Abstract
Objective
The capacity to generate new adipocytes from precursor cells is critical for maintaining metabolic health. Adipocyte precursor cells (APCs) constitute a heterogenous collection of cell types; however, the contribution of these various cell types to adipose tissue expansion in vivo remains unknown. The aim of the current study is to investigate the contribution of Dpp4+ progenitors to de novo adipogenesis.
Methods
Single cell analysis has identified several transcriptionally distinct subpopulations of APCs, including Dpp4+ progenitor cells concentrated in the connective tissue surrounding many organs, including white adipose tissue (WAT). Here, we generated a Dpp4CreER mouse model for in vivo lineage tracing of these cells and their downstream progeny in the setting of basal or high fat diet (HFD)-stimulated adipogenesis.
Results
Dpp4CreER mice enabled specific temporal labeling of Dpp4+ progenitor cells within their native connective tissue niche. Following a dietary chase period consisting of chow or HFD feeding for 18 weeks, Dpp4+ progenitors differentiated into mature adipocytes within the gonadal and subcutaneous WAT. HFD stimulated adipogenic contribution from Dpp4+ cells in the gonadal but not the subcutaneous depot. Flow cytometry analysis revealed that Dpp4+ progenitors give rise to DPP4(−)/ICAM1+ preadipocytes in vivo. HFD feeding did not perturb the flux of Dpp4+ cell conversion into ICAM1+ preadipocytes in gonadal WAT. Conversely, in subcutaneous WAT, HFD feeding/obesity led to an accumulation of ICAM1+ preadipocytes without a corresponding increase in mature adipocyte differentiation. Examination of non-classical murine visceral depots with relevance to humans, including omentum and retroperitoneal WAT, revealed robust contribution of Dpp4+ progenitors to de novo adipogenesis, which was further stimulated by HFD.
Conclusion
Our data demonstrate that Dpp4+ interstitial progenitor cells contribute to basal adipogenesis in all fat depots and are recruited to support de novo adipogenic expansion of visceral WAT in the setting of HFD-induced obesity.
Keywords: Mesenchymal stem cell, Adipose tissue, DPP4, ICAM1, Obesity, Preadipocyte
Highlights
-
•
Dpp4+ progenitor cells contribute to basal and HFD-induced visceral adipogenesis.
-
•
Dpp4+ progenitors give rise to ICAM1+/DPP4-committed preadipocytes in vivo.
-
•
Dpp4+ progenitors contribute to adipogenesis in the murine omentum and rpWAT.
-
•
Dpp4+ progenitors become stalled as Dpp4(−)/ICAM1+ cells in subcutaneous adipose.
1. Introduction
Obesity is a leading public health concern that is predicted to affect over 50 percent of adults in the United States by 2030 [1]. Obesity is characterized by the excessive accumulation of white adipose tissue (WAT), which serves as the primary reservoir for energy storage. Over time, chronic positive energy balance and obesity can overwhelm and impair the capacity of WAT to efficiently store excess nutritional energy. Insufficient amounts of WAT (as in lipodystrophy) or its diminished function (as in obesity) is a major driver of ectopic lipid deposition in other organs and systemic metabolic diseases [[2], [3], [4], [5]].
WAT expands via two mechanisms: increase in the size of existing adipocytes (hypertrophy), and recruitment of de novo differentiated adipocytes (hyperplasia). Studies in mice and humans indicate that hyperplastic adipocyte expansion via adipocyte precursor cell (APC) differentiation is metabolically favorable, often limiting or delaying the metabolic complications of obesity [[6], [7], [8], [9]]. By contrast, hypertrophic adipocyte growth is linked to adipose tissue fibrosis, inflammation, and metabolic dysfunction [10]. The anatomic location of adipose tissue expansion also influences metabolic outcomes [11,12]. In mice, hyperplastic de novo adipogenesis occurs mainly in the visceral depots (i.e. gonadal WAT), while both visceral and subcutaneous (i.e. inguinal WAT) depots engage in hypertrophic growth [[13], [14], [15]].
The capacity for hyperplastic expansion relies on the differentiation activity of APCs [5,16]. Loss of de novo adipogenesis in subcutaneous WAT results in the pathologic spill-over of free fatty acids into the visceral WAT [4,17,18], liver [19], and muscle [20,21], triggering metabolic derangement [17,22,23]. The importance of APC function is reinforced by the observation that de novo adipogenesis stimulated by PPARγ overexpression [7], thiazolidinedione treatment [24,25], or transplantation [[26], [27], [28]] leads to significant improvements in insulin sensitivity despite an increase in body weight. Investigation of the mechanisms governing APC biology is paramount to our understanding of the metabolic consequences of obesity.
APCs represent a heterogeneous pool of stromal and perivascular cells that are nearly universally marked by the cell surface protein PDGFRα [[29], [30], [31]]. Recent studies employing single-cell RNA-sequencing technology have revealed subpopulations of Pdgfrα+ cells, including Dpp4+/Pi16+ ‘progenitors’, Icam1+/Col15a1+ ‘preadipocytes’, and Cd142+ ‘adipose regulatory cells’, as well as additional cell types [[32], 33, [34], [35], [36], [37], [38]]. Dpp4+ cells possess enhanced proliferative capacity and multilineage differentiation potential [32,39]. By contrast, Icam1+ cells are highly committed to the adipose lineage and rapidly undergo adipocyte differentiation with minimal stimulus. Cell transplantation studies suggest a lineage hierarchy in which Dpp4+/Pi16+ progenitors give rise to Icam1+ preadipocytes and mature adipocytes [32].
Dpp4+/Pi16+ progenitors are enriched in an anatomic niche termed the Reticular Interstitium (RI), a relatively understudied connective tissue that envelops many organs [32,40]. Dpp4+/Pi16+ cells from a diverse set of organs express a conserved transcriptional profile, leading to their designation as ‘universal fibroblasts’ [41]. Pseudotemporal analysis of fibroblast populations throughout the body predicts that Dpp4+/Pi16+ universal fibroblasts give rise to specialized cell types commiserate with their resident tissue [41]; however, these lineage relationships have yet to be determined in vivo.
In this study, we developed a Dpp4CreER mouse model for lineage tracing of Dpp4+/Pi16+ progenitor cells within their native in vivo niche. We found that Dpp4+ progenitors give rise to ICAM1+ preadipocytes as well as mature adipocytes in both subcutaneous and visceral WAT, and that this process is stimulated in a depot-specific manner during the development of obesity. The relatively lower levels of adipogenesis in the subcutaneous WAT of obese animals was associated with a buildup of ICAM1+ preadipocytes derived from Dpp4+ progenitors, suggesting that HFD and/or obesity impairs preadipocyte commitment. Overall, these data show that ‘universal’ Dpp4+ progenitors contribute to the development of adipocytes in several major fat depots.
2. Results
2.1. Dpp4+ progenitor cells contribute to basal and HFD-induced visceral adipogenesis
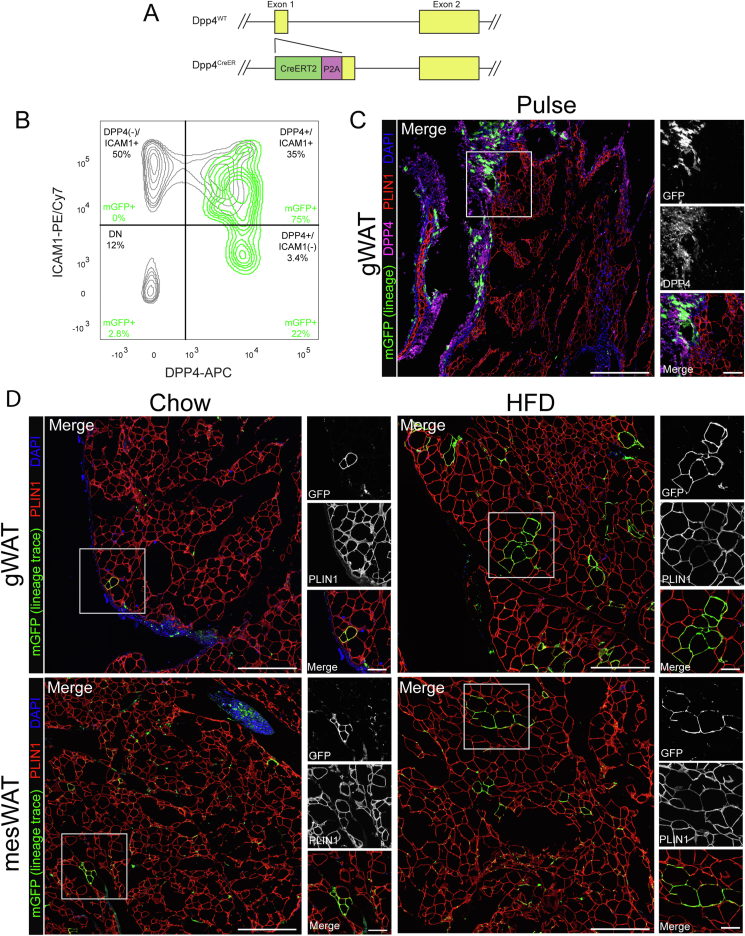
Dpp4+ progenitors possess the capacity to differentiate into mature adipocytes in vitro and after transplantation. However, the lineage contribution of this cell population has yet to be investigated in vivo. To determine the role of Dpp4+ mesenchymal progenitors in WAT growth, we developed a Dpp4CreER mouse model, enabling the indelible labeling of Dpp4+ progenitors and their progeny. The coding sequence for Cre-ERT2 was inserted into the first exon of Dpp4, separated by a self-cleaving P2A peptide (Figure 1A). This design allows for the expression of CRE-ERT2 under the control of the native Dpp4 gene regulatory elements. Dpp4CreER mice were crossbred with a R26mTmG fluorescent reporter mouse line for inducible lineage tracing.
Figure 1.
Dpp4+ progenitor cells contribute to basal and diet-induced visceral adipogenesis. (A) Schematic of the targeting strategy used to generate Dpp4CreER mice. (B) Flow cytometric analysis of SVF cells from gonadal WAT (gWAT) of Dpp4CreER; mGFP reporter mice immediately following tamoxifen injection (pulse timepoint). CD45(−)/DAPI(−) gated cells were plotted by expression of DPP4 vs ICAM1. mGFP + lineage-traced cells are shown as a green overlay on top of mGFP(−) cells depicted in gray. (C) Immunohistochemical (IHC) examination of gWAT from Dpp4CreER; mGFP mice at the pulse timepoint [mGFP: lineage traced cells (green), cells currently expressing DPP4 (magenta), PLIN1: mature adipocytes (red), DAPI: nuclei (blue); scale bar = 380 μM]. Individual antibody staining is shown in grayscale on the right-side inset panels (inset scale bar = 100 μM). (D) IHC staining of gWAT (top panels) and mesenteric (mesWAT, bottom panels) from Dpp4CreER; mGFP mice following 18 weeks of feeding chow (left) or HFD (right).
Tamoxifen treatment of these mice induces heritable labeling of Dpp4-expressing cells with membrane-GFP (mGFP). Flow cytometry analysis of mGFP+ cells immediately following tamoxifen injection (pulse timepoint) revealed specific labeling of DPP4+ cells in gonadal WAT (Figure 2B, mGFP labeled cells are depicted as a green overlay on top of unlabeled total stromal cells). There was no mGFP labeling of DPP4(−)/ICAM1+ preadipocytes at the pulse timepoint, demonstrating the specificity of this mouse model.
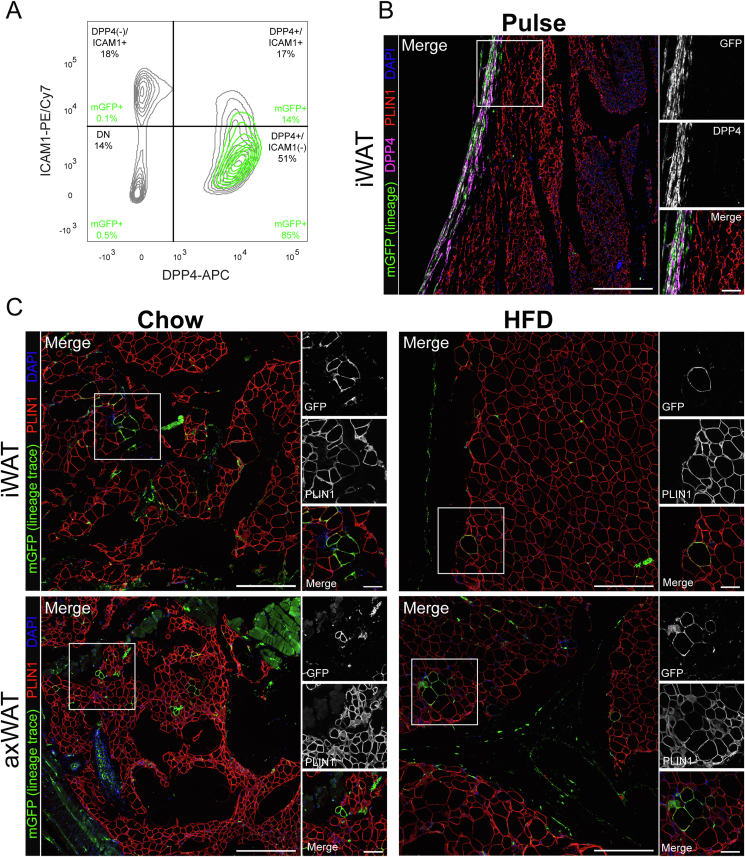
Figure 2.
Dpp4+ progenitor cells contribute to basal adipogenesis in subcutaneous depots. (A) Flow cytometric analysis of SVF cells (CD45(−)/DAPI(−)) from subcutaneous WAT of Dpp4CreER; mGFP mice at the pulse timepoint. mGFP+ lineage traced cells are shown as a green overlay on top of mGFP(−) cells, which are depicted in gray. (B) IHC staining of subcutaneous inguinal WAT (iWAT) at the pulse timepoint. [mGFP: lineage traced cells (green), cells currently expressing DPP4 (magenta), PLIN1: mature adipocytes (red), DAPI: nuclei (blue); scale bar = 380 μM]. Individual antibody staining is represented in grayscale on the right-side inset panels (inset scale bar = 100 μM). (C) IHC staining of Dpp4CreER iWAT (top panels) and axillary WAT (axWAT, bottom panels) following 18 weeks of feeding chow (left) or HFD.
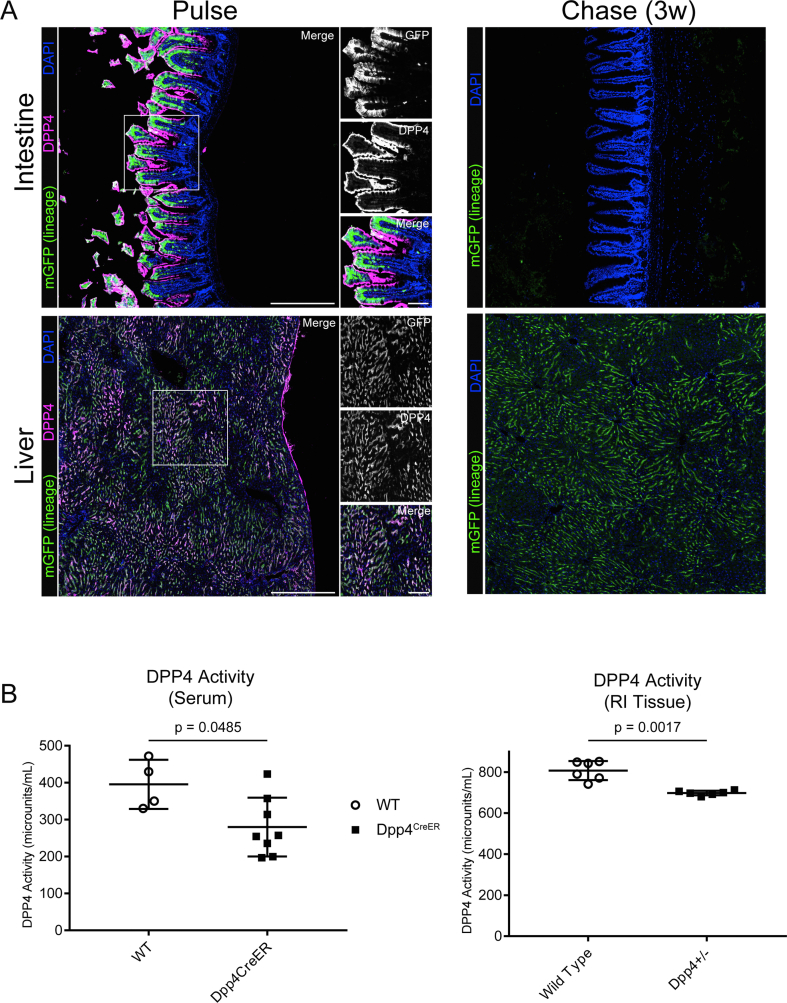
DPP4 is highly expressed in hepatocytes as well as mature epithelial cells of the intestinal villi, which we used as positive labeling controls. At the pulse timepoint, we observed robust mGFP labeling of hepatocytes and intestinal epithelial cells (Figure S1). Several weeks later, mGFP+ epithelial cells were absent from the intestinal villi, owing to continual cellular turnover and loss of previously labeled cells. These results demonstrate the specificity of the Dpp4CreER transgene as well as the temporal fidelity of the tamoxifen pulse.
Histological examination of gonadal WAT (gWAT) from Dpp4CreER; mGFP mice at the pulse timepoint showed numerous mGFP-labeled cells, which co-stained with anti-DPP4 antibody. The labeled cells displayed a spindle-shaped fibroblast morphology and were concentrated in the connective tissue at both the periphery of the adipose tissue depot and in the fibrous septae between adipose tissue lobes (Figure 1C). We did not observe mGFP labeling of any mature adipocytes at the pulse timepoint.
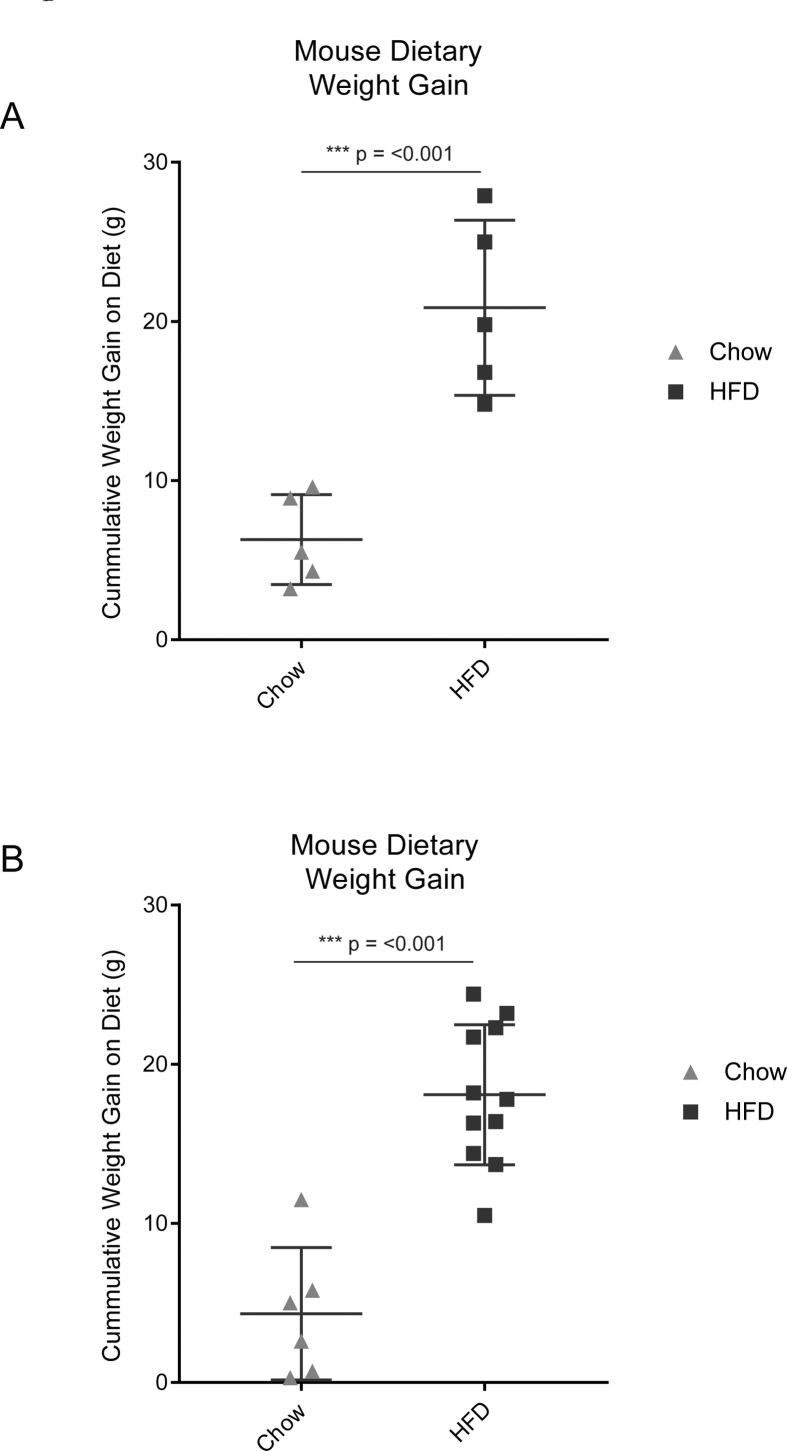
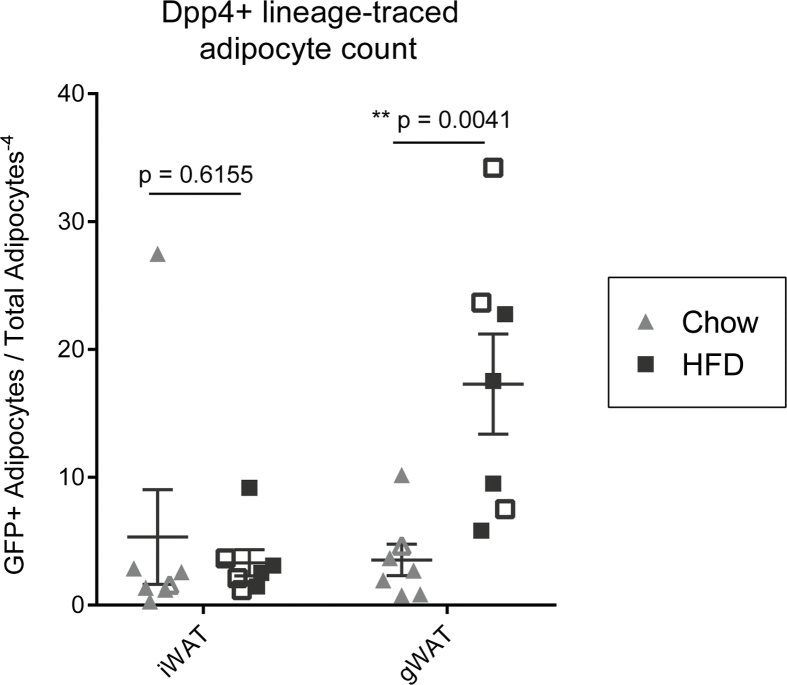
We next sought to determine the lineage contribution of Dpp4+ progenitors under conditions of chow diet (i.e. basal adipocyte turnover), and in the setting of HFD-stimulated adipose tissue expansion. Two weeks after tamoxifen injection, Dpp4CreER; mGFP mice were assigned to either chow or HFD feeding groups for an additional 18 weeks, leading to increases in body weight (Figure S2). Following the chase period in the chow fed group, we detected mGFP+ and Perilipin-1 (PLIN1)-expressing mature adipocytes, derived from Dpp4-lineage traced cells, in the gonadal and mesenteric WAT (Figure 1D, left panels). In the HFD group, we observed more numerous clusters of mGFP+ lineage traced mature adipocytes, which were often located near the periphery of the tissue (Figure 1D, right panels). Quantification of the histological images showed a significant obesity-stimulated increase in the number of mature adipocytes derived from Dpp4+ progenitors in the gWAT (Figure S3).
2.2. Dpp4+ progenitor cells contribute to basal adipogenesis in subcutaneous WAT
Flow cytometry analysis of mGFP+ labeled cells from the subcutaneous WAT of tamoxifen-injected Dpp4CreER mice at the pulse timepoint revealed highly specific labeling of DPP4+ cells (Figure 2A, green overlay). Histological examination of inguinal WAT (iWAT) at the pulse timepoint showed extensive mGFP labeling of DPP4+ cells within the reticular interstitium at the periphery of the tissue as well as fibrous septa between adipose tissue lobes (Figure 2B). No mGFP+ mature adipocytes were detected at the time of the tamoxifen pulse. Following an 18-week chase period with chow or HFD feeding, rare GFP+ adipocytes were detected within the inguinal and axillary WAT (Figure 2C, right panels). Quantitative analysis showed that HFD-induced obesity did not increase the number of Dpp4+ lineage-traced adipocytes in the subcutaneous iWAT depot (Figure 2C, right panels, Figure S3).
2.3. Dpp4+ progenitors give rise to ICAM1+/DPP4- committed preadipocytes in vivo
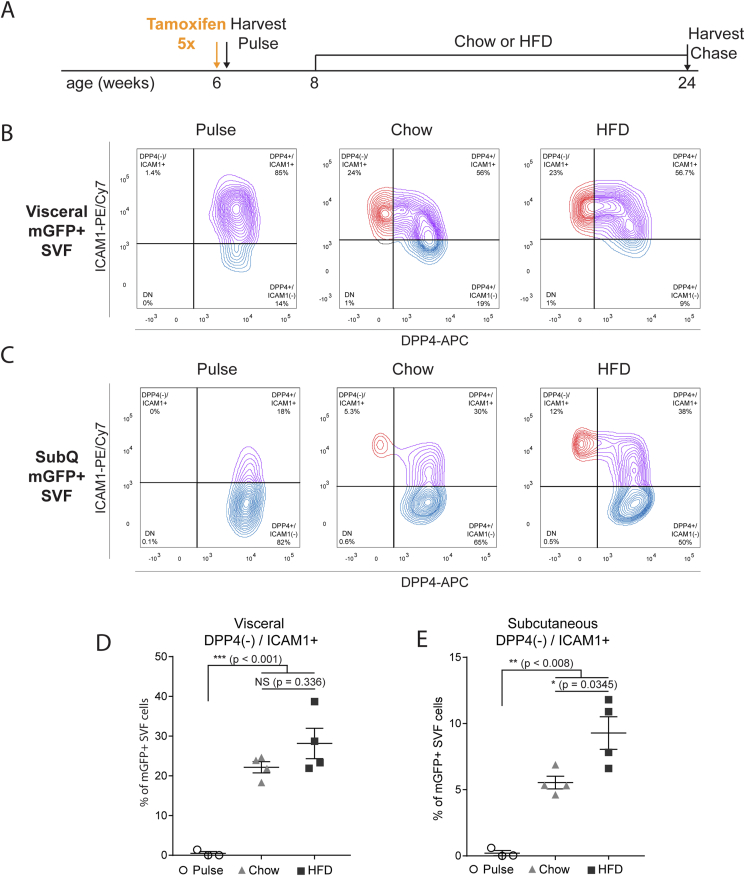
Previous studies have employed in silico pseudotemporal analysis [41], in vitro adipogenic differentiation, and transplantation models [32] to conceptualize an adipose lineage hierarchy in which Dpp4+ progenitors give rise to Dpp4(−)/Icam1+ preadipocytes. To investigate the lineage allocation of Dpp4+ progenitors in their native environment, we assessed the fate of mGFP-traced cells in mice under basal or HFD feeding conditions (Figure 3A). Immediately after tamoxifen injection (pulse timepoint), subcutaneous and visceral WAT depots were dissociated and examined by flow cytometry. After gating for CD45(−)/mGFP(+) cells, we observed mGFP-expressing DPP4+ cells in both the visceral and subcutaneous depots (Figure 3B and C, left panels, pulse timepoint), with almost no labeling of DPP4(−)/ICAM1+ preadipocytes. Following 18 weeks of chow or HFD, there was a significant shift in the lineage traced cells with the appearance of DPP4(−)/ICAM1+ preadipocytes (Figure 3B and C, right panels). Quantification of these data revealed a large increase in the number of DPP4(−)/ICAM1+ preadipocytes arising from DPP4+ cells during the dietary chase period. HFD-induced obesity did not shift the steady state proportion of lineage traced DPP4(−)/ICAM1+ preadipocytes in the visceral depot (Figure 3D). By contrast, we observed a marked buildup of DPP4(−)/ICAM1+ preadipocytes in the subcutaneous WAT of HFD-fed mice, as compared to chow-fed mice (Figure 3E).
Figure 3.
Dpp4+ progenitors give rise to ICAM1+/DPP4(−) preadipocytes in vivo. (A) Schematic depicting the lineage tracing timeline. Six-week-old Dpp4CreER; mGFP mice were injected with tamoxifen to produce heritable mGFP labeling of Dpp4+ cells. Lineage traced mice were placed on either chow or HFD at 8 weeks of age and harvested at 24 weeks. (B, C) Flow cytometric analysis of mGFP+ SVF cells from visceral (B) and subcutaneous (C) WAT of Dpp4CreER mice. CD45(−)/DAPI(−) SVF cells were gated on expression of mGFP to isolate lineage traced cells. Staining with anti-DPP4 and anti-ICAM1 at the pulse timepoint (left panel) and following 18 weeks of either chow diet (middle panel) or HFD (right panel). Data are representative of four biologic replicates. (D, E) Quantification of relative Dpp4+ lineage traced population abundance from visceral (D) and subcutaneous (E) WAT of Dpp4CreER; mGFP mice.
2.4. Dpp4+ progenitors contribute extensively to adipogenesis in the murine omentum
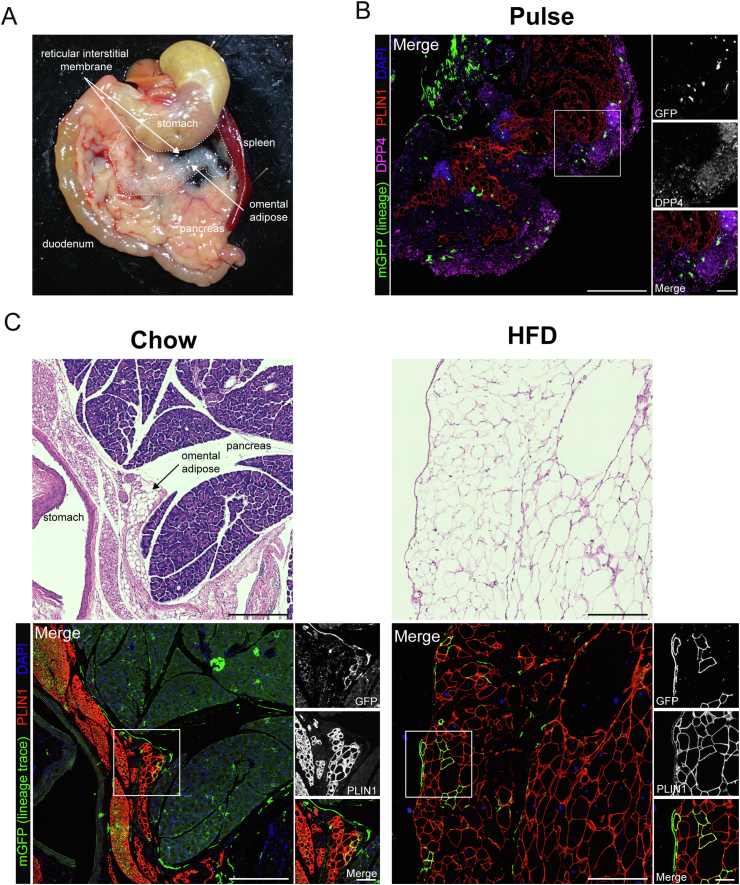
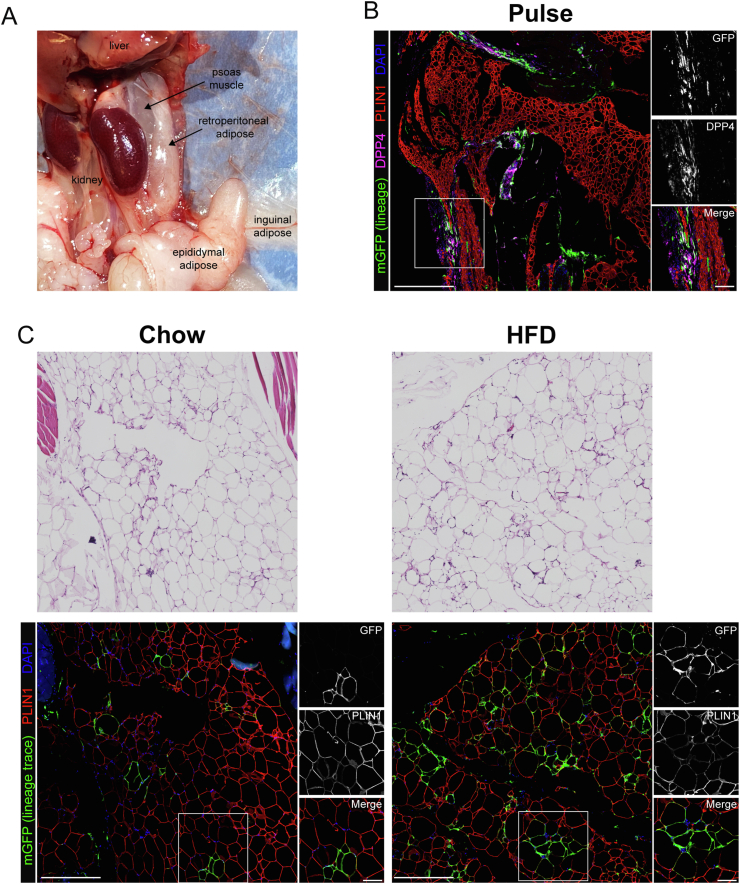
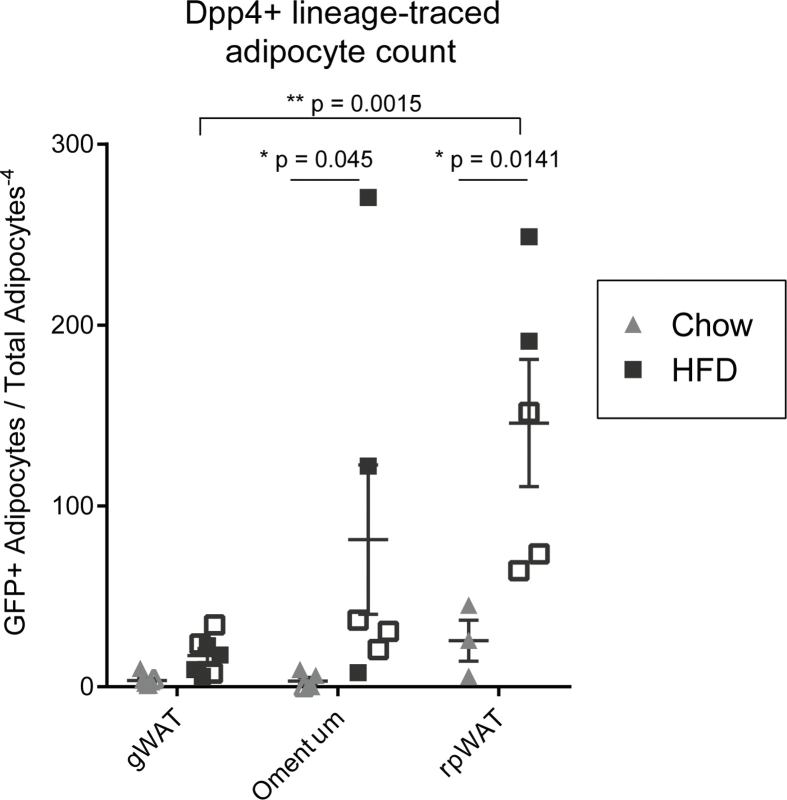
Murine gWAT has been the focus of extensive study due to its robust expansion in response to HFD and ease of surgical procurement. However, gWAT may not be the best proxy for human visceral adipose tissue given that there is no anatomically analogous tissue present in humans. We sought to expand our investigation to include non-classical visceral adipose tissue depots with human correlation, including the omental and retroperitoneal adipose tissue depots. In chow-fed lean mice, the greater omentum is composed of a translucent sheet of connective tissue containing small islands of adipose tissue, which bridges between the greater curvature of the stomach and the pancreas (Figure 4A) [42]. Following dissection and paraffin embedding, the translucent reticular interstitial membrane, which is studded with DPP4+ progenitor cells, appears bunched around the omental adipose tissue (Figure 4B, pulse timepoint). The omental depot undergoes massive expansion and lipid filling in the setting of HFD-induced obesity (Figure 4C, chow vs HFD H&E panels). Dpp4-lineage traced mature adipocytes were observed in the omentum of chow fed animals and numerous clusters mGFP + adipocytes emerged in response to HFD (Figure 4C, right panels). Quantitative analyses showed that HFD greatly increased the number of Dpp4 lineage-traced mature adipocytes in the omentum (Figure S4). We found an average of seven times more Dpp4-lineage traced adipocytes in the omentum of obese mice as compared to the gWAT, suggesting that DPP4+ progenitors play a particularly important role in the maintenance and expansion of omental adipose.
Figure 4.
Dpp4+ progenitors contribute extensively to adipogenesis in the murine omentum. (A) Gross histology showing the anatomic location of the mouse omentum. The murine greater omentum is a translucent sheet of collagen-filled connective tissue (reticular interstitium) that extends from the greater curve of the stomach and is draped over the pancreas and intestine. The inferior edge of the omentum is filled with adipose tissue (omental adipose). (B) IHC staining of omental adipose tissue from Dpp4CreER; mGFP mice at the pulse timepoint [mGFP: lineage-traced cells (green), DPP4: cells currently expressing DPP4 (magenta), PLIN1: mature adipocytes (red), DAPI: nuclei (blue); scale bar = 380 μM]. Individual antibody staining is represented in grayscale on the right-side inset panels (inset scale bar = 100 μM). (C) H&E and IHC staining of omental WAT from Dpp4CreER; mGFP mice under chow (left panels) or HFD (right panels) feeding conditions.
2.5. Dpp4+ progenitors contribute extensively to adipogenesis in retroperitoneal adipose tissue
Retroperitoneal WAT (rpWAT) is located within the retroperitoneal space against the dorsal surface of the abdomen. This depot contains a mixture of white and brown adipocytes and undergoes massive expansion and whitening in the setting of obesity. rpWAT is located within a thin connective tissue sheath, populated by DPP4+ cells, that tethers it to the underlying paraspinal muscles (Figure 5A). At the pulse timepoint, mGFP-labeled DPP4+ progenitor cells were found in the connective tissue surrounding the parenchyma of the rpWAT, as well as the intralobular septae (Figure 5A). Following the dietary chase period, we observed a large number of mGFP + lineage traced mature adipocytes in the rpWAT (Figure 5B). Like in omentum adipose tissue, DPP4+ cells constituted a significant source of newly formed adipocytes in rpWAT, and this lineage contribution was greatly stimulated in the setting of obesity (Figure S4).
Figure 5.
Dpp4+ progenitors contribute extensively to adipogenesis in retroperitoneal WAT. (A) Gross histology showing the anatomic location of the mouse retroperitoneal WAT. Murine retroperitoneal WAT is contained within a sheath of reticular interstitial connective tissue that affixes the depot within the retroperitoneal space against the psoas muscles. (B) IHC staining of retroperitoneal WAT from Dpp4CreER; mGFP mice at the pulse timepoint [mGFP: lineage traced cells (green), DPP4: cells currently expressing DPP4 (magenta), PLIN1: mature adipocytes (red), DAPI: nuclei (blue); scale bar = 380 μM]. Individual antibody staining is represented in grayscale on the right-side inset panels (inset scale bar = 100 μM). (C) H&E and IHC staining of Dpp4CreER retroperitoneal adipose under chow (left panels) or high fat diet (right panels) feeding conditions.
3. Discussion
The impairment of new adipocyte formation contributes significantly to the development of metabolic sequalae in obesity. The recent application of single-cell RNA transcriptomics enabled the unbiased identification of precursor subpopulations that may contribute to adipocyte differentiation in vivo. Numerous groups have consistently revealed the presence of several major APC populations in WAT, including those marked by Dpp4/Pi16 and Icam1/Col15a1 (reviewed in [[43], [44], [45]]). In silico pseudotemporal analysis and in vitro differentiation and transplantation experiments provide evidence of a lineage hierarchy in which Dpp4+ progenitors give rise to Icam1+ preadipocytes that can differentiate into mature adipocytes. However, these previous studies analyzed the differentiation capacity of isolated cell populations removed from their native context and niche components.
We applied genetic lineage tracing analysis to determine the contribution of Dpp4+ progenitor cells to de novo adipose tissue expansion in vivo. Our previous work identified Dpp4 as a highly specific marker gene of interstitial progenitor cells that enables their transcriptional, flow cytometric, and functional distinction from Icam1+/Col15a1+ preadipocytes [32]. Flow cytometry and histological analysis validated the labeling fidelity of our Dpp4CreER allele at the pulse timepoint. Our results conclusively show that Dpp4+ progenitors give rise to DPP4(−)/ICAM1+ preadipocytes, as well as mature adipocytes. Thus, these findings provide direct in vivo evidence of an adipogenic lineage hierarchy that contributes to adipose tissue renewal and growth.
Recent single cell analyses conducted across a broad range of murine and human mesenchymal tissues showed the ubiquitous presence of Dpp4+/Pi16+ progenitor/fibroblast cells [41]. Dpp4+/Pi16+ cells, named “universal fibroblasts”, are predicted to give rise to “specialized fibroblasts” as directed by local tissue requirements. The authors of this study generated a DptCreER lineage tracing mouse model to track the fate of labeled universal fibroblasts. Using a subcutaneous tumor model, the authors found that Dpt + fibroblasts gave rise to Lrrc15+ myofibroblast-like cells, confirming a lineage relationship between universal and specialized fibroblasts in the skin. However, Dpt is also highly expressed by Icam1+/Col15a1+ fibroblasts, which were co-labeled with the Dpp4+/Pi16+ cells in the study. Due to the broad expression profile of Dpt, the authors were unable to distinguish between the lineage contribution of Dpp4+/Pi16+ versus Icam1+/Col15a1+ cells. Our results provide additional support for the concept of DPP4+ cells functioning as universal fibro-progenitors and demonstrate that these cells give rise to preadipocytes (specialized fibroblasts) and adipocytes across several adipose tissue depots.
The adipogenic contribution of Dpp4+ progenitors depends on the anatomic location of the depot as well as the metabolic status of the organism. We found that Dpp4+ progenitors participated in basal adipocyte turnover of in all examined depots of chow fed animals. HFD stimulated a significant increase in Dpp4+ adipogenic differentiation in visceral depots, especially those arising within a dense layer of reticular interstitial connective tissue, including omental and retroperitoneal adipose.
Obesogenic diets have been shown to stimulate adipose tissue expansion via metabolically favorable hyperplasia of new adipocytes as well as pathologic hypertrophy of existing adipocytes. While both hyperplasia and hypertrophy are thought to occur in most human adipose depots [46,47], in mice, the subcutaneous adipose tissues (iWAT and axWAT) expand primarily via hypertrophy while the visceral WAT (gWAT, mesWAT, omentum and rpWAT) expands via both hyperplasia and hypertrophy [16]. Consistent with this hypothesis, we did not see a significant effect of obesogenic diet on Dpp4+ contribution to mature adipocytes in murine subcutaneous depots. When we examined the subcutaneous APC populations by flow cytometry, we found that HFD promoted an accumulation of DPP4(−)/ICAM1+ preadipocytes. These cells likely represent Dpp4+ progenitors that were induced to progress toward adipogenic commitment by HFD but became stalled as DPP4(−)/ICAM1+ cells. The proportion of Dpp4+ progenitors transiting through the DPP4(−)/ICAM1+ preadipocyte fate was unchanged by HFD in visceral adipose tissue, suggesting that preadipocytes in visceral depots possess higher differentiation activity. The cellular and environmental signaling pathways that underlie this depot-specific effect remain an intriguing avenue for future investigation.
Our results suggest that Dpp4+ cells do not play a major role in basal adipocyte turnover in the subcutaneous WAT. Dpp4(−)/Icam1+ cells, which are distributed throughout the parenchyma of the depot and are poised to undergo adipogenic differentiation, are likely responsible for mediating adipocyte turnover [32]. By contrast, our data suggest that the visceral WAT is more highly dependent on Dpp4+ interstitial progenitors to supply sufficient preadipocytes, especially when de novo adipogenesis is stimulated in the setting of obesity.
Dpp4+/Pi16+ cells exhibit many properties of progenitor/stem cells, including high proliferative capacity and multipotent differentiation potential, yet simultaneously express many gene pathways classically attributed to fibroblasts, including collagen production and ECM modification [34]. This expression profile is consistent with the anatomic location of DPP4+ cells, which cling to collagen/elastin fibers within the reticular interstitium, and likely contribute to the structural integrity of connective tissues [40]. Because murine subcutaneous WAT expands primarily via hypertrophy of existing adipocytes with very little de novo adipogenesis, Dpp4+ cells may not be called upon to support adipose tissue expansion. Rather, DPP4+ cells in the subcutaneous WAT may be primarily tasked with collagen remodeling to provide structural support for the enlarging adipose depot. As obesity progresses, it is plausible that this beneficial fibroblast-like activity of Dpp4+ cells may become maladaptive and promote tissue fibrosis.
In summary, our studies identify DPP4+ progenitor cells as an important source of committed preadipocytes and adipocytes, especially in human relevant visceral fat depots. Further studies are necessary to identify the genes and pathways controlling the activity of DPP4+ cells, including the mechanisms that control adipogenic commitment within adipose tissue. Understanding the regulation of these cells may reveal new avenues to promote adipocyte differentiation and metabolic health.
4. Methods
4.1. Animals
C57BL/6J WT [stock number (no.) 000664] and C57BL/6J:ROSAmT/mG (stock no. 007676) mouse lines were obtained from Jackson Laboratories. CD1 WT mice (stock no. 022) were obtained from Charles River. All animal procedures were performed under the guidance of the University of Pennsylvania Institutional Animal Care and Use Committee and strictly in accordance with the guidelines of the Institutional Animal Care and Use Committee (IACUC). For lineage tracing studies, tamoxifen (Sigma, T5648) was dissolved in corn oil (20 mg/ml) and administered by an intraperitoneal injection on five subsequent days at the indicated timepoint (0.1 μg tamoxifen per gram of mouse body weight). All animals were maintained at 25 °C and fed either regular chow diet (10% kcal fat) or high fat diet (60% kcal fat) for 18 weeks.
4.2. Generation of Dpp4CreER mouse strain
The Dpp4-CreERT2 mouse was generated by insertion of the coding sequence for CreERT2 followed by a self-cleaving P2A peptide (derived from derived from porcine teschovirus-1) into the start codon of the Dpp4 gene by CRISPR/Cas9 stimulated homologous recombination. In detail, a high-fidelity gRNA target was identified in close proximity to the start codon of Dpp4. A DNA vector was synthesized containing the coding sequence for CreERT2, with a carboxy-terminal P2A peptide, which was flanked by two 500 bp homology arms spanning the upstream and downstream regions of the Dpp4 locus on either side of the Cas9 cut site. The CreER-P2A construct was designed in-frame with the native Dpp4 gene product, such that CreERT2 and Dpp4 are simultaneously expressed under the control of the native Dpp4 promoter/enhancer elements. Single stranded template DNA (ssDNA) was generated by strand-specific exonuclease digestion and purified by gel electrophoresis. Ribonucleoprotein complexes (RNPs) were assembled by combination of recombinant Cas9 protein (Alt-R HiFi Cas9 Nuclease, IDT technologies), with synthesized guide and tracer RNAs (IDT technologies). ssDNA was mixed with the RNPs in pronuclear injection buffer (1 mM Tris HCl, pH 7.5; 0.1 mM EDTA) and injected into C57/Bl6 mouse zygotes at the 1 cell stage, followed by implantation into pseudo-pregnant female mice. Proper transgene insertion was confirmed by sanger sequencing of genomic PCR products spanning the entire homology region. DPP4 enzymatic activity was measured from mouse serum using the DPP4 Activity Assay Kit (Millipore Sigma # MAK088), which showed that Dpp4CreER mice retained 75% and 86% of wild-type DPP4 activity levels in serum, and RI tissue, respectively (Figure S1B).
4.3. Isolation of stromal cells from mouse adipose tissues
Visceral and subcutaneous WAT depots were surgically removed and processed using enzymatic and mechanical methods. Adipose tissues were manually minced and digested with collagenase type 1 (1.5 units/ml; Worthington) and dispase II (2.4 units/ml; Roche) in DMEM/F12 containing 1.0% fatty acid–free bovine serum albumin (Sigma). Mechanical dissociation was carried out in GenleMACS C-tubes using the genleMACS Octo Dissociated with Heaters (Miltenyi Biotec). The digestion was quenched with DMEM/F12 containing 10% FBS, and the dissociated cells were passed through a 100-μM filter and then subjected to centrifugation at 400×g for 10 min. The resulting supernatant containing mature adipocytes was aspirated, and the pellet, consisting of stromal vascular cells (SVCs), was resuspended in red blood cell lysis buffer (Biolegend) for 5 min at RT and then quenched in DMEM/F12 containing 10% FBS. The cells were then passed through a 40-μM filter and collected by centrifugation at 400×g for 5 min. The cells were recovered in FACS buffer (HBSS containing 1% FBS; Fisher).
4.4. Flow cytometry
SVCs from the visceral and subcutaneous depots were prepared as described above, resuspended in FACS buffer, and labeled with the following antibodies for 30 min at 4 °C: CD26 (DPP4)–fluorescein isothiocyanate (APC) (Biolegend, San Diego, CA; catalog no. 137806; 1:400), anti–mouse ICAM1–phycoerythrin (PE)/Cy7 (Biolegend catalog no. 116122; 1:200), anti–mouse CD45–allophycocyanin (APC)/Cy7 (Biolegend catalog no. 103116; 1:500). 4′,6-Diamidino-2-phenylindole (DAPI) (Roche catalog no. 10236276001; 1:10,000) was added for the final 5 min. The cells were washed three times with FACS buffer to remove unbound antibodies. The cells were analyzed using a BD LSRII flow cytometer (BD Biosciences). All compensation was performed at the time of acquisition in using compensation beads (BioLegend catalog no. A10497) for single-color staining and SVCs for negative staining and fluorescence (DAPI and mTomato). Analysis was performed using FlowJo 10.7. All gates were precisely defined using full-minus-one stained control cells.
4.5. Histology
Tissues were fixed with 4% paraformaldehyde via transcardiac perfusion, after which dissected adipose pads were allowed to fix overnight. The tissues were subsequently dehydrated through a series of ethanol washes and then embedded in paraffin for thin sectioning. Immunohistochemistry analysis was performed by following heat antigen retrieval methods, and samples were stained with the following antibodies: anti-GFP (goat, 1:500, Abcam #AB6673), anti-DPP4 (goat; 1:500; R&D Systems # AF954), anti-PLIN1 (rabbit, 1:500, Cell Signaling #3470). Paraffin processing, embedding, sectioning, hematoxylin/eosin and fluorescent staining were performed by Lan Cheng at the Institute for Diabetes, Obesity and Metabolism Histology Core at the University of Pennsylvania.
4.6. Quantification of adipocyte differentiation
Lineage-traced mature adipocytes were quantified on histology by manually counting GFP+/PLIN1+ double-positive adipocytes across an entire cross section of adipose tissue. Briefly, adipose tissues were thin sectioned in the maximal axis as described above and stained with anti-GFP and anti-PLIN1. Whole adipose tissue sections were imaged on a Keyence inverted fluorescence microscope (BZX-710) equipped with the following filters: DAPI (excitation, 360/40 nm; emission, 460/50 nm; Keyence, OP-87762), GFP (excitation, 470/40 nm; emission, 525/50 nm; Keyence, OP-87763) TRITC (excitation, 545/25 nm, emission 605/70 nm; Keyence OP-87764), and Cy5 (excitation, 620/60 nm, emission 700/75 nm; Keyence OP-87766). Adipose tissue sections were imaged in their entirety at 10x magnification using a tiled grid. Tiling and stitching were performed with Keyence BZ-X Viewer software. Quantification of labeled adipocytes was performed in ImageJ software using the “Cell Counter” plugin to count GFP+/PLIN1+ adipocytes. Total adipocyte count was performed using CellProfiler software (Broad Institute Inc) using the “IdentifyPrimaryObjects” module. Normalization for differences in adipose tissue slice size and cell number was accomplished by dividing the number of GFP + lineage traced adipocytes by the total PLIN1+ adipocyte count.
4.7. Statistical methods
Statistical methods were not used to predetermine sample size. The experiments were not randomized, and investigators were not blinded in experiments. All p-values are reported with adjustment for multiple comparisons. All flow and histological data are representative of at least three biological replicates. Most statistical tests were performed using an ANOVA followed by multiple comparisons. For quantification of flow cytometric data, tissue samples generated both DPP4+ and ICAM1+ cell populations, therefore two-way ANOVA was used to account for the matched nature of these data. Where indicated, biologic replicates (BRs) represent independent samples (i.e., cells derived from different mice). Statistical analyses were conducted using GraphPad Prism 7.0 (GraphPad Software, Inc).
Funding
This work was supported by NIH grants R01DK120982 and R01DK121801 to P.S., NIH grant K08DK122099 and the Measey Physician Scientist Fellowship to DM and P30-DK19525 (to Penn Diabetes Research Center).
Author contributions
D.M. and P.S. were responsible for conceptualization, investigation, data curation and analysis, methodology, validation, visualization, and writing and review. M.S. was involved in writing and review. M.S., S.T., and L.C. performed experiments and assisted with data analysis.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.molmet.2021.101357.
Contributor Information
Megan Stefkovich, Email: mstefko@pennmedicine.upenn.edu.
Sarah Traynor, Email: sarah.traynor@pennmedicine.upenn.edu.
Lan Cheng, Email: lancheng@pennmedicine.upenn.edu.
David Merrick, Email: david.merrick@pennmedicine.upenn.edu.
Patrick Seale, Email: sealep@pennmedicine.upenn.edu.
Conflict of interest
None.
Appendix ASupplementary data
The following are the Supplementary data to this article:
FigS1.
The Dpp4CreERmouse enables robust and specific labeling of DPP4+ cells in the intestine and liver. (A) Histological examination of the intestine and liver were conducted as a positive control for Dpp4CreER; mGFP labeling of cells known to express high levels of Dpp4. The pulse timepoint is depicted in the left panels and a 3-week chow diet chase timepoint on the right panels. DPP4+ intestinal epithelial cells are sloughed off and replaced by unlabeled cells at the chase timepoint. [mGFP: lineage traced cells (green), DPP4: cells currently expressing DPP4 (magenta), DAPI: nuclei (blue); scale bar = 380 μM]. Individual antibody staining is represented in grayscale on the right-side inset panels (inset scale bar = 100 μm). (B) Quantification of DPP4 enzymatic activity from serum (left panel) or dissected RI tissue (right panel) of wild type (WT) and Dpp4CreER mice.
FigS2.
Quantification of mouse weight gain during dietary chase. (A) Total body weight was quantified both before and after the 16-week dietary chase period for mice used in histologic experiments and (B) flow cytometry experiments.
FigS3.
Quantification of Dpp4+ lineage traced mature adipocytes in subcutaneous and gonadal WAT. GFP+/PLIN1+ mature adipocytes were quantified and normalized to the total number of PLIN1+ adipocytes per section. Open shapes = female mice, solid shapes = male mice.
FigS4.
Quantification of Dpp4+ lineage traced mature adipocytes in the omental and retroperitoneal depots. GFP+/PLIN1+ mature adipocytes were quantified and normalized the total number of PLIN1+ adipocytes per section. Gonadal WAT quantification was included for reference. Open shapes = female mice, solid shapes = male mice.
References
- 1.Wang Y., Beydoun M.A., Min J., Xue H., Kaminsky L.A., Cheskin L.J. Has the prevalence of overweight, obesity and central obesity levelled off in the United States? Trends, patterns, disparities, and future projections for the obesity epidemic. International Journal of Epidemiology. 2020;49(3):810–823. doi: 10.1093/ije/dyz273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hepler C., Gupta R.K. The expanding problem of adipose depot remodeling and postnatal adipocyte progenitor recruitment. Molecular and Cellular Endocrinology. 2017;445:95–108. doi: 10.1016/j.mce.2016.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ghaben A.L., Scherer P.E. Adipogenesis and metabolic health. Nature Reviews Molecular Cell Biology. 2019;20(4):242–258. doi: 10.1038/s41580-018-0093-z. [DOI] [PubMed] [Google Scholar]
- 4.Ravussin E., Smith S.R. Increased fat intake, impaired fat oxidation, and failure of fat cell proliferation result in ectopic fat storage, insulin resistance, and type 2 diabetes mellitus. Annals of the New York Academy of Sciences. 2002;967:363–378. doi: 10.1111/j.1749-6632.2002.tb04292.x. [DOI] [PubMed] [Google Scholar]
- 5.Isakson P., Hammarstedt A., Gustafson B., Smith U. Impaired preadipocyte differentiation in human abdominal obesity: role of Wnt, tumor necrosis factor-alpha, and inflammation. Diabetes. 2009;58(7):1550–1557. doi: 10.2337/db08-1770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vishvanath L., MacPherson K.A., Hepler C., Wang Q.A., Shao M., Spurgin S.B., et al. Pdgfrbeta+ mural preadipocytes contribute to adipocyte hyperplasia induced by high-fat-diet feeding and prolonged cold exposure in adult mice. Cell Metabolism. 2016;23(2):350–359. doi: 10.1016/j.cmet.2015.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shao M., Vishvanath L., Busbuso N.C., Hepler C., Shan B., Sharma A.X., et al. De novo adipocyte differentiation from Pdgfrbeta(+) preadipocytes protects against pathologic visceral adipose expansion in obesity. Nature Communications. 2018;9(1):890. doi: 10.1038/s41467-018-03196-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kusminski C.M., Holland W.L., Sun K., Park J., Spurgin S.B., Lin Y., et al. MitoNEET-driven alterations in adipocyte mitochondrial activity reveal a crucial adaptive process that preserves insulin sensitivity in obesity. Nature Medicine. 2012;18(10):1539–1549. doi: 10.1038/nm.2899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim J.Y., van de Wall E., Laplante M., Azzara A., Trujillo M.E., Hofmann S.M., et al. Obesity-associated improvements in metabolic profile through expansion of adipose tissue. Journal of Clinical Investigation. 2007;117(9):2621–2637. doi: 10.1172/JCI31021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hammarstedt A., Gogg S., Hedjazifar S., Nerstedt A., Smith U. Impaired adipogenesis and dysfunctional adipose tissue in human hypertrophic obesity. Physiological Reviews. 2018;98(4):1911–1941. doi: 10.1152/physrev.00034.2017. [DOI] [PubMed] [Google Scholar]
- 11.Karpe F., Pinnick K.E. Biology of upper-body and lower-body adipose tissue--link to whole-body phenotypes. Nature Reviews Endocrinology. 2015;11(2):90–100. doi: 10.1038/nrendo.2014.185. [DOI] [PubMed] [Google Scholar]
- 12.Manolopoulos K.N., Karpe F., Frayn K.N. Gluteofemoral body fat as a determinant of metabolic health. International Journal of Obesity. 2010;34(6):949–959. doi: 10.1038/ijo.2009.286. [DOI] [PubMed] [Google Scholar]
- 13.van Beek L., van Klinken J.B., Pronk A.C., van Dam A.D., Dirven E., Rensen P.C., et al. The limited storage capacity of gonadal adipose tissue directs the development of metabolic disorders in male C57Bl/6J mice. Diabetologia. 2015;58(7):1601–1609. doi: 10.1007/s00125-015-3594-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jeffery E., Church C.D., Holtrup B., Colman L., Rodeheffer M.S. Rapid depot-specific activation of adipocyte precursor cells at the onset of obesity. Nature Cell Biology. 2015;17(4):376–385. doi: 10.1038/ncb3122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang Q.A., Tao C., Gupta R.K., Scherer P.E. Tracking adipogenesis during white adipose tissue development, expansion and regeneration. Nature Medicine. 2013;19(10):1338–1344. doi: 10.1038/nm.3324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim S.M., Lun M., Wang M., Senyo S.E., Guillermier C., Patwari P., et al. Loss of white adipose hyperplastic potential is associated with enhanced susceptibility to insulin resistance. Cell Metabolism. 2014;20(6):1049–1058. doi: 10.1016/j.cmet.2014.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gyllenhammer L.E., Alderete T.L., Toledo-Corral C.M., Weigensberg M., Goran M.I. Saturation of subcutaneous adipose tissue expansion and accumulation of ectopic fat associated with metabolic dysfunction during late and post-pubertal growth. International Journal of Obesity. 2016;40(4):601–606. doi: 10.1038/ijo.2015.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Suganami T., Tanaka M., Ogawa Y. Adipose tissue inflammation and ectopic lipid accumulation. Endocrine Journal. 2012;59(10):849–857. doi: 10.1507/endocrj.ej12-0271. [DOI] [PubMed] [Google Scholar]
- 19.Yatsuya H., Nihashi T., Li Y., Hotta Y., Matsushita K., Muramatsu T., et al. Independent association of liver fat accumulation with insulin resistance. Obesity Research & Clinical Practice. 2014;8(4):e350–e355. doi: 10.1016/j.orcp.2013.08.002. [DOI] [PubMed] [Google Scholar]
- 20.Tardif N., Salles J., Guillet C., Tordjman J., Reggio S., Landrier J.F., et al. Muscle ectopic fat deposition contributes to anabolic resistance in obese sarcopenic old rats through eIF2alpha activation. Aging Cell. 2014;13(6):1001–1011. doi: 10.1111/acel.12263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Uezumi A., Fukada S., Yamamoto N., Takeda S., Tsuchida K. Mesenchymal progenitors distinct from satellite cells contribute to ectopic fat cell formation in skeletal muscle. Nature Cell Biology. 2010;12(2):143–152. doi: 10.1038/ncb2014. [DOI] [PubMed] [Google Scholar]
- 22.Gustafson B., Smith U. Regulation of white adipogenesis and its relation to ectopic fat accumulation and cardiovascular risk. Atherosclerosis. 2015;241(1):27–35. doi: 10.1016/j.atherosclerosis.2015.04.812. [DOI] [PubMed] [Google Scholar]
- 23.Shulman G.I. Ectopic fat in insulin resistance, dyslipidemia, and cardiometabolic disease. New England Journal of Medicine. 2014;371(12):1131–1141. doi: 10.1056/NEJMra1011035. [DOI] [PubMed] [Google Scholar]
- 24.Miyazaki Y., Mahankali A., Matsuda M., Mahankali S., Hardies J., Cusi K., et al. Effect of pioglitazone on abdominal fat distribution and insulin sensitivity in type 2 diabetic patients. Journal of Clinical Endocrinology & Metabolism. 2002;87(6):2784–2791. doi: 10.1210/jcem.87.6.8567. [DOI] [PubMed] [Google Scholar]
- 25.Tang W., Zeve D., Seo J., Jo A.Y., Graff J.M. Thiazolidinediones regulate adipose lineage dynamics. Cell Metabolism. 2011;14(1):116–122. doi: 10.1016/j.cmet.2011.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gavrilova O., Marcus-Samuels B., Graham D., Kim J.K., Shulman G.I., Castle A.L., et al. Surgical implantation of adipose tissue reverses diabetes in lipoatrophic mice. Journal of Clinical Investigation. 2000;105(3):271–278. doi: 10.1172/JCI7901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tran T.T., Yamamoto Y., Gesta S., Kahn C.R. Beneficial effects of subcutaneous fat transplantation on metabolism. Cell Metabolism. 2008;7(5):410–420. doi: 10.1016/j.cmet.2008.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Foster M.T., Softic S., Caldwell J., Kohli R., de Kloet A.D., Seeley R.J. Subcutaneous adipose tissue transplantation in diet-induced obese mice attenuates metabolic dysregulation while removal exacerbates it. Physics Reports. 2013;1(2) doi: 10.1002/phy2.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee Y.H., Petkova A.P., Mottillo E.P., Granneman J.G. In vivo identification of bipotential adipocyte progenitors recruited by β3-adrenoceptor activation and high-fat feeding. Cell Metabolism. 2012;15(4):480–491. doi: 10.1016/j.cmet.2012.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee Y.H., Petkova A.P., Granneman J.G. Identification of an adipogenic niche for adipose tissue remodeling and restoration. Cell Metabolism. 2013;18(3):355–367. doi: 10.1016/j.cmet.2013.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Han X., Zhang Z., He L., Zhu H., Li Y., Pu W., et al. A suite of new Dre recombinase drivers markedly expands the ability to perform intersectional genetic targeting. Cell Stem Cell. 2021;28(6):1160–1176.e1167. doi: 10.1016/j.stem.2021.01.007. [DOI] [PubMed] [Google Scholar]
- 32.Merrick D., Sakers A., Irgebay Z., Okada C., Calvert C., Morley M.P., et al. Identification of a mesenchymal progenitor cell hierarchy in adipose tissue. Science. 2019;364(6438) doi: 10.1126/science.aav2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Burl R.B., Ramseyer V.D., Rondini E.A., Pique-Regi R., Lee Y.H., Granneman J.G. Deconstructing adipogenesis induced by beta3-adrenergic receptor activation with single-cell expression profiling. Cell Metabolism. 2018;28(2):300–309. doi: 10.1016/j.cmet.2018.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hepler C., Shan B., Zhang Q., Henry G.H., Shao M., Vishvanath L., et al. Identification of functionally distinct fibro-inflammatory and adipogenic stromal subpopulations in visceral adipose tissue of adult mice. Elife. 2018;7 doi: 10.7554/eLife.39636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schwalie P.C., Dong H., Zachara M., Russeil J., Alpern D., Akchiche N., et al. A stromal cell population that inhibits adipogenesis in mammalian fat depots. Nature. 2019;559(7712):103–108. doi: 10.1038/s41586-018-0226-8. [DOI] [PubMed] [Google Scholar]
- 36.Angueira A.R., Sakers A.P., Holman C.D., Cheng L., Arbocco M.N., Shamsi F., et al. Defining the lineage of thermogenic perivascular adipose tissue. Nature Metabolism. 2021;3(4):469–484. doi: 10.1038/s42255-021-00380-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Oguri Y., Shinoda K., Kim H., Alba D.L., Bolus W.R., Wang Q., et al. CD81 controls beige fat progenitor cell growth and energy balance via FAK signaling. Cell. 2020;182(3):563–577.e520. doi: 10.1016/j.cell.2020.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Spallanzani R.G., Zemmour D., Xiao T., Jayewickreme T., Li C., Bryce P.J., et al. Distinct immunocyte-promoting and adipocyte-generating stromal components coordinate adipose tissue immune and metabolic tenors. Science Immunology. 2019;4(35) doi: 10.1126/sciimmunol.aaw3658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zachara M., Rainer P.Y., Russeil J.M., Hashimi H., Alpern D., Ferrero R., et al. Mammalian adipogenesis regulators (Aregs) exhibit robust non- and anti-adipogenic properties that arise with age and involve retinoic acid signalling. bioRxiv. 2021;2002:2024. 432431. [Google Scholar]
- 40.Benias P.C., Wells R.G., Sackey-Aboagye B., Klavan H., Reidy J., Buonocore D., et al. Structure and distribution of an unrecognized interstitium in human tissues. Scientific Reports. 2018;8(1):4947. doi: 10.1038/s41598-018-23062-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Buechler M.B., Pradhan R.N., Krishnamurty A.T., Cox C., Calviello A.K., Wang A.W., et al. Cross-tissue organization of the fibroblast lineage. Nature. 2021;593(7860):575–579. doi: 10.1038/s41586-021-03549-5. [DOI] [PubMed] [Google Scholar]
- 42.Wilkosz S., Ireland G., Khwaja N., Walker M., Butt R., de Giorgio-Miller A., et al. A comparative study of the structure of human and murine greater omentum. Anatomy and Embryology. 2005;209(3):251–261. doi: 10.1007/s00429-004-0446-6. [DOI] [PubMed] [Google Scholar]
- 43.Rondini E.A., Granneman J.G. Single cell approaches to address adipose tissue stromal cell heterogeneity. Biochemical Journal. 2020;477(3):583–600. doi: 10.1042/BCJ20190467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ferrero R., Rainer P., Deplancke B. Toward a consensus view of mammalian adipocyte stem and progenitor cell heterogeneity. Trends in Cell Biology. 2020;30(12):937–950. doi: 10.1016/j.tcb.2020.09.007. [DOI] [PubMed] [Google Scholar]
- 45.Vishvanath L., Gupta R.K. Contribution of adipogenesis to healthy adipose tissue expansion in obesity. Journal of Clinical Investigation. 2019;129(10):4022–4031. doi: 10.1172/JCI129191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Van Harmelen V., Röhrig K., Hauner H. Comparison of proliferation and differentiation capacity of human adipocyte precursor cells from the omental and subcutaneous adipose tissue depot of obese subjects. Metabolism. 2004;53(5):632–637. doi: 10.1016/j.metabol.2003.11.012. [DOI] [PubMed] [Google Scholar]
- 47.Belligoli A., Compagnin C., Sanna M., Favaretto F., Fabris R., Busetto L., et al. Characterization of subcutaneous and omental adipose tissue in patients with obesity and with different degrees of glucose impairment. Scientific Reports. 2019;9(1):11333. doi: 10.1038/s41598-019-47719-y. [DOI] [PMC free article] [PubMed] [Google Scholar]