Abstract
Recently, we found that more than 10% of the cases of acute non-A, non-B, non-C hepatitis in Taiwan were caused by a novel strain of hepatitis E virus (HEV). Since none of these patients had a history of travel to areas where HEV is endemic, the source of transmission remains unclear. The recent discovery of a swine HEV in herd pigs in the United States has led us to speculate that HEV may also circulate in herd pigs in Taiwan and may serve as a reservoir for HEV in Taiwan. Of 275 herd pigs obtained from 10 pig farms in Taiwan, 102 (37%) were seropositive for serum anti-HEV immunoglobulin G (IgG). A 185-bp genomic sequence within the ORF-2 of the HEV genome was amplified and cloned from serum samples of an anti-HEV positive pig and subsequently from serum samples of a patient with acute hepatitis E. Sequence comparison revealed that the swine and human isolates of HEV share 97.3% identity. Phylogenetic analyses further showed that the Taiwan swine and human isolates of HEV form a distinct branch divergent from all other known strains of HEV, including the U.S. swine strain. To examine the potential risk of cross-species transmission of swine HEV to humans, the seroprevalences of anti-HEV IgG in 30 swine handlers, 20 pork dealers, and 50 control subjects were assessed and were found to be 26.7, 15, and 8%, respectively (for swine handlers versus controls, P = 0.048). Our findings may help provide an understanding of the modes of HEV transmission and may also raise potential public health concerns for HEV zoonosis.
Hepatitis E virus (HEV) is the major causative pathogen of enterically transmitted non-A, non-B hepatitis (3, 4, 17, 33). HEV is a nonenveloped RNA virus. Its genome, about 7.5 kb in size, contains three partially overlapping open reading frames (ORFs) (18, 40). ORF-1 likely encodes nonstructural viral proteins (putative RNA helicase, protease, and RNA-dependent RNA polymerase). ORF-2 encodes the putative capsid protein, and ORF-3 encodes a cytoskeleton-associated phosphoprotein (14, 18, 40, 50).
HEV is transmitted primarily via a fecal-oral route, and hepatitis E occurs predominantly as outbreaks of waterborne epidemics in developing countries (3, 18). Hepatitis E is rarely diagnosed in developed countries and is usually regarded as imported (1, 8, 21, 38, 39). It has, however, long been an enigma that the seroprevalence of HEV antibodies (anti-HEV) is relatively high in areas where HEV is not endemic compared to the documented disease burden (7, 22, 25, 30, 32, 45).
Taiwan, an area of endemicity for viral hepatitis A and B, has never had an epidemic of hepatitis E. However, about 10 to 20% of the cases of acute hepatitis identified on this island are without a defined etiology. Recently, we found that more than 10% of our cases of acute non-A, non-B, non-C hepatitis were caused by acute infection with a novel strain of HEV (12). A partial sequence within the ORF-1 of this novel strain of HEV was successfully cloned from four of these patients. Phylogenetic studies indicated that all four of these isolates of HEV form a distinct branch divergent from all previously reported isolates worldwide (12). Since none of our patients had a disease-related history of travel to areas where HEV is endemic, it is quite likely that the transmission of HEV occurred in Taiwan. Therefore, it is important to determine the source of transmission of HEV in Taiwan. Recently, a novel strain of HEV from swine (the U.S. swine HEV strain) was isolated from herd pigs in the midwestern United States (26). Subsequently, two cases of acute hepatitis E reported in the United States were found to be caused by an HEV strain genetically very similar to the U.S. swine HEV strain (36). These findings suggest that swine HEV may be involved in cross-species infection between swine and humans. Since swine HEV and the U.S. human strains of HEV are so similar genetically and since Taiwan has a dense population of pigs, it was of interest to examine whether pigs in Taiwan are a potential reservoir for HEV transmission to humans. Herein we report evidence of HEV circulation in herd pigs in Taiwan and identification of a Taiwan strain of swine HEV genetically distinct from the U.S. swine HEV strain. Interestingly, sequence comparison and phylogenetic analyses indicated that the Taiwan swine and human isolates of HEV were distinct from other known strains of HEV but were very closely related to each other.
MATERIALS AND METHODS
Swine serum samples.
Serum samples were collected from 275 pigs on 10 pig farms in different geographic regions in Taiwan. Almost all the pigs tested in this study were older than 3 months. Serum samples from 10 specific-pathogen-free (SPF) pigs raised under laboratory conditions were included as negative controls for the enzyme-linked immunosorbent assay (ELISA) for detection of anti-HEV in swine sera.
ELISA for anti-HEV in swine.
The standard ELISA for anti-HEV in swine was performed essentially as described previously (26, 27). Briefly, a purified 55-kDa truncated form of the putative capsid protein derived from a human strain of HEV (Sar-55) was used as the antigen for the ELISA (42, 43). Peroxidase-labeled goat anti-swine immunoglobulin G (IgG) was used as the secondary antibody. All of the swine serum samples were assayed in duplicate. Preimmune and hyperimmune anti-HEV-positive sera as well as preinoculation and postinoculation sera from a pig experimentally infected with swine HEV were included as negative and positive controls, respectively (26, 27). In addition, serum samples from 10 SPF pigs were included as negative controls.
Human serum samples.
To examine the genetic relationship of the Taiwan swine isolate of HEV to the human HEV isolate causing sporadic cases of acute hepatitis E in Taiwan, acute-phase sera from a patient with acute hepatitis E were used for molecular cloning of a cognate region within the ORF-2 of the HEV genome. We had used the same serum sample to clone a partial sequence within the ORF-1 of HEV in a previous study (patient T821) (12). To assess the seroprevalence of anti-HEV IgG in pig handlers, serum samples were obtained from 30 individuals who had worked on swine farms for at least 5 years. Twenty individuals who had worked in pork companies and 50 age- and sex-matched (to the swine handlers) healthy individuals were included as control groups.
ELISA for anti-HEV IgG and anti-HEV IgM in human.
Serum was assayed for anti-HEV IgG and anti-HEV IgM in humans by using commercial diagnostic kits according to the manufacturer's instructions (Genelabs Diagnostica, Singapore) (48).
RNA extraction and reverse transcription-PCR (RT-PCR).
RNA was extracted from 100 μl of each serum sample by the single-step acid-phenol method and was converted to cDNA by a random primer method as described previously (11). The cDNA templates were subjected to amplification by a PCR standardized to detect HEV RNA. Two sets of primers derived from the putative capsid gene of the U.S. swine HEV strain (26) were used for the nested PCR. The first-round and second-round PCRs were performed similarly with 25 and 35 cycles, respectively, and the PCR parameters included denaturation for 40 s at 94°C, annealing for 1 min at 55°C, and extension for 1 min at 72°C. The amplified DNA products were analyzed by gel electrophoresis and were subsequently cloned into the pGEM-T vector according to the manufacturer's instructions (Promega, Madison, Wis.). The cloned PCR products were sequenced with an Automatic Sequencer Kit (ABI PRISM 337 DNA sequencer PRISM 337 Collection; Sequence Analysis 3.0; Perkin-Elmer). The primer sequences for the first-round PCR were as follows: sense primer, 5′-AACACACCTTACACTGGTGCAT-3′, and antisense primer, 5′-TGCCCTTGTCCTGCTGCGCATTCT-3′. Those for the second-round PCR were as follows: sense primer, 5′-CGTGTTTCCCGGTACACCAGCACA-3′, and antisense primer, 5′-CAGTTGGCCCCCGGCCGACGAA-3′.
Sequence analyses.
Pairwise sequence comparison and phylogenetic analyses were carried out with the aid of computer software (Clustal) with a weighted residue weight table (DNAstar, Inc., Madison, Wis.).
Nucleotide sequence accession numbers.
The GenBank accession numbers for the Taiwan swine and human isolates of HEV reported herein are AF077004 and AF077005, respectively.
RESULTS
Serological evidence that HEV is circulating in herd pigs in Taiwan.
Two hundred seventy-five serum samples obtained from pigs on 10 pig farms were assayed for the presence of anti-HEV IgG. As shown in Table 1, about 37.1% of the pigs tested were seropositive for anti-HEV IgG. However, the percentage of pigs seropositive for anti-HEV IgG varied among different farms: from as low as 0% (0 of 6 pigs) and 4.8% (2 of 42 pigs) to as high as 61.5% (16 of 26 pigs) and 66.7% (20 of 30 pigs). The variations were not considered to be a result of age differences, since almost all the pigs tested were older than 3 months. The specificity of the ELISA for assaying anti-HEV IgG in swine sera had been validated previously (26) and was further supported by seronegativity for anti-HEV in 10 SPF pigs raised under laboratory conditions (data not shown).
TABLE 1.
Prevalence of antibodies to HEV in herd pigs in Taiwan
| Farm no. | No. (%) of pigs
|
|
|---|---|---|
| Assayed | Positive for anti-HEV | |
| 1 | 6 | 0 (0) |
| 2 | 42 | 2 (4.8) |
| 3 | 27 | 3 (11.1) |
| 4 | 23 | 5 (21.7) |
| 5 | 16 | 5 (31.3) |
| 6 | 38 | 12 (31.6) |
| 7 | 12 | 6 (50) |
| 8 | 55 | 33 (60) |
| 9 | 26 | 16 (61.5) |
| 10 | 30 | 22 (66.7) |
| Total | 275 | 102 (37.1) |
Identification of a novel sequence of HEV from a pig serum sample positive for anti-HEV.
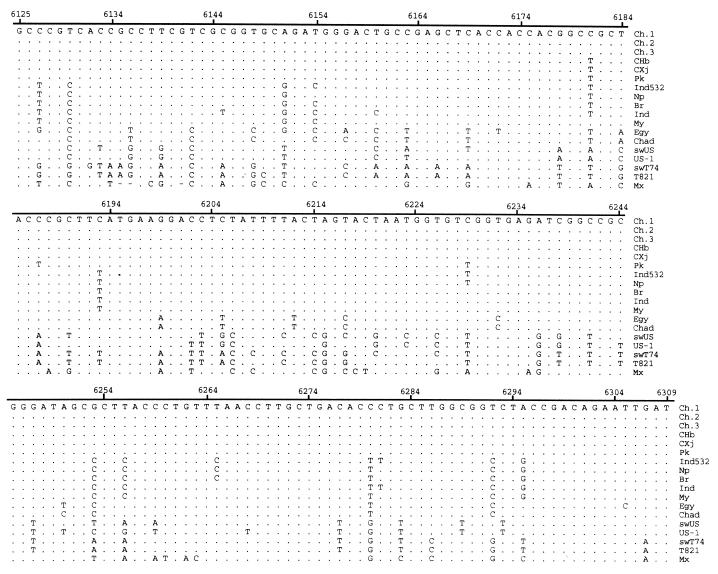
Fifty-six serum samples obtained from farms 9 and 10 were subjected to RT-PCR amplification to attempt to identify the sequence within the conserved ORF-2 region of HEV. Only 1 of the 38 anti-HEV-positive serum samples was positive by RT-PCR for HEV RNA, while all 18 anti-HEV-negative serum samples remained RT-PCR negative (data not shown). Cloning and sequencing analysis confirmed that the amplified PCR product was derived from the genome of a novel HEV strain. The nucleotide sequence of this novel strain of HEV (strain swT74) from swine was compared to the sequences of the corresponding regions of the U.S. swine HEV and other strains of human HEV (Fig. 1).
FIG. 1.
Nucleotide sequence alignments of a 185-bp genomic sequence within the ORF-2 region of HEV. Ch.1, Ch.2, Ch.3, CHb, and CXj are five isolates from China (GenBank accession nos., D11092, L25547, L25595, M94177, and L08816, respectively); Pk, Ind532, Np, Br, Ind, MY, Egy, and Chad are the isolates from Pakistan (accession no. M80581), India (accession no. U62621), Nepal (accession no. AF020607), Myanmar (accession no. M73218), India (accession no. X99441), Myanmar (accession no. D10330), Egypt (accession no. AF051351), and Chad (accession no. U62121), respectively; swUS and USP-1 are swine and human isolates, respectively, from the United States (accession nos. AF011921 and AF035437, respectively); swT74 (accession no. AF077004) is the swine isolate from Taiwan; T821 (accession no. AF077005) is the isolate from patient T821 (this report); Mx (accession no. M74506) is an isolate from Mexico. The nucleotide position is relative to the numbering system for the Burmese isolate (40). The nucleotide sequence of isolate Ch.1 is shown on top, and only nucleotide substitutions are indicated (uppercase) for other isolates.
Comparison of Taiwan swine HEV to Taiwan human HEV.
We have previously reported the identification of a novel strain of HEV from patients with acute non-A, non-B, non-C hepatitis in Taiwan (12). The cDNA fragments cloned from these human strains of HEV were within the ORF-1. In order to determine the genetic relationship between strains of swine HEV and human HEV in Taiwan, a patient with acute hepatitis E was selected. The patient was a 72-year-old retired farmer who had lived beside a pig farm in the northern region of Taiwan for decades. The patient had no history of travel abroad before the episode of acute hepatitis. The patient was admitted to the Chang Gung Memorial Hospital presenting with a typical clinical course of acute viral hepatitis. The peak levels of serum alanine aminotransferase (ALT) and total bilirubin were 1,126 U/liter and 22.5 mg%, respectively. The diagnosis of acute hepatitis E was based on elevated levels of ALT and total bilirubin along with the following findings: (i) negative results of testing for serum hepatitis A virus IgM antibodies, hepatitis B virus IgM core antibodies, hepatitis B virus surface antigen, hepatitis C virus antibodies, and hepatitis C virus RNA as determined by RT-PCR; (ii) seropositivity for HEV IgM and IgG antibodies; and (iii) positivity for HEV RNA in serum by an RT-PCR as described previously (12). Serum samples collected on day 10 after the onset of jaundice were used for the molecular cloning of a partial genomic fragment within the ORF-2 of HEV (Fig. 1). We had used the same serum sample to clone a partial sequence within the ORF-1 of HEV in a previous report (patient T821) (12). Comparison of the sequence of this ORF-2 region revealed that this Taiwan isolate of human HEV (T821) shared 97.3 and 98.4% sequence identities with the Taiwan swine HEV isolate (swT74) at the nucleotide level and the amino acid level, respectively (Table 2).
TABLE 2.
Identities of nucleotide sequences and amino acid sequences shown in Fig. 1
| Strain | % Identitya
|
|||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Ch.1 | Ch.2 | Ch.3 | CHb | CXj | Pk | Ind532 | Np | Br | Ind | My | Egy | Chad | swUS | US-1 | swT74 | T821 | Mx | |
| Ch.1 | 100.0 | 100.0 | 100.0 | 100.0 | 100.0 | 100.0 | 100.0 | 100.0 | 100.0 | 100.0 | 98.4 | 98.4 | 95.1 | 95.1 | 90.2 | 91.8 | 83.3 | |
| Ch.2 | 100.0 | 100.0 | 100.0 | 100.0 | 100.0 | 100.0 | 100.0 | 100.0 | 100.0 | 100.0 | 98.4 | 98.4 | 95.1 | 95.1 | 90.2 | 91.8 | 83.3 | |
| Ch.3 | 100.0 | 100.0 | 100.0 | 100.0 | 100.0 | 100.0 | 100.0 | 100.0 | 100.0 | 100.0 | 98.4 | 98.4 | 95.1 | 95.1 | 90.2 | 91.8 | 83.3 | |
| CHb | 99.5 | 99.5 | 99.5 | 100.0 | 100.0 | 100.0 | 100.0 | 100.0 | 100.0 | 100.0 | 98.4 | 98.4 | 95.1 | 95.1 | 90.2 | 91.8 | 83.3 | |
| CXj | 99.5 | 99.5 | 99.5 | 100.0 | 100.0 | 100.0 | 100.0 | 100.0 | 100.0 | 100.0 | 98.4 | 98.4 | 95.1 | 95.1 | 90.2 | 91.8 | 83.3 | |
| Pk | 98.4 | 98.4 | 98.4 | 98.9 | 98.9 | 100.0 | 100.0 | 100.0 | 100.0 | 100.0 | 98.4 | 98.4 | 95.1 | 95.1 | 90.2 | 91.8 | 83.3 | |
| Ind532 | 92.4 | 92.4 | 92.4 | 93.0 | 93.0 | 93.0 | 100.0 | 100.0 | 100.0 | 100.0 | 98.4 | 98.4 | 95.1 | 95.1 | 90.2 | 91.8 | 83.3 | |
| Np | 93.5 | 93.5 | 93.5 | 94.1 | 94.1 | 94.1 | 98.9 | 100.0 | 100.0 | 100.0 | 98.4 | 98.4 | 95.1 | 95.1 | 90.2 | 91.8 | 83.3 | |
| Br | 93.5 | 93.5 | 93.5 | 94.1 | 94.1 | 93.0 | 98.9 | 98.9 | 100.0 | 100.0 | 98.4 | 98.4 | 95.1 | 95.1 | 90.2 | 91.8 | 83.3 | |
| Ind | 92.4 | 92.4 | 92.4 | 93.0 | 93.0 | 91.9 | 97.8 | 96.8 | 97.8 | 100.0 | 98.4 | 98.4 | 95.1 | 95.1 | 90.2 | 91.8 | 83.3 | |
| My | 94.6 | 94.6 | 94.6 | 94.1 | 94.1 | 93.0 | 97.8 | 97.8 | 98.9 | 97.8 | 98.4 | 98.4 | 95.1 | 95.1 | 90.2 | 91.8 | 83.3 | |
| Egy | 87.0 | 87.0 | 87.0 | 87.5 | 87.6 | 86.5 | 87.6 | 87.6 | 88.6 | 88.6 | 88.6 | 100.0 | 93.4 | 93.4 | 88.5 | 90.2 | 81.7 | |
| Chad | 89.2 | 89.2 | 89.2 | 89.7 | 89.7 | 88.6 | 88.1 | 88.1 | 89.2 | 89.2 | 89.2 | 96.8 | 93.4 | 93.4 | 88.5 | 90.2 | 81.7 | |
| swUS | 78.4 | 78.4 | 78.4 | 78.4 | 78.4 | 78.9 | 74.6 | 75.1 | 75.1 | 74.6 | 75.7 | 74.6 | 76.8 | 100.0 | 95.1 | 96.7 | 88.3 | |
| US-1 | 80.0 | 80.0 | 80.0 | 80.0 | 80.0 | 80.5 | 76.8 | 77.3 | 77.3 | 76.8 | 77.8 | 75.7 | 77.3 | 92.4 | 95.1 | 96.7 | 88.3 | |
| swT74 | 69.2 | 69.2 | 69.2 | 69.7 | 69.7 | 70.3 | 68.6 | 69.2 | 69.2 | 68.6 | 69.2 | 67.0 | 68.6 | 80.0 | 77.3 | 98.4 | 88.3 | |
| T821 | 72.4 | 72.4 | 72.4 | 73.0 | 73.0 | 73.5 | 71.9 | 72.4 | 72.4 | 71.9 | 72.4 | 70.3 | 71.9 | 81.6 | 78.4 | 97.3 | 86.7 | |
| Mx | 70.9 | 70.9 | 70.9 | 70.9 | 70.9 | 70.3 | 68.7 | 69.8 | 69.8 | 69.2 | 70.9 | 71.9 | 69.2 | 69.8 | 66.5 | 71.4 | 70.3 | |
The percent identities below the diagonal space are for nucleotide sequences, and those above the diagonal space are for amino acid sequences.
Phylogenetic analyses of the swine and human strains of HEV isolated in Taiwan and other available strains of HEV worldwide.
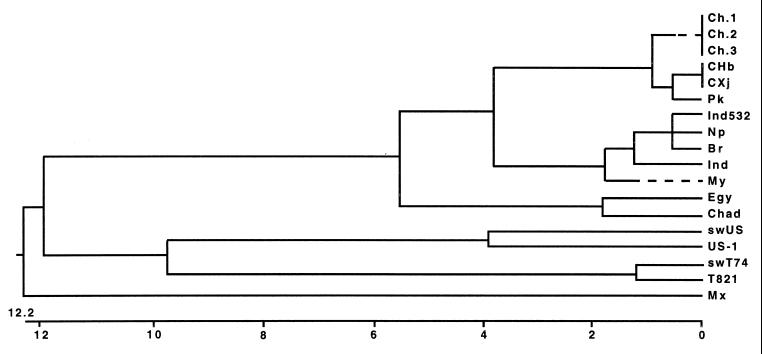
A pairwise comparison based on a 185-bp sequence within the ORF-2 was performed (Table 2). The Taiwan swine HEV strain (swT74), the Taiwan human HEV strain (T821), the U.S. swine HEV strain (swUS), a strain of HEV identified from a patient in the United States (US-1), and other strains of HEV isolated from other regions of the world were included in the comparison. As shown in Table 2, the nucleotide sequence identity between the U.S. swine isolate and US-1 was 92.4%, whereas the identity between the Taiwan swine isolate and the U.S. swine isolate was 80.0%, and that between the Taiwan swine isolate and US-1 was 77.3%. Clearly, the two swine strains of HEV isolated in different geographic regions were genetically distinct. The nucleotide sequences of the Taiwan swine and human isolates of HEV were 97.3% identical to each other, while they shared only about 70% nucleotide sequence identities with HEV strains isolated from China, Pakistan, India, Nepal, Burma, Egypt, and Chad. Phylogenetic analysis, as shown in Fig. 2, indicated that the Taiwan swine and Taiwan human isolates of HEV were clustered into a single branch, while the U.S. swine isolate and the U.S. human isolate (US-1) formed another distinct branch. The isolates from Asia including China, Pakistan, India, Nepal, and Myanmar formed the third branch, and the Mexican isolate alone represented the fourth branch (Fig. 2). The isolates from Africa (Egypt and Chad) were clustered with each other and were relatively related to the isolates from Asia, but they were quite divergent from the Taiwan, U.S., and Mexican isolates. As a result, both of the isolates from Egypt and Chad might be classified as a subbranch of the Asian strains. The phylogenetic tree based on the partial sequence within ORF-2 is very similar to the tree based on the partial sequence within ORF-1 presented in our previous report (12). However, the sequence within ORF-1 of the Taiwan swine isolate was not available for analysis in our previous study.
FIG. 2.
Phylogenetic tree based on a 185-bp sequence within the ORF-2 region of the HEV genome. The sources of these isolates are described in the text as well as in the legend to Fig. 1. The phylogenetic tree was constructed with the computer software Clustal with the weighted residue weight table (DNAstar) grounded on the sequence divergence. The length of each pair of branches represents the distance between sequence pairs. The scale beneath the tree measures the distance between sequences. Units indicate the number of substitution events.
Prevalence of serum anti-HEV IgG in pig handlers and in general population in Taiwan.
To examine whether pig handlers are at higher risk for HEV infection, 30 individuals who had been handling pigs for more than 5 years were assayed for the presence of anti-HEV. Anti-HEV IgG was detected in 26.7% (8 of 30) of these pig handlers. In comparison, 20 individuals who worked in pork companies but who had no direct contact with pigs were included, and 15% (3 of 20) were found to be positive for serum anti-HEV IgG. In addition, 50 age- and sex-matched (to the group of pig handlers) individuals were also selected as controls, and 8% (4 of 50) were found to be positive for serum anti-HEV IgG. As shown in Table 3, the differences in the seroprevalence rates among the three groups are not statistically significant, while the difference between the pig handlers and the controls was marginally significant (P = 0.048 by Fisher's exact test).
TABLE 3.
Rates of serum antibodies against HEV in persons handing pig or not handling pigs
| Group | No. of persons | Age (yr)a | No. (%) positive for anti-HEVb |
|---|---|---|---|
| Pig handlers | 30 | 42.8 ± 10 | 8 (26.7)c |
| Pork workers | 20 | 44 ± 8 | 3 (15) |
| Controls | 50 | 42.3 ± 4 | 4 (8)c |
Values are means ± standard errors.
Human serum anti-HEV IgG was assayed with an ELISA kit from Genelabs Diagnostica.
P = 0.048 by Fisher's exact test.
DISCUSSION
It has long been an enigma that the seroprevalence of anti-HEV is relatively high in countries where HEV is not endemic (7, 22, 25, 30, 41). Recently, an increasing number of patients with sporadic acute hepatitis E but with no history of travel to areas where HEV is endemic have been reported in industrialized countries (15, 16, 20, 37, 49, 51), suggesting that HEV infection in those patients is acquired domestically. The need to explain domestic infection of HEV in areas where HEV is not endemic has led to a hypothesis that animals may serve as reservoirs for the transmission of human hepatitis E (34). This hypothesis is further supported by the findings that anti-HEV has been detected in a number of wild animals including pigs, sheep, and rats in areas where HEV is endemic (5, 6) and that domestic pigs, monkeys, rats, and sheep were reportedly susceptible to infection with HEV (2, 13, 24, 45). Balayan et al. (2) reported that Russian domestic swine were susceptible to experimental infection with a Central Asian strain of HEV isolated from a human. However, the identity of the virus infecting pigs in that study was not known since the virus recovered from the pigs was not sequenced. Others have failed to experimentally infect pigs with two well-characterized epidemic strains of human HEV, including an Asian strain of human HEV (27, 31). Recently, a U.S. strain of human HEV isolated from a U.S. resident was shown to infect pigs, but this U.S. strain of human HEV is genetically very closely related to the U.S. strain of swine HEV (28). It is possible that the genetic makeup of a particular HEV strain may determine the host range. Experimental infections of rhesus monkeys and chimpanzees with swine HEV and of pigs with a human strain of HEV (28) provided experimental and genetic evidence for cross-species HEV infection. However, further experiments are needed to confirm that swine serve as a natural reservoir for transmission of human HEV.
We report herein on another isolate of swine HEV identified from a pig in Taiwan. The Taiwan swine HEV strain is genetically distinct from the swine HEV strain reported in the United States (26) and other known strains of HEV, but it is very closely related to the HEV strain isolated from humans with sporadic acute hepatitis E in Taiwan (12). Several lines of evidence suggest that the Taiwan swine isolate of HEV and the human strains of HEV causing sporadic acute hepatitis E in Taiwan may be variants of the same virus. (i) On the basis of a 185-bp sequence within the ORF-2 of the HEV genome, the Taiwan swine and human isolates of HEV are genetically very similar to each other (97.3 and 98.4% sequence identities at the nucleotide and amino acid levels, respectively). (ii) The phylogenetic tree based on the partial ORF-2 sequence (Fig. 2) is very similar to the one generated previously with a partial ORF-1 sequence, the sequence of which has not been determined for the Taiwan swine isolate (12). The similar genetic relationship among different isolates of HEV was obtained by separate phylogenetic analyses of the entire genome and of each of the putative viral genes, as well as the respective peptides of HEV (10), which further support our hypothesis. (iii) The human isolate of HEV from which the partial ORF-2 sequence was determined in this study emanated from the same serum sample (T821) that was used to clone the partial sequence within ORF-1 in our previous study (12). Therefore, the Taiwan swine isolate of HEV very likely forms a monophyletic group with the four human isolates of HEV described in our previous report (12). Further studies are warranted to characterize genetically and experimentally the novel swine HEV strain identified in Taiwan and to determine definitively the phylogenetic relationships among these novel HEV strains.
Since the identification of novel strains of HEV from two U.S. residents without a history of travel to areas where HEV is endemic, numerous HEV variants have recently been identified in areas where the disease is not endemic (9, 12, 20, 36, 37, 46, 47, 51). Novel HEV variants which are genetically distinct from the Burmese, Mexican, and U.S. genotypes have been identified from Taiwan (12), Italy (37, 51), Greece (37), Egypt (44), and China (46). However, the sequences of the new Chinese isolates that have been reported were not in the same region as those of the Taiwan isolates of human and swine HEV whose sequences have been reported (46), and the sequences of the Italian and Greek isolates are not yet available in the database (37, 51) to permit us to perform a phylogenetic analysis. Recently, by using bootstrap resampling of the multiple-sequence alignment, Schlauder et al. (37) reported a phylogenetic tree based on a partial sequence within ORF-1. They showed that the isolates from Taiwan were relatively more closely related to the isolates from Liaoning Province of China (namely, the new Chinese genotype) but were divergent from Burmese-like group (including isolates from Burma, Pakistan, India, Nepal, and western China), the Mexican isolate, the U.S. group (including two human isolates and one swine isolate from the United States), or the novel isolates from Europe (including two the Italian isolates and one Greek isolate). However, a definitive genetic relationship between the Taiwan swine HEV strain and the human isolates of HEV from Liaoning Province of China remains to be determined.
The seroprevalence of anti-HEV in swine in Taiwan was found to be relatively high, suggesting that HEV is widespread in the swine population in Taiwan. Similar results were reported for pigs in the United States (26). Therefore, it is reasonable to speculate that HEV may circulate in the swine population worldwide. This may pose a potential public health concern as a zoonosis for pig handlers as well as for the general population, since HEV is transmitted by the fecal-oral route. In addition, considering the immense interest in xenotransplantation of pig organs, swine HEV may also pose a potential risk for xenozoonosis.
As a first step toward addressing the question of whether swine HEV can be transmitted to humans, we assessed the prevalence of serum anti-HEV IgG in pig handlers, pork dealers, and the general population. Similar to previous reports (22), the prevalence of serum anti-HEV was relatively high even in the control groups (8% of the general population and 15% of pork dealers). Although the prevalence of serum anti-HEV was higher in pig handlers (26.7%), the difference in prevalence between pig handlers and the general population group is only marginally significant (for pig handlers versus the general population group, P = 0.048 by Fisher's exact test). These findings indicate (i) that HEV infection occasionally occurs in the general population. (ii) Transmission of swine HEV to humans does occur, but not frequently. In fact, only a few cases of acute hepatitis E have been identified in the United States, although most of the pigs there were infected with HEV. Considering that pigs usually become infected at an early age and the majority of adult pigs are seropositive for anti-HEV (26) and considering that shedding of virus in feces occurs only during the acute stage of infection, anti-HEV-positive adult pigs very unlikely shed virus in their feces or sera. In addition, because the virus is transmitted by the fecal-oral route, transmission of HEV is greatly dependent on the sanitary conditions under which the pig handlers work, which have been much improved in recent years in industrialized countries. These factors should significantly reduce the risk for transmission of HEV from swine to humans. (iii) Swine may not be the only animal reservoir for HEV. That hepatitis E has been reported in countries that do not traditionally raise pigs and that the high rate of seroprevalence of anti-HEV in the general population in areas where HEV is not endemic indicate the presence of other animal reservoirs for HEV. In fact, serum anti-HEV has been found in wild rats, monkeys, and sheep in areas where HEV is endemic (6). Domestic animals such as dogs, cats, and rats may also serve as reservoirs for transmission of human hepatitis E. Clearly, the natural history of HEV must be determined in order to effectively prevent HEV zoonosis.
Overt hepatitis E is usually associated with a higher serum bilirubin level (17, 29, 47) and a higher mortality rate than acute hepatitis caused by hepatitis A or B virus (18). However, most of the individuals positive for serum anti-HEV IgG do not have a history of overt hepatitis, suggesting that subclinical infection with HEV exists and occurs more frequently than overt hepatitis. Indeed, in pigs naturally infected with HEV and in pigs experimentally infected with swine HEV, only mild histological evidence of hepatitis was observed (26, 28). Similarly, only mild biochemical and histological evidence of hepatitis was observed in nonhuman primates experimentally infected with swine HEV (27). Variations in the clinical manifestations of viral hepatitis are usually dependent on multiple factors, such as the age of the patients and the immune response of the host. The genetic heterogeneity of the virus may also be an important determining factor. For example, infection with genotype 1b of the hepatitis C virus usually runs a graver course and has a poorer response to interferon therapy than infection with other genotypes (19, 23, 35). It is, therefore, very important to determine whether an avirulent strain of HEV is causing a subclinical infection or whether a virulent strain of HEV is causing severe hepatitis in humans. The Taiwan swine HEV strain does not appear to cause any clinical symptoms in swine (data not shown). However, a strain of HEV very similar to the Taiwan swine HEV strain caused overt hepatitis in humans in Taiwan (for example, patient T821). Furthermore, pigs inoculated with the US-2 human strain of HEV remained clinically normal (28). Nevertheless, these findings do not exclude the possibility that clinically divergent presentations of human HEV infection are a result of the genetic heterogeneity of the virus. Comparison of the sequences of HEV isolates from patients with or without overt hepatitis is necessary to address this question. However, it is almost impossible to isolate HEV from individuals without overt hepatitis, since HEV infection is transient and subclinical hepatitis goes unattended.
ACKNOWLEDGMENTS
We thank Suzanne U. Emerson and Robert H. Purcell for providing the purified HEV antigen for detection of swine anti-HEV. We also thank Pei-Ying Yang for excellent technical assistance and Su-Chen Chi for help in preparation of the manuscript.
This work was supported in part by a fund from the Chang Gung Memorial Hospital and a grant (DOH89-TD-1027) from the Department of Health, Executive Yuan, Taiwan.
REFERENCES
- 1.Bader T, Krawczynski K, Polish L, Favorov M. Hepatitis E in a U.S. traveler to Mexico. N Engl J Med. 1991;325:1659. doi: 10.1056/NEJM199112053252321. . (Letter.) [DOI] [PubMed] [Google Scholar]
- 2.Balayan M S, Usmanov P K, Zamayatina N A, Djumalieva D I, Karas F R. Brief report: experimental hepatitis E infection in domestic pigs. J Med Virol. 1990;32:58–59. doi: 10.1002/jmv.1890320110. [DOI] [PubMed] [Google Scholar]
- 3.Bradley D W, Krawczynski K, Cook E H, Jr, McCaustaland K, Humphrey C D, Spelbring J E, Myint H, Maynard J E. Enterically transmitted non-A, non-B hepatitis: serial passages of disease in cynomolgus macaques and tamarines and recovery of disease-associated 27- to 34-nm virus-like particles. Proc Natl Acad Sci USA. 1987;84:6277–6281. doi: 10.1073/pnas.84.17.6277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bradley D W. Enterically-transmitted non-A, non-B hepatitis. Br Med Bull. 1990;46:442–461. doi: 10.1093/oxfordjournals.bmb.a072409. [DOI] [PubMed] [Google Scholar]
- 5.Clayson E T, Innis B L, Myint K S A, Narupiti S, Vaughn D W, Giril S, Ranabhat P, Shrestha M P. Detection of hepatitis E virus infections among domestic swine in the Kathmandu Valley of Nepal. Am J Trop Med Hyg. 1995;53:228–232. doi: 10.4269/ajtmh.1995.53.228. [DOI] [PubMed] [Google Scholar]
- 6.Clayson E T, Snitbhan R, Ngarmpochana M, Vaughn D W, Shresth M P. Evidence of the hepatitis E virus (HEV) is a zoonotic virus: detection of natural infections among swine, rats and chickens in an area endemic for human disease. In: Busson Y, Coursaget P, Kane M, editors. Enterically transmitted hepatitis viruses. France: La Simarre, Joué-lès-Tours; 1996. pp. 329–335. [Google Scholar]
- 7.Dawson G J, Chau K H, Cabal C M, Yarbough P O, Reye G R, Mushahwar I K. Solid-phase enzyme-linked immunosorbent assay for hepatitis E virus IgG and IgM antibodies utilizing recombinant antigens and synthetic peptides. J Virol Methods. 1992;38:175–186. doi: 10.1016/0166-0934(92)90180-l. [DOI] [PubMed] [Google Scholar]
- 8.Dawson G J, Mushahwar I K, Chau K H. Detection of long-lasting antibody to hepatitis E virus in an U.S. traveler to Pakistan. Lancet. 1992;340:426–427. doi: 10.1016/0140-6736(92)91507-5. [DOI] [PubMed] [Google Scholar]
- 9.Erker J C, Desai S M, Schlauder G G, Dawsone G J, Mushahwa I K. A hepatitis virus variant from the United States: molecular characterization and transmission in cynomolgus macaques. J Gen Virol. 1999;80:681–690. doi: 10.1099/0022-1317-80-3-681. [DOI] [PubMed] [Google Scholar]
- 10.Gouvea V, Snellings N, Popek M J, Longer C F, Innis B L. Hepatitis E virus: complete genome sequence and phylogenetic analysis of a Nepali isolate. Virus Res. 1998;57:21–26. doi: 10.1016/s0168-1702(98)00079-3. [DOI] [PubMed] [Google Scholar]
- 11.Hsieh S Y, Yang P Y, Chen H C, Liaw Y F. Cloning and characterization of the 5′ terminal sequences of the RNA genome of GB virus C/hepatitis G virus. Proc Natl Acad Sci USA. 1997;94:3206–3210. doi: 10.1073/pnas.94.7.3206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hsieh S Y, Yang Y P, Ho Y P, Chu C M, Liaw Y F. Identification of a novel strain of hepatitis E virus responsible for sporadic acute hepatitis in Taiwan. J Med Virol. 1998;55:300–304. doi: 10.1002/(sici)1096-9071(199808)55:4<300::aid-jmv8>3.0.co;2-4. [DOI] [PubMed] [Google Scholar]
- 13.Jameel S, Durgapal H, Habibullah C M, Khuroo M S, Panda S K. Enteric non-A, non-B hepatitis: epidemics, animal transmission, and hepatitis E virus detection by the polymerase chain reaction. J Med Virol. 1992;37:263–270. doi: 10.1002/jmv.1890370405. [DOI] [PubMed] [Google Scholar]
- 14.Jameel S, Zafrullah M, Ozdener M H, Panda S K. Expression in animal cells and characterization of the hepatitis E virus structural proteins. J Virol. 1996;70:207–216. doi: 10.1128/jvi.70.1.207-216.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jardi R, Buti M, Rodriguez-Frias F, Esteban R. Hepatitis E infection in acute sporadic hepatitis in Spain. Lancet. 1993;341:1355–1356. doi: 10.1016/0140-6736(93)90874-g. [DOI] [PubMed] [Google Scholar]
- 16.Johansson P J, Mushahwar I K, Norkrans G, Weiland O, Nordenfelt E. Hepatitis E infection in patients with acute hepatitis non-A-D in Sweden. Scand J Infect Dis. 1995;27:543–546. doi: 10.3109/00365549509047064. [DOI] [PubMed] [Google Scholar]
- 17.Khuroo M S. Study of an epidemic of non-A, non-B hepatitis: possibility of another human hepatitis virus distinct from post-transfusion non-A, non-B type. Am J Med. 1980;68:818–823. doi: 10.1016/0002-9343(80)90200-4. [DOI] [PubMed] [Google Scholar]
- 18.Krawczynski K. Hepatitis E. Hepatology. 1993;17:932–941. [PubMed] [Google Scholar]
- 19.Kurosaki M, Enomoto N, Murakami T, Sakuma I, Ashina Y, Yamamoto C, Ikeda T, Tozuda S, Izumi N, Marumo F, Sato C. Analysis of genotype and amino acid residues 2209 to 2248 of the NS5A region of hepatitis C virus in relation to the response to interferon-beta therapy. Hepatology. 1997;25:750–753. doi: 10.1002/hep.510250343. [DOI] [PubMed] [Google Scholar]
- 20.Kwo P Y, Schlauder G G, Carpenter H A, Murphy P J, Rosenblatt J E, Dawson G J, Mase E E, Krawczynski K, Balan V. Acute hepatitis E by a new isolate acquired in the United States. Mayo Clin Proc. 1997;72:1133–1136. doi: 10.4065/72.12.1133. [DOI] [PubMed] [Google Scholar]
- 21.Lau J Y, Sallie R, Fang J W, Yarbough P O, Reyes G R, Portmann B C, Mieli-Vergani G, Willian R. Detection of hepatitis E virus genome and gene products in two patients with fulminant hepatitis E. J Hepatol. 1995;22:605–610. doi: 10.1016/0168-8278(95)80215-0. [DOI] [PubMed] [Google Scholar]
- 22.Lee S D, Wang Y J, Lu R H, Chan C Y, Lo K J, Moeckli R. Seroprevalence of antibody to hepatitis E virus among Chinese subjects in Taiwan. Hepatology. 1994;19:866–870. [PubMed] [Google Scholar]
- 23.Le Guen B, Squadrito G, Nalpas B, Berthelot P, Pol S, Berchot C. Hepatitis C virus genome complexity correlates with response to interferon therapy: a study in French patients with chronic hepatitis C. Hepatology. 1997;25:1250–1254. doi: 10.1002/hep.510250531. [DOI] [PubMed] [Google Scholar]
- 24.Maneerat Y, Clayson E T, Myint K S A, Young G D, Innis B L. Experimental infection of the laboratory rat with the hepatitis E virus. J Med Virol. 1996;48:121–128. doi: 10.1002/(SICI)1096-9071(199602)48:2<121::AID-JMV1>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- 25.Mast E E, Kuramoto I K, Favorov M O, Sehocning V R, Burkholder B T, Shapirs C N, Holland P V. Prevalence of risk factors for antibodies to hepatitis E virus seroreactivity among blood donors in northern California. J Infect Dis. 1997;176:34–40. doi: 10.1086/514037. [DOI] [PubMed] [Google Scholar]
- 26.Meng X J, Purcell R H, Halbur P G, Lehman J R, Webb D M, Tsareva T S, Haynes J S, Thacker B J, Emerson S U. A novel virus in swine in closely related to the human hepatitis E virus. Proc Natl Acad Sci USA. 1997;94:9860–9865. doi: 10.1073/pnas.94.18.9860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Meng X J, Halbur P G, Haynes J S, Tsareva T S, Bruna J D, Royer P L, Purcell R H, Emerson S U. Experimental infection of pigs with the newly identified swine hepatitis E virus (swine HEV), but not with human strains of HEV. Arch Virol. 1998;143:1405–1415. doi: 10.1007/s007050050384. [DOI] [PubMed] [Google Scholar]
- 28.Meng X J, Halbur P G, Shapiro M S, Govindarajan S, Bruna I D, Mushahwar I K, Purcell R H, Emerson S U. Genetic and experimental evidence for cross-species infection by swine hepatitis E virus. J Virol. 1998;72:9714–9721. doi: 10.1128/jvi.72.12.9714-9721.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Morrow R H, Smetana H F, Sai F T, Edgcomb J H. Unusual features of viral hepatitis in Accra, Ghana. Ann Intern Med. 1968;68:1250–1264. doi: 10.7326/0003-4819-68-6-1250. [DOI] [PubMed] [Google Scholar]
- 30.Paul D A, Knigge M F, Ritter A, Gutierrez R, Pilo-Matia T, Chau K H, Dawson J G. Determination of hepatitis E virus seroprevalence by using recombinant fusion proteins and synthetic peptides. J Infect Dis. 1994;169:801–806. doi: 10.1093/infdis/169.4.801. [DOI] [PubMed] [Google Scholar]
- 31.Platt K B, Yoon K J, Zimmerman S. Swine research report. Ames: Iowa State University; 1998. Susceptibility of swine to hepatitis E virus and its significance to human health; pp. 125–126. [Google Scholar]
- 32.Quiroga J A, Cotonat T, Castillo I, Carreno V. Hepatitis E virus seroprevalence in acute viral hepatitis in a developed country confirmed by a supplemental assay. J Med Virol. 1996;50:16–19. doi: 10.1002/(SICI)1096-9071(199609)50:1<16::AID-JMV4>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 33.Reyes G R, Purdy M A, Kim J P, Luk K C, Young L M, Fry K E, Bradley D W. Isolation of a cDNA from the virus responsible for enterically transmitted non-A, non-B hepatitis. Science. 1990;247:1335–1339. doi: 10.1126/science.2107574. [DOI] [PubMed] [Google Scholar]
- 34.Reyes G R. Overview of the epidemiology and biology of the hepatitis E virus. In: Willsen R A, editor. Viral Hepatitis. New York, N.Y: Marcel Dekker, Inc.; 1997. pp. 239–258. [Google Scholar]
- 35.Romeo R, Rumi M G, Del Ninno E, Colombo M. Hepatitis C genotype 1b and risk of hepatocellular carcinoma. Hepatology. 1997;26:1077–1081. doi: 10.1002/hep.510260443. [DOI] [PubMed] [Google Scholar]
- 36.Schlauder G G, Dawson G J, Erker J C, Kwo P Y, Knigge M F, Smalley D L, Rosenblatt J E, Desai S M, Mushahwar I K. The sequence and phylogenetic analysis of a novel hepatitis E virus isolated from a patient with acute hepatitis reported in the United States. J Gen Virol. 1998;79:447–456. doi: 10.1099/0022-1317-79-3-447. [DOI] [PubMed] [Google Scholar]
- 37.Schlauder G G, Desai S M, Zanetti A R, Tassopoulos N C, Mushahwar I K. Novel hepatitis E virus (HEV) isolates from Europe: evidence for additional genotypes of HEV. J Med Virol. 1999;57:243–251. doi: 10.1002/(sici)1096-9071(199903)57:3<243::aid-jmv6>3.0.co;2-r. [DOI] [PubMed] [Google Scholar]
- 38.Skidmore S J, Yarbough P O, Gabor K A, Tam A W, Reyes G R. Imported hepatitis E in UK. Lancet. 1991;337:1541. doi: 10.1016/0140-6736(91)93227-z. [DOI] [PubMed] [Google Scholar]
- 39.Smalley D L, Brewer S C, Dawson G J, Kyrk C, Waters B. Hepatitis E virus infection in an immigrant to the United States. South Med J. 1996;89:993–996. doi: 10.1097/00007611-199610000-00012. [DOI] [PubMed] [Google Scholar]
- 40.Tam A W, Smith M M, Guerra M E, Huang C C, Bradley D W, Fry K E, Reyes G R. Hepatitis E virus (HEV): molecular cloning and sequencing of the full-length viral genome. Virology. 1991;185:120–131. doi: 10.1016/0042-6822(91)90760-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Thomas D L, Yarbough P O, Vlahov D, Tsarev S A, Nelson K E, Saah A J, Purcell R H. Seroreactivity to hepatitis E virus in areas where the disease is not endemic. J Clin Microbiol. 1997;35:1244–1247. doi: 10.1128/jcm.35.5.1244-1247.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tsareva S A, Emerson S U, Reyes G R, Tsareva T S, Legters L J, Malik I A, Iqbal M, Purcell R H. Characterization of a prototype strain of hepatitis E virus. Proc Natl Acad Sci USA. 1992;89:559–563. doi: 10.1073/pnas.89.2.559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tsareva S A, Tsareva T S, Emerson S U, Kapikian A Z, Ticehurst J, London W, Purcell R H. ELISA for antibody to hepatitis E virus (HEV) based on complete open-reading frame-2 protein expressed in inset cells: identification of HEV infection in primates. J Infect Dis. 1993;168:369–378. doi: 10.1093/infdis/168.2.369. [DOI] [PubMed] [Google Scholar]
- 44.Tsareva S A, Binn L N, Gomatos P J, Arthure R R, Monier M K, van Cuyck-Gandr H, Longer C F, Innis B L. Phylogenetic analysis of hepatitis E virus isolates from Egypt. J Med Virol. 1999;57:68–74. doi: 10.1002/(sici)1096-9071(199901)57:1<68::aid-jmv10>3.0.co;2-e. [DOI] [PubMed] [Google Scholar]
- 45.Usmanov R K, Balayan M S, Dvoinkova O V, Alymbaeva D B, Zamiatina N A, Kazachkov I A, Belov V I. An experimental infection in lambs by the hepatitis E virus. Vopr Virusol. 1994;9:165–168. [PubMed] [Google Scholar]
- 46.Wang Y, Ling R, Erker J C, Zhang H, Li H, Desai S, Mushahwar I K, Harrison T I. A divergent genotype of hepatitis E virus in Chinese patients with acute hepatitis. J Gen Virol. 1999;80:169–177. doi: 10.1099/0022-1317-80-1-169. [DOI] [PubMed] [Google Scholar]
- 47.Wu J C, Sheen I J, Chiang T Y, Sheng W Y, Wand Y J, Chan C Y, Lee S D. The impact of traveling to endemic areas on the spread of hepatitis E virus infection: epidemiological and molecular analyses. Hepatology. 1998;27:1415–1420. doi: 10.1002/hep.510270532. [DOI] [PubMed] [Google Scholar]
- 48.Yarbough P O, Tam A W, Fry K E, Krawczynski I K, McCaustland K A, Bradley D W, Reyes G R. Hepatitis E virus: identification of type-common epitopes. J Virol. 1991;65:5790–5797. doi: 10.1128/jvi.65.11.5790-5797.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zaaijer H L, Mauser-Bunschoten F P, ten Veen J H, Kapprell H P, Kok M, van den Berg H M, Lelie P N. Hepatitis E virus antibodies among patients with hemophilia, blood donors, and hepatitis patients. J Med Virol. 1995;46:244–246. doi: 10.1002/jmv.1890460313. [DOI] [PubMed] [Google Scholar]
- 50.Zafrullah M, Ozadener M H, Panda S K, Jameel S. The ORF3 protein of hepatitis E virus is a phosphoprotein that associates with the cytoskeleton. J Virol. 1997;71:9045–9053. doi: 10.1128/jvi.71.12.9045-9053.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zanetti A R, Schlauder G G, Romano L, Tanzi E, Fabris P, Dawson G J, Mushahwar I K. Identification of a novel variant of hepatitis E virus in Italy. J Med Virol. 1999;57:356–360. doi: 10.1002/(sici)1096-9071(199904)57:4<356::aid-jmv5>3.0.co;2-d. [DOI] [PubMed] [Google Scholar]