Abstract

The analysis of glycans presents a significant challenge that arises from their isomeric heterogeneity. While high-resolution ion mobility spectrometry (IMS) has shown the ability to resolve subtly different glycan isomers, their unambiguous assignment remains difficult. Here, we demonstrate an infrared (IR) spectroscopic approach for identifying isomers in a glycan mixture. To display the feasibility of this approach, we have constructed a small database of cryogenic spectra of five lacto-N-fucopentaose (LNFP) and six disaccharide isomers and demonstrated that in the cases where they cannot be separated by IMS, we can use a cryogenic IR spectrum to identify the isomeric components of a mixture.
Introduction
Oligosaccharides, or glycans, play a fundamental role in a multitude of biological processes. For example, in the form of glycoproteins or glycolipids at the cell surface, they are involved in cell-to-cell communication and immune response.1,2 As free glycans in human milk, they influence the development of gut microbiota in infants.3,4 A first step in elucidating the function of a particular glycan is to identify its primary structure, including the stereochemistry of the glycosidic linkages. However, the inherent isomeric complexity of this class of molecules presents a formidable analytical challenge.5,6
A variety of techniques have been employed for the structural determination of isomeric glycans. While the speed and sensitivity of mass spectrometry (MS) make it ideal for most forms of biomolecular analysis, it is impossible for single-stage MS alone to distinguish between isomers. Tandem MS, most frequently using collision-induced dissociation (CID), can identify many isomeric glycans by their fragmentation patterns,7−11 however, it often has difficulty in distinguishing between the most subtly different isomeric forms. These limitations can be partially overcome by the addition of an orthogonal technique, such as ion mobility spectrometry (IMS), which separates gas-phase ions based on their collisional cross section (CCS).12−15 Recent developments in IMS methods, such as trapped ion mobility spectrometry (TIMS),16 cyclic IMS (cIMS),17 and structures for lossless ion manipulation (SLIM),18,19 offer significantly higher resolution than standard drift-tube IMS in many cases, enabling the separation of glycan isomers.20−23 While combining high-resolution IMS with tandem MS helps reduce the problem, the highly complex mobility profiles often leave isomer identification ambiguous.24
Another orthogonal technique that has been combined with MS to identify glycans is infrared (IR) spectroscopy.25−30 We have recently coupled cryogenic IR spectroscopy with IMS-MS and demonstrated how spectroscopic fingerprints can be used for glycan identification.14,26,31−34 This approach is particularly robust since a vibrational spectrum is a highly structure-specific property of the molecule itself that is relatively insensitive to experimental conditions. Any slight variations in the molecular structure will affect the vibrational mode frequencies, which can easily be detected.33,34
This study aims to demonstrate that cryogenic IR spectra can be used in a database approach for the unambiguous identification of glycan isomers in a mixture. Database approaches are used for identifying glycans on the basis of tandem MS16,35−38 as well as collision cross sections determined by ion mobility.39 In our approach, we create a spectroscopic database using high-resolution ion mobility to separate isomers and then measure an IR spectrum of the isomerically pure species. For the initial creation of the IR reference database, high ion mobility resolving power is necessary, but once these spectra are measured, any separation technique (liquid chromatography (LC), IMS, etc.) can be used in conjunction with IR spectroscopy for identification and quantification of the isomeric composition. Even components that are not fully separated by ion mobility or liquid chromatography can be identified by decomposition of the cryogenic IR spectra of the overlapping isomers, as long as the corresponding isomerically pure reference spectra have been added to the database.
To illustrate the feasibility of this approach, we use a set of five isomeric human milk oligosaccharides (HMOs), lacto-N-fucopentaoses (LNFPs), shown in Figure 1, from which we construct a small database of cryogenic IR spectra. We then use this database to determine the presence of these glycans in mixtures.
Figure 1.
SNFG representation of the studied human milk oligosaccharides.
Experimental Methods
Sample Preparation
Human milk oligosaccharides (HMOs), LNFP I, II, III, and V, were acquired from Carbosynth Ltd., and LNFP VI was purchased from Dextra Laboratories Ltd. An HMO mixture that includes 60% penta- and hexasaccharides was purchased from Carbosynth Ltd. and made into a 0.1 mg/mL solution. All samples were prepared in a 50:50 mixture of water/methanol and used without further purification.
Instrumentation
The experiments were performed on a home-built instrument,26,40 shown schematically in Figure 2, which incorporates structures for lossless ion manipulation (SLIM)18,41 for ion mobility separation and a cryogenic planar ion trap with a time-of-flight (TOF) mass spectrometer for IR spectroscopy.37
Figure 2.
Schematic of the experimental setup.
Briefly, oligosaccharide solutions with concentrations of 10–25 μM are introduced into the instrument through a heated stainless-steel capillary (170 °C) using a nanoelectrospray (nESI) source. Initially, the ions are guided through a set of ion funnels and accumulated in an ion funnel trap (IFT). Short intense ion packets (≈130 μs wide) are then released from the IFT and guided to the SLIM-IMS region filled with helium at 3 mbar, where molecules are separated according to their drift time.
Our SLIM-IMS device, which we described in detail previously,26,37,40 was designed following the work of Smith and co-workers.18,41 It comprises a pair of mirrored 15 × 15 cm2 printed circuit boards (see Figure 2), featuring a 1.5 m single-cycle serpentine path for ion mobility separation and a set of six individually accessible traps for storage of ions with selected mobility. Long path lengths are achieved by allowing the ions to cycle through the serpentine path multiple times.26,42
Following separation, mobility-selected ions are guided through differential pumping stages and a quadrupole mass filter toward the cryogenic planar ion trap maintained at 40 K, where they are cooled through collisions with a He/N2 (90:10) gas mixture and tagged by weakly binding one or two nitrogen molecules. At these low temperatures, different ions will have different tagging efficiencies, which we measure. The cold tagged ions are subsequently irradiated with a 50 ms slice from a continuous-wave mid-IR laser (IPG Photonics) and mass analyzed using the TOF mass spectrometer. Upon absorption of an IR photon, energy is redistributed among the vibrational modes of the ion, leading to dissociation of the nitrogen tag(s). We obtain an IR fingerprint spectrum of the tagged species by measuring the ratio between tagged and [tagged + untagged] ions at each laser wavenumber step. The power across the entire IR frequency range is maintained at 0.2 W. A typical spectrum takes 5 min to record. To acquire an arrival time distribution (ATD) of ions following SLIM-IMS separation, a channeltron detector is used after m/z selection by the quadruple mass filter.
Infrared spectral decomposition was performed using MATLAB. We used the fminsearch function to iteratively minimize the root-mean-square deviation (RMSD) between the spectrum of a mixture and a synthetic spectrum constructed from all normalized reference spectra. This spectral decomposition process yields an isomer ratio that corresponds to the best fit of the measured spectrum. The resulting isomer ratios are then linearly corrected by the relative tagging efficiencies at 40 K of all isomers to give a more accurate representation of the mixture composition.
Results and Discussion
Sodiated LNFP Isomers
Our initial studies focused on the isomers LNFP I, II, III, V, and VI in their singly sodiated charge state (m/z = 876.3 Th). A pure sample of each species was first analyzed to obtain a set of IR reference spectra (Figure S1, Supporting Information), which serve as identifying fingerprints. In the measured 3250–3700 cm–1 spectral region, which covers the free and hydrogen-bonded OH and NH stretch vibrations, each isomer shows a unique spectrum that is easily distinguishable from the others. The arrival time distribution of each isomer sample exhibits a single major peak, although the line shapes suggest additional unresolved features in most cases (Figure S2). This is likely due to the overlap of the reducing-end anomers, which are in equilibrium in solution before being electrosprayed but do not interconvert in the gas phase.43 In many cases, we can resolve these anomers and assign them, for example, by comparing with spectra of O-methylated species, which are locked in a particular configuration.34 Since we do not resolve the individual reducing-end anomers in this case, all reference spectra acquired for the [M + Na]+ charge state correspond to a mixture of these species.
Having measured the reference spectra, we then prepared an equimolar mixture containing all five isomers, introduced them into our instrument, and used SLIM-IMS to separate them. Figure 3a shows the ATD of the mixture after 7 cycles on the SLIM separation track (∼10.5 m drift length), which exhibits three major peaks. Figure 3b,c displays the IR spectra in colors corresponding to each of these arrival time peaks. Comparison of these spectra to our database (Figure S1) allowed us to attribute the second and third peaks in the ATD of Figure 3a to LNFP V and I (Figure 3b), respectively, and the first peak to an overlap of LNFP II, III, and VI (Figure 3c,d).
Figure 3.
(a) Arrival time distribution of the mixture including LNFP I, II, III, V, and VI ([M + Na]+ charge state) obtained after 7 SLIM cycles, ≈10.5 m drift length; (b) messenger tagging IR depletion spectra corresponding to individual peaks 2 (green) and 3 (blue) shown in panel (a) compared with the reference spectra identifying each (gray); (c) messenger tagging IR depletion spectra of the species found in peak 1 (red) shown in panel (a) compared with the best fit synthetic spectrum identifying the isomers present (gray); and (d) bar plot showing percentages of LNFP isomers in the best fit synthetic spectrum of panel (c).
LNFP Isomers in the [M + H + K]+2 Charge State
To address the issue of overlapping isomers in the ATD of the sodiated species, we also investigated the protonated–potassiated doubly charged species of the LNFP isomers (m/z = 446.6 Th). As before, we acquired a reference IR spectrum for each pure isomer (Figure S3), which serves as an identifying fingerprint. Unlike the sodiated species, some of the LNFP isomers in the [M + H + K]2+ charge state clearly display two major peaks in their ATD (Figure S2b), most probably originating from the two reducing-end anomers,17,20,34 which further complicates complete IMS separation of all isomers in the mixture. Examples of anomer-specific spectra are shown in Figures S4 and S5; these were also added to our small database.
Figure 4 shows the arrival time distribution of the mixture of all five LNFP isomers in the [M + H + K]2+ charge state after eight separation cycles (12 m), where the identity of individual drift peaks is assigned based on their drift times (Figure S2b). Even though LNFP II, III, and VI show partial separation, the overlap between other isomers is observed due to multiple drift peaks appearing for the individual compounds in the [M + H + K]2+ charge state. Thus, we cannot completely separate and identify all isomers of LNFPs present in this mixture using IMS alone, even when using both the singly sodiated and potassiated–protonated charge states. If we were to cycle the ions further to increase the resolution and separate the individual peaks, the ions would begin to overlap in the 1.5 m mobility separation region of our SLIM device.
Figure 4.
Arrival time distribution of LNFP I, II, III, V, and VI mixtures ([M + H + K]2+ charge state) obtained after eight SLIM cycles (12 m drift length). Each peak is denoted with its isomer content.
Isomer Identification Using Spectral Decomposition
A vibrational spectrum is an intrinsic property of a molecule, and given its sensitivity to the precise molecular structure, it provides a unique identifying fingerprint. If a reference spectrum of each of the constituents of a mixture is known, such as in the database for LNFP isomers presented in this work, it should be possible to identify the individual components in the IR spectrum of the mixture. In this case, high-resolution ion mobility serves to reduce the number of components in the composite spectrum, thus decreasing the complexity of the spectral decomposition.
To test our ability to identify species that we cannot fully separate by ion mobility, we investigated equimolar mixtures of those LNFP isomers that overlap most closely in their arrival time distributions: LNFP I and II in the [M + H + K]2+ charge state (Figure 4) and LNFP II, III, and VI in the [M + Na]+ charge state (peak 1 in Figure 3a).
Figure 5 shows the optimal synthetic spectrum (red) for the equimolar mixture of LNFP I and II in the [M + H + K]2+ charge state obtained from the MATLAB decomposition algorithm, compared to the measured spectrum (gray). Even though these species almost completely overlap in their ATD after a 12 m separation drift length, the algorithm correctly identifies them as major constituents of the mixture (43.9% LNFP I and 55.5% LNFP II), demonstrating the sensitivity of the cryogenic IR spectra to the slightest structural differences.
Figure 5.
(a) Messenger tagging IR depletion spectra of an equimolar mixture of LNFP I and II in the [M + H + K]2+ charge state (red) compared with the best fit synthetic spectrum (gray) and (b) bar plot showing percentages corresponding to the best fit synthetic spectrum.
As for the equimolar mixture in the [M + Na]+ charge state (Figure 3c,d), the decomposition algorithm successfully identifies the major isomers that overlap in the first peak of the ATD shown in Figure 3a. However, the percentages we obtain (26, 23.3, and 48.7%, for LNFP II, III, and VI, respectively) do not reflect the expected isomer ratios of the equimolar solution. The reasons for this are discussed below.
To further assess the performance of spectral decomposition, the reference cryogenic IR spectra of a series of six isomeric disaccharides (Figure S6) were measured in the [M + Na]+ charge state (Figure S7). Various mixtures of these disaccharides were then prepared, and the IR spectrum of each mixture without IMS separation was measured and analyzed. The decomposition algorithm successfully identified the constituent isomer makeup of each spectrum (Figures S8–S14), with a detection limit of 2.9% (2σ) in the mixtures we examined. This value was determined by considering all of the percentages of falsely identified isomers. This method is thus well suited for solving the problem of overlapping ion mobility peaks and can give reliable identification of the constituent isomer makeup in complex mixtures, although as discussed more fully below, our ability to perform a quantitative analysis is still limited.
Application to an Unknown Mixture
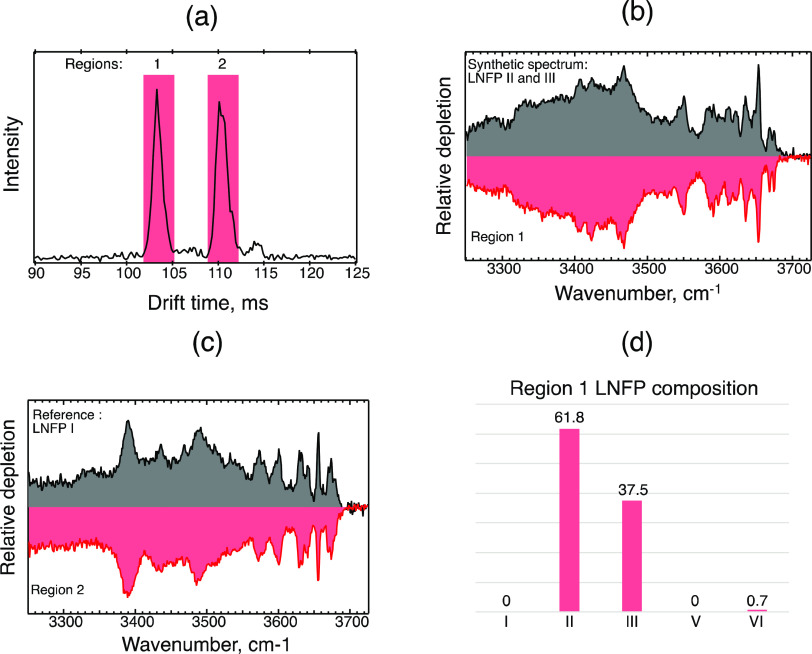
Having applied our database approach to mixtures of known concentration, we proceeded to analyze a sample obtained from Carbosynth Ltd., which contains an unknown mixture of HMOs including lacto-N-fucopentaoses. According to the supplier, the raw material for this sample was obtained from a panel of subjects and purified using liquid chromatography. Figure 6a shows the ATD of sodiated LNFP found in this sample after 7 cycles on our SLIM board. Visual comparison of this ATD to that in Figure 3a suggests the presence of at least one of LNFP II, III, and VI in the first peak, the possibility of LNFP V in the middle region, and LNFP I in the second, well-resolved peak. It is important to note that the ATDs shown in Figures 3 and 6 correspond to the same drift length, however, the drift times differ due to different separation conditions used in each experiment (different traveling wave pulse voltage and speed). A small peak appears in the ATD at about 114 ms, which is not present in Figure 3a. We believe this is an artifact, perhaps from a solvent cluster since it disappears when the drift gas is changed from He to N2 (Figure S15).
Figure 6.
(a) Arrival time distribution of LNFPs found in the Carbosynth HMO sample ([M + Na]+ charge state) obtained after 7 SLIM cycles (∼10.5 m drift length); (b) messenger tagging IR spectra of species found in the ATD region 1 (red), plotted against the best fit synthetic spectra identifying the isomers present (gray); (c) region 2 spectrum identifying LNFP I; and (d) bar plot showing percentages of LNFP isomers in the best fit synthetic spectrum of panel (b).
The cryogenic IR spectrum of the first ATD region was measured and analyzed using the spectral decomposition method as described above. Indeed, Figure 6b shows that the synthetic spectrum using the percentages returned by the decomposition algorithm (gray trace) is virtually identical to the measured spectrum (red trace), with its major components being LNFP II (61.8%) and III (37.5%). The algorithm also shows a minor contribution of LNFP VI (0.7%), which is below the limit of detection using this approach. Thus, we conclude that the first peak in the ATD of Figure 6a contains primarily LNFP II and III. The IR spectrum of region 2, shown in Figure 6c, indicates an exact match with LNFP I, thus confirming its presence. We also found that the spectrum of the ions arriving between the two main peaks in the ATD corresponds to that of LNFP V in our database (Figure S16).
Issues Related to Quantitation
While we have demonstrated that spectral decomposition is well suited for the identification of isomers present in the mixtures, the precise quantitation of the isomer content in the mixture solution is complicated by several issues that are intrinsic to ESI-MS analysis.44,45 These issues include possible differences in the ionization efficiencies and charge-state distributions of different isomers, as well as charge competition upon ESI.46 For example, the spectral decomposition of an equimolar mixture of LNFP I and II, as well as some of the binary mixtures of disaccharides that we studied, showed nearly equal ratios between each isomer, whereas in equimolar solutions with more than two isomers, at least one isomer was underestimated (Figures S10–S14). It is important to emphasize that this issue is not related to the spectral decomposition itself since this method accurately determines the gas-phase isomer ratios even though they might differ from the solution ratios due to the aforementioned effects.
To be able to determine the in-solution isomer composition quantitively, one can perform a derivatization step to tag glycans with a label, such as ProA or RapiFluor,47 which can help to mitigate the differences in the ionization efficiency and charge competition of different isomers. We are currently studying the spectroscopic fingerprints of labeled glycans and planning to further develop our approach for quantitative spectroscopic analysis of isomeric glycan mixtures.
Conclusions
In this work, we have demonstrated the principle that a library of cryogenic IR spectra can be used for identification of isomeric glycans in mixtures. We emphasize that high-resolution ion mobility is only necessary for obtaining isomerically pure IR fingerprints to build the database. Once the database is available, any moderate resolution IMS device could be used for partial separation and reduction of the number of overlapping species for reliable IR spectra decomposition and isomer identification. It is worth noting that under moderate separation conditions, it would be almost impossible to perform isomer identification using ion mobility alone or even in combination with fragmentation techniques since this would require unique fragment peaks for each isomer. In contrast, the linear nature of IR spectra and their high sensitivity to even minor structural variations between isomers make identification by spectral decomposition feasible. The identification process could be accelerated by shortening the IR scanning range, thereby reducing the IR spectral acquisition time. In addition, with an extensive database, machine learning methods could be introduced to identify unique regions of the isomer spectra, which would further reduce the scan range and acquisition time. The spectral measurement process can also be multiplexed, which will allow the acquisition of spectra of many species (including isomers) within a single laser scan.48
While our approach correctly identifies glycans from a mixture, issues remain in relating the relative gas-phase intensities that we observe to concentrations in solution prior to electrospray. Ongoing work using labeled glycans, which should have much more uniform ionization efficiencies, is currently underway.
Acknowledgments
The authors thank the European Research Council (Grant 788697-GLYCANAL) and the Swiss National Science Foundation (Grant 206021_177004) for their generous support of this work.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acs.analchem.1c03190.
Reference vibrational spectra and arrival time distributions (ATDs) for the LNFP isomers in different charge states; ATDs and anomer-resolved vibrational spectra of LNFP I and II; reference vibrational spectra of a set of disaccharides; and vibrational spectra of various glycan mixtures (PDF)
The authors declare no competing financial interest.
Supplementary Material
References
- Rudd P. M.; Elliott T.; Cresswell P.; Wilson I. A.; Dwek R. A. Glycosylation and the immune system. Science 2001, 291, 2370–2376. 10.1126/science.291.5512.2370. [DOI] [PubMed] [Google Scholar]
- Sriram V.; Willard C. A.; Liu J.; Brutkiewicz R. R. Importance of N-linked glycosylation in the functional expression of murine CD1d1. Immunology 2008, 123, 272–281. 10.1111/j.1365-2567.2007.02696.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akkerman R.; Faas M. M.; de Vos P. Non-digestible carbohydrates in infant formula as substitution for human milk oligosaccharide functions: Effects on microbiota and gut maturation. Crit. Rev. Food Sci. Nutr. 2019, 59, 1486–1497. 10.1080/10408398.2017.1414030. [DOI] [PubMed] [Google Scholar]
- Wiciński M.; Sawicka E.; Gebalski J.; Kubiak K.; Malinowski B. Human Milk Oligosaccharides: Health Benefits, Potential Applications in Infant Formulas, and Pharmacology. Nutrients 2020, 12, 266 10.3390/nu12010266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laine R. A. A calculation of all possible oligosaccharide isomers both branched and linear yields 1.05 ×10(12) structures for a reducing hexasaccharide: the Isomer Barrier to development of single-method saccharide sequencing or synthesis systems. Glycobiology 1994, 4, 759–767. 10.1093/glycob/4.6.759. [DOI] [PubMed] [Google Scholar]
- Mariño K.; Bones J.; Kattla J. J.; Rudd P. M. A systematic approach to protein glycosylation analysis: a path through the maze. Nat. Chem. Biol. 2010, 6, 713–723. 10.1038/nchembio.437. [DOI] [PubMed] [Google Scholar]
- Harvey D. J.; Royle L.; Radcliffe C. M.; Rudd P. M.; Dwek R. A. Structural and quantitative analysis of N-linked glycans by matrix-assisted laser desorption ionization and negative ion nanospray mass spectrometry. Anal. Biochem. 2008, 376, 44–60. 10.1016/j.ab.2008.01.025. [DOI] [PubMed] [Google Scholar]
- Zaia J. Mass spectrometry and glycomics. OMICS: J. Integr. Biol. 2010, 14, 401–418. 10.1089/omi.2009.0146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu H. C.; Liew C. Y.; Huang S. P.; Tsai S. T.; Ni C. K. Simple Method for De Novo Structural Determination of Underivatised Glucose Oligosaccharides. Sci. Rep. 2018, 8, 5562 10.1038/s41598-018-23903-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S. P.; Hsu H. C.; Liew C. Y.; Tsai S. T.; Ni C. K. Logically derived sequence tandem mass spectrometry for structural determination of Galactose oligosaccharides. Glycoconjugate J. 2021, 38, 177–189. 10.1007/s10719-020-09915-1. [DOI] [PubMed] [Google Scholar]
- Yu X.; Jiang Y.; Chen Y.; Huang Y.; Costello C. E.; Lin C. Detailed glycan structural characterization by electronic excitation dissociation. Anal. Chem. 2013, 85, 10017–10021. 10.1021/ac402886q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pu Y.; Ridgeway M. E.; Glaskin R. S.; Park M. A.; Costello C. E.; Lin C. Separation and Identification of Isomeric Glycans by Selected Accumulation-Trapped Ion Mobility Spectrometry-Electron Activated Dissociation Tandem Mass Spectrometry. Anal. Chem. 2016, 88, 3440–3443. 10.1021/acs.analchem.6b00041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu F.; Lee S.; Valentine S. J.; Reilly J. P.; Clemmer D. E. Mannose7 glycan isomer characterization by IMS-MS/MS analysis. J. Am. Soc. Mass Spectrom. 2012, 23, 2158–2166. 10.1007/s13361-012-0491-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann J.; Hahm H. S.; Seeberger P. H.; Pagel K. Identification of carbohydrate anomers using ion mobility-mass spectrometry. Nature 2015, 526, 241–244. 10.1038/nature15388. [DOI] [PubMed] [Google Scholar]
- Gaye M. M.; Nagy G.; Clemmer D. E.; Pohl N. L. Multidimensional Analysis of 16 Glucose Isomers by Ion Mobility Spectrometry. Anal. Chem. 2016, 88, 2335–2344. 10.1021/acs.analchem.5b04280. [DOI] [PubMed] [Google Scholar]
- Wei J.; Tang Y.; Ridgeway M. E.; Park M. A.; Costello C. E.; Lin C. Accurate Identification of Isomeric Glycans by Trapped Ion Mobility Spectrometry-Electronic Excitation Dissociation Tandem Mass Spectrometry. Anal. Chem. 2020, 92, 13211–13220. 10.1021/acs.analchem.0c02374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ujma J.; Ropartz D.; Giles K.; Richardson K.; Langridge D.; Wildgoose J.; Green M.; Pringle S. Cyclic Ion Mobility Mass Spectrometry Distinguishes Anomers and Open-Ring Forms of Pentasaccharides. J. Am. Soc. Mass Spectrom. 2019, 30, 1028–1037. 10.1007/s13361-019-02168-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng L.; Ibrahim Y. M.; Hamid A. M.; Garimella S. V.; Webb I. K.; Zheng X.; Prost S. A.; Sandoval J. A.; Norheim R. V.; Anderson G. A.; Tolmachev A. V.; Baker E. S.; Smith R. D. Ultra-High Resolution Ion Mobility Separations Utilizing Traveling Waves in a 13 m Serpentine Path Length Structures for Lossless Ion Manipulations Module. Anal. Chem. 2016, 88, 8957–8964. 10.1021/acs.analchem.6b01915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamid A. M.; Ibrahim Y. M.; Garimella S. V.; Webb I. K.; Deng L.; Chen T. C.; Anderson G. A.; Prost S. A.; Norheim R. V.; Tolmachev A. V.; Smith R. D. Characterization of Traveling Wave Ion Mobility Separations in Structures for Lossless Ion Manipulations. Anal. Chem. 2015, 87, 11301–11308. 10.1021/acs.analchem.5b02481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy G.; Attah I. K.; Garimella S. V. B.; Tang K.; Ibrahim Y. M.; Baker E. S.; Smith R. D. Unraveling the isomeric heterogeneity of glycans: ion mobility separations in structures for lossless ion manipulations. Chem. Commun. 2018, 54, 11701–11704. 10.1039/C8CC06966B. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arndt J. R.; Wormwood Moser K. L.; Van Aken G.; Doyle R. M.; Talamantes T.; DeBord D.; Maxon L.; Stafford G.; Fjeldsted J.; Miller B.; Sherman M. High-Resolution Ion-Mobility-Enabled Peptide Mapping for High-Throughput Critical Quality Attribute Monitoring. J. Am. Soc. Mass Spectrom. 2021, 2019–2032. 10.1021/jasms.0c00434. [DOI] [PubMed] [Google Scholar]
- Ropartz D.; Fanuel M.; Ujma J.; Palmer M.; Giles K.; Rogniaux H. Structure Determination of Large Isomeric Oligosaccharides of Natural Origin through Multipass and Multistage Cyclic Traveling-Wave Ion Mobility Mass Spectrometry. Anal. Chem. 2019, 91, 12030–12037. 10.1021/acs.analchem.9b03036. [DOI] [PubMed] [Google Scholar]
- Gray C. J.; Thomas B.; Upton R.; Migas L. G.; Eyers C. E.; Barran P. E.; Flitsch S. L. Applications of ion mobility mass spectrometry for high throughput, high resolution glycan analysis. Biochim. Biophys. Acta, Gen. Subj. 2016, 1860, 1688–1709. 10.1016/j.bbagen.2016.02.003. [DOI] [PubMed] [Google Scholar]
- Yalovenko N.; Yatsyna V.; Bansal P.; AbiKhodr A. H.; Rizzo T. R. Analyzing glycans cleaved from a biotherapeutic protein using ultrahigh-resolution ion mobility spectrometry together with cryogenic ion spectroscopy. Analyst 2020, 145, 6493–6499. 10.1039/D0AN01206H. [DOI] [PubMed] [Google Scholar]
- Mucha E.; Gonzalez Florez A. I.; Marianski M.; Thomas D. A.; Hoffmann W.; Struwe W. B.; Hahm H. S.; Gewinner S.; Schollkopf W.; Seeberger P. H.; von Helden G.; Pagel K. Glycan Fingerprinting via Cold-Ion Infrared Spectroscopy. Angew. Chem., Int. Ed. 2017, 56, 11248–11251. 10.1002/anie.201702896. [DOI] [PubMed] [Google Scholar]
- Ben Faleh A.; Warnke S.; Rizzo T. R. Combining Ultrahigh-Resolution Ion-Mobility Spectrometry with Cryogenic Infrared Spectroscopy for the Analysis of Glycan Mixtures. Anal. Chem. 2019, 91, 4876–4882. 10.1021/acs.analchem.9b00659. [DOI] [PubMed] [Google Scholar]
- Kirschbaum C.; Saied E. M.; Greis K.; Mucha E.; Gewinner S.; Schollkopf W.; Meijer G.; von Helden G.; Poad B. L. J.; Blanksby S. J.; Arenz C.; Pagel K. Resolving Sphingolipid Isomers Using Cryogenic Infrared Spectroscopy. Angew. Chem., Int. Ed. 2020, 59, 13638–13642. 10.1002/anie.202002459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Depraz Depland A.; Renois-Predelus G.; Schindler B.; Compagnon I. Identification of sialic acid linkage isomers in glycans using coupled InfraRed Multiple Photon Dissociation (IRMPD) spectroscopy and mass spectrometry. Int. J. Mass Spectrom. 2018, 434, 65–69. 10.1016/j.ijms.2018.09.005. [DOI] [Google Scholar]
- Martens J.; Berden G.; van Outersterp R. E.; Kluijtmans L. A. J.; Engelke U. F.; van Karnebeek C. D. M.; Wevers R. A.; Oomens J. Molecular identification in metabolomics using infrared ion spectroscopy. Sci. Rep. 2017, 7, 3363 10.1038/s41598-017-03387-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindler B.; Laloy-Borgna G.; Barnes L.; Allouche A. R.; Bouju E.; Dugas V.; Demesmay C.; Compagnon I. Online Separation and Identification of Isomers Using Infrared Multiple Photon Dissociation Ion Spectroscopy Coupled to Liquid Chromatography: Application to the Analysis of Disaccharides Regio-Isomers and Monosaccharide Anomers. Anal. Chem. 2018, 90, 11741–11745. 10.1021/acs.analchem.8b02801. [DOI] [PubMed] [Google Scholar]
- Masellis C.; Khanal N.; Kamrath M. Z.; Clemmer D. E.; Rizzo T. R. Cryogenic Vibrational Spectroscopy Provides Unique Fingerprints for Glycan Identification. J. Am. Soc. Mass Spectrom. 2017, 28, 2217–2222. 10.1007/s13361-017-1728-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khanal N.; Masellis C.; Kamrath M. Z.; Clemmer D. E.; Rizzo T. R. Cryogenic IR spectroscopy combined with ion mobility spectrometry for the analysis of human milk oligosaccharides. Analyst 2018, 143, 1846–1852. 10.1039/C8AN00230D. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellegrinelli R. P.; Yue L.; Carrascosa E.; Warnke S.; Ben Faleh A.; Rizzo T. R. How General Is Anomeric Retention during Collision-Induced Dissociation of Glycans?. J. Am. Chem. Soc. 2020, 142, 5948–5951. 10.1021/jacs.0c00264. [DOI] [PubMed] [Google Scholar]
- Warnke S.; Ben Faleh A.; Scutelnic V.; Rizzo T. R. Separation and Identification of Glycan Anomers Using Ultrahigh-Resolution Ion-Mobility Spectrometry and Cryogenic Ion Spectroscopy. J. Am. Soc. Mass Spectrom. 2019, 30, 2204–2211. 10.1007/s13361-019-02333-0. [DOI] [PubMed] [Google Scholar]
- Morrison K. A.; Clowers B. H. Differential Fragmentation of Mobility-Selected Glycans via Ultraviolet Photodissociation and Ion Mobility-Mass Spectrometry. J. Am. Soc. Mass Spectrom. 2017, 28, 1236–1241. 10.1007/s13361-017-1621-3. [DOI] [PubMed] [Google Scholar]
- Mank M.; Welsch P.; Heck A. J. R.; Stahl B. Label-free targeted LC-ESI-MS(2) analysis of human milk oligosaccharides (HMOS) and related human milk groups with enhanced structural selectivity. Anal. Bioanal. Chem. 2019, 411, 231–250. 10.1007/s00216-018-1434-7. [DOI] [PubMed] [Google Scholar]
- Bansal P.; Yatsyna V.; AbiKhodr A. H.; Warnke S.; Ben Faleh A.; Yalovenko N.; Wysocki V. H.; Rizzo T. R. Using SLIM-Based IMS-IMS Together with Cryogenic Infrared Spectroscopy for Glycan Analysis. Anal. Chem. 2020, 92, 9079–9085. 10.1021/acs.analchem.0c01265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remoroza C. A.; Liang Y.; Mak T. D.; Mirokhin Y.; Sheetlin S. L.; Yang X.; San Andres J. V.; Power M. L.; Stein S. E. Increasing the Coverage of a Mass Spectral Library of Milk Oligosaccharides Using a Hybrid-Search-Based Bootstrapping Method and Milks from a Wide Variety of Mammals. Anal. Chem. 2020, 92, 10316–10326. 10.1021/acs.analchem.0c00342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struwe W. B.; Pagel K.; Benesch J. L.; Harvey D. J.; Campbell M. P. GlycoMob: an ion mobility-mass spectrometry collision cross section database for glycomics. Glycoconjugate J. 2016, 33, 399–404. 10.1007/s10719-015-9613-7. [DOI] [PubMed] [Google Scholar]
- Warnke S.; Ben Faleh A.; Pellegrinelli R. P.; Yalovenko N.; Rizzo T. R. Combining ultra-high resolution ion mobility spectrometry with cryogenic IR spectroscopy for the study of biomolecular ions. Faraday Discuss. 2019, 217, 114–125. 10.1039/C8FD00180D. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamid A. M.; Garimella S. V. B.; Ibrahim Y. M.; Deng L.; Zheng X.; Webb I. K.; Anderson G. A.; Prost S. A.; Norheim R. V.; Tolmachev A. V.; Baker E. S.; Smith R. D. Achieving High Resolution Ion Mobility Separations Using Traveling Waves in Compact Multiturn Structures for Lossless Ion Manipulations. Anal. Chem. 2016, 88, 8949–8956. 10.1021/acs.analchem.6b01914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng L.; Webb I. K.; Garimella S. V. B.; Hamid A. M.; Zheng X.; Norheim R. V.; Prost S. A.; Anderson G. A.; Sandoval J. A.; Baker E. S.; Ibrahim Y. M.; Smith R. D. Serpentine Ultralong Path with Extended Routing (SUPER) High Resolution Traveling Wave Ion Mobility-MS using Structures for Lossless Ion Manipulations. Anal. Chem. 2017, 89, 4628–4634. 10.1021/acs.analchem.7b00185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraschetti C.; Guarcini L.; Zazza C.; Mannina L.; Circi S.; Piccirillo S.; Chiavarino B.; Filippi A. Real time evolution of unprotected protonated galactosamine probed by IRMPD spectroscopy. Phys. Chem. Chem. Phys. 2018, 20, 8737–8743. 10.1039/C7CP07642H. [DOI] [PubMed] [Google Scholar]
- Grünwald-Gruber C.; Thader A.; Maresch D.; Dalik T.; Altmann F. Determination of true ratios of different N-glycan structures in electrospray ionization mass spectrometry. Anal. Bioanal. Chem. 2017, 409, 2519–2530. 10.1007/s00216-017-0235-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirschbaum C.; Greis K.; Mucha E.; Kain L.; Deng S.; Zappe A.; Gewinner S.; Schollkopf W.; von Helden G.; Meijer G.; Savage P. B.; Marianski M.; Teyton L.; Pagel K. Unravelling the structural complexity of glycolipids with cryogenic infrared spectroscopy. Nat. Commun. 2021, 12, 1201 10.1038/s41467-021-21480-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang K.; Page J. S.; Smith R. D. Charge competition and the linear dynamic range of detection in electrospray ionization mass spectrometry. J. Am. Soc. Mass Spectrom. 2004, 15, 1416–1423. 10.1016/j.jasms.2004.04.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keser T.; Pavić T.; Lauc G.; Gornik O. Comparison of 2-Aminobenzamide, Procainamide and RapiFluor-MS as Derivatizing Agents for High-Throughput HILIC-UPLC-FLR-MS N-glycan Analysis. Front. Chem. 2018, 6, 324 10.3389/fchem.2018.00324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warnke S.; Ben Faleh A.; Rizzo T. R. Towards High-Throughput, Cryogenic, IR Fingerprinting of Mobili-ty-Separated Glycan Isomers. ACS Meas. Sci. Au 2021, 10.1021/acsmeasuresciau.1c00018. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.