Summary
Hematopoietic stem cells (HSCs) reside at the apex of the hematopoietic differentiation hierarchy and sustain multilineage hematopoiesis. Here, we show that the transcriptional regulator CITED2 is essential for life-long HSC maintenance. While hematopoietic-specific Cited2 deletion has a minor impact on steady-state hematopoiesis, Cited2-deficient HSCs are severely depleted in young mice and fail to expand upon aging. Moreover, although they home normally to the bone marrow, they fail to reconstitute hematopoiesis upon transplantation. Mechanistically, CITED2 is required for expression of key HSC regulators, including GATA2, MCL-1, and PTEN. Hematopoietic-specific expression of anti-apoptotic MCL-1 partially rescues the Cited2-deficient HSC pool and restores their reconstitution potential. To interrogate the Cited2→Pten pathway in HSCs, we generated Cited2;Pten compound heterozygous mice, which had a decreased number of HSCs that failed to reconstitute the HSC compartment. In addition, CITED2 represses multiple pathways whose elevated activity causes HSC exhaustion. Thus, CITED2 promotes pathways necessary for HSC maintenance and suppresses those detrimental to HSC integrity.
Keywords: hematopoietic stem cell, CITED2, MCL-1, PTEN, hematopoiesis
Highlights
-
•
Unperturbed hematopoiesis can be sustained long term while the HSC pool is depleted
-
•
CITED2 promotes HSC survival but not quiescence under homeostatic conditions
-
•
CITED2 maintains the HSC pool by controlling Mcl1 and Pten expression
Kranc, Guitart, and colleagues demonstrate that Cited2 deletion causes a progressive HSC loss under steady-state conditions without perturbing normal hematopoiesis. The authors show that CITED2 maintains HSCs by regulating the expression of Mcl1 and Pten. Finally, they indicate that CITED2 promotes multiple pathways necessary for HSC maintenance and suppresses those detrimental to HSC integrity to coordinate HSC function.
Introduction
Hematopoiesis is a dynamic and essential process, with the capacity to meet a large demand for differentiated blood cells (∼1011 cells per day in humans) (Sender and Milo, 2021). Hematopoiesis critically depends on a pool of bone marrow (BM)-resident adult hematopoietic stem cells (HSCs) at the apex of the hematopoietic differentiation hierarchy, which possess unique self-renewal capacity and multilineage differentiation potential (McCracken et al., 2016). The strict regulation of survival, quiescence, self-renewal, and differentiation in HSCs is essential for life-long maintenance of their pool. While much progress has been made in identifying individual pathways that suppress or promote these fates, the key regulators which coordinate these pathways to maintain the HSC pool both in the steady state and under conditions of physiologic stress remain poorly understood.
CITED2 (CBP/p300-interacting-transactivator-with-an ED-rich-tail 2) is a transcriptional regulator that co-activates or represses multiple transcription factors, including AP-2 (Bamforth et al., 2001), HIF-1alpha (Bhattacharya et al., 1999), PPAR-α (Tien et al., 2004), SMAD2/3 (Chou and Yang, 2006), and c-MYC (Chou et al., 2012) to regulate fundamental cellular processes, such as proliferation, metabolism, differentiation, migration, and autophagy. Consistent with its ubiquitous expression and pleiotropic impact on diverse transcription factors, CITED2 is essential for embryonic development, including fetal liver hematopoiesis (Bamforth et al., 2001, 2004; Chen et al., 2007; Weninger et al., 2005; Withington et al., 2006; Yin et al., 2002), ESC biology (Kranc et al., 2015; Li et al., 2012), adult tissue functions (Kim et al., 2018; Lee et al., 2009; Liu et al., 2019), cellular proliferation (Kranc et al., 2003), and cancer progression (Fernandes et al., 2020). Thus, CITED2 is an important regulator of diverse molecular, cellular, and developmental processes.
A growing body of evidence indicates that CITED2 is a key regulator of adult HSC biology (Du et al., 2012, 2014; Korthuis et al., 2015; Kranc et al., 2009). We previously reported that inducible Mx1-Cre-mediated deletion of Cited2 (in which Mx1-Cre is induced by poly(I:C)-stimulated IFN-α production) results in a rapid loss of HSCs via apoptosis and a resultant BM failure (Kranc et al., 2009). In this context, the significant loss of the HSC pool upon inducible Cited2 deletion is at least in part caused by upregulation of the p19ARF-p53 pathway, as genetic ablation of Cdkn2a (encoding p16INK4A and p19ARF) or Trp53 (encoding p53) rescues depletion of Cited2-deficient HSCs. Another study (Du et al., 2012) employing a different Cited2 floxed allele, also demonstrated that poly(I:C)-inducible Mx1-Cre-mediated Cited2 deletion results in loss of HSCs (by affecting their quiescence and survival), compromises their reconstitution potential, and leads to a rapid BM failure upon myelotoxic stress. In this study, loss of quiescence, but not increased apoptosis, upon inducible Cited2 deletion is mediated at least in part by HIF-1alpha, as Hif1a deletion partially restores impaired quiescence of HSCs lacking Cited2 and improves their ability to reconstitute the HSC compartment upon transplantation (Du et al., 2012). Additional analyses of Cited2-deficient HSCs indicated alterations in HSC metabolism, namely a decrease in glycolytic flux and an increase in mitochondrial activity (Du et al., 2014), a state associated with a decline in HSC function (Lawson et al., 2021; Wang et al., 2014). However, given that the previous studies outlined above involved poly(I:C) administration, which is known to induce interferon response, proinflammatory pathways, and subsequent over-proliferation in HSCs, the functional significance of CITED2 in maintenance of the HSC pool under steady-state conditions is yet to be investigated.
Here, we reveal that, under steady-state conditions, CITED2 is largely dispensable for unperturbed long-term multilineage hematopoiesis, but is critically required for the maintenance of the HSC pool and HSC function post-transplantation. We found that CITED2 represses pathways that inhibit HSC maintenance and promotes pathways required for HSC integrity. Notably, our functional genetic approaches found that key regulators of HSC maintenance, namely MCL-1 and PTEN, act downstream of CITED2 and mediate, at least in part, its critical role in regulating the HSC pool. Taken together, we propose that CITED2 coordinates multiple fundamental stem cell regulatory pathways to promote the maintenance of the HSC pool under steady-state conditions and upon transplantation.
Results
Cited2 deletion does not derail normal steady-state hematopoiesis
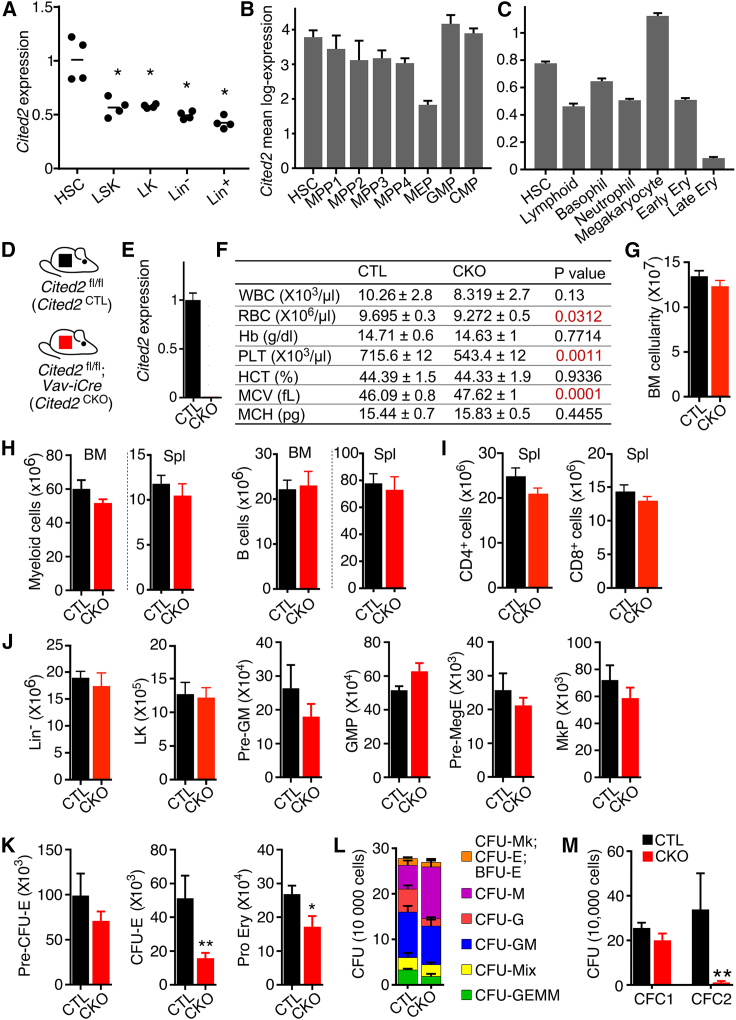
To determine the expression of Cited2 at different levels of the hematopoietic hierarchy, we sorted Lin–Sca-1+c-Kit+ (LSK) cells, LSKCD48−CD150+ HSCs, Lin−Sca-1−c-Kit+ (LK) myeloid progenitors, and more-mature Lin– and differentiated Lin+ hematopoietic cell populations, and performed qRT-PCR. Cited2 was expressed in all compartments, with significantly higher expression in the HSC population compared with myeloid progenitors and more mature hematopoietic cell populations (Figure 1A). Furthermore, to compare the expression of Cited2 in long-term HSCs (LSKCD34−CD135−), MPP1 (LSKCD34+CD135−CD150+CD48−), MPP2 (LSKCD34+CD135−CD150+CD48+), and MPP3 (LSKCD34+CD135−CD150−CD48+) populations, lymphoid-primed multipotent progenitors (LSKCD34+CD135+), which correspond to the MPP4 population, and CMP (LKCD34+FcγRII/IIIlow), GMP (LKCD34+FcγRII/IIIhigh), and MEP (LKCD34−FcγRII/IIIlow) compartments, we analyzed our SMART2-seq single-cell expression data in these populations (Nestorowa et al., 2016). Cited2 was rather uniformly expressed across these populations (Figure 1B), with the highest expression in the HSC, GMP, and CMP compartments, and lowest in the MEP population. Moreover, we employed our 10× Genomics single-cell RNA sequencing (RNA-seq) data set (Dahlin et al., 2018) to compare Cited2 expression between HSCs and committed progenitor cell compartments. Cited2 was expressed highly in HSCs and committed megakaryocytic progenitors, but decreased in committed lymphoid, basophilic, neutrophilic, and erythroid progenitor cells (Figure 1C). Thus, Cited2 is ubiquitously expressed in hematopoiesis, with robust high expression at the apex of the hematopoietic hierarchy, and lower expression in more differentiated cells.
Figure 1.
Hematopoiesis-specific Cited2 deletion does not derail multilineage hematopoiesis
(A) Cited2 mRNA levels in cells isolated from 8-week-old C57BL/6 mice (n = 4).
(B) Cited2 expression in different hematopoietic compartments determined using single-cell SMART2-seq.
(C) Cited2 expression in HSCs and committed progenitor cell compartments determined by 10× Genomics single-cell RNA-seq.
(D) Cited2fl/fl mice were bred to Vav-iCre mice to generate Cited2fl/fl;Vav-iCre (Cited2CKO) mice. Cited2fl/fl mice were used as controls (Cited2CTL).
(E) Cited2 expression in c-Kit+ cells from BM of Cited2CTL and Cited2CKO mice (n = 4).
(F) PB counts of 8- to 10-week-old mice (n = 14).
(G) Total BM cellularity (n = 6–9).
(H) Total number of differentiated myeloid (Gr-1+Mac-1+) and B cells (CD19+B220+) in BM and spleen (Spl) (n = 6–9).
(I) Total number of mature CD4+ and CD8+ T cells in spleens (n = 6–9).
(J) Total number of Lin− cells, LK cells, pre-GM (LKCD41−FcγRII/III−CD150−CD105−), GMP (LKCD41−FcγRII/III+), Pre-MegE (LKCD41−FcγRII/III−CD150−CD105+), MkP (LKCD41+CD150+) cells (n = 5).
(K) Total number of Pre-CFU-E (LKCD41−FcγRII/III−CD150+CD105+), CFU-E (LKCD41−FcγRII/III−CD150+CD105+CD71+Ter119−), and Pro Ery (LKCD41−FcγRII/III−CD150+CD105+CD71+Ter119+) cells (n = 5).
(L) Colony-forming unit (CFU) assay performed with 104 BM cells from 8- to 10-week-old mice. CFU-megakaryocyte (CFU-Mk), CFU-erythroid (CFU-E), burst-forming unit (BFU-E), CFU-granulocyte (CFU-G), CFU-monocyte/macrophage (CFU-M), CFU-granulocyte, and monocyte/macrophage (CFU-GM); at least three lineages (CFU-Mix), CFU with all four lineages, granulocyte, erythroid, monocyte/macrophage, and megakaryocyte (CFU-GEMM) (n = 7).
(M) Colony counts of primary and secondary cultures (n = 7). For (B), (C), and (F)–(M), data are mean ± SEM. ∗ p < 0.05, ∗∗p < 0.01
Given that the functional significance of Cited2 in unperturbed hematopoiesis remains poorly understood, we combined the Cited2fl allele (Kranc et al., 2009; MacDonald et al., 2008) with Vav-iCre (de Boer et al., 2003) to generate Cited2fl/fl;Vav-iCre (Cited2CKO) mice (Figure 1D), where Cited2 is deleted specifically from the hematopoietic system shortly after the emergence of HSCs. Consequently, Cited2 expression was completely lost in BM c-Kit+ cells isolated from Cited2CKO mice (Figure 1E). Surprisingly, Cited2 deletion had no impact on animal viability, and all animals survived to adulthood without any obvious defects. Peripheral blood (PB) analyses of 8- to 12-week-old Cited2CKO mice revealed unaffected WBC counts, with mild anemia and thrombocythemia (Figure 1F). Furthermore, Cited2CKO mice displayed normal BM cellularity (Figure 1G) and unaffected numbers of differentiated myeloid and B lymphoid cells in the BM and spleens (Figure 1H), as well as normal distribution of T cells in the thymi and spleens (Figures S1 and 1I). Cited2CKO mice also had normal myeloid and megakaryocytic progenitor cell numbers (Figure 1J). Notably, consistent with mild anemia (Figure 1F), mice lacking Cited2 had significantly decreased numbers of BM erythroid progenitors (Figure 1K). Finally, colony-forming cell (CFC) assays showed normal differentiation potential of Cited2CKO BM cells (Figures 1L and 1M). Importantly, however, Cited2-deficient cells failed to form secondary colonies after replating, suggesting that Cited2 is required for propagation or self-renewal of progenitor cells (Figure 1M). Taken together, while Cited2 deletion compromises erythroid progenitors and causes mild anemia, it is otherwise not essential for normal steady-state multilineage hematopoiesis in young adult mice.
Cited2 is required for the maintenance of the HSC pool under steady-state conditions and its expansion upon aging
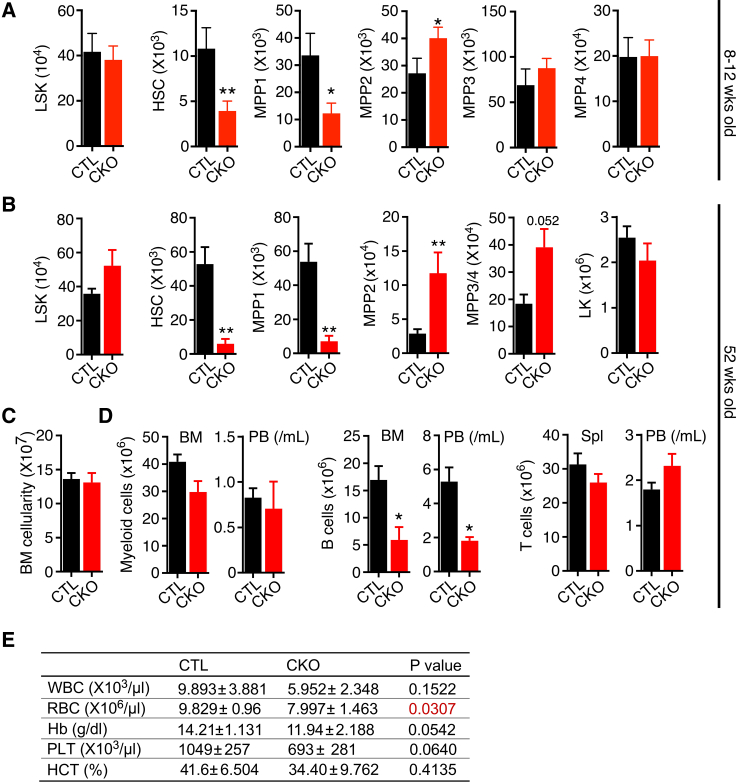
We next determined the impact of Cited2 deletion on HSCs and primitive progenitor cells. We found that the total number of LSK cells (the compartment comprising HSCs and functionally distinct lineage-biased multipotent progenitors [MPP1-4]) was not affected in young 8- to 12-week-old Cited2CKO mice (Figure 2A). Markedly, however, mice lacking Cited2 displayed significant depletion of HSCs and the most primitive progenitors (i.e., MPP1 population), with an increase in MPP2 population and unchanged numbers of the MPP3-4 populations. Thus, despite a select reduction in absolute HSC and MPP1 cell numbers, Cited2CKO mice sustain largely unaffected unperturbed steady-state multilineage hematopoiesis.
Figure 2.
Cited2 is required for the maintenance of the HSC pool and its expansion upon aging
(A) Total number of LSK cells, LSKCD48−CD150+Flt3−CD34− HSCs, LSKCD48−CD150+Flt3−CD34+ MPP1, LSKCD48+CD150+Flt3− MPP2, LSKCD48+CD150−Flt3− MPP3, and LSKCD48+CD150−Flt3+ MPP4 populations in BM of 8- to 10-week-old mice (n = 6–9).
(B) Total number of LSK cells, HSCs, MPP1, MPP2, MPP3/MPP4, and LK cells in BM in 52-week-old mice (n = 6).
(C) BM cellularity in 52-week-old mice (n = 6–9).
(D) Total number of differentiated myeloid cells (Gr-1+Mac-1+) and B cells (CD19+B220+) in BM, spleen (Spl) and PB in 52-week-old mice (n = 6–9).
(E) PB counts in 52-week-old Cited2CTL and Cited2CKO mice (n = 6–9). For (A)–(E), data are mean ± SEM. ∗p < 0.05, ∗∗p < 0.01.
Next, we investigated the impact of prolonged Cited2 deficiency on long-term HSC maintenance during unperturbed hematopoiesis. We aged mouse cohorts for 52 weeks and found that Cited2 deficiency had no impact on mouse survival (data not shown). Consistent with physiological HSC aging, during which the HSC pool undergoes expansion (Geiger et al., 2013), we found that aging Cited2CTL mice had approximately 5-fold more HSCs compared with 8- to 12-week-old Cited2CTL mice (Figures 2A and 2B). Strikingly however, Cited2CKO HSCs failed to expand upon aging and remained severely depleted compared with Cited2CTL HSCs (Figure 2B). Furthermore, while the MPP1 population also remained depleted, the numbers of MPP2 and MPP3/4 populations were increased in 52-week-old Cited2CKO mice. Aging Cited2CKO mice had normal BM cellularity and unaffected numbers of primitive and differentiated myeloid cells and T cells, while B cell numbers were decreased (Figures 2C and 2D). Apart from anemia and a significant drop in B cells, PB analyses showed no other major abnormalities (Figures 2D and 2E). Taken together, Cited2 is essential for both the maintenance of HSCs and their expansion during physiological aging, but remarkably, despite this, Cited2 is not critical for long-term multilineage hematopoiesis. Notably, these data also reveal the requirement for CITED2 in the maintenance of the B cell lineage, meriting further investigations.
Cited2 is essential for post-transplantation HSC functions
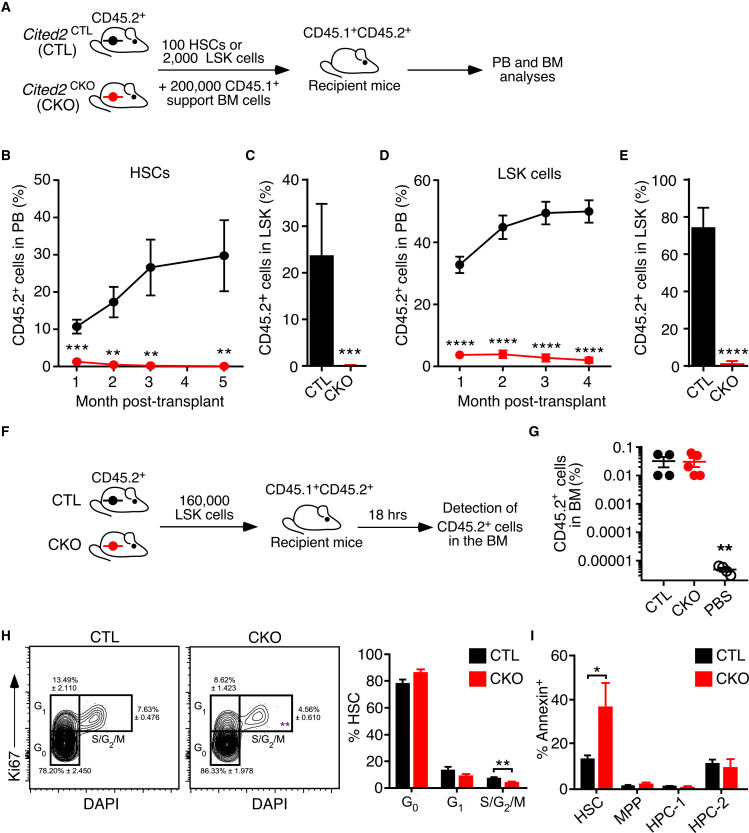
Given that Cited2CKO mice do not display any severe hematopoietic defects despite a substantial depletion of the HSC pool, we next investigated the multilineage reconstitution capacity of Cited2CKO HSCs. We competitively transplanted 100 HSCs from 8- to 12-week-old Cited2CKO and Cited2CTL mice into lethally irradiated recipients (Figure 3A). Cited2-deficient HSCs failed to reconstitute hematopoiesis (Figure 3B), and were unable to contribute to the LSK pool of recipient mice (Figure 3C). Given that HSCs lacking Cited2 have no reconstitution activity but that Cited2CKO mice are able to sustain hematopoiesis, we next asked whether the reconstitution activity is contained within the LSK compartment. We competitively transplanted LSK cells from 8- to 12-week-old Cited2CKO and Cited2CTL mice (together with 200,000 unfractionated CD45.1+ BM cells) and found that they also dramatically failed to reconstitute hematopoiesis (Figure 3D), and did not contribute to the LSK pool of the recipients (Figure 3E). Given that both Cited2-deficient HSCs and LSK cells fail to reconstitute hematopoiesis, we asked whether Cited2 loss impacts on the ability of HSCs to home to the BM. We transplanted LSK cells from Cited2CKO and Cited2CTL mice into irradiated recipients (Figure 3F), and found equal numbers of control and Cited2-deficient CD45.2+ cells 18 h after injection, indicating that LSK cells lacking Cited2 are able to home to the BM as efficiently as their Cited2CTL counterparts (Figure 3G). Therefore, although multilineage steady-state hematopoiesis is maintained in Cited2CKO mice, neither HSC nor LSK populations lacking Cited2 have the ability to repopulate hematopoiesis upon transplantation. Thus, although Cited2-deficient HSCs home to the BM, they critically require Cited2 to contribute to and sustain hematopoiesis upon transplantation.
Figure 3.
HSCs critically require Cited2 to reconstitute hematopoiesis upon transplantation
(A) Transplantation assay: 100 HSCs or 2,000 LSK cells sorted from Cited2CTL and Cited2CKO mice were transplanted into lethally irradiated recipients together with 200,000 support CD45.1+ total BM cells.
(B and C) Percentage of donor-derived CD45.2+ cells in (B) PB and (C) the LSK cell compartment of recipient mice following transplantation of 100 HSCs (n = 5 recipients per donor; 2 donors per genotype).
(D and E) Percentage of donor-derived CD45.2+ cells in (D) PB and (E) the LSK compartment of the recipient mice following transplantation of 2,000 LSK cells (n = 4 recipients per donor; 4 donors per genotype).
(F) Homing assay: 160,000 LSK cells were injected into irradiated CD45.1+-recipient mice and analyzed 18 h later.
(G) Percentage of donor-derived CD45.2+ cells in BM (n = 4–5).
(H) Percentage of HSCs from 8- to 10-week-old Cited2CKO and Cited2CTL mice in the G0 (DAPI−Ki67−), G1 (DAPI−Ki67+), and S/G2/M (DAPI+Ki67+) phases of the cell cycle (n = 3).
(I) Percentage of Annexin V+ cells in the HSC, MPP, HPC-1, and HPC-2 cell compartments in BM of Cited2CKO and Cited2CTL mice (n = 3). For (B)–(E) and (G)–(I), data are mean ± SEM. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001.
HSCs lacking Cited2 remain quiescent but display increased apoptotic rate
Depletion of HSCs and their failure to sustain hematopoiesis upon serial transplantation frequently result from a loss of HSC quiescence or increased apoptosis (Rossi et al., 2012). Thus, we investigated whether the reduction of HSCs and their reconstitution failure upon Cited2 deletion is associated with changes in these HSC fates. To determine the cell-cycle status of Cited2-deficient HSCs, we employed Ki67 and DAPI staining, which showed no differences in quiescence between Cited2CTL and Cited2CKO HSCs (Figure 3H). Notably, however, the percentage of actively cycling HSCs (i.e., those in S/G2/M phases) was decreased in the absence of Cited2 (Figure 3H). Furthermore, to determine the rate of cell death in Cited2-deficient HSCs, we used Annexin V staining. Cited2-deficient HSCs, but not primitive progenitors, displayed a significantly increased rate of apoptosis compared with their Cited2CTL counterparts (Figure 3I). Therefore, depletion of Cited2-deficient HSCs, and their inability to expand over time and reconstitute hematopoiesis upon transplantation likely result, at least in part, from a decrease in HSC cycling and an increase in their apoptosis.
Cited2 maintains HSCs by regulating Mcl1 expression
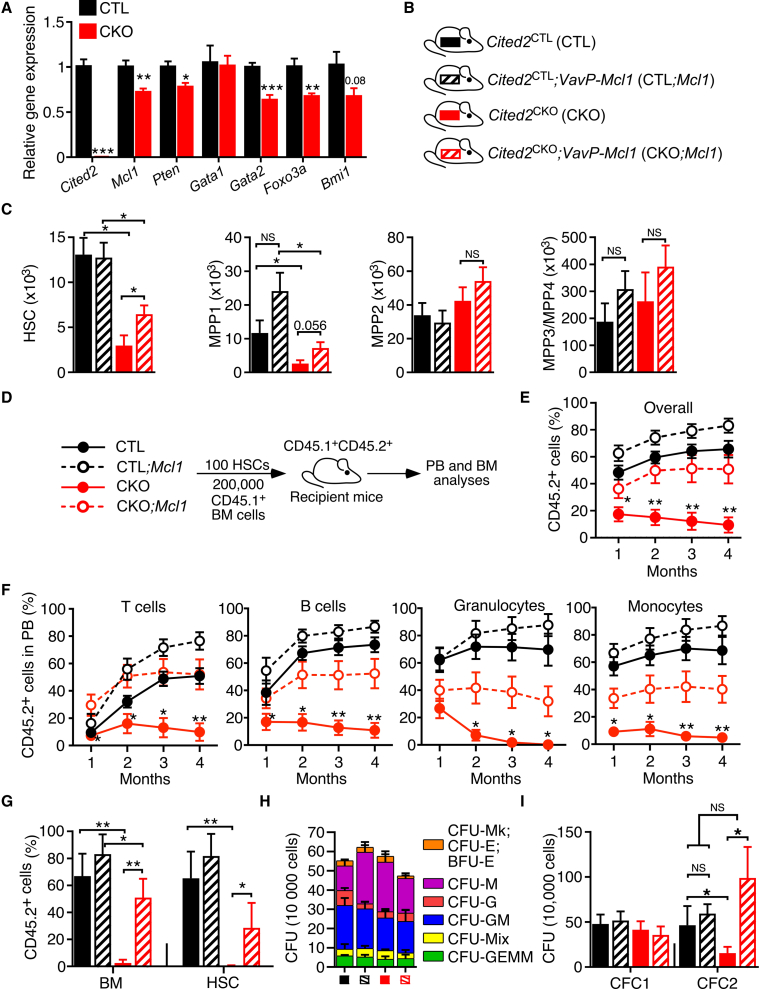
To understand the mechanisms through which Cited2 loss depletes the HSC pool in Cited2CKO mice and compromises HSC functions upon transplantation, we performed gene expression analyses in HSCs sorted from Cited2CKO and Cited2CTL mice. Interestingly, we found that the expression of key HSC regulators, including Mcl1, Pten, and Gata2 (Menendez-Gonzalez et al., 2019; Opferman et al., 2005; Yilmaz et al., 2006), was decreased in Cited2-deficient HSCs (Figure 4A), suggesting that CITED2 may control several pathways important for HSC maintenance.
Figure 4.
Mcl1 partially restores normal HSC maintenance and function of Cited2-deficient HSCs
(A) Expression of Cited2, Mcl1, Pten, Gata1, Gata2, Foxo3a, and Bmi1 in LSK cells sorted from 8-week-old Cited2CTL and Cited2CKO mice (n = 4).
(B) Schematic representation of experimental mouse cohorts; Cited2CTL, Cited2CTL;Mcl1, Cited2CKO, and Cited2CKO;Mcl1.
(C) Total number of HSC, MPP1, MPP2, and MPP3/MPP4 cell populations in BM of 8- to 10-week-old mice (n = 4–6 mice per genotype).
(D) Transplantation assay: 100 HSCs were transplanted into lethally irradiated recipients together with 200,000 unfractionated CD45.1+ BM cells.
(E) Percentage of donor-derived CD45.2+ cells in PB following transplantation (n = 4 recipients per donor; n = 2–3 donors).
(F) Percentage of donor-derived CD45.2+ cells overall in the monocyte, granulocyte, B cell, and T cell compartments of PB.
(G) Percentage of donor-derived CD45.2+ cells in total BM and HSC compartments of the recipient mice.
(H) CFU assay performed with 104 BM cells from 8- to 10-week-old mice (n = 5).
(I) Total CFC counts of primary and secondary cultures (n = 5). For (A), (C), and (E)–(I), data are mean ± SEM. ∗p < 0.05, ∗∗p < 0.01.
Myeloid cell leukemia 1 (MCL-1) is a pro-survival BCL-2 protein family member, whose Mx1-Cre-mediated deletion results in severe loss of HSCs and BM failure (Opferman et al., 2005), thus resembling the phenotype resulting from Mx1-Cre-mediated Cited2 deletion (Kranc et al., 2009). Moreover, MCL-1 is also required for self-renewal of human HSCs (Campbell et al., 2010a). Given that Mcl1 expression was decreased in Cited2-deficient HSCs, we sought to determine whether MCL-1 can rescue HSC defects resulting from Cited2 deletion in vivo. We bred Cited2CKO mice to VavP-Mcl1 transgenic mice (Campbell et al., 2010b), which overexpresses Mcl1 specifically within hematopoietic system, under the control of Vav regulatory elements (Figure 4B). Interestingly, Mcl1 overexpression in Cited2CKO mice (i.e., Cited2CKO;Mcl1 mice) resulted in a partial rescue of the HSC and MPP1 cell pools compared with Cited2CKO mice (Figure 4C). To investigate if Mcl1 overexpression can support Cited2-deficient HSCs to reconstitute hematopoiesis, we competitively transplanted 100 HSCs from Cited2CTL, Cited2CTL;Mcl1, Cited2CKO, and Cited2CKO;Mcl1 mice into lethally irradiated recipient mice (Figure 4D). Significantly, while Cited2-deficient HSCs failed to reconstitute recipient mice, HSCs from Cited2CKO;Mcl1 mice successfully repopulated recipients, comparable with the reconstitution potential of HSCs from Cited2CTL and Cited2CTL;Mcl1 mice (Figures 4E and 4F). No statistically significant differences were found in repopulation potential between HSCs from Cited2CTL and Cited2CTL;Mcl1 mice (Figures 4E and 4F). Consistent with these data, HSCs from Cited2CKO;Mcl1 mice contributed significantly more to the total BM and HSC compartments of recipient mice compared with Cited2CKO HSCs, which failed to efficiently reconstitute recipients (Figure 4G). Finally, BM cells from Cited2CTL, Cited2CTL;Mcl1, Cited2CKO, and Cited2CKO;Mcl1 mice efficiently generated primary colonies (Figure 4H), and while Cited2-deficient cells failed to replate secondary colonies, strikingly, Cited2CKO;Mcl1 cells efficiently produced comparable colony numbers to control cells (Figure 4I). Therefore, Mcl1 acts downstream of Cited2 in vivo and at least in part mediates CITED2 functions in sustaining the HSC pool under steady-state hematopoiesis and promoting their long-term reconstitution capacity.
Cited2 regulates Pten to maintain the HSC pool
PTEN is required for cell-autonomous HSC maintenance (Yilmaz et al., 2006; Zhang et al., 2006) and its deletion results in loss of adult HSC function through activation of mTORC2-dependent signaling (Magee et al., 2012). Given that Pten expression was decreased in Cited2-deficient HSCs (Figure 4A), we sought to genetically interrogate the putative CITED2→PTEN axis. We took advantage of the principle that if genes act in common pathways they should genetically interact, i.e., the compound heterozygosity should generate phenotypes not observed in mice heterozygous for a single gene of interest (Vidal et al., 2011). We investigated whether combined Pten and Cited2 heterozygosity causes loss of HSC functions compared with Pten or Cited2 heterozygosity alone. We generated Cited2+/fl;Pten+/fl;Vav-iCre (Cited2Het;PtenHet), Cited2Het, PtenHet, and control mice (Figure S2A). We found that Cited2Het and PtenHet mice had normal BM cellularity and largely unaffected numbers of Lin−, LK, and LSK cells within the BM (Figures S2B and S2C). However, while control, Cited2Het, and PtenHet mice had comparable numbers of HSCs, Cited2Het;PtenHet mice displayed subtly but significantly decreased numbers of HSCs. To assess the repopulation capacity of Cited2Het;PtenHet HSCs, we competitively transplanted HSCs of all relevant genotypes into lethally irradiated recipients (Figure S2D). While HSCs of all genotypes gave equal overall long-term reconstitution capacity in the PB compartment (Figure S2E), Cited2Het;PtenHet HSCs had a slightly decreased capacity to contribute to the BM reconstitution of the recipient mice (Figure S2F). Moreover, Cited2Het;PtenHet HSCs failed to contribute to the HSC compartments of the recipients (Figure S2G). Therefore, given that Cited2Het;PtenHet HSCs sustain hematopoiesis upon transplantation, but fail to efficiently repopulate the HSC compartment of the recipients, the CITED2→PTEN axis may contribute to long-term HSC maintenance but is unlikely to be essential for the reconstitution potential of HSCs.
Cited2 deletion dysregulates multiple pathways whose strict control is required for HSC integrity
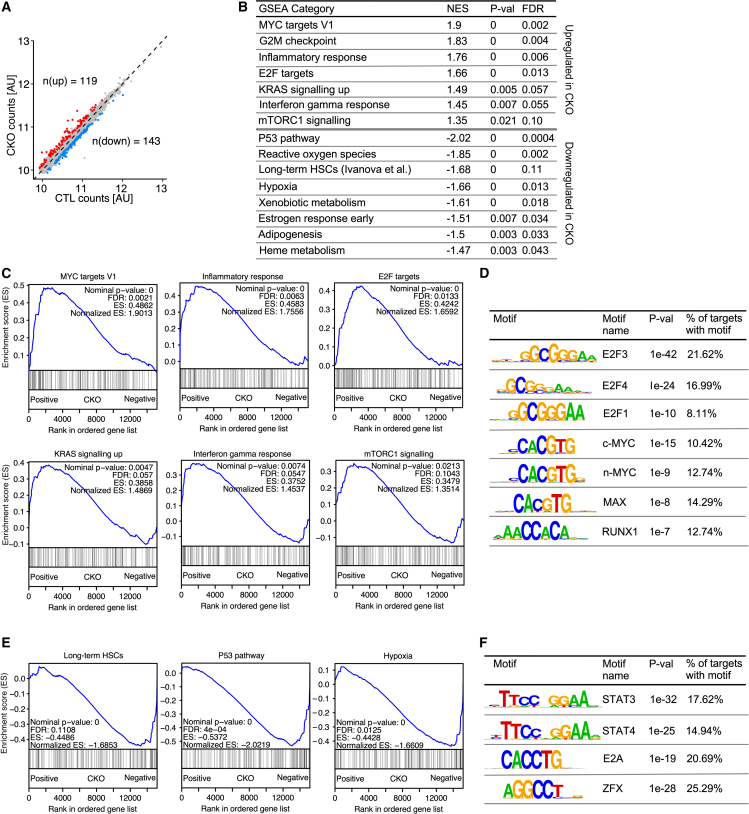
To further understand why Cited2-deficient HSCs undergo depletion and fail upon transplantation, we examined global gene expression in Cited2-deficient HSCs by RNA-seq. This analysis identified a number of dysregulated genes, with 119 upregulated and 143 downregulated genes (Figure 5A). Gene set enrichment analysis (GSEA) indicated a broad dysregulation of multiple pathways and processes, which was compatible with the loss of HSC function. Notably, we found that Cited2-deficient HSCs displayed activation of proinflammatory pathways, c-MYC and E2F targets, and K-RAS and mTORC1 signaling, whose elevated activity is known to lead to HSC exhaustion or loss of their reconstitution potential (Kim et al., 2017; Pietras, 2017; Sasine et al., 2018; Wilson et al., 2004; Yilmaz et al., 2006) (Figures 5B and 5C). Consistent with upregulation of E2F and c-MYC targets, analysis of proximal promoters of upregulated genes revealed the presence of E2F and c-MYC motifs, suggesting that CITED2 may repress E2Fs and c-MYC (Figure 5D). We also found that promoters of the upregulated genes were enriched for the motif of RUNX1 (Figure 5D), whose increased expression in adult HSCs results in loss of their reconstitution potential (Ichikawa et al., 2008).
Figure 5.
Cited2-deficient HSCs exhibit molecular signatures of functional HSC decline
(A) Expression scatterplot of Cited2CTL versus Cited2CKO HSCs from 8- to 12-week-old mice (n = 5). Transcripts significantly up- (red) and downregulated (blue) in Cited2CKO are highlighted (FDR < 0.05 and FC > 20%).
(B) GSEA showing hallmark pathways up- and downregulated in Cited2CKO HSCs.
(C) GSEA plots showing upregulated pathways in Cited2CKO HSCs.
(D) DNA motifs of transcription factors enriched in proximal promoters (from −200 to +100 bp from transcription start site) of genes upregulated in Cited2CKO HSCs.
(E) GSEA plots showing downregulated pathways in Cited2CKO HSCs.
(F) DNA motif enrichments in proximal promoters of genes that are downregulated in Cited2CKO HSCs.
Given that we observed activation of the mTORC1 signaling signature upon Cited2 deletion in HSCs (Figures 5B and 5C), we asked whether rapamycin, a known mTORC1 inhibitor (Guertin and Sabatini, 2007), can rescue the replating defect resulting from Cited2 deficiency. We serially plated BM cells from Cited2CTL and Cited2CKO mice into CFC assays, in the presence or absence of rapamycin. As expected, Cited2CTL and Cited2CKO cells generated primary colonies (Figure S3A). However, while Cited2CTL cultures efficiently produced secondary colonies, Cited2CKO cells displayed a replating defect, regardless of the presence or absence of rapamycin (Figure S3A). As such, we conclude that upregulation of the mTORC1 pathway alone in Cited2-deficient cells is insufficient to elicit the observed phenotypes. The identification and dissection of upregulated pathways upon Cited2 deletion, which act together to cause HSC depletion, merit future investigation.
We next focused on pathways that were downregulated in Cited2CKO HSCs. Interestingly, Cited2 loss led to an overall downregulation in genes whose expression is a hallmark of long-term HSC signature (Ivanova et al., 2002) (Figures 5B and 5E). Cited2 loss also led to a decreased signature of the p53 pathway (Figures 5B and 5E), whose inhibition is detrimental to HSC integrity and function (Liu et al., 2009). Cited2CKO HSCs displayed downregulation of the hypoxic signature (Figures 5B and 5E), implying their intrinsic inability to adapt to the physiologically hypoxic BM microenvironment where HSCs reside (Spencer et al., 2014). Moreover, we found that the reactive oxygen species (ROS) defense pathway was downregulated (Figure 5B). However, ROS levels were not elevated in Cited2CKO HSCs (Figure S3B), and treatment of Cited2CKO mice with the antioxidant N-acetyl-L-cysteine (NAC) did not rescue HSC depletion (Figures S3C and S3D), suggesting that ROS is unlikely to be responsible for the observed effects in HSCs lacking Cited2. Furthermore, in addition to the GSEA, our data revealed that several genes essential for HSC functions, including Prdm16, Men1, Rnh1, and Akt2 were downregulated in Cited2CKO HSCs (Figure S4) (Andina et al., 2019; Gudmundsson et al., 2020; Juntilla et al., 2010; Maillard et al., 2009). Finally, further interrogation of promoters of the downregulated genes revealed the presence of motifs for STAT3, STAT4, E2A, and ZFX transcription factors (Figure 5F), all of which are required for HSC maintenance and function (Galan-Caridad et al., 2007; Holmfeldt et al., 2016; Mantel et al., 2012; Semerad et al., 2009). Thus, CITED2 controls multiple fundamental pathways in HSCs to safeguard their integrity.
Discussion
Given that the functional role of CITED2 in HSC biology under unperturbed conditions remains poorly understood, here we conditionally deleted Cited2 specifically from the hematopoietic system. While steady-state hematopoiesis in young and aging mice lacking Cited2 was largely unaffected, HSCs were significantly depleted in young mice, at least in part via increased apoptosis, and failed to expand upon physiological aging. Importantly, these findings imply that HSCs under steady-state conditions require Cited2 to regulate the size of their pool but not to sustain long-term multilineage hematopoiesis. Furthermore, while Cited2-deficient HSCs successfully homed to the BM upon transplantation, they completely failed to reconstitute hematopoiesis, indicating that HSCs critically require Cited2 to function under stressful conditions imposed by transplantation. Finally, significantly, phenotypes elicited by Cited2 deficiency demonstrate that unperturbed multilineage hematopoiesis can be sustained long term while the HSC pool is drastically depleted, thus underscoring the remarkable plasticity of the hematopoietic system under steady-state conditions.
Previous studies using poly(I:C)-inducible Mx1-Cre concluded that CITED2 regulates HSC survival and quiescence by repressing INK4A/ARF and HIF-1alpha, respectively (Du et al., 2012; Kranc et al., 2009). However, cellular and molecular mechanisms via which CITED2 controls the HSC pool under steady-state conditions and HSC reconstitution potential upon transplantation have remained largely unexplored. Here, we found that, under steady-state conditions, Cited2-deficient HSCs maintain their quiescent state but undergo increased apoptosis. Notably, Cited2 is required for the expression of HSC survival and self-renewal regulator Mcl1 (Campbell et al., 2010b; Opferman et al., 2005), whose hematopoiesis-specific overexpression partially rescued depletion of Cited2-deficient HSCs, and their ability to reconstitute long-term multilineage hematopoiesis. We therefore propose that the CITED2→MCL-1 axis protects the integrity of the HSC pool by promoting HSC survival under the steady-state conditions and upon transplantation. Furthermore, Cited2 was necessary for normal expression of Pten, whose deletion results in a cell-autonomous HSC depletion and defective reconstitution potential (Yilmaz et al., 2006). Consistent with Pten downregulation, Cited2-deficient HSCs displayed increased signatures of PI3K/AKT and mTOR signaling, whose suppression by PTEN is essential for HSC maintenance (Lee et al., 2010; Yilmaz et al., 2006). These data are in concordance with the previous demonstration that Cited2 deletion in HSCs leads to an increased AKT activity (Du et al., 2014). Indeed, given that PTEN is a negative regulator of AKT, our results help to explain increased AKT activation upon Cited2 loss. Given that Cited2Het;PtenHet HSCs displayed normal multilineage reconstitution potential but poorly contributed to the HSC pool upon transplantation, and that rapamycin did not rescue defects resulting from Cited2 deficiency, it is likely that the CITED2→PTEN/PI3K/AKT/mTOR axis contributes to long-term HSC maintenance but is not solely critical for HSC functions.
Poly(I:C)-inducible Mx1-Cre deletion of Cited2 results in loss of quiescence of HSCs, a phenotype that is partially mediated by HIF-1alpha (Du et al., 2012, 2014). Notably, however, our data indicate that Vav-iCre-mediated deletion of Cited2 under homeostasis has no impact on HSC quiescence. Furthermore, our GSEA analyses showed that the hypoxia signature was in fact downregulated in Cited2-deficient HSCs. The discrepancies between our findings and those by Du et al. (2012, 2014) may be explained by different strategies of gene deletion, as poly(I:C) (used to induce Mx1-Cre-driven gene ablation) is known to transiently alter HSCs (i.e., induce cycling, increase frequency, and alter phenotype), unlike Vav-iCre, which is constitutively expressed, and as such more accurately allows for gene deletion under steady-state conditions. Thus, it is possible that concurrent poly(I:C) administration and Cited2 deletion in the Mx1-Cre-mediated model may exhibit exacerbated phenotypes not seen in the Vav-iCre model.
While in this study we focused on functional interrogation of the CITED2→MCL-1 and CITED2→PTEN axes, our work suggests that CITED2 is also likely to control other diverse pathways to coordinate HSC biology. Our analyses indicate that CITED2 represses multiple pathways downstream of c-MYC, E2F, and RUNX1 transcription factors and K-RAS and proinflammatory signaling pathways, whose upregulation has detrimental consequences for HSC integrity (Ichikawa et al., 2008; Kim et al., 2017; Pietras, 2017; Sasine et al., 2018; Wilson et al., 2004; Yilmaz et al., 2006). Finally, CITED2 is necessary for the expression of genes regulated by STAT3, STAT4, E2A, and ZFX transcription factors, which are essential for HSC maintenance (Galan-Caridad et al., 2007; Holmfeldt et al., 2016; Mantel et al., 2012; Semerad et al., 2009). Thus, given its dual action in gene transcription, we propose that CITED2 functions at the center of the transcriptional regulatory network to repress pathways detrimental to HSC integrity, and promote those necessary for HSC maintenance.
Experimental procedures
Mice
All mice were on the C57BL/6J genetic background. Cited2fl/fl (Kranc et al., 2009; MacDonald et al., 2008), VavP-Mcl1 (Campbell et al., 2010b), Ptenfl/fl (Yilmaz et al., 2006), and Vav-iCre (de Boer et al., 2003) mice were described previously. All transgenic and knockout mice were CD45.2+. Recipient mice were CD45.1+/CD45.2+. All experiments on animals were performed under UK Home Office authorization.
Flow cytometry
BM, spleen, and PB samples were stained and analyzed as described previously (Guitart et al., 2017). FACS analyses were performed using an LSRFortessa (BD). Cell sorting was performed on a FACSAria Fusion (BD). Data were analyzed using FlowJo.
A combination of anti-mouse antibodies purchased from BD Biosciences, BioLegend, and Life Technologies was used. The following BD Biosciences antibodies were used: Fc block (cat. no. 553142), anti-CD4; biotin conjugated (cat. no. 553649), anti-CD5; biotin conjugated (cat. no. 553019), anti-CD8a; biotin conjugated (cat. no. 553029), anti-CD11b; biotin conjugated (cat. no. 553309), anti-CD45R/B220; biotin conjugated (cat. no. 553086), anti-Ter119; biotin conjugated (cat. no. 553672), anti-Gr-1/Ly-6G/C; biotin conjugated (cat. no. 553125), anti-CD34; FITC conjugated (cat. no. 553733), and streptavidin; BV421 conjugated (cat. no. 563259). BioLegend antibodies used were anti-c-Kit/CD117; APC conjugated (cat. no. 105812), anti-c-Kit/CD117; APC-Cy7 conjugated (cat. no. 105826), anti-Sca-1; APC-Cy7 conjugated (cat. no. 122520), anti-Sca-1; PB conjugated (cat. no. 108125), anti-CD48; PE conjugated (cat. no. 103406), anti-CD150; PE-Cy7 conjugated (cat. no. 115914), anti-CD135; APC conjugated (cat. no. 135310), anti-CD135; PE conjugated (cat. no. 135305), anti-CD16/32; APC-Cy7 conjugated (cat. no. 101328), anti-CD41; APC conjugated (cat. no. 133914), anti-CD105; PE conjugated (cat. no. 120408), anti-CD127; BV421 conjugated (cat. no. 135023), anti-TER-119; FITC conjugated (cat. no. 116206), streptavidin; PerCp conjugated (cat. no. 405213), anti-CD19; APC-Cy7 conjugated (cat. no. 115530), anti-CD45R/B220; APCCy7 conjugated (cat. no. 103224), anti-CD11b; APC conjugated (cat. no. 101211), anti-Gr-1/Ly-6G/C; PE-Cy7 conjugated (cat. no. 108416), anti-CD4; PE conjugated (cat. no. 130310), anti-CD8a; PE conjugated (cat. no. 100708), anti-CD45.1; FITC conjugated (cat. no. 110706), anti-CD45.2; PB conjugated (cat. no. 109820), anti-Ki67; FITC conjugated (cat. no. 652410). Annexin V; FITC conjugated (cat. no. 640906), and 7-AAD (cat. no. 420403) were purchased from BioLegend. DAPI was purchased from Life Technologies (cat. no. D1306).
Administration of NAC
Mice received 30 mg/mL of NAC (Sigma) in drinking water for 4 weeks. The water bottles containing NAC were changed twice a week.
ROS detection
For detection of mitochondrial super oxide, 3 × 106 BM cells stained first for LSK were resuspended in X-Vivo 15 medium (without phenol red) supplemented with 10% FCS, loaded with 5 μM MitoSox red (Invitrogen) for 20 min at 37°C and analyzed using FACS.
CFC assays
CFC assays were performed using MethoCult M3434 (STEMCELL Technologies). Colonies were tallied at day 10. For CFC replating, CFC1 cells were washed with IMDM then seeded in M3434.
Transplantation assays
Lethal irradiation of CD45.1+/CD45.2+ recipient mice was achieved using a split dose of 11 Gy (two doses of 5.5 Gy administered 4 h apart) at an average rate of 0.58 Gy/min using a Cesium 137 GammaCell 40 irradiator. For transplantations 100 HSCs or 2,000 LSK sorted from BM of 8- to 10-week old adult mice mixed with 200,000 support CD45.1+ wild-type BM cells were injected into lethally irradiated CD45.1+/CD45.2+-recipient mice.
Homing assay
LSK cells sorted from CD45.2+ BM were injected into CD45.1+ lethally irradiated recipients (160,000 cells per mouse). After 18 h, recipients were sacrificed and BM CD45.2+ chimerism was analyzed by FACS.
qRT-PCR
Gene expression analyses were performed as described previously (Guitart et al., 2017). Differences in input cDNA were normalized with Actb expression.
Statistical analyses
Statistical analyses were performed using GraphPad Prism software. p values were calculated using a Mann-Whitney U test.
RNA-seq, GSEA, and DNA motif analysis
RNA-seq was performed on sorted HSCs from Cited2CKO and Cited2CTL animals (n = 5 per genotype). On average, 6,200 cells per sample were collected and 53.7 million single-ended 85-bp reads per sample were sequenced. Reads were aligned to the GRCm38 mouse genome using HISAT2 v.2.1.0 (Kim et al., 2015), and read counts were assessed per gene using the Rsubread package v.2.0.1 in R. Differential expression analysis was further performed using the Wald test in DESeq2 v.1.24, and genes were ranked according to moderated t statistics computed by DESeq2 for GSEA. The ranked gene list was compared with gene lists in the hallmark subset of the MSigDb database v.7.0 using the GSEA software tool v.3.0 (Subramanian et al., 2005).
For DNA motif analysis of proximal promoter regions, lists of up- and downregulated genes (FDR < 0.05, fold change > 20%) were mapped to gene loci using the basic set of ENCODE genomic annotation from Ensembl v.91. Next, proximal promoters were defined as the region from 200 bp upstream to 100 bp downstream of the transcription start site. Proximal promoter coordinates were shuffled within chromosomes using the bedtools shuffle tool with the –chrom flag to generate control DNA regions for motif analysis. Finally, DNA motifs overrepresented in promoters compared with control regions were identified by Homer v.4.10.
Author contributions
K.R.K., A.V.G., and H.L. designed the experiments and wrote the paper. A.V.G. and H.L. performed in vivo experiments, and data analyses and interpretation. L.N.L. and M.B. analyzed gene expression. K.J.C. and S.C. produced VavP-Mcl1 transgenic mice. A.T., J.D., A.V., A.B., C.M., E.G., C.M.-C., C.S., L.A., and J.C. helped with in vivo experiments, FACS, and data analyses. D.O., B.G., and N.P.R. provided significant expertise to this study. K.R.K. and A.V.G. contributed equally to this work.
Conflicts of interests
The authors declare no competing interests.
Acknowledgments
This research was funded by Cancer Research UK (CRUK) Senior Cancer Research Fellowship and a CRUK Program Grant awarded to K.R.K. (C29967/A14633 and C29967/A26787). K.R.K.'s laboratory is also supported by grants from The Barts Charity, The Kay Kendall Leukaemia Fund and Blood Cancer UK. A.V.G. is supported by Ligue contre le cancer. This research was also funded in part by the Wellcome (203151/Z/16/Z) and the Medical Research Council (MC_PC_17230). We thank Vladimir Benes and Jelena Pistolic (Genomics Core Facility, EMBL, Heidelberg) for performing RNA-seq.
Published: October 28, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.stemcr.2021.10.001.
Contributor Information
Amelie V. Guitart, Email: amelie.guitart@u-bordeaux.fr.
Kamil R. Kranc, Email: kamil.kranc@qmul.ac.uk.
Supplemental information
Data and code availability
The RNA-seq data are deposited in NCBI's Gene Expression Omnibus (accession no. GSE175372).
References
- Andina N.D., Sarangdhar M., Tardivel A., Bombaci G., Hallal M., Keller I., Allam R. Higher vertebrate specific gene ribonuclease inhibitor (RNH1) is essential for adult hematopoietic stem cell function and cell cycle regulation. Blood. 2019;134:273. [Google Scholar]
- Bamforth S.D., Braganca J., Eloranta J.J., Murdoch J.N., Marques F.I., Kranc K.R., Farza H., Henderson D.J., Hurst H.C., Bhattacharya S. Cardiac malformations, adrenal agenesis, neural crest defects and exencephaly in mice lacking Cited2, a new Tfap2 co-activator. Nat. Genet. 2001;29:469–474. doi: 10.1038/ng768. [DOI] [PubMed] [Google Scholar]
- Bamforth S.D., Braganca J., Farthing C.R., Schneider J.E., Broadbent C., Michell A.C., Clarke K., Neubauer S., Norris D., Brown N.A., et al. Cited2 controls left-right patterning and heart development through a Nodal-Pitx2c pathway. Nat. Genet. 2004;36:1189–1196. doi: 10.1038/ng1446. [DOI] [PubMed] [Google Scholar]
- Bhattacharya S., Michels C.L., Leung M.K., Arany Z.P., Kung A.L., Livingston D.M. Functional role of p35srj, a novel p300/CBP binding protein, during transactivation by HIF-1. Genes Dev. 1999;13:64–75. doi: 10.1101/gad.13.1.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell C.J., Lee J.B., Levadoux-Martin M., Wynder T., Xenocostas A., Leber B., Bhatia M. The human stem cell hierarchy is defined by a functional dependence on Mcl-1 for self-renewal capacity. Blood. 2010;116:1433–1442. doi: 10.1182/blood-2009-12-258095. [DOI] [PubMed] [Google Scholar]
- Campbell K.J., Bath M.L., Turner M.L., Vandenberg C.J., Bouillet P., Metcalf D., Scott C.L., Cory S. Elevated Mcl-1 perturbs lymphopoiesis, promotes transformation of hematopoietic stem/progenitor cells, and enhances drug resistance. Blood. 2010;116:3197–3207. doi: 10.1182/blood-2010-04-281071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Haviernik P., Bunting K.D., Yang Y.C. Cited2 is required for normal hematopoiesis in the murine fetal liver. Blood. 2007;110:2889–2898. doi: 10.1182/blood-2007-01-066316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou Y.T., Hsieh C.H., Chiou S.H., Hsu C.F., Kao Y.R., Lee C.C., Chung C.H., Wang Y.H., Hsu H.S., Pang S.T., et al. CITED2 functions as a molecular switch of cytokine-induced proliferation and quiescence. Cell Death Differ. 2012;19:2015–2028. doi: 10.1038/cdd.2012.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou Y.T., Yang Y.C. Post-transcriptional control of Cited2 by transforming growth factor beta. Regulation via Smads and Cited2 coding region. J. Biol. Chem. 2006;281:18451–18462. doi: 10.1074/jbc.M601720200. [DOI] [PubMed] [Google Scholar]
- Dahlin J.S., Hamey F.K., Pijuan-Sala B., Shepherd M., Lau W.W.Y., Nestorowa S., Weinreb C., Wolock S., Hannah R., Diamanti E., et al. A single-cell hematopoietic landscape resolves 8 lineage trajectories and defects in Kit mutant mice. Blood. 2018;131:e1–e11. doi: 10.1182/blood-2017-12-821413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Boer J., Williams A., Skavdis G., Harker N., Coles M., Tolaini M., Norton T., Williams K., Roderick K., Potocnik A.J., Kioussis D. Transgenic mice with hematopoietic and lymphoid specific expression of Cre. Eur. J. Immunol. 2003;33:314–325. doi: 10.1002/immu.200310005. [DOI] [PubMed] [Google Scholar]
- Du J., Chen Y., Li Q., Han X., Cheng C., Wang Z., Danielpour D., Dunwoodie S.L., Bunting K.D., Yang Y.C. HIF-1alpha deletion partially rescues defects of hematopoietic stem cell quiescence caused by Cited2 deficiency. Blood. 2012;119:2789–2798. doi: 10.1182/blood-2011-10-387902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du J., Li Q., Tang F., Puchowitz M.A., Fujioka H., Dunwoodie S.L., Danielpour D., Yang Y.C. Cited2 is required for the maintenance of glycolytic metabolism in adult hematopoietic stem cells. Stem Cells Dev. 2014;23:83–94. doi: 10.1089/scd.2013.0370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandes M.T., Calado S.M., Mendes-Silva L., Braganca J. CITED2 and the modulation of the hypoxic response in cancer. World J. Clin. Oncol. 2020;11:260–274. doi: 10.5306/wjco.v11.i5.260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galan-Caridad J.M., Harel S., Arenzana T.L., Hou Z.E., Doetsch F.K., Mirny L.A., Reizis B. Zfx controls the self-renewal of embryonic and hematopoietic stem cells. Cell. 2007;129:345–357. doi: 10.1016/j.cell.2007.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiger H., de Haan G., Florian M.C. The ageing haematopoietic stem cell compartment. Nat. Rev. Immunol. 2013;13:376–389. doi: 10.1038/nri3433. [DOI] [PubMed] [Google Scholar]
- Gudmundsson K.O., Nguyen N., Oakley K., Han Y., Gudmundsdottir B., Liu P., Tessarollo L., Jenkins N.A., Copeland N.G., Du Y. Prdm16 is a critical regulator of adult long-term hematopoietic stem cell quiescence. Proc. Natl. Acad. Sci. U S A. 2020;117:31945–31953. doi: 10.1073/pnas.2017626117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guertin D.A., Sabatini D.M. Defining the role of mTOR in cancer. Cancer Cell. 2007;12:9–22. doi: 10.1016/j.ccr.2007.05.008. [DOI] [PubMed] [Google Scholar]
- Guitart A.V., Panagopoulou T.I., Villacreces A., Vukovic M., Sepulveda C., Allen L., Carter R.N., van de Lagemaat L.N., Morgan M., Giles P., et al. Fumarate hydratase is a critical metabolic regulator of hematopoietic stem cell functions. J. Exp. Med. 2017;214:719–735. doi: 10.1084/jem.20161087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmfeldt P., Ganuza M., Marathe H., He B., Hall T., Kang G., Moen J., Pardieck J., Saulsberry A.C., Cico A., et al. Functional screen identifies regulators of murine hematopoietic stem cell repopulation. J. Exp. Med. 2016;213:433–449. doi: 10.1084/jem.20150806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichikawa M., Goyama S., Asai T., Kawazu M., Nakagawa M., Takeshita M., Chiba S., Ogawa S., Kurokawa M. AML1/Runx1 negatively regulates quiescent hematopoietic stem cells in adult hematopoiesis. J. Immunol. 2008;180:4402–4408. doi: 10.4049/jimmunol.180.7.4402. [DOI] [PubMed] [Google Scholar]
- Ivanova N.B., Dimos J.T., Schaniel C., Hackney J.A., Moore K.A., Lemischka I.R. A stem cell molecular signature. Science. 2002;298:601–604. doi: 10.1126/science.1073823. [DOI] [PubMed] [Google Scholar]
- Juntilla M.M., Patil V.D., Calamito M., Joshi R.P., Birnbaum M.J., Koretzky G.A. AKT1 and AKT2 maintain hematopoietic stem cell function by regulating reactive oxygen species. Blood. 2010;115:4030–4038. doi: 10.1182/blood-2009-09-241000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D., Langmead B., Salzberg S.L. HISAT: a fast spliced aligner with low memory requirements. Nat. Methods. 2015;12:357–360. doi: 10.1038/nmeth.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim E., Cheng Y., Bolton-Gillespie E., Cai X., Ma C., Tarangelo A., Le L., Jambhekar M., Raman P., Hayer K.E., et al. Rb family proteins enforce the homeostasis of quiescent hematopoietic stem cells by repressing Socs3 expression. J. Exp. Med. 2017;214:1901–1912. doi: 10.1084/jem.20160719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim G.D., Das R., Rao X., Zhong J., Deiuliis J.A., Ramirez-Bergeron D.L., Rajagopalan S., Mahabeleshwar G.H. CITED2 restrains proinflammatory macrophage activation and response. Mol. Cell Biol. 2018;38 doi: 10.1128/MCB.00452-17. e00452–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korthuis P.M., Berger G., Bakker B., Rozenveld-Geugien M., Jaques J., de Haan G., Schuringa J.J., Vellenga E., Schepers H. CITED2-mediated human hematopoietic stem cell maintenance is critical for acute myeloid leukemia. Leukemia. 2015;29:625–635. doi: 10.1038/leu.2014.259. [DOI] [PubMed] [Google Scholar]
- Kranc K.R., Bamforth S.D., Braganca J., Norbury C., van Lohuizen M., Bhattacharya S. Transcriptional coactivator Cited2 induces Bmi1 and Mel18 and controls fibroblast proliferation via Ink4a/ARF. Mol. Cell Biol. 2003;23:7658–7666. doi: 10.1128/MCB.23.21.7658-7666.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kranc K.R., Oliveira D.V., Armesilla-Diaz A., Pacheco-Leyva I., Catarina Matias A., Luisa Escapa A., Subramani C., Wheadon H., Trindade M., Nichols J., et al. Acute loss of Cited2 impairs Nanog expression and decreases self-renewal of mouse embryonic stem cells. Stem Cells. 2015;33:699–712. doi: 10.1002/stem.1889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kranc K.R., Schepers H., Rodrigues N.P., Bamforth S., Villadsen E., Ferry H., Bouriez-Jones T., Sigvardsson M., Bhattacharya S., Jacobsen S.E., Enver T. Cited2 is an essential regulator of adult hematopoietic stem cells. Cell Stem Cell. 2009;5:659–665. doi: 10.1016/j.stem.2009.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawson H., Sepulveda C., van de Lagemaat L.N., Durko J., Barile M., Tavosanis A., Georges E., Shmakova A., Timms P., Carter R.N., et al. JMJD6 promotes self-renewal and regenerative capacity of hematopoietic stem cells. Blood Adv. 2021;5:889–899. doi: 10.1182/bloodadvances.2020002702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.Y., Nakada D., Yilmaz O.H., Tothova Z., Joseph N.M., Lim M.S., Gilliland D.G., Morrison S.J. mTOR activation induces tumor suppressors that inhibit leukemogenesis and deplete hematopoietic stem cells after Pten deletion. Cell Stem Cell. 2010;7:593–605. doi: 10.1016/j.stem.2010.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.Y., Taub P.J., Wang L., Clark A., Zhu L.L., Maharam E.R., Leong D.J., Ramcharan M., Li Z., Liu Z., et al. Identification of CITED2 as a negative regulator of fracture healing. Biochem. Biophys. Res. Commun. 2009;387:641–645. doi: 10.1016/j.bbrc.2009.07.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q., Ramirez-Bergeron D.L., Dunwoodie S.L., Yang Y.C. Cited2 gene controls pluripotency and cardiomyocyte differentiation of murine embryonic stem cells through Oct4 gene. J. Biol. Chem. 2012;287:29088–29100. doi: 10.1074/jbc.M112.378034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L., He Z., Xu L., Lu L., Feng H., Leong D.J., Kim S.J., Hirsh D.M., Majeska R.J., Goldring M.B., et al. CITED2 mediates the mechanical loading-induced suppression of adipokines in the infrapatellar fat pad. Ann. N. Y Acad. Sci. 2019;1442:153–164. doi: 10.1111/nyas.14025. [DOI] [PubMed] [Google Scholar]
- Liu Y., Elf S.E., Miyata Y., Sashida G., Liu Y., Huang G., Di Giandomenico S., Lee J.M., Deblasio A., Menendez S., et al. p53 regulates hematopoietic stem cell quiescence. Cell Stem Cell. 2009;4:37–48. doi: 10.1016/j.stem.2008.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacDonald S.T., Bamforth S.D., Chen C.M., Farthing C.R., Franklyn A., Broadbent C., Schneider J.E., Saga Y., Lewandoski M., Bhattacharya S. Epiblastic Cited2 deficiency results in cardiac phenotypic heterogeneity and provides a mechanism for haploinsufficiency. Cardiovasc. Res. 2008;79:448–457. doi: 10.1093/cvr/cvn101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magee J.A., Ikenoue T., Nakada D., Lee J.Y., Guan K.L., Morrison S.J. Temporal changes in PTEN and mTORC2 regulation of hematopoietic stem cell self-renewal and leukemia suppression. Cell Stem Cell. 2012;11:415–428. doi: 10.1016/j.stem.2012.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maillard I., Chen Y.X., Friedman A., Yang Y., Tubbs A.T., Shestova O., Pear W.S., Hua X. Menin regulates the function of hematopoietic stem cells and lymphoid progenitors. Blood. 2009;113:1661–1669. doi: 10.1182/blood-2009-01-135012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mantel C., Messina-Graham S., Moh A., Cooper S., Hangoc G., Fu X.Y., Broxmeyer H.E. Mouse hematopoietic cell-targeted STAT3 deletion: stem/progenitor cell defects, mitochondrial dysfunction, ROS overproduction, and a rapid aging-like phenotype. Blood. 2012;120:2589–2599. doi: 10.1182/blood-2012-01-404004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCracken M.N., George B.M., Kao K.S., Marjon K.D., Raveh T., Weissman I.L. Normal and neoplastic stem cells. Cold Spring Harb Symp. Quant Biol. 2016;81:1–9. doi: 10.1101/sqb.2016.81.030965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menendez-Gonzalez J.B., Vukovic M., Abdelfattah A., Saleh L., Almotiri A., Thomas L.A., Agirre-Lizaso A., Azevedo A., Menezes A.C., Tornillo G., et al. Gata2 as a crucial regulator of stem cells in adult hematopoiesis and acute myeloid leukemia. Stem Cell Rep. 2019;13:291–306. doi: 10.1016/j.stemcr.2019.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nestorowa S., Hamey F.K., Pijuan Sala B., Diamanti E., Shepherd M., Laurenti E., Wilson N.K., Kent D.G., Gottgens B. A single-cell resolution map of mouse hematopoietic stem and progenitor cell differentiation. Blood. 2016;128 doi: 10.1182/blood-2016-05-716480. e20–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Opferman J.T., Iwasaki H., Ong C.C., Suh H., Mizuno S., Akashi K., Korsmeyer S.J. Obligate role of anti-apoptotic MCL-1 in the survival of hematopoietic stem cells. Science. 2005;307:1101–1104. doi: 10.1126/science.1106114. [DOI] [PubMed] [Google Scholar]
- Pietras E.M. Inflammation: a key regulator of hematopoietic stem cell fate in health and disease. Blood. 2017;130:1693–1698. doi: 10.1182/blood-2017-06-780882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi L., Lin K.K., Boles N.C., Yang L., King K.Y., Jeong M., Mayle A., Goodell M.A. Less is more: unveiling the functional core of hematopoietic stem cells through knockout mice. Cell Stem Cell. 2012;11:302–317. doi: 10.1016/j.stem.2012.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasine J.P., Himburg H.A., Termini C.M., Roos M., Tran E., Zhao L., Kan J., Li M., Zhang Y., de Barros S.C., et al. Wild-type Kras expands and exhausts hematopoietic stem cells. JCI Insight. 2018;3:e98197. doi: 10.1172/jci.insight.98197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semerad C.L., Mercer E.M., Inlay M.A., Weissman I.L., Murre C. E2A proteins maintain the hematopoietic stem cell pool and promote the maturation of myelolymphoid and myeloerythroid progenitors. Proc. Natl. Acad. Sci. U S A. 2009;106:1930–1935. doi: 10.1073/pnas.0808866106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sender R., Milo R. The distribution of cellular turnover in the human body. Nat. Med. 2021;27:45–48. doi: 10.1038/s41591-020-01182-9. [DOI] [PubMed] [Google Scholar]
- Spencer J.A., Ferraro F., Roussakis E., Klein A., Wu J., Runnels J.M., Zaher W., Mortensen L.J., Alt C., Turcotte R., et al. Direct measurement of local oxygen concentration in the bone marrow of live animals. Nature. 2014;508:269–273. doi: 10.1038/nature13034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A., Paulovich A., Pomeroy S.L., Golub T.R., Lander E.S., Mesirov J.P. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U S A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tien E.S., Davis J.W., Vanden Heuvel J.P. Identification of the CREB-binding protein/p300-interacting protein CITED2 as a peroxisome proliferator-activated receptor alpha coregulator. J. Biol. Chem. 2004;279:24053–24063. doi: 10.1074/jbc.M401489200. [DOI] [PubMed] [Google Scholar]
- Vidal M., Cusick M.E., Barabasi A.L. Interactome networks and human disease. Cell. 2011;144:986–998. doi: 10.1016/j.cell.2011.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y.H., Israelsen W.J., Lee D., Yu V.W., Jeanson N.T., Clish C.B., Cantley L.C., Vander Heiden M.G., Scadden D.T. Cell-state-specific metabolic dependency in hematopoiesis and leukemogenesis. Cell. 2014;158:1309–1323. doi: 10.1016/j.cell.2014.07.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weninger W.J., Floro K.L., Bennett M.B., Withington S.L., Preis J.I., Barbera J.P., Mohun T.J., Dunwoodie S.L. Cited2 is required both for heart morphogenesis and establishment of the left-right axis in mouse development. Development. 2005;132:1337–1348. doi: 10.1242/dev.01696. [DOI] [PubMed] [Google Scholar]
- Wilson A., Murphy M.J., Oskarsson T., Kaloulis K., Bettess M.D., Oser G.M., Pasche A.C., Knabenhans C., Macdonald H.R., Trumpp A. c-Myc controls the balance between hematopoietic stem cell self-renewal and differentiation. Genes Dev. 2004;18:2747–2763. doi: 10.1101/gad.313104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Withington S.L., Scott A.N., Saunders D.N., Lopes Floro K., Preis J.I., Michalicek J., Maclean K., Sparrow D.B., Barbera J.P., Dunwoodie S.L. Loss of Cited2 affects trophoblast formation and vascularization of the mouse placenta. Dev. Biol. 2006;294:67–82. doi: 10.1016/j.ydbio.2006.02.025. [DOI] [PubMed] [Google Scholar]
- Yilmaz O.H., Valdez R., Theisen B.K., Guo W., Ferguson D.O., Wu H., Morrison S.J. Pten dependence distinguishes haematopoietic stem cells from leukaemia-initiating cells. Nature. 2006;441:475–482. doi: 10.1038/nature04703. [DOI] [PubMed] [Google Scholar]
- Yin Z., Haynie J., Yang X., Han B., Kiatchoosakun S., Restivo J., Yuan S., Prabhakar N.R., Herrup K., Conlon R.A., et al. The essential role of Cited2, a negative regulator for HIF-1alpha, in heart development and neurulation. Proc. Natl. Acad. Sci. U S A. 2002;99:10488–10493. doi: 10.1073/pnas.162371799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Grindley J.C., Yin T., Jayasinghe S., He X.C., Ross J.T., Haug J.S., Rupp D., Porter-Westpfahl K.S., Wiedemann L.M., et al. PTEN maintains haematopoietic stem cells and acts in lineage choice and leukaemia prevention. Nature. 2006;441:518–522. doi: 10.1038/nature04747. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The RNA-seq data are deposited in NCBI's Gene Expression Omnibus (accession no. GSE175372).