Abstract
To develop a better understanding of the epidemiology and molecular biology of rifampin-resistant Mycobacterium tuberculosis strains in Australia, 50 clinical isolates (33 rifampin-resistant and 17 rifampin-sensitive strains) cultured between 1990 and 1997 were analyzed by a number of bacteriological and molecular techniques. Examination of the drug resistance profiles of the 33 rifampin-resistant isolates revealed that 91% were resistant to rifampin in combination with resistance to isoniazid, 88% were resistant to rifampin on first isolation, and 81% showed cross-resistance with rifabutin. On the basis of the demographic data provided for the patients infected with the rifampin-resistant strains, 90% of the patients were born overseas. Of these patients, 64% developed clinical symptoms within 5 years of residence in Australia. On a molecular level, analysis of the rpoB gene revealed that 97% of the rifampin-resistant isolates had missense mutations within a conserved region of the gene, and eight types of missense mutations were detected. Of the 31 rifampin-resistant isolates that were typed by restriction fragment length polymorphism (RFLP) analysis, 28 distinct patterns were obtained by RFLP analysis with IS6110, and three clusters of genetically related isolates were identified. All isolates within the clusters were from patients who were born overseas and who had the same country of origin. The results from this study provide an overview of the current situation of rifampin resistance in Australia and can serve as a basis for continued monitoring of drug-resistant M. tuberculosis strains isolated within the country.
Tuberculosis (TB) is a disease of major public health concern worldwide. Despite the availability of highly effective anti-TB drugs and widespread efforts to implement prevention programs, about a third of the world's population (1.7 billion people) are infected with Mycobacterium tuberculosis and are at risk of developing clinical TB (17). Each year, approximately 8 million new active cases and 3 million deaths associated with the disease are reported from around the world.
In the 1990s, control of TB has been further complicated by an increase in the incidence of drug-resistant strains of M. tuberculosis which have been found in both developing and industrialized countries (1). The most common causes of acquired drug resistance in M. tuberculosis are inadequate chemotherapeutic regimens and noncompliance by patients during therapy (20). In the United States, multidrug-resistant (MDR) strains of M. tuberculosis have been responsible for a large number of institutional outbreaks, resulting in more than 200 cases of clinical diseases, and over 80% of these cases have occurred in people infected with the human immunodeficiency virus (HIV) (6, 7). In this setting, infections with MDR M. tuberculosis are often associated with extraordinarily high mortality rates and rapid progression from diagnosis to death (7).
In Australia, the incidence of TB is low by international standards. Since 1989, the annual incidence of laboratory-confirmed cases has remained between 3 and 4 cases per 100,000 population (2–5), and the rate of drug resistance is less than 18%. The majority of the drug-resistant infections involved either streptomycin or isoniazid alone or in combination with other antibiotics excluding rifampin. In 1996, the rates of resistance to rifampin alone and to rifampin in combination with isoniazid were 0.1 and 2.0%, respectively (3). However, according to Australian Communicable Diseases Intelligence figures (8), approximately 75% of the new TB cases reported in 1996 were among patients born overseas, particularly those born in countries where drug resistance among M. tuberculosis strains is an increasing problem. If this trend continues, emergence of MDR M. tuberculosis is likely to become a major public health risk in Australia.
This paper represents the results of a study of 33 rifampin-resistant M. tuberculosis strains that were isolated in Australia between 1990 and 1997. Rifampin is an important antibiotic used in the treatment of TB, and efficacy of chemotherapy can be markedly reduced when infections are caused by M. tuberculosis strains that are rifampin resistant (22). The goals of the present study were (i) to evaluate the drug resistance profiles of these isolates, (ii) to analyze the demographics of the patients from whom the rifampin-resistant isolates were cultured, (iii) to analyze a region within the isolates' rpoB gene that had been associated with rifampin resistance (31), and (iv) to evaluate the genetic polymorphisms of these isolates.
MATERIALS AND METHODS
Bacterial samples.
A total of 50 clinical isolates of M. tuberculosis were selected for this study (Table 1). All isolates were selected on the basis of their rifampin susceptibility, which was determined by reference laboratories. Thirty-three rifampin-resistant isolates were selected from among the 50 clinical isolates received from the mycobacterial reference laboratories of the following institutions in Australia: Victorian Infectious Diseases Reference Laboratory, Melbourne, Victoria, Queensland Diagnostic and Reference Laboratory for Mycobacterial Diseases, The Prince Charles Hospital, Brisbane, Queensland; Institute of Clinical Pathology and Medical Research, Westmead, New South Wales; Western Australian Centre for Pathology and Medical Research, Perth, Western Australia; and Institute of Medical and Veterinary Science, Adelaide, South Australia. Seventeen rifampin-susceptible M. tuberculosis isolates were selected as controls from the reference laboratory at the Victorian Infectious Diseases Reference Laboratory. All patient demographic data were collected and were kindly provided by the reference laboratories and the Department of Human Services, Melbourne, Victoria.
TABLE 1.
Number of M. tuberculosis isolates selected from the five state mycobacterial reference laboratories in Australia
| State | No. of isolates
|
|
|---|---|---|
| Sensitive | Resistant | |
| Victoria | 17 | 17 |
| New South Wales | 0 | 8 |
| Queensland | 0 | 6 |
| South Australia | 0 | 1 |
| Western Australia | 0 | 1 |
| Total | 17 | 33 |
On the basis of reports from the Australian Mycobacterium Reference Laboratory Network (2, 3, 5), a total of 4,735 clinical isolates of M. tuberculosis were isolated in the country between 1990 and 1996. Of these, 73 isolates were resistant to rifampin. Since 29 of the 33 rifampin-resistant M. tuberculosis isolates selected in this study were cultured within the same period, this study has evaluated approximately 40% of all the rifampin-resistant M. tuberculosis isolates that have been cultured in Australia.
Drug susceptibility testing.
In the early 1990s, the resistance ratio method, as described by the Centers for Disease Control and Prevention, U.S. Department of Health and Human Services (16), was used by the mycobacterial references laboratories in Queensland, New South Wales, and Victoria to determine the first-line drug susceptibilities of all first-time M. tuberculosis isolates recovered from patients. Currently, all mycobacterial references laboratories in Australia use the BACTEC 460 (Becton Dickinson) radiometric method for drug susceptibility testing (28).
The Etest (AB BIODISK, Solna, Sweden) was used to determine the rifampin MICs for all the M. tuberculosis isolates selected. The test was performed by using a modification of the method described by Wanger and Mills (34). Briefly, McFarland no. 3 standard suspensions of M. tuberculosis were lawn cultured onto Middlebrook 7H10 agar plates (Difco, Detroit, Mich.) enriched with 10% Middlebrook oleic acid-albumin-dextrose-citrate supplement (Micro Diagnostics, Coorparoo, Queensland, Australia) and were incubated at 37°C in an atmosphere of 5% CO2. After 24 h, a rifampin Etest strip was placed onto the center of each plate and the plate was reincubated under the same conditions until sufficient growth occurred (2 to 4 weeks) to determine the MIC.
All M. tuberculosis isolates resistant to rifampin were tested to determine their susceptibility to the remaining first-line anti-TB antibiotics (isoniazid, ethambutol, and pyrazinamide) and to rifabutin. Susceptibility to isoniazid and ethambutol was tested by the resistance ratio method, and susceptibility to rifabutin was tested by the absolute concentration method. Both of these methods were performed as described by the Centers for Disease Control and Prevention (16). To determine the susceptibilities of the selected isolates to pyrazinamide, the pyrazinamidase screening method described by Wayne (35) was used. All isolates which demonstrated pyrazinamidase activity were considered susceptible to pyrazinamide.
PCR.
The oligonucleotide primers described by Telenti et al. (31), TR1 (5′-TACGGTCGGCGAGCTAT-3′) and TR2b (5′-TACGGCGTTTCGATGAACC-3′), were used to amplify a 411-bp fragment from a conserved region within the rpoB gene. Chromosomal DNA was amplified in a 50-μl mixture containing 20 mM Tris-HCl (pH 8.4), 50 mM KCl, 0.75 mM MgCl2, dimethyl sulfoxide, each deoxynucleoside triphosphate at a concentration of 200 μM, each primer at a concentration of 0.5 μM, and 1.25 U of Taq DNA polymerase (Gibco BRL, Gaithersburg, Md.). The reaction was performed in an automated thermal cycler (Hybaid, Middlesex, United Kingdom) by using a program consisting of an initial denaturation at 94°C for 3 min, followed by 25 cycles of denaturation for 1 min at 94°C, annealing for 1 min at 60°C, and extension for 1 min at 72°C and an additional extension step at 72°C for 5 min. The PCR products were then purified with Geneclean III (Bio 101, Inc., La Jolla, Calif.) according to the manufacturer's instructions.
DNA sequencing.
Sequencing of the PCR product was performed with the Sequenase DNA Sequencing Kit (Amersham) according to the manufacturer's instructions. Both DNA strands were sequenced in the presence of 10% dimethyl sulfoxide with either oligonucleotide primer TR9 (5′-TCGCCGCGATCAAGGAGT-3′) or oligonucleotide primer TR8 (5′-TGCACGTCGCGGACCTCC A-3′ (31).
RFLP typing.
Chromosomal DNA extraction and Southern blot hybridization were performed as described by Ross et al. (26). To determine the patterns of the rifampin-resistant M. tuberculosis isolates by restriction fragment length polymorphism (RFLP) analysis with IS6110 (RFLP-IS6110 patterns), chromosomal DNA was digested with restriction endonuclease PvuII and was hybridized with a probe prepared from a 247-bp PCR product of IS6110 (26). The chromosomal DNAs of M. tuberculosis isolates with similar IS6110-RFLP patterns were redigested with AluI and hybridized with a previously described polymorphic GC-rich repetitive sequence probe, pTBN12 (26). Both IS6110 and pTBN12 probes were labelled with digoxigenin with a nucleic acid labelling system supplied by Boehringer Mannheim GmbH (Mannheim, Germany).
Computer analysis.
Southern blots were scanned at a resolution of 0.2 mm by using Molecular Analyst software and an imaging densitometer (model GS-670; Bio-Rad Laboratories, Richmond, Calif.). The images created were saved as TIFF files and were analyzed with Gelcompar (version 4.0) software (Applied Maths, Ghent, Belgium). RFLP patterns were normalized with an EcoRI-HindIII-digested bacteriophage DNA molecular weight marker (Boehringer Mannheim), which was used as an external size marker. Banding patterns were compared with the Gelcompar software by the unweighted pair group method with arithmetic means clustering method and the Jaccard coefficient method and were verified visually.
Definition of clustering.
A cluster of M. tuberculosis isolates was defined as two or more isolates which either (i) exhibit the same number of copies of the IS6110 fragment with identical molecular weights and identical pTBN12 RFLP patterns or (ii) showed one additional or one missing IS6110 fragment and identical pTBN12 RFLP patterns. Genetic relatedness is considered a possibility when strains have a similarity coefficient of ≥75% (27).
RESULTS
Drug resistance profiles. (i) Rifampin MICs.
The Etest was used to determine the rifampin MICs for 30 rifampin-resistant M. tuberculosis isolates (3 isolates were nonviable) (Table 2). Of the M. tuberculosis isolates tested, the MICs for 2 (isolates R1 and R9) were 32 μg/ml and the MICs for 28 were ≥256 μg/ml. For all rifampin-susceptible isolates of M. tuberculosis selected as controls, rifampin MICs were ≤1 μg/ml.
TABLE 2.
Characteristics of rifampin-resistant M. tuberculosis strains isolated in Australia between 1990 and 1997
| Strain no. | Country of birth | First-line drug to which strain is resistanta | Rifabutin susceptibilityb | Rifampin MIC (μg/ml) | RFLP cluster | Mutated codon | Mutation |
|---|---|---|---|---|---|---|---|
| R1 | Timor | I, R | S | 32 | A | 522 | TCG→CAG |
| R2 | Timor | I, R | S | >256 | A | 522 | TCG→CAG |
| R3 | Timor | I, R, E | R | >256 | B | 526 | CAC→GAC |
| R4 | Timor | I, R, Pz | R | >256 | B | 526 | CAC→GAC |
| R5 | Timor | I, R, E (Int), Pz | R | Nonviable | B | 526 | CAC→GAC |
| R6 | Turkey | I, R | R | >256 | 526 | CAC→GAC | |
| R7 | Australia | R | R | >256 | 526 | CAC→TAC | |
| R8 | Ethiopia | I, R | R | >256 | 526 | CAC→TAC | |
| R9 | Vietnam | I, R, E | S | 32 | 526 | CAC→CTC | |
| R10 | Timor | I, R, Pz | S | >256 | 531 | TCG→TTG | |
| R11 | Vietnam | I, R, E | R | >256 | 531 | TCG→TTG | |
| R12 | Former Yugoslavia | I, R, E | R | >256 | 531 | TCG→TTG | |
| R13 | Vietnam | R | R | >256 | 531 | TCG→TTG | |
| R14 | Australia | I, R | R | >256 | 516 | GAC→GTC | |
| R15 | Turkey | I, R | S | >256 | 516 | GAC→GTC | |
| R16 | Vietnam | I, R, E | R | >256 | 572 | ATC→TTC | |
| R17 | Wales | I, R | S | >256 | |||
| R18 | Egypt | I, R, E | R | >256 | 526 | CAC→CGC | |
| R19 | Philippines | I, R | R | >256 | 531 | TCG→TTG | |
| R20 | Vietnam | I, R | Nonviable | Nonviable | 526 | CAC→TAC | |
| R21 | Unknown | I, R, E | R | >256 | 531 | TCG→TTG | |
| R22 | Korean | I, R, Pz | R | >256 | 531 | TCG→TTG | |
| R23 | Unknown | I, R | R | >256 | 531 | TCG→TTG | |
| R24 | Unknown | I, R, E | R | >256 | 516 | GAC→TAC | |
| R25 | Australia | I, R | R | >256 | 526 | CAC→TAC | |
| R26 | Philippines | I, R, E | Nonviable | Nonviable | 531 | TCG→TTG | |
| R27 | Laos | I, R | R | >256 | 531 | TCG→TTG | |
| R28 | Papua New Guinea | I, R | R | >256 | C | 531 | TCG→TTG |
| R29 | Vietnam | I, R | R | >256 | 531 | TCG→TTG | |
| R30 | Papua New Guinea | R | R | >256 | C | 531 | TCG→TTG |
| R31 | Philippines | I, R | R | >256 | 531 | TCG→TTG | |
| R32 | Korea | I, R, E | R | >256 | 531 | TCG→TTG | |
| R33 | Vietnam | I, R, E | R | >256 | 531 | TCG→TTG |
I, isoniazid; R, rifampin; E, ethambutol; Pz, pyrazinamide; Int, intermediate resistance.
S, sensitive; R, resistant.
Twenty-nine (88%) of the M. tuberculosis isolates studied were resistant to rifampin on first isolation (primary resistance) and four acquired rifampin resistance within 3 to 6 months of the patients' commencement of chemotherapy. For the M. tuberculosis isolates with acquired rifampin resistance, the rifampin MICs increased from <1 μg/ml on primary isolation to >256 μg/ml within 3 to 6 months.
(ii) First-line drug susceptibilities.
Analysis of the first-line drug susceptibility profiles for the 33 rifampin-resistant M. tuberculosis isolates revealed that only three were resistant to rifampin alone; all remaining isolates were also resistant to isoniazid (Table 2). Of the rifampin- and isoniazid-resistant isolates, 14 (42%) were susceptible to the remaining two first-line anti-TB drugs, 11 (33%) were also resistant to ethambutol, 4 (12%) were also resistant to pyrazinamide, and 1 (3%) isolate was resistant to all four of the first-line anti-TB drugs. Since M. tuberculosis is defined as MDR when it is resistant to both rifampin and isoniazid, approximately 91% of the rifampin-resistant M. tuberculosis isolates studied were MDR.
(iii) Rifabutin susceptibility.
To assess the extent of cross-resistance between rifampin and rifabutin, rifabutin susceptibility was determined for 30 of the 33 rifampin-resistant strains of M. tuberculosis (3 strains were nonviable) (Table 2). Of the rifampin-resistant M. tuberculosis isolates tested, 6 were susceptible to rifabutin at concentrations of 1.0 μg/ml, while 24 showed cross-resistance with rifabutin at concentrations of >2 μg/ml. The rifabutin susceptibility of one of the nonviable rifampin-resistant strains of M. tuberculosis had previously been reported to be resistance. Hence, approximately 81% of M. tuberculosis strains with high levels of resistance to rifampin demonstrated cross-resistance to rifabutin. All rifampin-susceptible M. tuberculosis control strains were also susceptible to rifabutin.
Patient demographics. (i) Age and sex distributions.
Of the 33 patients from whom the rifampin-resistant M. tuberculosis isolates were cultured, 18 were men and 15 were women. The male-to-female ratio was 1.2:1. With the exception of one person whose age was not known, these patients were between 18 and 70 years of age, and the median age was 40.3 years. The age distribution was as follows: 5 (16%) patients, between 18 and 24 years of age; 14 (47%) patients, between 25 and 45 years of age; and 13 (41%) patients, between 46 and 70 years of age. Of the female patients, 12 (80%) were less than 45 years old and 3 were greater than 50 years old. Of the male patients, 7 were less than 45 years old and 10 were over 45 years old.
(ii) HIV infection status.
The patient's HIV infection status is not usually provided to the laboratories. From the demographic details provided for the 33 patients, for only 1 was the HIV status disclosed, and that patient was HIV positive. The patient was a 42-year-old Caucasian male who had been born in Australia and who presented with disseminated TB. However, details of other illnesses and treatment for this patient were not available. The M. tuberculosis isolate cultured from the patient was resistant to rifampin, rifabutin, ciprofloxacin, clarithromycin, and cycloserine.
(iii) Country of origin.
The countries of origin were known for 30 patients, and 27 of them were born overseas (Table 2). The countries from which these patients migrated include Timor (n = 6), Papua New Guinea (n = 2), Philippines (n = 3), Korea (n = 2), Laos (n = 1), Vietnam (n = 7), Egypt (n = 1), Ethiopia (n = 1), Turkey (n = 2), Wales (n = 1), and the former Yugoslavia (n = 1). Among the patients born overseas, three were members of the same Timorese family. Of the three patients born in Australia, one was a 55-year old woman and two were men ages 42 and 63 years, respectively, at the time of diagnosis.
(iv) Period of residence in Australia prior to diagnosis.
The date of arrival in Australia was known for 25 of the 27 patients born overseas. The interval from the time of arrival in Australia to the time of isolation of M. tuberculosis for these 25 patients ranged between 0 months and 23 years. Of these patients, 10 received a diagnosis of TB within the same year of arriving in Australia (2 within the same month), 6 received a diagnosis of TB within 2 to 5 years, 4 received a diagnosis of TB within 6 to 10 years, and 5 received a diagnosis of TB after 11 or more years.
Genetic analysis. (i) rpoB gene analysis.
A 69-bp region of the rpoB genes from all M. tuberculosis strains included in the study was analyzed by DNA sequencing. All sequence data obtained were compared with the sequence obtained from the GenBank database (accession no. L27989). Of the 17 rifampin-sensitive M. tuberculosis isolates sequenced, no mutations were detected within the 69-bp segment. On the other hand, eight different types of mutations, consisting of either one or two nucleotide substitutions, were detected within the same 69-bp segment in 31 of the rifampin-resistant M. tuberculosis (Table 2).
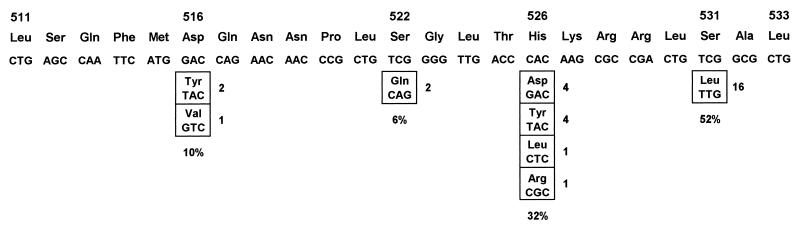
By using the rpoB codon numbering system described by Telenti et al. (31), all mutations were detected within codons 516, 522, 526, and 531 (Fig. 1). The most frequently encountered mutation was in codon 531 (52%), from TCG (Ser) to TTG (Leu). The next most frequently encountered mutation was in codon 526 (32%). The nucleotide substitutions found within this codon include CAC (His) to GAC (Asp), TAC (Tyr), CTC (Leu), or CGC (Arg). In strains R1 and R2, the substitution of two nucleotide in codon 522 from TCG (Ser) to CAG (Gln) has not been reported previously.
FIG. 1.
Nucleotide substitutions detected within the 69-bp region of the rpoB gene in rifampin-resistant M. tuberculosis isolates. The wild-type sequence is shown across the top, and the positions of point mutations and substituted amino acids are shown in the boxes below. The numbers on the right of the boxes are the number of strains with the same mutation. The frequency of mutations detected within the codons is indicated below the boxes. The codons were numbered on the basis of the system used by Telenti et al. (32).
Two rifampin-resistant M. tuberculosis isolates had no mutation within the 69-bp region. Of these two isolates, one (isolate R16) had a nucleotide substitution in codon 572 from ATC (Ile) to TTC (Phe), and one (isolate R17) had no mutation within the entire 411-bp fragment amplified.
(ii) RFLP analysis.
To assess the genetic polymorphism of the rifampin-resistant M. tuberculosis strains, DNA was digested with PvuII and was hybridized with the IS6110 probe. The RFLP patterns produced were then analyzed with the Gelcompar software program, and a dendrogram was constructed to illustrate the genetic relatedness among the isolates. Two rifampin-resistant isolates (isolates R5 and R20) were nonviable and so could not be typed by RFLP analysis.
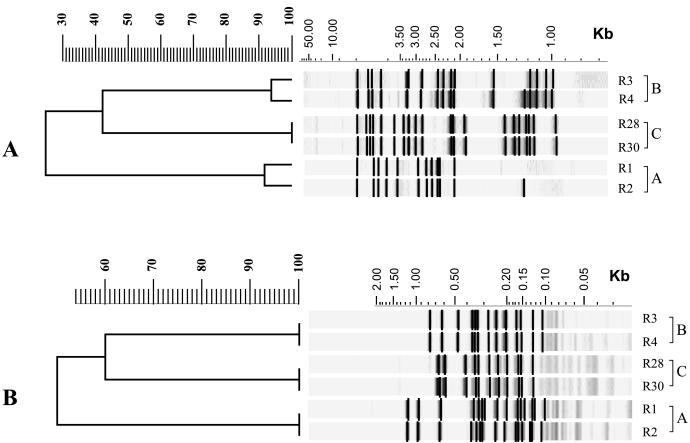
A total of 28 distinct RFLP patterns were obtained for the 31 rifampin-resistant isolates of M. tuberculosis tested (data not shown). Three groups of rifampin-resistant isolates exhibited very similar or identical IS6110-RFLP patterns (Fig. 2). To confirm their genetic relatedness, the chromosomal DNAs of the isolates were redigested with AluI and were hybridized with the pTBN12 probe. With this GC-rich repetitive sequence probe, all M. tuberculosis isolates within the groups (designated clusters A, B, and C) produced identical RFLP patterns.
FIG. 2.
Dendrograms of Southern blots produced with Gelcompar software showing the RFLP patterns and the percent similarities between the three clusters (designated clusters A, B, and C) of rifampin-resistant M. tuberculosis isolates that are epidemiologically related. The RFLP profiles were produced by digesting chromosomal DNA with PvuII and hybridization with the IS6110 probe (A) and by digesting chromosomal DNA with AluI and hybridization with the pTBN12 probe (B).
The isolates of M. tuberculosis in clusters A and B were initially cultured in Victoria. Cluster A consists of two isolates (isolates R1 and R2) with 92% similar IS6110-RFLP patterns. Direct comparison of the RFLP profiles revealed an extra 1.25-kb band in isolate R2 (Fig. 2). Both M. tuberculosis isolates were cultured from specimens from patients who originated in East Timor, and the same mutation was detected within codon 522 of their rpoB genes. Cluster B consists of three M. tuberculosis isolates (isolates R3, R4, and R5) cultured from specimens from members of the same Timorese family. However, chromosomal DNA was available for RFLP analysis of only isolates R3 and R4, and the IS6110-RFLP patterns produced were 94% similar according to the results obtained with the Gelcompar software. Direct comparison of the RFLP patterns revealed an extra 1.24-kb band in isolate R4 (Fig. 2). All three M. tuberculosis isolates had the same mutation within codon 526 of their rpoB genes. Cluster C consists of two M. tuberculosis isolates (isolates R28 and R30) initially cultured in Queensland (Fig. 2). It was the only cluster with isolates with identical IS6110-RFLP patterns. Both isolates were cultured from patients who originally came from Papua New Guinea, and the same mutation was detected in codon 531 of their rpoB genes.
DISCUSSION
Rifampin is the most potent sterilizing antibiotic used for the treatment of TB. It has bactericidal activity against extracellular tubercle bacilli as well as the active and the near dormant bacilli that may reside within macrophages. In the case of drug-susceptible strains of M. tuberculosis, the combined use of the first-line anti-TB drugs, rifampin, isoniazid, ethambutol, and pyrazinamide, will most often result in successful cures. Rifampin and isoniazid are the most active of the first-line anti-TB drugs, and M. tuberculosis strains that are resistant to both of these antibiotics are considered MDR (13). In general, multidrug resistance is acquired in two steps, with the first step being the development of isoniazid resistance rather than rifampin resistance (23), suggesting that rifampin resistance can be used as a surrogate marker for the detection of MDR M. tuberculosis (9, 31). Since 91% of the rifampin-resistant strains examined in this study were also resistant to isoniazid, this indicates that rifampin resistance is a good predictor for multidrug resistance in Australia.
Analysis of demographic data revealed that 27 of 33 (90%) patients infected with the rifampin-resistant M. tuberculosis strains were born outside Australia. Twenty-five of the 27 patients born overseas were infected with MDR M. tuberculosis strains, and the majority of them (19 of 30 strains) were from Southeast Asia. Of the six Timorese patients with drug-resistant TB, contact tracings have shown that three of them are members of the same family, suggesting intrafamily transfer, and RFLP analysis has confirmed that the isolates cultured from specimens from these patients (cluster B) are genetically related (Fig. 2). In addition, RFLP analysis revealed that two other rifampin-resistant isolates cultured from specimens from Timorese patients (cluster A) are genetically related to each other.
An important factor that was observed in this study is the short period of time that patients with drug-resistant TB had resided in Australia prior to diagnosis; 40% of the patients presented with drug-resistant TB within the same year as their arrival in Australia (two patients presented with TB within a month of their arrival in the country). Given the short time in the country prior to clinical diagnosis, it is highly probable that these patients were already infected with the drug-resistant strains on arrival. It is unfortunate that there are no adequate details on whether the patients had been given chemotherapy previously. Such data are important in determining whether the patients were initially infected with an MDR M. tuberculosis strain or whether the emergence of MDR M. tuberculosis was due to inadequate or inappropriate antibiotic treatment that resulted in acquisition of antibiotic resistance by the initial infecting isolate.
Of the 33 patients with rifampin-resistant TB studied, isolates from 29 (88%) were found to be resistant to rifampin on primary isolation. Although such patients represented only a very low proportion of the total number of the patients with TB nationwide, the possibility that drug-resistant strains were introduced into a highly susceptible population is a significant concern for public health. These data not only illustrate the need for a rapid and specific test for detection of MDR M. tuberculosis but also raise the question as to whether the present TB screening protocol for immigrants needs to be modified to include a wider group of people such as visitors who will reside in the country for long periods.
The HIV infection status of patients is not usually made available to the laboratories. On the basis of the data provided, only one case of rifampin-resistant TB was associated with HIV infection. Although the source of this drug-resistant M. tuberculosis strain is not known, to date there has been no evidence of further transmission of this strain into the HIV-infected population in Australia.
The molecular resistance mechanism for rifampin has been deciphered. On the basis of work conducted with Escherichia coli, rifampin resistance is the result of mutations within certain regions of the rpoB gene (clusters N, I, II, and III), which encodes the β-subunit of RNA polymerase (14, 18). Substitutions of amino acids within any of these regions may result in either conformational changes to the rifampin binding site or a loss of affinity for the drug. In effect, rifampin can no longer bind to the enzyme to inhibit its function, hence resulting in resistance. Recently, the sequence of the rpoB gene of M. tuberculosis has been determined, and different types of mutations associated with rifampin resistance have been reported (24, 31, 32). The majority of mutations were detected within a 69-bp segment of the gene which is equivalent to the cluster I region in E. coli (30).
Analysis of the conserved region within the rpoB gene of 33 rifampin-resistant M. tuberculosis isolates showed that 31 of them have missense mutations within the 69-bp segment of the gene, and a total of eight different types of mutations were detected within codons 516, 522, 526, and 531. These findings differed from those of previous studies in that the mutations are restricted to only four codon sites within the rpoB gene (15, 24, 31, 32). The lack of diversity could be due to the small number of rifampin-resistant strains that were studied and the limited population from which isolates were obtained. Interestingly, all rifampin-resistant M. tuberculosis isolates analyzed were resistant to high levels of rifampin (≥32 μg/ml), and this finding is consistent with previous reports in which amino acid substitutions within codons 516, 526, and 531 have been associated with high-level rifampin resistance in M. tuberculosis (24, 31, 32). Since 94% of the rifampin-resistant M. tuberculosis isolates studied have missense mutations within a confined segment of the rpoB gene, molecular techniques could be used to analyze this segment of the gene for the detection of mutations associated with rifampin resistance in M. tuberculosis. However, techniques such as DNA sequencing, PCR–single-strand conformation polymorphism analysis, and PCR-heteroduplex formation analysis, have been used in previous studies to analyze this region of the rpoB gene, but their use is not practical for routine diagnostic laboratories. Recently, a line probe assay (Inno-LiPA-Rif.TB) from Innogenetics, Ghent, Belgium, has been released commercially as a diagnostic test that can rapidly identify clinical isolates as a member of the M. tuberculosis complex and determine the presence of point mutations within the rpoB gene. Evaluations performed with this kit showed that it is highly sensitive and specific and is a good means of identifying the rifampin susceptibility status of clinical isolates (10, 21). However, it was shown in this study that a small percentage of rifampicin-resistant M. tuberculosis strains still do not possess mutations within the 69-bp segment of the rpoB gene, and these resistant strains would not have been detected by the line-probe assay or a similar assay. This may be due to a mutation outside the segment of the rpoB gene analyzed or an entirely different mechanism of resistance. While genetic analysis of the rpoB gene is useful for rapid detection of rifampin resistance in M. tuberculosis isolates, particularly in outbreak situations, susceptibility testing results obtained should always be confirmed by conventional drug sensitivity testing methods.
Rifabutin is a derivative of the rifamycin group of antibiotics and has been reported to have some activity against rifampin-resistant M. tuberculosis (19). It is used as a second-line anti-TB drug for the treatment of drug-resistant strains of M. tuberculosis. However, rifabutin and rifampin have similar modes of action (12), and cross-resistance between the two antibiotics has been shown to occur (11). In the present study, 81% of the isolates showed cross-resistance between rifampin and rifabutin. This finding is very similar to that from a study conducted by Heifets and Iseman (11), in which 88% of rifampin-resistant M. tuberculosis strains examined had cross-resistance to rifampin and rifabutin. In a recent study, Williams et al. (36) demonstrated that mutations within codons 526 and 531 of the rpoB gene could contribute to rifamycin cross-resistance in M. tuberculosis. However, no particular type of mutation detected within the rpoB gene of rifampin-resistant isolates could be associated with rifabutin resistance in the present study. In addition, rifampin MICs for two M. tuberculosis isolates (isolates R9 and R10) were high, and isolates with mutations in codons 526 and 531 of the rpoB gene remained susceptible to rifabutin in vitro, but the clinical efficacy of rifabutin treatment in this setting is untested.
RFLP analysis is a powerful technique that enables epidemiological studies to be performed on a molecular level. The IS6110 insertion sequence is a transposable element that is found only in species of the M. tuberculosis complex (33) and is widely used as a genetic marker for DNA fingerprinting of clinical M. tuberculosis isolates to monitor bacterial strain transmission and to confirm laboratory contaminations (25, 27, 29). In general, M. tuberculosis isolates that are genetically related demonstrate very similar or identical RFLP patterns, while nonrelated strains demonstrate significantly different patterns. By this technique, three clusters with very similar IS6110-RFLP patterns were identified among the rifampin-resistant M. tuberculosis isolates screened. Within each cluster, all the isolates were cultured from patients who were born overseas and who had the same country of origin. In addition, there was no evidence of transmission of MDR M. tuberculosis strains from individuals born overseas into the wider Australian population. The IS6110-RFLP patterns obtained for the rifampin-resistant strains of M. tuberculosis enabled a database to be established, and clinical isolates of M. tuberculosis with identical or very similar profiles could be deemed resistant to rifampin.
Rapid detection of rifampin resistance is very important for the timely and successful treatment of TB. The use of standard drug combination therapy against infections caused by MDR M. tuberculosis may result in prolonged or progressive disease and increased risk of transmission of drug-resistant strains of M. tuberculosis into the general community. The 33 rifampin-resistant M. tuberculosis isolates characterized in this study represented approximately 40% of the clinical rifampin-resistant M. tuberculosis isolates cultured nationwide between 1990 and 1997, and the data obtained can provide a general overview of the current situation of rifampin resistance in Australia. These results can also serve as a basis for continued monitoring of drug-resistant M. tuberculosis strains isolated in Australia.
ACKNOWLEDGMENTS
We thank Aina Sievers of the Victorian Infectious Diseases Reference Laboratory, Regina Lasaitis and William Chew of the Institute of Clinical Pathology and Medical Research, David Dawson of the Queensland Diagnostic and Reference Laboratory for Mycobacterial Diseases, The Prince Charles Hospital, Frank Haverkort of the Western Australian Centre for Pathology and Medical Research, Richard Lumb of the Institute of Medical and Veterinary Sciences, and the Department of Human Services of Victoria for providing the M. tuberculosis isolates and the patient demographics required for the study.
REFERENCES
- 1.Cohn D L, Bustreo F, Raviglione M C. Drug-resistant tuberculosis: review of the worldwide situation and the WHO/IUATLD Global Surveillance Project. Clin Infect Dis. 1997;24:S121–S130. doi: 10.1093/clinids/24.supplement_1.s121. [DOI] [PubMed] [Google Scholar]
- 2.Dawson D. Tuberculosis in Australia: bacteriologically confirmed cases and drug resistance, 1994 and 1995. Commun Dis Intell. 1997;21:245–249. doi: 10.33321/cdi.1997.21.48. [DOI] [PubMed] [Google Scholar]
- 3.Dawson D. Tuberculosis in Australia: bacteriologically confirmed cases and drug resistance, 1996. Commun Dis Intell. 1998;22:183–188. doi: 10.33321/cdi.1998.22.42. [DOI] [PubMed] [Google Scholar]
- 4.Dawson D, Anargyros P, Blacklock Z, Chew W, Dagnia H, Gow B, Jackson K, Sievers A. Tuberculosis in Australia: an analysis of cases identified in reference laboratories in 1986–88. Pathology. 1991;23:130–134. doi: 10.3109/00313029109060811. [DOI] [PubMed] [Google Scholar]
- 5.Dawson D J, Cheah D F, Chew W K, Haverkort F C, Lumb R, Sievers A S. Tuberculosis in Australia, 1989–1992. Bacteriologically confirmed cases and drug resistance. Med J Aust. 1995;162:287–290. doi: 10.5694/j.1326-5377.1995.tb139901.x. [DOI] [PubMed] [Google Scholar]
- 6.Dooley S W, Jarvis W R, Martone W J, Snider D E., Jr Multidrug-resistant tuberculosis. Ann Intern Med. 1992;117:257–259. doi: 10.7326/0003-4819-117-3-257. [DOI] [PubMed] [Google Scholar]
- 7.Frieden T R, Sherman L F, Maw K L, Fujiwara P I, Crawford J T, Nivin B, Sharp V, Hewlett D, Jr, Brudney K, Alland D, Kreisworth B N. A multi-institutional outbreak of highly drug-resistant tuberculosis: epidemiology and clinical outcomes. JAMA. 1996;276:1229–1235. [PubMed] [Google Scholar]
- 8.Gilroy N, Oliver G, Harvey B. Tuberculosis notifications in Australia, 1996. Commun Dis Intell. 1998;22:173–183. doi: 10.33321/cdi.1998.22.41. [DOI] [PubMed] [Google Scholar]
- 9.Goble M, Iseman M D, Madsen L A, Waite D, Ackerson L, Horsburgh C R., Jr Treatment of 171 patients with pulmonary tuberculosis resistant to isoniazid and rifampin. N Engl J Med. 1993;328:527–532. doi: 10.1056/NEJM199302253280802. [DOI] [PubMed] [Google Scholar]
- 10.Goyal M, Shaw R J, Banerjee D K, Coker R J, Robertson B D, Young D B. Rapid detection of multidrug-resistant tuberculosis. Eur Respir J. 1997;10:1120–1124. doi: 10.1183/09031936.97.10051120. [DOI] [PubMed] [Google Scholar]
- 11.Heifets L B, Iseman M D. Determination of in-vitro susceptibility of mycobacteria to ansamycin. Am Rev Respir Dis. 1985;132:710–711. doi: 10.1164/arrd.1985.132.3.710. [DOI] [PubMed] [Google Scholar]
- 12.Inderlied C B. Antimycobacterial agents: in-vitro susceptibility testing, spectrum of activity, mechanism of action and resistance, and assays for activity in biological fluids. In: Lorian V, editor. Antibiotics in laboratory medicine. 3rd ed. Baltimore, Md: The Williams & Wilkins Co.; 1991. pp. 134–197. [Google Scholar]
- 13.Jacobs R F. Multiple-drug-resistant tuberculosis. Clin Infect Dis. 1994;19:1–8. doi: 10.1093/clinids/19.1.1. [DOI] [PubMed] [Google Scholar]
- 14.Jin D J, Gross C A. Mapping and sequencing of mutations in the Escherichia coli rpoB gene that lead to rifampicin resistance. J Mol Biol. 1988;202:45–58. doi: 10.1016/0022-2836(88)90517-7. [DOI] [PubMed] [Google Scholar]
- 15.Kapur V, Li L L, Iordanescu S, Hamrick M R, Wanger A, Kreiswirth B N, Musser J M. Characterization by automated DNA sequencing of mutations in the gene (rpoB) encoding the RNA polymerase beta-subunit in rifampin-resistant Mycobacterium tuberculosis strains from New York City and Texas. J Clin Microbiol. 1994;32:1095–1098. doi: 10.1128/jcm.32.4.1095-1098.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kent P T, Kubica G P. Public health mycobacteriology: a guide for the level III laboratory. Atlanta, Ga: Centers for Disease Control; 1985. [Google Scholar]
- 17.Kochi A. The global tuberculosis situation and the new control strategy of the World Health Organization. Tubercle. 1991;72:1–6. doi: 10.1016/0041-3879(91)90017-m. [DOI] [PubMed] [Google Scholar]
- 18.Lisitsyn N, Sverdlov E, Moiseyeva E, Danilevskaya O, Nikiforov V. Mutation to rifampicin resistance at the beginning of the RNA polymerase β-subunit gene in Escherichia coli. Mol Gen Genet. 1984;196:173–174. doi: 10.1007/BF00334112. [DOI] [PubMed] [Google Scholar]
- 19.Luna-Herrera J, Reddy M V, Gangadharam P R. In-vitro and intracellular activity of rifabutin on drug-susceptible and multiple drug-resistant (MDR) tubercle bacilli. J Antimicrob Chemother. 1995;36:355–363. doi: 10.1093/jac/36.2.355. [DOI] [PubMed] [Google Scholar]
- 20.Mahmoudi A, Iseman M D. Pitfalls in the care of patients with tuberculosis: common errors and their association with the acquisition of drug resistance. JAMA. 1993;270:65–68. [PubMed] [Google Scholar]
- 21.Matsiota-Bernard P, Vrioni G, Marinsi E. Characterization of rpoB mutations in rifampicin-resistant clinical isolates of Mycobacterium tuberculosis isolates from Greece. J Clin Microbiol. 1998;36:20–23. doi: 10.1128/jcm.36.1.20-23.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mitchison D A, Nunn A J. Influence of initial drug resistance on the response to short-course chemotherapy of pulmonary tuberculosis. Am Rev Respir Dis. 1986;133:423–430. doi: 10.1164/arrd.1986.133.3.423. [DOI] [PubMed] [Google Scholar]
- 23.Nolan C M, Williams D L, Cave M D, Eisenach K D, el-Hajj H, Hooton T M, Thompson R L, Goldberg S V. Evolution of rifampin resistance in human immunodeficiency virus-associated tuberculosis. Am J Respir Crit Care Med. 1995;152:1067–1071. doi: 10.1164/ajrccm.152.3.7663785. [DOI] [PubMed] [Google Scholar]
- 24.Ohno H, Koga H, Kohno S, Tashiro T, Hara K. Relationship between rifampin MICs and rpoB mutations of Mycobacterium tuberculosis strains isolated in Japan. Antimicrob Agents Chemother. 1996;40:1053–1056. doi: 10.1128/aac.40.4.1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Otal I, Martin C, Vincent-Levy-Frebault V, Thierry D, Gicquel B. Restriction fragment length polymorphism analysis using IS6110 as an epidemiological marker in tuberculosis. J Clin Microbiol. 1991;29:1252–1254. doi: 10.1128/jcm.29.6.1252-1254.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ross B C, Raios K, Jackson K, Sievers A, Dwyer B. Differentiation of Mycobacterium tuberculosis strains by use of a nonradioactive Southern blot hybridization method. J Infect Dis. 1991;163:904–907. doi: 10.1093/infdis/163.4.904. [DOI] [PubMed] [Google Scholar]
- 27.Safi H, Aznar J, Palomares J C. Molecular epidemiology of Mycobacterium tuberculosis strains isolated during a 3-year period (1993 to 1995) in Seville, Spain. J Clin Microbiol. 1997;35:2472–2476. doi: 10.1128/jcm.35.10.2472-2476.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Siddiqi S H. Bactec® TB System, product and procedure manual, section IV. Sparks, Md: Becton Dickinson Diagnostic Instrument Systems; 1995. pp. 1–23. [Google Scholar]
- 29.Strassle A, Putnik J, Weber R, Fehr-Merhof A, Wust J, Pfyffer G E. Molecular epidemiology of Mycobacterium tuberculosis strains isolated from patients in a human immunodeficiency virus cohort in Switzerland. J Clin Microbiol. 1997;35:374–378. doi: 10.1128/jcm.35.2.374-378.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Taniguchi H, Aramaki H, Nikaido Y, Mizuguchi Y, Nakamura M, Koga T, Yoshida S-I. Rifampicin resistance and mutation of the rpoB gene in Mycobacterium tuberculosis. FEMS Microbiol Lett. 1996;144:103–108. doi: 10.1111/j.1574-6968.1996.tb08515.x. [DOI] [PubMed] [Google Scholar]
- 31.Telenti A, Imboden P, Marchesi F, Lowrie D, Cole S, Colston M J, Matter L, Schopfer K, Bodmer T. Detection of rifampicin-resistance mutations in Mycobacterium tuberculosis. Lancet. 1993;341:647–650. doi: 10.1016/0140-6736(93)90417-f. [DOI] [PubMed] [Google Scholar]
- 32.Telenti A, Imboden P, Marchesi F, Schmidheini T, Bodmer T. Direct, automated detection of rifampin-resistant Mycobacterium tuberculosis by polymerase chain reaction and single-strand conformation polymorphism analysis. Antimicrob Agents Chemother. 1993;37:2054–2058. doi: 10.1128/aac.37.10.2054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Thierry D, Cave M D, Eisenach K D, Crawford J T, Bates J H, Gicquel B, Guesdon J L. IS6110, an IS-like element of Mycobacterium tuberculosis complex. Nucleic Acids Res. 1990;18:188. doi: 10.1093/nar/18.1.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wanger A, Mills K. Testing of Mycobacterium tuberculosis susceptibility to ethambutol, isoniazid, rifampin, and streptomycin by using Etest. J Clin Microbiol. 1996;34:1672–1676. doi: 10.1128/jcm.34.7.1672-1676.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wayne L G. Simple pyrazinamidase and urease tests for routine identification of mycobacteria. Am Rev Respir Dis. 1974;109:147–151. doi: 10.1164/arrd.1974.109.1.147. [DOI] [PubMed] [Google Scholar]
- 36.Williams D L, Spring L, Collins L, Miller L P, Heifets L B, Gangadharam P R, Gillis T P. Contribution of rpoB mutations to development of rifamycin cross-resistance in Mycobacterium tuberculosis. Antimicrob Agents Chemother. 1998;42:1853–1857. doi: 10.1128/aac.42.7.1853. [DOI] [PMC free article] [PubMed] [Google Scholar]