Abstract
Temozolomide (TMZ), one of the few effective drugs used during adjuvant therapy, could effectively prolong the overall survival (OS) of glioma patients. In our previous study, the mRNA level of G Protein Subunit Alpha 13 (GNA13) was found to be inversely correlated with OS and was therefore identified as a potential biomarker for the prognosis of glioma. Henceforth, this study aims to identify the molecular mechanism of GNA13 in enhancing TMZ sensitization through bioinformatic analyses of GSE80729 and GSE43452 and other experiments. In glioma, overexpression of GNA13 downregulated PRKACA, which is a subunit of PKA, hence reducing phosphorylated RELA and MGMT. Since p-RELA and MGMT were proven to be closely associated with TMZ resistance, we therefore investigated whether thetwo signaling pathways, “GNA13/PRKACA/p-RELA”, and “GNA13/PRKACA/MGMT”, were involved in the molecular mechanism of GNA13 in TMZ sensitization. Our conclusion was that, GNA13 overexpression in glioma cells were more sensitive in TMZ treatment.
Keywords: GNA13, glioma, glioblastoma, temozolomide (TMZ), PRKACA, MGMT, RELA
Introduction
Glioma is one of the most malignant tumors of the central nervous system [1]. Surgical operation with Stupp protocol has proven to be the most effective strategy in prolonging OS of glioma patients.
To date, TMZ is still the most widely used chemotherapeutic agent in the treatment of glioblastoma multiforme (GBM) and astrocytoma. Despite its side effects of some of the gene mutation and epigenetic alteration, such as decoupling of DNA, that may cause damage to response signaling, TMZ is currently the best option of drug available for glioma treatment. Therefore, the acquisition of TMZ resistance would greatly affect the effectiveness of glioma treatment.
Guanine nucleotide-binding proteins, so-called G proteins, are a protein family functioning as molecular switches, which can transmit signals from various extracellular stimuli into the interior part of cells. Gα13, a subunit of G protein encoded by GNA13, is a member of the Gα12 family. It is regarded as the potential gene that is directly associated with cancer, mainly via controlling the cell cytoskeleton remodeling, migration and differentiation. Besides, GNA13 mutation can lead to cell dysfunction such as increased necrosis and growth inhibition in lymphoma [2] and promotion of G1/S cell cycle transition in gastric cancer [3]. Besides, several studies have suggested the possible correlation between GNA13 and chemotherapy. For example, the induced drug resistance in head and neck squamous cells where GNA13 is overexpressed [4]. In the retrospection of some Gα12 proteins, it was shown that they were associated with some key proteins in drug resistance of different tumors, like JNK, NF-κB, YAP and so on [5-7]. Nonetheless, the relationship between Gα13 and these proteins remains unclear despite being in the same subfamily.
The complicated pathways of resistance to alkylating agent include mismatch repair (MMR), base excision repair (BER), nucleotide excision repair (NER), translesion DNA synthesis (TLS) and homologous recombination (HR) [8]. Besides, O6-alkylguanine DNA alkyltransferase (AGT or MGMT) also plays a specific role in resisting alkylating mutagens, especially TMZ [9]. This protein removes the alkyl group from lesions through a stoichiometric reaction, thus denying it as a real enzyme [10]. MGMT is influenced by many factors in vivo, such as transcription factors, epigenetic alteration like methylation of the MGMT promoter region, phosphorylation, regulation by histone acetylation and microRNAs expression [11]. Other than that, MGMT is also affected by the PKA signaling pathway [12], MAPK/JNK signaling pathway [13] and NFκB [14]. Several studies pointed out that some of the factors influencing MGMT as mentioned above were partially associated with GNA13 in specific tumors for example, GNA13/NFκB in prostate cancer [15] and colorectal cancer [16]. Therefore, it is crucial to explore the possibility of such association between glioma cells and drug resistance. The detection of MGMT protein expression and methylation status of MGMT promoter is the most effective way in evaluating the efficiency of TMZ treatment.
Our previous bioinformatic analysis suggested the correlation between highly expressed GNA13 in glioma and good OS, making GNA13 a potential unfavorable prognostic biomarker [17]. However, the mechanism is yet to be identified. Herein, this study aims to elucidate the role of GNA13 in prolonging OS via TMZ sensitization.
The results of this study shows that the glioma cell line with GNA13 overexpression yielded lower half maximal inhibitory concentration (IC50) when compared to the vector group. There is no previous report that has suggested the role of G proteins in reducing the resistance of such a widely-used drug before. Besides, the previously described result may also wrongfully identify GNA13 as a marker of good prognosis. Thus, the strategy of bioinformatic analyses and experiments were deployed in this attempt to clarify the relationship between GNA13 and the complicated pathway of TMZ treatment. Functioning GNA13 can be found in the cell membrane. Due to its role as a switch to many cascade reactions, it is essential to find out the downstream effectors of GNA13 that are associated with the response of TMZ.
Materials and methods
Cell lines, reagents and transfection
The U251MG and U138MG cell lines were purchased from Jennio Biotech Co., Ltd (Guangzhou, China) and cultured in DMEM-basic (GIBCO,USA), supplemented with 10% Fetal Bovine Serum (FBS, GIBCO, USA) and 1% Penicillin/Streptomycin (PS). The U251MG and U138MG stable cell lines expressing pCMV3-Vector and pCMV3-GNA13-Flag respectively were cultured in DMEM-basic (GIBCO) supplemented with 10% FBS, 1% PS and 50 μg/ml Hygromycin (Dalian meilune biotechnology Co, LTD, China). The Human GNA13 ORF mammalian expression plasmid, C-FLAG tag (HG12512-CF) and pCMV3-C-FLAG negative Control Vector (CV012) were purchased from Sino Biological Inc. (Beijing, China). The Lipofectamine™ 3000 Transfection Reagent was purchased from Thermo Fisher Scientific (USA). For transfection, U251MG and U138MG cells were seeded in 35 mm dish and were plated at 8×105 cells per plate 24 hours prior to transfection. The transfection-mix was prepared using 2 μg of the cDNA plasmid. 6 hours after transfection, the media was replaced with culture media without antibiotic. After the first 48 hours upon transfection, media was replaced every 24 hours with culture media containing 100 μg/ml Hygromycin for 7 days and then maintained with culture media containing 50 μg/ml Hygromycin.
Extraction and reverse transcription of total RNA in U138MG and U251MG cell lines
Total RNA from the cells was extracted using Trizol according to the manufacturer’s protocol. The RNA was eluted in 20 μl of RNase-free water, and 1 μg of the extracted RNA was used as a template for complementary DNA synthesis. The gDNA erasing was done by using PrimeScript™ RT reagent Kit with gDNA Eraser (Perfect Real time) (RR047A, Takara Bio, Shiga, Japan). A 10 μl gDNA erasing mixture was made by adding 1 μl of gDNA Eraser, 2 μl of 5 times gDNA Eraser Buffer and 1 μg of the extracted RNA, then topped up to 10 ul with RNase-free water before performing at 4°C for 2 minutes. A 20 μl reverse transcription mixture was made by including 10 μl of gDNA erasing mixture as described previously, 1 μl of PrimeScript RT Enzyme Mix I, 1 μl of RT Enzyme Mix, 4 μl of 5 times PrimeScript Buffer 2 and 4 μl RNase-free water before performing at 37°C for 15 minutes, followed by incubation at 85°C for 5 seconds.
Total RNA of tumor samples from glioma patients
41 glioma samples and 2 normal brain tissues were obtained from glioma patients in Shantou Central Hospital from 2010 to 2018. The total RNA of these samples were extracted by using the PureLink RNA Mini Kit (12183018A, Thermo Fisher Scientific, USA) according to the quick reference provided by the manufacturer. Reverse transcription was done as the cell lines described in section 2.2.
Real-time quantitative PCR analysis
The cDNA products were analyzed by real-time PCR using LightCycler® 480 (Roche Diagnostics, Basel, Switzerland). Real-time PCR was performed in a 20 μl mixture that includes 0.4 μl of the diluted cDNA (1 μg) template, 10 μl of 2 times RealStar Green Fast mixture (Genstar, China), 0.8 μl of gene-specific primers (0.4 ul forward primer and 0.4 ul reverse primer) and 8.8 μl of RNase-free water. Genstar cycling conditions are as follows: an initial denaturation step at 95°C for 2 minutes, followed by 40 cycles including denaturation at 95°C for 15 seconds, annealing and extension at 60°C for 30 seconds; a melt curve analysis was performed from 60 to 95°C at 0.11°C increment per second. The reference gene β-actin was used as an internal control. Primers used for detecting the mRNAs of β-actin (forward: 5’-GCCGACAGGATGCAGAAGG-3’; reverse: 5’-GATGGAGGGGCCGGACTC-3’), GNA13 (forward: 5’-ACCATCTACAGCAACGTGATC-3’; reverse: 5’-TTGGTTTGAGTTGTCTCCCC-3’), PRKACA (forward: 5’-CAAGGACAACTCAAACTTATACATGG-3’; reverse: 5’-CAGATACTCAAAGGTCAGGACG-3’) and MGMT (forward: 5’-GCTGAATGCCTATTTCCACC-3’; reverse: 5’-CACTTCTCCGAATTTCACAACC-3’) were purchased from BGI (Hubei, China). The relative quantification of GNA13 expression was analyzed using the comparative 2-ΔΔCp method and data was visualized through the GraphPad Prism 7 software.
Immunoblot analysis
U251 and U138 cells were seeded in 35 mm dishes and grown in culture media containing 10% FBS for 24 hours. The cells were then washed with phosphate-buffered saline (PBS) and lysed in protein extraction buffer (50 M HEPES pH 7.5, 1 mM EDTA, 3 mM dithiothreitol, 10 mM MgSO4, 1% polyoxyethylene-10-lauryl ether) with the protease inhibitor cocktails (Thermo Fisher Scientific, USA). BCA Protein Assay Kit (Thermo Fisher Scientific, USA) was used to determine the protein concentrations of the lysates. 30 μg of the lysates were separated on a 10% polyacrylamide gel with SDS-PAGE before being transferred to a PVDF membrane. The membrane was then incubated with antibody against Flag-tag (MA1-91878, Thermo Fisher Scientific, USA), antibody against MGMT (#86039, CST, USA), antibody against NF-κB (#4767, CST, USA) and antibody against GAPDH (MA5-15738, Thermo Fisher Scientific, USA) overnight at 4°C, and subsequently with secondary antibody, goat-anti-mouse (31430, Thermo Fisher Scientific, USA). Finally, the membrane is visualized using chemiluminescence (ECL, Abcam, USA) in the ChemiDoc™ Imaging System.
Cell viability assay
To determine the cell viability after TMZ treatment, U251 and U138 cells were seeded in 96-well plates at densities of 1×104 per well, 24 hours before TMZ treatment. TMZ (S1237, Selleck, China) was added at a serial dilution of 10, 50, 100, 200 and 500 μM/mL. CCK-8 assays (AR1160-500, Boster, China) were performed 24 hours after treatment. The TMZ mixture was replaced with 100 μL DMEM-basic medium and 10 μL CCK-8 in each well for 1 hour at 37°C. As for modified CCK-8 assays as control, three blank wells were not seeded but only filled with 100 μL medium and 10 μL CCK-8. The absorbance at 450 nm was assessed by using a Multiskan FC microplate reader (Thermo Fisher Scientific, USA). All experiments were done in triplicates.
Apoptosis detection
The U251 and U138 cells were seeded in 35 mm dishes and 24-well plates, grown in culture media containing 10% FBS for 24 hours. All cells were then treated with 100 μM/mL TMZ for 24 hours. After that, cells were collected from the 35 mm dishes and stained with Annexin V-FITC/7-AAD Apoptosis Detection Kit (APK10448-F, Sino Biological, China) according to the manufacturer’s guideline. Flow cytometric was subsequently done using the BD Accuri™ C6 cytometric machine (Becton, Dickinson and Company, New York, USA). Cells from the 24-well plates were conjugated to the antibody against Flag-tag (MA1-91878, Thermo Fisher Scientific, USA) and the Alexa Fluor 488 donkey anti-rabbit IgG as secondary antibody (R37118, Thermo Fisher Scientific, USA). The nucleus was stained by DAPI.
Microarray data and differentially expressed genes (DEGs)
Gene expression profiles of GSE80729 and GSE43452 were downloaded from the Gene Expression Omnibus (GEO, https://www.ncbi.nlm.nih.gov/geo/). GSE80729 is a dataset for genome-wide analysis of BCL3 mediated chemotherapy resistance that contains 3 GSMs pairs of U87 siCtrl with TMZ treatment and U87 siCtrl with DMSO control, based on the GPL10558 platform (Illumina HumanHT-12 V4.0 expression beadchip); whereas GSE43452 is a dataset for gene profile of glioblastoma cells treated with y15 and TMZ that contains 2 GSMs pairs of U87 with 20 μM TMZ treatment and U87 untreated control, based on the same platform as GSE80729. Since these two GSE datasets originate from the same platform, quantile normalization could be accomplished. The analysis of DEGs was carried out with GEO2R, an online analysis tool designed for the GEO database based on R language. We applied GEO2R to identify the sample with expression patterns that are significantly different between the TMZ treated group and the control group. The differentially expressed genes (DEGs) were defined using a cut-off standard as Log2-fold threshold or an adjusted P value of <0.05. Also, R studio 1.1.4 was used to show the data in a volcano plot.
Integration of protein-protein interaction (PPI) network and module analysis
PPI information was downloaded from the Human Protein Reference Database and the Biological General Repository for Interaction Datasets (BioGRID) (released on 07/24/2017). Cytoscape software was then used to visualize the PPI network. The STITCH database is an online tool for showing and predicting protein-protein and chemical-protein interactions, including both direct association and indirect relationship. STITCH version 5.0 covers 9,643,763 proteins from 2031 organisms. It was used to assess the interactional relationships among the GNA13, DEGs and TMZ. The cut-off standard was defined at an interaction score of 0.4. GeNets is a unified web platform developed by Harvard and MIT that is specifically designed for network-based genomic analyses. It involves several nets of machines learning to analyze protein-protein interactions. Due to its high AUC of Inweb3 algorithm compared with other tools, GeNets was used for speculating the most probable connection between GNA13 and its PPI including functional association.
Functional and pathway enrichment analysis
Gene ontology (GO) annotation was integrated into the total DEGs PPI sub-networks for enriched GO “molecular function” (MF), “biological pathways” (BP), and “cellular component” (CC) terms of proteins using DAVID online tools (https://david.ncifcrf.gov/). DAVID provides annotated information of systematic and comprehensive biological function for high throughput gene expression. Meanwhile, the Kyoto Encyclopedia of Genes and Genomes (KEGG) is an online database for obtaining information about high-level functions and utilities of the biological system. Both GO and KEGG were used to analyze DEGs based on DAVID and the data was visualized using babble chart by R studio. A value of < 0.01 was considered statistically significant.
Verification of the correlation between GNA13 and the candidate gene
LinkedOmics (http://www.linkedomics.org/admin.php) is a public portal, containing multi-omics data from 32 TCGA cancer types [18]. The gene correlation analysis was done through Pearson’s correlation test in HiSeq RNA platform and Meth450 Methylation platform using the TCGA-GBMLGG dataset and the outcome was drawn with the online tools of LinkedOmics and R studio 3.5.1.
mRNA library construction
DNase I was used for digesting double-stranded and single-stranded DNA in total RNA. Magnetic beads were then purified to recover the reaction products. RNase H or Ribo-Zero method (human, mouse, plants) (Illumina, USA) was applied to remove the rRNA. Purified mRNA from previous steps was fragmented into small pieces. The first- and second-strand cDNA was generated in First Strand Reaction System by PCR. The reaction product was purified with magnetic beads. A-Tailing Mix and RNA Index Adapters were then added to carry out end repair. The cDNA fragments with adapters were subsequently amplified by PCR, and the products were purified by Ampure XP Beads. The quality of the library was validated on the Agilent Technologies 2100 bioanalyzer. After that, the double stranded PCR products were denatured via heating and circularized with the help of the splint oligo sequence. The single strand circle DNA (ssCir DNA) was formatted as the final library. The final library was amplified with phi29 (Thermo Fisher Scientific, USA) to create DNA nanoball (DNB) that contains more than 300 copies of one molecular. These DNBs were loaded for patterned nanoarray and 50 base-pair single-end reads were generated on the BGISEQ500 platform (BGI-Shenzhen, China). The analysis of the RNA-seq was visualized using the network platform of Dr. Tom Data Visualisation Solution on BGI (http://report.bgi.com).
Result
The expression of GNA13 in human glioma is inversely correlated with tumor grade and the sensitivity of TMZ
Pearson’s correlation analysis indicated an inverse correlation between the mRNA of tumor samples and tumor grade in TCGA datasets (Figure 1A) and GSE4412 (Figure 1B). qRT-PCR was used to detect the mRNA expression level of GNA13 in 43 samples collected from 41 patients at the Shantou Central Hospital between 2010 and 2018 (Figure 1C). Samples were divided into three categories according to tumor grades: normal, low and high. GNA13 is significantly downregulated in high-grade glioma as compared to low-grade glioma. Furthermore, the quantity of GNA13 mRNA in glioma is also lower than that in normal brain tissue. These data suggest that GNA13 may play an important role in the inhibition of glioma malignancy. This result also correlates to the data in TCGA GBMLGG datasets.
Figure 1.
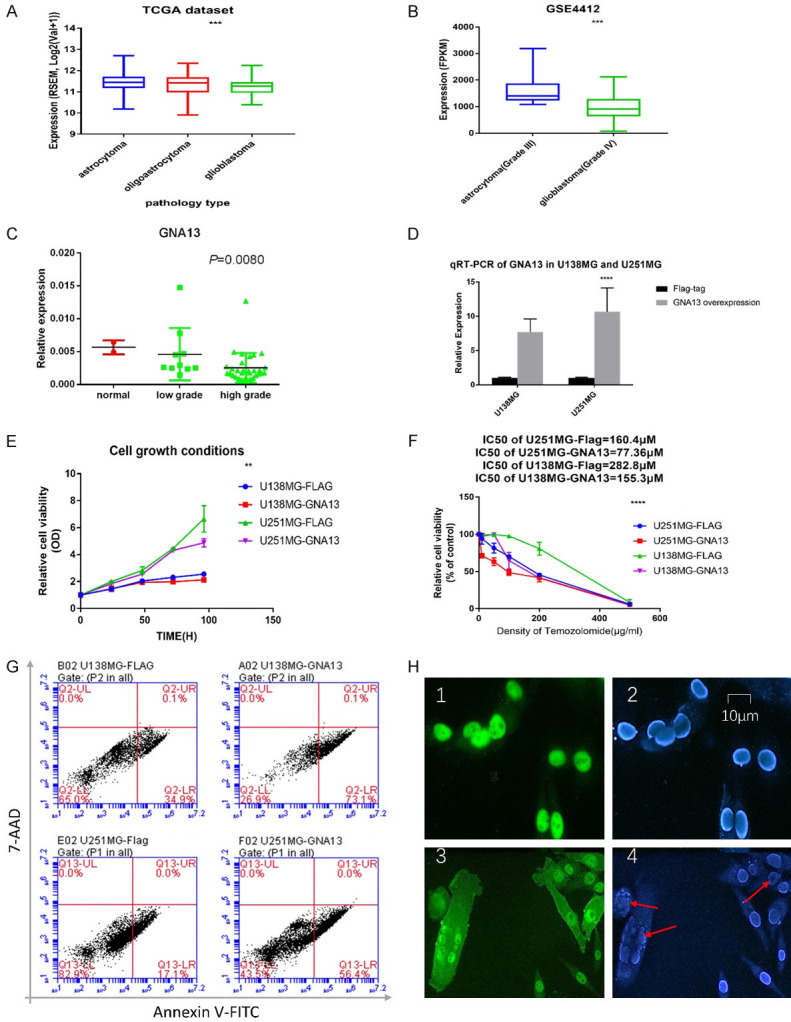
Basic bioinformation of GNA13 in datasets and glioma cell lines. A. The mRNA level of GNA13 is inversely correlated with tumor grade in TCGA datasets and. B. The mRNA level of GNA13 is inversely correlated with tumor grade in GSE4412. C. GNA13 is inversely correlated to glioma grade at mRNA level, as proven by qRT-PCR. D. The GNA13-overexpressed cell lines were confirmed through qRT-PCR. E. Cell growth inhibition is observed in GNA13-overexpressed cell lines when compared with the cell growth of vector cell lines, both U138MG and U251MG were significantly inhibited while GNA13 was overexpressed (P<0.01). F. The relative cell viability is significantly decreased in GNA13-overexpressed glioma cells after TMZ treatment (P<0.01). G. 24 hours after TMZ treatment, the proportion of cells at end stage apoptosis or dead was seen to be significantly increased in the GNA13-overexpressed group (P<0.01). X axis represented the conditions of Annexin V-FITC incubated shown in 575 nm light while Y axis represent the 7-AAD incubated shown in 488 nm light. H. 24 hours after TMZ treatment, the proportion of karyorrhexis occurrence was significantly increased in the GNA13-overexpressed group (P<0.01). Green represented the GNA13 expressed conditions, blue means the nucleus staining by DAPI and red arrow was used for emphasizing the karyorrhexis.
To investigate the possible role of GNA13 in TMZ sensitization, GNA13 was overexpressed in U251MG and U138MG cells, which was subsequently confirmed through immunoblotting and qRT-PCR (Figures 1D and 5D). The cell growth was inhibited in GNA13 overexpressed U138MG and U251mg cells (Figure 1E). The CCK-8 assay was used to verify TMZ cytotoxicity. Notably, the IC50 of both GNA13-overexpressed U251MG and U138MG cells are approximately 50% less than their respective vector controls (Figure 1F). Besides, the proportions of GNA13 overexpressed cell lines induced apoptosis or dead by TMZ were higher than the control group (Figure 1G, 1H).
Figure 5.
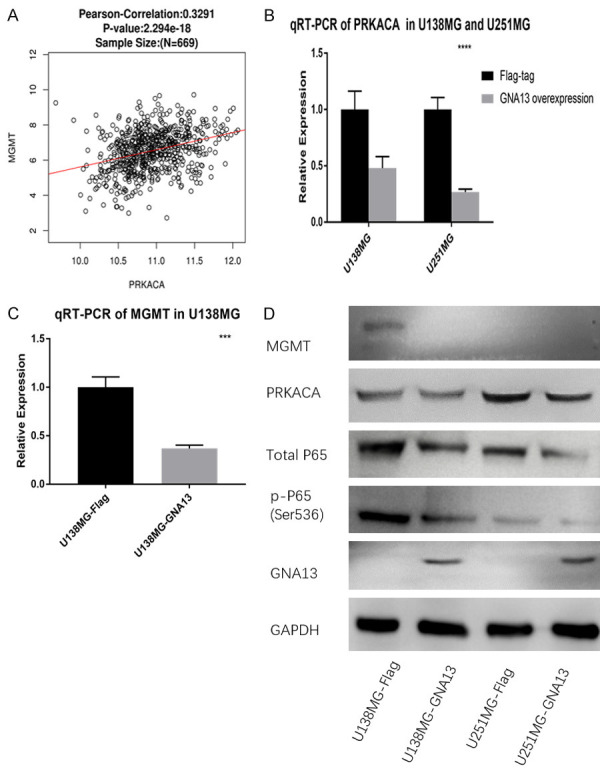
Cell lines and datasets evidence of GNA13 and TMZ sensitizations. A. Pearson’s correlation analysis shows that PRKACA is proportional to the expression of MGMT. B. qRT-PCR suggests that mRNA of PRKACA is downregulated in both GNA13-overexpressed cell lines of U251MG and U138MG. C. mRNA of MGMT decreases in GNA13-overexpressed U138MG cells. D. Result of immunoblot suggests that MGMT, total P65, p-P65 (Ser536) and PRKACA are significantly decreased in GNA13-overexpressed cells.
The 10 first neighbors of GNA13 are associated with the DEGs from the merged GEO dataset
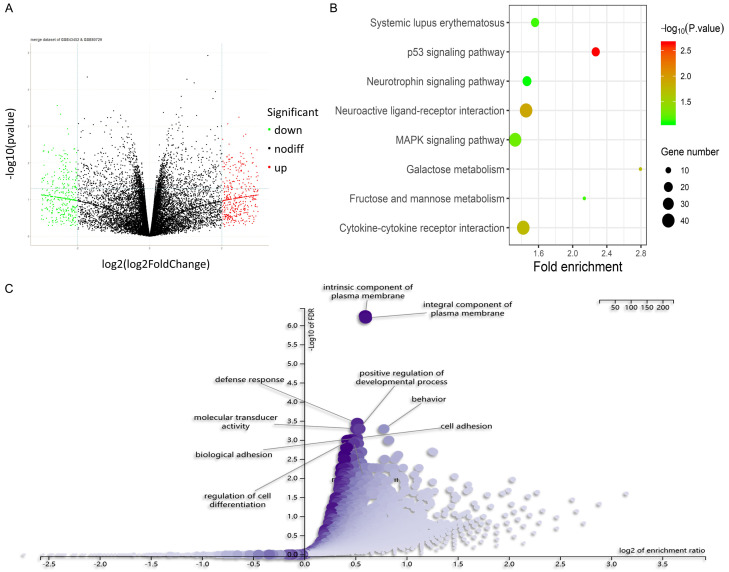
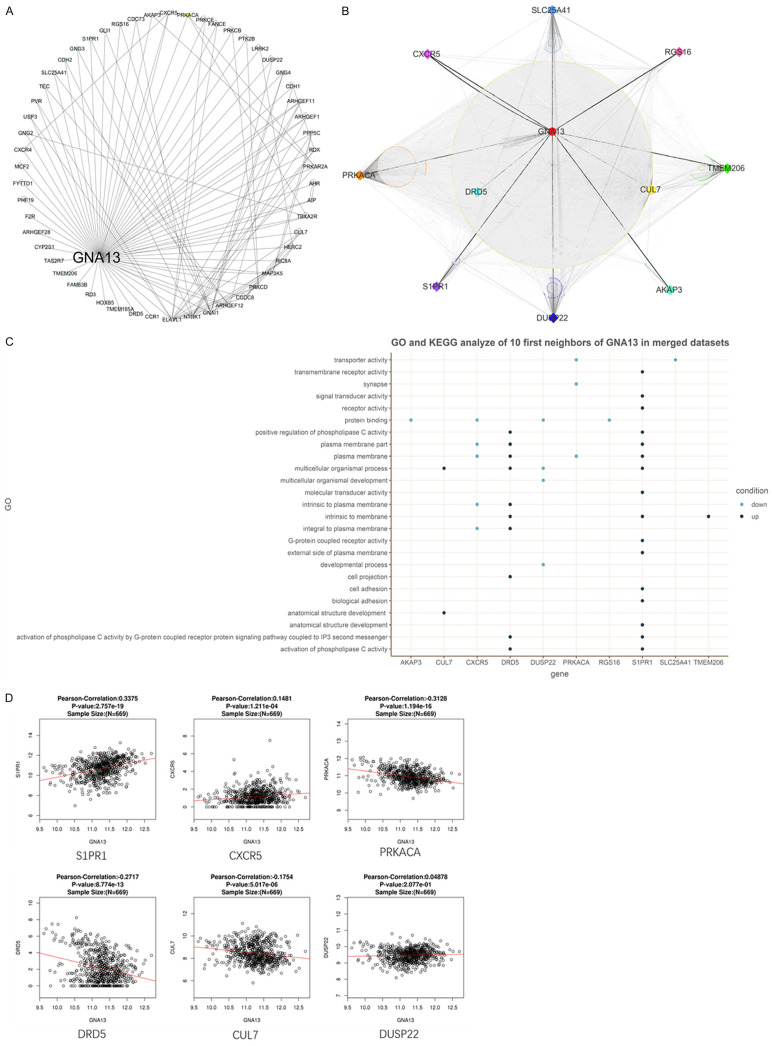
A total of 2121 DEGs were obtained from the merged microarray dataset of GSE80729 and GSE43452, in which 1038 were upregulated and 1083 were downregulated (Figure 2A-C). 56 nodes of GNA13 within 132 edges had been discovered via PPI analysis in human species (Figure 3A). The intersection of the said 56 nodes and the 2121 DEGs is shown as a group of 10 genes that includes CXCR5, DUSP22, CUL7, RGS16, AKAP3, TMEM206, S1PR1, SLC25A41, PRKACA and DRD5. These 11 genes (including GNA13) built a PPI network of 1107 nodes that have interacted with 19211 edges (Figure 3B). Based on the PPI information of humans, PRKACA comes into direct contact with the most neighbors (Figure 3C), indicating its possibly crucial role in reducing TMZ resistance in GNA13-overexpressed glioma. Moreover, PPI of these 10 genes indicated that there is no direct interaction among them. The GO and KEGG analyses aimed at identifying upregulated and downregulated genes were then done separately through DAVID (Figure 3C). According to the merged dataset, AKAP3, CXCR5, DUPS22, PRKACA, RGS16 and SCL25A41 were noticed to have been downregulated, most of which are cellular components and are involved in certain biological processes. On the other hand, CUL7, DRD5, S1PR1 and TMEM206 were noted to have been upregulated, some of which are cellular components and play important roles in both molecular functions and biological processes (Figure 3C). GO analysis suggested that the DEGs were enriched in signal transduction activity, plasma membrane and multicellular organismal process. Meanwhile, 8 of the first neighbors are either plasma membrane or part of the plasma membrane, which suggests that GNA13 may sensitize the cell response to TMZ by regulating the membrane structure (Figure 3C). KEGG analysis of the merged dataset shows that the upregulated genes are associated with the p53 signaling pathway, a finding that parallels the outcome of a previous study [19]. Focus is then set on CUL7, CXCR5, DRD5, PRKACA, CUL7 and DUSP22, all of which corresponded to the GO analysis of the merged datasets (Figure 3C).
Figure 2.
Preliminary bioinformatic analysis about TMZ resistance based on GSE data. A. A total of 2121 DEGs that were identified in the merged datasets and the logical relations of them in GSE80729 and GSE43452; 1695 of those were from GSE80729 while the other 426 were from GSE43452; 1038 were upregulated and 1083 were downregulated. B. The KEGG pathway analyses of DEGs in merged datasets indicated that the key genes responding to TMZ treatment are possibly associated with signal transduction activity, p53 signaling pathways and multicellular organismal process. C. The GO analyses of DEGs in merged datasets indicated that the key genes responding to TMZ treatment are possibly associated with plasma membrane.
Figure 3.
Basic informatic analysis of GNA13. A. A total of 56 first neighbors of GNA13 were identified in the PPI analysis of HRPD and BioGRID databases. B. The PPI network of 11 selected genes in HRPD and BioGRID databases. The black lines represent direct interactions among the 10 genes. The dark gray lines represent the interactions between the 10 genes and their first neighbors. The light gray lines represent the interactions among these first neighbors. PRKACA interacted with the most neighbors (directly and indirectly) among the 10 genes. C. The GO analysis of the merged datasets indicated that most of these 10 genes are plasma membrane (part). This analysis suggests that CUL7, CXCR5, DRD5, PRKACA, CUL7 and DUSP22 may be the downstream genes of GNA13 in regulating TMZ sensitization. D. Pearson’s correlation test pointed out that S1PR1 and CXCR5 are positively correlated to the expressed alteration of GNA13 while PRKACA, DRD5 and CUL7 are negatively correlated. DUSP22 shows no correlation with GNA13 in the GBMLGG dataset of TCGA.
Verification of the correlation between GNA13 and the 6 selected genes in TCGA GBMLGG datasets
Pearson’s correlation test was applied in the HiSeq RNA platform of the TCGA GBMLGG dataset. Significant value was set at P<0.05 to determine the correlation between GNA13 and the 6 selected genes. CXCR5 and S1PR1 were proven to correlate positively with GNA13; while CUL7, DRD5 and PRKACA exhibited negative correlation. No correlation was evident between GNA13 and DUSP22. PRKACA was downregulated both in the drug treatment GSE datasets and the GNA13 correlated conditions in glioma cells. These data seem to point out the possibility of GNA13 inducing TMZ sensitization through PRKACA (Figure 3D).
Verification of the correlation of GNA13 and the classical TMZ resistance pathway in TCGA GBMLGG datasets
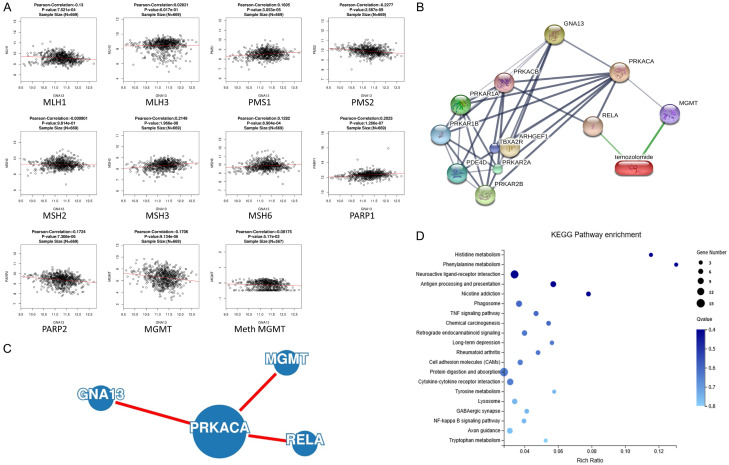
It was demonstrated that GNA13 might be positively correlated to MSH3, MSH6, PMS1 in MMR and PARP1 in BER; while exhibiting negative correlation with MLH1 and PMS2 in MMR, PARP2 in BER and MGMT. There is no correlation between GNA13 and both MLH3 and MSH2. GNA13 and methylated MGMT shows no correlation (P<0.05 as the significant value) in another Pearson’s correlation test based on the Meth450 platform in the TCGA GBMLGG dataset. Based on these data, it is deduced that GNA13 might induce the sensitization of TMZ by downregulating MGMT instead of MMR or BER. There seems to be no connection between GNA13 and the methylation of MGMT (Figure 4A).
Figure 4.
Bioinformatic analysis among GNA13 and TMZ resistance factors. A. Pearson’s correlation analysis shows that GNA13 is proportional to the expression of MGMT. B. The interactional network among GNA13, PRKACA and TMZ suggests that PRKACA can interact with MGMT and RELA, therefore it is responsive to TMZ stimulation. C. The PPI network among GNA13, PRKACA, RELA and MGMT as proven by one of the most effective PPI network tool, GeNets. D. RNA-seq indicated that 4 DEGs were enriched in NF-κB signaling pathway, which is one of the top 20 KEGG pathway enrichment analyses.
Verification of GNA13-associated pathway in TMZ sensitization through bioinformatic analysis
Interactive analysis was carried out through GeNets and STITCH. GNA13 is able to associate with PRKACA, which may in turn interact with MGMT. The GNA13-PRKACA-MGMT axon was shown in both datasets. Moreover, STITCH data suggests that there is no direct interaction between TMZ and neither GNA13 nor PRKACA. Besides, STITCH also indicated that PRKACA would interact with RELA simultaneously for TMZ sensitization (Figure 4B, 4C). Based on the RNA-seq analysis that includes both GNA13-overexpressed and vector cell lines of U251MG and U138MG, NF-κB signaling pathway is also identified as one of the key pathways.
Verification of the PRKACA, MGMT, p-RELA in glioma cell lines
Pearson’s correlation test between PRKACA and MGMT based on Hiseq mRNA platform of TCGA GBMLGG datasets also shows a positive correlation between GNA13 and PRKACA (Figure 5A). GNA13 overexpression leads to significant downregulation (approximately 50%) of PRKACA mRNA in both U138MG and U251MG cells, which was proven by qRT-PCR (Figure 5B, 5C). Although MGMT is not expressed in U251MG, qRT-PCR and immunoblotting of MGMT in U138MG cells demonstrated that MGMT will be downregulated if GNA13 is overexpressed. As a subunit of PKA, it can therefore be hypothesized that PRKACA would influence the phosphorylation status of RELA in glioma cell lines independent of MGMT expression. The result of RELA and pho-RELA (Ser536) immunoblotting shows that the expression of both genes were downregulated (Figure 5D).
Discussion
TMZ resistance is a critical issue that influences the OS and the quality of life since TMZ is one of the few effective drugs in glioma treatment. To date, various mechanisms including protein, non-coding RNAs and epigenetics have been suggested to be involved in TMZ resistance.
In this bioinformatic study, GNA13 was proven to be downregulated and indirectly inhibits MGMT activity via MGMT phosphorylation through PKA activation. Furthermore, GNA13 is able to downregulate the phosphorylation on Ser536 of RELA. Besides, GNA13 seems to be associated with the pathological grades of glioma as well, both in the TCGA GBMLGG datasets, GSE4412 and tumor samples. GNA13 is also highly associated with IDH1_p.R132G (P<0.01), which appears in 4% of IDH1-mutant glioma, indicating the potential role of GNA13 alteration in the early events of glioma.
PRKACA gene encodes the catalytic subunit α of protein kinase A, commonly abbreviated as PKA Cα (Protein kinase A catalytic subunit). Protein kinase A is an essential enzyme responsible for phosphorylating many intracellular molecules to regulate various cellular activities. In 2001, Niu et al. first observed that the interaction between Gα13 and AKAP110 can lead to the cyclic AMP-independent activation of protein kinase A [20]. They have proven that Gα13 could regulate many phosphorylation reactions in cells. Prior to this study, Srivenugopal et al. suggested that the activity of MGMT can be inhibited upon its phosphorylation [12], and PKA is one of the key proteins in this process. Therefore, it was speculated that GNA13 could downregulate the activity of MGMT by MGMT phosphorylation through PKA. By that, the interacting axon of ‘GNA13/PRKACA/p-MGMT’ is partly confirmed. Based on the outcome of bioinformatic analyses and other experiments in this study, it was observed that RELA phosphorylation is downregulated after GNA13 overexpression, thus validating the signaling pathway of ‘GNA13/PRKACA/p-RELA’.
However, there are limitations in this study. The experiment outcomes suggested that PRKACA is downregulated in GNA13-overexpressed glioma cell lines, which parallels the results of the informatic analysis in TCGA GBMLGG datasets and the merged dataset from GSE which has shown negative correlation. However, Manganello et al. reported that GNA13 can be phosphorylated at T203 [21]. This phenomenon indicated the possibility of a loop within the GNA13 and PKA system. Besides, there may be some other mechanisms that require activation by GNA13 and may subsequently lead to the reduction of RELA and JNK phosphorylation. Due to its potential negative correlation with the pathological grade of glioma, it is logical to speculate that GNA13 may be a molecular marker in predicting the low grade of glioma, and thus mistakenly being linked to the better prognosis sets after TMZ treatment in the same pathological grade.
Although the interacting axon has been elucidated, the relationship between GNA13 and the sensitization of TMZ may be far more complex than what is understood to date. GNA13 is known to activate NFκB and JNK/MAPK-AP-1 signaling pathways and therefore influencing the susceptibility of HNSCC cells to cisplatin treatment [4]. JNK was also reported to enhance TMZ resistance by regulating the MGMT expression [13]. These phenomena are different in glioma. The JNK expressed in GNA13-overexpressed U251 cells is opposite of that in GNA13-overexpressed U138 cells. Combined with the fact that mRNA level of PRKACA is downregulated in GNA13-overexpressed cell lines as proven, it remains unclear whether GNA13 can regulate MGMT phosphorylation through the JNK pathway in specific glioma cell lines.
Finally, TMZ sensitization cannot be determined by a single pathway. Proteins like STAT3 [22] and APE1 [23] were previously proven to play significant roles in TMZ resistance as well. They might also be involved in the GNA13-derived TMZ response. Previous studies have implied the possible relationship between GNA13 and STAT3 [24], but the mechanism is yet to be elucidated.
Acknowledgements
This study was funded by Guangdong Provincial Special Foundation for Innovation Strategy of Science and Technology (No. 180914164960240, No. 200114135896989). Shantou Science and Technology Bureau (180817094013077). This study was also funded by Natural Science Foundation of Guangdong Province (No. 2018A030313597).
Disclosure of conflict of interest
None.
References
- 1.Ahmed R, Oborski MJ, Hwang M, Lieberman FS, Mountz JM. Malignant gliomas: current perspectives in diagnosis, treatment, and early response assessment using advanced quantitative imaging methods. Cancer Manag Res. 2014;6:149–70. doi: 10.2147/CMAR.S54726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.O’Hayre M, Inoue A, Kufareva I, Wang Z, Mikelis CM, Drummond RA, Avino S, Finkel K, Kalim KW, DiPasquale G, Guo F, Aoki J, Zheng Y, Lionakis MS, Molinolo AA, Gutkind JS. Inactivating mutations in GNA13 and RHOA in Burkitt’s lymphoma and diffuse large B-cell lymphoma: a tumor suppressor function for the Galpha13/RhoA axis in B cells. Oncogene. 2016;35:3771–80. doi: 10.1038/onc.2015.442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang JX, Yun M, Xu Y, Chen JW, Weng HW, Zheng ZS, Chen C, Xie D, Ye S. GNA13 as a prognostic factor and mediator of gastric cancer progression. Oncotarget. 2016;7:4414–27. doi: 10.18632/oncotarget.6780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rasheed SAK, Leong HS, Lakshmanan M, Raju A, Dadlani D, Chong FT, Shannon NB, Rajarethinam R, Skanthakumar T, Tan EY, Hwang JSG, Lim KH, Tan DS, Ceppi P, Wang M, Tergaonkar V, Casey PJ, Iyer NG. GNA13 expression promotes drug resistance and tumor-initiating phenotypes in squamous cell cancers. Oncogene. 2018;37:1340–53. doi: 10.1038/s41388-017-0038-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kelley GG, Reks SE, Smrcka AV. Hormonal regulation of phospholipase Cepsilon through distinct and overlapping pathways involving G12 and Ras family G-proteins. Biochem J. 2004;378:129–39. doi: 10.1042/BJ20031370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Juneja J, Casey PJ. Role of G12 proteins in oncogenesis and metastasis. Br J Pharmacol. 2009;158:32–40. doi: 10.1111/j.1476-5381.2009.00180.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kelly P, Casey PJ, Meigs TE. Biologic functions of the G12 subfamily of heterotrimeric g proteins: growth, migration, and metastasis. Biochemistry. 2007;46:6677–87. doi: 10.1021/bi700235f. [DOI] [PubMed] [Google Scholar]
- 8.Lee SY. Temozolomide resistance in glioblastoma multiforme. Genes Dis. 2016;3:198–210. doi: 10.1016/j.gendis.2016.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Arora A, Somasundaram K. Glioblastoma vs temozolomide: can the red queen race be won? Cancer Biol Ther. 2019;20:1083–90. doi: 10.1080/15384047.2019.1599662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kaina B, Christmann M, Naumann S, Roos WP. MGMT: key node in the battle against genotoxicity, carcinogenicity and apoptosis induced by alkylating agents. DNA Repair (Amst) 2007;6:1079–99. doi: 10.1016/j.dnarep.2007.03.008. [DOI] [PubMed] [Google Scholar]
- 11.Cabrini G, Fabbri E, Lo Nigro C, Dechecchi MC, Gambari R. Regulation of expression of O6-methylguanine-DNA methyltransferase and the treatment of glioblastoma (review) Int J Oncol. 2015;47:417–28. doi: 10.3892/ijo.2015.3026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Srivenugopal KS, Mullapudi SR, Shou J, Hazra TK, Ali-Osman F. Protein phosphorylation is a regulatory mechanism for O6-alkylguanine-DNA alkyltransferase in human brain tumor cells. Cancer Res. 2000;60:282–7. [PubMed] [Google Scholar]
- 13.Okada M, Sato A, Shibuya K, Watanabe E, Seino S, Suzuki S, Seino M, Narita Y, Shibui S, Kayama T, Kitanaka C. JNK contributes to temozolomide resistance of stem-like glioblastoma cells via regulation of MGMT expression. Int J Oncol. 2014;44:591–9. doi: 10.3892/ijo.2013.2209. [DOI] [PubMed] [Google Scholar]
- 14.Caporali S, Levati L, Graziani G, Muzi A, Atzori MG, Bonmassar E, Palmieri G, Ascierto PA, D’Atri S. NF-kappaB is activated in response to temozolomide in an AKT-dependent manner and confers protection against the growth suppressive effect of the drug. J Transl Med. 2012;10:252. doi: 10.1186/1479-5876-10-252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lim WK, Chai X, Ghosh S, Ray D, Wang M, Rasheed SAK, Casey PJ. Galpha-13 induces CXC motif chemokine ligand 5 expression in prostate cancer cells by transactivating NF-kappaB. J Biol Chem. 2019;294:18192–206. doi: 10.1074/jbc.RA119.010018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang Z, Tan X, Luo J, Cui B, Lei S, Si Z, Shen L, Yao H. GNA13 promotes tumor growth and angiogenesis by upregulating CXC chemokines via the NF-kappaB signaling pathway in colorectal cancer cells. Cancer Med. 2018;7:5611–20. doi: 10.1002/cam4.1783. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 17.Liu M, Xu Z, Du Z, Wu B, Jin T, Xu K, Xu L, Li E, Xu H. The identification of key genes and pathways in glioma by bioinformatics analysis. J Immunol Res. 2017;2017:1278081. doi: 10.1155/2017/1278081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vasaikar SV, Straub P, Wang J, Zhang B. LinkedOmics: analyzing multi-omics data within and across 32 cancer types. Nucleic Acids Res. 2018;46:D956–D63. doi: 10.1093/nar/gkx1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chapuy B, Cheng H, Watahiki A, Ducar MD, Tan Y, Chen L, Roemer MG, Ouyang J, Christie AL, Zhang L, Gusenleitner D, Abo RP, Farinha P, von Bonin F, Thorner AR, Sun HH, Gascoyne RD, Pinkus GS, van Hummelen P, Wulf GG, Aster JC, Weinstock DM, Monti S, Rodig SJ, Wang Y, Shipp MA. Diffuse large B-cell lymphoma patient-derived xenograft models capture the molecular and biological heterogeneity of the disease. Blood. 2016;127:2203–13. doi: 10.1182/blood-2015-09-672352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Niu J, Vaiskunaite R, Suzuki N, Kozasa T, Carr DW, Dulin N, Voyno-Yasenetskaya TA. Interaction of heterotrimeric G13 protein with an A-kinase-anchoring protein 110 (AKAP110) mediates cAMP-independent PKA activation. Curr Biol. 2001;11:1686–90. doi: 10.1016/s0960-9822(01)00530-9. [DOI] [PubMed] [Google Scholar]
- 21.Manganello JM, Huang JS, Kozasa T, Voyno-Yasenetskaya TA, Le Breton GC. Protein kinase A-mediated phosphorylation of the Galpha13 switch I region alters the Galphabetagamma13-G protein-coupled receptor complex and inhibits Rho activation. J Biol Chem. 2003;278:124–30. doi: 10.1074/jbc.M209219200. [DOI] [PubMed] [Google Scholar]
- 22.Lee ES, Ko KK, Joe YA, Kang SG, Hong YK. Inhibition of STAT3 reverses drug resistance acquired in temozolomide-resistant human glioma cells. Oncol Lett. 2011;2:115–21. doi: 10.3892/ol.2010.210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Montaldi AP, Godoy PR, Sakamoto-Hojo ET. APE1/REF-1 down-regulation enhances the cytotoxic effects of temozolomide in a resistant glioblastoma cell line. Mutat Res Genet Toxicol Environ Mutagen. 2015;793:19–29. doi: 10.1016/j.mrgentox.2015.06.001. [DOI] [PubMed] [Google Scholar]
- 24.Tiacci E, Ladewig E, Schiavoni G, Penson A, Fortini E, Pettirossi V, Wang Y, Rosseto A, Venanzi A, Vlasevska S, Pacini R, Piattoni S, Tabarrini A, Pucciarini A, Bigerna B, Santi A, Gianni AM, Viviani S, Cabras A, Ascani S, Crescenzi B, Mecucci C, Pasqualucci L, Rabadan R, Falini B. Pervasive mutations of JAK-STAT pathway genes in classical Hodgkin lymphoma. Blood. 2018;131:2454–65. doi: 10.1182/blood-2017-11-814913. [DOI] [PMC free article] [PubMed] [Google Scholar]