Abstract
Background
Wall-associated kinase (WAK)/WAK-like (WAKL) is one of the subfamily of receptor like kinases (RLK). Although previous studies reported that WAK/WAKL played an important role in plant cell elongation, response to biotic and abiotic stresses, there are no systematic studies on RcWAK/RcWAKL in rose.
Results
In this study, we identified a total of 68 RcWAK/RcWAKL gene family members within rose (Rosa chinensis) genome. The RcWAKs contained the extracellular galacturonan-binding domain and calcium-binding epidermal growth factor (EGF)-like domain, as well as an intracellular kinase domains. The RcWAKLs are missing either calcium-binding EGF-like domain or the galacturonan-binding domain in their extracellular region. The phylogenetic analysis showed the RcWAK/RcWAKL gene family has been divided into five groups, and these RcWAK/RcWAKL genes were unevenly distributed on the 7 chromosomes of rose. 12 of RcWAK/RcWAKL genes were significantly up-regulated by Botrytis cinerea-inoculated rose petals, where RcWAK4 was the most strongly expressed. Virus induced gene silencing of RcWAK4 increased the rose petal sensitivity to B. cinerea. The results indicated RcWAK4 is involved in the resistance of rose petal against B. cinerea.
Conclusion
Our study provides useful information to further investigate the function of the RcWAK/RcWAKL gene family and breeding research for resistance to B. cinerea in rose.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12870-021-03307-9.
Keywords: Rosa chinensis, WAK, WAKL, Botrytis cinerea, grey mold disease, immune response
Background
Plant defense strategies of necrotrophic pathogens such as Botrytis cinerea are various. Pathogen-derived microbial-associated molecular patterns (MAMPs) and host damage-associated molecular patterns (DAMPs) as two immune signalings active plant immunity [1]. The plant cell wall is the first barrier to plant defense responses [2], and also plays an important role in maintaining cell morphology, regulating plant growth and development, and responding to biotic and abiotic stresses [3]. Pattern recognition receptors (PRRs) are a class of receptor kinases localized in the cell membrane that recognize pathogen invasion and transmit danger signals in plants [4]. Wall-associated kinase (WAK)/ WAK-like (WAKL) is a typical category of PRRs classified to receptor-like protein kinases (RLKs) [5].
WAKs/WAKLs are transmembrane proteins with an intracellular Ser/Thr kinase domain and an extracellular structure, contacting extracellular and cytoplasmic signals. Their extracellular structure contains a galacturonan-binding domain and/or a calcium-binding epidermal growth factor (EGF)-like domain for signal perception [6]. WAKs/WAKLs play important roles in regulating plant growth, development, and responding to environmental stresses. Expression of the AtWAK4 antisense gene in Arabidopsis results in impaired cell elongation and stunted lateral root development [7]. In addition, knocked out CaWAKL20 significantly increased pepper heat tolerance, whereas overexpression of CaWAKL20 in Arabidopsis reduced heat tolerance, indicating that CaWAKL20 negatively regulated pepper heat stress response [8].
Meanwhile, WAKs/WAKLs play a vital role in plant defense response to fungal diseases. Oligogalactouronides (OGs), as the cell wall polysaccharide degradation productions, are recognized by AtWAK1. And overexpression AtWAK1 could enhance resistance to B. cinerea in Arabidopsis [9, 10]. The high expression of ZmWAK, which was located in the head smut quantitative resistance locus (qHSP1), effectively suppressed the growth of Sporisorium reilianum in the mesocotyl [11]. Rice OsWAK14, OsWAK91 and OsWAK92 positively regulated the defense response to blast fungus (Magnaporthe oryzae), while OsWAK112d worked as a negative regulator of the defense response [12]. SlWAK1 is involved in the regulation of PRR-mediated immune response through FLS2/FLS3 complex in tomatoes [13].
Rose (Rosa sp.), as an important ornamental crop, accounts for more than 30% of the world's annual trade in cut flowers [14]. The long-distance transportation of cut rose flowers leads to water loss, senescence and postharvest diseases on cut flowers, resulting in serious economic losses. Gray mold disease, caused by B. cinerea, as one of the most serious postharvest fungal diseases of rose. The role WAKs/WAKLs family members in resistance against necrotrophic pathogens have been reported in Arabidopsis [9, 10]. However, whether WAKs/WAKLs are involved in rose resistance to B. cinerea is largely unknown.
Our previous study found that a large number of rose WAK/WAKL genes significantly up-regulated in response to B. cinerea inoculation [15]. In this study, we comprehensively characterized the of RcWAKs/RcWAKLs in rose genome, including their gene structural features, conserved domain, evolutionary relationships, and their expression pattern upon B. cinerea inoculation. This analysis suggested RcWAK4 may play a role in the response to B. cinerea infection. We finally used virus-induced gene silencing (VIGS) to confirm that RcWAK4 is involved in the rose defense against B. cinerea.
Results
Identification of RcWAK/ RcWAKL family members in rose
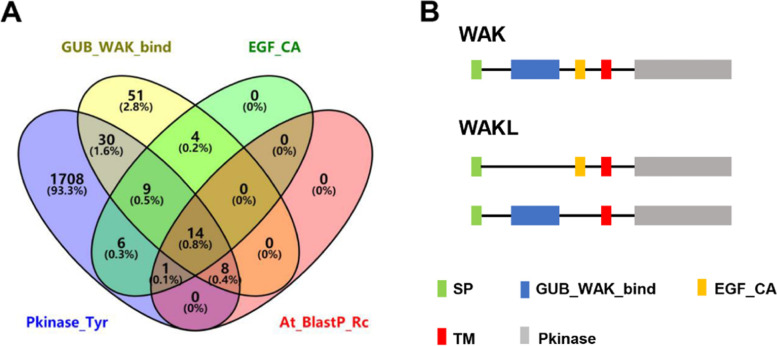
To identify the WAK/WAKL gene family in Rosa chinensis, 27 Arabidopsis WAK/WAKL family members were used as a reference to perform BlastP (E value<0.001) in the rose genome database (R. chinensis Homozygous Genome v2.0; available at https://lipmbrowsers.toulouse.inra.fr/pub/RchiOBHm-V2/). A total of 23 homologous RcWAK/RcWAKL were obtained after BlastP. Then, the HMM files of the calcium-binding EGF-like domain (EGF_CA, PF07645.15), galacturonan-binding domain (GUB_WAK_bind, PF13947.6) and kinase domain (Pkinase_Tyr, PF07714.17) from the Pfam database (http://pfam.xfam.org/) were used for hmmsearch. The EGF_CA, GUB_WAK_bind and Pkinase_Tyr hmmsearch lead to the identification of 34, 116 and 1723 candidate proteins, respectively (Fig. 1). Finally, we have verified a total of 68 non-redundant RcWAK/RcWAKL family genes in rose genome, where 23 candidate genes contained EGF_CA, GUB_WAK_bind and Phkinase_Tyr conserved domains, and considered as RcWAK genes. The other candidate genes were RcWAKLs, of which 38 contained GUB_WAK_bind and Phkinase_Tyr domain, the remaining 7 contained EGF_CA and Pkinase_Tyr domain (Fig. 1).
Fig. 1.
The predicted results of RcWAKs/RcWAKLs. A Venn diagram of the number of predicted RcWAKs/RcWAKLs or their conserved motifs by using Blastp or HMM search. At_BlastP_Rc represented the number of predicted RcWAKs/RcWAKLs by using Arabidopsis AtWAK/AtWAKL protein sequences to search for homologous proteins in Rosa chinensis, with E value<1e-3. GUB_WAK_bind, EGF_CA and PKinase_Ser/Thr represented number of proteins carring galacturonan-binding domain, calcium-binding EGF-like domain, or a kinase domain, respectively. B The Schematic representation of RcWAKs/RcWAKLs
The length of RcWAKs and RcWAKLs was extremely varied. The shortest is RcWAKL41, which encodes 369 amino acids. The longest is RcWAKL11, it encodes a protein of 891 amino acids. The average length of RcWAK/RcWAKL proteins was 678 amino acids. The accession number, chromosomal location, numbers of exon and intron, length of CDS and subgroup of RcWAK/RcWAKL gene family were listed in Table 1.
Table 1.
Predicted members of RcWAK/RcWAKL family in ‘Rosa chinensis’
| Gene | Accession number a | Chr.b | Position c | Intron | Exon | CDS (bp) | Amino Acids | Group |
|---|---|---|---|---|---|---|---|---|
| RcWAK1 | RchiOBHm_Chr1g0329511 | 1 | 19.17431 | 2 | 3 | 2328 | 775 | I |
| RcWAK2 | RchiOBHm_Chr1g0331391 | 1 | 21.25817 | 1 | 2 | 2346 | 781 | II |
| RcWAK3 | RchiOBHm_Chr1g0336911 | 1 | 28.55835 | 2 | 3 | 2364 | 787 | I |
| RcWAK4 | RchiOBHm_Chr2g0152591 | 2 | 69.97138 | 2 | 3 | 2190 | 729 | III |
| RcWAK5 | RchiOBHm_Chr2g0152901 | 2 | 70.29155 | 2 | 3 | 2223 | 740 | III |
| RcWAK6 | RchiOBHm_Chr2g0175101 | 2 | 87.45908 | 3 | 4 | 1794 | 597 | II |
| RcWAK7 | RchiOBHm_Chr5g0016551 | 5 | 11.37102 | 2 | 3 | 2157 | 718 | I |
| RcWAK8 | RchiOBHm_Chr5g0016611 | 5 | 11.43604 | 3 | 4 | 2280 | 759 | I |
| RcWAK9 | RchiOBHm_Chr5g0016871 | 5 | 11.56994 | 2 | 3 | 1848 | 615 | I |
| RcWAK10 | RchiOBHm_Chr5g0016931 | 5 | 11.65443 | 2 | 3 | 2331 | 776 | I |
| RcWAK11 | RchiOBHm_Chr5g0017041 | 5 | 11.72528 | 2 | 3 | 2094 | 697 | I |
| RcWAK12 | RchiOBHm_Chr5g0017091 | 5 | 11.76752 | 1 | 2 | 1545 | 514 | I |
| RcWAK13 | RchiOBHm_Chr5g0017291 | 5 | 11.98662 | 2 | 3 | 2337 | 778 | I |
| RcWAK14 | RchiOBHm_Chr5g0017321 | 5 | 12.00918 | 3 | 4 | 2490 | 829 | I |
| RcWAK15 | RchiOBHm_Chr5g0019001 | 5 | 13.45881 | 2 | 3 | 2325 | 774 | I |
| RcWAK16 | RchiOBHm_Chr5g0028941 | 5 | 22.72841 | 2 | 3 | 2325 | 774 | I |
| RcWAK17 | RchiOBHm_Chr5g0046061 | 5 | 42.10957 | 2 | 3 | 2286 | 761 | III |
| RcWAK18 | RchiOBHm_Chr5g0047341 | 5 | 43.74824 | 3 | 4 | 2244 | 747 | III |
| RcWAK19 | RchiOBHm_Chr5g0052141 | 5 | 53.75484 | 2 | 3 | 2256 | 751 | I |
| RcWAK20 | RchiOBHm_Chr5g0054801 | 5 | 57.50674 | 2 | 3 | 2304 | 767 | I |
| RcWAK21 | RchiOBHm_Chr6g0259291 | 6 | 14.53278 | 2 | 3 | 2352 | 783 | I |
| RcWAK22 | RchiOBHm_Chr6g0306911 | 6 | 65.42927 | 2 | 3 | 2262 | 753 | II |
| RcWAK23 | RchiOBHm_Chr7g0210751 | 7 | 28.16683 | 1 | 2 | 2352 | 783 | III |
| RcWAKL1 | RchiOBHm_Chr1g0328221 | 1 | 17.58247 | 1 | 2 | 1938 | 645 | V |
| RcWAKL2 | RchiOBHm_Chr1g0328281 | 1 | 17.62497 | 1 | 2 | 1851 | 616 | V |
| RcWAKL3 | RchiOBHm_Chr1g0328501 | 1 | 17.90567 | 5 | 6 | 1788 | 595 | V |
| RcWAKL4 | RchiOBHm_Chr1g0328701 | 1 | 18.10089 | 6 | 7 | 1464 | 487 | V |
| RcWAKL5 | RchiOBHm_Chr1g0333331 | 1 | 25.34515 | 4 | 5 | 2013 | 670 | V |
| RcWAKL6 | RchiOBHm_Chr1g0333351 | 1 | 25.40108 | 2 | 3 | 1893 | 630 | V |
| RcWAKL7 | RchiOBHm_Chr1g0333441 | 1 | 25.56581 | 2 | 3 | 1968 | 655 | V |
| RcWAKL8 | RchiOBHm_Chr1g0333481 | 1 | 25.62574 | 1 | 2 | 2175 | 724 | V |
| RcWAKL9 | RchiOBHm_Chr1g0333571 | 1 | 25.70152 | 2 | 3 | 1968 | 655 | V |
| RcWAKL10 | RchiOBHm_Chr1g0333611 | 1 | 25.73956 | 2 | 3 | 1332 | 443 | V |
| RcWAKL11 | RchiOBHm_Chr1g0361321 | 1 | 53.32277 | 11 | 12 | 2676 | 891 | IV |
| RcWAKL12 | RchiOBHm_Chr1g0368501 | 1 | 58.6087 | 2 | 3 | 1884 | 627 | IV |
| RcWAKL13 | RchiOBHm_Chr2g0117481 | 2 | 29.64339 | 2 | 3 | 2007 | 668 | III |
| RcWAKL14 | RchiOBHm_Chr2g0150651 | 2 | 68.33199 | 2 | 3 | 1137 | 378 | III |
| RcWAKL15 | RchiOBHm_Chr2g0150741 | 2 | 68.39579 | 2 | 3 | 2208 | 735 | III |
| RcWAKL16 | RchiOBHm_Chr2g0150781 | 2 | 68.45249 | 2 | 3 | 2262 | 753 | III |
| RcWAKL17 | RchiOBHm_Chr2g0152581 | 2 | 69.95926 | 3 | 4 | 2178 | 725 | III |
| RcWAKL18 | RchiOBHm_Chr2g0152641 | 2 | 70.0389 | 2 | 3 | 2217 | 738 | III |
| RcWAKL19 | RchiOBHm_Chr3g0449381 | 3 | 1.182279 | 2 | 3 | 1875 | 624 | V |
| RcWAKL20 | RchiOBHm_Chr3g0471461 | 3 | 17.30996 | 0 | 1 | 1950 | 649 | IV |
| RcWAKL21 | RchiOBHm_Chr3g0476711 | 3 | 22.69575 | 2 | 3 | 1509 | 502 | I |
| RcWAKL22 | RchiOBHm_Chr4g0405811 | 4 | 26.57302 | 1 | 2 | 1881 | 626 | V |
| RcWAKL23 | RchiOBHm_Chr4g0434711 | 4 | 58.40836 | 1 | 2 | 1899 | 632 | V |
| RcWAKL24 | RchiOBHm_Chr4g0445241 | 4 | 65.73948 | 0 | 1 | 1920 | 639 | IV |
| RcWAKL25 | RchiOBHm_Chr5g0016561 | 5 | 11.37718 | 3 | 4 | 2109 | 702 | I |
| RcWAKL26 | RchiOBHm_Chr5g0016581 | 5 | 11.39569 | 2 | 3 | 2247 | 748 | I |
| RcWAKL27 | RchiOBHm_Chr5g0016601 | 5 | 11.43082 | 2 | 3 | 2208 | 735 | I |
| RcWAKL28 | RchiOBHm_Chr5g0016641 | 5 | 11.46383 | 2 | 3 | 1821 | 606 | I |
| RcWAKL29 | RchiOBHm_Chr5g0016881 | 5 | 11.57806 | 2 | 3 | 2262 | 753 | I |
| RcWAKL30 | RchiOBHm_Chr5g0017021 | 5 | 11.71215 | 3 | 4 | 1566 | 521 | I |
| RcWAKL31 | RchiOBHm_Chr5g0017141 | 5 | 11.87352 | 2 | 3 | 2268 | 755 | I |
| RcWAKL32 | RchiOBHm_Chr5g0017281 | 5 | 11.98236 | 2 | 3 | 2253 | 750 | I |
| RcWAKL33 | RchiOBHm_Chr5g0017331 | 5 | 12.01933 | 2 | 3 | 2310 | 769 | I |
| RcWAKL34 | RchiOBHm_Chr5g0017401 | 5 | 12.06204 | 1 | 2 | 2148 | 715 | I |
| RcWAKL35 | RchiOBHm_Chr5g0019071 | 5 | 13.55579 | 2 | 3 | 1296 | 431 | I |
| RcWAKL36 | RchiOBHm_Chr5g0047581 | 5 | 44.05517 | 3 | 4 | 2238 | 745 | V |
| RcWAKL37 | RchiOBHm_Chr6g0273041 | 6 | 33.64626 | 4 | 5 | 2082 | 693 | IV |
| RcWAKL38 | RchiOBHm_Chr6g0292991 | 6 | 55.81773 | 3 | 4 | 1554 | 517 | II |
| RcWAKL39 | RchiOBHm_Chr7g0185911 | 7 | 5.968027 | 1 | 2 | 1860 | 619 | V |
| RcWAKL40 | RchiOBHm_Chr7g0185931 | 7 | 5.988849 | 2 | 3 | 1956 | 651 | V |
| RcWAKL41 | RchiOBHm_Chr7g0185941 | 7 | 5.993237 | 2 | 3 | 1110 | 369 | V |
| RcWAKL42 | RchiOBHm_Chr7g0185971 | 7 | 6.0123 | 3 | 4 | 1989 | 662 | IV |
| RcWAKL43 | RchiOBHm_Chr7g0186001 | 7 | 6.026964 | 3 | 4 | 1974 | 657 | IV |
| RcWAKL44 | RchiOBHm_Chr7g0209921 | 7 | 27.47616 | 3 | 4 | 1872 | 623 | V |
| RcWAKL45 | RchiOBHm_Chr7g0221871 | 7 | 42.97117 | 2 | 3 | 2328 | 775 | III |
aAvailable at https://lipm-browsers.toulouse.inra.fr/pub/RchiOBHm-V2/
bChromosome
cStarting position on chromosome (Mb)
Chromosomal locations of rose RcWAK/RcWAKL genes
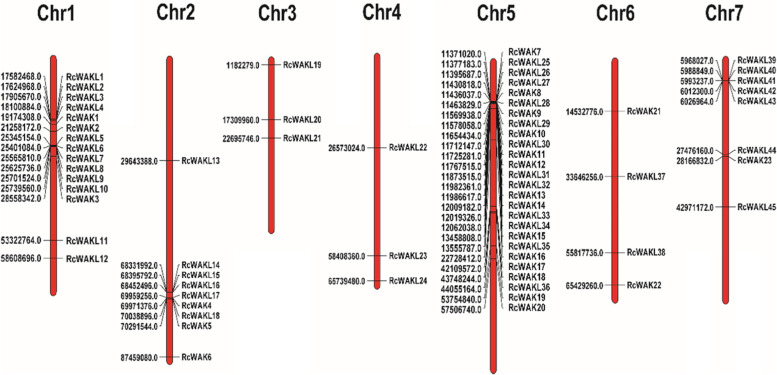
RcWAKs/RcWAKLs were mapped to seven chromosomes of the R. chinensis genome using Mapchat2.2. RcWAK1-RcWAK23 and RcWAKL1-RcWAKL45 were named according to their order on the chromosomes (Fig. 2). RcWAK/RcWAKL family genes were unevenly distributed on the seven chromosomes of rose (Table 1; Fig. 2). The high density of RcWAKs/RcWAKLs location was observed in several specific regions, such as six RcWAKLs (RcWAKL5-RcWAKL10) distributed at 25.34-25.74 Mb on chromosome 1 and fifteen RcWAKs/RcWAKLs distributed at 11.37-11.98 Mb on chromosome 5. In contrast, chromosomes 3, 4 and 6 contained only 3, 3 and 4 of RcWAKs/RcWAKLs, respectively. RcWAK members were located on chromosomes 1, 2, 5, 6 and 7, while RcWAKLs were located on all seven chromosomes. The unbalanced distribution of RcWAK/RcWAKL genes indicated genetic variation during the evolutionary process.
Fig. 2.
The localization of RcWAK/RcWAKL family members on ‘Rose chinensis’ chromosome. Numbers represented the gene starting position on chromosome
Phylogenetic analysis and structure analysis of rose RcWAK/RcWAKL genes
A phylogenetic analysis of RcWAKs/RcWAKLs was performed by using neighbor-joining method (Fig. 3). The subsequent intron-exon structure analysis and conserved domain analysis of RcWAKs/RcWAKLs were consistent with the results of the phylogenetic analysis (Fig. 3). RcWAKs/RcWAKLs contained mostly 2-3 introns, while RcWAKL20 and RcWAKL24 had no introns and RcWAKL11 contained 11 introns. Most of the genes in the same evolutionary branch exhibited similar exon-intron structures, such as RcWAKL21, RcWAKL35, RcWAK15, RcWAK3, RcWAK21, RcWAK1 and RcWAKL32 all contained 2 introns (Table 1; Fig. 3). However, there were a few exceptions, for example, RcWAKL43, RcWAKL42, RcWAKL37 and RcWAK24. More than that, the intron lengths of RcWAK/RcWAKL family members were spanning a wide range, from tens to tens of thousands of nucleotides. RcWAKL45 contained the longest intron (12,233bp), while RcWAKL38 had the shortest intron (59bp).
Fig. 3.
Phylogenetic analysis of RcWAK/RcWAKL family. A complete alignment was used to construct the tree with NJ method. The bootstrap value were symbolled on the branch node. RcWAK/RcWAKL gene conserved domain were analyzed by conserved domain database (CDD). Exon, intron and UTR were represented by yellow box, dark line and green box, respectively
In addition, we further analyzed the conserved domains of RcWAK/RcWAKL protein sequences by conserved domains database (CDD), and the results were similar to the previous results with Pfam database. All RcWAKs contained the highly conserved EGF_CA, GUB_WAK_bind and Pkinase, while RcWAKLs lacked the EGF_CA or GUB_WAK_bind domain. Meanwhile, the transmembrane (TM) structures and the signaling peptides (SP) were determined based on TMHMM Server (http://www.cbs.dtu.dk/services/TMHMM/) and SignalP (http://www.cbs.dtu.dk/services/SignalP/). Several RcWAKs/RcWAKLs were missing TM or SP in structure prediction. RcWAKL4 and RcWAKL23 missed both two structures (Fig. 3). Overall, RcWAKs/RcWAKLs are highly conserved in amino acid sequences from phylogenetic analysis, although some of RcWAKs/RcWAKLs lack one or two structures. RcWAKs/RcWAKLs from different evolutionary branches exist in diversity.
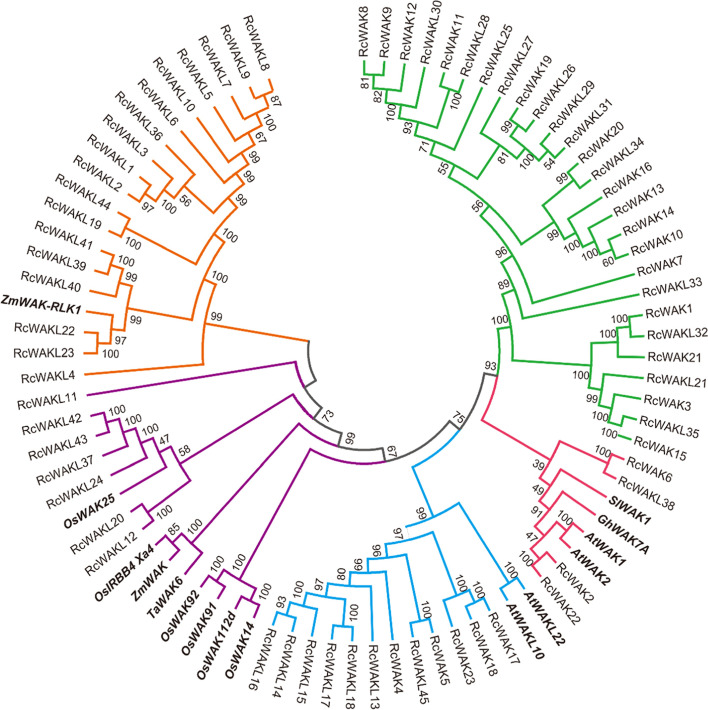
In recent years, an increasing number of studies have confirmed that WAK/WAKL family genes played a critical role in defense response. We compiled a total of 15 plants defense response-related WAK/WAKL family genes, including Arabidopsis, rice (Oryza sativa), tomato (Solanum lycopersicum), cotton (Gossypium hirsutum), maize (Zea mays L.), and wheat (Triticum aestivum) (Supplemental Table S2). To evaluate the evolutionary relationship between RcWAK/RcWAKL and the defense-related WAK/WAKL genes reported in different species, a phylogenetic tree was established using the neighbor-joining method (Fig. 4). The results showed that the proteins of the WAK/WAKL family were divided into five groups, which were labeled with different colors in Fig. 4. The groups II, III, IV and V contained WAK/WAKL from other species which were involved in plant resistance. We have also performed a comparative analysis of conserved domains in different groups. The result showed all of the five groups contained GUB_WAK_bind domain logo (PF13947) (Supplemental Fig. S1) and Pkinase_Tyr (PF07714.17) logo (Supplemental Fig. S3). However, the group V consisting of RcWAKLs was lack of EGF_CA logo, which was constituted with six cysteine (C) skeleton (Supplemental Fig. S2).
Fig. 4.
Phylogenetic analysis of the RcWAKs/RcWAKLs with defense-related WAKs/WAKLs from other plant species. Complete alignments of the rose and and the defense-related WAKs/WAKLs from other plant species, including Arabidopsis, cotton (Gossypium hirsutum), rice (Oryza sativa), tomato (Solanum lycopersicum), maize (Zea mays), and wheat (Triticum aestivum), were used to construct a phylogenetic tree using the Neighbor-Joining method. The bootstrap values are indicated on the nodes of the branches. The WAKs/WAKLs reported to be involved in plant disease resistance are marked in bold. Group I-V were labled in green, red, blue, purple and orange, respectively
Expression patterns of RcWAK/RcWAKL genes in rose petals induced by B. cinerea
Studies have shown that WAK/WAKL family members played a vital role in plant defense response to fungal pathogens. The WAKs/WAKLs which were up-regulated upon pathogen infection may involve in plant immunity. Our previous study established an RNA-seq analysis of B. cinerea inoculated rose petals at 30 hours post inoculation (hpi) and 48 hpi. On rose petals, B. cinerea conidia germinate at 24 hpi, and the early response to infection is occurred at 30 hpi, with petals having no visible lesion formation at this time point. 48 hpi corresponds to a later response, with the primary lesions starting to expand [15].
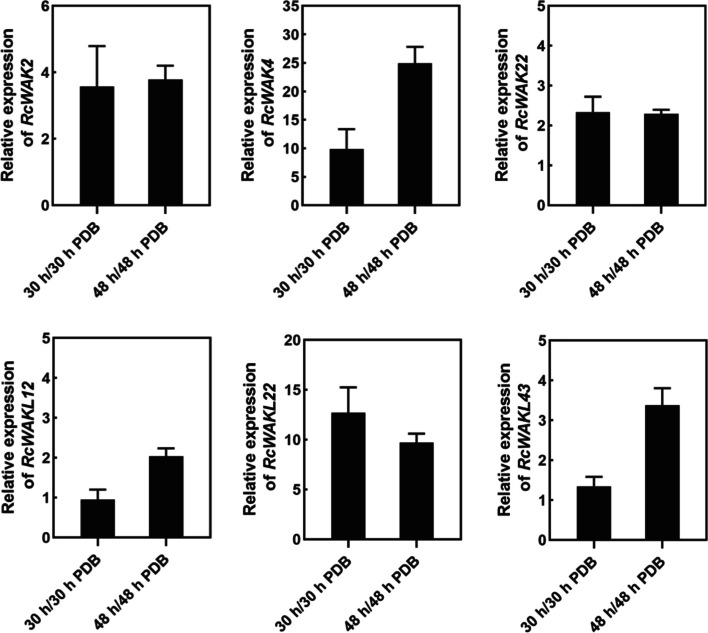
Twelve of RcWAK/RcWAKL genes were significantly up-regulated expression in RNA-seq data (Table 2). Among them, RcWAK2, RcWAK4, RcWAK8, RcWAK14, RcWAK16, RcWAKL6, RcWAKL22 and RcWAKL43 were induced in rose petals at 30 hpi, while RcWAK22, RcWAKL12, RcWAKL23, and RcWAKL43 were also induced at 48 hpi. To further clarify the expression profiles from the transcriptome data, RcWAK2, RcWAK4, RcWAK22, RcWAKL12, RcWAKL22 and RcWAKL43 expression patterns were confirmed by RT-qPCR (Fig. 5).
Table 2.
Expresion of the RcWAK/RcWAKL genes under B.cinerea infectiona
| Gene | Accession number | Group | log2Ratio 30hpi | log2Ratio 48hpi |
|---|---|---|---|---|
| RcWAK2 | RchiOBHm_Chr1g0331391 | II | 1.75 | 2.96 |
| RcWAK4 | RchiOBHm_Chr2g0152591 | III | 7.04 | - |
| RcWAK8 | RchiOBHm_Chr5g0016611 | I | 2.19 | 3.13 |
| RcWAK14 | RchiOBHm_Chr5g0017321 | I | 3.41 | 5.11 |
| RcWAK16 | RchiOBHm_Chr5g0028941 | I | 1.40 | 5.64 |
| RcWAK22 | RchiOBHm_Chr6g0306911 | II | - | 1.66 |
| RcWAKL6 | RchiOBHm_Chr1g0333351 | V | 1.56 | 4.32 |
| RcWAKL12 | RchiOBHm_Chr1g0368501 | IV | - | 1.53 |
| RcWAKL22 | RchiOBHm_Chr4g0405811 | V | 4.65 | 3.94 |
| RcWAKL23 | RchiOBHm_Chr4g0434711 | V | - | 3.69 |
| RcWAKL39 | RchiOBHm_Chr7g0185911 | V | - | 2.33 |
| RcWAKL43 | RchiOBHm_Chr7g0186001 | IV | 2.07 | 2.92 |
aThe log2 transformed expression profiles were obtained from the RNA-seq dataset
Fig. 5.
Validation of expression of selected RcWAK/RcWAKL genes using qPCR at 30 and 48 hours post inoculation. RcUBI2 was used as a reference gene. The primers used for each transcript were listed in Table S1. PDB, potato dextrose broth. ‘T’ represented the standard deviation
RcWAK4 participated in rose resistance to B. cinerea
To further illustrate the potential role of RcWAK/RcWAKL in rose petals against gray mold, we knocked down the expression of RcWAK4 using VIGS. RcWAK4 was the most significantly up-regulated RcWAKs in RNA-seq data (Table 2) and our qPCR also confirmed RcWAK4 was the most strongly induced RcWAKs upon B. cinerea inoculation (Fig. 5). Meanwhile, RcWAK4 belonged to the group III of RcWAKs/RcWAKLs and was close to AtWAKL10 and AtWAKL22, which were known to involve in plant disease resistance. Thus, RcWAK4 was considered an important candidate gene for rose resistance to B. cinerea.
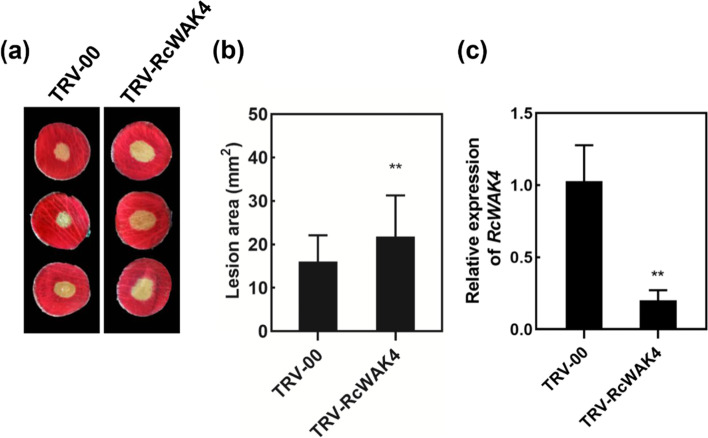
To clarify whether RcWAK4 was involved in rose petal defense response, we knocked down the expression of RcWAK4 in petals. To this end, we cloned the coding sequence fragment 309 bp of RcWAK4 into the TRV2 vector to generate TRV2-RcWAK4. Mixed agrobacterium cells carried TRV2-RcWAK4 and TRV1 in a 1:1 ratio and then vacuum infiltrated into rose petal discs for RcWAK4 silencing. The silenced petals were subsequently inoculated with B. cinerea. The RcWAK4-silencing petals exhibited a significant increase in lesion area compared with control petals inoculated with an empty TRV vector (TRV-00) (Fig. 6a and b). Finally, the silencing efficiency was confirmed by qPCR (Fig. 6c). These results suggested that RcWAK4 played an important role in rose petal resistance to B. cinerea.
Fig. 6.
Functional analysis of rose wall-associated kinase RcWAK4. a Decreased rose petal resistance to B. cinerea upon silencing of RcWAK4, shown at 60 hours post inoculation. b Statistical analysis of lesion area in rose petals. The value represented the mean of lesion area from at least 48 petal discs, Error bar=standard deviation, **P < 0.01. c Quantification of RcWAK4 expression in RcWAK4-silenced and control plants
Discussion
Plant resistance responses are beginning from the PRRs located on cell membrane recognizing to pathogenic signals, such as microbial-associated molecular patterns/damage-associated molecular patterns (MAMPs/DAMPs) [16, 17]. WAK/WAKL is a vital class of PRRs for necrotrophic pathogens (e.g., B. cinerea). Genome-wide identification of WAKs/WAKLs has been successively reported in Arabidopsis [18], rice (Oryza sativa) [19], cotton (Gossypium hirsutum) [20] and barley (Hordeum vulgare) [5]. However, there is no systematic analysis of the RcWAK/RcWAKL gene family, and the functions of the RcWAKs/RcWAKLs were largely unclear. In this study, we used ‘R. chinensis’ genome as a reference to perform a genome-wide analysis of RcWAK/RcWAKL gene family. We predicted the potential function of the RcWAK/RcWAKL gene family in rose through chromosome localization, exon-intron structural analysis, conserved domain, phylogenetic analysis, and gene expression pattern under B. cinerea infection.
The rose RcWAK/RcWAKL family contained 68 members (Table 1), while in Arabidopsis, rice, and barley were identified 26, 125, and 91 WAK/WAKL family members, respectively. The large disparity in the number of WAK/WAKL family members among different species may indicate that WAK/WAKL genes underwent different degrees of expansion during the evolution of plants. However, we have identified a total of 23 RcWAKs, they have a highly conservative domain with AtWAK1-AtWAK5 [18] and GhWAK1-GhWAK29 [20], including signaling peptide, EGF_CA, transmembrane and Pkinase (Fig. 3).
In phylogenetic analysis (Fig. 4), RcWAKs/RcWAKLs were divided into five groups. Evolutionary relationship analysis of barley also divided 91 HvWAK/HvWAKL into five groups [5]. The RcWAKL11, which was located in the group V, was evolutionarily distant from the other members in the same group (RcWAKL42, RcWAKL43, RcWAKL37, RcWAKL24, RcWAKL20 and RcWAKL12), probably due to more exons in the RcWAKL11 coding sequence. RcWAK members were distributed on chromosomes 1, 2, 5, 6 and 7, and evolutionary analysis showed that none of the RcWAK members were divided in group V (Figs. 2 and 4). Nevertheless, RcWAKLs were distributed on all seven chromosomes, but none of RcWAKLs appeared in group III in the evolutionary analysis. All four groups contained plant defenses associated WAKs/WAKLs except group I. While twelve of RcWAKs/RcWAKLs significantly up-regulated in the RNA-seq data (Table 2) induced by B. cinerea were distributed in all groups. The results implied that there was no group specificity on the role of WAK/WAKL in plant defense responses.
The expression pattern of RcWAKs/RcWAKLs induced by B. cinerea provided abundant candidate genes for its possible involvement in rose defense response to gray mold. The differential expression of RcWAK4 was more than 7-fold at the early stage after B. cinerea infection (Table 2), and the qPCR results were also consistent (Fig. 5). Transient silencing of RcWAK4 in rose petals exhibited increased susceptibility to B. cinerea (Fig. 6). In phylogenetic analysis, AtWAKL10 and AtWAKL22 were in the same group with RcWAK4 (Fig. 4). AtWAKL10 as a candidate guanylyl cyclase (GC), involved in Arabidopsis defense response [21]. Together, these results suggested that RcWAK4 as an important RcWAKs/RcWAKLs involved in the response of rose against B. cinerea.
Conclusions
A genome-wide characterization of RcWAK/RcWAKL family was performed in this study, mainly including chromosome localization, gene structure analysis, phylogenetic relationships, gene expression analysis induced by B. cinerea. We have identified a total of 68 non-redundant RcWAK/RcWAKL family members in the whole genome of R. chinensis. RcWAK4 was confirmed to be involved in rose petal resistance to gray mold by VIGS. These results provide a piece of essential information for further study RcWAK/RcWAKL family members in rose and may facilitate the further breeding of disease resistance roses.
Methods
Identification of the WAK/WAKL genes in rose genome
The complete rose (Rosa chinensis ‘Old blush’) genome was used as a reference and the data were downloaded from https://lipm-browsers.toulouse.inra.fr/pub/RchiOBHm-V2/ [22]. Firstly, we performed a BlastP homology search in rose protein library based on Arabidopsis WAK/WAKL family members. Secondly, based on the definition of WAK/WAKL, the HMM files of EGF_CA (PF07645.15), GUB_WAK_bind (PF13947.6) and PKinase_Ser/Thr (PF07714.17) were download from Pfam database (http://pfam.xfam.org/) for hmmsearch with E-value <1e-3. Finally, combining the predictions with SignalP (http://www.cbs.dtu.dk/services/SignalP/), TMHMM Server (http://www.cbs.dtu.dk/services/TMHMM/) and Conserved domain database (https://www.ncbi.nlm.nih.gov/cdd/?term=), the candidate RcWAKs/RcWAKLs containing both SP, EGF_CA or GUB_WAK_bind, transmembrane structures and PKinase structural domains were selected. Gene chromosome location distribution was mapped by Mapchat2.2 [23].
Gene structure and phylogenetic analyses
Extracted coding sequences region of RcWAK/RcWAKL genes, and the gene intron-exon structure was mapped using TBtools [24] with the genome-wide gene information file as reference. The conserved domain information of RcWAK/RcWAKL genes were obtained from Pfam database and Conserved domain database (CDD, https://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi). Full sequence alignment of RcWAK/RcWAKL was performed using the ClustalW method. Phylogenetic analysis of the comparison results was constructed using Neighbor-Joining of MEGA6.0 [25] with a bootstrap of 1000 replicates.
The WAK/WAKL from other species, containing Arabidopsis, rice (Oryza sativa), tomato (Solanum lycopersicum), cotton (Gossypium hirsutum), maize (Zea mays L.), and wheat (Triticum aestivum), which were related to plant defense response were used to phylogenetic analysis with RcWAKs/RcWAKLs. The protein sequences were collected from Genbank. All the WAKs/WAKLs were aligned by ClustalW before phylogenetic analysis with NJ of MEGA6.0 [24] (bootstrap test=1000 replicates).
The comparative analysis of conserved domains between different groups of RcWAKs/RcWAKLs. The group menbers of RcWAKs/RcWAKLs were aligned by ClustalW before performing sequences logo on WebLogo3 (http://weblogo.threeplusone.com/).
Expression data of RcWAKs/RcWAKLs in rose petal induced by B. cinerea
The RNA-seq data of rose petals infected by B. cinerea were obtained from the National Center for Biotechnology Information (NCBI) database with the accession number PRJNA414570 [15]. There no special permissions are necessary to collect these data from NCBI. Clean data were mapped to ‘Rosa chinensis’ genome, and reads per kb per million reads (RPKM) were used to value gene expression levels, and log2 (RPKM treatment/ RPKM control) was used to calculate the differential expression levels of genes.
The real time quantitative PCR were performed to confirm the RNA-seq results. To this end, cDNA synthesis were according to the RT Super Mix kit (Vazyme). qPCR was performed with TB Green Premix qRT-PCR kit (TAKARA) based on One Plus real-time PCR system (Applied Biosystems). RcUBI2 was used as the housekeeping gene, and the primers used in the experiment were shown in Supplemental Table S1.
Virus induced gene silencing (VIGS) and B. cinerea inoculation assays
The CDS sequence of RcWAK4 was used as a reference sequence to design specific primers VIGS-RcWAK4-F/R. The target was amplified using rose petal cDNA as a template to obtain 309 bp fragment. The fragment is constructed into the tobacco rattle virus (TRV) vector [26].
The VIGS and B. cinerea inoculation was performed as previously described [27]. Briefly, petals were detached from the outer whorls of the rose, which was at the opening stage 2 [28]. A 15-mm disk was punched from the center of each petal. Constructs expressing TRV1 and recombinant TRV2 cultured by Agrobacterium tumefaciens were mixed in a 1:1 ratio and vacuum-infiltrated into the petal disks. Six days after the TRV infection, the petal disks were inoculated with 2 μL B. cinerea with the concentration of 1x105 spores/mL. The VIGS was repeated at least three times using at least 48 discs. After the B. cinerea inoculation, the lesion area was measured by ImageJ, and a Student’s t-test was conducted to identify any significant differences. All the constructs and plant materials used in this study are freely available from the corresponding author, for research purposes. There no special permissions are necessary to collect item.
Supplementary Information
Additional file 1: Supplemental Table S1. List of primers used in this study
Additional file 2: Supplemental Table S2. Plant WAK/WAKL genes involved in disease resistance [29–35]
Additional file 3: Supplemental Figure S1. Comparative analysis of GUB_WAK domain between different groups of RcWAKs/RcWAKLs
Additional file 4: Supplemental Figure S2. Comparative analysis of EGF_CA domain between different groups of RcWAKs/RcWAKLs
Additional file 5: Supplemental Figure S3. Comparative analysis of Pkinase_Try domain between different groups of RcWAKs/RcWAKLs
Additional file 6: Supplemental Data. WAK/WAKL protein sequences
Acknowledgements
None.
Abbreviations
- hpi
hours post inoculation
- NJ
neighbor-joining
- RPKM
number of reads per kb per million reads
- HMM
Hidden Markov Model
- CDD
Conserved Domains Database
- VIGS
virus-induced gene silencing
Authors’ contributions
Z.Z., C.Z., W.D. and X.L. conceived and designed the experiments. X.L., Z.W., T.Y. and D.L. performed the experiments. X.L., Z.W. and S.Z. analyzed the data. Z.Z., W.D. and X.L. wrote the paper. All the authors have read and approved the final version of the manuscript.
Funding
This study was supported by the National Natural Science Foundation of China to Zhao Zhang (grant number 31772344) and to Changqing Zhang (grant number 31872147), and Hebei Provincial Key Natural Science Foundation and Key Basic Research Projects of Basic Research Program to Wenqi Dong (grant number 18962901D). The funders played no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Availability of data and materials
The protein sequences used and/or analyzed during the current study are available in ‘Rosa chinensis’ genome (https://lipm-browsers.toulouse.inra.fr/pub/RchiOBHm-V2/) and Genebank (https://www.ncbi.nlm.nih.gov/genbank/). The RNA-seq data used in the current study is available in NCBI database with the accession number PRJNA414570. The plant materials are available from the corresponding author on reasonable request.
Declarations
Ethics approval and consent to participate
Not applicable. Our research did not involve any human or animal subjects, material, or data. The plant materials used in this study are freely available for research purposes following institutional, national and international guidelines.
Consent for publication
Not applicable.
Competing interests
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Changqing Zhang, Email: chqzhang@cau.edu.cn.
Zhao Zhang, Email: zhangzhao@cau.edu.cn.
References
- 1.Broekgaarden C, Caarls L, Vos IA, Pieterse CM, Van Wees SC. Ethylene: traffic controller on hormonal crossroads to defense. Plant Physiol. 2015;169(4):2371–2379. doi: 10.1104/pp.15.01020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Underwood W. The plant cell wall: a dynamic barrier against pathogen invasion. Front Plant Sci. 2012;3:85. doi: 10.3389/fpls.2012.00085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kohorn BD, Kobayashi M, Johansen S, Riese J, Huang LF, Koch K, Fu S, Dotson A, Byers N. An Arabidopsis cell wall-associated kinase required for invertase activity and cell growth. Plant J. 2006;46(2):307–316. doi: 10.1111/j.1365-313X.2006.02695.x. [DOI] [PubMed] [Google Scholar]
- 4.Bacete L, Mélida H, Miedes E, Molina A. Plant cell wall-mediated immunity: cell wall changes trigger disease resistance responses. Plant J. 2018;93(4):614–636. doi: 10.1111/tpj.13807. [DOI] [PubMed] [Google Scholar]
- 5.Tripathi RK, Aguirre JA, Singh J. Genome-wide analysis of wall associated kinase (WAK) gene family in barley. Genomics. 2021;113(1):523–530. doi: 10.1016/j.ygeno.2020.09.045. [DOI] [PubMed] [Google Scholar]
- 6.He Z-H, Cheeseman I, He D, Kohorn BD. A cluster of five cell wall-associated receptor kinase genes, Wak1–5, are expressed in specific organs of Arabidopsis. Plant Mol Biol. 1999;39(6):1189–1196. doi: 10.1023/A:1006197318246. [DOI] [PubMed] [Google Scholar]
- 7.Lally D, Ingmire P, Tong H-Y, He Z-H. Antisense expression of a cell wall–associated protein kinase, WAK4, inhibits cell elongation and alters morphology. Plant Cell. 2001;13(6):1317–1332. doi: 10.1105/tpc.13.6.1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang H, Niu H, Liang M, Zhai Y, Huang W, Ding Q, Du Y, Lu M. A wall-associated kinase gene CaWAKL20 from pepper negatively modulates plant thermotolerance by reducing the expression of ABA-responsive genes. Front Plant Sci. 2019;10:591. doi: 10.3389/fpls.2019.00591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brutus A, Sicilia F, Macone A, Cervone F, De Lorenzo G. A domain swap approach reveals a role of the plant wall-associated kinase 1 (WAK1) as a receptor of oligogalacturonides. Proc Natl Acad Sci. 2010;107(20):9452–9457. doi: 10.1073/pnas.1000675107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ferrari S, Savatin DV, Sicilia F, Gramegna G, Cervone F, De Lorenzo G. Oligogalacturonides: plant damage-associated molecular patterns and regulators of growth and development. Front Plant Sci. 2013;4:49. doi: 10.3389/fpls.2013.00049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zuo W, Chao Q, Zhang N, Ye J, Tan G, Li B, Xing Y, Zhang B, Liu H, Fengler KA. A maize wall-associated kinase confers quantitative resistance to head smut. Nat Genet. 2015;47(2):151–157. doi: 10.1038/ng.3170. [DOI] [PubMed] [Google Scholar]
- 12.Delteil A, Gobbato E, Cayrol B, Estevan J, Michel-Romiti C, Dievart A, Kroj T, Morel J-B. Several wall-associated kinases participate positively and negatively in basal defense against rice blast fungus. BMC Plant Biol. 2016;16(1):1–10. doi: 10.1186/s12870-016-0711-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang N, Pombo MA, Rosli HG, Martin GB. Tomato wall-associated kinase SlWak1 depends on Fls2/Fls3 to promote apoplastic immune responses to Pseudomonas syringae. Plant Physiol. 2020;183(4):1869–1882. doi: 10.1104/pp.20.00144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liorzou M, Pernet A, Li S, Chastellier A, Thouroude T, Michel G, Malécot V, Gaillard S, Briée C, Foucher F. Nineteenth century French rose (Rosa sp.) germplasm shows a shift over time from a European to an Asian genetic background. J Exp Bot. 2016;67(15):4711–4725. doi: 10.1093/jxb/erw269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu X, Cao X, Shi S, Zhao N, Li D, Fang P, Chen X, Qi W, Zhang Z. Comparative RNA-Seq analysis reveals a critical role for brassinosteroids in rose (Rosa hybrida) petal defense against Botrytis cinerea infection. BMC Genet. 2018;19(1):62. doi: 10.1186/s12863-018-0668-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Newman M-A, Sundelin T, Nielsen JT, Erbs G. MAMP (microbe-associated molecular pattern) triggered immunity in plants. Front Plant Sci. 2013;4. [DOI] [PMC free article] [PubMed]
- 17.AbuQamar S, Moustafa K, Tran LS. Mechanisms and strategies of plant defense against Botrytis cinerea. Crit Rev Biotechnol. 2017;37(2):262–274. doi: 10.1080/07388551.2016.1271767. [DOI] [PubMed] [Google Scholar]
- 18.Verica JA, He Z-H. The cell wall-associated kinase (WAK) andWAK-like kinase gene family. Plant Physiol. 2002;129(2):455–459. doi: 10.1104/pp.011028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.de Oliveira LFV, Christoff AP, de Lima JC, de Ross BCF, Sachetto-Martins G, Margis-Pinheiro M, Margis R. The Wall-associated Kinase gene family in rice genomes. Plant Sci. 2014;229:181–192. doi: 10.1016/j.plantsci.2014.09.007. [DOI] [PubMed] [Google Scholar]
- 20.Dou L, Li Z, Shen Q, Shi H, Li H, Wang W, Zou C, Shang H, Li H, Xiao G. Genome-wide characterization of the WAK gene family and expression analysis under plant hormone treatment in cotton. BMC Genomics. 2021;22(1):1–17. doi: 10.1186/s12864-021-07378-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Meier S, Ruzvidzo O, Morse M, Donaldson L, Kwezi L, Gehring C. The Arabidopsis wall associated kinase-like 10 gene encodes a functional guanylyl cyclase and is co-expressed with pathogen defense related genes. PLoS One. 2010;5(1):e8904. doi: 10.1371/journal.pone.0008904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Raymond O, Gouzy J, Just J, Badouin H, Verdenaud M, Lemainque A, Vergne P, Moja S, Choisne N, Pont C. The Rosa genome provides new insights into the domestication of modern roses. Nat Genet. 2018;50(6):772–777. doi: 10.1038/s41588-018-0110-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Voorrips R. MapChart: software for the graphical presentation of linkage maps and QTLs. J Hered. 2002;93(1):77–78. doi: 10.1093/jhered/93.1.77. [DOI] [PubMed] [Google Scholar]
- 24.Chen C, Chen H, He Y, Xia R. TBtools, a toolkit for biologists integrating various HTS-data handling tools with a user-friendly interface. biRxiv. In.; 2018.
- 25.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 2013;30(12):2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu Y, Schiff M, Dinesh-Kumar S. Virus-induced gene silencing in tomato. Plant J. 2002;31(6):777–786. doi: 10.1046/j.1365-313X.2002.01394.x. [DOI] [PubMed] [Google Scholar]
- 27.Cao X, Yan H, Liu X, Li D, Sui M, Wu J, Yu H, Zhang Z. A detached petal disc assay and virus-induced gene silencing facilitate the study of Botrytis cinerea resistance in rose flowers. Hortic Res. 2019;6:136. doi: 10.1038/s41438-019-0219-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ma N, Cai L, Lu W, Tan H, Gao J. Exogenous ethylene influences flower opening of cut roses (Rosa hybrida) by regulating the genes encoding ethylene biosynthesis enzymes. Sci China Ser C Life Sci. 2005;48(5):434. doi: 10.1360/062004-37. [DOI] [PubMed] [Google Scholar]
- 29.Kohorn BD, Kohorn SL, Todorova T, Baptiste G, Stansky K, McCullough M. A dominant allele of Arabidopsis pectin-binding wall-associated kinase induces a stress response suppressed by MPK6 but not MPK3 mutations. Mol Plant. 2012;5(4):841–851. doi: 10.1093/mp/ssr096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Diener AC, Ausubel FM. RESISTANCE TO FUSARIUM OXYSPORUM 1, a dominant Arabidopsis disease-resistance gene, is not race specific. Genetics. 2005;171(1):305–321. doi: 10.1534/genetics.105.042218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang P, Zhou L, Jamieson P, Zhang L, Zhao Z, Babilonia K, Shao W, Wu L, Mustafa R, Amin I. The cotton wall-associated kinase GhWAK7A mediates responses to fungal wilt pathogens by complexing with the chitin sensory receptors. Plant Cell. 2020;32(12):3978–4001. doi: 10.1105/tpc.19.00950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Harkenrider M, Sharma R, De Vleesschauwer D, Tsao L, Zhang X, Chern M, Canlas P, Zuo S, Ronald PC. Overexpression of rice wall-associated kinase 25 (OsWAK25) alters resistance to bacterial and fungal pathogens. PLoS One. 2016;11(1):e0147310. doi: 10.1371/journal.pone.0147310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hu K, Cao J, Zhang J, Xia F, Ke Y, Zhang H, Xie W, Liu H, Cui Y, Cao Y. Improvement of multiple agronomic traits by a disease resistance gene via cell wall reinforcement. Nat Plants. 2017;3(3):1–9. doi: 10.1038/nplants.2017.9. [DOI] [PubMed] [Google Scholar]
- 34.Dmochowska-Boguta M, Kloc Y, Zielezinski A, Werecki P, Nadolska-Orczyk A, Karlowski WM, Orczyk W. TaWAK6 encoding wall-associated kinase is involved in wheat resistance to leaf rust similar to adult plant resistance. PLoS One. 2020;15(1):e0227713. doi: 10.1371/journal.pone.0227713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hurni S, Scheuermann D, Krattinger SG, Kessel B, Wicker T, Herren G, Fitze MN, Breen J, Presterl T, Ouzunova M. The maize disease resistance gene Htn1 against northern corn leaf blight encodes a wall-associated receptor-like kinase. Proc Natl Acad Sci. 2015;112(28):8780–8785. doi: 10.1073/pnas.1502522112. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Supplemental Table S1. List of primers used in this study
Additional file 2: Supplemental Table S2. Plant WAK/WAKL genes involved in disease resistance [29–35]
Additional file 3: Supplemental Figure S1. Comparative analysis of GUB_WAK domain between different groups of RcWAKs/RcWAKLs
Additional file 4: Supplemental Figure S2. Comparative analysis of EGF_CA domain between different groups of RcWAKs/RcWAKLs
Additional file 5: Supplemental Figure S3. Comparative analysis of Pkinase_Try domain between different groups of RcWAKs/RcWAKLs
Additional file 6: Supplemental Data. WAK/WAKL protein sequences
Data Availability Statement
The protein sequences used and/or analyzed during the current study are available in ‘Rosa chinensis’ genome (https://lipm-browsers.toulouse.inra.fr/pub/RchiOBHm-V2/) and Genebank (https://www.ncbi.nlm.nih.gov/genbank/). The RNA-seq data used in the current study is available in NCBI database with the accession number PRJNA414570. The plant materials are available from the corresponding author on reasonable request.