ABSTRACT
RNAi is a potent technique for the knockdown of target genes. However, its potential off-target effects limit the widespread applications in both reverse genetic analysis and genetic manipulation. Previous efforts have uncovered rules underlying specificity of siRNA-based silencing, which has broad applications in humans, but the basis for specificity of dsRNAs, which are better suited for use as insecticides, is poorly understood. Here, we investigated the rules governing dsRNA specificity. Mutational analyses showed that dsRNAs with >80% sequence identity with target genes triggered RNAi efficiently. dsRNAs with ≥16 bp segments of perfectly matched sequence or >26 bp segments of almost perfectly matched sequence with one or two mismatches scarcely distributed (single mismatches inserted between ≥5 bp matching segments or mismatched couplets inserted between ≥8 bp matching segments) also able to trigger RNAi. Using these parameters to predict off-target risk, dsRNAs can be designed to optimize specificity and efficiency, paving the way to the widespread, rational application of RNAi in pest control.
KEYWORDS: RNA interference, off-target effect, sequence identity, RNAi efficiency, dsRNA specificity, risk assessment
Introduction
RNA interference (RNAi) is an endogenous, post-transcriptional gene silencing mechanism highly conserved among eukaryotes [1–3]. RNAi has been harnessed as a powerful tool for functional genomic studies [4–6] and genetic manipulation [7–9]. However, research in mammalian cells revealed that not only can siRNA/dsRNA trigger non-specific off-target effects including immune (interferon) response [10], competition between siRNA and miRNA [11] and downstream effects including changes in expression of non-target genes [12], but also specific off-target effects because of siRNA hybridizing with unintended mRNA resulting in degradation of unrelated transcripts [13,14]. These issues impede the utilization of RNAi. Non-specific off-target effects usually occur at high treatment doses or in organisms with a high sensitivity to dsRNA. The specific off-target effects occur due to the sequence similarity between siRNA and target mRNA. They have the potential for confounding genetic analysis and present serious safety risks for genetic manipulation [15,16] and therefore attract more attention.
The specific off-target effects of siRNA have been well studied because siRNA is used as an RNAi trigger in medical applications. Through expression profiling analyses, Jackson, et al. [14] showed that as few as 11 contiguous nucleotides complementing with unintended transcripts are sufficient to induce knockdown of off-target genes. In D. melanogaster embryos, siRNA-directed ribonucleoprotein complex (RISC) uses as few as nine contiguous complementary nucleotides on the 3ʹ end of siRNA guide strand for binding to off-target mRNAs and induces their degradation [17]. Moreover, similar to miRNA, siRNA seed region (5ʹ end of guide strand between 2 and 8 nt) can effectively suppress translation of mRNAs opening the door for potential off-target effects [18–20]. Bioinformatics algorithms that filter potential off-target and non-target effects (or cross-species off-target effects, dsRNA inducing silencing genes in non-target organisms) have been developed [18,21,22].
Though dsRNA triggers RNAi after they are processed to siRNA, which induces silencing, because of the additional steps in processing and length, dsRNAs could have additional potential non-target effects [23,24]. For example, dsRNA longer than 30 bp triggers a host immune response in mammals [7,25], that is why it is not suitable for medical applications. Kulkarni, et al. [26] evaluated the specificity of long dsRNAs in D. melanogaster by high-throughput screens and suggested that as few as 19 nt-long perfect, cross-hybridizing sequences in a dsRNA likely be capable of silencing non-target genes. However, Poreddy, et al. [27] found the dsRNA shared 83% sequence identity and contained a > 21 bp-long contiguous complementary nucleotides, yet showed no off-target knockdown in two lepidopteran insects, perhaps due to their insensitive to RNAi. These studies also showed that dsRNA with sequences with identity ≥96% can induce significant off-target gene knockdown.
However, dsRNA is the best choice for RNAi-based pest control due to its suitability for genetic engineering as a plant-incorporated protectant (PIP) and its higher efficiency in insects compared to siRNA [8,28–30]. The essential insect genes associated with development, growth, or reproduction are suppressed by dsRNA, which can result in severe larval or pupal deformity and mortality [9,31,32]. RNAi has the potential to contribute to pest management to replace traditional chemical pesticides [33,34]. Indeed, transgenic pest-resistant crops expressing pesticidal dsRNA have already been approved for commercial use [35,36], and spray-induced gene silencing (SIGS) is being studied as an alternative method for delivering dsRNA to pests [37,38]. For applications of RNAi in pest management, non-target effects are considered very important to protect ecosystem services. However, uncertainties concerning dsRNA non-target risks present serious impediments to the broader development of dsRNA pesticides and their widespread use, particularly given the potential for silencing in non-target organisms, presenting a serious ecological risk [15,35]. Here, we investigated rules governing dsRNA specificity and provide information for designing specific and highly efficient dsRNA for use in pest management. Since off-target effects are elicited meanly from a complementary base pairing between siRNA derived from dsRNA and mRNA, we pursued this issue mainly by studying the correlation between dsRNA complementarity with target gene and RNAi knockdown efficiency.
Results
Off-target knockdown by dsRNA is correlated with sequence identity
Since the nucleotide matching of mRNA with siRNA derived from dsRNA or other sources is necessary to trigger RNAi, we investigated how the degree of sequence identity affects knockdown efficiency. First, we treated T. castaneum fifth instar larvae with a special dsRNA targeting CYP6BQ6, a member of the cytochrome P450 superfamily. The off-target occurrence was evaluated by checking the knockdown efficiency on homologous CYP genes with higher identity to the dsRNA sequence (>45%). The results showed that dsCYP6BQ6 silenced nine out of the 54 genes tested – CYP6BQ6 itself and eight off-target genes, all sharing ≥68% nucleotide sequence identity with dsCYP6BQ6 (Fig. 1A). We observed a modest positive correlation between knockdown efficiency and degree of sequence identity (Spearman correlation r = 0.6749 calculated with all the genes with ˃60% identity). These results suggest that nucleotide sequence identity between dsRNA and potential target genes contributes to knockdown efficiency, but the r- value indicated that there might be some other influencing factors.
Figure 1.
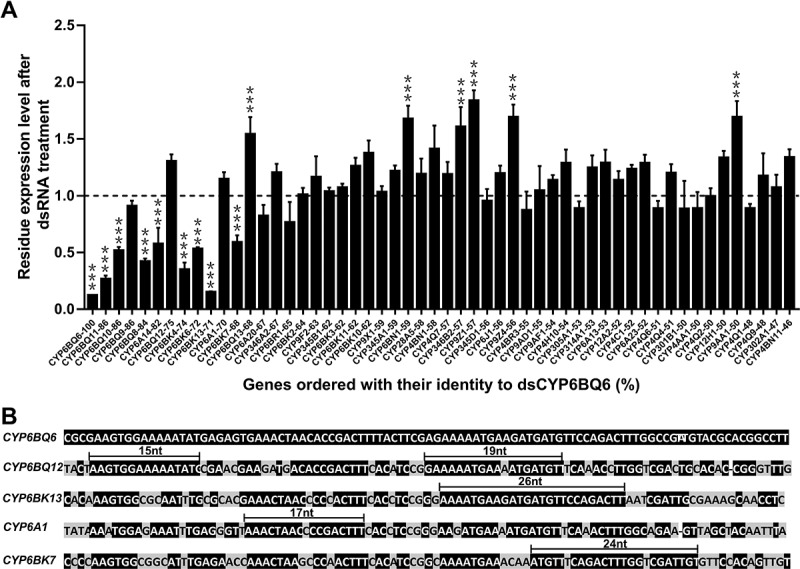
Knockdown efficiency of genes in T. castaneum triggered by 100 bp dsCYP6BQ6. (A) The number connected with a dash to the gene name represents the identity between the gene fragment and dsCYP6BQ6. The dsEGFP of the same length was used as control. Values are presented as mean±SE, n = 4 (*, p < 0.05; **, p < 0.01; ***, p < 0.001). The ‘n = 4’ represents four biological repeats, each biological repeat contains 10 larval individuals for RNAi experiments. (B) Alignments showing the complementarity between dsCYP6BQ6 and the corresponding area of the extremely sensitive or insensitive genes, CYP6BQ12, CYP6BK13, CYP6A1 and CYP6BK7, with the longest fragments of perfectly or almost perfectly matching sequence are indicated
Indeed, we found that two genes unexpectedly showed extreme sensitivity to RNAi irrespective of sequence identity between the dsRNA and the corresponding target gene fragment. CYP6BK13 and CYP6BK7, which share only 71% and 68% identity with dsCYP6BQ6, showed significant knockdown. Further sequence analysis found that CYP6BK13 and CYP6BK7 both possess long stretches with high sequence identity to dsCYP6BQ6 (Fig. 1B). For instance, CYP6BK13 contains 26 bp of contiguous matching bases. Since siRNAs generally range in length between 20 and 25 nt, this dsRNA is likely capable of producing 100% complementary siRNAs. CYP6BK7 has a 24 bp stretch of sequence similarity with just two single mismatched bases and thus is capable of generating siRNAs with >90% complementarity. All other tested genes that had lower sequence identity to full-length dsCYP6BQ6 and lacked long stretches of the complementary sequence showed insensitivity to RNAi. These results suggest that the length of the contiguous matching sequence, or partial sequence identity, could also be a key factor influencing the off-target effect of dsRNAs.
Otherwise, among the six genes shared high identity with dsCYP6BQ6 (>80%) we also found one unexpectedly refractory gene CYP6BQ9. When checking the expression level of the six genes, we found that the only unsusceptible gene CYP6BQ9 had the lowest expression level (Table S1). Thus, some other factors may influence the off-target effect of dsRNAs, including gene expression level and renewing rate of gene expression products. Previous reports have shown that genes for different functions differed in their susceptibility to RNAi [7].
Finally, some non-target genes were upregulated as generally found in RNAi experiments [27,39,40]. This phenomenon might reflect the dynamics of gene expression. Sure, some regulations might mediate by dsRNA treatment, but the reason might be diverse. We speculate compensation effect could be one for this experiment, because different CYP genes we checked may have a combined effect on some function, such as metabolism. Thus, when a gene is silenced, the others of the same function will be upregulated for metabolism compensation.
Mutational analysis uncovers factors governing dsRNA knockdown efficiency
To further understand how the sequence identity relates to knockdown efficiency, ruling out the interference influence mediated by different target genes, we used random mutagenesis to generate a series of dsRNAs with different identities to interfere the same target gene. Five different genes were selected for replications. The results showed that dsRNAs with 100% sequence identity to target genes induced 82% to 94% knockdown of target gene expression (Fig. 2A–E). However, as the sequence identity between dsRNA and target gene decreased, the knockdown efficiency also decreased, reaching zero when the sequence identity dipped below 80% (Fig. 2A–E & Table S2). The difference was found only in the shape of the index curves for different target genes (Fig. 2F). The most susceptible gene we tested, CYP4G7, was efficiently silenced by dsRNAs with >80% sequence identity, but knockdown efficiency dropped sharply to zero when dsRNA sequence identity dipped below 80% (Fig. 2A). In contrast, the most unsusceptible gene, CYP6BQ6, showing a more gradual decline in knockdown efficiency as dsRNA sequence identity decreased to ≤80% (Fig. 2E).
Figure 2.
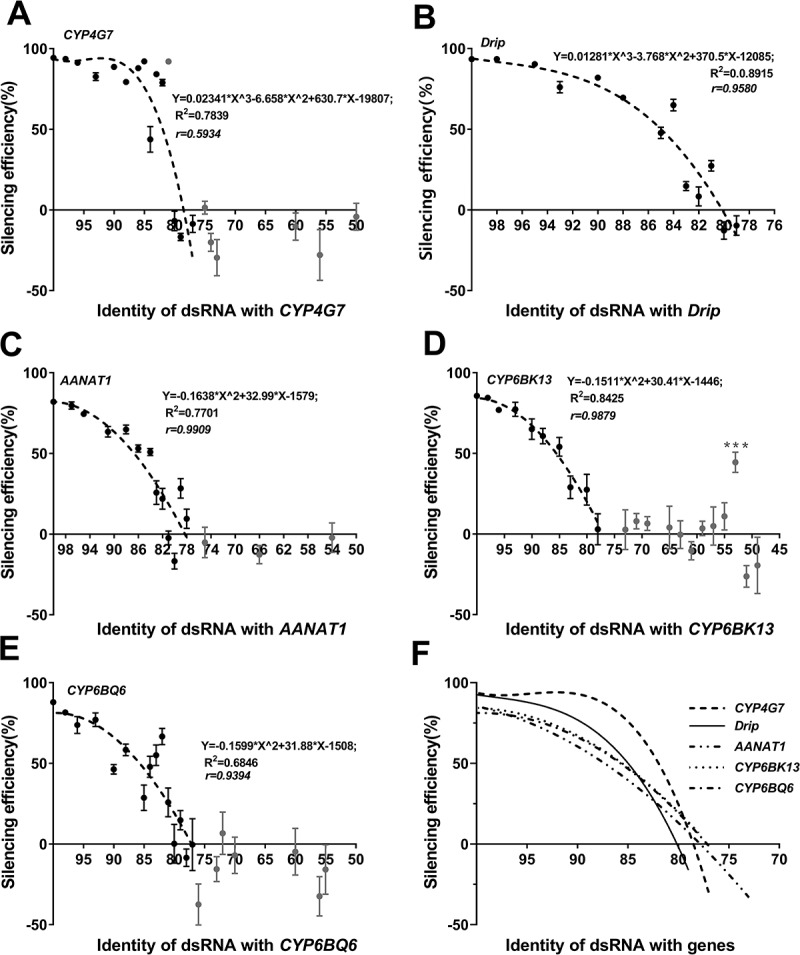
Knockdown efficiency of five genes in fifth instar T. castaneum larvae triggered by a series dsRNAs with varied identity. (A) CYP4G7; (B) Drip: D. melanogaster integral protein homologous; (C) AANAT1: Arylalkylamine N-acetyltransferase 1; (D) CYP6BK13; and (E) CYP6BQ6. The expression levels of these genes were 600, 272, 246, 189 and 54 times that of CPR18, respectively (Chen, et al., under review). The per cent depletions are presented as mean±SE, n = 4 (*, p < 0.05; **, p < 0.01; ***, p < 0.001). Bold grey dots (dsRNA identity <77%) were excluded from the curve modulations. (F) Sketch map used to compare the five index curves obtained
We also found two exceptions that dsRNAs can moderately silence genes with which they share low overall sequence identity (<80%) but possess a long stretch of sequence identity. DsCYP6BK13-53 sharing only 53% overall identity with CYP6BK13 caused 44.6% knockdown of target mRNA (Fig. 2D). DsAANAT1-79 sharing 79% overall identity with AANAT1, caused 28.3% knockdown of target mRNA (Fig. 2C). Sequence analysis showed that the randomly mutated bases were not distributed evenly in these two dsRNAs. DsCYP6BK13-53 possessed a 36 bp stretch of contiguous matching bases, and DsAANAT1-79 possessed a 26 bp stretch of almost perfectly matching bases. These data reproved that partial sequence identity could also be a key factor in the off-target effect of dsRNAs.
Determination of minimal contiguous sequence matching needed for efficient knockdown
To calibrate the minimal contiguous sequence matching requirements for silencing, we selected one of the most susceptible gene CYP4Q7 as target gene with another gene CYP6BK13 with medium susceptibility as replication and synthesized a series of chimeric 100 bp dsRNAs with target sequence stretches of various lengths inserted into an EGFP dsRNA (Fig. 3A). In this experiment, we tested different lengths of contiguously matching sequence (from 7 to 23 bp) and did not test data points of 8, 9 and 10 bp for simplifying the experiment, because we have determined the region for curve turning in the preparing experiment and these points were far from it. The RNAi experiments revealed a strong correlation between the length of a continuously complementary sequence and knockdown efficiency of target gene expression (R = 0.9273 and 0.9833), with the data fitting near-perfect logarithmic curves (Fig. 3B). Chimeric dsRNAs containing fewer than 15 bp of the continuously complementary sequence were not sufficient to trigger obvious RNAi effect even for the highly susceptible target gene, but knockdown efficiency increased sharply when the continuously complementary sequence exceeded 15 bp and reached high levels when the sequence identity was over 16 bp. The highly susceptible CYP4Q7 exhibited maximum knockdown efficiency at about 91%, while the medium susceptible CYP6BK13 showed lower knockdown efficiency at about 64%. All these results suggest that >15 bp or ≥16 bp of contiguously matching bases should be sufficient for dsRNAs to trigger RNAi (Table S3).
Figure 3.
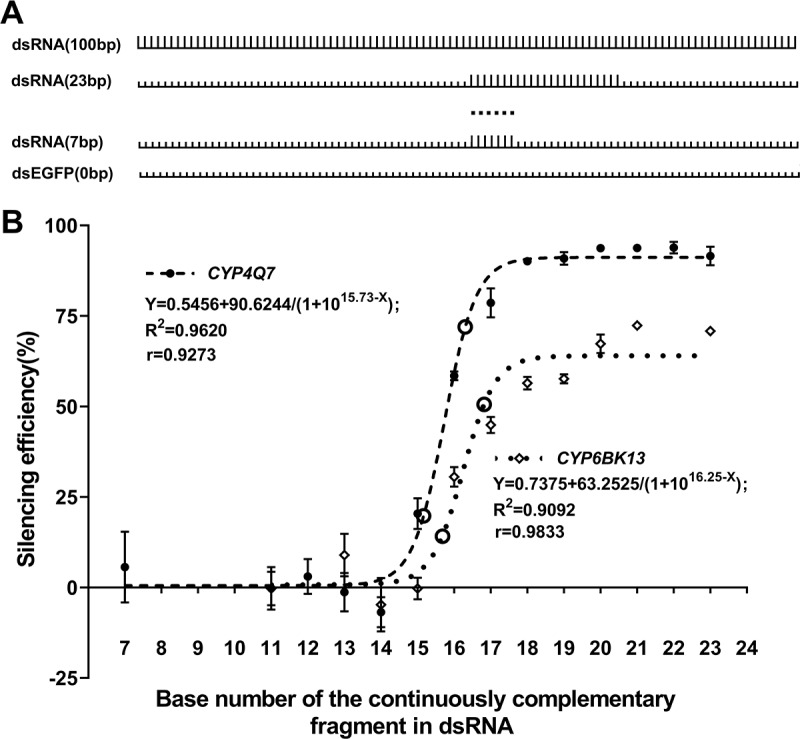
The scatter diagram showing knockdown efficiencies in T. castaneum triggered by a series of chimeric dsRNA containing a varied length fragment of contiguous matching bases. (A) Design of model of chimeric dsRNA. The short bars on the line represents EGFP sequence, and the tall bars on the line represent the target sequence. Numbers in the brackets represent the length of the target sequence. (B) Knockdown efficiency of CYP4Q7 and CYP6BK13 triggered by 100 bp chimeric dsRNA. Expression levels of these two genes were 3142.8 and 188.7 times that of CPR18, respectively. The circle represents the location of coordinate points with the largest slope change. Coordinate points (bp length, per cent depletion) for CYP4Q7 were (15.2, 19.8) and (16.3, 72.0), which were calculated using the formula deriv(derivn(0.5456 + 90.6244/(1 + 1015.73-x), x, 2), x) = 0. For CYP6BK13, they were (15.7, 14.2) and (16.8, 50.6) and the formula was deriv(derivn(0.7375 + 63.2525/(1 + 1016.25-x), x, 2), x) = 0
Defining minimal length criteria for imperfectly matched dsRNAs to induce off-targets
To more precisely define the sequence parameters governing RNAi triggering capacity, we test whether repeated small segments of contiguous bases linked by mismatches could efficiently trigger the RNAi effect. We selected four different target genes as target replications and synthesized a series of 100 bp dsRNAs with single-base mutations interspersed at different intervals (3 to 6 bp). RNAi experiments found that for the high susceptible gene CYP4Q7, dsRNAs containing ≥5 bp repeated stretches of contiguously matching bases linked by single mismatched bases were sufficient to trigger silencing. But for the medium susceptible genes, efficient dsRNA contained at least 6 bp of the repeated stretches (Table 1).
Table 1.
Knockdown efficiency of different genes in T. castaneum triggered by dsRNA containing evenly distributed single mismatching bases
| Gene name |
Expression levelsa | Knockdown efficiency (%) of the dsRNA with varied matching nucleotide ratio |
|||
|---|---|---|---|---|---|
| 6:1 | 5:1 | 4:1 | 3:1 | ||
| CYP4Q7 | 3142.8 | 40.6 ± 3.8b | 27.7±b | −5.3 ± 7.8 | −42.9 ± 7.5 |
| Drip | 272.4 | 21.8 ± 3.1b | 0.32 ± 3.9 | - | - |
| AANAT1 | 245.6 | 37.9 ± 2.4b | 0.8 ± 2.2 | −15.4 ± 7.2 | −37.0 ± 13.8 |
| CYP6BK13 | 188.7 | 18.2 ± 0.6b | 2.9 ± 9.5 | 6.0 ± 7.1 | 0.51 ± 7.4 |
aExpression levels were calculated using CPR18 as a reference gene.
bSignificant knockdown.
We then selected high susceptible target gene, CYP4Q7, and synthesized a series of 100 bp dsRNAs with two sequential nucleotide mismatches interspersed at different intervals between segments of perfectly matched sequences (Fig. 4A). The experiment data indicated that repeating segments of 8 contiguously matched bases interspersed with 2 bp long mismatches were sufficient to trigger silencing (Fig. 4B & Table S4).
Figure 4.
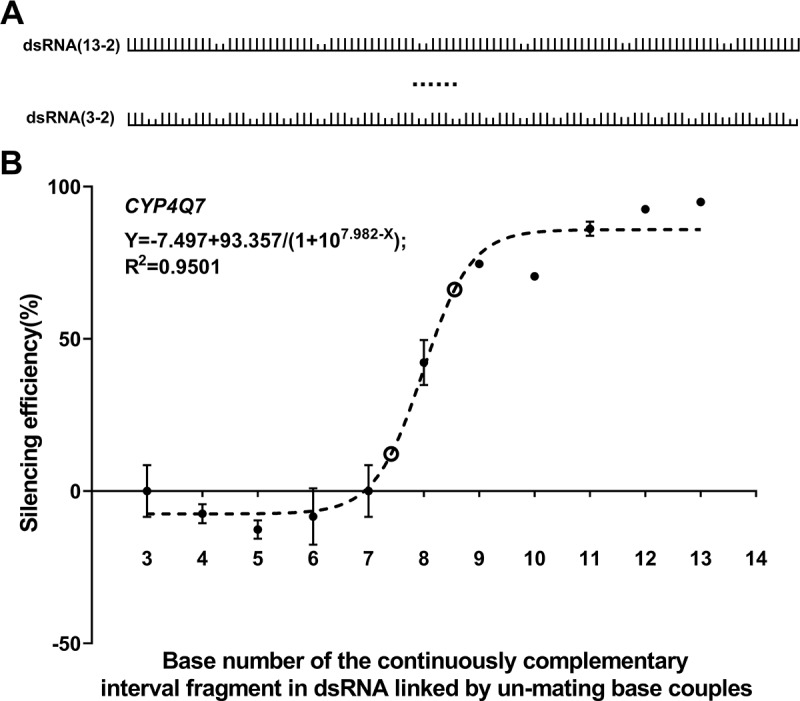
Knockdown induced by dsCYP4Q7 containing evenly distributed mismatching base couples in T. castaneum. (A) The distribution model of the mismatching base couples in the sequence. The tall bars standing on the line represent matching bases between the target gene and dsRNA, and the adjoin short bars standing on the line represent mismatching base couples. (B) Knockdown of CYP4Q7 gene triggered by dsRNA containing evenly distributed mismating base couples at varying intervals. The circle represents the location of coordinate points with the largest slope change. Coordinate points (bp length, per cent depletion) was (7.4, 12.2) and (8.6, 66.2), calculated using the formula deriv(derivn(−7.497 + 93.357/(1 + 107.982-x), x, 2), x) = 0
Since mutations do not distribute evenly in natural genes, we examined the dsRNAs with natural sequences and their random mutations for their off-target transcript knockdown and gene complementarity. The results found that all the dsRNAs able to induce obvious knockdown (>20%) of the genes with lower sequence identity (53%~83%) contained either ≥16 bp perfectly matching segments or ≥19 bp almost perfectly matching segments consisting of ≥5 bp matches linked by single mismatches and/or ≥8 bp matches linked by mismatched couplets (Table 2). Conversely, all the dsRNAs that were ineffective at silencing (≤20% knockdown) of the genes with higher sequence identity (86%~77%) contained ≤16 bp contiguous matching segments and ≤26 bp of almost perfectly matching segments or had a refractory target gene with extremely low expression levels (Table 3).
Table 2.
Comparison between sequence matching (dsRNA with target gene) and knockdown efficiency for dsRNAs inducing significant RNAi in T. castaneum.
| Interaction couple | Gene expression level |
dsRNA/ gene identity |
Longest segment |
Per cent knockdown | |
|---|---|---|---|---|---|
| Contiguous matching |
Imperfect matching |
||||
| dsMutant/CYP6BK13 | 188.7 | 53 | 36 | 4 | 44.5 |
| dsMutant/CYP6BQ6 | 53.7 | 82 | 29 | 19 | 66.6 |
| dsCYP6BQ6/CYP6BK13 | 188.7 | 71 | 26 | 9 | 84.9 |
| dsMutant/CYP4G7 | 600.1 | 83 | 26 | 11 | 84.1 |
| dsMutant/CYP4G7 | 600.1 | 81 | 26 | 14 | 92.0 |
| dsMutant/CYP4G7 | 600.1 | 82 | 22 | 12 | 78.9 |
| dsCYP6BQ6/CYP6BK4 | 75.1 | 74 | 18 | 36 | 65.1 |
| dsMutant/AANAT1 | 245.6 | 82 | 16 | 23 | 21.9 |
| dsCYP6BQ6/CYP6A14 | 212.7 | 82 | 15 | 50 | 42.6 |
| dsMutant/CYP6BQ6 | 53.7 | 83 | 14 | 36 | 54.9 |
| dsCYP6BQ6/CYP6BK6 | 67.5 | 72 | 10 | 30 | 46.9 |
| dsMutant/CYP6BK13 | 188.7 | 80 | 11 | 26 | 27.5 |
| dsMutant/AANAT1 | 245.6 | 79 | 11 | 26 | 28.3 |
| dsMutant/Drip | 272.4 | 81 | 13 | 24 | 27.3 |
| dsCYP6BQ6/CYP6BK7 | 229.8 | 68 | 10 | 24 | 41.2 |
| dsMutant/CYP6BQ6 | 53.7 | 81 | 12 | 20 | 25.8 |
| dsMutant/CYP6BK13 | 188.7 | 83 | 11 | 19 | 28.9 |
Table 3.
Comparison between sequence matching (dsRNA with target gene) and knockdown efficiency for dsRNAs inducing insignificant RNAi in T. castaneum.
| Interaction couple | Gene expression level |
dsRNA/gene identity | Longest segment |
Per cent knockdown | |
|---|---|---|---|---|---|
| Contiguous matching |
Imperfect matching |
||||
| dsCYP6BQ6/CYP6BQ9 | 6.3 | 86 | 22 | 74 | 9.2 |
| dsMutant/Drip | 272.4 | 79 | 8 | 8 | <0 |
| dsMutant/Drip | 272.4 | 80 | 10 | 15 | <0 |
| dsMutant/AANAT1 | 245.6 | 78 | 11 | 15 | 9.5 |
| dsMutant/CYP6BK13 | 188.7 | 78 | 9 | 16 | 2.9 |
| dsMutant/AANAT1 | 245.6 | 80 | 11 | 18 | <0 |
| dsMutant/CYP4G7 | 600.1 | 80 | 15 | 18 | <0 |
| dsMutant/CYP4G7 | 600.1 | 79 | 13 | 20 | <0 |
| dsMutant/CYP6BQ6 | 53.7 | 79 | 14 | 21 | 14.7 |
| dsMutant/CYP6BQ6 | 53.7 | 80 | 16 | 21 | 0.2 |
| dsMutant/CYP4G7 | 600.1 | 77 | 10 | 22 | <0 |
| dsMutant/Drip | 272.4 | 82 | 11 | 23 | 8.3 |
| dsMutant/CYP6BQ6 | 53.7 | 77 | 11 | 23 | <0 |
| dsMutant/CYP6BQ6 | 53.7 | 78 | 13 | 23 | <0 |
| dsMutant/CYP6BQ6 | 53.7 | 85 | 13 | 25 | 20.1 |
| dsMutant/AANAT1 | 245.6 | 81 | 9 | 26 | <0 |
| dsMutant/Drip | 272.4 | 83 | 10 | 26 | 14.8 |
Taken together, these data established minimal length criteria for sequence matching of dsRNAs with the off-target genes for triggering efficient RNAi effect. For dsRNAs containing contiguous segments of perfectly matching sequence, ≥16 bp stretches of sequence identity are needed to trigger off-target effects (with 15 bp being marginal). However, for dsRNAs with almost perfectly matching sequence to target genes, >26 bp stretches (single mismatches inserted between ≥5 bp matching segments or mismatched couplets inserted between ≥8 bp matching segments) were sufficient to trigger obvious off-target effects. When the length dropped below 19 bp, the knockdown possibility lost. dsRNAs with 19–26 bp of almost perfectly matching sequences could sometime trigger low levels of knockdown, which is generally insufficient to trigger phenotypical off-target effects [41]. Thus, for almost perfectly matching sequences, we consider 19–26 bp to be in the ‘warning zone’.
Evaluation of dsRNA off-target effects between insect species
To determine whether dsRNA non-target effect in other species follows the same rules for off-target effect in the same insect as those we established with T. castaneum, we synthesized dsRNAs targeting elongation factor 1 alpha (EF1) homologs in various insect species (Chilo suppressalis, Helicoverpa armigera, Spodoptera litura, Tribolium castaneum, Drosophila melanogaster, and Locusta migratoria) and treated insect larvae with conspecific dsRNA to determine target sensitivity, and with C. suppressalis dsEF1 (dsCsEF1) to determine off-target effects. EF1 homologs in all these insects share high sequence identity (83%~100%) with dsCsEF1 (Table 4) and possess long enough sequence fragments of perfectly or almost perfectly matching with dsCsEF1, so according to established off-target rules, we predicted that it would trigger significant RNAi effects in non-target insects. Indeed, the degree of target gene silencing showed good agreement with the predicted non-target effects. In T. castaneum, D. melanogaster cells, and L. migratoria significant RNAi effects of dsCsEF1 were observed. However, lepidopteran insects (C. suppressalis, H. armigera, S. litura) showed little to no silencing, either with perfectly or partially matched dsEF1. As lepidopterans have previously exhibited insensitivity to RNAi [7,42], it is likely that lepidopterans are refractory species that are difficult to target by RNAi.
Table 4.
Non-target effects in six insect species triggered by a dsRNA targeting Chilo suppressalis EF1.
| Insects | Target region identity with CsEF1 | Longest contiguous matching segment | Longest imperfect matching segment | Knockdown efficiency (%)a by perfectly matching dsEF1 | Knockdown of EF1 in non-target (%)b by dsCsEF1 | Off-target knockdown potential (%)c |
|---|---|---|---|---|---|---|
| Chilo suppressalis | 100 | 100 | - | 23.6 | - | - |
| Helicoverpa armigera | 94 | 35 | 83 | <0 | <0 | - |
| Spodoptera litura | 84 | 14 | 26 | <0 | <0 | - |
| Tribolium castaneum | 91 | 32 | 47 | 96.2 | 95.7 | 99.5 |
| Drosophila melanogaster | 90 | 26 | 74 | 84.9 | 62.4 | 73.5 |
| Locusta migratoria | 83 | 14 | 41 | 91.2 | 46.4 | 50.9 |
aKnockdown efficiency (%) was calculated by comparing EF1 mRNA levels in dsEF1 treated with those in control insects treated with dsEGFP.
bKnockdown of EF1 in non-target insects (%) was calculated by comparing EF1 mRNA levels in non-target insects treated with dsCsEF1 with those treated with dsEGFP.
cNon-target knockdown potential (%) was calculated by non-target knockdown efficiency/target knockdown efficiency.
In the RNAi experiments, each sample contains four biological repeats and each biological repeat contains five larval individuals for Chilo suppressalis, Helicoverpa armigera, Spodoptera litura, and Locusta migratoria.
Finally, since the ultimate goal is to use dsRNA to control pest populations, we further evaluated our capacity to predict dsRNA non-target effects using phenotypic effects as readout. We tested a plant-incorporated insecticide dsDvSnf7 targeting the maize pest Diabrotica virgifera virgifera for dsRNA induced non-target effects in T. castaneum with the dsCsEF1 as a positive control. The 240 bp target region of TcSnf7 and DvSnf7 share only 72% homology (Fig. 5A), which is lower than our predicted threshold (>80%) for effective silencing of non-target genes. Moreover, the longest segment of the almost perfectly matching sequence is 20 bp, which is in the ‘warning zone’ and below the critical length (≥26 bp) required for efficient silencing of the target gene. The results showed that T. castaneum Snf7 was highly sensitive to RNAi, with dsTcSnf7 inducing 83.6% transcript knockdown and 100% larval mortality in 7 days (Fig. 5C). In contrast, dsDvSnf7 induced only 24.2% non-target gene knockdown and failed to cause significant mortality (Fig. 5B). Thus, even in a related coleopteran species with high susceptibility to RNAi, dsDvSnf7 induced only a low level of transcript depletion and no obvious phenotypic change, indicating that our prediction is reliable and this dsRNA should be safe for other organisms. On the other hand, the positive control dsCsEF1, which shares 91% homology with T. castaneum EF1, was able to trigger 95.7% transcript depletion and 100% mortality, similar to dsTcEF1 (Fig. 5D).
Figure 5.
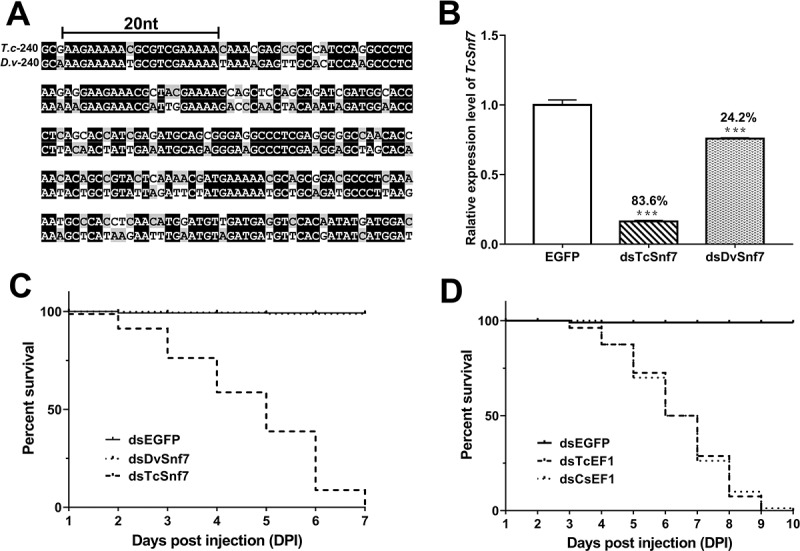
The non-target effects in T. castaneum induced by dsRNA synthesized using Diabrotica virgifera virgifera SNF7 gene fragment as a template (dsDvSNF7). (A) Alignment of sequences of SNF7 homologs from T. castaneum and D. virgifera. (B) The expression depletion of T. castaneum SNF7 triggered by dsDvSNF7 and dsTcSNF7. (C) Mortality of T. castaneum induced by dsDvSNF7 and dsTcSNF7 (Tc, T. castaneum; Dv, D. virgifera). (D) Mortality of T. castaneum induced by dsCsEF1 and dsTcEF1. Mean±SE (n = 4) are presented. *, p < 0.05; **, p < 0.01; ***, p < 0.001)
Taken together, all these results above demonstrate that the identity between dsRNA and non-target mRNA determines the occurrence of both off-target and non-target RNAi, and we can use these rules to design dsRNAs with different specificities to control non-target phenotypic effects.
Discussion
Our studies established clear rules that govern specific off-target effects by dsRNAs. We found that 100 bp dsRNAs containing ≥16 bp contiguous sequence matching with the off-target gene could trigger significant silencing. Previous work demonstrated that for siRNAs, ≥7 bp of contiguous sequence matching could suppress the translation of mRNA or degrade transcripts [7,13,17,26,43,44], while for miRNAs the minimal matching sequence was found to be ≥12 bp [45,46]. Thus, dsRNAs, which are much longer than either siRNAs or miRNAs, appear to require a longer contiguous matching sequence for efficient silencing. However, we found that unlike siRNA and miRNA, dsRNAs with long enough segments of imperfectly (almost perfectly) matching sequences could also trigger significant silencing, provided that the mismatch interval was no longer than 2 bp. DsRNAs with ≥5 bp repeated segments of contiguous matches linked by single mismatches, or ≥8 bp linked by mismatch couplets, were sufficient for inducing RNAi response. The exact minimal length might depend on the number of mismatches, but since this is difficult to test due to a large number of possible combinations, we used experimental data obtained with the dsRNAs prepared from natural or randomly mutated DNA templates to identify the minimal length of almost perfectly matched segments needed for efficient silencing. As expected, we determined this length to be ≥26 bp, with variable silencing efficiency between 19 and 26 bp (warning zone). For dsRNAs in the ‘warning zone’, accurate prediction of off-target effects will likely require further analysis of the influence of their mismatch intervals.
Obviously, though dsRNA and miRNA must be processed to siRNA to trigger RNAi, their off-target needs different numbers of contiguous matching bases in sequence. To make clear the mechanism underlying, it seems necessary to make sure what kinds of siRNA could be derived from a given dsRNA, which of them could function and there is any joint action. We tried to analyse our data by evaluating possible efficient siRNAs (Table S5). The derived siRNAs were supposed as 21 bp in length, and theoretically, 80 different siRNAs could be derived from 100 bp dsRNA. The siRNAs with the sequences containing ≥15 bp contiguous matching bases or ≤2 mismatching bases were thought as efficient according to our results obtained above. The analysis found that 100 bp dsRNA producing >18 different efficient siRNAs was certainly efficient, and those producing <4 was certainly inefficient, with a wide range of uncertainty (4 ~ 18). This work surely calls for further studies with more information, more reasonable parameters, and bioinformatics analysis. Recently, a sequence complementarity-based approach for evaluating off-target transcript knockdown has been reported [47].
Apart from sequence matching, factors concerning with RNAi sensitivity among species, tissues, and genes, such as differences in core RNAi machinery composition [48,49], amplification of RNAi signals in vivo [50], activity of dsRNA-degrading nucleases [51–53], dsRNA uptake and intracellular transport [54], and gene expression levels (Chen et al., 2020, under review), all may contribute to off-target RNAi in the same organism and non-target RNAi among different species. If the organism species or genes are refractory to RNAi, no off-target RNAi will happen. It was found in our study that off-target RNAi did not occur in lepidopteran insects tested; nevertheless, the identity between dsRNA and genes were positive (Table 4). Similarly for the gene CYP6BQ9 (Table 3), where little off-target RNAi happened (9% knockdown) because the gene expression level was very low, whereas dsRNA shared 86% identity with it and contained 22 bp perfect matching stretches and 74 bp almost perfectly matching stretches. Therefore, occurring probability of off-target effect mediated by dsRNA is highly decided by the sensitivity of organisms and related genes to RNAi and the off-target rules.
Finally, though not many insect species were tested for evaluation of the possible variation, which calls for further studies, our experiments in several species of insects showed that the dsRNA specificity rules we established in T. castaneum apply to non-target species, and the conserved RNAi mechanism among eukaryotes might fund the base for this situation. Thus, dsRNA used in the field needs to take cross-species off-target effects into account. At present, there is no perfect evaluation system for insecticide dsRNAs. Therefore, evaluation systems for understanding of the non-target effects and potential ecotoxicology of dsRNA treatments are urgently needed. Our study opens the way for the rational design of dsRNA for pest control. These studies also provide information that could help in the development of tests for the evaluation of dsRNAs off-target effects in non-target species. The information from these studies also could help to design dsRNAs that specifically target one or multiple genes in a single pest species or a closely related pest complex. Overall, our findings facilitate better interpretation of RNAi effects and the safe use of dsRNA agents in pest management.
Materials and Methods
Insects rearing and cells culture
The Tribolium castaneum was reared on whole wheat flour and dry yeast powder at 31 ± 1°C with 40% relative humidity [55]. The Locusta migratoria nymphs were reared on fresh wheat sprouts at 28 ± 1°C, 14:10 h (Light: Dark) photoperiod, 50% relative humidity [56]. The Spodoptera litura larvae were reared on an artificial diet [51] at 14:10 h (Light: Dark) photoperiod and 70 ± 10% relative humidity (RH) at 27 ± 1°C. The Chilo suppressalis larvae were reared on potted rice seedlings in a glass chamber at 28 ± 1°C and a 16:8 h (light: dark) photoperiod [57]. The Helicoverpa armigera larvae were reared on an artificial diet at 27 ± 1°C, 70 ± 10% relative humidity, 14:10 h (Light: Dark) photoperiod. The D. melanogaster S2 cells maintained in an incubator at 27°C in serum-free insect cell culture medium (HyClone, Logan, Utah, USA) and 10% heat-inactivated foetal bovine serum (HyClone, Logan, Utah, USA).
Genes sampling and sequence identity calculation
We selected Cytochrome P450 (CYP) gene superfamily for analysis of dsRNA off-target effect in T. castaneum. The large number of CYP family members with higher identity should make it easy to find the occurrence of dsRNA off-target effect. The members of the CYP gene family in T. castaneum have been identified previously [58]. The sequences of CYP genes were downloaded from the NCBI database and confirmed by PCR cloning and sequencing. Then, based on phylogenetic analysis and sequence alignment of the verified gene sequences, we selected the genes with high similarity to most others as target genes and 100 bp well-conserved gene fragments as the targeting regions.
We selected EF1 and Snf7 as target gene to evaluate off-target occurrence rules in different insects. EF1 is a housekeeping gene well expressed in different insects [59]. The sequences of EF1 genes from different insects were downloaded from the NCBI database and also confirmed by PCR cloning and sequencing. The primers were listed in Table S6. Snf7 in the maize pest Diabrotica virgifera virgifera is the target gene for the first commercially used plant-incorporated insecticide dsRNA, dsDvSnf7. The target region sequence of dsDvSnf7 was download from the published paper [60] and synthesized by GENEWIZ (Suzhou, China). Snf7 in T. castaneum was cloned with the primers designed with Primer Premier 5, which were listed in Table S6.
We calculated dsRNA sequence identities to genes with only the corresponding target region by using DNAMAN 8.0 following with sequence alignments by BioEdit 7.0.
RNA extraction, cDNA synthesis and RT-qPCR
The total RNA was extracted using Trizol (Thermo Fisher Scientific, Waltham, Mass, USA). One μg total RNA was used to prepare cDNA for determining relative mRNA levels using RT-qPCR with QuantStudio™ 6 (Thermo Fisher Scientific, USA). RT-qPCR reaction mixture contained 10 μL of TB Green Premix Ex Taq (Tli RNaseH Plus), 2 μL cDNA, 0.4 μL of each forward and reverse primer (10 μM), 0.4 μL of ROX dye II, and 6.8 μL nuclease-free water to a total volume of 20 μL. The thermal cycling protocol of RT-qPCR was: 95°C for 30 s, 40 cycles of 95°C for 5 s, and 60°C for 34 s. RT-qPCR data analysis was performed by the 2–ΔΔCT method. The primers used for RT-qPCR are listed in Table S6.
Because a lot of RT-qPCR experiments are needed to do in this paper, we firstly screen reliable reference genes with stable expression under our experimental conditions, injecting 5th instar larvae of red flour beetle with 100 bp different dsRNAs and checking gene expression level in their whole body, midgut, and carcase without midgut in 36 h. Thus, we evaluate the stability of the candidate reference genes with the larva treated with dsEGFP, dsDrip, dsCYP4G7, dsCYP4Q7, dsCYP6BK13, dsCYP6BQ6, and dsAANAT1 as various dsRNA treatments, with midgut and carcase without midgut as tissue treatments. Treated samples were collected in 36 h, the total RNA was extracted and the stability of eight candidate reference genes (Rp49, RpS3, EF1, Actin, GAPDH, SYN1, RPS18, and RPL13a) were evaluated by geNorm and NormFinder methods. RNA quality was checked by a NanoDrop ND-1000 spectrophotometer with the criterion values of OD260/OD280 between 1.80 and 2.00. The primers for RT-qPCR were selected with melting curves and amplification efficiency (E = 10^(−1/slope)-1) in accordance with the requirements (Table S7). The expression stability values (M) calculated by geNorm indicated that Rp49 and RpS3 were the best internal references with the minimum value 0.093244 (Fig. S1 & Table S8) and average pairwise variations V(2/3) 0.032 (<0.15 as shown in Figure S2). Another evaluation with NormFinder method indicated that Rp49 was the best internal reference gene with minimum value of 0.028 (Table S9). In addition, the best three reference genes (Rp49, EF1, and RpS3) selected by geNorm were all further verified by preparing RNAi experiments with randomly selected six genes. It was found that similar results were obtained when Rp49 and RpS3 were used as reference and variation was found when EF1 as reference (Fig. S3), indicating that both Rp49 and RpS3 were the suitable references. Due to lots of RNAi experiments and for saving time, only Rp49 was selected as the reference gene for T. castaneum. For other insects, we adopted the reported reference genes where the experiment is similar as ours, such as Rp49 for L. migratoria, Rp49 for D. melanogaster S2 cells, Actin for C. suppressalis and H. armigera, GAPDH for S. litura [51,56,61,62].
Synthesis of dsRNA, chimeric dsRNA and mutations
DNA template for dsRNA synthesize was amplified by PCR with cDNA and a pair of primers (Table S6) flanked by T7 promoter, and 2× Rapid Taq Master Mix (Vazyme, Nanjing, China). Thermal cycling protocol of PCR was: 95°C for 3 min, 34 cycles of 95°C for 30 s, 55°C for 30 s, and 72°C for 30 s, and 72°C for 10 min was used. The PCR products were purified using the Wizard® SV Gel and PCR Clean-Up System (Promega, Madison, WI, USA). dsRNAs were synthesized using a T7 RiboMAX system (Promega, Madison, WI, USA) following the manufacturer’s protocol. The quantity of synthesized dsRNA was measured using a NanoDrop ND-1000 spectrophotometer (Thermo Fisher Scientific, Waltham, Mass, USA), and kept at −80°C until further use.
Mutant dsRNAs were synthesized with the template sequences generated by a Python program named ‘change_function.py’. When we input a fragment of gene sequence and a number of mutant bases, the program will change the given gene sequence to a corresponding randomly mutated sequence with an expected designed identity. The mutated sequence fragments were synthesized at GENEWIZ (Suzhou, China) and cloned into plasmid pUC57. Then, the synthesized DNA templates were PCR amplified using T7 flanked primers and used as templates for synthesis of dsRNA. The chimeric dsRNA sequences were constructed by a gene-specific target sequence fragment with its both ends flanking by two pieces of EGFP sequence and marked as ‘EGFP-Target-EGFP’ (Fig. 3A). The mutated DNA template was synthesized by GENEWIZ (Suzhou, China).
RNAi methods
Based on the references and preparing experiment results, we designed 100 bp dsRNA targeting the coding region of a gene in our RNAi experiments. For Tribolium castaneum, 5th instar larvae were treated by injection with 200 ng dsRNA dissolved in 0.15 µL nuclease-free water. The control insects were treated with dsEGFP. Before injection, the insects were cleaned and placed on ice for 3–5 minutes to keep them comatose. The larvae were arranged in a row on slides, and the dsRNA solution was injected into sulcus between the second and third abdominal segments on the ventral side with a MicroSyringe Pump Controller (World Precision Instrument, USA) using a glass needle pulled from 4878 Glass Capillary (Sutter, USA) with the P-97 Flaming/Brown micropipette puller (Sutter, USA). The treated larvae were reared in 9 cm culture dishes with whole wheat flour and yeast powder. For L. migratoria, S. litura, and C. suppressalis, third instar nymphs or larvae were used as testing insects. Thus, 2 µg dsRNA in 1 μL solution was used for dsRNA injection using a sterile Hamilton™ 701 micro-injector (Hamilton, Switzerland). The dsRNA solution was injected into the abdomens of L. migratoria, S. litura, and C. suppressalis at a lateral site between the third and fourth abdomen segments.
For D. melanogaster S2 cell RNAi experiments, the cell 1 × 106 cells/mL were plated in each well of a sterilized 6-well plate. 60 µg dsRNA dissolved in 30 μL nuclease-free water was added to each well was added. After 1-h incubation, 2 mL medium mixed with 0.2 mL heat-inactivated foetal bovine serum was added to each well and the plates were incubated in a humidified incubator at 27°C for 36 h. Then, the total RNA was extracted using Trizol and mRNA expression levels were determined using RT-qPCR.
We performed preliminary experiments with T. castaneum, S. litura, L. migratoria, and D. melanogaster S2 cells to select the suitable observing time point by testing knockdown efficiency in a time course of 6, 12, 24, 36, 48, 72, and 96 h. In S. litura, we did not find any significant knockdown at all times tested. T. castaneum, L. migratoria, and D. melanogaster S2 cells showed the highest knockdown efficiency at 36 h after the application of dsRNA (Fig. S4 & Fig. S5 & Fig. S6). Thus, 36 h was designed as the observing time for our RNAi experiments.
We performed 3–4 biological replicates for each RNAi experiment. For insect samples, each biological replicate contained 5–10 larvae based on the size of insects. The relative mRNA levels were determined by RT-qPCR using the housekeeping gene Rp49 as internal normalization. The expression level of related genes was calculated as the times the mRNA levels over the gene CPR18, which is a stably lower expressed gene. The knockdown efficiency was calculated by the formula (expression level in the control group – expression level in the treated group)/expression level in control group ×100%.
Evaluation of the effect of dsRNA identity on off-target knockdown efficiency
For determination of off-target knockdown among genes with different sequence identities, dsRNA for a specific gene was synthesized and employed to knockdown expression of a group of related genes. For dsRNA could hardly complement the genes with little sequence identity and elicit off-target RNAi, the gene from a superfamily was selected to ensure there are enough homologous genes with high identity for off-target knockdown observation. We synthesized a 100 bp dsRNA specific for TcCYP6BQ6 from CYP supergene family and treated the fifth instar larvae of T. castaneum with this dsRNA. Then, we checked the knockdown efficiency of the target and 53 homologous genes of CYP family members with identity >45% by quantifying their mRNA levels before and after dsRNA treatment.
For eliminating the influence from different genes in the experiments described above, we also used a series of dsRNA of various identities to silence the same target gene. The sequence of a 100 bp fragment of the target gene was randomly mutated by a Python program ‘change_function.py’ to produce templets to synthesize a series of dsRNAs with a different identity to the target gene as described in Section ‘Synthesis of dsRNA, chimeric dsRNA and mutations’. For replications, we selected five genes with different expression levels as the targets to repeat this experiment.
Defining available sequence linkups for imperfectly matched dsRNAs to off-targets
Firstly, we identified the minimal length of a single fragment of contiguous matching sequence in the dsRNA needed for efficient RNAi. ‘‘In order to keep all test dsRNA with a varied contiguous matching sequence in the same molecular length, we used chimeric dsRNAs with the sequence mode ‘EGFP-Target-EGFP’ (see Fig. 3A). Secondly, we identified the minimal length of repeated contiguous matching sequences linked by single mismatching bases in the dsRNA, which could trigger efficient RNAi. For replications, we selected four genes as targets, synthesized dsRNA series for each gene by mutation of a single base with various intervals (3–6 contiguous matching bases). Thirdly, we identified the minimal length of repeated contiguous matching sequences linked by couple mismatching bases in the efficient dsRNA. Similarly, we selected a target gene sensitive to RNAi and synthesized dsRNA series for it by mutation of two neighbour bases with various intervals (3–13 contiguous matching bases, see Fig. 4A) for the identification RNAi experiment. All the results indicated that the efficient dsRNA contained longer repeated contiguous matching sequences linked by few mismatching bases. Thus, we named this kind of sequences as almost perfectly matching sequences.
After finding the valid composition of almost perfectly matching sequence for efficient dsRNA with artificially mutated dsRNA, we identified the shortest valid segments of the almost perfectly matched sequence by check the efficient dsRNA sharing relatively low identity with the acting gene and identified the longest invalid segments of the almost perfectly matched sequence by check the inefficient dsRNA sharing relatively high identity with the acting gene. We discriminated against the efficient dsRNAs with inefficient by 20% knockdown of the gene expression according to the preparing experiments where a turnover occurred. Basically, we used 80% to discriminate dsRNAs sharing high or low identity with the acting gene according to the results shown in Fig. 2. However, for data overlapping (±3%) to qualify the analysis, we sorted the inefficient dsRNAs sharing ≥77% identity and the efficient dsRNAs sharing ≤83% identity for analysis.
Evaluation of dsRNA off-target in non-target insects
For determination of dsRNA off-target effects in various insect species, we first selected elongation factor 1 alpha (EF1), a gene conserved among different insects, as target and six test insect species to determine off-target effects. We treated different insect larvae with conspecific dsRNA to determine target sensitivity and with C. suppressalis dsEF1 (dsCsEF1) to observe off-target effects. Then, we determined off-target effects in two coleopteran species using commercial dsDvSnf7 and with high off-target dsEF1 for comparison.
Statistical analysis
Correlation analysis of knockdown efficiency and dsRNA identities was performed by GraphPad Prism 7.0 (GraphPad Software Inc., La Jolla, CA, USA) using Spearman’s correlation coefficient. One-way ANOVA analysis followed by Dunnett’s multiple-comparisons test was performed by GraphPad Prism 7.0. Curve fitting was also processed by GraphPad Prism 7.0.
Supplementary Material
Funding Statement
The work was supported by the National Natural Science Foundation of China (31672053).
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Supplementary material
Supplemental data for this article can be accessed here.
References
- [1].Jin L, Song H, Tropea JE, et al. The molecular mechanism of dsRNA processing by a bacterial Dicer. Nucleic Acids Res. 2019. DOI: 10.1093/nar/gkz208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Gordon KHJ, Waterhouse PM.. RNAi for insect-proof plants. Nat Biotechnol. 2007;25:1231–1232. [DOI] [PubMed] [Google Scholar]
- [3].Tijsterman M, Plasterk RHA. Dicers at RISC: the mechanism of RNAi. Cell. 2004;117:1–3. [DOI] [PubMed] [Google Scholar]
- [4].Qiao HH, Wang F, Xu RG, et al. An efficient and multiple target transgenic RNAi technique with low toxicity in Drosophila. Nat Commun. 2018;9:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Boettcher M, McManus MT. Choosing the right tool for the job: rNAi, TALEN, or CRISPR. Molecular Cell. 2015;58: 575–585. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Yang D, Buchholz F, Huang Z, et al. Short RNA duplexes produced by hydrolysis with Escherichia coli RNase III mediate effective RNA interference in mammalian cells. PNAS. 2002;26:199–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Zhu KY, Palli SR. Mechanisms, applications, and challenges of insect RNA interference. Annu Rev Entomol. 2019;65:293–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Ramaseshadri P, Segers G, Flannagan R, et al. Physiological and cellular responses caused by RNAi- mediated suppression of Snf7 orthologue in western corn rootworm (Diabrotica virgifera virgifera) larvae. PLoS One. 2013;8:e54270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Baum JA, Bogaert T, Clinton W, et al. Control of coleopteran insect pests through RNA interference. Nat Biotechnol. 2007;25:1322–1326. [DOI] [PubMed] [Google Scholar]
- [10].Bridge AJ, Pebernard S, Ducraux A, et al. Induction of an interferon response by RNAi vectors in mammalian cells. Nat Genet. 2003;34:263–264. [DOI] [PubMed] [Google Scholar]
- [11].Khan AA, Betel D, Miller ML, et al. Transfection of small RNAs globally perturbs gene regulation by endogenous microRNAs. Nat Biotechnol. 2009;27:549–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Scacheri PC, Rozenblatt-Rosen O, Caplen NJ, et al. Short interfering RNAs can induce unexpected and divergent changes in the levels of untargeted proteins in mammalian cells. Proc Natl Acad Sci U S A. 2004;101:1892–1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Birmingham A, Anderson EM, Reynolds A, et al. 3ʹ UTR seed matches, but not overall identity, are associated with RNAi off-targets. Nat Methods. 2006;3:199–204. [DOI] [PubMed] [Google Scholar]
- [14].Jackson AL, Bartz SR, Schelter J, et al. Expression profiling reveals off-target gene regulation by RNAi. Nat Biotechnol. 2003;21:635–637. [DOI] [PubMed] [Google Scholar]
- [15].Papadopoulou N, Devos Y, Alvarez-Alfageme F, et al. Risk assessment considerations for genetically modified RNAi plants: EFSA’s activities and perspective. Front Plant Sci. 2020;11:1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Pan H, Yang X, Bidne K, et al. Dietary risk assessment of v-ATPase A dsRNAs on monarch butterfly larvae. Front Plant Sci. 2017;8:242–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Haley B, Zamore PD. Kinetic analysis of the RNAi enzyme complex. Nat Struct Molecular Biol. 2004;11: 599–606. . [DOI] [PubMed] [Google Scholar]
- [18].Bartoszewski R, Sikorski AF. Editorial focus: understanding off-target effects as the key to successful RNAi therapy. Cell Mol Biol Lett. 2019;24:1–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. [DOI] [PubMed] [Google Scholar]
- [20].Lim LP, Lau NC, Garrett-Engele P, et al. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433:769–773. [DOI] [PubMed] [Google Scholar]
- [21].Lück S, Kreszies T, Strickert M, et al. siRNA-Finder (si-Fi) software for RNAi-target design and off-target prediction. Front Plant Sci. 2019;10:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Perrimon N, Matter Arising M-PB. Off-targets and genome-scale RNAi screens in Drosophila. Fly (Austin). 2014;1:1–5. [DOI] [PubMed] [Google Scholar]
- [23].Hoehener C, Hug I, Nowacki M. Dicer-like enzymes with sequence cleavage preferences. Cell. 2018;173:234–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Guan R, Hu S, Li H, et al. The in vivo dsRNA cleavage has sequence preference in insects. Front Physiol. 2018;9:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Marques JT, Devosse T, Wang D, et al. A structural basis for discriminating between self and nonself double-stranded RNAs in mammalian cells. Nat Biotechnol. 2006;24:559–565. [DOI] [PubMed] [Google Scholar]
- [26].Kulkarni MM, Booker M, Silver SJ, et al. Evidence of off-target effects associated with long dsRNAs in Drosophila melanogaster cell-based assays. Nat Methods. 2006;3:833–838. [DOI] [PubMed] [Google Scholar]
- [27].Poreddy S, Li J, Baldwin IT. Plant-mediated RNAi silences midgut-expressed genes in congeneric lepidopteran insects in nature. BMC Plant Biol. 2017;17:199–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Kumar P, Pandit SS, Steppuhn A, et al. Natural history-driven, plant-mediated RNAi-based study reveals CYP6B46’s role in a nicotine-mediated antipredator herbivore defense. Proc Natl Acad Sci U S A. 2014;111:1245–1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Klumper W, Qaim M. A meta-analysis of the impacts of genetically modified crops. PLoS One. 2014;9:e111629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Gurusamy D, Howell JL, Chereddy S, et al. Transport of orally delivered dsRNA in southern green stink bug, Nezara viridula. Arch Insect Biochem Physiol. 2020;104:e21692. [DOI] [PubMed] [Google Scholar]
- [31].Christiaens O, Whyard S, Velez AM, et al. Double-stranded RNA technology to control insect pests: current status and challenges. Front Plant Sci. 2020;11:451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Fishilevich E, Velez AM, Storer NP, et al. RNAi as a management tool for the western corn rootworm, Diabrotica virgifera virgifera. Pest Manage Sci. 2016;72:1652–1663. . [DOI] [PubMed] [Google Scholar]
- [33].Cooper AM, Silver K, Zhang J, et al. Molecular mechanisms influencing efficiency of RNA interference in insects. Pest Manage Sci. 2019;75: 18–28. . [DOI] [PubMed] [Google Scholar]
- [34].Zotti M, Dos Santos EA, Cagliari D, et al. RNA interference technology in crop protection against arthropod pests, pathogens and nematodes. Pest Manage Sci. 2018;74: 1239–1250. . [DOI] [PubMed] [Google Scholar]
- [35].Bachman PM, Huizinga KM, Jensen PD, et al. Ecological risk assessment for DvSnf7 RNA: a plant-incorporated protectant with targeted activity against western corn rootworm. Regul Toxicol Pharmacol. 2016;81:77–88. [DOI] [PubMed] [Google Scholar]
- [36].Bolognesi R, Ramaseshadri P, Anderson J, et al. Characterizing the mechanism of action of double-stranded RNA activity against western corn rootworm (Diabrotica virgifera virgifera LeConte). PLoS One. 2012;7:e47534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Ghosh SKB, Hunter WB, Park AL, et al. Double-stranded RNA oral delivery methods to induce RNA interference in phloem and plant-sap-feeding hemipteran insects. Jove-J Vis Exp. 2018; 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Miguel KS, Scott JG. The next generation of insecticides: dsRNA is stable as a foliar-applied insecticide. Pest Manage Sci. 2016;72: 801–809. [DOI] [PubMed] [Google Scholar]
- [39].Miller SC, Miyata K, Brown SJ, et al. Dissecting systemic RNA interference in the red flour beetle Tribolium castaneum: parameters affecting the efficiency of RNAi. PLoS One. 2012;7:e47431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Wang JD, Wu M, Wang BJ, et al. Comparison of the RNA interference effects triggered by dsRNA and siRNA in Tribolium castaneum. Pest Manage Sci. 2013;69(7):781–786. . [DOI] [PubMed] [Google Scholar]
- [41].Zhang H, Li HC, Miao XX. Feasibility, limitation and possible solutions of RNAi-based technology for insect pest control. Insect Sci. 2013;20:15–30. [DOI] [PubMed] [Google Scholar]
- [42].Terenius O, Papanicolaou A, Garbutt JS, et al. RNA interference in Lepidoptera: an overview of successful and unsuccessful studies and implications for experimental design. J Insect Physiol. 2011;57:231–245. [DOI] [PubMed] [Google Scholar]
- [43].Tschuch C, Schulz A, Pscherer A, et al. Off-target effects of siRNA specific for GFP. BMC Mol Biol. 2008;9:1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Lin X, Ruan X, Anderson MG, et al. siRNA-mediated off-target gene silencing triggered by a 7 nt complementation. Nucleic Acids Res. 2005;33:4527–4535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Schnorrer F, Schonbauer C, Langer CC, et al. Systematic genetic analysis of muscle morphogenesis and function in Drosophila. Nature. 2010;464:287–291. [DOI] [PubMed] [Google Scholar]
- [46].Mummery-Widmer JL, Yamazaki M, Stoeger T, et al. Genome-wide analysis of Notch signalling in Drosophila by transgenic RNAi. Nature. 2009;458:987–992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Taning CNT, Gui S, De Schutter K, et al. A sequence complementarity-based approach for evaluating off-target transcript knockdown in Bombus terrestris, following ingestion of pest-specific dsRNA. J Pest Sci. 2020. DOI: 10.1007/s10340-020-01273-z. [DOI] [Google Scholar]
- [48].Yoon JS, Mogilicherla K, Gurusamy D, et al. Double-stranded RNA binding protein, Staufen, is required for the initiation of RNAi in coleopteran insects. Proc Natl Acad Sci U S A. 2018;115:8334–8339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Duan G, Saint RB, Helliwell CA, et al. C. elegans RNA-dependent RNA polymerases rrf-1 and ego-1 silence Drosophila transgenes by differing mechanisms. Cell Mol Life Sci. 2013;70::1469–1481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Cogoni C, Macino G. Gene silencing in Neurospora crassa requires a protein homologous to RNA dependent RNA polymerase. Nature. 1999;399:166–169. [DOI] [PubMed] [Google Scholar]
- [51].Peng YC, Wang KX, Zhu GH, et al. Identification and characterization of multiple dsRNases from a lepidopteran insect, the tobacco cutworm, Spodoptera litura (Lepidoptera: noctuidae). Pestic Biochem Physiol. 2019;162:86–95. [DOI] [PubMed] [Google Scholar]
- [52].Peng YC, Wang KX, Fu WX, et al. Biochemical comparison of dsRNA degrading nucleases in four different insects. Front Physiol. 2018;9:1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Shukla JN, Kalsi M, Sethi A, et al. Reduced stability and intracellular transport of dsRNA contribute to poor RNAi response in lepidopteran insects. RNA Biol. 2016;13:656–669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Lee H-C, Li L, Gu W, et al. Diverse pathways generate MicroRNA-like RNAs and dicer-independent small interfering RNAs in fungi. Mol Cell. 2010;38:803–814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Haliscak JP, Beeman RW. Status of malathion resistance in five genera of beetles infesting farm-stored corn, wheat, and oats in the United States. J Econ Entomol. 1983;76:717–722. [Google Scholar]
- [56].Song H, Zhang J, Li D, et al. A double-stranded RNA degrading enzyme reduces the efficiency of oral RNA interference in migratory locust. Insect Biochem Mol Biol. 2017;86:68–80. [DOI] [PubMed] [Google Scholar]
- [57].Peng YC, Sheng CW, Casida JE, et al. Ryanodine receptor genes of the rice stem borer, Chilo suppressalis: molecular cloning, alternative splicing and expression profiling. Pestic Biochem Physiol. 2017;135:69–77. [DOI] [PubMed] [Google Scholar]
- [58].Zhu F, Moural TW, Shah K, et al. Integrated analysis of cytochrome P450 gene superfamily in the red flour beetle, Tribolium castaneum. BMC Genomics. 2013;14:174–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Basu S, Pereira AE, Pinheiro DH, et al. Evaluation of reference genes for real-time quantitative PCR analysis in southern corn rootworm, Diabrotica undecimpunctata howardi (Barber). Sci Rep. 2019;9:10703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Petrick JS, Frierdich GE, Carleton SM, et al. Corn rootworm-active RNA DvSnf7: repeat dose oral toxicology assessment in support of human and mammalian safety. Regul Toxicol Pharmacol. 2016;81:57–68. [DOI] [PubMed] [Google Scholar]
- [61].Zhang X, Liu X, Ma J, et al. Silencing of cytochrome P450 CYP6B6 gene of cotton bollworm (Helicoverpa armigera) by RNAi. Bull Entomol Res. 2013;103:584–591. [DOI] [PubMed] [Google Scholar]
- [62].Zhao J, Xu L, Sun Y, et al. UDP-glycosyltransferase genes in the striped rice stem borer, Chilo suppressalis (Walker), and their contribution to chlorantraniliprole resistance. Int J Mol Sci. 2019;20:1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


