Abstract
Objective
To study the correlation between SNPs at phosphatidylinositol-4, 5-bisphosphate 3-kinase catalytic subunit alpha (PIK3CA) rs9838117 site, erb-b2 receptor tyrosine kinase 2 (ERBB2) rs1058808 site, and their interactions with environmental factors and the epithelial ovarian cancer (EOC) risk.
Methods
Sanger sequencing was used to analyze the genotypes of PIK3CA rs9838117 and ERBB2 rs1058808 site in 587 patients with epithelial ovarian cancer (EOC). Multi-factor dimensionality reduction (MDR) was applied to analyze the interaction between PIK3CA rs9838117 and ERBB2 rs1058808 site and the clinical data.
Results
The risk of EOC in T allele carriers at PIK3CA rs9838117 was 1.95 times (95%CI: 1.55–2.46, P<0.01) that of G allele carriers. The risk of EOC in G allele carriers at ERBB2 rs1058808 was as 0.64 times (95%CI: 0.54–0.75, P <0.01) as the risk for C allele carriers. In the interaction model between clinical data, PIK3CA rs9838117 site and ERBB2 rs1058808 SNP site, EOC risk in high-risk combination was 3.10 times (95%CI: 1.49–6.46, P <0.01) that of low-risk combination.
Conclusion
The SNPs at PIK3CA rs9838117 and ERBB2 rs1058808 loci were associated with the risk of EOC.
Supplementary Information
The online version contains supplementary material available at 10.1007/s10815-021-02177-2.
Keywords: Epithelial ovarian cancer, PIK3CA, ERBB2, Single nucleotide polymorphism, Susceptibility
Introduction
Ovarian cancer (OC) is the most lethal tumors of female reproductive system. Lacking of effective early screening methods, and the resistance to chemotherapy therapies lead to high mortality of patients with OC [1]. The occurrence of OC is caused by a variety of genes and environmental factors, thereby studying the combined effects of genes-genes and genes-environmental factors is critical for the prevention and treatment of OC [2–4].
The PIK3CA gene is located on human chromosome 3q26.32 and encodes the catalytic subunit of phosphatidylinositol 3-kinase α (PI3Kα). The mutation and aberrant activation of PIK3CA is one of the major genomic changes in ovarian cancer [5–7]. There are many single nucleotide polymorphisms (SNPs) sites on PIK3CA gene, of which the rs9838117 site is the single nucleotide variation (SNV) type of PIK3CA gene, and its position on the chromosome is 3:17895250. Previous study showed that SNP at rs9838117 locus is closely associated with the reduction in the incidence of radiation pneumonitis (RP) ≥ grade 3 [8]. Genome-wide copy number variation analysis showed that PIK3CA has a higher degree of copy number amplification in highly invasive/migrating ovarian cancer cell lines [9]. Correlation analysis indicated that SNPs at rs9838117 site on PIK3CA is associated with the occurrence of reflex pneumonia [8]. However, few researches have focused on the correlation between the rs9838117 SNPs of PIK3CA gene and the development of ovarian cancer.
ERBB2 gene, also named as HER2, is located on human chromosome 17q12. ERBB2 is one of the proto-oncogenes with the high mutation rate. Activation of ERBB2 contributes to the processes of cell mitosis, proliferation, survival, apoptosis and anti-apoptosis [10, 11]. There may be a correlation between the Val allele at Ile655Val locus of the ERBB2 and the increased risk of OC and poor prognosis of patients with OC [12]. A number of studies suggest that the rs1058808 locus of ERBB2 is closely associated with susceptibility to several types of solid tumors such as gastric cancer [13], cervical cancer [14] and osteosarcoma [15]. Analysis of functional polymorphism revealed that the G allele at rs1058808 site is related to the up-regulation of HER2 expression [14]. But the correlation between SNPs of ERBB2 rs1058808 site and the susceptibility to EOC remains unclear.
In the current study, with a case-control study, we analyzed the correlation between the SNPs of PIK3CA rs9838117 site and ERBB2 rs1058808 site and the risk of EOC, as well as the correlation between the interaction between these SNPs sites and the subjects’ clinical characteristics and the risk of EOC, providing reference for clinical prevention and treatment of EOC.
Materials and methods
Subjects
Subjects including 587 EOC patients were recruited from Longyan People Hospital and Weihai Central Hospital from January 2015 to October 2017. The age of enrolled OC patients was from 32 to 76 years old, with an average age of (58.97±8.71) years old. The 587 EOC patients were classified according to the American Joint Committee on Cancer (AJCC) [16] and the International Federation of Obstetricians and Gynecologists (FIGO) [17]. All the enrolled EOC patients were diagnosed by pathological diagnosis, and received surgical resection after routine pathological examination and immunohistochemistry. Patients with cancer history, patients with non-epithelial ovarian cancer and patients with immune system diseases were excluded from the present study. Another 650 women with no history of cancer were selected as the control group, aged from 34 to 85 years old, with an average age of (58.31±10.18) years old.
Genotyping analysis
Genomic DNA (gDNA) was prepared from peripheral blood using QIAamp DNA Blood Mini Kit (Qiagen, Hilden, Germany). DNA was quantified by NanoDrop 2000 ultraviolet spectrophotometer (Thermo Fisher Scientific, CA, USA) and stored at −80°C until use. DNA fragment containing PIK3CA rs9838117 locus and ERBB2 rs1058808 locus was amplified by PCR. The primers used for DNA amplification were shown as follows: PIK3CA rs9838117-F 5′-TGA TGG AGA AGG AAA AAG TGA TGG-3′, PIK3CA rs9838117-R 5′-TGT TGT GTC CAC ATT TCA AAA CAT-3′; ERBB2 rs1058808-F 5′-TGA ACC AGC CAG ATG TTC GG-3, ERBB2 rs1058808-R 5′-TCC CTG GGG AGA GAG TCT TG-3′. PCR was carried out in 50 μl reaction mixture containing 40 ng of gDNA, 4 μl of 2.5 mM dNTP, 10 μl polymerase buffer, 1 μl of 10 pM forward primer and reverse primer, and 0.5 U Prime STAR HS DNA polymerase (TaKaRa, Guangzhou, China). PCR amplification conditions were as follows: 94°C, 5 min; 98°C 20 s, 60°C 20 s, 72°C 2 min, 35 cycles; and 72°C 5 min. The genotype was determined according to the comparison with sequences in dbSNP (https://www.ncbi.nlm.nih.gov/snp/).
Statistical analysis
Continuous variables between groups were analyzed using t test and one-way analysis of variance. The χ2 test was used for statistical analysis of categorical variables. The consistency between genotype frequencies of the SNPs at PIK3CA rs9838117 site and ERBB2 gene rs1058808 site in the control group and Hardy-Weinberg equilibrium (HWE) was evaluated by χ2 test. Logistic regression analysis of odds ratio (OR) and 95% confidence interval (CI) was used to analyze the genotypes and allele frequencies of SNPs at PIK3CA rs9838117 locus and ERBB2 rs1058808, and to calculate the association between two genetic models (dominant model and sex model) and the risk of OC. They were also utilized to adjust for age, BMI, number live-born, smoking, drinking, ovarian cancer family history. The interaction between the PIK3CA rs9838117 locus and the ERBB2 rs1058808 locus and the clinical characteristics of the subjects was analyzed using multi-factor dimensionality reduction (MDR). All the statistical analysis was performed using SPSS 22.0 (SPSS, IL, USA), and P<0.05 indicated statistically significant.
Results
Clinical characteristics of subjects
The general clinical characteristics of 587 EOC patients and 650 control individuals were summarized in Table 1. There was no statistically significant difference in age, body mass index (BMI), number live-born, smoking, drinking between EOC patients and control group (P>0.05). But the proportion of EOC family history in EOC patients was obviously higher than that in the control group (P<0.05). According to the classification standers of FIGO, among the 587 patients with EOC, 104 cases (17.72%) were stage I, 121 cases (20.61%) were stage II, 293 cases (49.91%) were stage III, and 69 cases were stage IV (11.75%). Among the recruited EOC patients, 59 cases (10.05%) were in tumor grade G1, 168 cases (28.62%) were in G2, and the left 360 patients (61.33%) were in G3. There were 364 cases (62.01%) of EOC patients with lymphatic metastasis, and 223 cases (37.99%) of OC patients without lymphatic metastasis.
Table 1.
Comparison of clinical characteristics between OC patients and control group
| Variables | EOC (n=587) | Control (n=650) | P |
|---|---|---|---|
| Age (years, mean ± SD) | 58.97±8.71 | 58.31±10.18 | 0.22 |
| <60 | 336 (51.69%) | 384 (59.08%) | 0.51 |
| ≥60 | 251 (38.62%) | 266 (40.92%) | |
| BMI (kg/m2, n (%)) | 25.44±2.15 | 25.39±2.13 | 0.68 |
| <24 | 164 (27.94%) | 197 (30.31%) | 0.36 |
| ≥24 | 423 (72.06%) | 453 (69.69%) | |
| Number live-born (n (%)) | |||
| 0 | 47 (8.01%) | 45 (6.92%) | 0.31 |
| 1~2 | 364 (62.01%) | 430 (66.15%) | |
| ≥3 | 176 (29.98%) | 175 (26.92%) | |
| Smoking (n (%)) | |||
| Yes | 100 (17.04%) | 114 (17.54%) | 0.82 |
| No | 487 (82.96%) | 536 (82.46%) | |
| Drinking (n (%)) | |||
| Yes | 96 (16.35%) | 98 (15.08%) | 0.54 |
| No | 491 (83.65%) | 552 (84.92%) | |
| OC family history (n (%)) | |||
| Yes | 94 (16.01%) | 22 (3.38%) | <0.01 |
| No | 493 (83.99%) | 628 (96.62%) | |
| FIGO stage (n (%)) | |||
| I | 104 (17.72%) | ||
| II | 121 (20.61%) | ||
| III | 293 (49.91%) | ||
| IV | 69 (11.75%) | ||
| Tumor grade (n (%)) | |||
| G1 | 59 (10.05%) | ||
| G2 | 168 (28.62%) | ||
| G3 | 360 (61.33%) | ||
| Lymphatic metastasis (n (%)) | |||
| Yes | 364 (62.01%) | ||
| No | 223 (37.99%) | ||
BMI, body mass index; FIGO, International Federation of Gynecology and Obstetrics
Correlation between the polymorphism of PIK3CA and ERBB2 and the risk of EOC
The correlation between different genotypes, genetic models and allele frequencies of PIK3CA rs9838117 site and ERBB2 rs1058808 site and the risk of EOC were concluded in Table 2. The genotype frequencies of SNPs at PIK3CA rs9838117 locus and ERBB2 rs1058808 locus were in accordance with Hardy-Weinberg equilibrium (P>0.05). The TT genotype of PIK3CA rs9838117 locus, dominant model (GT+TT vs. GG), recessive model (TT vs. GG+GT) were associated with the increased risk of EOC (OR=6.54, 95%CI: 3.17–13.51, P<0.01; OR=1.68, 95%CI: 1.29–2.19, P<0.01; OR=6.20, 95%CI: 3.01–12.76, P<0.01). The risk of EOC in carriers of T allele at PIK3CA rs9838117 locus was 1.95 times that of carriers of G allele (95%CI: 1.55–2.46, P<0.01).
Table 2.
Correlation of genotype and allele frequency of PIK3CA rs9838117 locus and ERBB2 rs1058808 with the risk of EOC
| EOC (n=587) | Control (n=650) | HWE P | OR (95%CI) * | P | |
|---|---|---|---|---|---|
| rs9838117 | |||||
| GG | 419 (71.38%) | 525 (80.77%) | 0.37 | 1.00 (reference) | |
| GT | 121 (20.61%) | 116 (17.85%) | 1.31 (0.98–1.74) | 0.08 | |
| TT | 47 (8.01%) | 9 (1.38%) | 6.54 (3.17–13.51) | <0.01 | |
| GT+TT | 168 (28.62%) | 125 (19.23%) | 1.68 (1.29–2.19) | <0.01 | |
| GG+GT | 540 (91.99%) | 641 (98.62%) | 1.00 (reference) | ||
| TT | 47 (8.01%) | 9 (1.38%) | 6.20 (3.01–12.76) | <0.01 | |
| G | 959 (81.69%) | 1166 (89.69%) | 1.00 (reference) | ||
| T | 215 (18.31%) | 134 (10.31%) | 1.95 (1.55–2.46) | <0.01 | |
| rs1058808 | |||||
| CC | 262 (44.63%) | 241 (37.08%) | 0.15 | 1.00 (reference) | |
| CG | 298 (50.77%) | 295 (45.38%) | 0.93 (0.73–1.18) | 0.59 | |
| GG | 27 (4.60%) | 114 (17.54%) | 0.22 (0.14–0.34) | <0.01 | |
| CG+GG | 325 (55.37%) | 409 (62.92%) | 0.73 (0.58–0.92) | <0.01 | |
| CC+CG | 560 (95.40%) | 536 (82.46%) | 1.00 (reference) | ||
| GG | 27 (4.60%) | 114 (17.54%) | 0.23 (0.15–0.35) | <0.01 | |
| C | 822 (70.02%) | 777 (59.77%) | 1.00 (reference) | ||
| G | 352 (29.98%) | 523 (40.23%) | 0.64 (0.54–0.75) | <0.01 | |
HWE, Hardy-Weinberg equilibrium; OR, odds ratio; CI, confidence interval
*Adjust age, BMI, number live-born, smoking, drinking, ovarian cancer family history
The GG genotype, dominant model (CG+GG vs. CC), recessive model (GG vs. CC+CG) at ERBB2 rs1058808 locus were associated with the reduced risk of EOC (OR=0.22, 95%CI: 0.14–0.34, P<0.01; OR=0.73, 95%CI: 0.58–0.92, P<0.01; OR=0.23, 95%CI: 0.15–0.35, P<0.01). The risk of EOC in individuals with G allele at ERBB2 rs1058808 locus was as 0.64 times as the risk for carriers of C allele (95%CI: 0.54–0.75, P<0.01)
Stratified analysis
To investigate the correlation between different genotypes of PIK3CA gene rs9838117 site and ERBB2 gene rs1058808 site and the risk of ovarian cancer, enrolled subjects were classified based on age, BMI, number live-born, drinking history, smoking history and family history of ovarian cancer. As shown in Table 3, carriers of GT and TT genotypes of PIK3CA gene rs9838117 locus had higher risk of EOC than carriers of GG in people with the following features: less than 60 years old (OR=2.08, 95%CI: 1.48–2.95, P<0.01), BMI<24kg/m2 (OR=2.51, 95%CI: 1.51–4.19, P<0.01), BMI ≥24kg/m2 (OR=1.44, 95%CI: 1.06–1.97, P =0.03), number live-born 0–2 (OR=1.73, 95%CI: 1.26–2.36, P <0.01), smoking (OR=2.94, 95%CI: 1.52–5.69, P <0.01), no smoking history (OR=1.46, 95%CI: 1.09–1.95, P=0.01), drinking (OR=2.86, 95%CI: 1.47–5.57, P<0.01), no drinking history (OR=1.51, 95%CI: 1.13–2.02, P<0.01), no OC family history (OR=1.61, 95%CI: 1.22–2.13, P<0.01).
Table 3.
Stratified analysis of correlation between genotypes frequency at PIK3CA rs9838117 site and the risk of EOC
| EOC (n=587) | Control (n=650) | OR (95%CI) * | P | |
|---|---|---|---|---|
| Age | ||||
| <60 | ||||
| GG | 228 (67.86%) | 313 (81.51%) | 1.00 (reference) | |
| GT+TT | 108 (32.14%) | 71 (18.49%) | 2.08 (1.48–2.95) | <0.01 |
| ≥60 | ||||
| GG | 191 (76.10%) | 212 (79.70%) | 1.00 (reference) | |
| GT+TT | 60 (23.90%) | 54 (20.30%) | 1.23 (0.81–1.87) | 0.38 |
| BMI (kg/m2) | ||||
| <24 | ||||
| GG | 113 (68.90%) | 167 (84.77%) | 1.00 (reference) | |
| GT+TT | 51 (31.10%) | 30 (15.23%) | 2.51 (1.51–4.19) | <0.01 |
| ≥24 | ||||
| GG | 306 (72.34%) | 358 (79.03%) | 1.00 (reference) | |
| GT+TT | 117 (27.66%) | 95 (20.97%) | 1.44 (1.06–1.97) | 0.03 |
| Number live-born | ||||
| 0–2 | ||||
| GG | 294 (71.53%) | 386 (81.26%) | 1.00 (reference) | |
| GT+TT | 117 (28.47%) | 89 (18.74%) | 1.73 (1.26–2.36) | <0.01 |
| ≥3 | ||||
| GG | 125 (71.02%) | 139 (79.43%) | 1.00 (reference) | |
| GT+TT | 51 (28.98%) | 36 (20.57%) | 1.58 (0.97–2.57) | 0.09 |
| Smoking | ||||
| Yes | ||||
| GG | 66 (66.00%) | 97 (85.09%) | 1.00 (reference) | |
| GT+TT | 34 (34.00%) | 17 (14.91%) | 2.94 (1.52–5.69) | <0.01 |
| No | ||||
| GG | 353 (73.08%) | 428 (79.85%) | 1.00 (reference) | |
| GT+TT | 130 (26.92%) | 108 (20.15%) | 1.46 (1.09–1.95) | 0.01 |
| Drinking | ||||
| Yes | ||||
| GG | 60 (62.50%) | 81 (82.65%) | 1.00 (reference) | |
| GT+TT | 36 (37.50%) | 17 (17.35%) | 2.86 (1.47–5.57) | <0.01 |
| No | ||||
| GG | 359 (73.12%) | 444 (80.43%) | 1.00 (reference) | |
| GT+TT | 132 (26.88%) | 108 (19.57%) | 1.51 (1.13–2.02) | <0.01 |
| Ovarian cancer family history | ||||
| Yes | ||||
| GG | 64 (68.09%) | 19 (86.36%) | 1.00 (reference) | |
| GT+TT | 30 (31.91%) | 3 (13.64%) | 2.97 (0.82–10.81) | 0.15 |
| No | ||||
| GG | 355 (72.01%) | 506 (80.57%) | 1.00 (reference) | |
| GT+TT | 138 (27.99%) | 122 (19.43%) | 1.61 (1.22–2.13) | <0.01 |
BMI, body mass index; OR, odds ratio; CI, confidence interval
*Adjust age, BMI, number live-born, smoking, drinking, ovarian cancer family history
Individuals with CG and GG genotypes at ERBB2 gene rs1058808 site had a lower risk of EOC than CC genotype carriers in people classified as follows: age <60 years (OR=0.69, 95%CI: 0.51–0.93, P =0.02), number live-born 0–2 (OR=0.72, 95%CI: 0.55–0.94, P=0.02), no smoking (OR=0.61, 95%CI: 0.48–0.79, P<0.01), no drinking (OR=0.73, 95%CI: 0.57–0.94, P=0.02), no ovarian cancer family history (OR=0.72, 95%CI: 0.57–0.92, P<0.01) (Table 4).
Table 4.
Stratified analysis of correlation between genotypes frequency at ERBB2 rs1058808 site and the risk of EOC
| EOC (n=587) | Control (n=650) | OR (95%CI) * | P | |
|---|---|---|---|---|
| Age | ||||
| <60 | ||||
| CC | 156 (46.43%) | 144 (37.50%) | 1.00 (reference) | |
| CG+GG | 180 (53.37%) | 240 (62.50%) | 0.69 (0.51–0.93) | 0.02 |
| ≥60 | ||||
| CC | 106 (42.23%) | 97 (36.47%) | 1.00 (reference) | |
| CG+GG | 145 (57.77%) | 169 (63.53%) | 0.79 (0.55–1.12) | 0.21 |
| BMI (kg/m2) | ||||
| <24 | ||||
| CC | 77 (46.95%) | 69 (35.03%) | 1.00 (reference) | |
| CG+GG | 87 (53.05%) | 128 (64.97%) | 0.61 (0.40–0.93) | 0.03 |
| ≥24 | ||||
| CC | 185 (43.74%) | 172 (37.97%) | 1.00 (reference) | |
| CG+GG | 238 (56.26%) | 281 (62.03%) | 0.79 (0.60–1.03) | 0.10 |
| Number live-born | ||||
| 0–2 | ||||
| CC | 189 (45.99%) | 180 (37.89%) | 1.00 (reference) | |
| CG+GG | 222 (54.01%) | 295 (62.11%) | 0.72 (0.55–0.94) | 0.02 |
| ≥3 | ||||
| CC | 73 (41.48%) | 61 (34.86%) | 1.00 (reference) | |
| CG+GG | 103 (58.52%) | 114 (65.14%) | 0.76 (0.49–1.16) | 0.24 |
| Smoking | ||||
| Yes | ||||
| CC | 28 (28.00%) | 47 (41.23%) | 1.00 (reference) | |
| CG+GG | 72 (72.00%) | 67 (58.77%) | 1.80 (1.02–3.20) | 0.06 |
| No | ||||
| CC | 234 (48.05%) | 194 (36.19%) | 1.00 (reference) | |
| CG+GG | 253 (51.95%) | 342 (63.81%) | 0.61 (0.48–0.79) | <0.01 |
| Drinking | ||||
| Yes | ||||
| CC | 44 (45.83%) | 37 (37.76%) | 1.00 (reference) | |
| CG+GG | 52 (54.17%) | 61 (62.24%) | 0.72 (0.40–1.27) | 0.32 |
| No | ||||
| CC | 218 (44.40%) | 204 (36.96%) | 1.00 (reference) | |
| CG+GG | 273 (55.60%) | 348 (63.04%) | 0.73 (0.57–0.94) | 0.02 |
| Ovarian cancer family history | ||||
| Yes | ||||
| CC | 41 (43.62%) | 9 (40.91%) | 1.00 (reference) | |
| CG+GG | 53 (56.38%) | 13 (59.09%) | 0.90 (0.35–2.30) | 0.82 |
| No | ||||
| CC | 221 (44.83%) | 232 (36.94%) | 1.00 (reference) | |
| CG+GG | 272 (55.17%) | 396 (63.06%) | 0.72 (0.57–0.92) | <0.01 |
BMI, body mass index; OR, odds ratio; CI, confidence interval
*Adjust age, BMI, number live-born, smoking, drinking, ovarian cancer family history
Multi-factor dimensionality reduction (MDR) analysis of the interaction between SNP sites of PIK3CA gene and ERBB2 gene and clinical characteristic of subjects
The interaction between the PIK3CA rs9838117 locus and ERBB2 rs1058808 locus and the subject’s age, BMI, number live-born, smoking, drinking, and OC family history was shown in Fig. 1. The ERBB2 rs1058808 site had the strongest interaction with smoking, followed by PIK3CA gene rs9838117 site (Fig. 1). MDR analysis showed that the interaction model between age, BMI, number live-born, smoking, drinking, OC family history, PIK3CA gene rs9838117 locus, and ERBB2 gene rs1058808 locus was the best model for prediction of the risk of OC. The EOC risk of the “high-risk combination” was 3.10 times that of the “low-risk combination” (95%CI: 1.49–6.46, P<0.01). The training balanced accuracy was 0.7238, the testing balanced accuracy was 0.6377, and the cross-validation consistency was 10/10 (Table 5).
Fig. 1.
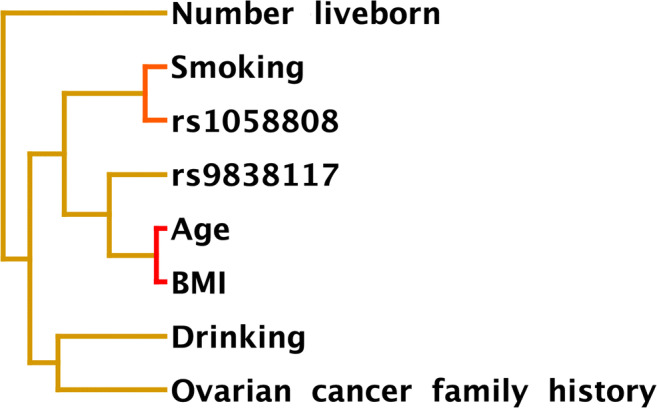
MDR analysis for the interaction between SNP sites of PIK3CA gene and ERBB2 gene and clinical data of subjects including age, BMI, number live-born, smoking, drinking, and OC family history. The distance was inversely proportional to the interaction
Table 5.
The best model prediction of the interaction between the PIK3CA gene rs9838117 and ERBB2 gene rs1058808 sites and the clinical data of subjects
| Model | Training balanced accuracy | Testing balanced accuracy | χ2 | OR (95%CI) | P | Cross-validation consistency |
|---|---|---|---|---|---|---|
| rs1058808 | 0.5666 | 0.5415 | 0.86 | 1.40 (0.69–2.87) | 0.35 | 6/10 |
| OC family history, rs9838117 | 0.5906 | 0.5500 | 1.53 | 1.64 (0.75–3.62) | 0.21 | 8/10 |
| Age, BMI, rs1058808 | 0.6092 | 0.5503 | 1.30 | 1.52 (0.74–3.14) | 0.25 | 3/10 |
| Age, BMI, rs9838117, rs1058808 | 0.6361 | 0.6233 | 7.55 | 2.77 (1.33–5.76) | <0.01 | 10/10 |
| Age, BMI, OC family history, rs9838117, rs1058808 | 0.6538 | 0.6277 | 8.05 | 2.84 (1.37–5.90) | <0.01 | 10/10 |
| Age, BMI, number live-born, drinking, rs9838117, rs1058808 | 0.6729 | 0.5913 | 4.12 | 2.09 (1.02–4.29) | 0.04 | 4/10 |
| Age, BMI, number live-born, smoking, drinking, rs9838117, rs1058808 | 0.6985 | 0.6124 | 6.23 | 2.50 (1.21–5.15) | 0.01 | 6/10 |
| Age, BMI, number live-born, smoking, drinking, ovarian cancer family history, rs9838117, rs1058808 | 0.7238 | 0.6377 | 9.36 | 3.10 (1.49–6.46) | <0.01 | 10/10 |
BMI, body mass index; OR, odds ratio; CI, confidence interval
Correlation of SNPs at PIK3CA rs9838117 and ERBB2 rs1058808 loci with the progression of EOC
The genotypes and allele frequency of rs9838117 locus of PIK3CA gene and rs1058808 locus of ERBB2 gene were not associated with FIGO stage and lymphatic metastasis in patients with EOC (P>0.05, Supplementary Table 1 to Table 3).
Discussion
In the current study, we found SNPs at PIK3CA rs9838117 site and ERBB2 gene rs1058808 locus correlated with the risk of EOC. In addition, the interaction between SNP sites of PIK3CA rs9838117 and ERBB2 rs1058808 loci and clinical data of subjects including age, BMI, number live-born, smoking, drinking, and OC family history was strongly associated with the risk of EOC.
EOC is a malignant tumor of the female reproductive system. Lacking of early diagnosis techniques and effective long-term treatment programs is the main reason for the high mortality rate of ovarian cancer [18, 19]. Therefore, there is an urgent need to find new tumor markers and therapeutic targets for clinical diagnosis and treatment of OC [20, 21]. In recent years, growing evidence supports that genetic polymorphisms are significantly associated with the occurrence and development of OC [3, 22, 23]. The influence of genetic polymorphisms of OC molecular markers on the occurrence and development of OC is also worthy of further researches.
PI3K/Akt signal pathway is the downstream transducing cascade of a variety of growth factors and cytokines. PI3K/Akt signal pathway closely correlates to the occurrence and development of tumors, and can promote cell proliferation, invasion, metastasis, and inhibits cell apoptosis [24–26]. The PIK3CA gene is the only gene with somatic mutations, and these mutations mostly appear in the regions coding the helical domain and kinase domain [27].
PIK3CA gene rs9838117 SNP site is located in the 3′ untranslated region (UTR). The present study showed that the T allele at PIK3CA rs9838117 site was associated with increased risk of OC. The possible reason is that the rs9838117 site may be at the targeted binding region of microRNA targeting PIK3CA, and allelic variation may affect the regulatory effect of microRNA on the expression of their target gene PIK3CA, which needed to be verified by in vitro assays.
The ERBB2 gene rs1058808 locus is also located in its 3′UTR region. Individuals carrying G allele at ERBB2 rs1058808 locus have a lower risk of EOC than carriers with C allele, suggesting that rs1058808 locus G allele may be a protective factor for EOC. Although there is no evidence that the SNPs at ERBB2 gene rs1058808 locus is associated with the risk of OC, there has been research that SNPs at ERBB2 rs1058808 locus is associated with the high risk of osteosarcoma [15]. But the study showed that G allele of ERBB2 gene rs1058808 locus is one high risk factor for osteosarcoma, which is inconsistent with the results of the present study [15]. This may be due to the large differences in the genetic background of the individuals enrolled in this study. The frequency of the G allele of the ERBB2 gene rs1058808 was 18% in the control group selected in the previous study, and was 40.23% in this proposed study. According to the data in the 1000 genome database, the frequency of the G allele at rs1058808 site in Han Chinese population in Beijing is 48.06%, which is close to the individuals selected in this study, indicating that the population selected herein is representative. In addition, the calculation of minimum sample size showed that the minimum sample size required for EOC patients and control groups in this study was 286 and 317, respectively, which were lower than the number of samples selected in the proposed study, which showed the objectivity of the obtained results.
Similarly, regarding the specific cause of the correlation between SNP at ERBB2 rs1058808 and OC, we speculated that the rs1058808 site may be at the binding site of the microRNA targeting ERBB2, and allelic variation may change the interaction between ERBB2 and microRNAs. At present, ERBB2 inhibitors have been used in clinical treatment, and they exert inhibitory effect on the proliferation and migration of ERBB2-positive OC cells. This finding implies that the association between genetic polymorphism and the risk of EOC may be reflected in the expressional level of ERBB2, which will be verified in in vitro cell model.
The occurrence of OC closely correlated with genes and environment, and the combined effects of genes and genes, genes and environment are related to the risk of ovarian cancer [28, 29]. The stratification of the clinical data of the subjects significantly affected the correlation between the SNPs at PIK3CA gene rs9838117 and ERBB2 rs1058808 loci and OC. Further MDR analysis displayed that the interaction model between age, BMI, number live-born, smoking, drinking, OC family history, PIK3CA rs9838117, and ERBB2 rs1058808 locus is the best model for the risk of OC prediction. This analysis further proved that the interaction of PIK3CA rs9838117 and ERBB2 rs1058808 sites with environmental factors has a significant correlation with the occurrence of EOC.
However, the present study still had the following deficiencies. In vitro analysis was required to further investigate the specific molecular mechanism of the correlation between the risk of OC and the SNPs at PIK3CA rs9838117 locus and the ERBB2 rs1058808 locus. Additionally, the effect of SNPs at PIK3CA rs9838117 locus and ERBB2 rs1058808 locus on gene expression remained to be elucidated in future. Moreover, this study lacks the functional investigation of the SNPs at PIK3CA rs9838117 locus and ERBB2 rs1058808 locus, which needs to be explored in future. Finally, in the present study, we did not include patient survival data, nor did we collect specific causes of death of EOC patients. In future studies, we need to supplement survival analysis data based on more detailed clinical characteristic.
Conclusion
In summary, SNPs of PIK3CA gene rs9838117 locus and ERBB2 gene rs1058808 locus were strongly associated with the risk of EOC. The underlying mechanism was still needed to be further explored.
Supplementary Information
(DOCX 24 kb)
Author contribution
Haibo Chen, Zhenyuan Zhai, Qinghai Xie, and Guiping Chen contributed to the experimental design, writing, review, and/or revision of the manuscript. Haibo Chen, Zhenyuan Zhai, and Yuanbin Lai contributed to the acquisition and analysis of data. Haibo Chen and Zhenyuan Zhai contributed equally to this work. All authors approved the submitted version.
Declarations
Ethics approval and consent to participate
This study was approved by the ethics committee of Longyan People Hospital, and all the subjects signed an informed consent form.
Conflict of interest
The authors declare no competing interests.
Footnotes
Haibo Chen and Zhenyuan Zhai are co-authors: authors contribute equally
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Voelker R. Ovarian cancer drug approved. JAMA. 2017;317(5):466. doi: 10.1001/jama.2017.0026. [DOI] [PubMed] [Google Scholar]
- 2.Jones MR, Kamara D, Karlan BY, Pharoah PDP, Gayther SA. Genetic epidemiology of ovarian cancer and prospects for polygenic risk prediction. Gynecol Oncol. 2017;147(3):705–713. doi: 10.1016/j.ygyno.2017.10.001. [DOI] [PubMed] [Google Scholar]
- 3.Kar SP, Berchuck A, Gayther SA, Goode EL, Moysich KB, Pearce CL, Ramus SJ, Schildkraut JM, Sellers TA, Pharoah PDP. Common genetic variation and susceptibility to ovarian cancer: current insights and future directions. Cancer Epidemiol Biomarkers Prev. 2018;27(4):395–404. doi: 10.1158/1055-9965.EPI-17-0315. [DOI] [PubMed] [Google Scholar]
- 4.Pearce CL, Rossing MA, Lee AW, Ness RB, Webb PM. for Australian Cancer S, et al.: Combined and interactive effects of environmental and GWAS-identified risk factors in ovarian cancer. Cancer Epidemiol Biomarkers Prev. 2013;22(5):880–890. doi: 10.1158/1055-9965.EPI-12-1030-T. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Astanehe A, Arenillas D, Wasserman WW, Leung PC, Dunn SE, Davies BR, et al. Mechanisms underlying p53 regulation of PIK3CA transcription in ovarian surface epithelium and in ovarian cancer. J Cell Sci. 2008;121(Pt 5):664–674. doi: 10.1242/jcs.013029. [DOI] [PubMed] [Google Scholar]
- 6.Woenckhaus J, Steger K, Sturm K, Munstedt K, Franke FE, Fenic I. Prognostic value of PIK3CA and phosphorylated AKT expression in ovarian cancer. Virchows Arch. 2007;450(4):387–395. doi: 10.1007/s00428-006-0358-3. [DOI] [PubMed] [Google Scholar]
- 7.Wang D, Li C, Zhang Y, Wang M, Jiang N, Xiang L, Li T, Roberts TM, Zhao JJ, Cheng H, Liu P. Combined inhibition of PI3K and PARP is effective in the treatment of ovarian cancer cells with wild-type PIK3CA genes. Gynecol Oncol. 2016;142(3):548–556. doi: 10.1016/j.ygyno.2016.07.092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tang Y, Liu B, Li J, Wu H, Yang J, Zhou X, Yi M, Li Q, Yu S, Yuan X. Genetic variants in PI3K/AKT pathway are associated with severe radiation pneumonitis in lung cancer patients treated with radiation therapy. Cancer Med. 2016;5(1):24–32. doi: 10.1002/cam4.564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li L, Bai H, Yang J, Cao D, Shen K. Genome-wide DNA copy number analysis in clonally expanded human ovarian cancer cells with distinct invasive/migratory capacities. Oncotarget. 2017;8(9):15136–15148. doi: 10.18632/oncotarget.14767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Krishnamurti U, Silverman JF. HER2 in breast cancer: a review and update. Adv Anat Pathol. 2014;21(2):100–107. doi: 10.1097/PAP.0000000000000015. [DOI] [PubMed] [Google Scholar]
- 11.Boku N. HER2-positive gastric cancer. Gastric Cancer. 2014;17(1):1–12. doi: 10.1007/s10120-013-0252-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Watrowski R, Castillo-Tong DC, Schuster E, Fischer MB, Speiser P, Zeillinger R. Association of HER2 codon 655 polymorphism with ovarian cancer. Tumour Biol. 2016;37(6):7239–7244. doi: 10.1007/s13277-015-4609-2. [DOI] [PubMed] [Google Scholar]
- 13.Vazquez-Ibarra KC, Bustos-Carpinteyro AR, Garcia-Ruvalcaba A, Magaaa-Torres MT, Gutierrez-Aguilar R, Marin-Contreras ME, et al. The ERBB2 gene polymorphisms rs2643194, rs2934971, and rs1058808 are associated with increased risk of gastric cancer. Braz J Med Biol Res. 2019;52(5):e8379. doi: 10.1590/1414-431x20198379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gao Y, Tang X, Cao J, Rong R, Yu Z, Liu Y, Lu Y, Liu X, Han L, Liu J, Zhang J, Xu M, Liu F. The effect of HER2 single nucleotide polymorphisms on cervical cancer susceptibility and survival in a Chinese population. J Cancer. 2019;10(2):378–387. doi: 10.7150/jca.27976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xin DJ, Shen GD, Song J. Single nucleotide polymorphisms of HER2 related to osteosarcoma susceptibility. Int J Clin Exp Pathol. 2015;8(8):9494–9499. [PMC free article] [PubMed] [Google Scholar]
- 16.Edge SB, Compton CC. The American Joint Committee on Cancer: the 7th edition of the AJCC cancer staging manual and the future of TNM. Annals of surgical oncology. 2010;17(6):1471–1474. doi: 10.1245/s10434-010-0985-4. [DOI] [PubMed] [Google Scholar]
- 17.Sheiner E, Kapur A, Retnakaran R, Hadar E, Poon LC, McIntyre HD, et al. FIGO (International Federation of Gynecology and Obstetrics) postpregnancy initiative: long-term maternal implications of pregnancy complications-follow-up considerations. Int J Gynaecol Obstet. 2019;147(Suppl 1):1–31. doi: 10.1002/ijgo.12926. [DOI] [PubMed] [Google Scholar]
- 18.Webb PM, Jordan SJ. Epidemiology of epithelial ovarian cancer. Best Pract Res Clin Obstet Gynaecol. 2017;41:3–14. doi: 10.1016/j.bpobgyn.2016.08.006. [DOI] [PubMed] [Google Scholar]
- 19.Rooth C. Ovarian cancer: risk factors, treatment and management. Br J Nurs. 2013;22(17):S23–S30. doi: 10.12968/bjon.2013.22.Sup17.S23. [DOI] [PubMed] [Google Scholar]
- 20.Pujade-Lauraine E. New treatments in ovarian cancer. Ann Oncol. 2017;28(suppl_8):viii57–viii60. doi: 10.1093/annonc/mdx442. [DOI] [PubMed] [Google Scholar]
- 21.Stewart C, Ralyea C, Lockwood S. Ovarian cancer: an integrated review. Semin Oncol Nurs. 2019;35(2):151–156. doi: 10.1016/j.soncn.2019.02.001. [DOI] [PubMed] [Google Scholar]
- 22.Pu D, Jiang SW, Wu J. Association between MTHFR gene polymorphism and the risk of ovarian cancer: a meta-analysis of the literature. Curr Pharm Des. 2014;20(11):1632–1638. doi: 10.2174/13816128113199990564. [DOI] [PubMed] [Google Scholar]
- 23.Koensgen D, Bruennert D, Ungureanu S, Sofroni D, Braicu EI, Sehouli J, Sümnig A, Delogu S, Zygmunt M, Goyal P, Evert M, Olek S, Biebler KE, Mustea A. Polymorphism of the IL-8 gene and the risk of ovarian cancer. Cytokine. 2015;71(2):334–338. doi: 10.1016/j.cyto.2014.07.254. [DOI] [PubMed] [Google Scholar]
- 24.Diaz-Serrano A, Angulo B, Dominguez C, Pazo-Cid R, Salud A, Jimenez-Fonseca P, et al. Genomic profiling of HER2-positive gastric cancer: PI3K/Akt/mTOR pathway as predictor of outcomes in HER2-positive advanced gastric cancer treated with trastuzumab. Oncologist. 2018;23(9):1092–1102. doi: 10.1634/theoncologist.2017-0379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lv B, Yang X, Lv S, Wang L, Fan K, Shi R, Wang F, Song H, Ma X, Tan X, Xu K, Xie J, Wang G, Feng M, Zhang L. Retraction note to: CXCR4 signaling induced epithelial-mesenchymal transition by PI3K/AKT and ERK pathways in glioblastoma. Mol Neurobiol. 2017;54(3):2380. doi: 10.1007/s12035-017-0464-z. [DOI] [PubMed] [Google Scholar]
- 26.Costa RLB, Han HS, Gradishar WJ. Targeting the PI3K/AKT/mTOR pathway in triple-negative breast cancer: a review. Breast Cancer Res Treat. 2018;169(3):397–406. doi: 10.1007/s10549-018-4697-y. [DOI] [PubMed] [Google Scholar]
- 27.Samuels Y, Wang Z, Bardelli A, Silliman N, Ptak J, Szabo S, Yan H, Gazdar A, Powell SM, Riggins GJ, Willson JK, Markowitz S, Kinzler KW, Vogelstein B, Velculescu VE. High frequency of mutations of the PIK3CA gene in human cancers. Science. 2004;304(5670):554. doi: 10.1126/science.1096502. [DOI] [PubMed] [Google Scholar]
- 28.Goodman MT, McDuffie K, Kolonel LN, Terada K, Donlon TA, Wilkens LR, et al. Case-control study of ovarian cancer and polymorphisms in genes involved in catecholestrogen formation and metabolism. Cancer Epidemiol Biomarkers Prev. 2001;10(3):209–216. [PubMed] [Google Scholar]
- 29.Lu ZH, Gu XJ, Shi KZ, Li X, Chen DD, Chen L. Association between genetic polymorphisms of inflammatory response genes and the risk of ovarian cancer. J Formos Med Assoc. 2016;115(1):31–37. doi: 10.1016/j.jfma.2015.01.002. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX 24 kb)


