Abstract
Bioluminescence, the emission of light catalysed by luciferases, has evolved in many taxa from bacteria to vertebrates and is predominant in the marine environment. It is now well established that in animals possessing a nervous system capable of integrating light stimuli, bioluminescence triggers various behavioural responses and plays a role in intra- or interspecific visual communication. The function of light emission in unicellular organisms is less clear and it is currently thought that it has evolved in an ecological framework, to be perceived by visual animals. For example, while it is thought that bioluminescence allows bacteria to be ingested by zooplankton or fish, providing them with favourable conditions for growth and dispersal, the luminous flashes emitted by dinoflagellates may have evolved as an anti-predation system against copepods. In this short review, we re-examine this paradigm in light of recent findings in microorganism photoreception, signal integration and complex behaviours. Numerous studies show that on the one hand, bacteria and protists, whether autotrophs or heterotrophs, possess a variety of photoreceptors capable of perceiving and integrating light stimuli of different wavelengths. Single-cell light-perception produces responses ranging from phototaxis to more complex behaviours. On the other hand, there is growing evidence that unicellular prokaryotes and eukaryotes can perform complex tasks ranging from habituation and decision-making to associative learning, despite lacking a nervous system. Here, we focus our analysis on two taxa, bacteria and dinoflagellates, whose bioluminescence is well studied. We propose the hypothesis that similar to visual animals, the interplay between light-emission and reception could play multiple roles in intra- and interspecific communication and participate in complex behaviour in the unicellular world.
Keywords: bioluminescence, luciferase, photoreceptors, lux operon, dinoflagellate, communication, signalling, symbiosis, rhizosphere
1. Introduction
In this short review, we examine the relationships between catalysed light-emission [1,2,3,4], light sensing and light behavioural responses in unicellular organisms with a view to exploring new functions of microorganism bioluminescence. During the past decades, many studies have shown that most unicellular species possess a great diversity of photoreceptors [5,6,7]. In particular, the presence of photoreceptors in non-phototrophic bacteria which do not use ambient light as an energy source was unexpected [8,9]. Photoreceptors allow the perception of light of different wavelengths and are responsible for various light-dependent behaviours in the unicellular world from simple phototaxis to more complex and collective behaviours such as hunting [10,11,12]. Through a great variety of receptors, prokaryotic and eukaryotic unicellular organisms are extremely sensitive to a variety of physical and chemical stimuli. In addition, to survive in their fluctuating environment, microorganisms must be able to integrate light and other stimuli to produce appropriate behavioural responses. Based on the observation of complex behaviours, learning phenomena and therefore the ability to memorise, some authors have evoked the “cognitive” properties of bacteria [13] or even the “consciousness” of organisms lacking nervous systems [14,15,16,17,18]. For example, it has been demonstrated that diverse single-cell eukaryotes such as the ciliates Paramecia and Stentor display complex behaviours such as decision-making or even associative learning [19,20,21,22]. At the molecular level, in addition to well documented cell signalling processes [23,24,25], the recent discovery of neural-like ribosomal protein networks that have the potential to transfer and integrate the information flow in transit during protein synthesis in the ribosomes [26,27,28] reinforced the seminal Bray’s hypothesis that protein networks could play a role analogous to nerve circuits in cells [29]. On the basis of this new conceptual framework, we propose that the interplay of light emission and reception in the unicellular world may, as in animals with nervous systems, also play a role in intra- or inter-species signalling.
1.1. Bioluminescence
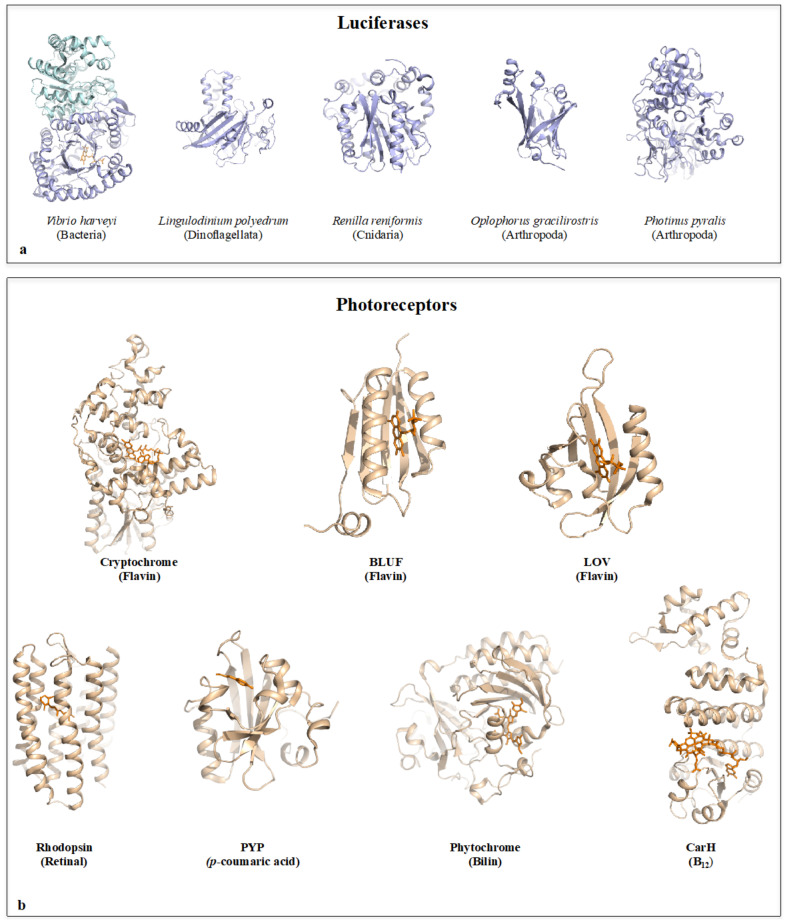
Bioluminescence, one of the most fascinating manifestations of life, is most often observed in the marine environment [1,2,3,4,30,31] and is shared by many taxa including bacteria [32], unicellular eukaryotes such as dinoflagellates [33,34], radiolarian [35,36] and vertebrates [37]. Light-emission is usually produced by the catalytic oxidation of various substrates by luciferase enzymes that have been isolated in many different taxa such as bacteria [32,38], dinoflagellates [39,40,41], cnidarian [42], arthropods [43,44,45,46,47,48] and fungi [49,50,51,52,53]. However, their molecular structures and mechanisms reveal that they display distinct protein folds and have evolved independently and convergently numerous times from non-luminous enzymes [54,55,56] (Figure 1a). The fact that many organisms have independently developed bioluminescence suggests that it has many functional advantages in the different ecosystems where it is observed. In 1983, Hastings proposed four functional categories of bioluminescence: (i) defense (ii) offense (iii) communication and (iv) metabolic (light emission is a by-product) [57]. Currently, it is widely accepted that the intra- or inter-species communication function of bioluminescence is reserved for visual animals with a nervous system capable of integrating and processing light stimuli: “In order for luminescence to be used for communicating information between two conspecifics, at least one must emit the light and the other must be able to detect and react to the light. In most known cases both emit and perceive light. Intraspecific communication is primarily the realm of organisms with well-developed nervous and visual systems and complicated behaviour. These include crustaceans, cephalopods, fishes and a few annelids” [58].
Figure 1.
Structures of luciferases and photoreceptors. (a) the structures of the luciferases are depicted with blue cartoons. At the periphery, the various types of photoreceptors are represented with beige cartoons. The corresponding pdb codes for the luciferases are: Vibrio harveyi: 3fgc, Lingulodinium polyedrum: 1vpr; Oplophorus gracilirostris: 5b0u; Photinus pyralis: 1lci; Renilla reniformis: 2psh. (b) The structures of the main classes of photoreceptors are represented by wheat cartoons. The pdb codes for the photoreceptors are: Cryptochrome: 1np7; BLUF: 1yrx; LOV: 1g28; Phytochrome: 2o9c; CarH: 5c8f; PYP: 2phy; sensory rhodopsin: 1jgj. The co-factors and pigments are indicated and represented with orange sticks if present in the structures of both luciferases and photoreceptors. The pigments are written in parentheses.
The functions of light-emission in the unicellular world are still subject to much speculation and remain to be explored. Surprisingly, the role of bioluminescence in communication between unicellular organisms lacking visual and nervous systems has, to our knowledge, never been mentioned. In bacteria, for example, while the lux operon has become one of the best characterised bioluminescent systems for almost half a century [38,59,60,61,62], the functions of bacterial luminescence remain an open question. It has been proposed that light-emission evolved from ancient detoxifying mechanisms [63,64], that it may play a role in stimulating DNA repair [65,66] or that it constitutes a visual attractant for zooplankton (copepods) and fishes that could provide them with a growth medium and dispersal [67]. Moreover, many bioluminescent bacteria have developed complex symbiotic relationships with animals in which the bacterial light emission may provide a reciprocal selective advantage [68,69,70,71]. In the protist world, it is generally accepted that dinoflagellate bioluminescence mainly evolved as an anti-predation system [34,72,73,74,75,76,77]. However, in the light of recent data on unicellular photoreception, we argue that many aspects of bacterial and dinoflagellate bioluminescence point to additional functions.
1.2. Perception and Responses to Light in the Unicellular World
The light-induced behavioural responses of microorganisms and fungi are mediated by a great variety of photoreceptors associated with diverse functions with gradual complexities [5,6,11,12,78,79,80,81,82,83,84,85,86,87]. While non-directional photoreception monitors variations in the light intensity, directional photoreception and low-resolution vision are used for orientation and habitat selection. High resolution vision constitutes the most elaborate system that enables complex behaviours such as prey detection and hunting [78,79,88]. In these behavioural responses to light, the photon activation of the light harvesting pigment is converted to an electric or chemical signal that is transmitted to the motor apparatus of the cells through modular cell signalling pathways such as the two component systems in bacteria [89,90,91,92] or phosphorylation cascades in eukaryotes [93,94]. Various systems have convergently evolved to provide accurate light-sensing such as the cellular micro-lenses of Synechocystis cyanobacteria used as micro-optics to sense light direction [95] or the elaborate eye-like ocelloids of certain dinoflagellates [96]. The best-characterised photoreceptors belong to different protein families and are associated with distinct chromophores [97] (Figure 1b). Despite their diversity, they share a similar property, which converts the ultrafast and local photo-activated changes in their pigment structure into long-lived changes in the receptor protein that are transmitted to other proteins in the signalling chain of the organisms [6,11]. The rhodopsin and photoactive yellow protein (PYP) use the photoisomerization of a C=C bond in retinal or the p-coumaric acid, respectively. Phytochromes use bilin, an open tetrapyrrole and the LOV proteins, cryptochromes and BLUF proteins use flavin mononucleotides (FMN) as a chromophore [98].
1.2.1. The Great Diversity of Bacterial and Eukaryotic Photoreceptors
Rhodopsin consisting of seven trans-membrane α-helices belongs to the vast family of G-protein-coupled receptors [99,100,101,102,103,104,105]. The light inducing isomerization of the 11-cis-retinal is coupled to a cascade of events that ultimately acts as a switch to relay the signal to G-proteins [106,107], guanylate kinases [101,108] or phosphodiesterase [102] that triggers various intracellular responses. Sensory rhodopsins that are responsible for light signalling and photo-behaviours have been found in archaea and bacteria [109,110,111,112].
The PYP adopt a Per-ARNT-Sim (PAS) domain [113,114,115,116,117,118], a protein sensor superfamily for a vast range of stimuli, from light sensing to ligand binding [119]. The trans to cis photoisomerization of its p-coumaric acid chromophore proceeds with a “volume-conserved” mechanism that has been followed by time-resolved crystallography [120,121]. This primary-event induces small structural changes and is associated with small structural rearrangements associated with charge delocalization around the chromophore [122].
LOV (Light, Oxygen or Voltage) domains are photosensors that also adopt the PAS domain structure and use the FMN as a chromophore in bacteria, archaea and certain eukaryotes (plants and fungi) [7,80,123,124,125,126,127,128,129,130]. They can be found associated with various effectors in bacteria such as histidine kinases, proteins involved in the synthesis of cyclic guanosine monophosphate (cGMP), STAS (Sulfate Transporter and Anti-Sigma factor antagonist) and helix-turn-helix and have been found to be associated with other sensor domains [131]. Several signalling mechanisms have been proposed based on their dark and light-activated structures such as helix Jα unwinding [132,133] or dimerization [134].
BLUF, bacterial and eukaryotic Blue Light receptor Using Flavine adenine dinucleotide (FAD) have a modular architecture comprising a 150 amino acids (aas) receptor domain with a ferredoxin-like fold that can be connected with many different effectors whose activity is modulated by the BLUF domain in response to light [135,136,137,138,139]. The BLUF domains contains a flavin adenine dinucleotide chromophore. They can be found as single photosensory domains involved in light-regulated protein interactions [140] or in multidomain proteins fused to light-regulated transcriptional effectors [141,142], phosphodiesterases [135] or adenylyl cyclases [143,144]. The BLUF domain binds to the FAD/FMN/RF pigment non-covalently for the properties of the isoalloxazine to absorb blue light. In BLUF, a photo-induced proton-coupled electron transfer (PCET) initiates a rearrangement of the hydrogen bonds around the flavin cofactor after illumination that is transmitted to the surface of the receptor and leads to the activation of the various effectors.
Cryptochromes adopt a Rossman-like fold similar to photolyases [145,146,147,148,149,150,151,152,153,154,155,156,157,158,159,160] involved in functions ranging from DNA repair to the blue-light regulation of growth, development and circadian rhythms [161]. Cryptochrome photoreception is based on blue light-induced interconversion between several redox states of flavin as a chromophore [162]. While the role of tryptophan triad is controversial [163], the light-induced oligomerisation of the plant cryptochrome is an accepted mechanism for light-regulation interactions with various signalling partners [164,165,166].
Phytochromes are modular multi-domain red/far-red photosensory proteins that share conserved PAS, GAF and PHY core photosensory domains that adopt a knotted structure [167] and bind covalently to a linear tetrapyrrole chromophore (bilin, phytochromobilin or biliverdin) in a broad range of organisms such as bacteria, protists and plants [168,169,170,171]. Photoconversion triggers structural changes in the dimer interactions and the refolding of a “tongue” loop modulates the activity of the C-terminal “output” domains [172,173].
Light-sensitive ion channels involved in phototaxis by depolarizing the plasma membrane have been found in some eukaryotic unicellular algae (chlorophytes) [174,175]. These channels, such as the rhodopsin channel (737 aas), evolved from bacterio-rhodopsin consisting of seven transmembrane segments covalently linked to a retinal chromophore [176]. Light absorption induces the isomerization of the retinal chromophore, which by turning causes a set of conformational changes leading to the opening of a pore that allows ions to pass through [177,178,179]. In Cryptophyte algae, there are also cation-conducting rhodopsin channels that specifically conduct anions [180,181]; this group of channels is more structurally related to haloarchaeal rhodopsins and has different functional properties [182]. Light-sensitive channels have also been found in nucleocytoplasmic large viruses that infect plankton [183].
However, it has recently been shown that other types of receptors such as the chemoreceptors Aer, Tar, Tsr, Tap and Trg also mediate bacterial responses to blue-light [184]. In addition, a new photosensor family uses coenzyme B12 for sensing blue and green-light and regulating gene expression [185,186,187,188,189,190,191,192]. A bacteriochlorophyll usually involved in photosynthesis with bacteria has been found as a photosensor in the retina of dragon-fish Malacosteus niger living in the deep-sea [193]. Moreover, ChrR regulators mediate blue-light responses through the intermediate stage of the production of ROS in V. cholera [194]. These recent findings have considerably extended the landscape of photoreception in the microbial world. Interestingly, some photoreceptors and luciferase share the same pigments and therefore their metabolic pathways. For example, the flavin and bilin chromophores are found in the active sites of luciferases of some bacteria and dinoflagellates to catalyse the photon emission. Conversely, as found in many bioluminescent organisms, the pigment of photoreceptors may be not synthetised by the organism but is acquired from the external environment [195]. To date, the ultimate stage of unicellular high-resolution vision is achieved by eye-spots or ocelloids organelles observed in some dinoflagellate or Chlamydomonas species [96,155,196,197,198,199,200].
1.2.2. Light and Unicellular Behaviours
Since the first observation of phototactic bacteria by Engelmann in 1883 [201], many studies have shown that light influences both the phototrophic and heterotrophic bacterial behaviours and physiology [8,9,11,202,203]. Phototaxis, the positive or negative displacement along a light gradient, displays various degrees of light-sensitivity among unicellular organisms including bacteria, archaea and eukaryotes [10,12,79,88,204,205,206]. Thus, very soon in the evolution, complex systems have emerged to accurately perceive and respond to light. Some phototactic archaea and cyanobacteria use photosensory rhodopsins to sense the intensity of light [112,207]. However, it has recently been shown that cyanobacteria represent the smallest and oldest organisms to use a kind of camera eye micro-optic for sensing light direction: individual Synechocystis cells directly sense the position of the light source through their own cells that act as micro-lenses [95,208] through cyanobacteriochromes [209,210]. Light can also induce collective behaviours in photosynthetic bacteria such as Rhodospirillum centenum [211] and other cyanobacteria [212,213].
In addition to phototaxis, light induces various physiological and behavioural responses in both photo and heterotrophic bacteria through their diverse photoreceptors [214]. This includes, for example, the light modulation of virulence, cell attachment, cell growth or biofilm formation [215,216,217,218,219]. Light plays an important role in modulating the bacterial behaviours and host–symbiont relationships in the rhizosphere [220]. Interestingly, the important effect of blue-light on bacteria has led to light-based anti-infective strategies being conceived [221,222,223,224]. However, particularly relevant for the present review is the finding that deep-sea bacteria that usually live in the non-photic zone possess photoreceptors and respond to light [225,226]. While it has been proposed that these deep-sea chemoreceptors may help to switch the behaviours between a planktonic and a benthic lifestyle, this finding may also support the hypothesis that bacteria perceive the bioluminescence of other deep-sea organisms, including themselves, which is the unique source of light in deep-sea sediments.
Light also modulates the phototaxis and circadian rhythms of multiple eukaryotic algae [83,161,227,228] but also induces various behavioural responses from other, non-photosynthetic protists such as dinoflagellates [198,229,230,231,232,233,234,235,236], ciliates [237,238,239]; affects biofilm formation by Chlorella vulgaris [240]; controls the morphogenesis in green plants and stramenopiles [241,242] and the sexual cycle in Chlamydomonas [243]; and influences the phototactic swimming in sponge larvae [244]. Interestingly, light also mediates cross-kingdom communication: it has recently been demonstrated that the green fluorescence of the cnidarian host attracts symbiotic algae [245] (Table 1).
Table 1.
Influence of light on dinoflagellate behaviours.
| Title | Light Response | Species | References |
|---|---|---|---|
| Diurnal rhythms | |||
| Bioluminescence is under circadian control | Gonyaulax polyedra | Sweeney and Hasting, 1957 [349] Hasting and Sweeney, 1958 [350] |
|
| Roenneberg and Hasting, 1988 [351] | |||
| Roenneberg and Deng, 1997 [352] | |||
| Roenneberg and Taylor, 1994 [353] | |||
| Morse et al., 1989 [354] | |||
| Krasnov et al. 1980 [339] | |||
|
Pyrodinium bahamense
Gonyaulax polyedra Pyrocystis lunula |
Biggley et al. 1969 [334] | ||
|
Mixed dinoflagellate communities:
Gonyaulax spp. Alexandrium spp. Ceratium fusus Pyrocystis spp. Protoperidinium spp. |
Marcinko et al., 2013 [355] | ||
| Multiple bioluminescent species | Backus et al. 1961 [356] Yentsh et al., 1964 [357] |
||
| Photoenhancement of bioluminescence | Ceratium fusus | Sullivan and Swift, 1995 [236] | |
| Photoinhibition of bioluminescence |
Alexandrium catenella, A. acatenella, A. tamerensis |
Esaias et al., 1973 [344] | |
| Gonyaulax polyedra | Hamman and Seliger, 1982 [345] | ||
| Ceratius fusus | Sullivan and Swift, 1994 [346] | ||
| Protoperidinium spp. | Buskey et al., 1992 [358] | ||
| Laser-induced bioluminescence | |||
| Artificial red flashes induce bioluminescence (wavelength 585 nm) |
Pyrocystis lunula | Hickman and Lynch, 1981; Hickman et al. 1982 [340,341] | |
| Detailed study of flash-induced bioluminescence | Pyrocystis lunula, Pyrocystis fusiformis, G. polyedra | Sweeney et al., 1983 [342] | |
| Gene expression induced by light | |||
| Xanthorhodopsin subgroup II: proton pump for energy supplement during light-limited photosynthesis | Prorocentrum donghaiense | Shi et al., 2015 [359] | |
| Oxyrrhis marina | Guo et al., 2014 [360] | ||
| A few genes are only transcriptionally regulated by light | Symbiodinium kawagutii | Zaheri et al., 2019 [361] | |
| Light-transcriptional control of 9.8% of the genes | Karenia brevis | Van Dolah et al., 2007 [362] | |
| Different photoresponses according to species | Dissodinium lunula, Pyrocystis fusiformis, Pyrocysti noctiluca | Swift and Meunier, 1976 [363] | |
|
Gonyaulax tamarensis
Heterocapsa trique |
Anderson and Stolzenbach, 1985 [364] | ||
| Motility, phototaxis | |||
| Swarming, diel vertical migration | Gonyaulax polyedra | Roenneberg et al., 1989 [365] | |
| Light induces “stop-responses” or “shock reaction” (cessation of movement) | Girodinium dorsum | Forward and Davenport, 1968; 1970 [348,366] | |
| Stop-response followed by positive phototaxis | Gymnodinium splendens | Forward, 1974 [233] Ekelund and Björn, 1987 [347] |
|
| Positive-Phototaxis (Comparison of phototaxis in dinoflagellate species with and without eyespots) |
Scrippsieiia hexapraecinguia
Peridlnium foliaceum Atexandrium hiranoi Gymnodinium mikimotoi |
Horiguchi et al. 1999 [367] | |
| Photoresponse of an heterotrophic dinoflagellate in three wavelengths: 450 nm, 525 nm and 680 nm) mediated by rhodopsin | Oxyrrhis marina | Hartz et al., 2011 [231] | |
| Modulation of phototactic and stop response by wavelengths | Girodinium dorsum | Hand et al., 1967 [368] | |
| Support the hypothesis of a two-pigment system in phototactic response | Girodinium dorsum | Forward, 1973 [232] | |
| Peridinium gatunense | Häder et al., 1990 [234] | ||
| Effect of light on symbiosic species | |||
| Blue-light has a dominant effect on the cell cycle | Symbiodinium spp. | Wang et al., 2008 [369] | |
| Physiological adaptation to light gradient in corals | Symbiodinium spp. | Wangpraseurt et al., 2016 [370] | |
| Green light facilitates symbiont capture by coral larvae | Symbiodinium spp. | Hollingsworth et al., 2005 [371] | |
| Symbiodinium species are specifically attracted by the green fluorescence emitted by its coral host | Symbiodinium spp. | Aihara et al., 2019 [245] | |
| Variable phototaxis responses according to different Symbiodinium species | Symbiodinium spp. | Yamashita, 2021 [372] | |
| Optical feedback loop involving dinoflagellates and coral in coral bleaching | Multiple species | Bollati et al., 2020 [373] | |
| Eye-spot and spectral sensitivity of phototaxis | Kryptoperidinium foliaceum | Moldrup and Garl, 2012 [230] | |
1.3. Communication, Collective Behaviours and “Cognition” in the Unicellular World
In his pioneer work “the psychic life of microorganisms” published in 1889 [246], Alfred Binet described the great behavioural richness of unicellular organisms that he observed under the microscope. He listed all the organs of locomotion and the sense organs that can be observed in protists and noticed their analogies with the corresponding organs in metazoans. He carefully described the movements, the complex and the species-specific behaviours of microorganisms and noticed, as Engelmann did in 1883 [201], the phototactic properties and sensitivity of bacteria to oxygen tension. A few decades later, in the United States, Jennings and Loeb also looked at and noticed the richness of the behaviour of what they called the lower organisms [247,248]. These works led to the realisation, which lasted for more than a century, that microorganisms, although lacking a nervous system, were capable of performing complex individual and collective behaviours. Today’s knowledge of the behavioural landscape of microorganisms goes far beyond what Binet could have imagined: microorganisms display sensitivities to diverse stimuli, they communicate with each other, they can change their behaviour according to their individual or collective way of life and are able to accomplish complex tasks such as associative learning and finding their way into a maze. The detailed knowledge of molecular sensors and signalling circuits now makes it possible to decipher how cells perceive, integrate and react to external and internal signals.
1.3.1. Bacterial Communication and “Cognition”
Firstly, many studies have shown that unicellular organisms communicate with each other using a wide variety of chemical or physical signals. The best known system is quorum sensing that allows bacteria and other microorganisms to inform each other about their density and trigger and synchronise collective behaviour [249,250,251,252,253,254]. However, while chemical communication has been the first to be characterised, intra- and inter-cellular communication can also be established in a physical way by electrical, electromagnetic and acoustic waves or mechanical contacts [255,256,257,258,259,260,261,262,263,264,265,266]. For example, a fascinating study showed that biofilms formed by Bacillus subtilis could attract and incorporate motile cells of different species through the ion channel-mediated electrical signalling [259]. Other studies have provided evidence that voltage-gated signalling mediates long-range communication between bacteria [260,267].
Secondly, studies on bacterial chemotaxis have led to the realisation that bacteria can make decisions based on the integration of multiple signals and led the pioneers of chemotaxis studies to propose an analogy between these phenomena and neurobiology in the early 1980s [268,269]. Chemotaxis is the ability to choose its orientation according to a source of attractive or repulsive substances [270]. It integrates the perception of external or internal signals through two-component system signalling [89]. In these systems, a “sensor” protein auto-phosphorylates in response to a specific signal coming from specific receptors and transfers the phosphoryl group to a “response regulator” protein that carries out a cellular response [271,272,273,274,275,276]. These studies soon revealed “cognitive” properties of bacteria, showing that they could indeed make decisions in complex situations [270,277], amplify signals, show habituation faculties and retain the memory of past situations [278,279]. Later, on the basis of genomic studies, Michael Galperin demonstrated that bacteria display distinct behavioural types according to their genomes and ecosystems [271,280]. Today, many articles converge towards the universality of the notion of “consciousness”, “cognition” or “intelligence” in microorganisms [13,15,16,17,18,281,282,283,284,285,286].
1.3.2. Complex Behaviours in Unicellular Eukaryotes
More elaborate behaviours that are close to that of metazoans have been observed in unicellular eukaryotes. Habituation, the simplest form of learning that consists of attenuating the response and eventually ceasing to respond to a repeated stimulus that has proven not to be harmful was initially characterised in a mollusc, the aplysia [287]. Habituation has been recently observed in organisms lacking a nervous system [288]. More elaborate, associative learning establishes a link between the joint appearances of different stimuli and associates them. This faculty is the basis of what is known as Pavlovian conditioning. In the middle of the 20th century, Beatrice Gelber demonstrated that, as with Pavlov’s dogs, Paramecia could be conditioned to link two different stimuli [21]. Although initially contested, these fascinating studies have recently been rehabilitated [22]. Since these pioneering studies, growing evidence has shown that they are not exclusive to beings with a nervous system and that complex behaviours such as associative learning and the solving of complex tasks are observed in unicellular organisms [19,20,22,289,290,291,292].
2. Hypothesis: Bioluminescence Signalling in the Unicellular World
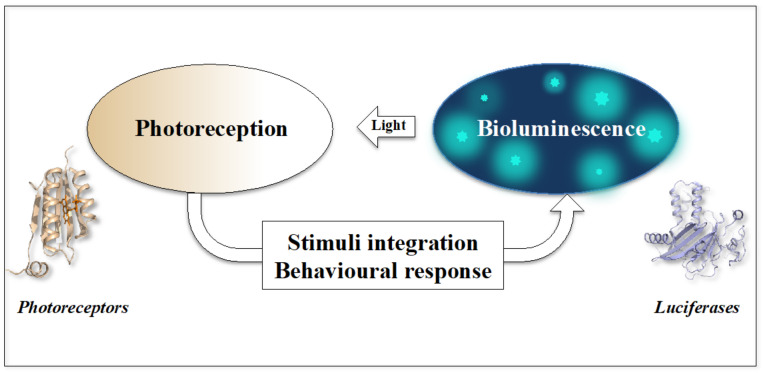
Whereas chemical communication has long dominated the literature about signalling between single-cell organisms, many studies have recently shown that cells can also exchange physical signals such as sounds, electrons or electromagnetic waves [255,256,257,258,264,266,293,294]. However, while the influence of ambient light on the behaviours of microorganisms is now well established, communication by light emitted by luciferases of bioluminescent unicellular organisms has not been previously addressed. Previous studies have brought experimental evidence that non-bioluminescent prokaryotic and eukaryotic unicellular organisms such E. coli and Paramecia communicate between themselves through the emission and reception of weak photons [264,295,296,297]. It is therefore likely that organisms that have evolved specific bioluminescent systems to emit more intense light and modulate light intensity communicate with each other or with other species through light. Thus, bioluminescence could also contribute to survival strategies involving light signalling within the unicellular world. Bioluminescence produced by bacterial or eukaryotic unicellular organisms is therefore expected to produce various kinds of intra- or inter-species behavioural responses in cells that possess photoreceptors or even more elaborate high-resolution vision such as eye spots or ocelloids [96,196,197]. Thus, we propose here that the interplay between bioluminescence and photoreception should be systematically explored from both an evolutionary and functional perspective such as, for example, in light-signalling within a single or among different species and taxa. Of course, bioluminescence is expected to exert an effect on other organisms in a particular ecological context protected from ambient-light: in the non-photic zones of the ocean, during the night, into the internal organs of animals or in the plant rhizosphere (Figure 2).
Figure 2.
Interplay between photoreception and bioluminescence in microorganisms. Schematic diagram of the feed-back loop between light emission and light reception. During the night, in the non-photic zone or in the rhizosphere (soil), light emission by luciferases (symbolised here by dinoflagellate luciferase) and light reception by photoreceptors (symbolised here by BLUF) has the potential to mediate intra- or inter-species light-communication.
2.1. Light Signalling in Bioluminescent Bacteria
Most of the bioluminescent bacterial species fall within the Gammaproteobacteria class and cluster phylogenetically in three families (Vibrionaceae, Shewanellaceae and Enterobacteriaceae) which all carry the canonical highly conserved lux operon [32,62]. Since its first identification more than 40 years ago, it has been shown that the lux operon adopts a “canonical” luxCDAB(F)E(G) organization that has been systematically observed in genomes of bioluminescent bacterial. The luxA and luxB genes encode the α and β luciferase subunits that emit light by the oxidation of FMNH2 and a long chain aldehyde [38,61]. LuxC, D and E proteins form a fatty acid reductase complex that synthesises the long chain aldehyde substrate. The recent discovery of new lux operon organisations in other bacterial taxa has opened new evolutionary and functional perspectives on bacterial bioluminescence [60].
Bioluminescent bacteria occupy a large variety of habitats and lifestyles [62]. It has long been thought that the genes involved in bioluminescence catalysis and control have co-evolved to adapt to various ecological niches [298,299,300]. However, many aspects of the evolution and functions of bacterial luminescence remain enigmatic. It is currently thought that the bacterial emission of light is essentially intended for the visual systems of their infected or symbiotic hosts (see above). However, recurrent features indicate that light plays a critical role in host–symbiont communication and in the formation of bacterial biofilms on host tissues by animal photoreception systems other than visual systems. For example, Vibrio fischeri establishes a complex symbiotic relationship with the squid Euprymna scolopes in which the bacterial luminescence is perceived by photoreceptors within the light organ of the host [70,301]. Thus, V. fischeri cells defective in light production are not retained for symbiosis and are expelled from the organ [302,303]. In addition, the control of bioluminescence by the regulation of the lux operon has also revealed some surprises. While it was initially thought that the lux operon is dependent on cell density through quorum sensing [251], it has recently been found that in some species, light-emission is independent of quorum sensing [304] or controlled by other factors depending on the environmental conditions [305,306,307,308,309,310].
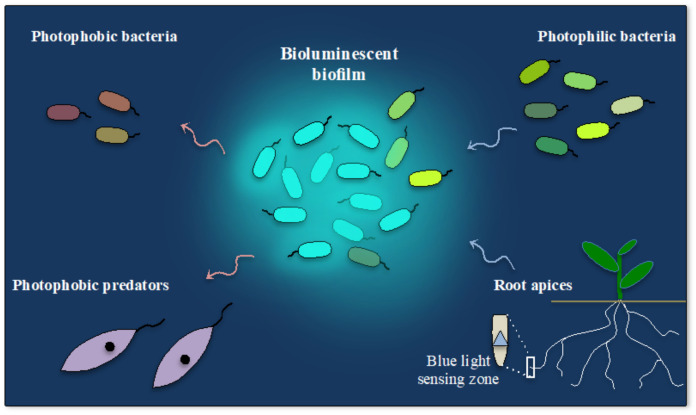
In Photorhabdus luminescens, the only terrestrial luminescent bacteria, light-emission is specifically associated with a specific symbiotic stage of its life cycle. P. luminescens switches between different phenotypic traits associated with a symbiotic life in the gut of the nematode Heterorhabditis (primary 1° cells), the infection of insect larvae (2° cells) and the plant root in the rhizosphere [311,312,313]. Only the primary cells (1°) that correspond to cell clumping and the symbiotic stage with the nematode is bioluminescent suggesting that light emission is required in communication with its host. However, little is known about its bioluminescence during the rhizosphere life stage, since this life form has only recently been discovered [313]. Knowing that many soil bacteria display photoreceptors, it is possible that the bioluminescence of P. luminescens plays a role in its association with the rhizosphere bacterial community. In addition, it has recently been found that other bacterial species possess non-canonical lux operons and are probably luminescent [60]. Some of these species are also frequently associated with the rhizosphere, thus reinforcing the idea that light may play an important role in cell-to-cell communication among the different species of the rhizosphere. Interestingly, several studies have shown that plant roots also possess multiple photoreceptors that sense different wavelengths including blue-light and display various behavioural and physiological responses to light [314]. This suggests that the bioluminescence of the bacteria that live in the rhizosphere may not only influence other root associated bacteria but also affect the growth and behaviour of root apices (Figure 3). Furthermore, the existence of several species of bioluminescent earthworms [315,316,317] reinforces the idea that the emission and perception of light in soils could play an ecological role that would be interesting to elucidate in more detail.
Figure 3.
Light signalling in the bacterial world. Schematic representation of the hypothesis that bioluminescent bacteria may shape the bacterial communities in their luminous biofilm (represented by a cyan halo). They can attract different species of photophilic bacteria (green) or repell photophobic bacteria (brown) and eukaryotes (violet). Thin, wavy arrows symbolise cell motility. Light emission may also influence the plant roots that possess blue-light photoreceptors (blue triangle) (see [314]).
In addition, the biofilm formation of bioluminescent bacteria isolated from the marine fish gut has recently been described [318]. This study shows that the bioluminescent bacteria Kosakonia cowanii forms cross-species biofilms with Gram-Positive coccoid bacteria. Does light emission play a role in this association in the gut? An important hint that supports our hypotheses is that bacteria from the deep-sea, and therefore living in the non-photic zone, possess photoreceptors that mediate light-behavioural responses. For example, it has been shown that some deep-sea species possess either PYP or bacteriophytochrome photoreceptors that regulate biofilm formation or growth [225,226]. While these studies have proposed that these photoreceptors contribute to the regulation of their benthic and pelagic lifestyle, they may also make bacteria sensitive to bioluminescence in the non-photic zone. This suggests that bioluminescence in the deep-sea could play a signalling role in the formation of biofilms or modulate the behaviours of other benthic bacterial species.
Since photons can be perceived by the multiple photoreceptors found throughout the tree kingdoms of life, bioluminescent bacteria may also have evolved a selective advantage through light-signalling with both prokaryotic and eukaryotic unicellular organisms. They could for example repel photophobic predators such as certain ciliates [239] but they could also contribute to the selection of the bacterial consortium around them: light may attract photophilic or repel photophobic species (Figure 3). Light emission and perception may therefore contribute to the shaping of bacterial communities and their symbiotic associations with various eukaryotic organisms. Thus, light signalling may be a general property of the bacterial collective way of life and could mediate their symbiotic associations with animal, fungi or plants [71,214,319]. Light communication could for example contribute to the formation of algae/bacteria blooms [320] such as the spectacular milky sea, whose bioluminescence can be observed from space [321,322] or bioluminescent blooms that occur in the deep-sea [323]. Thus, following on from Seliger’s and O’Kane’s works [299,300], it would also be interesting to study the co-evolution of genes involved in bioluminescence and photoreception in bacteria, or even the co-evolution of these systems between symbiotic organisms.
2.2. Do Dinoflagellates Converse by Light Signals?
Bioluminescent dinoflagellates, which are the major contributor to stimulated light-emission in the upper ocean, are currently the best-characterised luminescent unicellular eukaryotes, both on a molecular and ecological scale [33,34,74]. Among the 1555 species of dinoflagellates, there are ~70 bioluminescent species, including both autotrophic and heterotrophic species, occupying diverse oceanic niches [33,34,324]. The bioluminescence of dinoflagellates is part of numerous excitable responses to a variety of physical or chemical stimuli leading to complex individual or collective behaviours. Light emission occurs in specific organelles, the scintillons. They consist of dense vesicles of about 0.5–0.9 μm in diameter that contain the luciferase enzyme (LCF), the luciferin substrate and in some species, a luciferin binding protein (LBP) [325,326,327]. Mechanical stress on the cell membrane triggers an action potential on the vacuole membrane of their associated scintillons through a mechanotransduction pathway involving G-proteins and calcium signalling [328,329,330,331]. The action potential opens the voltage-gated proton channels and induces a rapid and transient acidification of the scintillons [332] where the catalytic oxidation of luciferin, a chlorophyll-derived open tetrapyrrole, by pH-regulated luciferases produces light flashes [40]. The emitted light has a wavelength in the range of 474–476 nm with a wide range of duration and intensities depending on the species. While the large dinoflagellate species such as P. noctiluca (~ 350 μm) or P. fusiformis (up to 1000 μm) can emit long (200–500 ms) and bright flashes (from 3.7 × 1010 to 6.5 × 1014 photon cell−1), smaller cells such as L. polyedra (~20–40 μm) emit brief (~100 ms) and dim flashes (from 3.1 × 107 to 1.2 × 108 photons cell−1) [324,333,334]. From membrane deformation to light emission, the overall bioluminescent response is rapid and can take only 12 to 20 μs [72,328]. The bioluminescence response threshold or flow sensitivity also depends on the size and morphological features of the different species [335].
Although these phenomena are well described from a mechanistic point of view, there are still many points to be clarified, in particular with regard to the ecological function of dinoflagellate bioluminescence. Overall, the current paradigm is that dinoflagellate bioluminescence has evolved as an anti-predation system. Three major hypotheses that are not exclusive have been considered and tested experimentally (reviewed in [33,34,58,324]): (i) the startle response, (ii) the burglar alarm and the (ii) aposematic warning. The first hypothesis proposes that the emission of light protects prey by frightening the predators. In the “burglar alarm” hypothesis, it is thought that the bioluminescence emitted in the presence of an herbivore (predator of the dinoflagellates) provides a selective advantage by attracting and allowing a carnivore (predator of the herbivore) to perceive to capture the herbivore, to the benefit of the dinoflagellate population. In the aposematic warning, light is used as a “grazing deterrent”, a lure that signals to predators that the prey may be dangerous, toxic or has unpalatable traits. While several experimental studies support some of these hypotheses [75,333,336] some characteristics of dinoflagellate bioluminescence may suggest that it could also have other functions. For example, some strains of Noctiluca scintillans off the west coast of the USA have lost their bioluminescence but are still able to form dense populations despite the presence of predators, thus questioning the ecological role of bioluminescence in this species [337]. Moreover, some studies have reported that many species of dinoflagellates can spontaneously emit light flashes, without being mechanically stimulated [334,338,339]. Furthermore, 30 years ago, three studies indicated that artificial light flashes on dinoflagellate cultures could induce their bioluminescence [340,341,342] (Table 1). These observations therefore call for a closer look at the inter-relationships between the ability of dinoflagellates to emit and perceive light and their possible functional consequences.
2.2.1. Photoreception and Light-Induced Behaviours in Dinoflagellates
In addition to their bioluminescent capabilities, dinoflagellates can perceive and distinguish between different light wavelengths and can exhibit a number of behavioural and physiological responses to light exposure (Table 1). Circadian rhythms [161,343], whose clock is generally light-regulated, control many functions such as bioluminescence, cell-cycle, collective behaviours and daily vertical migration along the water column. For example, bioluminescence is generally inhibited during the day and its intensity during the dark phase may depend on the amount of illumination during the previous day. However, a distinct phenomenon, the “photoinhibition” of bioluminescence has also been observed: this consists of the transient suppression of stimulated bioluminescence when cells are exposed to intense light [344,345,346]. Conversely, some studies have shown that artificial laser flashes may induce the light-emission of several dinoflagellate species such as P. lunula, P. fusiformis and G. polyedra [340,341,342]. Many species also display positive or negative phototactic behaviours at different wavelengths [12,230,232] (Table 1). The influence of light on the mobility of symbiotic species such as Symbiodinium sp. also plays an important role in symbiotic relationships with corals. It has been shown, for example, that some Symbiodinium species are specifically attracted to the colours emitted by the fluorescent proteins of their coral hosts. In contrast, a stop-response, the abrupt cessation of movement induced by intense light was also observed in Gyrodinium dorsum [347,348].
How do dinoflagellates perceive light? Similar to most of the planktonic organisms [78,374,375], dinoflagellates possess various kinds of photoreceptors and some species possess elaborate vision systems such as eye-spots or ocelloids [96,199,376]. Identified in many genomes of distantly related species, rhodopsin is considered ubiquitous in dinoflagellates [377,378,379,380,381,382]. For example, in the genome of O. marina, two forms of rhodopsins have been identified: proteorhodopsins related to bacteriorhodopsins [107,383,384], which probably function as proton pumps and sensory rhodopsins [360,385]. It has been proposed that proton pump proteorhodopsins contribute to energy resources and compensate photosynthesis in light deprived environments [105,359,386]. On the other hand, sensory rhodopsins [111,112] are more likely to have a function in photoperception and signalling. Interestingly, rhodopsins have been also found in the retinal body of Erythropsidinium eye-spots [376]. Knowing that rhodopsins found in the eye-spot membrane of Chlamydomonas and Volvox mediate phototaxis [387] or complex light-behavioural responses in Peranema [235], it is likely that they play a similar role in dinoflagellates. Supporting this idea, it has been shown that the outer cell membrane rhodopsins of the heterotrophic dinoflagellate O. marina, mediate the photosensory response to detect algal prey based on chlorophyll autofluorescence [231].
A cryptochrome blue-light receptor belonging to the CRY-DASH family has also been identified in Karena brevis [388]. While cryptochromes are generally known to be involved in circadian control [155,156], this family of photoreceptors may also play a role in the phototactic behaviour of sponges [244]. This opens interesting perspectives on other potential functions of cryptochromes in dinoflagellates.
Although phytochromes are generally absent from dinoflagellates, transcriptomic study has also found a phytochrome gene in the diatom symbiont of the dinotom Durinskia baltica [169,389,390]. The most spectacular vision systems observed in unicellular organisms are the eye-spots and ocelloids that have evolved in Erythropsidinium and Warnowia [96,376,391]. They have subcellular analogues to cornea, lens, iris and retina similar to vertebrate eyes that evolved from mitochondria and plastids. They are thought to provide a cellular high-resolution vision and allow the cells to react to the intensity and direction of light.
2.2.2. Excitability, Stimuli Integration and Complex Behaviours in Dinoflagellates
In addition to acute light-perception, growing evidence suggests that eukaryotic unicellular organisms including planktonic ones are able to integrate multiple chemical or physical stimuli to produce appropriate behavioural responses in their fluctuating environments. Many studies have reached the conclusion that the “behaviour evolved before nervous systems” [392] and the “neural aspects of biological systems were present in single-celled organisms” [393]. Interestingly, both bacteria and protists are excitable cells that perceive and propagate stimuli through voltage-gated ion channels similar to those of the animal nervous systems [267,394]. For example, a recent study showed that the diverse families of dinoflagellate Hv1 proton channels that were initially thought to control bioluminescence may also play diverse cellular roles [395]. Molecular networks in the cells have probably evolved to integrate the multiple stimuli and mediate complex behavioural responses such habituation, associative learning or decision-making, abilities suitable for the establishment of elaborate modes of communication [26,27,28,29,396].
Indeed, dinoflagellates exhibit a rich real-time behavioural and morphological repertoire to adapt to their diverse and fluctuating environmental conditions [397,398]. For example, in addition to the high-resolution vision systems described above [96,376], some dinoflagellate species have developed complex organs such as a piston for efficient propulsion [198] or nematocysts for hunting [399] that require complex “sensorimotor” and coordination molecular processes. In bioluminescent dinoflagellates, excitable light emission is stimulated by mechanical forces that trigger an “action potential” and a subsequent signalling cascade leading to the activation of luciferase in the scintillon [34,72,328,331,332,400]. Such excitable responses share both mechanistic and functional analogies with that of animals that possess nervous systems [401,402,403] and interestingly, the dinoflagellate responses are modulated by the neuromediators or inhibitors of voltage gated channels found in animal nervous systems [404,405]. In addition, it has been shown that some dinoflagellate species change their rheotactic behaviours, actively diversify their swimming strategies, form long multicellular chains or have the capacity to adopt compensating morphological countermeasures in response to variations in hydrodynamic environments [397,406,407,408]. Dinoflagellates also display collective and concerted behaviours such as the transition between life cycle stages during blooms or the formation of temporary cysts for escaping parasite infections [409,410]. Heterotrophic species such as Oxyrrhis marina display complex feeding behaviours such as prey discrimination and preference [411,412]. These studies that reveal the elaborate cognitive capacities of dinoflagellates are conducive to the consideration of new functions for their bioluminescence.
2.2.3. Photosensing of Bioluminescence: Emitting Light for Signalling
Knowing firstly that dinoflagellates possess various photoreceptors or even sometimes highly developed vision systems, secondly that some species have developed various types of responses to light stimuli and thirdly, that they can demonstrate highly elaborate individual and collective behaviours in the face of fluctuating situations in their environment, it seems likely that dinoflagellates could perceive, integrate and respond to the light stimuli emitted by their own bioluminescence, at night or in the non-photic zone (Figure 2). Interestingly, a recent study has shown that the photoreceptors of eukaryotic plankton communities are expressed predominantly at night, at a time that coincides with the maximum spontaneous (not externally stimulated) bioluminescence [413]. This study therefore supports the idea that these photoreceptors are dedicated to the perception of the bioluminescence that is most often under circadian control and emitted during the night [355,414,415].
On the other hand, some clues in the known properties of light emission in different dinoflagellate species may support this hypothesis. While it is now well known that the intense flashes produced by dinoflagellates are triggered by mechanical stress or flows to startle the copepod predators [75,76,328,329,330,405,416], it cannot be excluded that the modulation of flash intensities or durations may play more complex roles in intra- or inter-species communication. Moreover, some early works have shown that mechanical stress is not the only cause of light emission in dinoflagellates. For instance, dinoflagellates can spontaneously emit flashes or weak glowing without any apparent stress [334,338,339]. While it is thought that these spontaneous light emissions are stochastic, one cannot exclude the possibility that some cells voluntarily trigger these phenomena for a reason that is still unknown. Other works have also shown that dinoflagellate light-emission is not simply an all or nothing phenomenon: the intensity of the flashes can vary according to whether the induced flow is turbulent or laminar [417,418,419] or can depend on amplitude and the rate of cell wall deformations [420]. It is therefore possible that the intense flashes induced by stress contain more nuanced types of light emissions playing other functional roles which have not yet been investigated. The modulation of flash duration and intensity therefore also points towards the informational potential to communicate through light.
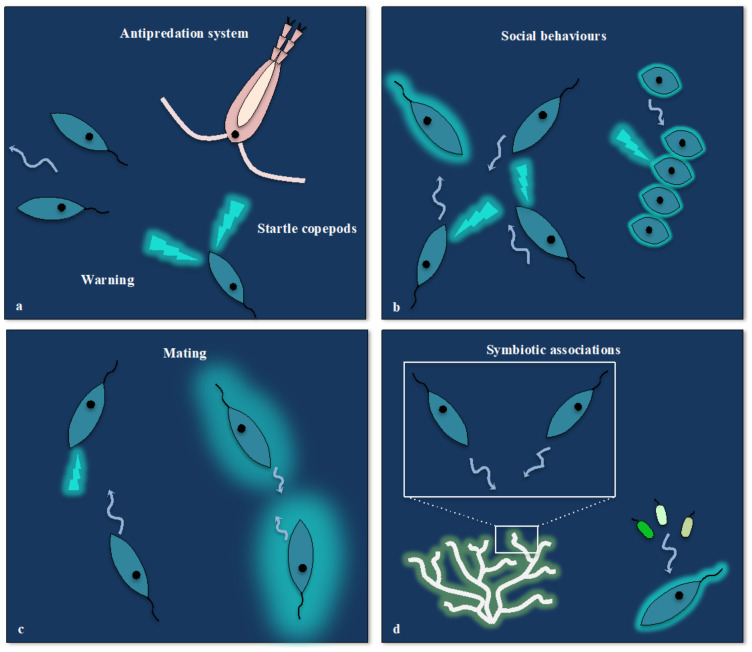
However, the emergence of communication during evolution is intimately linked to the concomitant development of a social life in various organisms. For example, communication through visual signals is widespread in the animals that display various degrees of social behaviours. Many properties of light such as colours, skin patterns, iridescence and bioluminescence have been developed by evolution for intra- or inter-species signalling [421,422,423,424,425,426,427,428]. While intra-species communication generally involves social interactions and mating, inter-species communication mediates symbiotic relationships or shapes ecosystem interactions. It turns out that many species of dinoflagellates have both a “social” and sexual life [429,430,431,432] that may require an animal-like “language”. One hypothesis could be, for example, that dinoflagellate bioluminescence displays some analogies with light-emission strategies used by various bioluminescent animal species to communicate [433,434,435,436,437,438,439,440,441]. Interestingly, complex collective behaviours involving communication for mating have been observed in other unicellular organisms such as ciliates [442,443,444]. It would therefore be interesting to investigate which types of light signals bioluminescent dinoflagellates emit “spontaneously” with modern technology using single-cell systems. In summary, dinoflagellates have the “cognitive abilities” and “social behaviour” that suggest that they could use light as a means of intra- and inter-species communication. Establishing a parallel between light emission in the macroscopic and microscopic world may bring some heuristic value for deciphering the still mysterious bioluminescence of unicellular planktonic organisms (Figure 4).
Figure 4.
Schematic representations of the multiple potential functions of dinoflagellate bioluminescence and photoreception. (a) The intense emitted flashes play an anti-predation role such as startling copepod predators. They can also alert other cells to danger and provide information about the location and the nature of predators (warning). (b) Flashes and glowing (cyan halo) can contribute to cell signalling in complex social behaviours such as blooms (left) or chain formation (right). (c) Glowing and the modulation of flash intensities and durations may contribute to communication for mating. (d) Light can also mediate symbiotic associations with coral (left) or bacteria (right).
3. Conclusions
Whereas chemical communication has long dominated the literature about signalling between microorganisms, many studies have shown that cells are also sensitive to physical signals such as sounds, electrons or electromagnetic waves. For example, ambient light is known to induce various responses in microorganisms through their large variety of photoreceptors. However, how bioluminescence is perceived and influences unicellular communication and behaviours remains to be explored. The functions of unicellular bioluminescence organisms are still enigmatic and it is thought that light-emission is mainly directed at visual animals, either as an anti-predator strategy, as for example in dinoflagellates, or in establishing symbiotic relationships in bacteria. In this short review, the interplay between bioluminescence and the photoreception of microorganisms has been examined in the light of numerous studies demonstrating their behavioural richness. We argue here that many aspects of bacterial and dinoflagellate bioluminescence point to additional functions and propose that similar to visual animals, feed-back loops between light-emission and reception could play multiple roles in intra- or inter-specific communication and participate in complex behaviour in the unicellular world. In addition, bioluminescence and photoreception could play a general role in the communication between symbiotic microorganisms and their plant or animal hosts.
Acknowledgments
We thank our colleagues T. Vannier, P. Hingamp, C. Vernette, S. Romac, K. Hartle-Mougiou, G. Gregori, C. Tamburini, S. Martini, P. Bonin, our students T. Antoine, E. Rozis and O. Di Valentin for interesting discussions and the reviewers’ advice for improving the quality of the manuscript.
Author Contributions
Conceptualization, Y.T. and M.L.; writing—original draft preparation, Y.T.; writing—review and editing, Y.T., M.L., M.V. and F.N.; supervision, Y.T., M.L., M.V. and F.N.; funding acquisition, Y.T. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by Modélisation du vivant–AAP 2021, CNRS and OCEANOMICS [ANR-11-BTBR-0008].
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Haddock S.H.D., Moline M.A., Case J.F. Bioluminescence in the Sea. Ann. Rev. Mar. Sci. 2010;2:443–493. doi: 10.1146/annurev-marine-120308-081028. [DOI] [PubMed] [Google Scholar]
- 2.Widder E.A. Bioluminescence in the Ocean: Origins of Biological, Chemical, and Ecological Diversity. Science. 2010;328:704–708. doi: 10.1126/science.1174269. [DOI] [PubMed] [Google Scholar]
- 3.Herring P.J. How to Survive in the Dark: Bioluminescence in the Deep Sea. Symp. Soc. Exp. Biol. 1985;39:323–350. [PubMed] [Google Scholar]
- 4.Wilson T., Hastings J.W. Bioluminescence. Annu. Rev. Cell Dev. Biol. 1998;14:197–230. doi: 10.1146/annurev.cellbio.14.1.197. [DOI] [PubMed] [Google Scholar]
- 5.Hegemann P. Algal Sensory Photoreceptors. Annu. Rev. Plant. Biol. 2008;59:167–189. doi: 10.1146/annurev.arplant.59.032607.092847. [DOI] [PubMed] [Google Scholar]
- 6.Kottke T., Xie A., Larsen D.S., Hoff W.D. Photoreceptors Take Charge: Emerging Principles for Light Sensing. Annu. Rev. Biophys. 2018;47:291–313. doi: 10.1146/annurev-biophys-070317-033047. [DOI] [PubMed] [Google Scholar]
- 7.Losi A., Mandalari C., Gärtner W. The Evolution and Functional Role of Flavin-Based Prokaryotic Photoreceptors. Photochem. Photobiol. 2015;91:1021–1031. doi: 10.1111/php.12489. [DOI] [PubMed] [Google Scholar]
- 8.Van der Horst M.A., Key J., Hellingwerf K.J. Photosensing in Chemotrophic, Non-Phototrophic Bacteria: Let There Be Light Sensing Too. Trends Microbiol. 2007;15:554–562. doi: 10.1016/j.tim.2007.09.009. [DOI] [PubMed] [Google Scholar]
- 9.Beattie G.A., Hatfield B.M., Dong H., McGrane R.S. Seeing the Light: The Roles of Red- and Blue-Light Sensing in Plant Microbes. Annu. Rev. Phytopathol. 2018;56:41–66. doi: 10.1146/annurev-phyto-080417-045931. [DOI] [PubMed] [Google Scholar]
- 10.Armitage J.P., Hellingwerf K.J. Light-Induced Behavioral Responses (‘Phototaxis’) in Prokaryotes. Photosynth Res. 2003;76:145–155. doi: 10.1023/A:1024974111818. [DOI] [PubMed] [Google Scholar]
- 11.Braatsch S., Klug G. Blue Light Perception in Bacteria. Photosynth. Res. 2004;79:45–57. doi: 10.1023/B:PRES.0000011924.89742.f9. [DOI] [PubMed] [Google Scholar]
- 12.Jékely G. Evolution of Phototaxis. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2009;364:2795–2808. doi: 10.1098/rstb.2009.0072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lyon P. The Cognitive Cell: Bacterial Behavior Reconsidered. Front. Microbiol. 2015;6:264. doi: 10.3389/fmicb.2015.00264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Baluška F., Miller W.B., Reber A.S. Biomolecular Basis of Cellular Consciousness via Subcellular Nanobrains. Int. J. Mol. Sci. 2021;22:2545. doi: 10.3390/ijms22052545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Baluška F., Levin M. On Having No Head: Cognition throughout Biological Systems. Front. Psychol. 2016;7:902. doi: 10.3389/fpsyg.2016.00902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Trewavas A.J., Baluška F. The Ubiquity of Consciousness. EMBO Rep. 2011;12:1221–1225. doi: 10.1038/embor.2011.218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Marijuán P.C., Navarro J., del Moral R. On Prokaryotic Intelligence: Strategies for Sensing the Environment. Biosystems. 2010;99:94–103. doi: 10.1016/j.biosystems.2009.09.004. [DOI] [PubMed] [Google Scholar]
- 18.Marijuán P.C., Navarro J., del Moral R. How the Living Is in the World: An Inquiry into the Informational Choreographies of Life. Prog. Biophys. Mol. Biol. 2015;119:469–480. doi: 10.1016/j.pbiomolbio.2015.07.002. [DOI] [PubMed] [Google Scholar]
- 19.Trinh M.K., Wayland M.T., Prabakaran S. Behavioural Analysis of Single-Cell Aneural Ciliate, Stentor Roeseli, Using Machine Learning Approaches. J. R. Soc. Interface. 2019;16:20190410. doi: 10.1098/rsif.2019.0410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dexter J.P., Prabakaran S., Gunawardena J. A Complex Hierarchy of Avoidance Behaviors in a Single-Cell Eukaryote. Curr. Biol. 2019;29:4323–4329.e2. doi: 10.1016/j.cub.2019.10.059. [DOI] [PubMed] [Google Scholar]
- 21.Gelber B. Food or Training Paramecium. Science. 1957;126:1340–1341. doi: 10.1126/science.126.3287.1340. [DOI] [PubMed] [Google Scholar]
- 22.Gershman S.J., Balbi P.E., Gallistel C.R., Gunawardena J. Reconsidering the Evidence for Learning in Single Cells. eLife. 2021;10:e61907. doi: 10.7554/eLife.61907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kholodenko B.N. Cell-Signalling Dynamics in Time and Space. Nat. Rev. Mol. Cell Biol. 2006;7:165–176. doi: 10.1038/nrm1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Azeloglu E.U., Iyengar R. Signaling Networks: Information Flow, Computation, and Decision Making. Cold Spring Harb. Perspect. Biol. 2015;7:a005934. doi: 10.1101/cshperspect.a005934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nair A., Chauhan P., Saha B., Kubatzky K.F. Conceptual Evolution of Cell Signaling. Int. J. Mol. Sci. 2019;20:3292. doi: 10.3390/ijms20133292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Timsit Y., Sergeant-Perthuis G., Bennequin D. Evolution of Ribosomal Protein Network Architectures. Sci. Rep. 2021;11:625. doi: 10.1038/s41598-020-80194-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Poirot O., Timsit Y. Neuron-Like Networks Between Ribosomal Proteins Within the Ribosome. Sci. Rep. 2016;6:26485. doi: 10.1038/srep26485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Timsit Y., Bennequin D. Nervous-Like Circuits in the Ribosome Facts, Hypotheses and Perspectives. Int. J. Mol. Sci. 2019;20:2911. doi: 10.3390/ijms20122911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bray D. Protein Molecules as Computational Elements in Living Cells. Nature. 1995;376:307–312. doi: 10.1038/376307a0. [DOI] [PubMed] [Google Scholar]
- 30.Widder E. Bioluminescence and the Pelagic Visual Environment. Mar. Freshwater Behav. Physiol. 2002;35:1–26. doi: 10.1080/10236240290025581. [DOI] [Google Scholar]
- 31.Young R.E. Oceanic Bioluminescence: An Overview of General Functions. Bull. Mar. Sci. 1983;33:829–845. [Google Scholar]
- 32.Meighen E.A. Genetics of Bacterial Bioluminescence. Annu. Rev. Genet. 1994;28:117–139. doi: 10.1146/annurev.ge.28.120194.001001. [DOI] [PubMed] [Google Scholar]
- 33.Marcinko C.L.J., Painter S.C., Martin A.P., Allen J.T. A Review of the Measurement and Modelling of Dinoflagellate Bioluminescence. Prog. Oceanogr. 2013;109:117–129. doi: 10.1016/j.pocean.2012.10.008. [DOI] [Google Scholar]
- 34.Valiadi M., Iglesias-Rodriguez D. Understanding Bioluminescence in Dinoflagellates-How Far Have We Come? Microorganisms. 2013;1:3–25. doi: 10.3390/microorganisms1010003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Herring P.J. Some Features of the Bioluminescence of the Radiolarian Thalassicolla sp. Mar. Biol. 1979;53:213–216. doi: 10.1007/BF00952428. [DOI] [Google Scholar]
- 36.Latz M.I., Frank T.M., Case J.F., Swift E., Bidigare R.R. Bioluminescence of Colonial Radiolaria in the Western Sargasso Sea. J. Exp. Mar. Biol. Ecol. 1987;109:25–38. doi: 10.1016/0022-0981(87)90183-3. [DOI] [Google Scholar]
- 37.Mallefet J., Stevens D.W., Duchatelet L. Bioluminescence of the Largest Luminous Vertebrate, the Kitefin Shark, Dalatias Licha: First Insights and Comparative Aspects. Front. Mar. Sci. 2021;8:153. doi: 10.3389/fmars.2021.633582. [DOI] [Google Scholar]
- 38.Baldwin T.O., Christopher J.A., Raushel F.M., Sinclair J.F., Ziegler M.M., Fisher A.J., Rayment I. Structure of Bacterial Luciferase. Curr. Opin. Struct. Biol. 1995;5:798–809. doi: 10.1016/0959-440X(95)80014-X. [DOI] [PubMed] [Google Scholar]
- 39.Liu L., Hastings J.W. Two Different Domains of the Luciferase Gene in the Heterotrophic Dinoflagellate Noctiluca Scintillans Occur as Two Separate Genes in Photosynthetic Species. Proc. Natl. Acad. Sci. USA. 2007;104:696–701. doi: 10.1073/pnas.0607816103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schultz L.W., Liu L., Cegielski M., Hastings J.W. Crystal Structure of a PH-Regulated Luciferase Catalyzing the Bioluminescent Oxidation of an Open Tetrapyrrole. Proc. Natl. Acad. Sci. USA. 2005;102:1378–1383. doi: 10.1073/pnas.0409335102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nakamura H., Kishi Y., Shimomura O., Morse D., Hastings J.W. Structure of Dinoflagellate Luciferin and Its Enzymic and Nonenzymic Air-Oxidation Products. J. Am. Chem. Soc. 1989;111:7607–7611. doi: 10.1021/ja00201a050. [DOI] [Google Scholar]
- 42.Loening A.M., Fenn T.D., Gambhir S.S. Crystal Structures of the Luciferase and Green Fluorescent Protein from Renilla Reniformis. J. Mol. Biol. 2007;374:1017–1028. doi: 10.1016/j.jmb.2007.09.078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Conti E., Franks N.P., Brick P. Crystal Structure of Firefly Luciferase Throws Light on a Superfamily of Adenylate-Forming Enzymes. Structure. 1996;4:287–298. doi: 10.1016/S0969-2126(96)00033-0. [DOI] [PubMed] [Google Scholar]
- 44.Tomabechi Y., Hosoya T., Ehara H., Sekine S.-I., Shirouzu M., Inouye S. Crystal Structure of NanoKAZ: The Mutated 19 KDa Component of Oplophorus Luciferase Catalyzing the Bioluminescent Reaction with Coelenterazine. Biochem Biophys Res. Commun. 2016;470:88–93. doi: 10.1016/j.bbrc.2015.12.123. [DOI] [PubMed] [Google Scholar]
- 45.Viviani V.R. The Origin, Diversity, and Structure Function Relationships of Insect Luciferases. Cell Mol. Life Sci. 2002;59:1833–1850. doi: 10.1007/PL00012509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Larionova M.D., Markova S.V., Vysotski E.S. Bioluminescent and Structural Features of Native Folded Gaussia Luciferase. J. Photochem. Photobiol. B. 2018;183:309–317. doi: 10.1016/j.jphotobiol.2018.04.050. [DOI] [PubMed] [Google Scholar]
- 47.Markova S.V., Larionova M.D., Vysotski E.S. Shining Light on the Secreted Luciferases of Marine Copepods: Current Knowledge and Applications. Photochem. Photobiol. 2019;95:705–721. doi: 10.1111/php.13077. [DOI] [PubMed] [Google Scholar]
- 48.Wu N., Kobayashi N., Tsuda K., Unzai S., Saotome T., Kuroda Y., Yamazaki T. Solution Structure of Gaussia Luciferase with Five Disulfide Bonds and Identification of a Putative Coelenterazine Binding Cavity by Heteronuclear NMR. Sci. Rep. 2020;10:20069. doi: 10.1038/s41598-020-76486-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Desjardin D.E., Oliveira A.G., Stevani C.V. Fungi Bioluminescence Revisited. Photochem. Photobiol. Sci. 2008;7:170–182. doi: 10.1039/b713328f. [DOI] [PubMed] [Google Scholar]
- 50.Oliveira A.G., Desjardin D.E., Perry B.A., Stevani C.V. Evidence That a Single Bioluminescent System Is Shared by All Known Bioluminescent Fungal Lineages. Photochem. Photobiol. Sci. 2012;11:848–852. doi: 10.1039/c2pp25032b. [DOI] [PubMed] [Google Scholar]
- 51.Ke H.-M., Lee H.-H., Lin C.-Y.I., Liu Y.-C., Lu M.R., Hsieh J.-W.A., Chang C.-C., Wu P.-H., Lu M.J., Li J.-Y., et al. Mycena Genomes Resolve the Evolution of Fungal Bioluminescence. Proc. Natl. Acad. Sci. USA. 2020;117:31267–31277. doi: 10.1073/pnas.2010761117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kotlobay A.A., Sarkisyan K.S., Mokrushina Y.A., Marcet-Houben M., Serebrovskaya E.O., Markina N.M., Gonzalez Somermeyer L., Gorokhovatsky A.Y., Vvedensky A., Purtov K.V., et al. Genetically Encodable Bioluminescent System from Fungi. Proc. Natl. Acad. Sci. USA. 2018;115:12728–12732. doi: 10.1073/pnas.1803615115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kaskova Z.M., Dörr F.A., Petushkov V.N., Purtov K.V., Tsarkova A.S., Rodionova N.S., Mineev K.S., Guglya E.B., Kotlobay A., Baleeva N.S., et al. Mechanism and Color Modulation of Fungal Bioluminescence. Sci. Adv. 2017;3:e1602847. doi: 10.1126/sciadv.1602847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lau E.S., Oakley T.H. Multi-Level Convergence of Complex Traits and the Evolution of Bioluminescence. Biol. Rev. Camb. Philos. Soc. 2021;96:673–691. doi: 10.1111/brv.12672. [DOI] [PubMed] [Google Scholar]
- 55.Delroisse J., Ullrich-Lüter E., Blaue S., Ortega-Martinez O., Eeckhaut I., Flammang P., Mallefet J. A Puzzling Homology: A Brittle Star Using a Putative Cnidarian-Type Luciferase for Bioluminescence. Open Biol. 2017;7:160300. doi: 10.1098/rsob.160300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Waldenmaier H.E., Oliveira A.G., Stevani C.V. Thoughts on the Diversity of Convergent Evolution of Bioluminescence on Earth. Int. J. Astrobiol. 2012;11:335–343. doi: 10.1017/S1473550412000146. [DOI] [Google Scholar]
- 57.Hastings J.W. Biological Diversity, Chemical Mechanisms, and the Evolutionary Origins of Bioluminescent Systems. J. Mol. Evol. 1983;19:309–321. doi: 10.1007/BF02101634. [DOI] [PubMed] [Google Scholar]
- 58.Morin J.G. Coastal Bioluminescence: Patterns and Functions. Bull. Mar. Sci. 1983;33:787–817. [Google Scholar]
- 59.Sitnikov D.M., Schineller J.B., Baldwin T.O. Transcriptional Regulation of Bioluminesence Genes from Vibrio Fischeri. Mol. Microbiol. 1995;17:801–812. doi: 10.1111/j.1365-2958.1995.mmi_17050801.x. [DOI] [PubMed] [Google Scholar]
- 60.Vannier T., Hingamp P., Turrel F., Tanet L., Lescot M., Timsit Y. Diversity and Evolution of Bacterial Bioluminescence Genes in the Global Ocean. NAR Genom. Bioinform. 2020;2:lqaa018. doi: 10.1093/nargab/lqaa018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Brodl E., Winkler A., Macheroux P. Molecular Mechanisms of Bacterial Bioluminescence. Comput. Struct. Biotechnol. J. 2018;16:551–564. doi: 10.1016/j.csbj.2018.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Dunlap P. Biochemistry and Genetics of Bacterial Bioluminescence. Adv. Biochem. Eng. Biotechnol. 2014;144:37–64. doi: 10.1007/978-3-662-43385-0_2. [DOI] [PubMed] [Google Scholar]
- 63.Rees J.F., de Wergifosse B., Noiset O., Dubuisson M., Janssens B., Thompson E.M. The Origins of Marine Bioluminescence: Turning Oxygen Defence Mechanisms into Deep-Sea Communication Tools. J. Exp. Biol. 1998;201:1211–1221. doi: 10.1242/jeb.201.8.1211. [DOI] [PubMed] [Google Scholar]
- 64.Timmins G.S., Jackson S.K., Swartz H.M. The Evolution of Bioluminescent Oxygen Consumption as an Ancient Oxygen Detoxification Mechanism. J. Mol. Evol. 2001;52:321–332. doi: 10.1007/s002390010162. [DOI] [PubMed] [Google Scholar]
- 65.Czyz A., Plata K., Wegrzyn G. Stimulation of DNA Repair as an Evolutionary Drive for Bacterial Luminescence. Luminescence. 2003;18:140–144. doi: 10.1002/bio.715. [DOI] [PubMed] [Google Scholar]
- 66.Kozakiewicz J., Gajewska M., Lyzeń R., Czyz A., Wegrzyn G. Bioluminescence-Mediated Stimulation of Photoreactivation in Bacteria. FEMS Microbiol. Lett. 2005;250:105–110. doi: 10.1016/j.femsle.2005.06.047. [DOI] [PubMed] [Google Scholar]
- 67.Zarubin M., Belkin S., Ionescu M., Genin A. Bacterial Bioluminescence as a Lure for Marine Zooplankton and Fish. Proc. Natl. Acad. Sci. USA. 2012;109:853–857. doi: 10.1073/pnas.1116683109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Aschtgen M.-S., Brennan C.A., Nikolakakis K., Cohen S., McFall-Ngai M., Ruby E.G. Insights into Flagellar Function and Mechanism from the Squid-Vibrio Symbiosis. NPJ Biofilms Microbiomes. 2019;5:32. doi: 10.1038/s41522-019-0106-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tanet L., Martini S., Casalot L., Tamburini C. Reviews and Syntheses: Bacterial Bioluminescence–Ecology and Impact in the Biological Carbon Pump. Biogeosciences. 2020;17:3757–3778. doi: 10.5194/bg-17-3757-2020. [DOI] [Google Scholar]
- 70.McFall-Ngai M. Divining the Essence of Symbiosis: Insights from the Squid-Vibrio Model. PLoS Biol. 2014;12:e1001783. doi: 10.1371/journal.pbio.1001783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ruby E.G. Symbiotic Conversations Are Revealed under Genetic Interrogation. Nat. Rev. Microbiol. 2008;6:752–762. doi: 10.1038/nrmicro1958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Eckert R., Sibaoka T. The Flash-Triggering Action Potential of the Luminescent Dinoflagellate Noctiluca. J. Gen. Physiol. 1968;52:258–282. doi: 10.1085/jgp.52.2.258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Prevett A., Lindström J., Xu J., Karlson B., Selander E. Grazer-Induced Bioluminescence Gives Dinoflagellates a Competitive Edge. Curr. Biol. 2019;29:R564–R565. doi: 10.1016/j.cub.2019.05.019. [DOI] [PubMed] [Google Scholar]
- 74.Fajardo C., De Donato M., Rodulfo H., Martinez-Rodriguez G., Costas B., Mancera J.M., Fernandez-Acero F.J. New Perspectives Related to the Bioluminescent System in Dinoflagellates: Pyrocystis Lunula, a Case Study. Int. J. Mol. Sci. 2020;21:1784. doi: 10.3390/ijms21051784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hanley K.A., Widder E.A. Bioluminescence in Dinoflagellates: Evidence That the Adaptive Value of Bioluminescence in Dinoflagellates Is Concentration Dependent. Photochem. Photobiol. 2017;93:519–530. doi: 10.1111/php.12713. [DOI] [PubMed] [Google Scholar]
- 76.Lindström J., Grebner W., Rigby K., Selander E. Effects of Predator Lipids on Dinoflagellate Defence Mechanisms—Increased Bioluminescence Capacity. Sci. Rep. 2017;7:13104. doi: 10.1038/s41598-017-13293-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Valiadi M., Debora Iglesias-Rodriguez M., Amorim A. Distribution and genetic diversity of the luciferase gene within marine dinoflagellates(1) J. Phycol. 2012;48:826–836. doi: 10.1111/j.1529-8817.2012.01144.x. [DOI] [PubMed] [Google Scholar]
- 78.Colley N.J., Nilsson D.-E. Photoreception in Phytoplankton. Integr. Comp. Biol. 2016;56:764–775. doi: 10.1093/icb/icw037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Häder D.P. Photosensory Behavior in Procaryotes. Microbiol. Rev. 1987;51:1–21. doi: 10.1128/mr.51.1.1-21.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Herrou J., Crosson S. Function, Structure and Mechanism of Bacterial Photosensory LOV Proteins. Nat. Rev. Microbiol. 2011;9:713–723. doi: 10.1038/nrmicro2622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ho M.-Y., Soulier N.T., Canniffe D.P., Shen G., Bryant D.A. Light Regulation of Pigment and Photosystem Biosynthesis in Cyanobacteria. Curr. Opin. Plant. Biol. 2017;37:24–33. doi: 10.1016/j.pbi.2017.03.006. [DOI] [PubMed] [Google Scholar]
- 82.Ikeuchi M., Ishizuka T. Cyanobacteriochromes: A New Superfamily of Tetrapyrrole-Binding Photoreceptors in Cyanobacteria. Photochem. Photobiol. Sci. 2008;7:1159–1167. doi: 10.1039/b802660m. [DOI] [PubMed] [Google Scholar]
- 83.Ueki N., Matsunaga S., Inouye I., Hallmann A. How 5000 Independent Rowers Coordinate Their Strokes in Order to Row into the Sunlight: Phototaxis in the Multicellular Green Alga Volvox. BMC Biol. 2010;8:103. doi: 10.1186/1741-7007-8-103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Möglich A. Signal Transduction in Photoreceptor Histidine Kinases. Protein Sci. 2019;28:1923–1946. doi: 10.1002/pro.3705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Singh A.H., Doerks T., Letunic I., Raes J., Bork P. Discovering Functional Novelty in Metagenomes: Examples from Light-Mediated Processes. J. Bacteriol. 2009;191:32–41. doi: 10.1128/JB.01084-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Yu Z., Fischer R. Light Sensing and Responses in Fungi. Nat. Rev. Microbiol. 2019;17:25–36. doi: 10.1038/s41579-018-0109-x. [DOI] [PubMed] [Google Scholar]
- 87.Porter M.L. Beyond the Eye: Molecular Evolution of Extraocular Photoreception. Integr. Comp. Biol. 2016;56:842–852. doi: 10.1093/icb/icw052. [DOI] [PubMed] [Google Scholar]
- 88.Wilde A., Mullineaux C.W. Light-Controlled Motility in Prokaryotes and the Problem of Directional Light Perception. FEMS Microbiol. Rev. 2017;41:900–922. doi: 10.1093/femsre/fux045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Bijlsma J.J.E., Groisman E.A. Making Informed Decisions: Regulatory Interactions between Two-Component Systems. Trends Microbiol. 2003;11:359–366. doi: 10.1016/S0966-842X(03)00176-8. [DOI] [PubMed] [Google Scholar]
- 90.Swartz T.E., Tseng T.-S., Frederickson M.A., Paris G., Comerci D.J., Rajashekara G., Kim J.-G., Mudgett M.B., Splitter G.A., Ugalde R.A., et al. Blue-Light-Activated Histidine Kinases: Two-Component Sensors in Bacteria. Science. 2007;317:1090–1093. doi: 10.1126/science.1144306. [DOI] [PubMed] [Google Scholar]
- 91.Cheung J., Hendrickson W.A. Sensor Domains of Two-Component Regulatory Systems. Curr. Opin. Microbiol. 2010;13:116–123. doi: 10.1016/j.mib.2010.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Jacob-Dubuisson F., Mechaly A., Betton J.-M., Antoine R. Structural Insights into the Signalling Mechanisms of Two-Component Systems. Nat. Rev. Microbiol. 2018;16:585–593. doi: 10.1038/s41579-018-0055-7. [DOI] [PubMed] [Google Scholar]
- 93.Jin J., Pawson T. Modular Evolution of Phosphorylation-Based Signalling Systems. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2012;367:2540–2555. doi: 10.1098/rstb.2012.0106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Lemmon M.A., Schlessinger J. Cell Signaling by Receptor Tyrosine Kinases. Cell. 2010;141:1117–1134. doi: 10.1016/j.cell.2010.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Schuergers N., Lenn T., Kampmann R., Meissner M.V., Esteves T., Temerinac-Ott M., Korvink J.G., Lowe A.R., Mullineaux C.W., Wilde A. Cyanobacteria Use Micro-Optics to Sense Light Direction. eLife. 2016;5:e12620. doi: 10.7554/eLife.12620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Gavelis G.S., Hayakawa S., White R.A., Gojobori T., Suttle C.A., Keeling P.J., Leander B.S. Eye-like Ocelloids Are Built from Different Endosymbiotically Acquired Components. Nature. 2015;523:204–207. doi: 10.1038/nature14593. [DOI] [PubMed] [Google Scholar]
- 97.Möglich A., Yang X., Ayers R.A., Moffat K. Structure and Function of Plant Photoreceptors. Annu. Rev. Plant. Biol. 2010;61:21–47. doi: 10.1146/annurev-arplant-042809-112259. [DOI] [PubMed] [Google Scholar]
- 98.Conrad K.S., Manahan C.C., Crane B.R. Photochemistry of Flavoprotein Light Sensors. Nat. Chem. Biol. 2014;10:801–809. doi: 10.1038/nchembio.1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Fuhrman J.A., Schwalbach M.S., Stingl U. Proteorhodopsins: An Array of Physiological Roles? Nat. Rev. Microbiol. 2008;6:488–494. doi: 10.1038/nrmicro1893. [DOI] [PubMed] [Google Scholar]
- 100.Finkel O.M., Béjà O., Belkin S. Global Abundance of Microbial Rhodopsins. ISME J. 2013;7:448–451. doi: 10.1038/ismej.2012.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Avelar G.M., Schumacher R.I., Zaini P.A., Leonard G., Richards T.A., Gomes S.L. A Rhodopsin-Guanylyl Cyclase Gene Fusion Functions in Visual Perception in a Fungus. Curr. Biol. 2014;24:1234–1240. doi: 10.1016/j.cub.2014.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Ikuta T., Shihoya W., Sugiura M., Yoshida K., Watari M., Tokano T., Yamashita K., Katayama K., Tsunoda S.P., Uchihashi T., et al. Structural Insights into the Mechanism of Rhodopsin Phosphodiesterase. Nat. Commun. 2020;11:5605. doi: 10.1038/s41467-020-19376-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Gallot-Lavallée L., Archibald J.M. Evolutionary Biology: Viral Rhodopsins Illuminate Algal Evolution. Curr. Biol. 2020;30:R1469–R1471. doi: 10.1016/j.cub.2020.10.080. [DOI] [PubMed] [Google Scholar]
- 104.Oda K., Vierock J., Oishi S., Rodriguez-Rozada S., Taniguchi R., Yamashita K., Wiegert J.S., Nishizawa T., Hegemann P., Nureki O. Crystal Structure of the Red Light-Activated Channelrhodopsin Chrimson. Nat. Commun. 2018;9:3949. doi: 10.1038/s41467-018-06421-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Ma M., Shi X., Lin S. Heterologous Expression and Cell Membrane Localization of Dinoflagellate Opsins (Rhodopsin Proteins) in Mammalian Cells. Mar. Life Sci. Technol. 2020;2:302–308. doi: 10.1007/s42995-020-00043-1. [DOI] [Google Scholar]
- 106.Palczewski K., Kumasaka T., Hori T., Behnke C.A., Motoshima H., Fox B.A., Le Trong I., Teller D.C., Okada T., Stenkamp R.E., et al. Crystal Structure of Rhodopsin: A G Protein-Coupled Receptor. Science. 2000;289:739–745. doi: 10.1126/science.289.5480.739. [DOI] [PubMed] [Google Scholar]
- 107.Ernst O.P., Lodowski D.T., Elstner M., Hegemann P., Brown L.S., Kandori H. Microbial and Animal Rhodopsins: Structures, Functions, and Molecular Mechanisms. Chem. Rev. 2014;114:126–163. doi: 10.1021/cr4003769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Butryn A., Raza H., Rada H., Moraes I., Owens R.J., Orville A.M. Molecular Basis for GTP Recognition by Light-Activated Guanylate Cyclase RhGC. FEBS J. 2020;287:2797–2807. doi: 10.1111/febs.15167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Spudich E.N., Takahashi T., Spudich J.L. Sensory Rhodopsins I and II Modulate a Methylation/Demethylation System in Halobacterium Halobium Phototaxis. Proc. Natl. Acad. Sci. USA. 1989;86:7746–7750. doi: 10.1073/pnas.86.20.7746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Luecke H., Schobert B., Lanyi J.K., Spudich E.N., Spudich J.L. Crystal Structure of Sensory Rhodopsin II at 2.4 Angstroms: Insights into Color Tuning and Transducer Interaction. Science. 2001;293:1499–1503. doi: 10.1126/science.1062977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Spudich J.L., Luecke H. Sensory Rhodopsin II: Functional Insights from Structure. Curr. Opin. Struct. Biol. 2002;12:540–546. doi: 10.1016/S0959-440X(02)00359-7. [DOI] [PubMed] [Google Scholar]
- 112.Vogeley L., Sineshchekov O.A., Trivedi V.D., Sasaki J., Spudich J.L., Luecke H. Anabaena Sensory Rhodopsin: A Photochromic Color Sensor at 2.0 A. Science. 2004;306:1390–1393. doi: 10.1126/science.1103943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Borgstahl G.E.O., Williams D.R., Getzoff E.D. 1.4.ANG. Structure of Photoactive Yellow Protein, a Cytosolic Photoreceptor: Unusual Fold, Active Site, and Chromophore. Biochemistry. 1995;34:6278–6287. doi: 10.1021/bi00019a004. [DOI] [PubMed] [Google Scholar]
- 114.Pellequer J.L., Wager-Smith K.A., Kay S.A., Getzoff E.D. Photoactive Yellow Protein: A Structural Prototype for the Three-Dimensional Fold of the PAS Domain Superfamily. Proc. Natl. Acad. Sci. USA. 1998;95:5884–5890. doi: 10.1073/pnas.95.11.5884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Mix L.T., Hara M., Rathod R., Kumauchi M., Hoff W.D., Larsen D.S. Noncanonical Photocycle Initiation Dynamics of the Photoactive Yellow Protein (PYP) Domain of the PYP-Phytochrome-Related (Ppr) Photoreceptor. J. Phys. Chem. Lett. 2016;7:5212–5218. doi: 10.1021/acs.jpclett.6b02253. [DOI] [PubMed] [Google Scholar]
- 116.Jiang Z., Swem L.R., Rushing B.G., Devanathan S., Tollin G., Bauer C.E. Bacterial Photoreceptor with Similarity to Photoactive Yellow Protein and Plant Phytochromes. Science. 1999;285:406–409. doi: 10.1126/science.285.5426.406. [DOI] [PubMed] [Google Scholar]
- 117.Wang J. Visualization of H Atoms in the X-Ray Crystal Structure of Photoactive Yellow Protein: Does It Contain Low-Barrier Hydrogen Bonds? Protein Sci. 2019;28:1966–1972. doi: 10.1002/pro.3716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Kumauchi M., Hara M.T., Stalcup P., Xie A., Hoff W.D. Identification of Six New Photoactive Yellow Proteins—Diversity and Structure-Function Relationships in a Bacterial Blue Light Photoreceptor. Photochem. Photobiol. 2008;84:956–969. doi: 10.1111/j.1751-1097.2008.00335.x. [DOI] [PubMed] [Google Scholar]
- 119.Taylor B.L., Zhulin I.B. PAS Domains: Internal Sensors of Oxygen, Redox Potential, and Light. Microbiol. Mol. Biol. Rev. 1999;63:479–506. doi: 10.1128/MMBR.63.2.479-506.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Schmidt M. A Short History of Structure Based Research on the Photocycle of Photoactive Yellow Protein. Struct. Dyn. 2017;4:032201. doi: 10.1063/1.4974172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Jung Y.O., Lee J.H., Kim J., Schmidt M., Moffat K., Srajer V., Ihee H. Volume-Conserving Trans-Cis Isomerization Pathways in Photoactive Yellow Protein Visualized by Picosecond X-Ray Crystallography. Nat. Chem. 2013;5:212–220. doi: 10.1038/nchem.1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Sigala P.A., Tsuchida M.A., Herschlag D. Hydrogen Bond Dynamics in the Active Site of Photoactive Yellow Protein. Proc. Natl. Acad. Sci. USA. 2009;106:9232–9237. doi: 10.1073/pnas.0900168106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Glantz S.T., Carpenter E.J., Melkonian M., Gardner K.H., Boyden E.S., Wong G.K.-S., Chow B.Y. Functional and Topological Diversity of LOV Domain Photoreceptors. Proc. Natl. Acad. Sci. USA. 2016;113:E1442–E1451. doi: 10.1073/pnas.1509428113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Pathak G.P., Losi A., Gärtner W. Metagenome-Based Screening Reveals Worldwide Distribution of LOV-Domain Proteins. Photochem. Photobiol. 2012;88:107–118. doi: 10.1111/j.1751-1097.2011.01024.x. [DOI] [PubMed] [Google Scholar]
- 125.Pathak G.P., Ehrenreich A., Losi A., Streit W.R., Gärtner W. Novel Blue Light-Sensitive Proteins from a Metagenomic Approach. Environ. Microbiol. 2009;11:2388–2399. doi: 10.1111/j.1462-2920.2009.01967.x. [DOI] [PubMed] [Google Scholar]
- 126.Krauss U., Minh B.Q., Losi A., Gärtner W., Eggert T., von Haeseler A., Jaeger K.-E. Distribution and Phylogeny of Light-Oxygen-Voltage-Blue-Light-Signaling Proteins in the Three Kingdoms of Life. J. Bacteriol. 2009;191:7234–7242. doi: 10.1128/JB.00923-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Mandalari C., Losi A., Gärtner W. Distance-Tree Analysis, Distribution and Co-Presence of Bilin- and Flavin-Binding Prokaryotic Photoreceptors for Visible Light. Photochem. Photobiol. Sci. 2013;12:1144–1157. doi: 10.1039/c3pp25404f. [DOI] [PubMed] [Google Scholar]
- 128.Takahashi F. Blue-Light-Regulated Transcription Factor, Aureochrome, in Photosynthetic Stramenopiles. J. Plant. Res. 2016;129:189–197. doi: 10.1007/s10265-016-0784-5. [DOI] [PubMed] [Google Scholar]
- 129.Kroth P.G., Wilhelm C., Kottke T. An Update on Aureochromes: Phylogeny-Mechanism-Function. J. Plant. Physiol. 2017;217:20–26. doi: 10.1016/j.jplph.2017.06.010. [DOI] [PubMed] [Google Scholar]
- 130.Matiiv A.B., Chekunova E.M. Aureochromes-Blue Light Receptors. Biochemistry. 2018;83:662–673. doi: 10.1134/S0006297918060044. [DOI] [PubMed] [Google Scholar]
- 131.Weber A.M., Kaiser J., Ziegler T., Pilsl S., Renzl C., Sixt L., Pietruschka G., Moniot S., Kakoti A., Juraschitz M., et al. A Blue Light Receptor That Mediates RNA Binding and Translational Regulation. Nat. Chem. Biol. 2019;15:1085–1092. doi: 10.1038/s41589-019-0346-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Harper S.M., Neil L.C., Gardner K.H. Structural Basis of a Phototropin Light Switch. Science. 2003;301:1541–1544. doi: 10.1126/science.1086810. [DOI] [PubMed] [Google Scholar]
- 133.Okajima K. Molecular Mechanism of Phototropin Light Signaling. J. Plant. Res. 2016;129:149–157. doi: 10.1007/s10265-016-0783-6. [DOI] [PubMed] [Google Scholar]
- 134.Möglich A., Moffat K. Structural Basis for Light-Dependent Signaling in the Dimeric LOV Domain of the Photosensor YtvA. J. Mol. Biol. 2007;373:112–126. doi: 10.1016/j.jmb.2007.07.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Barends T.R.M., Hartmann E., Griese J.J., Beitlich T., Kirienko N.V., Ryjenkov D.A., Reinstein J., Shoeman R.L., Gomelsky M., Schlichting I. Structure and Mechanism of a Bacterial Light-Regulated Cyclic Nucleotide Phosphodiesterase. Nature. 2009;459:1015–1018. doi: 10.1038/nature07966. [DOI] [PubMed] [Google Scholar]
- 136.Kennis J.T.M., Mathes T. Molecular Eyes: Proteins That Transform Light into Biological Information. Interface Focus. 2013;3:20130005. doi: 10.1098/rsfs.2013.0005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Fujisawa T., Masuda S. Light-Induced Chromophore and Protein Responses and Mechanical Signal Transduction of BLUF Proteins. Biophys. Rev. 2018;10:327–337. doi: 10.1007/s12551-017-0355-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Park S.-Y., Tame J.R.H. Seeing the Light with BLUF Proteins. Biophys. Rev. 2017;9:169–176. doi: 10.1007/s12551-017-0258-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Kutomi O., Yamamoto R., Hirose K., Mizuno K., Nakagiri Y., Imai H., Noga A., Obbineni J.M., Zimmermann N., Nakajima M., et al. A Dynein-Associated Photoreceptor Protein Prevents Ciliary Acclimation to Blue Light. Sci. Adv. 2021;7:eabf3621. doi: 10.1126/sciadv.abf3621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Jung A., Domratcheva T., Tarutina M., Wu Q., Ko W.-H., Shoeman R.L., Gomelsky M., Gardner K.H., Schlichting I. Structure of a Bacterial BLUF Photoreceptor: Insights into Blue Light-Mediated Signal Transduction. Proc. Natl. Acad. Sci. USA. 2005;102:12350–12355. doi: 10.1073/pnas.0500722102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Masuda S., Bauer C.E. AppA Is a Blue Light Photoreceptor That Antirepresses Photosynthesis Gene Expression in Rhodobacter Sphaeroides. Cell. 2002;110:613–623. doi: 10.1016/S0092-8674(02)00876-0. [DOI] [PubMed] [Google Scholar]
- 142.Winkler A., Heintz U., Lindner R., Reinstein J., Shoeman R.L., Schlichting I. A Ternary AppA-PpsR-DNA Complex Mediates Light Regulation of Photosynthesis-Related Gene Expression. Nat. Struct. Mol. Biol. 2013;20:859–867. doi: 10.1038/nsmb.2597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Ohki M., Sugiyama K., Kawai F., Tanaka H., Nihei Y., Unzai S., Takebe M., Matsunaga S., Adachi S.-I., Shibayama N., et al. Structural Insight into Photoactivation of an Adenylate Cyclase from a Photosynthetic Cyanobacterium. Proc. Natl. Acad. Sci. USA. 2016;113:6659–6664. doi: 10.1073/pnas.1517520113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Lindner R., Hartmann E., Tarnawski M., Winkler A., Frey D., Reinstein J., Meinhart A., Schlichting I. Photoactivation Mechanism of a Bacterial Light-Regulated Adenylyl Cyclase. J. Mol. Biol. 2017;429:1336–1351. doi: 10.1016/j.jmb.2017.03.020. [DOI] [PubMed] [Google Scholar]
- 145.Brautigam C.A., Smith B.S., Ma Z., Palnitkar M., Tomchick D.R., Machius M., Deisenhofer J. Structure of the Photolyase-like Domain of Cryptochrome 1 from Arabidopsis Thaliana. Proc. Natl. Acad. Sci. USA. 2004;101:12142–12147. doi: 10.1073/pnas.0404851101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Mei Q., Dvornyk V. Evolutionary History of the Photolyase/Cryptochrome Superfamily in Eukaryotes. PLoS ONE. 2015;10:e0135940. doi: 10.1371/journal.pone.0135940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Brudler R., Hitomi K., Daiyasu H., Toh H., Kucho K., Ishiura M., Kanehisa M., Roberts V.A., Todo T., Tainer J.A., et al. Identification of a New Cryptochrome Class. Structure, Function, and Evolution. Mol. Cell. 2003;11:59–67. doi: 10.1016/S1097-2765(03)00008-X. [DOI] [PubMed] [Google Scholar]
- 148.Ahmad M. Photocycle and Signaling Mechanisms of Plant Cryptochromes. Curr. Opin. Plant. Biol. 2016;33:108–115. doi: 10.1016/j.pbi.2016.06.013. [DOI] [PubMed] [Google Scholar]
- 149.Cashmore A.R., Jarillo J.A., Wu Y.J., Liu D. Cryptochromes: Blue Light Receptors for Plants and Animals. Science. 1999;284:760–765. doi: 10.1126/science.284.5415.760. [DOI] [PubMed] [Google Scholar]
- 150.Essen L.-O., Franz S., Banerjee A. Structural and Evolutionary Aspects of Algal Blue Light Receptors of the Cryptochrome and Aureochrome Type. J. Plant. Physiol. 2017;217:27–37. doi: 10.1016/j.jplph.2017.07.005. [DOI] [PubMed] [Google Scholar]
- 151.Fortunato A.E., Annunziata R., Jaubert M., Bouly J.-P., Falciatore A. Dealing with Light: The Widespread and Multitasking Cryptochrome/Photolyase Family in Photosynthetic Organisms. J. Plant. Physiol. 2015;172:42–54. doi: 10.1016/j.jplph.2014.06.011. [DOI] [PubMed] [Google Scholar]
- 152.Franz S., Ignatz E., Wenzel S., Zielosko H., Putu E.P.G.N., Maestre-Reyna M., Tsai M.-D., Yamamoto J., Mittag M., Essen L.-O. Structure of the Bifunctional Cryptochrome ACRY from Chlamydomonas Reinhardtii. Nucleic Acids Res. 2018;46:8010–8022. doi: 10.1093/nar/gky621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Haug M.F., Gesemann M., Lazović V., Neuhauss S.C.F. Eumetazoan Cryptochrome Phylogeny and Evolution. Genome Biol. Evol. 2015;7:601–619. doi: 10.1093/gbe/evv010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Kimø S.M., Friis I., Solov’yov I.A. Atomistic Insights into Cryptochrome Interprotein Interactions. Biophys. J. 2018;115:616–628. doi: 10.1016/j.bpj.2018.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Kottke T., Oldemeyer S., Wenzel S., Zou Y., Mittag M. Cryptochrome Photoreceptors in Green Algae: Unexpected Versatility of Mechanisms and Functions. J. Plant. Physiol. 2017;217:4–14. doi: 10.1016/j.jplph.2017.05.021. [DOI] [PubMed] [Google Scholar]
- 156.Chaves I., Pokorny R., Byrdin M., Hoang N., Ritz T., Brettel K., Essen L.-O., van der Horst G.T.J., Batschauer A., Ahmad M. The Cryptochromes: Blue Light Photoreceptors in Plants and Animals. Annu. Rev. Plant. Biol. 2011;62:335–364. doi: 10.1146/annurev-arplant-042110-103759. [DOI] [PubMed] [Google Scholar]
- 157.Heijde M., Zabulon G., Corellou F., Ishikawa T., Brazard J., Usman A., Sanchez F., Plaza P., Martin M., Falciatore A., et al. Characterization of Two Members of the Cryptochrome/Photolyase Family from Ostreococcus Tauri Provides Insights into the Origin and Evolution of Cryptochromes. Plant. Cell Environ. 2010;33:1614–1626. doi: 10.1111/j.1365-3040.2010.02168.x. [DOI] [PubMed] [Google Scholar]
- 158.Müller M., Carell T. Structural Biology of DNA Photolyases and Cryptochromes. Curr. Opin. Struct. Biol. 2009;19:277–285. doi: 10.1016/j.sbi.2009.05.003. [DOI] [PubMed] [Google Scholar]
- 159.Lin C., Todo T. The Cryptochromes. Genome Biol. 2005;6:220. doi: 10.1186/gb-2005-6-5-220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Solov’yov I.A., Domratcheva T., Moughal Shahi A.R., Schulten K. Decrypting Cryptochrome: Revealing the Molecular Identity of the Photoactivation Reaction. J. Am. Chem. Soc. 2012;134:18046–18052. doi: 10.1021/ja3074819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Saini R., Jaskolski M., Davis S.J. Circadian Oscillator Proteins across the Kingdoms of Life: Structural Aspects. BMC Biol. 2019;17:13. doi: 10.1186/s12915-018-0623-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Zoltowski B.D. Resolving Cryptic Aspects of Cryptochrome Signaling. Proc. Natl. Acad. Sci. USA. 2015;112:8811–8812. doi: 10.1073/pnas.1511092112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Gao J., Wang X., Zhang M., Bian M., Deng W., Zuo Z., Yang Z., Zhong D., Lin C. Trp Triad-Dependent Rapid Photoreduction Is Not Required for the Function of Arabidopsis CRY1. Proc. Natl. Acad. Sci. USA. 2015;112:9135–9140. doi: 10.1073/pnas.1504404112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Wang Q., Lin C. A Structural View of Plant CRY2 Photoactivation and Inactivation. Nat. Struct. Mol. Biol. 2020;27:401–403. doi: 10.1038/s41594-020-0432-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Wang Q., Lin C. Mechanisms of Cryptochrome-Mediated Photoresponses in Plants. Annu. Rev. Plant. Biol. 2020;71:103–129. doi: 10.1146/annurev-arplant-050718-100300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Palayam M., Ganapathy J., Guercio A.M., Tal L., Deck S.L., Shabek N. Structural Insights into Photoactivation of Plant Cryptochrome-2. Commun Biol. 2021;4:28. doi: 10.1038/s42003-020-01531-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Wagner J.R., Brunzelle J.S., Forest K.T., Vierstra R.D. A Light-Sensing Knot Revealed by the Structure of the Chromophore-Binding Domain of Phytochrome. Nature. 2005;438:325–331. doi: 10.1038/nature04118. [DOI] [PubMed] [Google Scholar]
- 168.Essen L.-O., Mailliet J., Hughes J. The Structure of a Complete Phytochrome Sensory Module in the Pr Ground State. Proc. Natl. Acad. Sci. USA. 2008;105:14709–14714. doi: 10.1073/pnas.0806477105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Rockwell N.C., Lagarias J.C. Phytochrome Evolution in 3D: Deletion, Duplication, and Diversification. New Phytol. 2020;225:2283–2300. doi: 10.1111/nph.16240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Nagano S. From Photon to Signal in Phytochromes: Similarities and Differences between Prokaryotic and Plant Phytochromes. J. Plant. Res. 2016;129:123–135. doi: 10.1007/s10265-016-0789-0. [DOI] [PubMed] [Google Scholar]
- 171.Woitowich N.C., Halavaty A.S., Waltz P., Kupitz C., Valera J., Tracy G., Gallagher K.D., Claesson E., Nakane T., Pandey S., et al. Structural Basis for Light Control of Cell Development Revealed by Crystal Structures of a Myxobacterial Phytochrome. IUCrJ. 2018;5:619–634. doi: 10.1107/S2052252518010631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Burgie E.S., Zhang J., Vierstra R.D. Crystal Structure of Deinococcus Phytochrome in the Photoactivated State Reveals a Cascade of Structural Rearrangements during Photoconversion. Structure. 2016;24:448–457. doi: 10.1016/j.str.2016.01.001. [DOI] [PubMed] [Google Scholar]
- 173.Takala H., Björling A., Berntsson O., Lehtivuori H., Niebling S., Hoernke M., Kosheleva I., Henning R., Menzel A., Ihalainen J.A., et al. Signal Amplification and Transduction in Phytochrome Photosensors. Nature. 2014;509:245–248. doi: 10.1038/nature13310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Nagel G., Ollig D., Fuhrmann M., Kateriya S., Musti A.M., Bamberg E., Hegemann P. Channelrhodopsin-1: A Light-Gated Proton Channel in Green Algae. Science. 2002;296:2395–2398. doi: 10.1126/science.1072068. [DOI] [PubMed] [Google Scholar]
- 175.Deisseroth K., Hegemann P. The Form and Function of Channelrhodopsin. Science. 2017;357:6356. doi: 10.1126/science.aan5544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Kato H.E. Structure-Function Relationship of Channelrhodopsins. Adv. Exp. Med. Biol. 2021;1293:35–53. doi: 10.1007/978-981-15-8763-4_3. [DOI] [PubMed] [Google Scholar]
- 177.Schneider F., Grimm C., Hegemann P. Biophysics of Channelrhodopsin. Annu. Rev. Biophys. 2015;44:167–186. doi: 10.1146/annurev-biophys-060414-034014. [DOI] [PubMed] [Google Scholar]
- 178.Lórenz-Fonfría V.A., Heberle J. Channelrhodopsin Unchained: Structure and Mechanism of a Light-Gated Cation Channel. Biochim. Biophys. Acta. 2014;1837:626–642. doi: 10.1016/j.bbabio.2013.10.014. [DOI] [PubMed] [Google Scholar]
- 179.Oda K., Nomura T., Nakane T., Yamashita K., Inoue K., Ito S., Vierock J., Hirata K., Maturana A.D., Katayama K., et al. Time-Resolved Serial Femtosecond Crystallography Reveals Early Structural Changes in Channelrhodopsin. eLife. 2021;10:e62389. doi: 10.7554/eLife.62389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Govorunova E.G., Sineshchekov O.A., Janz R., Liu X., Spudich J.L. Natural Light-Gated Anion Channels: A Family of Microbial Rhodopsins for Advanced Optogenetics. Science. 2015;349:647–650. doi: 10.1126/science.aaa7484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Kim Y.S., Kato H.E., Yamashita K., Ito S., Inoue K., Ramakrishnan C., Fenno L.E., Evans K.E., Paggi J.M., Dror R.O., et al. Crystal Structure of the Natural Anion-Conducting Channelrhodopsin GtACR1. Nature. 2018;561:343–348. doi: 10.1038/s41586-018-0511-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Sineshchekov O.A., Govorunova E.G., Li H., Spudich J.L. Bacteriorhodopsin-like Channelrhodopsins: Alternative Mechanism for Control of Cation Conductance. Proc. Natl. Acad. Sci. USA. 2017;114:E9512–E9519. doi: 10.1073/pnas.1710702114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Zabelskii D., Alekseev A., Kovalev K., Rankovic V., Balandin T., Soloviov D., Bratanov D., Savelyeva E., Podolyak E., Volkov D., et al. Viral Rhodopsins 1 Arean Unique Family of Light-Gated Cation Channels. Nat. Commun. 2020;11:5707. doi: 10.1038/s41467-020-19457-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Perlova T., Gruebele M., Chemla Y.R. Blue Light Is a Universal Signal for Escherichia Coli Chemoreceptors. J. Bacteriol. 2019;201 doi: 10.1128/JB.00762-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Jost M., Fernández-Zapata J., Polanco M.C., Ortiz-Guerrero J.M., Chen P.Y.-T., Kang G., Padmanabhan S., Elías-Arnanz M., Drennan C.L. Structural Basis for Gene Regulation by a B12-Dependent Photoreceptor. Nature. 2015;526:536–541. doi: 10.1038/nature14950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Takano H., Mise K., Hagiwara K., Hirata N., Watanabe S., Toriyabe M., Shiratori-Takano H., Ueda K. Role and Function of LitR, an Adenosyl B12-Bound Light-Sensitive Regulator of Bacillus Megaterium QM B1551, in Regulation of Carotenoid Production. J. Bacteriol. 2015;197:2301–2315. doi: 10.1128/JB.02528-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Ortiz-Guerrero J.M., Polanco M.C., Murillo F.J., Padmanabhan S., Elías-Arnanz M. Light-Dependent Gene Regulation by a Coenzyme B12-Based Photoreceptor. Proc. Natl. Acad. Sci. USA. 2011;108:7565–7570. doi: 10.1073/pnas.1018972108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Padmanabhan S., Jost M., Drennan C.L., Elías-Arnanz M. A New Facet of Vitamin B12: Gene Regulation by Cobalamin-Based Photoreceptors. Annu. Rev. Biochem. 2017;86:485–514. doi: 10.1146/annurev-biochem-061516-044500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Sumi S., Suzuki Y., Matsuki T., Yamamoto T., Tsuruta Y., Mise K., Kawamura T., Ito Y., Shimada Y., Watanabe E., et al. Light-Inducible Carotenoid Production Controlled by a MarR-Type Regulator in Corynebacterium Glutamicum. Sci. Rep. 2019;9:13136. doi: 10.1038/s41598-019-49384-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Fernández-Zapata J., Pérez-Castaño R., Aranda J., Colizzi F., Polanco M.C., Orozco M., Padmanabhan S., Elías-Arnanz M. Plasticity in Oligomerization, Operator Architecture, and DNA Binding in the Mode of Action of a Bacterial B12-Based Photoreceptor. J. Biol. Chem. 2018;293:17888–17905. doi: 10.1074/jbc.RA118.004838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Bridwell-Rabb J., Drennan C.L. Vitamin B12 in the Spotlight Again. Curr. Opin. Chem. Biol. 2017;37:63–70. doi: 10.1016/j.cbpa.2017.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Sumi S., Shiratori-Takano H., Ueda K., Takano H. Role and Function of Class III LitR, a Photosensor Homolog from Burkholderia Multivorans. J. Bacteriol. 2018;200:e00285-18. doi: 10.1128/JB.00285-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Douglas R.H., Genner M.J., Hudson A.G., Partridge J.C., Wagner H.-J. Localisation and Origin of the Bacteriochlorophyll-Derived Photosensitizer in the Retina of the Deep-Sea Dragon Fish Malacosteus Niger. Sci. Rep. 2016;6:39395. doi: 10.1038/srep39395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Tardu M., Bulut S., Kavakli I.H. MerR and ChrR Mediate Blue Light Induced Photo-Oxidative Stress Response at the Transcriptional Level in Vibrio Cholerae. Sci. Rep. 2017;7:40817. doi: 10.1038/srep40817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Nakajima Y., Kojima K., Kashiyama Y., Doi S., Nakai R., Sudo Y., Kogure K., Yoshizawa S. Bacterium Lacking a Known Gene for Retinal Biosynthesis Constructs Functional Rhodopsins. Microbes Environ. 2020;35 doi: 10.1264/jsme2.ME20085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Francis D. On the Eyespot of the Dinoflagellate, Nematodinium. J. Exp. Biol. 1967;47:495–501. doi: 10.1242/jeb.47.3.495. [DOI] [PubMed] [Google Scholar]
- 197.Kreimer G. The Green Algal Eyespot Apparatus: A Primordial Visual System and More? Curr. Genet. 2009;55:19–43. doi: 10.1007/s00294-008-0224-8. [DOI] [PubMed] [Google Scholar]
- 198.Gómez F. The Function of the Ocelloid and Piston in the Dinoflagellate Erythropsidinium (Gymnodiniales, Dinophyceae) J. Phycol. 2017;53:629–641. doi: 10.1111/jpy.12525. [DOI] [PubMed] [Google Scholar]
- 199.Gehring W.J. The Evolution of Vision. Wiley Interdiscip. Rev. Dev. Biol. 2014;3:1–40. doi: 10.1002/wdev.96. [DOI] [PubMed] [Google Scholar]
- 200.Losi A., Gärtner W. The Evolution of Flavin-Binding Photoreceptors: An Ancient Chromophore Serving Trendy Blue-Light Sensors. Annu. Rev. Plant. Biol. 2012;63:49–72. doi: 10.1146/annurev-arplant-042811-105538. [DOI] [PubMed] [Google Scholar]
- 201.Drews G. Contributions of Theodor Wilhelm Engelmann on Phototaxis, Chemotaxis, and Photosynthesis. Photosynth Res. 2005;83:25–34. doi: 10.1007/s11120-004-6313-8. [DOI] [PubMed] [Google Scholar]
- 202.McBride M.J. Shining a Light on an Opportunistic Pathogen. J. Bacteriol. 2010;192:6325–6326. doi: 10.1128/JB.01141-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Lubner C.E. Bacteria “Read” Light To Gain a Competitive Advantage. J. Bacteriol. 2019;201:e00082-19. doi: 10.1128/JB.00082-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Hoff W.D., van der Horst M.A., Nudel C.B., Hellingwerf K.J. Prokaryotic Phototaxis. Methods Mol. Biol. 2009;571:25–49. doi: 10.1007/978-1-60761-198-1_2. [DOI] [PubMed] [Google Scholar]
- 205.Taylor B.L., Miller J.B., Warrick H.M., Koshland D.E. Electron Acceptor Taxis and Blue Light Effect on Bacterial Chemotaxis. J. Bacteriol. 1979;140:567–573. doi: 10.1128/jb.140.2.567-573.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Sprenger W.W., Hoff W.D., Armitage J.P., Hellingwerf K.J. The Eubacterium Ectothiorhodospira Halophila Is Negatively Phototactic, with a Wavelength Dependence That Fits the Absorption Spectrum of the Photoactive Yellow Protein. J. Bacteriol. 1993;175:3096–3104. doi: 10.1128/jb.175.10.3096-3104.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Hoff W.D., Jung K.H., Spudich J.L. Molecular Mechanism of Photosignaling by Archaeal Sensory Rhodopsins. Annu. Rev. Biophys. Biomol. Struct. 1997;26:223–258. doi: 10.1146/annurev.biophys.26.1.223. [DOI] [PubMed] [Google Scholar]
- 208.Schuergers N., Mullineaux C.W., Wilde A. Cyanobacteria in Motion. Curr. Opin. Plant. Biol. 2017;37:109–115. doi: 10.1016/j.pbi.2017.03.018. [DOI] [PubMed] [Google Scholar]
- 209.Hardman S.J.O., Heyes D.J., Sazanovich I.V., Scrutton N.S. Photocycle of Cyanobacteriochrome TePixJ. Biochemistry. 2020;59:2909–2915. doi: 10.1021/acs.biochem.0c00382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Burgie E.S., Walker J.M., Phillips G.N., Vierstra R.D. A Photo-Labile Thioether Linkage to Phycoviolobilin Provides the Foundation for the Blue/Green Photocycles in DXCF-Cyanobacteriochromes. Structure. 2013;21:88–97. doi: 10.1016/j.str.2012.11.001. [DOI] [PubMed] [Google Scholar]
- 211.Ragatz L., Jiang Z.Y., Bauer C.E., Gest H. Macroscopic Phototactic Behavior of the Purple Photosynthetic Bacterium Rhodospirillum Centenum. Arch. Microbiol. 1995;163:1–6. doi: 10.1007/BF00262196. [DOI] [PubMed] [Google Scholar]
- 212.Menon S.N., Varuni P., Menon G.I. Information Integration and Collective Motility in Phototactic Cyanobacteria. PLoS Comput. Biol. 2020;16:e1007807. doi: 10.1371/journal.pcbi.1007807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Varuni P., Menon S.N., Menon G.I. Phototaxis as a Collective Phenomenon in Cyanobacterial Colonies. Sci. Rep. 2017;7:17799. doi: 10.1038/s41598-017-18160-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Losi A., Mandalari C., Gärtner W. From Plant Infectivity to Growth Patterns: The Role of Blue-Light Sensing in the Prokaryotic World. Plants. 2014;3:70–94. doi: 10.3390/plants3010070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Verma R.K., Biswas A., Kakkar A., Lomada S.K., Pradhan B.B., Chatterjee S. A Bacteriophytochrome Mediates Interplay between Light Sensing and the Second Messenger Cyclic Di-GMP to Control Social Behavior and Virulence. Cell Rep. 2020;32:108202. doi: 10.1016/j.celrep.2020.108202. [DOI] [PubMed] [Google Scholar]
- 216.Purcell E.B., Siegal-Gaskins D., Rawling D.C., Fiebig A., Crosson S. A Photosensory Two-Component System Regulates Bacterial Cell Attachment. Proc. Natl. Acad. Sci. USA. 2007;104:18241–18246. doi: 10.1073/pnas.0705887104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Mussi M.A., Gaddy J.A., Cabruja M., Arivett B.A., Viale A.M., Rasia R., Actis L.A. The Opportunistic Human Pathogen Acinetobacter Baumannii Senses and Responds to Light. J. Bacteriol. 2010;192:6336–6345. doi: 10.1128/JB.00917-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Santamaría-Hernando S., Cerna-Vargas J.P., Martínez-García P.M., de Francisco-de Polanco S., Nebreda S., Rodríguez-Palenzuela P., Rodríguez-Herva J.J., López-Solanilla E. Blue-Light Perception by Epiphytic Pseudomonas Syringae Drives Chemoreceptor Expression, Enabling Efficient Plant Infection. Mol. Plant. Pathol. 2020;21:1606–1619. doi: 10.1111/mpp.13001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Maresca J.A., Keffer J.L., Hempel P.P., Polson S.W., Shevchenko O., Bhavsar J., Powell D., Miller K.J., Singh A., Hahn M.W. Light Modulates the Physiology of Nonphototrophic Actinobacteria. J. Bacteriol. 2019;201 doi: 10.1128/JB.00740-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Losi A., Gärtner W. A Light Life Together: Photosensing in the Plant Microbiota. Photochem. Photobiol. Sci. 2021;20:451–473. doi: 10.1007/s43630-021-00029-7. [DOI] [PubMed] [Google Scholar]
- 221.Yin R., Dai T., Avci P., Jorge A.E.S., de Melo W.C.M.A., Vecchio D., Huang Y.-Y., Gupta A., Hamblin M.R. Light Based Anti-Infectives: Ultraviolet C Irradiation, Photodynamic Therapy, Blue Light, and Beyond. Curr. Opin. Pharmacol. 2013;13:731–762. doi: 10.1016/j.coph.2013.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Dai T., Gupta A., Murray C.K., Vrahas M.S., Tegos G.P., Hamblin M.R. Blue Light for Infectious Diseases: Propionibacterium Acnes, Helicobacter Pylori, and Beyond? Drug Resist. Updates. 2012;15:223–236. doi: 10.1016/j.drup.2012.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Angarano V., Smet C., Akkermans S., Watt C., Chieffi A., Van Impe J.F.M. Visible Light as an Antimicrobial Strategy for Inactivation of Pseudomonas Fluorescens and Staphylococcus Epidermidis Biofilms. Antibiotics. 2020;9:171. doi: 10.3390/antibiotics9040171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Hadi J., Wu S., Brightwell G. Antimicrobial Blue Light versus Pathogenic Bacteria: Mechanism, Application in the Food Industry, Hurdle Technologies and Potential Resistance. Foods. 2020;9:1895. doi: 10.3390/foods9121895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Van der Horst M.A., Stalcup T.P., Kaledhonkar S., Kumauchi M., Hara M., Xie A., Hellingwerf K.J., Hoff W.D. Locked Chromophore Analogs Reveal That Photoactive Yellow Protein Regulates Biofilm Formation in the Deep Sea Bacterium Idiomarina Loihiensis. J. Am. Chem. Soc. 2009;131:17443–17451. doi: 10.1021/ja9057103. [DOI] [PubMed] [Google Scholar]
- 226.Liu G., Shan Y., Zheng R., Liu R., Sun C. Growth Promotion of a Deep-Sea Bacterium by Sensing Infrared Light through a Bacteriophytochrome Photoreceptor. Environ. Microbiol. 2021;23:4466–4477. doi: 10.1111/1462-2920.15639. [DOI] [PubMed] [Google Scholar]
- 227.Foster K.W., Smyth R.D. Light Antennas in Phototactic Algae. Microbiol. Rev. 1980;44:572–630. doi: 10.1128/mr.44.4.572-630.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Lenci F., Colombetti G. Photobehavior of Microorganisms: A Biophysical Approach. Annu. Rev. Biophys. Bioeng. 1978;7:341–361. doi: 10.1146/annurev.bb.07.060178.002013. [DOI] [PubMed] [Google Scholar]
- 229.Moldrup M., Moestrup Ø., Hansen P.J. Loss of Phototaxis and Degeneration of an Eyespot in Long-Term Algal Cultures: Evidence from Ultrastructure and Behaviour in the Dinoflagellate Kryptoperidinium Foliaceum. J. Eukaryot. Microbiol. 2013;60:327–334. doi: 10.1111/jeu.12036. [DOI] [PubMed] [Google Scholar]
- 230.Moldrup M., Garm A. Spectral Sensitivity of Phototaxis in the Dinoflagellate Kryptoperidinium Foliaceum and Their Reaction to Physical Encounters. J. Exp. Biol. 2012;215:2342–2346. doi: 10.1242/jeb.066886. [DOI] [PubMed] [Google Scholar]
- 231.Hartz A.J., Sherr B.F., Sherr E.B. Photoresponse in the Heterotrophic Marine Dinoflagellate Oxyrrhis Marina. J. Eukaryot. Microbiol. 2011;58:171–177. doi: 10.1111/j.1550-7408.2011.00529.x. [DOI] [PubMed] [Google Scholar]
- 232.Forward R.B. Phototaxis in a Dinoflagellate: Action Spectra as Evidence for a Two-Pigment System. Planta. 1973;111:167–178. doi: 10.1007/BF00386277. [DOI] [PubMed] [Google Scholar]
- 233.Forward R.B. Phototaxis by the Dinoflagellate Gymnodinium Splendens Lebour. J. Protozool. 1974;21:312–315. doi: 10.1111/j.1550-7408.1974.tb03659.x. [DOI] [PubMed] [Google Scholar]
- 234.Häder D.P., Häder M., Liu S.M., Ullrich W. Effects of Solar Radiation on Photoorientation, Motility and Pigmentation in a Freshwater Peridinium. Biosystems. 1990;23:335–343. doi: 10.1016/0303-2647(90)90015-S. [DOI] [PubMed] [Google Scholar]
- 235.Saranak J., Foster K.W. Photoreceptor for Curling Behavior in Peranema Trichophorum and Evolution of Eukaryotic Rhodopsins. Eukaryot. Cell. 2005;4:1605–1612. doi: 10.1128/EC.4.10.1605-1612.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Sullivan J.M., Swift E. Photoenhancement of Bioluminescence Capacity in Natural and Laboratory Populations of the Autotrophic Dinoflagellate Ceratium Fusus (Ehrenb.) Dujardin. J. Geophys. Res. Oceans. 1995;100:6565–6574. doi: 10.1029/94JC01511. [DOI] [Google Scholar]
- 237.Iseki M., Matsunaga S., Murakami A., Ohno K., Shiga K., Yoshida K., Sugai M., Takahashi T., Hori T., Watanabe M. A Blue-Light-Activated Adenylyl Cyclase Mediates Photoavoidance in Euglena Gracilis. Nature. 2002;415:1047–1051. doi: 10.1038/4151047a. [DOI] [PubMed] [Google Scholar]
- 238.Häder D.-P., Iseki M. Photomovement in Euglena. Adv. Exp Med. Biol. 2017;979:207–235. doi: 10.1007/978-3-319-54910-1_11. [DOI] [PubMed] [Google Scholar]
- 239.Sobierajska K., Fabczak H., Fabczak S. Photosensory Transduction in Unicellular Eukaryotes: A Comparison between Related Ciliates Blepharisma Japonicum and Stentor Coeruleus and Photoreceptor Cells of Higher Organisms. J. Photochem. Photobiol. B. 2006;83:163–171. doi: 10.1016/j.jphotobiol.2006.01.005. [DOI] [PubMed] [Google Scholar]
- 240.Hultberg M., Asp H., Marttila S., Bergstrand K.-J., Gustafsson S. Biofilm Formation by Chlorella Vulgaris Is Affected by Light Quality. Curr. Microbiol. 2014;69:699–702. doi: 10.1007/s00284-014-0645-1. [DOI] [PubMed] [Google Scholar]
- 241.Suetsugu N., Wada M. Evolution of Three LOV Blue Light Receptor Families in Green Plants and Photosynthetic Stramenopiles: Phototropin, ZTL/FKF1/LKP2 and Aureochrome. Plant. Cell Physiol. 2013;54:8–23. doi: 10.1093/pcp/pcs165. [DOI] [PubMed] [Google Scholar]
- 242.Higa T., Suetsugu N., Wada M. Plant Nuclear Photorelocation Movement. J. Exp. Bot. 2014;65:2873–2881. doi: 10.1093/jxb/ert414. [DOI] [PubMed] [Google Scholar]
- 243.Zou Y., Wenzel S., Müller N., Prager K., Jung E.-M., Kothe E., Kottke T., Mittag M. An Animal-Like Cryptochrome Controls the Chlamydomonas Sexual Cycle. Plant. Physiol. 2017;174:1334–1347. doi: 10.1104/pp.17.00493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Rivera A.S., Ozturk N., Fahey B., Plachetzki D.C., Degnan B.M., Sancar A., Oakley T.H. Blue-Light-Receptive Cryptochrome Is Expressed in a Sponge Eye Lacking Neurons and Opsin. J. Exp. Biol. 2012;215:1278–1286. doi: 10.1242/jeb.067140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Aihara Y., Maruyama S., Baird A.H., Iguchi A., Takahashi S., Minagawa J. Green Fluorescence from Cnidarian Hosts Attracts Symbiotic Algae. Proc. Natl. Acad. Sci. USA. 2019;116:2118–2123. doi: 10.1073/pnas.1812257116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Binet A. The Psychic Life of Micro-Organisms. A Study in Experimental Psychology. The Open Court Publishing Company, London Agents Kegan Paul Trench, Trubner & Co. LTD; London, UK: 1888. [Google Scholar]
- 247.Jennings H.S., Herbert S. Behavior of the Lower Organisms. Columbia University Press; New York, NY, USA: 1906. [Google Scholar]
- 248.Loeb J. Harvard University Press; Cambridge, MA, USA: 2013. The Mechanistic Conception of Life. [Google Scholar]
- 249.Mukherjee S., Bassler B.L. Bacterial Quorum Sensing in Complex and Dynamically Changing Environments. Nat. Rev. Microbiol. 2019;17:371–382. doi: 10.1038/s41579-019-0186-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Shiner E.K., Rumbaugh K.P., Williams S.C. Inter-Kingdom Signaling: Deciphering the Language of Acyl Homoserine Lactones. FEMS Microbiol. Rev. 2005;29:935–947. doi: 10.1016/j.femsre.2005.03.001. [DOI] [PubMed] [Google Scholar]
- 251.Reading N.C., Sperandio V. Quorum Sensing: The Many Languages of Bacteria. FEMS Microbiol. Lett. 2006;254:1–11. doi: 10.1111/j.1574-6968.2005.00001.x. [DOI] [PubMed] [Google Scholar]
- 252.Joint I., Tait K., Wheeler G. Cross-Kingdom Signalling: Exploitation of Bacterial Quorum Sensing Molecules by the Green Seaweed Ulva. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2007;362:1223–1233. doi: 10.1098/rstb.2007.2047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Henke J.M., Bassler B.L. Bacterial Social Engagements. Trends Cell Biol. 2004;14:648–656. doi: 10.1016/j.tcb.2004.09.012. [DOI] [PubMed] [Google Scholar]
- 254.Mukherjee S., Jemielita M., Stergioula V., Tikhonov M., Bassler B.L. Photosensing and Quorum Sensing Are Integrated to Control Pseudomonas Aeruginosa Collective Behaviors. PLoS Biol. 2019;17:e3000579. doi: 10.1371/journal.pbio.3000579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Reguera G. When Microbial Conversations Get Physical. Trends Microbiol. 2011;19:105–113. doi: 10.1016/j.tim.2010.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Cifra M., Fields J.Z., Farhadi A. Electromagnetic Cellular Interactions. Prog. Biophys. Mol. Biol. 2011;105:223–246. doi: 10.1016/j.pbiomolbio.2010.07.003. [DOI] [PubMed] [Google Scholar]
- 257.Kučera O., Cifra M. Cell-to-Cell Signaling through Light: Just a Ghost of Chance? Cell Commun. Signal. 2013;11:87. doi: 10.1186/1478-811X-11-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Scholkmann F., Fels D., Cifra M. Non-Chemical and Non-Contact Cell-to-Cell Communication: A Short Review. Am. J. Transl. Res. 2013;5:586–593. [PMC free article] [PubMed] [Google Scholar]
- 259.Humphries J., Xiong L., Liu J., Prindle A., Yuan F., Arjes H.A., Tsimring L., Süel G.M. Species-Independent Attraction to Biofilms through Electrical Signaling. Cell. 2017;168:200–209.e12. doi: 10.1016/j.cell.2016.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Larkin J.W., Zhai X., Kikuchi K., Redford S.E., Prindle A., Liu J., Greenfield S., Walczak A.M., Garcia-Ojalvo J., Mugler A., et al. Signal Percolation within a Bacterial Community. Cell Syst. 2018;7:137–145.e3. doi: 10.1016/j.cels.2018.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Mitchell R.J., Lee S.K., Kim T., Ghim C.-M. Microbial Linguistics: Perspectives and Applications of Microbial Cell-to-Cell Communication. BMB Rep. 2011;44:1–10. doi: 10.5483/BMBRep.2011.44.1.1. [DOI] [PubMed] [Google Scholar]
- 262.Ben Jacob E., Becker I., Shapira Y., Levine H. Bacterial Linguistic Communication and Social Intelligence. Trends Microbiol. 2004;12:366–372. doi: 10.1016/j.tim.2004.06.006. [DOI] [PubMed] [Google Scholar]
- 263.Majumdar S., Pal S. Information Transmission in Microbial and Fungal Communication: From Classical to Quantum. J. Cell Commun. Signal. 2018;12:491–502. doi: 10.1007/s12079-018-0462-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Fels D. Endogenous Physical Regulation of Population Density in the Freshwater Protozoan Paramecium Caudatum. Sci. Rep. 2017;7:13800. doi: 10.1038/s41598-017-14231-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Bassler B.L., Losick R. Bacterially Speaking. Cell. 2006;125:237–246. doi: 10.1016/j.cell.2006.04.001. [DOI] [PubMed] [Google Scholar]
- 266.Combarnous Y., Nguyen T.M.D. Cell Communications among Microorganisms, Plants, and Animals: Origin, Evolution, and Interplays. Int. J. Mol. Sci. 2020;21:8052. doi: 10.3390/ijms21218052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Bruni G.N., Weekley R.A., Dodd B.J.T., Kralj J.M. Voltage-Gated Calcium Flux Mediates Escherichia Coli Mechanosensation. Proc. Natl. Acad. Sci. USA. 2017;114:9445–9450. doi: 10.1073/pnas.1703084114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Adler J. Bacterial Chemotaxis and Molecular Neurobiology. Cold Spring Harb. Symp. Quant. Biol. 1983;48:803–804. doi: 10.1101/SQB.1983.048.01.082. [DOI] [PubMed] [Google Scholar]
- 269.Koshland D.E. Bacterial Chemotaxis in Relation to Neurobiology. Annu. Rev. Neurosci. 1980;3:43–75. doi: 10.1146/annurev.ne.03.030180.000355. [DOI] [PubMed] [Google Scholar]
- 270.Adler J. Motile Escherichia Coli Migrate in Bands That Are Influenced by Oxygen and Organic Nutrients. Science. 1966;153:708–716. doi: 10.1126/science.153.3737.708. [DOI] [PubMed] [Google Scholar]
- 271.Galperin M.Y. What Bacteria Want. Environ. Microbiol. 2018;20:4221–4229. doi: 10.1111/1462-2920.14398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.Baker M.D., Wolanin P.M., Stock J.B. Signal Transduction in Bacterial Chemotaxis. Bioessays. 2006;28:9–22. doi: 10.1002/bies.20343. [DOI] [PubMed] [Google Scholar]
- 273.Micali G., Endres R.G. Bacterial Chemotaxis: Information Processing, Thermodynamics, and Behavior. Curr. Opin. Microbiol. 2016;30:8–15. doi: 10.1016/j.mib.2015.12.001. [DOI] [PubMed] [Google Scholar]
- 274.Hellingwerf K.J. Bacterial Observations: A Rudimentary Form of Intelligence? Trends Microbiol. 2005;13:152–158. doi: 10.1016/j.tim.2005.02.001. [DOI] [PubMed] [Google Scholar]
- 275.Hazelbauer G.L., Falke J.J., Parkinson J.S. Bacterial Chemoreceptors: High-Performance Signaling in Networked Arrays. Trends Biochem. Sci. 2008;33:9–19. doi: 10.1016/j.tibs.2007.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276.Falke J.J., Piasta K.N. Architecture and Signal Transduction Mechanism of the Bacterial Chemosensory Array: Progress, Controversies, and Challenges. Curr. Opin. Struct. Biol. 2014;29:85–94. doi: 10.1016/j.sbi.2014.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277.Adler J., Tso W.W. “Decision”-Making in Bacteria: Chemotactic Response of Escherichia Coli to Conflicting Stimuli. Science. 1974;184:1292–1294. doi: 10.1126/science.184.4143.1292. [DOI] [PubMed] [Google Scholar]
- 278.Koshland D.E. A Response Regulator Model in a Simple Sensory System. Science. 1977;196:1055–1063. doi: 10.1126/science.870969. [DOI] [PubMed] [Google Scholar]
- 279.Koshland D.E., Goldbeter A., Stock J.B. Amplification and Adaptation in Regulatory and Sensory Systems. Science. 1982;217:220–225. doi: 10.1126/science.7089556. [DOI] [PubMed] [Google Scholar]
- 280.Galperin M.Y. A Census of Membrane-Bound and Intracellular Signal Transduction Proteins in Bacteria: Bacterial IQ, Extroverts and Introverts. BMC Microbiol. 2005;5:35. doi: 10.1186/1471-2180-5-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 281.Baluška F., Miller W.B. Senomic View of the Cell: Senome versus Genome. Commun. Integr. Biol. 2018;11:1–9. doi: 10.1080/19420889.2018.1489184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 282.Ben-Jacob E., Levine H. Self-Engineering Capabilities of Bacteria. J. R. Soc. Interface. 2006;3:197–214. doi: 10.1098/rsif.2005.0089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 283.Keijzer F. Moving and Sensing without Input and Output: Early Nervous Systems and the Origins of the Animal Sensorimotor Organization. Biol. Philos. 2015;30:311–331. doi: 10.1007/s10539-015-9483-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 284.Pattee H.H. Cell Phenomenology: The First Phenomenon. Prog. Biophys. Mol. Biol. 2015;119:461–468. doi: 10.1016/j.pbiomolbio.2015.06.010. [DOI] [PubMed] [Google Scholar]
- 285.Richardson K. Heritability Lost; Intelligence Found. Intelligence Is Integral to the Adaptation and Survival of All Organisms Faced with Changing Environments. EMBO Rep. 2012;13:591–595. doi: 10.1038/embor.2012.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 286.Hein A.M., Carrara F., Brumley D.R., Stocker R., Levin S.A. Natural Search Algorithms as a Bridge between Organisms, Evolution, and Ecology. Proc. Natl. Acad. Sci. USA. 2016;113:9413–9420. doi: 10.1073/pnas.1606195113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 287.Kandel E., Abel T. Neuropeptides, Adenylyl Cyclase, and Memory Storage. Science. 1995;268:825–826. doi: 10.1126/science.7754367. [DOI] [PubMed] [Google Scholar]
- 288.Dussutour A. Learning in Single Cell Organisms. Biochem. Biophys. Res. Commun. 2021;564:92–102. doi: 10.1016/j.bbrc.2021.02.018. [DOI] [PubMed] [Google Scholar]
- 289.Marshall W.F. Cellular Cognition: Sequential Logic in a Giant Protist. Curr. Biol. 2019;29:R1303–R1305. doi: 10.1016/j.cub.2019.10.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 290.Beekman M., Latty T. Brainless but Multi-Headed: Decision Making by the Acellular Slime Mould Physarum Polycephalum. J. Mol. Biol. 2015;427:3734–3743. doi: 10.1016/j.jmb.2015.07.007. [DOI] [PubMed] [Google Scholar]
- 291.De la Fuente I.M., Bringas C., Malaina I., Fedetz M., Carrasco-Pujante J., Morales M., Knafo S., Martínez L., Pérez-Samartín A., López J.I., et al. Evidence of Conditioned Behavior in Amoebae. Nat. Commun. 2019;10:3690. doi: 10.1038/s41467-019-11677-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 292.Burgos J.E. Is a Nervous System Necessary for Learning? Perspect. Behav. Sci. 2018;41:343–368. doi: 10.1007/s40614-018-00179-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 293.Majumdar S., Pal S. Cross- Species Communication in Bacterial World. J. Cell Commun. Signal. 2017;11:187–190. doi: 10.1007/s12079-017-0383-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 294.Manna S., Ghanty C., Baindara P., Barik T.K., Mandal S.M. Electrochemical Communication in Biofilm of Bacterial Community. J. Basic Microbiol. 2020;60:819–827. doi: 10.1002/jobm.202000340. [DOI] [PubMed] [Google Scholar]
- 295.Fels D. Cellular Communication through Light. PLoS ONE. 2009;4:e5086. doi: 10.1371/annotation/8d99ccc5-cc76-44f4-b468-d63e42e0b9e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 296.Trushin M.V. Studies on Distant Regulation of Bacterial Growth and Light Emission. Microbiology. 2003;149:363–368. doi: 10.1099/mic.0.25825-0. [DOI] [PubMed] [Google Scholar]
- 297.Trushin M.V. Light-Mediated “Conversation” among Microorganisms. Microbiol. Res. 2004;159:1–10. doi: 10.1016/j.micres.2003.11.001. [DOI] [PubMed] [Google Scholar]
- 298.Seliger H.H. The Origin of Bioluminescence. Photochem. Photobiol. 1975;21:355–361. doi: 10.1111/j.1751-1097.1975.tb06684.x. [DOI] [PubMed] [Google Scholar]
- 299.Seliger H.H. The Evolution of Bioluminescence in Bacteria. Photochem. Photobiol. 1987;45:291–297. doi: 10.1111/j.1751-1097.1987.tb05377.x. [DOI] [Google Scholar]
- 300.O’Kane D.J., Prasher D.C. Evolutionary Origins of Bacterial Bioluminescence. Mol. Microbiol. 1992;6:443–449. doi: 10.1111/j.1365-2958.1992.tb01488.x. [DOI] [PubMed] [Google Scholar]
- 301.Miyashiro T., Ruby E.G. Shedding Light on Bioluminescence Regulation in Vibrio Fischeri. Mol. Microbiol. 2012;84:795–806. doi: 10.1111/j.1365-2958.2012.08065.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 302.Visick K.L., Foster J., Doino J., McFall-Ngai M., Ruby E.G. Vibrio Fischeri Lux Genes Play an Important Role in Colonization and Development of the Host Light Organ. J. Bacteriol. 2000;182:4578–4586. doi: 10.1128/JB.182.16.4578-4586.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 303.Tong D., Rozas N.S., Oakley T.H., Mitchell J., Colley N.J., McFall-Ngai M.J. Evidence for Light Perception in a Bioluminescent Organ. Proc. Natl. Acad. Sci. USA. 2009;106:9836–9841. doi: 10.1073/pnas.0904571106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 304.Tanet L., Tamburini C., Baumas C., Garel M., Simon G., Casalot L. Bacterial Bioluminescence: Light Emission in Photobacterium Phosphoreum Is Not Under Quorum-Sensing Control. Front. Microbiol. 2019;10:365. doi: 10.3389/fmicb.2019.00365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 305.Norsworthy A.N., Visick K.L. Gimme Shelter: How Vibrio Fischeri Successfully Navigates an Animal’s Multiple Environments. Front. Microbiol. 2013;4:356. doi: 10.3389/fmicb.2013.00356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 306.Septer Alecia N., Lyell Noreen L., Stabb Eric V. The Iron-Dependent Regulator Fur Controls Pheromone Signaling Systems and Luminescence in the Squid Symbiont Vibrio Fischeri ES114. Appl. Environ. Microbiol. 2013;79:1826–1834. doi: 10.1128/AEM.03079-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 307.Septer A.N., Bose J.L., Lipzen A., Martin J., Whistler C., Stabb E.V. Bright Luminescence of Vibrio Fischeri Aconitase Mutants Reveals a Connection between Citrate and the Gac/Csr Regulatory System. Mol. Microbiol. 2015;95:283–296. doi: 10.1111/mmi.12864. [DOI] [PubMed] [Google Scholar]
- 308.Septer A.N., Bose J.L., Dunn A.K., Stabb E.V. FNR-Mediated Regulation of Bioluminescence and Anaerobic Respiration in the Light-Organ Symbiont Vibrio Fischeri. FEMS Microbiol. Lett. 2010;306:72–81. doi: 10.1111/j.1574-6968.2010.01938.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 309.Tabei Y., Ogawa A., Era M., Ninomiya J., Morita H. Influence of Cations and Anions on the Induction of Cell Density-Independent Luminescence in Photorhabdus Luminescens. J. Basic Microbiol. 2013;53:268–276. doi: 10.1002/jobm.201100568. [DOI] [PubMed] [Google Scholar]
- 310.Tabei Y., Era M., Ogawa A., Morita H. Effects of Magnesium Sulfate on the Luminescence of Vibrio Fischeri under Nutrient-Starved Conditions. Biosci. Biotechnol. Biochem. 2011;75:1073–1078. doi: 10.1271/bbb.100880. [DOI] [PubMed] [Google Scholar]
- 311.Clarke D.J. Photorhabdus: A Tale of Contrasting Interactions. Microbiology. 2020;166:335–348. doi: 10.1099/mic.0.000907. [DOI] [PubMed] [Google Scholar]
- 312.Eckstein S., Heermann R. Regulation of Phenotypic Switching and Heterogeneity in Photorhabdus Luminescens Cell Populations. J. Mol. Biol. 2019;431:4559–4568. doi: 10.1016/j.jmb.2019.04.015. [DOI] [PubMed] [Google Scholar]
- 313.Regaiolo A., Dominelli N., Andresen K., Heermann R. The Biocontrol Agent and Insect Pathogen Photorhabdus Luminescens Interacts with Plant Roots. Appl. Environ. Microbiol. 2020;86 doi: 10.1128/AEM.00891-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 314.Mo M., Yokawa K., Wan Y., Baluška F. How and Why Do Root Apices Sense Light under the Soil Surface? Front. Plant. Sci. 2015;6:775. doi: 10.3389/fpls.2015.00775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 315.Wampler J.E., Jamieson B.G.M. Earthworm Bioluminescence: Comparative Physiology and Biochemistry. Compar. Biochem. Physiol. Part. B Compar. Biochem. 1980;66:43–50. doi: 10.1016/0305-0491(80)90081-4. [DOI] [Google Scholar]
- 316.Rodionova N.S., Rota E., Tsarkova A.S., Petushkov V.N. Progress in the Study of Bioluminescent Earthworms. Photochem. Photobiol. 2017;93:416–428. doi: 10.1111/php.12709. [DOI] [PubMed] [Google Scholar]
- 317.Verdes A., Gruber D.F. Glowing Worms: Biological, Chemical, and Functional Diversity of Bioluminescent Annelids. Integr. Comp. Biol. 2017;57:18–32. doi: 10.1093/icb/icx017. [DOI] [PubMed] [Google Scholar]
- 318.Burtseva O., Baulina O., Zaytseva A., Fedorenko T., Chekanov K., Lobakova E. In Vitro Biofilm Formation by Bioluminescent Bacteria Isolated from the Marine Fish Gut. Microb Ecol. 2021;81:932–940. doi: 10.1007/s00248-020-01652-0. [DOI] [PubMed] [Google Scholar]
- 319.Hirsch, null; McFall-Ngai, null Fundamental Concepts in Symbiotic Interactions: Light and Dark, Day and Night, Squid and Legume. J. Plant. Growth Regul. 2000;19:113–130. doi: 10.1007/s003440000025. [DOI] [PubMed] [Google Scholar]
- 320.Grimes D.J., Ford T.E., Colwell R.R., Baker-Austin C., Martinez-Urtaza J., Subramaniam A., Capone D.G. Viewing Marine Bacteria, Their Activity and Response to Environmental Drivers from Orbit: Satellite Remote Sensing of Bacteria. Microb Ecol. 2014;67:489–500. doi: 10.1007/s00248-013-0363-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 321.Miller S.D., Haddock S.H.D., Elvidge C.D., Lee T.F. Detection of a Bioluminescent Milky Sea from Space. Proc. Natl. Acad. Sci. USA. 2005;102:14181–14184. doi: 10.1073/pnas.0507253102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 322.Lapota D., Galt C., Losee J.R., Huddell H.D., Orzech J.K., Nealson K.H. Observations and Measurements of Planktonic Bioluminescence in and Around a Milky Sea. Naval Ocean Systems Center; San Diego, CA, USA: 1988. [Google Scholar]
- 323.Tamburini C., Canals M., Durrieu de Madron X., Houpert L., Lefèvre D., Martini S., D’Ortenzio F., Robert A., Testor P., Aguilar J.A., et al. Deep-Sea Bioluminescence Blooms after Dense Water Formation at the Ocean Surface. PLoS ONE. 2013;8:e67523. doi: 10.1371/journal.pone.0067523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 324.Cusick K.D., Widder E.A. Bioluminescence and Toxicity as Driving Factors in Harmful Algal Blooms: Ecological Functions and Genetic Variability. Harmful Algae. 2020;98:101850. doi: 10.1016/j.hal.2020.101850. [DOI] [PubMed] [Google Scholar]
- 325.DeSa R., Hastings J.W. The Characterization of Scintillons: Bioluminescent Particles from the Marine Dinoflagellate, Gonyaulax Polyedra. J. Gen. Physiol. 1968;51:105–122. doi: 10.1085/jgp.51.1.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 326.Johnson C.H., Inoué S., Flint A., Hastings J.W. Compartmentalization of Algal Bioluminescence: Autofluorescence of Bioluminescent Particles in the Dinoflagellate Gonyaulax as Studied with Image-Intensified Video Microscopy and Flow Cytometry. J. Cell Biol. 1985;100:1435–1446. doi: 10.1083/jcb.100.5.1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 327.Nicolas M.T., Sweeney B.M., Hastings J.W. The Ultrastructural Localization of Luciferase in Three Bioluminescent Dinoflagellates, Two Species of Pyrocystis, and Noctiluca, Using Anti-Luciferase and Immunogold Labelling. J. Cell Sci. 1987;87:189–196. doi: 10.1242/jcs.87.1.189. [DOI] [PubMed] [Google Scholar]
- 328.Latz M.I., Bovard M., VanDelinder V., Segre E., Rohr J., Groisman A. Bioluminescent Response of Individual Dinoflagellate Cells to Hydrodynamic Stress Measured with Millisecond Resolution in a Microfluidic Device. J. Exp. Biol. 2008;211:2865–2875. doi: 10.1242/jeb.011890. [DOI] [PubMed] [Google Scholar]
- 329.Chen A.K., Latz M.I., Sobolewski P., Frangos J.A. Evidence for the Role of G-Proteins in Flow Stimulation of Dinoflagellate Bioluminescence. Am. J. Physiol.-Regul. Integr. Compar. Physiol. 2007;292:R2020–R2027. doi: 10.1152/ajpregu.00649.2006. [DOI] [PubMed] [Google Scholar]
- 330.Von Dassow P., Latz M.I. The Role of Ca2+ in Stimulated Bioluminescence of the Dinoflagellate Lingulodinium Polyedrum. J. Exp. Biol. 2002;205:2971–2986. doi: 10.1242/jeb.205.19.2971. [DOI] [PubMed] [Google Scholar]
- 331.Fogel M., Hastings J.W. Bioluminescence: Mechanism and Mode of Control of Scintillon Activity. Proc. Natl. Acad. Sci. USA. 1972;69:690–693. doi: 10.1073/pnas.69.3.690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 332.Smith S.M.E., Morgan D., Musset B., Cherny V.V., Place A.R., Hastings J.W., Decoursey T.E. Voltage-Gated Proton Channel in a Dinoflagellate. Proc. Natl. Acad. Sci. USA. 2011;108:18162–18167. doi: 10.1073/pnas.1115405108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 333.Cusick K.D., Widder E.A. Intensity Differences in Bioluminescent Dinoflagellates Impact Foraging Efficiency in a Nocturnal Predator. Bull. Mar. Sci. 2014;90:797–811. doi: 10.5343/bms.2013.1059. [DOI] [Google Scholar]
- 334.Biggley W.H., Swift E., Buchanan R.J., Seliger H.H. Stimulable and Spontaneous Bioluminescence in the Marine Dinoflagellates, Pyrodinium Bahamense, Gonyaulax Polyedra, and Pyrocystis Lunula. J. Gen. Physiol. 1969;54:96–122. doi: 10.1085/jgp.54.1.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 335.Latz M.I., Nauen J.C., Rohr J. Bioluminescence Response of Four Species of Dinoflagellates to Fully Developed Pipe Flow. J. Plankton Res. 2004;26:1529–1546. doi: 10.1093/plankt/fbh141. [DOI] [Google Scholar]
- 336.Esaias W.E., Curl H.C. Effect of Dinoflagellate Bioluminescence on Copepod Ingestion Rates. Limnol. Oceanogr. 1972;17:901–906. doi: 10.4319/lo.1972.17.6.0901. [DOI] [Google Scholar]
- 337.Valiadi M., de Rond T., Amorim A., Gittins J.R., Gubili C., Moore B.S., Iglesias-Rodriguez M.D., Latz M.I. Molecular and Biochemical Basis for the Loss of Bioluminescence in the Dinoflagellate Noctiluca Scintillans along the West Coast of the U.S.A. Limnol. Oceanogr. 2019;64:2709–2724. doi: 10.1002/lno.11309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 338.Latz M.I., Lee A.O. Spontaneous and Stimulated Bioluminescence of the Dinoflagellate Ceratocorys Horrzda (Peridiniales)1. J. Phycol. 1995;31:120–132. doi: 10.1111/j.0022-3646.1995.00120.x. [DOI] [Google Scholar]
- 339.Krasnow R., Dunlap J.C., Taylor W., Hastings J.W., Vetterling W., Gooch V. Circadian Spontaneous Bioluminescent Glow and Flashing of Gonyaulax Polyedra. J. Comparat. Physiol. 1980;138:19–26. doi: 10.1007/BF00688730. [DOI] [Google Scholar]
- 340.Daniel Hickman G., Edmonds J.A., Lynch R.V. Laser-Induced Marine Bioluminescence Measurements and the Potential for Airborne Remote Sensing. Remote Sens. Environ. 1984;15:77–89. doi: 10.1016/0034-4257(84)90053-1. [DOI] [Google Scholar]
- 341.Hickman G.D., Richard V., Lynch I.I.I. Laser-Induced Bioluminescence; Proceedings of the Los Alamos Conference on Optics ’81, International Society for Optics and Photonics; Los Alamos, NM, USA. 30 December 1981; pp. 263–268. [Google Scholar]
- 342.Sweeney B.M., Fork D.C., Satoh K. Stimulation of Bioluminescence in Dinoflagellates by Red Light*. Photochem. Photobiol. 1983;37:457–465. doi: 10.1111/j.1751-1097.1983.tb04499.x. [DOI] [Google Scholar]
- 343.Roenneberg T., Merrow M. Circadian Systems: Different Levels of Complexity. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2001;356:1687–1696. doi: 10.1098/rstb.2001.0969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 344.Esaias W.E., Curl H.C., Seliger H.H. Action Spectrum for a Low Intensity, Rapid Photoinhibition of Mechanically Stimulable Bioluminescence in the Marine Dinoflagellates Gonyaulax Catenella, G. Acatenella, and G. Tamarensis. J. Cell Physiol. 1973;82:363–372. doi: 10.1002/jcp.1040820306. [DOI] [PubMed] [Google Scholar]
- 345.Hamman J.P., Seliger H.H. The Chemical Mimicking of the Mechanical Stimulation, Photoinhibition, and Recovery from Photoinhibition of Bioluminescence in Marine Dinoflagellate, Gonyaulax Polyedra. J. Cell Physiol. 1982;111:315–319. doi: 10.1002/jcp.1041110314. [DOI] [PubMed] [Google Scholar]
- 346.Sullivan J.M., Swift E. Photoinhibition of Mechanically Stimulable Bioluminescence in the Autotrophic Dinoflagellate Ceratium Fusus (Pyrrophyta)1. J. Phycol. 1994;30:627–633. doi: 10.1111/j.0022-3646.1994.00627.x. [DOI] [Google Scholar]
- 347.Ekelund N.G.A., Björn L.O. Photophobic Stop-Response in a Dinoflagellate: Modulation by Preirradiation. Physiol. Plantarum. 1987;70:394–398. doi: 10.1111/j.1399-3054.1987.tb02834.x. [DOI] [Google Scholar]
- 348.Forward R., Davenport D. Red and Far-Red Light Effects on a Short-Term Behavioral Response of a Dinoflagellate. Science. 1968;161:1028–1029. doi: 10.1126/science.161.3845.1028. [DOI] [PubMed] [Google Scholar]
- 349.Sweeney B.M., Hastings J.W. Characteristics of the Diurnal Rhythm of Luminescence in Gonyaulax Polyedra. J. Cell. Compar. Physiol. 1957;49:115–128. doi: 10.1002/jcp.1030490107. [DOI] [Google Scholar]
- 350.Hastings J.W., Sweeney B.M. A Persistent Diurnal Rhythm of Luminescence in Gonyaulax Polyedra. Biol. Bull. 1958;115:440–458. doi: 10.2307/1539108. [DOI] [Google Scholar]
- 351.Roenneberg T., Hastings J.W. Two Photoreceptors Control the Circadian Clock of a Unicellular Alga. Naturwissenschaften. 1988;75:206–207. doi: 10.1007/BF00735584. [DOI] [PubMed] [Google Scholar]
- 352.Roenneberg T., Deng T.-S. Photobiology of the Gonyaulax Circadian System. I. Different Phase Response Curves for Red and Blue Light. Planta. 1997;202:494–501. doi: 10.1007/s004250050154. [DOI] [Google Scholar]
- 353.Roenneberg T., Taylor W. Light-Induced Phase Responses in Gonyaulax Are Drastically Altered by Creatine. J. Biol. Rhythms. 1994;9:1–12. doi: 10.1177/074873049400900101. [DOI] [PubMed] [Google Scholar]
- 354.Morse D., Milos P.M., Roux E., Hastings J.W. Circadian Regulation of Bioluminescence in Gonyaulax Involves Translational Control. Proc. Natl. Acad. Sci. USA. 1989;86:172–176. doi: 10.1073/pnas.86.1.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 355.Marcinko C.L.J., Allen J.T., Poulton A.J., Painter S.C., Martin A.P. Diurnal Variations of Dinoflagellate Bioluminescence within the Open-Ocean North-East Atlantic. J. Plankton Res. 2013;35:177–190. doi: 10.1093/plankt/fbs081. [DOI] [Google Scholar]
- 356.Backus R.H., Yentsch C.S., Wing A. Bioluminescence in the Surface Waters of the Sea. Nature. 1961;192:518–521. doi: 10.1038/192518a0. [DOI] [Google Scholar]
- 357.Yentsch C.S., Backus R.H., Wing A. Factors Affecting the Vertical Distribution of Bioluminescence in the Euphotic Zone1. Limnol. Oceanogr. 1964;9:519–524. doi: 10.4319/lo.1964.9.4.0519. [DOI] [Google Scholar]
- 358.Buskey E.J., Strom S., Coulter C. Biolumiscence of Heterotrophic Dinoflagellates from Texas Coastal Waters. J. Exp. Mar. Biol. Ecol. 1992;159:37–49. doi: 10.1016/0022-0981(92)90256-A. [DOI] [Google Scholar]
- 359.Shi X., Li L., Guo C., Lin X., Li M., Lin S. Rhodopsin Gene Expression Regulated by the Light Dark Cycle, Light Spectrum and Light Intensity in the Dinoflagellate Prorocentrum. Front. Microbiol. 2015;6:555. doi: 10.3389/fmicb.2015.00555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 360.Guo Z., Zhang H., Lin S. Light-Promoted Rhodopsin Expression and Starvation Survival in the Marine Dinoflagellate Oxyrrhis Marina. PLoS ONE. 2014;9:e114941. doi: 10.1371/journal.pone.0114941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 361.Zaheri B., Dagenais-Bellefeuille S., Song B., Morse D. Assessing Transcriptional Responses to Light by the Dinoflagellate Symbiodinium. Microorganisms. 2019;7:261. doi: 10.3390/microorganisms7080261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 362.Van Dolah F.M., Lidie K.B., Morey J.S., Brunelle S.A., Ryan J.C., Monroe E.A., Haynes B.L. Microarray Analysis of Diurnal- and Circadian-Regulated Genes in the Florida Red-Tide Dinoflagellate Karenia Brevis (Dinophyceae) J. Phycol. 2007;43:741–752. doi: 10.1111/j.1529-8817.2007.00354.x. [DOI] [Google Scholar]
- 363.Swift E., Meunier V. Effects of Light Intensity on Division Rate, Stimulable Bioluminescence and Cell Size of the Oceanic Dinoflagellates Dissodinium Lunula, Pyrocystis Fusiformis and Pyrocystis Noctiluca. J. Phycol. 1976;12:14–22. [Google Scholar]
- 364.Anderson D.M., Stolzenbach K.D. Selective Retention of Two Dinoflagellates in a Well-Mixed Estuarine Embayment: The Importance of Diel Vertical Migration and Surface Avoidance. Mar. Ecol. Progress Ser. 1985;25:39–50. doi: 10.3354/meps025039. [DOI] [Google Scholar]
- 365.Roenneberg T., Colfax G.N., Hastings J.W. A Circadian Rhythm of Population Behavior in Gonyaulax Polyedra. J. Biol. Rhythms. 1989;4:201–216. doi: 10.1177/074873048900400208. [DOI] [PubMed] [Google Scholar]
- 366.Forward R.B., Davenport D. The Circadian Rhythm of a Behavioral Photoresponse in the Dinoflagellate Gyrodinium Dorsum. Planta. 1970;92:259–266. doi: 10.1007/BF00388560. [DOI] [PubMed] [Google Scholar]
- 367.Horiguchi T., Kawai H., Kubota M., Takahashi T., Watanabe M. Phototactic Responses of Four Marine Dinoflagellates with Different Types of Eyespot and Chloroplast. Phycol. Res. 1999;47:101–107. doi: 10.1111/j.1440-1835.1999.tb00290.x. [DOI] [Google Scholar]
- 368.Hand W.G., Forward R., Davenport D. Short-Term Photic Regulation of a Receptor Mechanism in a Dinoflagellate. Biol. Bull. 1967;133:150–165. doi: 10.2307/1539800. [DOI] [Google Scholar]
- 369.Wang L.-H., Liu Y.-H., Ju Y.-M., Hsiao Y.-Y., Fang L.-S., Chen C.-S. Cell Cycle Propagation Is Driven by Light–Dark Stimulation in a Cultured Symbiotic Dinoflagellate Isolated from Corals. Coral Reefs. 2008;27:823. doi: 10.1007/s00338-008-0434-z. [DOI] [Google Scholar]
- 370.Wangpraseurt D., Pernice M., Guagliardo P., Kilburn M.R., Clode P.L., Polerecky L., Kühl M. Light Microenvironment and Single-Cell Gradients of Carbon Fixation in Tissues of Symbiont-Bearing Corals. ISME J. 2016;10:788–792. doi: 10.1038/ismej.2015.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 371.Hollingsworth L.L., Kinzie R.A., Lewis T.D., Krupp D.A., Leong J.-A.C. Phototaxis of Motile Zooxanthellae to Green Light May Facilitate Symbiont Capture by Coral Larvae. Coral Reefs. 2005;24:523. doi: 10.1007/s00338-005-0063-8. [DOI] [Google Scholar]
- 372.Yamashita H., Koike K., Shinzato C., Jimbo M., Suzuki G. Can Acropora Tenuis Larvae Attract Native Symbiodiniaceae Cells by Green Fluorescence at the Initial Establishment of Symbiosis? PLoS ONE. 2021;16:e0252514. doi: 10.1371/journal.pone.0252514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 373.Bollati E., D’Angelo C., Alderdice R., Pratchett M., Ziegler M., Wiedenmann J. Optical Feedback Loop Involving Dinoflagellate Symbiont and Scleractinian Host Drives Colorful Coral Bleaching. Curr. Biol. 2020;30:2433–2445.e3. doi: 10.1016/j.cub.2020.04.055. [DOI] [PubMed] [Google Scholar]
- 374.Jaubert M., Bouly J.-P., Ribera d’Alcalà M., Falciatore A. Light Sensing and Responses in Marine Microalgae. Curr. Opin. Plant. Biol. 2017;37:70–77. doi: 10.1016/j.pbi.2017.03.005. [DOI] [PubMed] [Google Scholar]
- 375.Duanmu D., Rockwell N.C., Lagarias J.C. Algal Light Sensing and Photoacclimation in Aquatic Environments. Plant. Cell Environ. 2017;40:2558–2570. doi: 10.1111/pce.12943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 376.Hayakawa S., Takaku Y., Hwang J.S., Horiguchi T., Suga H., Gehring W., Ikeo K., Gojobori T. Function and Evolutionary Origin of Unicellular Camera-Type Eye Structure. PLoS ONE. 2015;10:e0118415. doi: 10.1371/journal.pone.0118415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 377.Stephens T.G., González-Pech R.A., Cheng Y., Mohamed A.R., Burt D.W., Bhattacharya D., Ragan M.A., Chan C.X. Genomes of the Dinoflagellate Polarella Glacialis Encode Tandemly Repeated Single-Exon Genes with Adaptive Functions. BMC Biol. 2020;18:1–21. doi: 10.1186/s12915-020-00782-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 378.Lin S. Genomic Understanding of Dinoflagellates. Res. Microbiol. 2011;162:551–569. doi: 10.1016/j.resmic.2011.04.006. [DOI] [PubMed] [Google Scholar]
- 379.Fajardo C., Amil-Ruiz F., Fuentes-Almagro C., De Donato M., Martinez-Rodriguez G., Escobar-Niño A., Carrasco R., Mancera J.M., Fernandez-Acero F.J. An “Omic” Approach to Pyrocystis Lunula: New Insights Related with This Bioluminescent Dinoflagellate. J. Proteomics. 2019;209:103502. doi: 10.1016/j.jprot.2019.103502. [DOI] [PubMed] [Google Scholar]
- 380.Rhiel E., Westermann M., Steiniger F., Hoischen C. The Proteorhodopsins of the Dinoflagellate Oxyrrhis Marina: Ultrastructure and Localization by Immunofluorescence Light Microscopy and Immunoelectron Microscopy. Protoplasma. 2020;257:1531–1541. doi: 10.1007/s00709-020-01530-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 381.Rhiel E., Ammermann S. The Influence of Light Quality and Prey Organisms on the Amounts of Rhodopsins of the Dinoflagellate Oxyrrhis Marina. Nova Hedwigia. 2021;112:307–321. doi: 10.1127/nova_hedwigia/2021/0631. [DOI] [Google Scholar]
- 382.Lowe C.D., Mello L.V., Samatar N., Martin L.E., Montagnes D.J.S., Watts P.C. The Transcriptome of the Novel Dinoflagellate Oxyrrhis Marina (Alveolata: Dinophyceae): Response to Salinity Examined by 454 Sequencing. BMC Genom. 2011;12:519. doi: 10.1186/1471-2164-12-519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 383.Kandori H. History and Perspectives of Ion-Transporting Rhodopsins. Adv. Exp. Med. Biol. 2021;1293:3–19. doi: 10.1007/978-981-15-8763-4_1. [DOI] [PubMed] [Google Scholar]
- 384.Kandori H. Ion-Pumping Microbial Rhodopsins. Front. Mol. Biosci. 2015;2:52. doi: 10.3389/fmolb.2015.00052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 385.Slamovits C.H., Okamoto N., Burri L., James E.R., Keeling P.J. A Bacterial Proteorhodopsin Proton Pump in Marine Eukaryotes. Nat. Commun. 2011;2:183. doi: 10.1038/ncomms1188. [DOI] [PubMed] [Google Scholar]
- 386.Janke C., Scholz F., Becker-Baldus J., Glaubitz C., Wood P.G., Bamberg E., Wachtveitl J., Bamann C. Photocycle and Vectorial Proton Transfer in a Rhodopsin from the Eukaryote Oxyrrhis Marina. Biochemistry. 2013;52:2750–2763. doi: 10.1021/bi301412n. [DOI] [PubMed] [Google Scholar]
- 387.Sineshchekov O.A., Jung K.-H., Spudich J.L. Two Rhodopsins Mediate Phototaxis to Low- and High-Intensity Light in Chlamydomonas Reinhardtii. Proc. Natl. Acad. Sci. USA. 2002;99:8689–8694. doi: 10.1073/pnas.122243399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 388.Brunelle S.A., Hazard E.S., Sotka E.E., Dolah F.M.V. Characterization of a Dinoflagellate Cryptochrome Blue-Light Receptor with a Possible Role in Circadian Control of the Cell Cycle1. J. Phycol. 2007;43:509–518. doi: 10.1111/j.1529-8817.2007.00339.x. [DOI] [Google Scholar]
- 389.Rockwell N.C., Lagarias J.C. Phytochrome Diversification in Cyanobacteria and Eukaryotic Algae. Curr. Opin. Plant. Biol. 2017;37:87–93. doi: 10.1016/j.pbi.2017.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 390.Hehenberger E., Burki F., Kolisko M., Keeling P.J. Functional Relationship between a Dinoflagellate Host and Its Diatom Endosymbiont. Mol. Biol. Evol. 2016;33:2376–2390. doi: 10.1093/molbev/msw109. [DOI] [PubMed] [Google Scholar]
- 391.Greuet C. [Ultrastructural organization of the ocelloide of Nematodinium. Phylogenetic aspect of the evolution of Warnowiidae Lindemann dinoflagellates photoreceptor (author’s transl)] Cytobiologie. 1978;17:114–136. [PubMed] [Google Scholar]
- 392.Jékely G. Origin and Early Evolution of Neural Circuits for the Control of Ciliary Locomotion. Proc. Biol. Sci. 2011;278:914–922. doi: 10.1098/rspb.2010.2027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 393.Baluska F., Mancuso S. Deep Evolutionary Origins of Neurobiology: Turning the Essence of “neural” Upside-Down. Commun. Integr. Biol. 2009;2:60–65. doi: 10.4161/cib.2.1.7620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 394.Wan K.Y., Jékely G. Origins of Eukaryotic Excitability. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2021;376:20190758. doi: 10.1098/rstb.2019.0758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 395.Kigundu G., Cooper J.L., Smith S.M.E. Hv 1 Proton Channels in Dinoflagellates: Not Just for Bioluminescence? J. Eukaryot. Microbiol. 2018;65:928–933. doi: 10.1111/jeu.12627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 396.Bray D. Molecular Networks: The Top-down View. Science. 2003;301:1864–1865. doi: 10.1126/science.1089118. [DOI] [PubMed] [Google Scholar]
- 397.Smayda T.J. Adaptations and Selection of Harmful and Other Dinoflagellate Species in Upwelling Systems. 2. Motility and Migratory Behaviour. Progress Oceanogr. 2010;85:71–91. doi: 10.1016/j.pocean.2010.02.005. [DOI] [Google Scholar]
- 398.Gómez F. A Quantitative Review of the Lifestyle, Habitat and Trophic Diversity of Dinoflagellates (Dinoflagellata, Alveolata) Syst. Biodivers. 2012;10:267–275. doi: 10.1080/14772000.2012.721021. [DOI] [Google Scholar]
- 399.Gavelis G.S., Wakeman K.C., Tillmann U., Ripken C., Mitarai S., Herranz M., Özbek S., Holstein T., Keeling P.J., Leander B.S. Microbial Arms Race: Ballistic “Nematocysts” in Dinoflagellates Represent a New Extreme in Organelle Complexity. Sci. Adv. 2017;3:e1602552. doi: 10.1126/sciadv.1602552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 400.Stires J.C., Latz M.I. Contribution of the Cytoskeleton to Mechanosensitivity Reported by Dinoflagellate Bioluminescence. Cytoskeleton. 2018;75:12–21. doi: 10.1002/cm.21392. [DOI] [PubMed] [Google Scholar]
- 401.Van Haastert P.J., Janssens P.M., Erneux C. Sensory Transduction in Eukaryotes. A Comparison between Dictyostelium and Vertebrate Cells. Eur. J. Biochem. 1991;195:289–303. doi: 10.1111/j.1432-1033.1991.tb15706.x. [DOI] [PubMed] [Google Scholar]
- 402.Christensen S.T., Leick V., Rasmussen L., Wheatley D.N. Signaling in Unicellular Eukaryotes. Int. Rev. Cytol. 1998;177:181–253. doi: 10.1016/s0074-7696(08)62233-0. [DOI] [PubMed] [Google Scholar]
- 403.Van Duijn M. Phylogenetic Origins of Biological Cognition: Convergent Patterns in the Early Evolution of Learning. Interface Focus. 2017;7:20160158. doi: 10.1098/rsfs.2016.0158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 404.Forward R.B. Effects of Neurochemicals upon a Dinoflagellate Photoresponse. J. Protozool. 1977;24:401–405. doi: 10.1111/j.1550-7408.1977.tb04760.x. [DOI] [PubMed] [Google Scholar]
- 405.Jin K., Klima J.C., Deane G., Stokes M.D., Latz M.I. Pharmacological Investigation of the Bioluminescence Signaling Pathway of the Dinoflagellate Lingulodinium Polyedrum: Evidence for the Role of Stretch-Activated Ion Channels. J. Phycol. 2013;49:733–745. doi: 10.1111/jpy.12084. [DOI] [PubMed] [Google Scholar]
- 406.Li S.-W., Lin P.-H., Ho T.-Y., Hsieh C.-H., Sun C.-L. Change in Rheotactic Behavior Patterns of Dinoflagellates in Response to Different Microfluidic Environments. Sci. Rep. 2021;11:11105. doi: 10.1038/s41598-021-90622-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 407.Sengupta A., Carrara F., Stocker R. Phytoplankton Can Actively Diversify Their Migration Strategy in Response to Turbulent Cues. Nature. 2017;543:555–558. doi: 10.1038/nature21415. [DOI] [PubMed] [Google Scholar]
- 408.Lovecchio S., Climent E., Stocker R., Durham W.M. Chain Formation Can Enhance the Vertical Migration of Phytoplankton through Turbulence. Sci. Adv. 2019;5:eaaw7879. doi: 10.1126/sciadv.aaw7879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 409.Brosnahan M.L., Velo-Suárez L., Ralston D.K., Fox S.E., Sehein T.R., Shalapyonok A., Sosik H.M., Olson R.J., Anderson D.M. Rapid Growth and Concerted Sexual Transitions by a Bloom of the Harmful Dinoflagellate Alexandrium Fundyense (Dinophyceae) Limnol. Oceanogr. 2015;60:2059–2078. doi: 10.1002/lno.10155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 410.Toth G.B., Norén F., Selander E., Pavia H. Marine Dinoflagellates Show Induced Life-History Shifts to Escape Parasite Infection in Response to Water-Borne Signals. Proc. Biol. Sci. 2004;271:733–738. doi: 10.1098/rspb.2003.2654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 411.Wootton E.C., Zubkov M.V., Jones D.H., Jones R.H., Martel C.M., Thornton C.A., Roberts E.C. Biochemical Prey Recognition by Planktonic Protozoa. Environ. Microbiol. 2007;9:216–222. doi: 10.1111/j.1462-2920.2006.01130.x. [DOI] [PubMed] [Google Scholar]
- 412.Guo Z., Zhang H., Liu S., Lin S. Biology of the Marine Heterotrophic Dinoflagellate Oxyrrhis Marina: Current Status and Future Directions. Microorganisms. 2013;1:33–57. doi: 10.3390/microorganisms1010033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 413.Coesel S.N., Durham B.P., Groussman R.D., Hu S.K., Caron D.A., Morales R.L., Ribalet F., Armbrust E.V. Diel Transcriptional Oscillations of Light-Sensitive Regulatory Elements in Open-Ocean Eukaryotic Plankton Communities. Proc. Natl. Acad. Sci. USA. 2021;118:e2011038118. doi: 10.1073/pnas.2011038118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 414.Colepicolo P., Roenneberg T., Morse D., Taylor W.R., Hastings J.W. Circadian Regulation of Bioluminescence in the Dinoflagellate Pyrocystis Lunula1. J. Phycol. 1993;29:173–179. doi: 10.1111/j.0022-3646.1993.00173.x. [DOI] [Google Scholar]
- 415.Hastings J.W. The Gonyaulax Clock at 50: Translational Control of Circadian Expression. Cold Spring Harb. Symp. Quant. Biol. 2007;72:141–144. doi: 10.1101/sqb.2007.72.026. [DOI] [PubMed] [Google Scholar]
- 416.Deane G.B., Stokes M.D., Latz M.I. Bubble Stimulation Efficiency of Dinoflagellate Bioluminescence. Luminescence. 2016;31:270–280. doi: 10.1002/bio.2957. [DOI] [PubMed] [Google Scholar]
- 417.Cussatlegras A.-S., Le Gal P. Variability in the Bioluminescence Response of the Dinoflagellate Pyrocystis Lunula. J. Exp. Mar. Biol. Ecol. 2007;343:74–81. doi: 10.1016/j.jembe.2006.11.009. [DOI] [Google Scholar]
- 418.Rohr J., Hyman M., Fallon S., Latz M.I. Bioluminescence Flow Visualization in the Ocean: An Initial Strategy Based on Laboratory Experiments. Deep Sea Res. Part I Oceanogr. Res. Papers. 2002;49:2009–2033. doi: 10.1016/S0967-0637(02)00116-4. [DOI] [Google Scholar]
- 419.Cussatlegras A.-S., Le Gal P. Bioluminescence of the Dinoflagellate Pyrocystis Noctiluca Induced by Laminar and Turbulent Couette Flow. J. Exp. Mar. Biol. Ecol. 2004;310:227–246. doi: 10.1016/j.jembe.2004.04.012. [DOI] [Google Scholar]
- 420.Jalaal M., Schramma N., Dode A., de Maleprade H., Raufaste C., Goldstein R.E. Stress-Induced Dinoflagellate Bioluminescence at the Single Cell Level. Phys. Rev. Lett. 2020;125:028102. doi: 10.1103/PhysRevLett.125.028102. [DOI] [PubMed] [Google Scholar]
- 421.Rosenthal G.G., Ryan M.J. Visual and Acoustic Communication in Non-Human Animals: A Comparison. J. Biosci. 2000;25:285–290. doi: 10.1007/BF02703937. [DOI] [PubMed] [Google Scholar]
- 422.Gagnon Y.L., Templin R.M., How M.J., Marshall N.J. Circularly Polarized Light as a Communication Signal in Mantis Shrimps. Curr. Biol. 2015;25:3074–3078. doi: 10.1016/j.cub.2015.10.047. [DOI] [PubMed] [Google Scholar]
- 423.Burford B.P., Robison B.H. Bioluminescent Backlighting Illuminates the Complex Visual Signals of a Social Squid in the Deep Sea. Proc. Natl. Acad. Sci. USA. 2020;117:8524–8531. doi: 10.1073/pnas.1920875117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 424.Stuart-Fox D., Ospina-Rozo L., Ng L., Franklin A.M. The Paradox of Iridescent Signals. Trends Ecol. Evol. 2021;36:187–195. doi: 10.1016/j.tree.2020.10.009. [DOI] [PubMed] [Google Scholar]
- 425.Doucet S.M., Meadows M.G. Iridescence: A Functional Perspective. J. R. Soc. Interface. 2009;6((Suppl. 2)):S115–S132. doi: 10.1098/rsif.2008.0395.focus. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 426.Mäthger L.M., Denton E.J., Marshall N.J., Hanlon R.T. Mechanisms and Behavioural Functions of Structural Coloration in Cephalopods. J. R. Soc. Interface. 2009;6:S149–S163. doi: 10.1098/rsif.2008.0366.focus. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 427.Macel M.-L., Ristoratore F., Locascio A., Spagnuolo A., Sordino P., D’Aniello S. Sea as a Color Palette: The Ecology and Evolution of Fluorescence. Zool. Lett. 2020;6:9. doi: 10.1186/s40851-020-00161-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 428.Hauser F.E., Chang B.S. Insights into Visual Pigment Adaptation and Diversity from Model Ecological and Evolutionary Systems. Curr. Opin. Genet. Dev. 2017;47:110–120. doi: 10.1016/j.gde.2017.09.005. [DOI] [PubMed] [Google Scholar]
- 429.Salgado P., Figueroa R.I., Ramilo I., Bravo I. The Life History of the Toxic Marine Dinoflagellate Protoceratium Reticulatum (Gonyaulacales) in Culture. Harmful Algae. 2017;68:67–81. doi: 10.1016/j.hal.2017.07.008. [DOI] [PubMed] [Google Scholar]
- 430.Shah S., Chen Y., Bhattacharya D., Chan C.X. Sex in Symbiodiniaceae Dinoflagellates: Genomic Evidence for Independent Loss of the Canonical Synaptonemal Complex. Sci. Rep. 2020;10:9792. doi: 10.1038/s41598-020-66429-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 431.Figueroa R.I., Dapena C., Bravo I., Cuadrado A. The Hidden Sexuality of Alexandrium Minutum: An Example of Overlooked Sex in Dinoflagellates. PLoS ONE. 2015;10:e0142667. doi: 10.1371/journal.pone.0142667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 432.Berdieva M., Kalinina V., Lomert E., Knyazev N., Skarlato S. Life Cycle Stages and Evidence of Sexual Reproduction in the Marine Dinoflagellate Prorocentrum Minimum (Dinophyceae, Prorocentrales) J. Phycol. 2020;56:941–952. doi: 10.1111/jpy.12989. [DOI] [PubMed] [Google Scholar]
- 433.Moiseff A., Copeland J. Firefly Synchrony: A Behavioral Strategy to Minimize Visual Clutter. Science. 2010;329:181. doi: 10.1126/science.1190421. [DOI] [PubMed] [Google Scholar]
- 434.Lewis S.M., Cratsley C.K. Flash Signal Evolution, Mate Choice, and Predation in Fireflies. Annu. Rev. Entomol. 2008;53:293–321. doi: 10.1146/annurev.ento.53.103106.093346. [DOI] [PubMed] [Google Scholar]
- 435.Goodheart J.A., Minsky G., Brynjegard-Bialik M.N., Drummond M.S., Munoz J.D., Fallon T.R., Schultz D.T., Weng J.-K., Torres E., Oakley T.H. Laboratory Culture of the California Sea Firefly Vargula Tsujii (Ostracoda: Cypridinidae): Developing a Model System for the Evolution of Marine Bioluminescence. Sci. Rep. 2020;10:10443. doi: 10.1038/s41598-020-67209-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 436.Hensley N.M., Ellis E.A., Gerrish G.A., Torres E., Frawley J.P., Oakley T.H., Rivers T.J. Phenotypic Evolution Shaped by Current Enzyme Function in the Bioluminescent Courtship Signals of Sea Fireflies. Proc. Biol. Sci. 2019;286:20182621. doi: 10.1098/rspb.2018.2621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 437.Rivers T.J., Morin J.G. The Relative Cost of Using Luminescence for Sex and Defense: Light Budgets in Cypridinid Ostracods. J. Exp. Biol. 2012;215:2860–2868. doi: 10.1242/jeb.072017. [DOI] [PubMed] [Google Scholar]
- 438.Rivers T.J., Morin J.G. Complex Sexual Courtship Displays by Luminescent Male Marine Ostracods. J. Exp. Biol. 2008;211:2252–2262. doi: 10.1242/jeb.011130. [DOI] [PubMed] [Google Scholar]
- 439.Herring P.J. Species Abundance, Sexual Encounter and Bioluminescent Signalling in the Deep Sea. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2000;355:1273–1276. doi: 10.1098/rstb.2000.0682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 440.Schweikert L.E., Davis A.L., Johnsen S., Bracken-Grissom H.D. Visual Perception of Light Organ Patterns in Deep-Sea Shrimps and Implications for Conspecific Recognition. Ecol. Evol. 2020;10:9503–9513. doi: 10.1002/ece3.6643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 441.López-Palafox T., Macías-Ordóñez R., Cordero C.R. The Size of Signal Detection and Emission Organs in a Synchronous Firefly: Sexual Dimorphism, Allometry and Assortative Mating. PeerJ. 2020;8:e10127. doi: 10.7717/peerj.10127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 442.Clark K.B. Origins of Learned Reciprocity in Solitary Ciliates Searching Grouped “courting” Assurances at Quantum Efficiencies. Biosystems. 2010;99:27–41. doi: 10.1016/j.biosystems.2009.08.005. [DOI] [PubMed] [Google Scholar]
- 443.Clark K.B. Social Biases Determine Spatiotemporal Sparseness of Ciliate Mating Heuristics. Commun. Integr. Biol. 2012;5:3–11. doi: 10.4161/cib.18337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 444.Görtz H.D., Kuhlmann H.W., Möllenbeck M., Tiedtke A., Kusch J., Schmidt H.J., Miyake A. Intra- and Intercellular Communication Systems in Ciliates. Naturwissenschaften. 1999;86:422–434. doi: 10.1007/s001140050646. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.