Abstract
Cyclical inactivation of B-type cyclins has been proposed to be required for alternating DNA replication and mitosis. Destruction box-dependent Clb5p degradation is strongly increased in mitotic cells, and constitutive overexpression of Clb5p lacking the destruction box resulted in rapid accumulation of inviable cells, frequently multiply budded, with DNA contents ranging from unreplicated to apparently fully replicated. Loss of viability correlated with retention of nuclear Clb5p at the time of nuclear division. CLB2-Δdb overexpression that was quantitatively comparable to CLB5-Δdb overexpression with respect to Clb protein production and Clb-associated kinase activity resulted in a distinct phenotype: reversible mitotic arrest with uniformly replicated DNA. Simultaneous overexpression of CLB2-Δdb and CLB5-Δdb overexpressers similarly resulted in a uniform arrest with replicated DNA, and this arrest was significantly more reversible than that observed with CLB5-Δdb overexpression alone. These results suggest that Clb2p and not Clb5p can efficiently block mitotic completion. We speculate that CLB5-Δdb overexpression may be lethal, because persistence of high nuclear Clb5p-associated kinase throughout mitosis leads to failure to load origins of replication, thus preventing DNA replication in the succeeding cell cycle.
Cyclin-dependent kinase activity drives the eukaryotic cell cycle. In Saccharomyces cerevisiae, three G1, or CLN, cyclins and six B-type, or CLB, cyclins bind and activate the cyclin-dependent kinase Cdc28p. CLB function is required for initiation of DNA replication, spindle formation, and initiation of mitosis. With respect to DNA replication and mitosis, the main role of the CLN cyclins is to allow activation of Clbp-Cdc28p kinase, although the CLN cyclins have additional cell cycle roles (6, 26).
It is likely that all of the CLB cyclins are descendants of a single B-type cyclin-like ancestor, and it has been proposed (28) that a single B-type cyclin regulated both DNA replication and mitosis in a primordial eukaryotic cell. Multiple B-type cyclins derived from gene duplication have diverged in function. Functional divergence could simply reflect different timing of accumulation of functionally interchangeable cyclins; alternatively, specific cyclin coding sequences could have become intrinsically specialized for particular cell cycle roles. Recently, we showed that Clb5p is intrinsically specialized for activation of replication in comparison to Clb2p (8).
Clbp-Cdc28p kinase drives some essential step(s) in replication, including the binding of Cdc45p and replication protein A (RPA) to the prereplicative complex (PRC) (44, 50). The PRC is formed by Cdc6p-dependent loading of minichromosome maintenance (MCM) proteins onto the origin recognition complex at origins of replication. PRC formation occurs in the absence of Cdk activity. B-type cyclin-associated kinase activity is thought to limit DNA replication to once per cell cycle by blocking loading of MCM proteins onto origins until B cyclin-Cdk inactivation at the end of mitosis (reviewed in references 26 and 29). There is conflict over whether inactivation of anaphase-promoting complex (APC) components allows rereplication in a single cell cycle even in the presence of Clbp-Cdc28p kinase (17, 18, 32).
High Clb2-associated kinase activity blocks exit from mitosis: cells expressing high levels of Clb2p arrest with long spindles and separated chromosomes before cytokinesis (43). Thus, there is an additional requirement for the level of Clb-associated kinases to fall for the cell cycle to cycle. It is unclear if all Clb-associated kinases are efficient at inhibition of mitotic exit.
If Clb-associated kinases have both positive and negative roles in the cell cycle, their accumulation and degradation must be accurately regulated. Clb2p degradation is restricted to late mitosis after chromosome separation and the subsequent G1 period before initiation of the succeeding cell cycle. This is probably due to the requirement for Cdh1p to associate with the APC to allow Clb2p degradation. Cdh1p is inactive due to Cdk-mediated phosphorylation and is activated late in the cell cycle by the Cdc14p phosphatase (20, 41, 47). Clb5p is not under the control of Cdh1p, and relatively little cell cycle regulation of Clb5p degradation has been observed (39), although its degradation was reported to be destruction box dependent and dependent on components of the APC that are also required for Clb2p ubiquitination and degradation (19). Clb5p and the anaphase inhibitor Pds1p may both be targets of Cdc20p-directed APC degradation, because it has been observed that deleting CLB5 rescues cdc20 pds1 strains which would otherwise arrest in late anaphase, and deleting cdc20 stabilizes Clb5p (40).
Here we report on cell cycle dependence of Clb5p degradation. We also compare the effects of expression of stabilized Clb5p and Clb2p on cell cycle progression, to examine the issue of intrinsic Clb specialization in driving cell cycle events.
MATERIALS AND METHODS
Yeast strains.
All strains are isogenic with 15Dau (MATa leu2 ura3 trp1 his2 ade1) (33). Strains were constructed and analyzed by standard genetic methods. DNA transformations were done by the lithium acetate method.
Plasmids.
All plasmids were derived from CE119, a YCP50-based GAL1::CLB5 construct (GAL1::CLB5 URA3 CEN4 ARS1 Ampr) (11, 30). CE119-4, a hemagglutinin (HA)-tagged version of CE119, and the GAL1::CLB5-Δdb-HA construct DB4 were described previously (8). A point mutation, S399P, presumably generated during the PCR-based construction of CE119 (11), was present in CE119-4 and in the GAL1::CLB5-Δdb-HA construct. The S399P point mutation is relatively innocuous; it slightly reduces Clb5p function, but CLB5-S399P under control of the CLB5 promoter fully rescues the clb5 replication defect (12) and clb3,4,5,6 lethality (37), and GAL1::CLB5-Δdb-S399P is lethal with a phenotype similar to that of the S399S version (data not shown). The experiments in Fig. 1, 2, 4, and 5 were performed with S399P versions of GAL1::CLB5. The remaining experiments were performed with constructs in which the S399P mutation was repaired by subcloning the wild-type fragment to make CE119-4R and DB4R. We assume that this does not significantly affect the results. The destruction box-dependent instability of Clb5p documented in Fig. 2 has been found to be similar to those of S399S and S399P versions of Clb5p (compare Fig. 2B and 3A).
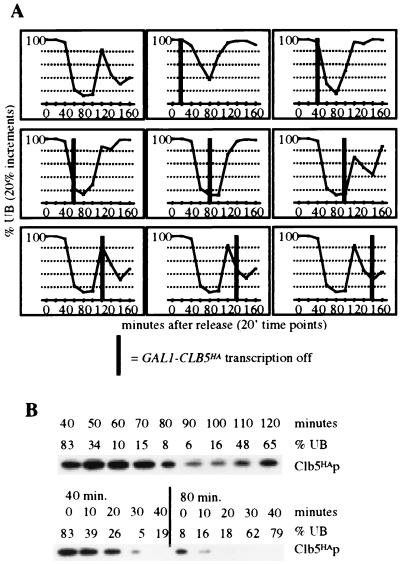
FIG. 1.
Cell cycle-regulated changes in Clb5p stability. 17-48-1 (cln1 cln2 cln3 GAL1::CLB5HA) cultures were synchronized with a cln block, by incubation in raffinose for 2.5 h to turn off GAL1::CLB5HA, followed by galactose addition to 3% to release the block. GAL::CLB5HA transcription was then turned off again at various times by the addition of glucose to a final concentration of 2%. (A) Graphs plotting the percent unbudded cells (% UB) against the time after release. The black bar represents the time at which glucose was added to the culture. (B) Clb5HAp immunoblots and percent unbudded cells in a synchronized culture (top) and in synchronized cultures with GAL1::CLB5HA transcription turned off either 40 or 80 min after release (bottom). Clb5HAp is undetectable in raffinose-arrested cultures of this strain (data not shown).
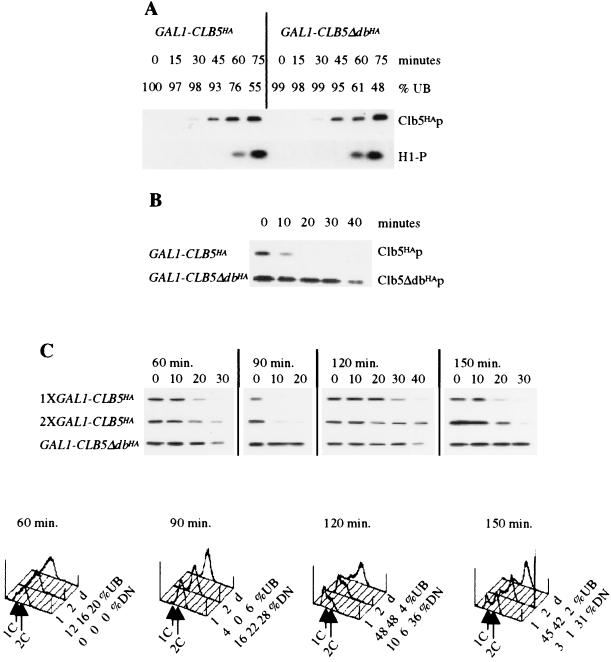
FIG. 2.
A destruction box-dependent decrease in Clb5p stability coincides with nuclear division. All strains have the indicated construct integrated in 1255-5C (wild type). Cultures for each experiment were grown overnight to log phase in YEP medium containing 3% raffinose. (A) Immunoblots for Clb5HAp and Clb5dbHAp (Clb5HAp), histone H1 kinase blots (H1-P), and percent unbudded cells (% UB) after release from an α-factor block in raffinose medium into galactose medium lacking α-factor (YPGal). (B) Clb5HAp and Clb5dbHAp immunoblots, as indicated, from asynchronous cultures after GAL1-driven transcription was turned off by the addition of 2% glucose. (C) Following synchronization with α-factor, cultures were released into YPGal. The GAL1-driven transcription was turned off by the addition of glucose to a final concentration of 2% (time zero) at 60, 90, 120, and 150 min after release. Immunoblots for Clb5HAp from one (1×) or two (2×) copies of GAL1::CLB5HA and for Clb5dbHAp from one copy of GAL1::CLB5dbHA (top) are shown, as are DNA content profiles generated by FACS analysis, percent unbudded cells, and percent of divided nuclei (% DN) as assayed by the presence of two DAPI-stained spots for the 1× GAL1::CLB5HA (1), 2× GAL1::CLB5HA (2), and GAL1::CLB5dbHA (d) cultures (bottom).
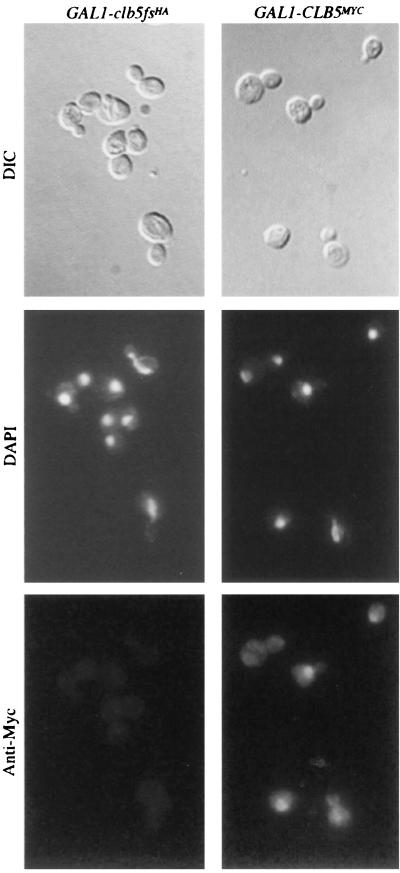
FIG. 4.
Loss of Clb5p nuclear localization in mitotic cells. Photographs of cell bodies (differential interference contrast [DIC]), nuclei visualized by DAPI staining (DAPI), and Myc-tagged protein detected by indirect immunofluorescence (Anti-Myc) from 1255-5C (wild type) strains carrying integrated copies of GAL1-CLB5MYC or GAL1-clb5fsHA. Samples were taken from cycling cultures.
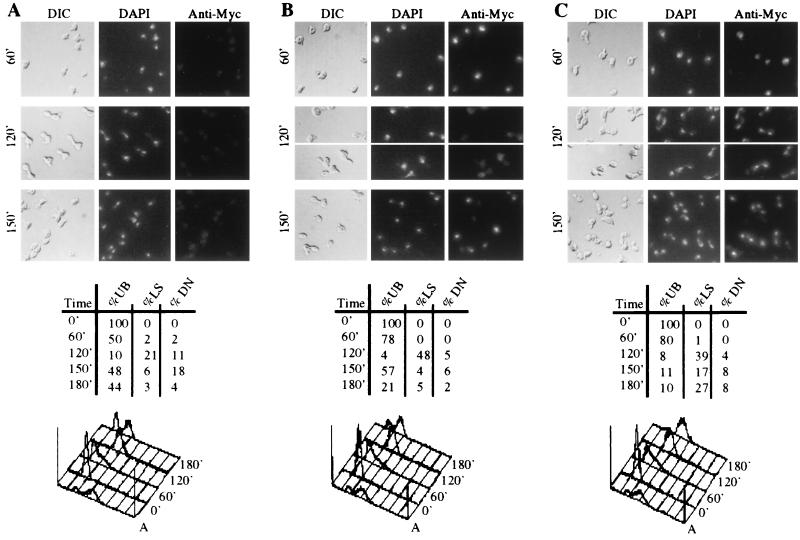
FIG. 5.
Loss of nuclear Clb5p is destruction box dependent. Subcellular localization of Myc-tagged protein and markers for cell cycle progression for synchronized cultures of 1255-5C (wild type) cells containing (A) GAL1-clb5fsHA, (B) GAL1-CLB5MYC, or (C) GAL1-CLB5dbMYC. All cultures were grown and synchronized as in Fig. 1. The photographs show cell bodies (differential interference contrast [DIC]), nuclei visualized by DAPI staining (DAPI), and Myc-tagged protein detected by indirect immunofluorescence (Anti-Myc) at 60 (60′), 120 (120′), and 150 (150′) min after release from α-factor. Data on cell cycle progression in each culture includes the percent unbudded cells (%UB), the percent long spindles (%LS), the percent divided nuclei (%DN), and DNA content profiles. Spindles were visualized by indirect immunofluorescence, nuclei were visualized by DAPI staining, and DNA content profiles were generated by FACS analysis.
FIG. 3.
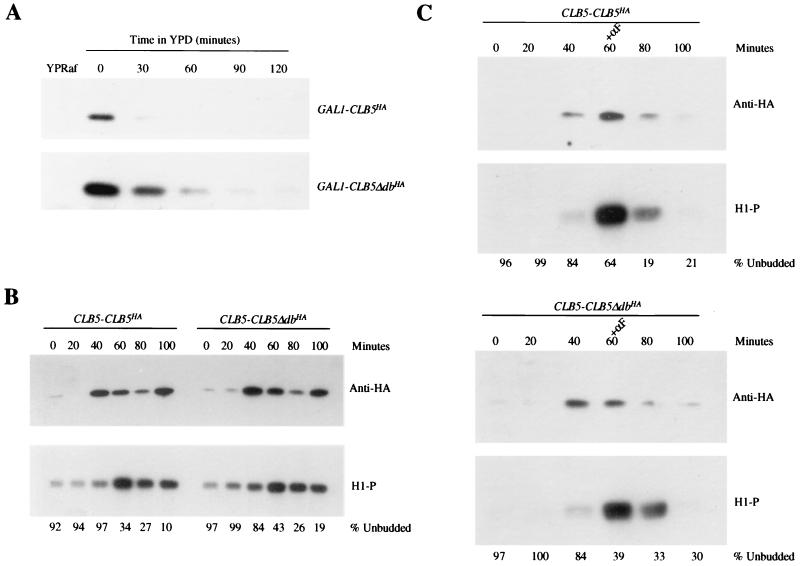
Instability of Clb5Δdb protein when expressed from GAL1 and CLB5 promoters. All strains have the indicated constructs integrated in 1255-5C (wild type). (A) Cultures were grown overnight to log phase in YEP medium containing 3% raffinose (YPRaf). Following 3 h of incubation with 3% galactose to induce the GAL1 promoter, 2% glucose was added to repress expression of GAL1::CLB5HA and GAL1::CLB5ΔdbHA. Protein levels were monitored by Western blotting against the HA tag. (B and C) Strains containing CLB5HA under the control of its endogenous promoter (CLB5-CLB5HA and CLB5-CLB5ΔdbHA) were synchronized in G1 with α-factor. Strains were either released into yeast extract-peptone-dextrose (YPD) (B) or released into YPD and rearrested with α-factor (+αF) 60 min later (C). Synchrony was gauged by counting the percentage of unbudded cells. CLB5HA expression was monitored by immunoblots (anti-HA) and associated histone H1 kinase blots (H1-P).
The control GAL1-clb5fsHA construct is a product of the GAL1-CLB5dbHA construction in which a single nucleotide deletion introduced a frameshift 19 nucleotides into the CLB5HA sequence. GAL1::CLB2-HA was constructed by gap repair of AflII-digested CE119-4R with XhoI-BamHI-digested C52-H4 or C52-HDB1 plasmids containing CLB5::CLB2-HA or CLB5::CLB2-Δdb-HA (8). The GAL1-CLB5MYC and GAL1-CLB5dbMYC plasmids were constructed by exchanging the NotI fragment containing the HA tag for a NotI fragment containing nine repeats of the MYC epitope tag. Two copies of the MYC cassette were inserted in the GAL1-CLB5MYC plasmid, and one copy was inserted in the GAL1-CLB5dbMYC plasmid. The Myc-tagged GAL1-CLB5 constructs all contained the S399P mutation. Integrating derivatives of these plasmids were constructed by digestion with SmaI and HpaI and religation to eliminate the CEN4 sequence and targeted for integrated at ARS1 by BglII digestion. For the GAL1::CLB2 plasmids, integration was targeted to CLB2 by XbaI digestion.
Growth conditions.
Cells were grown at 30°C in (yeast extract-peptone) YEP medium. Arrest of cultures with α-factor was done by incubating log-phase YEP-raffinose (3%) (YEPRaf) cultures with α-factor at a final concentration of 0.1 μM for 2 h. To release the arrest, cells were collected by centrifugation, washed once in 30°C prewarmed YEPRaf, and then resuspended in YEPRaf with 3% galactose.
Protein analysis.
Total protein extraction, anti-HA immunoprecipitation, and histone H1 kinase assays were performed as described previously (22).
FACS analysis and indirect immunofluorescence.
Fluorescence-activated cell sorter analysis (FACS) was performed as described previously (12). For immunofluorescence, cells from a log-phase culture were fixed by rotation at 30°C in 3.7% formaldehyde. After fixation, the cells were pelleted, and the formaldehyde solution was decanted. The pellet was resuspended in 1× phosphate-buffered saline (PBS), sonicated for 12 s, and washed once in PBS and once in sorbitol-citrate buffer (17.4 g of K2HPO4 [anhydrous], 7 g of citric acid, 218.6 g of sorbitol, 2 ml of 1 M dithiothreitol [DTT] in 1 liter of water). The cell walls were digested in sorbitol-citrate buffer with 1 mM DTT, 0.01% zymolyase 20T (wt/vol), 10% glusulase in a 30°C water bath for 2 h with occasional gentle mixing then were washed three times in sorbitol-citrate buffer. Cells were deposited in wells on poly-l-lysine-treated slides (Sigma) for 3 min at room temperature. Cells were fixed to the slide by completely aspirating off the buffer, dehydrating the slide for 5 min in methanol and 5 min in acetone, and quickly air drying the slide. Cells were rehydrated in blocking solution (PBS, 0.2% Tween 20, 2% [wt/vol] nonfat dry milk) at room temperature for 30 min and incubated with the primary antibody (9E10 mouse anti-Myc monoclonal antibody [Santa-Cruz Biotechnology] or YOL1-32 rat antitubulin monoclonal antibody [33]) in blocking solution at a 1:200 dilution for 2 h at room temperature in a humidified chamber. Cells were washed four times briefly and three times for 5 min in blocking solution before adding the secondary antibody (antimouse fluorescein isothiocyanate [FITC]-conjugated polyclonal antibody for Myc, antirat FITC-conjugated polyclonal antibody for tubulin [Jackson ImmunoResearch Laboratories]) at a 1:200 dilution in blocking solution for 2 h at room temperature in the dark in a humidified chamber. Cells were washed as before and then washed three times briefly with PBS before being mounted under a slide cover with mounting medium (10% PBS in glycerol with 22.5 ng of 4′,6′-diamidino-2-phenylindole [DAPI] per ml and 1 mg of phenylenediamine per ml). DAPI staining in samples not processed for immunofluorescence was done as previously described (45). Fluorescence was visualized on a Zeiss Axiophot microscope, and images were captured with a Sony digital photo camera (DKC-5000) by using Photoshop software. Images were manipulated with Photoshop software. Comparable images were treated identically by Photoshop manipulations.
RESULTS
Clb5p degradation is cell cycle regulated.
We constructed a cln1 cln2 cln3 GAL1::CLB5-HA strain, in which CLB5-HA substitutes for the CLN G1 cyclins in driving cell cycle initiation (12, 30). This strain was synchronized by raffinose block in G1, followed by release into the cell cycle with galactose addition to induce expression of the GAL1 promoter. Using timed glucose addition to repress GAL1::CLB5-HA expression, we saw a sharp increase in Clb5p instability approximately coincident with the time of nuclear division (Fig. 1). Consistent with this, shutoff of the GAL1 promoter at any time before 100 min (approximately the time of nuclear division) resulted in failure to bud in the next cell cycle (Fig. 1), consistent with a drop in Clb5p to a nonfunctional level during division (12, 30).
To confirm these results in a wild-type background and to address the role of the Clb5p destruction box in Clb5p degradation, we constructed wild-type strains containing integrated GAL1::CLB5-HA or GAL1::CLB5-Δdb-HA (lacking amino acids 56 to 64) (8). Addition of galactose to cultures of these strains yielded comparable initial accumulation of Clb5p and Clb5Δdbp protein and associated kinase (Fig. 2A). Glucose addition to such galactose-induced cultures to inactivate GAL1-driven transcription showed that the destruction box-containing protein decayed much faster than the destruction box-deleted protein (Fig. 2B). A longer time course, however, revealed that Clb5p lacking its destruction box was still somewhat unstable (Fig. 3A). In addition, at best, minor stabilization of Clb5Δdbp was observed when it was expressed from the endogenous CLB5 promoter (Fig. 3B and C), consistent with a hypothesis that additional regions of Clb5p may contribute to its targeted degradation. Involvement of a Skp1-Cdc53-F box (SCF)-ubiquitinating activity in Clb5p degradation was suggested previously (4).
We employed timed glucose addition to shut off GAL1::CLB5-HA transcription in cells synchronized by α-factor block-release, in which the cultures were blocked in raffinose plus α-factor medium and released into galactose medium lacking α-factor (Fig. 2C). We did this with cells with one or two copies of GAL1::CLB5-HA or one copy of GAL1::CLB5-Δdb-HA. Clb5p was moderately stable in cells completing DNA replication (glucose addition 60 min after release), but by 90 min after release, Clb5p became highly unstable. After division (120 min), Clb5p became highly stable, and stability decreased again later in the second cell cycle. Thus, Clb5p stability is cyclically regulated, with peak instability in dividing cells, consistent with the results in Fig. 1. The destruction box-deleted Clb5-Δdb protein was degraded similarly slowly at each time point. We controlled for increased Clb5p levels due to destruction box deletion with the two-copy GAL1::CLB5-HA integrant; the pattern of instability with this strain was similar to that with the one-copy strain (Fig. 2C). Thus, we observed destruction box-dependent and cell-cycle-dependent Clb5p degradation, with peak instability in dividing cells. The GAL1::CLB5-Δdb-HA strain did not complete nuclear division during the time course (Fig. 2C), and an accumulation of cells with divided nuclei was observed. A further examination of the effects of GAL1::CLB5-Δdb-HA is presented below.
Clb5p, but not Clb5Δdbp, is degraded in the nucleus at mitosis.
We replaced the HA tag with a Myc tag to allow immunofluorescent detection of Clb5p (Fig. 4). (In our hands, the HA tag is not suitable for immunofluorescent detection.) In unbudded cells, small budded cells, and most large budded cells with an undivided nucleus, Clb5p is concentrated in the nucleus. In large budded cells with an undivided DNA mass near or spanning the bud neck and in large budded cells with two DNA signals, Clb5p is distributed diffusely throughout the cell. This pattern indicates that Clb5p accumulates in the nucleus before budding and during DNA replication, but during mitosis, Clb5p nuclear abundance is strikingly reduced.
We compared the localization patterns of Myc-tagged protein throughout the cell cycle in GAL1::CLB5-MYC, GAL1::CLB5Δdb-MYC, and control strains blocked in raffinose plus α-factor medium and released into galactose medium lacking α-factor. The synchrony between samples was similar during DNA replication and entry into nuclear division, but the GAL1::CLB5Δdb-MYC culture did not complete nuclear division during the time course (Fig. 5). The accumulation of binucleate cells in this α-factor synchrony protocol is reduced in GAL1::CLB5Δdb-MYC strains compared to that observed with GAL1::CLB5Δdb-HA (Fig. 2C). We do not know the reason for this, but it correlates with increased stability of the Myc-tagged Clb5-Δdbp compared to the HA-tagged protein (R. Waesch, unpublished data). Clb5p was concentrated in the nucleus of most cells while DNA replication occurred, was reduced to very low levels in the nucleus of most cells when long spindles became abundant, and then became concentrated in the nucleus of most cells again after cytokinesis (Fig. 5), consistent with the results with asynchronous culture (Fig. 4). Clb5-Δdbp remained concentrated in the nucleus throughout the time course, including in cells with DAPI staining, similar to that observed in cells with long spindles (Fig. 5 and data not shown). Our localization results using overexpressed Clb5p agree with those recently reported for endogenously expressed Clb5p (40). Our results also suggest that the observed nuclear persistence of Clb5-Δdbp around the time of division is destruction box dependent. Removal of Clb5p from the nucleus is likely to be due to degradation resulting from Cdc20p-dependent targeting of the APC to Clb5p (40).
Removal of Clb5p regulation.
CLB5 is controlled in three ways: it is transcriptionally induced early in the cell cycle (12), its associated kinase activity is inhibited by Sic1p (36), and it is degraded in a destruction box-dependent manner (14, 19 [see above]). To address the biological significance of proteolytic control, we deleted the CLB5 destruction box, and we also eliminated transcriptional control by placing CLB5 under the strong GAL1 promoter.
GAL1::CLB5 expression was not lethal, but expression of GAL1::CLB5-Δdb was (8). GAL1::CLB5 expression results in the accumulation of cells with a 2C DNA content (see Fig. 8). GAL1::CLB5 expression becomes lethal in a sic1 background (data not shown). Deletion of the SWE1 inhibitory kinase (5) had little additional effect on these phenotypes (data not shown).
FIG. 8.
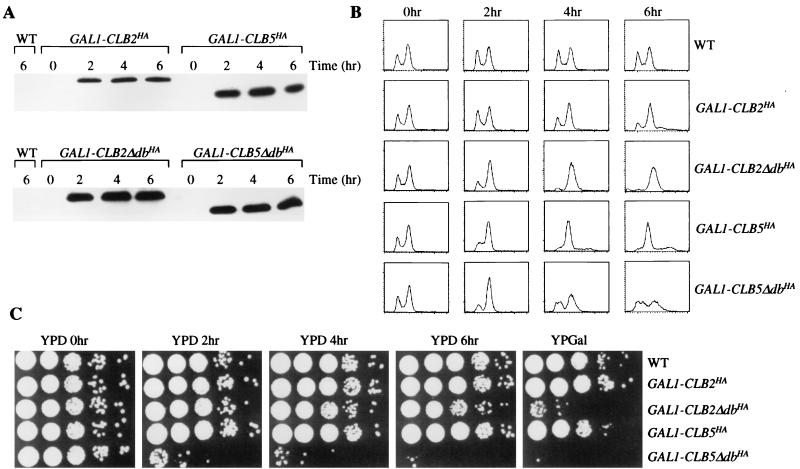
Overexpression of Clb5ΔdbHAp results in a lethal phenotype unlike the reversible arrest caused by GAL1::CLB2ΔdbHA. All strains have the indicated construct integrated in 1255-5C (wild type [WT]). Wild-type, GAL1::CLB2HA, GAL1::CLB2ΔdbHA, GAL1::CLB5HA, and GAL1::CLB5ΔdbHA strains were grown overnight in YEPRaf. The GAL1 promoters were then induced by the addition of galactose to a final concentration of 3%, and samples were taken at 2-h intervals while the cultures were incubating at 30°C. The samples were analyzed for protein levels by an anti-HA Western blot (A), DNA content by FACS analysis (B), and viability by 10-fold serial dilutions on yeast extract-peptone-dextrose (YPD) and galactose medium lacking α-factor (YPGal) (C). Samples from all time points resulted in identical YPGal plating efficiencies, and therefore only a representative time point is shown.
In contrast, CLB5-Δdb expressed from its own promoter was not lethal (8), even in the absence of SIC1 (data not shown). Therefore, loss of transcriptional control (of periodicity, levels or both), in addition to loss of either proteolytic or Sic1p control, was required to elevate Clb5p activity sufficiently to achieve lethality. Clb5p-Δdb expressed from the CLB5 promoter was moderately if at all stabilized in cell cycle time courses, in contrast to the strong stabilization observed with GAL1::CLB5-Δdb (Fig. 2 and 3). It may be that overexpression of Clb5-Δdb is required to saturate some means of Clb5p degradation that is independent of the identified destruction box. These observations on expression of CLB5-Δdb pose a paradox with respect to the results of Shirayama et al. (40). If Cdc20-dependent degradation of Clb5p is destruction box dependent, and if failure of Cdc20-dependent degradation of Clb5p (expressed from its own promoter) blocks mitotic exit, then CLB5-Δdb expression should similarly result in significant persistence of Clb5p and a block to mitotic exit. There may be additional unidentified destruction boxes in Clb5p (S. Holloway, unpublished data), or there may be Cdc20-dependent but destruction box-independent means of Clb5p degradation. We are exploring these possibilities.
Since GAL1::CLB5-Δdb and GAL1::CLB5 yield nearly comparable levels of protein through most of the cell cycle, it seemed likely that the lethality of GAL1::CLB5-Δdb may require specific persistence of Clb5p through mitosis, when Clb5p but not Clb5-Δdbp is degraded. Since Sic1p protein accumulation is induced during late mitosis, this may explain the lethality of GAL1::CLB5 in sic1 strains: a low level of residual Clb5p escaping degradation may require Sic1p inhibition to avoid lethality due to active Clb5p complexes persisting through mitosis. Overexpressed Clb5-Δdbp may saturate the available Sic1p.
Confirming that high levels of Clb5p activity are required to induce lethality, the introduction of a double mutation (K253A, E282A) that partially interferes with Cdc28p kinase activation into the GAL1::CLB5-Δdb strain relieved lethality (8). Deletion of SIC1 made expression of GAL1::CLB5-KA,EA-Δdb lethal (data not shown), again confirming that destruction box-dependent degradation and Sic1p inhibition can coregulate Clb5p activity, probably specifically in mitosis. GAL1::CLB5-KA,EA was not lethal even in the simultaneous absence of sic1 and swe1 (data not shown). The KA,EA mutation does not significantly affect degradation rates of Clb5p with or without its destruction box (data not shown).
Characterization of GAL1::CLB5-Δdb lethality.
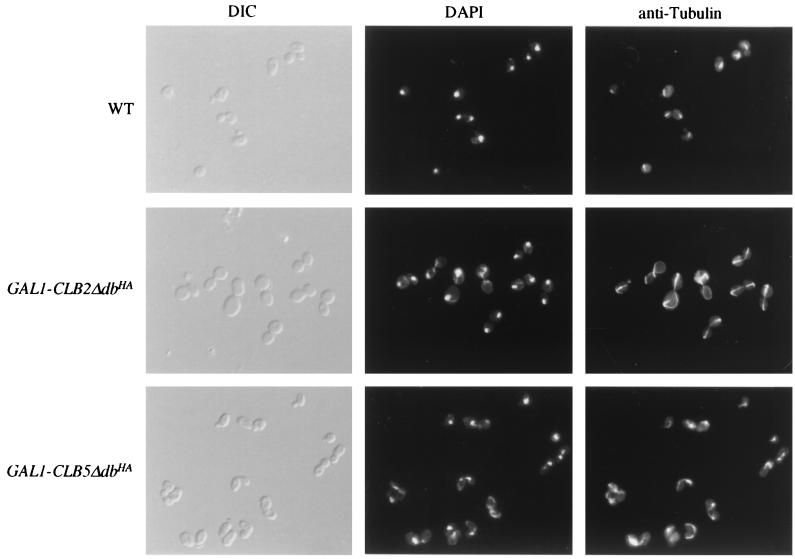
GAL1::CLB5-Δdb cells grown in raffinose medium arrest after several hours of galactose induction. Arrest is associated with lethality in that the plating efficiency of these cells on glucose medium drops approximately 1,000-fold by 4 to 6 h of galactose incubation (see Fig. 8C and 9C). After approximately 2 h of expression, the GAL1::CLB5-Δdb cells appear to delay as binucleate large budded cells with a 2C DNA content, resembling a GAL1::CLB2-Δdb-induced arrest (data not shown), but cells escape this mitotic block over the succeeding 2 h. By 4 to 6 h, the cells are heterogeneous with respect to DNA content: approximately half of the cells have a 2C (replicated) DNA content, and almost half have only 1C (Fig. 8B and 9B). There is usually some accumulation of cells that appear to have intermediate DNA contents. The cells are predominately large budded, and frequently they rebud after 4 h of galactose induction (Table 1). Tubulin staining by indirect immunofluorescence indicates that most of the cells are arresting with postmitotic spindles (Fig. 6). The DNA signals (DAPI) are heterogeneous in strength, compared to those of wild-type controls, possibly the result of an unequal distribution into the mother and daughter buds (Fig. 6). Combined with the FACS analysis showing a significant population of cells with approximately 1C DNA content, these results suggest that some cells have undergone abortive mitosis despite failure of DNA replication (31).
FIG. 9.
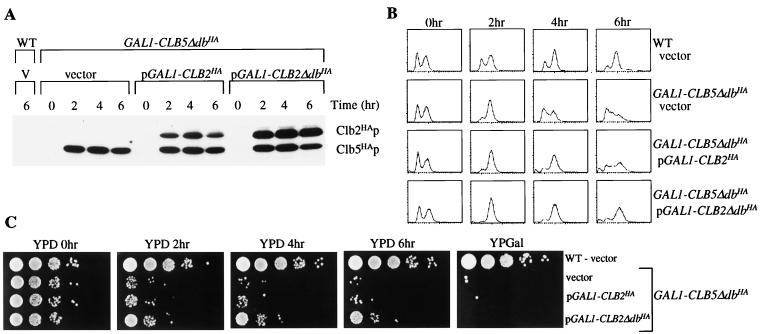
Overexpression of Clb2ΔdbHAp in a GAL1::CLB5ΔdbHA strain blocks cells in mitosis and partially suppresses lethality due to GAL1::CLB5ΔdbHA. Wild-type (WT) 1255-5C transformed with vector (V) and 1255-5C with integrated GAL1::CLB5ΔdbHA (transformed with either vector, pGAL1::CLB2HA, or pGAL1::CLB2ΔdbHA) were grown overnight in synthetic complete raffinose-uracil medium. The cells were harvested by centrifugation and resuspended in galactose medium lacking α-factor (YPGal) to induce the GAL1 promoter. Samples were taken at 2-h intervals while the cultures were incubating at 30°C. The samples were analyzed for protein levels by an anti-HA Western blot (A), DNA content by FACS analysis (B), and viability by 10-fold serial dilutions on yeast extract-peptone-dextrose (YPD) and YPGal (C). Samples from all time points resulted in identical YPGal plating efficiencies, and therefore only a representative time point is shown.
TABLE 1.
Budding and rebudding percentages resulting from GAL1-induced CLB2 and CLB5 constructs
| Genotype | % of cellsa
|
|||
|---|---|---|---|---|
| Expt 1
|
Expt 2
|
|||
| Budded | Rebudded | Budded | Rebudded | |
| Set I | ||||
| Wild type | 67 | 0 | 68 | 0 |
| GAL1::CLB2 | 59 | 0 | 60 | 0 |
| GAL1::CLB2-Δdb | 88 | 0 | 90 | 0 |
| GAL1::CLB5 | 67 | 2 | 68 | 2 |
| GAL1::CLB5-Δdb | 56 | 39 | 56 | 42 |
| Set II | ||||
| Wild type + vector | 54 | 2 | 55 | 5 |
| GAL1::CLB5-Δdb + vector | 68 | 27 | 69 | 28 |
| GAL1::CLB5-Δdb + pGAL1::CLB2 | 64 | 22 | 48 | 24 |
| GAL1::CLB5-Δdb + pGAL1::CLB2-Δdb | 54 | 4 | 53 | 12 |
Percentages of budded and rebudded cells were determined by counting 200 cells for each sample in two separate experiments. The samples were processed by the methods described for Fig. 7 (set I) and 8 (set II) following 6 h of galactose induction. Budded cells represent only singly budded cells, and rebudded cells represent all variations of multiply budded cells (typically either three or four cell bodies).
FIG. 6.
Antitubulin and DAPI staining of cells arrested due to GAL1::CLB2ΔdbHA and GAL1::CLB5ΔdbHA expression. Wild-type (WT), GAL1::CLB2ΔdbHA, and GAL1::CLB5ΔdbHA strains (integrated in 1255-5C) were grown overnight in YPRaf. The GAL1 promoters were then induced by the addition of galactose to a final concentration of 3%. Following 4 h of incubation at 30°C, the cells were fixed and processed for DAPI and antitubulin staining. DIC, differential interference contrast.
We interpret this phenotype as indicating that Clb5-Δdbp overexpression delays completion of mitosis, but many cells nevertheless ultimately divide in the presence of Clb5-Δdbp. These cells do not efficiently replicate DNA after division, accounting for the accumulation of cells with 1C DNA content. The GAL1::CLB5-Δdb rebudding phenotype (Table 1) may be due to the initiation of a G1 cell cycle program without properly completing the later stages of mitosis and cytokinesis. The rebudding phenotype was significantly reduced when the cells were processed for indirect immunofluorescence. This most likely results from digestion of the cell wall during sample preparation and could indicate that cytokinesis (but not cell separation) is largely complete in many of the apparently rebudded cells.
Comparison between the GAL1::CLB5-Δdb and GAL1::CLB2-Δdb phenotypes.
GAL1::CLB2-Δdb was reported to cause uniform arrest in late mitosis with replicated DNA (43), unlike the GAL1::CLB5-Δdb phenotype we observed; these cells also did not rebud, unlike the GAL1::CLB5-Δdb cells (Table 1). These differences could be due to intrinsic differences in the ability of Clb2p and Clb5p to block exit from mitosis; alternatively, levels of expression could differ between the two GAL1::CLB-Δdb constructs. Therefore, we replaced the CLB5 coding sequence with the CLB2 coding sequence in the GAL1::CLB5-Δdb construct. We found that these constructs yielded comparable levels of Clb protein and associated kinase activity (Fig. 7). As reported previously (43), GAL1::CLB2-Δdb expression results in late mitotic arrest, with few or no cells with unreplicated DNA detected (Fig. 8) and almost all cells displaying elongated spindles with the replicated DNA separated into the mother cell and the bud (Fig. 6). This phenotype was similar with or without the HA tag on GAL1::CLB2-Δdb (data not shown). In addition, the rapid induction of inviability by GAL1::CLB5-Δdb was not observed with GAL1::CLB2-Δdb (Fig. 8). The multiple-budding phenotype of GAL1::CLB5-Δdb was also not observed with GAL1::CLB2-Δdb (Table 1). Thus, even at comparable levels of expression, the phenotypes due to overexpression of stabilized Clb2p and Clb5p are different.
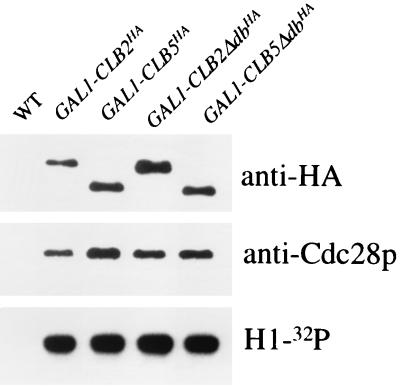
FIG. 7.
Expression of Clb2p and Clb5p from the GAL1 promoter results in comparable levels of protein, associated Cdc28p, and in vitro kinase activity. All strains have the indicated construct integrated in 1255-5C (wild type [WT]). Strains were grown overnight in YEPRaf. After 4 h of induction of the GAL1 promoter with 3% galactose at 30°C, the cultures were processed for immunoprecipitation of the HA-tagged proteins, followed by an in vitro kinase assay of the immunoprecipitates. Protein samples were analyzed by Western blotting with antibodies against either the HA tag or Cdc28p. 32P-phosphorylated histone-H1 (H1-32P) was detected by autoradiography.
We also carried out experiments in which strains were synchronized in G1 by using α-factor and then released into galactose medium to induce GAL1::CLB5-Δdb or GAL1::CLB2-Δdb. For reasons that we do not understand, in this protocol, GAL1::CLB5-Δdb expression resulted in a long preanaphase delay with apparently fully replicated DNA (data not shown); such a delay was not detected in the experiments (Fig. 8) in which galactose was added to asynchronous cultures. This difference makes it hard to directly compare results between the two protocols. Despite this, a significant population of the GAL1::CLB5-Δdb cells eventually escaped this block and divided their nuclei, and, overall, about 30% of the cells then rebudded (data not shown). In contrast, the GAL1::CLB2-Δdb cells arrested stably late in mitosis with divided nuclei and without rebudding, as was seen in the experiments where galactose was added to asynchronous cultures (Fig. 6 and 8). These results suggest that a new cell cycle is being initiated despite the presence of overexpressed Clb5-Δdbp, but that this is blocked by overexpressed Clb2-Δdbp, consistent with the results when galactose was added to asynchronous cultures (Fig. 8).
Clb5p is inefficient at blocking mitotic exit.
Clb2p is probably the major B-type cyclin active in mitosis (26, 27, 43). Previous studies have shown that Clb2p-associated kinase activity must be eliminated for completion of division and for proper loading of DNA replication origins during G1 (10, 15, 43). Clb2p can be stabilized by Clb2p-Cdc28p or Clnp-Cdc28p (1). If Clb5p-Cdc28p also stabilizes Clb2p, then some aspects of the phenotype due to overexpression of CLB5Δdb could be indirect, due to stabilized Clb2p-associated kinase activity. To test this, we constructed GAL1-CLB5 clb2::LEU2 strains, with or without the destruction box and with or without the KA,EA mutation in CLB5. clb2 deletion enhanced inviability due to CLB5, since GAL1::CLB5 and GAL1::CLB5-KA,EA-Δdb were lethal in a clb2 background. clb2 deletion also enhanced the speed of induction of irreversible lethality due to GAL1::CLB5-Δdb, although this effect was somewhat variable (data not shown). In almost all cases, GAL1::CLB5-Δdb induction in clb2 strains resulted in a significantly greater accumulation of cells with 1C DNA content than was observed in CLB2 strains (data not shown).
These results suggested that Clb2p might be restraining mitotic exit in the CLB5-Δdb overexpressers. We constructed a strain expressing both GAL1::CLB5-Δdb and GAL1::CLB2-Δdb. The strain arrested as large budded cells with a 2C DNA content (Fig. 9) where the replicated DNA was separated into the mother and daughter cell bodies (data not shown), typical of a GAL1::CLB2-Δdb-induced arrest. This is consistent with the idea that the accumulation of 1C DNA content in cells with GAL1::CLB5-Δdb alone requires mitosis, which may be blocked by GAL1::CLB2-Δdb expression. Similarly, GAL1::CLB2-Δdb expression significantly reduces the number of cells displaying the characteristic GAL1::CLB5-Δdb rebudding phenotype (Table 1), consistent with the inhibitory effects of Clb2p on cell polarization and bud emergence reported previously (2, 23). Although the reversibility of the arrest in the GAL1::CLB5-Δdb/GAL1::CLB2-Δdb strain was reduced from that observed in GAL1::CLB2-Δdb cells, viability was increased 10-fold from strains expressing GAL1::CLB5-Δdb alone (Fig. 9). These results support the idea that Clb2p can restrain mitotic exit in GAL1::CLB5-Δdb expressers and that the severity of GAL1::CLB5-Δdb-induced lethality is correlated with an accumulation of cells containing a 1C DNA content. Accumulation of 1C cells may not be the sole cause of irreversible arrest, because GAL1::CLB5-Δdb-expressing cells arrest with a heterogeneous population of DNA content. This is difficult to interpret fully, though, because these cells may ultimately divide upon plating for the viability assay.
DISCUSSION
Clb5p degradation.
We find that Clb5p degradation is cell cycle regulated, and Clb5p is most unstable in dividing cells. These differences in stability correlate with loss of detectable nuclear accumulation of Clb5p in dividing cells (although significant cytoplasmic signal remains). We speculate that the specific loss of Clb5p from the nucleus may be due to nucleus-localized degradation, but further work is required to eliminate other possibilities (for example, that Clb5p is exported from the nucleus during mitosis and is independently rendered less stable at this time).
Clb5-Δdbp expressed from the CLB5 promoter has at best minor phenotypes, even in the absence of sic1, and destruction box deletion from endogenously expressed Clb5p also results in, at most, minor stabilization of the protein, monitored with HA-tagged Clb5p in synchronous culture (Fig. 3 and data not shown). Destruction box-independent degradation of Clb5p may be responsible, or there may be other unidentified destruction boxes in Clb5p. In either case, the overexpression of Clb5-Δdbp from the GAL1 promoter may swamp out these alternative means of degrading Clb5p.
Cyclin specificity.
The six CLB B-type cyclins have distinct roles in vivo, although they overlap significantly in function. Distinct in vivo roles could be due to time of expression during the cell cycle or to intrinsic specialization of the different CLB coding sequences. Previously, we showed that Clb5p was much more potent at inducing DNA replication than Clb2p, even when time of expression and protein accumulation were made comparable, by placing both under control of the CLB5 promoter (8). Here, we examine the cell cycle-inhibitory activity of Clb5p compared to that of Clb2p. To make this comparison, we eliminated differential transcriptional control by using the GAL1 promoter and eliminated differential proteolytic control by removing the proteins' destruction boxes. We find that while Clb2p is able to block exit from mitosis, as reported previously, Clb5p is relatively weak at this activity, even when strongly overexpressed. Even the limited apparent capacity of Clb5p to inhibit mitotic exit is likely to be dependent on endogenous Clb2p. Clb5p could recruit endogenous Clb2p for this role by stabilizing it, perhaps by phosphorylation of Cdh1p (1, 49). In addition, the potency of existing Clb2p may be increased due to Clb5p-dependent phosphorylation of Sic1p (46). The idea that Clb5p restrains mitotic exit by phosphorylation of Cdh1p and Sic1p was also suggested by Shirayama et al. (40).
Accumulation of cells with unreplicated DNA in GAL1::CLB5-Δdb cells may be due to failure to license replication origins due to high Clb5p-associated kinase (9), combined with the permissiveness of high Clb5p-associated kinase for completion of mitosis. The consequence may be that cells divide without being able to initiate DNA replication after division. If cells under these conditions pass the “point of no return” (31) when endogenous Clb-associated kinases block reloading of replication origins, then even after shutoff of GAL1::CLB5-Δdb, irreversible lethality is predicted. It is also possible that some of these cells undergo anaphase without DNA replication, leading to unequal segregation of the haploid DNA content (31), a clearly lethal event.
The molecular basis for differences in cyclin specificity is unknown. The results reported here and previously (8) do not suggest that differential accumulation of the protein or associated kinase is responsible, although subtle differences in timing or levels are hard to rule out unambiguously. A candidate substrate-targeting domain (the hydrophobic patch [8, 34]) could function differently between Clb5p and Clb2p, potentially leading to differential substrate targeting. This region contributes to but is not essential for lethality of GAL1::CLB5-Δdb (8). The analogous region in Clb2p may be required for efficient blocking of mitotic exit in GAL1::CLB5-Δdb, GAL1::CLB2-Δdb overexpressers in the assay shown in Fig. 9 (data not shown). Failure to detect a strong requirement for this region for some effects of Clb-Δdb overexpression may be due to masking the role for the region due to high expression levels. The region is required for efficient lethality of Clb5p in the absence of cdc20 and pds1 and is also required for efficient Clb2p mitotic function at lower levels of expression (7).
S and M cyclins and the organization of the yeast cell cycle.
CLB5 and CLB6 have been called S-phase cyclins and CLB1 and CLB2 have been called M-phase cyclins, due to their time of expression and evident function as deduced from null phenotypes (12, 15, 26, 27, 37, 43). In fission yeast, cdc13 appears to be the predominant M-phase cyclin, and cig2 (with help from cig1) may be the major S-phase cyclin. Despite this differentiation of function among B-type cyclins, models have been proposed for both budding yeast and fission yeast in which a single generic B-type cyclin-dependent kinase activity could be sufficient to control both the S and M phases in proper alternation (13, 26, 42). These models suggest that functional differences in the roles of different B-type cyclins (as deduced from null phenotypes) are due solely to differential accumulation due to transcriptional or proteolytic controls. At least for budding yeast, this appears to be an oversimplification. Previously we showed that Clb5p is intrinsically specialized for induction of DNA replication, functioning much better at this activity than Clb2p (8). The results reported here suggest that Clb2p has a specific function of restraining mitotic exit, which is weak or absent in Clb5p. This difference is not due to defects in Clb5p protein accumulation or kinase activation, and thus the difference is likely to be intrinsic to the protein. Thus, at least for these two activities, one early in the cell cycle and one at the end, the yeast cell cycle is driven by early accumulation of S cyclins, including Clb5p, and late accumulation of M cyclins, including Clb2p, which are intrinsically specialized for appropriate roles.
cdc20 pds1 cells are blocked late in mitosis (25), and this block has been attributed to failure of Cdc20p-dependent Clb5p degradation (40). These observations lead to the prediction that expression of stabilized Clb5p in mitotic cells should permanently block mitotic exit. In contrast, upon expression of GAL1::CLB5-Δdb, we observe only a transient mitotic delay followed by completion of mitosis (with concomitant loss in cell viability). From our results, it appears likely that restraint of mitotic exit in cdc20 pds1 cells (25), while genetically due to Clb5p (40), may be more directly due to Clb5p-dependent activation of mitotic cyclins, including Clb2p. We do not know why this effect is only transient under our conditions.
In addition to regulating mitotic exit, Clb activity must somehow lead to mitotic entry (for example, by activation of Cdc20p [21] leading to Pds1p degradation [48] and/or by some direct activation of spindle function). A specific role for Clb5p in some aspects of spindle function has been suggested based on the phenotype of clb3,4,5,6 strains (37) and on analysis of clb5 cdc28-4 diploid strains (38). Thus, some aspects of spindle function may be more efficiently driven by early-expressed cyclins such as Clb5p than by late-expressed cyclins such as Clb2p.
Clb1,2,3,4p activity has been implicated in down-regulation of expression of SBF-regulated genes, such as the G1 cyclins CLN1 and CLN2 (3), while CLB5 probably lacks this activity, and at least under some circumstances may actually drive expression of this class of genes (24, 30). This may partially explain the ability of GAL1::CLB5-Δdb but not GAL1::CLB2-Δdb cells to undergo extra rounds of budding (see above), since Cln1p and Cln2p are probably major activators of bud emergence (23). This could provide another example of restriction of late functions to late-expressed Clb proteins by intrinsic specialization.
Thus, the available data suggest that the budding yeast cell cycle is largely segregated into early functions promoted by S-phase cyclins and late functions promoted by M-phase cyclins. This segregation correlates with time of expression of these cyclins and also with the intrinsic functional capacities of these cyclins.
The linkage between these sets of functions is unknown, but it is an intriguing possibility that Clb5p may be important for stabilizing Clb2p through Cdh1p phosphorylation (1, 40, 49). Since Cdh1p is not involved in Clb5p degradation (35), this could allow early accumulation of Clb5p and performance of Clb5p-specific early cell cycle functions, followed by accumulation of Clb2p. Transcriptional positive feedback control of CLB2 (3) could enhance temporal segregation of accumulation of different cyclins.
Fission yeasts are able to proliferate fairly normally with only one of the three identified B-type cyclins (cdc13), suggesting that this system may be significantly simpler than the budding yeast system. (It is worth noting, though, that the fission yeast sequencing project, while still incomplete, has identified a fourth B-type cyclin [EMBL locus SPBC16E9; accession no. Z99759.1] whose involvement in S phase and mitosis has not been explored to our knowledge.) Even in budding yeast, sufficient overexpression of the CLB1 B-type cyclin is sufficient for viability in the absence of CLB2-6 (16), indicating that it there is no absolute requirement for differentially targeted B-type cyclins for cell cycle progression. This is consistent with the evolutionary speculation (28) that the primordial eukaryotic cell cycle was driven by a single B-type cyclin capable of driving events in DNA replication and in mitosis. In metazoans, it is likely that cyclins A and E are specialized for induction of S phase and that B-type cyclins are specialized for induction of mitosis, and this situation may be closer to the budding yeast system when cyclins are expressed at endogenous levels.
REFERENCES
- 1.Amon A. Regulation of B-type cyclin proteolysis by Cdc28-associated kinases in budding yeast. EMBO J. 1997;16:2693–2702. doi: 10.1093/emboj/16.10.2693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amon A, Irniger S, Nasmyth K. Closing the cell cycle circle in yeast: G2 cyclin proteolysis initiated at mitosis persists until the activation of G1 cyclins in the next cycle. Cell. 1994;77:1037–1050. doi: 10.1016/0092-8674(94)90443-x. [DOI] [PubMed] [Google Scholar]
- 3.Amon A, Tyers M, Futcher B, Nasmyth K. Mechanisms that help the yeast cell cycle clock tick: G2 cyclins transcriptionally activate G2 cyclins and repress G1 cyclins. Cell. 1993;74:993–1007. doi: 10.1016/0092-8674(93)90722-3. [DOI] [PubMed] [Google Scholar]
- 4.Bai C, Sen P, Hofmann K, Ma L, Goebl M, Harper J W, Elledge S J. SKP1 connects cell cycle regulators to the ubiquitin proteolysis machinery through a novel motif, the F-box. Cell. 1996;86:263–274. doi: 10.1016/s0092-8674(00)80098-7. [DOI] [PubMed] [Google Scholar]
- 5.Booher R N, Deshaies R J, Kirschner M W. Properties of Saccharomyces cerevisiae wee1 and its differential regulation of p34CDC28 in response to G1 and G2 cyclins. EMBO J. 1993;12:3417–3426. doi: 10.1002/j.1460-2075.1993.tb06016.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cross F R. Starting the cell cycle: what's the point? Curr Opin Cell Biol. 1995;7:790–797. doi: 10.1016/0955-0674(95)80062-x. [DOI] [PubMed] [Google Scholar]
- 7.Cross F R, Jacobson M D. Conservation and function of a potential substrate-binding domain in the yeast Clb5 B-type cyclin. Mol Cell Biol. 2000;20:4782–4790. doi: 10.1128/mcb.20.13.4782-4790.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cross F R, Yuste-Rojas M, Gray S, Jacobson M D. Specialization and targeting of B-type cyclins. Mol Cell. 1999;4:11–19. doi: 10.1016/s1097-2765(00)80183-5. [DOI] [PubMed] [Google Scholar]
- 9.Dahmann C, Diffley J F X, Nasmyth K A. S-phase-promoting cyclin-dependent kinases prevent re-replication by inhibiting the transition of replication origins to a pre-replicative state. Curr Biol. 1995;5:1257–1269. doi: 10.1016/s0960-9822(95)00252-1. [DOI] [PubMed] [Google Scholar]
- 10.Detweiler C S, Li J J. Ectopic induction of Clb2 in early G1 phase is sufficient to block prereplicative complex formation in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1998;95:2384–2389. doi: 10.1073/pnas.95.5.2384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Epstein C B. Ph.D. thesis. New York, N.Y: Rockefeller University; 1992. [Google Scholar]
- 12.Epstein C B, Cross F R. CLB5: a novel B cyclin from budding yeast with a role in S phase. Genes Dev. 1992;6:1695–1706. doi: 10.1101/gad.6.9.1695. [DOI] [PubMed] [Google Scholar]
- 13.Fisher D L, Nurse P. A single fission yeast mitotic cyclin B p34CDC2 kinase promotes both S-phase and mitosis in the absence of G1 cyclins. EMBO J. 1996;15:850–860. [PMC free article] [PubMed] [Google Scholar]
- 14.Germain D, Hendley J, Futcher B. DNA damage inhibits proteolysis of the B-type cyclin Clb5 in S. cerevisiae. J Cell Sci. 1997;110:1813–1820. doi: 10.1242/jcs.110.15.1813. [DOI] [PubMed] [Google Scholar]
- 15.Ghiara J B, Richardson H E, Sugimoto K, Henze M, Lew D J, Wittenberg C, Reed S I. A cyclin B homolog in S. cerevisiae: chronic activation of the Cdc28 protein kinase by cyclin prevents exit from mitosis. Cell. 1991;65:163–174. doi: 10.1016/0092-8674(91)90417-w. [DOI] [PubMed] [Google Scholar]
- 16.Haase S B, Reed S I. Evidence that a free-running oscillator drives G1 events in the budding yeast cell cycle. Nature. 1999;401:394–397. doi: 10.1038/43927. [DOI] [PubMed] [Google Scholar]
- 17.Heichman K A, Roberts J M. CDC16 controls initiation at chromosome replication origins. Mol Cell. 1998;1:457–463. doi: 10.1016/s1097-2765(00)80046-5. [DOI] [PubMed] [Google Scholar]
- 18.Heichman K A, Roberts J M. The yeast CDC16 and CDC27 genes restrict DNA replication to once per cell cycle. Cell. 1996;85:39–48. doi: 10.1016/s0092-8674(00)81080-6. [DOI] [PubMed] [Google Scholar]
- 19.Irniger S, Nasmyth K. The anaphase-promoting complex is required in G1 arrested cells to inhibit B-type cyclin accumulation and to prevent uncontrolled entry into S-phase. J Cell Sci. 1997;110:1523–1531. doi: 10.1242/jcs.110.13.1523. [DOI] [PubMed] [Google Scholar]
- 20.Jaspersen S L, Charles J F, Morgan D O. Inhibitory phosphorylation of the APC regulator Hct1 is controlled by the kinase Cdc28 and the phosphatase Cdc14. Curr Biol. 1999;9:227–236. doi: 10.1016/s0960-9822(99)80111-0. [DOI] [PubMed] [Google Scholar]
- 21.Kotani S, Tanaka H, Yasuda H, Todokoro K. Regulation of APC activity by phosphorylation and regulatory factors. J Cell Biol. 1999;146:791–800. doi: 10.1083/jcb.146.4.791. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 22.Levine K, Huang K, Cross F R. Saccharomyces cerevisiae G1 cyclins differ in their intrinsic functional specificities. Mol Cell Biol. 1996;16:6794–6803. doi: 10.1128/mcb.16.12.6794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lew D J, Reed S I. Morphogenesis in the yeast cell cycle: regulation by Cdc28 and cyclins. J Cell Biol. 1993;120:1305–1320. doi: 10.1083/jcb.120.6.1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li X, Cai M. Recovery of the yeast cell cycle from heat shock-induced G(1) arrest involves a positive regulation of G(1) cyclin expression by the s phase cyclin Clb5. J Biol Chem. 1999;274:24220–24231. doi: 10.1074/jbc.274.34.24220. [DOI] [PubMed] [Google Scholar]
- 25.Lim H H, Goh P Y, Surana U. Cdc20 is essential for the cyclosome-mediated proteolysis of both Pds1 and Clb2 during M phase in budding yeast. Curr Biol. 1998;8:231–234. doi: 10.1016/s0960-9822(98)70088-0. [DOI] [PubMed] [Google Scholar]
- 26.Nasmyth K. At the heart of the budding yeast cell cycle. Trends Genet. 1996;12:405. doi: 10.1016/0168-9525(96)10041-x. [DOI] [PubMed] [Google Scholar]
- 27.Nasmyth K. Control of the yeast cell cycle by the Cdc28 protein kinase. Curr Opin Cell Biol. 1993;5:166–179. doi: 10.1016/0955-0674(93)90099-c. [DOI] [PubMed] [Google Scholar]
- 28.Nasmyth K. Evolution of the cell cycle. Philos Trans R Soc Lond B Biol Sci. 1995;349:271–281. doi: 10.1098/rstb.1995.0113. [DOI] [PubMed] [Google Scholar]
- 29.Newlon C S. Putting it all together: building a prereplicative complex. Cell. 1997;91:717–720. doi: 10.1016/s0092-8674(00)80459-6. [DOI] [PubMed] [Google Scholar]
- 30.Oehlen L J W M, Jeoung D-I, Cross F R. Cyclin-specific START events and the G1-phase specificity of arrest by mating factor in budding yeast. Mol Gen Genet. 1998;258:183–198. doi: 10.1007/s004380050722. [DOI] [PubMed] [Google Scholar]
- 31.Piatti S, Böhm T, Cocker J H, Diffley J F X, Nasmyth K. Activation of S-phase-promoting CDKs in late G1 defines a “point of no return” after which Cdc6 synthesis cannot promote DNA replication in yeast. Genes Dev. 1996;10:1516–1531. doi: 10.1101/gad.10.12.1516. [DOI] [PubMed] [Google Scholar]
- 32.Pichler S, Piatti S, Nasmyth K. Is the yeast anaphase promoting complex needed to prevent re-replication during G2 and M phases? EMBO J. 1997;16:5988–5997. doi: 10.1093/emboj/16.19.5988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Richardson H E, Wittenberg C, Cross F, Reed S I. An essential G1 function for cyclin-like proteins in yeast. Cell. 1989;59:1127–1133. doi: 10.1016/0092-8674(89)90768-x. [DOI] [PubMed] [Google Scholar]
- 34.Schulman B A, Lindstrom D L, Harlow E. Substrate recruitment to cyclin-dependent kinase 2 by a multipurpose docking site on cyclin A. Proc Natl Acad Sci USA. 1998;95:10453–10458. doi: 10.1073/pnas.95.18.10453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schwab M, Lutum A S, Seufert W. Yeast Hct1 is a regulator of Clb2 cyclin proteolysis. Cell. 1997;90:683–693. doi: 10.1016/s0092-8674(00)80529-2. [DOI] [PubMed] [Google Scholar]
- 36.Schwob E, Böhm T, Mendenhall M D, Nasmyth K. The B-type cyclin kinase inhibitor p40SIC1 controls the G1 to S transition in S. cerevisiae. Cell. 1994;79:233–244. doi: 10.1016/0092-8674(94)90193-7. [DOI] [PubMed] [Google Scholar]
- 37.Schwob E, Nasmyth K. CLB5 and CLB6, a new pair of B cyclins involved in DNA replication in Saccharomyces cerevisiae. Genes Dev. 1993;7:1160–1175. doi: 10.1101/gad.7.7a.1160. [DOI] [PubMed] [Google Scholar]
- 38.Segal M, Clarke D J, Reed S I. Clb5-associated kinase activity is required early in the spindle pathway for correct preanaphase nuclear positioning in Saccharomyces cerevisiae. J Cell Biol. 1998;143:135–145. doi: 10.1083/jcb.143.1.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Seufert W, Futcher B, Jentsch S. Role of a ubiquitin-conjugating enzyme in degradation of S- and M-phase cyclins. Nature. 1995;373:78–81. doi: 10.1038/373078a0. [DOI] [PubMed] [Google Scholar]
- 40.Shirayama M, Toth A, Galova M, Nasmyth K. APCCdc20 promotes exit from mitosis by destroying the anaphase inhibitor Pds1 and cyclin Clb5. Nature. 1999;402:203–207. doi: 10.1038/46080. [DOI] [PubMed] [Google Scholar]
- 41.Shou W, Seol J H, Shevchenko A, Baskerville C, Moazed D, Chen Z W, Jang J, Shevchenko A, Charbonneau H, Deshaies R J. Exit from mitosis is triggered by Tem1-dependent release of the protein phosphatase Cdc14 from nucleolar RENT complex. Cell. 1999;97:233–244. doi: 10.1016/s0092-8674(00)80733-3. [DOI] [PubMed] [Google Scholar]
- 42.Stern B, Nurse P. A quantitative model for the cdc2 control of S phase and mitosis in fission yeast. Trends Genet. 1996;12:345–350. [PubMed] [Google Scholar]
- 43.Surana U, Amon A, Dowzer C, McGrew J, Byers B, Nasmyth K. Destruction of the CDC28/CLB mitotic kinase is not required for the metaphase to anaphase transition in budding yeast. EMBO J. 1993;12:1969–1978. doi: 10.1002/j.1460-2075.1993.tb05846.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tanaka T, Knapp D, Nasmyth K. Loading of an MCM protein onto DNA replication origins is regulated by Cdc6p and CDKs. Cell. 1997;90:649–660. doi: 10.1016/s0092-8674(00)80526-7. [DOI] [PubMed] [Google Scholar]
- 45.Vallen E A, Cross F R. Mutations in RAD27 define a potential link between G1 cyclins and DNA replication. Mol Cell Biol. 1995;15:4291–4302. doi: 10.1128/mcb.15.8.4291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Verma R, Annan R S, Huddleston M J, Carr S A, Reynard G, Deshaies R J. Phosphorylation of Sic1p by G1 Cdk required for its degradation and entry into S phase. Science. 1997;278:455–460. doi: 10.1126/science.278.5337.455. [DOI] [PubMed] [Google Scholar]
- 47.Visintin R, Craig K, Hwang E S, Prinz S, Tyers M, Amon A. The phosphatase Cdc14 triggers mitotic exit by reversal of Cdk-dependent phosphorylation. Mol Cell. 1998;2:709–718. doi: 10.1016/s1097-2765(00)80286-5. [DOI] [PubMed] [Google Scholar]
- 48.Zachariae W, Nasmyth K. Whose end is destruction: cell division and the anaphase-promoting complex. Genes Dev. 1999;13:2039–2058. doi: 10.1101/gad.13.16.2039. [DOI] [PubMed] [Google Scholar]
- 49.Zachariae W, Schwab M, Nasmyth K, Seufert W. Control of cyclin ubiquitination by CDK-regulated binding of Hct1 to the anaphase promoting complex. Science. 1998;282:1721–1724. doi: 10.1126/science.282.5394.1721. [DOI] [PubMed] [Google Scholar]
- 50.Zou L, Stillman B. Formation of a preinitiation complex by S-phase cyclin CDK-dependent loading of Cdc45p onto chromatin. Science. 1998;280:593–596. doi: 10.1126/science.280.5363.593. [DOI] [PubMed] [Google Scholar]