Summary
For those people with cystic fibrosis carrying rare CFTR mutations not responding to currently available therapies, there is an unmet need for relevant tissue models for therapy development. Here, we describe a new testing platform that employs patient-specific induced pluripotent stem cells (iPSCs) differentiated to lung progenitor cells that can be studied using a dynamic, high-throughput fluorescence-based assay of CFTR channel activity. Our proof-of-concept studies support the potential use of this platform, together with a Canadian bioresource that contains iPSC lines and matched nasal cultures from people with rare mutations, to advance patient-oriented therapy development. Interventions identified in the high-throughput, stem cell-based model and validated in primary nasal cultures from the same person have the potential to be advanced as therapies.
Keywords: high-throughput phenotypic platform, therapy testing, CF-causing nonsense mutations, pluripotent stem cells, apical chloride conductance assay, complementary assays of primary and iPSC derived tissues, precision medicine
Highlights
-
•
A Canadian resource (CFIT) has CF donor-matched iPSCs and nasal epithelial cells
-
•
Lung progenitor cells (LPCs) differentiated from iPSCs express CFTR
-
•
LPCs from people with rare CFTR mutations enable high-throughput therapy testing
-
•
Matching nasal cultures can validate patient-specific drug responses in LPCs
Bear and colleagues show that lung progenitor cells (LPCs) differentiated from cystic fibrosis (CF) iPSCs recapitulate the primary defects conferred by different types of CF mutations. LPCs enable high-throughput testing of interventions, and pilot studies show that responses in LPCs can be validated in patient-matched primary nasal epithelial cultures, confirming the potential utility of LPCs in precision CF therapy development.
Introduction
Mutations in the CFTR gene can result in the disease called cystic fibrosis (CF). The major mutation, called F508del, leads to CFTR protein misassembly, mistrafficking, and altered function as a phosphorylation-regulated chloride channel (Cheng et al., 1990; Lukacs and Verkman, 2012; Qu and Thomas, 1996). Combinations of small-molecule modulators called correctors, which rescue CFTR protein misassembly, together with potentiators that augment CFTR channel activation have been shown to be effective in improving lung function in individuals harboring the major mutation, F508del (Middleton et al., 2019; Taylor-Cousar et al., 2017; Wainwright et al., 2015). However, not all individuals with this mutation show the same level of clinical improvement, supporting the ongoing need for novel therapy development (Heijerman et al., 2019; Middleton et al., 2019). In addition, there are multiple, rarer CF-causing mutations, such as nonsense and splicing mutations for which there are no approved therapies. Given the variability in transcriptional and translational efficiencies across patient-specific tissues, the field would be well served by tissue models that reflect this variability and are scalable to high-throughput formats to facilitate such therapy development.
Traditionally, in vitro studies of CF therapies have employed bronchial epithelial tissue cultures from explanted CF lungs obtained at the time of transplant. Such cultures were, and continue to be, very instructive with respect to understanding the mechanism of modulator activity and evaluating their efficacy in relevant tissues. The functional consequences of modulator treatment are typically measured as changes in ion conductance in the Ussing chamber apparatus (Guerra et al., 2020; Van Goor et al., 2011; Veit et al., 2018). Although informative, these methods are low throughput and not readily conducive to comparative studies of multiple interventions simultaneously. In addition, as the cell donor has undergone a lung transplant, the potential for directing individualized therapy is limited. Further, for rare mutations, reduced availability of explanted tissue harboring the appropriate genotype impedes drug development.
In lieu of bronchial epithelial cells, the development of other tissue models of CF, including nasal epithelium and rectal organoids (Berkers et al., 2019; Brewington et al., 2018; de Winter-de Groot et al., 2020; Dekkers et al., 2013; Graeber et al., 2020; Pranke et al., 2017; Ramalho et al., 2021), greatly enhanced studies of the in vitro efficacy of CFTR modulators in patient-specific tissues. Influential papers by Dekkers et al. (Berkers et al., 2019; Fawcett et al., 2021; Ramalho et al., 2021; Silva et al., 2020, 2021) highlighted the potential for the rectal organoid model to facilitate drug development and precision medicine for CF. Stem cells in the rectal mucosa provide a renewable source of this tissue, and biobanks of intestinal organoids from CF populations have been created around the world to enhance CF therapy development. In organoids, CFTR channel function is measured indirectly as forskolin-induced organoid swelling. The correlation between patient-specific organoid responses and the clinical biomarkers forced expiratory volume in 1 s (FEV1) and sweat chloride concentrations suggests that organoid swelling can potentially be used to predict individual therapeutic outcomes (Berkers et al., 2019; de Winter-de Groot et al., 2020). However, the variability in organoid shape and size can lead to variable swelling responses that may limit its application to high-throughput screening and therapy discovery (Vonk et al., 2020). Therefore, there is a need for additional tissue models that directly report CFTR channel function and are potentially amenable to higher-throughput phenotypic assays.
The use of primary, patient-derived nasal cultures in preclinical studies of CFTR modulators has been highly informative (Keegan and Brewington, 2021; Laselva et al., 2021a, 2021b; Oren et al., 2021; Park et al., 2020; Phuan et al., 2021; Veit et al., 2020). Interestingly, Amaral's group demonstrated a correlation between CFTR rescue by CFTR modulators in primary nasal epithelial cells and those in rectal organoids from the same individual (Silva et al., 2021). There are several published accounts of the use of nasal cultures in validating the relative efficacy of the new modulator combination in TRIKAFTA for rescuing F508del-CFTR and certain rare mutations (Laselva et al., 2021a; Veit et al., 2020). Interestingly, our group and others observed that patients with the same genotype can exhibit different in vitro drug responses (de Wilde et al., 2019; Laselva et al., 2021b; Molinski et al., 2017; Phuan et al., 2018; Wu et al., 2019). The molecular basis for interpatient variations in in vitro response size remains unknown, but is likely a harbinger of the extent of variability in clinical response size (Berkers et al., 2019; Graeber et al., 2020; Merkert et al., 2019; Pranke et al., 2017). In fact, patient-specific in vitro responses to interventions measured in differentiated nasal epithelial cultures in the Ussing chamber have been correlated with clinical effect size (Pranke et al., 2017, 2019).
Although primary nasal cultures have the potential to reflect patient-specific responses to therapeutic interventions, there are limitations to their use for repeated drug testing. While multiple methods have been developed to expand brushed nasal cells prior to differentiation at the air-liquid interface (ALI) in order to enhance testing capacity using this model (Awatade et al., 2021), the functional expression of CFTR appears to worsen with progressive population doublings (Brewington et al., 2018). This property is limiting to combinatorial studies of investigational compounds in these cultures. Further, Ussing chamber studies of nasal epithelial cultures are low throughput and time consuming; hence, there is a need to develop a higher-throughput testing platform of patient-specific tissue that enables therapy development, particularly for those CF-causing mutations like W1282X for which no therapies exist.
Numerous protocols have been developed for the differentiation of CF-patient-derived induced pluripotent stem cells (iPSCs) to CF-affected lung (Firth et al., 2014; Hawkins et al., 2017; Huang et al., 2014; McCauley et al., 2017; Wong et al., 2015), thereby providing the potential for a renewable source of patient-derived tissue. Although these methods likely achieve varying proportions of mature lung cells (Kerschner et al., 2020), functional rescue of F508del-CFTR chloride channel activity by CFTR modulators was consistently shown for lung tissue differentiated at the air-liquid interface using the Ussing chamber assay (Crane et al., 2015; Hawkins et al., 2021; Wong et al., 2012, 2015). These findings raise the possibility that the functional expression of CFTR is not dependent on differentiation to the fully mature lung epithelium.
Merkert and colleagues (Merkert et al., 2019) engineered iPSCs to stably express the halide-sensitive yellow fluorescent protein (YFP) for the purpose of developing a fluorescence-based assay of CFTR function. These engineered iPSCs were differentiated to intestinal epithelium in a 2D format, enabling higher-throughput fluorescence-based studies of the functional expression of F508del-CFTR before and after CFTR modulators. However, this platform required genetic engineering to introduce the halide-sensitive YFP, and the assay was relatively insensitive, requiring augmentation of drug responses by incubation at low, nonphysiological temperatures (Merkert et al., 2019). In the future, increased sensitivity in phenotypic screens will be particularly important for discovery of interventions that target rare CF mutations such as the nonsense mutation W1282X.
The goal of the current project was to develop a sensitive, fluorescence-based, medium-to-high-throughput assay of CFTR channel function for testing interventions targeting the rare CF-causing mutations. We asked if lung progenitor epithelium grown on 96 well plates suitable for fluorescence-based assays of channel activity expresses functional CFTR. Further, we were prompted to determine if this platform was sensitive enough to measure CFTR function in cultures generated from patient-derived iPSCs harboring the rare mutation W1282X. Finally, using the Canadian Program for Individualized CF Therapy (CFIT) bioresource of matched, patient-specific iPSC lines and nasal epithelial cultures (https://lab.research.sickkids.ca/cfit/cystic-fibrosis-patients-families-researchers/cell-resources-available/), we tested the correlation between drug responses measured in iPSC-derived cultures and responses determined in the same patient’s primary nasal epithelial cultures. A correlation in these pilot experiments could prompt development of a new, two-stage preclinical pipeline for developing personalized medicine for people with CF caused by rare mutations.
Results
Differentiation of iPSCs to lung progenitor cells in high-throughput format
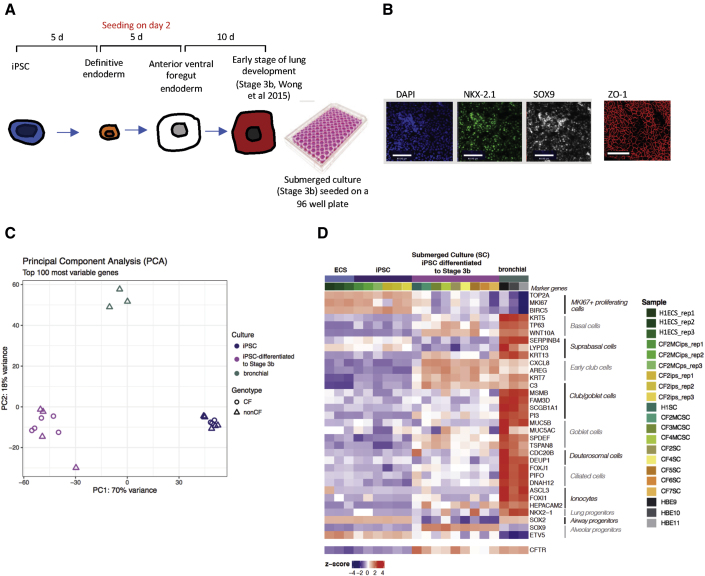
We employed the protocol developed by Wong et al. (Wong et al., 2015; Figure 1A) to differentiate non-CF and CF iPSCs to tissue in the early stages of lung development in a 96 well plate format. These cultures remained submerged under differentiation medium; hence, we refer to them as “submerged cultures.” As shown in immunofluorescence images (Figure 1B), embryonic stem cells (CA1) differentiated to this stage express NKX2.1 and SOX9 markers of lung tip progenitors (Hawkins et al., 2017; Nikolic et al., 2017). ZO-1 expression supports the claim that the cultures are epithelial and polarized.
Figure 1.
Differentiation of patient-derived iPSCs to lung progenitor cells
(A) Schematic of differentiation protocol and timeline. Human iPSCs were directed to definitive endoderm and passaged onto 96 well plates during the anterior foregut endoderm differentiation. The submerged cultures were differentiated for 10 more days into lung progenitor cells (stage 3b; Wong et al., 2015).
(B) Immunofluorescence images of submerged cultures. Scale bar, 80 μm. Most cells stained positive for TTF1 (NKX2-1) and SOX9. Negative controls are shown in Figure S1.
(C) Principal-component analysis (PCA) comparing iPSC lines, submerged lung progenitor cultures differentiated from iPSC lines, and primary bronchial cultures. Both CF and non-CF (including mutation-corrected) iPSC lines were studied.
(D) Heatmap of gene expression clustered according to cell type using marker genes (Deprez et al., 2020). The columns correspond to different donors and whether lines are CF or non-CF (including mutation corrected [MC]). Columns are also clustered as an embryonic stem cell line (H1), iPSC lines, lung progenitors differentiated from iPSC lines as submerged cultures (SC), and primary human bronchial epithelial (HBE) cultures. Relative CFTR expression across cultures is shown in the bottom row of the heatmap.
We conducted transcriptomic analyses to further characterize the properties of submerged cultures generated from iPSCs. Bulk RNA sequencing (RNA-seq) was performed for: (1) an embryonic stem cell line, H1, with three replicates, and two undifferentiated iPSC lines, each with three replicate cultures; (2) nine iPSC lines differentiated under “submerged” conditions to create progenitor lung cells (four non-CF and five CF); and (3) non-CF differentiated bronchial airway cultures (n = 3) as positive controls (NIH Tissue Core, University of Iowa) (Figures 1C and 1D). Principal-component analysis (PCA) showed that the submerged cultures were intermediate (as determined by PC1) between the iPSCs and differentiated mature bronchial epithelium with respect to gene expression of the 100 genes displaying the greatest variability across samples (Figure 1C). Further, at least with these samples, mutant CFTR genotypes (with or without a CF-causing mutation) did not associate with different cell sub-populations.
The heatmap visualization of RNA-seq analysis of the same cultures is shown in Figure 1D. The map shows the relative change in expression of genes, clustered according to developmental stage or cell type with differentiation from iPSC cultures to day 20 as shown in Figure 1A. First, we noticed that there was considerable variation among the patient-specific lines with respect to gene expression associated with early lung development. Although there was a variable increase in NKX2.1, the increase in SOX9 expression was more consistent with differentiation to stage 3b under submerged conditions. With reference to the gene expression changes reported using the protocol for direct lung differentiation published by Hawkins et al. (Hawkins et al., 2017), the submerged cultures generated in the current study resemble D15 cultures as described by Hawkins et al. (2017) or lung tip progenitors as described by Nikolic and colleagues (Nikolic et al., 2017). Although the expression of genes typically associated with early club cells is uniformly upregulated in submerged cultures relative to iPSCs, expression of genes typically expressed in mature cell types (i.e., SCGB1A1 and MUC5AC) is low and variable. Consistent with the RT-PCR analyses, CFTR expression, while variable across patient-specific cultures, is generally high and comparable to levels measured in mature non-CF bronchial cultures differentiated at the air-liquid interface. Altogether, these transcriptomic studies suggest that the submerged cultures differentiated on 96 well plates model multipotent, progenitor lung tissue. And, we cannot rule out the contribution of nonlung epithelial cells. However, the relatively high levels of CFTR expression support their potential application in middle-to-high-throughput studies of mutation-targeted therapies.
Submerged progenitor lung cultures express functional CFTR
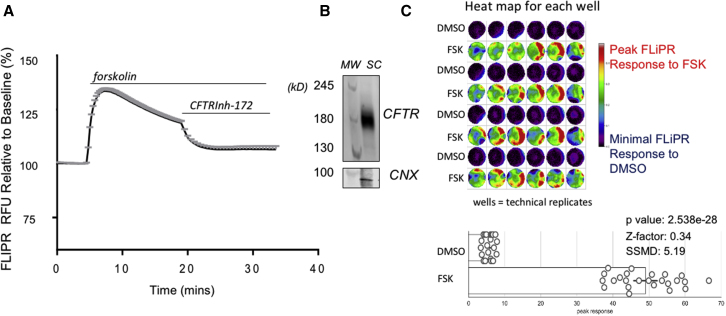
Cyclic-AMP-activated wild-type (WT) CFTR channel function was detected for lung progenitor cultures differentiated in wells of a 96 well plate, using a fluorescence-based assay of membrane potential changes (FLIPR) (Ahmadi et al., 2017). The trace in Figure 2A shows representative forskolin-mediated membrane depolarization and increase in FLIPR in the presence of an outward chloride gradient (mean change ± SEM for four wells). This response was inhibited by the CFTR inhibitor, CFTRInh-172, as expected for CFTR-mediated channel activity. Expression of the mature CFTR protein as a 180 kDa polypeptide also known as Band C. was confirmed for cultures grown in this format (Figure 2B).
Figure 2.
Lung progenitor cultures express WT CFTR and channel activity in fluorescence-based assay
(A) Representative FLIPR trace (mean ± SEM) responses of four wells plated with submerged cultures stimulated by 10 μM forskolin and inhibited by 10 μM CFTRInh-172. This cell line was derived from donor CF2MC for which the F508del mutation was corrected to WT. The method for mutation editing is described in Eckford et al. (2019), and characterization of the CF2MC line is given in Document S1. The naming of lines is consistent with Figure 1.
(B) Western blot shows molecular weight (MW) markers and mature CFTR expression in submerged culture of the same line, (180 kDa). Calnexin (CNX) was used as loading control.
(C) Top, heatmap shows representative data for a single plate (48 wells), used for establishing assay statistics (p value, Z-prime factor, and SSMD). Multiple wells were seeded with iPSCs differentiated to submerged lung progenitor cells. Alternating rows (each containing six wells) show well scans of CFTR channel activation after agonist (forskolin, FSK) or vehicle control (DMSO) addition. The response size is color coded as shown in the side bar, with red representing the highest response and purple-black, the lowest response. Bottom, bars show data from all of the above wells treated with control (DMSO) or FSK. Assay statistics are superimposed.
The heatmap in Figure 2C shows well scans of FLIPR dye fluorescence change with depolarization caused by forskolin or the DMSO vehicle added to alternating rows of six wells (i.e., technical replicates). Red shows the highest responses and purple-black, the lowest responses. Interestingly, for certain wells, there are focal hotspots of activity, reflecting heterogeneity of the differentiated culture. The open circles in the bar graph of Figure 2C show the mean peak CFTR channel activity after DMSO or forskolin measured by FLIPR for a single well in a 96 well plate. This FLIPR assay of CFTR-mediated chloride conductance in lung progenitor cultures exhibits reproducibility in a 96 well plate format with a Z-prime factor of 0.34 and an SSMD (strictly standardized mean difference) score of 5.19. These metrics support the claim that this is a good to excellent assay platform for CFTR channel function (Gubler, 2009; Zhang, 2007).
Lung progenitor tissue differentiated from CF iPSCs models primary defects caused by F508del
Having shown that the FLIPR assay of lung progenitor cells differentiated from iPSCs is suitable for a high-throughput assay of WT CFTR, we applied it to study the mutation F508del-CFTR and its modulation by small molecules. In Figure 3, we show that the primary defects caused by the major mutation F508del are recapitulated in iPSC-derived lung progenitor cultures generated from two different patient donors who are homozygous for this mutation. The donor identification numbers are consistent those shown in Figure 1. For lung progenitor cultures from both CF donors, CF2 and CF4, the mean residual forskolin-activated CFTR channel activities (5.67 ± 2.16 [SD], n = 4, and 7.25 ± 0.91 [SD], n = 4) were significantly reduced relative to that exhibited by non-CF culture (37.50 ± 8.02 [SD], n = 4) as shown in Figures 3 and 2, respectively. As expected, the abundance of mature band C was also reduced for the major mutation in iPSC-derived lung progenitor epithelium in the absence of small-molecule modulators (Figure S2).
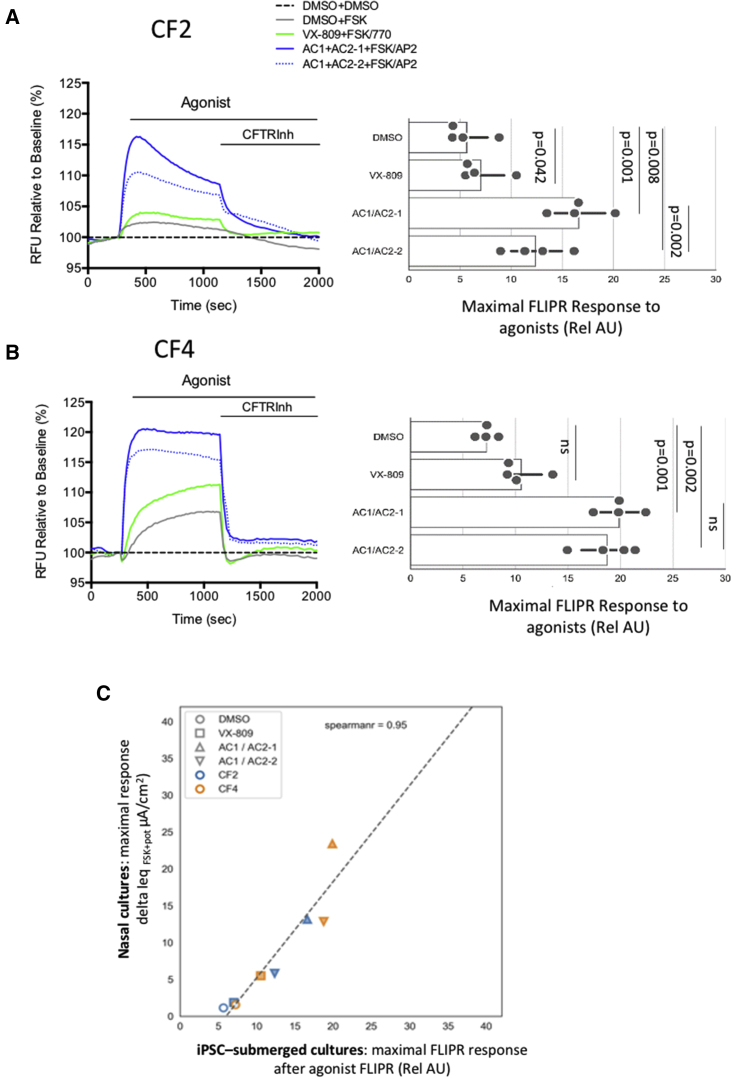
Figure 3.
Submerged lung progenitor cultures generated from iPSCs from two different donors homozygous for F508del exhibit robust responses to modulators
(A) Left: representative FLIPR traces of cultures derived from iPSCs with F508del mutation after chronic rescue (48 h) with DMSO (0.1%) or small molecules as defined in the key with concentrations indicated in Table S1. After a 5 min baseline, the cells were stimulated with DMSO or FSK ± 1 μM VX-770 (or 1.5 μM AP2). Right: bar graph shows peak response from each modulator combination. Each solid circle represents mean peak response of 4 wells in a 96 well plate for four plates of lung progenitor culture generated from a single differentiation of iPSC line CF2. The horizontal line in each bar, indicates the range amongst the mean measurements. The naming of cell lines is consistent with Figure 1.
(B) Left: FLIPR traces showing responses to small-molecule modulators on epithelium differentiated from an iPSC line derived from donor CF4. Right: bars show reproducibility of the FLIPR assay. Each solid dot represents the mean peak response for 4 technical replicates (wells) of a 96 well plate, and there were four plates generated from a single differentiation of iPSC line CF4. The horizontal line in each bar, indicates the range amongst the mean measurements. Significant differences between treatment groups were determined using an ordinary one-way ANOVA, multiple comparisons test (Prism version 9.2).
(C) Correlation between mean donor-specific activations measured using FLIPR (mean values ± interventions, extracted from bars above) and mean donor-specific changes measured in the Ussing chamber, delta Ieq (μA/cm2), after forskolin and treatment as reported in Laselva et al. (2018).
Treatment of lung progenitor cultures from donor CF2 with small-molecule modulator combinations led to reproducible responses in the FLIPR assay (Figure 3A). Relative to the vehicle (DMSO) control, treatment with the corrector, lumacaftor, i.e., VX-809, led to a partial rescue of the functional expression for CF2 cultures in the presence of the potentiator (VX-770). As expected, there were robust functional responses to the investigational modulator combinations AC1 + AC2-1 and AC1 + AC2-2 and the potentiator AP2 using the FLIPR assay on lung progenitor cultures (Figure 3A). Moreover, we found that the combination of AC1 + AC2-1 rescued the mature form of F508del-CFTR (band C) (Figure S2). The efficacy of these modulator combinations was published previously for studies on primary nasal epithelial cultures (Laselva et al., 2020a, 2020b, 2021b).
We compared modulator responses in lung progenitor cultures generated from donor CF4, another individual who is homozygous for F508del. While the ranking of modulator efficacies was similar for the two donors CF2 and CF4 (Figures 3A, 3B, and S2), the responses to the modulator combination AC1 + AC2-2 was higher in cultures from donor CF4 than from donor CF2 (p = 0.022). Together, these findings suggest that donor-specific differences in in vitro response to modulators can be measured using this platform.
In Figure 3C, we show that the responses to modulators observed for epithelial cultures generated from CF2 and CF4 correlate (r = 0.95) with the in vitro responses previously reported for well-differentiated primary nasal cultures from the same donors (Laselva et al., 2018). The bioelectric responses to modulators were previously measured for primary nasal cultures in Ussing chambers, considered the “gold standard” for measuring CFTR channel function. Interestingly, as in the FLIPR assay of lung progenitor cultures, the fully differentiated nasal epithelial cultures generated from donor CF4 consistently exhibited a higher functional response to the combinations AC1 + AC2-1 and AC1 + AC2-2 than the nasal cultures generated from donor CF2 (p = 0.022). Hence the patient-specific responses to modulators of the major mutation (F508del) observed in the high-throughput FLIPR-based assay recapitulated those observed in the gold-standard, but low-throughput, Ussing chamber assay.
Lung progenitor epithelium differentiated from CF iPSCs models primary defects caused by W1282X and reports differential responses to investigational rescue compounds
Having shown that the FLIPR assay of epithelial cultures differentiated from iPSC lines faithfully recapitulates therapeutic responses to modulator drugs targeting the major mutation F508del, we then tested the fidelity with which this novel platform can be used to measure patient-specific responses to investigational compounds targeting the rare nonsense mutation W1282X. The format for these phenotypic assays is shown in Figure 4A. Here, we show a heatmap for the FLIPR assay of submerged epithelial cultures. Wells (24) seeded with submerged cultures containing WT CFTR (CF2 mutation corrected [MC]) are displayed on the left. Wells (32) seeded with lung progenitor epithelium differentiated from iPSCs generated from a donor who is homozygous for the rare nonsense mutation W1282X (CF7) are shown on the right. The color scale on the right of the wells corresponds to the peak FLIPR fluorescence after activation and potentiation of CFTR channel activity. The wells seeded with non-CF (WT) epithelial tissue exhibit robust activation by forskolin, and the peak corresponds to red. On the other hand, the color of the wells containing cultures from a donor homozygous for the nonsense mutation varied (from dark blue to red, reflecting low to high responses) according to the treatment provided.
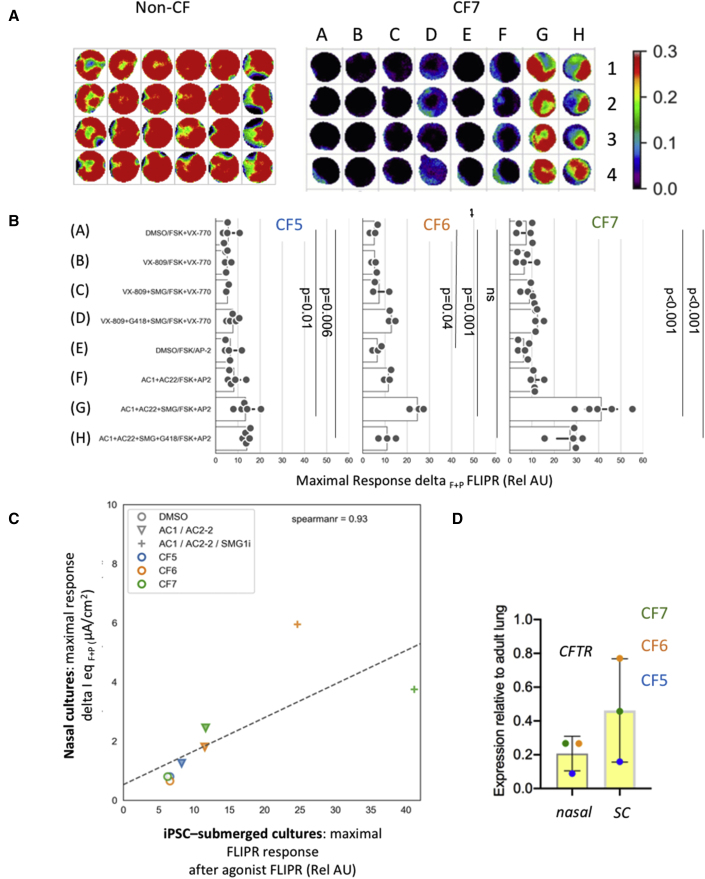
Figure 4.
Lung progenitor cultures generated from iPSCs from three different donors with the nonsense mutation W1282X exhibit differential phenotypic responses to modulators
(A) Heatmap of peak responses generated from non-CF culture stimulated with 10 μM forskolin (FSK) (left) and CF culture (W1282X) treated with a combination of small molecules (A–H, concentrations defined in Table S1; see also Video S1 for movie showing dynamic CFTR activation in the assay).
(B) Bar graph showing the peak response of W1282X CFTR treated with DMSO (0.1%) or small molecules (defined on y axis with concentrations indicated in Table S1). After a 5 min baseline, the cells were stimulated with DMSO or FSK ± 1 μM VX-770 (VX) or 1.5 μM AP2 (AC). Mean peak responses (of 4 wells in a 96 well plate) after agonist and potentiator are shown for each small-molecule combination as a solid circle. The horizontal line in each bar, indicates the range amongst the mean measurements. Three patients homozygous for W1282X were studied (CF5, 6, and 7). For CF5, 5 plates generated from four differentiations from iPSCs were studied. For CF6, 3 plates from one differentiation were studied, and for CF7, 5 plates from two differentiations were studied. CFTR modulators (VX-809 or AC1 + AC2-2), in combination with SMG1i, were effective in increasing the abundance of the truncated W1282X protein in lung progenitor cultures (Figure S4).
(C) Correlation plot between FLIPR peak response and Ussing chamber studies (data from Laselva et al., 2020a).
(D) Basal W1282X-CFTR transcript (left) expression in primary nasal epithelial cultures (nasal) and submerged iPSC-derived lung progenitor cultures (SC) from three donors. The vertical line in each bar shows the range in CFTR expression amongst the three donors.
A panel of small-molecule interventions was tested for efficacy in rescuing the functional expression of W1282X in three donors, all homozygous for this mutation. These interventions were chosen based on previous experiments in a bronchial cell line (Valley et al., 2019) and our previous bioelectric studies of primary nasal epithelial cultures (Laselva et al., 2020a). Based on these previous studies, we expect that only those cultures receiving SMG1i, a small-molecule inhibitor of nonsense-mediated decay (NMD), together with protein modulators of assembly and function (Valley et al., 2019) would enable functional rescue.
As shown in the bar graphs in Figure 4B, the responses observed for each patient-specific culture are reproducible. Interestingly, there were variable responses to the same panel of modulators across the three donors. For example, for donor CF5, the magnitude of the response to the modulator combinations is muted relative to the cultures derived from CF6 and CF7. As expected, low responses observed for CF5 were fully reversed by mutation correction of the same line, as shown in Figure S3.
With respect to the tested interventions, significant functional rescue was observed only in lung progenitor cultures treated with the NMD inhibitor, SMG1i, in combination with correctors and potentiator of the truncated W1282 CFTR protein. Moreover, as previously demonstrated in primary nasal epithelial cells (Laselva et al., 2020a), we showed band C rescue of W1282 CFTR only in the presence of SMG1i compound in combination with CFTR modulators (Figure S4). The migration of the mutant CFTR protein corresponds to that expected for the fully glycosylated truncated protein (Haggie et al., 2017). Interestingly, the premature termination codon (PTC) readthrough agent (G418) was not effective in promoting the full-length polypeptide, nor was it effective in augmenting functional rescue of W1282X mediated by SMG1i, as suggested in some but not all studies of primary cultures harboring this nonsense mutation (Laselva et al., 2020a). Hence, we show that combinatorial interventions targeting W1282 CFTR can be tested in patient-derived cultures using the FLIPR-based high-throughput assay. Further, we show that NMD is limiting modulator activity in these cultures and, finally, that different responses are observed for patient-specific cultures.
As for the studies of F508del CFTR, we observed a strong correlation (r = 0.93) between the patient-specific responses to interventions measured by FLIPR in submerged lung and the responses measured in primary nasal epithelial cultures in the Ussing chamber (data from Laselva et al., 2020a; Figure 4C). Unfortunately, due to constraints related to the limited expansion of primary nasal epithelial cultures, the correlation for all conditions, for all three donors, could not be assessed. However, in these pilot studies, it is clear that the best functional responses in both iPSC-derived lung progenitor and primary nasal epithelial cultures were achieved for donors CF6 and CF7 relative to CF5. Interestingly, the residual mutant CFTR transcript abundance (expressed relative to adult lung, shown in Figure 4D) also tended to be higher for both CF6 and CF7 relative to CF5, in both the iPSC-derived lung progenitor cultures and the matched nasal cultures.
We then asked if this fluorescence-based assay of drug responses in iPSC-derived lung progenitor cultures recapitulates patient-specific responses measured in primary nasal epithelial cultures from an individual with a different rare mutation, one that causes aberrant splicing of CFTR, c.3700A > G. Our previous studies showed that the combination of lumacaftor plus ivacaftor was inefficient at causing functional rescue of the mutant protein p.Ile1234_Arg1239del-CFTR generated by aberrant splicing in c.3700A > G, in patient-derived nasal cultures (Molinski et al., 2017). On the other hand, in a subsequent report, we showed that the combination of novel modulators AC1 + AC2-2 did rescue this mutant protein (Laselva et al., 2020b). Interestingly, in these pilot studies, we show that the fluorescence-based assay of lung progenitor cells differentiated from iPSCs from the same person recapitulates the differential drug responses reported using their primary nasal epithelial cultures (Figure S5).
Hence, these new, higher-throughput methods for studying lung progenitor cells differentiated from iPSCs have the potential to predict mutation- and patient-specific responses to therapeutic interventions measured in primary cultures.
Discussion
In these studies, we showed that fluorescence-based channel activity assays of lung progenitor cells generated from CF iPSCs have the potential to support therapy development for individuals with CF. Such cultures can be grown in a 96 well format to enable robust phenotypic screening, comparing approved and emerging therapeutic interventions. Further, our proof-of-concept studies show that functional responses seen in this novel assay associate with patient-specific responses measured in primary nasal epithelial cultures, a model previously shown to correlate with clinical outcomes (Pranke et al., 2017, 2019). Therefore, this new phenotypic platform has the potential to aid in precision medicine development for those individuals with CF caused by rare mutations and who currently lack effective therapeutic options.
Given the good to excellent statistical properties of the FLIPR assay and the availability of CF iPSC lines through the CFIT bioresource (Eckford et al., 2019), we suggest that patient-specific iPSCs may be used as a prescreen of multiple interventions prior to validation on mature, patient-specific nasal cultures. We suggest that this iPSC-based platform be employed as a tool to evaluate the range of patient-specific responses to existing and emerging therapies targeting unique CF-causing mutations. This platform could constitute a preclinical trial and ranking of the relative efficacy of multiple candidates in tissues from a number of CF-affected individuals harboring the same mutation. Then, the best therapy as defined using the stem cell-based platform can be validated in each individual using their own primary nasal epithelial cells prior to future recommendations regarding its advancement to clinical use (Figure 5). This therapy development pipeline is enabled by the Canadian CFIT bioresource containing matched iPSC lines and nasal epithelium cultures from people with defined CFTR genotypes. These lines and cultures are available to international researchers (Eckford et al., 2019).
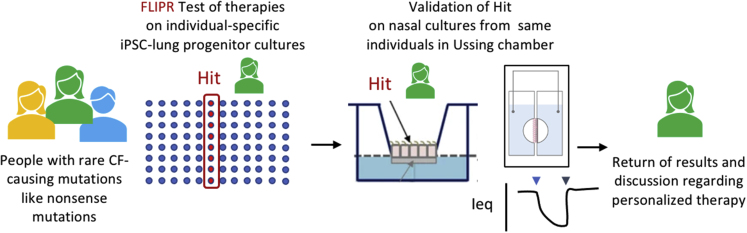
Figure 5.
Open access bioresource (CFIT, https://lab.research.sickkids.ca/cfit/cystic-fibrosis-patients-families-researchers/cell-resources-available/) contains matched iPSCs and nasal epithelial cultures from multiple people with the same CFTR genotype to enable two-step preclinical trials that account for individual variation and select the best intervention for each individual with a rare CF-causing mutation.
Although an extremely active area of research, there are no approved treatments targeting the primary defects caused by nonsense mutations in CFTR. Individuals who are homozygous for nonsense mutations like W1282X are rare worldwide (188 patients, http://cftr2.org) and, as a result, primary airway tissue models for therapy testing are limited. The identification of therapies targeting the nonsense mutation W1282X would benefit from the phenotypic platform described in this paper. Small molecules that facilitate readthrough of PTCs to enable production of full-length CFTR protein are undergoing development. However, only one such compound, called ELOX-2, is currently in clinical trial for subjects carrying the CF-causing nonsense mutation G542X (Crawford et al., 2021): https://www.cff.org/Trials/Pipeline. A complementary approach may be to suppress NMD using small-molecule inhibitors (Keenan et al., 2019; Laselva et al., 2020a; Valley et al., 2019), although such inhibitors may have nonspecific, deleterious effects. Further, such NMD inhibitors will need to be combined with protein modulators in order to correct the primary defects in W1282 CFTR function. Hence, combinatorial treatments aimed at promoting PTC readthrough, inhibition of NMD, and modulation of the stability and function of the mutant protein will need to be tested to identify the most effective interventions. In summary, we anticipate that the iPSC phenotypic platform that we describe here will facilitate such urgently needed therapy development by facilitating comparative analysis of multiple therapeutic strategies.
The FLIPR assay of CFTR channel activity described here is amenable to middle-to-high-throughput studies (Ahmadi et al., 2017; Zomer-van Ommen et al., 2018). The membrane-potential-sensitive FLIPR dye can be applied to any cell line or tissue, and this feature distinguishes it from the halide-sensitive YFP as employed by the Martin group in studies of intestinal tissue generated from iPSCs (Merkert et al., 2019). This group employed iPSCs stably expressing halide-sensitive YFP and differentiated to intestinal epithelium for their drug discovery studies. A major advantage of the FLIPR system over the YFP assay is that it enables evaluation of the kinetics of channel activation, a fundamental property of interest to biophysicists. A movie of the kinetics of CFTR activation measured by FLIPR is provided as supplemental material (Video S1). The YFP quenching assay measures the number of active channels, rather than their kinetics. Finally, the FLIPR assay is more sensitive to anion conductance changes than the YFP-based assay. For example, the modest functional rescue of F508del-CFTR channel expression with modulator compounds such as ORKAMBI can be readily detected at physiological temperatures using FLIPR, in contrast to the YFP-based assay, which requires augmentation of the small-molecule effects by low-temperature rescue.
A caveat of our work relates to the relatively early stage of lung differentiation modeled by the submerged cultures employed for the phenotypic screen. The submerged cultures grown on 96 well plates model multipotent lung progenitor epithelium and there is a deficit of mature cell types, including ciliated, club, and goblet cells and ionocytes. Interestingly, despite this early stage of differentiation, the expression of CFTR is high, comparable to that observed in primary bronchial epithelium grown at the air-liquid interface. However, it is likely that the regulation of CFTR protein synthesis, trafficking, and channel activity may be cell-type specific. Hence, this necessitates that findings reported via the high-throughput assay on submerged cultures be validated using that person’s primary nasal epithelial cultures. Our proof-of-concept studies and access to the Canadian CFIT bioresource of patient-matched iPSCs and nasal cultures support the value and feasibility of this drug development pipeline.
Experimental procedures
Differentiation to submerged lung progenitor culture
Human iPSCs were obtained from the CFIT program (Eckford et al., 2019) and were generated from blood samples collected with informed consent, under oversight of the Research Ethics Board of the Hospital for Sick Children (approval #1000044783). The submerged lung progenitor cultures were generated from iPSCs as previously described (Wong et al., 2015). Additional details are given in the supplemental information.
Apical chloride conductance assay for CFTR function
The apical chloride conductance (ACC) assay was used to assess CFTR-mediated changes in membrane depolarization using methods as previously described (Ahmadi et al., 2017; Erwood et al., 2020). Additional details are given in the supplemental information.
Analysis and heatmap generation
Experiments were exported as multi-frame TIFF images of which every frame recorded the entire plate. Pixels outside of well areas were filtered out using the initial signal. All fluorescence pixels (500–1,000) from inside the wells contributed to the heatmaps shown in Figure 2. All trajectories were normalized to the last point of the baseline intensity to define the peak response for each pixel. Heatmap representation was generated from the peak response of each pixel and the mean response trace of wells was generated by averaging the corresponding pixel traces.
Statistical analyses
Unpaired two-tailed t test was performed on data with two datasets. Ordinary one-way ANOVA with Tukey's multiple comparison test was performed on all data with more than two datasets. Correlations between datasets were assessed using the “Spearman's test”. p < 0.05 was considered statistically significant. Statistical analyses were performed using GraphPad Prism 9.2.
Real-time quantitative PCR
As previously described (Cao et al., 2020), total mRNA was extracted with an RNeasy Plus Micro Kit and used for reverse transcription as well as real-time PCR expression analysis. A detailed description is given in the supplemental information and primer sequences are listed in Table S2.
RNA sequencing and analysis
RNA samples were extracted and submitted for bulk RNA-seq. SAMtools was used for sequence alignment and featureCounts was used for raw counts. Downstream analysis and PCA were generated using R package DESeq2 (v.1.24.0). See the supplemental information for a description.
Immunofluorescence
Samples were fixed in 4% paraformaldehyde and permeabilized in 0.05% Triton X-100. After being blocked, the samples were labeled with primary antibodies and then secondary antibodies. Images were acquired on the SP8/TED confocal microscope. A detailed description is available in the supplemental information.
Western blotting
Samples were resuspended, and soluble fractions were analyzed by SDS-PAGE on 6% Tris-glycine gels. Proteins were transferred and detected with antibodies, and relative levels of CFTR protein were quantified with Image Studio Lite (see the supplemental information).
Data and code availability
Raw sequence data are available through the NCBI sequence read archive (https://www.ncbi.nlm.nih.gov/sra) with the BioProject: PRJNA721455.
Author contribution
J.X.J. designed and performed experiments, analyzed data, and revised the manuscript. L.W. designed and performed experiments. O.L. performed experiments and revised the manuscript. I.U. performed data analysis and interpretation. Z.B. developed analysis methods. T.G. performed experiments. Z.N. performed experiments. S.X. performed data collection. M.D.P. performed data collection. P.D.W.E. performed oversight and reviewed and edited the manuscript. F.R. provided scientific insight and reviewed and edited the manuscript. T.M. provided scientific insight and reviewed and edited the manuscript. J.P. performed data analysis and interpretation and reviewed the manuscript. A.P.W. performed data analysis and interpretation and reviewed the manuscript. C.E.B. conceived and designed the work and drafted and revised the manuscript.
Conflicts of interest
The authors declare no competing interests.
Acknowledgments
The iPSCs and primary nasal cell cultures were obtained through the CF Canada-SickKids Program for Individualized CF Therapy (CFIT). SMG1i was obtained through Cystic Fibrosis Foundation Therapeutics and the small molecules AC1, AC2-1, AC2-2, and AP2 were provided by AbbVie. We thank Ashvani Singh for reviewing the manuscript. We acknowledge the insightful discussions with Saumel Ahmadi as we started this project. This work was supported by the CFIT program with funding provided by CF Canada and the SickKids Foundation, by the government of Canada through Genome Canada and the Ontario Genomics Institute (OGI-148), and a grant to C.E.B. from Medicine by Design. J.P. and I.U. were supported by a grant from the Natural Sciences and Engineering Research Council (RGPIN-2019-06852). This work was also supported by the Cystic Fibrosis Foundation (OOC 590131). This study was supported by a grant from the government of Ontario. T.J.M. was also supported by funding from Emily's Entourage.
Published: October 21, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.stemcr.2021.09.020.
Supplemental information
References
- Ahmadi S., Bozoky Z., Di Paola M., Xia S., Li C., Wong A.P., Wellhauser L., Molinski S.V., Ip W., Ouyang H., et al. Phenotypic profiling of CFTR modulators in patient-derived respiratory epithelia. NPJ Genom Med. 2017;2:12. doi: 10.1038/s41525-017-0015-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Awatade N.T., Wong S.L., Capraro A., Pandzic E., Slapetova I., Zhong L., Turgutoglu N., Fawcett L.K., Whan R.M., Jaffe A., et al. Significant functional differences in differentiated Conditionally Reprogrammed (CRC)- and Feeder-free Dual SMAD inhibited-expanded human nasal epithelial cells. J. Cyst Fibros. 2021;20:364–371. doi: 10.1016/j.jcf.2020.12.019. [DOI] [PubMed] [Google Scholar]
- Berkers G., van Mourik P., Vonk A.M., Kruisselbrink E., Dekkers J.F., de Winter-de Groot K.M., Arets H.G.M., Marck-van der Wilt R.E.P., Dijkema J.S., Vanderschuren M.M., et al. Rectal organoids enable personalized treatment of cystic fibrosis. Cell Rep. 2019;26:1701–1708.e1703. doi: 10.1016/j.celrep.2019.01.068. [DOI] [PubMed] [Google Scholar]
- Brewington J.J., Filbrandt E.T., LaRosa F.J., 3rd, Moncivaiz J.D., Ostmann A.J., Strecker L.M., Clancy J.P. Brushed nasal epithelial cells are a surrogate for bronchial epithelial CFTR studies. JCI Insight. 2018;3:13. doi: 10.1172/jci.insight.99385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao H., Ouyang H., Laselva O., Bartlett C., Zhou Z.P., Duan C., Gunawardena T., Avolio J., Bear C.E., Gonska T., et al. A helper-dependent adenoviral vector rescues CFTR to wild-type functional levels in cystic fibrosis epithelial cells harbouring class I mutations. Eur. Respir. J. 2020;56:5. doi: 10.1183/13993003.00205-2020. [DOI] [PubMed] [Google Scholar]
- Cheng S.H., Gregory R.J., Marshall J., Paul S., Souza D.W., White G.A., O'Riordan C.R., Smith A.E. Defective intracellular transport and processing of CFTR is the molecular basis of most cystic fibrosis. Cell. 1990;63:827–834. doi: 10.1016/0092-8674(90)90148-8. [DOI] [PubMed] [Google Scholar]
- Crane A.M., Kramer P., Bui J.H., Chung W.J., Li X.S., Gonzalez-Garay M.L., Hawkins F., Liao W., Mora D., Choi S., et al. Targeted correction and restored function of the CFTR gene in cystic fibrosis induced pluripotent stem cells. Stem Cell Rep. 2015;4:569–577. doi: 10.1016/j.stemcr.2015.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford D.K., Mullenders J., Pott J., Boj S.F., Landskroner-Eiger S., Goddeeris M.M. Targeting G542X CFTR nonsense alleles with ELX-02 restores CFTR function in human-derived intestinal organoids. J. Cyst Fibros. 2021;20:436–442. doi: 10.1016/j.jcf.2021.01.009. [DOI] [PubMed] [Google Scholar]
- de Wilde G., Gees M., Musch S., Verdonck K., Jans M., Wesse A.S., Singh A.K., Hwang T.C., Christophe T., Pizzonero M., et al. Identification of GLPG/ABBV-2737, a novel class of corrector, which exerts functional synergy with other CFTR modulators. Front Pharmacol. 2019;10:514. doi: 10.3389/fphar.2019.00514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Winter-de Groot K.M., Berkers G., Marck-van der Wilt R.E.P., van der Meer R., Vonk A., Dekkers J.F., Geerdink M., Michel S., Kruisselbrink E., Vries R., et al. Forskolin-induced swelling of intestinal organoids correlates with disease severity in adults with cystic fibrosis and homozygous F508del mutations. J. Cyst Fibros. 2020;19:614–619. doi: 10.1016/j.jcf.2019.10.022. [DOI] [PubMed] [Google Scholar]
- Dekkers J.F., Wiegerinck C.L., de Jonge H.R., Bronsveld I., Janssens H.M., de Winter-de Groot K.M., Brandsma A.M., de Jong N.W., Bijvelds M.J., Scholte B.J., et al. A functional CFTR assay using primary cystic fibrosis intestinal orga noids. Nat. Med. 2013;19:939–945. doi: 10.1038/nm.3201. [DOI] [PubMed] [Google Scholar]
- Deprez M., Zaragosi L.E., Truchi M., Becavin C., Ruiz Garcia S., Arguel M.J., Plaisant M., Magnone V., Lebrigand K., Abelanet S., et al. A single-cell atlas of the human healthy airways. Am. J. Respir. Crit. Care Med. 2020;202:1636–1645. doi: 10.1164/rccm.201911-2199OC. [DOI] [PubMed] [Google Scholar]
- Eckford P.D.W., McCormack J., Munsie L., He G., Stanojevic S., Pereira S.L., Ho K., Avolio J., Bartlett C., Yang J.Y., et al. The CF Canada-Sick Kids Program in individual CF therapy: a resource for the advancement of personalized medicine in CF. J. Cyst Fibros. 2019;18:35–43. doi: 10.1016/j.jcf.2018.03.013. [DOI] [PubMed] [Google Scholar]
- Erwood S., Laselva O., Bily T.M.I., Brewer R.A., Rutherford A.H., Bear C.E., Ivakine E.A. Allele-specific prevention of nonsense-mediated decay in cystic fibrosis using homology-independent genome editing. Mol. Ther. Methods Clin. Dev. 2020;17:1118–1128. doi: 10.1016/j.omtm.2020.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fawcett L.K., Wakefield C.E., Sivam S., Middleton P.G., Wark P., Widger J., Jaffe A., Waters S.A. Avatar acceptability: views from the Australian Cystic Fibrosis community on the use of personalised organoid technology to guide treatment decisions. ERJ Open Res. 2021;7 doi: 10.1183/23120541.00448-2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Firth A.L., Dargitz C.T., Qualls S.J., Menon T., Wright R., Singer O., Gage F.H., Khanna A., Verma I.M. Generation of multiciliated cells in functional airway epithelia from human induced pluripotent stem cells. Proc. Natl. Acad. Sci. U S A. 2014;111:E1723–E1730. doi: 10.1073/pnas.1403470111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graeber S.Y., van Mourik P., Vonk A.M., Kruisselbrink E., Hirtz S., van der Ent C.K., Mall M.A., Beekman J.M. Comparison of organoid swelling and in vivo biomarkers of CFTR function to determine effects of lumacaftor-ivacaftor in patients with cystic fibrosis homozygous for the F508del mutation. Am. J. Respir. Crit. Care Med. 2020;202:1589–1592. doi: 10.1164/rccm.202004-1200LE. [DOI] [PubMed] [Google Scholar]
- Gubler H. Assay data quality assessment. Methods Mol. Biol. 2009;552:79–95. doi: 10.1007/978-1-60327-317-6_6. [DOI] [PubMed] [Google Scholar]
- Guerra L., Favia M., Di Gioia S., Laselva O., Bisogno A., Casavola V., Colombo C., Conese M. The preclinical discovery and development of the combination of ivacaftor + tezacaftor used to treat cystic fibrosis. Expert Opin. Drug Discov. 2020;15:873–891. doi: 10.1080/17460441.2020.1750592. [DOI] [PubMed] [Google Scholar]
- Haggie P.M., Phuan P.W., Tan J.A., Xu H., Avramescu R.G., Perdomo D., Zlock L., Nielson D.W., Finkbeiner W.E., Lukacs G.L., et al. Correctors and potentiators rescue function of the truncated W1282X-cystic fibrosis transmembrane regulator (CFTR) translation product. J. Biol. Chem. 2017;292:771–785. doi: 10.1074/jbc.M116.764720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawkins F., Kramer P., Jacob A., Driver I., Thomas D.C., McCauley K.B., Skvir N., Crane A.M., Kurmann A.A., Hollenberg A.N., et al. Prospective isolation of NKX2-1-expressing human lung progenitors derived from pluripotent stem cells. J. Clin. Invest. 2017;127:2277–2294. doi: 10.1172/JCI89950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawkins F.J., Suzuki S., Beermann M.L., Barilla C., Wang R., Villacorta-Martin C., Berical A., Jean J.C., Le Suer J., Matte T., et al. Derivation of airway basal stem cells from human pluripotent stem cells. Cell Stem Cell. 2021;28:79–95.e78. doi: 10.1016/j.stem.2020.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heijerman H.G.M., McKone E.F., Downey D.G., Van Braeckel E., Rowe S.M., Tullis E., Mall M.A., Welter J.J., Ramsey B.W., McKee C.M., et al. Efficacy and safety of the elexacaftor plus tezacaftor plus ivacaftor combination regimen in people with cystic fibrosis homozygous for the F508del mutation: a double-blind, randomised, phase 3 trial. Lancet. 2019;394:1940–1948. doi: 10.1016/S0140-6736(19)32597-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S.X., Islam M.N., O'Neill J., Hu Z., Yang Y.G., Chen Y.W., Mumau M., Green M.D., Vunjak-Novakovic G., Bhattacharya J., et al. Efficient generation of lung and airway epithelial cells from human pluripotent stem cells. Nat. Biotechnol. 2014;32:84–91. doi: 10.1038/nbt.2754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keegan D.E., Brewington J.J. Nasal epithelial cell-based models for individualized study in cystic fibrosis. Int. J. Mol. Sci. 2021;22:4448. doi: 10.3390/ijms22094448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keenan M.M., Huang L., Jordan N.J., Wong E., Cheng Y., Valley H.C., Mahiou J., Liang F., Bihler H., Mense M., et al. Nonsense-mediated RNA decay pathway inhibition restores expression and function of W1282X CFTR. Am. J. Respir. Cell Mol Biol. 2019;61:290–300. doi: 10.1165/rcmb.2018-0316OC. [DOI] [PubMed] [Google Scholar]
- Kerschner J.L., Paranjapye A., Yin S., Skander D.L., Bebek G., Leir S.H., Harris A. A functional genomics approach to investigate the differentiation of iPSCs into lung epithelium at air-liquid interface. J. Cell Mol. Med. 2020;24:9853–9870. doi: 10.1111/jcmm.15568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laselva O., Bartlett C., Gunawardena T.N.A., Ouyang H., Eckford P.D.W., Moraes T.J., Bear C.E., Gonska T. Rescue of multiple class II CFTR mutations by elexacaftor+tezacaftor+ivacaftor mediated in part by the dual activities of elexacaftor as both corrector and potentiator. Eur. Respir. J. 2021;57:2002774. doi: 10.1183/13993003.02774-2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laselva O., Bartlett C., Popa A., Ouyang H., Gunawardena T.N.A., Gonska T., Moraes T.J., Bear C.E. Emerging preclinical modulators developed for F508del-CFTR have the potential to be effective for ORKAMBI resistant processing mutants. J. Cyst Fibros. 2021;20:106–119. doi: 10.1016/j.jcf.2020.07.015. [DOI] [PubMed] [Google Scholar]
- Laselva O., Bartlett C., Popa A., Ip W., Ouyang H., Moraes J.T., Gonska T., Bear C.E. 15th ECFS Basic Science Conference (Loutraki, Greece) 2018. Profiling CFTR modulators in nasal epithelial cultures reveals differential, patient-specific responses. [Google Scholar]
- Laselva O., Eckford P.D., Bartlett C., Ouyang H., Gunawardena T.N., Gonska T., Moraes T.J., Bear C.E. Functional rescue of c.3846G>A (W1282X) in patient-derived nasal cultures achieved by inhibition of nonsense mediated decay and protein modulators with complementary mechanisms of action. J. Cyst Fibros. 2020;19:717–727. doi: 10.1016/j.jcf.2019.12.001. [DOI] [PubMed] [Google Scholar]
- Laselva O., McCormack J., Bartlett C., Ip W., Gunawardena T.N.A., Ouyang H., Eckford P.D.W., Gonska T., Moraes T.J., Bear C.E. Preclinical studies of a rare CF-causing mutation in the second nucleotide binding domain (c.3700A>G) show robust functional rescue in primary nasal cultures by novel CFTR modulators. J. Pers Med. 2020;10:209. doi: 10.3390/jpm10040209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukacs G.L., Verkman A.S. CFTR: folding, misfolding and correcting the DeltaF508 conformational defect. Trends Mol. Med. 2012;18:81–91. doi: 10.1016/j.molmed.2011.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCauley K.B., Hawkins F., Serra M., Thomas D.C., Jacob A., Kotton D.N. Efficient derivation of functional human airway epithelium from pluripotent stem cells via temporal regulation of WNT signaling. Cell Stem Cell. 2017;20:844–857.e846. doi: 10.1016/j.stem.2017.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merkert S., Schubert M., Olmer R., Engels L., Radetzki S., Veltman M., Scholte B.J., Zollner J., Pedemonte N., Galietta L.J.V., et al. High-throughput screening for modulators of CFTR activity based on genetically engineered cystic fibrosis disease-specific iPSCs. Stem Cell Rep. 2019;12:1389–1403. doi: 10.1016/j.stemcr.2019.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Middleton P.G., Mall M.A., Drevinek P., Lands L.C., McKone E.F., Polineni D., Ramsey B.W., Taylor-Cousar J.L., Tullis E., Vermeulen F., et al. Elexacaftor-Tezacaftor-Ivacaftor for Cystic Fibrosis with a Single Phe508del Allele. N. Engl. J. Med. 2019;381:1809–1819. doi: 10.1056/NEJMoa1908639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molinski S.V., Ahmadi S., Ip W., Ouyang H., Villella A., Miller J.P., Lee P.S., Kulleperuma K., Du K., Di Paola M., et al. Orkambi(R) and amplifier co-therapy improves function from a rare CFTR mutation in gene-edited cells and patient tissue. EMBO Mol. Med. 2017;9:1224–1243. doi: 10.15252/emmm.201607137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikolic M.Z., Caritg O., Jeng Q., Johnson J.A., Sun D., Howell K.J., Brady J.L., Laresgoiti U., Allen G., Butler R., et al. Human embryonic lung epithelial tips are multipotent progenitors that can be expanded in vitro as long-term self-renewing organoids. eLife. 2017;6:e26575. doi: 10.7554/eLife.26575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oren Y.S., Irony-Tur Sinai M., Golec A., Barchad-Avitzur O., Mutyam V., Li Y., Hong J., Ozeri-Galai E., Hatton A., Leibson C., et al. Antisense oligonucleotide-based drug development for Cystic Fibrosis patients carrying the 3849+10 kb C-to-T splicing mutation. J. Cyst Fibros. 2021:S1569–S1993. doi: 10.1016/j.jcf.2021.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J.K., Shrivastava A., Zhang C., Pollok B.A., Finkbeiner W.E., Gibb E.R., Ly N.P., Illek B. Functional profiling of CFTR-directed therapeutics using pediatric patient-derived nasal epithelial cell models. Front Pediatr. 2020;8:536. doi: 10.3389/fped.2020.00536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phuan P.W., Haggie P.M., Tan J.A., Rivera A.A., Finkbeiner W.E., Nielson D.W., Thomas M.M., Janahi I.A., Verkman A.S. CFTR modulator therapy for cystic fibrosis caused by the rare c.3700A>G mutation. J. Cyst Fibros. 2021;20:452–459. doi: 10.1016/j.jcf.2020.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phuan P.W., Son J.H., Tan J.A., Li C., Musante I., Zlock L., Nielson D.W., Finkbeiner W.E., Kurth M.J., Galietta L.J., et al. Combination potentiator ('co-potentiator') therapy for CF caused by CFTR mutants, including N1303K, that are poorly responsive to single potentiators. J. Cyst Fibros. 2018;17:595–606. doi: 10.1016/j.jcf.2018.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pranke I., Hatton A., Masson A., Flament T., Le Bourgeois M., Chedevergne F., Bailly C., Urbach V., Hinzpeter A., Edelman A., et al. Might brushed nasal cells be a surrogate for CFTR modulator clinical response? Am. J. Respir. Crit. Care Med. 2019;199:123–126. doi: 10.1164/rccm.201808-1436LE. [DOI] [PubMed] [Google Scholar]
- Pranke I.M., Hatton A., Simonin J., Jais J.P., Le Pimpec-Barthes F., Carsin A., Bonnette P., Fayon M., Stremler-Le Bel N., Grenet D., et al. Correction of CFTR function in nasal epithelial cells from cystic fibrosis patients predicts improvement of respiratory function by CFTR modulators. Sci. Rep. 2017;7:7375. doi: 10.1038/s41598-017-07504-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu B.H., Thomas P.J. Alteration of the cystic fibrosis transmembrane conductance regulator folding pathway. J. Biol. Chem. 1996;271:7261–7264. doi: 10.1074/jbc.271.13.7261. [DOI] [PubMed] [Google Scholar]
- Ramalho A.S., Furstova E., Vonk A.M., Ferrante M., Verfaillie C., Dupont L., Boon M., Proesmans M., Beekman J.M., Sarouk I., et al. Correction of CFTR function in intestinal organoids to guide treatment of cystic fibrosis. Eur. Respir. J. 2021;57:1902426. doi: 10.1183/13993003.02426-2019. [DOI] [PubMed] [Google Scholar]
- Silva I.A.L., Dousova T., Ramalho S., Centeio R., Clarke L.A., Railean V., Botelho H.M., Holubova A., Valaskova I., Yeh J.T., et al. Organoids as a personalized medicine tool for ultra-rare mutations in cystic fibrosis: the case of S955P and 1717-2A>G. Biochim. Biophys. Acta Mol. Basis Dis. 2020;1866:165905. doi: 10.1016/j.bbadis.2020.165905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva I.A.L., Railean V., Duarte A., Amaral M.D. Personalized medicine based on nasal epithelial cells: comparative studies with rectal biopsies and intestinal organoids. J. Pers Med. 2021;11:421. doi: 10.3390/jpm11050421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor-Cousar J.L., Munck A., McKone E.F., van der Ent C.K., Moeller A., Simard C., Wang L.T., Ingenito E.P., McKee C., Lu Y., et al. Tezacaftor-ivacaftor in patients with cystic fibrosis homozygous for Phe508del. N. Engl. J. Med. 2017;377:2013–2023. doi: 10.1056/NEJMoa1709846. [DOI] [PubMed] [Google Scholar]
- Valley H.C., Bukis K.M., Bell A., Cheng Y., Wong E., Jordan N.J., Allaire N.E., Sivachenko A., Liang F., Bihler H., et al. Isogenic cell models of cystic fibrosis-causing variants in natively expressing pulmonary epithelial cells. J. Cyst Fibros. 2019;18:476–483. doi: 10.1016/j.jcf.2018.12.001. [DOI] [PubMed] [Google Scholar]
- Van Goor F., Hadida S., Grootenhuis P.D., Burton B., Stack J.H., Straley K.S., Decker C.J., Miller M., McCartney J., Olson E.R., et al. Correction of the F508del-CFTR protein processing defect in vitro by the investigational drug VX-809. Proc. Natl. Acad. Sci. U S A. 2011;108:18843–18848. doi: 10.1073/pnas.1105787108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veit G., Roldan A., Hancock M.A., Da Fonte D.F., Xu H., Hussein M., Frenkiel S., Matouk E., Velkov T., Lukacs G.L. Allosteric folding correction of F508del and rare CFTR mutants by elexacaftor-tezacaftor-ivacaftor (Trikafta) combination. JCI Insight. 2020;5:e139983. doi: 10.1172/jci.insight.139983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veit G., Xu H., Dreano E., Avramescu R.G., Bagdany M., Beitel L.K., Roldan A., Hancock M.A., Lay C., Li W., et al. Structure-guided combination therapy to potently improve the function of mutant CFTRs. Nat. Med. 2018;24:1732–1742. doi: 10.1038/s41591-018-0200-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vonk A.M., van Mourik P., Ramalho A.S., Silva I.A.L., Statia M., Kruisselbrink E., Suen S.W.F., Dekkers J.F., Vleggaar F.P., Houwen R.H.J., et al. Protocol for application, standardization and validation of the forskolin-induced swelling assay in cystic fibrosis human colon organoids. STAR Protoc. 2020;1:100019. doi: 10.1016/j.xpro.2020.100019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wainwright C.E., Elborn J.S., Ramsey B.W. Lumacaftor-ivacaftor in patients with cystic fibrosis homozygous for Phe508del CFTR. N. Engl. J. Med. 2015;373:1783–1784. doi: 10.1056/NEJMc1510466. [DOI] [PubMed] [Google Scholar]
- Wong A.P., Bear C.E., Chin S., Pasceri P., Thompson T.O., Huan L.J., Ratjen F., Ellis J., Rossant J. Directed differentiation of human pluripotent stem cells into mature airway epithelia expressing functional CFTR protein. Nat. Biotechnol. 2012;30:876–882. doi: 10.1038/nbt.2328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong A.P., Chin S., Xia S., Garner J., Bear C.E., Rossant J. Efficient generation of functional CFTR-expressing airway epithelial cells from human pluripotent stem cells. Nat. Protoc. 2015;10:363–381. doi: 10.1038/nprot.2015.021. [DOI] [PubMed] [Google Scholar]
- Wu Y.S., Jiang J., Ahmadi S., Lew A., Laselva O., Xia S., Bartlett C., Ip W., Wellhauser L., Ouyang H., et al. ORKAMBI-mediated rescue of mucociliary clearance in cystic fibrosis primary respiratory cultures is enhanced by arginine uptake, Arginase inhibition, and promotion of nitric oxide signaling to the cystic fibrosis transmembrane conductance regulator channel. Mol. Pharmacol. 2019;96:515–525. doi: 10.1124/mol.119.117143. [DOI] [PubMed] [Google Scholar]
- Zhang X.D. A pair of new statistical parameters for quality control in RNA interference high-throughput screening assays. Genomics. 2007;89:552–561. doi: 10.1016/j.ygeno.2006.12.014. [DOI] [PubMed] [Google Scholar]
- Zomer-van Ommen D.D., de Poel E., Kruisselbrink E., Oppelaar H., Vonk A.M., Janssens H.M., van der Ent C.K., Hagemeijer M.C., Beekman J.M. Comparison of ex vivo and in vitro intestinal cystic fibrosis models to measure CFTR-dependent ion channel activity. J. Cyst Fibros. 2018;17:316–324. doi: 10.1016/j.jcf.2018.02.007. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw sequence data are available through the NCBI sequence read archive (https://www.ncbi.nlm.nih.gov/sra) with the BioProject: PRJNA721455.