Abstract
The regulatory factor X (RFX) complex, which contains RFXANK(B), RFXAP, and RFX5, binds to X and S boxes in major histocompatibility complex class II (MHC II) promoters. In the bare lymphocyte syndrome (BLS), which is a human severe combined immunodeficiency, MHC II promoters are neither occupied nor transcribed. Thus, the absence of any one subunit prevents the formation of the RFX complex. Nevertheless, except for a weak binding between RFX5 and RFXAP, no other interactions between RFX proteins have been described. In this study, we demonstrate that RFXANK(B) binds to RFXAP to form a scaffold for the assembly of the RFX complex, which then binds to DNA. Moreover, mutant RFXANK(B) and RFXAP proteins from complementation groups B and D of BLS, respectively, cannot support this interaction. Our data elucidate an intriguing medical situation, where a genetic disease targets two different surfaces that are required for the nucleation of a multisubunit DNA-protein complex.
By presenting processed antigens to CD4+ lymphocytes, major histocompatibility complex class II (MHC II) determinants play a critical role in the immune response (6). They not only are expressed constitutively on thymic epithelial cells, mature B lymphocytes, and dendritic cells but can be induced on many other cells by gamma interferon (2–4, 11, 18). Three different MHC II isotypes are found in humans. They are called the human leukocyte antigens DR, DP, and DQ and form heterodimers of α and β chains (6). The expression of MHC II genes is regulated principally at the level of transcription (2–4, 11, 18). MHC II genes and genes involved in antigen processing and presentation (invariant chain, Ii; DMA and DMB) are transcribed from compact promoters containing conserved upstream sequences (CUS) from positions −135 to −60 (DRA promoter) and variable proximal sequences that lack a TATA box (2–4, 11, 14, 18). CUS have been subdivided further into S, pyrimidine tract, X, X2, and Y boxes, which interact with many different proteins and protein complexes that mediate constitutive and inducible expression of MHC II genes (2–4, 11, 14, 18).
These complexes are composed of trans-acting factors. The regulatory factor X (RFX) complex binds to X and S boxes. It is composed of three subunits. RFX5 is a 75-kDa protein that contains a DNA-binding domain (25). RFXAP is a 41-kDa protein that interacts weakly with RFX5 in vivo (8). The third subunit, RFXANK or RFX(B) (henceforth RFXANK), is a 33-kDa protein that contains three ankyrin repeats (20, 21). Genes that code for these proteins are mutated in complementation groups B (RFXANK), C (RFX5), and D (RFXAP) of the bare lymphocyte syndrome (BLS), which is an autosomal recessive immunodeficiency characterized by the congenital absence of MHC II molecules on B cells (18). Complementation group A is caused by mutations in the class II transactivator (CIITA) (26). The mutated gene in the complementation group E is still unknown but is expected to code for a protein involved in the organization of chromatin (7).
Complementation group B, where a functional RFXANK protein is not expressed, contains 15 affected individuals (17). The mutation has been mapped precisely in four patients. In three patients (represented by cell lines Abdullah, Nacera, and Bequit), a deletion of 26 nucleotides (nt) results in the loss of the splice acceptor site and the first nucleotide of exon 6. mRNA from the fourth patient (BLS-1) contains a 58-nt deletion that removes the last 23 nt and the splice donor site of exon 6 (20, 21). Until recently, complementation group D was represented only by the 6.1.6 cell line, which was generated in vitro (13). However, eight patients from six unrelated families were identified later and found to have mutated RFXAP genes (9, 27).
Despite extensive investigations, the assembly of the RFX complex remains a mystery. Only a weak interaction between RFX5 and RFXAP has been reported (8). In this study, we asked whether additional and specific binding could be observed between these two proteins and the newly identified RFXANK. To this end, we expressed wild-type and mutant RFX proteins in vitro and in vivo and performed structural and functional studies. Indeed, a specific and direct interaction could be demonstrated between RFXANK and RFXAP, which was abrogated in complementation groups B and D of BLS. Studies of protein-protein and DNA-protein interactions also revealed that RFXANK and RFXAP nucleate the RFX complex in the absence of DNA.
MATERIALS AND METHODS
Cell culture.
6.1.6 is an immunoselected clonal variant of the T5-1 B-cell line, which does not express MHC class II determinants due to a mutation in the RFXAP gene (8, 13). 6.1.6 cells were maintained in RPMI 1640 medium, and COS cells were grown in Dulbecco's modified Eagle's medium. Media were supplemented with 10% fetal bovine serum, 100 mM l-glutamine, and 50 μg each of penicillin and streptomycin per ml.
Plasmid constructions.
RFXANK cDNA was generated by PCR (forward primer 5′-ATGGAGCTTACCCAGCCTGCA-3′; reverse primer 5′-TCACTCAGGGTCAGCGGGCAC-3′) using a B-cell cDNA library as a template. The amplified product was ligated in pCR3.1 TA vector (Invitrogen, Carlsbad, Calif.) and confirmed by DNA sequencing. RFXANK was excised from pCR3.1 and ligated into a BamHI-EcoRI-cleaved pcDNA3.1 vector (Invitrogen) and modified pEFBOS vectors in frame with an N-terminal Myc epitope tag (1). Mutant RFXANK (mutRFXANK) cDNA was acquired from the cell line Bequit by reverse transcription (RT)-PCR amplification and was a generous gift from Jeremy Boss (21). Glutathione S-transferase (GST)–RFXANK, GST-mutRFXANK, and a GST fusion with the first 90 residues of RFXANK were generated by ligating various RFXANK cDNAs in frame with the coding region of the GST gene in pGEX-2TK (Amersham-Pharmacia Biotech, Piscataway, N.J.). The chimeric Gal4-RFXANK protein was engineered by fusing the RFXANK cDNA in frame with the Gal4 DNA-binding domain (residues from positions 1 to 147) in pSG424 (23). The wild-type RFXAP and various mutant RFXAP proteins [RFXAP(69-272), RFXAP(1-151), and RFXAP(1-243)] were expressed from cDNAs that were PCR amplified from pT7T3-RFXAP (J. D. Fontes, unpublished data) and the primer pairs F (5′-CGTTCGGGATCCGCCACCATGGAGGCGCAGGGTGTA-3′)-R (5′-ACGTATGAATTCTCACATTGATGTTCCTGG-3′), F69 (5′-CGTCGTGGATCCGCCACCATGAAGCCCGTTAGGTACCTG-3′)-R, F-R151 (5′-CGTCGTGAATTCCTAGCTCGTGGTCTCGCTGCA-3′), and F-R243 (5′-CGTTGCGAATTCCTATTCTGGACTTCTTAGTAA-3′), respectively. The amplified products were ligated into BamHI-EcoRI-cleaved pcDNA3.1 vector. Hemagglutinin (HA) epitope-tagged wild-type RFXAP protein was generated by PCR and inserted into BamHI-EcoRI sites of the modified pEFBOS vector. HA epitope-tagged RFXAP and mutant RFXAP(1-243) cDNAs were ligated into EcoRI-HindIII and HindIII-XbaI sites of pSVSPORT1 (Invitrogen), respectively. RFX5 cDNA was introduced into HindIII-XbaI sites of pcDNA3.1. All cDNAs were confirmed by DNA sequencing. The Myc epitope-tagged full-length RFXAP and mutant RFXAP(1-243) cDNAs were ligated into pSV12 vector (Fontes, unpublished data) in frame with the activation domain (residues from positions 413 to 490) of VP16 from the herpes simplex virus (5), located 5′ from the multiple cloning site. Gal4-VP16, RFX5-VP16, and pG5bCAT plasmid constructs were described previously (12, 15, 22).
Transient transfections and CAT assays.
For chloramphenicol acetyltransferase (CAT) enzymatic assays, COS cells were seeded into 50-mm-diameter petri dishes 18 h prior to transfection. Cells were transfected with a total of 2 μg of plasmid DNA using Lipofectamine reagent as instructed by the manufacturer (Gibco-BRL, Rockville, Md.). For immunoprecipitations, COS cells were seeded into 100-mm-diameter petri dishes and transfected with a total of 6 μg of plasmid DNA. 6.1.6 cells were transfected by electroporation as previously described (15) with a total of 45 μg of plasmid DNA. A cytomegalovirus–β-galactosidase reporter plasmid (Gibco-BRL) was used to monitor transfection efficiency. Cells were harvested 72 h posttransfection, and CAT activity was analyzed as described elsewhere (16).
Immunoprecipitation and Western blotting.
At 48 h posttransfection, COS cells were harvested in 1 ml of lysis buffer (1% [vol/vol] NP-40, 10 mM Tris-HCl [pH 7.4], 150 mM NaCl, 2 mM EDTA, 0.1% protease inhibitors) for 45 min at 4°C, and the amounts of the solubilized proteins were measured (bicinchoninic acid protein assay; Pierce, Rockford, Ill.). Protein A-Sepharose (Amersham-Pharmacia Biotech)-precleared proteins were subjected to immunoprecipitation using a rabbit polyclonal anti-c-Myc antibody (A-14; Santa Cruz Biotechnology, Santa Cruz, Calif.). Immune complexes were recovered by binding to protein A-Sepharose beads during the overnight rotation at 4°C, resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) on an SDS–10% polyacrylamide gel and transferred to nitrocellulose membranes by a semidry technique. The membranes were immunostained with a mouse monoclonal anti-HA antibody (1:2,000; Boehringer Mannheim, Indianapolis, Ind.) followed by horseradish peroxidase-conjugated goat anti-mouse immunoglobulin G secondary antibody (1:2,000; Gibco-BRL, Rockville, Md.). Blots were developed by chemiluminescence assay (NEN Life Science Products, Boston, Mass.).
In vitro transcription and translation.
Plasmids containing RFXANK, mutRFXANK, RFXAP, RFX5, RFXAP(69-272), RFXAP(1-151), and RFXAP(1-243) cDNAs were transcribed and translated in vitro using the TnT T7 coupled reticulocyte lysate system (Promega, Madison, Wis.) as instructed by the manufacturer in the presence or absence of 35S-labeled cysteine (NEN).
In vitro binding assays.
GST fusion proteins were produced in Escherichia coli BL21(DE3)pLysS competent cells (Novagen, Madison, Wis.) during 4 h of induction with 1 mM isopropyl-β-d-thiogalactopyranoside and purified from the total cell lysates with glutathione-Sepharose beads (Amersham-Pharmacia Biotech). For the GST pull-down assay, 10 μg of GST or GST fusion protein was mixed with 10 μl of in vitro-translated proteins in 300 μl of binding buffer (50 mM Tris-HCl [pH 8.0], 5% glycerol, 0.5 mM EDTA, 5 mM MgCl2, 1% bovine serum albumin, 500 mM NaCl, 1% Triton X-100, 0.5% NP-40). After 4 h at 4°C, GST-coupled beads were washed five times with binding buffer. Bound proteins were eluted by boiling in the SDS sample buffer. Proteins were resolved by SDS-PAGE on a 10% gel and revealed by autoradiography.
Electrophoretic mobility shift assay (EMSA).
The following oligonucleotides were used in this study. The SX oligonucleotide contains sequences from positions −144 to −69 in the DRA promoter. The SRE oligonucleotide contains the c-fos serum response element (19). Oligonucleotides were prepared by annealing of two synthesized, complementary strands as described before (15). Binding buffer contained 12% glycerol, 12 mM HEPES (pH 7.9), 60 mM KCl, 5 mM MgCl2, 0.12 mM EDTA, 0.3 mM phenylmethylsulfonyl fluoride, and 0.3 mM dithiothreitol. Each reaction mixture contained 1 to 2 μg of salmon sperm DNA, 10,000 to 20,000 cpm of 32P-labeled SX oligonucleotide, and 3 μl of each protein. Proteins were incubated for 5 min at 4°C in the presence or absence of competitor oligonucleotide before 32P-labeled SX oligonucleotides were added. Binding was then allowed to proceed for 30 min at room temperature. DNA-protein complexes were separated on a 4% nondenaturing polyacrylamide gel, which was run in 0.25× Tris-borate-EDTA buffer for 3 h at 4°C and at 200 V. Gels were then dried and analyzed by autoradiography.
RESULTS
RFXAP, but not RFX5, binds to RFXANK in vitro.
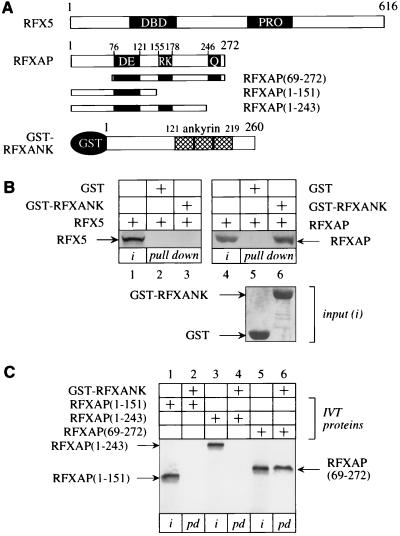
To examine direct protein-protein interactions within the RFX complex, the GST-RFXANK fusion protein was expressed in E. coli, and wild-type RFXAP and RFX5 proteins were transcribed and translated in vitro using the rabbit reticulocyte lysate (Fig. 1A). First, the GST-RFXANK fusion protein was tested for its ability to rescue RFXANK-deficient Bequit cells, where it restored the expression of MHC II determinants as analyzed by fluorescence-activated cell sorting (data not shown). Second, RFX5 and RFXAP were combined with the GST-RFXANK fusion protein in a GST pull-down assay (Fig. 1B). Under stringent conditions, RFX5 bound neither to GST alone nor to the GST-RFXANK fusion protein (Fig. 1B, lanes 2 and 3). In sharp contrast, RFXAP bound to the GST- RFXANK fusion protein but did not interact with GST alone, demonstrating that the interaction between RFXANK and RFXAP was specific (Fig. 1B, compare lanes 5 and 6). The input amounts of all proteins were equivalent (Fig. 1B, lanes 1 and 4 and bottom panel). We conclude that RFXANK binds to RFXAP in vitro.
FIG. 1.
Specific binding between RFXANK and RFXAP in vitro. (A) Schematic representation of the proteins used in the GST pull-down assay. RFX5 contains 616 residues and two well-defined domains. The DNA-binding (DBD) and the proline-rich (PRO) domains are depicted as black rectangles. RFXAP contains 272 residues and three structural domains, which are rich in acidic (DE), basic (RK), and glutamine (Q) residues. Mutant RFXAP(69-272) protein is truncated at its N terminus. Mutant RFXAP(1-151) and RFXAP(1-243) proteins are truncated at their C termini, where both glutamine and basic or just the glutamine region, respectively, was deleted. The GST-RFXANK fusion protein links GST to 260 residues from RFXANK, which contains three ankyrin repeats (striped bars). (B) RFXANK binds to RFXAP but not to RFX5 in vitro. 35S-labeled RFX5 (left) and RFXAP (right) proteins were incubated with GST alone or with the GST-RFXANK fusion protein and selected on glutathione-Sepharose beads. Bound proteins were separated by SDS-PAGE and revealed by autoradiography. Lanes 1 and 4, 25% of input (i) labeled proteins; lanes 2, 3, 5, and 6, results of binding assay. Pluses above the autoradiographs indicate the presence of different proteins in the assay; 25% of the input (i) GST alone (lane 1) and the GST-RFXANK fusion protein (lane 2) were equivalent and are presented in a Coomassie blue-stained SDS-polyacrylamide gel [input (i)]. (C) The C terminus of RFXAP is required for the binding to RFXANK in vitro. Three different labeled mutant in vitro-translated (IVT) RFXAP proteins (A) were incubated with the GST-RFXANK fusion protein and analyzed as described above. Lanes 1, 3, and 5, 25% of the input (i) mutant RFXAP proteins; lanes 2, 4, and 6; pull-down (pd) results. Gels are labeled as in panel B.
To determine which part of RFXAP could interact with RFXANK, several deletion mutants of RFXAP (Fig. 1A) were transcribed and translated in vitro using the rabbit reticulocyte lysate and examined for the ability to bind to the GST-RFXANK fusion protein. Mutant RFXAP(1-151) and RFXAP(1-243) proteins, which contained acidic or acidic and basic domains, respectively, did not interact with the GST-RFXANK fusion protein (Fig. 1C, lanes 2 and 4). Only the mutant RFXAP(69-272) protein, which retained the glutamine-rich domain and the C terminus of RFXAP (Fig. 1A), bound to the GST-RFXANK fusion protein (Fig. 1C, lane 6). This interaction was determined by comparing the amount of the bound protein with the total amount of the mutant RFXAP(69-272) protein in the reaction (Fig. 1C, compare lanes 5 and 6). Thus, the C terminus of RFXAP, which includes the glutamine-rich domain, binds to RFXANK.
RFXANK and RFXAP bind to each other in vivo.
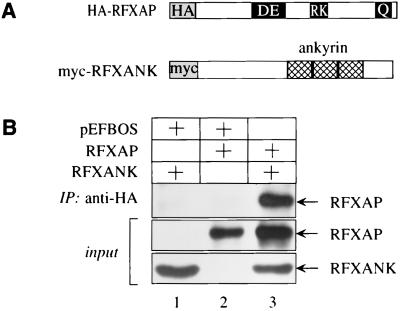
To examine whether the interaction between RFXANK and RFXAP could also be observed in vivo, these proteins were coexpressed in cells. COS cells were transfected with plasmids which directed expression of the N-terminally Myc epitope-tagged RFXANK protein, N-terminally HA epitope-tagged RFXAP protein, or both (Fig. 2A). Total cell lysates were incubated with the anti-Myc antibody, and immunoprecipitates were examined for the presence of RFXAP by Western blotting with the anti-HA antibody; 10% of total cell lysates were also examined for the expression of both proteins (Fig. 2B, input). The immunoprecipitation was first optimized for the specific interaction being studied. When either of the two plasmids alone was transfected into COS cells, no RFXAP was detected in the immunoprecipitates (Fig. 2B, lanes 1 and 2). However, when these two proteins were coexpressed, RFXAP was detected abundantly with the anti-HA antibody (Fig. 2B, lane 3). We conclude that RFXANK also binds to RFXAP in vivo.
FIG. 2.
RFXANK and RFXAP interact in COS cells. (A) Schematic representation of the proteins used in immunoprecipitations. N termini of RFXANK and RFXAP were linked to epitope tags (Myc and HA, respectively). Proteins are labeled as in Fig. 1A. (B) RFXANK and RFXAP interact in COS cells. Epitope-tagged proteins were expressed alone (lanes 1 and 2) or in combination (lane 3) in COS cells. Total cell lysates were immunoprecipitated with the anti-Myc antibody and protein A-Sepharose beads and examined for the presence of RFXAP by Western blotting with the anti-HA antibody; 10% of total cell lysates were analyzed for the presence of RFXANK and RFXAP (input).
RFXANK and RFXAP also interact in a mammalian two-hybrid system.
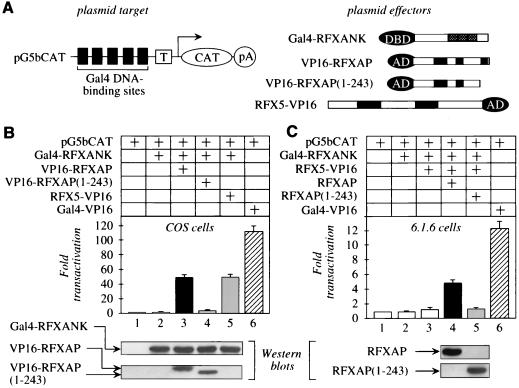
To characterize further the interaction between RFXAP and RFXANK and their roles in the formation of the RFX complex, mammalian two-hybrid binding assays were performed with COS cells (Fig. 3A). The plasmid target pG5bCAT contained five Gal4 DNA-binding sites upstream of a TATA box linked to the CAT reporter gene (Fig. 3A, top left). Protein effectors consisted of hybrid bait and prey proteins (Fig. 3A, top right). The former contained the DNA-binding domain (residues 1 to 147) of yeast Gal4 linked to the N terminus of RFXANK (Gal4-RFXANK). Prey proteins contained the activation domain (residues 413 to 490) of VP16 linked to the N terminus of RFXAP (VP16-RFXAP) or the C terminus of RFX5 (RFX5-VP16). The COS cells used in our assays do not express CIITA, which is a master switch for the expression of MHC II genes (11). In contrast, they contain all other proteins that are required for MHC II transcription including the RFX complex (10).
FIG. 3.
RFXANK and RFXAP also interact in a mammalian two-hybrid assay. (A) Schematic representation of the plasmid target and protein effectors used in the mammalian two-hybrid assay. pG5bCAT contained five Gal4 DNA-binding sites upstream of the TATA box (T) linked to the CAT reporter gene (CAT), which terminates in a poly(A) signal (pA). Protein effectors consisted of a hybrid bait (Gal4-RFXANK) as well as wild-type and mutant prey [VP16-RFXAP, VP16-RFXAP(1-243), and RFX5-VP16] proteins. All proteins except the hybrid RFX5-VP16 protein contained epitope tags (Myc) at their N termini. DBD, DNA-binding domain; AD, activation domain. (B) The interaction between RFXANK and RFXAP activates transcription from pG5bCAT. pG5bCAT and the hybrid Gal4-RFXANK protein were coexpressed (lane 2) with the hybrid VP16-RFXAP as well as mutant VP16-RFXAP(1-243) and RFX5-VP16 proteins (lanes 3 to 5) in COS cells. The hybrid Gal4-VP16 protein (striped bar) was used as the positive control. The total amount of transfected plasmid DNA was held constant at 2 μg. Fold transactivation represents values from experiments with coexpressed protein effectors over those obtained with pG5bCAT alone (lane 1). CAT enzymatic activities represent the mean value of three independent experiments performed in triplicate with indicated standard errors of the mean. The expression of the chimeras was monitored with the anti-Myc antibody and Western blotting (bottom). Expression of the hybrid RFX5-VP16 protein was detected with the anti-VP16 antibody (data not shown). (C) RFXAP is also required to activate transcription from pG5bCAT in 6.1.6 cells. Experiments were performed as for panel B except that the hybrid RFX5-VP16 protein was used as the prey and 6.1.6 cells were electroporated. CAT enzymatic activity increased only when the wild-type RFXAP protein was expressed in these cells (black bar). In sharp contrast, there was no effect when the mutant RFXAP(1-243) protein, which lacked the C terminus and resembled the endogenous RFXAP protein in 6.1.6 cells, was added (gray bar). The hybrid Gal4-VP16 protein (striped bar) represented the positive control. Western blots were performed with the anti-Myc antibody [for the hybrid mutant RFXAP(1-243) protein] or the anti-HA antibody (for RFXAP). Expression of the Gal4-RFXANK and RFX5-VP16 hybrid proteins paralleled data in panel B (data not shown).
Compared to pG5bCAT alone, coexpression of the hybrid Gal4-RFXANK protein and pG5bCAT did not increase the CAT enzymatic activity (Fig. 3B, compare lanes 1 and 2). However, when the hybrid VP16-RFXAP protein was added, levels of CAT activity increased 48-fold (lane 3). In sharp contrast, when the hybrid mutant VP16-RFXAP(1-243) protein was used, only basal levels of transactivation were observed, indicating that a critical interaction within the RFX complex was absent (lane 4). Western blots demonstrated that the levels of expression of the hybrid Gal4-RFXANK, VP16-RFXAP, and mutant VP16-RFXAP(1-243) proteins were equivalent (Fig. 3B, bottom). The Gal4-VP16 fusion protein, which bound directly to pG5bCAT and was used as the positive control in all transfections, increased CAT activity 100-fold over basal levels (compare lanes 1 and 6). Thus, levels of transactivation with RFXANK and RFXAP were 50% of maximal levels that can be achieved. They reflect the binding between these two proteins that was observed in vitro and in vivo (Fig. 1 and 2). We conclude that not only are the 30 C-terminal residues of RFXAP necessary for its interaction with RFXANK but RFX5 cannot promote formation of the RFX complex in the absence of this heterodimer.
To characterize further the role that RFXANK and RFXAP play in assembly of the RFX complex, pG5bCAT was coexpressed with the hybrid Gal4-RFXANK and RFX5-VP16 proteins, which also increased CAT activity 50-fold above basal levels (Fig. 3B, lane 5). Not only did the hybrid RFX5-VP16 protein activate transcription to the same levels as the hybrid VP16-RFXAP protein, but the coexpression of VP16-RFXAP and RFX5-VP16 fusion proteins alone with pG5bCAT had no effect (data not shown). Since no binding was observed between RFXANK and RFX5 in vitro (Fig. 1B), the endogenous RFXAP must bridge and connect RFXANK and RFX5 in cells. Thus, the RFX complex, tethered by RFXANK to pG5bCAT and requiring RFXAP, binds to RFX5 and presents the activation domain of VP16 to the transcription complex.
This bridge was confirmed in 6.1.6 cells that do not express a functional RFXAP protein. When pG5bCAT and the hybrid Gal4-RFXANK protein alone (Fig. 3C, lane 2) or in combination with the hybrid RFX5-VP16 protein (lane 3) were coexpressed in these cells, CAT activity did not increase significantly over basal levels (lane 1). However, when the wild-type RFXAP protein was included, the activation of pG5bCAT increased fivefold (lane 4). Moreover, this effect was abolished with the mutant RFXAP(1-243) protein (compare lanes 4 and 5). Of note, ratios of fold transactivation between the RFX complex formed on the hybrid Gal4-RFXANK protein and the Gal4-VP16 fusion protein were similar in COS and 6.1.6 cells (compare lanes 5 and 6 in Fig. 3B with lanes 4 and 6 in Fig. 3C). These results confirm the central role that the binding of RFXANK to RFXAP plays in formation of the RFX complex.
mutRFXANK does not bind to RFXAP.
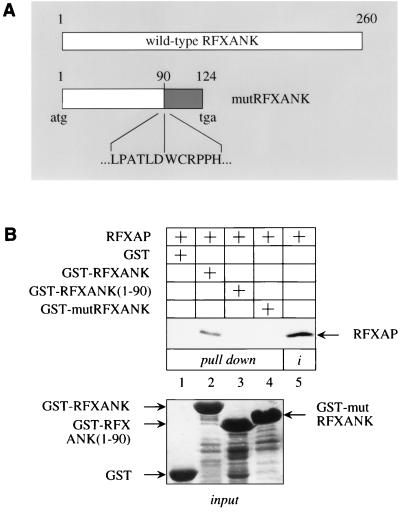
Our data indicated the importance of the binding between RFXANK and RFXAP within the RFX complex. This finding suggested that mutations in one of these two proteins might prevent their interaction and thus the transcription of MHC II genes in BLS. Several types of mutations, which include insertions, substitutions, and deletions, were found in RFXAP genes from different BLS patients. They all have alterations from nt 116 to 540 in the RFXAP cDNA and lead to truncated RFXAP proteins (9, 27). Since none of these mutant RFXAP proteins from complementation group D contain the C terminus of the wild-type protein, which is required for the binding to RFXANK, they were not examined further. Rather, mutRFXANK from Bequit cells was tested for its ability to interact with RFXAP. The mutRFXANK cDNA was amplified by RT-PCR and cloned (20, 21). A deletion of 26 nt changes the splicing pattern and removes exons 5 and 6 in the mature transcripts. Thus, the first 90 residues in mutRFXANK are conserved, followed by a frameshift that creates a truncated protein of 124 residues (Fig. 4A).
FIG. 4.
mutRFXANK does not bind to RFXAP. (A) Schematic representation comparing mutRFXANK from BLS (Bequit and Nacera) with the wild-type RFXANK protein. A 26-bp deletion at the boundary between intron 5 and exon 6 in the RFXANK gene directs the synthesis of a protein that contains 124 residues, of which 90 are from RFXANK, followed by 34 irrelevant residues and a premature stop codon (tga). The white portion of the mutRFXANK depicts the first 90 residues that are unchanged, ending in LPATLD. The frameshift starts a new sequence, colored in gray. Residues from position 91 on (WCRPPH…) are not present in the wild-type protein. (B) mutRFXANK does not bind to RFXAP in vitro. GST pull-down assays were performed with three different GST fusion proteins [GST-RFXANK, GST-RFXANK(1-90) and GST-mutRFXANK]. Results are presented in lanes 1 to 4; lane 5 contains 25% of the input labeled RFXAP (i); 25% of the input GST-fusion proteins are also presented in the bottom panel.
mutRFXANK was linked to GST and expressed in E. coli. To help characterize the interaction between mutRFXANK and RFXAP, the mutant GST-RFXANK(1-90) fusion protein was also synthesized. Neither chimera bound to RFXAP in the GST pull-down assay (Fig. 4A, lanes 3 and 4). As in Fig. 1B, the GST-RFXANK fusion protein bound to RFXAP (Fig. 4A, lane 2). Inputs of all GST fusion proteins were equivalent (Fig. 4B, input). These data indicate that the inability of mutRFXANK to bind to RFXAP blocks the formation of the RFX complex in BLS. Additionally, this interaction cannot occur without the C terminus of RFXAP.
Mutant RFXAP and RFXANK proteins do not assemble into the RFX complex in the presence of DNA.
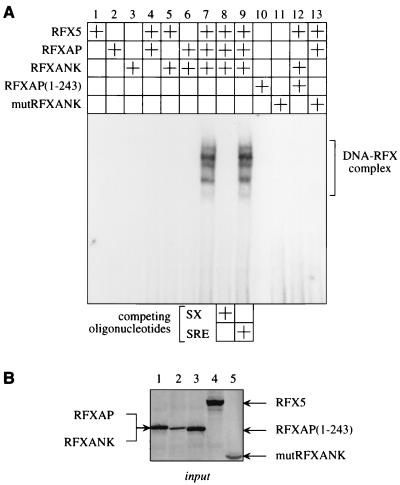
Data from our binding assays in vitro and in vivo indicated that the mutant RFXAP and RFXANK proteins cannot support the formation of the RFX complex. Thus, there existed the possibility that DNA could help to assemble the RFX complex. To examine whether individual subunits or only the complete RFX complex can assemble on the X and S boxes from the DRA promoter, EMSAs were performed. Different combinations of wild-type and mutant RFX proteins were transcribed and translated in vitro using the rabbit reticulocyte lysate system and mixed with the 32P-labeled SX oligonucleotide. Indeed, the RFX complex bound to DNA (Fig. 5A, lane 7). The formation of this DNA-RFX complex required RFXANK, RFXAP, and RFX5 and was not observed with individual subunits (lanes 1 to 3) or combinations of any two proteins (lanes 4 to 6). The competition with the unlabeled SX oligonucleotide completely abolished the formation of the RFX complex on the labeled SX oligonucleotide (lane 8), and the unlabeled non-specific SRE oligonucleotide had no effect (lane 9). Thus, the RFX complex binds specifically to DNA. Moreover, mutRFXANK and mutant RFXAP(1-243) proteins could not interact with DNA alone (lanes 10 and 11). Finally, they did not support the formation of the DNA-RFX complex when combined with other wild-type subunits (lanes 12 and 13). We conclude that protein-protein interactions between RFXANK, RFXAP, and RFX5 are independent of DNA and that only the complete RFX complex binds to DNA.
FIG. 5.
Mutations in RFXANK and RFXAP genes in BLS prevent the assembly of the RFX complex on DNA. (A) Analyses of interactions between the three RFX proteins and DNA. Wild-type RFXANK, RFXAP, and RFX5 proteins, mutant RFXAP(1-243) protein, and mutRFXANK were transcribed and translated in vitro using the rabbit reticulocyte lysate system. Different combinations of proteins were incubated with the 32P-labeled oligonucleotide containing S and X boxes from the DRA promoter (SX oligonucleotide). Two different unlabeled competitor oligonucleotides were used, the specific SX oligonucleotide and the nonspecific SRE oligonucleotide (lanes 8 and 9). The rabbit reticulocyte lysate alone did not bind to the SX oligonucleotide (data not shown). (B) Amounts of proteins used in EMSA. Presented are inputs of the 35S-labeled proteins that were transcribed and translated in vitro using the rabbit reticulocyte lysate in parallel with their unlabeled counterparts: RFXAP (lane 1), RFXANK (lane 2), RFXAP(1-243) (lane 3), RFX5 (lane 4), and mutRFXANK (lane 5).
DISCUSSION
In this study, we observed a direct interaction between RFXANK and RFXAP and explained why the RFX complex cannot form in patients from complementation groups B and D of BLS. The binding of RFXANK to RFXAP was demonstrated in vitro and in vivo, in the latter case by two independent systems which included immunoprecipitations and mammalian two-hybrid assays. Using several different mutants of these proteins, we also demonstrated that the interaction was specific; i.e., it depended completely on their C termini. Functional assays, performed in two different cell lines, one of which lacked RFXAP, indicated that the assembly of the RFX complex requires all three subunits. It also occurs in the absence of DNA. Finally, we studied the RFX complex assembly on X and S boxes from the DRA promoter and observed that all three subunits must be present in their wild-type forms for productive DNA-protein interactions.
In vitro binding assays led us to concentrate on the interaction between RFXAP and RFXANK. However, in the mammalian two-hybrid system, we also demonstrated that RFX5 is brought into the RFX complex. Interestingly, since no binding between RFXANK and RFX5 could be demonstrated and the binding between RFXAP and RFX5 is weak, RFXANK and RFXAP must form a combinatorial surface that binds to RFX5. To examine this issue, pG5bCAT was transactivated equivalently by the Gal4-RFXANK and VP16-RFXAP fusion proteins as well as by the Gal4-RFXANK, RFXAP, and RFX5-VP16 fusion proteins. Importantly, the exogenous wild-type RFXAP protein was absolutely essential for the function of this tripartite complex in 6.1.6 cells, which contain mutated RFXAP genes. Thus, RFXAP forms a bridge between RFXANK and RFX5 and connects all three subunits. Another interesting feature of this study is that mutant RFX proteins can be expressed and are stable in cells. They could be detected by Western blotting via their epitope tags. That they have not been detected in BLS cells is most likely due to the lack of epitope-specific antibodies against RFXANK, RFXAP, and RFX5. Thus, although these mutant proteins should be expressed in BLS patients, they cannot support the function of their wild-type counterparts.
To date, the binding between RFXAP and RFXANK is the strongest interaction within the RFX complex. It occurs in the presence of only two RFX subunits. However, these two proteins need RFX5 to bind to DNA. Since a weak interaction was also demonstrated between RFX5 and CIITA, the same protein could help to attract CIITA to MHC II promoters. However, RFX5 cannot bind to DNA in its full-length form (25), suggesting that its conformation has to be changed. This modification could occur following its binding to RFXAP (8) and could be strengthened by RFXANK. Taken together, these data explain why MHC II promoters in patients from complementation groups B, C, and D of BLS are bare (14). Furthermore, mutant RFXAP proteins from BLS, which all lack the C terminus, cannot bind to RFXANK. Likewise, mutRFXANK cannot bind to RFXAP. It is fascinating that two different complementation groups of a human genetic disease meet at the same protein-protein interaction. In other words, the mutation of either protein from complementation groups B and D of BLS prevents its binding to the other, indicating that the very first step in the formation of the RFX complex is blocked.
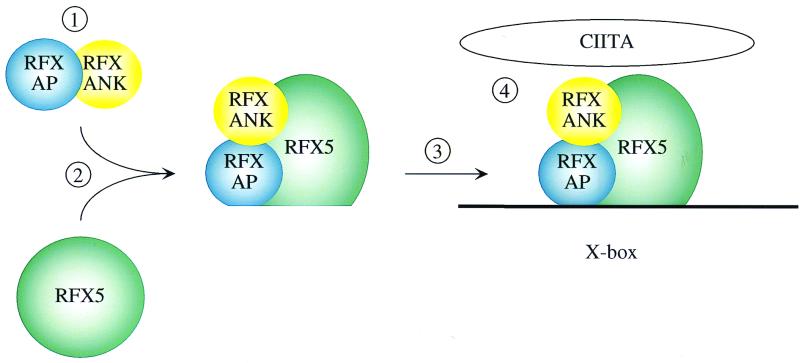
Our data suggest the following model for the formation of the DNA-RFX complex (Fig. 6). RFXANK and RFXAP assemble first and represent a scaffold that attracts RFX5. Upon binding, the conformational change of RFX5 exposes its DNA-binding domain, which anchors the RFX complex to X and S boxes. The final shape of the RFX complex also allows RFXAP to make extensive contacts with DNA (28). Another part of the RFX5 protein touches CIITA, which is attracted to MHC II promoters by a combinatorial surface formed on CUS. With the availability of nuclear factor Y and RFX complexes as well as CIITA, the structural and functional assembly of these protein-protein and DNA-protein complexes can now be performed to elucidate the complex transcription of MHC II genes.
FIG. 6.
A model for the assembly of the RFX complex. RFXANK and RFXAP bind to each other and form a heterodimer (step 1) that subsequently interacts with RFX5 (step 2). All proteins at this stage are depicted as circles. Upon binding, the conformation of RFX5 changes (step 2) in a way that enables the RFX complex to bind to DNA (step 3) and to recruit other proteins that are required for the transcription of MHC II genes, especially CIITA (step 4).
ACKNOWLEDGMENTS
We thank Paula Zupanc-Ecimovic for secretarial assistance; Jeremy Boss for reagents; and Satoshi Kanazawa, Giovanna Tosi, and other members of the laboratory for help with experiments and comments on the manuscript.
This work was supported by a grant from the Nora Eccles Treadwell Foundation.
REFERENCES
- 1.Alberts A S, Treisman R. Activation of RhoA and SAPK/JNK signalling pathways by the RhoA-specific exchange factor mNET1. EMBO J. 1998;17:4075–4085. doi: 10.1093/emboj/17.14.4075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Benoist C, Mathis D. Regulation of major histocompatibility complex class-II genes: X, Y and other letters of the alphabet. Annu Rev Immunol. 1990;8:681–715. doi: 10.1146/annurev.iy.08.040190.003341. [DOI] [PubMed] [Google Scholar]
- 3.Boss J M. Regulation of transcription of MHC class II genes. Curr Opin Immunol. 1997;9:107–113. doi: 10.1016/s0952-7915(97)80166-5. [DOI] [PubMed] [Google Scholar]
- 4.Cogswell J P, Zeleznik-Le N, Ting J P. Transcriptional regulation of the HLA-DRA gene. Crit Rev Immunol. 1991;11:87–112. [PubMed] [Google Scholar]
- 5.Cress W D, Triezenberg S J. Critical structural elements of the VP16 transcriptional activation domain. Science. 1991;251:87–90. doi: 10.1126/science.1846049. [DOI] [PubMed] [Google Scholar]
- 6.Cresswell P. Assembly, transport, and function of MHC class II molecules. Annu Rev Immunol. 1994;12:259–293. doi: 10.1146/annurev.iy.12.040194.001355. [DOI] [PubMed] [Google Scholar]
- 7.Douhan J, III, Hauber I, Eibl M M, Glimcher L H. Genetic evidence for a new type of major histocompatibility complex class II combined immunodeficiency characterized by a dyscoordinate regulation of HLA-D alpha and beta chains. J Exp Med. 1996;183:1063–1069. doi: 10.1084/jem.183.3.1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Durand B, Sperisen P, Emery P, Barras E, Zufferey M, Mach B, Reith W. RFXAP, a novel subunit of the RFX DNA binding complex is mutated in MHC class II deficiency. EMBO J. 1997;16:1045–1055. doi: 10.1093/emboj/16.5.1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fondaneche M C, Villard J, Wiszniewski W, Jouanguy E, Etzioni A, Le Deist F, Peijnenburg A, Casanova J L, Reith W, Mach B, Fischer A, Lisowska-Grospierre B. Genetic and molecular definition of complementation group D in MHC class II deficiency. Hum Mol Genet. 1998;7:879–885. doi: 10.1093/hmg/7.5.879. [DOI] [PubMed] [Google Scholar]
- 10.Fontes J D, Jabrane-Ferrat N, Peterlin B M. Assembly of functional regulatory complexes on MHC class II promoters in vivo. J Mol Biol. 1997;270:336–345. doi: 10.1006/jmbi.1997.1121. [DOI] [PubMed] [Google Scholar]
- 11.Fontes J D, Kanazawa S, Nekrep N, Peterlin B M. The class II transactivator CIITA is a transcriptional integrator. Microbes Infect. 1999;1:863–869. doi: 10.1016/s1286-4579(99)00232-4. [DOI] [PubMed] [Google Scholar]
- 12.Ghosh S, Selby M J, Peterlin B M. Synergism between Tat and VP16 in trans-activation of HIV-1 LTR. J Mol Biol. 1993;234:610–619. doi: 10.1006/jmbi.1993.1615. [DOI] [PubMed] [Google Scholar]
- 13.Gladstone P, Pious D. Stable variants affecting B cell alloantigens in human lymphoid cells. Nature. 1978;271:459–461. doi: 10.1038/271459a0. [DOI] [PubMed] [Google Scholar]
- 14.Glimcher L H, Kara C J. Sequences and factors: a guide to MHC class-II transcription. Annu Rev Immunol. 1992;10:13–49. doi: 10.1146/annurev.iy.10.040192.000305. [DOI] [PubMed] [Google Scholar]
- 15.Jabrane-Ferrat N, Fontes J D, Boss J M, Peterlin B M. Complex architecture of major histocompatibility complex class II promoters: reiterated motifs and conserved protein-protein interactions. Mol Cell Biol. 1996;16:4683–4690. doi: 10.1128/mcb.16.9.4683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jabrane-Ferrat N, Peterlin B M. Ets-1 activates the DRA promoter in B cells. Mol Cell Biol. 1994;14:7314–7321. doi: 10.1128/mcb.14.11.7314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lisowska-Grospierre B, Fondaneche M C, Rols M P, Griscelli C, Fischer A. Two complementation groups account for most cases of inherited MHC class II deficiency. Hum Mol Genet. 1994;3:953–958. doi: 10.1093/hmg/3.6.953. [DOI] [PubMed] [Google Scholar]
- 18.Mach B, Steimle V, Martinez-Soria E, Reith W. Regulation of MHC class II genes: lessons from a disease. Annu Rev Immunol. 1996;14:301–331. doi: 10.1146/annurev.immunol.14.1.301. [DOI] [PubMed] [Google Scholar]
- 19.Marais R, Wynne J, Treisman R. The SRF accessory protein Elk-1 contains a growth factor-regulated transcriptional activation domain. Cell. 1993;73:381–393. doi: 10.1016/0092-8674(93)90237-k. [DOI] [PubMed] [Google Scholar]
- 20.Masternak K, Barras E, Zufferey M, Conrad B, Corthals G, Aebersold R, Sanchez J C, Hochstrasser D F, Mach B, Reith W. A gene encoding a novel RFX-associated transactivator is mutated in the majority of MHC class II deficiency patients. Nat Genet. 1998;20:273–277. doi: 10.1038/3081. [DOI] [PubMed] [Google Scholar]
- 21.Nagarajan U M, Louis-Plence P, DeSandro A, Nilsen R, Bushey A, Boss J M. RFX-B is the gene responsible for the most common cause of the bare lymphocyte syndrome, an MHC class II immunodeficiency. Immunity. 1999;10:153–162. doi: 10.1016/s1074-7613(00)80016-3. . (Erratum, 10:399.) [DOI] [PubMed] [Google Scholar]
- 22.Sadowski I, Ma J, Triezenberg S, Ptashne M. GAL4-VP16 is an unusually potent transcriptional activator. Nature. 1988;335:563–564. doi: 10.1038/335563a0. [DOI] [PubMed] [Google Scholar]
- 23.Sadowski I, Ptashne M. A vector for expressing GAL4(1-147) fusions in mammalian cells. Nucleic Acids Res. 1989;17:7539. doi: 10.1093/nar/17.18.7539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Scholl T, Mahanta S K, Strominger J L. Specific complex formation between the type II bare lymphocyte syndrome-associated transactivators CIITA and RFX5. Proc Natl Acad Sci USA. 1997;94:6330–6334. doi: 10.1073/pnas.94.12.6330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Steimle V, Durand B, Barras E, Zufferey M, Hadam M R, Mach B, Reith W. A novel DNA-binding regulatory factor is mutated in primary MHC class II deficiency (bare lymphocyte syndrome) Genes Dev. 1995;9:1021–1032. doi: 10.1101/gad.9.9.1021. [DOI] [PubMed] [Google Scholar]
- 26.Steimle V, Otten L A, Zufferey M, Mach B. Complementation cloning of an MHC class II transactivator mutated in hereditary MHC class II deficiency (or bare lymphocyte syndrome) Cell. 1993;75:135–146. [PubMed] [Google Scholar]
- 27.Villard J, Mach B, Reith W. MHC class II deficiency: definition of a new complementation group. Immunobiology. 1997;198:264–272. doi: 10.1016/S0171-2985(97)80046-0. [DOI] [PubMed] [Google Scholar]
- 28.Westerheide S D, Boss J M. Orientation and positional mapping of the subunits of the multicomponent transcription factors RFX and X2BP to the major histocompatibility complex class II transcriptional enhancer. Nucleic Acids Res. 1999;27:1635–1641. doi: 10.1093/nar/27.7.1635. [DOI] [PMC free article] [PubMed] [Google Scholar]