Abstract
Aged individuals, particularly males, display an impaired level of antibody response compared with their younger counterparts, yet the molecular mechanisms responsible for the discrepancy are not well understood. We hypothesize that some of this difference may be linked to B cell somatic hypermutation (SHM) targeting including error-prone DNA repair activities that are crucial to antibody diversification. To examine the effects of aging on SHM targeting, we analyzed B cell immunoglobulin repertoire sequences from 27 healthy male and female human subjects aged 20 to 89. By studying mutation patterns based on 985,069 mutations obtained from 123,415 sequences, we found that the SHM mutability hierarchies on micro-sequence motifs (i.e., SHM hot/cold spots) are mostly consistent between different age and sex groups. However, we observed a lower frequency in mutations involving Phase II SHM DNA repair activities in older males, but not in females. We also observed from a separate study a decreased expression level of DNA mismatch repair genes involved in SHM in older individuals compared with younger individuals, with larger fold changes in males than in females. Finally, we showed that the balance between Phase I vs. Phase II SHM activities impacts the resulting immunoglobulin phenotypes. Our results showed that the SHM process is altered in some older individuals, providing insights into observed clinical differences in immunologic responses between different age and sex groups.
- SHM hot- and cold-spot biases are largely consistent between different age and sex groups
- A shift in SHM targeting is observed between young and older male subjects
- This shift in SHM targeting influences the Ig amino acid composition
INTRODUCTION
Age and sex are important sources of variation in immune responses. Older individuals tend to be more vulnerable to bacterial and viral infections, more prone to developing cancer and autoimmune diseases, and less likely to respond to vaccinations (1–3). The age effects on the immune system, however, are not uniform between sexes (4). Females tend to display a more robust immunologic response to infections and vaccinations, yet females are also at a higher risk of developing autoimmune diseases (5–8). Though the age and sex biases have long been observed, the molecular mechanisms causing these differences remain unclear. Gaining an understanding of the molecular mechanisms is crucial for designing more targeted therapies and vaccinations for different age and sex groups.
Among many immune cells that experience immunosenescence, B cells are of great interest due to their central role in humoral immunity. B cells produce antibodies that defend us from a wide range of extracellular pathogens, and bind to cells to elicit antibody-dependent cytotoxicity. Moreover, most vaccines for viral and bacterial infections to date target B cells and utilize their memory response to prevent future infections. Previous studies have shown that though the quantity of antibodies does not alter significantly between young and aged individuals, older individuals display a decrease in the affinity, specificity and diversity of the antibodies they produce (9–12). Such observations prompted us to examine the mechanisms governing the antibody diversification process.
To generate potent B cell receptors (BCRs), which are the membrane-bound form of antibodies, the immunoglobulin (Ig) loci in B cells undergo a number of diversification steps, including V(D)J recombination and somatic hypermutation (SHM) (13). SHM, a process that occurs in germinal centers (GCs) in secondary lymphoid organs, diversifies B cell receptors (BCRs) by introducing point mutations into the immunoglobulin genes at a high rate (14). B cells with mutations leading to a high affinity to antigens survive and clonally expand in a selection process. SHM is initiated when the enzyme activation-induced cytidine deaminase (AID) converts cytosine (C) to uracil (U) in the Ig gene in B cells. Following this, three scenarios may occur. In Phase Ia, a simple transcription leads to a C-to-T or a G-to-A transition on the complement strand. In Phase Ib, the activation of UNG-induced short-patch base-excision repair (BER) pathway introduces mutation at the targeted C or G site (hereafter written as C/G site). Phase II results in the activation of long-patch BER and mismatch repair (MMR) pathways, leading to mutations on neighboring bases that can be on any nucleotide, including A and T site (hereafter written as A/T site). Although stochastic, SHM preferentially targets certain DNA motifs (hot spots) while avoiding others (cold spots) (15). A previous study shows a decrease in AID expression level in older individuals, suggesting that a lower degree of SHM initiation may contribute to the compromised immunologic responses in these individuals (16). However, there has been a lack of investigation on sex biases or the impact of aging on SHM targeting, which dictates the types of mutations resulting from SHM.
With the advancement of high-throughput technologies, we can now profile immune repertoires using Adaptive Immune Receptor Repertoire sequencing (AIRR-seq) at an unprecedented depth (17, 18). Such technologies involve sequencing the immunoglobulin loci in DNA or RNA, enabling us to examine mutation patterns in B cell receptors in great detail. For example, it has been observed that aging is associated with longer CDR3 regions (the most diverse region in B cell immunoglobulin genes), a trend toward the accumulation of more highly mutated IgM and IgG Ig genes, larger persistent clones in B cells in peripheral blood, and diminished intralineage diversification (19–21). Previously, we used such datasets to study SHM targeting patterns across individuals and species at a high resolution (22, 23). We hypothesized that such approaches may also provide insights into the effects of age and sex in the SHM targeting process.
In this study, we aimed to compare and contrast the SHM targeting patterns between different age and sex groups. We analyzed high-throughput B cell immunoglobulin sequences from peripheral blood of 27 healthy individuals from two age groups: younger individuals (20–31 years old) and older individuals (61–89 years old) (19). As a validation, we examined mutations in Ig genes in whole-genome RNA-seq data from 147 individuals aged 20 to 70 in the Genotype-Tissue Expression (GTEx) project (24). We examined mutations involved in Phase I vs. Phase II SHM and found a significant decrease in Phase II-induced mutations in older males. Such observations lead to the hypothesis that older individuals, particularly males, experience alterations in SHM activities, which may contribute to their lower level of immunologic responses.
MATERIALS AND METHODS
Repertoire Sequencing Data Processing and Clonotype Assignment
The high-throughput Ig repertoire AIRR-seq dataset was published in (19) by Wang et al. Raw high-throughput sequencing reads were quality controlled, assembled, and filtered using pRESTO (25). IMGT/High-VQUEST was used to assign germline V(D)J segments and determine functionality (26). Change-O command line tool was used to partition sequences into clonal groups (27). V(D)J sequences were assigned into clones based on identical IGHV gene, IGHJ gene, and junction length, with a weighted intraclonal distance threshold of 10 using the substitution probabilities previously described (28). Mutations were defined as nucleotides that were different from the germline sequence. A sliding window approach was used to filter out sequences with highly clustered mutations, defined by 6 or more nucleotide differences in 10 consecutive nucleotides. Sequences with less than 5% mutation frequency were filtered out to reduce the effect of sequencing errors in the analysis.
Targeting and Substitution Models
The 1-mer substitution model was generated using the shazam R package version 0.1.1 in the Change-O suite (22, 27). In brief, substitution matrix is defined by calculating the frequency of substitutions for each nucleotide. The 5-mer targeting model used for the construction of S (synonymous) 5-mer models is previously described (22).
GTEx RNA-seq dataset
The raw whole-genome RNA-seq data was collected by the Genotype-Tissue Expression (GTEx) project (v6) (24). The IgH locus was isolated and mapped to the germline Ig genes published by IMGT via IgBLAST (29). Reads with more than 65 nts (out of 75 nts) aligned with a mutation frequency between 75% and 95% were used for mutation analyses. Mutations from different tissues were aggregated for each individual. The A/T mutation frequency was normalized by the total number of A and T nucleotides in the germline sequences. Individuals with greater than 0.8 or less than 0.2 A/T mutation frequency were considered outliers and excluded from the analysis.
Gene Expression Analysis
Gene expression data of healthy individuals of different age and sex groups was obtained from GEO GSE65442 (30). Subject metadata were accessed from ImmPort SDY404. The baseline data of B cells prior to vaccination from the 2011–2012 season, which has at least 4 individuals in each age and sex group, was selected for the analysis. The gene expression profiles were quantile normalized using R package limma. The genes involved in SHM repair activities, including AICDA, EXO1, MSH2, MSH6, PMS2, POLH, POLI, POLK, POLL, POLQ, REV1, UNG, UNG2, were considered in the initial analysis (13). Probes with low detection signals, i.e., those with less than 90% of samples having a gene expression detection p-value less than 0.01, were eliminated from downstream analyses.
Criteria for Frailty
Operational definition of frailty is based on the following criteria: a) low grip strength, b) low energy, c) slowed walking speed, d) low physical activity, e) unintentional weight loss. Individuals with three to five symptoms were defined as frail.
RESULTS
B cell immunoglobulin repertoire sequencing data
To assess the effect of aging on SHM targeting, we analyzed a publicly available B cell immunoglobulin repertoire sequencing (AIRR-Seq) data from 27 healthy individuals (hereafter referred to as the “AIRR-seq” dataset) (19). The study includes ten young adults (20–31 yr old), five of whom are females, and seventeen older adults (61–89 yr old), twelve of whom are females. Within each age group, male and female subjects were of comparable ages (Young: p = 0.65, Older: p = 0.83; Welch’s t-test).
High-throughput next generation sequencing was used to profile the Ig sequences. For each individual, three replicates were obtained from different sequencing methods: genomic data sequenced at Stanford Genomic Center (DNA-GC), genomic data sequenced on the Roche 454 platform (DNA-Roche), and cDNA data from RNAs (RNA). We analyzed each replicate separately to prevent potential biases that arise from sequencing methods. A total of 123,415 unique sequences passed quality control (Materials and Methods) and were used for the downstream analyses (Table 1). IMGT was used to determine the functionality of the Ig sequences (26). Non-functional sequences were defined as sequences containing out-of-frame junction region and were presumably not having participated in selection. To identify independent SHM events, we partitioned the sequences into groups that were likely to be clonally-related (Materials and Methods), and identified a set of distinct mutations from each clone. We collected a total of 985,069 somatic hypermutations, where mutations were defined as the nucleotides that were different from the inferred germline sequences in the V region. From the RNA replicate where isotype information is available, we found that the majority of the mutations were obtained from sequences of the IgG, IgA and IgM isotypes. To study intrinsic SHM targeting patterns in the absence of selection pressure, we analyzed two types of mutations from each replicate: 1) silent mutations (i.e., mutations that do not alter the amino acid sequence) from functional sequences, and 2) both silent and replacement mutations from non-functional sequences (i.e., sequences that do not encode proteins).
Table 1:
Sequencing data in the AIRR-seq dataset
| No. of Unique Sequences | No. of Clones | No. of Mutations | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
|
| ||||||||||
| SAMPLE | Processed | Functional | Non-functional | Total | Functional | Non-functional | Functional | Non-functional | Functional Silent | Non-functional Silent |
| DNA-GC | 24,901 | 20,092 | 3,858 | 17,615 | 13,708 | 3,216 | 141,409 | 34,319 | 53,760 | 10,486 |
| DNA-Roche | 27,951 | 22,720 | 4,197 | 19,600 | 15,387 | 3,478 | 163,958 | 38,038 | 61,953 | 11,440 |
| RNA | 70,563 | 66,719 | 3,301 | 40,987 | 37,775 | 3,026 | 562,764 | 44,581 | 208,108 | 15,906 |
|
| ||||||||||
| TOTAL | 123,415 | 109,531 | 11,356 | 78,202 | 66,870 | 9,720 | 868,131 | 116,938 | 323,821 | 37,832 |
Aging does not influence SHM targeting hierarchy on micro-sequence motifs
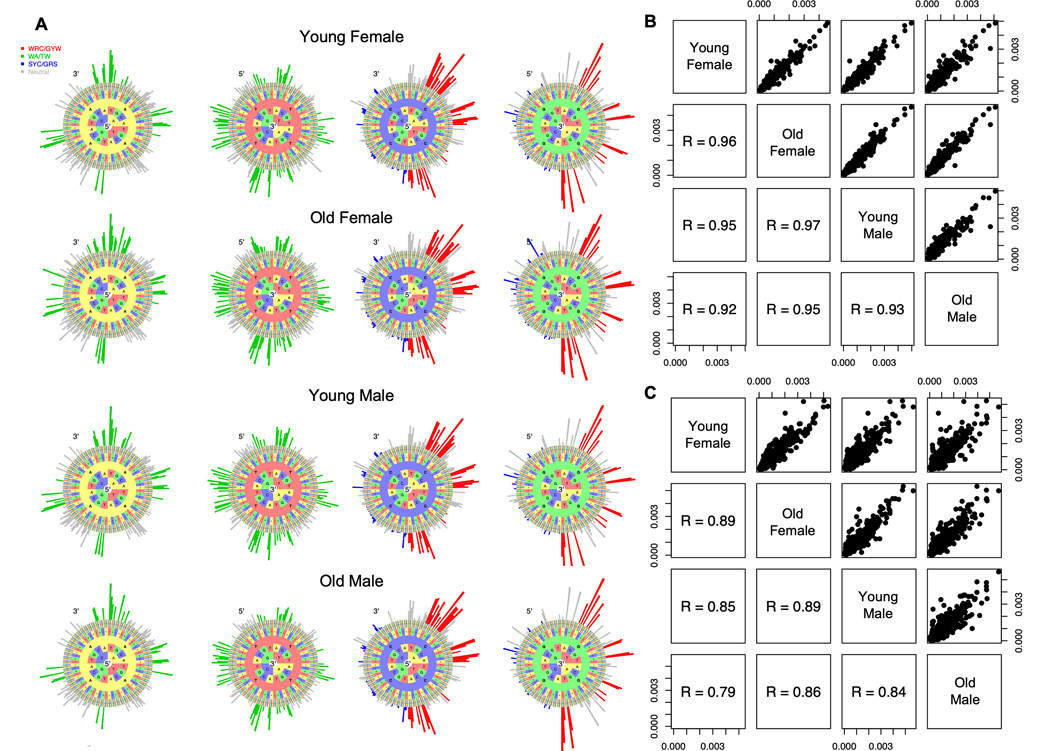
To assess whether aging influences the intrinsic biases of SHM targeting (i.e. hot/cold spots), we built targeting models based on micro-sequence motifs. To capture the classic SHM hotspot (WRCY/RGYW) on both the forward and reverse strands, the model was based on 5-mer motifs including 2 bases up- and 2 bases down-stream of the mutated base (Materials and Methods) (22). The mutability of each 5-mer is defined as the probability of the central base being targeted by SHM relative to all other motifs. Since 5-mer mutability hierarchies are highly similar across individuals (22), we built a single targeting model for each age and sex group. We then used “hedgehog” plots to visualize the targeting biases (i.e., hot/cold-spot motifs) (Figure 1A). In all age and sex groups, we observed high mutability in classic hot spots and low mutability in classic cold spots, consistent with our expectations. We compared the targeting models between age and sex groups, and found that the mutability hierarchies were highly consistent between different ages and sexes (Spearman ρ > 0.92 in silent mutations in functional sequences, and ρ > 0.79 in both silent and replacement mutations in non-functional sequences in the RNA replicate) (Figure 1B, C). The results suggest that age and sex do not influence the hierarchy of SHM targeting on micro-sequence motifs.
Figure 1: SHM mutability hierarchy on micro-sequence motifs is consistent across age and sex groups.
A) SHM targeting model for each age and sex group. Hedgehog plots depict the relative targeting of 5-mer motifs centered on the bases A, T, C, and G, with each center base in an individual circle. Classic hot spots are colored in red (WRC/GYW) and green (WA/TW), whereas cold spots are colored in blue (SYC/GRS). B) Scatterplot of 5-mer mutabilities between each age and sex group built on silent mutations in functional sequences. C) Scatterplot of 5-mer mutabilities between each age and sex group built on all mutations in non-functional sequences. Each dot represents the mutability value of a 5-mer motif. R denotes Spearman correlation coefficient.
Aging is associated with a shift in SHM repair activities in males
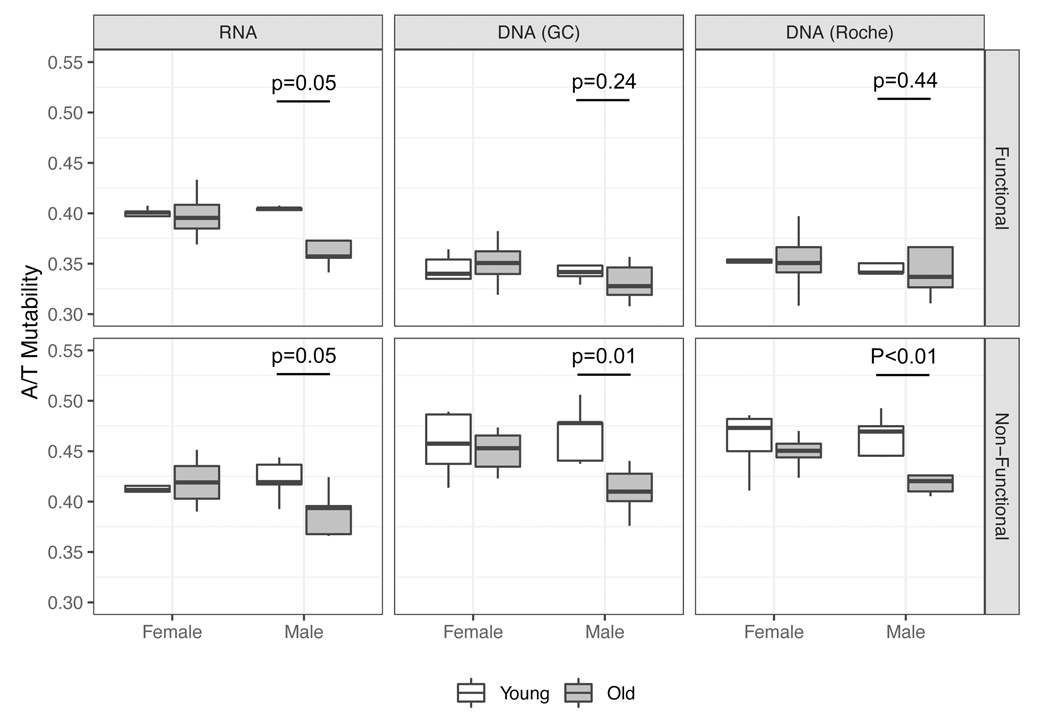
In SHM, C/G nucleotides are targeted in both Phase I and Phase II, while A/T nucleotides are targeted in Phase II only. To detect whether there is a shift in SHM phases between age and sex groups, we examined the balance between mutations at C/G nucleotides vs. mutations at A/T nucleotides. For each individual, we computed the proportion of mutations that occur on A/T bases, normalized by the number of each base in the germline sequences (hereafter written as A/T mutability). Across all technical replicates, we observed a significant decrease in A/T mutability in older males in non-functional sequences, with at least four of the five older males having lower mutations on A/T than all of the younger males (Figure 2). We also observed a decrease in A/T mutability in older males in silent mutations from functional sequences, and the difference was statistically significant in the RNA replicate. In the male group, there may be a continuous decrease in A/T mutability with age (ρ < −0.70, p < 0.03, Pearson product-moment correlation in the RNA replicate) (Figure S1), though the level of continuity is unclear in the 31 to 60-year age group due to the lack of subjects. In females, the older individuals have comparable mutation patterns as the younger individuals. The older males also exhibit lower A/T mutabilities compared with older females across all replicates.
Figure 2: Older males display a lower level of A/T mutability in SHM.
A/T mutability was calculated by normalizing frequency of unselected mutations occurring on A or T nucleotides by frequency of A or T bases in germline immunoglobulin sequences for each individual in the AIRR-seq dataset. Unselected mutations were obtained from silent mutations in functional sequences (Functional) or all mutations in non-functional sequences (Non-functional). Boxes represent the median and interquartile range of A/T mutabilities in each age and sex group. The analysis was performed in all three technical replicates: RNA (left panels), DNA-GC (middle panels), DNA-Roche (right panels). p-values were obtained from Welch’s t-test.
We used an RNA-seq dataset published by the Genotype Tissue Expression (GTEx) project to validate the results (hereafter referred to as the “GTEx RNA-seq” dataset) (24). This dataset contains RNA sequences from the entire transcriptome across many cell types, and therefore includes a limited number of mutated B cell immunoglobulin transcripts. We extracted the IgH locus and mapped the cDNA sequences to germline Ig genes using IgBLAST (Materials and Methods). The resulting data contains 11,312 mutations from 147 individuals. To maximize the number of mutations we can obtain per individual, sequences from different tissue sites were aggregated. We again calculated the A/T mutability by normalizing the proportion of mutations on A/T bases by the number of A/T nucleotides in the germline genes. Although the data contains a large amount of variation due to a small number of mutations that can be extracted from such type of data, we observed a similar trend as before: both sex groups display a decrease in A/T mutability with age, but the males have a much more prominent decrease (Pearson ρ = −0.24, p = 0.023 in males vs. Pearson ρ = −0.13, p = 0.363 in females) (Figure S2). By applying linear models, we observed that A/T mutabilities in males decrease at a slightly faster rate (a mutability value of 0.5826 at age 0, decreasing by 0.0018 per year) compared with their female counterparts (a mutability value of 0.5291 at age 0, decreasing by 0.0011 per year). The decrease in A/T (vs. C/G) mutability in older males suggested that these individuals have fewer mutations involved in Phase II SHM, which are induced by the activation of the long-patch base excision repair and MSH2/MSH6 DNA mismatch repair pathway that drives mutation spreading from the original AID-targeted C-to-U lesion to neighboring bases.
To determine whether there is also a shift between Phase Ia and Ib SHM repair pathways (i.e., simple replication vs. UNG-induced short-patch base excision repair at the AID-targeted C/G bases), we examined the frequency of C-to-T and G-to-A mutations (Phase Ia mutations) with respect to all mutations on C or G bases (Phase I mutations) in the AIRR-Seq dataset. To observe this, we computed the frequency of each substitution (Figure S3A), and normalized them by the total number of substitutions from that base (Figure S3B). While there appears to be an insignificant elevation of Phase Ia mutations before normalization, this elevation disappears after normalization. We tested the hypothesis on the GTEx RNA-seq dataset, and again did not see any correlation between age and Phase Ia mutation frequency. Thus, our analysis does not support a shift between Phase Ia and Phase Ib with age in these datasets.
Older males display a decrease in gene expression levels in molecules involved in SHM
The observation of a decreased mutation frequency occurring on A and T bases may suggest that the DNA repair mechanisms involved in SHM are altered in some older individuals. To assess whether DNA repair molecules involved in SHM have a different gene expression level, we analyzed the gene expressions profiles of peripheral blood B cells collected from another cohort of healthy individuals (see Materials and Methods; hereafter referred to as the “gene expression” dataset). This cohort included 16 younger (including 7 male) and 14 older (including 4 male) subjects. B cells were sorted from blood samples and the gene expression levels of 47,260 probes were measured (30).
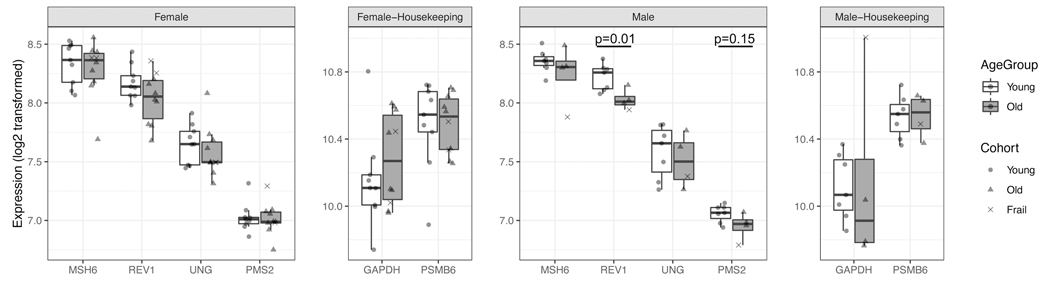
We examined the genes involved in SHM that were measured with significant detection level on the microarray (detection p < 0.01 for >90% of the samples), including MSH6, PMS2, REV1 and UNG. We compared the expression levels between the younger and older individuals in each sex group, and found that these genes consistently displayed lower expression values in older males compared with younger males (Figure 3). In particular, REV1 exhibits the largest gap between age groups (p = 0.01, Welch’s t-test). In females, the expressions were comparable between age groups, though the older individuals had slightly lower expressions in most of the genes. Next, we examined whether the decrease in SHM repair activities was linked to health state. We divided the older individuals into “healthy” and “frail” categories based on the criteria described in Materials and Methods. There were a total of 11 healthy and 3 frail older individuals. Interestingly, the frail male showed lowest expression in multiple genes among the male group, though such trend was not observed in the female group. These findings of gene expression changes in SHM molecules further demonstrate that older males may experience alterations in SHM targeting activities.
Figure 3: Older males display a decrease in the expression of DNA repair genes involved in SHM.
Log transformed expressions of SHM-repair genes with high detection levels in the gene expression dataset (described in Materials and Methods). Housekeeping genes were included as a control. Boxes represent the median and interquartile range of gene expressions in each age and sex group. p-values were obtained from Welch’s t-test.
Balance between phase I vs. phase II SHM repair activities impacts immunoglobulin phenotype
We hypothesized that an alteration in somatic hypermutation targeting at the nucleotide level would affect the resulting Ig phenotypes, and consequently contribute to a different immunological response. To detect the impact of a shift in A/T mutability on Ig amino acid composition, we examined the characteristics of amino acid substitutions by computationally translating the Ig nucleotide sequences into amino acids. We hypothesized that a decrease in A/T mutability would result in a decrease in the amino acid substitutions that occur due to mutations at A/T bases. Therefore, we examined a list of amino acid substitutions that would occur from a single nucleotide change unambiguously occurring at an A/T site (Table 2). For example, the change from phenylalanine to serine was included in the list because the nucleotide T must be converted to C in order to induce this amino acid exchange in a single mutational event. On the other hand, the change from serine to threonine was not included because it could be induced by mutations on T or G. There are 57 amino acid substitutions that match the criteria, with the most common ones being tyrosine (Y) to phenylalanine (F) or histidine (H), lysine (K) to arginine (R), valine (V) to alanine (A), and serine (S) to glycine (G).
Table 2:
Amino acid substitutions that are influenced by mutations on A/T nucleotides
| From | F | L | I | M | V | S | T | Y | N | K | D | E | C | W |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| To | I | S | F | L | A | P | P | H | Y | Q | V | V | R | R |
| V | W | L | V | D | A | A | N | H | E | A | A | G | G | |
| S | V | T | E | G | D | D | I | G | G | |||||
| Y | T | K | G | F | I | M | ||||||||
| C | N | R | S | T | T | |||||||||
| K | C | S | R | |||||||||||
| S | ||||||||||||||
| R |
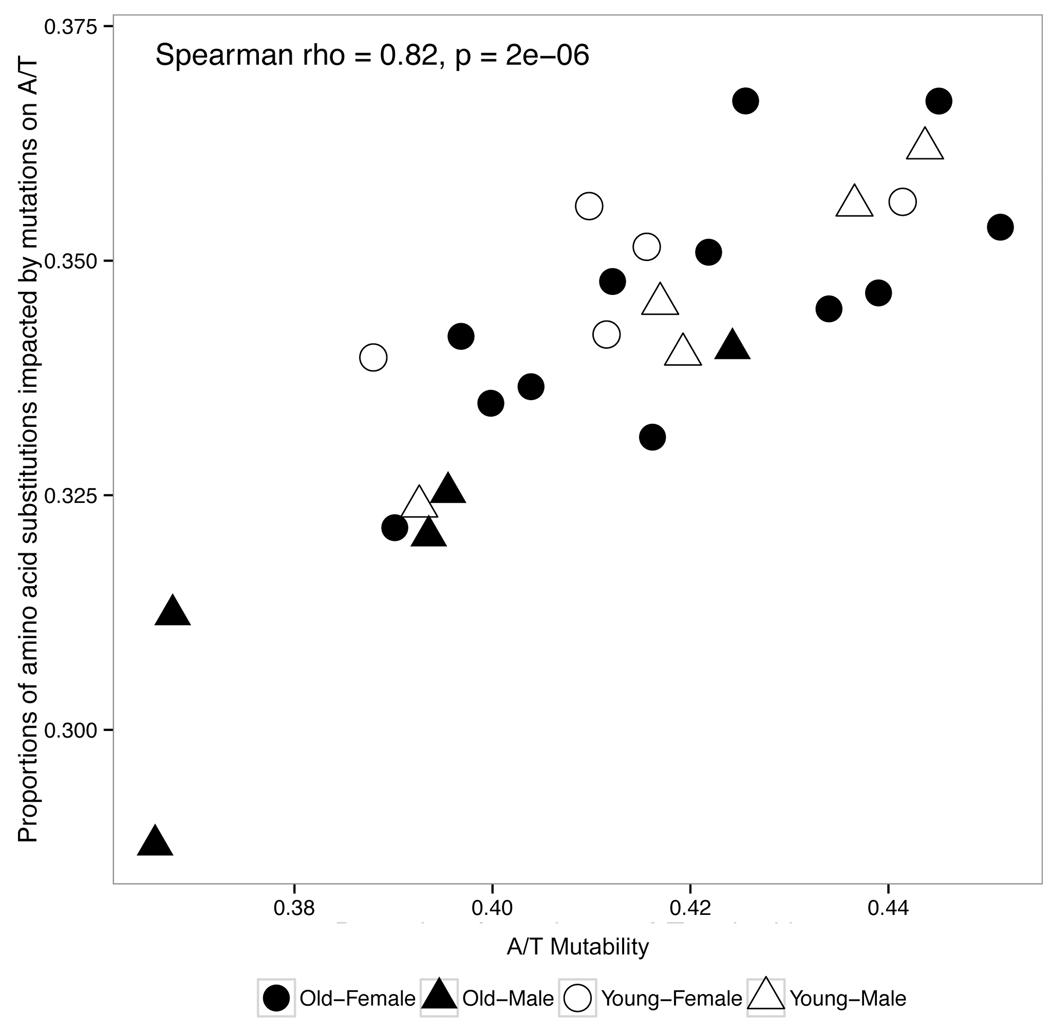
For each individual, we calculated the proportion of such amino acid substitutions on functional sequences (i.e., sequences that encode BCRs), and compared it to the A/T mutability in non-functional sequences, which reflects the level of SHM targeting activities not influenced by selection pressure. We found that a lower A/T mutability is significantly correlated with a lower frequency of amino acid substitutions that result from mutations at A/T sites for all replicates in the AIRR-Seq dataset (RNA: Spearman ρ = 0.82, p = 2e-6) (Figure 4 and Figure S4). We observed that younger individuals cluster at the top right corner, which represents a higher A/T mutability and a higher frequency of amino acid substitutions due to such mutations at the nucleotide level. Older males and some older females occupy the bottom right corner, indicating a decrease in the proportion in amino acid substitutions involving A/T mutations. These results indicate that a shift in Phase I vs. Phase II mutations affect the resulting immunoglobulin phenotype even if they are under selection pressure. As different amino acids have distinct properties (i.e., sizes, charges and hydrophobicity), alterations in Ig amino acid composition would potentially impact immunologic responses, such as causing changes in Ig binding affinity.
Figure 4: Changes in somatic hypermutation targeting affects immunoglobulin phenotype.
Comparison between A/T mutability found in non-functional sequences (reflecting intrinsic SHM targeting biases) and the proportion of amino acid substitutions that are exclusively caused by A/T nucleotide changes in functional sequences (reflecting properties of amino acid sequences encoding functional BCRs) in the AIRR-seq dataset. Each point represents an individual; their corresponding age group is indicated by shade and sex group is indicated by shape. The analysis was performed in all three technical replicates: RNA (shown here), DNA-GC and DNA-Roche (Figure S4).
DISCUSSION
Elucidating age and sex discrepancies in immune responses at the molecular level can provide mechanistic insights to the differences in immune responses observed in clinical settings. Though it is well established that males and females of different age groups have different immune responses, the mechanisms responsible for the lack of antibody diversity and potency for certain groups are yet to be clarified. In this study, we analyzed high-throughput sequencing and gene expression datasets to study an antibody diversification process in different age and sex groups. We analyzed a large number of mutations gathered from high-throughput Ig sequencing data, and used 1-mer and 5-mer micro-sequence targeting frameworks to show a shift in A/T vs. C/G targeting in older males. We used RNA-seq data from 147 individuals to validate the patterns observed. We also used gene expression profiles of an independent study to show that males have decreased levels of expression in several genes crucial to SHM targeting activities. Finally, we showed that a shift in the balance between Phase I and Phase II SHM at the nucleotide level measured in the absence of selection pressure influences the amino acid compositions of functional immunoglobulins, potentially altering immunologic responses.
It has been observed that the antibodies generated by older individuals have a lower level of affinity, specificity and diversity. However, the molecular processes responsible for such declines have not been elucidated. Previous repertoire analyses have observed alterations in repertoire properties in older individuals, such as CDR3 lengths and clonal sizes. Here we studied DNA repair pathways involved in SHM, processes upstream of these observations, and found that the mutations involved in different phases of SHM repair pathways are altered in older individuals. Using one of the cohorts analyzed here (19), we previously found a decrease in WA hot-spot targeting with age (31). However, unlike the sex-specific differences observed here in targeting of A/T more generally, these hot-spot differences were similar in males and females. Our finding that males exhibit much more prominent alterations in these pathways with age is consistent with the clinical finding that older men have a weaker response to vaccination and are more likely to develop infections or cancer. One explanation based on this study would be that the Phase II SHM process is less efficient in older males, resulting in a less optimal antibody repertoire in response to antigens.
Aside from aging, other factors may also affect SHM targeting, such as genetic factors and health state. Despite the fact that many factors can impact one’s immunity, we found significant correlations between A/T mutability and age in males. It is interesting that the frail male showed the largest decrease in the major SHM repair molecules; this might suggest that a lack of SHM repair activities is associated with weakened immune response. Future studies should examine the role of sex-specific molecules in the SHM process. For example, a study revealed the immunosuppressive function of testosterone in response to influenza vaccination, indicating that sex hormones could play a role (32).
It is interesting to note that many DNA repair enzymes and polymerases are involved in both antibody diversification and DNA damage repair in other parts of the genome. A previous study observed a correlation between microsatellite stability, particularly PMS2 expression, and types of mutations in B cell repertoires (33). Here, we observed a slight reduction of PMS2 expression in older males, which may reflect their conclusion on the influence of mismatch repair activity on the types of mutations generated through SHM. Future studies should examine the pleiotropic effects of DNA repair molecules. For example, individuals with deficiency in DNA repair genes may not only have an impaired antibody repertoire, but also a higher predisposition to diseases caused by DNA damages, such as cancer.
A limitation of the gene expression dataset is that the B cells were collected from peripheral blood instead of the germinal center where SHM takes place. Because of this, we found that some genes encoding important molecules in SHM, such as AID and pol-η, were not highly expressed, limiting our ability to study all genes involved in SHM. Additionally, other machineries, aside from gene expression of SHM-related molecules, may also influence SHM targeting. This prompts future studies to examine B cells in germinal centers to see what other DNA repair pathways contribute to the aging effects that we observed. A limitation of the GTEx RNA-seq dataset is the small number of B cell Ig genes one can extract from genome-wide RNA-seq data on all cell types in a tissue. The AIRR-seq dataset has many more mutations per individual, though has fewer subjects. However, the fact that both AIRR-Seq and GTEx RNA-seq analyses yielded consistent findings improves our confidence in the sex biased aging effects in SHM targeting. In the future, larger cohorts of individuals should be recruited to characterize the extent to which age and sex can influence SHM targeting activities, and which genes experience the most alteration in expression levels with age. Nevertheless, this study showed for the first time that age and sex could influence the balance between the two phases of SHM targeting, which is crucial to antibody diversification.
In conclusion, our analyses on BCR sequencing and B cell gene expression data showed alterations in SHM mutation patterns and gene expressions between age and sex groups. These results suggest an uneven rate of alteration in SHM DNA repair machineries between males and females across lifetime, providing insights into designing more targeted treatment and vaccination strategies for different age and sex groups.
Supplementary Material
ACKNOLEDGEMENTS
The authors thank the Kleinstein lab members for their feedback and technical assistance and Dr. Scott Boyd for sharing data. The authors thank the Yale University Biomedical High Performance Computing Center and the Broad Institute Computing Center for use of their computing resources.
This work was supported by grants from the National Institute of Health R03AI092379 and R01AI104739 to SHK and a Natural Sciences and Engineering Research Council of Canada postgraduate fellowship (NSERC PGS-M) to AC. The Yale University Biomedical High Performance Computing Center is funded by National Institutes of Health grants RR19895 and RR029676-01. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
LIST OF ABBREVIATIONS
- SHM
somatic hypermutation
- AID
activation-induced cytidine deaminase
- Ig
Immunoglobulin
- GC
germinal center
- BER
base-excision repair
- MMR
mismatch repair
References:
- 1.Weinberger B, Herndler‐Brandstetter D, Schwanninger A, Weiskopf D, and Grubeck‐Loebenstein B. 2008. Biology of Immune Responses to Vaccines in Elderly Persons. Clin. Infect. Dis. 46: 1078–1084. [DOI] [PubMed] [Google Scholar]
- 2.Castle SC 2000. Clinical relevance of age-related immune dysfunction. Clin. Infect. Dis. 31: 578–85. [DOI] [PubMed] [Google Scholar]
- 3.Castelo-Branco C, and Soveral I. 2014. The immune system and aging: A review. Gynecol. Endocrinol. 30: 16–22. [DOI] [PubMed] [Google Scholar]
- 4.Márquez EJ, Chung C-H, Marches R, Rossi RJ, Nehar-Belaid D, Eroglu A, Mellert DJ, Kuchel GA, Banchereau J, and Ucar D. 2020. Sexual-dimorphism in human immune system aging. Nat. Commun. 11: 751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beeson PB 1994. Age and sex associations of 40 autoimmune diseases. Am. J. Med. 96: 457–62. [DOI] [PubMed] [Google Scholar]
- 6.Klein SL, Marriott I, and Fish EN. 2015. Sex-based differences in immune function and responses to vaccination. Trans. R. Soc. Trop. Med. Hyg. 109: 9–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fish EN 2008. The X-files in immunity: sex-based differences predispose immune responses. Nat. Rev. Immunol. 8: 737–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gubbels Bupp MR 2015. Sex, the aging immune system, and chronic disease. Cell. Immunol. 294: 102–10. [DOI] [PubMed] [Google Scholar]
- 9.Dunn-Walters DK 2016. The ageing human B cell repertoire: a failure of selection? Clin. Exp. Immunol. 183: 50–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dunn-Walters DK, Banerjee M, and Mehr R. 2003. Effects of age on antibody affinity maturation. Biochem. Soc. Trans. 31: 447–8. [DOI] [PubMed] [Google Scholar]
- 11.Weiskopf D, Weinberger B, and Grubeck-Loebenstein B. 2009. The aging of the immune system. Transpl. Int. 22: 1041–50. [DOI] [PubMed] [Google Scholar]
- 12.LeMaoult J, Szabo P, and Weksler ME. 1997. Effect of age on humoral immunity, selection of the B-cell repertoire and B-cell development. Immunol. Rev. 160: 115–26. [DOI] [PubMed] [Google Scholar]
- 13.Peled JU, Kuang FL, Iglesias-Ussel MD, Roa S, Kalis SL, Goodman MF, and Scharff MD. 2008. The biochemistry of somatic hypermutation. Annu. Rev. Immunol. 26: 481–511. [DOI] [PubMed] [Google Scholar]
- 14.McKean D, Huppi K, Bell M, Staudt L, Gerhard W, and Weigert M. 1984. Generation of antibody diversity in the immune response of BALB/c mice to influenza virus hemagglutinin. Proc. Natl. Acad. Sci. U. S. A. 81: 3180–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rogozin IB, and a Kolchanov N. 1992. Somatic hypermutagenesis in immunoglobulin genes. II. Influence of neighbouring base sequences on mutagenesis. Biochim. Biophys. Acta 1171: 11–8. [DOI] [PubMed] [Google Scholar]
- 16.Frasca D, Landin AM, Lechner SC, Ryan JG, Schwartz R, Riley RL, and Blomberg BB. 2008. Aging down-regulates the transcription factor E2A, activation-induced cytidine deaminase, and Ig class switch in human B cells. J. Immunol. 180: 5283–90. [DOI] [PubMed] [Google Scholar]
- 17.Benichou J, Ben-Hamo R, Louzoun Y, and Efroni S. 2012. Rep-Seq: uncovering the immunological repertoire through next-generation sequencing. Immunology 135: 183–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rubelt F, Busse CE, Bukhari SAC, Bürckert J, Mariotti-Ferrandiz E, Cowell LG, Watson CT, Marthandan N, Faison WJ, Hershberg U, Laserson U, Corrie BD, Davis MM, Peters B, Lefranc M, Scott JK, Breden F, AIRR Community ET Luning Prak, and S. H. Kleinstein. 2017. Adaptive Immune Receptor Repertoire Community recommendations for sharing immune-repertoire sequencing data. Nat. Immunol. 18: 1274–1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang C, Liu Y, Xu LT, Jackson KJL, Roskin KM, Pham TD, Laserson J, Marshall EL, Seo K, Lee J-Y, Furman D, Koller D, Dekker CL, Davis MM, Fire AZ, and Boyd SD. 2014. Effects of aging, cytomegalovirus infection, and EBV infection on human B cell repertoires. J. Immunol. 192: 603–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jiang N, He J, Weinstein JA, Penland L, Sasaki S, He X, Dekker CL, Zheng N-Y, Huang M, Sullivan M, Wilson PC, Greenberg HB, Davis MM, Fisher DS, and Quake SR. 2013. Lineage structure of the human antibody repertoire in response to influenza vaccination. Sci. Transl. Med. 5: 171ra19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.de Bourcy CFA, Angel CJL, Vollmers C, Dekker CL, Davis MM, and Quake SR. 2017. Phylogenetic analysis of the human antibody repertoire reveals quantitative signatures of immune senescence and aging. Proc. Natl. Acad. Sci. U. S. A. 114: 1105–1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yaari G, Vander Heiden JA, Uduman M, Gadala-Maria D, Gupta N, Joel JN, O’Connor KC, Hafler DA, Laserson U, Vigneault F, and Kleinstein SH. 2013. Models of somatic hypermutation targeting and substitution based on synonymous mutations from high-throughput immunoglobulin sequencing data. Front. Immunol. 4: 358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cui A, Di Niro R, Vander Heiden JA, Briggs AW, Adams K, Gilbert T, OConnor KC, Vigneault F, Shlomchik MJ, and Kleinstein SH. 2016. A Model of Somatic Hypermutation Targeting in Mice Based on High-Throughput Ig Sequencing Data. J. Immunol. 197: 3566–3574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Consortium GTEx. 2013. The Genotype-Tissue Expression (GTEx) project. Nat. Genet. 45: 580–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vander Heiden J a, Yaari G, Uduman M, Stern JNH, O’Connor KC, a Hafler D, Vigneault F, and Kleinstein SH. 2014. pRESTO: a toolkit for processing high-throughput sequencing raw reads of lymphocyte receptor repertoires. Bioinformatics 30: 1930–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Alamyar E, Duroux P, Lefranc M-P, and Giudicelli V. 2012. IMGT(®) tools for the nucleotide analysis of immunoglobulin (IG) and T cell receptor (TR) V-(D)-J repertoires, polymorphisms, and IG mutations: IMGT/V-QUEST and IMGT/HighV-QUEST for NGS. Methods Mol. Biol. 882: 569–604. [DOI] [PubMed] [Google Scholar]
- 27.Gupta NT, Vander Heiden J. a., Uduman M, Gadala-Maria D, Yaari G, and Kleinstein SH. 2015. Change-O: a toolkit for analyzing large-scale B cell immunoglobulin repertoire sequencing data. Bioinformatics 31: 3356–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Smith DS, Creadon G, Jena PK, Portanova JP, Kotzin BL, and Wysocki LJ. 1996. Di- and trinucleotide target preferences of somatic mutagenesis in normal and autoreactive B cells. J. Immunol. 156: 2642–52. [PubMed] [Google Scholar]
- 29.Ye J, Ma N, Madden TL, and Ostell JM. 2013. IgBLAST: an immunoglobulin variable domain sequence analysis tool. Nucleic Acids Res. 41: W34–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Avey S, Mohanty S, Chawla DG, Meng H, Bandaranayake T, Ueda I, Zapata HJ, Park K, Blevins TP, Tsang S, Belshe RB, Kaech SM, Shaw AC, and Kleinstein SH. 2020. Seasonal Variability and Shared Molecular Signatures of Inactivated Influenza Vaccination in Young and Older Adults. J. Immunol. 204: 1661–1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hoehn KB, Vander Heiden JA, Zhou JQ, Lunter G, Pybus OG, and Kleinstein SH. 2019. Repertoire-wide phylogenetic models of B cell molecular evolution reveal evolutionary signatures of aging and vaccination. Proc. Natl. Acad. Sci. U. S. A. 116: 22664–22672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Furman D, Hejblum BP, Simon N, Jojic V, Dekker CL, Thiébaut R, Tibshirani RJ, and Davis MM. 2014. Systems analysis of sex differences reveals an immunosuppressive role for testosterone in the response to influenza vaccination. Proc. Natl. Acad. Sci. U. S. A. 111: 869–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rosner K, Winter DB, Kasmer C, Skovgaard GL, Tarone RE, Bohr VA, and Gearhart PJ. 2001. Impact of age on hypermutation of immunoglobulin variable genes in humans. J. Clin. Immunol. 21: 102–15. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.