ABSTRACT
Human/humanized IgG4 antibodies have reduced effector function relative to IgG1 antibodies, which is desirable for certain therapeutic purposes. However, the developability and biophysical properties for IgG4 antibodies are not well understood. This work focuses on the head-to-head comparison of key biophysical properties, such as self-interaction and viscosity, for 14 human/humanized, and chimeric IgG1 and IgG4 S228P monoclonal antibody pairs that contain the identical variable regions. Experimental measurements showed that the IgG4 S228P antibodies have similar or higher self-interaction and viscosity than that of IgG1 antibodies in 20 mM sodium acetate, pH 5.5. We report sequence and structural drivers for the increased viscosity and self-interaction detected in IgG4 S228P antibodies through a combination of experimental data and computational models. Further, we applied and extended a previously established computational model for IgG1 antibodies to predict the self-interaction and viscosity behavior for each antibody pair, providing insight into the structural characteristics and differences of these two isotypes. Interestingly, we observed that the IgG4 S228P swapped variants, where the CH3 domain was swapped for that of an IgG1, showed reduced self-interaction behavior. These domain swapped IgG4 S228P molecules also showed reduced viscosity from experiment and coarse-grained simulations. We also observed that experimental diffusion interaction parameter (kD) values have a high correlation with computational diffusivity prediction for both IgG1 and IgG4 S228P isotypes.
Abbreviations: , constant region Hamaker constant; , variable region Hamaker constant; CDRs, Complementarity-determining regions; CG, Coarse-grained model; CH1, Constant heavy chain 1; CH2 Constant heavy chain 2; CH3 Constant heavy chain 3; chgCH3 Effective charge on the CH3 region; CL Constant light chain; cP, Centipoise; DLS, Dynamic light scattering; Fab, Fragment antigen-binding; Fc, Fragment crystallizable; Fv, Variable domaing; (r) Radial distribution function; H1 CDR1 of Heavy Chain; H2 CDR2 of Heavy Chain; H3 CDR3 of Heavy Chain; HVI, High viscosity index; IgG1 human immunoglobulin of IgG1 subclass; IgG4 human immunoglobulin of IgG4 subclass; kD, Diffusion interaction parameter; L1 CDR1 of Light Chain; L2 CDR2 of Light Chain; L3 CDR3 of Light Chain; mAb, Monoclonal antibody; MD, Molecular dynamics; PPI Protein–protein interactions; SCM, Spatial charge map; UP-SEC, Ultra-high-performance size-exclusion chromatography; VH, Variable domain of Heavy Chain; VL, Variable domain of Light Chain
KEYWORDS: Monoclonal antibody self-interaction, monoclonal antibody viscosity, diffusion interaction parameter, IgG1, IgG4 S228P, IgG4P, developability, computational models
Introduction
The immunoglobulin G (IgG) is the most abundant class of immunoglobulins in circulation as well as the most used class as therapeutic proteins.1–3 The IgG class has four isotypes: IgG1, IgG2, IgG3, and IgG4.2 In 2021, antibody drugs approved in the United States and European Union include ~60% hIgG1, ~9% hIgG2, ~17% hIgG4, and ~15% other isotypes or formats (Fab, domain, mouse, hybrid molecules).4 Selection of isotype often depends on the required interaction with the human immune system. Typically, IgG4 antibodies have reduced effector function relative to the IgG1.2 The lack of effector functions, such as antibody-dependent cell-mediated cytotoxicity and complement-dependent cytotoxicity, is desirable for therapeutic purposes when the objective is to block certain receptors or deliver a toxic payload.5
Although IgG1 and IgG4 share high sequence similarity at the amino acid level, these two isotypes differ structurally. The length and flexibility of the hinge region vary among the IgG isotypes.2 The hinge region of IgG4 (12 amino acids) is shorter and less flexible than that of IgG1 (15 amino acids). The interchain disulfide bonds between light chain and heavy chain for IgG1 and IgG4 differ in their position.6 The disulfide bonds form between light chain C-terminus (C214) and upper hinge region (C220) for IgG1 and N-terminal CH1 (C131) for IgG4, respectively. Wildtype IgG4 is also known to undergo antibody-binding fragment (Fab)-arm exchange where a half molecule (one heavy and one light chain) may dissociate and form a whole antibody with other half molecules.7,8 A mutation for IgG4 (S228P), IgG4P, on the hinge region was reported to abolish Fab-arm exchange.9,10 However, the reverse mutation, P228S, for IgG1 did not induce Fab-arm exchange, suggesting that the core hinge alone is not responsible for this dynamic process.11 A residue (K409) on the CH3 domain for IgG1 has also been reported to be essential for stabilizing the half molecules.11 Despite these intriguing features of IgG4, the underlying mechanisms and biophysical properties compared to their IgG1 counterparts remain unclear.
The stability of monoclonal antibodies (mAbs) is crucial for new drug development in different stages, including manufacturing, storage, and delivery.12 However, many therapeutic proteins can present developability and instability challenges.11–15 These include solubility, aggregation, depressed colloidal properties, stability, and viscosity. It has been reported that IgG4s have lower thermostability than IgG1 due in part to a different pattern in the disulfide bond network between heavy chain and light chain of the two isotypes. The thermostability of IgG4 Fab regions can be improved to be closer to that of an IgG1 Fab by manipulating the IgG4’s disulfide bond arrangement to mimic that of IgG1 to reduce disulfide bond heterogeneity.16,17 In addition, Neergaard et al. compared the stability of IgG1 and IgG4P with the same variable domain at high protein concentration and suggested that the stability profile (aggregation tendency, thermostability, chemical stability) of an antibody is likely dependent on the IgG framework on the constant region and that IgG1s are more stable than IgG4s.18 Moreover, Heads et al. reported that the net charge state of variable domains relative to the net charge state of the constant domains is predominantly responsible for the different native state aggregation behavior of IgG1 and IgG4P mAbs.19 An independent study performed on the panel of 152 mAbs (72 IgG1 and 80 IgG4 antibody molecules) showed that IgG4P mAbs had overall less optimal developability properties than IgG1 mAbs across several assays (aggregation formation upon low pH stress, AC-SINS, Tm/Tagg). However, a caveat from this study was that most of these mAbs that were examined on human IgG1 and IgG4 backbones had different variable domain sequences.20
The diffusion interaction parameter (kD) has been reported to show some correlation and be predictive of viscosity behavior.20 A study on the viscosity of IgG1 and IgG4 (which included one IgG1 and two IgG4s) showed that solution viscosity can be predicted by quantifying protein–protein interactions (PPI) such as kD measured by dynamic light scattering (DLS).21 Another study found that kD is effective in predicting poor solution behavior (high viscosity and opalescence) with a set of 59 mAbs in low ionic buffer.22 However, Yadav et al. showed a limitation of using kD measured in diluted solution to predict high concentration behavior.13 Wang et al. showed examples of antibody solutions that had low viscosities at high concentrations >150 mg/ml across different pH conditions while presenting strong negative second virial coefficient B2 values.23 In a different study, it is reported that under a wide range of pH and ionic strength, big changes in net PPI did not always lead to significant changes in viscosities, putting into question the validity of using kD as a predictor of viscosity.24 A systematic comparison of a larger number of antibody pairs is required to corroborate the interplay between sequence, structure, self-interactions, and viscosity characteristics of IgG1s versus IgG4s.
In this work, we focused on the head-to-head comparison of self-interaction (kD) measured by DLS and viscosity behaviors of 14 IgG1 and IgG4P mAb pairs, having identical variable domain for each antibody pair. We implemented experimental and computational methods to study and predict these self-interaction and viscosity behaviors. All 14 IgG4P mAbs in our study showed an increase in self-interaction as compared to their corresponding IgG1 mAbs. Three computational models, namely, spatial charge map (SCM),25 a decision tree model previously described,26 and a coarse-grained (CG) model,27 were applied in an attempt to predict self-interaction and viscosity behaviors of these IgG1/IgG4 pairs. The microstructure of clusters by the radial distribution function was also analyzed to gain insight into domain interactions. Although these models were previously developed based on the IgG1 isotype, this work aims to validate and extend these models to the IgG4P isotype. We found that experimental self-interaction (kD) correlated with computational diffusivity prediction and the computational model also showed that IgG4P antibodies have similar or higher viscosity than their IgG1 counterparts, which agrees with experimental results. In addition, our domain swapping experimental studies reveal a critical role of the CH3 domain, which, when combined with the net charge of the complementarity-determining regions (CDRs), drives the self-interaction of some IgG4P antibodies. This is further supported by the finding from computational structure analysis that the CH3 region of the IgG4P molecules interact more favorably with the variable regions compared to that of the IgG1 molecules. Predictive models that can inform on the potential risk of increased self-interaction and elevated viscosity are of utmost value to enable candidate selection, drive sequence optimization via protein engineering, or lead to the decision to use a mutated human IgG1 isotype with reduced effector function and favorable biophysical properties instead of an IgG4 when effector function reduction is required for biological activity. Mutated IgG1 mAbs for reduced effector function (e.g., with L234A/L235A mutations) maintain for the most part identical biophysical properties and viscosity profiles than native IgG1 (unpublished data).
Results
Experimental viscosity and kD of IgG1 and IgG4P mAbs
Fourteen approved monoclonal antibody sequences (sequences obtained from patent and peer-reviewed literature in supplemental Table 1) were produced recombinantly in Expi-CHO transient system as human/humanized IgG1 and IgG4P isotypes and purified via protein A chromatography alone or followed by polishing steps whenever necessary as described in Materials and Methods. The 14 antibody pairs were dialyzed in 20 mM sodium acetate, pH 5.5 and concentrated to 50, 100, 150, and 175 mg/ml for viscosity measurements. Possible aggregation at the various protein concentrations was monitored by ultra-high-performance size-exclusion chromatography (UP-SEC) and was <2% at all concentrations for all the pairs and showed no signs of visible turbidity (data not shown) unless otherwise specified.
Table 1.
Diffusion interaction parameter (kD) and viscosity at 150 mg/ml of human IgG1 and human IgG4P antibodies. The viscosity is averaged from different shear rates. The zero-shear viscosity is calculated by extrapolating to zero shear rate using a polynomial fit. All mAbs except cetuximab exhibited Newtonian behavior for viscosity
| mAbs | human IgG1 |
human IgG4P |
||||
|---|---|---|---|---|---|---|
| kD (up to 20 mg/ml) | Viscosity at 150 mg/ml (cP) (Averaged experimental data) | zero-shear viscosity at 150 mg/ml (cP) (Extrapolated) | kD (up to 20 mg/ml) | viscosity at 150 mg/ml (cP) (Averaged experimental data) | zero-shear viscosity at 150 mg/ml (cP) (Extrapolated) | |
| Ganitumab | 38.2 | 10.1 | 10.9 | 4.73 | 19.2.0 | 19.1 |
| Adalimumab | 14.95 | 11.8 | 12.8 | −2.89 | 23.4 | 23.7 |
| Cetuximab | −10.6 | 40.9 | 55.7 | −26.49 | 205.6 | 264.6 |
| Tremelimumab | 8.8 | 14.3 | 14.2 | 4.34 | 17.2 | 17.3 |
| Ipilimumab | 8.6 | 17.8 | 18 | 4.95 | 13.5 | 13.7 |
| Basiliximab | 25.05 | 8 | 8.6 | 2.94 | 20.8 | 20.8 |
| Natalizumab | 13.67 | 11.1 | 11.3 | −0.001 | 11.1 | 12.2 |
| Vesencumab | 23.57 | 11 | 12 | 4.65 | 33.9 | 34.1 |
| Atezolizumab | 11.56 | 21.8 | 22.3 | −7.17 | 49.9 | 53 |
| Trastuzumab | 14.68 | 9.2 | 9.3 | −4.45 | 8.7 | 8.8 |
| TGN1412 | 16.42 | 6.1 | 6.1 | −1.68 | 11.8 | 11.8 |
| Omalizumab | −13.37 | 30.3 (100 mg/ml) | 32.5 (100 mg/ml) | −31.3 (up to 8 mg/ml) | 1.11 (20 mg/ml) | |
| Bevacizumab | 3.17 | 6.8 | 6.7 | −11.82 | 12.9 | 12.8 |
| Golimumab | 9.31 | 7.5 | 7.6 | −3.88 | 8.6 | 8.7 |
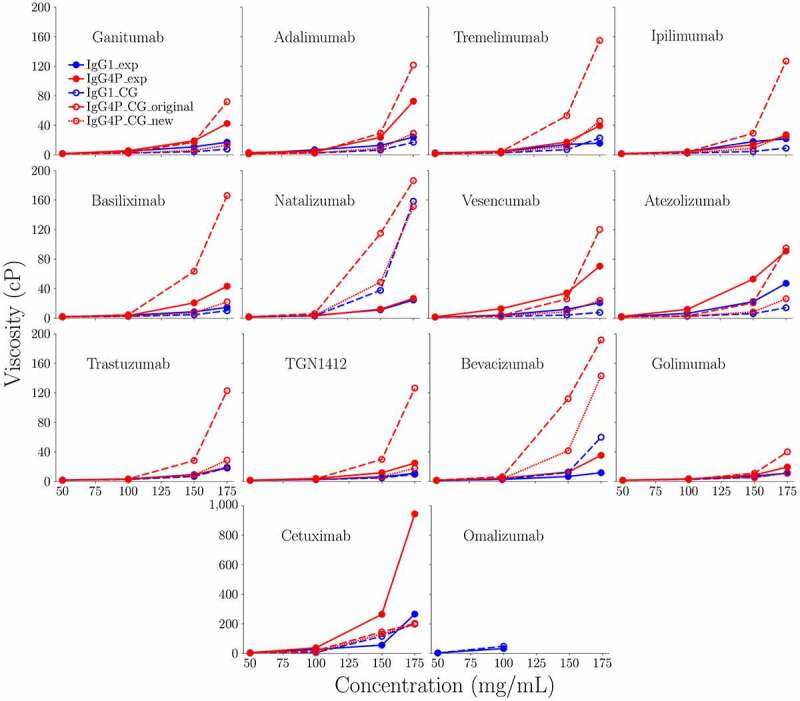
Viscosities were measured from 50 to 175 mg/ml at various shear rates (Figure S1). Viscosity values were also extrapolated to zero shear-rate at concentrations up to 150 mg/ml to directly compare with values obtained from simulation for the 14 IgG1/IgG4P mAb pairs, which are reported in Table 1 and in Figure 1. Experimental kD values were also measured at protein concentrations ranging from 1 to 20 mg/ml in 20 mM sodium acetate, pH 5.5 (Table 1 and Figure S2). An increased negative kD value would correspond to an increased tendency for self-interaction whereas positive kD values would indicate low self-interaction.
Figure 1.
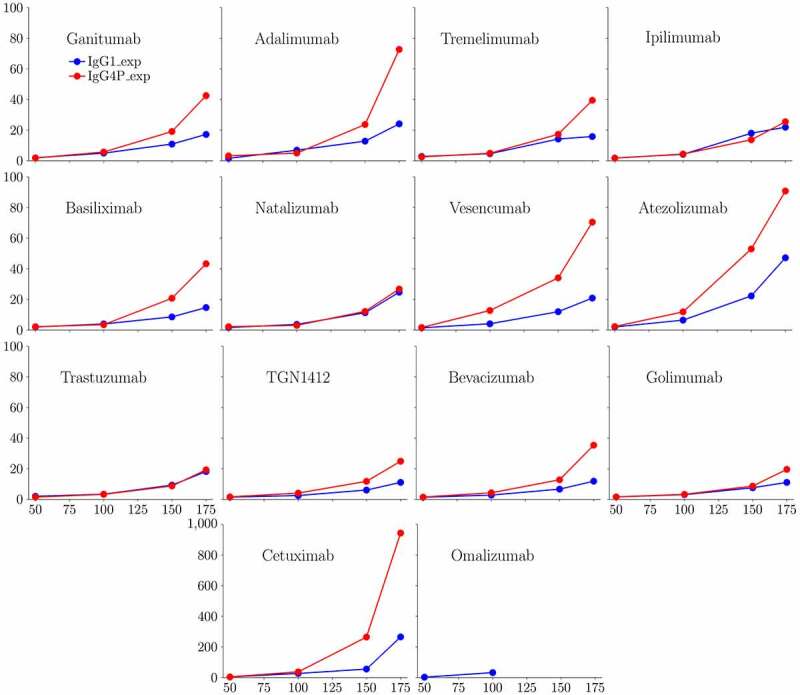
Concentration dependence of the zero-shear rate (extrapolated) viscosity of 14 IgG1 and IgG4P mAbs at 50, 100, 150, and 175 mg/ml in 20 mM sodium acetate at pH 5.5. The zero-shear viscosity was calculated by extrapolating viscosity at different shear-rate to zero. Omalizumab IgG1 cannot be concentrated >100 mg/ml and omalizumab IgG4P cannot be concentrated past 20 mg/ml
Experimental results showed that IgG1 mAbs exhibit viscosity values from ~6 to 40 cP, while IgG4P mAbs exhibit noticeably higher viscosity values ranging from ~8 to 200 cP at 150 mg/ml. Eight of 14 IgG4P mAbs had significantly elevated viscosity (~2-5-fold increase at 150 mg/ml) and noticeable increased self-interaction (kD by DLS) as IgG4P compared to IgG1. These mAbs are ganitumab, adalimumab, cetuximab, basiliximab, vesencumab, atezolizumab, TGN1412, and bevacizumab. In one case, omalizumab, the IgG4P antibody could not even be concentrated past 20 mg/ml due to increased turbidity and viscosity, while omalizumab IgG1 could be concentrated to 100 mg/ml (but not to higher concentrations due to increased turbidity and viscosity). The high viscosity of omalizumab has been reported previously by others.28
Diffusion interaction parameter (kD) and correlations with viscosity measurements
As shown in Table 1, kD was measured by DLS at 1–20 mg/ml in 20 mM sodium acetate, pH 5.5. Figure 2 shows the correlation of experimental viscosity at 150 mg/mL with measured kD values for IgG1 and IgG4P. The correlation coefficients are 0.62 and 0.8 for IgG1 and IgG4P, respectively. However, the correlation coefficients drop to 0.13 and 0.05 for IgG1 and IgG4P, respectively, when excluding the measurements for cetuximab, which has a significantly higher viscosity than the other antibodies in the study. This indicates that kD does not always quantitatively correlate with experimental viscosity but is a useful qualitative tool to identify low and high viscosity trends.
Figure 2.
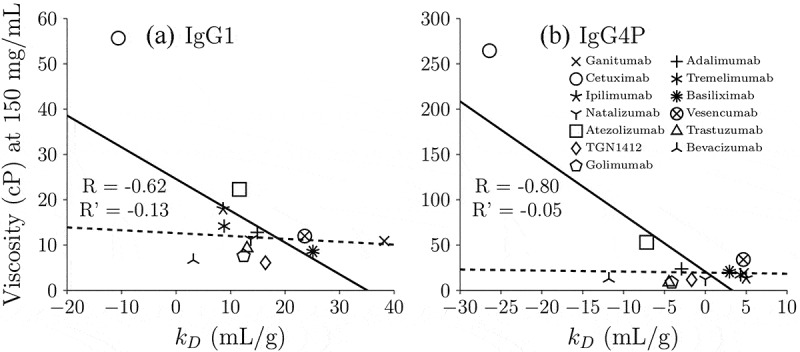
Correlation of experimental zero-shear viscosity at 150 mg/mL with measured kD values for (a) IgG1 and (b) IgG4P. The solid lines indicate linear correlation of all 13 mAbs (including cetuximab and excluding omalizumab since it could not be concentrated 150 mg/ml)) and R is its linear correlation coefficient. Cetuximab is the only high viscosity mAb for which we were able to measure viscosity at 150 mg/ml concentrations for both IgG1 and IgG4P. The dashed lines indicate linear correlation excluding cetuximab, which has the highest viscosity (55.7 cP for IgG1 and 264.6 cP for IgG4P) and R’ is its linear correlation coefficient
Charge distribution of IgG1 and IgG4P mAbs
Protein net charge and charge distribution on the surface are believed to be a dominating factor of antibody viscosity.29 Experimental determination of the effective net charge on the individual domain is challenging; therefore, net charge calculation based on sequence and structure are often used. Table 2 lists the net charge on different regions of IgG1 and IgG4P mAbs based on sequence and molecular structure. The regions include the six CDRs individually in the heavy and light chains, total CDR (sum of the 6 CDR regions), VH, CH1, hinge, CH2, CH3, VL, CL, variable fragment (Fv), fragment crystallizable (Fc), and full-length mAb. The constant regions of IgG4P mAbs (CH1, CH2, CH3, and Fc) have significantly less positive charge compared to the corresponding constant regions of IgG1 mAbs. Of note is the significant charge difference between the Fc domain of IgG4P and IgG1 mAbs (2 e for IgG4P vs. 12 e for IgG1) and between the CH3 region of IgG4P and IgG1 mAbs (a net negative charge of −1 e for IgG4P versus +2 e for IgG1).
Table 2.
Charges (e) of different regions based on sequence and molecular structure in 20 mM sodium acetate, pH 5.5. The protonation state of histidine is calculated from PROPKA 3.0 using the homology model. H1, H2, H3, L1, L2, and L3 indicate different CDR regions. CDR indicates the sum of the six CDR regions
| mAbs | H1 | H2 | H3 | L1 | L2 | L3 | CDR | VH | CH1 | hinge | CH2 | CH3 | VL | CL | Fv | Fc | mAb |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Ganitumab IgG1 | 0 | 1 | −1 | 1 | 1 | 1 | 3 | 4 | 6 | 1 | 4 | 2 | 1 | 1 | 5 | 12 | 38 |
| Ganitumab IgG4P | 0 | 1 | −1 | 1 | 1 | 1 | 3 | 4 | 5 | 0 | 2 | −1 | 1 | 1 | 5 | 2 | 24 |
| Adalimumab IgG1 | −2 | 1 | −1 | 2 | 0 | 2 | 2 | −2 | 5 | 1 | 4 | 2 | 5 | 1 | 3 | 12 | 32 |
| Adalimumab IgG4P | −2 | 1 | −1 | 2 | 0 | 2 | 2 | −2 | 4 | 0 | 2 | −1 | 5 | 1 | 3 | 2 | 18 |
| Cetuximab IgG1 | 0 | 0 | −2 | 1 | −1 | 0 | −2 | 2 | 5 | 1 | 4 | 2 | −1 | 0 | 1 | 12 | 26 |
| Cetuximab IgG4P | 0 | 0 | −2 | 1 | −1 | 0 | −2 | 2 | 4 | 0 | 2 | −1 | −1 | 0 | 1 | 2 | 12 |
| Tremelimumab IgG1 | 0 | −1 | −1 | 0 | 0 | 0 | −2 | 3 | 5 | 1 | 4 | 2 | 1 | 1 | 4 | 12 | 34 |
| Tremelimumab IgG4P | 0 | −1 | −1 | 0 | 0 | 0 | −2 | 3 | 4 | 0 | 2 | −1 | 1 | 1 | 4 | 2 | 20 |
| Ipilimumab IgG1 | 0 | −1 | −1 | 1 | 1 | 0 | 0 | 3 | 6 | 1 | 4 | 2 | 1 | 1 | 4 | 12 | 36 |
| Ipilimumab IgG4P | 0 | −1 | −1 | 1 | 1 | 0 | 0 | 3 | 5 | 0 | 2 | −1 | 1 | 1 | 4 | 2 | 22 |
| Basiliximab IgG1 | 1 | −1 | −2 | 1 | 0 | 1 | 0 | 3 | 5 | 1 | 4 | 2 | 4 | 1 | 7 | 12 | 40 |
| Basiliximab IgG4P | 1 | −1 | −2 | 1 | 0 | 1 | 0 | 3 | 4 | 0 | 2 | −1 | 4 | 1 | 7 | 2 | 26 |
| Natalizumab IgG1 | 0 | −1 | −2 | 1 | 0 | −1 | −3 | 1 | 4 | 1 | 4 | 2 | 1 | 1 | 2 | 12 | 28 |
| Natalizumab IgG4P | 0 | −1 | −2 | 1 | 0 | −1 | −3 | 1 | 4 | 0 | 2 | −1 | 1 | 1 | 2 | 2 | 16 |
| Vesencumab IgG1 | 0 | 0 | 0 | 1 | 1 | 0 | 2 | 1 | 5 | 1 | 4 | 2 | 3 | 1 | 4 | 12 | 34 |
| Vesencumab IgG4P | 0 | 0 | 0 | 1 | 1 | 0 | 2 | 1 | 4 | 0 | 2 | −1 | 3 | 1 | 4 | 2 | 20 |
| Atezolizumab IgG1 | −1 | 0 | 1 | 0 | 0 | 0 | 0 | 1 | 5 | 1 | 4 | 2 | 1 | 1 | 2 | 12 | 30 |
| Atezolizumab IgG4P | −1 | 0 | 1 | 0 | 0 | 0 | 0 | 1 | 4 | 0 | 2 | −1 | 1 | 1 | 2 | 2 | 16 |
| Trastuzumab IgG1 | 0 | 0 | −2 | 0 | 0 | 0 | −2 | 1 | 6 | 1 | 4 | 2 | 2 | 1 | 3 | 12 | 34 |
| Trastuzumab IgG4P | 0 | 0 | −2 | 0 | 0 | 0 | −2 | 1 | 5 | 0 | 2 | −1 | 2 | 1 | 3 | 2 | 20 |
| TGN1412 IgG1 | 0 | 0 | −1 | 1 | 2 | 0 | 2 | 2 | 5 | 1 | 4 | 2 | 4 | 1 | 6 | 12 | 38 |
| TGN1412 IgG4P | 0 | 0 | −1 | 1 | 2 | 0 | 2 | 2 | 4 | 0 | 2 | −1 | 4 | 1 | 6 | 2 | 24 |
| Bevacizumab IgG1 | 0 | −1 | 0 | −1 | 1 | 0 | −1 | 1 | 5 | 1 | 4 | 2 | 1 | 1 | 2 | 12 | 30 |
| Bevacizumab IgG4P | 0 | −1 | 0 | −1 | 1 | 0 | −1 | 1 | 4 | 0 | 2 | −1 | 1 | 1 | 2 | 2 | 16 |
| Omalizumab IgG1 | 0 | −1 | 2 | −2 | −1 | −1 | −3 | 3 | 5 | 1 | 4 | 2 | −3 | 1 | 0 | 12 | 26 |
| Omalizumab IgG4P | 0 | −1 | 2 | −2 | −1 | −1 | −3 | 3 | 4 | 0 | 2 | −1 | −3 | 1 | 0 | 2 | 12 |
| Golimumab IgG1 | 0 | −1 | −1 | 1 | 0 | 1 | 0 | 3 | 6 | 1 | 4 | 2 | 1 | 1 | 4 | 12 | 36 |
| Golimumab IgG4P | 0 | −1 | −1 | 1 | 0 | 1 | 0 | 3 | 5 | 0 | 2 | −1 | 1 | 1 | 4 | 2 | 22 |
Prediction of IgG1 and IgG4P viscosity by SCM
A spatial charge map (SCM) is a tool to predict low and high viscosity based on the negative charge distribution on the surface obtained from molecular dynamics (MD) simulation.25 SCM was developed based on the assumption that most conserved regions of mAbs at common formulation conditions carry a net positive charge. If there are negative charge patches on the Fv region, they will produce favorable electrostatic interactions with other positively charged regions. This tool was previously tested on 19 IgG1 mAbs.25 The performance of IgG4P mAbs has not been validated with a large dataset. Table 3 shows a summary of the key parameters used in this study for IgG1 and IgG4P mAbs (isoelectric point, molecular weight, extinction coefficient, net charge, SCM, and HVI). Figure 3 displays the correlation of SCM with experimental viscosity at 150 mg/mL for IgG1 and IgG4P mAbs. Antibodies with SCM scores of above 1000 are predicted to have high viscosity. The cutoff value for low/high viscosity is 30 cP. Among the 14 IgG1 mAbs, 11 IgG1 mAbs were correctly classified as low viscosity mAbs (ganitumab, tremelimumab, ipilimumab, basiliximab, natalizumab, vesencumab, atezolizumab, trastuzumab, TGN1412, bevacizumab, and golimumab) as their SCM scores were below 1000, and 2 IgG1 mAbs were correctly classified as high viscosity mAbs (cetuximab and omalizumab). Among the 14 IgG4P mAbs, 9 IgG4P mAbs were correctly classified as low viscosity mAbs (ganitumab, tremelimumab, ipilimumab, basiliximab, natalizumab, trastuzumab, TGN1412, bevacizumab, and golimumab) and 2 IgG4P mAbs were correctly classified as high viscosity mAbs (cetuximab and omalizumab). The prediction failed in the case of one IgG1 (adalimumab) and three IgG4P mAbs (adalimumab, vesencumab, and atezolizumab), with a 7.1% and 21.4% failure rate for IgG1 and IgG4P variants, respectively.
Table 3.
Summary of the key parameters used in this study. The molecular weight (MW) has a unit of Da. The extinction coefficient (EC) has a unit of ml/mg/cm. The net charge is based on the antibody’s full-length sequence and molecular structure (using Propka) and has a unit of e. SCM represents the spatial charge map score. HVI represents the high viscosity index
| mAbs | pI | MW | EC | net charge | SCM | HVI |
|---|---|---|---|---|---|---|
| Ganitumab IgG1 | 8.75 | 145455.4 | 1.606 | 38 | 806.5 | 15.15 |
| Ganitumab IgG4P | 7.58 | 145144.7 | 1.609 | 24 | 759.9 | 15.15 |
| Adalimumab IgG1 | 8.72 | 145190.9 | 1.463 | 32 | 1316.6 | 16.67 |
| Adalimumab IgG4P | 7.55 | 144880.3 | 1.466 | 18 | 1325 | 16.67 |
| Cetuximab IgG1 | 8.54 | 145188.7 | 1.498 | 26 | 1127.9 | 20.35 |
| Cetuximab IgG4P | 7.25 | 144878 | 1.501 | 12 | 1046.3 | 20.35 |
| Tremelimumab IgG1 | 8.93 | 146572.4 | 1.551 | 34 | 704.2 | 18.97 |
| Tremelimumab IgG4P | 8.23 | 146261.8 | 1.554 | 20 | 723.6 | 18.97 |
| Ipilimumab IgG1 | 8.46 | 144911.7 | 1.57 | 36 | 754 | 15.49 |
| Ipilimumab IgG4P | 8.34 | 144768.2 | 1.544 | 22 | 769.2 | 15.49 |
| Basiliximab IgG1 | 9.12 | 143522.9 | 1.619 | 40 | 640.8 | 25.11 |
| Basiliximab IgG4P | 8.72 | 143212.2 | 1.623 | 26 | 637.1 | 25.11 |
| Natalizumab IgG1 | 8.68 | 146344.3 | 1.533 | 28 | 815.5 | 20.52 |
| Natalizumab IgG4P | 7.55 | 146033.7 | 1.536 | 16 | 809.3 | 20.52 |
| Vesencumab IgG1 | 8.97 | 144910.7 | 1.411 | 34 | 661.3 | 14.78 |
| Vesencumab IgG4P | 8.25 | 144600.1 | 1.414 | 20 | 693 | 14.78 |
| Atezolizumab IgG1 | 8.53 | 144377.9 | 1.624 | 30 | 759.6 | 13.33 |
| Atezolizumab IgG4P | 7.28 | 144067.3 | 1.627 | 16 | 774.3 | 13.33 |
| Trastuzumab IgG1 | 8.41 | 150000 | 1.43 | 34 | 700.2 | 17.18 |
| Trastuzumab IgG4P | 7.91 | 144792.2 | 1.488 | 20 | 690.6 | 17.18 |
| TGN1412 IgG1 | 8.79 | 145689.5 | 1.554 | 38 | 844.6 | 21.15 |
| TGN1412 IgG4P | 7.91 | 145378.8 | 1.558 | 24 | 808.3 | 21.15 |
| Bevacizumab IgG1 | 8.25 | 146243.8 | 1.664 | 30 | 987.3 | 19.1 |
| Bevacizumab IgG4P | 7.05 | 145933.2 | 1.668 | 16 | 997 | 19.1 |
| Omalizumab IgG1 | 7.36 | 146215.4 | 1.569 | 26 | 1193.7 | 21.6 |
| Omalizumab IgG4P | 6.63 | 145904.7 | 1.572 | 12 | 1086.8 | 21.6 |
| Golimumab IgG1 | 8.98 | 146688.6 | 1.489 | 36 | 807.2 | 13.2 |
| Golimumab IgG4P | 8.32 | 146378 | 1.492 | 22 | 785.2 | 13.2 |
Figure 3.
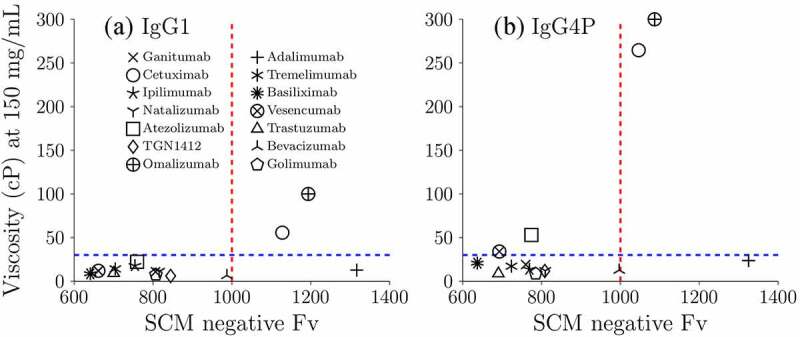
The correlation of SCM with experimental zero-shear viscosity at 150 mg/mL for (a) IgG1 and (b) IgG4P mAbs. The blue dashed line indicates the cutoff value (30 cP) of low and high viscosity mAbs from experimental measurements. The red dashed line indicates the cutoff value of predicted low and high viscosity mAbs. When SCM > 1000, the mAbs are predicted to have high viscosity. The experimental viscosities of omalizumab are hypothetical. Omalizumab has been reported to exhibit high viscosity. SCM model cannot predict precise viscosity values, but provide some directional information on potential low or high viscosity mAbs
Classification of IgG1 and IgG4P viscosity by the decision tree model
The decision tree model depends on two parameters, namely, mAb net charge and the high viscosity index (HVI), to classify low and high viscosity mAbs. It was trained based on 21 IgG1, 4 IgG2, and 2 IgG4 mAbs formulated in 10 mM histidine-HCl at pH 6.0.26 The training set is slightly different from the solution conditions in this study as all mAbs here are formulated in 20 mM sodium acetate pH 5.5 and are pairs of mAbs with the same variable domains on IgG1 and IgG4P isotypes. For this purpose, we define high viscosity mAbs as having viscosity values >30 cP at 150 mg/ml.
We believe the sequence-based charge might overestimate the net charge difference between IgG1s and IgG4Ps, requiring different criteria for IgG4Ps. To determine the best lower and upper charge bound range for IgG4Ps, a range of upper and lower charge bounds were tested (Figure 5). It is assumed that the lower bound is still 22 e lower than the upper bound. For example, when the upper bound is +32 e, the lower bound is +10 e. Finally, we found that, when the upper bound is +20 e, having the corresponding lower bound −2 e gives the best accuracy. Figure 4 (C) shows the classification of the IgG4P mAbs with new splitting criteria. Ten of 14 IgG4P mAbs were now correctly classified and only four mAbs were misclassified (vesencumab, atezolizumab, natalizumab, and bevacizumab). It should be noted that this is a training accuracy using the 14 IgG4P mAbs in this study. More elaborate datasets will be needed to test the revised model.
Figure 5.
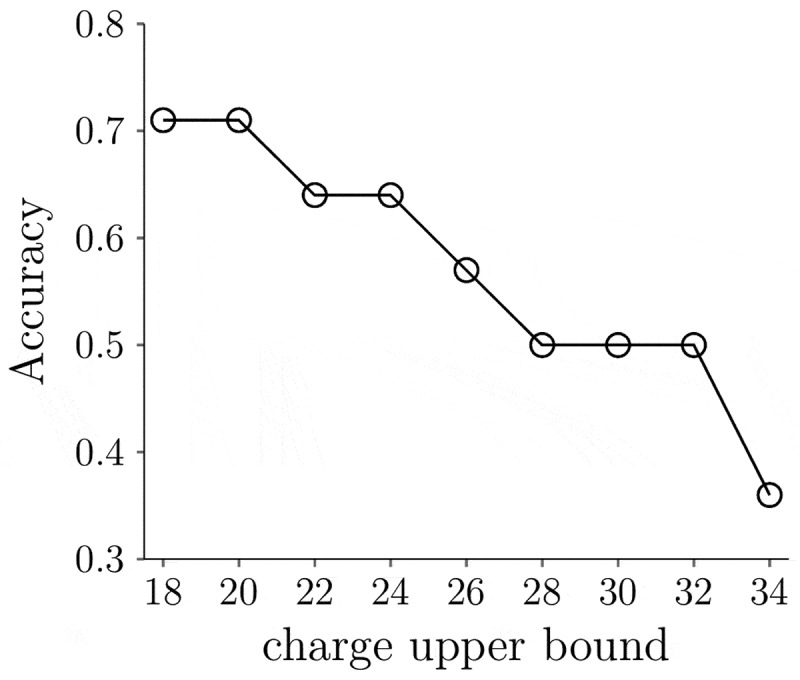
The classification accuracy of the decision tree model for IgG4P mAbs using different charge upper and lower bound criteria. The charge lower bound is 22 e lower than the upper bound. The accuracy is defined as the number of correctly classified mAbs divided by the total number of mAbs analyzed (14)
Figure 4.
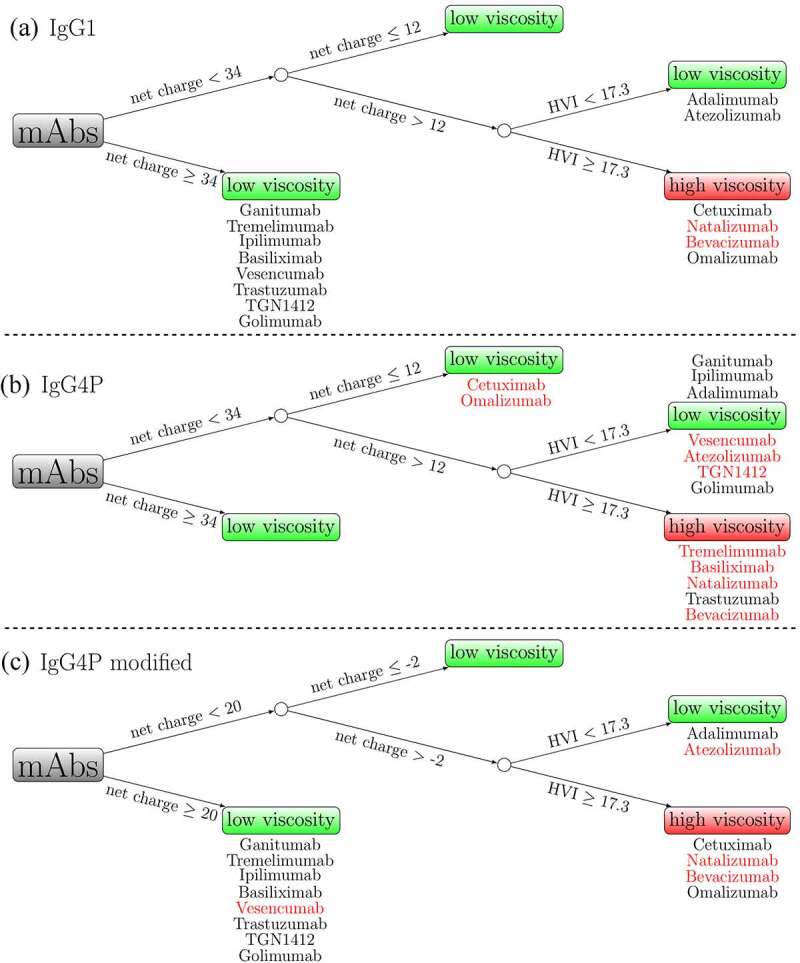
Classification of low and high viscosity mAbs based on a decision tree model for (a) IgG1 (b) IgG4P (c) IgG4P with modified splitting criteria. MAbs with black colors indicate correct classification, while mAbs with red colors indicate incorrect classification. The accuracy is based on the experimental zero-shear viscosity at 150 mg/ml using a cutoff value of 30 cP
Calculation of IgG1 and IgG4P viscosity by the CG model
In addition to low and high viscosity classifiers, we also implemented a recently developed CG model combined with hydrodynamic calculations to calculate viscosity.27 This model was developed and validated based on 20 IgG1 mAbs up to 150 mg/ml in 10 mM histidine-HCl at pH 6.0.27 We attempted to validate and extend this model to the IgG4P isotype. Figure 6 illustrates the concentration dependence of the viscosity from CG models with the experimental viscosity data for IgG1 and IgG4P mAbs extrapolated to zero shear rate. We investigated structural details in an attempt to provide information on the domain interactions involved in viscosity. Figure 7 shows the radial distribution function of VH–VL, VH–VH, VL–VL, VH–CH3, and VL–CH3 pairs for IgG1 and IgG4P mAbs. In short, the radial distribution function (g(r)) describes how density varies as a function of distance (r) between the various antibody domains. It was computed as described in the Materials and Methods section. Although the CG simulation shows that the viscosity of IgG4P mAbs is higher than that of IgG1 mAbs, consistent with experimental data, the simulation tends to overestimate the predicted viscosity of IgG4P mAbs at high concentrations. The radial distribution function shows that the CH3 region interacts more strongly with the variable regions for IgG4P mAbs compared to that of IgG1 mAbs.
Figure 6.
Concentration dependence of the zero-shear viscosity from CG models with the experimental data for IgG1 and IgG4P mAbs. Blue circles indicate experimental viscosity measurements of IgG1. Red squares indicate experimental viscosity measurements of IgG4P. Blue lines indicate CG results of IgG1. Red solid lines indicate CG results of IgG4P with kcal/mol and chgCH3 = −1.0 e. Red dashed lines indicate CG results of IgG4P with kcal/mol and chgCH3 = −0.8 e
Figure 7.
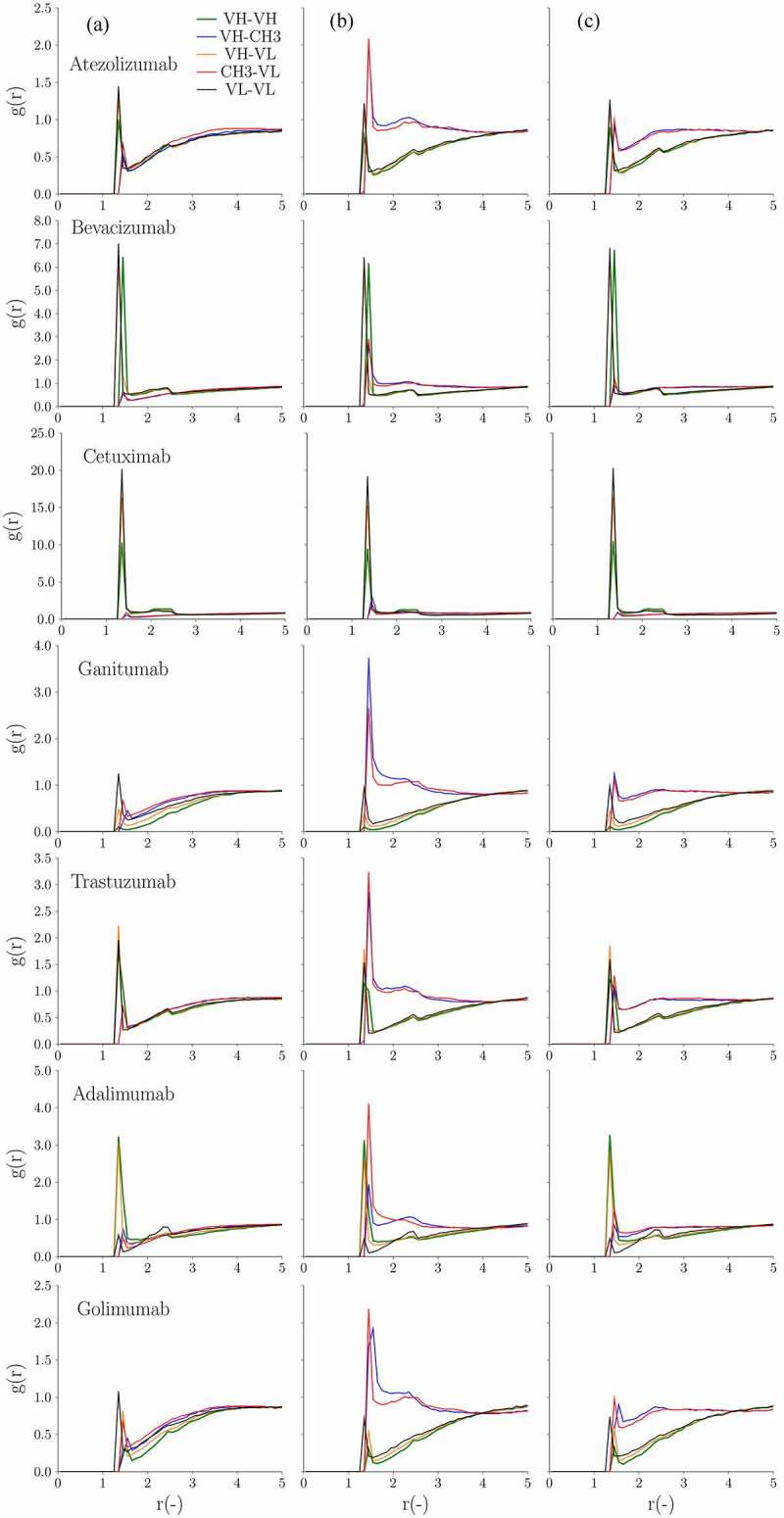
(A) Radial distribution (g(r)) at 150 mg/ml as a function of the distance (r(-)) in between the various mAb domains (VH-VH, VH-CH3, VH-VL, CH3-VL and VL-VL) for the 14 antibody pairs. From left to right are (a) IgG1, (b) IgG4P with kcal/mol and chgCH3 = −1.0 e and (c) IgG4P with kcal/mol and chgCH3 = −0.8 e, respectively. (B) Radial distribution (g(r)) at 150 mg/ml as a function of the distance (r(-)) in between the various mAb domains (VH-VH, VH-CH3, VH-VL, CH3-VL and VL-VL) for the 14 antibody pairs. From left to right are (a) IgG1, (b) IgG4P with kcal/mol and chgCH3 = −1.0 e and (c) IgG4P with kcal/mol and chgCH3 = −0.8 e, respectively
Figure 7.
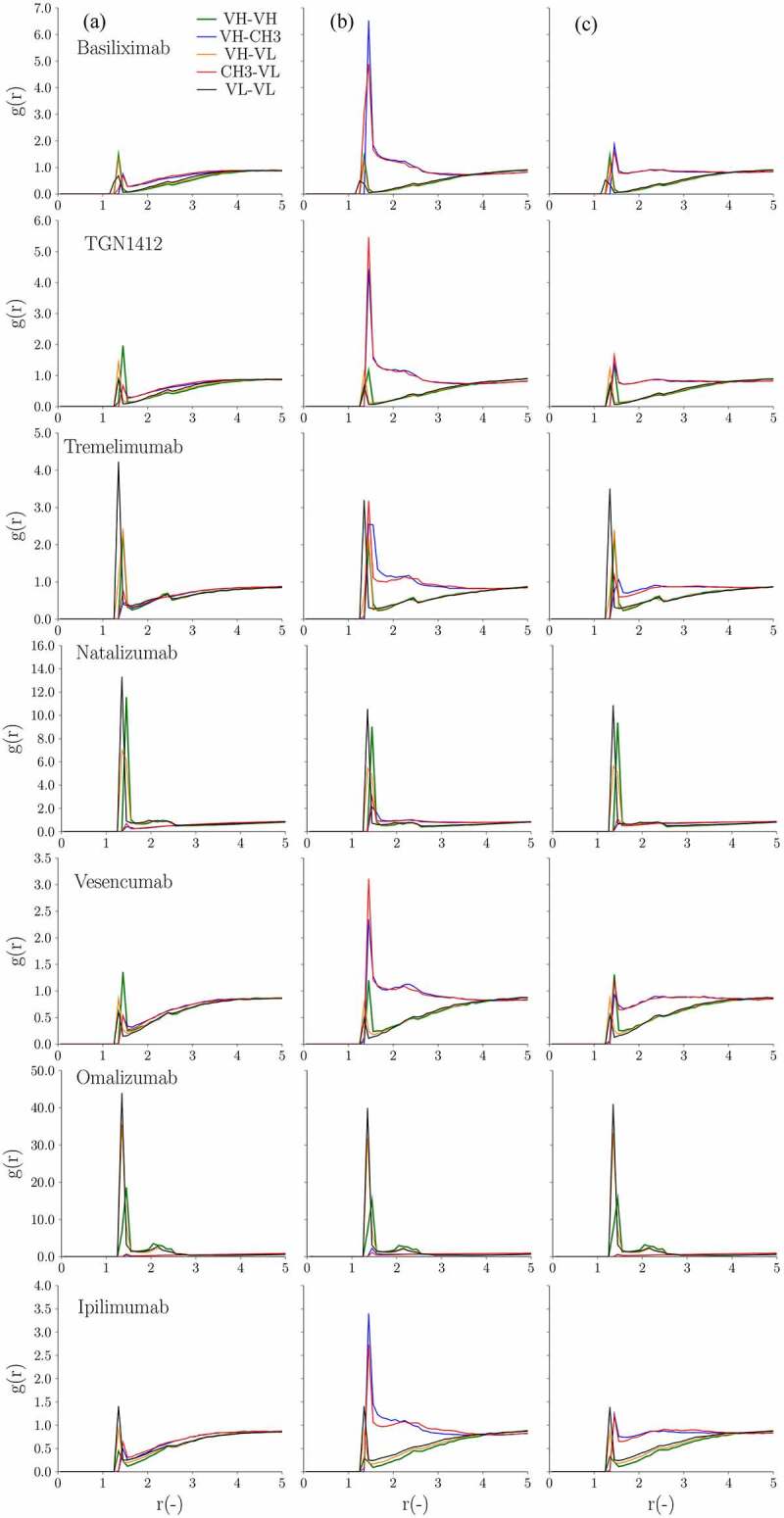
Continued
Determination of the best parameters for the IgG4P CG model
The CG model represents an antibody using 12 beads. The intermolecular interactions are governed by long-range electrostatic interactions and short-range van der Waals interactions. The electrostatic interactions are determined by the charge pairs on the beads. The magnitude of van der Waals interactions is determined by the Hamaker constants on the beads. In our previous work, the Hamaker constants were divided into variable and constant region contributions.22 The constant region Hamaker constants ( were determined by using 20 IgG1 mAbs from a previous study.22 It was hypothesized that it would not be directly applicable to the IgG4P isotype. In addition, the negative charge on the CH3 region of the IgG4P isotype tends to overestimate the electrostatic interactions present in the CG model. The effective charge on the CH3 region (chgCH3) in the IgG4P needs to be modified to properly characterize the various interactions. In order to determine the best parameter sets, we performed a parameter screening for from 0.12 kcal/mol to 0.18 kcal/mol with a step size of 0.02 kcal/mol and for chgCH3 from −0.2 e to −0.8 e with a step size of 0.2 e to minimize the total deviation of viscosity between experiment and simulation of IgG4P. The deviation is defined as . The viscosity difference between experiment and simulation of IgG4P isotype not only comes from the constant regions, but also comes from the variable regions. The parameterization here is mainly for the constant region of IgG4P. Some of the deviation between CG prediction and experiment could come from the variable regions.
In order to exclude the contribution from the variable regions affecting the parameterization of the IgG4P constant regions, we decided to use IgG1 as a reference to remove the effect from the variable regions when both IgG1 and IgG4P share the same variable domains. To account for this effect, the deviation is offset by the viscosity difference between IgG1 experiment and simulation. Table 4 contains the result of the total deviation of 8 IgG4P mAbs (ganitumab, tremelimumab, ipilimumab, basiliximab, trastuzumab, TGN1412, adalimumab, and golimumab) at 150 mg/ml and 175 mg/ml. The four misclassified IgG4P mAbs from the machine learning model were excluded (atezolizumab, natalizumab, bevacizumab, and vesencumab) because their viscosity difference comes primarily from the variable regions. Cetuximab IgG4P was also excluded because its high viscosity reached the limit of the CG simulation. The parameter set for IgG4P which showed improved accuracy is kcal/mol and chgCH3 = −0.8 e.
Table 4.
Total deviation of viscosity at 150 and 175 mg/ml for eight IgG4P mAbs (ganitumab, trastuzumab (Herceptin®), Adalimumab, golimumab, basiliximab, TGN1412, tremelimumab, and ipilimumab) as a function of and chgCH3. The deviation for each IgG4P mAb is defined as . The deviation is offset by the viscosity difference between experiment and computation of IgG1 mAbs to account for the effect of variable regions on the viscosity. The total deviation is defined as
| Total deviation (cP) | (kcal/mol) |
||||
|---|---|---|---|---|---|
| 0.12 | 0.14 | 0.16 | 0.18 | ||
| chgCH3 (e) | −0.2 | 180.8 | 183.4 | 183.5 | 181 |
| −0.4 | 164.5 | 166.5 | 165.3 | 165.1 | |
| −0.6 | 163.1 | 160.8 | 160 | 163.6 | |
| −0.8 | 153.3 | 154.9 | 164.6 | 184.4 | |
Correlation of kD values with diffusivities from the CG model
The diffusivities calculated from CG models have been reported to correlate well with measured kD values.30 Figure 8 shows that the linear correlation coefficients of the normalized diffusivities with the measured kD are 0.81, 0.63, and 0.85 for IgG1, IgG4P ( kcal/mol and chgCH3 = −1.0 e), and IgG4P ( kcal/mol and chgCH3 = −0.8 e), respectively. The modified parameters for the IgG4P CG model significantly improved the correlation. For the high viscosity mAbs, cetuximab and omalizumab, the normalized self-diffusivities are <0.62 for both IgG1 and IgG4 despite different measured kD values.
Figure 8.
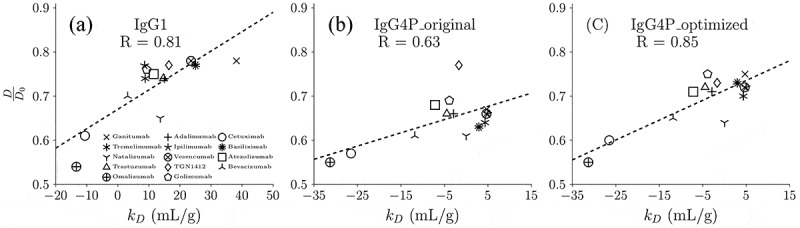
Correlation of self-diffusivities (d) from CG simulation with measured kD values measured by DLS (1–20 mg/ml) for (a) IgG1 with kcal/mol, (b) IgG4P with kcal/mol and chgCH3 = −1.0 e (original parameters) and (c) IgG4P with kcal/mol and chgCH3 = −0.8 e (optimized parameters). The self-diffusivities are calculated at 50 mg/ml. D0 is the single antibody diffusivity in free space. The dashed lines indicate linear correlation and R is the linear correlation coefficient
Viscosity and kD measurements of selected CH3 IgG1-IgG4 swapped variants
Given the significant charge differences between the CH3 domain of IgG1 (+2 e) and IgG4P (−1 e) mAbs (Table 2), we hypothesized that the CH3 domain could play a role in the difference seen in viscosity and kD (self-interactions) for IgG1 and IgG4P mAbs. To test this hypothesis, we selected four mAbs exhibiting different viscosities (cetuximab, trastuzumab, atezolizumab, and golimumab) for a domain swapping experiment, in which the CH3 domain of IgG4P is replaced with the CH3 domain of IgG1 (sequences in supplemental Table 1). The kD and viscosities of the selected IgG4P CH3 swapped variants were measured (Figures S1 and S2). Cetuximab was chosen as it is the highest viscosity mAb in our study for which we have kD and viscosity data for both IgG1 and IgG4P isotypes. Trastuzumab and golimumab were selected as examples of antibodies of low viscosity having similar values as IgG1 (9.1 cP and 7.5 cP, respectively) and as IgG4P mAbs (8.7 cP and 8.5 cP, respectively). Atezolizumab was selected because it is an antibody with moderate viscosity as an IgG1 (21.8 cP) that is further increased as an IgG4P (49.9 cP).
For these four IgG1/IgG4P selected pairs, kD was more negative for the IgG4P isotypes than the IgG1 isotypes indicating a stronger self-interaction. Table 5 shows the viscosity at 150 mg/mL and kD measurements of the four IgG4P CH3 swapped mutants as compared to their IgG1 and IgG4P counterparts. The kD values of the IgG4P swapped mutants are much closer to that of the IgG1 mAbs (Table 5). Among the four selected mAbs, trastuzumab and golimumab, which had similar viscosity values between IgG1 and IgG4P, also had similar values as an IgG4P CH3 swapped mutant. On the other hand, the viscosity of cetuximab IgG4P (high viscosity mAb) CH3 swapped mutant was reduced significantly compared to the IgG4P counterpart (72.6 cP for the CH3 swapped versus 205.6 cP for IgG4P), but was still 31.5 cp higher than that of at the IgG1 (40.9 cP) counterpart. Atezolizumab, which has a moderate viscosity as an IgG1 (21.8 cP) and elevated viscosity as an IgG4P (49.9 cP), show a viscosity profile lower than that of IgG1 and IgG4P as a CH3 IgG4P swapped variant (13.81 cP). The kD of the swapped mutant (−3.81 ml/g) was also in between the kD of IgG1 and IgG4P (11.56 and −7.17 ml/g respectively).
Table 5.
Diffusion interaction parameter (kD) (0–20 mg/ml) and experimental viscosity of IgG1, IgG4P, and IgG4P CH3 swapped mutants. The viscosity is averaged from different shear rates. The zero-shear viscosity is calculated by extrapolating to zero shear rate using a polynomial fit. All mAbs except Cetuximab exhibited a Newtonian behavior for viscosity
| mAbs | IgG1 |
IgG4P |
IgG4P CH3 swapped mutants |
||||||
|---|---|---|---|---|---|---|---|---|---|
| kD (ml/g) | Experimental viscosity (cP) at 150 mg/ml (Averaged) | zero-shear viscosity (cP) at 150 mg/ml (Extrapolated) | kD (ml/g) | Experimental viscosity(cP) at 150 mg/ml (Averaged) | zero-shear viscosity(cP) at 150 mg/ml (Extrapolated) | kD (ml/g) | Experimental viscosity (cP) at 150 mg/ml (Averaged) | zero-shear viscosity (cP) at 150 mg/ml |
|
| (Extrapolated) | |||||||||
| Cetuximab | −10.6 | 40.9 | 55.7 | −26.49 | 205.6 | 264.6 | −12.24 | 72.6 | 75 |
| Atezolizumab | 11.56 | 21.8 | 22.3 | −7.17 | 49.9 | 53 | −3.81 | 13.81 | 14.01 |
| Trastuzumab | 14.68 | 9.2 | 9.3 | −4.45 | 8.7 | 8.8 | 4.77 | 7.7 | 7.9 |
| Golimumab | 9.31 | 7.5 | 7.6 | −3.88 | 8.6 | 8.7 | 8.5 | 7.3 | 8.9 |
For all four mAbs, the IgG4P had a stronger self-interaction (more negative measured kD) compared to its IgG1 counterpart. Interestingly, all IgG4P CH3 swapped variants tested had a measured kD close to the kD of the IgG1 (golimumab) or between the kD of the IgG1 and the IgG4P (cetuximab, atezolizumab, and trastuzumab). Figure 9 shows the concentration-dependent viscosity of these four IgG1, IgG4P, and IgG4P CH3 swapped mutants from experimental results and CG simulation at 50, 100, 150, and 175 mg/ml. For IgG4P CG simulation, the improved parameter sets were used. For trastuzumab and golimumab, the three IgG4 CH3 swapped mutants exhibit similar measured viscosity behavior as their IgG1and IgG4P counterparts (also described in Table 5). For cetuximab and atezolizumab, the IgG4P CH3 swapped mutants showed significant viscosity reduction compared to their IgG4P counterparts (also shown in Table 5). Figure 10 shows the radial distribution function of IgG4 swapped mutants and the comparison with the corresponding IgG1 and IgG4P for trastuzumab, golimumab, cetuximab, and atezolizumab. For the low viscosity IgG4P mAbs (trastuzumab and golimumab), which show similar viscosity (~7–9 cP at 150 mg/ml), the peak heights/intensities for VH–CH3 and CH3–VL regions are comparable to the peak heights for VH–VH, VH–VL, and VL–VL regions, despite IgG4P having slightly higher VH–CH3 and CH3–VL peaks compared to those of IgG1 and IgG4P CH3 swapped mutants. On the contrary, the high viscosity mAb (cetuximab) shows markedly high VH–VH, VH–VL, and VL–VL peaks, and exhibits an intermediate viscosity value between IgG1 (40.9 cP) and IgG4P (205 cP) as an IgG4P CH3 swapped mutant (~72 cP).
Figure 9.
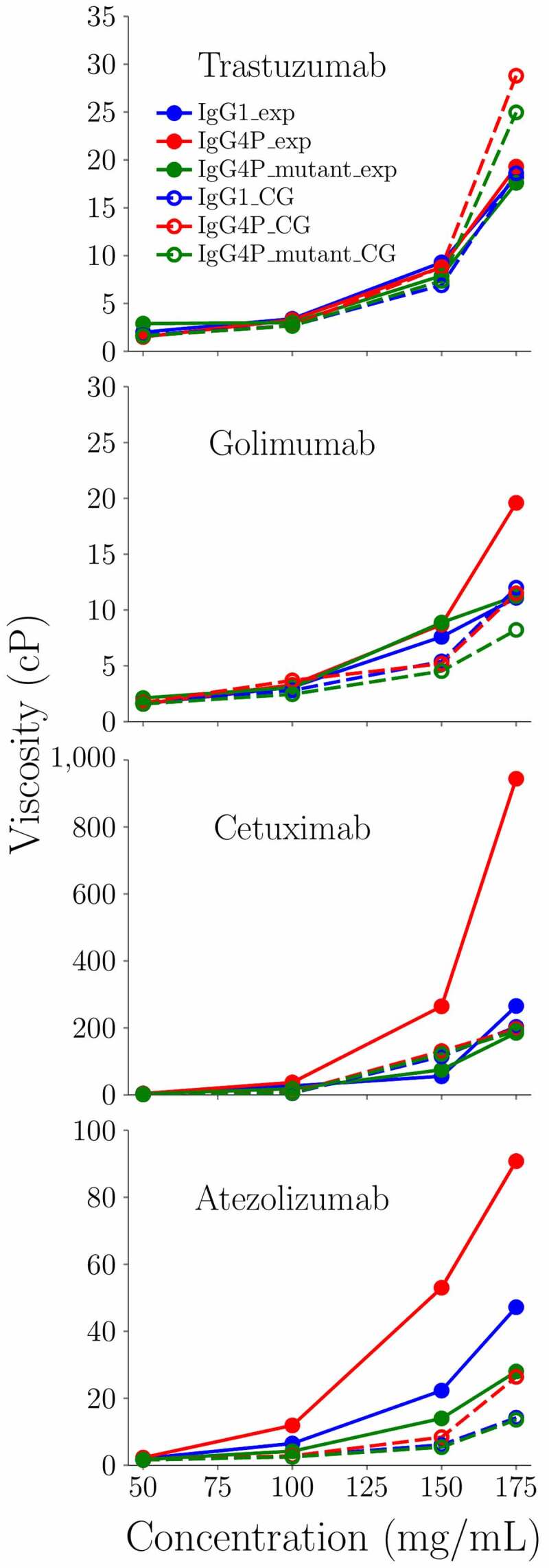
Concentration dependence of the zero-shear viscosity from CG models with the experimental viscosity data for IgG1 and IgG4P mAbs. Blue circles indicate experimental measurement of IgG1. Red squares indicate experimental measurement of IgG4P. Green triangles indicate experimental measurement of IgG4P CH3 swapped mutants. Blue lines indicate CG results of IgG1. Red dashed lines indicate CG results of IgG4P with kcal/mol and chgCH3 = −0.8 e. Green dotted dashed lines indicate CG results of IgG4P CH3 swapped mutants
Figure 10.
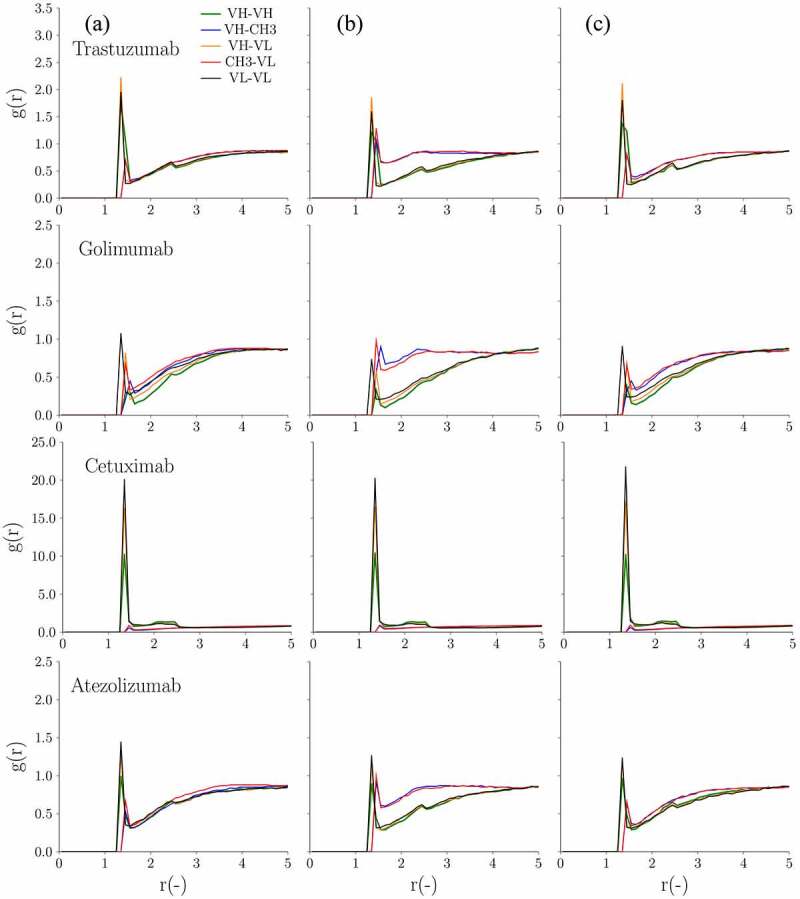
Radial distribution as a function of the distance in between the various mAb domains at 150 mg/ml for the 4 selected antibody pairs. From left to right are (a) IgG1, (b) IgG4P with kcal/mol and chgCH3 = −0.8 e, and (c) IgG4P CH3 swapped mutants, respectively
Discussion
IgG1 and IgG4P antibodies are known to have differences in sequence, structure, and key biophysical characterization parameters, such as isoelectric point, disulfide bond networking, thermostability, and colloidal properties, including self-interaction and viscosity2,16–19,31. However, previous studies have neither dissected mechanisms leading to some of these differences nor have isolated the contribution of the constant domains. Here, we focused on self-interactions and viscosity differences between IgG1 and IgG4P mAbs and used computational approaches, such as charge distribution, high viscosity index classification, and CG with hydrodynamic calculation, to predict self-interaction and viscosity in antibodies and to achieve an understanding of sequence and structural characteristics within the constant domains that affect these behaviors in paired IgG1 and IgG4 isotypes. We selected 14 IgG1/IgG4P antibody pairs having low to high viscosity as IgG1. We defined low viscosity mAbs as those that exhibit viscosity <30 cP at 150 mg/ml with the IgG1 isotype. The viscosity differential behavior between IgG1 and IgG4P can be divided into three categories: 1) low or moderately elevated viscosity as IgG1 and significantly higher viscosity as IgG4P (e.g., ganitumab, adalimumab, tremelimumab, basiliximab, vesencumab, atezolizumab, TGN1412, and bevacizumab); 2) low or moderately elevated viscosity as IgG1 and similar viscosity as IgG4P (e.g., ipilimumab, natalizumab, trastuzumab, and golimumab); and 3) high viscosity as IgG1 and significantly higher viscosity as IgG4P (e.g., cetuximab, 40.9 cP at 150 mg/ml, and omalizumab, 30.3 cP at 100 mg/ml). Cetuximab IgG4P showed a 4- to 5-fold viscosity increase compared to its IgG1 counterpart at 150 mg/ml (>200 cP) and omalizumab IgG4P could not be concentrated beyond 20 mg/ml in 20 mM sodium acetate, pH 5.5 buffer due to precipitation, consistent with previous reports that omalizumab is a viscous mAb.28 Overall, the IgG4P mAbs in this study exhibited higher or similar viscosity compared to their IgG1 counterparts at 150 mg/ml. To complement the data acquired with viscosity measurements, we also measured the diffusion interaction parameter (kD) by DLS for the 14 IgG1/IgG4P antibody pairs.
The validity of using kD measured from low protein concentrations to predict high concentration viscosity remains controversial. Woldeyes et al. reported that kD did not correlate well with viscosity at high concentration.24 However, Kingsbury et al. pointed out that, although kD may be limited in providing quantitative regressions for viscosity-mAb concentration relationships, consensus in the field is that it has great directional value in identifying poorly and well-behaved mAbs from a self-interaction standpoint.22 From this study, we demonstrate that directional kD and viscosity are in alignment with the most elevated viscosity mAbs having a strongly negative kD (indicative of strong self-interaction), whereas kD only has a modest correlation with the viscosity of low viscosity mAbs, highlighting the utility of kD measurements to identify high viscosity mAbs.
Significant differences are observed in the charges of IgG1 and IgG4P mAbs in 20 mM sodium acetate pH 5.5 buffer. The net charge of IgG1 mAbs range from +26 e to +40 e with the two high viscosity IgG1 mAbs in our study, cetuximab and omalizumab, having the lowest net charge (+26). IgG4P mAbs have significantly lower net charges compared to their IgG1 mAbs counterparts (a difference of 14 e). This major difference in net charge comes from the Fc region, composed of two CH2 and two CH3 domains, which has a difference of 10 e between IgG1 Fc (12 e) and IgG4 Fc (2 e). The net charges on the CH2 regions are +4 e and +2 e for the IgG1 and IgG4P mAbs, respectively. The net charges on the CH3 regions are significantly different, with +2 e and −1 e for the IgG1 and IgG4P mAbs, respectively, likely due to differences in the pI of the CH3 domains. It has been reported previously that at a similar pH (pH 5.8) the pI of IgG1 CH3 is ~7 and the pI of IgG4P is ~5.32
We propose that the lower CH3 net charges could result in weaker repulsive interactions between IgG4P mAbs, which is consistent with the higher peaks (strong interaction) observed between the CH3 and the variable region seen in the radial distribution function analysis. In particular, the negative charges on the CH3 regions of IgG4P mAbs could produce favorable electrostatic interactions with other positively charged regions. This would explain, at least in part, why most IgG4P mAbs displayed higher viscosity at high concentrations than their IgG1 mAbs counterparts. However, these differences in net charge do not explain by themselves why some IgG4P mAbs had similar viscosity behaviors with their IgG1 mAb counterparts. For example, the net charges of ipilimumab, natalizumab, trastuzumab, and golimumab IgG4P mAbs, which showed comparable viscosity for IgG1 and IgG4P isotypes, range from +16 e to +22 e. The net charges of other IgG4P mAbs, which exhibit viscosity difference between IgG1 and IgG4 especially above 100 mg/ml, range from +12 e to +26 e. More complex molecular interactions and phenomena are likely involved.
The SCM analysis showed that for IgG1 mAbs, the prediction for high and low viscosity is mostly accurate with only one false positive case (adalimumab). The negative charge patches of adalimumab on the heavy chains are spatially close while distant from other positive charge patches, resulting in a large SCM score. Since the variable regions are the same, the dynamic average of SCM scores of IgG1 and IgG4P mAbs would be very similar. In addition to one false positive case, there are two false negative cases for the IgG4P mAbs. It is noted that vesencumab IgG4P is only slightly above the high viscosity threshold (30 cP). One of the underlying premises of the SCM model is that most of the constant regions carry net positive charges, and an increase the in extent and magnitude of the exposed negative electrostatic patch on the Fv region will result in increases in viscosity. The model was initially developed based only on IgG1 mAbs. However, the constant region of IgG4 mAbs not only has lower net positive charges but the CH3 domain also carries net negative charges. Under our buffer condition, the Fv domains of the IgG4 mAbs are positively charged. The overall positively charged Fv domain may be dominant over negatively charged patches and form strong electrostatic interactions with the negatively charged CH3 domain, resulting in elevated viscosity.
Twelve of 14 IgG1 mAbs were correctly classified using the decision tree model. The misclassified mAbs (natalizumab and bevacizumab) are both false positives. Based on the earlier work, the number of serine and glutamic acid residues on the Fv regions are significantly higher for high viscosity mAbs.26 On average, there are 34 serines and 10 glutamic acids for high viscosity mAbs.26 From sequence analysis, although natalizumab has a large HVI value, it only has 24 serines on the Fv regions. The significant difference in the amino acid composition compared to the high viscosity mAbs in the previous work could be the reason for the misclassification. Moreover, bevacizumab was also misclassified by the decision tree model in the previous work.26
The accuracy of the classification of the viscosity for the IgG4P mAbs using our classification previously established for IgG1 mAbs was 36%, only 5 of 14 cases were correctly classified. There are two features in the decision tree model, HVI and mAbs net charge. HVI only depends on the Fv sequence, and, in this case, both IgG1s and IgG4Ps have the same Fv sequences. Therefore, we did not choose to modify HVI. Instead, we found that one major difference between IgG1s and IgG4Ps is the charge on the constant region; therefore, we only revised the mAbs net charge criterion for the IgG4P decision tree model. Yang et al. also reported that experimentally measured charge differences are much smaller than the calculated charge differences.33 We understood that the splitting criteria previously used for IgG1 mAbs was not applicable for IgG4P mAbs and the net charge criteria of the decision tree model for the IgG4P mAbs needed to be adjusted. The splitting results of IgG4P mAbs based on the new criteria resemble the results of IgG1 mAbs. Only vesencumab and atezolizumab IgG4P mAbs exceed the high viscosity threshold. Future application of this decision tree model may require separation of the mAbs based on their isotypes first and then implementation of the corresponding splitting criteria.
For IgG1 mAbs, the CG model predicts the concentration dependence of most low viscosity mAbs quite well, but overestimates the viscosity of natalizumab IgG1 at >100 mg/ml, bevacizumab IgG1 at 175 mg/mL, and atezolizumab IgG1 at >100 mg/ml. For IgG4P mAbs, the CG model generally predicts higher viscosity than for IgG1 mAbs. An exception is the high viscosity cetuximab for which the CG model predicted very similar results for IgG1 and IgG4P mAbs due to the strong Fv–Fv interactions that dominate in the cluster formation. In this specific case, the constant regions only had a marginal effect on viscosity.
The analysis of the radial distribution function for VH–VL, VH–VH, VL–VL, VH–CH3, and VL–CH3 showed differences in domain interactions for IgG1 and IgG4P isotypes. For IgG1 mAbs (with both low and high viscosity), VH–VL, VH–VH, and VL–VL (Fv–Fv) domains generally have higher peaks than those of VH–CH3 and VL–CH3 (Fv–CH3), which suggests a strong interaction between Fv–Fv domains. However, we observed two different patterns for IgG4P mAbs. For low viscosity IgG4P mAbs, in contrast to IgG1 mAbs, VH–CH3, and VL–CH3 pairs were found to have significantly higher peaks than those of VH–VL, VH–VH, and VL–VL pairs ( kcal/mol), indicating a stronger Fv–CH3 interaction. The strong interaction observed between CH3 and Fv could be due to the electrostatic interaction between the negatively charged IgG4P CH3 domain with the positively charged regions of VH and VL. On the contrary, high viscosity IgG4P mAbs (omalizumab and cetuximab) showed very similar profiles as their IgG1 counterparts, i.e., stronger VL–VL, VH–VH, and VH–VL domain interactions (large values) and reduced Fv–CH3 interactions. ( kcal/mol) due to negatively charged Fv. The negatively charged Fv domain would reduce the interaction between Fv and CH3.
Due to the challenges of experimentally measuring charges of isolated antibody domains, computational models lack the data needed to improve prediction accuracy, which is likely the reason why the model overestimates sequence-based charges and electrostatic interaction predictions. To improve our models for the IgG4 isotype, we adjusted the values previously described for IgG128 and the charge on the CH3 beads/domains (chgCH3) for IgG4P. The optimal we found was 0.12 kcal/mol and chgCH3 = −0.8 e. Overall, the computationally predicted viscosity values of IgG4P using the optimized parameters ( kcal/mol and chgCH3 = −0.8 e) was much closer to the experimental measurements. We showed that the VH-CH3 and VL-CH3 peaks of IgG4P with kcal/mol and chgCH3 = −0.8 e are lower than those of IgG4P with kcal/mol and chgCH3 = −1.0 e, indicating weaker interactions between these pairs with the new parameter sets.
Given the observed relatively strong interaction between Fv and CH3 in IgG4P in the radial distribution function and the negative charge of the CH3 domain, we hypothesized that CH3 could be a key region for the viscosity and self-interaction difference between IgG1 and IgG4P antibodies. We designed CH3 domain swapped IgG4P mutants, replacing the IgG4 CH3 with the IgG1 CH3 regions, for four selected IgG1/IgG4P pairs having differential viscosity behaviors. These selected mAbs were trastuzumab and golimumab (low viscosity as IgG1 and IgG4P), cetuximab (high viscosity mAb as IgG1, more viscous as IgG4P) and atezolizumab (low to moderate viscosity as IgG1, more viscous as IgG4P). The results showed the CH3 IgG4P swapped mutants have similar (or within same range) kD values as their IgG1 counterparts, highlighting the CH3 region as a key contributor to the difference in the self-interaction observed between IgG1 and IgG4P mAbs.
We compared the effect of IgG4 CH3 swapped mutants on the low, medium, and high viscosity mAbs using experimental and computational approaches. As anticipated, trastuzumab and golimumab showed similar viscosity behavior as IgG1, IgG4P, and IgG4 CH3 swapped mutants. CG simulation also showed that IgG1, IgG4P, and IgG4 CH3 swapped mutant have similar viscosity for trastuzumab and golimumab. However, the CG simulation has a slight overestimation of the viscosity at 175 mg/ml for trastuzumab, and a slight underestimation of the viscosity >150 mg/ml for golimumab compared to experimental results. For cetuximab, the viscosity of the IgG4P CH3 swapped mutant showed significant reduction compared to that of IgG4P isotype and exhibited similar viscosity behavior as that of IgG1 isotype. This clearly demonstrates that the viscosity difference between IgG1 and IgG4 was primarily driven by the CH3 region for cetuximab. However, since the CG simulation overpredicts the Fv–Fv interactions, as described above, the difference in the constant region only has a minimal effect on the predicted viscosity. For atezolizumab, the viscosity of the IgG4P CH3 swapped mutant showed reduced viscosity that is lower than the IgG1 and IgG4P mAbs. The CG simulation qualitatively and correctly predicted that the IgG1 and IgG4P CH3 swapped mutant exhibit lower viscosity than that of IgG4P isotype.
The difference in the radial distribution function profile indicates that the regions in the mAbs involved in self-interactions and viscosity (i.e., the microstructures of the cluster formation) of IgG1 and IgG4P are different. For IgG1 mAbs, the clusters are formed predominantly by connecting the Fv–Fv regions (head-to-head). For IgG4P mAbs, Fv–Fc interactions (head-to-tail) are also playing a role and can even be dominant for antibodies with weak Fv–Fv interactions. It is noted that the CG model is different from the SCM model and should not be compared directly. SCM is based on an all-atom representation, and it takes into account different charge distribution on the surface. It also is based on the assumption that negative charge patches can interact with positive charge patches on other regions and viscosity behavior is predominantly governed by charge effect. On the other hand, the CG model does not have high-resolution information for charge distribution, but includes the effect of short-range interactions.
Previous publications have described that head-to-head interactions lead to extended linear clusters, while head-to-tail interactions lead to compact and branched clusters.27,34 Given the same interaction energies, the extended cluster would have higher viscosity than that of branched clusters.34 Furthermore, IgG4P mAbs that exhibit lower net charge could lead to enhanced self-association. The theoretical model assumes that, given the same interaction energies, extended clusters could have higher viscosity; however, in the real situation, the interaction energies between IgG4Ps are more favorable compared to that of IgG1s. Here, we postulate that the balance between the interaction energies between Fv–Fv and Fv and the constant domain of the Fc (with the CH3 domain playing a critical role) account for the difference between the IgG1 and IgG4P viscosity and self-interaction behavior.
To our knowledge, this work is the first to provide an explanation for differences in colloidal properties (viscosity, self-interactions) between IgG1 and IgG4P mAbs and reveal the CH3 domain’s significant contributions in conjunction with the nature of the variable domains (amino acid composition, net charge) in driving viscosity in mAbs. This is also the first report of a computational model to predict viscosity and self-interaction of IgG4P mAbs.
Materials and Methods
Expression and Purification (gene synthesis, transfection, and purification)
IgG1 and IgG4P monoclonal antibodies were generated from transiently transfected in CHO-Expi Expression System (Thermo Fisher Scientific, Waltham, MA) according to the manufacturer’s protocol. Briefly, the cells were grown and maintained in ExpiCHO Expression Medium (Thermo Fisher Scientific). The cultures were harvested on day 7, the cells were pelleted by centrifugation, and the supernatant was passed through a 0.2-micron filter. Antibodies were purified from culture supernatant by affinity chromatography using a Protein A coupled resin (MabSelect™ SuRe™ (GE Healthcare Life Sciences, Pittsburgh PA). The mAb was eluted from the resin with 20 mM sodium acetate at pH 3.5 and immediately neutralized with 0.333 M Tris, 1 M sodium acetate, pH 8.0 and buffer exchanged into 20 mM sodium acetate pH 5.5.
Aggregation levels were assessed by UP-SEC. When necessary, a secondary polishing purification step such as preparative SEC or cation-exchange chromatography (CEX) (Capto S resin, GE Healthcare) was used to achieve a purity of >98% by UP-SEC.
Size-exclusion chromatography HPLC
To determine aggregation by UP-SEC, 5 μg of purified antibody was injected onto a BEH200 (Waters Acquity BEH200 SEC, 1.7 μm, 4.6 × 150 mm) size exclusion column that was equilibrated with 100 mM sodium phosphate, 200 mM sodium chloride, 0.02% sodium azide pH 7, at 0.5 mL/minute using a Waters H-Class UPLC. Chromatograms were collected at both 215 and 280 nm wavelengths, and integration of the absorption at 280 nm (wavelength) trace was performed using EMPOWER 2 (Waters).
Diffusion interaction parameter (kD) by DLS assessment
All DLS studies were performed at 25°C in glass bottom 96-well plates using a DynaPro Plate Reader II (Wyatt, Santa Barbara, Ca). High concentration samples were diluted with the buffers of interest (20 mM sodium acetate pH 5.5) to obtain a concentration of 20 mg/mL, filtered through 0.22 µm filters, and diluted in filtered buffers (with desired pH and ionic strength) to obtain lower concentration samples (2, 5, 10, and 15 mg/mL), which were then added to the microplate. The kD was determined by a linear fit of the measured (mutual) diffusion coefficients as a function of concentration.
Viscosity assessment
Samples were evaluated for viscosity. Viscosities were measured using the VROC Initium (RheoSense, San Ramon, CA) viscometer. The technology can accurately measure the pressure drop by using an array of pressure sensors as the sample flow through a micro-channel, which can be used to calculate the viscosity.
Viscosities at different shear rates (shear rate sweep) were measured in 20 mM sodium acetate pH 5.5 at 50 mg/ml, 100 mg/ml, 150 mg/ml, and 175 mg/ml. All viscosity measurements were carried out at 25°C. First, the estimated viscosity was measured at a shear rate such that the pressure is 50% of Pmax of the B05 chip (NC1226294, RheoSense, San Ramon, CA). Twenty-six ul of samples was used for this measurement. The estimated viscosity was then used to generate the protocol for shear rate sweep experiment. The range of accessible shear rates for a certain viscosity was limited by the maximum flow rate of the chip. For these measurements, 26–50 ul of sample was used depending on the estimated viscosity of the samples. E02 (NC1577288, RheoSense) chip was used if the estimated viscosity is higher than 200 cP. Concentrated samples were filtered using a 0.2 um Polyvinylidene fluoride (PVDF) membrane prior to loading into glass vials. For each sample, viscosity values at each shear rate were average and then were plotted as a function of a series of shear rates. The viscosity at zero shear rate was then extrapolated using a polynomial fit.
Homology Modeling of mAbs
The sequences of the mAbs used in this study are available in Supplemental Table 1. The mAb molecules were constructed followed the protocol proposed by Brandt et al.35 The Fab structures were obtained from either available crystal structures or homology models built from RosettaAntibody.36–38 The structures of the Fab regions were superimposed on a template structure of a full-length IgG1 and IgG4 models. The IgG1 template was obtained from the KOL/Padlan structure.39,40 For the IgG4 model, the Fc regions (PDB:4C54) were superimposed on the KOL/Padlan IgG1 structure.41,42 The glycan structure was G0F. The homology models were energy-minimized to remove steric clashes in protein structures using NAMD2.43
All-Atom Molecular Dynamics Simulation
MD simulations were performed using all-atom structures with explicit solvent using the TIP3P water model.44 Simulation boxes were set up using VMD45 to place a single antibody in a water box extending 12 Å beyond the protein surface. Simulations were performed at 300 K and 1 atm in the NPT ensemble, using the NAMD243 software package and the CHARMM36m force field.46 The system pH was set to 5.5 to match the experimental pH by adjusting the protonation states of the histidine residues using the PROPKA3 protocol.47 Electrostatic interactions were treated with the Particle Mesh Ewald method and van der Waals interactions were calculated using a switching distance of 10 Å and a cutoff of 12 Å. The integration time step was set to 2 fs. Each mAb system was pre-equilibrated for 10 ns, followed by 50 ns production runs.
Computational Models of Viscosity Behavior
The spatial charge map (SCM) calculates a score based on the negative charges of the exposed residues in the Fv region.25 The decision tree model uses two features, net charge of full-length mAbs and HVI, to classify high and low viscosity mAbs.26
| (1) |
The CG model was described elsewhere.27 Briefly, a 12-bead CG model derived from all-atom MD simulations was used to construct the structure of all the mAbs. The intramolecular interaction parameters were obtained from the dynamic averages of the MD simulations. The charges on the beads are the sum of all the atomic charges on the corresponding domains. The radii of the beads were calculated from the average radius of gyration from MD simulations. The CG models were treated as colloidal particles as applied in a previous work.48 Brownian dynamics simulation was used to describe the dynamic behaviors of the CG beads. The electrostatic interactions between the beads were approximated as a form of the Yukawa potential. The dispersion interactions were described as van der Waals interactions.27 The magnitude of interactions depends on the Hamaker constant, . The Hamaker constants are divided into and to account for the variable regions and constant regions, respectively. The variable region equals to kcal/mol. The constant region equals to 0.2 kcal/mol for IgG1 mAbs. The total viscosity is contributed from hydrodynamic, interparticle, and solvent interactions. In addition, the maximum viscosity that the CG model can reach is ~200 cP due to the finite size effects as described elsewhere.27 The details are referred to the previous work.27 The calculation of self-diffusivity (D) follows a previous study.48 The single antibody diffusivity in free space (D0) was calculated by extrapolating the concentration-dependence of self-diffusivity to zero concentration.
Calculation of radial distribution function g(r)
The radial distribution function g(r) is calculated by
where r is the distance between two beads. N is the number of beads. is the average number density of the beads. is the Dirac delta function. <> is the ensemble average. Bead pairs belong to the same molecule are excluded from the calculation.
Supplementary Material
Acknowledgments
This work is supported by Merck Sharp and Dohme. The authors would like to thank Jeanne Baker and members of the protein expression group, Ellen Chien and members of the protein purification group, Marc Bailly, Daniela Tomazela and members of the protein characterization and mass spectrometry groups from the Merck Sharp and Dohme South San Francisco Protein Sciences Department within Discovery Biologics and the Merck Sharp and Dohme postdoctoral program.
Funding Statement
The authors reported that there is no funding associated with the work featured in this article.
Disclosure statement
No potential conflict of interest was reported by the authors.
Declarations of Interest
During the execution of this work, Gaurav Ghag, Yao Yu, Veronica Juan, and Laurence Fayadat-Dilman were employees of subsidiaries of Merck & Co., Inc., Kenilworth, NJ, USA and stockholders of Merck & Co., Inc., Kenilworth, NJ, USA.
Supplementary material
Supplemental data for this article can be accessed on the publisher’s website
References
- 1.Schur PH. IgG subclasses. A Historical Perspective Monographs iAllergy. 1988;23:1–19. [PubMed] [Google Scholar]
- 2.Vidarsson G, Dekkers G, Rispens T.. IgG Subclasses and Allotypes: from Structure to Effector Functions. Front Immunol. 2014;5. doi: 10.3389/fimmu.2014.00520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Grilo AL, Mantalaris A. The Increasingly Human and Profitable Monoclonal Antibody Market. Trends Biotechnol. 2019;37(1):9–16. doi: 10.1016/j.tibtech.2018.05.014. [DOI] [PubMed] [Google Scholar]
- 4.The Antibody Society . Therapeutic monoclonal antibodies approved or in review in the EU or US. [accessed 2021. Mar 21]. https://www.antibodysociety.org/antibody-therapeutics-product-data/
- 5.Labrijn AF, Aalberse RC, Schuurman J. When binding is enough: nonactivating antibody formats. Curr Opin Immunol. 2008;20(4):479–85. doi: 10.1016/j.coi.2008.05.010. [DOI] [PubMed] [Google Scholar]
- 6.Frangione B, Milstein C, Pink JRL. Immunoglobulins: structural Studies of Immunoglobulin G. Nature. 1969;221(5176):145–48. doi: 10.1038/221145a0. [DOI] [PubMed] [Google Scholar]
- 7.van der Neut Kolfschoten M, Schuurman J, Losen M, Bleeker WK, Martínez-Martínez P, Vermeulen E, den Bleker TH, Wiegman L, Vink T, Aarden LA, et al. Anti-inflammatory activity of human IgG4 antibodies by dynamic Fab arm exchange. Science. 2007;317(5844):1554–57. doi: 10.1126/science.1144603. [DOI] [PubMed] [Google Scholar]
- 8.Rispens T, Ooijevaar-de Heer P, Bende O, Aalberse RC. Mechanism of immunoglobulin G4 Fab-arm exchange. J Am Chem Soc. 2011;133(26):10302–11. doi: 10.1021/ja203638y. [DOI] [PubMed] [Google Scholar]
- 9.Angal S, Dj K, Mw B, Turner A, ADG L, Roberts G, Pedley B, Jr A. A single amino acid substitution abolishes the heterogeneity of chimeric mouse/human (IgG4) antibody. Mol Immunol. 1993;30(1):105–08. doi: 10.1016/0161-5890(93)90432-B. [DOI] [PubMed] [Google Scholar]
- 10.Silva J-P, Vetterlein O, Jose J, Peters S, The KH. S228P mutation prevents in vivo and in vitro IgG4 Fab-arm exchange as demonstrated using a combination of novel quantitative immunoassays and physiological matrix preparation. J Biol Chem. 2015;290(9):5462–69. doi: 10.1074/jbc.M114.600973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Labrijn AF, Rispens T, Meesters J, Rose RJ, Bleker THD, Loverix S, ETJ B, van den, Neijssen J, Vink T, Lasters I, et al. Species-Specific Determinants in the IgG CH3 Domain Enable Fab-Arm Exchange by Affecting the Noncovalent CH3–CH3 Interaction Strength. J Immunol. 2011;187(6):3238–46. doi: 10.4049/jimmunol.1003336. [DOI] [PubMed] [Google Scholar]
- 12.Le Basle Y, Chennell P, Tokhadze N, Astier A, Sautou V. Physicochemical Stability of Monoclonal Antibodies: a Review. J Pharm Sci. 2020;109(1):169–90. doi: 10.1016/j.xphs.2019.08.009. [DOI] [PubMed] [Google Scholar]
- 13.Yadav S, Shire SJ, Kalonia DS. Factors affecting the viscosity in high concentration solutions of different monoclonal antibodies. J Pharm Sci. 2010;99(12):4812–29. doi: 10.1002/jps.22190. [DOI] [PubMed] [Google Scholar]
- 14.Roberts CJ. Therapeutic protein aggregation: mechanisms, design, and control. Trends Biotechnol. 2014;32(7):372–80. doi: 10.1016/j.tibtech.2014.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pindrus M, Shire SJ, Kelley RF, Demeule B, Wong R, Xu Y, Yadav S. Solubility Challenges in High Concentration Monoclonal Antibody Formulations: relationship with Amino Acid Sequence and Intermolecular Interactions. Mol Pharm. 2015;12(11):3896–907. doi: 10.1021/acs.molpharmaceut.5b00336. [DOI] [PubMed] [Google Scholar]
- 16.Peters SJ, Smales CM, Henry AJ, Stephens PE, West S, Humphreys DP. Engineering an improved IgG4 molecule with reduced disulfide bond heterogeneity and increased Fab domain thermal stability. J Biol Chem. 2012;287(29):24525–33. doi: 10.1074/jbc.M112.369744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Heads JT, Adams R, D’Hooghe LE, Page MJT, Humphreys DP, Popplewell AG, Lawson AD, Henry AJ. Relative stabilities of IgG1 and IgG4 Fab domains: influence of the light–heavy interchain disulfide bond architecture. Protein Science. 2012;21(9):1315–22. doi: 10.1002/pro.2118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Neergaard MS, Nielsen AD, Parshad H. Van De Weert M. Stability of monoclonal antibodies at high-concentration: head-to-head comparison of the IgG1 and IgG4 subclass. J Pharm Sci. 2014;103(1):115–27. doi: 10.1002/jps.23788. [DOI] [PubMed] [Google Scholar]
- 19.Heads JT, Lamb R, Kelm S, Adams R, Elliott P, Tyson K, Topia S, West S, Nan R, Turner A, et al. Electrostatic interactions modulate the differential aggregation propensities of IgG1 and IgG4P antibodies and inform charged residue substitutions for improved developability. Protein. 2019;32(6):277–88. doi: 10.1093/protein/gzz046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bailly M, Mieczkowski C, Juan V, Metwally E, Tomazela D, Baker J, Uchida M, Kofman E, Raoufi F, Motlagh S, et al. Predicting Antibody Developability Profiles Through Early Stage Discovery Screening. mAbs. 2020;12(1):1743053. doi: 10.1080/19420862.2020.1743053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Neergaard MS, Kalonia DS, Parshad H, Nielsen AD, Møller EH. van de Weert M. Viscosity of high concentration protein formulations of monoclonal antibodies of the IgG1 and IgG4 subclass - prediction of viscosity through protein-protein interaction measurements. Eur J Pharmaceu Scie. 2013;49(3):400–10. doi: 10.1016/j.ejps.2013.04.019. [DOI] [PubMed] [Google Scholar]
- 22.Kingsbury JS, Saini A, Auclair SM, Fu L, Lantz MM, Halloran KT, Calero-Rubio C, Schwenger W, Airiau CY, Zhang J, et al. A single molecular descriptor to predict solution behavior of therapeutic antibodies. Sci Adv. 2020;6(32):eabb0372. doi: 10.1126/sciadv.abb0372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang W, Lilyestrom W, Yu HZY, Scherer T. S Cluster Size and Quinary Structure Determine the Rheological Effects of Antibody Self-Association at High Concentrations. Pharm Sci. 2019. Jan;108(1):142–54. [DOI] [PubMed] [Google Scholar]
- 24.Woldeyes MA, Qi W, Razinkov VI, Furst EM, Roberts CJ. How Well Do Low- and High-Concentration Protein Interactions Predict Solution Viscosities of Monoclonal Antibodies? J Pharm Sci. 2019;108(1):142–54. doi: 10.1016/j.xphs.2018.07.007. [DOI] [PubMed] [Google Scholar]
- 25.Agrawal NJ, Helk B, Kumar S, Mody N, Sathish HA, Samra HS, Buck PM, Li L, Trout BL. Computational tool for the early screening of monoclonal antibodies for their viscosities. mAbs. 2015;8(1):43–48. doi: 10.1080/19420862.2015.1099773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lai P-K, Fernando A, Cloutier TK, Gokarn Y, Zhang J, Schwenger W, Chari R, Calero-Rubio C, Trout BL. Machine Learning Applied to Determine the Molecular Descriptors Responsible for the Viscosity Behavior of Concentrated Therapeutic Antibodies. Mol Pharm. 2021;18(3):1167–75. doi: 10.1021/acs.molpharmaceut.0c01073. [DOI] [PubMed] [Google Scholar]
- 27.Lai P-K, Swan JW, Trout BL. Calculation of Therapeutic Antibody Viscosity with Coarse-Grained Models, Hydrodynamic Calculations and Machine Learning-Based Parameters. mAbs. 2021;13(1):e1907882. doi: 10.1080/19420862.2021.1907882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Belliveau PP, Lahoz MR. Evaluation of omalizumab from a health plan perspective. Journal of Managed Care Pharmacy: JMCP. 2005;11(9):735–45. doi: 10.18553/jmcp.2005.11.9.735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tomar DS, Kumar S, Singh SK, Goswami S, Li L. Molecular basis of high viscosity in concentrated antibody solutions: strategies for high concentration drug product development. mAbs. 2016;8(2):216–28. doi: 10.1080/19420862.2015.1128606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Izadi S, Patapoff TW, Walters BT. Multiscale Coarse-Grained Approach to Investigate Self-Association of Antibodies. Biophys J. 2020;118(11):2741–54. doi: 10.1016/j.bpj.2020.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Laptoš T, Omersel J. The importance of handling high-value biologicals: physico-chemical instability and immunogenicity of monoclonal antibodies. Exp Ther Med. 2018;15(4):3161–68. doi: 10.3892/etm.2018.5821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li L, Kumar S, Buck PM, Burns C, Lavoie J, Singh SK, Warne NW, Nichols P, Luksha N, Boardman D. Concentration dependent viscosity of monoclonal antibody solutions: explaining experimental behavior in terms of molecular properties. Pharm Res. 2014;31(11):3161–78. doi: 10.1007/s11095-014-1409-0. [DOI] [PubMed] [Google Scholar]
- 33.Yang D, Kroe-Barrett R, Singh S, Laue T. IgG Charge: practical and Biological Implications. Antibodies. 2019;8(1):24 doi: 10.3390/antib8010024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ramallo N, Paudel S, Schmit J. Cluster Formation and Entanglement in the Rheology of Antibody Solutions. J Phys Chem B. 2019;123(18):3916–23. doi: 10.1021/acs.jpcb.9b01511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brandt JP, Patapoff TW, Aragon SR. Construction, MD simulation, and hydrodynamic validation of an all-atom model of a monoclonal IgG antibody. Biophys J. 2010;99(3):905–13. doi: 10.1016/j.bpj.2010.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sivasubramanian A, Sircar A, Chaudhury S, Gray JJ. Toward high-resolution homology modeling of antibody Fv regions and application to antibody-antigen docking. Proteins. 2009;74(2):497–514. doi: 10.1002/prot.22309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Weitzner BD, Kuroda D, Marze N, Xu J, Gray JJ. Blind prediction performance of RosettaAntibody 3.0: grafting, relaxation, kinematic loop modeling, and full CDR optimization. Proteins: Structure, Function, and Bioinformatics. 2014;82(8):1611–23. doi: 10.1002/prot.24534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Weitzner BD, Jeliazkov JR, Lyskov S, Marze N, Kuroda D, Frick R, Adolf-Bryfogle J, Biswas N, Dunbrack RL, Gray JJ. Modeling and docking of antibody structures with Rosetta. Nat Protoc. 2017;12(2):401–16. doi: 10.1038/nprot.2016.180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Padlan EA. Anatomy of the antibody molecule. Mol Immunol. 1994;31(3):169–217. doi: 10.1016/0161-5890(94)90001-9. [DOI] [PubMed] [Google Scholar]
- 40.Boehm MK, Woof JM, Kerr MA, Perkins SJ, Fab T. and Fc fragments of IgA1 exhibit a different arrangement from that in IgG: a study by X-ray and neutron solution scattering and homology modelling. J Mol Biol. 1999;286(5):1421–47. doi: 10.1006/jmbi.1998.2556. [DOI] [PubMed] [Google Scholar]
- 41.Teplyakov A, Zhao Y, Malia TJ, Obmolova G, Gilliland GL. IgG2 Fc structure and the dynamic features of the IgG CH2–CH3 interface. Mol Immunol. 2013;56(1–2):131–39. doi: 10.1016/j.molimm.2013.03.018. [DOI] [PubMed] [Google Scholar]
- 42.Davies AM, Rispens T, Ooijevaar-de Heer P, Gould HJ, Jefferis R, Aalberse RC, Sutton BJ. Structural determinants of unique properties of human IgG4-Fc. J Mol Biol. 2014;426(3):630–44. doi: 10.1016/j.jmb.2013.10.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Phillips JC, Hardy DJ, Maia JDC, Stone JE, Ribeiro JV, Bernardi RC, Buch R, Fiorin G, Hénin J, Jiang W, et al. Scalable molecular dynamics on CPU and GPU architectures with NAMD. J Chem Phys. 2020;153(4):044130. doi: 10.1063/5.0014475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML. Comparison of simple potential functions for simulating liquid water. J Chem Phys. 1983;79(2):926–35. doi: 10.1063/1.445869. [DOI] [Google Scholar]
- 45.Humphrey W, Dalke A, Schulten K. VMD: visual molecular dynamics. J Mol Graph. 1996;14(1):33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 46.Hung JJ, Dear BJ, Karouta CA, Chowdhury AA, Godfrin PD, Bollinger JA, Nieto MP, Wilks LR, Shay TY, Ramachandran K, et al. Protein–Protein Interactions of Highly Concentrated Monoclonal Antibody Solutions via Static Light Scattering and Influence on the Viscosity. J Phys Chem B. 2019;123(4):739–55. doi: 10.1021/acs.jpcb.8b09527. [DOI] [PubMed] [Google Scholar]
- 47.Olsson MHM, Søndergaard CR, Rostkowski M, Jensen JH. PROPKA3: consistent Treatment of Internal and Surface Residues in Empirical pKa Predictions. J Chem Theory Comput. 2011;7(2):525–37. doi: 10.1021/ct100578z. [DOI] [PubMed] [Google Scholar]
- 48.Wang G, Varga Z, Hofmann J, Zarraga IE, Swan JW. Structure and Relaxation in Solutions of Monoclonal Antibodies. J Phys Chem B. 2018;122(11):2867–80. doi: 10.1021/acs.jpcb.7b11053. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


