Abstract
The recent evolution of the SARS-like Coronavirus has ravaged the world. The deadly virus has claimed over millions of lives across the world and hence highlights the need to develop effective therapeutic drugs to contain the disease posed by this parasite. In this study, the inhibitory potential of fifty (50) dietary polyphenols against Coronavirus (SARS-CoV-2) main protease (Mpro) was conducted using the Autodock Vina Molecular docking tool. In the virtual screening process, the binding affinity of Remdesivir (-7.7 kcal/mol) currently used to treat COVID-19 patients was set as the cut-off value to screen out less probable inhibitors. Ellagic acid, Kievitone, and Punicalin were the only promising ligands with binding affinities (-8.9 kcal/mol, -8.0 kcal/mol and -7.9 kcal/mol respectively) lower than the set cut-off value. Furthermore, we validated Ellagic acid and Kievitone efficacy by subjecting them to molecular dynamics simulation and further stability was assessed at the molecular mechanics and quantum levels. The overall analysis indicates both compounds demonstrate higher stability and inhibitory potential to bind to the crucial His41 and Cys145 catalytic dyad of Mpro than the standard drug. However, further analysis of punicalin after evaluating its docking score was not conducted as the ligand pharmacokinetics properties suggests it could pose serious adverse effect to the health of participants in clinical trials. Hence, we employed a more safe approach by filtering out the compound during this study. Conclusively, while Ellagic acid and kievitone polyphenolic compounds have been demonstrated to be promising under this in silico research, further studies are needed to substantiate their clinical relevance.
Keywords: Molecular docking, Molecular dynamics, Quantum mechanics, Molecular mechanics, SARS-COV2 Mpro inhibitors
Abbreviations: Mpro, main protease; ADMET, absorption, distribution, metabolism, excretion, and toxicity; HOMO, highest occupied molecular orbital; LUMO, lowest unoccupied molecular orbital; ORF, open reading frame; GROMACS, GROningen MAchine for chemical simulations; MM-PBSA, molecular mechanics Poisson–Boltzmann Surface Area
Graphical abstract
1. Introduction
In this in silico-based research, we used ADMET profile, physicochemical properties analysis and molecular docking simulation to investigate some class of dietary polyphenols that could inhibit the viral growth of SAR-CoV-2 through the blocking of SARS-CoV-2 main protease 3CL (Mpro). We further subjected the selected compounds to 20 ns all atoms molecular dynamics simulation to investigate their stability at the Mpro active pocket. Molecular mechanics MMPBSA method was used to calculate the free energy of the residues involved in the interaction of the selected compounds with Mpro while Quantum mechanical-Density Functional Theory (HOMO and LUMO) was exploited to calculate the energy donating and accepting potential of these compounds. Our aim is to investigate the compounds that could emerge and developed as potent nutraceuticals for the management/treatment of SARS-CoV-2 mediated COVID-19.
Several reports of respiratory illness have been reported from Wuhan, Hubei province of China in December 2019, with confirmed evidence that the infections were caused by a novel coronavirus which was subsequently named SARS-COV-2 by World Health Organization [12,16,23,52]. The pathogenesis of SARS-COV-2 leads to a medical complication named COVID-19. This pathogen is a positive-sense single-stranded RNA virus from the genus β-coronavirus. The outbreak of this disease has become a pandemic to the world and a serious health challenge to the public health organization, owing to an historical record of about two million death cases. It was primarily believed to have been the first host to infect human, thorough investigation has established SARS-COV-2 to be closely related to SARS-CoV which occurs in 2003 and MERS-CoV in 2012 [3,5,17,38]. Out of the different therapeutic drugs and vaccines that have been proposed to reduce high increase or spread of this coronavirus disease, only the broad spectrum nucleotide analogue prodrug remdesivir (Velour) was the first and only approved drug by the FDA for the treatment of SAR-CoV-2-induced COVID-19 for hospitalized adults and children of age 12 years and above weighing at least 40 kg. In a research where 541 patients were assigned to Remdesivir and 521 to placebo, it was reported that Remdesivir was superior to placebo through the shortening of recovery time in COVID-19 infected hospitalized adults with evidence of lower respiratory tract infection [5], therefore, an urgent need to find effective natural anti‐SARS‐CoV‐2 measures for the treatment/management of COVID‐19 to alleviate the current global public health threat is the order of the day in the scientific world.
Persistent investigation of SARS-COV-2 genomics organization reveals that the viral genome has approximately 30 kb with at least 14 open reading frames which is released into the cytoplasm [21,30]. The ORF1a and ORF1ab encodes all non-structural proteins, the remnant genes encode for the structural proteins; spike (S) protein, envelope (E) protein, membrane (M) protein and nucleocapsid (N) protein, and nine accessory factors which together function in the virion formation. Mostly, the nonstructural proteins are important for the replication and transcriptions of the virus and it has been observed that a slippery sequence UUUAAAC is present at the junction between ORF1a and ORF1ab in all coronaviruses. Translation begins at the end of the slippery sequence through a RNA-mediated ribosome frame shift. The ORF1a encodes the papain-like (PLpro) and 3CL-protease (Mpro) which cleaves the replicates polypeptide to encode 16 non-structural proteins (NSPs) [15]. It has been reported that 3CL-protease proteolytic activity is greater than the Papain-like protease by cleaving larger number (11) of sites within the polyprotein, hence it is regarded as the major proteolytic enzyme of SARS-COVs [19,48]. The individual homo-dimeric protomers of SARS-COV2 Cysteine-like protease (Mpro) consist of three different domains: domain I consist of sequences between 8-101 amino acid residues, domain II is found at the region of 102–184 residues and sequence between 201-303 is made up of domain III [26]. The domain I and II which contains the catalytic dyad (His41 and Cys145) at the formed cleft [9,26,43] are made up of β-sheet while the dominant structural motifs of domain III is α-helix [26]. Crucially, Cys145 residue act as a nucleophile and has been identified as the major key player in the proteolytic activity of Mpro [26]. Hence, targeting this residue has been identified as a major therapeutic strategy in the prevention of polyprotein processing and Viral maturation exhibited by the Mpro [53]. Interestingly, we have recently reported the essence of targeting relevant amino acid residue on protein targets in computer-aided drug discovery [7]. In 2020, Jin and collaborators employed the use of Computer-aided drug design (CADD) and X-ray crystallographic technique to elucidate the structure of Mpro bound with a mechanism-based inhibitor (N3) [26]. This discovery as thus paves the way for researchers to investigate potential Mpro inhibitors from natural products/phytochemicals and clinically approved unrelated COVID-19 on Mpro (for drug repurposing) and other protein targets using in silico approach [[1],2,10,18,40].
Polyphenols majorly synthesized from phenylpropanoid pathways by plants are one of the phytochemicals which has been scientifically supported by nutritional and medicinal evidences as established functional foods or nutraceutical that makes the lines between foods and drugs to fade. They are polymeric unit of phenyl with more than a double phenyl rings with single or additional hydroxyl substituents. Food sources such as black tea, walnut, apples, blueberries, vegetables, almonds, spinach, tomatoes, parsley, leguminous plants, soy beans, cummoet, pomegranate, cherries, persimmons, rooibus tea, flex-seed etc., housed different subclasses of polyphenols in different proportions [4,8]. Long consumption of diet rich in polyphenol potentially prevent against infections and non-communicable diseases such as lungs damage, neurodegenerative diseases, osteoporosis, pancreatitis and type-2 diabetes. Several scientific examinations have established that various polyphenol have anti-inflammatory, antioxidant, antiviral activities and also have effect in numerous cellular processes. Ellagitannins possess antiviral activities in particular against HIV infections [39] and manifest inhibitory effects on Epstein-Barr virus, HSV-1 and HSV-2. Ellagic acid and Kievitone have been found to possess antioxidant, anti-cancer and antiviral activities [20,22,44]. Pre-existing drugs have been used by scientist in both ligand-based and structural-based method to control the spread of COVID-19, however some of these drugs have been thus far had inappreciable effects on SARS-COV-2 which could be due to the suggestion that the viral proteins may have actively acquire mutations by single nucleotide polymorphisms (SNPs) and thus escape for being targeted by antiviral drugs. However, the use of natural product such as polyphenols may leverage this insurgence and could be a promiscuous therapeutic target against SARS-COV-2 proteins [51].
2. Materials and methods
2.1. Preparation of target protein
The crystal structure of the SARS-COV-2 Main protease PDB ID: 6lu7 [26] was harvested from the RCSB Protein Data Bank (http://www.rcsb.org). We validated the protein by checking the presence of missing hydrogens, side-chain abnormalities, iMproper bonds etc. The mechanism-based inhibitor (N3) which was co-crystallized with Mpro was removed from the protein before executing our docking protocols. The protein structures were inserted in Autodock Vina and the pdbqt format for each protein was generated [49].
2.2. Determination of (6LU7) Mpro active sites
Binding pocket, ligands interactions and all amino acid residues in the active site of SARS-CoV-2 Mpro were determined with Computed Atlas for Surface Topography of Proteins (CASTp) (http://sts.bioe.uic.edu/castp/index.html?2011) and Biovia Discovery Studio (2019). Obtained data were compared and validated with previously reported experimental data for SARS-CoV-2 proteins.
2.3. Ligands selection and preparation for docking
We retrieve the canonical smiles for 3-hydroxyflavone, Apigenin, Baicelin, Caffeic acid, Catechin, Curcumin, Cyanidin, Diadezin, Delphinidin, Ellagic acid, Epiafzellichin, Epicatechin, Epigallocatechin, Ferulic acid, Gallic acid, Gallocatechin, Genisterin, Glycitein, Hesperetin, Hesperidin, Isorharmetin, Kampferol, Kievitone, Lariciresinol, Malvidin, Medioresinol, myricetin, Naringenin, Naringin, Narirutin, Neohesparindin, Nobiletin, p-Coumatic acid, Pelargiridin, Peonidin, Petinidin, Pinoresinol, Protocatechuic, Punicalin, Punicalagin, Quercetin, Resveratrol, Rutinoside, Secisolariciresinol, Sesamin, Sinapic acid, Syrigaresinol, Tangeretin, Vallic acid and Woogin acid and also the standard (Remdesivir) from PUBCHEM (http://pubchem.ncbi.nlm.nih.gov). Cactus Translator Server (https://cactus.nci.nih.gov/translate/) was employed to generate pdb format for each compound canonical smiles. The compounds were optimized and inserted into Autodock Vina to obtain their respective pdbqt formats [49].
2.4. Molecular docking
Further analyses were carried out using Autodock 4.2 for identifying specific interaction involved in binding of molecules to the target receptor. Subsequently, all the proteins were preprocessed and Gesteiger charges were added to it using AutoDock MGL tools 1.5.6. A receptor grid box was generated after the active sites had been defined. Grid box dimension of 60×60×60 for 3cl Mpro with spacing of 0.375 Å centering on residue Cys145 and His41 was defined. The protein-ligand interactions were visualized by Discovery Studio and Pymol [11].
2.5. ADMET and drug-likeness analysis
An efficient and widely used computational server-Admetsar2 (https://lmmd.ecust.edu.cn/admetsar2/) was entrusted to perform ligand-based studies of the selected phytochemicals to ascertain the compound that may have the potential of qualifying to be a druggable lead compounds with the aim to reduce the viral pathogenesis. Diverse parameters under numerous rules were used to screen the pharmacokinetics and drug-likeness of these compounds [45,54].
2.6. Molecular dynamics simulation
Prior to the 20 ns production run of the three complexes (including the standard), GROMACS (GROningen MAchine for Chemical Simulations) Molecular dynamics operating environment [50] was used to prepare the files for the simple dynamics normal control group “6LU7” and for the complexes which include 6LU7-Ela, 6LU7-Kie, and 6LU7-Rem. We used CHARMS-36 force field and TIP3P GROMACS recommended water model for the protein topology. CGENFF web server tool was used to prepare the “.str” file for the ligand topology after which the topology files of the complexes were appropriately updated using appropriate python codes to manually include ligands topology. We ionized and neutralized the systems and then solvated them using the simple point charge-216 explicit water model (spc216.gro). Energy minimization was run for 100 ps (picoseconds) using steepest descent algorithm to establish a stable conformation for the system after which we equilibrated the system using Verlet algorithm from 0-310 K at 100 ps, 2fs (femtoseconds) time step for NVT, while Berenson algorithm at 2fs time step for 100 ps was used for NPT equilibration. The system was treated using the “trjconv” module to centralize and compact the protein while we also made sure the atoms does not jump out of the PBC (Periodic Boundary Condition). RMSD (Root Mean Square Deviation) was calculated and reported as mean ± SD using the 2004 frames for each 20 ns production step run for each complex using “gmx analyze” GROMACS module after which the H-bonds were also calculated using “xmgrace” module.
2.7. Molecular mechanics (MM-PBSA)
The Molecular Mechanics Poisson–Boltzmann surface area (MM-PBSA) method [31], implemented in the g_mmpbsa module [32] of GROMACS, was used to compute the relative binding free energy of each complexes for the last 5 ns trajectories. Xmgrace was then used to plot the graph spectrum of the individual contributing energy of the residues for the three complexes.
The Binding energies consist of the van der Waal forces, electrostatic interactions and the solvation energy while this solvation energy comprises of both polar and non-polar energies. The equation is defined below as:
Where BE = Binding Energy, VDW =Van Der Waal, PSE = Polar Solvation Energy, and SASA=Solvent Accessible Surface Area Energy
2.8. Density functional theory (quantum mechanics)
The top two compounds (hits) from the docking screening and the standard (Remdesivir) were subjected to quantum mechanical calculation using density functional theory. The Gaussian 09 W program [13] was used for the calculations by optimizing the compounds' geometries at DFT/B3LYP/6–31G(d'p’) levels. The frontiers orbital energies, the highest occupied molecular orbital (HOMO), the lowest occupied molecular orbital (LUMO), energy gap, and the molecular electrostatic potential were computed in order to understand the electron acceptor and electron donor properties of the compounds. These also provide information about the chemical reactivity and stability of the compounds.
3. Results and discussion
3.1. Molecular docking experiment
Right here for the first time, we report the anti-SARS-CoV-2 potential of two polyphenols (Ellagic acid and Kievitone). We study the inhibitory potential of 50 dietary polyphenols against the Mpro target (6LU7) of SARS-COV2 using Autodock Vina. The binding affinity result of the 50 dietary polyphenols has been depicted in supplementary data (Table 1 ). The computational thermodynamic scoring function of the widely used Autodock Vina program reveals three top-ranking compounds-Ellagic acid, Kievitone and Punicalin with significant binding affinity of −8.9 kcal/mol (4 conventional H-bond and 3 hydrophobic bonds), −8.0 kcal/mol (8 H-bond and 3 hydrophobic bonds) and −7.9 kcal/mol (6 H-bonds and 1 hydrophobic interaction) respectively (see Table 1 and Fig. 1 ). Other compounds were filtered out based on the binding affinity of the standard drug-Remdesivir (−7.7 kcal/mol) which was used as cut-off to screen out less potent compounds (Table 1). Since lowest binding energy ligand represents highest binding affinity compound [47], Ellagic acid could be considered as the best docked compound in this study.
Table 1.
indicates the binding affinity, Inhibition constant, hydrogen bonds and hydrophobic bonds interaction of dietary polyphenols hits against SARS-COV-2 main protease (6LU7).
| Compounds | Targeted Proteins 6LU7 |
||
|---|---|---|---|
| Binding Affinities | Hydrogen Bonds interactions | Hydrophobic Bonds Interactions | |
| Ellagic acid | −8.9 kcal/mol | His41, Asn142, Gly143, Glu166 | Met49, Cys145 |
| Kievitone | −8.0 kcal/mol | Asp187, Cys143, asn142, Glu166, His172, his163, Ser144 | His41, Met49, Met165 |
| Punicalin | −7.9 kcal/mol | Thr26, Leu141, Gly143, His164, Met165, Asp187, Arg188, | Cys145 |
| Remdesivir | −7.7 kcal/mol | Glu166, Ser144, gly143, Cys145, asn142, His163 | His41, His164, Gln165 |
Fig. 1.
3D structure of SARS-COV2 Mpro and A. Ellagic acid B. Kievitone C. Punicalin D. Remdesivir conventional hydrogen bonds (Green), carbon hydrogen bonds (cyan) and hydrophobic (pink) 2D interactions with SARS-COV-2 Main protease residues.
Moreover, our result of conformational sampling of Ellagic acid and Kievitone against the Mpro molecular target also indicates that the phytocompounds binds efficiently with domain I and II of the receptor active site including the His41 and Cys145 catalytic dyad (Fig. 1). Although Punicalin only interacts with Cys145, however, the ligand depicted similar mode of binding when docked with the receptor. Interestingly, similar mode of binding of these phytochemicals was also observed in the standard drug (Remdesivir) which was also found interacting with the crucial catalytic dyad of Mpro (Fig. 1). Several investigations which support our findings have been reported in the studies of Gurung and collaborators, Das and colleagues, Joshi and Co-authors and Islam et al. [10,18,24,27]). We believe the competitive binding of our hit compounds with His41 and Cys145 could be a potent therapeutic measure that could inhibit the SARS-COV2 Mpro catalytic activity.
3.2. ADMET and drug-likeness
Computational drug-likeness test is done to facilitate the creation of new drug molecules, the molecules of which must comply with the criteria of the following five Lipinski's rule: molecular weight must not be greater than 500, number of hydrogen bond donors must not be greater than 5, number of hydrogen bond acceptors must not be more than 10 and lipophilicity (expressed in LogP) must not be greater than 5 [34]. Potential inhibitors with more than one violation of these rules are considered as molecules with poor absorption and low permeation properties [6]. The result of this study shown in Table 2 reveals the potential of Ellagic acid and Kievitone as an excellent drug-like compound since both compounds do not violate any of Lipinski rules. In contrast, Punicalin do not comply with Lipinski drug-likeness with three (3) violations of the physiochemical properties of the rule of five. Hence, there is high propensity that Ellagic acid and Kievitone could behave as a better drug-like molecule than the standard which disobeyed three properties from the rule of five.
Table 2.
Indicates the physicochemical properties of selected hit compounds and the standard used in this virtual screening.
| Compounds | Lipinski's Rule of 5 |
||||
|---|---|---|---|---|---|
| Mol. wt. | Hydrogen donor | Hydrogen Acceptor | Log P | No. of Violation | |
| Ellagic acid | 302.19 | 4 | 8 | 0.79 | 0 |
| Kievitone, | 356.37 | 4 | 6 | 2.24 | 0 |
| Punicalin | 782.53 | 13 | 22 | −0.07 | 3 |
| Remdesivir | 602.58 | 4 | 12 | 3.24 | 2 |
In silico ADMET analysis is used to assess the pharmacokinetic and toxicological properties of hit compounds before experimental procedures [25]. ADMET evaluation of compounds is crucial in determining whether a potential lead candidate is capable of passing through the clinical trials stage. Hence, it is imperative to perform ADMET test early during the drug discovery process in order to filter out small molecule inhibitors with unacceptable and poor pharmacological profile. Our result of pharmacokinetic predictions of Ellagic acid, Kievitone, Punicalin and the standard drug is represented in Table 3 .
Table 3.
Represents the ADMET properties of lead compounds selected from the bulk of dietary polyphenols used in this virtual screening. C-1= Ellagic Acid; C-2= Kievitone; C-3 = Punicalin C-4= Remdesivir.
| Absorption&Distribution | C-1 | C-2 | C-3 | C-4 |
|---|---|---|---|---|
| BBB (+/-) | – | – | – | + |
| p-glycoprotein substrate | – | – | – | + |
| HIA+ | + | + | + | + |
| Metabolism | ||||
| CYP450 2C19 Inhibitor |
– | + | – | – |
| CYP450 1A2 Inhibitor |
– | + | – | – |
| CYP450 3A4 Inhibitor |
– | – | – | – |
| CYP450 2C9 Inhibitor |
– | + | – | – |
| CYP450 2D6 Inhibitor |
– | – | – | – |
| Excretion | ||||
| Biodegradation | – | – | – | – |
| Toxicity | ||||
| AMES Mutagenesis | – | – | + | – |
| Acute Oral Toxicity | II | III | III | III |
| hERG Inhibition | – | – | + | – |
| Carcinogenicity | – | – | – | – |
| Hepatotoxicity | + | + | + | + |
Human intestinal absorption (HIA), substrate of P-glycoprotein and permeation through the blood-brain barrier (BBB) have been identified for the prediction of absorption and distribution properties of our drug candidates (Table 3). The positive value of HIA depicted by the selected compounds including the standard may indicate that they can be easily absorbed in the intestine after administration (Table 3). In the distribution section, Ellagic acid, Kievitone and Punicalin have negative values for BBB test which means they are unable to permeate through the blood-brain barrier and hence the central nervous system is protected from their action. In contrast, Remdesivir show a positive value for BBB which indicate its wider distribution and potential neurological effect. Furthermore, Ellagic acid and Punicalin were predicted to be non-substrate of P-glycoprotein which suggest both compounds could have good distribution properties. However, Kievitone and Remdesivir could act as substrate of p-glycoprotein and hence their distribution may be limited by the mediation of p-glycoprotein efflux [46].
In the metabolism part, the families of Cytp450 have been identified as an important pharmacological parameter. According to Lynch, the inhibition of these protein families may result in drug-drug interaction and bio-accumulation [36]. According to our result, the only compound found inhibiting one or more of these metabolic enzymes is Kievitone.
The excretion of all drug candidates was also assessed in our ADMET study by carrying out the biodegradation test. Interestingly, all compounds including the standard could be easily excreted as they return a negative value for biodegradation.
Finally, the pharmacodynamic profile of the selected drug candidates was also assessed. It is important to note that the failure of drugs to pass through the clinical trials is majorly because of their non-acceptable toxicological profile and hence we have put into consideration the Ames mutagenicity, Acute oral toxicity, hERG inhibition, carcinogenicity and hepatotoxicity testing in this study (Table 3). All the drug candidates and the standard drug show negative values for carcinogenicity test which may explain why the compounds may not be carcinogenic. However, it is disappointing that all the drug candidates are hepatotoxic. However, since clinically approved Remdesivir was also predicted to be hepatotoxic, it is likely that the negative consequence of this property is minimal to the liver cells. The hERG and Ames mutagenicity are crucial pharmacological parameters for evaluating if a drug-like compound could cause cardiac arrhythmia [37] and capable of mutating the DNA respectively. Ellagic acid, Kievitone and Remdesivir excel in these parameters while Punicalin with its positive values for hERG and Ames mutagenicity test may affect the rhythm of potassium channels in the heart (cardiac arrhythmia) and may induce carcinogenicity respectively. Although, Ellagic acid is the only predicted orally toxic compound in this section. However, it is evidenced that the drug candidate stands out among the selected compounds in terms of pharmacokinetic and pharmacodynamic profile. In contrast, the mutagenicity and inhibition of hERG protein by Punicalin may lead to fatal health consequence and hence we have employed a more safety measure by filtering out this phytochemical compound in our study.
3.3. Molecular dynamics simulation
Molecular dynamic is an in silico approach which is used to monitor the behavior and dynamics of binary protein-ligand complex at the atomistic level. Unlike molecular docking, MD simulation represents a more realistic delineation of the complex biological system by putting the flexibility of proteins, neutralizing ions and water environment into consideration. In this study, Ellagic acid (Ela), Kievitone (Kie) and Remdesivir (Rem) behavior with the Mpro target (6LU7) was simulated by subjecting them in Molecular dynamics environment. The reason for dropping Punicalin (the 3rd ranked inhibitor) for MD simulation is based on its poor pharmacodynamics and non-compliance with the rule of five (see Tables 3 and 2 respectively) which could be detrimental to the health of clinical trial volunteers. In order to investigate and compare the stability of 6LU7-Ela, 6LU7-Kie with the standard (6LU7-Rem), the graph spectrums of the RMSD, RMSF, H-bond and ROG of the complexes have been superimposed as depicted in Fig. 2, Fig. 3, Fig. 4, Fig. 5 .
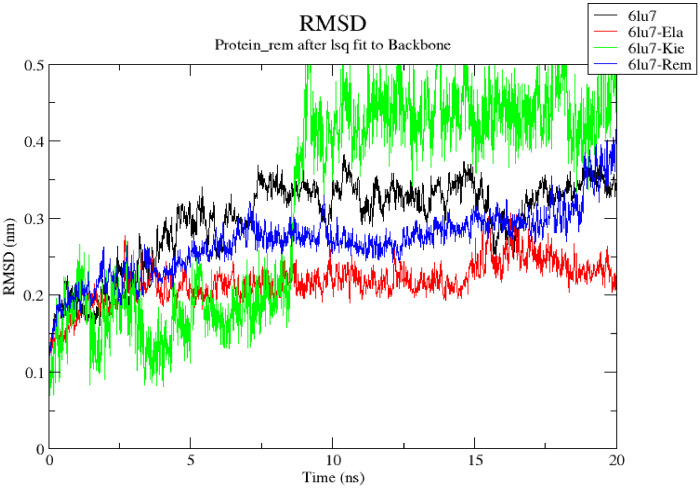
Fig. 2.
Superimposed RMSD spectrum of 6LU7 (Apoprotein), 6LU7-Ela, 6LU7-Kie and 6LU7-Rem.
Fig. 3.
Superimposed RMSF spectrum of 6LU7 (Apoprotein), 6LU7-Ela, 6LU7-Kie and 6LU7-Rem.
Fig. 4.
Represents the intermolecular H-bond spectrum of 6LU7-Ela, 6LU7-Kie and 6LU7-Rem.
Fig. 5.
ROG Spectrum of 6LU7, 6LU7-Ela, 6LU7-Kie and 6LU7-Rem.
3.3.1. Root mean square deviation (RMSD)
The RMSD backbone is used to assess the structural stability of protein-ligand complex and lower value of the ligand bound protein compared to the unbound form indicates a stable system [35]. The average RMSD values for 6LU7 (Apoprotein), 6LU7-Ela, 6LU7-Kie and 6LU7-Rem are 0.2981 ± 0.0552 nm, 0.2171 ± 0.0267 nm, 0.3230 ± 0.1366and 0.2692 ± 0.0433 respectively. From this result, the RMSD value of 6LU7-Ela is lower than the apoprotein and the standard (6LU7-Rem) hence it could be regarded as the most stable complex. The spectrum of the RMSD graph also suggests minimal perturbation in the dynamical conformation of the 6LU7-Ela complex (Fig. 2). This finding may explain why Ellagic acid could not deviate away from its initial docked pose and the ruggedness to consistently bind to Mpro catalytic dyad residues. In the case of Kievitone dynamics with Mpro receptor, there is abrupt fluctuation of the RMSD spectrum between 0 and 10 ns. However, after 10 ns, stability of the complex was noticed as equilibrium was attained till the end of the 20 ns production run (Fig. 2).
3.3.2. Root mean square fluctuation (RMSF)
RMSF graph of the individual residues of the unbound and bound protein were also plotted to assess the flexibility of the amino acids. It is known that regions with very high RMSF values correspond mostly to the loop region which could also be involved in ligand binding and conformational alterations [33]. Interestingly, His41 (0.0787 nm) and Cys145 (0.0546 nm) catalytic dyad which are found at the loop region and believe to be critical for inhibition of Mpro were among the most stable residues of the apoprotein. Moreover, the binding of Ellagic acid, Kievitone and Remdesivir with 6LU7 return RMSF score of 0.1010 nm, 0.1141 nm and 0.0770 nm respectively for His41. The RMSF score of Cys145 for 6LU7-Ela, 6LU7-Kie and 6LU7-Rem are 0.0632 nm, 0.0583 nm and 0.0738 nm respectively (Fig. 3). Since there were no significant variations in the RMSF values of these complexes when compared with apoprotein, we conclude that the catalytic dyad residues remain stabilized. Also, the average RMSF score of 6LU7-Ela (0.1147 ± 0.0501 nm) is lower than the apoprotein, 6LU7-Kie (0.10301 ± 0.1012 nm) and 6LU7-Rem (0.1417 ± 0.0621) (see Fig. 3). Hence, it could be regarded as complex with the most stable residues.
3.3.3. H-bond
It is well known that intermolecular H-bond is important for maintaining the stability of protein-ligand interaction. The higher the number of H-bond formed between a complex, the stronger their interaction [41]. In this research, 6LU7-Ela averaged intermolecular H-bond formation of 1.81 for the entire 20 ns simulation run which is higher than 6LU7-Kie (1.04) and the standard (1.501) (Fig. 4). The H-bond result may explain why Ellagic acid demonstrate higher binding affinity and better stability with Mpro than both the standard (Remdesivir) and Kievitone.
3.3.4. Radius of gyration (ROG)
The compactness of protein in a dynamical system can be monitored by computing their ROG values. A stably folded system could be measured by equilibration of its ROG spectrum throughout the simulation period while a non-compact system may be identified when there is abrupt fluctuation of ROG spectrum [29].
The low mean ROG values of 6LU7, 6LU7-Ela, 6LU7-Kie and 6LU7-Rem (2.2267 −2544 nm) indicate minimal fluctuation of the complexes (Fig. 5). Although, the unbound 6LU7 protein is the most compact structure according to its ROG spectrum, the minimum and maximum values of 6LU7-Ela (2.21002 nm- 2.3001 nm) and 6LU7-Kie (2.20016 nm- 2.23816 nm) may explain a reasonable well folded system. Importantly, the RMSD spectrum of both complexes suggest a better compact structure when compared with the standard which shows minimum and maximum ROG values of 2.2116 nm and 2.3913 nm respectively.
3.4. Molecular mechanics (MM-PBSA)
The individual decomposition energy of the residues and overall binding energies of the complexes have been represented in Fig. 6 and Table 5 respectively. From the result presented in Table 5, it is crystal clear that the major driving force responsible for the spontaneity and high binding energy of the complexes is the Van der waal's interaction (VDW). In contrast, the polar solvation energy observed in the three complexes show unfavorable binding of the protein-ligand interactions. The overall result of the mm-pbsa binding energy suggests that Kievitone (−60.480±24.834 kj/mol) and Ellagic acid (−47.487±11.028 kj/mol) may be more potent inhibitor than Remdesivir(−28.911±10.9kj/mol). Finally, we plotted the graph spectrum of the individual contributing energy of the residues to assess their cruciality in the binding of the complexes. From the docked pose, His41, Met49, Cys145, Gly143, Asn142 and Glu166 were found interacting with Ellagic acid and their contributing mm-pbsa energy (in kj/mol) are 0.4035, −1.1554, −2.4237, 0.1686, 0.2228, −1.3238 respectively. Also, the contributing energy of the amino acids found interacting with Kievitone have binding energies of −0.1159 (Tyr54),2.3352 (His 41), −1.5219 (Met49), −2.1422 (Cys145), −2.796 (Met165),16.6994 (Glu166), 2.4762 (Ser144), −0.3695 (His163),0.097 (His172),−5.6508Asn142, and −0.580 (Asp187) while those binding with Remdesivir have decomposition energies of −0.6983 (His41), −0.0369 (Cys145), −4.908 (Met165), 2.9349 (Glu166), 0.1885 (Asn142), −0.7251 (His164), −0.2039 (His163), −0.202 (His172),−0.175 (Leu141), 0.055 (Gly143) and −0.079 (Ser144). From this result, it is evident that the atomic interaction of Ellagic acid and Kievitone to Cys145 is largely responsible for their significant binding energy. Interestingly, our analysis also indicates the ability of the two drug candidates to effectively inhibit SARS-COV2 main protease by binding more stably to the crucial Cys145 residue [26] than the standard.
Fig. 6.
MM-PBSA Binding free energy contribution of individual residue of 6LU7 in complex with A.Ela B.Kie C.Rem.
Table 5.
MMPBSA free energies of the protein-ligand complexes.
| Compounds | VdW | Electrostatic | PSA | SASA | Total Binding Energy (Kj/mol) |
|---|---|---|---|---|---|
| Ellagic acid | −83.866±11.128 | −34.071±7.832 | 78.891±9.889 | −9.441±0.999 | −47.487±11.028 |
| Remdesivir | −122.849±12.780 | −81.955±24.436 | 190.929±28.989 | −15.036±1.110 | −28.911±10.98 |
| Kievitone | −126.286±40.575 | −9.836±8.023 | 92.032±37.945 | −16.390±4.363 | −60.480±24.834 |
3.5. Quantum mechanics (DFT)
The molecular frontier orbitals are well-established parameters to provide information about the compounds' reactivity and stability. The highest occupied molecular orbital (HOMO) and the lowest occupied molecular orbital (LUMO) depict the electrons donating and receiving the compounds' ability. The energy gap, which is the difference between the LUMO and the HOMO, describes the kinetic stability and the intramolecular charge transfer. Also, compounds with significant energy gaps are referred to have low chemical reactivity and high kinetic stability. In contrast, compounds with low energy gaps are more reactive with low stability [29]. The energy gap of the lead compounds increase accordingly: Ellagic acid (0.149 a.u) < Kevitone (0.161 a.u) < Remdesivir (0.175 a.u) (Table 4 and Fig 7 ). Notwithstanding, the chemical hardness and softness were also computed from the frontier orbitals. Compounds with high chemical reactivity and low stability are considered to be soft (σ) [14], and compounds with low reactivity and high stability are referred to as hard (η) [28]. The Ellagic acid with an energy gap of 0.149 a.u has the highest chemical reactivity with less stability when compared with Kievitone and Remdesivir with an energy gap of 0.161 a.u and 0.175 a.u, respectively. While the most stable compound is Remdesivir based on the energy gap value.
Table 4.
The Highest occupied molecular orbital (HOMO) energy, Lowest occupied molecular orbital (LUMO) energy.
| Frontiers Orbitals | Ellagic acid (a.u) | Kievitone (a.u) | Remdesivir (a.u) |
|---|---|---|---|
| HOMO | −0.237 | −0.224 | −0.242 |
| LUMO | −0.088 | −0.063 | −0.066 |
| Gap (∆E) | 0.149 | 0.161 | 0.175 |
| Chemical hardness(η=(ELUMO-EHOMO)/2) | 0.074 | 0.080 | 0.087 |
| Softness (σ=1/η) | 13.513 | 12.500 | 11.494 |
Fig. 7.
The Highest occupied molecular orbital (HOMO) energy, Lowest occupied molecular orbital (LUMO) energy, and the molecular electrostatic potential (MEP).
4. Conclusion
In this study, we employed a computational approach with the use of molecular docking, molecular dynamics, and Quantum mechanics to investigate the inhibitory potential of dietary polyphenols against SARS-COV-2 Mpro at the molecular and atomistic levels. What we noticed from our findings is that two of the screened ligands (Ellagic acid and Kievitone) could be potent competitive inhibitors of the coronavirus main protease. It is apparent from molecular dynamics and MMPBSA analysis that the binding of Ellagic acid to His41 and Cys145 catalytic dyad of the receptor (from the docked pose) forms a more stable system when compared to the standard drug. Our findings also revealed that Kievitone is capable of disrupting the proteolytic activity of Mpro and the analysis of MMPBSA and molecular orbital theory suggest that the drug candidate is slightly stable than Ellagic acid. We, therefore, propose that Ellagic acid and Kievitone be further developed as drug candidates for the treatment of COVID-19. Nonetheless, while these findings may be appreciable and appealing under in silico research, further experimental and clinical studies are needed to buttress their preclinical and clinical relevance respectively. In addition, functional foods endowed with kievitone and ellagic acid might be functional in the management/treatment of COVID-19 disease. Although a global concern about making polyphenols functional foods is their high susceptibility towards metabolism which may impede their bioavailability index, this could be addressed by packing them into nanocarriers or through an encapsulation mechanism for efficient delivery to their target site.
Funding information
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
CRediT authorship contribution statement
Temitope Isaac Adelusi: Conceptualization, Supervision, Writing – original draft, Formal analysis. Abdul-Quddus Kehinde Oyedele: Writing – original draft, Investigation, Formal analysis, Validation. Ojo Emmanuel Monday: Writing – original draft, Methodology, Investigation. Ibrahim Damilare Boyenle: Investigation, Validation. Mukhtar Oluwaseun Idris: Software, Methodology. Abdeen Tunde Ogunlana: Methodology, Investigation, Visualization. Ashiru Mojeed Ayoola: Writing – review & editing. John Olabode Fatoki: Supervision. Oladipo Elijah Kolawole: Writing – review & editing. Kehinde Busuyi David: Validation. Akintola Adebola Olayemi: Writing – review & editing.
Declaration of Competing Interest
Authors declare no area of competing interest.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.molstruc.2021.131879.
Appendix. Supplementary materials
References
- 1.Adelusi T.I., Abdul-Hammed M., Ojo E.M., Oyedele Q.K., Boyenle I.D., Adedotun I.O., Olaoba O.T., Folorunsho A.A., Kolawole O.E. Molecular docking assessment of clinically approved antiviral drugs against Mpro, spike glycoprotein and angiotensin converting enzyme-2 revealed probable anti-SARS-CoV-2 potential. Trop. J. Nat. Prod. Res. 2021;5(4) [Google Scholar]
- 2.Adelusi T.I., Misbaudeen A., Idris M.O., Oyedele Q.K., Adedotun I.O. Molecular dynamics, quantum mechanics and docking studies of some Keap1 inhibitor—an insight into the atomistic mechanisms of their antioxidant potential. Heliyon. 2021:e07317. doi: 10.1016/j.heliyon.2021.e07317. ISSN 2405 8440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Azhar E.I., El-Kafrawy S.A., Farraj S.A., Hassan A.M., MS Al-Saeed, Hashem A.M., Madani T.A. Evidence for camel-to-human transmission of MERS coronavirus. N. Engl. J. Med. 2014;370(26):2499–2505. doi: 10.1056/NEJMoa1401505. Jun 26Epub 2014 Jun 4. PMID: 24896817. [DOI] [PubMed] [Google Scholar]
- 4.Basheer L., Kerem Z. Interactions between CYP3A4 and dietary polyphenols. Oxid. Med. Cell Longev. 2015 doi: 10.1155/2015/854015. 2015Epub 2015 Jun 9. PMID: 26180597; PMCID: PMC4477257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beigel J.H., Tomashek K.M., Dodd L.E., Mehta A.K., Zingman B.S., Kalil A.C., Hohmann E., Chu H.Y., Luetkemeyer A., Kline S., Lopez de Castilla D., Finberg R.W., Dierberg K., Tapson V., Hsieh L., Patterson T.F., Paredes R., Sweeney D.A., Short W.R., Touloumi G., Lye D.C., Ohmagari N., Oh M.D., Ruiz-Palacios G.M., Benfield T., Fätkenheuer G., Kortepeter M.G., Atmar R.L., Creech C.B., Lundgren J., Babiker A.G., Pett S., Neaton J.D., Burgess T.H., Bonnett T., Green M., Makowski M., Osinusi A., Nayak S., Lane H.C. ACTT-1 study group members. Remdesivir for the treatment of Covid-19—final report. N. Engl. J. Med. 2020;383(19):1813–1826. doi: 10.1056/NEJMoa2007764. Nov 5Epub 2020 Oct 8. PMID: 32445440; PMCID: PMC7262788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Benet L.Z., Hosey C.M., Ursu O., Oprea T.I. BDDCS, the Rule of 5 and drugability. Adv. Drug. Deliv. Rev. 2016;101:89–98. doi: 10.1016/j.addr.2016.05.007. Jun 1Epub 2016 May 13. PMID: 27182629; PMCID: PMC4910824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boyenle I.D., Divine U.C., Adeyemi R., Ayinde K.S., Olaoba O.T., Apu C., Du L., Lu Q., Yin X., Adelusi T.I. Direct Keap1-kelch inhibitors as potential drug candidates for oxidative stress-orchestrated diseases: a review on In silico perspective. Pharmacol. Res. 2021;167 doi: 10.1016/j.phrs.2021.105577. MayEpub 2021 Mar 24. PMID: 33774182. [DOI] [PubMed] [Google Scholar]
- 8.Corcoran M.P., McKay D.L., Blumberg J.B. Flavonoid basics: chemistry, sources, mechanisms of action, and safety. J. Nutr. Gerontol. Geriatr. 2012;31(3):176–189. doi: 10.1080/21551197.2012.698219. PMID: 22888837. [DOI] [PubMed] [Google Scholar]
- 9.Dai W., Zhang B., Jiang X.M., Su H., Li J., Zhao Y., Xie X., Jin Z., Peng J., Liu F., Li C., Li Y., Bai F., Wang H., Cheng X., Cen X., Hu S., Yang X., Wang J., Liu X., Xiao G., Jiang H., Rao Z., Zhang L.K., Xu Y., Yang H., Liu H. Structure-based design of antiviral drug candidates targeting the SARS-CoV-2 main protease. Science. 2020;368(6497):1331–1335. doi: 10.1126/science.abb4489. Jun 19Epub 2020 Apr 22. PMID: 32321856; PMCID: PMC7179937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Das S., Sarmah S., Lyndem S., Singha Roy A. An investigation into the identification of potential inhibitors of SARS-CoV-2 main protease using molecular docking study. J. Biomol. Struct. Dyn. 2021;39(9):3347–3357. doi: 10.1080/07391102.2020.1763201. JunEpub 2020 May 13. PMID: 32362245; PMCID: PMC7232884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Delano W. Pymol: an open-source molecular graphics tool. CCP4 Newsl. Protein Crystallogr. 2002;40:44–53. [Google Scholar]
- 12.Dey S.K., Rahman M.M., Siddiqi U.R., Howlader A. Analyzing the epidemiological outbreak of COVID-19: a visual exploratory data analysis approach. J. Med. Virol. 2020;92(6):632–638. doi: 10.1002/jmv.25743. JunEpub 2020 Mar 11. PMID: 32124990; PMCID: PMC7228278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Frisch M.J., Trucks G.W., Schlegel H.B., Scuseria G.E., Robb M.A., Cheeseman J.R., Scalmani G., Barone V., Mennucci B., Petersson G.A., Nakatsuji H., Caricato M., Li X., Hratchian H.P., Izmaylov A.F., Bloino J., Zheng G., Sonnenberg J.L., Hada M., Ehara M., Toyota K., Fukuda R., Hasegawa J., Ishida M., Nakajima T., Honda Y., Kitao O., Nakai H., Vreven T., Montgomery J.A., Jr., Peralta J.E., Ogliaro F., Bearpark M., Heyd J.J., Brothers E., Kudin K.N., Staroverov V.N., Kobayashi R., Normand J., Raghavachari K., Rendell A., Burant J.C., Iyengar S.S., Tomasi J., Cossi M., Rega N., Millam J.M., Klene M., Knox J.E., Cross J.B., Bakken V., Adamo C., Jaramillo J., Gomperts R., Stratmann R.E., Yazyev O., Austin A.J., Cammi R., Pomelli C., Ochterski J.W., Martin R.L., Morokuma K., Zakrzewski V.G., Voth G.A., Salvador P., Dannenberg J.J., Dapprich S., Daniels A.D., Farkas Ö., Foresman J.B., Ortiz J.V., Cioslowski J., Fox D.J. Gaussian, Inc.; Wallingford CT: 2009. Gaussian 09, Revision E.01. [Google Scholar]
- 14.Gázquez J.L. Chemical Hardness. Springer; 1993. Hardness and softness in density functional theory; pp. 27–43. [Google Scholar]
- 15.Grum-Tokars V., Ratia K., Begaye A., Baker S.C., Mesecar A.D. Evaluating the 3C-like protease activity of SARS-Coronavirus: recommendations for standardized assays for drug discovery. Virus Res. 2008;133(1):63–73. doi: 10.1016/j.virusres.2007.02.015. AprEpub 2007 Mar 29. PMID: 17397958; PMCID: PMC4036818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guan W.J., Ni Z.Y., Hu Y., Liang W.H., Ou C.Q., He J.X., Liu L., Shan H., Lei C.L., Hui D.S.C., Du B., Li L.J., Zeng G., Yuen K.Y., Chen R.C., Tang C.L., Wang T., Chen P.Y., Xiang J., Li S.Y., Wang J.L., Liang Z.J., Peng Y.X., Wei L., Liu Y., Hu Y.H., Peng P., Wang J.M., Liu J.Y., Chen Z., Li G., Zheng Z.J., Qiu S.Q., Luo J., Ye C.J., Zhu S.Y., Zhong N.S. China medical treatment expert group for Covid-19. Clinical characteristics of coronavirus disease 2019 in China. N. Engl. J. Med. 2020;382(18):1708–1720. doi: 10.1056/NEJMoa2002032. Apr 30Epub 2020 Feb 28. PMID: 32109013; PMCID: PMC7092819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guan Y., Zheng B.J., He Y.Q., Liu X.L., Zhuang Z.X., Cheung C.L., Luo S.W., Li P.H., Zhang L.J., Guan Y.J., Butt K.M., Wong K.L., Chan K.W., Lim W., Shortridge K.F., Yuen K.Y., Peiris J.S., Poon L.L. Isolation and characterization of viruses related to the SARS coronavirus from animals in southern China. Science. 2003;302(5643):276–278. doi: 10.1126/science.1087139. Oct 10Epub 2003 Sep 4. PMID: 12958366. [DOI] [PubMed] [Google Scholar]
- 18.Gurung A.B., Ali M.A., Lee J., Farah M.A., Al-Anazi K.M. Unravelling lead antiviral phytochemicals for the inhibition of SARS-CoV-2 Mpro enzyme through in silico approach. Life Sci. 2020;255 doi: 10.1016/j.lfs.2020.117831. Aug 15Epub 2020 May 22. PMID: 32450166; PMCID: PMC7243810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Harcourt B.H., Jukneliene D., Kanjanahaluethai A., Bechill J., Severson K.M., Smith C.M., Rota P.A., Baker S.C. Identification of severe acute respiratory syndrome coronavirus replicase products and characterization of papain-like protease activity. J. Virol. 2004;78(24):13600–13612. doi: 10.1128/JVI.78.24.13600-13612.2004. DecPMID: 15564471; PMCID: PMC533933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hayes J.E., Allen P., Brunton N., et al. Phenolic composition and in vitro antioxidant capacity of four commercial phytochemical products: olive leaf extract (Olea europaea L.), lutein, sesamol and ellagic acid. Food Chem. 2011;126(3):948–955. https//doi.org/10.1016/j.foodchem.2010.11.092. [Google Scholar]
- 21.Hillen H.S., Kokic G., Farnung L., Dienemann C., Tegunov D., Cramer P. Structure of replicating SARS-CoV-2 polymerase. Nature. 2020;584(7819):154–156. doi: 10.1038/s41586-020-2368-8. AugEpub 2020 May 21. PMID: 32438371. [DOI] [PubMed] [Google Scholar]
- 22.Hoffman R. Potent inhibition of breast cancer cell lines by the isoflavonoid kievitone: comparison with genistein. Biochem. Biophys. Res. Commun. 1995;211(2):600–606. doi: 10.1006/bbrc.1995.1855. Jun 15PMID: 7794275. [DOI] [PubMed] [Google Scholar]
- 23.Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., Zhang L., Fan G., Xu J., Gu X., Cheng Z., Yu T., Xia J., Wei Y., Wu W., Xie X., Yin W., Li H., Liu M., Xiao Y., Gao H., Guo L., Xie J., Wang G., Jiang R., Gao Z., Jin Q., Wang J., Cao B. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395(10223):497–506. doi: 10.1016/S0140-6736(20)30183-5. Feb 15Epub 2020 Jan 24. Erratum in: Lancet. 2020 Jan 30; PMID: 31986264; PMCID: PMC7159299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Islam R., Parves M.R., Paul A.S., Uddin N., Rahman M.S., Mamun A.A., Hossain M.N., Ali M.A., Halim M.A. A molecular modeling approach to identify effective antiviral phytochemicals against the main protease of SARS-CoV-2. J. Biomol. Struct. Dyn. 2021;39(9):3213–3224. doi: 10.1080/07391102.2020.1761883. JunEpub 2020 May 12. PMID: 32340562; PMCID: PMC7232885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jayaraj J.M., Reteti E., Kesavan C., Muthusamy K. Structural insights on vitamin D receptor and screening of new potent agonist molecules: structure and ligand-based approach. J. Biomol. Struct. Dyn. 2020:1–12. doi: 10.1080/07391102.2020.1775122. Jun 11Epub ahead of print. PMID: 32462983. [DOI] [PubMed] [Google Scholar]
- 26.Jin Z., Du X., Xu Y., Deng Y., Liu M., Zhao Y., Zhang B., Li X., Zhang L., Peng C., Duan Y., Yu J., Wang L., Yang K., Liu F., Jiang R., Yang X., You T., Liu X., Yang X., Bai F., Liu H., Liu X., Guddat L.W., Xu W., Xiao G., Qin C., Shi Z., Jiang H., Rao Z., Yang H. Structure of Mpro from SARS-CoV-2 and discovery of its inhibitors. Nature. 2020;582(7811):289–293. doi: 10.1038/s41586-020-2223-y. JunEpub 2020 Apr 9. PMID: 32272481. [DOI] [PubMed] [Google Scholar]
- 27.Joshi T., Joshi T., Sharma P., Mathpal S., Pundir H., Bhatt V., Chandra S. In silico screening of natural compounds against COVID-19 by targeting Mpro and ACE2 using molecular docking. Eur. Rev. Med. Pharmacol. Sci. 2020;24(8):4529–4536. doi: 10.26355/eurrev_202004_21036. AprPMID: 32373991. [DOI] [PubMed] [Google Scholar]
- 28.Kaya S., Kaya C. A new equation for calculation of chemical hardness of groups and molecules. Mol. Phys. 2015;113(11):1311–1319. doi: 10.1080/00268976.2014.991771. [DOI] [Google Scholar]
- 29.Khoutoul M., Djedouani A., Lamsayah M., Abrigach F., Touzani R. Liquid-liquid extraction of metal ions, DFT and TD-DFT analysis for some pyrane derivatives with high selectivity for Fe (II) and Pb (II) Sep. Sci. Technol. 2016;51(7):1112–1123. doi: 10.1080/01496395.2015.1107583. [DOI] [Google Scholar]
- 30.Kim D., Lee J.Y., Yang J.S., Kim J.W., Kim V.N., Chang H. The architecture of SARS-CoV-2 transcriptome. Cell. 2020;181(4):914–921. doi: 10.1016/j.cell.2020.04.011. May 14e10Epub 2020 Apr 23. PMID: 32330414; PMCID: PMC7179501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kollman P.A., Massova I., Reyes C., Kuhn B., Huo S., Chong L., Lee M., Lee T., Duan Y., Wang W., Donini O., Cieplak P., Srinivasan J., Case D.A., Cheatham T.E., 3rd Calculating structures and free energies of complex molecules: combining molecular mechanics and continuum models. Acc. Chem. Res. 2000;33(12):889–897. doi: 10.1021/ar000033j. DecPMID: 11123888. [DOI] [PubMed] [Google Scholar]
- 32.Kumari R., Kumar R. Open Source Drug Discovery Consortium, Lynn A. g_mmpbsa–a GROMACS tool for high-throughput MM-PBSA calculations. J. Chem. Inf. Model. 2014;54(7):1951–1962. doi: 10.1021/ci500020m. Jul 28Epub 2014 Jun 19. PMID: 24850022. [DOI] [PubMed] [Google Scholar]
- 33.Lee G.R., Shin W.H., Park H.B., Shin S., Seok C. Conformational sampling of flexible ligand-binding protein loops. Bull. Korean Chem. Soc. 2012;33(3):770–774. doi: 10.5012/bkcs.2012.33.3.770. [DOI] [Google Scholar]
- 34.Lipinski C.A., Lombardo F., Dominy B.W., Feeney P.J. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv. Drug. Deliv. Rev. 2001;46(1–3):3–26. doi: 10.1016/s0169-409x(00)00129-0. Mar 1PMID: 11259830. [DOI] [PubMed] [Google Scholar]
- 35.Liu K., Watanabe E., Kokubo H. Exploring the stability of ligand binding modes to proteins by molecular dynamics simulations. J. Comput. Aided Mol. Des. 2017;31(2):201–211. doi: 10.1007/s10822-016-0005-2. FebEpub 2017 Jan 10. PMID: 28074360. [DOI] [PubMed] [Google Scholar]
- 36.Lynch T., Price A. The effect of cytochrome P450 metabolism on drug response, interactions, and adverse effects. Am. Fam. Physician. 2007;76(3):391–396. Aug 1PMID: 17708140. [PubMed] [Google Scholar]
- 37.Sanguinetti M.C., Jiang C., Curran M.E., Keating M.T. A mechanistic link between an inherited and an acquired cardiac arrhythmia: HERG encodes the IKr potassium channel. Cell. 1995;81(2):299–307. doi: 10.1016/0092-8674(95)90340-2. Apr 21PMID: 7736582. [DOI] [PubMed] [Google Scholar]
- 38.Ki M. MERS outbreak in Korea: hospital-to-hospital transmission. Epidemiol. Health. 2015;37 doi: 10.4178/epih/e2015033. 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Martino V., Morales J., Martínez-Irujo J.J., Font M., Monge A., Coussio J. Two ellagitannins from the leaves of Terminalia triflora with inhibitory activity on HIV-1 reverse transcriptase. Phytother. Res. 2004;18(8):667–669. doi: 10.1002/ptr.1504. AugPMID: 15472920. [DOI] [PubMed] [Google Scholar]
- 40.Mazzini S., Musso L., Dallavalle S., Artali R. Putative SARS-CoV-2 Mpro inhibitors from an in-house library of natural and nature-inspired products: a virtual screening and molecular docking study. Molecules. 2020;25(16):3745. doi: 10.3390/molecules25163745. Aug 17PMID: 32824454; PMCID: PMC7463876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Menéndez C.A., Accordino S.R., Gerbino D.C., Appignanesi G.A. Hydrogen bond dynamic propensity studies for protein binding and drug design. PLoS One. 2016;11(10) doi: 10.1371/journal.pone.0165767. Oct 28PMID: 27792778; PMCID: PMC5085089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Osman E.E.A., Toogood P.L., Neamati N. COVID-19: living through another pandemic. ACS Infect. Dis. 2020;6(7):1548–1552. doi: 10.1021/acsinfecdis.0c00224. Jul 10Epub 2020 May 10. PMID: 32388976; PMCID: PMC7216760. [DOI] [PubMed] [Google Scholar]
- 44.Park S.W., Kwon M.J., Yoo J.Y., Choi H.J., Ahn Y.J. Antiviral activity and possible mode of action of ellagic acid identified in Lagerstroemia speciosa leaves toward human rhinoviruses. BMC Complement. Altern. Med. 2014;14:171. doi: 10.1186/1472-6882-14-171. May 26PMID: 24885569; PMCID: PMC4052798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pires D.E., Blundell T.L., Ascher D.B. pkCSM: predicting small-molecule pharmacokinetic and toxicity properties using graph-based signatures. J. Med. Chem. 2015;58(9) doi: 10.1021/acs.jmedchem.5b00104. May 144066-72Epub 2015 Apr 22. PMID: 25860834; PMCID: PMC4434528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Prachayasittikul V., Prachayasittikul V. P-glycoprotein transporter in drug development. EXCLI J. 2016;15:113–118. doi: 10.17179/excli2015-768. Feb 12PMID: 27047321; PMCID: PMC4817426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Simon L., Imane A., Srinivasan K.K., Pathak L., Daoud I. In silico drug-designing studies on flavanoids as anticolon cancer agents: pharmacophore mapping, molecular docking, and Monte Carlo method-based QSAR modeling. Interdiscip. Sci. 2017;9(3):445–458. doi: 10.1007/s12539-016-0169-4. SepEpub 2016 Apr 8. PMID: 27059855. [DOI] [PubMed] [Google Scholar]
- 48.Thiel V., Ivanov K.A., Putics Á., Hertzig T., Schelle B., Bayer S., Weißbrich B., Snijder E.J., Rabenau H., Doerr H.W., Gorbalenya A.E., Ziebuhr J. Mechanisms and enzymes involved in SARS coronavirus genome expression. J. Gen. Virol. 2003;84(Pt 9):2305–2315. doi: 10.1099/vir.0.19424-0. SepPMID: 12917450. [DOI] [PubMed] [Google Scholar]
- 49.Trott O., Olson A.J. AutoDock Vina: iMproving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010;31(2):455–461. doi: 10.1002/jcc.21334. Jan 30PMID: 19499576; PMCID: PMC3041641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Van Der Spoel D., Lindahl E., Hess B., Groenhof G., Mark A.E., Berendsen H.J. GROMACS: fast, flexible, and free. J. Comput. Chem. 2005;26(16):1701–1718. doi: 10.1002/jcc.20291. DecPMID: 16211538. [DOI] [PubMed] [Google Scholar]
- 51.Vankadari N. Overwhelming mutations or SNPs of SARS-CoV-2: a point of caution. Gene. 2020;752 doi: 10.1016/j.gene.2020.144792. Aug 20Epub 2020 May 20. PMID: 32445924; PMCID: PMC7239005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang D., Hu B., Hu C., Zhu F., Liu X., Zhang J., Wang B., Xiang H., Cheng Z., Xiong Y., Zhao Y., Li Y., Wang X., Peng Z. Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus-infected pneumonia in Wuhan, China. JAMA. 2020;323(11):1061–1069. doi: 10.1001/jama.2020.1585. Mar 17Erratum in: JAMA. 2021 Mar 16;325(11):1113. PMID: 32031570; PMCID: PMC7042881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yan L., Velikanov M., Flook P., Zheng W., Szalma S., Kahn S. Assessment of putative protein targets derived from the SARS genome. FEBS Lett. 2003;554(3):257–263. doi: 10.1016/s0014-5793(03)01115-3. Nov 20PMID: 14623076; PMCID: PMC7159027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yang H., Lou C., Sun L., Li J., Cai Y., Wang Z., Li W., Liu G., Tang Y. admetSAR 2.0: web-service for prediction and optimization of chemical ADMET properties. Bioinformatics. 2019;35(6):1067–1069. doi: 10.1093/bioinformatics/bty707. Mar 15PMID: 30165565. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.