Abstract
Objective:
Subthalamic deep brain stimulation (DBS) is an established therapy for Parkinson’s disease. Connectomic DBS modeling is a burgeoning subfield of research aimed at characterizing the axonal connections activated by DBS. This article describes our approach and methods for evolving the StimVision software platform to meet the technical demands of connectomic DBS modeling in the subthalamic region.
Materials and Methods:
StimVision v2 was developed with Visualization Toolkit (VTK) libraries and integrates four major components: 1) medical image visualization, 2) axonal pathway visualization, 3) electrode positioning, and 4) stimulation calculation.
Results:
StimVision v2 implemented two key technological advances for connectomic DBS analyses in the subthalamic region. First was the application of anatomical axonal pathway models to patient-specific DBS models. Second was the application of a novel driving-force method to estimate the response of those axonal pathways to DBS. Example simulations with directional DBS electrodes and clinically defined therapeutic DBS settings are presented to demonstrate the general outputs of StimVision v2 models.
Conclusions:
StimVision v2 provides the opportunity to evaluate patient-specific axonal pathway activation from subthalamic DBS using anatomically detailed pathway models and electrically detailed electric field distributions with interactive adjustment of the DBS electrode position and stimulation parameter settings.
Keywords: Axonal pathways, basal ganglia, driving-force, electrode, hyperdirect, subthalamic nucleus
INTRODUCTION
Deep brain stimulation (DBS) of the subthalamic region is an established treatment for the motor symptoms of Parkinson’s disease (1). The clinical efficacy of subthalamic DBS is often dramatic, with off-medication on-stimulation improvements of 50% or greater in clinical rating scale scores (2). However, some patients fail to achieve that targeted level of therapeutic benefit. Therefore, scientific and clinical questions exist on the source of that therapeutic variability. Candidate topics of interest include: patient selection (3), electrode placement (4), stimulation parameter selection (5), and the neural response to stimulation (6). One tool that has been used to help address some of those questions is patient-specific DBS modeling (7). Computer models that integrate anatomical information from magnetic resonance imaging (MRI) data, and electrical information from the stimulation parameter settings, are now commonly used in clinical DBS research to help identify relationships between stimulation and behavioral responses (8,9).
Over the last decade, patient-specific DBS models have steadily evolved to incorporate greater levels of anatomical and electrical detail (10). On the anatomical side, the clinical imaging data available for a given patient is often inadequate for detailed analyses, so DBS models are typically augmented by the fitting of brain atlases to the patient (11,12). The fitted brain atlas provides estimates of anatomical structures that are not clearly visible on the MRI, which is often the case for the subthalamic nucleus (STN). On the electrical side, the estimated effect of DBS on neural activity can be calculated with a wide range of methods, using algorithms that range from simple to complex (8). However, as the scientific and clinical questions on subthalamic DBS have become more elaborate, the use of biophysically detailed models of stimulation is likely warranted (13).
One aspect of patient-specific DBS modeling that has recently received great attention is the incorporation of axonal pathway models into the anatomical rendering of the patient (14). This is because the primary neural response to DBS is thought to be axonal activation (15). As such, DBS research has gravitated toward attempting to understand the brain network connections that are being directly modulated by therapeutic stimulation. This burgeoning subfield, known as connectomic DBS modeling, is currently dominated by the use of diffusion-weighted imaging and tractography algorithms to estimate the structural connectivity of the brain (8,9). However, tractography is known to suffer from substantial limitations in its ability to generate anatomically realistic pathway representations (16). Therefore, alternative strategies are evolving to define human axonal pathway models, based on known connections in the brain and annotated by expert neuroanatomists, to overcome some of the limitations of tractography (17–19).
StimVision is the integrated software tool we developed for prospective testing of connectomic DBS hypotheses in clinical research studies (20). The initial application for StimVision was in subcallosal cingulate DBS, which has thus far been very successful at helping to improve DBS outcomes in treatment-resistant depression patients (21). However, the initial version of StimVision relied on tractography and simplistic DBS modeling methodologies, both of which make that tool likely to only be applicable in the study of cylindrical contact DBS electrodes directly implanted into large white matter pathways. Generalized use of connectomic DBS modeling concepts in other areas of the brain (i.e., basal ganglia), or other electrode designs (i.e., directional contact DBS electrodes), require greater technical detail to generate results with scientific credibility (8). As such, this article describes our approach and methods for evolving the StimVision platform to meet the demands of connectomic DBS modeling in the subthalamic region.
MATERIALS AND METHODS
Ethics Statement
StimVision is an academic DBS research tool and does not have any form of government body regulatory approval. As such, any use of StimVision is strictly limited to Institutional Review Board (IRB) approved research studies at individual academic institutions. The collection and analysis of all patient data used for this article was approved by the Case Western Reserve University and/or University Hospitals IRB.
StimVision Overview
StimVision integrates multiple computational functions and software tools into a common visualization framework for DBS research (20). StimVision v2 is an evolution of that platform, specifically designed for clinical research on subthalamic DBS. Preoperatively, StimVision v2 can be used to facilitate the identification of an electrode implant location that theoretically stimulates a targeted collection of subthalamic axonal pathways in an individual patient. Postoperatively, StimVision v2 can be used to evaluate the subthalamic axonal pathways that are theoretically activated for a specific stimulation parameter setting. To accomplish these tasks, StimVision v2 consists of four major components (Figs. 1 and 2): 1) medical image visualization, 2) axonal pathway visualization, 3) electrode positioning, and 4) stimulation calculation. A key aspect of the software is the ability to integrate these four components within the context of the patient-specific stereotactic neurosurgical coordinate system (or frame space). This enables the model predictions to be directly merged, or compared, with the actual neurosurgical equipment used in clinical practice.
Figure 1.
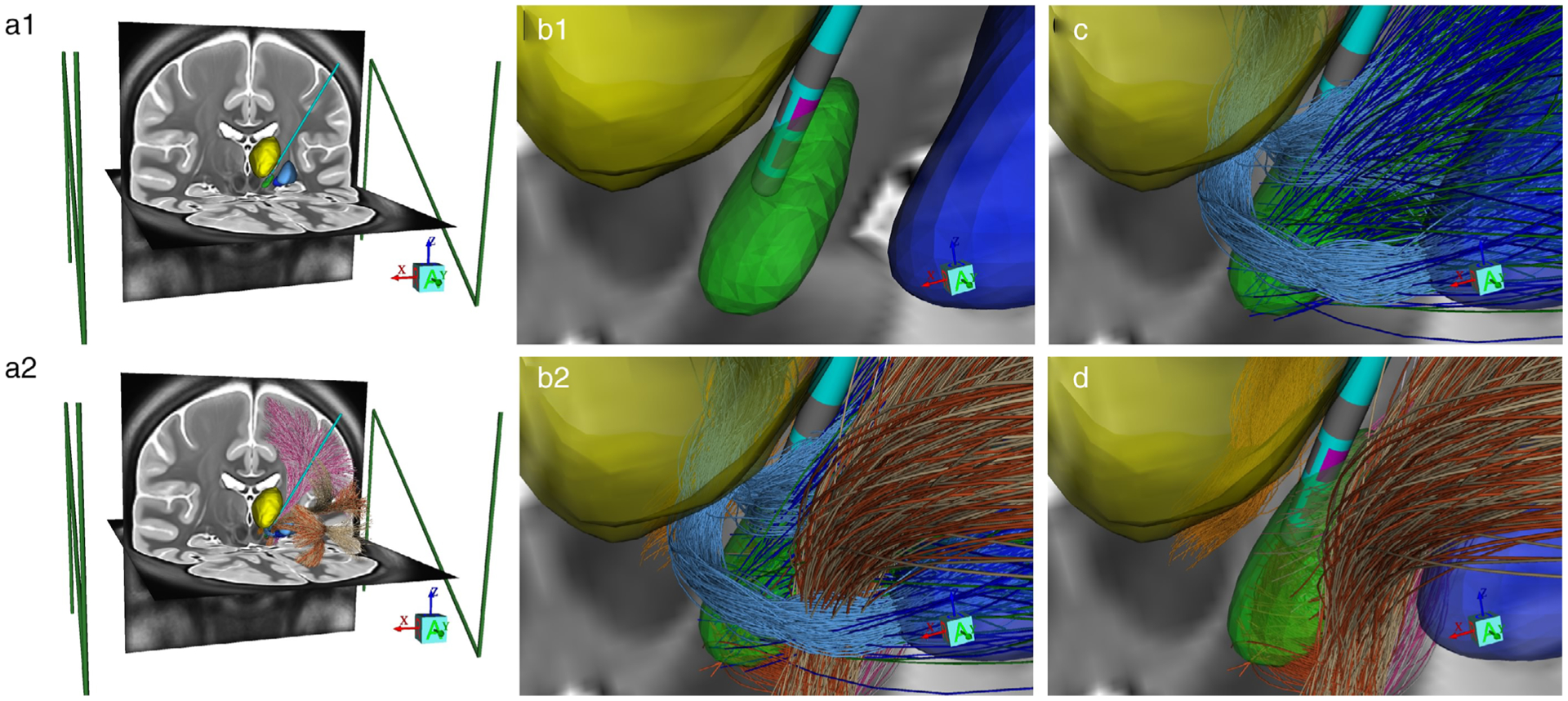
StimVision v2 anatomical model. a1. Integration of MRI data, anatomical nuclei, and the stereotactic frame coordinate system (green fiducial markers). The CIT168 brain atlas is used in this example (22). a2. Addition of the Petersen et al. (19) axonal pathway models. b1. Zoomed in view of the subthalamic region with a BSN 2202 lead placed in the STN (green volume), surrounded by the thalamus (yellow volume) and globus pallidus (blue volume). Contact 5 (pink) is active. b2. Addition of the Petersen et al. (19) axonal pathway models. c. Basal ganglia pathways: pallidothalamic (light blue), subthalamopallidal (green), pallidosubthalamic fibers (dark blue). d. Cerebellothalamic (orange) and corticofugal pathways: IC fibers of passage (white—IC motor; tan—IC PFC) and HDP (pink—HDP motor; burnt orange—HDP PFC).
Figure 2.
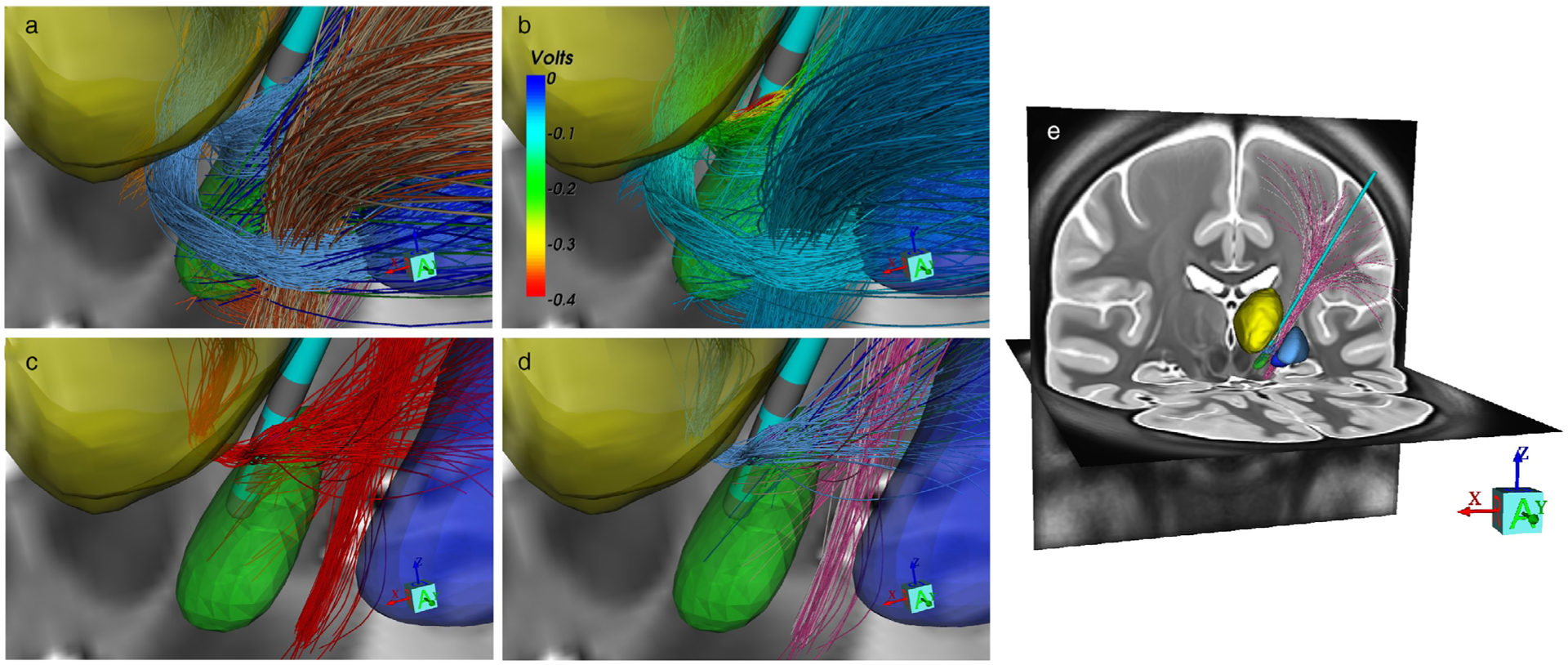
StimVision v2 activation model. a. Petersen et al. (19) pathway models. Nuclei and pathway color codes provided in Figure 1. b. Extracellular voltage distribution applied to each axon model from −1 mA (cathodic) 60 μsec stimulation delivered through contact 5. c. Activated pathways (red), as calculated by the Howell et al. (13) driving-force algorithm. d. Activated pathways displayed with their corresponding anatomical pathway color. e. Whole brain view of the activated connections with cortex.
Two key technological advances were necessary for the StimVision platform to facilitate connectomic DBS analyses in the subthalamic region. First was the creation of anatomically realistic axonal pathway models for many of the important pathways within the subthalamic region (19) (Fig. 1). Second was the transition away from volume of tissue activated (VTA) models (8), and implementation of a driving-force (DF) method to better estimate the response of complex axonal pathways to DBS (13) (Fig. 2).
Software Architecture
StimVision v2 is written in tool command language with a graphical user interface widget toolkit (tcl/Tk) (http://www.tcl.tk/) with Visualization Toolkit (VTK) libraries (http://www.vtk.org/). Tcl/Tk is a precompiled scripting language and VTK is an object-based visualization application programming interface that uses OpenGL (http://www.opengl.org/) for rendering. The software is cross-platform and can run on Linux, Windows, Mac OS, and Unix. The user interacts with the software through a tcl/Tk graphical user interface (GUI) which enables the import and display of images, electrode models, stimulation models, and axonal pathways using a VTK display window.
Medical Image Data
The first steps in the creation of a StimVision v2 patient-specific subthalamic DBS model are the acquisition, coregistration, and loading of the patient imaging data. StimVision is designed to read in coregistered imaging data sets in NIfTI format (http://nifti.nimh.nih.gov/). These images could include any range of different data sets available for the patient (e.g., T1-weighted [T1w] MRI, T2-weighted MRI, computed tomography [CT], etc.). The example patient-specific StimVision v2 models displayed in the figures were simply retrospective stimulation examples, so we only needed the postoperative T1w MRI to construct those models. Those images were acquired approximately one month after the DBS surgery on a 1.5T Siemens scanner with 0.8 mm isotropic resolution. However, StimVision v2 models that are used for prospective studies would likely include preoperative and postoperative imaging data sets in the model development process (see below).
Axonal Pathways
StimVision v2 is designed to work with the CIT168 human MRI brain atlas (22) and the Petersen et al. (19) axonal pathway models (Fig. 1). The Petersen et al. (19) anatomist defined axonal pathways were created within the CIT168 space and are coregistered with the 3D volumes of 16 subcortical nuclei included in the CIT168 atlas. For StimVision v2, we elected to condense the Petersen et al. (19) results into nine general pathways: 1) subthalamopallidal, 2) pallidosubthalamic, 3) pallidothalamic, 4) cerebellothalamic, 5) medial lemniscus, 6) internal capsule (motor), 7) internal capsule (prefrontal cortex [PFC]), 8) hyperdirect (motor), and 9) hyperdirect (PFC). Each of these pathways in StimVision v2 consists of 500 individual streamlines that mimic the anatomically defined 3D trajectory of the pathway (Fig. 1).
Model Coregistration
All patient images (MRI, CT, etc.) are converted from the clinical DICOM format to NIfTI format using dcm2nii, which is distributed with MRIcron (http://neuro.debian.net/pkgs/mricron.html). The intrapatient converted NIfTI images are then coregistered in Advanced Normalization Tools (ANTs) (http://picsl.upenn.edu/software/ants/). The preoperative surgical targeting T1w MRI is typically used to represent the anatomical foundation for each patient model. The StimVision v2 patient-specific model is then developed in layers from that foundation. The first step in that process is the coregistration of the CIT168 brain to the patient. The nonlinear transformation matrix and warp field necessary for this step is created via ANTs using symmetric normalization (SyN) (23). The patient’s T1w image is the “fixed image” and the CIT168 T1w image is the “moving image.” That resulting transformation matrix and warp field are then also applied to the polygonal data of the 3D anatomical nuclei (22) and axonal pathway streamlines (19), thereby placing those model components into patient-specific space.
DBS Electrode
Once the imaging data (e.g., preop MRI and frame images) and anatomical models (3D nuclei and pathways) are all coregistered in StimVision v2, the neurosurgical stereotactic coordinate system can be established. This requires selection of the appropriate frame fiducial model and fitting of that model to the fiducials visible in the frame image. StimVision v2 is currently capable of representing the Leksell (Elekta Instruments) and CRW (Integra LifeSciences) frame systems. A virtual model of the frame fiducials is displayed over the patient imaging data and the GUI provides options to position the frame model into correspondence with the image frame fiducials. Once the frame model is appropriately positioned in StimVision v2, the stereotactic coordinates of the anterior and posterior commissures can be checked against the commercial neurosurgical navigation system used in the operating room as a validation step.
Establishing the stereotactic coordinate system in StimVision v2 enables interactive positioning of the DBS electrode within the patient-specific brain volume such that any selected position can be directly compared with the surgical plan. With a target point defined, the entry point and trajectory are defined by the surgeon to avoid passing through sulci, ventricles, and vessels. These decisions dictate the arc and ring angles of the trajectory. If necessary, fine adjustment of the X, Y, Z position of the electrode tip, as well as the arc and ring angles, can be further evaluated with submillimeter adjustment in the StimVision v2 GUI.
Postoperatively, the electrode localization image (MRI or CT) can be coregistered with the patient model, thereby providing definition of the final electrode position in the brain volume. This enables direct comparison of the surgical plan with the actual surgical placement.
DBS Volume Conductor
Currently, most DBS models rely on VTA calculations to estimate the spatial extent of stimulation around the implanted electrode (24,25). However, recent comparisons of the neural activation predictions generated by DBS suggest that VTA models have substantial limitations when applied in the subthalamic region (8). A key step that is necessary for implementing more detailed models of the neural response to stimulation is the explicit calculation of the voltage distribution (Ve) generated in the brain by the DBS electrode (13) (Fig. 2). Therefore, StimVision v2 includes precomplied solutions for the Ve generated by four different types of commercial DBS lead designs: 1) Medtronic 3387 (MDT 3387), 2) Medtronic 3389 (MDT 3389), 3) Abbott 6172 (ABT 6172), and 4) Boston Scientific 2202 (BSN 2202). These DBS volume conductors (VCs) were constructed as finite element models in COMSOL Multiphysics (https://www.comsol.com/). The anatomical foundation for the VC models was the Multimodal Imaging-Based Detailed Anatomical (MIDA) model of the human head and neck (26). The VCs in StimVision v2 follow the methodology of Howell and McIntyre (27), using the MIDA11 template. However, to provide a greater degree of generality to the simulations, the gray matter and white matter components of the MIDA11 VC model were combined into a single isotropic medium of 0.2 S/m for the brain parenchyma. We called this generic head model MIDA10 for use in StimVision v2. The brain parenchyma simplification allows the electrode position in a patient-specific StimVision v2 model to be able to vary within the subthalamic region, and be interactively moved by the user, while still enabling a reasonably accurate representation of Ve for the DBS simulations in StimVision v2.
To create the StimVision v2 VC solutions, the DBS electrode was placed in the STN of the MIDA10 head model and the lead was surrounded by a 0.5 mm encapsulation layer (σ = 0.1 S/m). We calculated basis solutions of Ve for each electrode contact of each electrode design in a monopolar configuration using a single unitary current source. StimVision v2 currently includes precompiled basis solutions for four different commercial DBS leads (i.e., four basis Ve solutions for the MDT 3387 or MDT 3389 leads, and eight basis Ve solutions for the ABT 6172 or BSN 2202 leads). These monopolar basis solutions are then linearly combined to construct the Ve generated by any given stimulation parameter setting (i.e., multipolar and/or multisource configurations at any given stimulus amplitude) using superposition.
DBS Pathway Activation
The final steps in the development of a StimVision v2 patient-specific subthalamic DBS model are the calculation of axonal pathway activation as a function of the simulated electrode location(s) and stimulation parameter setting(s) (Fig. 2). This process relies on the Howell et al. (13) driving-force (DF) algorithm. DF algorithms use the steady-state polarization response of passive axon cable models to an applied stimulus to estimate activation thresholds (28). DF algorithms are well suited to DBS research applications attempting to estimate axonal pathway activation because they explicitly account for both the DBS electric field and the anatomical trajectory of the axon models. This becomes important in the subthalamic region where there are many different axonal pathways, projecting in many different directions, with each axon following its own tortuous trajectory. Therefore, the Petersen et al. (19) subthalamic pathway models provide the anatomical basis for the axon trajectories (Fig. 1), and the Ve solution along those trajectories provide the electrical inputs to the Howell DF algorithm for axonal activation estimation (13) (Fig. 2).
Each axonal streamline, within each pathway, is made up of a series of points along its trajectory. The 3D location of each of those points is established within the patient-specific anatomical model as part of the nonlinear transformations of the CIT168 space into the patient space. The DBS VC solution of Ve is also made up of points of data that enable interpolation of the profile of extracellular voltages along the trajectory of each individual streamline (Vstreamline) (Fig. 2b). This array of Vstreamline inputs for each axon model allows the Howell DF algorithm to calculate whether the DBS stimulus is suprathreshold or subthreshold for action potential activation (Fig. 2c). The overall result of the StimVision v2 simulation is then the collection of activated axon models, from all the different pathways, for the specific electrode type, electrode location, and stimulation parameter setting that is being evaluated (Fig. 2d).
The output of the Howell DF algorithm varies nonlinearly depending on the polarity of the stimulus, its pulse duration, the diameter of the axon model being evaluated, and the second spatial difference of Vstreamline along the axon trajectory (13). Each axon streamline in each pathway in the StimVision v2 model is currently defined as 2 μm in diameter, except for the corticofugal fibers making up the internal capsule and hyperdirect pathway (HDP) which are defined as 5.7 μm in diameter (29). The hyperdirect collateral branches to the STN are defined as 2 μm in diameter (10). Hyper-direct axons are considered activated if either the collateral branch or the corticofugal fiber is deemed suprathreshold by the Howell DF algorithm.
RESULTS
StimVision v2 provides the ability to evaluate axonal pathway activation from subthalamic DBS using anatomically detailed pathways and electrically detailed Ve distributions within a software tool that facilitates interactive adjustment of the stimulation. We propose that these features become useful when attempting to understand how different stimulation locations within the subthalamic region theoretically effect the activation of different axonal pathways. Such understanding may then be useful in defining the therapeutic neural targets of DBS, as well as the clinical evaluation of hypotheses that stimulation of a certain pathway is related to a specific therapeutic effect or side-effect.
Figure 3 provides an example of the kinds of results that can be generated by StimVision v2. A BSN 2202 lead was positioned in the posterior STN of the CIT168 brain atlas using a typical surgical trajectory. We then used the model to calculate pathway activation recruitment curves. Directional contacts located at the dorsal border of the STN (contacts 5, 6, 7), commonly assumed to be a clinically optimal location for subthalamic DBS, activated a wide range of different pathways with different degrees of coactivation. Stimulation through the anterior contact (5) primarily recruited the pallidothalamic pathway and HDP (Fig. 3a). However, activation of the HDP was accompanied by activation of internal capsule (IC) fibers of passage. Stimulation through the posterior contact (6) exhibited a similar trend in HDP and IC activation, while also gradually recruiting the pallidosubthalamic pathway (Fig. 3b). Stimulation through the medial contact (7) generated selective activation of the pallidothalamic pathway (Fig. 3c). Moving to the ventral ring of directional contacts, and selecting the posterior contact (3) for stimulation, provided an opportunity to more selectively activate the HDP, which is commonly assumed to be a key therapeutic target of subthalamic DBS (30) (Fig. 3d).
Figure 3.
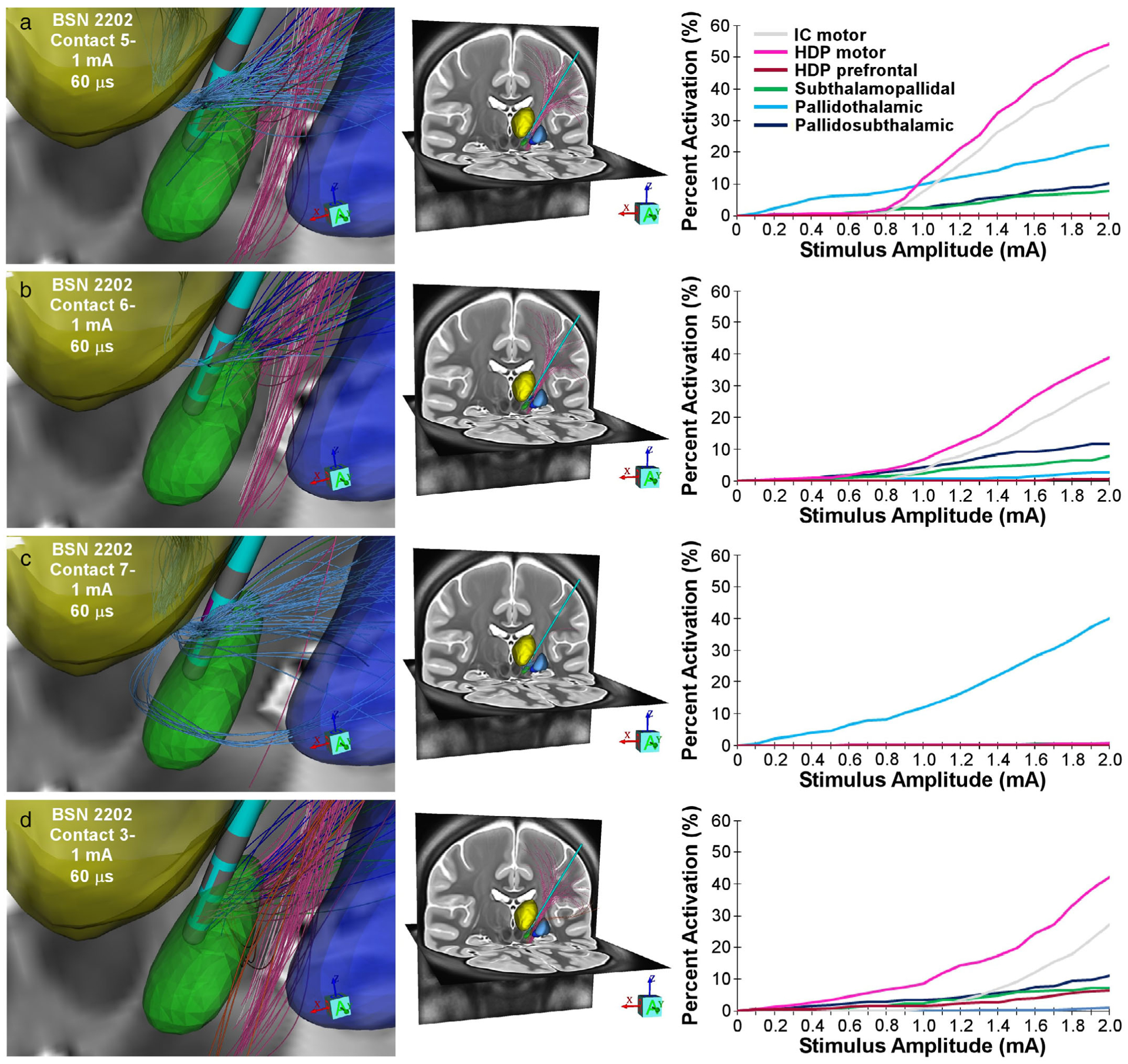
Pathway recruitment with directional stimulation. a. Stimulation through contact 5 (anterior facing) using 60 μsec cathodic stimulation. Left image shows the activated pathways at 1 mA with their corresponding anatomical pathway color. Middle image shows a whole brain view of those activated connections with cortex. The right image shows pathway recruitment curves for contact 5. b. Stimulation through contact 6 (posterior facing) using 60 μsec cathodic stimulation. c. Stimulation through contact 7 (medial facing) using 60 μsec cathodic stimulation. d. Stimulation through contact 3 (posterior facing) using 60 μsec cathodic stimulation. Nuclei and pathway color codes provided in Figure 1.
Figure 4 provides three examples of StimVision v2 patient-specific DBS models at their clinically defined therapeutic stimulation settings. Subjects with an excellent, good, and mediocre response to DBS were randomly selected and displayed for comparison. These example models demonstrate that many different pathways are likely activated by therapeutic DBS. Detailed statistical analyses with large numbers of subthalamic DBS patients will be forthcoming in subsequent reports. Nonetheless, the results suggest that it is unlikely that a single locus of stimulation, or activation of a single pathway, could be both necessary and sufficient for generic therapeutic benefit from subthalamic DBS. For example, we seldom see intrapatient bilateral consistency in the activated pathways, and we commonly note large variances in the interpatient pathway activation percentages (Fig. 4).
Figure 4.
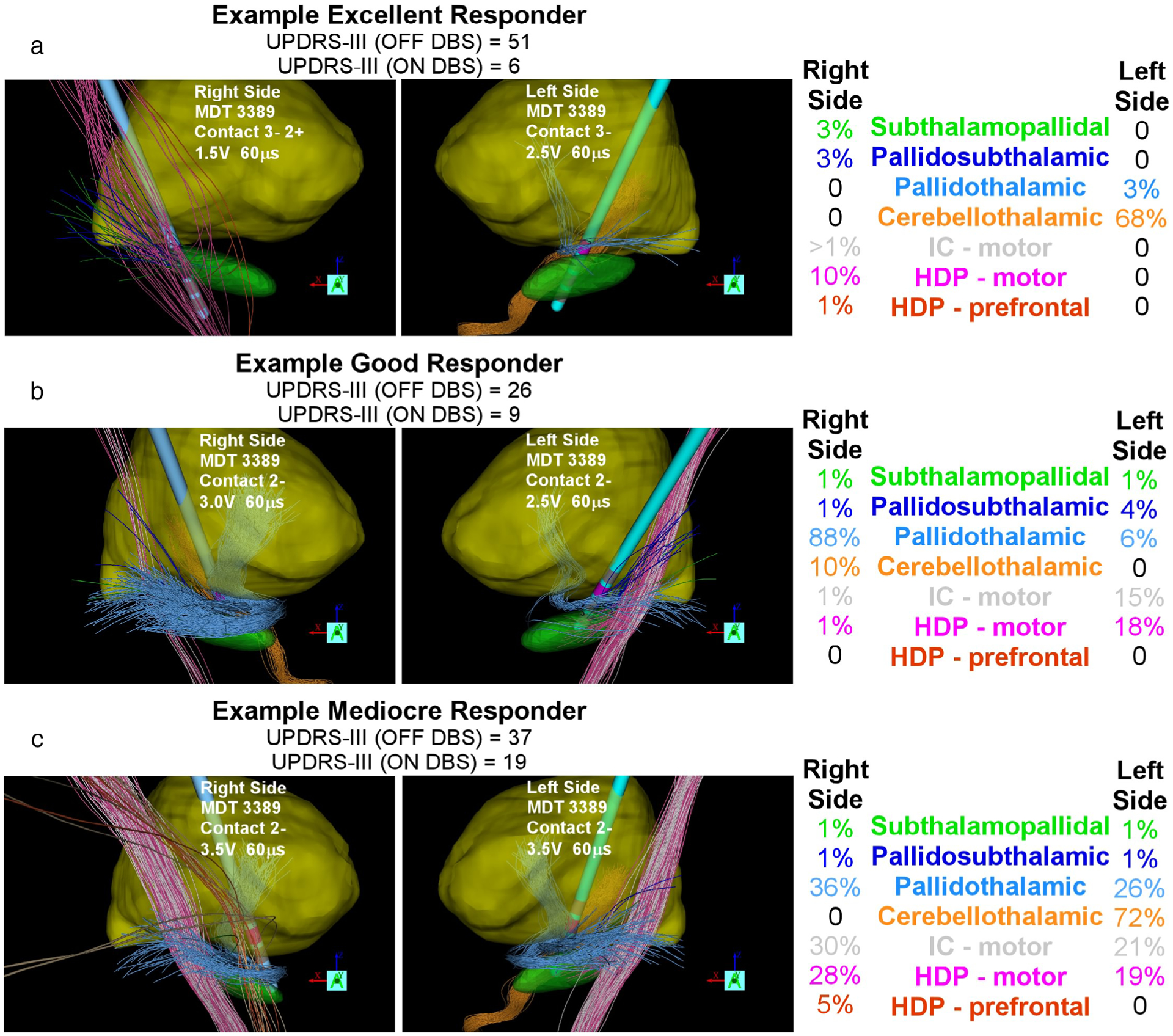
Therapeutic pathway activation. Example patient-specific DBS models and pathway activation estimates at their clinical DBS settings. a. Right and left hemispheres of an excellent responder. b. Right and left hemispheres of a good responder. c. Right and left hemispheres of a mediocre responder. Nuclei and pathway color codes provided in Figure 1. Unified Parkinson’s Disease Rating Scale—Motor (UPDRS-III).
A popular concept in the clinical DBS literature is to conduct DBS modeling analyses with electrode locations from populations of subjects transformed into an atlas space (9). While that approach has certain advantages, methodological issues also introduce uncertainty into the analyses. Figure 5 provides a visual demonstration of some of the issues associated with transforming a patient-specific DBS model into an atlas space and then performing connectomic DBS modeling. The first step in that process is to position the patient-specific DBS lead in the atlas space. Modern techniques for this step rely on nonlinear transformations (31). However, the size and shape of the DBS lead, as well as the relative position of the electrode contacts to the neuroanatomy, can be distorted in this process (Fig. 5b) (32). As such, arbitrary decisions then need to be made on the placement of the DBS electrode in the atlas space (Fig. 5c). Once the DBS electrode position is defined, an activation estimate is needed to calculate the DBS connectivity. Most connectomic DBS modeling analyses currently rely on the intersection of axon streamlines with a VTA model (Fig. 5d). However, the results of those simulations are dependent upon both the VTA model and axonal pathway models used in the analysis (8). For example, generalized whole brain tractography results (9) in the subthalamic region are highly biased by the internal capsule, which overwhelms opportunities to represent activation other pathways (Fig. 5e,f). Therefore, the technical details of the methods used in connectomic DBS modeling play an important role in dictating the results that are generated.
Figure 5.
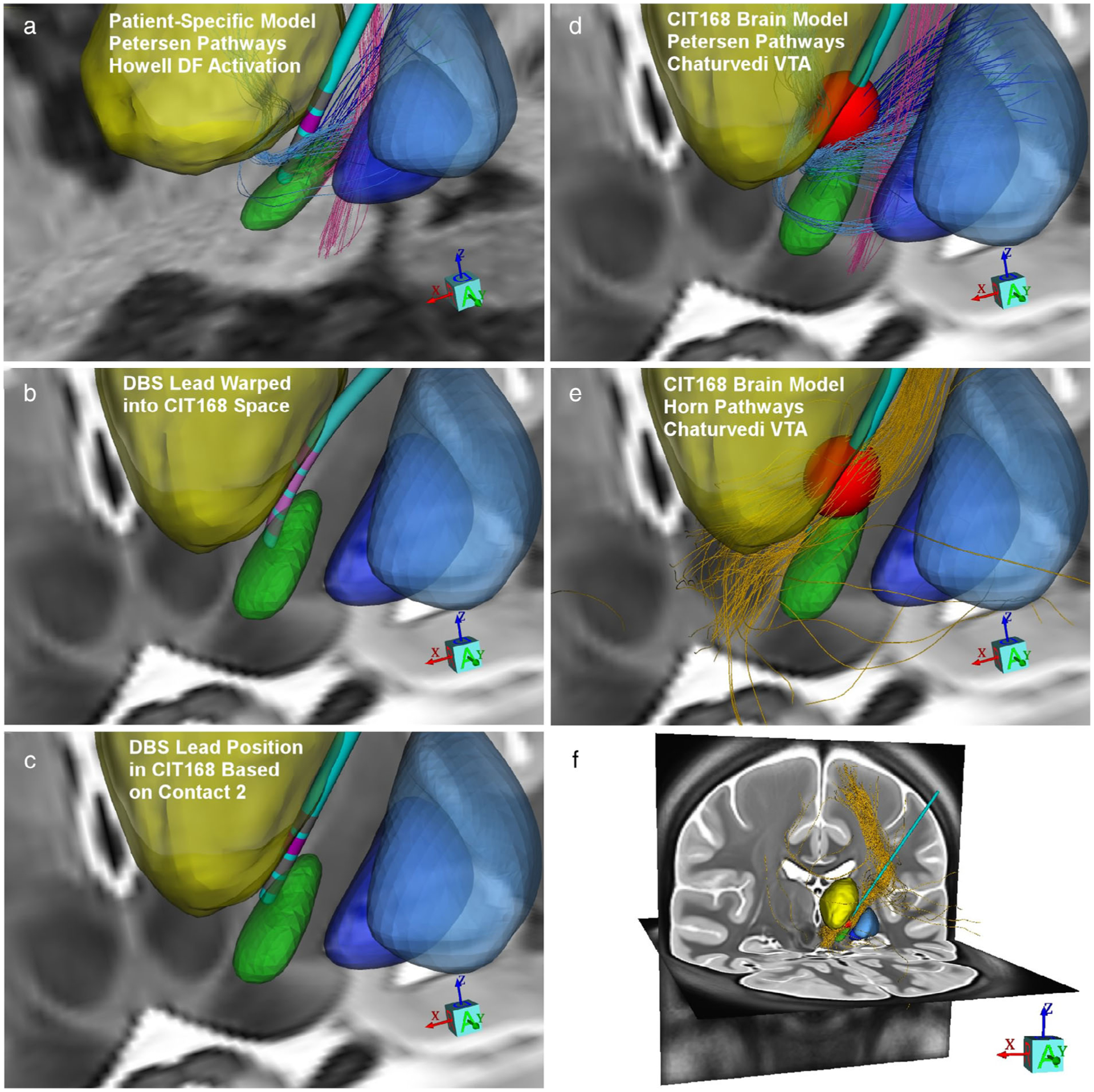
DBS model comparison. a. Example patient-specific DBS model from Figure 4b (left side of the brain). b. Inverse transform of the patient-specific DBS lead warped into CIT168 brain atlas space. c. DBS electrode model position based on the contact used for stimulation (contact 2). d. Petersen et al. (19) axonal pathways coursing through a Chaturvedi et al. (24) VTA model for the same stimulation parameters as panel a. e. Horn et al. (9) group connectome pathways (gold streamlines) coursing through the same VTA. f. Whole brain view of the group connectome activated pathways.
DISCUSSION
The concept of developing patient-specific models of DBS to identify correlations between stimulation parameter settings, modulation of anatomical features (e.g., nuclei or axonal pathways), and changes in clinical outcome measures, has a long history (7,33–35). Early attempts at using patient-specific models to study subthalamic DBS suggested that the STN per se was not the only therapeutic target within this very complex region of the brain (36,37). In turn, the prospective use of model-defined target stimulation volumes that encompassed the dorsal-posterior STN, as well as the white matter dorsal to the STN, were able to generate therapeutic outcomes that rivaled traditional clinical practice (38). However, the technical detail and computational resources necessary to implement those early patient-specific DBS models were considered prohibitive for clinical application. Yet from a scientific perspective, those early models were already substantial biophysical simplifications of what was known about DBS. As such, the field of patient-specific DBS modeling began to branch in two general directions. Clinical researchers moved forward by developing simple DBS models that ignored the biophysical details, but could be more easily applied to large populations of DBS subjects (39,40). While scientific researchers moved forward by developing ever more detailed biophysical models of DBS to better quantifying the neural response to stimulation, but those complex models negated opportunities to perform large-scale clinical analyses (10,41). The goal of StimVision v2 is to provide a new middle ground for the development of detailed patient-specific models of subthalamic DBS that can be applied to the study of large numbers of DBS patients.
StimVision v2
StimVision v2 integrates three advances in DBS modeling methodology, which based on our experience with scientifically detailed DBS models, should improve the anatomical and electrical accuracy of the simulations. First, was the incorporation of the Petersen et al. (19) axonal pathway models. The vast majority of patient-specific DBS models generated over the last decade have incorporated axonal pathway models derived from tractography (14). However, tractography results are indirect simulations of connectivity, derived from low resolution data, and notorious for anatomical inaccuracy (16). Our work in Petersen et al. (19) and Gunalan et al. (8) demonstrated glaring issues with using even the most advanced tractographic approaches to study subthalamic DBS. In addition, tractography results require extensive postprocessing to cull false positive results and annotate the realistic anatomical connections (18). In contrast, nonlinear fits of anatomical pathway models to de novo subjects can generate excellent alignment to patient-specific white matter (19). As such, anatomical pathway models are faster to implement, easier to work with, and provide greater anatomical detail than tractography. In addition, the use of a pathway atlas provides a more consistent and rigorous platform to compare pathway activation results across subjects.
The second advance was the inclusion of precompiled DBS Ve solutions that better account for the return path of current flow during “monopolar” DBS (42). The DBS VC models that are typically used in VTA modeling suffer from an overestimation of stimulus spread (8,27). This is because the boundary conditions of simplified VC models ignore the lowest resistivity return path, primarily made up of cerebrospinal fluids (CSFs) in the head and neck, to the implanted pulse generator (IPG). This causes the simulated DBS electric field to decay too slowly in the modeled brain tissue and the predicted neural response thresholds to become artificially low. The precompiled DBS Ve solutions in StimVision v2 explicitly include the CSF return paths to the IPG. These generic DBS Ve solutions represent our best suggestion for a compromise between electrical realism and computational simplicity. As such, they provide a reasonable option for calculating the neural response to DBS within an interactive software tool. However, if more detailed patient-specific DBS VC representations are available for a given subject, those Ve solutions can also be used within the StimVision v2 framework.
The third advance was the application of a driving-force (DF) predictor function to more accurately quantify the neural response to stimulation (13). Field-cable models represent the scientific “gold standard” for simulating neural activation from DBS, but they are technically demanding and computationally intensive (43). Therefore, DBS VTA models were created to provide a generalized estimate of stimulus spread (7). However, the simplifications associated with DBS VTA models result in substantial errors when applied to complex neural populations (8). DF models represent a more detailed approach than VTA models, and require additional processing steps, but are more accurate because they integrate data from both the DBS electric field and the trajectory of each neural process to calculate activation (13). Therefore, the StimVision v2 combination of anatomical axonal pathway models, electrode-specific DBS Ve solutions, and a DF predictor function enables interactive placement of the DBS electrode within the subthalamic region, and subsequent calculation of the neural response to DBS, all within a reasonable amount of time on a laptop computer. For example, it currently takes ~2 min to calculate the response of the 4500 axon models representing the nine different pathways on a modern MacBook Pro. Versions under development suggest that this time can be reduced to just a few seconds with optimization of the code. Comparatively, solutions for 4500 field-cable models in NEURON (https://neuron.yale.edu/neuron/) would require hundreds of minutes.
StimVision v2 Limitations
StimVision v2 subthalamic DBS models still suffer from substantial limitations when compared to scientifically detailed DBS models (8). The first major limitation is that predefined anatomical axonal pathway models may not be able to capture the true interpatient variability of the subthalamic white matter. The fit of the anatomical pathway models to the patient are dependent upon the nonlinear fit of the CIT168 brain atlas to the patient. In our experience, SyN fits with ANTs are typically excellent, assuming a high quality T1w image of the patient (23). However, it should be noted that even advanced nonlinear warps can be off by several millimeters in the subthalamic region for some subjects (11,12,32). Alternatively, patient-specific tractography results may be favored by some investigators. However, coregistration of the diffusion-weighted imaging data into the patient model introduces its own set of errors (44), which our experience suggests is larger than the errors associated with nonlinear warps of anatomical pathway models to the patient.
The second major limitation is that the precompiled DBS Ve solutions in StimVision v2 ignore the complexities of brain tissue anisotropy. This simplification was implemented to enable interactive movement of the DBS electrode within the patient-specific anatomical volume. This limitation primarily effects internal capsule pathway activation calculations, as the strong tissue anisotropy associated with those pathways acts to limit current spread in the lateral direction (15). As such, when using the precompiled VC models, StimVision v2 models will tend to overestimate internal capsule activation (Fig. 4b,c). Nonetheless, for scientific research studies, detailed patient-specific VC models can be loaded into StimVision for more accurate simulations.
The third major limitation is that StimVision v2 uses DF methods to predict pathway activation instead of field-cable methods. This simplification dramatically speeds up the model calculations, compared to field-cable simulations, and provides more accurate predictions than VTA models (13). However, VTA models provide effectively instantaneous prediction of pathway activation, which can be very useful during interactive DBS surgical planning (20). Therefore, we still use VTA models (or a simple ~2 mm radius activation sphere) to provide an initial estimate of pathway activation during interactive DBS electrode positioning. Then once a general target electrode location has been identified, we use DF calculations to verify that the pathway activation profile is consistent with the surgical targeting goals, and adjust as needed. However, for all retrospective or postoperative DBS analyses, where the electrode location is defined by patient-specific imaging data, we have abandoned VTA models in favor of DF calculations.
Caveats of Connectomic DBS Modeling
While connectomic DBS modeling is currently a popular concept, the technical details and computational methods that are necessary to generate patient-specific DBS models with anatomical, biophysical, and connectomic validity remain unknown (8). Nonetheless, wide spread studies are actively attempting to identify correlations between DBS pathway activation and clinical outcomes (9). As such, there is a need to evaluate some of the assumptions in connectomic DBS modeling and better define standards for its use in clinical research.
One important assumption of connectomic DBS modeling is that the behavioral metric being evaluated is directly modulated by a unique locus of stimulation in the brain, or a specific axonal pathway, that can be generalized across subjects. This assumption may be reasonable for a simple and quantifiable behavioral metric (e.g., reduction of tremor via DBS of the cerebellothalamic pathway), but is likely to be unreasonable for a complex clinical outcome score. The patient-specific DBS modeling results of Figure 4 provide some insight into why recent attempts to correlate UPDRS-III scores with simple DBS models have had such limited success (45,46). One issue is the diversity of axonal pathways that are simultaneously activated during therapeutic DBS. Another issue is the use of bilateral UPDRS-III scores for the correlations when DBS pathway activation is not bilaterally similar. These unaccounted for issues contribute variance to the analyses, and are likely factors in the weak correlations. As such, scientific studies with patient-specific DBS models should be performed as unilateral analyses (i.e., opposite side DBS turned off), with the DBS activation estimates tagged to a focused and quantitative metric that is associated with the corresponding hemibody (37).
Another important assumption in connectomic DBS modeling is that the geometric uncertainty associated with the overall model is smaller than the volume of brain tissue over which the statistical analyses are being performed. Connectomic DBS models have three general sources of uncertainty (anatomical, biophysical, and connectivity) that need to be considered when designing a study and evaluating the model predictions. The first general source of uncertainty is anatomical, which is the error associated with defining the position of the DBS electrode in the patient-specific brain model. This error has at least four sources: 1) stability of the electrode position as defined by the postoperative image (i.e., brain shift), 2) ambiguity in defining the DBS electrode position within the postoperative image (i.e., electrode artifact), 3) coregistration of the postoperative image to the patient brain model, and 4) fit of the brain atlas anatomical volumes to the patient imaging data. These errors are difficult to quantify, but at least 1 mm of anatomical uncertainty should be expected in the model (12,36). If the investigators then attempt to re-map that patient-specific model into an atlas space for the DBS analyses, another ~1 mm of anatomical uncertainty needs to be considered (31,32) (Fig. 5b). The second general source of uncertainty is the biophysics. Estimates of DBS activation can vary substantially depending on the methods employed (8). The most common approach is to use VTA models, which have at least 1 mm of biophysical uncertainty (i.e., error associated with the radius of the VTA) in their activation predictions (13) (Fig. 5d). The third general source of uncertainty is the connectivity. DBS connectivity estimates are highly dependent upon the axonal pathway models used in the analysis (9,19) (Fig. 5e). Geometric errors associated with using different pathway model representations are currently unknown; however, at least 1 mm of connectivity uncertainty (i.e., anatomical accuracy of the pathway model as it passes by the DBS electrode) likely needs to be considered.
Given the very small region of interest for subthalamic DBS (i.e., ~6 × 6 × 6 mm3) (36,37), the uncertainties associated with DBS modeling call into question the legitimacy of performing correlative analyses between generic UPDRS-III scores and simple VTA models mapped into an atlas space (45,46). Therein lies our motivation to improve the anatomical and electrical accuracy of connectomic DBS modeling with the technical advances implemented in StimVision v2. In parallel, we propose that far more scientific research, attention to detail, and validation studies are needed to define best practices for this burgeoning field. In our opinion, these standards need to be defined prior to promoting the use of connectomic DBS modeling by investigators that lack technical understanding of the models and assumptions they are employing.
CONCLUSION
StimVision v2 is a connectomic DBS modeling tool that integrates recent advances in scientific DBS modeling, while making reasonable computing compromises that should facilitate the analysis of larger numbers of research subjects. While StimVision v2 can be used for retrospective analyses, it is primarily designed for use in prospective studies aimed at testing connectomic DBS hypotheses. For example, is activation of pathway X causally associated with behavioral metric Y? Answering those types of questions is best done via prospective definition of patient-specific DBS electrode placements and/or parameter settings that theoretically provide selective activation of the desired pathway. Those model-defined DBS settings can then be experimentally evaluated in dedicated clinical research studies using quantitative behavioral metrics that are explicitly designed to test the hypothesis at hand.
Acknowledgement
This work was supported by grants from the National Institutes of Health (R01 NS105690; P50 NS098573; R01 NS085188).
Footnotes
Conflict of Interest: Cameron C. McIntyre is a paid consultant for Boston Scientific Neuromodulation, receives royalties from Hologram Consultants, Neuros Medical, Qr8 Health, and is a shareholder in the following companies: Hologram Consultants, Surgical Information Sciences, CereGate, Autonomic Technologies, Cardionomic, Enspire DBS. Ms. Noecker reports personal fees from Hologram Consultants, outside the submitted work. Dr. Howell reports personal fees from Abbott Laboratories, outside the submitted work. The remaining authors have no conflicts of interest to disclose.
REFERENCES
- 1.Krack P, Volkmann J, Tinkhauser G, Deuschl G. Deep brain stimulation in movement disorders: from experimental surgery to evidence-based therapy. Mov Disord 2019;34:1795–1810. [DOI] [PubMed] [Google Scholar]
- 2.Schuepbach WM, Rau J, Knudsen K et al. Neurostimulation for Parkinson’s disease with early motor complications. N Engl J Med 2013;368:610–622. [DOI] [PubMed] [Google Scholar]
- 3.Bronstein JM, Tagliati M, Alterman RL et al. Deep brain stimulation for Parkinson disease: an expert consensus and review of key issues. Arch Neurol 2011;68:165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hamel W, Köppen JA, Alesch F et al. Targeting of the subthalamic nucleus for deep brain stimulation: a survey among Parkinson disease specialists. World Neurosurg 2017;99:41–46. [DOI] [PubMed] [Google Scholar]
- 5.Volkmann J, Moro E, Pahwa R. Basic algorithms for the programming of deep brain stimulation in Parkinson’s disease. Mov Disord 2006;21:S284–S289. [DOI] [PubMed] [Google Scholar]
- 6.McIntyre CC, Anderson RW. Deep brain stimulation mechanisms: the control of network activity via neurochemistry modulation. J Neurochem 2016;139:338–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Butson CR, Cooper SE, Henderson JM, McIntyre CC. Patient-specific analysis of the volume of tissue activated during deep brain stimulation. Neuroimage 2007; 34:661–670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gunalan K, Howell B, McIntyre CC. Quantifying axonal responses in patient-specific models of subthalamic deep brain stimulation. Neuroimage 2018;172: 263–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Horn A, Li N, Dembek TA et al. Lead-DBS v2: towards a comprehensive pipeline for deep brain stimulation imaging. Neuroimage 2019;184:293–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gunalan K, Chaturvedi A, Howell B et al. Creating and parameterizing patient-specific deep brain stimulation pathway-activation models using the hyperdirect pathway as an example. PLoS One 2017;12:e0176132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ewert S, Horn A, Finkel F, Li N, Kühn AA, Herrington TM. Optimization and comparative evaluation of nonlinear deformation algorithms for atlas-based segmentation of DBS target nuclei. Neuroimage 2019;184:586–598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim J, Duchin Y, Shamir RR et al. Automatic localization of the subthalamic nucleus on patient-specific clinical MRI by incorporating 7 T MRI and machine learning: application in deep brain stimulation. Hum Brain Mapp 2019;40: 679–698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Howell B, Gunalan K, McIntyre CC. A driving-force predictor for estimating pathway activation in patient-specific models of deep brain stimulation. Neuromodulation 2019;22:403–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chaturvedi A, Butson CR, Lempka SF, Cooper SE, McIntyre CC. Patient-specific models of deep brain stimulation: influence of field model complexity on neural activation predictions. Brain Stimul 2010;3:65–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McIntyre CC, Mori S, Sherman DL, Thakor NV, Vitek JL. Electric field and stimulating influence generated by deep brain stimulation of the subthalamic nucleus. Clin Neurophysiol 2004;115:589–595. [DOI] [PubMed] [Google Scholar]
- 16.Schilling KG, Nath V, Hansen C et al. Limits to anatomical accuracy of diffusion tractography using modern approaches. Neuroimage 2019;185:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schmitz D, Muenzing SEA, Schober M et al. Derivation of fiber orientations from oblique views through human brain sections in 3D-polarized light imaging. Front Neuroanat 2018;12:75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yeh FC, Panesar S, Fernandes D et al. Population-averaged atlas of the macro-scale human structural connectome and its network topology. Neuroimage 2018; 178:57–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Petersen MV, Mlakar J, Haber SN et al. Holographic reconstruction of axonal pathways in the human brain. Neuron 2019;104:1056–1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Noecker AM, Choi KS, Riva-Posse P, Gross RE, Mayberg HS, McIntyre CC. StimVision software: examples and applications in subcallosal cingulate deep brain stimulation for depression. Neuromodulation 2018;21:191–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Riva-Posse P, Choi KS, Holtzheimer PE et al. A connectomic approach for subcallosal cingulate deep brain stimulation surgery: prospective targeting in treatment-resistant depression. Mol Psychiatry 2018;23:843–849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pauli WM, Nili AN, Tyszka JM. A high-resolution probabilistic in vivo atlas of human subcortical brain nuclei. Sci Data 2018;5:180063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Klein A, Andersson J, Ardekani BA et al. Evaluation of 14 nonlinear deformation algorithms applied to human brain MRI registration. Neuroimage 2009;46:786–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chaturvedi A, Luján JL, McIntyre CC. Artificial neural network based characterization of the volume of tissue activated during deep brain stimulation. J Neural Eng 2013;10:056023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Astrom M, Diczfalusy E, Martens H, Wardell K. Relationship between neural activation and electric field distribution during deep brain stimulation. IEEE Trans Biomed Eng 2015;62:664–672. [DOI] [PubMed] [Google Scholar]
- 26.Iacono MI, Neufeld E, Akinnagbe E et al. MIDA: a multimodal imaging-based detailed anatomical model of the human head and neck. PLoS One 2015;10: e0124126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Howell B, McIntyre CC. Role of soft-tissue heterogeneity in computational models of deep brain stimulation. Brain Stimul 2017;10:46–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Warman EN, Grill WM, Durand D. Modeling the effects of electric fields on nerve fibers: determination of excitation thresholds. IEEE Trans Biomed Eng 1992;39: 1244–1254. [DOI] [PubMed] [Google Scholar]
- 29.McIntyre CC, Richardson AG, Grill WM. Modeling the excitability of mammalian nerve fibers: influence of afterpotentials on the recovery cycle. J Neurophysiol 2002;87:995–1006. [DOI] [PubMed] [Google Scholar]
- 30.Macerollo A, Zrinzo L, Akram H, Foltynie T, Limousin P. Subthalamic nucleus deep brain stimulation for Parkinson’s disease: current trends and future directions. Expert Rev Med Devices 2020;17:1063–1074. [DOI] [PubMed] [Google Scholar]
- 31.D’Haese PF, Pallavaram S, Kao C, Neimat JS, Konrad PE, Dawant BM. Effect of data normalization on the creation of neuro-probabilistic atlases. Stereotact Funct Neurosurg 2013;91:148–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nowacki A, Nguyen TA, Tinkhauser G et al. Accuracy of different three-dimensional subcortical human brain atlases for DBS-lead localisation. Neuroimage Clin 2018;20: 868–874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.D’Haese PF, Cetinkaya E, Konrad PE, Kao C, Dawant BM. Computer-aided placement of deep brain stimulators: from planning to intraoperative guidance. IEEE Trans Med Imaging 2005;24:1469–1478. [DOI] [PubMed] [Google Scholar]
- 34.Nowinski WL, Belov D, Pollak P, Benabid AL. Statistical analysis of 168 bilateral subthalamic nucleus implantations by means of the probabilistic functional atlas. Neurosurgery 2005;57:319–330. [DOI] [PubMed] [Google Scholar]
- 35.Guo T, Finnis KW, Parrent AG, Peters TM. Visualization and navigation system development and application for stereotactic deep-brain neurosurgeries. Comput Aided Surg 2006;11:231–239. [DOI] [PubMed] [Google Scholar]
- 36.Maks CB, Butson CR, Walter BL, Vitek JL, McIntyre CC. Deep brain stimulation activation volumes and their association with neurophysiological mapping and therapeutic outcomes. J Neurol Neurosurg Psychiatry 2009;80:659–666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Butson CR, Cooper SE, Henderson JM, Wolgamuth B, McIntyre CC. Probabilistic analysis of activation volumes generated during deep brain stimulation. Neuroimage 2011;54:2096–2104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Frankemolle AM, Wu J, Noecker AM et al. Reversing cognitive-motor impairments in Parkinson’s disease patients using a computational modelling approach to deep brain stimulation programming. Brain 2010;133:746–761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vanegas-Arroyave N, Lauro PM, Huang L et al. Tractography patterns of subthalamic nucleus deep brain stimulation. Brain 2016;139:1200–1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Akram H, Sotiropoulos SN, Jbabdi S et al. Subthalamic deep brain stimulation sweet spots and hyperdirect cortical connectivity in Parkinson’s disease. Neuroimage 2017;158:332–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Slopsema JP, Peña E, Patriat R et al. Clinical deep brain stimulation strategies for orientation-selective pathway activation. J Neural Eng 2018;15:056029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Walckiers G, Fuchs B, Thiran JP, Mosig JR, Pollo C. Influence of the implanted pulse generator as reference electrode in finite element model of monopolar deep brain stimulation. J Neurosci Methods 2010;186:90–96. [DOI] [PubMed] [Google Scholar]
- 43.McNeal DR. Analysis of a model for excitation of myelinated nerve. IEEE Trans Biomed Eng 1976;23:329–337. [DOI] [PubMed] [Google Scholar]
- 44.Choi KS, Noecker AM, Riva-Posse P et al. Impact of brain shift on subcallosal cingulate deep brain stimulation. Brain Stimul 2018;11:445–453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dembek TA, Roediger J, Horn A et al. Probabilistic sweet spots predict motor outcome for deep brain stimulation in Parkinson disease. Ann Neurol 2019;86:527–538. [DOI] [PubMed] [Google Scholar]
- 46.Treu S, Strange B, Oxenford S et al. Deep brain stimulation: imaging on a group level. Neuroimage 2020;219:117018. [DOI] [PubMed] [Google Scholar]


