ESHRE). Hum Reprod Open. 2020:1–17. [DOI] [PMC free article] [PubMed]
- 2.ESHRE Guideline Group on Ovarian Stimulation. Bosch E, Broer S, Griesinger G, Grynberg M, Humaidan P, Kolibianakis E, Kunicki M, Marca A, Lainas G, Clef NL, Massin N, Mastenbroek S, Polyzos N, Sunkara SK, Timeva T, Töyli M, Urbancsek J, Vermeulen N, Broekmans F. Erratum: ESHRE guideline: ovarian stimulation for IVF/ICSI. Hum Reprod Open. 2020;2020(4):hoaa067. doi: 10.1093/hropen/hoaa067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kaiser UB. The pathogenesis of the ovarian hyperstimulation syndrome. N Engl J Med. 2003;349:729–732. doi: 10.1056/NEJMp038106. [DOI] [PubMed] [Google Scholar]
- 4.Delvigne A, Rozenberg S. Epidemiology and prevention of ovarian hyperstimulation syndrome (OHSS): a review. Hum Reprod Update. 2002;8:559–577. doi: 10.1093/humupd/8.6.559. [DOI] [PubMed] [Google Scholar]
- 5.Nenonen H, Lindgren I, Prahl A, Trzybulska D, Kharraziha I, Hultén M, et al. The N680S variant in the follicle-stimulating hormone receptor gene identifies hyperresponders to controlled ovarian stimulation. Pharmacogenet Genomics. 2019;29:114–120. doi: 10.1097/FPC.0000000000000374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Drakopoulos P, Blockeel C, Stoop D, Camus M, de Vos M, Tournaye H, Polyzos NP. Conventional ovarian stimulation and single embryo transfer for IVF/ICSI. How many oocytes do we need to maximize cumulative live birth rates after utilization of all fresh and frozen embryos? Hum Reprod. 2016;31:370–376. doi: 10.1093/humrep/dev316. [DOI] [PubMed] [Google Scholar]
- 7.Lainas T, Sfontouris I, Venetis C, Lainas G, Zorzovilis I, Tarlatzis B, et al. Live birth rates after modified natural cycle compared with high-dose FSH stimulation using GnRH antagonists in poor responders. Hum Reprod. 2015;30:2321–2330. doi: 10.1093/humrep/dev198. [DOI] [PubMed] [Google Scholar]
- 8.Lledo B, Ortiz JA, Llacer J, Bernabeu R. Pharmacogenetics of ovarian response. Pharmacogenomics. 2014;15(6):885–893. doi: 10.2217/pgs.14.49. [DOI] [PubMed] [Google Scholar]
- 9.Aittomaki K, Lucena JL, Pakarinen P. Mutation in the follicle-stimulating hormone receptor gene causes hereditary hypergonadotropic ovarian failure. Cell. 1995;82(6):959–968. doi: 10.1016/0092-8674(95)90275-9. [DOI] [PubMed] [Google Scholar]
- 10.Gromoll J, Simoni M, Nordhoff V, Behre HM, De Geyter C, Nieschlag E. Functional and clinical consequences of mutations in the FSH receptor. Mol Cell Endocrinol. 1996;125:177–182. doi: 10.1016/s0303-7207(96)03949-4. [DOI] [PubMed] [Google Scholar]
- 11.Levallet J, Pakarinen P, Huhtaniemi IT. Follicle-stimulating hormone ligand and receptor mutations, and gonadal dysfunction. Arch Med Res. 1999;30:486–494. doi: 10.1016/s0188-0128(99)00058-5. [DOI] [PubMed] [Google Scholar]
- 12.Lussiana C, Guani B, Mari C, Restagno G, Massobrio M, Revelli A. Mutations and polymorphisms of the FSH receptor (FSHR) gene: clinical implications in female fecundity and molecular biology of FSHR protein and gene. Obstet Gynecol Surv. 2008;63:785–789. doi: 10.1097/OGX.0b013e31818957eb. [DOI] [PubMed] [Google Scholar]
- 13.Simoni M, Gromoll J, Nieschlag E. The follicle-stimulating hormone receptor: biochemistry, molecular biology, physiology, and pathophysiology. Endocr Rev. 1997;18:739–773. doi: 10.1210/edrv.18.6.0320. [DOI] [PubMed] [Google Scholar]
- 14.Simoni M, Nieschlag E, Gromoll J. Isoforms and single nucleotide polymorphisms of the FSH receptor gene: implications for human reproduction. Hum Reprod Update. 2002;8:413–421. doi: 10.1093/humupd/8.5.413. [DOI] [PubMed] [Google Scholar]
- 15.Laan M, Grigorova M, Huhtaniemi IT. Pharmacogenetics of follicle-stimulating hormone action. Curr Opin Endocrinol Diabetes Obes. 2012;19:220–227. doi: 10.1097/MED.0b013e3283534b11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Loutradis D, Patsoula E, Minas V. FSH receptor gene polymorphisms have a role for different ovarian response to stimulation in patients entering IVF/ICSI-ET programs. J Assist Reprod Genet. 2006;23:177–184. doi: 10.1007/s10815-005-9015-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pabalan N, Trevisan CM, Peluso C, Jarjanazi H, Christofolini DM, Barbosa CP, Bianco B. Evaluating influence of the genotypes in the follicle-stimulating hormone receptor (FSHR) Ser680Asn (rs6166) polymorphism on poor and hyper-responders to ovarian stimulation: a meta-analysis. J Ovarian Res. 2014;7:285. doi: 10.1186/s13048-014-0122-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.García-Jiménez G, Zariñán T, Rodríguez-Valentín R, Mejía-Domínguez N, Gutiérrez-Sagal R, Hernández-Montes G, et al. Frequency of the T307A, N680S, and -29G>A single-nucleotide polymorphisms in the follicle-stimulating hormone receptor in Mexican subjects of Hispanic ancestry. Reprod Biol Endocrinol. 2018;16:100. doi: 10.1186/s12958-018-0420-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ilgaz N, Aydos OS, Karadag A, Taspinar M, Eryilmaz O, Sunguroglu A. Impact of follicle-stimulating hormone receptor variants in female infertility. J Assist Reprod Genet. 2015;32:1659–1668. doi: 10.1007/s10815-015-0572-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lledo B, Guerrero J, Turienzo A, Ortiz JA, Morales R, Ten J. Effect of follicle-stimulating hormone receptor N680S polymorphism on the efficacy of follicle-stimulating hormone stimulation on donor ovarian response. Pharmacogenet Genomics. 2013;23:262–268. doi: 10.1097/FPC.0b013e32835fe813. [DOI] [PubMed] [Google Scholar]
- 21.Tong Y, Liao WX, Roy AC, Ng SC. Absence of mutations in the coding regions of follicle-stimulating hormone receptor gene in Singapore Chinese women with premature ovarian failure and polycystic ovary syndrome. Horm Metab Res. 2001;33:221–226. doi: 10.1055/s-2001-14941. [DOI] [PubMed] [Google Scholar]
- 22.Perez Mayorga M, Gromoll J, Behre HM, Gassner C, Nieschlag E, Simoni M. Ovarian response to follicle-stimulating hormone (FSH) stimulation depends on the FSH receptor genotype. J Clin Endocrinol Metab. 2000;85:3365–3369. doi: 10.1210/jcem.85.9.6789. [DOI] [PubMed] [Google Scholar]
- 23.Trevisan C, Peluso C, Cordts E, De Oliveira R, Christofolini DM, Barbosa C, Bianco B. Ala307Thr and Asn680Ser Polymorphisms of FSHR Gene in Human Reproduction Outcomes. Cell Physiol Biochem. 2014;34:1527–1535. doi: 10.1159/000366356. [DOI] [PubMed] [Google Scholar]
- 24.Tang H, Yan Y, Wang T, Zhang T, Shi W, Fan R, Yao Y, Zhai S. Effect of follicle-stimulating hormone receptor Asn680Ser polymorphism on the outcomes of controlled ovarian hyperstimulation: an updated meta-analysis of 16 cohort studies. J Assist Reprod Genet. 2015;32:1801–1810. doi: 10.1007/s10815-015-0600-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lindgren I, Nenonen H, Henic E, Bungum L, Prah A, Bungum M, Leijonhufvud I, Huhtaniemi I, Andersen C, Giwercman Y. Gonadotropin receptor variants are linked to cumulative live birth rate after in vitro fertilization. J Assist Reprod Genet. 2019;36:29–38. doi: 10.1007/s10815-018-1318-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Čuš M, Vlaisavljević V, Repnik K, PotoČnik U, KovaČiČ B. Could polymorphisms of some hormonal receptor genes, involved in folliculogenesis help in predicting patient response to controlled ovarian stimulation? J Assist Reprod Genet. 2019;36:47–55. doi: 10.1007/s10815-018-1357-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yan Y, Gong Z, Zhang L, Li Y, Li X, Zhu L, et al. Association of follicle- stimulating hormone receptor polymorphisms with ovarian response in Chinese women: a prospective clinical study. PLoS One. 2013;8:e78138. doi: 10.1371/journal.pone.0078138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Achrekar S, Modi D, Desai S, Mangoli V, Mangoli R, Mahale S. Follicle-stimulating hormone receptor polymorphism (Thr307Ala) is associated with variable ovarian response and ovarian hyperstimulation syndrome in Indian women. Fertil Steril. 2009;91(2):432–439. doi: 10.1016/j.fertnstert.2007.11.093. [DOI] [PubMed] [Google Scholar]
- 29.Sheikhha MH, Eftekhar M, Kalantar SM. Investigating the association between polymorphism of follicle-stimulating hormone receptor gene and ovarian response in controlled ovarian hyperstimulation. J Hum Reprod Sci. 2011;4:86–90. doi: 10.4103/0974-1208.86089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Olijve W, De Boer W, Mulders JW, Van Wezenbeek PM. Molecular biology and biochemistry of human recombinant follicle stimulating hormone (Puregon) Mol Hum Reprod. 1996;2:371–382. doi: 10.1093/molehr/2.5.371. [DOI] [PubMed] [Google Scholar]
- 31.O'Shaughnessy PJ, Dudley K, Rajapaksha WR. Expression of follicle stimulating hormone-receptor mRNA during gonadal development. Mol Cell Endocrinol. 1996;125:169–175. doi: 10.1016/s0303-7207(96)03957-3. [DOI] [PubMed] [Google Scholar]
- 32.Menon KM, Clouser CL, Nair AK. Gonadotropin receptors: role of post-translational modifications and post-transcriptional regulation. Endocrine. 2005;26:249–257. doi: 10.1385/ENDO:26:3:249. [DOI] [PubMed] [Google Scholar]
- 33.Sower SA, Moriyama S, Kasahara M, Takahashi A, Nozaki M, Uchida K, Dahlstrom JM, Kawauchi H. Identification of sea lamprey GTHbeta-like cDNA and its evolutionary implications. Gen Comp Endocrinol. 2006;148:22–32. doi: 10.1016/j.ygcen.2005.11.009. [DOI] [PubMed] [Google Scholar]
- 34.De Castro F, Ruiz R, Montoro L, Pérez-Hernández D, Sánchez-Casas Padilla E, Real LM, et al. Role of follicle-stimulating hormone receptor Ser680Asn polymorphism in the efficacy of follicle-stimulating hormone. Fertil Steril. 2003;80:571–576. doi: 10.1016/s0015-0282(03)00795-7. [DOI] [PubMed] [Google Scholar]
- 35.Griesinger G. Diedrich1 K, Devroey P, Kolibianakis EM. GnRH agonist for triggering final oocyte maturation in the GnRH antagonist ovarian hyperstimulation protocol: a systematic review and meta-analysis. Hum Reprod Update. 2006;12:159–168. doi: 10.1093/humupd/dmi045. [DOI] [PubMed] [Google Scholar]
- 36.Danhua Pu WJ, Liu J. Comparisons of GnRH antagonist versus GnRH agonist protocol in poor ovarian responders undergoing IVF. Hum Reprod. 2011;26:2742–2749. doi: 10.1093/humrep/der240. [DOI] [PubMed] [Google Scholar]
- 37.Demirol A, Gurgan T. Comparison of microdose flare-up and antagonist multiple-dose protocols for poor-responder patients: a randomized study. Fertil Steril. 2009;92:481–485. doi: 10.1016/j.fertnstert.2008.07.006. [DOI] [PubMed] [Google Scholar]
- 38.Ferraretti AP, Goossens V, Kupka M, Bhattacharya S, de Mouzon J, Castilla JA, Erb K, Korsak V, Nyboe Andersen A, The European IVF-monitoring (EIM) Consortium, for The European Society of Human Reproduction and Embryology (ESHRE) Strohmer H, Bogaerts K, Kyurkchiev S, Vrcic H, Pelekanos M, Rezabek K, Erb K, Gissler M, Royere D, Bühler K, Tarlatzis BC, Kosztolanyi G, Bjorgvinsson H, Mocanu E, Scaravelli G, Lokshin V, Arajs M, Gudleviciene Z, Lazarevski S, Moshin V, Simic TM, Hazekamp JT, Kurzawa R, Calhaz–Jorge C, Rugescu I, Korsak V, Radunovic N, Tomazevic T, Hernandez JH, Karlström PO, Weder M, Lambalk C, Veselovsky V, Baranowski R. European IVF-Monitoring (EIM) Consortium for the European Society of Human Reproduction and Embryology (ESHRE). Assisted reproductive technology in Europe, 2009: results generated from European registers by ESHRE. Hum Reprod. 2013;28:2318–2331. doi: 10.1093/humrep/det278. [DOI] [PubMed] [Google Scholar]
- 39.Ferraretti AP, La Marca A, Fauser BC, Tarlatzis B, Nargund G, Gianaroli L, On behalf of the ESHRE working group on Poor Ovarian Response Definition ESHRE consensus on the definition of “poor response” to ovarian stimulation for in vitro fertilization: the Bologna criteria. Hum Reprod. 2011;26:1616–1624. doi: 10.1093/humrep/der092. [DOI] [PubMed] [Google Scholar]
- 40.Ferraretti AP, Gianaroli L. The Bologna criteria for the definition of poor ovarian responders: is there a need for revision? Hum Reprod. 2014;29:1842–1845. doi: 10.1093/humrep/deu139. [DOI] [PubMed] [Google Scholar]
- 41.Patrizio P, Vaiarelli A, Setti P, Tobler K, Shoham G, Leong M, et al. How to define, diagnose and treat poor responders? Responses from a worldwide survey of IVF clinics. Reprod BioMed Online. 2015;30:581–592. doi: 10.1016/j.rbmo.2015.03.002. [DOI] [PubMed] [Google Scholar]
- 42.Abu-Musa A, Haahr T, Humaidan P. Novel Physiology and Definition of Poor Ovarian Response; Clinical Recommendations. Int J Mol Sci. 2020;21(6):2110. doi: 10.3390/ijms21062110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Marcuello A, Martínez-Redondo D, Dahmani Y, Casajús JA, Ruiz-Pesini E, Montoya J, López-Pérez MJ, Díez-Sánchez C. Human mitochondrial variants influence on oxygen consumption. Mitochondrion. 2009;9:27–30. doi: 10.1016/j.mito.2008.10.002. [DOI] [PubMed] [Google Scholar]
- 44.Sudo S, Kudo M, Wada S, Sato O, Hsueh A, Fujimoto S. Genetic and functional analyses of polymorphisms in the human FSH receptor gene. Mol Hum Reprod. 2002;8:893–899. doi: 10.1093/molehr/8.10.893. [DOI] [PubMed] [Google Scholar]
- 45.Akaike H. Fitting autoregressive models for prediction. Ann Inst Stat Math. 1969;21:243–247. [Google Scholar]
- 46.Zilaitiene B, Dirzauskas M, Verkauskiene R, Ostrauskas R, Gromoll J, Nieschlag E. The impact of FSH receptor polymorphism on time-to-pregnancy: a cross-sectional single-Centre study. BMC Pregnancy Childbirth. 2018;18:272. doi: 10.1186/s12884-018-1910-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sasson R, Dantes A, Tajima K, Amsterdam A. Novel genes modulated by FSH in normal and immortalized FSH-responsive cells: new insights into the mechanism of FSH action. FASEB J. 2003;17:1256–1266. doi: 10.1096/fj.02-0740com. [DOI] [PubMed] [Google Scholar]
- 48.Fan QR, Hendrickson WA. Structural biology of glycoprotein hormones and their receptors. Endocrine. 2005;26:179–188. doi: 10.1385/endo:26:3:179. [DOI] [PubMed] [Google Scholar]
- 49.Fan QR, Hendrickson WA. Structure of human follicle-stimulating hormone in complex with its receptor. Nature. 2005;433:269–277. doi: 10.1038/nature03206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rajashekar L, Krishna D, Patil M. Polycystic ovaries and infertility: Our experience. J Hum Reprod Sci. 2008;1:65–72. doi: 10.4103/0974-1208.44113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Frydman R. Poor responders: still a problem. Fertil Steril. 2011;96:1057. doi: 10.1016/j.fertnstert.2011.09.051. [DOI] [PubMed] [Google Scholar]
- 52.Wunsch A, Sonntag B, Simoni M. Polymorphism of the FSH receptor and ovarian response to FSH. Ann Endocrinol. 2007;68:160–166. doi: 10.1016/j.ando.2007.04.006. [DOI] [PubMed] [Google Scholar]
- 53.Huang W, Cao Y, Shi L. Effects of FSHR polymorphisms on premature ovarian insufficiency in human beings: a meta-analysis. Reprod Biol Endocrinol. 2019;17:80. doi: 10.1186/s12958-019-0528-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.König T, Van der Lee J, Schats R, Lambalk C. The relationship between FSH receptor polymorphism status and IVF cycle outcome: a retrospective observational study. Reprod BioMed Online. 2019;39(2):231–240. doi: 10.1016/j.rbmo.2019.05.018. [DOI] [PubMed] [Google Scholar]
- 55.Greb RR, Grieshaber K, Gromoll J, Sonntag B, Nieschlag E, Kiesel L, Simoni M. A common single nucleotide polymorphism in exon 10 of the human follicle stimulating hormone receptor is a major determinant of length and hormonal dynamics of the menstrual cycle. J Clin Endocrinol Metab. 2005;90:4866–4872. doi: 10.1210/jc.2004-2268. [DOI] [PubMed] [Google Scholar]
- 56.Livshyts G, Podlesnaja S, Kravchenko S, Sudoma I, Livshits L. A distribution of two SNPs in exon 10 of the FSHR gene among the women with a diminished ovarian reserve in Ukraine. J Assist Reprod Genet. 2009;26:29–34. doi: 10.1007/s10815-008-9279-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Moron FJ, Ruiz A. Pharmacogenetics of controlled ovarian hyperstimulation: Time to corroborate the clinical utility of FSH receptor genetic markers. Pharmacogenomics. 2010;11:1613–1618. doi: 10.2217/pgs.10.156. [DOI] [PubMed] [Google Scholar]
- 58.Singhasena W, Pantasri T, Piromlertamorn W, Samchimchom S, Vutyavanich T. Follicle-stimulating hormone receptor gene polymorphism in chronic anovulatory women, with or without polycystic ovary syndrome: a cross-sectional study. Reprod Biol Endocrinol. 2014;12:86. doi: 10.1186/1477-7827-12-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sangeeta R. Ashish, Kumari P, Singh A, Singh R. Correlation of follicle-stimulating hormone receptor gene Asn 680 Ser (rs6166) polymorphism with female infertility. J Family Med Prim Care. 2019;8(10):3356–3361. doi: 10.4103/jfmpc.jfmpc_685_19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Klinkert E, Te Velde E, Weima S, van Zandvoort P, Hanssen R, Nilsson P, et al. FSH receptor genotype is associated with pregnancy but not with ovarian response in IVF. Reprod BioMed Online. 2006;13:687–695. doi: 10.1016/s1472-6483(10)60660-8. [DOI] [PubMed] [Google Scholar]
- 61.Kuijper E, Blankenstein M, Luttikhof L, Roek S, Overbeek A, Hompes P, et al. Frequency distribution of polymorphisms in the FSH receptor gene in infertility patients of different ethnicity. Reprod BioMed Online. 2010;20:588–593. doi: 10.1016/j.rbmo.2010.02.004. [DOI] [PubMed] [Google Scholar]
- 62.Laven J, Mulders A, Suryandari D, Gromoll J, Nieschlag E, Fauser B, Simoni M. Follicle-stimulating hormone receptor polymorphisms in women with normogonadotropic anovulatory infertility. Fertil Steril. 2003;80:986–992. doi: 10.1016/s0015-0282(03)01115-4. [DOI] [PubMed] [Google Scholar]
- 63.Behre H, Greb R, Mempel A, Sonntag B, Kiesel L, Kaltwasser P, et al. Significance of a common single nucleotide polymorphism in exon 10 of the follicle-stimulating hormone (FSH) receptor gene for the ovarian response to FSH: a pharmacogenetic approach to controlled ovarian hyperstimulation. Pharmacogenet Genomics. 2005;15:451–456. doi: 10.1097/01.fpc.0000167330.92786.5e. [DOI] [PubMed] [Google Scholar]
- 64.Boudjenah R, Molina-Gomes D, Torre A, Bergere M, Bailly M, Boitrelle F, Taieb S, Wainer R, Benahmed M, de Mazancourt P, Selva J, Vialard F. Genetic polymorphisms influence the ovarian response to rFSH stimulation in patients undergoing in vitro fertilization programs with ICSI. PLoS One. 2012;7:e38700. doi: 10.1371/journal.pone.0038700. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX 17 kb)
(DOCX 14 kb)
(DOCX 17 kb)
Data Availability Statement
All data are available upon request.