Abstract
The four human PIWI-LIKE gene family members PIWI-LIKE 1–4 play a pivotal role in stem cell maintenance and transposon repression in the human germline. Therefore, dysregulation of these genes negatively influences the genetic stability of the respective germ cell and subsequent development and maturation. Recently, we demonstrated that a lower PIWI-LIKE 2 mRNA expression in ejaculated spermatozoa is more frequent in men with oligozoospermia. In this study, we analysed how PIWI-LIKE 1–4 mRNA expression in ejaculated spermatozoa predicts ART outcome. From 160 IVF or ICSI cycles, portions of swim-up spermatozoa used for fertilization were collected, and the total RNA was isolated. PIWI-LIKE 1–4 mRNA expression was measured by qPCR using TaqMan probes with GAPDH as a reference gene. PIWI-LIKE 1 and 2 transcript levels in the spermatozoa of the swim-up fraction were positively correlated to each other (rS = 0.78; p < 0.001). Moreover, lower PIWI-LIKE 2 mRNA levels, as well as lower PIWI-LIKE 1 mRNA levels, in these spermatozoa were positively associated with a fertilization rate ≥ 50% in the respective ART cycles (p = 0.02 and p = 0.0499, Mann–Whitney U-Test). When separately analysing IVF and ICSI cycles, PIWI-LIKE 1 and 2 transcript levels were only significantly associated to increased fertilization rates in IVF, yet not in ICSI cycles. Spermatozoal PIWI-LIKE 3 and 4 transcript levels were not significantly associated to fertilization rates in ART cycles. In conclusion, lower levels of spermatozoal PIWI-LIKE 1 and 2 mRNA levels are positively associated with a higher fertilization rate in IVF cycles.
Keywords: male infertility, PIWI-LIKE genes, spermatozoa, ART, fertilization rate
1. Introduction
Infertility is a major health problem worldwide, with an estimated 48.5 million couples suffering from unwilling childlessness [1]. Male infertility contributes to roughly half of these cases, and impacts not only the reproductive function, but is also linked to the overall health of the affected male [2,3]. In most cases of male infertility, disturbances of the spermatogenesis result in the reduction of semen quality, measurable in total sperm count and the proportions of motile sperm and sperm with typical morphology, but may also result in a disturbed DNA condensation or in higher DNA fragmentation. Many couples with infertile men are referred to assisted reproduction therapy (IVF or ICSI), by which the disturbances in the sperm variables are encompassed by the in vitro selection of optimal spermatozoa. However, the selection of human spermatozoa based on criteria such as viability, motility, and morphology does not necessarily reflect in the subsequent sperm fertilizing capacity [4,5,6].
Depending on the technique chosen and the number of oocytes retrieved, the mean fertilization rate, which is a predictor of a subsequent pregnancy, is around 50–70% in ART cycles in Germany. Nevertheless, in some cases, fertilization rates remain below 25%. These decreased fertilization rates may be due to abnormal DNA condensation or even to fragmentation during spermatogenesis. The DNA fragmentation is described by the DNA fragmentation index (DFI), which can be analysed by various different testing systems [7,8]. Several studies have shown an increased DFI to be associated with a significantly lower fertilization rate [9,10]; however, there are also reports indicating no relation between DFI and embryologic outcomes [11,12,13]. Nevertheless, nowadays DFI measurements are performed routinely in many ART clinics to predict the fertilization capacity of the ejaculate [14,15]. Given the fact that these measurements have only a limited prognostic impact, further early markers of a lowered fertilization competence of the ejaculate are warranted as a diagnostic tool, but also as a therapeutic target, which may in future allow the selection of the optimal spermatozoa.
During spermatogenesis, a plethora of molecular factors are engaged in the maturation of a round, metabolic active, differentiation-competent spermatogonia to a mature spermatozoa. It is assumed that around 2000 genes play a role in the spermatogenic differentiation or the processing in the epididymis [16], many of them being associated with male infertility in mouse knock-out models. One striking example is the PIWI-LIKE gene family, a subclade of the Argonaute gene family. PIWI-LIKE genes are highly conserved among various species, consisting for example of four human family members (PIWI-LIKE 1–4; alternatively, HIWI, HILI, HIWI3, and HIWI2 [17]) or three murine homologs (Miwi, Mili, Miwi2 [18]). When generating Miwi-knockout mice, Deng and Lin observed that Miwinull male mice are sterile, while Miwinull female mice remain fertile. They also demonstrated that Miwinull mice exhibited an arrest of spermatogenesis on the round spermatid level, and that no mature sperm is found in the epididymides of these mice [19]. Similarly, Milinull and Miwi2null male mice are also sterile, with Milinull mice exhibiting a spermatogenic block in the early pachytene stage [20] and Miwi2null mice reaching spermatogenic arrest even in the zygotene stage of the first meiosis [21]. Yet, the impact of the human PIWI-LIKE genes on male fertility is still under debate.
Previously, we demonstrated that a dysregulated PIWI-LIKE 1 or 2 expression is associated with altered semen variables [22]. However, currently no data exist on the connection between dysregulated spermatozoal PIWI-LIKE gene expression as a surrogate of suboptimal spermatogenesis and the fertilization rates achieved in ART cycles using these spermatozoa. Therefore, the primary aim of this study was to measure PIWI-LIKE 1–4 mRNA expression in the swim-up fraction of ejaculated spermatozoa used for ART and to correlate these parameters with the corresponding fertilization rate.
2. Results
2.1. Distribution of PIWI-LIKE Gene Transcripts in the Spermatozoa
Firstly, we quantitatively assessed PIWI-LIKE transcript levels in the swim-up fraction and in the cell pellet. PIWI-LIKE gene transcripts were measurable in both fractions of the patients’ samples to a different extent. While PIWI-LIKE 1 expression could be quantified in 96.2% (swim-up fractions) and 98.2% (cell pellets) of the patients’ samples, PIWI-LIKE 2 was measurable in 74.9% and 75.2% of the samples, respectively. PIWI-LIKE 4 was only detectable in around one third of the patients’ samples (32.7% and 36.4%, respectively), while PIWI-LIKE 3 was more frequently detected in the cell pellet fraction (39.4% of the samples) than in the swim-up fraction (17.5%).
Comparing the transcript levels of PIWI-LIKE 1–4 in the different fractions, we observed a significantly higher expression of PIWI-LIKE 1 mRNA in the cell pellet than in the swim-up phase (median expression 0.00133 in the swim-up fraction vs. 0.00399 in the cell pellet, p < 0.001; Mann–Whitney U test, see Table 1). On the other hand, PIWI-LIKE 2 expression was not significantly different between the spermatozoa of both fractions (see Table 1).
Table 1.
Distribution of PIWI-LIKE gene transcripts in spermatozoa from the swim-up and the cell pellet fraction.
| Median Expression (ΔCq) | p (MWU) | n | ||
|---|---|---|---|---|
| PIWI-LIKE 1 | swim-up fraction | 0.00113 | <0.001 | 160 |
| cell pellet | 0.00399 | |||
| PIWI-LIKE 2 | swim-up fraction | 0.00001 | n.s. | 160 |
| cell pellet | 0.000012 | |||
| PIWI-LIKE 3 | swim-up fraction | 0.00 | <0.001 | 160 |
| cell pellet | 0.00 | |||
| PIWI-LIKE 4 | swim-up fraction | 0.00 | n.s. | 160 |
| cell pellet | 0.00 |
Abbreviation: MWU–Mann–Whitney U-test.
In bivariate correlation analyses (according to Spearman-Rho), the mRNA expression of PIWI-LIKE 1 (rS = 0.57; p < 0.001), PIWI-LIKE 2 (rS = 0.70; p < 0.001), PIWI-LIKE 3 (rS = 0.22; p = 0.006), and PIWI-LIKE 4 (rS = 0.37; p < 0.001) in the spermatozoa of the swim-up fraction and the cell pellet correlated to each other highly significantly. Furthermore, the PIWI-LIKE 1 and 2 transcript levels were highly significant, associated in the spermatozoa of the swim-up, as well as of the cell fraction (rS = 0.78 and rS = 0.49, respectively; p < 0.001). Additionally, PIWI-LIKE 4 transcript levels in spermatozoa of the swim-up fraction was inversely correlated to the PIWI-LIKE 1 and 2 levels (rS = −0.25 and rS = −0.28, respectively, p < 0.001).
2.2. Lower PIWI-LIKE 1 and 2 mRNA Expression Is Associated with Higher Fertilization Rates in ART
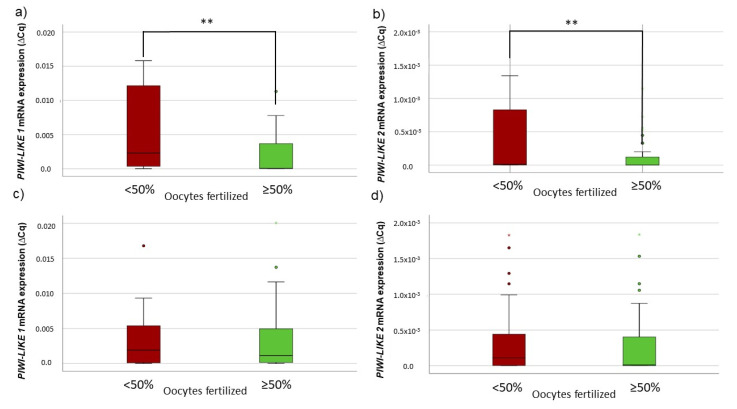
We compared the PIWI-LIKE 1–4 transcript levels in the spermatozoa of the swim-up fraction used for ART depending on the fertilization rates in the respective cycle. The median fertilization rate of 50% was chosen as a cut-off. In the swim-up spermatozoa fraction of patients with fertilization rates ≥ 50%, a significantly lower PIWI-LIKE 1 or lower PIWI-LIKE 2 mRNA level was observed (p = 0.0499 and 0.017, respectively, Mann–Whitney U-Test, see Figure 1a,b).
Figure 1.
Association between fertilization success and (a) PIWI-LIKE 1, (b) PIWI-LIKE 2, (c) PIWI-LIKE 3, or (d) PIWI-LIKE 4 spermatozoal mRNA levels in 160 ART cycles. * p < 0.05.
Furthermore, in cycles with ≥ 8 inseminated oocytes, the correlation between a lower PIWI-LIKE 1 or 2 mRNA expression and a fertilization rate ≥50% was even more pronounced (p = 0.019 or p = 0.009, respectively, Mann–Whitney U-test). Spermatozoal PIWI-LIKE 3 and PIWI-LIKE 4 mRNA levels did not show any correlations to fertilization rate.
2.3. PIWI-LIKE 1 and 2 mRNA Expression Is a Predictor for Fertilization in IVF, but Not ICSI Cycles
When subdividing the whole study cohort according to the ART treatment performed, only in the IVF group a lowered PIWI-LIKE 1 or 2 mRNA level was significantly associated with a higher fertilization success (p = 0.002 and p = 0.019, respectively, Mann–Whitney U-test; see Figure 2a,b). In comparison, in the ICSI group, no significant association between the spermatozoal PIWI-LIKE mRNA expression and the fertilization results was seen (see Figure 2c,d). Of note, only four (7.0%) of male patients in the IVF group exhibited mild impairments semen values, while 70 (72.9%) of the patients in the ICSI group showed abnormal sperm values.
Figure 2.
Association between fertilization success and spermatozoal PIWI-LIKE 1 spermatozoal mRNA levels in (a) IVF or (c) ICSI cycles and association between fertilization success and spermatozoal PIWI-LIKE 2 mRNA levels in (b) IVF or (d) ICSI cycles. ** p <0.01.
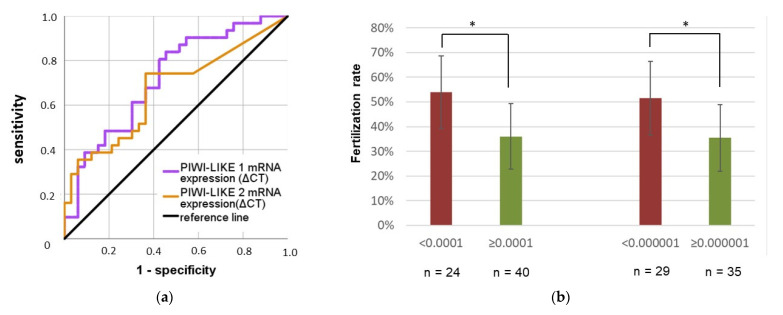
In ROC analyses, both PIWI-LIKE 1 and 2 transcript levels were fair predictors of a higher fertilization rate in IVF cycles (see Figure 3a). Spermatozoal PIWI-LIKE 1 mRNA levels exhibited an AUC of 0.72 (asymptotic p = 0.002), while PIWI-LIKE 2 mRNA expression had an AUC of 0.67 (asymptotic p = 0.021).
Figure 3.
Potential of PIWI-LIKE 1 and 2 spermatozoal mRNA level to distinguish between high and low-fertilization patient groups applying (a) ROC analyses and (b) different cut-off rates for PIWI-LIKE 1 and 2. * p < 0.05.
Based on the ROC analyses, we calculated a Youden optimized cut-off value for the spermatozoal PIWI-LIKE 1 and PIWI-LIKE 2 transcript levels. Below a spermatozoal PIWI-LIKE 1 mRNA level of 0.0001 (ΔCq), the mean fertilization rate was significantly higher than above this cut-off (53.9% vs. 36.0%; p = 0.011, Mann–Whitney U-test). Regarding spermatozoal PIWI-LIKE 2 mRNA levels, below 0.00001 (ΔCq), the mean fertilization rate was significantly increased (51.5% vs. 35.5%; p = 0.022; Mann–Whitney U-Test, see Figure 3b).
3. Discussion
In this study, we observed, for the first time, an association between low spermatozoal PIWI-LIKE 1 and 2 transcript level and a higher fertilization rate in IVF. As PIWI-LIKE gene expression was directly assessed in the motile spermatozoa from the swim-up fraction used in the respective ART cycle, an influence of an altered expression of the PIWI-LIKE genes during spermatogenesis and reproductive success may be hypothesized. Furthermore, the results of this study may pave the road to the inclusion of PIWI-LIKE gene expression as an additional marker for the clinical decision between IVF or ICSI as treatment of choice, if confirmed in larger independent studies.
We were able to quantify the PIWI-LIKE mRNA expressions of the four human family members in the spermatozoa of the swim-up and the primarily immotile cell fraction to a different extent. While PIWI-LIKE 1 and 2 mRNA were detectable in over 95% and in around 75% of the samples, PIWI-LIKE 3 and 4 transcripts were present in a fewer number of samples. This is concordant with our previous study, where we could detect PIWI-LIKE 1 in all studied samples, and PIWI-LIKE 2 in around 50% of the samples, while PIWI-LIKE 3 and PIWI-LIKE 4 were only measurable in less than 20% of the samples [22]. Furthermore, PIWI-LIKE 4 expression is inversely correlated to the expression of PIWI-LIKE 1/2. These findings are in concordance with the known expression sequence of PIWI proteins in the mammalian system, with PIWI-LIKE 4 being expressed in early stages, while PIWI-LIKE 2 is expressed over the main course of spermatogenesis, and finally being replaced by PIWI-LIKE 1 expression in the mid-pachytene stage (reviewed in [23]). In concordance with our results, Hempfling and colleagues demonstrated the mRNA expression of PIWI-LIKE 1 and 2 in the vast majority of the testis tissue samples displaying normal or impaired spermatogenesis [24].
Although decreased PIWI-LIKE 2 expression was associated to impaired semen parameters [22] in our previous study, our current study shows a relation between lower PIWI-LIKE 2 expression and higher fertilization success. The contradictory results regarding PIWI-LIKE 2 and sperm parameters compared to our first study could be explained by the assessment of spermatozoa from native ejaculate in the initial study versus the analysis of spermatozoa solely from the swim-up phase in the present study. However, both studies strongly suggest that PIWI-LIKE gene expression serves as a predictor for semen quality.
We observed a significant association between a lower PIWI-LIKE 1 or 2 transcript expression in the spermatozoa used for IVF and increased fertilization rates. To the best of our knowledge, this is the first time that PIWI-LIKE gene expression could be linked to fertilization success. Cui and colleagues demonstrated that the increased spermatozoal expressions of the piwi-interacting RNAs piR-31704, piR-39888, and piR-40349 are linked with a fertilization success >70% in 186 ICSI cycles [25]. A study from Denomme and colleagues showed that a group of ten miRNAs were identified as significantly altered in association with poor blastocyst development, with corresponding alterations to target genes involved in embryonic genome activation, blastocyst implantation, and DNA methylation [26]. The complex interplay of the PIWIL/piRNA machinery with gametogenesis and fertilization is still not elucidated yet.
The impact of the PIWI-LIKE proteins on human spermatogenesis is still unclear. In 2017, Gou and colleagues described several heterozygous mutations in the D-box region of HIWI/PIWI-LIKE 1 gene of three out of 413 patients with idiopathic azoospermia, thereby impairing the ubiquitination of PIWI-LIKE 1 [27]. Additionally, the authors demonstrated in mice that the resulting stabilization and accumulation of PIWI-LIKE 1 induces abnormal chromatin compaction due to an impaired histone–protamine exchange and thus leading to male sterility [27]. However, recently Oud and colleagues showed in a larger study cohort of 2740 patients with azoospermia or severe oligozoospermia, that HIWI D-box variations are less frequent than proposed by Gou and colleagues, and the that loss of function of PIWI-LIKE 1 is unlikely to cause fertility impairments when haploin-sufficient [28]. In our study, we did not perform mutational screenings on the PIWI-LIKE genes, therefore statements about the relationship of mutated PIWI-LIKE genes and male infertility cannot be drawn from our experimental setting.
Earlier studies showed that spermatozoal RNAs are also delivered to the oocyte and potentially contribute to early embryo development [29,30]. Although murine PIWI-LIKE 2 was detected alongside other nuage structure proteins in primordial follicular oocytes [31], less is known about the role of PIWI-LIKE genes in human oocytes. It can be hypothesized, that PIWI-LIKE transcript delivery via the spermatozoon may assist in retrotransposon control during early embryonic development [32].
A limitation of our study is that we do not have insight into all maternal factors involved in the fertilization process. Fertilization is a multifactorial process that involves maternal and paternal interplay. Success and failure of oocyte fertilization cannot be referred solely to one single gene family in men but remain multifactorial.
Furthermore, the evaluation of mRNA in spermatozoa as a predictor for male fertilization capacity is still experimental. The correlation between RT-qPCR data and fertilization success does not exactly reflect the mRNA content of the single spermatozoon that induces the fertilization of one particular oocyte.
4. Materials and Methods
4.1. Ethical Approval
The ethics committee of the Medical Faculty of the Martin Luther University Halle-Wittenberg approved the study. All patients gave written informed consent.
4.2. Study Population
Patients were included in this study if they were 18–50 years old and signed a written informed consent. Exclusion criteria were azoospermia, known genetic causes of infertility such as cystic fibrosis or Y chromosomal microdeletions, and testosterone or other drug abuse. ART cycles with at least one oocyte inseminated were included in this study. A total of 130 patients were included in this study, with 67 exhibiting normal semen values, while, in 63 patients, one or more sperm parameters were below the reference ranges defined in the WHO laboratory manual for the examination and processing of human semen, 5th edition. Of the patients with abnormal sperm values, 26 patients exhibited an oligozoospermia, 7 asthenozoospermia, and 2 teratozoospermia. Eleven patients had an oligoteratozoospermia, 7 patients oligoasthenozoospermia and 10 patients oligoasthenoteratozoospermia. For more information on semen values see Table 2.
Table 2.
Demographical and clinical data on the patients’ cohort. The minimal and maximal values are given in brackets.
| Parameter | Male Partner | Female Partner |
|---|---|---|
| age (y) | 37 (25–52) | 34 (26–41) |
| weight (kg) | 88.4 (58–150) | 67.1 (43–123) |
| BMI (kg/m2) | 26.9 (18.3–41.6) | 24.0 (16.5–40.2) |
| days of abstinence | 4 (2–21) | |
| semen volume (mL) | 2.8 (0.1–10.4) | |
| sperm count (total sperm/ejaculate) | 70.1 (0.1–717.6) | |
| progressive motility (%) | 50 % (0–90%) | |
| % morphologically normal spermatozoa | 8% (0–24%) |
In total, 160 ART cycles were performed at the Center for Reproductive Medicine and Andrology of the University Hospital Halle (Saale), either by in vitro fertilization (IVF; n = 64) or by intracytoplasmatic sperm injection (ICSI; n = 96). In 19 treatment cycles (11.9%), no fertilization was achieved (IVF: 11 cyles; 17.2%; ICSI: 8 cycles; 8.3%). In median, there were 9 oocytes retrieved per cycle, of which 8 could be inseminated. Median fertilization rate was 50%, and median clinical pregnancy rate was 26.3%. For detailed cycle information, see Table 3.
Table 3.
Clinical data on the ART cycles. The minimal and maximal values are given in brackets.
| Total (n = 160) | IVF (n = 64) | ICSI (n = 96) | |
|---|---|---|---|
| Cycles with male indication for ART | 26 | 0 | 26 |
| Cycles with female indication for ART | 61 | 56 | 5 |
| Cycles with both partner indication for ART | 73 | 8 | 65 |
| Oocytes retrieved (n) | 9 (1–36) | 10 (1–25) | 8 (1–36) |
| Inseminated (n) | 8 (1–25) | 10 (1–25) | 6 (1–19) |
| Fertilized (n) | 4 (0–14) | 4 (0–14) | 3 (0–10) |
| Fertilization rate (%) | 50 (0–100) | 50 (0–100) | 58 (0–100) |
| Transferred embryos (n) | 2 (0–3) | 2 (0–3) | 2 (0–3) |
| Clinical pregnancy rate (n) | 26.3 | 26.6 | 26.0 |
4.3. Specimen Preparation and RNA Isolation
After collection, the ejaculate was stored 30 min at 37 °C for liquefaction. Sperm concentration and motility were assessed as described in the WHO manual, 5th edition, with at least 2 × 200 spermatozoa counted for each parameter. After ejaculate analysis, the specimens were centrifuged (1000× g, 10 min) on a density gradient (Sil Select, FertiPro, Berneem, Belgium) to exclude cell debris and non-spermatozoal cells. The cell pellet was washed with Gamete Buffer (Cook Medical, Bloomington, IN, USA), subsequently overlaid with Gamete Buffer and incubated for 30 min at 37 °C. ART (either IVF or ICSI) was performed with spermatozoa from the swim-up fraction. Directly after the insemination, the remaining swim-up fraction and cell pellet were separated, washed with sterile PBS (Invitrogen, Carlsbad, CA, USA), and immediately stored at −80 °C until RNA isolation.
Total RNA was isolated using the RNeasy Micro Kit (Qiagen, Hilden, Germany). Briefly, cell pellets of either the swim-up or the cellular fraction were lysed with RLT buffer + 1% ß-mercaptoethanol (Sigma-Aldrich, St. Louis, MO, USA), and RNA was precipitated with 70% ethanol (Sigma-Aldrich, St. Louis, MO, USA). The total RNA was bound to a silica spin column and treated with DNase I (Sigma-Aldrich, St. Louis, MO, USA) for 10 min to remove residual DNA contaminations. After three rounds of washing with the appropriate buffers, the RNA was eluted in 14 μL RNAse-free water. RNA concentration and purity were assessed by absorption spectrometry (Eppendorf, Hamburg, Germany).
4.4. cDNA Synthesis and Quantitative Real-Time PCR
Spermatozoal RNA was transcribed to cDNA with the RevertAid First Strand cDNA Synthesis Kit (Thermo Fisher, Waltham, MA, USA) according to the manufacturers protocol using a mixture of random hexamer primers and OligodT primers. Quantitative PCR of the PIWI-LIKE genes was applied with TaqMan Primer (Applied biosystems; Waltham, MA, USA) on a MyIQ-Cycler (BioRad, Hercules, CA, USA). GAPDH expression was used as internal reference, and ΔCq values were calculated according to Livak and Schmittgen [33].
4.5. Statistical Analyses
Statistical analyses were performed using SPSS (Version 25, SPSS Inc., Chicago, IL, USA). Bivariate correlation analyses according to Spearman-Rho and non-parametric tests (Mann–Whitney U-Test) were conducted for the evaluation of the association between human PIWI-LIKE 1–4 mRNA expression, ejaculate variables, and fertilization rates. p < 0.05 was considered significant.
5. Conclusions
In conclusion, when analyzing the spermatozoal PIWI-LIKE gene transcript levels in the swim-up phase of sperm used for ART, lower PIWI-LIKE 1 and 2 transcript levels were associated with increased fertilization rates in IVF cycles. Based on these results, it could be speculated that spermatozoal PIWI-LIKE 1 and 2 might be a surrogate marker for proper sperm development and fertilization capacity of sperm used for IVF treatment, and that, therefore, patients with higher PIWI-LIKE 1 or 2 transcript levels in swim-up sperm should be offered ICSI despite having relatively good classical sperm values.
Acknowledgments
The authors would like to thank Sabine Moosdorf, Gabriele Wiener and Constanze Kloss for providing excellent technical assistance and performing the semen analysis.
Author Contributions
Conceptualization, T.G. and H.M.B.; Formal analysis, T.G. and M.G.; Funding acquisition, T.G. and H.M.B.; Investigation, T.G., M.G. and D.H.; Methodology, T.G., M.G. and D.H.; Project administration, T.G. and H.M.B.; Resources, G.S.; Visualization, T.G. and M.G.; Writing—original draft, T.G. and M.G.; Writing—review and editing, T.G., M.G., G.S. and H.M.B. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
The Ethics Committee of the Medical Faculty of the Martin Luther University Halle-Wittenberg approved the study (approval number 218/14.04.10/2.; date of approval: 19 April 2010).
Informed Consent Statement
Written informed consent has been obtained from the patients.
Data Availability Statement
Original data are available from the authors by reasonable behalf.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Agarwal A., Mulgund A., Hamada A., Chyatte M.R. A unique view on male infertility around the globe. Reprod. Biol. Endocrinol. 2015;13:37. doi: 10.1186/s12958-015-0032-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jensen T.K., Lindahl-Jacobsen R., Christensen K., Nielsen N.C., Bostofte E. Good Semen Quality and Life Expectancy: A Cohort Study of 43,277 Men. Am. J. Epidemiol. 2009;170:559–565. doi: 10.1093/aje/kwp168. [DOI] [PubMed] [Google Scholar]
- 3.Glazer C.H., Eisenberg M.L., Tøttenborg S.S., Giwercman A., Flachs E.M., Bräuner E., Vassard D., Pinborg A., Schmidt L., Bonde J.P. Male factor infertility and risk of death: A nationwide record-linkage study. Hum. Reprod. 2019;34:2266–2273. doi: 10.1093/humrep/dez189. [DOI] [PubMed] [Google Scholar]
- 4.Thilagavathi J., Venkatesh S., Kumar R., Dada R. Segregation of sperm subpopulations in normozoospermic infertile men. Syst. Biol. Reprod. Med. 2012;58:313–318. doi: 10.3109/19396368.2012.706361. [DOI] [PubMed] [Google Scholar]
- 5.Albani E., Castellano S., Gurrieri B., Arruzzolo L., Negri L., Borroni E.M., Levi-Setti P.E. Male age: Negative impact on sperm DNA fragmentation. Aging. 2019;11:2749–2761. doi: 10.18632/aging.101946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Poenicke K., Grunewald S., Glander H., Paasch U., Kamini A.R., Agarwal A., Srinivas M. Andrology Laboratory Manual. Jaypee Brothers Pvt Ltd.; New Delhi, India: 2010. Sperm Selection in Assisted Reproductive Techniques; pp. 173–187. [DOI] [Google Scholar]
- 7.Dutta S., Henkel R., Agarwal A. Comparative analysis of tests used to assess sperm chromatin integrity and DNA fragmentation. Andrologia. 2020;53:e13718. doi: 10.1111/and.13718. [DOI] [PubMed] [Google Scholar]
- 8.Evenson D.P., Wixon R. Clinical aspects of sperm DNA fragmentation detection and male infertility. Theriogenology. 2006;65:979–991. doi: 10.1016/j.theriogenology.2005.09.011. [DOI] [PubMed] [Google Scholar]
- 9.Tang L., Rao M., Yang W., Yao Y., Luo Q., Lu L., Wang L., Zhao S. Predictive value of the sperm DNA fragmentation index for low or failed IVF fertilization in men with mild-to-moderate asthenozoospermia. J. Gynecol. Obstet. Hum. Reprod. 2020;50:101868. doi: 10.1016/j.jogoh.2020.101868. [DOI] [PubMed] [Google Scholar]
- 10.Oleszczuk K., Giwercman A., Bungum M. Sperm chromatin structure assay in prediction of in vitro fertilization outcome. Andrology. 2016;4:290–296. doi: 10.1111/andr.12153. [DOI] [PubMed] [Google Scholar]
- 11.Lin M.-H., Lee R.K.-K., Li S.-H., Lu C.-H., Sun F.-J., Hwu Y.-M. Sperm chromatin structure assay parameters are not related to fertilization rates, embryo quality, and pregnancy rates in in vitro fertilization and intracytoplasmic sperm injection, but might be related to spontaneous abortion rates. Fertil. Steril. 2008;90:352–359. doi: 10.1016/j.fertnstert.2007.06.018. [DOI] [PubMed] [Google Scholar]
- 12.Green K.A., Patounakis G., Dougherty M.P., Werner M.D., Scott R.T., Franasiak J.M. Sperm DNA fragmentation on the day of fertilization is not associated with embryologic or clinical outcomes after IVF/ICSI. J. Assist. Reprod. Genet. 2019;37:71–76. doi: 10.1007/s10815-019-01632-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Speyer B.E., Pizzey A.R., Ranieri M., Joshi R., Delhanty J.D.A., Serhal P. Fall in implantation rates following ICSI with sperm with high DNA fragmentation. Hum. Reprod. 2010;25:1609–1618. doi: 10.1093/humrep/deq116. [DOI] [PubMed] [Google Scholar]
- 14.Deng C., Li T., Xie Y., Guo Y., Yang Q., Liang X., Deng C., Liu G. Sperm DNA fragmentation index influences assisted reproductive technology outcome: A systematic review and meta-analysis combined with a retrospective cohort study. Andrologia. 2019;51:e13263. doi: 10.1111/and.13263. [DOI] [PubMed] [Google Scholar]
- 15.Rex A.S., Wu C., Aagaard J., Fedder J. Implementation of an in-house flow cytometric analysis of DNA fragmentation in spermatozoa. Asian J. Androl. 2020;22:246. doi: 10.4103/aja.aja_51_19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hamada A.J., Esteves S.C., Agarwal A. A comprehensive review of genetics and genetic testing in azoospermia. Clinics. 2013;68:39–60. doi: 10.6061/clinics/2013(Sup01)06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sasaki T., Shiohama A., Minoshima S., Shimizu N. Identification of eight members of the Argonaute family in the human genome☆. Genomics. 2003;82:323–330. doi: 10.1016/S0888-7543(03)00129-0. [DOI] [PubMed] [Google Scholar]
- 18.Kuramochi-Miyagawa S., Kimura T., Yomogida K., Kuroiwa A., Tadokoro Y., Fujita Y., Sato M., Matsuda Y., Nakano T. Two mouse piwi-related genes: Miwi and mili. Mech. Dev. 2001;108:121–133. doi: 10.1016/S0925-4773(01)00499-3. [DOI] [PubMed] [Google Scholar]
- 19.Deng W., Lin H. miwi, a Murine Homolog of piwi, Encodes a Cytoplasmic Protein Essential for Spermatogenesis. Dev. Cell. 2002;2:819–830. doi: 10.1016/S1534-5807(02)00165-X. [DOI] [PubMed] [Google Scholar]
- 20.Kuramochi-Miyagawa S., Kimura T., Ijiri T.W., Isobe T., Asada N., Fujita Y., Ikawa M., Iwai N., Okabe M., Deng W., et al. Mili, a mammalian member ofpiwifamily gene, is essential for spermatogenesis. Development. 2004;131:839–849. doi: 10.1242/dev.00973. [DOI] [PubMed] [Google Scholar]
- 21.Carmell M.A., Girard A., van de Kant H.J., Bourc’His D., Bestor T.H., de Rooij D., Hannon G.J. MIWI2 Is Essential for Spermatogenesis and Repression of Transposons in the Mouse Male Germline. Dev. Cell. 2007;12:503–514. doi: 10.1016/j.devcel.2007.03.001. [DOI] [PubMed] [Google Scholar]
- 22.Behre H., Giebler M., Greither T., Müller L., Mösinger C. Altered PIWI-LIKE 1 and PIWI-LIKE 2 mRNA expression in ejaculated spermatozoa of men with impaired sperm characteristics. Asian J. Androl. 2018;20:260–264. doi: 10.4103/aja.aja_58_17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bak C.W., Yoon T.-K., Choi Y. Functions of PIWI proteins in spermatogenesis. Clin. Exp. Reprod. Med. 2011;38:61–67. doi: 10.5653/cerm.2011.38.2.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hempfling A.L., Lim S.L., Adelson D., Evans J., O’Connor A.E., Qu Z.P., Kliesch S., Weidner W., O’Bryan M.K., Bergmann M. Expression patterns of HENMT1 and PIWIL1 in human testis: Implications for transposon expression. Reproduction. 2017;154:363–374. doi: 10.1530/REP-16-0586. [DOI] [PubMed] [Google Scholar]
- 25.Cui L., Fang L., Shi B., Qiu S., Ye Y. Spermatozoa Expression of piR-31704, piR-39888, and piR-40349 and Their Correlation to Sperm Concentration and Fertilization Rate after ICSI. Reprod. Sci. 2017;25:733–739. doi: 10.1177/1933719117725822. [DOI] [PubMed] [Google Scholar]
- 26.Denomme M.M., McCallie B.R., Parks J.C., Schoolcraft W.B., Katz-Jaffe M.G. Alterations in the sperm histone-retained epigenome are associated with unexplained male factor infertility and poor blastocyst development in donor oocyte IVF cycles. Hum. Reprod. 2017;32:2443–2455. doi: 10.1093/humrep/dex317. [DOI] [PubMed] [Google Scholar]
- 27.Gou L.-T., Kang J.-Y., Dai P., Wang X., Li F., Zhao S., Zhang M., Hua M.-M., Lu Y., Zhu Y., et al. Ubiquitination-Deficient Mutations in Human Piwi Cause Male Infertility by Impairing Histone-to-Protamine Exchange during Spermiogenesis. Cell. 2017;169:1090–1104.e13. doi: 10.1016/j.cell.2017.04.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Oud M., Volozonoka L., Friedrich C., Kliesch S., Nagirnaja L., Gilissen C., O’Bryan M., McLachlan R., Aston K., Tüttelmann F., et al. Lack of evidence for a role of PIWIL1 variants in human male infertility. Cell. 2021;184:1941–1942. doi: 10.1016/j.cell.2021.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ostermeier G.C., Goodrich R.J., Moldenhauer J.S., Diamond M.P., Krawetz S.A. A suite of novel human spermatozoal RNAs. J. Androl. 2005;26:70–74. [PubMed] [Google Scholar]
- 30.Jodar M., Selvaraju S., Sendler E., Diamond M., Krawetz S.A., Reproductive Medicine Network The presence, role and clinical use of spermatozoal RNAs. Hum. Reprod. Update. 2013;19:604–624. doi: 10.1093/humupd/dmt031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lim A.K., Lorthongpanich C., Chew T.G., Tan C.W.G., Shue Y.T., Balu S., Gounko N., Kuramochi-Miyagawa S., Matzuk M.M., Chuma S., et al. The nuage mediates retrotransposon silencing in mouse primordial ovarian follicles. Development. 2013;140:3819–3825. doi: 10.1242/dev.099184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kohlrausch F.B., Berteli T.S., Wang F., Navarro P.A., Keefe D.L. Control of LINE-1 Expression Maintains Genome Integrity in Germline and Early Embryo Development. Reprod. Sci. 2021:1–13. doi: 10.1007/s43032-021-00461-1. [DOI] [PubMed] [Google Scholar]
- 33.Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Original data are available from the authors by reasonable behalf.