Abstract
Simple Summary
The glycosyltransferase β1,4-N-acetylgalactosaminyltransferae 2 (B4GALNT2), product of the B4GALNT2 gene is responsible for the biosynthesis of the carbohydrate antigen Sda. Both the enzyme and its cognate antigen display a restricted pattern of tissue expression and modulation in colorectal, gastric, and mammary cancers. In colorectal cancer, B4GALNT2 is generally downregulated, but patients displaying higher expression survive longer. The sialyl Lewisa and sialyl Lewisx antigens are associated with malignancy. Their biosynthesis and that of Sda are mutually exclusive. Forced expression of B4GALNT2 in colorectal cancer cell lines modulates the transcriptome towards lower malignancy, reducing stemness. These effects are independent of B4GALNT2-induced sLea/sLex inhibition. Thus, B4GALNT2 is a marker of better prognosis and a cancer-restraining enzyme in colorectal cancer, with a therapeutic potential.
Abstract
Terminal carbohydrate structures are particularly relevant in oncology because they can serve as cancer markers and alter the phenotype of cancer cells. The Sda antigen and the sialyl Lewisx and sialyl Lewisa (sLex and sLea) antigens are terminal structures whose biosynthesis is mutually exclusive. In this review, we describe the main features of the Sda antigen in cancer and its relationship with sLex/a antigens. Information was obtained from an extensive literature search and from The Cancer Genome Atlas (TCGA) public database. The Sda biosynthetic enzyme B4GALNT2 undergoes downregulation in colorectal (CRC) and stomach cancer, while it is ectopically expressed by a minority of breast cancer (BRCA) patients. High expression of B4GALNT2 is associated with better prognosis and a less malignant gene expression profile in CRC, while the opposite occurs in BRCA. The regulation of B4GALNT2 expression in CRC is multifactorial, involving gene methylation and miRNA expression. Forced expression of B4GALNT2 inhibited sLea/sLex and reduced malignancy and stemness in cells constitutively expressing sLex/a antigens. However, consistent effects were observed upon B4GALNT2 forced expression and in cells not expressing sLex/a antigens. Thus, B4GALNT2 and the Sda antigen exert a tumor-restraining activity in CRC and probably other gastrointestinal cancers, independently of sLex/a antigens.
Keywords: glycosylation, colorectal cancer, Sda antigen, Sialyl Lewis antigens, glycosyltransferases, B4GALNT2, gene expression control, transcriptomic analysis
1. Introduction
The impact of glycosylation on cell behavior largely depends on the terminal portions of glycoconjugates. Examples of terminal structures include α2,3- and α2,6-linked sialic acids, polysialic acid, the AB0 blood group, and other fucosylated structures such as the sialyl Lewisx (sLex) and the sialyl Lewisa (sLea) antigens [1,2,3]. The Sda antigen is a terminal carbohydrate structure expressed on erythrocytes, in secretions [4], and in a few organs [5] by the vast majority of Caucasians. The Sda antigen was discovered independently by two groups in 1967 [6,7] and found to be inherited as a dominant character. Although the percentage of Sda-negative individuals was found to be 4%, the percentage of individuals with “natural” anti Sda antibodies in serum was much lower. In urine and kidney, the major carrier of Sda is the urinary Tamm–Horsfall glycoprotein. In this review, we will focus on the role of Sda and of its biosynthetic enzyme B4GALNT2 in cancer and its relationship with sLex and sLea antigens. However, it is worth mentioning that Sda plays a role in a wide variety of biological systems, including the attenuation of the phenotype in mouse models of muscular dystrophy [8,9] and bleeding disorders [10,11], in the equilibrium of the gut microbiota [12,13], in the prolificacy of sheep [14], in the xenotransplantation of pig organs [15], and in the inhibition of influenza virus infection [16,17].
2. The Sda Antigen: Structure and Biosynthesis
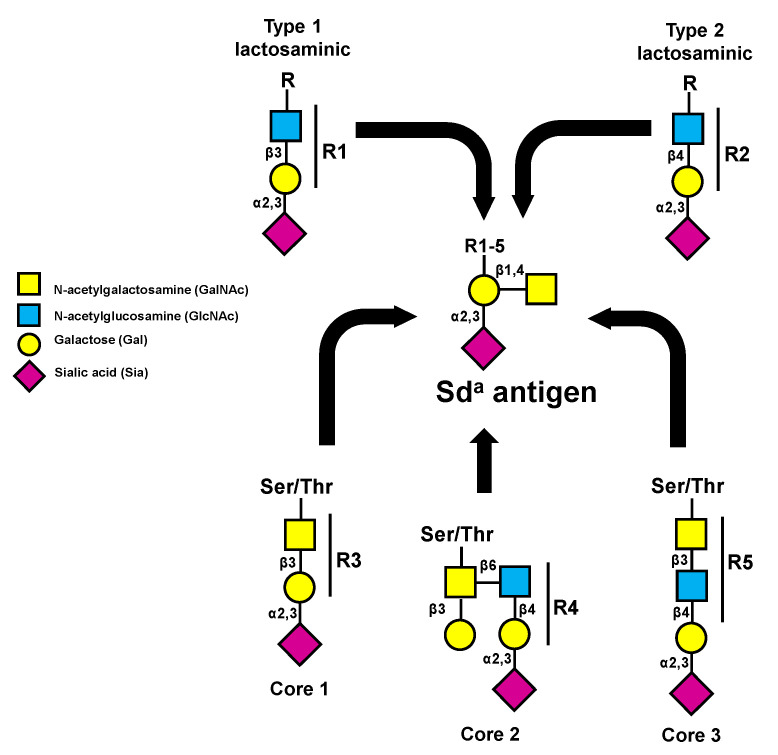
The structure of the Sda antigen was elucidated in the 1980s and found to have a basic composition of a α2,3-sialylated galactose to which a β1,4 GalNAc is linked [18]. The underlying carbohydrate structure is variable, including type 1 (Galβ1,3GlcNAc) and type 2 (Galβ1,4GlcNAc) lactosaminic chains, as well as the core 1, core 2, and core 3 structures of O-linked chains (Figure 1) [19]. In addition, the glycolipid sialylparagloboside can be decorated by the Sda antigen [20].
Figure 1.
Structure of the different carbohydrate chains that can be terminated by the Sda antigen. These structures can be present on N- or O-linked chains. The addition of β1,4-linked GalNAc is always mediated by B4GALNT2.
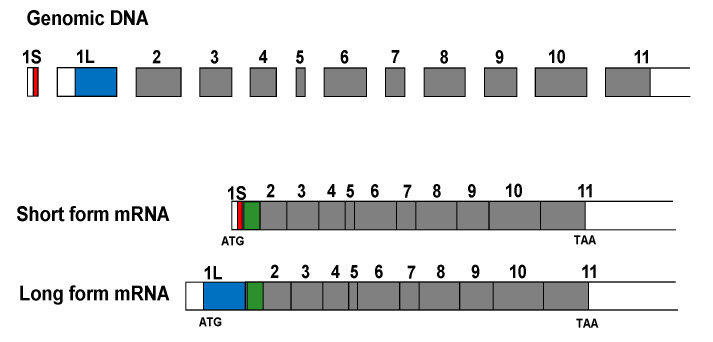
The addition of β1,4-linked GalNAc to α3-sialylated sugar chains is mediated by a single enzyme, product of the B4GALNT2 gene. After the first identification of this enzymatic activity in Guinea pig kidney [21], the mouse B4GALNT2 cDNA was cloned through a transient expression cloning approach [22]. Successively, the human cDNA was cloned from the colon cancer cell line Caco 2 by two different groups [23,24]. The human B4GALNT2 gene maps on 17q21.33 and encompasses at least 12 coding exons. The presence of multiple transcripts diverging in their 5′- and 3′-UTR, some of which are up to 9000 nucleotides long, and the occurrence of at least two alternative first exons were documented [23,24]. The transcripts were mainly expressed in the colon and to a lower extent in ileum, stomach, and kidney. The alternative presence of the 253-base-pair-long exon 1L or of the 38-base-pair-long exon 1S leads to two transcript variants containing different translational start sites. Consequently, the human B4GALNT2 gene can originate at least two different transmembrane peptides, diverging in their amino-terminal portion: a 566-amino acid long form and a 506-amino acid short form (Figure 2).
Figure 2.
Structure of the genomic DNA and the two major B4GALNT2 transcripts. Both the alternative exons 1S and 1L contain a translational start codon. Their coding portion is indicated in red and blue, respectively. Coding portions common to the two transcripts are in grey. The transmembrane portion is in green. Exons are drawn approximately to scale.
Both isoforms display Golgi localization, although the long form is present also in post-Golgi vesicles and the plasma membrane [25]. The short form appears to be enzymatically more active than the long form [26] and is by far the dominant form in both normal and cancer colon [27]. The genetic bases of B4GALNT2 deficiency linked to the Sda-negative phenotype have been recently elucidated and found to be mainly associated with missense mutations in the C-terminal portion of the enzyme [28]. Although the most frequent mutation appears to be Cys466Arg, other mutant alleles, including one affecting mRNA splicing, have been described [28]. In spite of the high similarity between the Sda antigen and the sugar chain of ganglioside GM2, B4GALNT2 is unable to synthesize the ganglioside GM2, which is instead the product of B4GALNT1. On the other hand, GM2 synthase B4GALNT1 is unable to synthesize the Sda antigen.
A role for DNA methylation in B4GALNT2 gene regulation was suggested by the presence in the genomic regions upstream of exons 1L and 1S of the features of a CpG island. A paper showed that the B4GALNT2 transcript was detectable by RT-PCR in some gastrointestinal cell lines and not in others, but only after 40 amplification cycles [29]. The promoter region of the cell lines not expressing B4GALNT2 was found to be methylated, and gene expression was restored by treatment with the DNA methylation inhibitor 5’aza 2’-deoxycytidine [29]. Another study [30] found methylation of the B4GALNT2 gene in the majority of gastric and colon cancer cell lines. A weak expression of the B4GALNT2 transcript and of the Sda antigen was induced by treatment of cell lines with anti-DNA-methylation agents [30]. Recently, it has been shown that the transcription factors ETS1 and, to a lesser extent, SP1 are required for B4GALNT2 transcription, although neither of the two is responsible for its differential expression in CRC [31].
3. Sda/B4GALNT2 in Development, Differentiation, and Cancer
3.1. Development and Differentiation
Ontogenic regulation of B4GALNT2 was formerly suggested by the observation that the Guinea pig kidney enzyme showed a five-fold increase after birth [32], while in rat colon, the enzyme was nearly undetectable at birth, but its level rapidly raised after weaning [33]. Consistently, the Sda antigen was not detected in human fetal colonic mucins [34]. The dependence of Sda/B4GALNT2 on cell differentiation was unclear. In fact, while the enzyme activity of both Sda [35] and B4GALNT2 [33] was higher in the poorly differentiated cells of the colonic crypt, B4GALNT2 expression increased upon differentiation of the human colon cancer Caco2 cells [36].
3.2. Cancer
Cancer-dependent modulation of B4GALNT2 was reported by numerous studies in CRC and by a few in gastric cancer. A possible involvement of B4GALNT2 in the biology and clinic of breast cancer (BRCA) has only recently emerged. These studies involved the analysis of clinical specimens, as well as of cell lines.
3.2.1. Clinical Studies
Traditionally, the clinical impact of cancer-modulated glycosyltransferases was investigated in cohorts of patients specifically recruited for the study by research Institutions. Although this approach has provided invaluable contributions to the field, it is strongly limited by the relatively small number of specimens that can be collected and analyzed. This limitation has been circumvented by public databases reporting genomic, transcriptomic, epigenetic, and clinical data of hundreds or thousands of cancer patients. The most comprehensive of these databases is probably The Cancer Genome Atlas (TCGA) (https://portal.gdc.cancer.gov/, accessed on 14 August 2021).
Colon Cancer
The dramatic downregulation of B4GALNT2 expression in colorectal cancer tissues was formerly reported in 1989 [37], successively confirmed [26] and found to be largely due to a reduced mRNA expression [38]. Moreover, the expression of the Sda antigen was reduced in colon cancer, compared with normal mucosa, paralleling the expression of the enzyme [39]. Recently, an association of B4GALNT2 mRNA with ulcerative colitis, an inflammatory pre-neoplastic condition [40], has been reported. The B4GALNT2 transcript was found to be more expressed in long-duration, compared with short-duration ulcerative colitis cases.
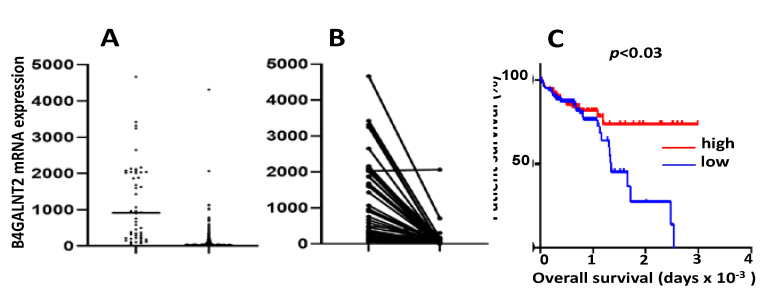
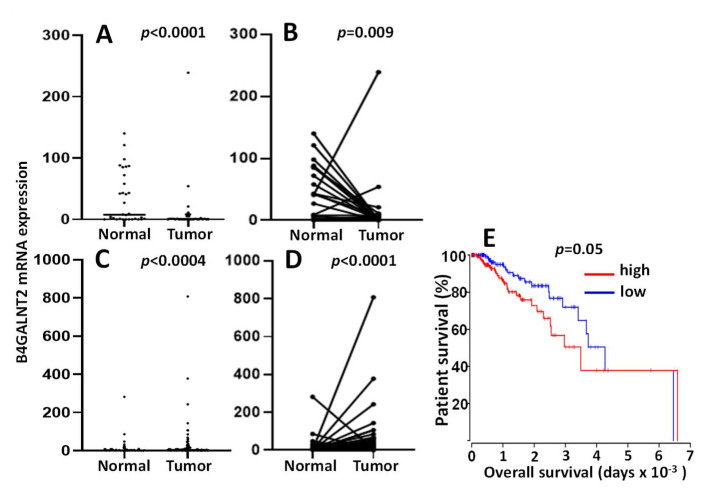
TCGA data confirm the general downregulation of B4GALNT2 in cancer tissues (Figure 3A) [41]. Although the vast majority of cancer samples lacked detectable levels of the transcript, several cancer cases displayed a level of expression comparable with that of many normal tissues. Paired comparison in 49 normal mucosa/cancer pairs (Figure 3B) revealed that only one sample displayed the same high expression in both the normal mucosa and the cancer. However, as previously observed in both European [26,37] and Japanese [38] studies, a small percentage of individuals expressing very low B4GALNT2 levels in normal colon was present also in TCGA cohort. These individuals are likely Sda-negative, although missense point mutations found to be responsible for several Sda-negative cases [28] cannot be responsible for low-mRNA expression data.
Figure 3.
B4GALNT2 mRNA in TCGA COADREAD (colon and rectal adenocarcinoma) cohort. (A): unpaired distribution of B4GALNT2 expression in 49 normal samples and 626 cancer samples. Statistical analysis was conducted by the Mann–Whitney test. (B): B4GALNT2 distribution in paired normal and tumor tissues of 49 patients. Statistical analysis was conducted by the Wilcoxon test. (C): Kaplan–Meier survival curves of patients belonging to the groups of high expressers (15th upper percentile, red) or no expressers (15th lower percentile, blue) of B4GALNT2 mRNA, as deduced from the Oncolnc.org website.
According to these data, B4GALNT2 mRNA level was nearly undetectable in the majority of cancer samples, but a remarkable number of cases retained a relatively high level of expression. These high-B4GALNT2 expressers (HBE) often displayed a non-mucinous, microsatellite-stable phenotype and a better response to therapy [41]. However, the most remarkable clinical finding emerging from TCGA data was the close association of high B4GALNT2 expression with long overall survival (Figure 3C). Patients stratified according to the B4GALNT2 transcription level displayed different gene expression profiles. In fact, numerous genes showed different levels of expression in HBE, compared with low B4GALNT2 expressers (LBE). In general, the gene expression profile of HBE is strongly oriented towards an attenuation of the neoplastic phenotype and the maintenance of functions associated with a normal epithelium, such as mucus secretion and solute transportation. Interestingly, the glycosylation machinery is also differentially expressed in HBE and LBE. In fact, several glycosyltransferases involved in the biosynthesis of O-linked chains, gangliosides, Lewis antigens, and galectins are more highly expressed in HBE, while ST6GAL sialyltransferases are more highly expressed in LBE [42].
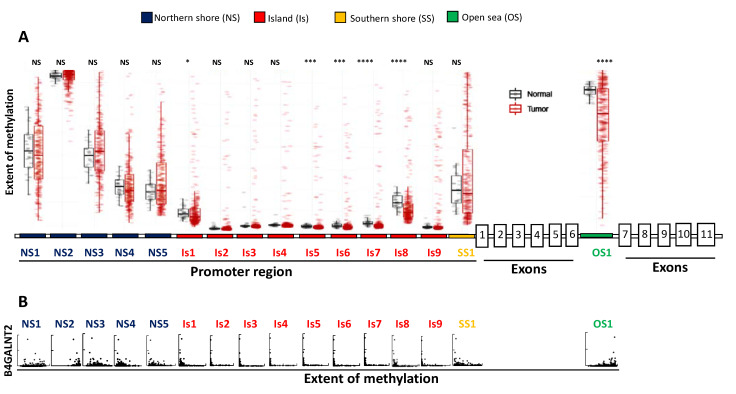
The mechanisms regulating B4GALNT2 expression in normal and cancer tissues are probably multifactorial and partially epigenetic. TCGA data allow a detailed analysis of the relationship between B4GALNT2 expression and methylation of individual methylation sites located in the CpG island, as well as in its upstream and downstream flanking regions known as “northern shore (NS)” and “southern shore (SS)”, respectively [42] (Figure 4). In addition, the methylation of an “open sea (OS)” site located in the intron between exons 6 and 7 was also reported. In general, methylation of the island and of the northern and southern shore positions in colon cancer is closely associated with low or no B4GALNT2 transcription. However, the methylation level in these positions is similar in the corresponding normal tissues (Figure 4A), ruling out the possibility that a differential methylation of these sites can be responsible for the downregulation of B4GALNT2 in cancer. The most important regulatory role appears to be played by the OS1 position, whose methylation is lower in cancer. As a common condition, all HBE displayed high methylation of the NS2 and OS1 sites and lack of methylation of all other positions (Figure 4B). However, not all cases sharing this condition are HBE, suggesting this is a necessary but not sufficient condition.
Figure 4.
TCGA data on the methylation of the B4GALNT2 gene in normal and CRC tissues. (A): extent of methylation in normal and CRC tissues of CpG positions in the NS (blue, five positions), CpG island (Is, red, nine positions), SS (yellow, one position), and OS (green, one position). The promoter region and the coding exons have not been drawn to scale. (B): B4GALNT2 mRNA expression plotted vs. the extent of methylation in cancer tissues. * p < 0.05; *** p < 0.001; **** p < 0.0001.
TCGA also provides information on the level of expression of numerous miRNA. Bioinformatics analysis has revealed that some miRNAs are potentially able to target B4GALNT2 [42]. TCGA data showed a clear tendency towards an inverse relationship between some of these miRNAs and B4GALNT2 in colon cancer tissues. However, hsa-miR-204-5p, that targets a sequence located in the 3’-UTR, about 120 bp downstream of the translational stop codon, displayed the best inverse relationship with B4GALNT2 expression. In fact, all HBE displayed no hsa-miR-204-5p expression, while all high-hsa-miR-204-5p expressers displayed no B4GALNT2 expression. However, the existence of many hsa-miR-204-5p non-expressers failing to express B4GALNT2 revealed that the lack of hsa-miR-204-5p is also a necessary but not sufficient condition for high levels of B4GALNT2.
In conclusion, the regulation of B4GALNT2 in normal and cancer colon is complex and multifactorial. The differential expression of specific transcription factors has not yet been investigated but is likely to play a major role in B4GALNT2 regulation.
Gastric Cancer
Using monoclonal antibodies specific for the Sda antigen and/or for the ganglioside GM2, it was found that the two structures displayed opposite regulation in normal gastric mucosa and gastric cancer; the former was downregulated, while the latter was upregulated in cancer [43]. The downregulation of B4GALNT2 in gastric cancer was documented successively [38]. A detailed structural analysis in sera identified eight carbohydrate structures expressing the Sda antigen on the various underlying structures depicted in Figure 1 [44]. Interestingly, in sera of healthy people the Sda antigen was expressed only on O-linked Core 1 structures. On the other hand, in sera of a few gastric (and a few pancreatic) cancer patients, these Sda structures were found to be carried by other underlying chains [44]. The importance of these markers in cancer management requires further investigation.
According to TCGA data, B4GALNT2 is expressed in the gastric mucosa by only a few cases, while it is virtually not expressed in gastric cancer, with few exceptions (Figure 5A,B). Overall, the level of B4GALNT2 expression in the gastric mucosa is about 50-fold lower than in the colon. In normal stomach, B4GALNT2 activity and Sda antigen expression were found to be associated with chief cells, the stomach cells releasing pepsinogen [43]. Methylation of the promoter was shown to be relevant for B4GALNT2 expression by the two previously mentioned studies [29,30] as well as in gastric cancer cell lines. However, the low level of expression in gastric cancer tissues did not allow investigating the relationship between B4GALNT2 expression and methylation or clinical features using TCGA data.
Figure 5.
B4GALNT2 mRNA in stomach adenocarcinoma (STAD) and breast cancer (BRCA) TCGA cohorts. In (A) and (C) plots, the unpaired distribution of B4GALNT2 expression in normal tissue and cancer samples of stomach (A) and breast (C) cancer patients is reported. In (B) and (D) graphs, B4GALNT2 distribution in paired normal and tumor tissues of stomach (B) and breast (D) cancer patients is reported. Stomach: 35 normal and 415 tumor samples; breast: 107 normal and 1100 cancer patients. Statistical analysis conducted by the Mann–Whitney test in A and C and by the Wilcoxon test in (B) and (D). (E): Kaplan–Meier survival curves of patients belonging to the groups of high expressers (15th upper percentile, red) or no expressers (15th lower percentile, blue) of mRNA B4GALNT2 as deduced from the Oncolnc.org website.
Breast Cancer
Little is known about B4GALNT2 in BRCA. A recent study reported a systemic upregulation of B4GALNT2 in an experimental model of breast cancer [45]. Consistently, TCGA data analysis showed that in normal breast tissues as well as in the majority of BRCA tissues, the level of B4GALNT2 mRNA expression was nearly undetectable (Figure 5C,D), although a minority of the cancer cases displayed a level of activity similar with that of colon. Considering the virtual absence of B4GALNT2 in normal breast tissues, it seems appropriate to define the B4GALNT2 expression by a minority of BRCA cases as ectopic. Notably, these high-B4GALNT2 expressers displayed a significantly shorter overall survival (Figure 5E). No obvious relationship was evident with clinical parameters, including receptor (estrogens, progesterone, or HER2) status. However, genes displaying over- or under-expression in the HBE cohorts provided a molecular signature strongly oriented towards malignancy (Table 1). This view was confirmed by a recent study showing growth promotion induced by B4GALNT2 in triple-negative breast cancer cell lines [46].
Table 1.
Cancer-relevant genes up- or downregulated in HBE expressers.
| Gene Symbol | Ratio | Function | PMID | |
|---|---|---|---|---|
| KRT20 | 494 | Cytokeratin 20, associated with worse prognosis | 22493626 | |
| CEACAM6 | 11 | Member of the CEA family. Associated with worse prognosis | 24186057 | |
| CRISP3 | 10 | High expression correlates with malignancy | 30609035 | |
| ABCC2 | 10 | Multidrug resistance-associated protein 2 | 26499806 | |
| LRRC31 | 9 | Inhibitor of DNA repair | 33005030 | |
| MUC21 | 8.5 | Associated with incohesive growth of lung cancer | 31301084 | |
| CEACAM5 | 8 | Member of the CEA family. Driver of breast cancer metastasis | 29736411 | |
| TRIM72 | 8 | Lower expression predicts recurrence in colon cancer | 30852740 | |
| PNMT | 7.8 | Co-amplified with ERBB2 | 12727839 | |
| CXCL17 | 7 | Promotes proliferation and invasion of breast cancer cells | 28943434 | |
| AMER3 | −8.7 | Enhances β-catenin signaling in CRC | 24251807 | |
| KCNC1 | −8.8 | Its inhibition is associated with poor survival in seminoma | 34105734 | |
| INSM1 | −9.3 | In SCLC, low expression associated with better prognosis | 32118626 | |
| SLIT1 | −9 | Suppresses breast cancer growth | 18829537 | |
| RUNDC3A | −10.6 | High expression correlates with shorter survival in rectal cancer | 29050227 | |
| HDC | −13 | Increased expression associated with better survival | 31748740 | |
| ECEL1 | −14 | Associated with good prognosis in neuroblastoma | 12632073 | |
| CHRNA9 | −19 | High expression associated with poor survival in BRCA | 20953833 | |
| PCSK1 | −23 | Reduces growth of breast cancer cells | 11241161 | |
| NELL1 | −27 | Down-regulated in kidney cancer | 25726761 |
Top 20 genes showing significantly (p ≤ 0.05 by Student’s t test) higher (10 genes) or lower (10 genes) expression in the 15% upper percentile of B4GALNT2 expression. The red or green color indicates that the change has a putatively cancer-promoting (red) or cancer-restraining (green) effect, according to the literature search. The “PubMed Identification Number” (PMID) of the most relevant paper is indicated.
4. Sialyl Lewis Antigens
sLex and sLea are well known cancer-associated fucosylated carbohydrate structures [47]. Although they are normally expressed by a variety of tissues, including leukocytes, their ectopic expression by several cancers is associated with malignancy [48]. The sLea tetrasaccharide is the epitope of the CA19.9 antigen, widely used in clinical practice [49,50,51].
4.1. Structure and Biosynthesis in Normal and Cancer Colon
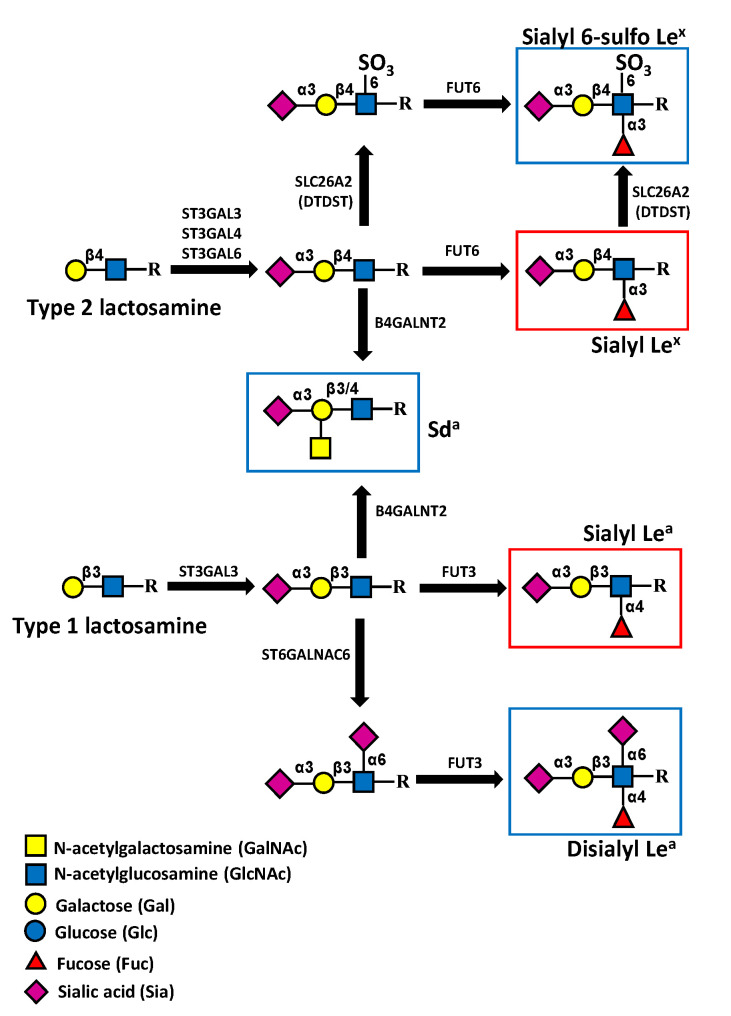
The terminal steps of the biosynthesis of the sLex and sLea antigens are mediated by α2,3 sialyltransferases and successively by α1,3/4 fucosyltransferases [52]. While type 1 chains can be α2,3 sialylated only by ST3GAL3, type 2 chains can be α2,3 sialylated by ST3GAL3, ST3GAL4, and ST3GAL6. The successive addition of fucose in α1,4 linkage to type 1 chains is catalyzed only by FUT3, while that in α1,3 linkage to type 2 chains in colon cancer is mediated mainly, if not exclusively, by FUT6 [53]. The usage of these different enzymes is strongly cell-type specific.
In CRC, the aberrant expression of sLex/a antigens appears to be particularly relevant [54,55,56,57,58,59,60] (recently reviewed in [61]). However, the molecular bases of their overexpression in this malignancy are unclear. Although several studies have shown that some of the above-mentioned α2,3 sialyltransferases [62,63,64,65,66,67] and α1,3/4 fucosyltransferases [53,68,69,70] have the ability to regulate sLex/a biosynthesis in experimental systems, the overexpression of these antigens in CRC is not due to the increased expression of their cognate sialyl- and/or fucosyltransferases [53,71]. In fact, the level of expression of sLex/a antigens does not reflect the level of expression of these terminal glycosyltransferases in normal and CRC tissues. In normal colon, sLex is expressed by only a few specimens, and the level of antigen expression is much lower than that in cancer samples [53], but the expression level of FUT6 is quite similar in normal and cancer colon tissues [53,71]. Glycosyltransferases synthesizing subterminal carbohydrate structures can play crucial roles. In particular, the GlcNAc transferases B3GNT5 and B3GNT7 appear to play opposite roles. The first one, which is involved in the biosynthesis of the underlying type 1 and type 2 chains, promotes sLex/a overexpression [72,73], whereas the second one, which extends sulfated polylactosaminic chains, inhibits sLex/a biosynthesis [74]. Overexpression of sialidases, in particular NEU4, which acts preferentially on mucins and is downregulated in CRC, has been shown to play a role in keeping low levels of sLex/a in normal colon and, consequently, higher levels in CRC [75]. Among the mechanisms that have been claimed to be responsible for the upregulation of sLex/a in CRC, the biosynthesis in normal colonic tissues of carbohydrate structures whose expression prevents that of sLex/a antigens is particularly relevant. An example is provided by the antigen sialyl 6-sulfo Lex, highly expressed in normal colon, which is replaced in CRC by sLex, due to reduced sulfation in the latter (Figure 6) [76]. The process is mainly regulated by the epigenetic silencing of the sulfate transporter DTDST, the product of the SLC26A2 gene [77]. Another example is provided by the alternative biosynthesis of di-sialyl Lewisa or sLea antigens in normal mucosa and CRC, respectively [78]. The former, mainly expressed by normal mucosa, is synthesized by the sequential action of sialyltransferase ST6GALNAC6 and fucosyltransferase FUT3 (Figure 6). However, when ST6GALNAC6 is downregulated, as occurs in CRC, only the sLea antigen is synthesized. It has been recently reported that biglycan (encoded by the BGN gene) is responsible for the epigenetic silencing of the SLC26A2 and ST6GALNA6 genes in CRC through an inflammatory pathway described below [79,80]. A third example of biosynthetic competition between different antigens is provided by sLex/a and Sda. Both derive from a common precursor, i.e., an α2,3-sialylated type 1 or type 2 chain. The addition of β1,4-linked GalNAc to Gal hinders the addition of fucose to GlcNAc and vice versa [26,81]. In fact, a structural analysis of mucins from normal colon specimens revealed a large preponderance of oligosaccharides frequently terminated by the Sda epitope [82], while sLex structures were minor components. Interestingly, no structures carrying both the Sda and the sLex determinants were detected. We have previously proposed that the right question to answer is not “why is sLex high in colon cancer?” but rather: “why is sLex low in normal colon?” The reduced B4GALNT2 expression in CRC is part of the answer. In fact, a significant relationship in normal colonic mucosa between the sLex level and the FUT6/B4GALNT2 ratio has been reported [27], supporting the notion that in normal colon, sLex is poorly expressed because of a high level of B4GALNT2.
Figure 6.
Sialyl Lewis-related antigens and Sda antigen in colonic tissues. In normal colon, the expression of sialyl 6-sulfo Lex antigen (upper) predominates over that of sLex. However, the reduced expression of enzymes responsible for its biosynthesis, such as SLC26A2 (formerly DTDST) in colon cancer, leads to the overexpression of sLex. The biosynthesis of the di-sialyl Lea antigen (lower), which predominates in normal colon, proceeds from α2,6 sialylation, catalyzed by ST6GALNAC6, followed by α1,4-fucosylation mediated by FUT3. The downregulation of ST6GALNAC6 in colon cancer leads to sLea overexpression. The addition of β1,4-linked GalNAc mediated by B4GALNT2 leads to the expression of the Sda, inhibiting that of sLex and sLea. Cancer-associated structures are boxed in red, while structures associated with normal colon are boxed in blue.
4.2. Role in Malignancy
sLex/a structures act as ligands for cell adhesion molecules of the selectin family, playing a fundamental role in leukocyte extravasation [83]. However, when overexpressed in cancer cells, they contribute to metastasis formation [84], particularly because they allow the interaction of circulating cancer cells with selectins expressed on endothelial cells [85,86,87,88,89]. This notion has been clearly demonstrated in vivo by showing that a CRC cell line injected subcutaneously in immunodeficient mice formed a much lower number of spontaneous lung metastases in E- and P-selectin-deficient mice [90]. In addition, E- and P-selectins were found to be crucial for peritoneal metastasization of pancreatic cancer cells [91] and bone metastasis of breast cancer cells [92]. Recently, it has been shown that sLe antigens are involved in bone metastasis formation by mediating adhesion to the bone niche [93]. Inflammation is a crucial feature associated with cancer, and cancer frequently develops on pre-existing inflammatory conditions. In inflammatory bowel diseases, sLex/a antigens expressed by mucosal cells on the CD44v6 molecule sustain inflammation by acting as ligands for transmigrating neutrophils [94]. Siglecs are sialic acid-binding molecules with a general anti-inflammatory and immunosuppressive activity [95]. The above-mentioned antigens sialyl 6-sulfo Lex and di-sialyl Lewisa, expressed by normal mucosa, are ligands for Siglec-7. Consequently, their downregulation in CRC contributes to the cancer-associated inflammatory status [79,80]. Biglycan, overexpressed in CRC, is a good ligand for Toll-like receptor 4 (TLR4) [73,74]. This leads to the activation in CRC of the TLR4/NFkB inflammatory pathway, resulting in the previously mentioned epigenetic silencing of the SLC26A2 and ST6GALNA6 genes, responsible for sialyl 6-sulfo Lex and di-sialyl Lewisa biosynthesis [79,80].
Apart from the increased adhesion to selectin-expressing vessels, several studies indicate that sLex/a structures increase motility, proliferation, and malignancy through selectin-independent mechanisms [87,88,96,97,98]. The colon cancer cell lines SW480 and SW620, which were derived from a primary colon cancer and a lymph node metastasis of the same patient, respectively, provide a good model of cells with different malignancy but a very similar genetic background. Forced expression of FUT6 in these cells resulted in profound changes of gene expression towards increased malignancy [97]. However, some genes were modulated only in one of the two cell lines, while others in both, despite the close relationship between the two cell lines. For example, transcription of a group of genes strongly involved in DNA replication, including those encoding telomerase (TERT), thymidylate synthase (TYMS), and DNA polymerase ε4 (POLE4), was stimulated by FUT6 in SW620 but not in SW480 [97]. Furthermore, the impact of FUT6/sLex on the phenotype of the two cell lines was different. In fact, the clonogenic ability in soft agar and the capacity to heal a wound were increased only in SW620 but not in SW480 [97].
5. Phenotypic Effects of B4GALNT2 on Cancer Phenotype
In vitro experiments have shown that forced expression of B4GALNT2 in colon and gastric cancer cell lines strongly reduced sLex/a expression [26,81]. In addition, B4GALNT2-expressing cells showed reduced metastatic ability [81,99]. More recently, we have shown that in the colon cancer cell line LS174T that constitutively expresses sLex/a, B4GALNT2 reduced stemness-associated features, in particular the ability to grow in poor adherence. Notably, B4GALNT2 strongly modulated the transcriptional activity towards an attenuation of the neoplastic phenotype in this cell line [41]. It has long been thought that the tumor-restraining activity exerted by B4GALNT2/Sda was dependent on sLex/a inhibition, rather than on de novo expression of Sda. However, this view has been challenged by the observation that in SW480 and SW620 cells (lacking sLex if not transfected with FUT6), forced expression of B4GALNT2 [97] downregulated all malignancy-associated phenotypes (including cell proliferation, growth in poor adherence, wound healing ability, and stemness marker expression) in SW620, but only those associated with stemness in SW480 [97]. Thus, attenuation of the stemness-associated malignant phenotype by B4GALNT2/Sda appears to be a common feature in CRC cells and is independent of sLex inhibition.
6. Discussion
In this review, we have described the mutual relationship of Sda and sLex/a antigens, showing how the terminal portions of glycoconjugates can play profound but sometimes opposite effects on cancer biology. The data summarized in this review reinforce or challenge established paradigms. First, the ectopic expression of sLex/a antigens in CRC would be due, together with other factors, to the downregulation of B4GALNT2 rather than to the upregulation of the sLex/a biosynthetic machinery (graphical abstract). Second, the Sda synthase B4GALNT2 is associated with better prognosis in CRC but with worse prognosis in BRCA. Moreover, the gene expression profile of HBE in CRC and BRCA is opposite. This supports the notion that the impact of a glycosyltransferase on cancer is strongly tissue-dependent. Third, Sda/B4GALNT2 inhibits numerous properties of malignancy in CRC cells, in particular those associated with stemness. However, this effect is not dependent on sLex/a inhibition, as previously thought. It remains to be established whether cell lines from other organs of the gastrointestinal tract, as well as from BRCA, display this effect. Fourth, the regulation of a glycosyltransferases can be very complex and multifactorial. B4GALNT2 is regulated by DNA methylation, although the crucial sites appear to be located outside the canonical CpG island. miRNAs also play a role. However, appropriate patterns of methylation and miRNA expression appear to be a necessary but not sufficient condition for high expression. It is possible to hypothesize that a differential expression of key transcription factors plays a pivotal role during the complete or partial shut-down of B4GALNT2 transcription associated with CRC transformation. The identification of these transcription factors deserves investigation. Fifth, glycosyltransferases impact the gene expression profile and the phenotype of cell lines. Consistent with clinical data, overexpression of B4GALNT2 attenuates malignancy, while overexpression of FUT6/sLex exacerbates malignancy, although in a strongly cell-type specific manner. While the use of sLe antigens as cancer markers has been debated since the 1980s and is already applied in the clinic (CA19.9), the use of Sda and/or B4GALNT2 as prognostic markers would be novel. The detection of the Sda antigen is limited by the lack of a commercially available antigen and by its poor detection on formalin-fixed/paraffin-embedded tissues (F. Dall’Olio, unpublished observation). On the contrary, the precise quantification in clinical samples of B4GALNT2 mRNA by RNA-seq techniques at a cost compatible with clinical routine, would be a realistic perspective.
7. Conclusions
In conclusion, B4GALNT2 and its cognate carbohydrate antigen Sda play a relevant role in CRC, not only for their association with better prognosis, but also because their expression is causally related to reduced malignancy in CRC experimental systems. For these reasons, their use as clinical markers deserves consideration. The reduced malignancy induced by the forced expression of B4GALNT2 in cancer cells by viral vectors [99] also suggests its potential as a therapeutic agent.
Abbreviations
| BRCA | breast cancer |
| COADREAD | colon adenocarcinoma rectal adenocarcinoma |
| CRC | colorectal cancer |
| DTDST | diastrophic dysplasia sulfate transporter |
| HBE | high-B4GALNT2 expressers |
| LBE | low-B4GALNT2 expressers |
| NS | northern shore |
| OS | open sea |
| RT-PCR | reverse transcriptase-polymerase chain reaction |
| sLea | sialyl Lewisa |
| sLex | sialyl Lewisx |
| SCLC | small cell lung cancer |
| SS | southern shore |
| STAD | stomach adenocarcinoma |
| TCGA | The Cancer Genome Atlas |
| TLR4 | Toll-like receptor 4 |
Author Contributions
Conceptualization, F.D.; writing—original draft preparation, F.D., M.P. and N.M.; writing—review and editing, N.M., M.P. and F.D.; supervision, F.D. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the University of Bologna and by the “Pallotti” Legacy for Cancer.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Dall’Olio F., Malagolini N., Trinchera M., Chiricolo M. Mechanisms of cancer-associated glycosylation changes. Front. Biosci. 2012;17:670–699. doi: 10.2741/3951. [DOI] [PubMed] [Google Scholar]
- 2.Dall’Olio F., Malagolini N., Trinchera M., Chiricolo M. Sialosignaling: Sialyltransferases as engines of self-fueling loops in cancer progression. Biochim. Biophys. Acta. 2014;1840:2752–2764. doi: 10.1016/j.bbagen.2014.06.006. [DOI] [PubMed] [Google Scholar]
- 3.Pinho S.S., Reis C.A. Glycosylation in cancer: Mechanisms and clinical implications. Nat. Rev. Cancer. 2015;15:540–555. doi: 10.1038/nrc3982. [DOI] [PubMed] [Google Scholar]
- 4.Morton J.A., Pickles M.M., Terry A.M. The Sda blood group antigen in tissues and body fluids. Vox Sang. 1970;19:472–482. doi: 10.1111/j.1423-0410.1970.tb01779.x. [DOI] [PubMed] [Google Scholar]
- 5.Morton J.A., Pickles M.M., Vanhegan R.I. The Sda antigen in the human kidney and colon. Immunol. Investig. 1988;17:217–224. doi: 10.3109/08820138809052961. [DOI] [PubMed] [Google Scholar]
- 6.Renton P.H., Howell P., Ikin E.W., Giles C.M., Goldsmith K.L. Anti Sda: A new blood group antibody. Vox Sang. 1967;13:493–501. doi: 10.1111/j.1423-0410.1967.tb03796.x. [DOI] [Google Scholar]
- 7.Macvie S.I., Morton J.A., Pickles M.M. The reactions and inheritance of a new blood group antigen. Vox Sang. 1967;13:485–492. [Google Scholar]
- 8.Cramer M.L., Xu R., Martin P.T. Soluble Heparin Binding EGF-like Growth Factor (HB-EGF) is a regulator of GALGT2 expression and GALGT2-dependent muscle and neuromuscular phenotypes. Mol. Cell Biol. 2019;39:e00140-19. doi: 10.1128/MCB.00140-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xu R., Singhal N., Serinagaoglu Y., Chandrasekharan K., Joshi M., Bauer J.A., Janssen P.M., Martin P.T. Deletion of Galgt2 (B4Galnt2) Reduces Muscle Growth in Response to Acute Injury and Increases Muscle Inflammation and Pathology in Dystrophin-Deficient Mice. Am. J. Pathol. 2015;185:2668–2684. doi: 10.1016/j.ajpath.2015.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vallier M., Abou C.M., Hindersin L., Linnenbrink M., Traulsen A., Baines J.F. Evaluating the maintenance of disease-associated variation at the blood group-related gene B4galnt2 in house mice. BMC Evol. Biol. 2017;17:187. doi: 10.1186/s12862-017-1035-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Linnenbrink M., Johnsen J.M., Montero I., Brzezinski C.R., Harr B., Baines J.F. Long-term balancing selection at the blood group-related gene B4galnt2 in the genus Mus (Rodentia; Muridae) Mol. Biol. Evol. 2011;28:2999–3003. doi: 10.1093/molbev/msr150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Staubach F., Kunzel S., Baines A.C., Yee A., McGee B.M., Backhed F., Baines J.F., Johnsen J.M. Expression of the blood-group-related glycosyltransferase B4galnt2 influences the intestinal microbiota in mice. ISME J. 2012;6:1345–1355. doi: 10.1038/ismej.2011.204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Galeev A., Suwandi A., Cepic A., Basu M., Baines J.F., Grassl G.A. The role of the blood group-related glycosyltransferases FUT2 and B4GALNT2 in susceptibility to infectious disease. Int. J. Med. Microbiol. 2021;311:151487. doi: 10.1016/j.ijmm.2021.151487. [DOI] [PubMed] [Google Scholar]
- 14.Ben J.S., Ruesche J., Sarry J., Woloszyn F., Lassoued N., Fabre S. The high prolificacy of D’man sheep is associated with the segregation of the FecL(L) mutation in the B4GALNT2 gene. Reprod. Domest. Anim. 2018;54:531–537. doi: 10.1111/rda.13391. [DOI] [PubMed] [Google Scholar]
- 15.Byrne G., Ahmad-Villiers S., Du Z., McGregor C. B4GALNT2 and xenotransplantation: A newly appreciated xenogeneic antigen. Xenotransplantation. 2018;25:e12394. doi: 10.1111/xen.12394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Heaton B.E., Kennedy E.M., Dumm R.E., Harding A.T., Sacco M.T., Sachs D., Heaton N.S. A CRISPR Activation Screen Identifies a Pan-avian Influenza Virus Inhibitory Host Factor. Cell Rep. 2017;20:1503–1512. doi: 10.1016/j.celrep.2017.07.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wong H.H., Fung K., Nicholls J.M. MDCK-B4GalNT2 cells disclose a a2,3-sialic acid requirement for the 2009 pandemic H1N1 A/California/04/2009 and NA aid entry of A/WSN/33. Emerg. Microbes. Infect. 2019;8:1428–1437. doi: 10.1080/22221751.2019.1665971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Donald A.S., Yates A.D., Soh C.P., Morgan W.T., Watkins W.M. A blood group Sda-active pentasaccharide isolated from Tamm-Horsfall urinary glycoprotein. Biochem. Biophys. Res. Commun. 1983;115:625–631. doi: 10.1016/S0006-291X(83)80190-9. [DOI] [PubMed] [Google Scholar]
- 19.Dall’Olio F., Malagolini N., Chiricolo M., Trinchera M., Harduin-Lepers A. The expanding roles of the Sda/Cad carbohydrate antigen and its cognate glycosyltransferase B4GALNT2. Biochim. Biophys. Acta. 2014;1840:443–453. doi: 10.1016/j.bbagen.2013.09.036. [DOI] [PubMed] [Google Scholar]
- 20.Blanchard D., Piller F., Gillard B., Marcus D., Cartron J.P. Identification of a novel ganglioside on erythrocytes with blood group Cad specificity. J. Biol. Chem. 1985;260:7813–7816. doi: 10.1016/S0021-9258(17)39523-6. [DOI] [PubMed] [Google Scholar]
- 21.Serafini-Cessi F., Dall’Olio F. Guinea-pig kidney b-N-acetylgalactosaminyltransferase towards Tamm- Horsfall glycoprotein. Requirement of sialic acid in the acceptor for transferase activity. Biochem. J. 1983;215:483–489. doi: 10.1042/bj2150483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Smith P.L., Lowe J.B. Molecular cloning of a murine N-acetylgalactosamine transferase cDNA that determines expression of the T lymphocyte-specific CT oligosaccharide differentiation antigen. J. Biol. Chem. 1994;269:15162–15171. doi: 10.1016/S0021-9258(17)36587-0. [DOI] [PubMed] [Google Scholar]
- 23.Lo Presti L., Cabuy E., Chiricolo M., Dall’Olio F. Molecular Cloning of the Human b1,4 N-Acetylgalactosaminyltransferase Responsible for the Biosynthesis of the Sda Histo-Blood Group Antigen: The Sequence Predicts a Very Long Cytoplasmic Domain. J. Biochem. 2003;134:675–682. doi: 10.1093/jb/mvg192. [DOI] [PubMed] [Google Scholar]
- 24.Montiel M.D., Krzewinski-Recchi M.A., Delannoy P., Harduin-Lepers A. Molecular cloning, gene organization and expression of the human UDP-GalNAc:Neu5Aca2-3Galb-R b1,4-N-acetylgalactosaminyltransferase responsible for the biosynthesis of the blood group Sda/Cad antigen: Evidence for an unusual extended cytoplasmic domain. Biochem. J. 2003;373:369–379. doi: 10.1042/bj20021892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Groux-Degroote S., Schulz C., Cogez V., Noel M., Portier L., Vicogne D., Solorzano C., Dall’Olio F., Steenackers A., Mortuaire M., et al. The extended cytoplasmic tail of the human B4GALNT2 is critical for its Golgi targeting and post-Golgi sorting. FEBS J. 2018;285:3442–3463. doi: 10.1111/febs.14621. [DOI] [PubMed] [Google Scholar]
- 26.Malagolini N., Santini D., Chiricolo M., Dall’Olio F. Biosynthesis and expression of the Sda and sialyl Lewis x antigens in normal and cancer colon. Glycobiology. 2007;17:688–697. doi: 10.1093/glycob/cwm040. [DOI] [PubMed] [Google Scholar]
- 27.Groux-Degroote S., Wavelet C., Krzewinski-Recchi M.A., Portier L., Mortuaire M., Mihalache A., Trinchera M., Delannoy P., Malagolini N., Chiricolo M., et al. B4GALNT2 gene expression controls the biosynthesis of Sda and sialyl Lewis X antigens in healthy and cancer human gastrointestinal tract. Int. J. Biochem. Cell Biol. 2014;53:442–449. doi: 10.1016/j.biocel.2014.06.009. [DOI] [PubMed] [Google Scholar]
- 28.Stenfelt L., Hellberg A., Moller M., Thornton N., Larson G., Olsson M.L. Missense mutations in the C-terminal portion of the B4GALNT2-encoded glycosyltransferase underlying the Sda phenotype. Biochem. Biophys. Rep. 2019;19:100659. doi: 10.1016/j.bbrep.2019.100659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang H.R., Hsieh C.Y., Twu Y.C., Yu L.C. Expression of the human Sda b-1,4-N-acetylgalactosaminyltransferase II gene is dependent on the promoter methylation status. Glycobiology. 2008;18:104–113. doi: 10.1093/glycob/cwm120. [DOI] [PubMed] [Google Scholar]
- 30.Kawamura Y.I., Toyota M., Kawashima R., Hagiwara T., Suzuki H., Imai K., Shinomura Y., Tokino T., Kannagi R., Dohi T. DNA hypermethylation contributes to incomplete synthesis of carbohydrate determinants in gastrointestinal cancer. Gastroenterology. 2008;135:142–151. doi: 10.1053/j.gastro.2008.03.031. [DOI] [PubMed] [Google Scholar]
- 31.Wavelet-Vermuse C., Groux-Degroote S., Vicogne D., Cogez V., Venturi G., Trinchera M., Brysbaert G., Krzewinski-Recchi M.A., Bachir E.H., Schulz C., et al. Analysis of the proximal promoter of the human colon-specific B4GALNT2 (Sda synthase) gene: B4GALNT2 is transcriptionally regulated by ETS1. Biochim. Biophys. Acta Gene Regul. Mech. 2021;1864:194747. doi: 10.1016/j.bbagrm.2021.194747. [DOI] [PubMed] [Google Scholar]
- 32.Dall’Olio F., Malagolini N., Serafini-Cessi F. Tissue distribution and age-dependent expression of b-4-N- acetylgalactosaminyl-transferase in guinea-pig. Biosci. Rep. 1987;7:925–932. doi: 10.1007/BF01122125. [DOI] [PubMed] [Google Scholar]
- 33.Dall’Olio F., Malagolini N., Di Stefano G., Ciambella M., Serafini-Cessi F. Postnatal development of rat colon epithelial cells is associated with changes in the expression of the b 1,4-N-acetylgalactosaminyltransferase involved in the synthesis of Sda antigen and of a 2,6-sialyltransferase activity towards N-acetyllactosamine. Biochem. J. 1990;270:519–524. doi: 10.1042/bj2700519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Robbe-Masselot C., Maes E., Rousset M., Michalski J.C., Capon C. Glycosylation of human fetal mucins: A similar repertoire of O-glycans along the intestinal tract. Glycoconj. J. 2009;26:397–413. doi: 10.1007/s10719-008-9186-9. [DOI] [PubMed] [Google Scholar]
- 35.Lefrancois L. Carbohydrate differentiation antigens of murine T cells: Expression on intestinal lymphocytes and intestinal epithelium. J. Immunol. 1987;138:3375–3384. [PubMed] [Google Scholar]
- 36.Malagolini N., Dall’Olio F., Serafini-Cessi F. UDP-GalNAc:NeuAc a 2,3Gal b-R (GalNAc to Gal) b 1,4-N- acetylgalactosaminyltransferase responsible for the Sda specificity in human colon carcinoma CaCo-2 cell line. Biochem. Biophys. Res. Commun. 1991;180:681–686. doi: 10.1016/S0006-291X(05)81119-2. [DOI] [PubMed] [Google Scholar]
- 37.Malagolini N., Dall’Olio F., Di Stefano G., Minni F., Marrano D., Serafini-Cessi F. Expression of UDP-GalNAc:NeuAc a2,3Gal b-R beta 1,4(GalNAc to Gal) N-acetylgalactosaminyltransferase involved in the synthesis of Sda antigen in human large intestine and colorectal carcinomas. Cancer Res. 1989;49:6466–6470. [PubMed] [Google Scholar]
- 38.Dohi T., Yuyama Y., Natori Y., Smith P.L., Lowe J.B., Oshima M. Detection of N-acetylgalactosaminyltransferase mRNA which determines expression of Sda blood group carbohydrate structure in human gastrointestinal mucosa and cancer. Int. J. Cancer. 1996;67:626–631. doi: 10.1002/(SICI)1097-0215(19960904)67:5<626::AID-IJC6>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 39.Robbe-Masselot C., Herrmann A., Maes E., Carlstedt I., Michalski J.C., Capon C. Expression of a core 3 disialyl-Lex hexasaccharide in human colorectal cancers: A potential marker of malignant transformation in colon. J. Proteome. Res. 2009;8:702–711. doi: 10.1021/pr800740j. [DOI] [PubMed] [Google Scholar]
- 40.Low E.N.D., Mokhtar N.M., Wong Z., Raja Ali R.A. Colonic Mucosal Transcriptomic Changes in Patients with Long-Duration Ulcerative Colitis Revealed Colitis-Associated Cancer Pathways. J. Crohns. Colitis. 2019;13:755–763. doi: 10.1093/ecco-jcc/jjz002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pucci M., Gomes Ferreira I., Orlandani M., Malagolini N., Ferracin M., Dall’Olio F. High Expression of the Sda Synthase B4GALNT2 Associates with Good Prognosis and Attenuates Stemness in Colon Cancer. Cells. 2020;9:948. doi: 10.3390/cells9040948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pucci M., Malagolini N., Dall’Olio F. Glycosyltransferase B4GALNT2 as a Predictor of Good Prognosis in Colon Cancer: Lessons from Databases. Int. J. Mol. Sci. 2021;22:4331. doi: 10.3390/ijms22094331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dohi T., Ohta S., Hanai N., Yamaguchi K., Oshima M. Sialylpentaosylceramide detected with anti-GM2 monoclonal antibody. Structural characterization and complementary expression with GM2 in gastric cancer and normal gastric mucosa. J. Biol. Chem. 1990;265:7880–7885. doi: 10.1016/S0021-9258(19)39013-1. [DOI] [PubMed] [Google Scholar]
- 44.Tanaka-Okamoto M., Hanzawa K., Mukai M., Takahashi H., Ohue M., Miyamoto Y. Identification of internally sialylated carbohydrate tumor marker candidates, including Sda/CAD antigens, by focused glycomic analyses utilizing the substrate specificity of neuraminidase. Glycobiology. 2018;28:247–260. doi: 10.1093/glycob/cwy010. [DOI] [PubMed] [Google Scholar]
- 45.Qusa M.H., Abdelwahed K.S., Siddique A.B., El Sayed K.A. Comparative Gene Signature of (-)-Oleocanthal Formulation Treatments in Heterogeneous Triple Negative Breast Tumor Models: Oncological Therapeutic Target Insights. Nutrients. 2021;13:1706. doi: 10.3390/nu13051706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yu P., Zhu L., Cui K., Du Y., Zhang C., Ma W., Guo J. B4GALNT2 Gene Promotes Proliferation, and Invasiveness and Migration Abilities of Model Triple Negative Breast Cancer (TNBC) Cells by Interacting With HLA-B Protein. Front Oncol. 2021;11:722828. doi: 10.3389/fonc.2021.722828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Holst S., Wuhrer M., Rombouts Y. Glycosylation characteristics of colorectal cancer. Adv. Cancer Res. 2015;126:203–256. doi: 10.1016/bs.acr.2014.11.004. [DOI] [PubMed] [Google Scholar]
- 48.Blanas A., Sahasrabudhe N.M., Rodriguez E., van Kooyk Y., van Vliet S.J. Fucosylated Antigens in Cancer: An Alliance toward Tumor Progression, Metastasis, and Resistance to Chemotherapy. Front Oncol. 2018;8:39. doi: 10.3389/fonc.2018.00039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Aronica A., Avagliano L., Caretti A., Tosi D., Bulfamante G.P., Trinchera M. Unexpected distribution of CA19.9 and other type 1 chain Lewis antigens in normal and cancer tissues of colon and pancreas: Importance of the detection method and role of glycosyltransferase regulation. Biochim. Biophys. Acta Gen. Subj. 2017;1861:3210–3220. doi: 10.1016/j.bbagen.2016.08.005. [DOI] [PubMed] [Google Scholar]
- 50.Mare L., Caretti A., Albertini R., Trinchera M. CA19.9 antigen circulating in the serum of colon cancer patients: Where is it from? Int. J. Biochem. Cell Biol. 2013;45:792–797. doi: 10.1016/j.biocel.2013.01.004. [DOI] [PubMed] [Google Scholar]
- 51.Trinchera M., Aronica A., Dall’Olio F. Selectin Ligands Sialyl-Lewis a and Sialyl-Lewis x in Gastrointestinal Cancers. Biology. 2017;6:16. doi: 10.3390/biology6010016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ferreira J.A., Magalhaes A., Gomes J., Peixoto A., Gaiteiro C., Fernandes E., Santos L.L., Reis C.A. Protein glycosylation in gastric and colorectal cancers: Toward cancer detection and targeted therapeutics. Cancer Lett. 2017;387:32–45. doi: 10.1016/j.canlet.2016.01.044. [DOI] [PubMed] [Google Scholar]
- 53.Trinchera M., Malagolini N., Chiricolo M., Santini D., Minni F., Caretti A., Dall’Olio F. The biosynthesis of the selectin-ligand sialyl Lewis x in colorectal cancer tissues is regulated by fucosyltransferase VI and can be inhibited by an RNA interference-based approach. Int. J. Biochem. Cell Biol. 2011;43:130–139. doi: 10.1016/j.biocel.2010.10.004. [DOI] [PubMed] [Google Scholar]
- 54.Balog C.I., Stavenhagen K., Fung W.L., Koeleman C.A., McDonnell L.A., Verhoeven A., Mesker W.E., Tollenaar R.A., Deelder A.M., Wuhrer M. N-glycosylation of Colorectal Cancer Tissues: A liquid chromatography and mass spectrometry-based investigation. Mol. Cell Proteom. 2012;11:571–585. doi: 10.1074/mcp.M111.011601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Holst S., Stavenhagen K., Balog C.I., Koeleman C.A., McDonnell L.M., Mayboroda O.A., Verhoeven A., Mesker W.E., Tollenaar R.A., Deelder A.M., et al. Investigations on aberrant glycosylation of glycosphingolipids in colorectal cancer tissues using liquid chromatography and matrix-assisted laser desorption time-of-flight mass spectrometry (MALDI-TOF-MS) Mol. Cell Proteom. 2013;12:3081–3093. doi: 10.1074/mcp.M113.030387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Holst S., Deuss A.J., van Pelt G.W., van Vliet S.J., Garcia-Vallejo J.J., Koeleman C.A., Deelder A.M., Mesker W.E., Tollenaar R.A., Rombouts Y., et al. N-glycosylation Profiling of Colorectal Cancer Cell Lines Reveals Association of Fucosylation with Differentiation and Caudal Type Homebox 1 (CDX1)/Villin mRNA Expression. Mol. Cell Proteom. 2016;15:124–140. doi: 10.1074/mcp.M115.051235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Madunic K., Zhang T., Mayboroda O.A., Holst S., Stavenhagen K., Jin C., Karlsson N.G., Lageveen-Kammeijer G.S.M., Wuhrer M. Colorectal cancer cell lines show striking diversity of their O-glycome reflecting the cellular differentiation phenotype. Cell Mol. Life Sci. 2021;78:337–350. doi: 10.1007/s00018-020-03504-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kaprio T., Satomaa T., Heiskanen A., Hokke C.H., Deelder A.M., Mustonen H., Hagstrom J., Carpen O., Saarinen J., Haglund C. N-glycomic Profiling as a Tool to Separate Rectal Adenomas from Carcinomas. Mol. Cell Proteom. 2015;14:277–288. doi: 10.1074/mcp.M114.041632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kikuchi D., Saito M., Saito K., Watanabe Y., Matsumoto Y., Kanke Y., Onozawa H., Hayase S., Sakamoto W., Ishigame T., et al. Upregulated solute carrier family 37 member 1 in colorectal cancer is associated with poor patient outcome and metastasis. Oncol. Lett. 2018;15:2065–2072. doi: 10.3892/ol.2017.7559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yamadera M., Shinto E., Tsuda H., Kajiwara Y., Naito Y., Hase K., Yamamoto J., Ueno H. Sialyl Lewis(x) expression at the invasive front as a predictive marker of liver recurrence in stage II colorectal cancer. Oncol. Lett. 2018;15:221–228. doi: 10.3892/ol.2017.7340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pothuraju R., Krishn S.R., Gautam S.K., Pai P., Ganguly K., Chaudhary S., Rachagani S., Kaur S., Batra S.K. Mechanistic and Functional Shades of Mucins and Associated Glycans in Colon Cancer. Cancers. 2020;12:649. doi: 10.3390/cancers12030649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Carvalho A.S., Harduin-Lepers A., Magalhaes A., Machado E., Mendes N., Costa L.T., Matthiesen R., Almeida R., Costa J., Reis C.A. Differential expression of a-2,3-sialyltransferases and a-1,3/4-fucosyltransferases regulates the levels of sialyl Lewis a and sialyl Lewis x in gastrointestinal carcinoma cells. Int. J. Biochem. Cell Biol. 2010;42:80–89. doi: 10.1016/j.biocel.2009.09.010. [DOI] [PubMed] [Google Scholar]
- 63.Perez-Garay M., Arteta B., Pages L., De Llorens R., de Bolos C., Vidal-Vanaclocha F., Peracaula R. a2,3-sialyltransferase ST3Gal III modulates pancreatic cancer cell motility and adhesion in vitro and enhances its metastatic potential in vivo. PLoS ONE. 2010;5:e12524. doi: 10.1371/journal.pone.0012524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Dimitroff C.J., Pera P., Dall’Olio F., Matta K.L., Chandrasekaran E.V., Lau J.T., Bernacki R.J. Cell surface n-acetylneuraminic acid a2,3-galactoside-dependent intercellular adhesion of human colon cancer cells. Biochem. Biophys. Res. Commun. 1999;256:631–636. doi: 10.1006/bbrc.1999.0388. [DOI] [PubMed] [Google Scholar]
- 65.Gomes C., Osorio H., Pinto M.T., Campos D., Oliveira M.J., Reis C.A. Expression of ST3GAL4 leads to SLex expression and induces c-Met activation and an invasive phenotype in gastric carcinoma cells. PLoS ONE. 2013;8:e66737. doi: 10.1371/journal.pone.0066737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Perez-Garay M., Arteta B., Llop E., Cobler L., Pages L., Ortiz R., Ferri M.J., de Bolos C., Figueras J., De Llorens R., et al. a2,3-Sialyltransferase ST3Gal IV promotes migration and metastasis in pancreatic adenocarcinoma cells and tends to be highly expressed in pancreatic adenocarcinoma tissues. Int. J. Biochem. Cell Biol. 2013;45:1748–1757. doi: 10.1016/j.biocel.2013.05.015. [DOI] [PubMed] [Google Scholar]
- 67.Colomb F., Krzewinski-Recchi M.A., El Machhour F., Mensier E., Jaillard S., Steenackers A., Harduin-Lepers A., Lafitte J.J., Delannoy P., Groux-Degroote S. TNF regulates sialyl-Lewisx and 6-sulfo-sialyl-Lewisx expression in human lung through up-regulation of ST3GAL4 transcript isoform BX. Biochimie. 2012;94:2045–2053. doi: 10.1016/j.biochi.2012.05.030. [DOI] [PubMed] [Google Scholar]
- 68.Hiller K.M., Mayben J.P., Bendt K.M., Manousos G.A., Senger K., Cameron H.S., Weston B.W. Transfection of a1,3 fucosyltransferase antisense sequences impairs the proliferative and tumorigenic ability of human colon carcinoma cells. Mol. Carcinog. 2000;27:280–288. doi: 10.1002/(SICI)1098-2744(200004)27:4<280::AID-MC6>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 69.Weston B.W., Hiller K.M., Mayben J.P., Manousos G.A., Bendt K.M., Liu R., Cusack J.C., Jr. Expression of human a1,3 fucosyltransferase antisense sequences inhibits selectin-mediated adhesion and liver metastasis of colon carcinoma cells. Cancer Res. 1999;59:2127–2135. [PubMed] [Google Scholar]
- 70.Pan S., Liu Y., Liu Q., Xiao Y., Liu B., Ren X., Qi X., Zhou H., Zeng C., Jia L. HOTAIR/miR-326/FUT6 axis facilitates colorectal cancer progression through regulating fucosylation of CD44 via PI3K/AKT/mTOR pathway. Biochim. Biophys. Acta Mol. Cell Res. 2019;1866:750–760. doi: 10.1016/j.bbamcr.2019.02.004. [DOI] [PubMed] [Google Scholar]
- 71.Kudo T., Ikehara Y., Togayachi A., Morozumi K., Watanabe M., Nakamura M., Nishihara S., Narimatsu H. Up-regulation of a set of glycosyltransferase genes in human colorectal cancer. Lab. Investig. 1998;78:797–811. [PubMed] [Google Scholar]
- 72.Holmes E.H., Hakomori S., Ostrander G.K. Synthesis of type 1 and 2 lacto series glycolipid antigens in human colonic adenocarcinoma and derived cell lines is due to activation of a normally unexpressed b1,3N-acetylglucosaminyltransferase. J. Biol. Chem. 1987;262:15649–15658. doi: 10.1016/S0021-9258(18)47776-9. [DOI] [PubMed] [Google Scholar]
- 73.Marcos N.T., Magalhaes A., Ferreira B., Oliveira M.J., Carvalho A.S., Mendes N., Gilmartin T., Head S.R., Figueiredo C., David L., et al. Helicobacter pylori induces b3GnT5 in human gastric cell lines, modulating expression of the SabA ligand sialyl-Lewis x. J. Clin. Investig. 2008;118:2325–2336. doi: 10.1172/JCI34324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lu C.H., Wu W.Y., Lai Y.J., Yang C.M., Yu L.C. Suppression of B3GNT7 gene expression in colon adenocarcinoma and its potential effect in the metastasis of colon cancer cells. Glycobiology. 2014;24:359–367. doi: 10.1093/glycob/cwu002. [DOI] [PubMed] [Google Scholar]
- 75.Shiozaki K., Yamaguchi K., Takahashi K., Moriya S., Miyagi T. Regulation of Sialyl Lewis Antigen Expression in Colon Cancer Cells by Sialidase NEU4. J. Biol. Chem. 2011;286:21052–21061. doi: 10.1074/jbc.M111.231191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Izawa M., Kumamoto K., Mitsuoka C., Kanamori C., Kanamori A., Ohmori K., Ishida H., Nakamura S., Kurata-Miura K., Sasaki K., et al. Expression of sialyl 6-sulfo Lewis X is inversely correlated with conventional sialyl Lewis X expression in human colorectal cancer. Cancer Res. 2000;60:1410–1416. [PubMed] [Google Scholar]
- 77.Yusa A., Miyazaki K., Kimura N., Izawa M., Kannagi R. Epigenetic silencing of the sulfate transporter gene DTDST induces sialyl Lewisx expression and accelerates proliferation of colon cancer cells. Cancer Res. 2010;70:4064–4073. doi: 10.1158/0008-5472.CAN-09-2383. [DOI] [PubMed] [Google Scholar]
- 78.Miyazaki K., Ohmori K., Izawa M., Koike T., Kumamoto K., Furukawa K., Ando T., Kiso M., Yamaji T., Hashimoto Y., et al. Loss of disialyl Lewisa the ligand for lymphocyte inhibitory receptor sialic acid-binding immunoglobulin-like lectin-7 (Siglec-7) associated with increased sialyl Lewis a expression on human colon cancers. Cancer Res. 2004;64:4498–4505. doi: 10.1158/0008-5472.CAN-03-3614. [DOI] [PubMed] [Google Scholar]
- 79.Huang H.C., Chao C.C., Wu P.H., Chung H.Y., Lee H.Y., Suen C.S., Hwang M.J., Cai B.H., Kannagi R. Epigenetic silencing of the synthesis of immunosuppressive Siglec ligand glycans by NF-kappaB/EZH2/YY1 axis in early-stage colon cancers. Biochim. Biophys. Acta Gene Regul. Mech. 2019;1862:173–183. doi: 10.1016/j.bbagrm.2019.01.002. [DOI] [PubMed] [Google Scholar]
- 80.Huang H.C., Cai B.H., Suen C.S., Lee H.Y., Hwang M.J., Liu F.T., Kannagi R. BGN/TLR4/NF-B Mediates Epigenetic Silencing of Immunosuppressive Siglec Ligands in Colon Cancer Cells. Cells. 2020;9:397. doi: 10.3390/cells9020397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kawamura Y.I., Kawashima R., Fukunaga R., Hirai K., Toyama-Sorimachi N., Tokuhara M., Shimizu T., Dohi T. Introduction of Sda carbohydrate antigen in gastrointestinal cancer cells eliminates selectin ligands and inhibits metastasis. Cancer Res. 2005;65:6220–6227. doi: 10.1158/0008-5472.CAN-05-0639. [DOI] [PubMed] [Google Scholar]
- 82.Capon C., Maes E., Michalski J.C., Leffler H., Kim Y.S. Sda-antigen-like structures carried on core 3 are prominent features of glycans from the mucin of normal human descending colon. Biochem. J. 2001;358:657–664. doi: 10.1042/bj3580657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Foxall C., Watson S.R., Dowbenko D., Fennie C., Lasky L.A., Kiso M., Hasegawa A., Asa D., Brandley B.K. The three members of the selectin receptor family recognize a common carbohydrate epitope, the sialyl Lewisx oligosaccharide. J. Cell Biol. 1992;117:895–902. doi: 10.1083/jcb.117.4.895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Liang J.X., Liang Y., Gao W. Clinicopathological and prognostic significance of sialyl Lewis X overexpression in patients with cancer: A meta-analysis. OncoTargets Ther. 2016;9:3113–3125. doi: 10.2147/OTT.S102389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Nakamori S., Kameyama M., Imaoka S., Furukawa H., Ishikawa O., Sasaki Y., Izumi Y., Irimura T. Involvement of carbohydrate antigen sialyl Lewisx in colorectal cancer metastasis. Dis. Colon Rectum. 1997;40:420–431. doi: 10.1007/BF02258386. [DOI] [PubMed] [Google Scholar]
- 86.Terraneo L., Avagliano L., Caretti A., Bianciardi P., Tosi D., Bulfamante G.P., Samaja M., Trinchera M. Expression of carbohydrate-antigen sialyl-Lewis a on colon cancer cells promotes xenograft growth and angiogenesis in nude mice. Int. J. Biochem. Cell Biol. 2013;45:2796–2800. doi: 10.1016/j.biocel.2013.10.003. [DOI] [PubMed] [Google Scholar]
- 87.Guerrero P.E., Miro L., Wong B.S., Massaguer A., Martinez-Bosch N., Llorens R., Navarro P., Konstantopoulos K., Llop E., Peracaula R. Knockdown of a2,3-Sialyltransferases Impairs Pancreatic Cancer Cell Migration, Invasion and E-selectin-Dependent Adhesion. Int. J. Mol. Sci. 2020;21:6239. doi: 10.3390/ijms21176239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Carrascal M.A., Silva M., Ramalho J.S., Pen C., Martins M., Pascoal C., Amaral C., Serrano I., Oliveira M.J., Sackstein R., et al. Inhibition of fucosylation in human invasive ductal carcinoma reduces E-selectin ligand expression, cell proliferation, and ERK1/2 and p38 MAPK activation. Mol. Oncol. 2018;12:579–593. doi: 10.1002/1878-0261.12163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Yoshihama N., Yamaguchi K., Chigita S., Mine M., Abe M., Ishii K., Kobayashi Y., Akimoto N., Mori Y., Sugiura T. A Novel Function of CD82/KAI1 in Sialyl Lewis Antigen-Mediated Adhesion of Cancer Cells: Evidence for an Anti-Metastasis Effect by Down-Regulation of Sialyl Lewis Antigens. PLoS ONE. 2015;10:e0124743. doi: 10.1371/journal.pone.0124743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kohler S., Ullrich S., Richter U., Schumacher U. E-/P-selectins and colon carcinoma metastasis: First in vivo evidence for their crucial role in a clinically relevant model of spontaneous metastasis formation in the lung. Br. J. Cancer. 2010;102:602–609. doi: 10.1038/sj.bjc.6605492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Gebauer F., Wicklein D., Stubke K., Nehmann N., Schmidt A., Salamon J., Peldschus K., Nentwich M.F., Adam G., Tolstonog G., et al. Selectin binding is essential for peritoneal carcinomatosis in a xenograft model of human pancreatic adenocarcinoma in pfp--/rag2--mice. Gut. 2013;62:741–750. doi: 10.1136/gutjnl-2011-300629. [DOI] [PubMed] [Google Scholar]
- 92.Stubke K., Wicklein D., Herich L., Schumacher U., Nehmann N. Selectin-deficiency reduces the number of spontaneous metastases in a xenograft model of human breast cancer. Cancer Lett. 2012;321:89–99. doi: 10.1016/j.canlet.2012.02.019. [DOI] [PubMed] [Google Scholar]
- 93.Esposito M., Mondal N., Greco T.M., Wei Y., Spadazzi C., Lin S.C., Zheng H., Cheung C., Magnani J.L., Lin S.H., et al. Bone vascular niche E-selectin induces mesenchymal-epithelial transition and Wnt activation in cancer cells to promote bone metastasis. Nat. Cell Biol. 2019;21:627–639. doi: 10.1038/s41556-019-0309-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Kelm M., Quiros M., Azcutia V., Boerner K., Cummings R.D., Nusrat A., Brazil J.C., Parkos C.A. Targeting epithelium-expressed sialyl Lewis glycans improves colonic mucosal wound healing and protects against colitis. JCI Insight. 2020;5:135843. doi: 10.1172/jci.insight.135843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Bordon Y. Inflammation: Live long and prosper with Siglecs. Nat. Rev. Immunol. 2015;15:266–267. doi: 10.1038/nri3851. [DOI] [PubMed] [Google Scholar]
- 96.Deschepper F.M., Zoppi R., Pirro M., Hensbergen P.J., Dall’Olio F., Kotsias M., Gardner R.A., Spencer D.I.R., Videira P.A. L1CAM as an E-selectin Ligand in Colon Cancer. Int. J. Mol. Sci. 2020;21:8286. doi: 10.3390/ijms21218286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Pucci M., Gomes F.I., Malagolini N., Ferracin M., Dall’Olio F. The Sda Synthase B4GALNT2 Reduces Malignancy and Stemness in Colon Cancer Cell Lines Independently of Sialyl Lewis X Inhibition. Int. J. Mol. Sci. 2020;21:6558. doi: 10.3390/ijms21186558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Zhan L., Chen L., Chen Z. Knockdown of FUT3 disrupts the proliferation, migration, tumorigenesis and TGF-beta induced EMT in pancreatic cancer cells. Oncol. Lett. 2018;16:924–930. doi: 10.3892/ol.2018.8738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Kawamura Y.I., Adachi Y., Curiel D.T., Kawashima R., Kannagi R., Nishimoto N., Dohi T. Therapeutic adenoviral gene transfer of a glycosyltransferase for prevention of peritoneal dissemination and metastasis of gastric cancer. Cancer Gene Ther. 2014;21:427–433. doi: 10.1038/cgt.2014.46. [DOI] [PubMed] [Google Scholar]