Abstract
Purpose
To identify the genetic causes for acephalic spermatozoa syndrome.
Methods
Whole-exome sequencing was performed on the proband from a non-consanguineous to identify pathogenic mutations for acephalic spermatozoa syndrome. Quantitative real-time polymerase chain reaction and whole genome sequencing were subjected to detect deletion. The functional effect of the identified splicing mutation was investigated by minigene assay. Western blot and immunofluorescence were performed to detect the expression level and localization of mutant TSGA10 protein.
Results
Here, we identified a novel heterozygous splicing mutation in TSGA10 (NM_025244: c.1108-1G > T), while we confirmed that there was a de novo large deletion in the proband. The splicing mutation led to the skipping of the exon15 of TSGA10, which resulted in a truncated protein (p. A370Efs*293). Therefore, we speculated that the splicing mutation might affect transcription and translation without the dosage compensation of a normal allele, which possesses a large deletion including intact TSGA10. Western blot and immunofluorescence demonstrated that the very low expression level of truncated TSGA10 protein led the proband to present the acephalic spermatozoa phenotype.
Conclusion
Our finding expands the spectrum of pathogenic TSGA10 mutations that are responsible for ASS and male infertility. It is also important to remind us of paying attention to the compound heterozygous deletion in patients from non-consanguineous families, so that we can provide more precise genetic counseling for patients.
Keywords: Male infertility, Teratozoospermia, Acephalic spermatozoa syndrome, TSGA10, Spermatogenesis
Introduction
Although not a life-threatening condition, with increasing incidence of reproductive disorders, infertility will be the third major disease threatening human health in the twenty-first century according to the WHO [1]. About 50% of infertility can be ascribed to males [2]. Acephalic spermatozoa syndrome (ASS) is a severe condition associated with male infertility. The typical ASS patients’ sperm consisted of a large number of headless sperm tails, a few tailless sperm heads, and sperm with abnormal head–tail connection [3]. Since ASS was first discovered in 1977, only a small number of genetic factors, such as SUN5 [4–10], PMFBP1 [9, 11–13], BRDT [14], HOOK1 [15], DNAH6 [16], TSGA10 [9, 17, 18], and CEP112 [19] mutations, have been proven to be associated with it, and there were still many cases that could not be explained by the reported genes and mutations, which remained to be elucidated.
TSGA10 (testis specific, 10) is expressed exclusively in testis [20]. It encoded a protein of 82 KDa, including a 27-KDa N-terminus localized in the principal piece, and a 55-KDa C-terminus accumulated in the midpiece of sperm to the centrosome and basal body [21]. Previous studies revealed that TSGA10 played an important role in the assembly of centriole in the head–tail connection, the arrangement of mitochondrial sheath, and the development of embryo [17, 18, 22, 23]. However, there were only several studies about TSGA10 reported since 2017, the first time linked it with ASS [9, 17, 18].
In the present study, we identified a novel splicing mutation (NM_025244: c.1108-1G > T) in TSGA10 and a de novo large deletion contained intact TSGA10. This is the first case reported as a heterozygous deletion in TSGA10. Our finding expanded the spectrum of pathogenic TSGA10 mutations responsible for ASS and male infertility.
Materials and methods
Human subjects
The proband was recruited from Reproductive Medicine Center, Department of Obstetrics and Gynecology, the First Affiliated Hospital of Anhui Medical University. The proband’s sperm met with a certain diagnostic standard of ASS according to the World Health Organization 2010 guidelines [1]. We obtained informed consent from all the participants, and the study was approved by the Ethics Committee of Anhui Medical University.
Papanicolaou staining
Papanicolaou staining was performed according to the World Health Organization standards for human semen examination and processing [1] to assess sperm morphology.
WES and co-segregation analysis
Genomic DNA was extracted following the protocol of the QIAamp DNA blood midi kit (Qiagen, Hilden, Germany). We performed WES on the proband and filtered out the most promising candidate mutations using the methods described in our previous studies [4, 11], except that we took the compound heterozygous mutations into consideration due to the proband from a non-consanguineous family.
The suspected mutation in TSGA10 was subjected to co-segregation analysis by Sanger sequencing using the primers shown in Table 1. As the discrepancy of the law of co-segregation, we speculated on the existence of the compound heterozygous deletion.
Table 1.
TSGA10 primers used for PCR
| Target | Forward | Reverse |
|---|---|---|
| Primers used for co-segregation analysis | ||
| exon15: c.1108-1G > T | TGCTTATTTGGGTCATTTGCC | ACCTGTCGTTCTGAGTATACCA |
| Primers used for real-time PCR | ||
| Exon6 | AAGTCCAAGACGCCCATCAC | ACTGTGCCATCAAAGAGACCTC |
| Exon7 | TGGCAGAAATTCAGGGTAATG | TTGCAGGTTTAAGCAGTCTCAT |
| Exon8 | ATTACCCGACTTCGACG | CTTTCCAACCTTTAGCCTC |
| Exon9 | AGAAGGCTCACCTGGAA | CCATTGAGTGTTTATAGCCA |
| Exon10 | TCCTGTCAGTCTTGCCAT | CTTTTTGTCTGCCAAGTTC |
| Exon11 | CTTTCTGATACTCAGCGACACC | GTTAAGGGCTTGTTTCCAAGTT |
| Exon12 | GGAGAAGTTCAAAAAGTACGATA | GCCAGATCATTGAGGGTTC |
| Exon13 | ATCATTGCTGAGATGGAACA | CGTATCATTAAGCCCCTTG |
| Exon14 | CTTGTGAGAATTTGTTGCTCTG | CTTTCCCTGGCGATCTGG |
| Exon15 | TGGAGGTTAACAAGCTGAAGAA | CTTTGATTCACCTGTCGTTCTG |
| Exon16 | CAGGAATCTGAGAACCG | CAGCAGTGATAAGTTCCAG |
| Exon17 | AGGTCTTACAAGTCCCAG | CAAGTTTAATACAGAGTTCCC |
| Exon18 | TGAACTCCTGAGGAGTCAGATG | TTTTCTGCCAAACAAAGGTG |
| Exon19 | GAGTAGAGATGTGGCCCAG | AGAACTAAAACCTCTTGTTACCTA |
| Exon20 | GCCGTACAAGAACTTCGC | GATGGTGAGCACGTTCTG |
| Exon21 | CTTACGTTTTGCTGAAGTGATACAC | TTGACCTTTCTCAGGGATGTG |
| Primers used to confirm the breakpoint | ||
| Breakpoint | ACTCAGGTGATGATTTTATGGG | ACCCTCTAACTTGCTCTGCCC |
| Primers used for constructing the minigene vectors | ||
| Exon14 | CTATAGGGAGACCCAAGCTTACAGTGTACTGAGGCCCTAA | GAAAACACATAAGCCCAATAGCATTCCATTTAAAAATTATGTGGCCTA |
| Exon15 | TAGGCCACATAATTTTTAAATGGAATGCTATTGGGCTTATGTGTTTTC | GAAAACAGGTCATCCTGTAATTAACATCCAGGCATCTTTCTGAACA |
| Exon16 | TGTTCAGAAAGATGCCTGGATGTTAATTACAGGATGACCTGTTTTC | ACGTCATATGGATAGGATCCTTGCTCAACCTCTCTGTTG |
| Primers used for site directed mutagenesis | ||
| Mu1108-1 | TTTTTTCGTATGCACTGTCCAAAAAATTGAATGACACTC | TGGACAGTGCATACGAAAAAAAGATAGGTATATTTGACTATG |
| Primers used for detecting splicing isoform | ||
| Exon14-16 | ACAGTGTACTGAGGCCCTAA | TTGCTCAACCTCTCTGTTG |
qPCR and WGS
To verify our speculation, the expression level of TSGA10 in proband and his parents were analyzed by qPCR. Primers for qPCR are shown in Table 1. qPCR was performed using TB Green Premix Ex Taq II (Tli RNase H Plus) (Takara Biomedical Technology, Beijing, China) in StepOnePlus™ Real-Time PCR System with Tower (Applied Biosystems, Foster City, CA). Forty cycles of denaturation at 95℃ for 5 s, annealing at 60℃ for 30 s were performed. Results were analyzed by the 2−ΔΔCT method. The expression of GAPDH served as reference gene and the proband’s father, as a heterozygote, served as the normal control. Each reaction was repeated three times.
WGS was performed on the proband to find the definite deletion range and fracture site further. Briefly, eligible DNA was broken into fractions followed with repaired and amplified. Then, the library was constructed by ligating the fractions. Finally, high qualitative reads were compared with the human genome and the unique reads were extracted and analyzed. Polymerase chain reaction (PCR) and Sanger sequencing were followed to confirm the breakpoint and the primers are also shown in Table 1.
Minigene assay
The c.1108-1G > T mutation was located at the acceptor splice site of intron 14. Due to the large size of intron, we were unable to clone the full sequence from exon 14 to exon 16 into the minigene vector. Thus, we integrated exon14, exon 15, exon 16, and the sequences 150–200 bp before and after every exon by PCR and cloned the integrated fragment into a pcDNA3.1 plasmid using the method of homologous recombination. The mutation was generated by Mut Express® II Fast Mutagenesis Kit V2 (Vazyme Biotech, Nanjing, China). The wild type and mutant plasmids were transfected into HEK293T cells, respectively. After incubation for 36 h, total RNA was extracted using trizol and subjected to reverse transcription-polymerase chain reaction (RT-PCR) with HyperScriptTM III RT SuperMix for qPCR with gDNA Remover (NovaBio, Shanghai, China) following the manufacturer’s instructions. PCR was performed to amplify and compare the TSGA10 transcript from the wild type and mutant plasmids. The primers for constructing the minigene vectors and for detecting alternative splice sites are listed in Table 1.
Western blot and immunofluorescence
Western blot and immunofluorescence were performed to detect the expression level and localization of mutant TSGA10 protein compared with the normal, in accordance with the methods described previously [4, 11, 13]. The detailed information of antibodies we used displayed in Table 2.
Table 2.
Antibodies used in Western blot and immunofluorescence
| Primary/secondary antibodies | Host species | Company | Cat/Lot no | Application | Dilution |
|---|---|---|---|---|---|
| TSGA10 Antibody | Rabbit | Proteintech | 12,593–1-AP | WB | 1:2000 |
| IF | 1:200 | ||||
| Acetyl-α-Tubulin Rabbit mAb | Rabbit | ABclonal | 5335 T | WB | 1:2000 |
| IF | 1:1000 | ||||
| Alexa Fluor® 488—Conjugated Goat anti-Mouse IgG(H + L) | Goat | ThermoFisher | 1,834,802 | IF | 1:400 |
| Alexa Fluor® 555—Conjugated Goat anti-Rabbit IgG(H + L) | Goat | ThermoFisher | 1,774,719 | IF | 1:400 |
| Goat anti-Rabbit IgG (H + L) Secondary Antibody, HRP | Goat | ThermoFisher | 31,460 | WB | 1:5000 |
| Goat anti-Mouse IgG (H + L) Secondary Antibody, HRP | Goat | Thermofisher | 31,430 | WB | 1:5000 |
Results
Clinical findings
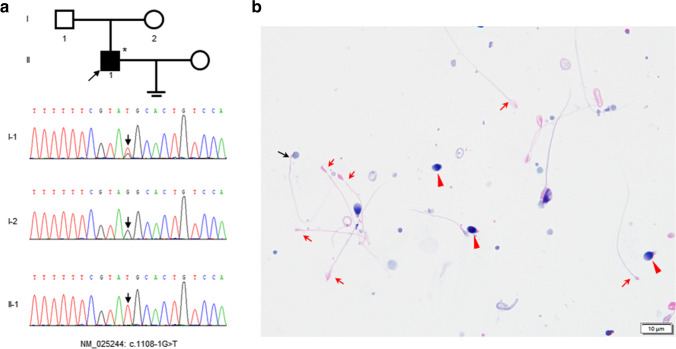
We recruited an ASS patient from a non-consanguineous family (Fig. 1a). The proband did not have any bad living habits or any adverse chemical contact history. Examination of the patient revealed normal development of male external genitalia, normal bilateral testicular size, and no abnormality upon palpation. The chromosomal karyotype of the patient was normal 46, XY. The hormonal levels of the proband were normal. The characteristics of the patient’s semen are shown in Table 3. Notably, it exhibited that spermatozoa of proband with normal morphology comprised 1% of the population, while ~ 98% were headless tails through Papanicolaou staining (Fig. 1b).
Fig. 1.
The family pedigree of the proband and papanicolaou staining of his sperm. a The family tree shows a patient with acephalic spermatozoa syndrome in a non-consanguineous pedigree. The black square shows the affected individuals with TSGA10 mutation and an arrow indicates the proband. The arrows in the chromatograms show the position of the mutation. The asterisk stands for the proband subjected WES. b Papanicolaou staining of the proband’s sperm. The sperm morphology was primarily acephalic (red arrow). Besides, there also were a few tailless heads (red arrowhead) and a small proportion of intact spermatozoa with abnormal head–tail junction (black arrow) can be observed. Scale bars: 10 μm
Table 3.
Statistical analysis of the semen parameters
| Proband | First ejaculate | Second ejaculate | Reference value |
|---|---|---|---|
| Volume (ml) | 2.6 | 2.7 | ≥ 2 |
| Concentration (× 106/ml) | 0.6 | 1.3 | ≥ 20 |
| Motility a + b (%) | 0 | 0 | ≥ 50 |
| Percentages of different morphologic spermatozoa(%) | |||
| Normally formed | 1 | 1 | ≥ 23% |
| Abnormal head–tail junction | 0.2 | 0.1 | / |
| Decaudated | 0.7 | 1 | / |
| Acephalic | 98.1 | 98.9 | / |
Genetic analysis
According to previous work suggested a genetic origin, WES was performed on the proband to identify the genetic defect. After screening with all our criteria, 41 mutations were noted (Table 4). The splicing mutation in TSGA10 is viewed as the most possible pathogenesis of the proband on the basis of previous studies. However, our results of Sanger sequencing demonstrated that the proband (II-1) was homozygous for the splicing mutation; his father (I-1) was heterozygous, while his mother (I-2) was a wild type, which were not in conformity with co-segregation (Fig. 1a). Therefore, we speculated that there was the possibility of deletion in TSGA10.
Table 4.
Screening strategy to identify the causal genes by WES
| Screening procedure | The proband (II-1) |
|---|---|
| Total variants | 153,484 |
| Filter rare variants (not reported or frequency < 0.1%) in public databasesa | 2491 |
| Coding (nonsynonymous exonic, indel or splice site variant) | 363 |
| Predicted to be highly deleterious | 41 |
| Relevancy for phenotypeb | 1 |
a represent the following three databases: 1000 Genomes variant database, Genome Aggregation Database, and Exome Aggregation Consortium; b represents mutations associated with phenotype, including expression in testis or not reported
Identification of the large deletion
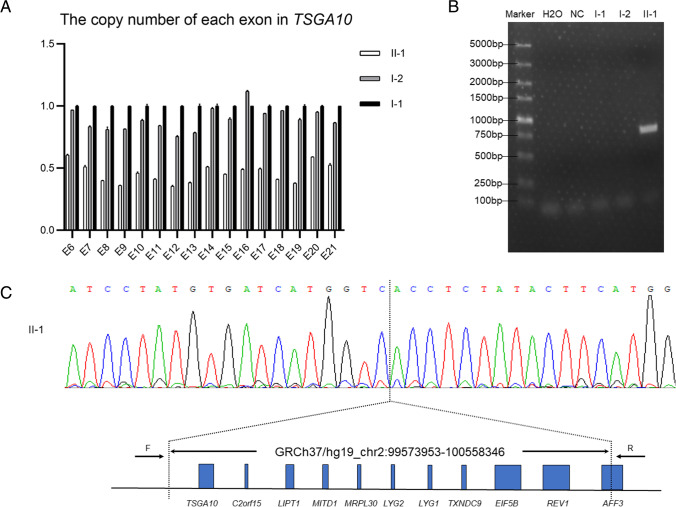
To confirm whether the deletion exists, we performed qPCR analysis on DNA from the proband and his parents to detect possible heterozygous deletions in TSGA10. Heterozygous deletion encompassing all of the exons of TSGA10 was identified in the proband compared with his parents, suggesting the deletion of the whole TSGA10 (Fig. 2a). To confirm qPCR results and identify the deletion range, WGS was performed on the proband and eight copy number variants (CNV) were identified (Table 5). Among them, a large heterozygous deletion, involved eleven genes, contained intact TSGA10, was detected (Fig. 2c). Moreover, the deletion range was ascertained to uncover 984,394 bp (chr2:99,573,953–100,558,346) and the breakpoint was identified by PCR using the specific primers designed on the basis of the WGS result (Fig. 2b, c).
Fig. 2.
Identification of the large deletion. a Detected heterozygous deletions in exons of TSGA10 in the proband and his parents with qPCR. b The DNA fragment containing the breakpoint was amplified with PCR using the special primers, shown in the Table 1, designed based on the WGS. c The sequencing result of the DNA fragment above shows an 984,394 bp deletion spanning the eleven genes, contained the entire TSGA10 gene
Table 5.
CNVs detected by WGS
| Chr | Start | End | Length | Type | Ratio | Cutoff | Gene |
|---|---|---|---|---|---|---|---|
| 2 | 99572631 | 100558590 | 985959 | Del*1 | 0.45 | > 100 kb | REV1 (NM_001037872, NM_016316); AFF3 (NM_001025108, NM_002285); TSGA10 (NM_182911, NM_025244); C2orf15 (NM_144706); LIPT1 (NM_015929, NM_145197, NM_145198, NM_145199, NM_001204830); MITD1 (NM_138798); MRPL30 (NM_145212); LYG2 (NM_175735); LYG1 (NM_174898); TXNDC9 (NM_005783); EIF5B (NM_015904) |
| 4 | 70121496 | 70235854 | 114358 | Del*1 | 0.63 | > 100 kb | UGT2B28(NM_001207004, NM_053039) |
| 8 | 39,229,215 | 39387492 | 158277 | Del*1 | 0.43 | > 100 kb | NULL |
| 6 | 11392006 | 11454521 | 62515 | Dup*3 | 1.37 | / | NULL |
| 7 | 102110025 | 102207359 | 97334 | Dup*3 | 1.86 | / | LRWD1(NM_152892); POLR2J(NM_006234); POLR2J3(NM_001097615); SPDYE2(NM_001031618); SPDYE2L(NM_001166339) |
| 9 | 210770 | 269060 | 58290 | Dup*3 | 1.17 | / | C9orf66 (NM_152569); DOCK8(NM_203447); |
| 17 | 34485533 | 34579126 | 93593 | Del*1 | 0.84 | / | TBC1D3B(NM_001001417); CCL3L3(NM_001001437); CCL3L1(NM_021006); CCL4L1(NM_001001435); CCL4L2(NM_207007) |
| 19 | 53931213 | 54006794 | 75581 | Dup*3 | 1.42 | / | ZNF761(NM_001008401); ZNF813(NM_001004301) |
Del*1 represents one copy and Dup*3 represents three copies
The influence of the splicing mutation in TSGA10 in vitro
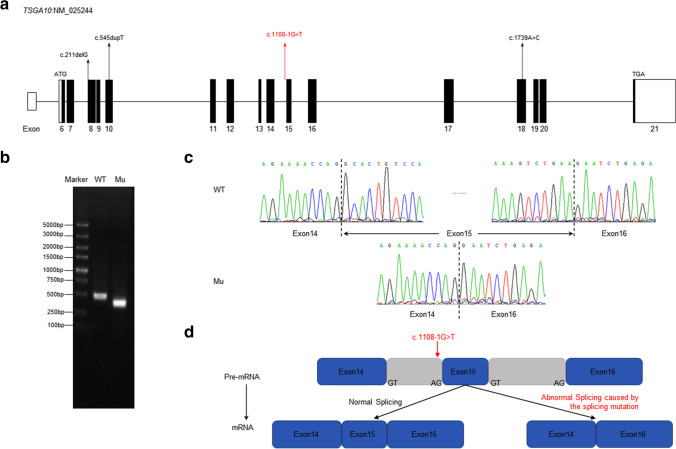
The splicing mutation in TSGA10 is located in the canonical receptor site of intron 14 (Fig. 3a, d). In order to study the effect of the splicing mutation NM_025244 (TSGA10) c.1108-1G > T, we performed a minigene assay. As agarose gel electrophoresis showed, we got two different sizes of RT-PCR productions from the wild type and the splicing mutation minigenes, respectively (Fig. 3b). The following Sanger sequencing indicated the abnormal splicing isoform of the mutation compared with the wild type (Fig. 3c). We found that the splicing mutation led to the skipping of the exon15 of TSGA10, which resulted in a frameshift and a predicted truncated protein (p. A370Efs*293) (Fig. 3d). The losing residue affected by the splicing mutation is conserved among different species (Fig. 4a).
Fig. 3.
The location and conservation of the mutant residue in TSGA10. a The location of mutations in the TSGA10 exons. Mutations described in the previous study are marked in black and the novel splicing mutation identified in our study is marked in red. b Agarose gel electrophoresis illustrating the effect of the TSGA10 c.1108-1G > T mutation. Compared with the wild type lane, the mutation lane showed a smaller band. c Identified the abnormal splicing isoform of the mutation. By RT-PCR and Sanger sequencing, the results showed that the splice-site mutation in TSGA10 lead to the skipping of exon15. d A possible scheme illustrating of the effect of the TSGA10 c.1108-1G > T mutation. In this case, the mutation skips the canonical receptor site leading to the entire deletion of exon15 in cDNA and resulting in abnormal transcription and translation
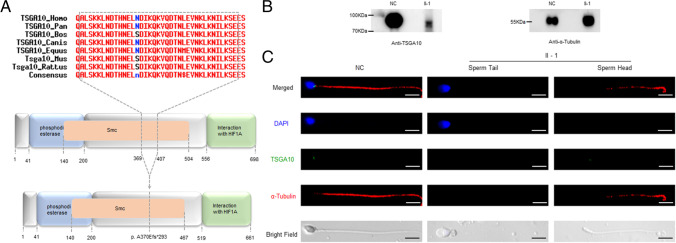
Fig. 4.
Expression and localization of TSGA10 in the proband’s sperm compared with the normal. a Domains in the TSGA10, the predicted mutant TSGA10. The full-length protein is 698 amino acids (aa). Domain with putative phosphodiesterase activity, aa 41 to 200 (blue box); domain that can interact with HIF1A, aa 556 to 689 (green box); the maintenance of the chromosome (Smc) domain, aa 140 to 504 (orange box). The p. A370Efs*293 mutation is located at the Smc domain of the TSGA10 protein and the truncated protein harbors a 293aa frameshift sequence. The 37aa, which was truncated, presented high-conserved among different species. b Western blot was performed on the proband’s sperm, using the normal sperm as a control. c The immunostaining of the proband’s sperm compared with the normal. The nucleus and tail of the sperm are stained with DAPI (blue) and α-Tubulin (red), respectively. The localization of TSGA10 in sperm from the proband and normal control were demonstrated by immunofluorescence staining with TSGA10 antibody (green). Scale bars: 5 μm
Expression and localization of TSGA10
In order to verify the effect of the mutation and deletion, Western blot and immunofluorescence assays were performed to detect the expression level and localization of mutant TSGA10 protein in the proband’s sperm compared with the normal control. Western blot analysis demonstrated that TSGA10 was expressed in the control as its theoretical molecular weight, but was lower and decayed significantly from the proband’s sperm (Fig. 4b), which was consistent with the result of minigene assay. By immunostaining the spermatozoa from a normal control, TSGA10 was found to localize at the head–tail junction of spermatozoa (Fig. 4c). However, no staining was observed in the proband’s sperm (Fig. 4c).
Discussion
TSGA10 is a testis-specific protein, which is located to the principal piece of sperm, to the centrosome and basal body [20, 21]. Previous studies revealed that TSGA10 played an important role in the assembly of centriole in the head–tail connection, the arrangement of mitochondrial sheath, and the development of embryo [17, 18, 22, 23]. So far, TSGA10 mutations, as the definitive genetic etiology, have been associated with about 3.1% cases which had reported [4–19].
Here, we identified a novel splicing mutation (c.1108-1G > T) in TSGA10 in an ASS patient from a non-consanguineous family (Fig. 1a). At the same time, we found a large deletion in the proband involved eleven genes, contained intact TSGA10, by qPCR and WGS due to the violation of co-segregation principle (Fig. 2). The other ten genes, except for TSGA10, were never reported to be associated with male infertility so far. Besides, we made a sketchy exploration of this large deletion. On the basis of the sequences of the deletion, we found that it existed line interspersed nuclear elements on the bilateral sides of the deletion, which may cause non-sister chromatids mismatch and unequal exchange to result in the deletion. However, the detailed pathogenic mechanism needs to be explained further.
By the minigene assay, we found that the splicing mutation led to the skipping of the exon15 of TSGA10 (Fig. 3c, d), and a truncated protein (p. A370Efs*293), which impaired the function of structural maintenance of the chromosome (Smc) domain of TSGA10 (Fig. 4a). Consistent with the minigene assay, Western blot showed a truncated TSGA10 protein (about 78KDa) in the proband, which was lower than its theoretical molecular weight and decayed significantly (Fig. 4b). Immunostaining was not observed in the proband’s sperm, compared with the normal, which staining was located in the head–tail junction of sperm (Fig. 4c). According to the result of Western bolt, the signal might not be detected, causing by its appreciable decay. Therefore, we considered that the ASS of the proband caused by the novel splicing mutation and deletion in TSGA10.
We herein summarized known data on the gene TSGA10 contribution of genes to acephalic spermatozoa (Fig. 3a) and broadened the spectrum of genetic causes. Furthermore, the identification of heterozygous deletion suggested that pathogenic genes we known might contribute more than we found in patients with ASS because genomic deletions cannot be screened by WES. All our studies were devoted to potentially facilitating diagnoses and promoting therapeutic development.
Acknowledgements
We would like to sincerely thank the patient and his family for participation.
Author contribution
F.Z., Y.C., and F.W. conceived and designed the experiments. J.Z., Z.D., and X.Z. collected the samples. M.X., Y.W., W.X., and N.Z. performed the experiments. F.Z., Y.C., F.W., M.X., Y.W., and X.S. analyzed the data. Y.W. and M.X. wrote the article. All authors approved the final article.
Funding
National Natural Science Foundation of China (82071701 to F. Z.; 81792641 to F. W.), Natural Science Foundation of Anhui Province (1908085J28 to F. Z.), Key R&D program of Anhui Province (201904a07020050 to F. Z.), the Non-profit Central Research Institute Fund of Chinese Academy of Medical Sciences (2019PT310002), and Scientific Research Foundation of the Institute for Translational Medicine of Anhui Province (SRFITMAP 2017zhyx29 and ZHYX2020A001) supported this study.
Data availability
The data underlying this article are available in the article.
Code availability
Not applicable
Declarations
Ethics approval
This study was approved by the Ethics Committee of Anhui Medical University. Written informed consent was obtained from each participant.
Consent to participate
Obtained.
Consent for publication
Obtained.
Conflict of interest
The authors declare no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Mingfei Xiang, Yu Wang and Weilong Xu contributed equally to this work.
Contributor Information
Fengsong Wang, Email: fengsongw@ahmu.edu.cn.
Yunxia Cao, Email: caoyunxia6@126.com.
Fuxi Zhu, Email: fxzhu@ahmu.edu.cn.
References
- 1.World Health Organization. WHO Laboratory Manual for the Examination and Processing of Human Semen, 5th edn. Cambridge: Cambridge University Press, 2010;37–44:65–67.
- 2.Mascarenhas MN, Flaxman SR, Boerma T, Vanderpoel S, Stevens GA. National, regional, and global trends in infertility prevalence since 1990: a systematic analysis of 277 health surveys. PLoS Med. 2012;9(12):e1001356. doi: 10.1371/journal.pmed.1001356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chemes HE, Carizza C, Scarinci F, Brugo S, Neuspiller N, Schwarsztein L. Lack of a head in human spermatozoa from sterile patients: a syndrome associated with impaired fertilization. Fertil Steril. 1987;47(2):310–316. doi: 10.1016/s0015-0282(16)50011-9. [DOI] [PubMed] [Google Scholar]
- 4.Zhu F, Wang F, Yang X, Zhang J, Wu H, Zhang Z, et al. Biallelic SUN5 mutations cause autosomal-recessive acephalic spermatozoa syndrome. Am J Hum Genet. 2016;99(4):942–949. doi: 10.1016/j.ajhg.2016.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Elkhatib RA, Paci M, Longepied G, Saias-Magnan J, Courbiere B, Guichaoua MR, et al. Homozygous deletion of SUN5 in three men with decapitated spermatozoa. Hum Mol Genet. 2017;26(16):3167–3171. doi: 10.1093/hmg/ddx200. [DOI] [PubMed] [Google Scholar]
- 6.Sha YW, Xu X, Ji ZY, Lin SB, Wang X, Qiu PP, et al. Genetic contribution of SUN5 mutations to acephalic spermatozoa in Fujian China. Gene. 2018;647:221–225. doi: 10.1016/j.gene.2018.01.035. [DOI] [PubMed] [Google Scholar]
- 7.Fang J, Zhang J, Zhu F, Yang X, Cui Y, Liu J. Patients with acephalic spermatozoa syndrome linked to SUN5 mutations have a favorable pregnancy outcome from ICSI. Hum Reprod. 2018;33(3):372–377. doi: 10.1093/humrep/dex382. [DOI] [PubMed] [Google Scholar]
- 8.Shang Y, Yan J, Tang W, Liu C, Xiao S, Guo Y, et al. Mechanistic insights into acephalic spermatozoa syndrome-associated mutations in the human SUN5 gene. J Biol Chem. 2018;293(7):2395–2407. doi: 10.1074/jbc.RA117.000861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu G, Wang N, Zhang H, Yin S, Dai H, Lin G, et al. Novel mutations in PMFBP1, TSGA10 and SUN5: Expanding the spectrum of mutations that may cause acephalic spermatozoa. Clin Genet. 2020;97(6):938–939. doi: 10.1111/cge.13747. [DOI] [PubMed] [Google Scholar]
- 10.Zhang D, Huang WJ, Chen GY, Dong LH, Tang Y, Zhang H, et al. Pathogenesis of acephalic spermatozoa syndrome caused by SUN5 variant. Mol Hum Reprod. 2021;27(5):gaab028. doi: 10.1093/molehr/gaab028. [DOI] [PubMed] [Google Scholar]
- 11.Zhu F, Liu C, Wang F, Yang X, Zhang J, Wu H, et al. Mutations in PMFBP1 cause acephalic spermatozoa syndrome. Am J Hum Genet. 2018;103(2):188–199. doi: 10.1016/j.ajhg.2018.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sha YW, Wang X, Xu X, Ding L, Liu WS, Li P, et al. Biallelic mutations in PMFBP1 cause acephalic spermatozoa. Clin Genet. 2019;95(2):277–286. doi: 10.1111/cge.13461. [DOI] [PubMed] [Google Scholar]
- 13.Lu M, Kong S, Xiang M, Wang Y, Zhang J, Duan Z, et al. A novel homozygous missense mutation of PMFBP1 causes acephalic spermatozoa syndrome. J Assist Reprod Genet. 2021;38(4):949–955. doi: 10.1007/s10815-021-02075-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li L, Sha Y, Wang X, Li P, Wang J, Kee K, et al. Whole-exome sequencing identified a homozygous BRDT mutation in a patient with acephalic spermatozoa. Oncotarget. 2017;8(12):19914–19922. doi: 10.18632/oncotarget.15251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen H, Zhu Y, Zhu Z, Zhi E, Lu K, Wang X, et al. Detection of heterozygous mutation in hook microtubule-tethering protein 1 in three patients with decapitated and decaudated spermatozoa syndrome. J Med Genet. 2018;55(3):150–157. doi: 10.1136/jmedgenet-2016-104404. [DOI] [PubMed] [Google Scholar]
- 16.Li L, Sha YW, Xu X, Mei LB, Qiu PP, Ji ZY, et al. DNAH6 is a novel candidate gene associated with sperm head anomaly. Andrologia. 2018 doi: 10.1111/and.12953. [DOI] [PubMed] [Google Scholar]
- 17.Sha YW, Sha YK, Ji ZY, Mei LB, Ding L, Zhang Q, et al. TSGA10 is a novel candidate gene associated with acephalic spermatozoa. Clin Genet. 2018;93(4):776–783. doi: 10.1111/cge.13140. [DOI] [PubMed] [Google Scholar]
- 18.Ye Y, Wei X, Sha Y, Li N, Yan X, Cheng L, et al. Loss-of-function mutation in TSGA10 causes acephalic spermatozoa phenotype in human. Mol Genet Genomic Med. 2020;8(7):e1284. doi: 10.1002/mgg3.1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sha Y, Wang X, Yuan J, Zhu X, Su Z, Zhang X, et al. Loss-of-function mutations in centrosomal protein 112 is associated with human acephalic spermatozoa phenotype. Clin Genet. 2020;97(2):321–328. doi: 10.1111/cge.13662. [DOI] [PubMed] [Google Scholar]
- 20.Modarressi MH, Cameron J, Taylor KE, Wolfe J. Identification and characterisation of a novel gene, TSGA10, expressed in testis. Gene. 2001;262(1–2):249–255. doi: 10.1016/s0378-1119(00)00519-9. [DOI] [PubMed] [Google Scholar]
- 21.Modarressi MH, Behnam B, Cheng M, Taylor KE, Wolfe J, van der Hoorn FA. Tsga10 encodes a 65-kilodalton protein that is processed to the 27-kilodalton fibrous sheath protein. Biol Reprod. 2004;70(3):608–615. doi: 10.1095/biolreprod.103.021170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Luo G, Hou M, Wang B, Liu Z, Liu W, Han T, et al. Tsga10 is essential for arrangement of mitochondrial sheath and male fertility in mice. Andrology. 2021;9(1):368–375. doi: 10.1111/andr.12889. [DOI] [PubMed] [Google Scholar]
- 23.Behnam B, Modarressi MH, Conti V, Taylor KE, Puliti A, Wolfe J. Expression of Tsga10 sperm tail protein in embryogenesis and neural development: from cilium to cell division. Biochem Biophys Res Commun. 2006;344(4):1102–1110. doi: 10.1016/j.bbrc.2006.03.240. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data underlying this article are available in the article.
Not applicable