Abstract
Background
The Wnt receptors ROR1 and ROR2 are generating increased interest as cancer therapeutic targets but remain understudied in pancreatic ductal adenocarcinoma (PDAC). Compared to canonical Wnt/ β-catenin signalling, the role of noncanonical Wnt signalling in PDAC remains largely unknown. Only one study has investigated the prognostic significance of the noncanonical Wnt signalling receptor, ROR2 in PDAC. No studies have investigated the prognostic role of ROR1 in PDAC.
Methods
Here, we performed analysis of ROR1 and ROR2 mRNA expression in three publicly available datasets ICGC-PACA-AU (n = 81), TCGA-PAAD (n = 150) and CPTAC-PDAC (n = 137). ROR1 and ROR2 protein expression from the CPTAC-PDAC discovery cohort were also analysed. Immunohistochemistry (IHC) using the validated anti ROR1 monoclonal antibody (4A5) was performed on the Australian Pancreatic Cancer Genome Initiative (APGI) cohort of PDAC samples (n = 152). Association between ROR1 cytoplasmic staining intensity and clinicopathological parameters including stage, grade and overall survival (OS) was investigated.
Results
High ROR1 mRNA expression levels correlated with a favourable OS outcome in all of the ICGC-PACA-AU, TCGA-PAAD and CPTAC-PDAC cohorts. ROR1 protein expression was not associated with stage, grade or OS in the APGI cohort.
Conclusion
ROR1 and ROR2 have potential as prognostic markers when measured at the mRNA level in PDAC. Our IHC cohort did not support ROR1 protein expression in predicting OS, and highlighted the discrepancy of prognostic biomarkers when measured by MS, IHC and RNAseq.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12885-021-08952-9.
Background
Pancreatic cancer represents the 11th most common cancer worldwide with 458,918 new cases and 432,242 deaths recorded in 2018 [1]. It is also one of the most lethal malignancies, with 5-year survival rate less than 10% [2]. Risk factors associated with the disease include smoking [3], family history [4], germline mutations [5] and genetic mutations [6]. Despite the increased awareness around risk factors, the worldwide incidence and mortality of pancreatic cancer are both predicted to increase in coming years [7].
Pancreatic ductal adenocarcinoma (PDAC) is the most common type of pancreatic cancer that accounts for more than 80% of total cases. The majority of PDAC cases originate from microscopic precursor lesions called pancreatic intraepithelial neoplasia (PanINs) that are challenging to detect by current clinical imaging techniques [8]. As such, PDAC is typically diagnosed at an advanced stage (Stage III or IV) and presents an extremely poor prognosis compared to the less common pancreatic neuroendocrine tumour [9]. Current treatment options for patients are limited, especially for those with locally advanced or metastatic disease who are not eligible for surgery. In terms of chemotherapy, gemcitabine plus Abraxane has become the standard treatment based on moderate improvement outcomes compared to gemcitabine alone (median overall survival 8.5 months compared to 6.7 months, p < 0.001) from a phase III clinical trial [10]. Unfortunately, combination therapy with other agents shown to be effective in other cancers have not shown promising results in PDAC [11, 12]. In addition, immunotherapies, either alone or in combination with chemoradiotherapy or targeted therapy failed to show much progress in PDAC [13–15]. There is an urgent need of identifying biomarkers which could be targeted therapeutically for the malignancy.
Wnt signalling is one of the key pathways involved in cell differentiation, polarity, migration, adhesion and invasion. Encompassing both canonical (β-catenin dependent) and non-canonical (β-catenin independent) arms, the pathway has been implicated in a range of cancers. In PDAC, activation of the β-catenin dependent Wnt pathway has been indicated by cytoplasmic and nuclear localization of β-catenin [16], and suggested to play a role in progression of pancreatic cancer [17]. Mutations in RNF43, a gene associated with Wnt/ β-catenin pathway regulation have also been reported in PDAC [18, 19].
In contrast, the role of the β-catenin independent Wnt signalling arm remains largely unexplored in PDAC. The receptor tyrosine kinase-like orphan receptors ROR1 and ROR2 regulate β-catenin independent signalling and have been linked to carcinogenesis and metastasis in several malignancies [20–23]. A single study in PDAC (n = 162, ROR2 polyclonal antibody #LS-C99126, LifeSpan BioSciences Inc., USA) found high ROR2 protein expression was significantly associated with malignant characteristics and reduced overall survival [24]. An early study into ROR1 protein expression across a range of tumour types reported “moderate” or “high” expression in 83% of pancreatic cancer samples (n = 48) using the high-affinity 4A5 monoclonal antibody against ROR1 [25]. A separate study (using a separate C-terminus targeting monoclonal antibody) detected ROR1 protein at low levels in 15% of pancreatic adenocarcinoma samples (n = 38) [26]. Both studies also highlighted the presence of ROR1 expression in normal pancreatic islet cells.
The aim of this study was to clarify expression of ROR1 or ROR2 in PDAC through the interrogation of publicly available datasets and by performing immunohistochemistry (IHC) with a high-affinity monoclonal ROR1 antibody (4A5) on an independent and well-characterised cohort of PDAC [25, 27]. As there are numerous ROR1/2 targeting therapies in development [28, 29] and clinical trials [30–32] in progress, PDAC patients may benefit from this approach in the future.
Methods
Public datasets
The mRNA expression and accompanying clinicopathological data were acquired via three public platforms: International Cancer Genome Consortium (ICGC, https://icgc.org/),the Cancer Genome Atlas (TCGA, http://cancergenome.nih.gov/) and the Clinical Proteomic Tumor Analysis Consortium (CPTAC, https://cptac-data-portal.georgetown.edu/). Specifically, the normalised mRNA expression generated by next generation sequencing (NGS) was extracted for the ICGC-Pancreatic Cancer - Australia (ICGC-PACA-AU) cohort from the supplementary material of the corresponding publication [33]. Only cases with tumour purity above 40% indicated by the Qpurity score were included. For the TCGA-pancreatic adenocarcinoma (TCGA-PAAD) cohort, normalised gene expression of NGS was downloaded from the UCSC Xena platform [34] on 3rd May, 2020. The normalised gene expression data of the CPTAC cohort was obtained from the Genomic Data Commons (GDC) Data Portal (https://portal.gdc.cancer.gov/) on 2nd Sep, 2021.
In addition, the mass-spectrometry (MS) based proteomic data of the PDAC Discovery Study used in this publication were generated by the National Cancer Institute (NCI) CPTAC. We extracted relative abundance level of ROR1 and ROR2 protein (unshared log ratio) from the CPTAC data portal (https://pdc.cancer.gov/pdc/) on 2nd Sep, 2021.For all of the cohorts, only PDAC cases were included in the analysis. The characteristics of each cohort was shown in Table 1.
Table 1.
Characteristics of the public datasets used in the study
| ICGC-PACA-AU [33] | TCGA-PAAD [18] | CPTAC-PDAC [35] | ||
|---|---|---|---|---|
| Total (n)a | 81 | 150 | 137 | |
| Stage | I | 6 | 13 | 4 |
| II | 70 | 128 | 76 | |
| III | 1 | 4 | 36 | |
| IV | 4 | 4 | 7 | |
| Tumour grade | G1 | 1 | 20 | 9 |
| G2 | 45 | 85 | 97 | |
| G3 | 31 | 42 | 30 | |
| G4 | 2 | 1 | 1 | |
| Detection platform | Illumina HiSeq 2000 | Illumina HiSeq 2000/2500 |
Illumina HiSeq (RNAseq) Thermo EASY-nLC 1200 UHPLC System (mass-spectrometry) |
|
aCases included in the analysis
Clinical cohort
Tumour tissue microarrays (TMAs) and accompanying clinicopathological data of the Pancreatic Ductal Carcinoma Validation set (n = 334) were acquired from the Australian Pancreatic Cancer Genome Initiative (APGI). Immunohistochemistry (IHC) staining of ROR1 (1:50, #564464, BD Biosciences, USA) was optimised by the Kinghorn Cancer Centre Histopathology Laboratory at the Garvan Institute of Medical Research using the Leica Bond RX system (Leica Microsystems, USA). Negative control TMA slides (omission of primary antibody) were stained in parallel. The intensity of ROR1 staining was graded as 0 (absence), 1 (weak) and 2 (intense). Only the cases with three intact TMA cores (n = 157) were included, with the highest score set as the final score. We excluded another 5 from the final analysis for non-Pancreatic ductal adenocarcinoma (PDA) subtype, and ended up with the analysis cohort (n = 152). Demographic and clinicopathological parameters of the 152 patients were summarised in Table 2.
Table 2.
Demographic and clinicopathological characteristics of the tumour samples in the APGI cohort
| Number of cases | Percentage (%) | ||
|---|---|---|---|
| Gender | Female | 76 | 50.0 |
| Male | 76 | 50.0 | |
| Tumour grade | 1 | 8 | 5.3 |
| 2 | 105 | 69.1 | |
| 3 | 39 | 25.7 | |
| FIGO stage (2009) | I | 11 | 7.2 |
| II | 128 | 84.2 | |
| III | 2 | 1.3 | |
| IV | 10 | 6.6 | |
| Recurrence status | Unknown | 8 | 8.1 |
| Confirmed | 91 | 91.9 | |
| Vital status | Alive | 18 | 11.8 |
| Deceased of PCa | 134 | 88.2 | |
aPC- pancreatic cancer
Statistical analysis
Both mRNA and protein expression levels were log2 transformed for all the tests. Statistical significance of expression level between tumour grades and stages was carried out using unpaired t-test in public datasets or Chi-square test in clinical cohort. Pearson’s correlation was performed to investigate relationship between mRNA and protein expression. Kaplan Meier curves were produced for univariate overall survival (OS) analyses. Optimal cutpoint for variables was determined by the maximally selected rank statistics from the ‘maxstat’ package in R. The log-rank test was used to evaluate the significance of the association. Cox multivariate regression including gender, stage and tumour grade was also applied on the OS. All the analyses were performed using R (v3.6.3). Figures were provided in R (v3.6.3) and GraphPad Prism (v7.02). Significance was set at p < 0.05.
Results
High ROR2 mRNA expression is associated with better overall survival in two independent PDAC cohorts
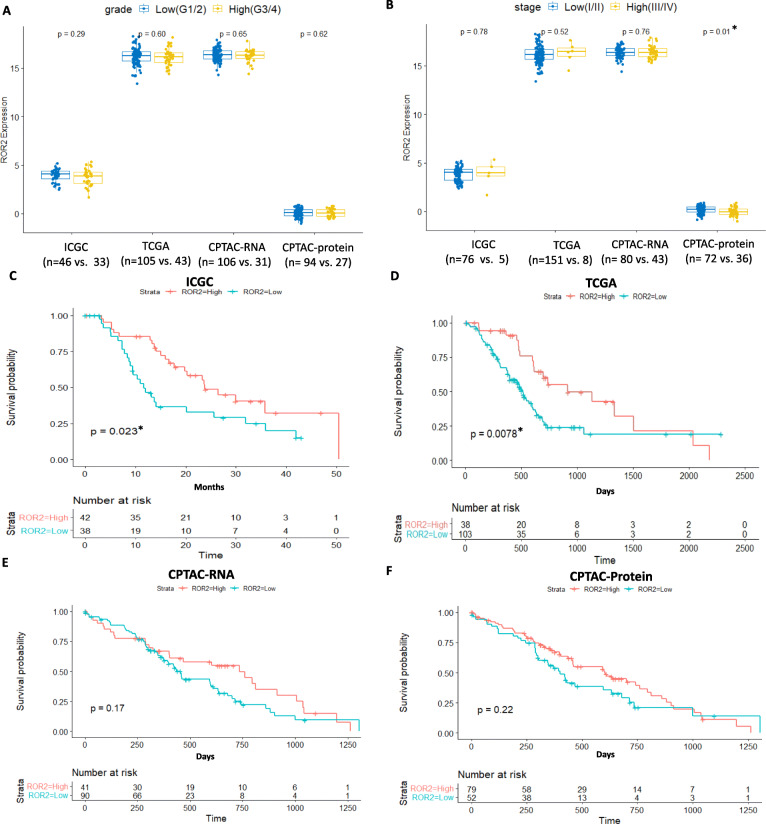
We correlated mRNA and protein expression of ROR2 with tumour grade, stage and overall survival (OS) in three independent pancreatic cancer cohorts (ICGC-PACA-AU, TCGA-PAAD and CPTAC-PDAC). No significant difference was observed for ROR2 mRNA expression between tumour stages or grades in either cohort (Fig. 1A,B). Lower ROR2 protein expression level was observed in high stage compared to low stage patients (p = 0.010, Fig. 1B). High ROR2 mRNA expression level was significantly associated with longer OS in both the ICGC and TCGA cohorts (p = 0.023 and 0.008 respectively, Fig. 1C,D), but not significant in the CPTAC-PDAC cohort (p = 0.170, Fig. 1E). ROR2 protein level was not significantly associated with overall survival based on the proteomic data of CPTAC-PDAC cohort (Fig. 1F).
Fig. 1.
Association between ROR2 mRNA/protein expression level and clinicopathological parameters in the ICGC-PACA-AU,TCGA-PAAD and CPTAC-PDAC cohorts. A. No significant difference in ROR2 expression was observed between low and high tumour grade. B. ROR2 mRNA expression was not associated with stage, ROR2 protein expression was significantly different between low and high stage (p = 0.010). C. High ROR2 mRNA expression level was associated with better overall survival (p = 0.023) in the ICGC-PACA-AU cohort. D. High ROR2 mRNA expression level was associated with better overall survival (p = 0.008) in the TCGA-PAAD cohort. E. ROR2 mRNA was not associated with overall survival in the CPTAC-PDAC cohort. F. ROR2 protein was not associate with overall survival in the CPTAC-PDAC cohort.*Significance at p < 0.05 level
High ROR1 mRNA expression is associated with better overall survival in three independent PDAC cohorts
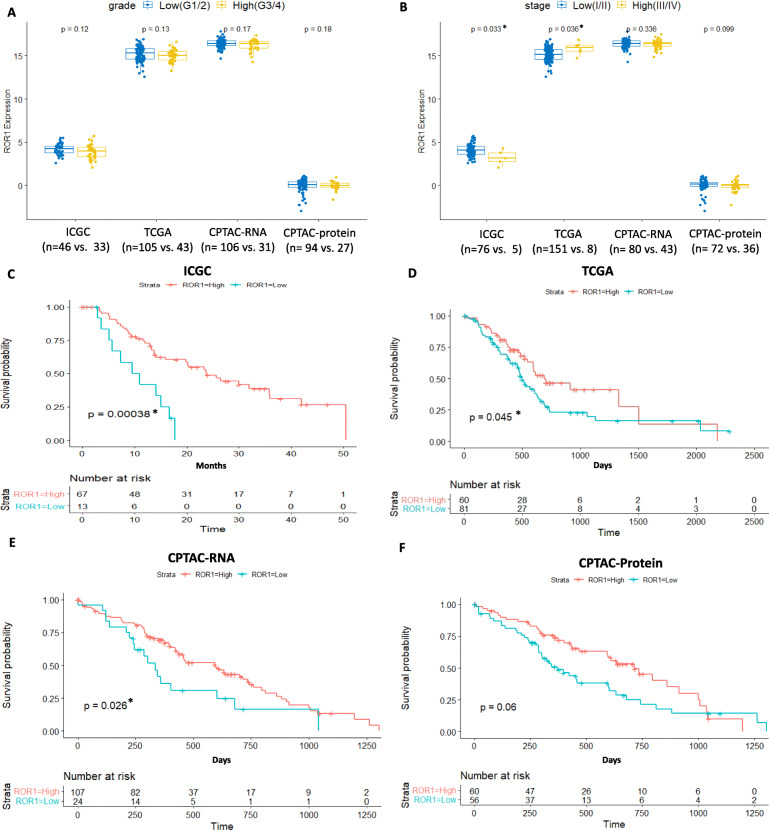
We next correlated ROR1 mRNA expression with grade, stage, and OS in the cohorts. No association was seen between ROR1 mRNA or protein expression and grade in either cohort (Fig. 2A). ROR1 mRNA expression was lower in high stage (stage III or IV) versus low stage (stage I or II) in the ICGC cohort (Fig. 2B, p = 0.033) while the opposite trend was observed in the TCGA cohort (Fig. 2B, p = 0.036). However, both of the cohorts are limited in the number of high stage cases (n = 5 in ICGC and n = 8 in TCGA), which compromised the statistical power of the analyses. No significant correlation between ROR1 mRNA or protein level and tumour stage was observed in the CPTAC-PDAC cohort in which more late-stage samples were included (Fig. 2B). High ROR1 mRNA expression level was significantly associated with longer OS in all the three cohorts (p < 0.001, p = 0.045 and p = 0.026 respectively, Fig. 2C,D,E). However, the overall survival was not significantly associated with ROR1 protein expression level based on the CPTAC-PDAC cohort (Fig. 2F).
Fig. 2.
Association between ROR1 mRNA/protein expression level and clinicopathological parameters in the ICGC-PACA-AU, TCGA-PAAD and CPTAC-PDAC cohorts. A. No significant difference of ROR1 expression was observed between low and high tumour grade. B. ROR1 expression was associated with stage in ICGC-PACA-AU and TCGA-PAAD cohorts in opposite directions (p = 0.033 and 0.036 respectively). No significant correlation between ROR1 mRNA and stage was observed in CPTAC-PDAC cohort. C. High ROR1 mRNA expression level was associated with better overall survival (p < 0.001) in the ICGC-PACA-AU cohort. D. High ROR1 mRNA expression was associated with better overall survival (p = 0.045) in the TCGA-PAAD cohort. E. High ROR1 mRNA expression was associated with better overall survival (p = 0.026) in the CPTAC-PDAC cohort. F. ROR1 protein expression was not associated with overall survival in the CPTAC-PDAC cohort. *Significance at p < 0.05 level
ROR1 is not correlated with OS or clinicopathological parameters in PDAC at the protein level
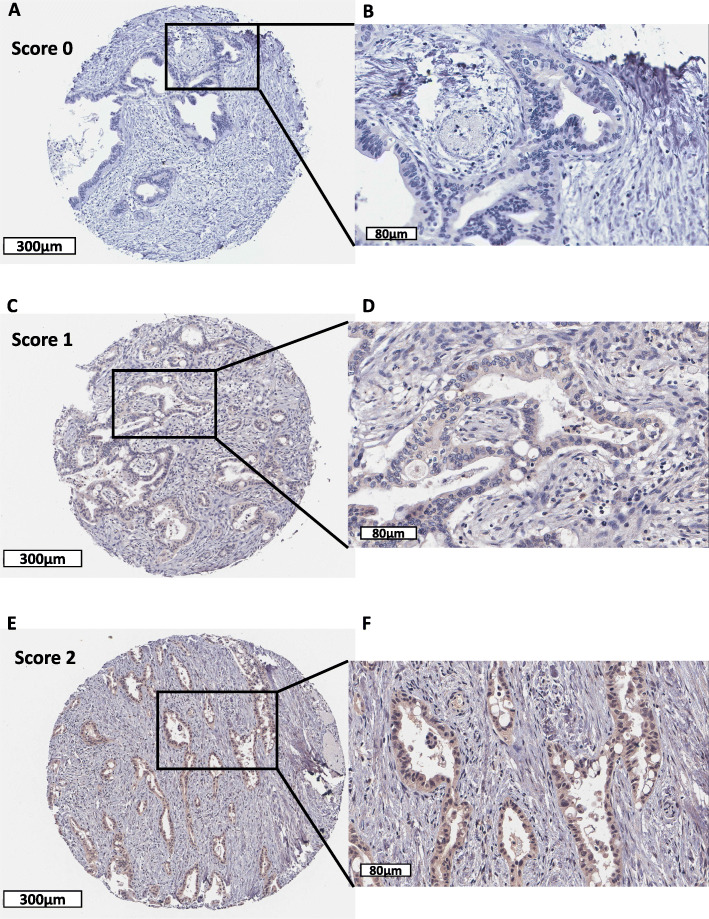
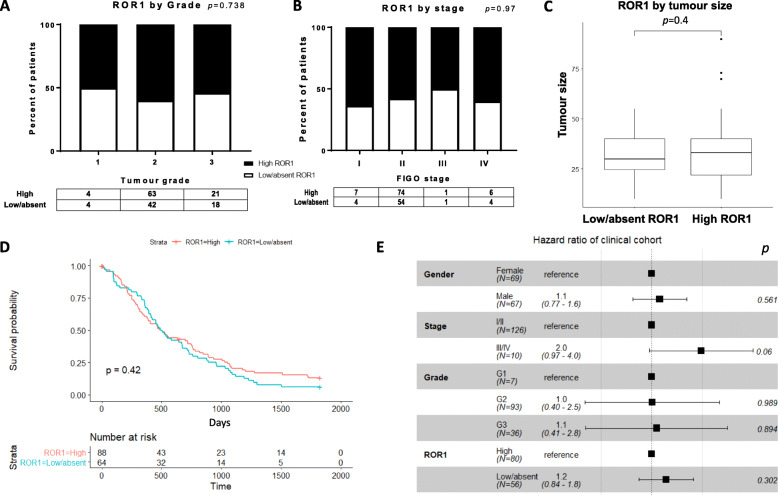
To investigate the role of ROR1 in PDAC at the translational level, we performed immunohistochemical staining of ROR1 in a well-defined cohort of 152 Australian PDAC tissue samples and correlated cytoplasmic expression with clinicopathological parameters and OS. Overall, the cohort showed a broad range of expression levels for ROR1 in PDAC tumour tissues (Fig. 3). ROR1 protein expression was not associated with tumour grade (Fig. 4A), stage (Fig. 4B) or tumour size (Fig. 4C), though it should be noted that the cohort is heavily skewed towards grade 2 and stage 2 tumours, with low numbers of samples from other grades and stages. No significant difference of OS was observed between high and low/absent ROR1 expression in terms of single or multivariable survival analysis (Fig. 4D,E).
Fig. 3.
Representative immunohistochemistry images of ROR1 staining. Representative images of score 0 (absence; A,B), 1 (low; C,D) and 2 (high; E,F) for ROR1 expression measured by immunohistochemistry. Original magnification × 8 (scale bar = 300 μm) for A, C, E; magnification × 30 (scale bar = 80 μm) for B, D, F
Fig. 4.
Correlation between ROR1 protein expression and tumour grade, stage, tumour size and overall survival in the APGI clinical cohort. A-C. Expression of ROR1 stratified by tumour grade, stage and tumour size. D. Kaplan-Meier analysis for ROR1 stratified by low/absent (score 0,1) and high (score 2). E. Multivariable overall survival analysis of ROR1 incorporating gender, tumour stage and grade
Discussion
The Wnt signalling receptors ROR1 and ROR2 are primarily expressed during embryogenesis and are downregulated in most adult tissues [36]. However, aberrant expression of ROR1 has been observed in several haematological malignancies and solid tumours including breast, ovarian and lung cancers [20, 22, 26, 37]. This tumour specific expression has made ROR1 a promising cancer drug target, leading to several ROR1 targeting therapies being developed. ROR1 targeting chimeric antigen receptors (CAR) T cell therapy has shown antitumour efficacy in vivo [38] and a humanised monoclonal antibody developed against ROR1 Fz and Ig-like domain, cirmtuzumab has demonstrated safety and efficacy in several Phase I/II clinical trials for patients with chronic lymphocytic leukemia (CLL), mantle cell lymphoma (MCL) (NCT03088878) and Her2-negative breast cancer (NCT02776917) [30, 31].
Two previous studies using independent ROR1 antibodies (4A5 and 6D4 monoclonal antibodies) have investigated ROR1 protein expression in small cohorts of pancreatic cancer tissues [25, 26]. While neither study investigated an association with overall survival, the detection of ROR1 protein expression prompted interest in pursuing ROR1 therapeutics in pancreatic cancer treatment. A small molecule KAN0439834 targeting the intracellular ROR1 TK domain induced significant apoptosis of ROR1 expressing PC cell lines [39], with additive effects observed in combination with erlotinib or ibrutinib. This raises the possibility of using ROR1 targeting therapies in future pancreatic cancer treatment. However, it should be noted that ROR1 protein expression was also detected in normal pancreatic tissue in previous studies [25, 26]. Off-target toxicity therefore needs to be considered when treating PDAC patients with ROR1-targeting therapies.
Ours is the first study to investigate an association between ROR1 protein expression and overall survival in pancreatic cancer patients. Previous studies of ROR1 expression via IHC reported cytoplasmic staining, even though ROR1 is a membrane receptor [25, 26]. We also detected ROR1 predominantly as cytoplasmic staining with the 4A5 antibody (same antibody used in [25]) in this cohort. It has been suggested that membranous staining is predominant if cells are fixed in paraformaldehyde while cytoplasmic expression more likely appears in formalin-fixed tissue [25]. ROR1 cytoplasmic staining was scored in tumour cells only, as no or low ROR1 was detected in surrounding stromal tissue. We observed no difference in overall survival in PDAC patients with low or high ROR1 expression in this cohort. We also saw no association with grade or stage in our cohort. However, it should be noted that this cohort (like most pancreatic cancer cohorts) is built from surgically resectable PDAC cases, which represents < 20% of PDAC patients. Tissue that makes its way into tumour biobanks and publicly available cohorts is often biased towards patients with a better overall prognosis. The majority of patients in our cohort were low grade (grade 1, 2 = 74%) and low stage (stage I, stage II = 92%).
At the transcriptional level, expression of ROR1 mRNA did associate with overall survival in three independent PDAC cohorts. All of the TCGA-PAAD, ICGC-PACA-AU and CPTAC-PDAC cohorts showed low ROR1 mRNA level was associated with worse OS compared to high ROR1 mRNA level. While two cohorts (TCGA and ICGC) showed divergent associations between ROR1 mRNA expression and stage, it should also be noted that both public cohorts consisted of only few high stage cases, which decreased the statistical power when analysing association between biomarker expression and stage in PDAC. As noted above, this is a consequence of cohort bias as only resectable tumours were biobanked. No significant correlation between ROR1 mRNA level and stage was observed in the CPTAC cohort which included a certain proportion of advanced staged cases (43 Stage III and IV out of total 137 cases). Apart from the limitation of resected PDAC samples, other differences in the cohorts could contribute to the discrepancy. One key difference could be the purity of tumour tissue biobanked and included in the separate mRNA expression analysis. PDAC tissue biopsies are typically composed of a significant proportion of stromal components, with low tumour cellularity commonly observed [18, 40]. Stromal and other cell contamination is likely to introduce noise that interferes with the gene expression analysis, particularly in the case of ROR1 where expression has been observed in healthy cells of the pancreas [25, 26]. One advantage of the ICGC cohort is the inclusion of tumour cellularity information for analysis. Tumour cellularity, the relative portion of tumour and normal cells of each sample in the cohort was estimated using the statistical model qpure [41]. In our analysis, we did not include any sample with epithelial content less than 40% from the ICGC cohort, which presented a more robust outcome than the TCGA cohort (median neoplastic cellularity 33% estimated with ABSOLUTE algorithm [18]). If we excluded the samples with tumour cellularity less than 40% in the TCGA cohort, we would lose statistical power for analysis dramatically due to a small sample size (n = 46). For the CPTAC cohort, all the cases included in this study had more than 40% cellularity. Other divergences among the cohorts were population origin, sample size, tumour grade distribution (Table 1), and mRNA expression cut-off.
Despite these limitations to individual cohort, it is unclear exactly why the prognostic significance of ROR1 expression at the mRNA level was not observed in ROR1 protein. A weak correlation between mRNA and protein expression (MS-based) was observed for ROR1 in the cases we included in CPTAC PDAC discovery cohort (R = 0.25, p = 0.005, Supplementary Fig.S1 A). Previous studies have reported a lack of direct relation between mRNA and protein expression [42, 43]. For ROR1, post-translational modifications such as glycosylation and mono-ubiquitination were observed extensively in CLL cells, which regulated the localization and stabilisation of ROR1 protein [44]. In addition, truncated ROR1 which lost the extracellular matrix and transmembrane regions was detected in central nervous system, leukemias and a variety of human cancers [45]. It is possible that individual isoforms of ROR1 could be undetected using the ROR1 monoclonal antibody we used in this cohort. Further studies are warranted to unravel the mechanisms behind the discrepancy between mRNA and protein level of ROR1 in PDAC.
Compared to ROR1, ROR2 has been less investigated as a cancer drug target, likely due to its complex dual role in carcinogenesis [46]. ROR2 has been reported to play either an oncogenic or tumour suppressor role based on different tumour context and appears more tightly linked to metastasis than proliferation [47–49]. Unlike ROR1, there are currently fewer ROR2 targeted drugs in clinical trials. A conditionally active biologic (CAB)-ROR2- antibody drug conjugate (ADC) named BA3021 was developed to selectively bind to ROR2 in the context of tumour microenvironment and has been reported to inhibit growth of ROR2 positive cell lines and xenografts of several cancers [50]. This drug is currently under Phase I/II trials for solid tumour, non-small cell lung cancer, triple negative breast cancer and soft tissue sarcoma (NCT03504488).
Our analysis of public datasets showed low ROR2 mRNA expression level correlated with worse OS in PDAC in the ICGC and TCGA cohorts (Fig. 1 C,D), which was contrary to the IHC staining results from a previous study [24]. For the ROR2 mRNA and protein level of the CPTAC cohort, no significant association was observed with OS (Fig. 1 E,F). That specific study reported that high ROR2 protein expression, indicated by both stromal and cytoplasmic staining, was correlated with worse OS and stage in a cohort of 162 PDAC cases. They used a polyclonal antibody against ROR2, and it should be noted that the validity of a number of ROR2 antibodies has been shown to be questionable [51]. We strongly recommend an independent assessment of ROR2 protein expression in a pancreatic cancer cohort to clarify this question.
In conclusion, ROR1 appears to be promising biomarkers in predicting survival outcome of PDAC patients at the mRNA expression level. However its use as protein biomarkers remains unclear. Our IHC analysis in a large well-defined PDAC cohort as well as the proteomic data of the CPTAC PDAC discovery cohort does not support the use of ROR1 as a prognostic biomarker at protein level in PDAC and highlights the discrepancy of prognostic biomarkers when measured by MS, IHC and RNAseq. The mechanism underlying the insufficient correlation between mRNA and protein level of ROR1 and ROR2 in PDAC remains unclear and warrants future investigation.
Supplementary Information
Additional file 1 : Supplementary Fig.S1. Correlation between mRNA and protein expression of ROR1 and ROR2 in CPTAC PDAC discovery cohort. A. ROR1 mRNA and protein expression was positively correlated (Pearson’s R=0.25, p=0.005). B. ROR2 mRNA and protein expression was positively correlated (Pearson’s R=0.48, p<0.001).
Acknowledgements
We would like to acknowledge Australian Pancreatic Cancer Genome Initiative (APGI) for providing biospecimens and clinicopathological data and the Kinghorn Cancer Centre Histopathology services for performing IHC. Australian Pancreatic Cancer Genome Initiative (APGI): Garvan Institute of Medical Research Amber L. Johns1, Anthony J Gill1, 5, Lorraine A. Chantrill1,22, Paul Timpson1, Angela Chou1,5, Marina Pajic1, Tanya Dwarte1, David Herrmann1, Claire Vennin1, Thomas R Cox1, Brooke Pereira1, Shona Ritchie1, Daniel A Reed1, Cecilia R Chambers1, Xanthe Metcalf1, Max Nobis1. QIMR Berghofer Medical Research Institute Nicola Waddell2, John V. Pearson2, Ann-Marie Patch2, Katia Nones2, Felicity Newell2, Pamela Mukhopadhyay2, Venkateswar Addala2, Stephen Kazakoff2, Oliver Holmes2, Conrad Leonard2, Scott Wood2. University of Melbourne, Centre for Cancer Research Sean M. Grimmond3, Oliver Hofmann3. Royal North Shore Hospital Jaswinder S. Samra5, Nick Pavlakis5, Jennifer Arena5, Hilda A. High5. Bankstown Hospital Ray Asghari6, Neil D. Merrett6, Amitabha Das6. Liverpool Hospital Peter H. Cosman7, Kasim Ismail7. St Vincent’s Hospital Alina Stoita8, David Williams8, Allan Spigellman8. Westmead Hospital Vincent W. Lam9, Duncan McLeod9, Judy Kirk9. Royal Prince Alfred Hospital, Chris O’Brien Lifehouse James G. Kench10, Peter Grimison10, Charbel Sandroussi10, Annabel Goodwin7,10. Prince of Wales Hospital R. Scott Mead1,11, Katherine Tucker11, Lesley Andrews11. Fiona Stanley Hospital Michael Texler12, Cindy Forrest12, Mo Ballal12,13, David Fletcher12. St John of God Healthcare Maria Beilin13, Kynan Feeney13 Krishna Epari13 Sanjay Mukhedkar13. Epworth HealthCare Nikolajs Zeps23. Royal Adelaide Hospital Nan Q Nguyen14, Andrew R. Ruszkiewicz14, Chris Worthley14. Flinders Medical Centre John Chen15, Mark E. Brooke-Smith15, Virginia Papangelis15. Envoi Pathology Andrew D. Clouston16. Princess Alexandra Hospital Andrew P. Barbour17, Thomas J. O’Rourke17, Jonathan W. Fawcett17, Kellee Slater17, Michael Hatzifotis17, Peter Hodgkinson17. Austin Hospital Mehrdad Nikfarjam18. Johns Hopkins Medical Institutes James R. Eshleman19, Ralph H. Hruban19, Christopher L. Wolfgang19. ARC-Net Centre for Applied Research on Cancer Aldo Scarpa20, Rita T. Lawlor20, Vincenzo Corbo20, Claudio Bassi20. University of Glasgow Andrew V Biankin 21, Nigel B. Jamieson21 David K. Chang1, 21, Stephan B. Dreyer21. 1The Kinghorn Cancer Centre, Garvan Institute of Medical Research, 370 Victoria Street, Darlinghurst, Sydney, New South Wales 2010, Australia. 2QIMR Berghofer Medical Research Institute, 300 Herston Rd., Herston, Queensland 4006, Australia. 3University of Melbourne, Centre for Cancer Research, Victorian Comprehensive Cancer Centre, 305 Grattan Street, Melbourne, Victoria 3000, Australia. 4 Institute for Molecular Bioscience, University of QLD, St Lucia, Queensland 4072, Australia.5Royal North Shore Hospital, Westbourne Street, St Leonards, New South Wales 2065, Australia.6Bankstown Hospital, Eldridge Road, Bankstown, New South Wales 2200, Australia.7Liverpool Hospital, Elizabeth Street, Liverpool, New South Wales 2170, Australia.8 St Vincent’s Hospital, 390 Victoria Street, Darlinghurst, New South Wales, 2010 Australia. 9Westmead Hospital, Hawkesbury and Darcy Roads, Westmead, New South Wales 2145, Australia.10Royal Prince Alfred Hospital, Missenden Road, Camperdown, New South Wales 2050, Australia.11Prince of Wales Hospital, Barker Street, Randwick, New South Wales 2031, Australia. 12Fremantle Hospital, Alma Street, Fremantle, Western Australia 6959, Australia.13 St John of God Healthcare, 12 Salvado Road, Subiaco, Western Australia 6008, Australia.14 Royal Adelaide Hospital, North Terrace, Adelaide, South Australia 5000, Australia.15 Flinders Medical Centre, Flinders Drive, Bedford Park, South Australia 5042, Australia.16 Envoi Pathology, 1/49 Butterfield Street, Herston, Queensland 4006, Australia.17 Princess Alexandra Hospital, 199 Ipswich Rd., Woolloongabba QLD 4102. 18 Austin Hospital, 145 Studley Road, Heidelberg, Victoria 3084, Australia.19 Johns Hopkins Medical Institute, 600 North Wolfe Street, Baltimore, Maryland 21287, USA.20 ARC-NET Center for Applied Research on Cancer, University of Verona, Via dell’Artigliere, 19 37129 Verona, Province of Verona, Italy.21 Wolfson Wohl Cancer Research Centre, Institute of Cancer Sciences, University of Glasgow, Garscube Estate, Switchback Road, Bearsden, Glasgow, Scotland G61 1BD, United Kingdom.22 Wollongong Hospital, Illawarra and Shoalhaven Local Health District, Loftus Street, Wollongong NSW 2500.23 Epworth HealthCare, 89 Bridge Rd., Richmond VIC 3121, Australia.
Authors’ contributions
Conceptualization, D.L., G.S., P.P. and C.E.F.; methodology, D.L. APGI and G.S.; resources, APGI; formal analysis, D.L.; writing—D.L.; writing—review and editing, G.S., P.P. and C.E.F.; funding acquisition, C.E.F. All authors have reviewed and approved this manuscript.
Funding
We acknowledge funding from NHMRC (Ideas Grant, APP2002707, Phillips, Sharbeen; Project grant, APP1144108, Phillips), Cancer Australia/Cancer Council (APP1126736, Phillips) and Cancer-Institute NSW Career Development Fellowship (CDF181166, Sharbeen), Tour de Cure Senior Research Grant (RSP-235-19/20) and Tour de Cure Pioneering Research Grant (RSP-255-19/20, Sharbeen, Phillips).
Availability of data and materials
The datasets analysed during the current study are available in the International Cancer Genome Consortium (ICGC, https://icgc.org/), the Cancer Genome Atlas (TCGA, http://cancergenome.nih.gov/) and the National Cancer Institute’s Clinical Proteomic Tumor Analysis Consortium (CPTAC, https://pdc.cancer.gov/pdc/).
Declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Caroline E. Ford, Email: caroline.ford@unsw.edu.au
Australian Pancreatic Cancer Genome Initiative:
Amber L. Johns, Anthony J. Gill, Lorraine A. Chantrill, Paul Timpson, Angela Chou, Marina Pajic, Tanya Dwarte, David Herrmann, Claire Vennin, Thomas R. Cox, Brooke Pereira, Shona Ritchie, Daniel A. Reed, Cecilia R. Chambers, Xanthe Metcalf, Max Nobis, Nicola Waddell, John V. Pearson, Ann-Marie Patch, Katia Nones, Felicity Newell, Pamela Mukhopadhyay, Venkateswar Addala, Stephen Kazakoff, Oliver Holmes, Conrad Leonard, Scott Wood, Sean M. Grimmond, Oliver Hofmann, Jaswinder S. Samra, Nick Pavlakis, Jennifer Arena, Hilda A. High, Ray Asghari, Neil D. Merrett, Amitabha Das, Peter H. Cosman, Kasim Ismail, Alina Stoita, David Williams, Allan Spigellman, Vincent W. Lam, Duncan McLeod, Judy Kirk, James G. Kench, Peter Grimison, Charbel Sandroussi, Annabel Goodwin, R. Scott Mead, Katherine Tucker, Lesley Andrews, Michael Texler, Cindy Forrest, Mo Ballal, David Fletcher, Maria Beilin, Kynan Feeney, Krishna Epari, Sanjay Mukhedkar, Nikolajs Zeps, Nan Q. Nguyen, Andrew R. Ruszkiewicz, Chris Worthley, John Chen, Mark E. Brooke-Smith, Virginia Papangelis, Andrew D. Clouston, Andrew P. Barbour, Thomas J. O’Rourke, Jonathan W. Fawcett, Kellee Slater, Michael Hatzifotis, Peter Hodgkinson, Mehrdad Nikfarjam, James R. Eshleman, Ralph H. Hruban, Christopher L. Wolfgang, Aldo Scarpa, Rita T. Lawlor, Vincenzo Corbo, Claudio Bassi, Andrew V. Biankin, Nigel B. Jamieson, David K. Chang, and Stephan B. Dreyer
References
- 1.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin. 2019;69(1):7–34. doi: 10.3322/caac.21551. [DOI] [PubMed] [Google Scholar]
- 3.Bosetti C, Lucenteforte E, Silverman DT, Petersen G, Bracci PM, Ji BT, Negri E, Li D, Risch HA, Olson SH, Gallinger S. Cigarette smoking and pancreatic cancer: an analysis from the International Pancreatic Cancer Case-Control Consortium (Panc4). Annals of oncology. 2012;23(7):1880-8. [DOI] [PMC free article] [PubMed]
- 4.Klein AP, Brune KA, Petersen GM, Goggins M, Tersmette AC, Offerhaus GJA, Griffin C, Cameron JL, Yeo CJ, Kern S, Hruban RH. Prospective risk of pancreatic cancer in familial pancreatic cancer kindreds. Cancer Res. 2004;64(7):2634–2638. doi: 10.1158/0008-5472.CAN-03-3823. [DOI] [PubMed] [Google Scholar]
- 5.Jones S, Hruban RH, Kamiyama M, Borges M, Zhang X, Parsons DW, et al. Exomic sequencing identifies PALB2 as a pancreatic cancer susceptibility gene. Science. 2009;324(5924):217. doi: 10.1126/science.1171202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hu ZI, Shia J, Stadler ZK, Varghese AM, Capanu M, Salo-Mullen E, Lowery MA, Diaz LA, Jr, Mandelker D, Yu KH, Zervoudakis A, Kelsen DP, Iacobuzio-Donahue CA, Klimstra DS, Saltz LB, Sahin IH, O'Reilly EM. Evaluating mismatch repair deficiency in pancreatic adenocarcinoma: challenges and recommendations. Clin Cancer Res. 2018;24(6):1326–1336. doi: 10.1158/1078-0432.CCR-17-3099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rahib L, Smith BD, Aizenberg R, Rosenzweig AB, Fleshman JM, Matrisian LM. Projecting cancer incidence and deaths to 2030: the unexpected burden of thyroid, liver, and pancreas cancers in the United States. Cancer Res. 2014;74(11):2913–2921. doi: 10.1158/0008-5472.CAN-14-0155. [DOI] [PubMed] [Google Scholar]
- 8.Collisson EA, Bailey P, Chang DK, Biankin AV. Molecular subtypes of pancreatic cancer. Nat Rev Gastroenterol Hepatol. 2019;16(4):207–220. doi: 10.1038/s41575-019-0109-y. [DOI] [PubMed] [Google Scholar]
- 9.Rawla P, Sunkara T, Gaduputi V. Epidemiology of pancreatic cancer: global trends, etiology and risk factors. World J Oncol. 2019;10(1):10–27. doi: 10.14740/wjon1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Von Hoff DD, Ervin T, Arena FP, Chiorean EG, Infante J, Moore M, et al. Increased survival in pancreatic cancer with nab-paclitaxel plus gemcitabine. N Engl J Med. 2013;369(18):1691–1703. doi: 10.1056/NEJMoa1304369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Moore MJ, Goldstein D, Hamm J, Figer A, Hecht JR, Gallinger S, Au HJ, Murawa P, Walde D, Wolff RA, Campos D, Lim R, Ding K, Clark G, Voskoglou-Nomikos T, Ptasynski M, Parulekar W, National Cancer Institute of Canada Clinical Trials Group Erlotinib plus gemcitabine compared with gemcitabine alone in patients with advanced pancreatic cancer: a phase III trial of the National Cancer Institute of Canada clinical trials group. J Clin Oncol. 2007;25(15):1960–1966. doi: 10.1200/JCO.2006.07.9525. [DOI] [PubMed] [Google Scholar]
- 12.Collisson EA, Sadanandam A, Olson P, Gibb WJ, Truitt M, Gu S, Cooc J, Weinkle J, Kim GE, Jakkula L, Feiler HS, Ko AH, Olshen AB, Danenberg KL, Tempero MA, Spellman PT, Hanahan D, Gray JW. Subtypes of pancreatic ductal adenocarcinoma and their differing responses to therapy. Nat Med. 2011;17(4):500–503. doi: 10.1038/nm.2344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Royal RE, Levy C, Turner K, Mathur A, Hughes M, Kammula US, et al. Phase 2 trial of single agent Ipilimumab (anti-CTLA-4) for locally advanced or metastatic pancreatic adenocarcinoma. J Immunother (Hagerstown, Md: 1997) 2010;33(8):828. doi: 10.1097/CJI.0b013e3181eec14c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Le DT, Lutz E, Uram JN, Sugar EA, Onners B, Solt S, et al. Evaluation of ipilimumab in combination with allogeneic pancreatic tumor cells transfected with a GM-CSF gene in previously treated pancreatic cancer. J Immunother (Hagerstown, Md: 1997) 2013;36(7):382. doi: 10.1097/CJI.0b013e31829fb7a2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.O’Reilly EM, Oh D-Y, Dhani N, Renouf DJ, Lee MA, Sun W, Fisher G, Hezel A, Chang SC, Vlahovic G, Takahashi O, Yang Y, Fitts D, Philip PA. Durvalumab with or without tremelimumab for patients with metastatic pancreatic ductal adenocarcinoma: a phase 2 randomized clinical trial. JAMA Oncol. 2019;5(10):1431–1438. doi: 10.1001/jamaoncol.2019.1588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Murtaugh LC, Law AC, Dor Y, Melton DA. β-Catenin is essential for pancreatic acinar but not islet development. Development. 2005;132(21):4663–4674. doi: 10.1242/dev.02063. [DOI] [PubMed] [Google Scholar]
- 17.Lowy AM, Fenoglio-Preiser C, Kim OJ, Kordich J, Gomez A, Knight J, James L, Groden J. Dysregulation of β-catenin expression correlates with tumor differentiation in pancreatic duct adenocarcinoma. Ann Surg Oncol. 2003;10(3):284–290. doi: 10.1245/ASO.2003.05.003. [DOI] [PubMed] [Google Scholar]
- 18.Raphael BJ, Hruban RH, Aguirre AJ, Moffitt RA, Yeh JJ, Stewart C, et al. Integrated genomic characterization of pancreatic ductal adenocarcinoma. Cancer Cell. 2017;32(2):185–203. e13. doi: 10.1016/j.ccell.2017.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jiang X, Hao H-X, Growney JD, Woolfenden S, Bottiglio C, Ng N, Lu B, Hsieh MH, Bagdasarian L, Meyer R, Smith TR, Avello M, Charlat O, Xie Y, Porter JA, Pan S, Liu J, McLaughlin ME, Cong F. Inactivating mutations of RNF43 confer Wnt dependency in pancreatic ductal adenocarcinoma. Proc Natl Acad Sci. 2013;110(31):12649–12654. doi: 10.1073/pnas.1307218110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.DaneshManesh AH, Mikaelsson E, Jeddi-Tehrani M, Bayat AA, Ghods R, Ostadkarampour M, et al. Ror1, a cell surface receptor tyrosine kinase is expressed in chronic lymphocytic leukemia and may serve as a putative target for therapy. Int J Cancer. 2008;123(5):1190–1195. doi: 10.1002/ijc.23587. [DOI] [PubMed] [Google Scholar]
- 21.Zhang S, Chen L, Cui B, Chuang H-Y, Yu J, Wang-Rodriguez J, Tang L, Chen G, Basak GW, Kipps TJ. ROR1 is expressed in human breast cancer and associated with enhanced tumor-cell growth. PLoS One. 2012;7(3):e31127. doi: 10.1371/journal.pone.0031127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Henry C, Emmanuel C, Lambie N, Loo C, Kan B, Kennedy C, et al. Distinct patterns of stromal and tumor expression of ROR1 and ROR2 in histological subtypes of epithelial ovarian cancer. Transl Oncol. 2017;10(3):346–356. doi: 10.1016/j.tranon.2017.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu D, Gunther K, Enriquez LA, Daniels B, O’Mara TA, Tang K, Spurdle AB, Ford CE. ROR1 is upregulated in endometrial cancer and represents a novel therapeutic target. Sci Rep. 2020;10(1):13906. doi: 10.1038/s41598-020-70924-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huang J, Fan X, Wang X, Lu Y, Zhu H, Wang W, Zhang S, Wang Z. High ROR2 expression in tumor cells and stroma is correlated with poor prognosis in pancreatic ductal adenocarcinoma. Sci Rep. 2015;5(1):12991. doi: 10.1038/srep12991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang S, Chen L, Wang-Rodriguez J, Zhang L, Cui B, Frankel W, Wu R, Kipps TJ. The onco-embryonic antigen ROR1 is expressed by a variety of human cancers. Am J Pathol. 2012;181(6):1903–1910. doi: 10.1016/j.ajpath.2012.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Balakrishnan A, Goodpaster T, Randolph-Habecker J, Hoffstrom BG, Jalikis FG, Koch LK, Berger C, Kosasih PL, Rajan A, Sommermeyer D, Porter PL, Riddell SR. Analysis of ROR1 protein expression in human cancer and normal tissues. Clin Cancer Res. 2017;23(12):3061–3071. doi: 10.1158/1078-0432.CCR-16-2083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fukuda T, Chen L, Endo T, Tang L, Lu D, Castro JE, Widhopf GF, Rassenti LZ, Cantwell MJ, Prussak CE, Carson DA, Kipps TJ. Antisera induced by infusions of autologous ad-CD154-leukemia B cells identify ROR1 as an oncofetal antigen and receptor for Wnt5a. Proc Natl Acad Sci. 2008;105(8):3047–3052. doi: 10.1073/pnas.0712148105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sheetz JB, Mathea S, Karvonen H, Malhotra K, Chatterjee D, Niininen W, et al. Structural Insights into Pseudokinase Domains of Receptor Tyrosine Kinases. Mol Cell. 2020;79, 390(3):–405.e7. [DOI] [PMC free article] [PubMed]
- 29.Hojjat-Farsangi M, Daneshmanesh A, Khan A, Shetye J, Mozaffari F, Kharaziha P, et al. First-in-class oral small molecule inhibitor of the tyrosine kinase ROR1 (KAN0439834) induced significant apoptosis of chronic lymphocytic leukemia cells. Leukemia. 2018;32(10):2291–2295. doi: 10.1038/s41375-018-0113-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Choi MY, Widhopf GF, II, Ghia EM, Kidwell RL, Hasan MK, Yu J, et al. Phase I trial: cirmtuzumab inhibits ROR1 signaling and stemness signatures in patients with chronic lymphocytic leukemia. Cell Stem Cell. 2018;22(6):951–9. e3. doi: 10.1016/j.stem.2018.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Choi MY, Wierda WG, Lee HJ, Tzachanis D, Ianopulos X, Jezior D, et al. Phase 1/2 trial of cirmtuzumab and ibrutinib: planned analysis of phase 1 CLL cohorts. Am Soc Clin Oncol. 2019. 10.1200/jco.2019.37.15_suppl.7527.
- 32.Antibody–drug conjugates for cancer score with ROR1. Nat Biotechnol. 2021;39:10. 10.1038/s41587-020-00798-z. [DOI] [PubMed]
- 33.Bailey P, Chang DK, Nones K, Johns AL, Patch A-M, Gingras M-C, et al. Genomic analyses identify molecular subtypes of pancreatic cancer. Nature. 2016;531(7592):47–52. doi: 10.1038/nature16965. [DOI] [PubMed] [Google Scholar]
- 34.Goldman M, Craft B, Brooks A, Zhu J, Haussler D. The UCSC Xena platform for cancer genomics data visualization and interpretation. BioRxiv. 2018;326470. 10.1101/326470.
- 35.Edwards NJ, Oberti M, Thangudu RR, Cai S, McGarvey PB, Jacob S, et al. The CPTAC data portal: a resource for cancer proteomics research. J Proteome Res. 2015;14(6):2707–2713. doi: 10.1021/pr501254j. [DOI] [PubMed] [Google Scholar]
- 36.Al-Shawi R, Ashton SV, Underwood C, Simons JP. Expression of the Ror1 and Ror2 receptor tyrosine kinase genes during mouse development. Dev Genes Evol. 2001;211(4). 10.1007/s004270100140. [DOI] [PubMed]
- 37.Baskar S, Kwong KY, Hofer T, Levy JM, Kennedy MG, Lee E, Staudt LM, Wilson WH, Wiestner A, Rader C. Unique cell surface expression of receptor tyrosine kinase ROR1 in human B-cell chronic lymphocytic leukemia. Clin Cancer Res. 2008;14(2):396–404. doi: 10.1158/1078-0432.CCR-07-1823. [DOI] [PubMed] [Google Scholar]
- 38.Hudecek M, Lupo-Stanghellini M-T, Kosasih PL, Sommermeyer D, Jensen MC, Rader C, Riddell SR. Receptor affinity and extracellular domain modifications affect tumor recognition by ROR1-specific chimeric antigen receptor T cells. Clin Cancer Res. 2013;19(12):3153–3164. doi: 10.1158/1078-0432.CCR-13-0330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Daneshmanesh AH, Hojjat-Farsangi M, Ghaderi A, Moshfegh A, Hansson L, Schultz J, Vågberg J, Byström S, Olsson E, Olin T, Österborg A, Mellstedt H. A receptor tyrosine kinase ROR1 inhibitor (KAN0439834) induced significant apoptosis of pancreatic cells which was enhanced by erlotinib and ibrutinib. PLoS One. 2018;13(6):e0198038. doi: 10.1371/journal.pone.0198038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Moffitt RA, Marayati R, Flate EL, Volmar KE, Loeza SGH, Hoadley KA, Rashid NU, Williams LA, Eaton SC, Chung AH, Smyla JK, Anderson JM, Kim HJ, Bentrem DJ, Talamonti MS, Iacobuzio-Donahue CA, Hollingsworth MA, Yeh JJ. Virtual microdissection identifies distinct tumor-and stroma-specific subtypes of pancreatic ductal adenocarcinoma. Nat Genet. 2015;47(10):1168–1178. doi: 10.1038/ng.3398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Song S, Nones K, Miller D, Harliwong I, Kassahn KS, Pinese M, et al. qpure: A tool to estimate tumor cellularity from genome-wide single-nucleotide polymorphism profiles. PLoS One. 2012;7(9):e45835. doi: 10.1371/journal.pone.0045835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Têtu B, Popa I, Bairati I, L'esperance S, Bachvarova M, Plante M, et al. Immunohistochemical analysis of possible chemoresistance markers identified by micro-arrays on serous ovarian carcinomas. Mod Pathol. 2008;21(8):1002–1010. doi: 10.1038/modpathol.2008.80. [DOI] [PubMed] [Google Scholar]
- 43.Greenbaum D, Colangelo C, Williams K, Gerstein M. Comparing protein abundance and mRNA expression levels on a genomic scale. Genome Biol. 2003;4(9):1–8. doi: 10.1186/gb-2003-4-9-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kaucká M, Krejčí P, Plevová K, Pavlová Š, Prochazkova J, Janovská P, et al. Post-translational modifications regulate signalling by Ror1. Acta Physiol. 2011;203(3):351–362. doi: 10.1111/j.1748-1716.2011.02306.x. [DOI] [PubMed] [Google Scholar]
- 45.Reddy UR, Phatak S, Pleasure D. Human neural tissues express a truncated Ror1 receptor tyrosine kinase, lacking both extracellular and transmembrane domains. Oncogene. 1996;13(7):1555–1559. [PubMed] [Google Scholar]
- 46.Ford CE, Qian Ma SS, Quadir A, Ward RL. The dual role of the novel Wnt receptor tyrosine kinase, ROR2, in human carcinogenesis. Int J Cancer. 2013;133(4):779–787. doi: 10.1002/ijc.27984. [DOI] [PubMed] [Google Scholar]
- 47.Tseng J-C, Huang S-H, Lin C-Y, Wang B-J, Huang S-F, Shen Y-Y, et al. ROR2 suppresses metastasis of prostate cancer via regulation of miR-199a-5p–PIAS3–AKT2 signaling axis. Cell Death Dis. 2020;11(5):1–11. doi: 10.1038/s41419-020-2587-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bayerlová M, Menck K, Klemm F, Wolff A, Pukrop T, Binder C, Beißbarth T, Bleckmann A. Ror2 signaling and its relevance in breast cancer progression. Front Oncol. 2017;7:135. doi: 10.3389/fonc.2017.00135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Xu J, Shi J, Tang W, Jiang P, Guo M, Zhang B, Ma G. ROR2 promotes the epithelial-mesenchymal transition by regulating MAPK/p38 signaling pathway in breast cancer. J Cell Biochem. 2020;121(10):4142–4153. doi: 10.1002/jcb.29666. [DOI] [PubMed] [Google Scholar]
- 50.Sharp LL, Chang C, Frey G, Wang J, Liu H, Xing C, et al. Anti-tumor efficacy of BA3021, a novel conditionally active biologic (CAB) anti-ROR2 ADC. Chicago: AACR; 2018.
- 51.Ma SS, Henry CE, Llamosas E, Higgins R, Daniels B, Hesson LB, et al. Validation of specificity of antibodies for immunohistochemistry: the case of ROR2. Virchows Arch. 2017;470(1):99–108. doi: 10.1007/s00428-016-2019-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1 : Supplementary Fig.S1. Correlation between mRNA and protein expression of ROR1 and ROR2 in CPTAC PDAC discovery cohort. A. ROR1 mRNA and protein expression was positively correlated (Pearson’s R=0.25, p=0.005). B. ROR2 mRNA and protein expression was positively correlated (Pearson’s R=0.48, p<0.001).
Data Availability Statement
The datasets analysed during the current study are available in the International Cancer Genome Consortium (ICGC, https://icgc.org/), the Cancer Genome Atlas (TCGA, http://cancergenome.nih.gov/) and the National Cancer Institute’s Clinical Proteomic Tumor Analysis Consortium (CPTAC, https://pdc.cancer.gov/pdc/).