Abstract
The increasing incidence of aspergillosis, a life-threatening infection in immunocompromised patients, emphasizes the need to improve the currently limited diagnostic tools. We developed a two-step PCR assay that specifically amplifies a region of the 18S rRNA gene that is highly conserved in Aspergillus species. A number of primers with the least homology to equivalent human or Candida gene sequences were screened for the pairs that gave the highest sensitivity and specificity. No cross-reaction with the wide range of fungal and bacterial pathogens so far tested was observed. This assay allows direct and rapid detection of down to 10 fg of Aspergillus DNA corresponding to 1 to 5 CFU per ml of blood. A total of 315 blood and bronchoalveolar lavage samples from 140 subjects, including 93 patients at risk for invasive fungal disease, were screened. The result was a 100% correlation between positive histology, culture, or high-resolution computed tomography findings and PCR results. The test specificity was 89%. Our data point to the considerable potential clinical value of this simple, specific, rapid, and inexpensive PCR assay for improving the means of early diagnosis of systemic aspergillosis in high-risk patients.
The incidence of life-threatening systemic fungal infections has been increasing in recent years, and the increasing incidence has been correlated with increasing numbers of immunocompromised patients (1). Patients at the greatest risk are those with prolonged periods of neutropenia after intensive immunosuppressive chemotherapy, for example, during treatment for acute leukemia or after bone marrow transplantation (31, 39). Particularly on the increase are invasive infections with Aspergillus species, resulting in high mortality rates or, if the patient survives, causing high levels of morbidity that often limit further antileukemic therapies. Antifungal prophylaxis, especially prophylaxis against molds, is controversial and is not generally practiced (3, 10). Moreover, amphotericin B, at present the “gold standard” of antifungal therapy, is toxic.
In contrast to other infections, only limited conventional diagnostic tools with poor sensitivity and reliability are available for early detection of invasive aspergillosis (13, 21, 50), with the systemic infection frequently being diagnosed late or confirmed only at autopsy (7, 12, 20, 35, 36). The tests commonly used in the initial period of infection during neutropenia are often insufficient not only for accurate early diagnosis but also for monitoring of the subsequent course of invasive aspergillosis (2, 6, 11, 21–23, 25–27, 32, 34, 42, 43, 46–49). In view of the low specificity and sensitivity rates of these methods, the diagnosis of an invasive aspergillosis can be proven conclusively only by positive histology or culture results. However, establishing cultures from blood and bronchoalveolar lavage (BAL) samples is often unsuccessful due to the low yields of CFU, and in the case of immunocompromised high-risk patients who are febrile, neutropenic, thrombocytopenic, and often seriously ill, tissue biopsy specimens, in general, are not available (15, 37, 53).
The diagnosis of invasive aspergillosis by molecular methods such as Southern blot analysis has been performed successfully with lung and liver material from animal models (41). The method showed a high degree of sensitivity but is limited by the necessity of performing invasive tissue biopsy. When applied to clinical samples which were obtained noninvasively, such as blood and BAL samples, hybridization methods were unsuccessful (41). As successfully accomplished for other pathogenic organisms that are difficult to detect (e.g., human immunodeficiency virus, cytomegalovirus, Borrelia burgdorferi, and Toxoplasma gondii) (9, 16, 24, 33, 40), more sensitive and rapid detection assays have been established by use of the PCR method, particularly following the identification and sequencing of multicopy gene templates in a range of fungi and other organisms. PCR assays for the detection of fungal nucleic acids may be the optimal diagnostic approach because they are potentially more sensitive than current culture-based methods and may be designed to encompass the desired range of genera and specimen types. Previous studies evaluating PCR-mediated detection of Aspergillus species showed significantly improved sensitivity but involved assays with different methods and objectives, partly to optimize culture assays (9, 28) and partly for typing in epidemiological studies (4, 5, 18). Therefore, the results of different groups are not consistent or comparable. By using PCR primers specific for the multicopy 28S rRNA gene, very small amounts (down to 1 pg) of genomic DNA from Aspergillus fumigatus have been detected, the sensitivity being increased to 100 fg by a subsequent Southern blot assay (41). Studies with clinical samples, e.g., blood or BAL samples, were mostly done retrospectively and with small numbers of patients (8, 16, 29, 30, 41, 44, 45, 51, 52).
Melchers et al. (30) first described a PCR assay for the detection of DNA from an Aspergillus sp. in immunocompromised patients with primers based on the coding sequence of the 18S rRNA gene which is highly conserved and which is amplified some hundredfold in the Aspergillus genome. PCR products were obtained from BAL samples of immunocompromised and neutropenic patients, while no amplicons were obtained from immunocompetent individuals. However, another report pointed to the high risk of BAL specimen contamination by Aspergillus conidia (8) resulting in approximately 25% false-positive results. Yamakami et al. (52) first described the use of a two-step PCR to detect Aspergillus spp. in blood with increased specificity and sensitivity, but that study was performed with only a small number of patients. Einsele et al. (17) described a PCR assay with subsequent Southern blot analysis which allowed the detection of fungal pathogens (including Aspergillus spp.) in blood samples. A recent publication described a panfungal PCR assay for amplification of a variety of fungal pathogens in human blood specimens, with the specific fungal species or genera being identified by subsequent Southern blot hybridization (45).
In order to achieve an improved, specific, and rapid means of detection of Aspergillus spp. in clinical specimens, we developed a two-step PCR assay for peripheral blood and BAL samples. Two optimal pairs of oligonucleotide primers derived from sequences of the 18S rRNA gene which are specific for Aspergillus spp. were selected from among a number of different candidates. The assay was evaluated for its sensitivity and specificity in vitro and was used to analyze clinical samples of immunocompromised patients. The results of the assay were also compared with the results of conventional diagnostic methods.
MATERIALS AND METHODS
Strains and growth conditions.
Prior to extraction of DNA, cultures of A. fumigatus were grown in Sabouraud agar for 72 h at 30°C. The cell density of the fungal suspensions (conidia) was determined microscopically by counting the cell number in a Neubauer cell chamber. Different titers of Aspergillus cell suspensions were used to spike EDTA-anticoagulated blood of healthy donors, and DNA was extracted by the method described below. These dilution experiments were done in triplicate. Extraction of DNA from the bacteria cultures was performed by ultrasonication of the pelleted bacteria and subsequent phenol-chloroform extraction as described by Sambrook et al. (38). Total DNA was used to determine the sensitivity of the PCR assay.
Strains tested for species specificity of PCR assay.
The following fungal strains were used in the study: A. fumigatus DSM 819, A. fumigatus CS, Aspergillus flavus DSM 1959, Aspergillus terreus DSM 1958, Aspergillus niger DSM 63263, A. niger CS, Aspergillus versicolor DSM 1943, Aspergillus clavatus DSM 3410, Candida albicans DSM 1386, C. albicans ATCC 44808, Candida tropicalis DSM 1346, Candida tropicalis DSM 5991, Candida krusei DSM 70079, Candida glabrata DSM 70614, Candida parapsilosis DSM 70126, Emericella nidulans DSM 820, Penicillium chrysogenum DSM 844, Penicillium expansum DSM 1282, Penicillium funiculosum DSM 1944, Aureobasidium pullulans DSM 2404, Paecilomyces variotii DSM 1961, Rhizopus oryzae DSM 854, Fusarium proliferatum DSM 848, Botrytis cinerea DSM 877, Scopulariopsis brevicaulis DSM 1218, and Neurospora crassa DSM 1257.
The following bacterial strains were used in the study: Streptococcus sanguis DSM 20068, Streptococcus mitis DSM 20568, Streptococcus pneumoniae DSM 20566, Staphylococcus aureus DSM 799, S. aureus ATCC 29213, S. aureus ATCC 25923, Staphylococcus epidermidis DSM 709, Enterococcus faecalis DSM 2570, Enterococcus faecium DSM 2146, Escherichia coli DSM 787, E. coli DSM 5923, E. coli ATCC 25921, Pseudomonas aeruginosa ATCC 27853, Klebsiella pneumoniae DSM 681, Enterobacter cloacae DSM 6234, Serratia marcescens DSM 1636, Proteus mirabilis DSM 788, and Haemophilus influenzae DSM 4690.
The fungal and bacterial test strains were obtained from the Deutsche Sammlung von Mikroorganismen und Zellkulturen GmbH (DSM), Braunschweig, Germany, from the American Type Culture Collection (ATCC), Manassas, Va., or from the Institute for Medical Microbiology and Hygiene, Klinikum Mannheim, University of Heidelberg, Heidelberg, Germany.
Clinical samples and DNA preparation.
The primary diseases of the immunocompromised patients group were hematological malignancies: acute myeloid or lymphoblastic leukemias, chronic myelogenous leukemia blast crisis, advanced myelodysplastic syndrome (refractory anemia-excess of blasts, refractory anemia-excess of blasts, transformation) while under antileukemic therapy, advanced or relapsed high- or intermediate-grade malignant lymphoma, advanced or relapsed Hodgkin's disease, or advanced chronic lymphocytic leukemia. Human immunodeficiency virus-positive patients were not included. Criteria for inclusion of immunocompromised patients were neutropenia (granulocyte count < 1.0 × 109/liter), fever (body temperature > 38.3°C), unresponsiveness to the first-line antibacterial treatment, and/or newly arisen nonspecific pulmonary infiltrates proven by conventional chest radiography.
Nonimmunocompromised patients had fever and lung infiltrates caused by complicated community-acquired pneumonia and/or suspect tumor findings by conventional chest radiography.
High-resolution computed tomography (HR-CT) scans of the lung were performed by standardized techniques by the Department of Clinical and Diagnostic Radiology, Klinikuum Mannheim, University of Heidelberg; small angiotropic round infiltrates, halo signs around infiltrates, or wedge-shaped small peripheral lung infarctions were judged as typical early pulmonary aspergillosis findings, the “air crescent sign” was considered a sign of a later disease stage. The HR-CT scans were performed, and the findings were analyzed by a panel of radiologically experienced staff.
Blood samples were obtained under sterile conditions by venipuncture, usually simultaneously with blood samples for culture for microbiological examination, in a sterile vessel containing potassium EDTA to a final concentration of 1.6 mg of EDTA per ml of blood. The sample volume was 5 to 7 ml.
A total of 3 to 5 ml of peripheral blood was mixed with 5 volumes of erythrocyte lysis buffer (0.155 M NH4Cl, 0.01 M NH4HCO3, 0.1 mM EDTA [pH 7.4]), and the mixture was incubated for 10 min at 4°C. After lysis of erythrocytes, the sample was centrifuged at 300 × g for 10 min. The supernatant was discarded, and the leukocytes were washed once with 1× phosphate-buffered saline (10× phosphate-buffered saline is 1.4 M NaCl, 50 mM KCl, 90 mM Na2PO4 · 2H2O, and 20 mM KH2PO4 [pH 7.4]) and recentrifuged.
Bronchoscopy was performed according to the guidelines of the Deutsche Gesellschaft für Pneumonologie (14), and BAL samples were obtained in a sterile vessel without conservation medium. The mean sample volume was 10 ml; the total volume of BAL samples taken from one patient at a time (ca. 100 ml) was aliquoted into 10-ml volumes, and these were placed in appropriate sterile vessels.
BAL samples were transferred to 1.5-ml tubes, the tubes were centrifuged at 13,000 rpm for 5 min (bench minifuge; Heraeus, and the supernatant was removed. Sedimented cell material from both blood and BAL specimens was processed as follows: the leukocyte pellet was resuspended in 300 μl of 1× phosphate-buffered saline and the mixture was incubated with 100 to 125 U of lyticase (50,000 U; Sigma) for 30 min at 37°C to achieve degradation of fungal cells. Residual human and fungal cell material was treated with 500 to 1,000 μg of proteinase K (Boehringer Mannheim, Mannheim, Germany) and 0.5% sodium dodecyl sulfate (Sigma) at 55°C for 1 h. Residual cell material was then lysed by incubation with an additional 100 μl of 2× Aspergillus extraction buffer (400 mM Tris-Cl, 1 M NaCl, 20 mM EDTA, 2% sodium dodecyl sulfate) for 30 min at 65°C. The purification of fungal and human DNA was performed by conventional phenol-chloroform extraction (38). The DNA was precipitated by the addition of 0.7 volume of isopropanol, pelleted, and washed once with 70% ethanol and air dried. The DNA concentration was assessed by spectrophotometry at 260 and 280 nm.
Oligonucleotide primers, primer sequences, and PCR assay.
The alignment of the three DNA sequences was performed with the program Geneworks (Intelligenetics, Inc.) by using standard algorithms. Primers were designed to have sequences homologous to those of various Aspergillus spp. but not to include the human 18S rRNA gene or the 16S rRNA genes of Candida spp. or other pathogenic microorganisms. Therefore, selection of the primer sequences was based on a close check for sequences with matching homologies in current DNA databases (GenBank, release June 1998) with a DNA alignment program (Blast).
By using a nested, two-step PCR technique, 15 different primers (Table 1) were tested, and the optimum two pairs (primers AFU5S and AFU5AS and primers AFU7S and AFU7AS) were chosen for all subsequent PCR assays. As an internal control, a 138-bp PCR fragment encoded by the human glucose-6-phosphate dehydrogenase gene (GenBank accession no. X55448) was amplified with primers G6PD1S and 1AS (Table 1) in each clinical sample.
TABLE 1.
Primer sequences and location
| Primer | DNA sequence (5′–3′)a | Locationb | Species specificity |
|---|---|---|---|
| AFU2S | ATG TCT AAG TAT AAG CAA TTT A | 17–38 | Aspergillus |
| AFU2AS | CTG TTA TTG CCG CGC ACT TCC A | 1366–1387 | Aspergillus |
| AFU3S | GCG AGT ACT GGT CCG GCT GGA | 628–648 | Aspergillus |
| AFU3AS | CCA GCG GCC CGC AAA TGC GG AC | 1314–1335 | Aspergillus |
| AFU4S | TAC TTA GAC ATG CAT GGC TTA A | 6–27 | Aspergillus |
| AFU4AS | TAG AGG AAG TAA AAG TCG TAA | 1704–1724 | Aspergillus |
| AFU5S | AGG GCC AGC GAG TAC ATC ACC TTG | 1436–1459 | Aspergillus |
| AFU5AS | GG G (AG)GT CGT TGC CAA C(CT)C (CT)CC TGA | 1648–1771 | Aspergillus |
| AFU7S | CGG CCC TTA AAT AGC CCG | 1296–1313 | Aspergillus |
| AFU7AS | GA CCG GGT TTG ACC AAC TTT | 1681–1700 | Aspergillus |
| AFU8S | GTC CGC ATT TGC GGG CCG CT | 1314–1333 | Aspergillus |
| AFU8AS | TGC CAA CTC CCC TGA GCC AG | 1643–1662 | Aspergillus |
| AFU9S | GCA CGC GCG CTA CAC TGA CAG GGC | 1417–1440 | Universal |
| AFU9AS | GGC CTC ACC GAG CCA TTC AAT CGG | 1613–1636 | Universal |
| AFU10AS | GCG ACG GGC GGT GTG TAC AAA GGG | 1584–1607 | Universal |
| G6PD1S | CAG CGT CAT GGC AGA GCA GGT GGC | 3344–3367 | Human |
| G6PD1AS | GGA GAT ACT CAC CGA TGC ACC CAT | 3459–3482 | Human |
Nucleotides in parentheses are degenerate.
PCR.
Per 25-μl PCR mixture, approximately 50 to 150 ng of total DNA was used as the template. The standard PCR mixture contained 0.5 U of Taq DNA polymerase, 6.25 nmol of the deoxynucleoside triphosphates, 10 pmol of primer (first step, primer AFU7S-AFU7AS; second step, primer AFU5S-AFU5AS). In preliminary studies the optimum reaction conditions were established by testing different DNA, primer, enzyme, and deoxynucleoside triphosphate concentrations as well as a range of cycling conditions. PCR was performed in a thermal cycler (Perkin-Elmer Cetus), as follows: for the first PCR, 2 min at 94°C and then 23 cycles of 40 s at 94°C, 1 min at 65°C, and 1 min at 72°C with a terminal step of 5 min at 72°C and then the mixture was held at 4°C; for the second PCR, 2 min at 94°C and then 35 cycles of 40 s at 94°C, 1 min at 65°C, and 1 min at 72°C, with a terminal step of 5 min at 72°C, and then the mixture was held at 4°C. For the second PCR, approximately 1 to 2 μl of the first-round PCR product was used. The PCR products were separated by 2.5% agarose gel electrophoresis, stained with ethidium bromide, and visualized with UV light. Control samples included all the constituents in the reaction mixture except genomic DNA. As negative and positive PCR controls, DNA from the human cell line T47D or dilute samples of A. fumigatus, respectively, were used as templates.
RESULTS
Strategy for primer selection.
The alignment of the 18S rRNA genes of Aspergillus spp., humans, and Candida spp. revealed several regions of significant divergence which were the basis for selection of the primers. In order to establish a PCR assay specific for several Aspergillus species of clinical importance the 18S rRNA gene (rDNA) sequence of A. fumigatus (GenBank accession no. AB008401) was aligned with the human 18S rDNA sequence (GenBank accession no. M10098) and the 16S rDNA sequence of C. albicans (GenBank accession no. X53497), another ubiquitious microorganism of major clinical importance. Fifteen different primers, comprising 7 sequences upstream and 8 sequences downstream from various divergent regions of the Aspergillus species gene which showed the least homology with the human or Candida genes, were selected for the evaluation and the optimization of the PCR assay.
Three different strategies were used. (i) A nested PCR with Aspergillus-specific primers in both PCR steps was used. Most of these primer combinations were relatively insensitive, detecting 1 pg of Aspergillus DNA. The pairs described below, however, were more sensitive. (ii) A nested PCR with one primer (universal primer) that had broad specificity and that matched most of the 18S rDNA sequences analyzed in combination with a primer specific for Aspergillus in the first step and a primer pair specific for only Aspergillus in the second step was used. None of these primer combinations reached a sensitivity sufficient to amplify a detectable DNA fragment from less than 1 pg of Aspergillus DNA, or they proved to be unspecific, amplifying human and Candida DNA in the second step. (iii) The intergenic region of the 18s rRNA gene of A. fumigatus was amplified with the primer pair AFU4S-AFU4AS to generate specific intergenic regions. This assay did not amplify any detectable PCR products, probably because the fragment length was too long for productive amplification.
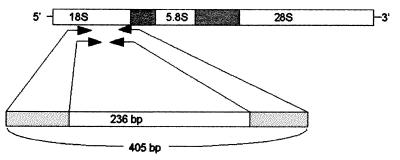
The two primer pairs that produced PCR products with the highest sensitivity and species specificity were the Aspergillus-specific primers AFU7S and AFU7AS, which amplified a fragment of 405 bp, followed by AFU5S and AFU5AS, which produced an internal fragment of 236 bp (Fig. 1 and 2; primer sequences in Table 1). These primer-binding sites are located in the 3′ part of the 18S rRNA gene and in variable region V7-V9 (AFU7AS) or V8-V9 (AFU5AS), with no sequence overlap between the primers used in the first and second PCRs to reduce contamination problems (Fig. 2).
FIG. 1.
Locations of primer pairs AFU5S-AFU5AS and AFU7S-AFU7AS used in the two-step PCR to detect Aspergillus DNA. The primers are derived from the 18S rRNA gene of Aspergillus spp. The first PCR step (with AFU7S-AFU7AS) results in amplification of a 405-bp fragment, and the second step (with AFU5S-AFU5AS) amplifies an internal fragment of 236 bp.
FIG. 2.
Alignment of DNAs of 18S rRNA genes of A. fumigatus (GenBank accession no. AB008401) and humans (GenBank accession no. M10098) and the corresponding 16S rRNA gene of C. albicans (GenBank accession no. X53497). The locations of primer pairs AFU5S-AFU5AS and AFU7S-AFU7AS used in the two-step PCR to detect Aspergillus DNA are indicated by arrows. Homologous regions are boxed.
Sensitivity of two-step PCR.
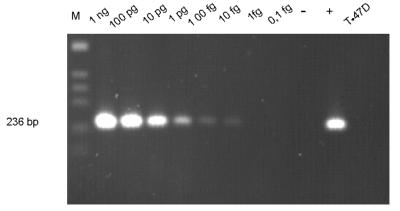
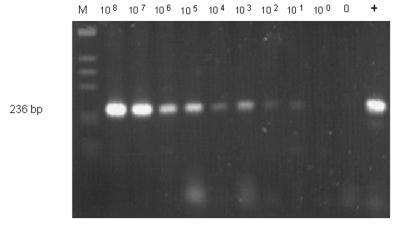
The sensitivity of the PCR assay with the optimal two pairs of nested oligonucleotides AFU7S-AFU7AS and AFU5S-AFU5AS was determined by dilution of A. fumigatus template in human leukocyte DNA. Signals derived from as little as 10 fg of A. fumigatus DNA could still be clearly detected by ethidium bromide staining of agarose gels (Fig. 3), whereas PCR with human or C. albicans DNA alone did not produce any detectable amplified DNA. The detection limit of this assay corresponds to 1 to 5 CFU/ml of blood, as additionally confirmed by spiking blood samples from healthy donors with serial dilutions of fungal conidia and performing similar two-step PCR amplification assays (Fig. 4).
FIG. 3.
Determination of the sensitivity of the two-step PCR assay with purified A. fumigatus DNA diluted in human DNA (50 ng). The signal derived from 10 fg of Aspergillus template DNA was clearly detectable by ethidium bromide staining of an agarose gel. As a positive control, only DNA extracted from A. fumigatus (10 pg) was used in a single PCR amplification of the 236-bp fragment with the second primer pair (lane +). A negative reagent control amplification without addition of DNA (lane −) as well as purified DNA from a human cell line (T47D) resulted in no bands. The 123-bp ladder (Gibco BRL) was used as molecular size marker (lane M).
FIG. 4.
Peripheral blood samples from healthy donors were spiked with defined numbers of conidia from A. fumigatus. The signal derived from 101 to 100 CFU per ml of blood were still detectable by ethidium bromide staining of an agarose gel. As a positive control, DNA extracted from A. fumigatus (10 pg) was used for single-step PCR amplification of the 236-bp fragment (lane +). Negative reagent control amplification without conidia (0) resulted in no bands. The 123-bp ladder (Gibco BRL) was used as a molecular size marker (M).
Genus and species specificity of PCR assay.
The products obtained after the second PCR from an A. fumigatus test strain were cloned and sequenced, confirming that the DNA sequences exactly matched the corresponding sequence of the A. fumigatus 18S rRNA gene from a database (GenBank). To further test the PCR specificity, template DNA extracted from a wide range of fungal and bacterial pathogens, listed in Materials and Methods, was investigated with the two primer pairs. PCR products could be obtained only from DNAs of cultures of Aspergillus spp. (A. fumigatus, A. flavus, A. niger, A. terreus, A. clavatus, A. versicolor, and the Aspergillus-like species E. nidulans), whereas the PCRs with the other fungal and bacterial test strains and DNA from a human cell line were negative. In the cases in which the PCR products were obtained from clinical samples (see below), three blood samples were chosen for cloning and sequencing of the amplified fragment. These samples had results identical to those for the control PCR product clones from the A. fumigatus test strain: the DNA sequences matched the sequence of the A. fumigatus 18S rRNA gene from the database for all samples (data not shown). These results indicate the high species specificity and sequence stringency of this two-step PCR assay.
Detection of Aspergillus DNA in clinical samples.
A total of 315 samples (250 blood samples and 65 BAL samples) from 140 subjects including 47 healthy individuals or nonimmunocompromised patients and 93 febrile neutropenic patients with malignant hematologic diseases (predominantly patients with acute leukemias) at high risk for invasive fungal disease were screened by the two-step PCR assay. Several specimens from all patients were tested (average, 3.0 specimens). Typical PCR results for clinical samples (peripheral blood) showed either no amplification product or a single defined band of 236 bp, corresponding exactly to the results for the Aspergillus controls.
A positive DNA sample containing 100 fg of Aspergillus DNA mixed with 50 ng of human DNA was used in PCRs with samples from each set of patients to check for proper amplification. If no signal was obtained for this positive control, the PCR was repeated.
Consecutive blood samples from 22 neutropenic leukemia patients had positive PCR signals, and aspergillosis was proven in 7 of these patients by the results of histology and/or culture. Histologic tissue samples were obtained from two patients by lung biopsy and from two subjects at autopsy. The three positive culture results that were obtained were exclusively for BAL specimens, thus highlighting the difficulties associated with obtaining positive culture results, particularly for blood samples.
No convincing clinical evidence of an Aspergillus infection could be found for only one of the leukemic patients (patient 13; Table 2), and only a single positive PCR result was obtained for one blood sample from that patient. By monitoring the clinical course of this febrile neutropenic patient, in whom a small lung infiltration was detected by chest X ray, we could neither prove nor exclude invasive aspergillosis. Antimycotic and antibacterial therapy and a prompt rise in granulocyte counts resulted in defervescence within a few days; no relapse of the pneumonia occurred during the next subsequent neutropenic phase. Subsequent positive PCR results were not obtained until 8 months later, following bone marrow transplantation, and invasive aspergillosis was also proven at this time by a positive result for a tracheal lavage specimen culture.
TABLE 2.
Positive PCR results for febrile neutropenic patients with lung infiltrates and proven (by histology and/or by culture) or probable (by HR-CT findings) invasive aspergillosisa
| Patient no. (age [yr]) | Diagnosis | Diagnostic tests | Sample with positive PCR findings | Outcome |
|---|---|---|---|---|
| 1 (54) | AML | HR-CT | B | Died (IA) |
| 2 (36) | AML | HR-CT, histology (chest surgery) | B | Died (IA) (relapse after BMT) |
| 3 (62) | ALL | Culture | B, BAL | Died (IA) |
| 4 (60) | AML | Histology (autopsy) | BAL | Died (IA) |
| 5 (61) | AML | HR-CT | B | Died (leukemia) |
| 6 (75) | AML | HR-CT | B | Died (leukemia) |
| 7 (75) | HD | HR-CT | B, BAL | Alive |
| 8 (42) | AML | HR-CT, histology (liver puncture) | B | Alive |
| 9 (50) | AML | HR-CT, histology (chest surgery), antigen assay | B | Alive |
| 10 (38) | AML | HR-CT, antigen assay | B, BAL | Died (leukemia) |
| 11 (30) | AML | HR-CT | B | Died (leukemia) |
| 12 (32) | SAA (post BMT) | HR-CT, culture, antigen assay | B, BAL | Alive |
| 13 (25) | ALL | Chest X ray | B | Died (IA) |
| 14 (67) | AML | HR-CT, histology (liver puncture) | B, BAL | Died (IA) |
| 15 (61) | AML | Histology (autopsy) | B, BAL | Died (leukemia) |
| 16 (64) | AML | HR-CT | BAL | Died (leukemia) |
| 17 (56) | AML | HR-CT | B | Died (leukemia) |
| 18 (69) | ALL | HR-CT | B | Died (leukemia) |
| 19 (20) | ALL | HR-CT | B | Died (BMT) |
| 20 (39) | CML-BC | HR-CT | B | Died (leukemia) |
| 21 (62) | CML-BC | HR-CT, culture | B, BAL | Died (leukemia) |
| 22 (56) | AML | HR-CT | B | Died (leukemia) |
Abbreviations: AML, acute myeloid leukemia; IA, invasive aspergillosis; HD, Hodgkin's disease; SAA, severe aplastic anemia; BMT, bone marrow transplantation; ALL, acute lymphoblastic leukemia; B, blood; CML-BC, chronic myelogenous leukemia blast crisis.
The remaining 14 (of the 22) patients had characteristic HR-CT findings that suggested invasive aspergillosis. (For almost all patients, blood and BAL specimens were collected before HR-CT scans were performed.) Therefore, the correlation between the positive histopathology results, positive culture results, or positive HR-CT findings and the PCR results was 100%. None of our patients had positive HR-CT findings and a negative PCR result. Among the samples from nonimmunocompromised patients and healthy volunteers, the PCR assay gave one single positive result with a blood sample (2.1%) and four positive results with BAL samples (8.5%) from different individuals.
DISCUSSION
PCR has been shown to be a highly sensitive diagnostic tool for the detection of infectious fungi in diverse specimens, but current PCR assays applied in clinical diagnostics have considerable drawbacks due to possible contamination with ubiquitious fungal conidia, coamplification of human DNA, or the necessity for additional time-consuming hybridization steps. Our aim was to establish a new, highly sensitive, and specific PCR assay for the rapid detection of the full range of human pathogenic Aspergillus species in blood and BAL samples. This would result in a test with increased sensitivity and increased predictive value.
Pioneering work in this direction (41) was initiated after multicopy gene targets, such as the 28S rRNA gene, had been identified and sequenced. The first PCR assay for the detection of Aspergillus DNA in immunocompromised patients to be described (30) used primers derived from the multicopy 18S rRNA gene, a gene of numerous microorganisms that has now been sequenced. More recent reports described PCR techniques with primers designed to target conserved 18S rRNA sequences common to a variety of fungal pathogens (17, 45). However, additional Southern blot analyses with longer probes were also necessary to achieve the detection sensitivity (17) for the identification of the specific fungal species or genera present (45).
Nested PCR assays improved the detection sensitivity. In the first description of such a two-step PCR for the detection of Aspergillus spp. in serum samples of patients, Yamakami et al. (52) reported that a PCR with two sets of 18S rRNA primers had considerably improved sensitivity compared to that of a PCR assay with a single set of primers. This could be further improved by subsequent Southern blot hybridization. These data also highlighted for the first time the use of PCR for the detection of Aspergillus DNA in blood samples, which can be obtained by a less noninvasive procedure that is associated with fewer risks than the procedure used to collect BAL specimens, which cannot be performed repeatedly with immunocompromised patients.
We also used a two-step PCR procedure with carefully selected primers to increase the sensitivity and specificity for Aspergillus detection, thereby eliminating a subsequent hybridization step otherwise obligatory for sensitive detection or verification of the amplified PCR products. Use of a two-step PCR increases the probability of contamination. Therefore, negative controls (human DNA and water) were used in each set of PCRs. If these controls gave a positive result, the set of PCRs was repeated. The multicopy 18S rRNA gene is highly conserved in all species but includes variable regions that are conserved among most Aspergillus species. On the basis of comparisons with sequences in a data bank, we chose oligonucleotides from the variable regions that specifically matched only the 18S rRNA gene from various Aspergillus species to avoid problems arising from coamplification of human or bacterial DNA or contamination with other fungus-derived DNA. A total of 15 different primers were screened for their sensitivities and specificities, and the optimal two pairs of nested primers were chosen for use in all subsequent assays. The negative test results for a wide range of fungal pathogens, including several Penicillium spp., and bacterial pathogens or a human cell line indicated the high species and genus specificity of this PCR assay. PCR products were obtained only from DNA from Aspergillus cultures (A. fumigatus, A. flavus, A. niger, A. terreus, A. clavatus, A. versicolor) and the Aspergillus-like species E. nidulans. In order to exclude possible contamination with Aspergillus conidia, a problem associated with BAL specimens (8), and also because of the clinically relevant ease of with which repeated blood samples can be obtained, peripheral blood from volunteer donors and patients was preferentially analyzed.
The detection threshold of the PCR assay described here was about 10 fg of template DNA, or between 1 and 5 CFU per ml of blood (Fig. 3 and 4). This demonstrates a sensitivity that may allow this PCR assay to be useful for the detection of small amounts of pathogen and demonstrates that this PCR assay may be a valid diagnostic tool. In addition, elimination of a subsequent hybridization step greatly simplifies and speeds up the diagnostic assay. Contamination with ubiquituous fungal spores may be a disadvantage of this PCR assay. The conditions and the origins of contamination are various and cannot be methodically excluded completely. The contamination rate that occurs during processing of DNA and PCRs was low under our optimized conditions. To assess the clinical implications and applicability of the assay, we screened clinical samples from 47 healthy individuals or nonimmunocompromised patients and 93 febrile neutropenic patients with malignant hematologic diseases, predominantly acute leukemias, at high risk for invasive fungal disease. No convincing evidence of an Aspergillus infection could be found in one leukemic patient who provided one blood sample that had a positive PCR result. Conversely, all 21 patients with clinically proven invasive aspergillosis (positive histopathology results, positive culture results, or positive HR-CT scans) had positive PCR results, demonstrating a test sensitivity of 100%.
Interestingly, only three positive culture results were obtained, and all of these were with BAL specimens, highlighting the difficulties associated with obtaining positive culture results, particularly with blood samples. In contrast, PCR results for all blood samples were consistent with those for BAL samples or positive clinical test results, demonstrating the sensitivity and potential clinical value of this PCR assay. For the group of nonimmunocompromised patients, the results indicate a test specificity of 89% due to a low contamination rate of one blood sample and four BAL samples. Preliminary data (unpublished data) from follow-up PCR assays correlated with the clinical course of the infected patients, indicating that this assay may provide a better means of monitoring antimycotic therapy. It has already been reported that the persistence of PCR signals reveals a trend toward a worse, possibly fatal outcome (17).
Due to the low incidence of proven invasive aspergillosis in immunocompromised patients, the prospective clinical validation of this assay requires large numbers of patients at high risk for fungal infection. The clinical validation of the assay in a prospective multicenter study to define the predictive value of the assay and its experimental validation with a histologically defined animal model of aspergillosis are in progress.
In summary, a specific two-step PCR assay for the rapid detection of the full range of human pathogenic Aspergillus species in both BAL and blood samples was established, facilitating improved early diagnosis and better monitoring of systemic aspergillosis during therapy in high-risk patients.
A patent of the assay is pending (23a).
ACKNOWLEDGMENTS
We thank H. Hof and M. Kretschmar, Institute for Medical Microbiology and Hygiene, Klinikum Mannheim, University of Heidelberg, Germany, for helpful support, especially with bacterial and fungal test strains, and H. Niachos for technical assistance. We thank A. Arthur-Goettig for critically reading the manuscript.
This work was supported in part by a grant from the Forschungsfonds der Fakultät für Klinische Medizin Mannheim der Universität Heidelberg (project nos. 62 and 2062).
REFERENCES
- 1.Anaissie E J, Bodey G P, Rinaldi M G. Emerging fungal pathogens. Eur J Clin Microbiol Infect Dis. 1989;8:323–330. doi: 10.1007/BF01963467. [DOI] [PubMed] [Google Scholar]
- 2.Aquino S L, Kee S T, Warnock M L, Gamsu G. Pulmonary aspergillosis: imaging findings with pathologic correlation. Am J Roentgenol. 1994;163:811–815. doi: 10.2214/ajr.163.4.8092014. [DOI] [PubMed] [Google Scholar]
- 3.Arning M, Aul C. Prophylaxis against mycosis in neutropenic patients. Mycoses. 1994;37(Suppl. 2):70–76. [PubMed] [Google Scholar]
- 4.Aufavre-Brown A, Cohen J, Holden D W. Use of randomly amplified polymorphic DNA markers to distinguish isolates of Aspergillus fumigatus. J Clin Microbiol. 1992;30:2991–2993. doi: 10.1128/jcm.30.11.2991-2993.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Birch M, Nolard N, Shankland G S, Denning D W. DNA typing of epidemiologically related isolates of Aspergillus fumigatus. Epidemiol Infect. 1995;114:161–168. doi: 10.1017/s0950268800052018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blum U, Windfuhr M, Buitrago-Tellez C, Sigmund G, Herbst E W, Langer M. Invasive pulmonary aspergillosis. MRI, CT and plain radiographic findings and their contribution for early diagnosis. Chest. 1994;106:1156–1161. doi: 10.1378/chest.106.4.1156. [DOI] [PubMed] [Google Scholar]
- 7.Bodey G P, Bueltmann B, Duguid W, Gibbs D, Hanak H, Hotchi M, Mall G, Martino P, Meunier F, Milliken S, Naoe S, Okudaira M, Scevola D, van't Wout J. Fungal infections in cancer patients: an international autopsy survey. Eur J Clin Microbiol Infect Dis. 1992;11:99–109. doi: 10.1007/BF01967060. [DOI] [PubMed] [Google Scholar]
- 8.Bretagne S, Costa J-M, Marmorat-Khuong A, Poron F, Cordonnier C, Vidaud M, Fleury-Feith J. Detection of Aspergillus species DNA in bronchoalveolar lavage samples by competitive PCR. J Clin Microbiol. 1995;33:1164–1168. doi: 10.1128/jcm.33.5.1164-1168.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Burg J L, Grover M, Pouletty P, Boothroyd J C. Direct and sensitive detection of a pathogenic protozoon, Toxoplasma gondii, by polymerase chain reaction. J Clin Microbiol. 1989;27:1787–1792. doi: 10.1128/jcm.27.8.1787-1792.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cafferkey M T. Chemoprophylaxis of invasive pulmonary aspergillosis. J Antimicrob Chemother. 1994;33:917–924. doi: 10.1093/jac/33.5.917. [DOI] [PubMed] [Google Scholar]
- 11.Caillot D, Casasnovas O, Bernard A, Couaillier J F, Durand C, Cuisenier B, Solary E, Piard F, Petrella T, Bonnin A, Couillault G, Dumas M, Guy H. Improved management of invasive pulmonary aspergillosis in neutropenic patients using early thoracic computed tomography scan and surgery. J Clin Oncol. 1997;15:139–147. doi: 10.1200/JCO.1997.15.1.139. [DOI] [PubMed] [Google Scholar]
- 12.Chandrasekar P H, Weinmann A, Shearer C. Autopsy-identified infections among bone marrow transplant recipients: a clinico-pathologic study of 56 patients. Bone Marrow Transplant. 1995;16:675–681. [PubMed] [Google Scholar]
- 13.Denning D W. Treatment of invasive aspergillosis. J Infect. 1994;28:25–33. doi: 10.1016/s0163-4453(94)95941-2. [DOI] [PubMed] [Google Scholar]
- 14.Deutsche Gesellschaft für Pneumologie. Empfehlungen zur diagnostischen bronchoalveolären Lavage. Pneumologie. 1994;48:311–323. [PubMed] [Google Scholar]
- 15.Duthie R, Denning D W. Aspergillus fungemia: report of two cases and review. Clin Infect Dis. 1995;20:598–605. doi: 10.1093/clinids/20.3.598. [DOI] [PubMed] [Google Scholar]
- 16.Einsele H, Steidle H, Vallbracht A, Saal J G, Ehninger G, Müller C A. Early occurrence of human cytomegalovirus infection after bone marrow transplantation as demonstrated by the polymerase chain reaction technique. Blood. 1991;77:1104–1110. [PubMed] [Google Scholar]
- 17.Einsele H, Hebart H, Roller G, Löffler J, Rothenhöfer I, Müller C A, Bowden R A, van Burik J-A, Engelhard D, Kanz L, Schumacher U. Detection and identification of fungal pathogens in blood using molecular probes. J Clin Microbiol. 1997;35:1353–1360. doi: 10.1128/jcm.35.6.1353-1360.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Girardin H, Sarfati J, Traore F, Dupouy-Camet J, Derouin F, Latge J P. Molecular epidemiology of nosocomial invasive aspergillosis. J Clin Microbiol. 1994;32:684–690. doi: 10.1128/jcm.32.3.684-690.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Glass N L, Donaldson G C. Development of primer sets designed for use with the PCR to amplify conserved genes from filamentous ascomycetes. Appl Environ Microbiol. 1995;61:1323–1330. doi: 10.1128/aem.61.4.1323-1330.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Groll A H, Shah P M, Mentzel C, Schneider M, Just-Nübling G, Huebner K. Trends in the postmortem epidemiology of invasive fungal infections at a university hospital. J Infect. 1996;33:23–32. doi: 10.1016/s0163-4453(96)92700-0. [DOI] [PubMed] [Google Scholar]
- 21.Guiot H F L, Fibbe W E, van't Wout J W. Risk factors for fungal infection in patients with malignant hematologic disorders: implications for empirical therapy and prophylaxis. Clin Infect Dis. 1994;18:525–532. doi: 10.1093/clinids/18.4.525. [DOI] [PubMed] [Google Scholar]
- 22.Haynes K, Rogers T R. Retrospective evaluation of a latex agglutination test for diagnosis of invasive aspergillosis in immunocompromised patients. Eur J Clin Microbiol Infect Dis. 1994;13:670–674. doi: 10.1007/BF01973998. [DOI] [PubMed] [Google Scholar]
- 23.Hearn V M, Pinel C, Blachier S, Ambroise-Thomas P, Grillot R. Specific antibody detection in invasive aspergillosis by analytical isoelectrofocusing and immunoblotting methods. J Clin Microbiol. 1995;33:982–986. doi: 10.1128/jcm.33.4.982-986.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23a.Hehlmann R, Skladny H, Buchheidt D. Verfahren zum Nachweis von Aspergillus-Nukleinsäuren in einer Körperprobe, Nukleinsäure-Sonden und Diagnosekits zur Durchführung des Verfahrens. Patent no. 198 06 274.5. German Patent Office; 16 February 1999. [Google Scholar]
- 24.Huppertz H-I, Schmidt H, Karch H. Detection of Borrelia burgdorferi by nested polymerase chain reaction in cerebrospinal fluid and urine of children with neuroborreliosis. Eur J Pediatr. 1993;152:414–418. doi: 10.1007/BF01955900. [DOI] [PubMed] [Google Scholar]
- 25.Kappe R, Schulze-Berge A, Sonntag H G. Evaluation of eight antibody tests and one antigen test for the diagnosis of invasive aspergillosis. Mycoses. 1996;39:13–23. doi: 10.1111/j.1439-0507.1996.tb00078.x. [DOI] [PubMed] [Google Scholar]
- 26.Kuhlmann J E, Fishman E K, Burch P A, Karp J E, Zerhouni E A, Siegelman S S. Invasive pulmonary aspergillosis in acute leukemia: the contribution of CT to early diagnosis and aggressive management. Chest. 1987;92:95–99. doi: 10.1378/chest.92.1.95. [DOI] [PubMed] [Google Scholar]
- 27.Logan P M, Primack S L, Miller R R, Muller N L. Invasive aspergillosis of the airways: radiographic, CT and pathologic findings. Radiology. 1994;193:383–388. doi: 10.1148/radiology.193.2.7972747. [DOI] [PubMed] [Google Scholar]
- 28.Makimura K, Murayama S Y, Yamaguchi H. Specific detection of Aspergillus and Penicillium species from respiratory specimens by PCR. Jpn J Med Sci Biol. 1994;47:141–156. doi: 10.7883/yoken1952.47.141. [DOI] [PubMed] [Google Scholar]
- 29.Makimura K, Murayama S Y, Yamaguchi H. Detection of a wide range of medically important fungi by the polymerase chain reaction. J Med Microbiol. 1994;40:358–364. doi: 10.1099/00222615-40-5-358. [DOI] [PubMed] [Google Scholar]
- 30.Melchers W J, Verweij P E, van den Hurk P, van Belkum A, De Pauw B E, Hoogkamp-Korstanje J A, Meis J F. General primer-mediated PCR for detection of Aspergillus species. J Clin Microbiol. 1994;32:1710–1717. doi: 10.1128/jcm.32.7.1710-1717.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Morrison V A, Haake R J, Weisdorf D J. The spectrum of non-Candida fungal infections following bone marrow transplantation. Medicine. 1993;72:78–89. doi: 10.1097/00005792-199303000-00002. [DOI] [PubMed] [Google Scholar]
- 32.Obayashi T, Yoshida M, Mori T, Goto H, Yasuoka A, Iwasaki H, Teshima H, Kohno S, Horiuchi A, Ito A. Plasma(1-3)-beta-d-glucan measurement in diagnosis of invasive deep mycosis and fungal febrile episodes. Lancet. 1994;345:17–20. doi: 10.1016/s0140-6736(95)91152-9. [DOI] [PubMed] [Google Scholar]
- 33.Ou C Y, Kwok S, Mitchell W, Mack D H, Sninsky J J, Krebs J W, Feorino P, Warfield G, Schochetman G. DNA amplification for direct detection of HIV-1 in DNA of peripheral blood mononuclear cells. Science. 1988;239:295–297. doi: 10.1126/science.3336784. [DOI] [PubMed] [Google Scholar]
- 34.Patterson T F, Miniter P, Patterson J E, Rappaport J M, Andriole V T. Aspergillus antigen detection in the diagnosis on invasive aspergillosis. J Infect Dis. 1995;171:1553–1558. doi: 10.1093/infdis/171.6.1553. [DOI] [PubMed] [Google Scholar]
- 35.Pfaffenbach B, Donhuijsen K, Pahnke J, Bug R, Adamek R J, Wegener M, Ricken D. Systemische Pilzinfektionen bei hämatologischen Neoplasien. Eine Autopsiestudie an 1053 Patienten. Med Klin. 1994;89:299–304. [PubMed] [Google Scholar]
- 36.Rinaldi M G. Problems in the diagnosis of invasive fungal diseases. Rev Infect Dis. 1991;13:493–498. doi: 10.1093/clinids/13.3.493. [DOI] [PubMed] [Google Scholar]
- 37.Robinson L A, Reed E C, Galbraith T A, Alonso A, Moulton A L, Fleming W H. Pulmonary resection for invasive Aspergillus infections in immunocompromised patients. J Thorac Cardiovasc Surg. 1995;109:1182–1196. doi: 10.1016/S0022-5223(95)70202-4. [DOI] [PubMed] [Google Scholar]
- 38.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 39.Saugier-Veber P, Devergie A, Sulahian A, et al. Epidemiology and diagnosis of invasive pulmonary aspergillosis in bone marrow transplant patients. Results of a 5 year retrospective study. Bone Marrow Transplant. 1993;12:121–124. [PubMed] [Google Scholar]
- 40.Shibata D, Martin J W, Appleman M D, Causey D M, Leedom J M, Arnheim N. Detection of cytomegalovirus DNA in peripheral blood of patients infected with human immunodeficiency virus. J Infect Dis. 1988;158:1185–1192. doi: 10.1093/infdis/158.6.1185. [DOI] [PubMed] [Google Scholar]
- 41.Spreadbury C, Holden D, Aufavre-Brown A, Bainbridge B, Cohen J. Detection of Aspergillus fumigatus by polymerase chain reaction. J Clin Microbiol. 1993;31:615–621. doi: 10.1128/jcm.31.3.615-621.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stynen D, Goris A, Sarfati J, Latge J P. A new sensitive sandwich enzyme-linked immunosorbent assay to detect galactofuran in patients with invasive aspergillosis. J Clin Microbiol. 1995;33:497–500. doi: 10.1128/jcm.33.2.497-500.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Swanink C M A, Meis J F G M, Rijs A J M M, Donnelly J P, Verweij P E. Specificity of a sandwich enzyme linked immunosorbent assay for detecting Aspergillus galactomannan. J Clin Microbiol. 1997;35:257–260. doi: 10.1128/jcm.35.1.257-260.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tang C M, Holden D, Aufauvre-Brown A, Cohen J. Detection of Aspergillus species by the polymerase chain reaction and its evaluation in bronchoalveolar lavage fluid. Am Rev Respir Dis. 1993;148:1313–1317. doi: 10.1164/ajrccm/148.5.1313. [DOI] [PubMed] [Google Scholar]
- 45.van Burik J-A, Myerson D, Schreckhise R W, Bowden R A. Panfungal PCR assay for detection of fungal infection in human blood specimens. J Clin Microbiol. 1998;36:1169–1175. doi: 10.1128/jcm.36.5.1169-1175.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Verweij P E, Rijs A J, De Pauw B E, Horrevorts A M, Hoogkamp-Korstanje J A, Meis J F. Clinical evaluation and reproducibility of the Pastorex Aspergillus antigen latex test for diagnosing invasive aspergillosis. J Clin Pathol. 1995;48:474–476. doi: 10.1136/jcp.48.5.474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Verweij P E, Latge J P, Rijs A J, Melchers W J, de Pauw B E, Hoogkamp-Korstanje J A, Meis J F. Comparison of antigen detection and PCR assay using bronchoalveolar lavage fluid for diagnosing invasive pulmonary aspergillosis in patients receiving treatment for hematological malignancies. J Clin Microbiol. 1995;33:3150–3153. doi: 10.1128/jcm.33.12.3150-3153.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.von Eiff M, Zuhlsdorf M, Roos N, Hesse M, Schulten R, van de Loo J. Pulmonary fungal infections in patients with hematological malignancies: diagnostic approaches. Ann Hematol. 1995;70:135–141. doi: 10.1007/BF01682033. [DOI] [PubMed] [Google Scholar]
- 49.Walsh T J, Garrett K, Feuerstein E, Girton M, Allende M, Bacher J, Francesconi A, Schaufele R, Pizzo P A. Therapeutic monitoring of experimental invasive pulmonary aspergillosis by ultrafast computerized tomography: a novel, noninvasive method for measuring responses to antifungal therapy. Antimicrob Agents Chemother. 1995;39:1065–1069. doi: 10.1128/aac.39.5.1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Walsh T J, Lee J, Lecciones J, Rubin M, Butler K, Francis P, Weinberger M, Roilides E, Marshall D, Gress J, Pizzo P A. Empiric therapy with amphotericin B in febrile granulocytopenic patients. Rev Infect Dis. 1991;13:496–503. doi: 10.1093/clinids/13.3.496. [DOI] [PubMed] [Google Scholar]
- 51.Walsh T J, Francesconi A, Kasai M, Chanock S J. PCR and single-strand conformational polymorphism for recognition of medically important opportunistic fungi. J Clin Microbiol. 1995;33:3216–3220. doi: 10.1128/jcm.33.12.3216-3220.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yamakami Y, Hashimoto A, Tokimatsu I, Nasu M. PCR detection of DNA specific for Aspergillus species in serum of patients with invasive aspergillosis. J Clin Microbiol. 1996;34:2464–2468. doi: 10.1128/jcm.34.10.2464-2468.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yu V L, Muder R R, Poorsattar A. Significance of isolation of Aspergillus from the respiratory tract in diagnosis of invasive pulmonary aspergillosis. Am J Med. 1986;81:249–254. doi: 10.1016/0002-9343(86)90259-7. [DOI] [PubMed] [Google Scholar]