Abstract
Enteropathogenic Escherichia coli (EPEC) and enterohemorrhagic E. coli (EHEC) constitute a significant risk to human health worldwide. A hallmark of both pathogens is their ability to produce characteristic attaching-and-effacing (A/E) lesions in intestinal epithelial cells. Genes encoding A/E lesion formation map to a chromosomal pathogenicity island termed the locus of enterocyte effacement (LEE). Intimin, an LEE-encoded bacterial adhesion molecule, mediates the intimate bacterium-host cell interaction characteristic of A/E lesions. On the basis of characterization of the C-terminal 280-amino-acid cell binding domain of intimin (Int280661–939), four distinct Int280 types (types α, β, γ, and δ) have been identified. Importantly, Int280α and Int280β antisera specifically recognized their respective intimin types. Using a conserved region of the intimin molecule (Int388–667) and primers synthesized to generate the recombinant Int388–667, we have now generated universal intimin antiserum and PCR primers that are reactive with the different intimin types expressed by both human and animal A/E lesion-forming strains. Use of immunogold electron microscopy to visualize intimin on the surfaces of EPEC and EHEC strains revealed, in general, a uniform distribution on the bacterial cell surface. However, a filamentous staining pattern was observed with a few strains expressing intimin γ. Cloning of the intimin eae gene from one such strain (strain ICC57) into strain CVD206, an EPEC strain which harbors a null deletion in eae, produced a uniform intimin staining pattern indicating that, if the filamentous staining pattern defines a filamentous form of intimin γ, it is dependent upon the genetic background of the strain and is not a feature of the intimin molecule.
Enteropathogenic Escherichia coli (EPEC) is a common cause of diarrhea, particularly among young infants in developing countries (for a review, see reference 28). Infection with EPEC is associated with a microscopic lesion of intestinal epithelial cells, the attaching-and-effacing (A/E) lesion (27), which is characterized by destruction of host cell microvilli and intimate attachment of bacteria to cup-like pedestals at the apical cell membrane (21, 30). A/E lesions are also induced by other enterobacteria, including enterohemorrhagic E. coli (EHEC), the causative agent of bloody and nonbloody diarrhea as well as of hemolytic-uremic syndrome in humans (for a review, see reference 28); Hafnia alvei, isolated from children with diarrhea (2); Citrobacter rodentium, the causative agent of transmissible colonic hyperplasia in laboratory mice (29); and rabbit-specific EPEC (REPEC) including rabbit diarrheagenic E. coli (RDEC-1), which cause diarrhea in rabbits (3).
Several genes (and their associated proteins) have been implicated in A/E lesion formation. All of these map to a pathogenicity island termed the locus of enterocyte effacement, or the LEE region (26). The LEE region encodes a type III secretion system (13); three associated EPEC-secreted proteins, proteins EspA (6), EspB (18), and EspD (23), required for protein translocation, signal transduction in host cells, and A/E lesion formation (for a review, see reference 9); an outer membrane adhesin, intimin (14); and a translocated intimin receptor, Tir (17). In addition to the LEE pathogenicity island, some EPEC strains also possess large EPEC adherence factor (EAF) virulence plasmids which encode a bundle-forming pilus important in colonization (4) and per regulatory genes (11). Strains which possess or lack EAF plasmids have been termed typical and atypical EPEC, respectively (15); EHEC also lack EAF plasmids.
Study of the intimin family of proteins has shown that their cell binding activity is localized to the C-terminal 280 amino acids (Int280661–939) (7) and that within this domain lies a 76-amino-acid loop formed by a disulfide bridge between two cysteines at positions 860 and 937 (16). This loop is required for intimin-mediated intimate attachment and invasion into cultured mammalian cells (8). In a human intestinal organ culture model of infection, intimin was essential for colonization of the mucosa and A/E lesion formation (12). Immunoglobulin A (IgA) antibodies to different EPEC intimins were shown to be present in colostrum from mothers in Brazil (25).
Recently, using antisera made against Int280, we have shown that the expression of intimin in EAF plasmid-positive EPEC strains is regulated by the EAF plasmid-encoded per locus and is influenced by growth phase and temperature (19). Moreover, using the anti-Int280 serum and PCR to investigate antigenic variation and classify the cell binding domain of intimin expressed by A/E lesion-forming bacterial pathogens, we identified four distinct intimin subtypes: intimin α, intimin β, intimin γ, and intimin δ (1). Importantly, intimin α was specifically expressed by a group of EPEC strains, all of which belong to one evolutionary branch of EPEC known as EPEC clone 1 (31), and H. alvei (1), while intimin β was mainly associated with EPEC and EHEC strains belonging to their respective clones 2, C. rodentium, and RDEC-1. Intimin γ was associated with EHEC O157:H7, EPEC O55:H7, and O55:H−, while intimin δ was expressed only by EPEC O86:H34. In this study we aimed to develop an intimin antiserum which is reactive with all the different intimin types and which can therefore be used to detect A/E lesion-forming E. coli. Here we report on the production of universal, broad-spectrum intimin primers and antiserum based on a domain which is conserved in all intimin types.
MATERIALS AND METHODS
Bacterial strains.
The bacterial strains used in this study included E. coli BL21; clinical EPEC isolate E2348/68 (O127:H6) (24), its EAF plasmid-cured derivative JPN15 (14), and its eae deletion mutant CVD206 (5); clinical isolate B171 (O111:H−) and its EAF plasmid-cured derivative B171-4 (10); clinical isolate ICC57 (O55:H7) (this study); and the other clinical human and animal isolates listed in Table 1. Other strains included in this study included H. alvei, enteroaggregative E. coli, enterotoxigenic E. coli, enteroinvasive E. coli, diffuse-adhering E. coli, E. coli K-12 HB101, E. coli K-12 harboring cloned Yersinia invasin [HB101(pRI203)], Salmonella typhimurium, Salmonella muenchen, Shigella flexneri, Yersinia enterocolitica, Yersinia pseudotuberculosis, Listeria monocytogenes, and Edwardsiella tarda. Bacterial strains were grown in L broth (1% tryptone, 0.5% yeast extract, 0.5% NaCl [pH 7.5]) or L agar (L broth containing 1.5% Bacto Agar) (Difco Laboratories). The medium was supplemented with 100 μg of ampicillin per ml or 30 μg kanamycin per ml where appropriate. For immunodetection of intimin in whole-cell extracts, stationary L-broth cultures were diluted 1:100 in Dulbecco's modified Eagle's medium (DMEM) and were incubated at 37°C for 3 h (1).
TABLE 1.
Characteristics of strains used in the study and Western blotting and PCR results for the strains
| Serotype | Intimin type | Host | No. of strains positive/ total no. tested
|
|
|---|---|---|---|---|
| Western blotting with Int388–667 | PCR with Int388–667 | |||
| O142:H6 | α | Human | 4/4 | 4/4 |
| O142:H34 | α | Human | 4/4 | 4/4 |
| O55:H6 | α | Human | 8/8 | 8/8 |
| O127:H6 | α | Human | 6/7 | 6/7 |
| O119:H6 | β | Human | 8/8 | 8/8 |
| O119:H2 | β | Human | 9/10 | 9/10 |
| O111:H2 | β | Human | 8/8 | 8/8 |
| O111:H− | β | Human | 1/1 | 1/1 |
| O128:H2 | β | Human | 4/5 | 4/5 |
| O26:H11 | β | Human | 1/1 | 1/1 |
| O26:H− | β | Human | 1/1 | 1/1 |
| O126:H− | β | Human | 2/2 | 2/2 |
| O114:H− | β | Human | 1/1 | 1/1 |
| O55:H7 | γ | Human | 2/3 | 2/3 |
| O157:H7 | γ | Human | 1/1 | 1/1 |
| O55:H− | γ | Human | 3/3 | 3/3 |
| O86:H11 | δ | Human | 1/1 | 1/1 |
| O86:H34 | δ | Human | 3/3 | 3/3 |
| O127:H40 | γ | Human | 4/4 | 4/4 |
| O157:H7 | γ | Cow | 1/1 | 1/1 |
| O157:H− | γ | Cow | 1/1 | 1/1 |
| O26 | β | Cow | 5/5 | 4/5 |
| O157:H− | γ | Sheep | 1/1 | 1/1 |
| O157:H7 | γ | Sheep | 1/1 | 1/1 |
| O26:H11 | β | Pig | 1/1 | 1/1 |
| O45 | β | Pigeon | NDa | 1/1 |
| O75 | β | Pigeon | ND | 1/1 |
| O15 | β | Pigeon | ND | 1/1 |
| O18 | β | Pigeon | ND | 1/1 |
| O128 | β | Pigeon | 1/1 | 1/1 |
| O2 | NCb | Osterich | 1/1 | 1/1 |
| NTc | β | Chick | 1/1 | 1/1 |
| O45 | β | Pig | ND | 1/1 |
| O49 | δ | Dog | ND | 1/1 |
| NT | α | Dog | 1/1 | 1/1 |
| NT | β | Buffalo | 1/1 | 1/1 |
| O26 | β | Roe deer | 1/1 | 2/2 |
| O110:H6 | NC | Bird | 3/3 | 3/3 |
| Osp:H6 | NC | Bird | 1/1 | 1/1 |
| O131:H− | NC | Bird | 1/1 | 1/1 |
| O63:H6 | NC | Bird | 1/1 | 1/1 |
| O109:H2 | NC | Rabbit | 1/1 | 1/1 |
| O128:H2 | NC | Rabbit | 1/1 | 1/1 |
| O8 | NC | Rabbit | 1/1 | 1/1 |
| O20:H7 | NC | Rabbit | 1/1 | 1/1 |
| O132:H2 | NC | Rabbit | 1/1 | 1/1 |
| O153:H7 | NC | Rabbit | 1/1 | 1/1 |
| O15:H− | NC | Rabbit | 1/1 | 1/1 |
| O103 | β | Rabbit | 4/4 | 5/5 |
| RDEC-1 | β | Rabbit | 1/1 | 0/1 |
| C. rodentium | β | Mouse | 1/1 | 0/1 |
ND, not done.
NC, Not classified.
NT, not typed.
Preparation of broad-spectrum intimin antiserum.
In order to produce an intimin antiserum reactive with all the intimin types, the fragment encoding the 280 amino acids upstream of the cell binding domain, residues Gly388 to Lys667, of eae from EPEC E2348/69 was amplified (see below) and was subcloned into the EcoRI and HindIII sites of pET28a (Novagen Biotechnology), and the recombinant plasmids were transformed into E. coli BL21. The pET28a vector, which encodes a kanamycin resistance marker, directs expression of cloned genes from an inducible T7 promoter as His tag fusions. Induced cultures were sonicated, and the soluble fraction was collected and purified on nickel columns as described previously (19). The purities of the polypeptide preparations were confirmed by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) analysis (see below). Female Sandy half-lop rabbits were immunized subcutaneously with 50 to 100 μg of the purified intimin antigen in complete Freund's adjuvant. The animals were boosted twice with the same antigen in incomplete Freund's adjuvant at 3-week intervals before exsanguination.
PAGE.
PAGE in the presence of SDS was performed as described previously (1, 22). Protein samples and bacterial extracts to be separated were diluted in an equal volume of 2× sample buffer (2% [wt/vol] SDS, 2% [vol/vol] 2-mercaptoethanol, 20% glycerol, and 0.01% [wt/vol] bromophenol blue in 0.0065 M Tris [pH 6.8]) and were boiled for 5 min prior to loading onto 7.5 to 10% gels. Molecular weights were estimated with Rainbow molecular markers (Amersham). Following electrophoresis, the separated proteins were visualized by staining the gel with Coomassie stain or were transferred to a nitrocellulose membrane.
Western blotting (immunoblotting).
Proteins separated by SDS-PAGE were transferred electrophoretically onto nitrocellulose membranes (Schleicher & Schuell) and were immunoblotted at 80 V for 90 min as described previously (1). The membranes were blocked overnight in 3% bovine serum albumin (BSA), washed three times with phosphate-buffered saline (PBS) containing 0.05% Tween 20 (PBST), and then reacted for 2 h with the Int388–667 serum diluted 1:1,000 in PBST containing 0.1% BSA. After three washes with PBST the bound antibodies were reacted with horseradish peroxidase-conjugated swine anti-rabbit serum (1:1,000 dilution; DAKO) and the membranes were developed with hydrogen peroxide and 3′,3′-diaminobenzidine (Sigma).
PCR.
PCR was used to amplify a segment of the eae gene encoding the conserved intimin domain. Thirty amplification cycles of 95°C for 20 sec, 45°C for 1 min, and 74°C for 1 min were used. A total of 25 pmol of each of the primers (primers Int-Fc [5′-CCG GAA TTC GGG ATC GAT TAC CGT CAT] and Int-Rc [5′-CCC AAG CTT TTA TTT ATC AGC CTT AAT CTC]) and 1.5 U of Taq DNA polymerase (Appligene, Durham, United Kingdom) were used. For each reaction, 1 μl of the diluted overnight cultures was transferred to a 0.5-ml tube containing the PCR mixture, and primers and the tubes were incubated at 95°C for 5 min prior to the PCR cycling. Ten microliters from each reaction mixture was analyzed by agarose gel electrophoresis.
Immunogold labelling of bacterial cells.
For immunogold labelling of bacteria, stationary-phase L-broth cultures of representative strains were diluted 1:100 in DMEM and were grown at 37°C for 4 h. A total of 10 μl of samples of washed bacterial suspensions was applied to carbon-coated grids for 5 min, excess liquid was removed, and the grids were immediately placed face down on drops of anti-Int388–667 serum (diluted 1:40 in PBS containing 0.2% BSA–PBS-BSA) for 30 min. After thorough washing in PBS-BSA, the grids were placed on drops of 10-nm gold-labelled goat anti-rabbit serum (diluted 1:20; British BioCell International) for 30 min. After further washing with PBS-BSA and distilled water, the grids were air dried and were examined in a Jeol 1200EX electron microscope operated at 80 kV.
Immunofluorescence labelling of bacterial cells.
Immunofluorescence staining was performed with bacteria adhering to HEp-2 cells following a 3-h incubation of HEp-2 cell monolayers with overnight cultures (1, 19). Formalin-fixed and washed, infected cell monolayers were incubated with anti Int388–667 antiserum (diluted 1:40) for 45 min. After three 5-min washes with PBS-BSA, the monolayers were stained with fluorescein isothiocyanate-conjugated goat anti-rabbit IgG (diluted 1:20; Sigma) for 45 min. When fluorescence actin staining (FAS) tests were to be performed, the HEp-2 cell preparations were permeabilized with Triton X-100 and were stained for cellular actin with a 5-μg/ml solution of tetramethyl rhodamine isocyanate-phalloidin (Sigma) (20). The preparations were washed three times with PBS, mounted in glycerol-PBS, and examined by incident light fluorescence with a Leitz Dialux microscope. Fluorescence and phase-contrast images of the same field were recorded.
Cloning of eae from ICC57.
Strain ICC57 chromosomal DNA, purified from 1-ml overnight bacterial cultures with the QIAamp tissue kit (QIAGEN), was used as a template. PCR was performed with the Gene Amp XL PCR kit (Perkin-Elmer) and primers orfU-F (5′-TTA TCT GAC ACT AAT GAC GAA TAT ATG ATG) and eae-R (5′-CCC AAG CTT TTA TTC TAC ACA AAC) primers with 28 cycles of 94°C for 1 min and 60°C for 10 min. The eae gene was cloned into the pGEM-T vector (Promega), which encodes an ampicillin resistance marker, to produce plasmid pICC57, which was transformed into CVD206. Recombinant plasmids were screened by PCR with the internal eae primers used as described above.
RESULTS
Production of a broad-spectrum intimin antiserum.
We have previously produced intimin antisera directed against the carboxy-terminal cell binding domains of intimin α and intimin β (1). Since these reagents were reactive only with specific subsets of A/E lesion-forming strains, the aim of this study was to generate a universal intimin antiserum reactive with all intimin types. For that purpose we cloned into pET28a, following DNA amplification, the 280 amino acids upstream of the cell binding domain (residues Gly388 to Lys667) of eae from EPEC E2348/69 (which encodes intimin α); this region of intimin is highly conserved in all the different intimins sequenced to date. The Int388–667 polypeptide was overexpressed in E. coli BL21, and the protein was purified on a nickel column and was used to raise rabbit polyclonal antiserum. The specificity of the antiserum was confirmed with the wild-type strain (strain E2348/69) and its eae-negative derivative (strain CVD206) on Western blots and by immunogold labelling electron microscopy (Fig. 1 and 2).
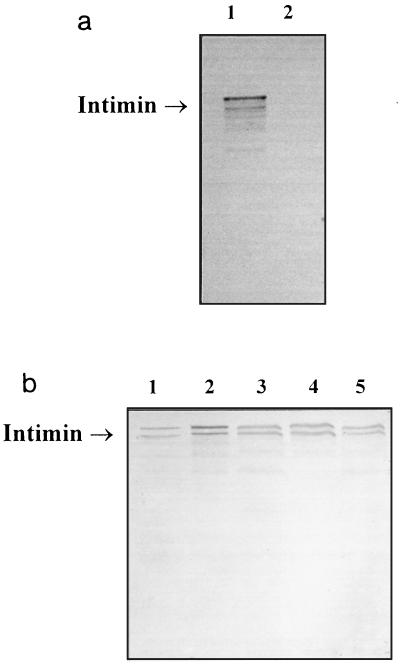
FIG. 1.
Western blot analysis of Int388–667 antiserum against various A/E lesion-forming E. coli strains. (a) The antiserum detected intimin α from strain E2348/69 (lane 1), while no reactivity was observed with eae-negative strain CVD206, which was used as a negative control (lane 2). (b) Similar levels of intimin expression are shown by representative human isolates: O111:H2 (intimin β; lane 1), O119:H6 (intimin β; lane 2), O55:H7 (intimin γ; lane 3), O55:H− (intimin γ; lane 4), and O157:H7 (intimin γ; lane 5).
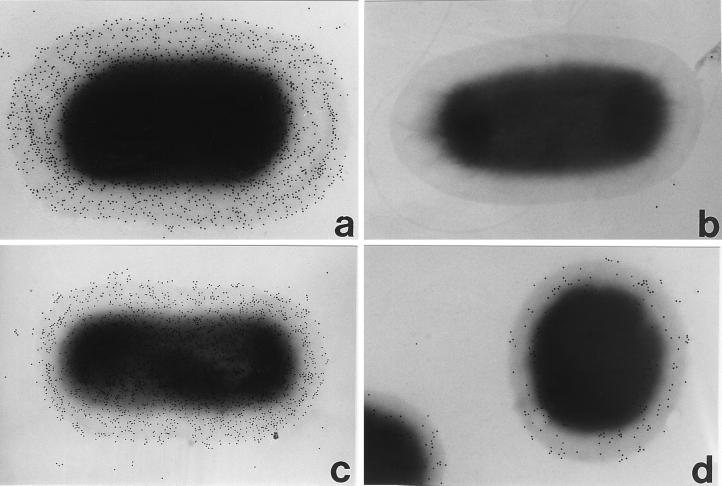
FIG. 2.
Immunogold labelling of intimin. The specificity of the Int388–667 antibody was confirmed by labelling wild-type EPEC strain E2348/69 (a) and its intimin-negative derivative, CVD206 (b). The antiserum labelled all strains that express intimins α, β, and δ, although there was some strain-to-strain variation in the level of intimin expression illustrated here, with two strains expressing intimins α (c) and β (d), respectively. Magnification, ×30,000.
Detection of intimin and eae by Western blotting and PCR.
In order to determine the reactivity of the Int388–667 antiserum with the different intimin types, clinical human and animal A/E lesion-forming isolates were examined by Western blotting (Table 1). The bacterial strains were grown to mid-logarithmic growth phase in DMEM at 37°C, growth conditions that have been established to be optimal for intimin expression (1). Following a 3-h incubation, whole-cell lysates representing equal numbers of bacteria were subjected to Western blot analysis. Seventy-one of the 75 (94.7%) human A/E lesion-forming E. coli isolates (Table 1) and H. alvei (data not shown) and all the representative animal A/E lesion-forming E. coli isolates (Table 1) reacted with the Int388–667 antiserum. Similar levels of reactivity were observed with intimins from wild-type EPEC and EHEC strains irrespective of whether they possessed the EPEC EAF plasmid encoding the positive regulator per; lower levels of reactivity were detected for wild-type strains cured of the EAF plasmid, including strains B171-4 (data not shown) and JPN15 (19). In addition to CVD206, no reactivity was detected with enterotoxigenic E. coli, enteroinvasive E. coli, diffuse-adhering E. coli, enteroaggregative E. coli E. coli K-12 HB101, E. coli HB101 expressing the Yersinia invasin [HB101(pIR203)], or any of the non-E. coli strains listed in Materials and Methods. These results demonstrate that the Int388–667 antiserum can react with any of the intimin types and can be used as a broad-spectrum (universal) intimin reagent.
We also screened the 75 human isolates and 42 animal strains with the PCR primers used, on the basis of the conserved eae region, to amplify Int388–667. This produced positive reactivities with the same 71 human isolates that were positive by Western blot analysis and with 39 of the 42 animal strains (Table 1; Fig. 3). Negative PCR results were obtained with all the non-EPEC E. coli isolates and the non-E. coli strains (Fig. 3 and data not shown).
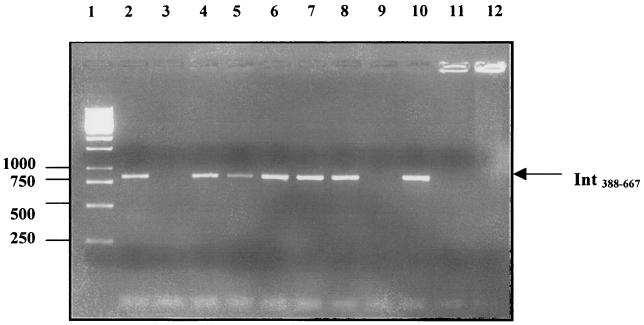
FIG. 3.
Detection of different intimin types with the universal intimin primers. A specific PCR product (840 bp) was generated from a human O127 EPEC isolate expressing intimin α (lane 2), cow O26 isolates expressing intimin β (lanes 4 and 8), sheep O157:H7 and O157:H− isolates expressing intimin γ (lanes 5 and 6, respectively), a cow O157:H7 isolate expressing intimin γ (lane 7), and a pig O26 isolate expressing intimin β (lane 10). No PCR product was obtained with CVD206 (lane 3), a cow O26 isolate expressing intimin β (lane 9), L. monocytogenes (lane 11), or Y. pseudotuberculosis (lane 12). Molecular size markers (1-kb ladder) were loaded in lane 1.
Detection of surface intimin expression by immunogold electron microscopy.
In order to determine if the Int388–667 region is exposed on the bacterial cell surface and accessible for binding of the antiserum, we used live EPEC bacteria and immunogold electron microscopy. By reacting the Int388–667 antiserum with 11 strains (belonging to serogroups O26, O55, O86, O119, O125, O127, O128, O142) expressing either intimin α, β, or δ, we revealed a uniform surface distribution of intimin but some interbacterial variation in the number of gold particles associated with individual bacteria; some strains showed a high density of gold particles per unit area, and others showed a lower density (Fig. 2). This is in contrast to the uniform level of reactivity seen on Western blots and may indicate variable access of the antibody to surface-expressed intimin.
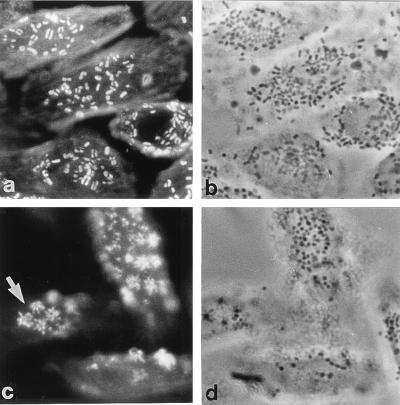
Because we had no intimin γ antiserum, the distribution of this intimin type on the surface of the bacteria has not yet been shown. In this study we have used the universal intimin antiserum to address this issue. Strains that express intimin γ (11 isolates belonging to serogroups O55 and O157) generally displayed a uniform surface distribution, although in a few strains belonging to serotypes O55:H7 and O55:H−, a distinct filamentous pattern of intimin staining was seen; filamentous staining appeared to be associated with rigid rod-like fimbrial structures (Fig. 4). This filamentous staining pattern was also evident on the same strains during infection of HEp-2 cells when examined by immunofluorescence (data not shown), although these bacteria did produce a normal FAS reaction (Fig. 5).
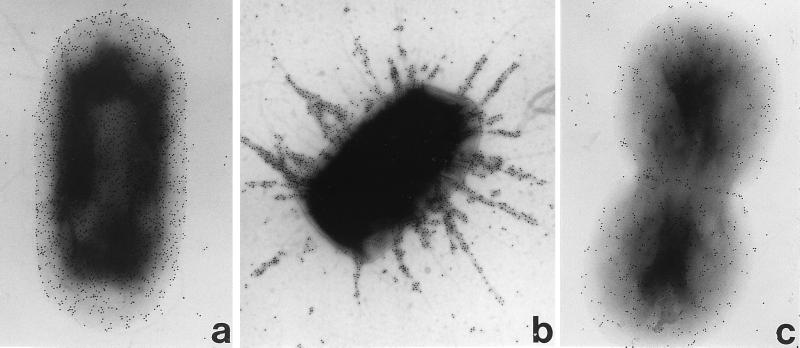
FIG. 4.
Immunogold labelling of intimin γ The antiserum labelled all strains expressing intimin γ, and most strains including EHEC 85-170 (O157:H7) (a) displayed a uniform distribution of surface intimin, although strains ICC39 (O55:H−) and ICC57 (O55:H7) (b) showed a filamentous pattern of intimin staining. Intimin γ from strain ICC57 cloned into CVD206 [strain CVD206(pICC57)] displayed a uniform surface intimin distribution (c). Magnification, ×30,000.
FIG. 5.
Corresponding actin fluorescence (a and c) and phase-contrast micrographs (b and d) showing HEp-2 cells infected with EPEC ICC57 (a and b) and CVD206(pICC57) (c and d) for 3 h. Both strains produced intense spots of actin fluorescence at sites of bacterial attachment (positive FAS test) indicative of A/E lesion formation. However, strain CVD206(pICC57) was atypical in that it produced a stellate distribution of actin accretion (c, arrow).
Expression of eae from ICC57 in CVD206.
In order to determine if production of a filamentous form of intimin γ is a feature associated with the primary sequence of this intimin, eae from ICC57 was cloned in the pGEM-T vector. The recombinant plasmid (pICC57) was used to transform CVD206. CVD206 expressing recombinant intimin α [CVD206(pCVD438)] was used as a control. Western blotting and Int388–667 antiserum have shown similar levels of intimin expression in the two CVD206 strains (data not shown). Immunogold labelling of CVD206(pICC57) revealed, as in CVD206(pCVD438), a uniform distribution of the intimin polypeptide on the bacterial cell surface (Fig. 4), although this strain did give an atypical FAS reaction and featured a stellate distribution of actin accretion beneath each adherent bacterium (Fig. 5).
DISCUSSION
The aim of this study was to develop a broad-spectrum intimin antiserum reactive with all of the intimin types expressed by A/E lesion-forming microbial pathogens. For that purpose we used a conserved region of intimin (Int388–667) located upstream of the cell binding domain. Reacting the antiserum with Western blots of eae-positive strains revealed that the antiserum recognized all intimin types. The observation that similar levels of reactivity were observed with all the EPEC and EHEC isolates indicates that atypical EPEC and EHEC strains possess a regulatory system for intimin expression but that it is different from those of typical EPEC strains which possess the Per regulatory proteins. Immunogold labelling confirmed that the lower levels of reactivity observed in the case of the EAF-positive EPEC strains that had been cured of their plasmids were, as observed previously (19), due to the fact that a much smaller fraction of bacteria expressed intimin instead of the fact that all bacteria had reduced levels of intimin expression. However, when examining large numbers of isolates, a few showed lower densities of gold particles per unit surface area, which was possibly due to the reduced accessibility of intimin on the bacterial cell surface. Thus, although the antiserum described in this study is clearly suitable as a diagnostic tool on Western blots, the variability in intimin expression or accessibility on the bacterial cell surface indicates that it might not provide in all cases a sensitive tool for diagnostic methods based on immunocapture procedures. We also found that the PCR primers synthesized to generate Int388–667 gave a specific PCR product with as the template eae-positive strains belonging to different EPEC and EHEC serogroups isolated from a variety of animal species. Accordingly, the Int388–667 antiserum and PCR primers described here provide universal reagents that can be used to detect a broad range of A/E lesion-forming strains.
In our previous investigations (1) we were unable to determine the distribution of intimin γ on the bacterial cell surface because no specific antiserum was available and the lack of cross-reactivity between intimin α and intimin β antisera and intimin γ polypeptide. In this investigation we used the broad-spectrum intimin antiserum to detect surface expression of intimin γ. An unexpected finding was the production by a few EPEC O55:H− and O55:H7 strains of a filamentous form of intimin γ, while on other O55:H7, O157:H7, and O127:H40 (which was recently classified as expressing intimin γ similar to O55:H7; GenBank accession no. AJ132982) strains a uniform intimin distribution was shown over the bacterial cell surface. While we cannot rule out the possible artifactual nature of the intimin staining in these few strains, we have no rational explanation as to why this antiserum would nonspecifically label filamentous bacterial surface structures. Importantly, though, EPEC strains expressing filamentous intimin produced a FAS reaction indistinguishable from that of the nonfilamentous intimin-producing isolates, indicating intimate intimin-mediated adhesion and A/E lesion formation (20).
Cloning of pICC57, a recombinant plasmid harboring eae from ICC57, a filamentous intimin-producing strain, into CVD206 (eae mutant derivative of E2348/69 encoding intimin α) restored A/E lesion formation activity, albeit with atypical actin accretion, and intimin γ appeared to be uniformly distributed over the cell surface, similar to the pattern observed in CVD206(pCVD438). Thus, the production of a filamentous form of intimin would appear to be dependent on the genetic background of the bacterial strain and not an intrinsic feature of intimin γ.
Previous studies have shown that in EPEC, intimin expression is upregulated by Per during bacterial growth and, following A/E lesion formation, is downregulated (19). EHEC and atypical EPEC isolates (15) lack EAF plasmids, per regulatory genes, and per homologues but appear to regulate intimin because intimin, which is not expressed when bacteria are grown in L broth, is expressed when they are grown in DMEM. The development of a universal intimin antiserum will now allow the expression and regulation of intimin in EHEC and atypical EPEC strains producing intimin γ to be investigated.
ACKNOWLEDGMENTS
We thank Stephen Reece for classifying the intimin from O127:H40. We also thank Luiz R. Trabulsi, James B. Kaper, Eric Oswald, Lothar Wieler, Henrik Chart, Roy Robins-Browne, and Josee Harel for providing E. coli strains.
V.H.'s visit to London was supported by a “short-term studentship for research abroad” from the Austrian Ministry of Science and Research. This work was supported by grants from the Wellcome Trust.
REFERENCES
- 1.Adu-Bobie J, Frankel G, Bain C, Goncaleves A G, Trabulsi L R, Douce G, Knutton S, Dougan G. Detection of intimin α, β, γ, and δ, four intimin derivatives expressed by attaching and effacing microbial pathogens. J Clin Microbiol. 1998;36:662–668. doi: 10.1128/jcm.36.3.662-668.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Albert M J, Faruque S M, Ansaruzzaman M, Islam M M, Haider K, Alam K, Kabir I, Robins-Browne R. Sharing of virulence-associated properties at the phenotypic and genetic levels between enteropathogenic Escherichia coli and Hafnia alvei. J Med Microbiol. 1992;37:310–314. doi: 10.1099/00222615-37-5-310. [DOI] [PubMed] [Google Scholar]
- 3.Cantey J R, Blake R K. Diarrhea due to Escherichia coli in the rabbit, a novel mechanism. J Infect Dis. 1977;135:454–462. doi: 10.1093/infdis/135.3.454. [DOI] [PubMed] [Google Scholar]
- 4.Donnenberg M S, Giron J A, Nataro J P, Kaper J B. A plasmid-encoded type IV fimbrial gene of enteropathogenic Escherichia coli associated with localized adherence. Mol Microbiol. 1992;6:3427–3437. doi: 10.1111/j.1365-2958.1992.tb02210.x. [DOI] [PubMed] [Google Scholar]
- 5.Donnenberg M S, Kaper J B. Construction of an eae deletion mutant of enteropathogenic Escherichia coli by using a positive-selection suicide vector. Infect Immun. 1991;59:4310–4317. doi: 10.1128/iai.59.12.4310-4317.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Donnenberg M S, Yu J, Kaper J B. A second chromosomal gene necessary for intimate attachment of enteropathogenic Escherichia coli to epithelial cells. J Bacteriol. 1993;175:4670–4680. doi: 10.1128/jb.175.15.4670-4680.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Frankel G, Candy D C, Everest P, Dougan G. Characterization of the C-terminal domains of intimin-like proteins of enteropathogenic and enterohemorrhagic Escherichia coli, Citrobacter freundii, and Hafnia alvei. Infect Immun. 1994;62:1835–1842. doi: 10.1128/iai.62.5.1835-1842.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Frankel G, Philips A D, Novakova M, Batchelor M, Hicks S, Dougan G. Generation of Escherichia coli intimin-derivatives with differing biological activities using site-directed mutagenesis of the intimin C-terminus domain. Mol Microbiol. 1998;29:559–570. doi: 10.1046/j.1365-2958.1998.00950.x. [DOI] [PubMed] [Google Scholar]
- 9.Frankel G, Phillips A D, Rosenshine I, Dougan G, Kaper J B, Knutton S. Enteropathogenic and enterohaemorrhagic Escherichia coli: more subversive elements. Mol Microbiol. 1998;30:911–921. doi: 10.1046/j.1365-2958.1998.01144.x. [DOI] [PubMed] [Google Scholar]
- 10.Giron J A, Ho A S, Schoolnik G K. An inducible bundle-forming pilus of enteropathogenic Escherichia coli. Science. 1991;254:710–713. doi: 10.1126/science.1683004. [DOI] [PubMed] [Google Scholar]
- 11.Gomez-Duarte O G, Kaper J B. A plasmid-encoded regulatory region activates chromosomal eaeA expression in enteropathogenic Escherichia coli. Infect Immun. 1995;63:1767–1776. doi: 10.1128/iai.63.5.1767-1776.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hicks S, Frankel G, Kaper J B, Dougan G, Phillips A D. Role of intimin and bundle-forming pili in enteropathogenic Escherichia coli adhesion to pediatric intestine in vitro. Infect Immun. 1998;66:1570–1578. doi: 10.1128/iai.66.4.1570-1578.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jarvis K G, Giron J A, Jerse A E, McDaniel T K, Donnenberg M S, Kaper J B. Enteropathogenic Escherichia coli contains a putative type III secretion system necessary for the export of proteins involved in attaching and effacing lesion formation. Proc Natl Acad Sci USA. 1995;92:7996–8000. doi: 10.1073/pnas.92.17.7996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jerse A E, Yu J, Tall B D, Kaper J B. A genetic locus of enteropathogenic Escherichia coli necessary for the production of attaching and effacing lesions on tissue culture cells. Proc Natl Acad Sci USA. 1990;87:7839–7843. doi: 10.1073/pnas.87.20.7839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kaper J B. Defining EPEC. Rev Microbiol Sao Paulo. 1996;27(Suppl. 1):130–133. [Google Scholar]
- 16.Kelly G, Prasannan S, Daniell S, Flemming K, Frankel G, Dougan G, Connerton I, Matthews S. Intimin from enteropathogenic E. coli belongs to a new family of bacterial adhesion molecules containing C-type lectin- and tandem immunoglobulin-like domains. Nat Struct Biol. 1999;6:313–318. doi: 10.1038/7545. [DOI] [PubMed] [Google Scholar]
- 17.Kenny B, DeVinney R, Stein M, Reinscheid D J, Frey E A, Finlay B B. Enteropathogenic E. coli (EPEC) transfers its receptor for intimate adherence into mammalian cells. Cell. 1997;91:511–520. doi: 10.1016/s0092-8674(00)80437-7. [DOI] [PubMed] [Google Scholar]
- 18.Kenny B, Lai L C, Finlay B B, Donnenberg M S. EspA, a protein secreted by enteropathogenic Escherichia coli, is required to induce signals in epithelial cells. Mol Microbiol. 1996;20:313–323. doi: 10.1111/j.1365-2958.1996.tb02619.x. [DOI] [PubMed] [Google Scholar]
- 19.Knutton S, Adu-Bobie J, Bain C, Phillips A D, Dougan G, Frankel G. Down regulation of intimin expression during attaching and effacing enteropathogenic Escherichia coli adhesion. Infect Immun. 1997;65:1644–1652. doi: 10.1128/iai.65.5.1644-1652.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Knutton S, Baldwin T, Williams P H, McNeish A S. Actin accumulation at sites of bacterial adhesion to tissue culture cells: basis of a new diagnostic test for enteropathogenic and enterohemorrhagic Escherichia coli. Infect Immun. 1989;57:1290–1298. doi: 10.1128/iai.57.4.1290-1298.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Knutton S, Lloyd D R, McNeish A S. Adhesion of enteropathogenic Escherichia coli to human intestinal enterocytes and cultured human intestinal mucosa. Infect Immun. 1987;55:69–77. doi: 10.1128/iai.55.1.69-77.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 23.Lai L C, Wainwright L A, Stone K D, Donnenberg M S. A third secreted protein that is encoded by the enteropathogenic Escherichia coli pathogenicity island is required for transduction of signals and for attaching and effacing activities in host cells. Infect Immun. 1997;65:2211–2217. doi: 10.1128/iai.65.6.2211-2217.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Levine M M, Berquist E J, Nalin D R, Waterman D H, Hornick R B, Young C R, Stoman S, Rowe B. Escherichia coli that cause diarrhoea but do not produce heat-labile or heat-stable enterotoxins and are non-invasive. Lancet. 1978;i:119–122. doi: 10.1016/s0140-6736(78)90299-4. [DOI] [PubMed] [Google Scholar]
- 25.Loureiro I, Frankel G, Adu-Bobie J, Dougan G, Trabulsi L R, Carneiro-Sampaio M M S. Human colostrum contains IgA antibodies reactive to enteropathogenic Escherichia coli-virulence-associated proteins: intimin, BfpA, EspA and EspB. J Pediatr Gastroenterol Nutr. 1998;27:166–171. doi: 10.1097/00005176-199808000-00007. [DOI] [PubMed] [Google Scholar]
- 26.McDaniel T K, Jarvis K G, Donnenberg M S, Kaper J B. A genetic locus of enterocyte effacement conserved among diverse enterobacterial pathogens. Proc Natl Acad Sci USA. 1995;92:1664–1668. doi: 10.1073/pnas.92.5.1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moon H W, Whipp S C, Argenzio R A, Levine M M, Giannella R A. Attaching and effacing activities of rabbit and human enteropathogenic Escherichia coli in pig and rabbit intestines. Infect Immun. 1983;41:1340–1351. doi: 10.1128/iai.41.3.1340-1351.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nataro J, Kaper J B. Diarrheagenic Escherichia coli. Clin Microbiol Rev. 1998;11:142–210. doi: 10.1128/cmr.11.1.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schauer D B, Falkow S. Attaching and effacing locus of a Citrobacter freundii biotype that causes transmissible murine colonic hyperplasia. Infect Immun. 1993;61:2486–2492. doi: 10.1128/iai.61.6.2486-2492.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ulshen M H, Rollo J L. Pathogenesis of Escherichia coli gastroenteritis in man—another mechanism. N Engl J Med. 1980;302:99–101. doi: 10.1056/NEJM198001103020207. [DOI] [PubMed] [Google Scholar]
- 31.Whittam T S, McGraw E A. Clonal analysis of EPEC serogroups. Rev Microbiol Sao Paulo. 1996;27(Suppl. 1):7–16. [Google Scholar]