Abstract
Gliomas are the most prevalent primary malignant central nervous system tumors among all tumors occurring in the brain and spinal cord. The poor outcome of glioma requires the discovery of novel biomarkers with potential therapeutic value. Somatostatin receptor subtype 2 (SSTR2) represents a diagnostic biomarker and potential therapeutic target in many cancers, such as meningioma and neuroendocrine tumors (NETs). However, the relationship of SSTR2 and glioma was unclear. Therefore, this study aimed to investigate the expression of SSTR2 and assess its prognostic and potential therapeutic value in a large cohort of patients with WHO grade I to IV glioma from a single Chinese center. Immunohistochemical analysis revealed that SSTR2 was highly expressed in 23.84% (72 of 302) of glioma (I-IV grade) samples. Among all glioma subtypes, high SSTR2 expression was detected mainly in oligodendroglioma, anaplastic oligodendroglioma, and astrocytoma, whereas SSTR2 was expressed at a low level, or not at all, in glioblastoma. Western blotting also confirmed the low expression of SSTR2 in glioblastoma cell lines. Statistical analysis showed that SSTR2 protein expression correlated significantly with WHO grade, the location of the tumor, epilepsy syndrome, mitosis (PHH3), proliferation index (Ki-67), IDH and 1p/19q-codeleted status. Kaplan-Meier analysis indicated that SSTR2 high expression was a good prognostic factor in glioma. In summary, this study demonstrated that SSTR2 might be a valuable prognostic factor and therapeutic target in certain glioma subtypes.
Keywords: SSTR2, glioma, immunohistochemistry, IDH
Introduction
Gliomas are the most common primary malignancy in central nervous system (CNS). High grade glioma is characterized by excessive proliferation and relentless invasion as well as profuse infiltration into the brain parenchyma. In spite of aggressive treatment, including surgical resection, a combination of radiotherapy and adjuvant chemotherapy with temozolomide (TMZ), patients with glioma usually have a dismal outcome, and the median survival of GBM remains 12-14 months [1]. Therefore, it is necessary to discover novel biomarkers to improve the current diagnostic and possible treatment strategy.
Somatostatin receptors (SSTRs), members of the seven-transmembrane G protein-coupled receptor family, are widely expressed in both normal tissues and tumor tissues. SSTRs have been classified into five subtypes (SSTR1-5), which exert their antisecretory and antiproliferative effects through interactions with somatostatin analogs [2]. Among these receptor subtypes, SSTR2 is the most investigated because of its predominance. In neuroendocrine tumors (NETs), in which SSTRs are commonly expressed, the detection of SSTR2 expression has both diagnostic and therapeutic potential. Besides NETs, SSTR2 immunostaining can be detected in other types of tumor cells [3]. The expression of SSTRs has also been reported to be a diagnostic factor [4,5] and to be associated with favorable outcome in various tumors [6-8]. In our clinical practice, we found that SSTR2 also can be detected in some glioma cases. However, the previous studies were controversial on expression of SSTR2 in gliomas. This may be due to limited number of tumor samples, and they did not differentiate glioma histological and molecular subtypes [9-13].
The aim of the present study was to assess the expression of SSTR2 in a large cohort of Chinese patients with glioma (grade I-IV), determine its prognostic value, assess the correlation between SSTR2 expression and clinicopathological parameters and explore the underlying mechanism via a bioinformatic approach, in order to provide a foundation for later studies and therapeutic strategy.
Materials and methods
Clinical data
A total of 302 paraffin-embedded glioma samples were diagnosed histopathologically from 2009-2016 at the Sun Yat-Sen University Cancer Center. The clinical materials were used for research purposes after obtaining the patient’s written consent. This study was approved by the Institutional Research Ethics Committee of the Sun Yat-Sen University Cancer Center. All cases were defined according to the 2016 WHO Classification of CNS Tumors.
SSTR2 immunohistochemistry and evaluation on a tissue microarray (TMA)
Immunohistochemical detection of SSTR2 protein expression was performed on 5-μm thick TMA sections using a Ventana Benchmark XT (Ventana Medical Systems SA, Illkirch, France). The SSTR2 monoclonal antibody (clone UMB1, ab134152, Abcam, Cambridge, MA, USA) was used at 1/400 dilution. A Benchmark Ventana autostainer was used for detection and TMA slides were simultaneously immunostained to avoid inter-manipulation variability.
The results of staining were scored independently by two pathologists and the diagnoses were confirmed by histological phenotypes and molecular alterations. SSTR2 is localized in the cell membrane or membrane/cytoplasm (Figure 1). The expression of SSTR2 was scored according to the proportion of positivity and the staining intensity of the tumor cells. The percentage of positively stained tumor cells was scored as follows: 0 (no positive tumor cells), 1 (1-24% positive tumor cells), 2 (25-49% positive tumor cells), 3 (50-74% positive tumor cells), and 4 (≥ 75% positive tumor cells). The intensity of tumor cell staining was graded as follows: 0 (no staining), 1 (weak staining, light yellow), 2 (moderate staining, yellow brown), and 3 (strong staining, brown). The staining index (SI) was calculated as the product of the staining intensity score and the percentage of positive tumor cells (SI = percentage of positively stained tumor cells × staining intensity), which resulting in scores of 0, 1, 2, 3, 4, 6, 8, 9, 12. Cutoff values to define the high- and low-expression of SSTR2 were chosen using the median method. An SI score of ≥ 6 was used to define tumors with high expression of SSTR2, and an SI ≤ 4 was defined as low SSTR2 expression.
Figure 1.
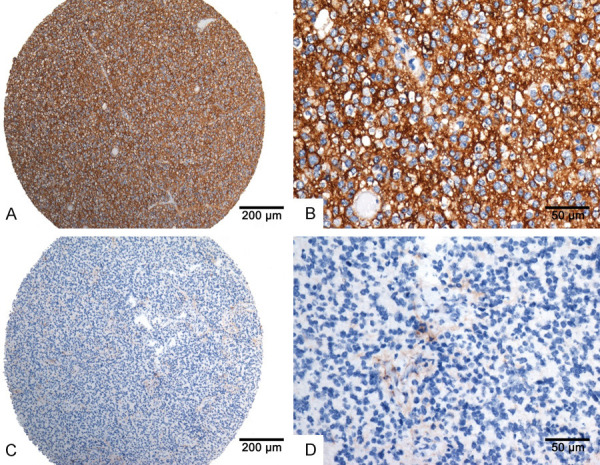
The protein expression of SSTR2 in paraffin-embedded glioma tissues. A and B. Strong staining of SSTR2 in oligodendroglioma; C and D. No staining in GBM.
Detection of other molecular alterations
To assess the association between SSTR2 expression and several established biomarkers, immunohistochemistry for p53 and Ki67, and fluorescent in situ hybridization for 1p/19q-codeletion status, IDH status, PHH3 (encoding phosphohistone H3), ATRX (encoding alpha thalassemia/mental retardation syndrome X-Linked), MGMT (encoding O-6-Methylguanine-DNA Methyltransferase), and TERT (encoding telomerase reverse transcriptase) promoter status were performed for all cases. The interpretation standard has been described previously [14].
Cell culture and western blotting
Normal human astrocyte (HA) cell line and human glioblastoma cell lines, including SF767, LN229, A172, U87, U251, DBTRG and T98G were cultured in Dulbecco’s Modified Eagle’s medium (DMEM) (Invitrogen, Carlsbad, CA, USA) containing 10% fetal bovine serum (Gibco-BRL, Gaithersburg, MD, USA). Cells were harvested and lysed in SDS lysis buffer. Protein concentrations were measured using the BCA protein assay kit (Beyotime, Shanghai, China) according to the manufacturer’s instructions. Equal amounts of protein samples (25 μg) were separated by electrophoresis on 10% SDS polyacrylamide gels, and then transferred to PVDF membrane (Millipore, Bedford, MA, USA). The membrane were blocked with 5% nonfat milk in Tris-buffered saline containing 0.1% Tween-20 (TBST) for 1 h at room temperature and was cut into two pieces in the location of 70 KD. Membranes were incubated with anti-SSTR2 antibody (1:500, Abcam, Cambridge, UK) overnight at 4°C, and then with horseradish peroxidase-conjugated anti-rabbit IgG antibodies (1:20000, Zhongshan Goldenbridge, Beijing, China) for 1 h at room temperature. A Vinculin antibody (1:1000, Santa cruz, CA, USA) was used as an internal reference. Immunoreactive bands were detected using an ECL kit (Beyotime, Shanghai, China) and the expression levels of protein were visualized using ChemiDoc Touching Imaging System (Bio-Rad, CA, USA).
Statistical analysis
All statistical analyses were performed using SPSS 25.0 (IBM Corp., Armonk, NY, USA) and GraphPad Prism version 8.0 (GraphPad Inc., La Jolla, CA, USA). Chi-square and Fisher’s exact tests were used to analyze the correlation between SSTR2 expression and clinicopathological characteristics. Quantitative variables were compared using Student’s t test and analysis of variance (ANOVA). Univariate and multivariate Cox-regression analyses were used to analyze the significance of various variables for survival. The type of Cox regression model chosen was the Forward: LR method. Survival curves were plotted using the Kaplan-Meier method and compared using the logrank test. Overall survival was calculated from the date of diagnosis to the date of death or the last known follow-up. All statistical tests were two-sided, and the threshold for statistical significance was P < 0.05.
Bioinformatic analyses
We utilized The Cancer Genome Atlas (TCGA) and the Chinese Glioma Genome Atlas (CGGA) to identify the differentially expressed genes (DEGs) between high SSTR2 expressing glioma samples and low SSTR2 expressing glioma samples. An adjusted P < 0.05 and |log2fold change (FC)| > 2 were chosen as the cut-off value. The Venn Diagram packages of R were applied to generate the Venn diagram to visualize the identified DEGs. To explore the underlying biological function and corresponding pathways of these significant DEGs, we used the DAVID database to perform functional enrichment analysis, including gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analyses. A P-value < 0.05 was considered a significant enrichment. Furthermore, the Search Tool for the Retrieval of Interacting Genes Database (STRING) (https://www.string-db.org/) was used to assess protein-protein interaction (PPI), and Cytoscape software (Cytoscape Consortium, San Diego, CA, USA) was used to visualize the results. A confidence score > 0.7 was regarded as significant. Finally, survival analysis related to key genes was performed via the TCGA dataset.
Results
The expression levels of SSTR2 in glioma
We firstly studied the mRNA expression of SSTR2 in CGGA and TCGA datasets. Cutoff values to define the high- and low-expression of SSTR2 were chosen using the median method. In both CGGA dataset and TCGA dataset, high mRNA expression were mainly detected in oligodendroglioma, anaplastic oligodendroglioma and astrocytoma, whereas high mRNA expression rate was lowest in glioblastoma (see Table S1). Statistically significant difference could be seen between glioblastoma and anaplastic oligodendroglioma (P < 0.001), glioblastoma and oligodendroglioma (P < 0.001), glioblastoma and diffuse astrocytoma (P < 0.001), anaplastic oligodendroglioma and anaplastic astrocytoma (P = 0.008), anaplastic astrocytoma and astrocytoma (P = 0.006), anaplastic astrocytoma and oligodendroglioma (P = 0.006), astrocytoma and oligodendroglioma (P < 0.001). In TGGA dataset, t test showed statistically significant difference between every two glioma subtype groups according to SSTR2 expression.
Then, we studied the protein expression of SSTR2 by IHC in 302 glioma samples, including 14 cases of pilocytic astrocytoma, 22 cases of oligodendroglioma (14 IDH-mutant, 1p/19q-codeleted, and 8 Not Otherwise Specified (NOS)), 59 cases of diffuse astrocytoma (41 IDH-mutant, 18 IDH-wild-type), 32 cases of anaplastic oligodendroglioma (27 IDH-mutant, 1p/19q-codeleted, and 5 NOS), 44 cases of anaplastic astrocytoma (19 IDH-mutant, 25 IDH-wildtype), and 131 cases of glioblastoma (21 IDH-mutant, 110 IDH-wildtype). The patients’ clinicopathological characteristics are shown in Table 1.
Table 1.
Clinicopathological characteristics of patient and the correlation between SSTR2 expression and clinical and molecular pathological characteristics of patients with glioma
| Characteristics | Total (n = 302) | SSTR2 Low (≤ 4) expression (%) | SSTR2 high (≥ 6) expression (%) | chi-square test P value |
|---|---|---|---|---|
| Sex | 0.668 | |||
| male | 178 | 134 (75.28) | 44 (24.71) | |
| female | 124 | 96 (77.42) | 28 (22.58) | |
| Age (y) | 0.152 | |||
| < 55 | 233 | 173 (74.25) | 60 (25.75) | |
| ≥ 55 | 69 | 57 (82.61) | 12 (17.39) | |
| WHO grade | < 0.001 | |||
| I | 14 | 11 (78.57) | 3 (21.43) | |
| II | 81 | 43 (53.09) | 38 (46.91) | |
| III | 76 | 56 (73.68) | 20 (26.31) | |
| IV | 131 | 120 (91.60) | 11 (8.40) | |
| epilepsy | 0.001 | |||
| no | 219 | 178 (81.28) | 41 (18.72) | |
| yes | 83 | 52 (62.65) | 31 (37.35) | |
| Location | 0.037 | |||
| subtentorial | 17 | 17 (100) | 0 (0) | |
| supratentorial | 285 | 213 (74.73) | 72 (25.26) | |
| Ki-67 | < 0.001 | |||
| < 10% | 74 | 45 (60.81) | 29 (39.19) | |
| ≥ 10% | 228 | 185 (81.14) | 43 (18.86) | |
| P53 | 0.18 | |||
| < 10% | 110 | 79 (71.82) | 31 (28.18) | |
| ≥ 10% | 192 | 151 (78.65) | 41 (21.35) | |
| PHH3 | < 0.001 | |||
| < 5 | 122 | 77 (63.11) | 45 (36.89) | |
| ≥ 5 | 180 | 153 (85.00) | 27 (15.00) | |
| ATRX | 0.571 | |||
| positive | 159 | 119 (74.84) | 40 (25.16) | |
| negative | 143 | 111 (77.62) | 32 (22.38) | |
| IDH1 | < 0.001 | |||
| Wild-type | 176 | 156 (88.64) | 20 (11.36) | |
| Mutated | 126 | 74 (58.73) | 52 (41.27) | |
| 1p/19q | < 0.001 | |||
| non-codeleted | 254 | 204 (80.31) | 50 (19.69) | |
| codeleted | 48 | 26 (54.17) | 22 (45.83) | |
| MGMTp | 0.856 | |||
| methylated | 144 | 109 (75.69) | 35 (24.31) | |
| unmethylated | 158 | 121 (76.58) | 37 (23.42) | |
| TERTp | 0.262 | |||
| Mutated | 142 | 104 (73.24) | 38 (26.76) | |
| Wild-type | 160 | 126 (78.75) | 34 (21.25) | |
Immunohistochemical analysis revealed that SSTR2 immunostaining could be detected in 49.00% (148/302) of the studied samples and high SSTR2 expression was present in 23.84% (72 of 302) of them. Among them, SSTR2 expression varied between glioma subtypes. High SSTR2 expression was mainly detected in oligodendrogliomas (12/22, 54.55%), astrocytomas (26/59, 44.07%), and anaplastic oligodendrogliomas (11/32, 34.38%) (Table 2). However, the majority of glioblastomas was associated with low SSTR2 expression, or was even negative for SSTR2 immunostaining.
Table 2.
SSTR2 expression in glioma subtypes
| Diagnosis | SSTR2 expression | SSTR2 expression | Total | ||
|---|---|---|---|---|---|
|
|
|
||||
| negative | positive | low | high | ||
| glioblastoma | 83 (63.36%) | 48 (36.64%) | 120 (91.60%) | 11 (8.40%) | 131 |
| anaplastic oligodendroglioma | 10 (31.25%) | 22 (68.75%) | 21 (65.63%) | 11 (34.38%) | 32 |
| oligodendroglioma | 4 (18.19%) | 18 (81.81%) | 10 (45.45%) | 12 (54.55%) | 22 |
| astrocytoma | 26 (44.97%) | 33 (55.93%) | 33 (55.93%) | 26 (44.07%) | 59 |
| anaplastic astrocytoma | 20 (45.45%) | 24 (54.55%) | 35 (79.55%) | 9 (20.45%) | 44 |
| pilocytic astrocytoma | 11 (78.57%) | 3 (21.43%) | 11 (78.57%) | 3 (21.43%) | 14 |
Western blotting analysis also confirmed the relatively low SSTR2 expression in glioblastoma cell lines (Figure 2). Among all the glioblastoma cell lines, SSTR2 protein expression was relatively higher in U251 and U87 cell lines than the others, while it can hardly be detected in T98G, DBTRG and SF767 cell lines.
Figure 2.
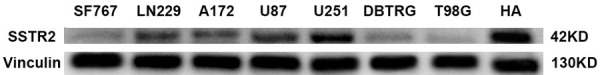
SSTR2 protein expression in human astrocyte (HA) and seven GBM cell lines, including SF767, LN229, A172, U87, U251, DBTRG and T98G. Protein levels were evaluated by western blotting. In general, SSTR2 protein expression in glioblastoma cell lines was lower than that in HA (astrocyte cell line). However, SSTR2 protein expression varies between different glioblastoma cell lines. Among the above glioblastoma cell lines, SSTR2 protein expression was relatively higher in U251 and U87 cell lines than the others, while it can hardly be detected in T98G, DBTRG and SF767 cell lines.
An ANOVA test revealed statistically significant difference between oligodendroglioma and glioblastoma (P = 0.002), anaplastic oligodendroglioma and glioblastoma (P = 0.007), and diffuse astrocytoma and glioblastoma (P < 0.001) according to SSTR2 protein expression (Figure 3).
Figure 3.
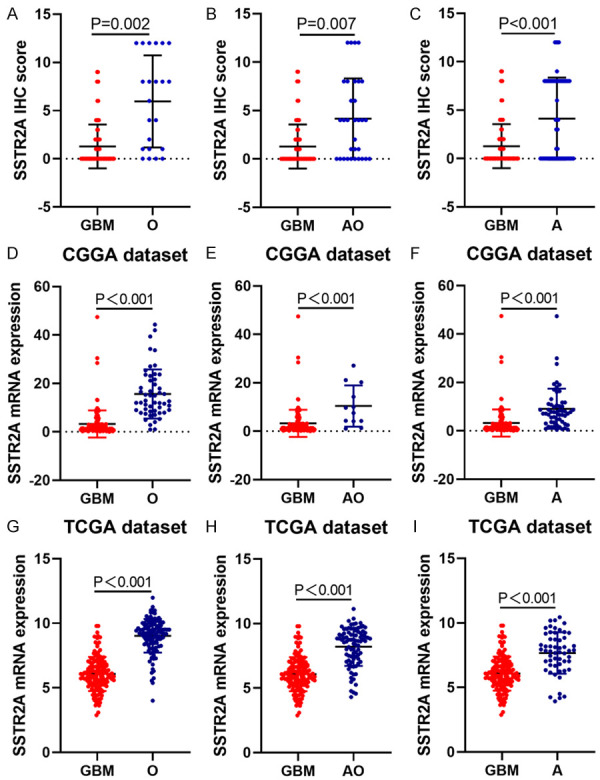
The expression pattern of SSTR2 in different histological subtypes of glioma. An ANOVA test revealed the statistical significance of the SSTR2 expression differences between oligodendroglioma and glioblastoma, anaplastic oligodendroglioma and glioblastoma, and diffuse astrocytoma and glioblastoma. A-C. Our data; D-F. CGGA RNAseq; G-I. TCGA RNAseq; GBM: glioblastoma multiforme; AO: anaplastic oligodendroglioma; O: oligodendroglioma; A: astrocytoma.
Correlation between SSTR2 expression and clinicopathological parameters in gliomas
T tests indicated that the SSTR2 expression detected in grade II to III glioma (lower grade glioma) tissues was higher than that in grade IV tissues (glioblastoma multiforme (GBM)) (P < 0.001), whereas its level did not correlate negatively with increasing WHO grade. Besides, SSTR2 expression in patients with epilepsy syndrome was higher than that in the patients without epilepsy syndrome (P < 0.001), and the expression of SSTR2 in supratentorial glioma was higher than that in subtentorial glioma (P = 0.006). Regarding molecular alterations, statistical analysis demonstrated that higher SSTR2 expression was detected in glioma with IDH mutations (P < 0.001), low Ki-67 expression (P = 0.024), low PHH3 expression (P < 0.001), and 1p/19q-codeletion (P < 0.001) (Figure 4). Further analysis showed that SSTR2 expression was markedly associated with WHO grade (P < 0.001), location (P = 0.037), epilepsy syndrome (P = 0.001), IDH mutation (P < 0.001), PHH3 (P < 0.001), 1p/19q-codeletion (P < 0.001), and Ki-67 (P < 0.001) (Table 1).
Figure 4.
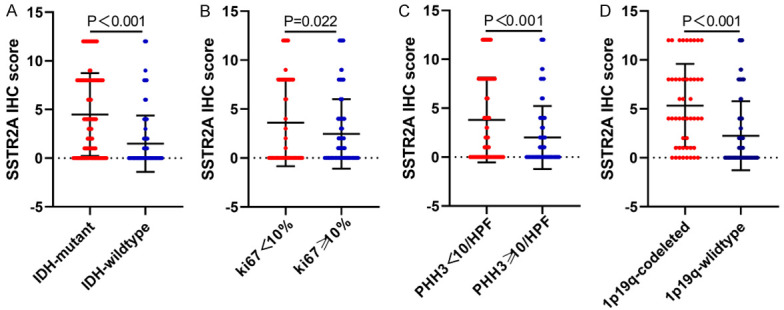
T test for SSTR2 expression and molecular alterations. Significantly higher expression of SSTR2 was observed in glioma tissue with IDH mutations (A), a Ki-67 label index < 10% (B), PHH3 < 5/10 HPF (C), and 1p/19q codeleted (D).
High SSTR2 expression was associated with a favorable prognosis in glioma, but was not an independent prognostic factor
We investigated the correlation between SSTR2 expression and patient survival using Kaplan-Meier analysis with the log-rank test. The survival plots in Figure 5 showed that high levels of SSTR2 protein correlated positively with longer overall survival (OS) in grade II to IV glioma patients (P < 0.001) as well as in grade III to IV glioma (P = 0.042). We further validated the prognostic value of SSTR2 in the CGGA and TCGA datasets. In the stratification analysis, SSTR2 was also a good prognostic maker in grade II glioma (P = 0.002) and grade II to III glioma (lower grade glioma, P = 0.001). However, no significance in OS for patients with grade III glioma (P = 0.288) and grade IV glioma/GBM (P = 0.931) was observed according to SSTR2 expression levels. In addition, compared with that of low level SSTR2 expression, patients with the IDH-wild-type subtype glioma with high SSTR2 expression had significantly longer OS, while no significance was observed in patients with the IDH-mutant subtype (see Figure S1). Taken together, patients with glioma with high SSTR2 expression were more frequently associated with better outcome than those with low or negative expression.
Figure 5.
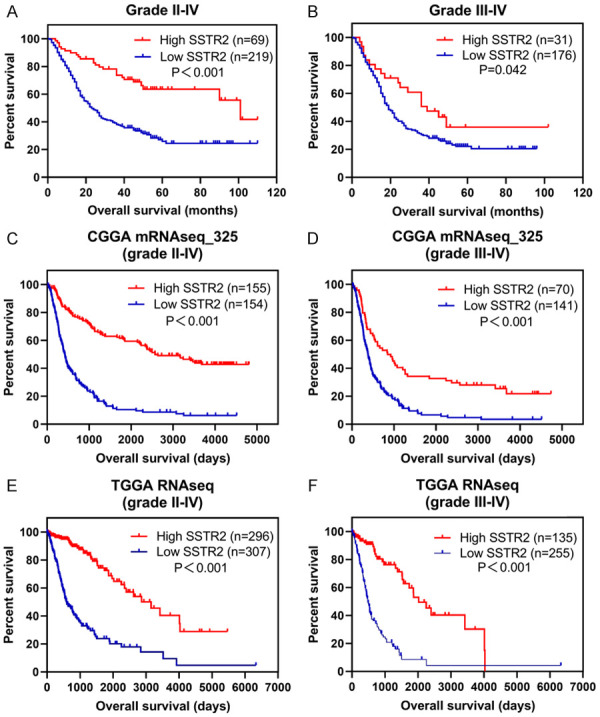
Kaplan-Meier curves for univariate analysis. High SSTR2 expression correlated with better prognosis in both of grade II to IV glioma (A, our data, P < 0.001; C, CGGA RNAseq, P < 0.001; E, TCGA RNAseq, P < 0.001; log-rank test) and grade III to IV glioma (B, our data, P = 0.042; D, CGGA RNAseq, P < 0.001; F, TCGA RNAseq, P < 0.001; log-rank test).
We used univariate Cox-regression analysis to determine the clinical significance of various prognostic factors that might influence the survival in patients with glioma. The results indicated that SSTR2 expression, age (≥ 55), WHO grade, tumor location, epilepsy syndrome, operative approaches, adjuvant chemotherapy, adjuvant radiation, Ki-67 label index, PHH3, IDH status, and the 1p/19q-codeletion were significant prognostic factors. However, multivariate Cox-regression analysis revealed that only operative approaches, Ki-67 index, IDH status, and 1p/19q-codeletion were independent prognostic factors for patients with glioma (Table 3).
Table 3.
Univariate and multivariate analysis of prognostic factors for patients with glioma
| Variable | Univariate analysis | Multivariate analysis | ||||
|---|---|---|---|---|---|---|
|
|
|
|||||
| Regression coefficient (SE) | Hazard ratio (HR) | P-value | HR | 95% Confidence interval | P-value | |
| Sex(male vs. female) | 0.156 | 1.338 | 0.061 | |||
| Age (≥ 55 vs. < 55) | 0.165 | 2.135 | < 0.001 | |||
| WHO grade | 0.105 | 2.352 | < 0.001 | 1.28 | 0.983-1.668 | 0.067 |
| Location | 0.363 | 2.374 | 0.017 | |||
| epilepsy | 0.183 | 0.552 | 0.001 | |||
| Resection | 0.156 | 0.638 | 0.004 | 0.603 | 0.443-0.822 | 0.001 |
| Adjuvant chemotherapy | 0.233 | 2.365 | < 0.001 | |||
| Adjuvant radiation | 0.3 | 2.796 | 0.001 | |||
| SSTR2 expression | 0.025 | 0.873 | < 0.0011 | |||
| Ki67 | 0.239 | 3.501 | < 0.001 | 2.427 | 1.345-4.380 | 0.003 |
| P53 | 0.162 | 1.22 | 0.221 | |||
| PHH3 | 0.173 | 2.756 | < 0.001 | |||
| IDH1 mut | 0.179 | 0.202 | < 0.001 | 0.338 | 0.227-0.505 | < 0.001 |
| 1p19q codeleted | 0.343 | 0.165 | < 0.001 | 0.306 | 0.148-0.633 | 0.001 |
| MGMTmet | 0.151 | 0.876 | 0.381 | |||
| ATRX | 0.153 | 1.225 | 0.185 | |||
| TERT | 0.151 | 1.136 | 0.399 | |||
Predicted functions and pathways of SSTR2 in gliomas
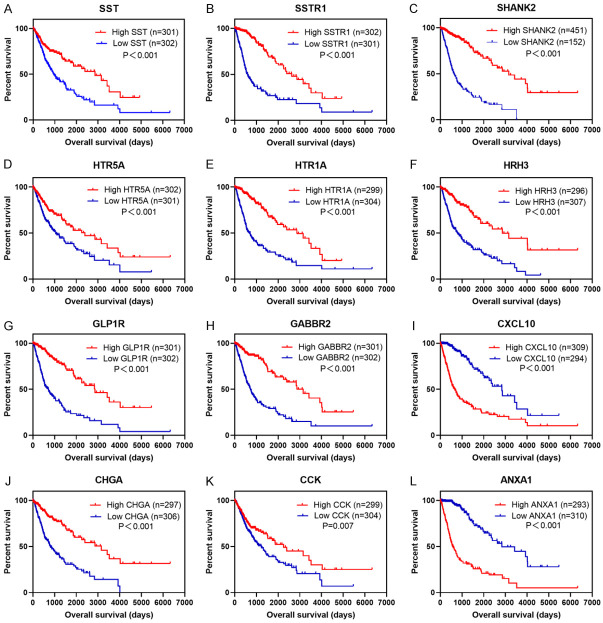
The TCGA and CGGA datasets were analyzed to identify genes that were differentially expressed in high SSTR2 expressing glioma samples compared with that in low SSTR2 expressing glioma samples. The TCGA dataset included 601 glioma cases and CGGA dataset included 608 glioma cases. We sorted each database according to the median expression level of SSTR2, and divided them into two groups (high and low). There were 485 DEGs (283 upregulated and 202 downregulated) in the TCGA dataset and 492 DEGs (323 upregulated and 169 downregulated) in the CGGA dataset that were differentially expressed between high SSTR2 expressing glioma samples and low SSTR2 expressing glioma samples. Further analysis of these DEGs using a Venn diagram revealed that 272 DEGs were observed consistently in both datasets (Figure 6). To further investigate the biological functions of the 272 DEGs, GO and KEGG pathway enrichment analyses were performed. In the GO enrichment analysis, the DEGs were mainly enriched in the nervous system, including synapse assembly, chemical synaptic transmission, and calcium iron transmembrane transport. In the KEGG analysis, the DEGs were mostly enriched in neuroactive ligand-receptor interaction, calcium signaling pathway, cAMP signaling pathway, and PI3K-Akt signaling pathway (Figure 6). Among the 272 DEGs, PPI analysis identified 12 genes whose protein products interact directly with SSTR2: SSTR1, SST (somatostatin), SHANK2 (SH3 and multiple ankyrin repeat domains 2), HTR5A (5-hydroxytryptamine receptor 5A), HTR1A, HRH3 (histamine receptor H3), GLP1R (glucagon like peptide 1 receptor), GABBR2 (gamma-aminobutyric acid type B receptor subunit 2), CXCL10 (C-X-C motif chemokine ligand 10), CHGA (chromogranin A), CCK (cholecystokinin), and ANXA1 (annexin A1) (Figure 6). Survival analysis of these genes in the TCGA dataset revealed that most of them are good prognostic markers in glioma, except CXCL10 and ANXA1 (Figure 7).
Figure 6.
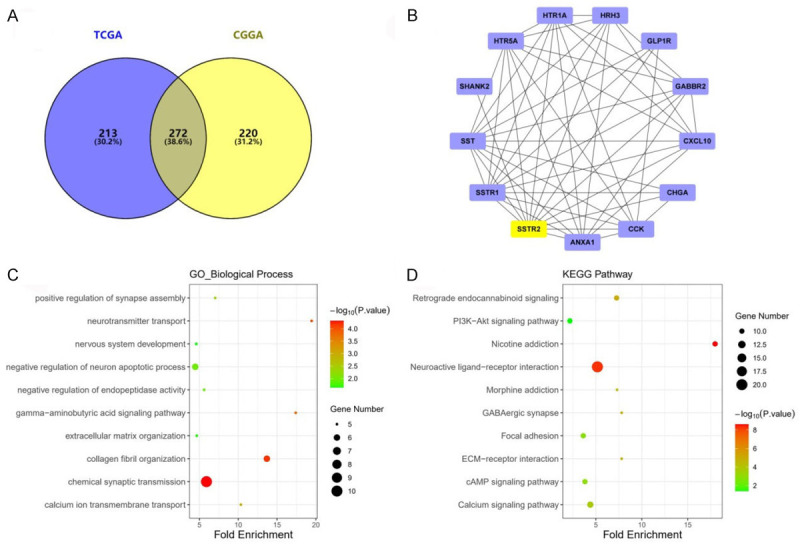
Bioinformatic analysis of the SSTR2-related differentially expressed genes (DEGs) in gliomas. A. Venn diagram showing the DEGs, including 485 in the TCGA, 429 in the CGGA ,and 272 belonging to both the TCGA and the CGGA. B. Protein-protein interaction (PPI) network of SSTR2-directly related genes. C. GO pathway enrichment analysis of DEGs. D. KEGG pathway enrichment analysis of DEGs.
Figure 7.
Survival analysis of DEGs in grade II-IV glioma in the TCGA dataset. Survival analysis of DEGs in the TCGA dataset revealed that the SST (A), SSTR1 (B), SHANK2 (C), HTR5A (D), HTR1A (E), HRH3 (F), GLP1R (G), GABBR2 (H), CHGA (J), and CCK (K) genes are good prognostic markers for glioma. While the high expression of CXCL10 (I) and ANXA1 (L) are correlated with poor outcome.
Discussion
Gliomas are the most prevalent primary CNS tumors. According to the 2016 WHO (World Health Organization) Classification of Tumors of the CNS, the novel diagnostic criteria take into account certain molecular alterations, such as IDH (encoding isocitrate dehydrogenase) mutation status and chromosome 1p/19q codeletion [15]. However, despite detailed molecular profiling and aggressive treatment, the prognosis of GBM remains dismal [1]. Consequently, it is necessary to discover biomarkers that possess prognostic and therapeutic potential in glioma.
SSTR2 is a promising candidate. In tumors of the CNS, owing to the strong and diffuse expression of SSTR2 in almost all cases of meningioma, the immunohistochemical detection of SSTR2 has been used as a specific marker of meningioma. A study reported that SSTR2 is the most sensitive (95.2%) and specific (92%) marker in the diagnosis of meningioma and achieved the maximum sensitivity (100%) when combined with an epithelial membrane antigen [5]. However, previous studies on SSTR2 expression in glioma have been controversial. A study conducted in Switzerland found that SSTR2 expression was predominantly detected in the majority of astrocytoma, two out of four oligodendrogliomas, and one out of twenty glioblastomas using autoradiographic assays [9]. Dutour et al. reported that SSTR2 mRNA could be detected in six out of nine glioma samples (the highest expression was detected in one glioblastoma) using northern blotting analysis, whereas immunostaining for SSTR2 was only conducted on representative glioma samples that were rich in mRNA [10]. A similar result was also observed in a study that included 50 glioma samples: SSTR2 immunoreactivity was found in 44% of glioblastomas (14/32), but only in 10% of anaplastic astrocytomas (1/10) and not in diffuse astrocytomas (0/8) [11]. In contrast to the first study mentioned above, the latter two studies demonstrated high SSTR2 expression in glioblastoma and low SSTR2 expression in grade II-III gliomas. Moreover, two studies analyzed SSTR2 expression in a relatively large-scale glioma sample. Kiviniemi et al. reported that SSTR2 expression was associated significantly with oligodendroglioma (79%), while it was less frequent in astrocytomas (25%) and negative in most glioblastomas (13%). The study also indicated that positive SSTR2 expression was related significantly to longer OS in patients with grade II-III glioma [12]. Similar observations were obtained by Appay et al. [13]. Various factors might contribute to the above controversial results, including the limited number of tumor samples, different diagnostic criteria, and different detection and analytical methods. However, most studies did not analyze the clinicopathological characteristics and the association between SSTR2 expression and other molecular alterations. Among them, only one study performed a stratification analysis of survival curves according to WHO grade [12].
In the present study, we investigated the clinicopathological significance of SSTR2 in a large cohort of Chinese patients with glioma (WHO grade I to IV). In line with the conclusion of Kiviniemi et al. and Appay et al. [14,15], we found that SSTR2 immunostaining correlated significantly with oligodendroglioma and anaplastic oligodendroglioma, whereas the majority of glioblastomas were associated with low or no SSTR2 expression by immunostaining. Western blotting analysis also indicated that SSTR2 expression was generally lower in glioblastoma cell lines than that in human astrocyte cell lines (Figure 2). The difference between the present study and the previous studies is that our statistical analysis showed that SSTR2 expression was also frequent in astrocytoma, and no statistical significance was observed between astrocytoma and oligodendroglioma and anaplastic oligodendroglioma according to SSTR2 expression. Besides, this study is the first to analyze the relationship between SSTR2 expression and several established molecular alterations.
Kaplan-Meier survival analysis revealed that patients with grade II-IV glioma with high SSTR2 expression showed a clear survival benefit compared with that for gliomas with low SSTR2 expression (P < 0.001). There are several potential explanations for this finding. First, high SSTR2 expression correlated significantly with oligodendrogliomas, and oligodendrogliomas are generally associated with better outcome, thus SSTR2 expression may also relate to better survival. Further stratification analysis of the OS of the histological subtypes showed that the survival benefit might be related to the association between SSTR2 and astrocytoma, because no significant difference in OS was observed within other histological subtypes according to SSTR2 status (see Figure S2), which also indicated that patients with astrocytoma should have their SSTR2 level detected in the future. The second probable reason was that a significantly higher expression of SSTR2 was observed in glioma tissue with IDH mutation, Ki-67 label index < 10%, PHH3 < 5/10HFP (high power field), and 1p/19q co-deleted which are generally associated with a better outcome. Although the independent prognostic factors defined by multivariate Cox-regression analysis did not include SSTR2 expression, better prognostic trends were noted for patients with grade II to IV glioma, and we had validated the prognostic value of SSTR2 in the CGGA and TCGA data. Our last speculation is that the molecular mechanisms underlying the antitumor activity of SSTR2 contributed to a better outcome. Our bioinformatic analysis revealed several SSTR2-related genes and signaling pathways in glioma. Most of the related genes were significantly associated with better survival in patients with glioma, as validated by the TCGA dataset. GO and KEGG analysis indicated that DEGs were significantly enriched in cAMP signaling pathways, the Calcium signaling pathway, and the PI3K-Akt signaling pathway, all of which have been reported as pathways associated with the anti-proliferation or antisecretory behavior induced by SSTR2 [16-18].
Due to the antisecretory and antiproliferative effects of SSTR2, patients with tumors that express SSTR2 can benefit from its clinical use. In NETs, several clinical trials had demonstrated some somatostatin analogues and peptide receptor radionuclide therapy (PRRT) based on SSTR2 enabled patients with NETs to have better survival times [19-21]. When it comes to glioma, a number of studies have reported promising therapeutic effectiveness of the peptide receptor radionuclide 90Y-DOTA-tyr3-Octreotide (90Y-DOTATOC) in II-IV grade glioma [22-24]. In our results, SSTR2 was expressed at the highest frequency in oligodendrogliomas (12/22, 54.55%) and at the lowest frequency in glioblastoma (11/131, 8.40%), indicating that SSTR2 targeted therapy could be promising for patients with oligodendroglioma. Likewise, high SSTR2 expression was detected in some patients with glioblastoma, which might be therapeutically meaningful, and further stressed the necessity of SSTR2 detection in patients with glioma. Nevertheless, defining groups of patients with glioma who will most likely benefit from this technically demanding procedure remains a challenge. Kiviniemi et al. reported the limited value of positron emission tomography-computed tomography (PET/CT) imaging with intravenously injected 68Ga-DOTA peptide targeting SSTR2 in defining suitable patients with high-grade glioma for peptide receptor radionuclide therapy (PRRT) [25]. However, these therapeutic strategies need further study.
In conclusion, our study demonstrated that high SSTR2 expression was mainly detected in oligodendroglioma, astrocytoma and anaplastic oligodendroglioma, whereas the expression of SSTR2 in glioblastoma was at a low level or barely detectable. High SSTR2 expression was associated with better outcome in patients with grade II to IV glioma. Consequently, SSTR2 may represent a predictor of patient outcome and could serve as a potential therapeutic target for glioma.
Disclosure of conflict of interest
None.
Supporting Information
References
- 1.Gately L, McLachlan SA, Dowling A, Philip J. Life beyond a diagnosis of glioblastoma: a systematic review of the literature. J Cancer Surviv. 2017;11:447–452. doi: 10.1007/s11764-017-0602-7. [DOI] [PubMed] [Google Scholar]
- 2.Theodoropoulou M, Stalla GK. Somatostatin receptors: from signaling to clinical practice. Front Neuroendocrinol. 2013;34:228–252. doi: 10.1016/j.yfrne.2013.07.005. [DOI] [PubMed] [Google Scholar]
- 3.Barbieri F, Bajetto A, Pattarozzi A, Gatti M, Würth R, Thellung S, Corsaro A, Villa V, Nizzari M, Florio T. Peptide receptor targeting in cancer: the somatostatin paradigm. Int J Pept. 2013;2013:926295. doi: 10.1155/2013/926295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tao LL, Huang YH, Chen YL, Yu GY, Yin WH. SSTR2a is a useful diagnostic marker for follicular dendritic cells and their related tumors. Am J Surg Pathol. 2019;43:374–381. doi: 10.1097/PAS.0000000000001205. [DOI] [PubMed] [Google Scholar]
- 5.Boulagnon-Rombi C, Fleury C, Fichel C, Lefour S, Marchal Bressenot A, Gauchotte G. Immunohistochemical approach to the differential diagnosis of meningiomas and their mimics. J Neuropathol Exp Neurol. 2017;76:289–298. doi: 10.1093/jnen/nlx008. [DOI] [PubMed] [Google Scholar]
- 6.Liu Y, Jiang L, Mu Y. Somatostatin receptor subtypes 2 and 5 are associated with better survival in operable hepatitis B-related hepatocellular carcinoma following octreotide long-acting release treatment. Oncol Lett. 2013;6:821–828. doi: 10.3892/ol.2013.1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Abdel-Rahman O, Lamarca A, Valle JW, Hubner RA. Somatostatin receptor expression in hepatocellular carcinoma: prognostic and therapeutic considerations. Endocr Relat Cancer. 2014;21:R485–493. doi: 10.1530/ERC-14-0389. [DOI] [PubMed] [Google Scholar]
- 8.Okuwaki K, Kida M, Mikami T, Yamauchi H, Imaizumi H, Miyazawa S, Iwai T, Takezawa M, Saegusa M, Watanabe M, Koizumi W. Clinicopathologic characteristics of pancreatic neuroendocrine tumors and relation of somatostatin receptor type 2A to outcomes. Cancer. 2013;119:4094–4102. doi: 10.1002/cncr.28341. [DOI] [PubMed] [Google Scholar]
- 9.Reubi JC, Lang W, Maurer R, Koper JW, Lamberts SW. Distribution and biochemical characterization of somatostatin receptors in tumors of the human central nervous system. Cancer Res. 1987;47:5758–5764. [PubMed] [Google Scholar]
- 10.Dutour A, Kumar U, Panetta R, Ouafik L, Fina F, Sasi R, Patel YC. Expression of somatostatin receptor subtypes in human brain tumors. Int J Cancer. 1998;76:620–627. doi: 10.1002/(sici)1097-0215(19980529)76:5<620::aid-ijc2>3.0.co;2-s. [DOI] [PubMed] [Google Scholar]
- 11.Mawrin C, Schulz S, Pauli SU, Treuheit T, Diete S, Dietzmann K, Firsching R, Schulz S, Höllt V. Differential expression of sst1, sst2A, and sst3 somatostatin receptor proteins in low-grade and high-grade astrocytomas. J Neuropathol Exp Neurol. 2004;63:13–19. doi: 10.1093/jnen/63.1.13. [DOI] [PubMed] [Google Scholar]
- 12.Kiviniemi A, Gardberg M, Kivinen K, Posti JP, Vuorinen V, Sipilä J, Rahi M, Sankinen M, Minn H. Somatostatin receptor 2A in gliomas: association with oligodendrogliomas and favourable outcome. Oncotarget. 2017;8:49123–49132. doi: 10.18632/oncotarget.17097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Appay R, Tabouret E, Touat M, Carpentier C, Colin C, Ducray F, Idbaih A, Mokhtari K, Uro-Coste E, Dehais C, Figarella-Branger D. Somatostatin receptor 2A protein expression characterizes anaplastic oligodendrogliomas with favorable outcome. Acta Neuropathol Commun. 2018;6:89. doi: 10.1186/s40478-018-0594-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hu WM, Wang F, Xi SY, Zhang X, Lai JP, Wu HY, Liu LL, Sai K, Zeng J. Practice of the new integrated molecular diagnostics in gliomas: experiences and new findings in a single Chinese center. J Cancer. 2020;11:1371–1382. doi: 10.7150/jca.38603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen R, Smith-Cohn M, Cohen AL, Colman H. Glioma subclassifications and their clinical significance. Neurotherapeutics. 2017;14:284–297. doi: 10.1007/s13311-017-0519-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tentler JJ, Hadcock JR, Gutierrez-Hartmann A. Somatostatin acts by inhibiting the cyclic 3’,5’-adenosine monophosphate (cAMP)/protein kinase A pathway, cAMP response element-binding protein (CREB) phosphorylation, and CREB transcription potency. Mol Endocrinol. 1997;11:859–866. doi: 10.1210/mend.11.7.9943. [DOI] [PubMed] [Google Scholar]
- 17.Stueven AK, Kayser A, Wetz C, Amthauer H, Wree A, Tacke F, Wiedenmann B, Roderburg C, Jann H. Somatostatin analogues in the treatment of neuroendocrine tumors: past, present and future. Int J Mol Sci. 2019;20:3049. doi: 10.3390/ijms20123049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bousquet C, Guillermet-Guibert J, Saint-Laurent N, Archer-Lahlou E, Lopez F, Fanjul M, Ferrand A, Fourmy D, Pichereaux C, Monsarrat B, Pradayrol L, Estève JP, Susini C. Direct binding of p85 to sst2 somatostatin receptor reveals a novel mechanism for inhibiting PI3K pathway. EMBO J. 2006;25:3943–3954. doi: 10.1038/sj.emboj.7601279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rinke A, Müller HH, Schade-Brittinger C, Klose KJ, Barth P, Wied M, Mayer C, Aminossadati B, Pape UF, Bläker M, Harder J, Arnold C, Gress T, Arnold R. Placebo-controlled, double-blind, prospective, randomized study on the effect of octreotide LAR in the control of tumor growth in patients with metastatic neuroendocrine midgut tumors: a report from the PROMID study group. J. Clin. Oncol. 2009;27:4656–4663. doi: 10.1200/JCO.2009.22.8510. [DOI] [PubMed] [Google Scholar]
- 20.Caplin ME, Pavel M, Ćwikła JB, Phan AT, Raderer M, Sedláčková E, Cadiot G, Wolin EM, Capdevila J, Wall L, Rindi G, Langley A, Martinez S, Blumberg J, Ruszniewski P. Lanreotide in metastatic enteropancreatic neuroendocrine tumors. N Engl J Med. 2014;371:224–233. doi: 10.1056/NEJMoa1316158. [DOI] [PubMed] [Google Scholar]
- 21.Bushnell DL Jr, O’Dorisio TM, O’Dorisio MS, Menda Y, Hicks RJ, Van Cutsem E, Baulieu JL, Borson-Chazot F, Anthony L, Benson AB, Oberg K, Grossman AB, Connolly M, Bouterfa H, Li Y, Kacena KA, LaFrance N, Pauwels SA. 90Y-edotreotide for metastatic carcinoid refractory to octreotide. J. Clin. Oncol. 2010;28:1652–1659. doi: 10.1200/JCO.2009.22.8585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Merlo A, Hausmann O, Wasner M, Steiner P, Otte A, Jermann E, Freitag P, Reubi JC, Müller-Brand J, Gratzl O, Mäcke HR. Locoregional regulatory peptide receptor targeting with the diffusible somatostatin analogue 90Y-labeled DOTA0-D-Phe1-Tyr3-octreotide (DOTATOC): a pilot study in human gliomas. Clin Cancer Res. 1999;5:1025–1033. [PubMed] [Google Scholar]
- 23.Heute D, Kostron H, von Guggenberg E, Ingorokva S, Gabriel M, Dobrozemsky G, Stockhammer G, Virgolini IJ. Response of recurrent high-grade glioma to treatment with (90)Y-DOTATOC. J Nucl Med. 2010;51:397–400. doi: 10.2967/jnumed.109.072819. [DOI] [PubMed] [Google Scholar]
- 24.Schumacher T, Hofer S, Eichhorn K, Wasner M, Zimmerer S, Freitag P, Probst A, Gratzl O, Reubi JC, Maecke R, Mueller-Brand J, Merlo A. Local injection of the 90Y-labelled peptidic vector DOTATOC to control gliomas of WHO grades II and III: an extended pilot study. Eur J Nucl Med Mol Imaging. 2002;29:486–493. doi: 10.1007/s00259-001-0717-x. [DOI] [PubMed] [Google Scholar]
- 25.Kiviniemi A, Gardberg M, Frantzén J, Pesola M, Vuorinen V, Parkkola R, Tolvanen T, Suilamo S, Johansson J, Luoto P, Kemppainen J, Roivainen A, Minn H. Somatostatin receptor subtype 2 in high-grade gliomas: PET/CT with (68)Ga-DOTA-peptides, correlation to prognostic markers, and implications for targeted radiotherapy. EJNMMI Res. 2015;5:25. doi: 10.1186/s13550-015-0106-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.