ABSTRACT
MicroRNAs (miRNAs) play critical roles in gene expression and numerous human diseases. The success of miRNA biogenesis is largely determined by the primary miRNA (pri-miRNA) processing by the DROSHA-DGCR8 complex, called Microprocessor. Here, we analysed the high-throughput pri-miRNA processing assays and secondary structures of pri-miRNAs to investigate the roles of bulges in the pri-miRNA processing. We found that bulges in multiple places control both the cleavage efficiency and accuracy of pri-miRNA processing. These bulges were shown to act on Microprocessor via its catalytic subunit, DROSHA, and function in a position and strand-dependent manner. Interestingly, we discovered that the enriched and conserved bulges, called midB, can correct DROSHA orientation on pri-miRNAs, thereby enhancing production of miRNAs. The revealed functions of the bulges help improve our understanding of pri-miRNA processing and suggest their potential roles in miRNA biogenesis regulation.
KEYWORDS: miRNA biogenesis, Microprocessor, DROSHA, DGCR8, Bulges
Introduction
MicroRNAs (miRNAs) are small single-stranded RNAs of 21–22 nucleotides (nt) that regulate the expression of genes involved in various cellular processes. miRNAs interact with and guide a protein called Ago to find target mRNAs, after which the miRNAs often form 7 base pairs with them. This base-paired region of the miRNA is called a seed sequence, ranging from the 2nd to the 8th nt from the 5ʹ-end of the miRNA. The miRNA-Ago complex makes up the core of the miRNA-mediated RNA silencing complex (miRISC), which induces the inhibition of translation and/or the degradation of mRNA [1–4]. Each miRNA is a part of a stem-loop RNA called a primary miRNA (pri-miRNA), synthesized in the nucleus by RNA polymerase II. The extraction of a miRNA sequence from a pri-miRNA requires two sequential double cleavages catalysed by the RNase III enzymes called Microprocessor (in the nucleus), and DICER (in the cytoplasm), as well as strand-selection, which is controlled by Ago [2–6]. Microprocessor initially processes the pri-miRNA to generate pre-miRNA, and this is then cleaved by DICER to produce a miRNA duplex. One strand of the miRNA duplex is eventually selected by Ago, and this acts as a mature miRNA functioning in gene silencing [2–6].
Microprocessor is a trimetric complex consisting of an RNase III enzyme called DROSHA and a dimer of the RNA-binding protein, DGCR8 [3,6–11] (Fig. S1A). Microprocessor interacts with multiple RNA elements within pri-miRNAs in order to ensure that its double cleavage occurs both accurately and efficiently [12–20]. These RNA elements can be classified into two groups, primary RNA sequencing motifs and secondary RNA structures.
An example of an essential secondary RNA structure is the ssRNA/dsRNA junction. DROSHA recognizes this junction, which helps the enzyme find its cleavage site located 11 base pairs (bp) away [10,14,17,18,20] (Fig. S1B). Since pri-miRNAs have an ssRNA/dsRNA junction at each end of the stem, DROSHA might cleave them at their basal junction, resulting in a productive cleavage, or at their apical junction, resulting in an unproductive cleavage. However, only the productive cleavages produce miRNAs, and so many mechanisms are required to enhance the productive cleavage capability and reduce the unproductive cleavage ability of DROSHA (Fig. S1B). For example, the mGHG motif is a secondary RNA structure element located in the lower stem. It contains a mismatch in position 8 from the basal junction and two Gs in positions 7 and 9 on the 3p-strand [13,17]. This motif interacts with the double-stranded RNA-binding domain (dsRBD) of DROSHA and determines its cleavage efficiency and accuracy [13,17,20]. The asymmetric internal loop (AIL), located in the lower stem, negatively affects the double cleavages of Microprocessor on the 5p- and 3p-strands by inhibiting its cleavage on the 3p-strand of pri-miRNAs [21]. In addition, there is midMW_79 (containing mismatches and wobble base pairs), which is located 7–9 nt from the cleavage sites in the middle region of the upper stem [19]. This influences the action of DROSHA by facilitating cleavage at 8 nt from its location [19]. Another midMW_1012, which is located in positions 10–12 from the cleavage sites, is enriched in human pri-miRNAs, blocks the unproductive cleavages by DROSHA, and thus enhances the productive cleavages [19]. A seedMW contains mismatches and wobble base pairs in positions 4–8 nt from the cleavage sites, which inhibit DROSHA cleavage. Although seedMW rarely appears in human pri-miRNAs, it might arise due to single nucleotide polymorphisms (SNPs) or RNA modifications [19]. For example, ADAR-edited pri-mir-142 has a seedMW, and thus it expresses fewer miRNAs when compared with non-edited pri-mir-142 [19,22].
Three sequencing motifs have also been identified: UG, CNNC, and UGU [12]. The UG motif is located at the basal junction, and it attracts and interacts with DROSHA at this site [12,14] (Fig. S1B). The CNNC motif binds to a splicing factor, SRSF3, which in turn recruits DROSHA to the basal junction [12,15,16] (Fig. S1B). Therefore, the UG and CNNC motifs both facilitate the productive cleavage of DROSHA at the basal junction. The UGU motif is located at the apical junction strengthening the interaction between DGCR8 and the loop. In this way, it prevents DROSHA from being dislocated at the apical junction and cleaving pri-miRNA at the unproductive sites [12,14,23,24] (Fig. S1B).
In this study, we investigated the effect of bulges, another type of secondary structure, on pri-miRNA processing. It was previously reported that bulges in positions 3–7 nt from the cleavage sites reduced the efficiency of pri-miRNA processing for pri-mir-16-1 and pri-mir-125b-2 [25]. However, it is still unknown if these bulges affected the DROSHA or DGCR8 subunit of the Microprocessor complex, and if the bulge on the 5p-strand might affect processing differently from that on the 3p-strand. In addition, it is still unclear how bulges in other positions of the stem might affect the efficiency and/or cleavage sites of processing. Here, we analysed pri-miRNA secondary structures, high-throughput pri-miRNA processing of DROSHA, and performed in vitro cleavage assays for numerous pri-miRNA variants, which contained bulges on the 5p- or 3p-strand in different positions of the upper stem of pri-miRNAs. We revealed that bulges in the upper stem control both the cleavage efficiency and accuracy of Microprocessor action. We also demonstrated that the function of each bulge is dependent on its relative position to the cleavage sites and its localization on the 5p- or 3p-strand. In addition, we validated the effects of these bulges on the biogenesis of miRNA in human cells and demonstrated their impact on the DROSHA subunit. These findings indicate the crucial role of the bulges in controlling pri-miRNA processing and extend our understanding of the cleavage mechanism of Microprocessor by demonstrating the regulation of miRNA expression via bulges in the upper stem.
Results
The midB_1013 blocks unproductive cleavages by Microprocessor
We estimated the frequency of the bulges in the different positions on the 5p- or 3p-strand of human pri-miRNAs (Fig. 1A). Interestingly, we found that on the 5p-strand but not on the 3p-strand, bulges frequently appeared in positions 11–13 when compared with the other positions. We extended a similar analysis to pri-miRNAs of 14 other organisms and again discovered a high enrichment of 5p-strand bulges in the 11–13 positions (Fig. 1A). This bulge feature seems highly conserved in many organisms for many pri-miRNAs, such as pri-mir-30a, pri-mir-34a, and pri-mir-181a-1 (Fig. 1B).
Figure 1.
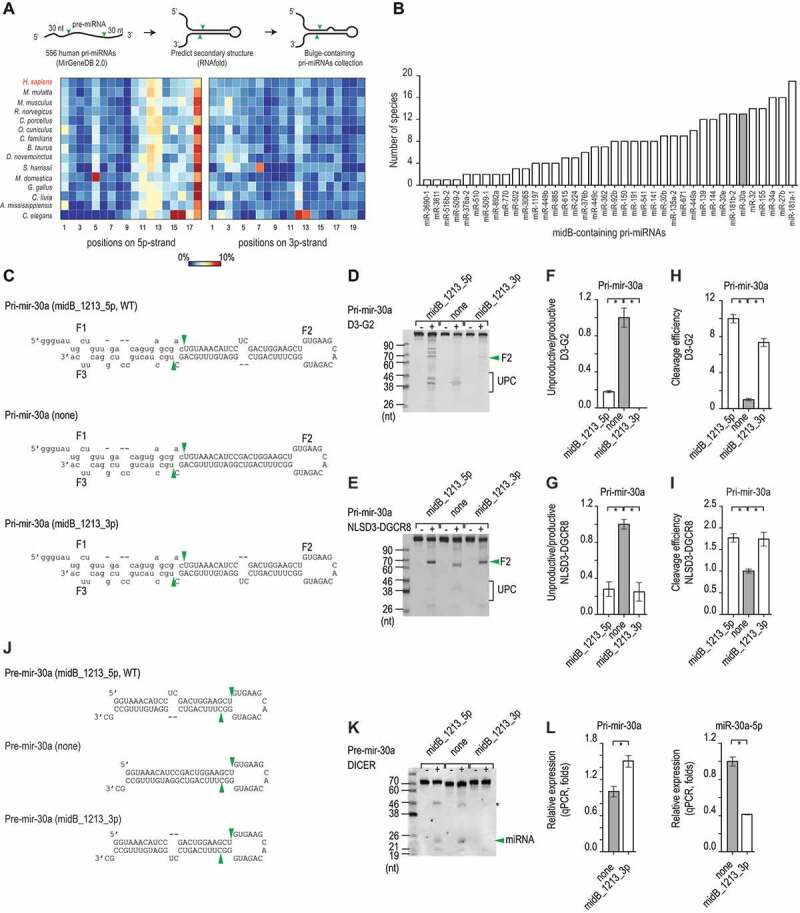
The 5p-strand bulges block the unproductive cleavage. (A) The percentage of pri-miRNAs that contain bulges in different positions in humans and various other organisms was estimated. The numbers indicate the positions of the nucleotides in the upper stem from the Microprocessor cleavage site, and the colour bar indicates the percentage ranging from 0% (blue) to 10% (red). (B) The number of species that share each midB_containing pri-miRNAs. (C) The sequences and secondary structures of the three pri-mir-30a variants. The productive cleavage sites of Microprocessor are indicated by the green arrowheads. The pre-mir-30a sequence is shown as capital letters. (D and E) Gel images showing the in vitro processing of the pri-mir-30a variants by D3-G2 (D) or NLSD3-DGCR8 (E). A 4 pmol aliquot of each pri-mir-30a variant was incubated with 16 pmol D3-G2 or 8 pmol NLSD3-DGCR8 at 37°C for 120 min. UPC: unproductive cleavage products. (F − I) Bar charts to show the unproductive/productive ratios or cleavage efficiency (i.e. the productive/original substrate ratio) estimated from three repeated processing assays. The ratios for each midB variant were normalized to that of the ‘none’ variant. The data are presented as mean +/− SEM values and the asterisks (*) indicate statistically significant results (p < 0.05) when compared with the two-sided t-test. The unproductive/productive ratios obtained for D3-G2 cleavage of pri-mir-30a, indicated that midB_1213_5p versus none: p = 0.002, and midB_1213_3p versus none: p = 0.001. The cleavage efficiency of this assay showed that midB_1213_5p versus none: p = 5.6e-5, and midB_1213_3p versus none: p = 1.6e-4. The unproductive/productive ratios obtained for NLSD3-DGCR8 cleavage of pri-mir-30a indicated that midB_1213_5p versus none: p = 0.002, and midB_1213_3p versus none: p = 0.003. The cleavage efficiency of this assay showed that midB_1213_5p versus none : p = 0.002, and midB_1213_3p versus none: p = 0.01. (J) The sequences and secondary structures of the pre-mir-30a variants. The DICER cleavage sites are indicated by the green arrowheads. (K) A representative gel image to show the in vitro cleavage of the pre-mir-30a variants by DICER. A 5 pmol aliquot of pre-mir-30a was incubated with 1.75 pmol DICER at 37°C for 120 min. The green arrowhead indicates the miRNA, and the asterisk (*) indicates the unidentified RNA fragments. (L) The relative level of pri-mir-30a (left) and miR-30a (right) expression in HCT116 cells transfected with the pri-mir-30a pcDNA3.0 plasmids followed by qPCR. The expression levels of pri-mir-30a and miR-30a were normalized to those of pri-mir-1226 and miR-1226, respectively. These normalized expression levels were then normalized again to the level of expression of pri-mir-30a or miR-30a (indicated by ‘none’ on both graphs). The data are presented as the mean +/− SEM, and the asterisks (*) indicate statistically significant (p < 0.05) results when compared with the two-sided t-test. For pri-mir-30a, midB_1213_3p versus none: p = 0.046, and for miR-30a, midB_1213_3p versus none: p = 0.007
Next, we selected pri-mir-30a, which has a 2-nt bulge in positions 12–13 (WT, midB_1213_5p), and synthesized three variants: midB_1213_5p (WT), midB_1213_3p and none (base-paired) (Fig. 1C). We found that both the 5p- and 3p-strand bulges reduced the number of unproductive cleavages and increased the number of productive cleavages of DROSHA (D3-G2) and Microprocessor (NLSD3-DGCR8) on pri-mir-30a (Fig. 1D–I). This suggests that these bulges control pri-miRNA processing by Microprocessor via the DROSHA subunit. Consistently, we demonstrated that bulges on either the 5p-strand (midB_1012_5p) or the 3p-strand (midB_1012_3p) significantly reduced unproductive cleavage of DROSHA on several of the pri-miRNAs tested, including pri-mir-576, pri-mir-30d, and pri-mir-128-1 (Fig. S1C–H, Table S1). Furthermore, we also observed a similar effect for midB on increasing the cleavage efficiency of DROSHA for pri-mir-576, pri-mir-30d, and pri-mir-128-1 (Fig. S1I–K, Table S1). The 3-nt bulges seemed to have a stronger effect than the 1-nt bugles (Fig. S1L and S1M, Table S1). These data indicated that the bulges in the positions 10–13 blocked a wrong orientation of DROSHA at the apical junction. The 10–13 bulges are located in the positions −1 to −3 from the cleavage site on 5p-strand as DROSHA orients at the apical junction. Here, we also found that the bulges in the positions −1 to −2 at the basal junction decreased the productive cleavage of DROSHA on pri-mir-576 (Fig. S1N, Table S1). We also demonstrated that the bulges in the positions −1 to −2 diminished the single cleavage of DROSHA mutants, DROSHA-TN1 and DROSHA-TN2, that could cleave pri-miRNAs at either 5p-strand or 3p-strand. This indicates that midB affects both cleavages of DROSHA on the 5p- and 3p-strands.
Although both 5p- and 3p-strand midB_1213 enhanced the productive cleavage of Microprocessor, only 5p-strand midB_1213 is enriched in pri-miRNA. Therefore, we reasoned that the 3p-strand bulge might support pri-miRNA processing by Microprocessor, but it might also entangle a downstream step in the miRNA biogenesis pathway, the action of DICER. We then tested the cleavage activity of DICER on three pre-mir-30a variants containing a 5p-strand bulge, a 3p-strand bulge, or no bulge (Fig. 1J). We found that DICER cleaved the 5p-strand bulge and the no-bulge (none) pre-mir-30a similarly, but it cleaved the 3p-strand bulge pre-mir-30a inefficiently (Fig. 1K). We also showed that DICER cleaved 3p-strand bulge-containing pre-mir-671 and pre-mir-30e less efficiently than 5p-strand bulge-containing pre-miRNAs (Fig. S1O and S1P). We expressed two pri-mir-30a variants (Fig. 1J) in human cells, and found that in human cells, the 3p-strand bulge pri-mir-30a (midB_1213_3p) expressed less miR-30a than the no-bulge pri-mir-30a (none) (Fig. 1L). These data support 3p-strand bulge reduced miRNA expression by inhibiting DICER, explaining why it is not enriched in human pri-miRNAs.
The midB_89 on the 3p-strand influences the cleavage sites of Microprocessor
The effect of bulges in the midB_1013 is similar to that of mismatches and wobble base pairs found in the previous study [19]. Therefore, we examined the effect of bulges in different positions on pri-miRNAs by analysing our previous high-throughput (HT) pri-miRNA processing assays using the randomized pri-mir-342 sequences and the purified Microprocessor complex (D3TN1-G2) (Fig. S2A and S2B). We collected many 3p-strand bulge-containing pri-mir-342 variants, but only a few 5p-strand bulge containing pri-mir-342 variants in the HT assays. Since Microprocessor cleaved pri-mir-342 at two different positions (CL-1 and CL0), we estimated CL-1/CL0 ratio for pri-miRNAs containing bulges in various positions on the 3p-strand. Interestingly, we found that bulges in position 8 on the 3p-strand induced Microprocessor to cleave pri-mir-342 more at the CL-1 position, when compared with the base-paired pri-mir-342 variants (Fig. 2A). The effect of these bulges was similar to that of mismatches and wobble base pairs in the same position (Fig. 2A) [19], which also stimulated the CL-1 cleavages.
Figure 2.
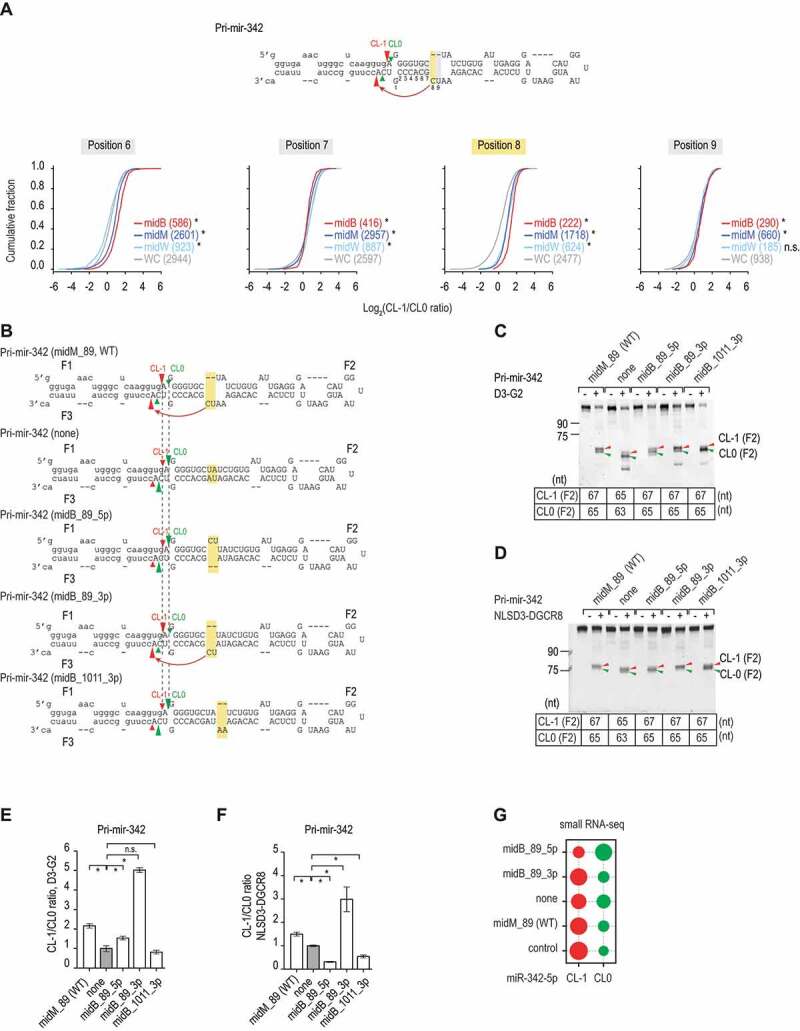
The 3p-strand bulges affect the cleavage sites. (A) The ratios of the CL-1/CL0 cleavage were estimated for the pri-mir-342 variants, which contained bulges (midB), mismatches (midM), wobble base pairs (midW), or Watson-Cricks base pairs (WC) on the 3p-strand in positions 6–9 from the 5p-strand cleavage site. The pri-mir-342 variants were generated and cleaved by DROSHA, as described in our previous study [19] and Fig. S2A and S2B. The red, dark blue, light blue, and grey curves indicate the midB, midM, midW, and WC variants, respectively. The p-values were calculated by a two-sided Wilcoxon rank-sum test, for position 8, midM versus WC: p = 5.3e-117, midW versus WC: p = 1.8e-56, midB versus WC: p = 1.1e-57. (B) Diagrams showing the RNA sequences and secondary structures of the pri-mir-342 variants. The CL-1 and CL0 cleavage sites of Microprocessor are indicated by the red and green arrowheads, respectively. The mutated regions are highlighted in yellow. The pre-mir-342 sequence is shown in capital letters. (C and D) Gel images showing the size of bands after the pri-mir-342 variants were processed by D3-G2 or NLSD3-DGCR8. A 5 pmol aliquot of each pri-mir-342 variant was incubated with 10 pmol D3-G2 (C) or 6 pmol NLSD3-DGCR8 (D) at 37°C for 120 min. The sizes of the F2 bands resulting from the CL-1 and CL0 cleavages for each variant are shown in the tables below the gel images. (E and F) Bar charts showing the mean +/− SEM CL-1/CL0 ratios from the three repeated cleavage assays shown in C and D, respectively, for each pri-mir-342 variant. The band densities of each F2 fragment were quantified by Image Lab v.6.0.1. The CL-1/CL0 ratio of each variant was normalized to that of pri-mir-342 (none). The asterisk (*) and (n.s.) indicate statistically significant (p < 0.05) and no significant differences, respectively, when data were compared with the two-sided t-test. For the D3-G2 cleavage (E), midM_89 versus none: p = 0.003, midB_89_5p versus none: p = 0.039, midB_89_3p versus none: p = 3.2e-5, and midB_1011_3p versus none: p = 0.356. For the NLSD3-DGCR8 cleavage, midM_89 versus none: p = 0.006, midB_89_5p versus none: p = 1.0e-4, midB_89_3p versus none: p = 0.020, and midB_1011_3p versus none: p = 0.004. (G) The relative abundance of CL-1 or CL0 miR-342 in HCT116 cells transfected with pcDNA3.0 plasmids expressing each of the pri-mir-342 variants. HCT116 cells transfected with the pcDNA3.0 alone were used as a control. The relative abundance of each miRNA is indicated by the size of the circle
We then generated several pri-mir-342 variants, which contained bulges in different positions on either the 5p- or 3p-strand. Consistent with our previous study, mismatches in the 8–9 positions (midM_89) significantly induced DROSHA and Microprocessor to cleave pri-mir-342 at CL-1 (compare midM_89 with none in Fig. 2B–F). Here, we also observed that bulges in positions 8–9 on the 3p-strand (midB_89_3p), but not on the 5p-strand (midB_89_5p), increased CL-1 cleavage (Fig. 2B–F). In addition, a bulge in positions 10–11 on the 3p-strand had no effect on the CL-1 cleavage (Fig. 2B–F). These data together indicate that RNA bulges can affect the cleavage sites of Microprocessor in a position- and 3p-strand-dependent manner. Like midM_89, midB_89_3p generated CL0 pre-mir-342, which was 2 nt shorter than CL-1 pre-mir-342 (Fig. 2C,Fig. 2D) [19], suggesting that midB_89_3p might shift both cleavage sites on the 5p- and 3p-strands each 1 nt.
We then transfected four plasmids, which expressed different pri-mir-342 variants, into human HCT116 cells, and we conducted a small RNA sequencing analysis for these transfected cells. Consistent with our in vitro pri-miRNA processing data, we demonstrated that midB_89_3p and midM_89 pri-mir-342s produced more of the CL-1 miR-342 isoform, whereas midB_89_5p and none pri-mir-342s generated more of the CL0 miR-342 isoform in human cells (Fig. 2G).
The seedB inhibits the productive cleavage of Microprocessor
We then generated different variants of several pri-miRNAs, including pri-mir-16-1, pri-mir-576, pri-mir-30a, and pri-mir-885, which contained bulges in the seed region of the miRNAs in positions 7–8 on either the 5p- or 3p-strand (Fig. 3A,Fig. 3B, Fig. S3A and S3B). Here, we found that 3p-strand bulges (seedB_3p) suppressed the enzymatic activity of Microprocessor more than 5p-strand bulges (seedB_5p) in the same positions (Fig. 3C–J, Fig. S3C and S3D). We then expressed two different variants of pri-mir-16-1 in human cells and quantified the expression of miR-16-1 by qPCR. We showed that bulge-containing pri-miRNAs expressed less miR-16-1 than the no-bulge pri-mir-16-1 (WT) (Fig. 3K). In addition, we found that seedB reduced the cleavage activity of DROSHA on either 5p- or 3p-strand by diminishing the single cleavage of DROSHA-TN1 or TN2 (Fig. S3E).
Figure 3.
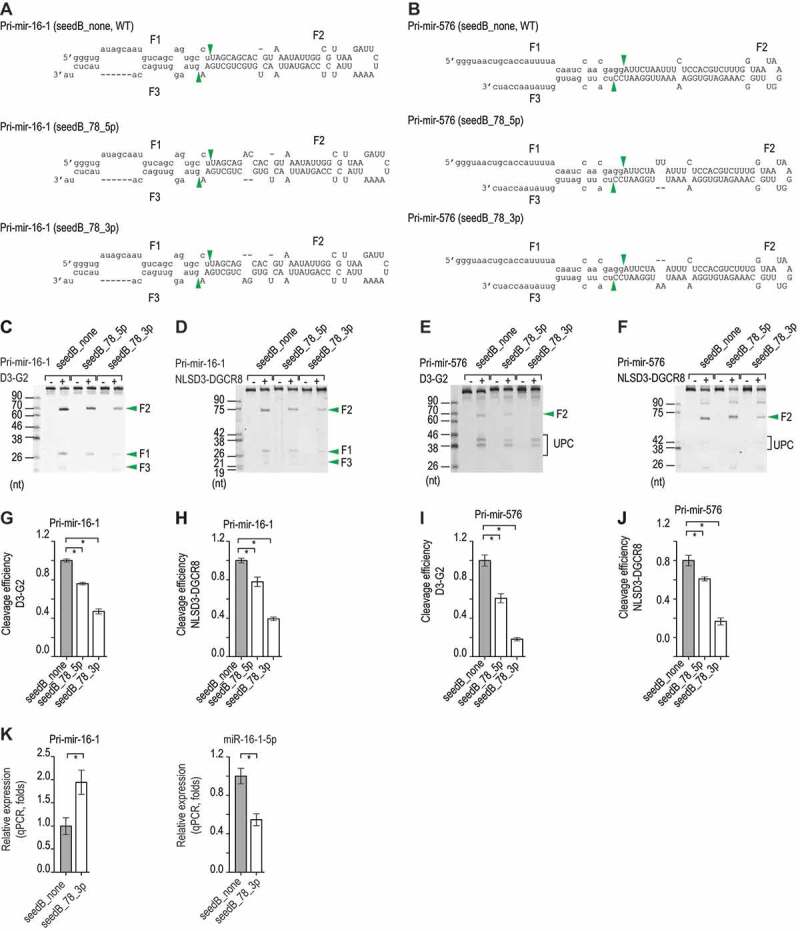
The 3p-strand bulges inhibit the productive cleavage. (A and B) The sequences and secondary structures of the pri-mir-16-1 and pri-mir-576 variants. The pre-mir-16-1 and pre-mir-576 sequences are presented as capital letters, and the productive cleavage sites of Microprocessor are indicated by the green arrowheads. (C–F) Representative images of gels showing the in vitro processing of the pri-mir-16-1 (C, D) and pri-mir-576 (E, F) variants by D3-G2 (C, E) or NLSD3-DGCR8 (D, F). A 5 pmol aliquot of the pri-miRNA substrate was incubated with 5 pmol D3-G2 or 4 pmol NLSD3-DGCR8 at 37°C for 120 min. UPC: unproductive cleavage products. (G–J) The cleavage efficiency of Microprocessor for pri-mir-16-1 and pri-mir-576 was estimated for the three repeated experiments shown in (C–F). The band densities of the F2 fragments or the original substrates were measured by Image Lab v.6.0.1. The cleavage efficiency was estimated as a ratio of F2/original substrate. These ratios (estimated for each pri-miRNA variant) were normalized to that of its ‘none’ variant. Data are presented as mean +/− SEM and the asterisks (*) indicate statistically significant differences (p < 0.05) when comparing the data with the two-sided t-test. For (G), seedB_78_5p versus seedB_none: p = 2.9e-4, and seedB_78_3p versus seedB_none: p = 6.9e-5. For (H), seedB_78_5p versus seedB_none: p = 0.016, and seedB_78_3p versus seedB_none: p = 3.9e-5. For (I), seedB_78_5p versus seedB_none: p = 0.004, and seedB_78_3p versus seedB_none: p = 6.2e-5, and for (J), seedB_78_5p versus seedB_none: p = 0.031, and seedB_78_3p versus seedB_none: p = 6.4e-4. (K) The relative level of expression of pri-mir-16-1 (left) and miR-16-1 (right) in HCT116 cells transfected with the pri-mir -16-1 pcDNA3.0 plasmid followed by qPCR. The expression levels of pri-mir-16-1 and miR-16-1 were normalized to those of pri-mir-1226 and miR-1226, respectively. These normalized expression levels were normalized to those of pri-mir-16-1 (left) or miR-16-1 (right; both labelled as seedB_none). The data are presented as mean +/− SEM, and the asterisks (*) and (n.s.) indicate statistically significant (p < 0.05) and no significant differences, respectively when data were compared with the two-sided t-test. For the left graph, seedB_78_3p versus seedB_none: p = 0.007, whereas for the right graph, seedB_78_3p versus seedB_none: p = 0.002
Coordination of the bulges and the primary sequencing motifs
We examined the combined effects of the bulges and each of the three known motifs, UG, UGU, and mGHG, which also govern the cleavage of Microprocessor. We found that midB_89_3p and UG showed a synergistic effect and enhanced the CL-1 cleavage (Fig. 4A,Fig. 4C, Fig. S4A). The mGHG motif promoted the cleavage of DROSHA at CL0, whereas midB_89_3p stimulated DROSHA to cleave more at CL-1 (Fig. 4B,Fig. 4C, Fig. S4A). As a result, the introduction of midB_89_3p to mGHG-containing pri-mir-342 reduced the cleavage efficiency of DROSHA at CL0 but could not stimulate DROSHA to cleave pri-mir-342 at CL-1 (Fig. 4B,Fig. 4C, Fig. S4A). This indicates that mGHG has a more dominant role in determining the cleavage sites of DROSHA than midB_89_3p. The addition of the natural UGU motif in the apical loop, which is known not to interact with DROSHA [14], mildly increased the CL-1/CL0 ratio of DROSHA between 1.2–1.7 folds for pri-mir-342 midB_89_3p and midM_89 substrates (Fig. 4D,Fig. 4E, Fig. S4A). However, by interacting with DGCR8, the UGU motif induced the CL-1 cleavage of the NLSD3-DGCR8 complex (Fig. 4F,Fig. 4G, Fig. S4A). A combination of midB_89_3p and UGU further enhanced the CL-1/CL0 ratio of the NLSD3-DGCR8 complex (2.3–3.7 folds), when compared with either UGU or midB_89_3p alone (Fig. 4F,Fig. 4G, Fig. S4A). This indicates that midB_89_3p and UGU might be coordinated in determining the cleavage sites of Microprocessor.
Figure 4.
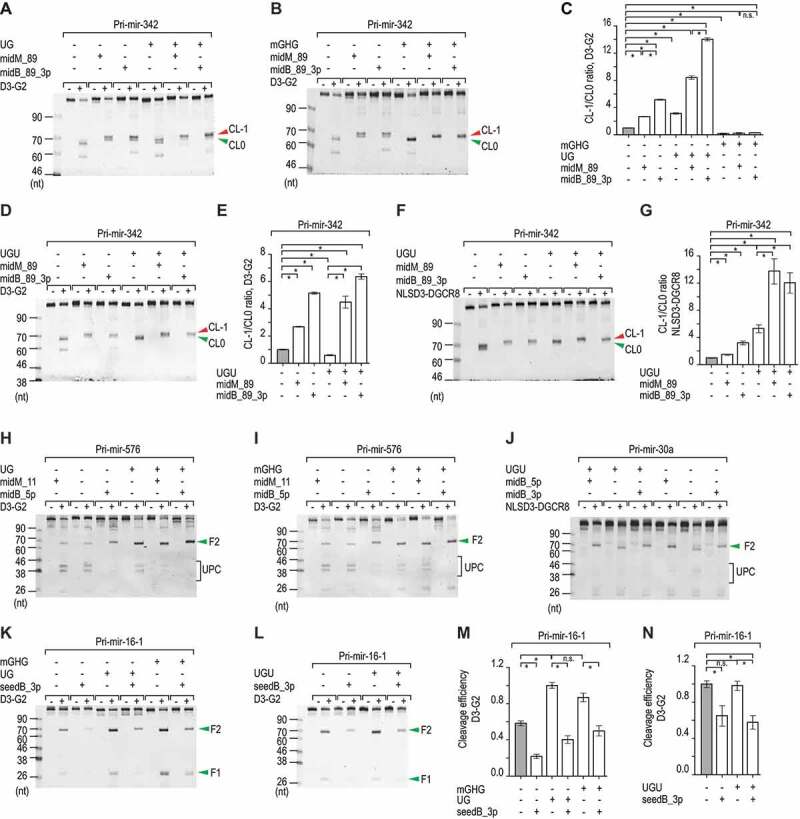
Coordination of bulges and known motifs. (A, B, D, F, H–L) Representative gel images showing the in vitro processing of the pri-mir-342, pri-mir-576, pri-mir-30a, and pri-mir-16-1 variants by D3-G2 or NLSD3-DGCR8. A 5 pmol aliquot of each pri-miRNA variant was incubated with 4–20 pmol D3-G2 or 4–8 pmol NLSD3-DGCR8 at 37°C for 120 min. UPC: unproductive cleavage products. (C, E and G) Bar charts showing the mean +/− SEM CL-1/CL0 ratios estimated from three repeated experiments for the pri-mir-342 variants. The band densities of the F2 fragments resulting from the CL-1 or CL0 cleavages for each variant were quantified by Image Lab v.6.0.1, and the CL-1/CL0 ratio of each variant was normalized to that of pri-mir-342 (midM_89). The asterisks (*) and (n.s.) indicate statistically significant (p < 0.05) and no significant differences, respectively, when the data were compared using the two-sided t-test. For (C), none versus midB_89: p = 9.6e-7, none versus UG+none: p = 6.4e-5, midB_89_3p versus UG+midB_89: p = 2.8e-6, none versus mGHG+none: p = 4.9e-4, midB_89_3p versus mGHG+midB_89_3p: p = 5.0e-7. For (E), none versus UGU+none: p = 0.002, midB_89_3p versus UGU+midB_89_3p: p = 0.004. For (G), none versus UGU+none: p = 0.002, midB_89_3p versus UGU+midB_89_3p: p = 0.005. (M and N) Bar charts to show the cleavage efficiency of Microprocessor, which was estimated from the three repeated processing experiments conducted for the pri-mir-16-1 variants. The band densities of the F2 fragments or the original substrates were quantified by Image Lab v.6.0.1. The cleavage efficiency was estimated as the ratio of the F2/original substrate. The ratio was normalized to that of the ‘UG’ variant (M) or to the ‘none’ variant (N). Data are presented as mean +/− SEM, and the asterisk (*) and (n.s.) indicate statistically significant (p < 0.05) and no significant differences, respectively when the data were compared using the two-sided t-test. For (M), none versus seedB_3p: p = 4.7e-4, UG+none versus UG+seedB_3p: p = 3.5e-4, and mGHG+none versus mGHG+seedB_3p: p = 0.007. For (N), none versus seedB_3p: p = 0.04, and UGU+none versus UGU+seedB_3p: p = 0.012
We next demonstrated that the midB_1013_5p reduced the unproductive cleavages of DROSHA on pri-mir-576 (Fig. 4H,Fig. 4I, Fig. S4B), which were introduced with UG or mGHG. The midB_1013_5p also blocked the unproductive cleavage of DROSHA on pri-mir-30a (Fig. 4J, Fig. S4C), which contained the UGU motif. A combination of the UGU motif and the midB_1013_5p or 3p resulted in fewer unproductive cleavages of the NLSD3-DGCR8 complex than the UGU motif, midB_1013_5p, or 3p alone (Fig. 4J). This suggests that UGU and midB_1013 work together to reduce the unproductive cleavage of Microprocessor on pri-mir-30a optimally.
Finally, we showed that seedB_3p could function to reduce the productive cleavage of Microprocessor on pri-miRNA regardless of other UG, UGU, and GHG motifs (Fig. 4K–N, Fig. S4D).
Discussion
We previously demonstrated that mismatches and wobble base pairs in a region 4–13 nt from the cleavage sites control the cleavage accuracy and efficiency of Microprocessor [19]. In this study, we showed that bulges in a similar region in the upper stem also impact on the cleavage activity of Microprocessor. We named the bulges (B), mismatches (M), and wobble base pairs (W) located 4–8 (seed region), 7–9 nt, and 10–13 nt from the cleavage sites as seedBMW, midBMW_79, and midBMW_1013, respectively (Fig. 5A–C). Our findings in previous [19] and current studies not only explain the functions of these BMW elements in pri-miRNA processing, but also provide a foundation for interpreting the effects of any events, such as SNPs, somatic mutations and RNA-editing or modifications, which add or remove these BMW elements in pri-miRNAs, on miRNA biogenesis. We recently found that the diseases-related SNP (rs2910164) in pri-mir-146a creates a non-canonical mGHG motif at the apical junction and thus enhances the unproductive cleavage of DROSHA [26]. By performing pri-miRNA processing assays, we explain fundamental mechanisms of how this SNP alters miRNA biogenesis. Therefore, it is interesting to investigate how the changes in the BMW elements, which might be caused by SNPs, somatic mutations, and RNA modifications, affect miRNA biogenesis via pri-miRNA processing.
Figure 5.
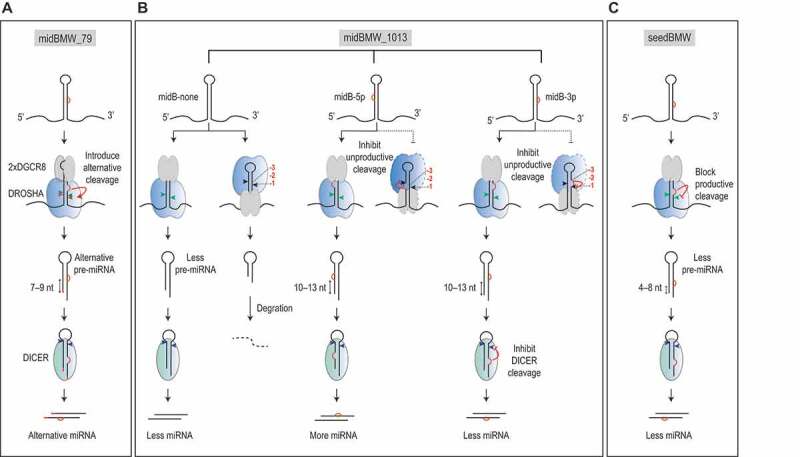
Models for controlling pri-miRNA processing by bulges, mismatches, and wobble base pairs. (A) Bulges on the 3p-strand, and both mismatches and wobble base pairs in positions 7–9 from the cleavage sites, facilitate the occurrence of alternative cleavages. These RNA elements appear in some pri-miRNAs such as pri-mir-342 (this study) and pri-mir-200b.[19], determining the cleavage sites of Microprocessor on these RNAs. These are called midBMW_79. (B) Bulges, mismatches, and wobble base pairs in the positions 10–13 from the cleavage sites, block unproductive cleavages, and thus enhance productive cleavages by Microprocessor. Although bulges on both the 5p-strand and 3p-strand reduce the unproductive cleavage of Microprocessor, and thus enhance the pre-miRNA production, the bulges on the 3p-strand reduce the cleavage of pre-miRNA by DICER. On the 5p-strand, bulges, mismatches, and wobble base pairs in the 10–13 region are enriched and conserved in many pri-miRNAs, and they are called midBMW_1013. (C) Bulges, mismatches, and wobble base pairs in the positions 4–8 from the cleavage sites, repress the number of productive cleavages by Microprocessor. Bulges on the 3p-strand have more substantial inhibitory effect than those on the 5p-strand. These RNA elements are called seedBMW. SeedBMW might occur in pri-miRNAs due to RNA-editing or SNPs[19]
We showed that the action of the BMW elements on pri-miRNA processing is position and strand-dependent. Thus, the seedBMW impedes the productive cleavages, whereas midBMW_79 influences the cleavage sites (Fig. 5A), and midBMW_1013 blocks the unproductive cleavages of Microprocessor (Fig. 5B). Since DROSHA might not cover the positions 10–13, we previously regarded midMW_1013 as mismatches and wobbles in the positions −1 to −3, 1 to 3 nt from the cleavage site of DROSHA on the 5p-strand as DROSHA oriented at the apical junction. We indeed demonstrated that the mismatches and wobbles in the positions −1 to −3 at the basal junction also decreased the cleavage activity of DROSHA. Here, we observed that the bulges in the positions −1 to −2 reduced the cleavage activity of DROSHA (Fig. S1N, Table S1). These suggest that BMWs in the positions −1 to −3 have a similar impact on the cleavage activity of DROSHA. In addition, we also showed that the BMW elements act on pri-miRNA processing in a strand-dependent manner. Although midBMW_1013 has a similar effect regardless of the location of the bulges (this study) or wobble base pairs [19] on the 5p- or 3p-strands (Fig. 5B), the actions of midBMW_79 and seedBMW are somewhat asymmetric (Fig. 5A,Fig. 5C). For example, for both midBMW_79 and seedBMW, bulges (this study) or wobble base pairs (G-U) [19] on the 3p-strand have a stronger effect than those on the 5p-strand (Fig. 5A,Fig. 5C).
In this study, we also found that bulges in midB_1013 have a distinct effect on Microprocessor and DICER. Although both 5p- and 3p-strand midB_1013 efficiently block the unproductive cleavage of DROSHA, thereby positively affecting the pre-miRNA production, 3p-strand midB_1013 inhibits DICER cleavage, and thus reduces the amount of miRNA product (Fig. 5B). These opposing effects of midB_1013_3p on Microprocessor and DICER for miRNA production might explain why bulges are not enriched in the positions 10–13 on the 3p-strand of pri-miRNAs from humans or other organisms. Thus, evolution might specifically select pri-miRNAs that have 5p-strand bulges enriched in midB_1013.
Here, we studied the effect of bulges on the D3-G2 complex, which contains an RNA-binding affinity only from DROSHA, and NLSD3-DGCR8, which contains RNA-binding affinity from both DROSHA and DGCR8. We found that the effects of seedBMW or midBMW are similar between these two types of complexes. This indicates that these bulges affect Microprocessor via DROSHA. Analysing the solved DROSHA-RNA structures and mutagenesis study would help us understand further how bulges function in pri-miRNA processing via affecting the DROSHA-RNA interaction.
Materials and methods
Protein purification
Recombinant proteins, NLSD3-DGCR8 and D3-G2, were prepared as described previously [19]. In brief, pXab-D3 (or pXab-NLSD3) was co-transfected with pXG-G2 (or pXG-DGCR8) into HEK293E cells. Three days after transfection, the transfected cells were collected by centrifugation and resuspended in lysis buffer (20 mM Tris-HCl [pH 7.5], 500 mM NaCl, 2 μg/ml RNase A, 4 mM β-mercaptoethanol and protease inhibitor cocktail (Thermo Fisher Scientific). The lysed mixture was subjected to sonication and centrifugation to obtain a clear cell lysate. The proteins were then purified from the cell lysate using two columns, an IMAC nickel resin and a UNOsphere Q resin (both from Bio-Rad). The fractions eluted from the UNOsphere Q column, which contained the highest protein concentration, were aliquoted and stored at −80°C until use. The D3TN1 and D3TN2 proteins had the mutation in the positions 1045 and 1222, respectively [9]. The D3TN1-G2 and D3TN2-G2 proteins were purified similarly as D3-G2.
The open reading frame of DICER was cloned in the pXG vector, after which the DICER proteins were purified using a similar protocol described for Microprocessor.
The information of the plasmids used for protein expression is shown in Table S2.
Pri-miRNA and pre-miRNA processing
Pri-miRNA and pre-miRNA were processed by Microprocessor and DICER, respectively, in 10 μl standard cleavage buffer (50 mM Tris-HCl (pH 7.5), 150 mM NaCl, 0.2 μg/μl BSA, 10% glycerol, 2 mM MgCl2 and 1 mM DTT) at 37°C. The amounts of the pri-miRNAs and Microprocessor used and the incubation times applied are shown in the Figures and Figure legends. The processing reaction was stopped by the addition of 10 μl loading buffer (8 M urea, 1 mM EDTA, 20 mM Tris-HCl (pH 8.0) and 0.05% (w/v) bromophenol blue). The stopped reaction mixture was then treated with 20 μg proteinase K (Thermo Fisher Scientific) for 15 min at 37°C, then for 15 min at 50°C, and finally for 5 min at 95°C. The proteinase K-treated mixture was then chilled on ice and loaded onto the urea-PAGE, which had been pre-run for 1 h at 300 V. The urea-PAGE electrophoresis was also run at 300 V, this time for 40 min, after which the gel was stained with SYBRTM Green II RNA gel stain (Invitrogen). The RNAs in the gel were visualized and photographed with a Bio-Rad Gel Doc XR+ system, and the images acquired were analysed by Image Lab v6.0.1.
The information of primers and DNA templates for pri-miRNA synthesis is shown in Table S3.
Analysis of the high through-put pri-mir-342 processing assays
The read counts of the pri-mir-342 library were obtained from Gene Expression Omnibus (Accession number: GSE142140) [19]. The structure prediction and selection of variants were as described in a previous study [19]. The pri-mir-342 variants were classified into four groups: mismatches, bulges, wobbles, and Watson-Cricks base pairs. The CL-1/CL0 ratio for each group was estimated as described previously [19].
Analysis of the bulges in the upper stem of pri-miRNAs
The pri-miRNA sequences of different species were acquired from MirGeneDB release 2.0 [27]. Each pri-miRNA contained 30-nt flanking segments and a pre-miRNA sequence. Only pri-miRNAs that were also deposited in miRBase release 22 [28] were selected, after which they were folded with RNAfold [29]. For a comprehensive comparison, the predicted structures of pri-miRNAs containing multiple loops were removed. We detected bulges along the upper stem of pri-miRNAs and determined their positions from the cleavage sites of DROSHA. The bulge frequency in each position was estimated as a ratio of a number of pri-miRNAs containing bulges at this position to the total number of all pri-miRNAs used in this analysis. The bulge frequency was estimated for the 5p- and 3p-strands separately.
Small RNA library preparation and sequencing
The pcDNA3.0 plasmids encoding pri-mir-342 or its variants (1.5 μg) (Table S4) and 0.5 μg pcDNA3.0 encoding pri-mir-1226 were co-transfected into HCT116 cells pre-seeded in one well of a 6-well plate using lipofectamine 3000 (Thermo Fisher Scientific). One and a half days after transfection, the total RNAs were extracted from the transfected cells using TRIzol reagent (Invitrogen). Fifteen μg of each total RNA was loaded into a 15% urea-PAGE, and the small RNAs within the 19–24 nt region were gel-purified. The purified small RNAs were ligated to the 3ʹ-adapter (4N-RA3,/5rApp/NN NNT GGA ATT CTC GGG TGC CAA GG/3ddC/) using T4 RNA ligase 2, truncated KQ (NEB). The 3ʹ-adapter-ligated RNAs were then separated from the free 3ʹ-adapter and unligated small RNAs by gel-purification. The purified 3ʹ-adapter RNAs were then ligated with a 5ʹ-adapter (RA5-4N, GUU CAG AGU UCU ACA GUC CGA CGA UCN NNN) using T4 RNA ligase 1 (NEB). Next, the 3ʹ- and 5ʹ-adapter-ligated RNAs were reverse-transcribed using SuperScript™ IV reverse transcriptase with the R-RA3 primer, which generated cDNAs. The resulting cDNAs were amplified using PCR with RP1 and RPI primers (Illumina), which produced DNA libraries. A RPI primer with a unique index sequence was used for each small RNA sample.
The DNA libraries of the small RNAs were run using Illumina Nextseq 500. The sequencing data were deposited in NCBI (Accession number: GSE158060). The raw paired-end reads were trimmed to remove 3p- and 5p-adapters using cutadapt (cutadapt -a TGGAATTCTCGGGTGCCAAGG -A GATCGTCGGACTGTAGAACTCTGAAC) [30]. The resulting trimmed reads were joined using fastq-join. Low quality reads and duplicated reads were discarded using fastq_quality_filter (-q 30 -p 90) and fastx_collapser (FASTX-Toolkit, http://hannonlab.cshl.edu/fastx_toolkit/index.html, version 0.0.13), respectively. The 4-nt randomized barcodes on both ends of the reads were then trimmed, after which the reads were mapped to the sequence of the transfected pri-mir-342 or its variants using BWA [31]. Since the 3p-end annotation of miRNAs is not accurate due to the modifications, we used the 5p miRNAs for cleavage site analysis.
qPCR
The miRNAs or U6 spliceosomal RNA were reverse transcribed in a reaction mixture containing 50 ng total RNA using Superscript IV reverse transcriptase and the stem-loop RT primers, which were designed according to the reported method [32]. The pri-miRNAs were reverse transcribed from 1 µg total RNA with the Superscript IV reverse transcriptase and the specific RT primers for each pri-miRNA. qPCRs were performed using the SYBR® green master mix (Bio-Rad). All the primers used for reverse transcription and qPCR are shown in Table S5.
Supplementary Material
Acknowledgments
We thank our lab members for their discussion and technical assistance.
Funding Statement
This work was supported by the Croucher Foundation [CIA17SC03]. T.D.N. is a recipient of the Hong Kong PhD Fellowship Scheme.
Disclosure statement
No potential conflict of interest was reported by the authors.
Supplementary material
Supplemental data for this article can be accessed here.
References
- [1].Iwasaki S, Tomari Y.. Argonaute-mediated translational repression (and activation). Fly (Austin). 2009;3:205–208. [PubMed] [Google Scholar]
- [2].Jonas S, Izaurralde E. Towards a molecular understanding of microRNA-mediated gene silencing. Nat Rev Genet. 2015;16:421–433. [DOI] [PubMed] [Google Scholar]
- [3].Bartel DP. Metazoan MicroRNAs. Cell. 2018;173:20–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Gebert LFR, MacRae IJ. Regulation of microRNA function in animals. Nat Rev Mol Cell Biol. 2019;20:21–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Kawamata T, Making TY. RISC. Trends Biochem Sci. 2010;35:368–376. [DOI] [PubMed] [Google Scholar]
- [6].Ha M, Kim VN. Regulation of microRNA biogenesis. Nat Rev Mol Cell Biol. 2014;15:509–524. [DOI] [PubMed] [Google Scholar]
- [7].Denli AM, Tops BBJ, Plasterk RHA, et al. Processing of primary microRNAs by the microprocessor complex. Nature. 2004;432:231–235. [DOI] [PubMed] [Google Scholar]
- [8].Gregory RI, Yan KP, Amuthan G, et al. The Microprocessor complex mediates the genesis of microRNAs. Nature. 2004;432:235–240. [DOI] [PubMed] [Google Scholar]
- [9].Han J, Lee Y, Yeom KH, et al. The Drosha-DGCR8 complex in primary microRNA processing. Genes Dev. 2004;18:3016–3027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Nguyen HM, Nguyen TD, Nguyen TL, et al. Orientation of human microprocessor on primary MicroRNAs. Biochemistry. 2019;58:189–198. [DOI] [PubMed] [Google Scholar]
- [11].Landthaler M, Yalcin A, Tuschl T. The human DiGeorge syndrome critical region gene 8 and its D. melanogaster homolog are required for miRNA biogenesis. Curr Biol. 2004;14:2162–2167. [DOI] [PubMed] [Google Scholar]
- [12].Auyeung VC, Ulitsky I, McGeary SE, et al. Beyond secondary structure: primary-sequence determinants license Pri-miRNA hairpins for processing. Cell. 2013;152:844–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Fang W, Bartel DP. The menu of features that define primary MicroRNAs and enable de novo design of MicroRNA genes. Mol Cell. 2015;60:131–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Nguyen TA, Jo MH, Choi YG, et al. Functional anatomy of the human microprocessor. Cell. 2015;161:1374–1387. [DOI] [PubMed] [Google Scholar]
- [15].Fernandez N, Cordiner RA, Young RS, et al. Genetic variation and RNA structure regulate microRNA biogenesis. Nat Commun. 2017;8:15114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Kim K, Duc Nguyen T, Li S, et al. SRSF3 recruits DROSHA to the basal junction of primary microRNAs. Rna. 2018;24:892–898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Kwon SC, Baek SC, Choi YG, et al. Molecular basis for the single-nucleotide precision of primary microRNA processing. Mol Cell. 2019;73:505–518.e5. [DOI] [PubMed] [Google Scholar]
- [18].Jin W, Wang J, Liu CP, et al. Structural basis for pri-miRNA recognition by Drosha. Mol Cell. 2020;78:423–433.e5. [DOI] [PubMed] [Google Scholar]
- [19].Li S, Nguyen TD, Nguyen TL, et al. Mismatched and wobble base pairs govern primary microRNA processing by human microprocessor. Nat Commun. 2020;11:1926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Partin AC, Zhang K, Jeong BC, et al. Cryo-EM structures of human drosha and DGCR8 in complex with primary MicroRNA. Mol Cell. 2020;78:411–422.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Nguyen TL, Nguyen TD, Bao S, et al. The internal loops in the lower stem of primary microRNA transcripts facilitate single cleavage of human Microprocessor. Nucleic Acids Res. 2020;48:2579–2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Yang W, Chendrimada TP, Wang Q, et al. Modulation of microRNA processing and expression through RNA editing by ADAR deaminases. Nat Struct Mol Biol. 2006;13:13–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Nguyen TA, Park J, Dang TL, et al. Microprocessor depends on hemin to recognize the apical loop of primary microRNA. Nucleic Acids Res. 2018;46:5726–5736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Dang TL, Le CT, Le MN, et al. Select amino acids in DGCR8 are essential for the UGU-pri-miRNA interaction and processing. Commun Biol. 2020;3:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Roden C, Gaillard J, Kanoria S, et al. Novel determinants of mammalian primary microRNA processing revealed by systematic evaluation of hairpin-containing transcripts and human genetic variation. Genome Res. 2017;27:374–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Le CT, Nguyen TL, Nguyen TD, et al. Human disease-associated single nucleotide polymorphism changes the orientation of DROSHA on pri-mir-146a. Rna. 2020;26:1777–1786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Fromm B, Domanska D, Høye E, et al. MirGeneDB 2.0: the metazoan microRNA complement. Nucleic Acids Res. 2020;48:D132–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Kozomara A, Birgaoanu M, Griffiths-Jones S. MiRBase: from microRNA sequences to function. Nucleic Acids Res. 2019;47:D155–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Lorenz R, Bernhart SH, Höner Zu Siederdissen C, et al. ViennaRNA Package 2.0. Algorithms Mol Biol. 2011;6:26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 2011;17:10. [Google Scholar]
- [31].Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Chen C, Ridzon DA, Broomer AJ, et al. Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic Acids Res. 2005;33:e179. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


