Abstract
The C and D box-containing (box C/D) small nucleolar RNAs (snoRNAs) function in the nucleolytic processing and 2′-O-methylation of precursor rRNA. In vertebrates, most box C/D snoRNAs are processed from debranched pre-mRNA introns by exonucleolytic activities. Elements directing accurate snoRNA excision are located within the snoRNA itself; they comprise the conserved C and D boxes and an adjoining 5′,3′-terminal stem. Although the terminal stem has been demonstrated to be essential for snoRNA accumulation, many snoRNAs lack a terminal helix. To identify the cis-acting elements supporting the accumulation of intron-encoded box C/D snoRNAs devoid of a terminal stem, we have investigated the in vivo processing of the human U46 snoRNA and an artificial snoRNA from the human β-globin pre-mRNA. We demonstrate that internal and/or external stem structures located within the snoRNA or in the intronic flanking sequences support the accumulation of mammalian box C/D snoRNAs lacking a canonical terminal stem. In the intronic precursor RNA, transiently formed external and/or stable internal base-pairing interactions fold the C and D boxes together and therefore facilitate the binding of snoRNP proteins. Since the external intronic stems are degraded during snoRNA processing, we propose that the C and D boxes alone can provide metabolic stability for the mature snoRNA.
The nucleolus contains a large number of small nucleolar RNAs (snoRNAs), which function in the nucleolytic processing and nucleotide modification of precursor rRNAs (pre-rRNAs) (reviewed in references 34, 48, and 49). The vast majority of snoRNAs fall into two families, which can be distinguished on the basis of common sequence boxes. Many snoRNAs share the conserved C (consensus sequence: RUGAUGA) and D (CUGA) boxes (50; reviewed in reference 34), and the rest of the snoRNAs possess the H (AnAnnA) and ACA motifs (4, 16). A few C and D box-containing (box C/D) snoRNAs (U3, U8, U14, and U22) and an H and ACA motif-containing (box H/ACA) snoRNA (snR30) are required for the production of mature-sized rRNAs (21, 22, 30, 35, 40, 46, 53). However, the majority of box C/D and box H/ACA snoRNAs direct site-specific 2′-O-ribose methylation (10, 26, 32, 51) and pseudouridylation (17, 37) of rRNAs, respectively.
Genes coding for snoRNAs have been found in diverse genomic contexts (reviewed in references 34, 49, and 59). While the majority of vertebrate snoRNAs are synthesized as parts of pre-mRNA introns, most yeast and plant snoRNAs are transcribed as monocistronic or polycistronic precursor snoRNAs (pre-snoRNAs) from independent transcription units. In each case, mature snoRNAs are released from the primary transcript by nucleolytic processing. The intronic snoRNAs are processed from the removed and debranched host introns (23, 25, 39, 41) or, less frequently, from endonucleolytically released intronic fragments (7, 15, 42, 55) by exonucleolytic activities (2, 9, 11, 23–25, 41, 52). From yeast polycistronic pre-snoRNA transcripts, the RNase III (Rnt1p) endoribonuclease releases individual pre-snoRNA fragments and, again, exonucleolytic trimming forms the correct 5′ and 3′ termini of the snoRNA (12, 13, 43). The yeast Rat1p and Xrn1p 5′→3′ exonucleases play an essential role in the 5′-end formation of both intronic and polycistronic snoRNAs (41, 43, 55). Maturation of some yeast snoRNAs by trimming of short 3′-terminal trailer sequences specifically requires the Rrp6p 3′→5′ exonuclease that is a component of the yeast exosome (1, 2).
The exonucleolytic processing of intronic and polycistronic snoRNAs is directed by cis-acting elements which have been found within the snoRNA itself. Consistent with this finding, snoRNAs placed into nonnatural sequence contexts are accurately processed both in vitro and in vivo (5, 16, 17, 25–27, 44, 57, 60). The conserved sequence boxes and the neighboring stem structures constitute the signal elements directing the correct snoRNA formation (4, 6, 9, 16, 20, 44, 57, 60). Processing and accumulation of the intron-encoded and polycistronic box C/D snoRNAs depend on the C and D boxes and an adjacent 4- or 5-bp helix that folds together the 5′ and 3′ ends of the snoRNAs. This terminal structure, called the box C/D core motif (57, 60), functions as a binding site for snoRNP core proteins (8, 28, 33, 58). Most probably, snoRNA proteins bound to the pre-snoRNA protect the snoRNA sequences from the processing exonucleases and therefore delineate the correct snoRNA termini and provide metabolic stability for the mature snoRNA.
Several laboratories have demonstrated that a base-paired 5′,3′-terminal stem is absolutely required for the processing and accumulation of box C/D snoRNAs (6, 9, 20, 31, 57, 60). However, mysteriously enough, many intron-encoded box C/D snoRNAs expressed in mammalian cells (26, 54) or polycistronic snoRNAs accumulating in yeast (32) lack the canonical 5′,3′-terminal helix. Elements supporting the processing and accumulation of this large group of box C/D snoRNAs remain largely speculative. In this study, we demonstrate that processing of mammalian box C/D snoRNAs lacking a 5′,3′-terminal stem is supported by external intronic stem structures that are fully or partially degraded during the exonucleolytic processing of these snoRNAs.
MATERIALS AND METHODS
General procedures and oligos.
Unless stated otherwise, all techniques used for manipulating DNA, RNA, and oligodeoxynucleotides (oligos) were performed according to standard laboratory protocols (45). The following oligos were used in this study: 1, ATAATCGATGTAGGGTGATGAAAAAGAATCCTTAGGCG; 2, ATAACGCGTAGTCAGTGTAACTATGACAAG; 3, ATAATCGATGGAGGCAAGTAGGGTGATGAAAAAGAATCC; 4, ATAACGCGTGGAGGCAACAGTCAGTGTAAC; 5, ATAATCGATGGCGCCTCGCGTC ATAGTCAG; 6, ATACTCGAGACCAAGGAGAGCAAGGCAAGTGAG GG; 7, ATAATCGATCTCTATTCGTAGGGTGATGAAAAAGAATCC; 8, ATAACGCGTCTCTATTCCAGTCAGTGTAACTATG; 9, CGATGCAACAGCATGATGATCGACCGC; 10, GGTAAGATCTCTGATCAATGATGACATATGGTCAAG; 11, TCCATTTACCTGACTGTTGCA; 12, TTGATCAGAGATCTTACCGCGGTCGATCATCATGCTGTTGCAT; 13, CGCGTGCAACAGTCAGGTAAATGGACTTGACCATATGTCATCA; 14, ATAATCGATTCGAGACATGATGATCGACCGCGG; 15, ATAACGCGTGCAACAGTCAGATAAATGG; 16, ATAATCGATCAAGAGACATGATGATCG; 17, ATAATCGATTGCAAGAGACATGATGATGCG; 18, ATAATCGATCGTGCAAGAGACATGATGATCG; 19, ATAATCGATCGCGTGCAAGAGACATGATGATCGACCGCGG; 20, ATAATCGATTCGACACATGATGATCGACCGCGG; 21, CGCGTGCAACAGTCAGGTAAATGACCGCGGTCA; 22, TATGACCGCGGTCATTTACCTGACTGTTGCA; and 23, GGCCACAACCACGCCTAAGG.
Construction of plasmids for transfection of COS7 cells.
Construction of the pGLCXM mammalian expression vector has been described elsewhere (25). In brief, the human β-globin gene carrying three novel restriction sites (ClaI, XhoI, and MluI) in its second intron was placed under the control of the cytomegalovirus (CMV) promoter. To generate pGL-U46(0/0), the coding region of human U46 snoRNA was PCR amplified using oligos 1 and 2 as 5′- and 3′-end-specific primers, respectively. The amplified DNA fragment was digested with ClaI and MluI and inserted into the same sites of the pGLCXM expression construct. A similar approach was used to generate pGL-U46(8/9) and pGL-U46(93/54). Fragments of the human S8 ribosomal protein gene carrying the U46 gene and its 8- or 93-nucleotide upstream and 9- or 54-nucleotide downstream flanking sequences were amplified using oligos 3 and 5 and oligos 4 and 6, respectively. The amplified (8-U46-9) and (93-U46-54) fragments were inserted into the ClaI/MluI and ClaI/XhoI sites of pGLCXM, respectively. To obtain pGL-U46(8/9) and pGL-U46(8/9) (underlining indicates the destroyed part of the construct [see Results]), the U46 gene was PCR amplified using oligos 7 and 4 and oligos 3 and 8 as primers, respectively. The amplified fragments were cloned into the ClaI/MluI sites of pGLCXM. The same strategy was used to generate pGL-U46(8/9), except that oligos 7 and 8 were used as 5′- and 3′-end-specific PCR primers, respectively.
To generate pGL-X1, five synthetic oligos (oligos 9 to 13) were annealed, mixed with ClaI- and MluI-digested plasmid pGLCXM, and treated with T4 DNA ligase. Prior to annealing, oligos 10, 11, and 13 had been phosphorylated with T4 polynucleotide kinase. The resulting plasmid, pGL-X1, was used as a template for PCR amplification with oligos 14 and 15 as 5′- and 3′-end-specific primers, respectively. The resulting DNA fragment was inserted into the ClaI/MluI sites of pGLCXM, yielding pGL-X1(0). The same approach was used to construct pGL-X1(3), pGL-X1(5), pGL-X1(7), pGL-X1(9), and pGL-X1(5t), except that in these PCRs pGL-X1(0) was used as a template and oligos 16, 17, 18, 19, and 20 were used as 5′-end-specific primers, respectively. To generate pGL-X1(0)-(9) and pGL-X1(5t)-(9), the NdeI-MluI fragments of pGL-X1(0) and pGL-X1(5t) encompassing the 3′-terminal regions of the X1(0) and X1(5t) artificial snoRNAs, respectively, were replaced with a synthetic DNA fragment obtained after annealing of oligos 21 and 22. The identity of all constructions was verified by sequence analyses.
RNA analyses.
RNase A/T1 mapping was performed as described earlier (19, 25). Complementary RNA probes were synthesized in vitro with T7 RNA polymerase. All probes were purified on 5% denaturing polyacrylamide gels. To generate templates for the preparation of antisense RNA probes, the HindIII-EcoRI fragments of the pGL-U46, pGL-U46(8/9), pGL-U46(93/54), pGL-U46(8/9), pGL-U46(8/9), pGL-U46(8/9), pGL-X1, pGL-X1(0), pGL-X1(3), pGL-X1(5), pGL-X1(7), pGL-X1(9), pGL-X1(5t), pGL-X1(0)-(9), and pGL-X1(5t)-(9) expression constructs were cloned into the HindIII/EcoRI sites of pBluescribe KS(−) (Stratagene). The resulting recombinant plasmids were linearized with HindIII and used as templates for T7 RNA polymerase transcription. The 5′ terminus of the human U46 snoRNA was determined by primer extension analysis with 5 μg of template RNA extracted from a nuclear fraction of human HeLa cells (50) and 5 pmol of terminally labeled oligo 23 as a primer. Computer folding of human and mouse intronic sequences was performed by using the RNAdraw program (http://rnadraw.base8.se/).
RESULTS
Intronic flanking sequences are required for the accumulation of U46 intron-encoded snoRNA.
Contrary to the fact that a short 5′,3′-terminal helix structure has been demonstrated to be fundamental to the accumulation of both intron-encoded and polycistronic box C/D snoRNAs, a large number of naturally occurring box C/D snoRNAs lack a terminal stem (see above). To solve this apparent contradiction, we have set about identifying the cis-acting elements that direct the processing of the human U46 box C/D snoRNA, which has been reported to lack a canonical 5′,3′-terminal stem (26). Primer extension (Fig. 1A) and oligo ligation-PCR amplification (Fig. 1B) experiments demonstrated that the C and D boxes of the mature U46 snoRNA are preceded by five 5′-terminal and followed by two 3′-terminal nucleotides which are apparently unable to form a canonical terminal stem. The human U46 snoRNA is encoded within the second intron of the S8 ribosomal protein gene (26). To dissect elements essential for the accumulation of U46 snoRNA, fragments of the human S8 gene encompassing the coding region of U46 snoRNA with or without intronic flanking sequences were inserted into the second intron of the human β-globin gene (Fig. 2A). The resulting constructs were placed under the control of the CMV promoter and transfected into simian COS7 cells (25). The accumulation of U46 snoRNA and the spliced globin mRNA was monitored by RNase A/T1 mapping using antisense RNA probes specific for each expression construct (Fig. 2B). Control mapping with RNAs obtained from human HeLa cells (Fig. 2B, lanes 2, 6, and 10) and nontransfected COS7 cells (lanes 3, 7, and 11) revealed that the human and simian U46 snoRNAs can be readily distinguished based on their different migration properties in a denaturing polyacrylamide gel. RNA fragments protected by the simian U46 snoRNA migrated slightly faster than those protected by the human U46 snoRNA.
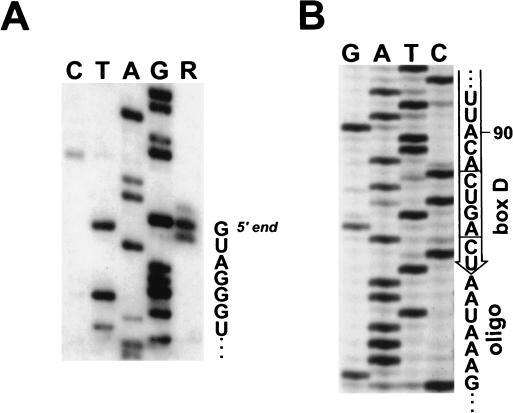
FIG. 1.
Characterization of the 5′ and 3′ termini of human U46 snoRNA. (A) Primer extension analysis. A terminally labeled oligo complementary to the human U46 snoRNA from positions 21 to 40 was annealed to HeLa cell snoRNAs and extended by use of avian myeloblastosis virus reverse transcriptase (lane R). Lanes G, A, T, and C represent dideoxy sequencing reactions performed on a recombinant plasmid carrying the human U46 gene. The extended DNA products were fractionated on a 6% sequencing gel. (B) Determination of the 3′ end of the human U46 snoRNA by the T4 RNA ligase-PCR procedure. Sequence analysis of three cDNA clones obtained from independent PCRs resulted in the same 3′-terminal snoRNA sequences. Sequences of the oligoribonucleotide ligated to the snoRNA are also indicated.
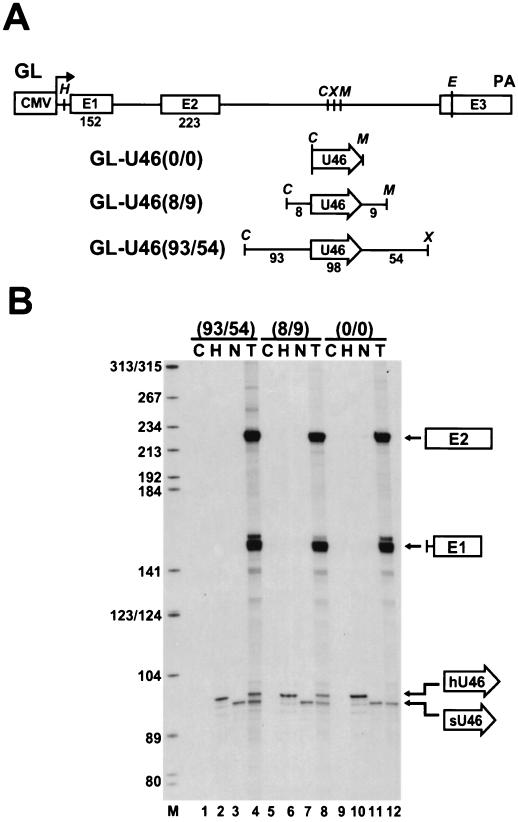
FIG. 2.
Processing of the U46 snoRNA from the second intron of the human β-globin pre-mRNA expressed in COS7 cells. (A) Schematic representation of the expression constructs used for transfection of simian COS7 cells. The CMV promoter, the exon regions (E1 to E3), and the polyadenylation (PA) site of the human β-globin gene are indicated. The relevant restriction sites are shown (C, ClaI; E, EcoRI; H, HindIII; M, MluI; X, XhoI). The human U46 intronic snoRNA (open arrow)—with or without its authentic flanking sequences—was inserted into the ClaI/MluI or ClaI/XhoI sites of the globin (GL) expression construct. (B) RNase A/T1 mapping of U46 snoRNA accumulation. RNAs isolated from human HeLa (H) cells and from transfected (T) or nontransfected (N) COS7 cells were mapped with sequence-specific RNA probes as indicated above the lanes; C, control mapping with Escherichia coli tRNA. RNA fragments protected by the first (E1) and second (E2) exons of the spliced globin mRNA and the human (hU46) and simian (sU46) U46 snoRNAs are indicated. Lane M, size markers in nucleotides.
Mapping of RNAs obtained from COS7 cells transfected with the GL-U46(93/54) (Fig. 2B, lane 4) and GL-U46(8/9) (lane 8) constructs yielded a protected RNA identical in size to the human U46 snoRNA, in addition to the endogenous simian U46-specific fragment. However, no human U46-specific sequences were detected in RNAs extracted from COS7 cells transfected with the GL-U46(0/0) construct, which carried only the coding region of the human U46 gene. The human β-globin mRNA was correctly and efficiently processed from the GL-U46(0/0) transcript (Fig. 2B, lane 12), as it was from the GL-U46(93/54) and GL-U46(8/9) transcripts (lanes 4 and 8). Thus, we concluded that the 8-nucleotide upstream and 9-nucleotide downstream flanking sequences of the U46 snoRNA carry indispensable information for snoRNA processing.
An intronic stem structure is required for the accumulation of U46 snoRNA.
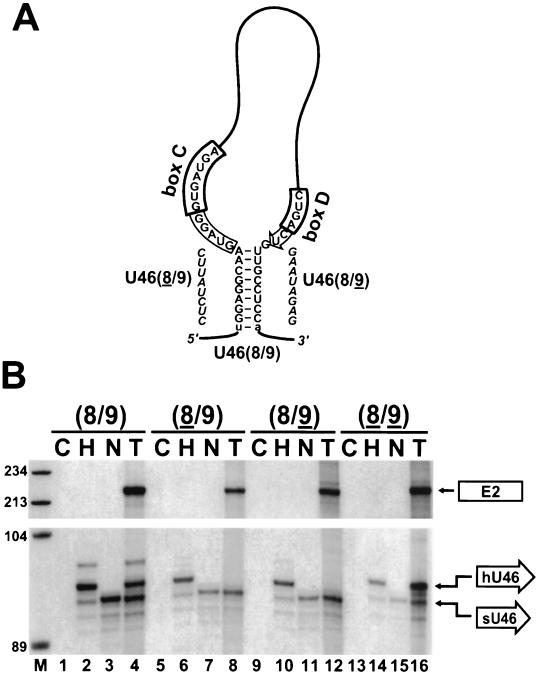
We noticed that the upstream and downstream intronic sequences flanking the human U46 snoRNA in the S8 pre-mRNA possess the potential to form an 8-bp perfect double helix (Fig. 3A). In the GL-U46(8/9) transcript, this interaction is accidentally elongated with an additional U-A base pair originating from the terminal nucleotides of the ClaI and MluI restriction sites used for the insertion of U46-containing fragments of the S8 gene. A possible function of the 5′,3′-terminal stem might be the folding of C and D boxes into close proximity with each other (57, 60). For processing of the human U46 snoRNA, instead of the canonical 5′,3′-terminal stem, an intronic helix may juxtapose the C and D boxes. To test this hypothesis, the putative intronic stem of the GL-U46(8/9) construct was destroyed by substitution of either the 5′ or the 3′ side of the helix (Fig. 3A). Upon transfection of the resulting GL-U46(8/9) and GL-U46(8/9) constructs into COS7 cells, the expression of the U46 snoRNA and the globin mRNA was tested by RNase mapping (Fig. 3B). The GL-U46(8/9) and GL-U46(8/9) pre-mRNAs were correctly and efficiently spliced, but no detectable amount of U46 snoRNA was processed from the excised introns of these transcripts (Fig. 3B, lanes 8 and 12). When the two stem mutations were combined and, therefore, the intronic stem structure of U46 was reestablished, the accumulation of faithfully processed U46 snoRNA was restored (Fig. 3B, lane 16). These results demonstrate that the formation of an external helix structure is essential for the production of U46 snoRNA and that the nucleotide composition of the intronic stem structure does not influence the efficiency or fidelity of the processing reaction.
FIG. 3.
Processing of U46 snoRNA with altered external stems. (A) Proposed secondary structure of the 5′,3′-terminal region of human U46 snoRNA in the GL-U46(8/9) pre-mRNA. Nucleotides derived from the human S8 pre-mRNA are in uppercase letters. Lowercase letters indicate nucleotides originating from the ClaI/MluI sites of the GL expression construct. Sequences retained in the mature U46 snoRNA are boxed. The C and D box motifs are highlighted. Sequences used to destroy the external stem of U46 in the GL-U46(8/9) and GL-U46(8/9) constructs are in italics. (B) RNase A/T1 mapping of U46 accumulation. The GL-U46(8/9), GL-U46(8/9), GL-U46(8/9), and GL-U46(8/9) constructs were transfected into COS7 cells. RNAs isolated from transfected (T) or nontransfected (N) COS cells were mapped with sequence-specific RNA probes as indicated above the lanes; H and C, control mappings with HeLa cell or E. coli RNAs, respectively. Lane M, size markers in nucleotides. See the legend to Fig. 2 for designations.
The accumulation of an intronic artificial box C/D snoRNA is supported by an external stem structure.
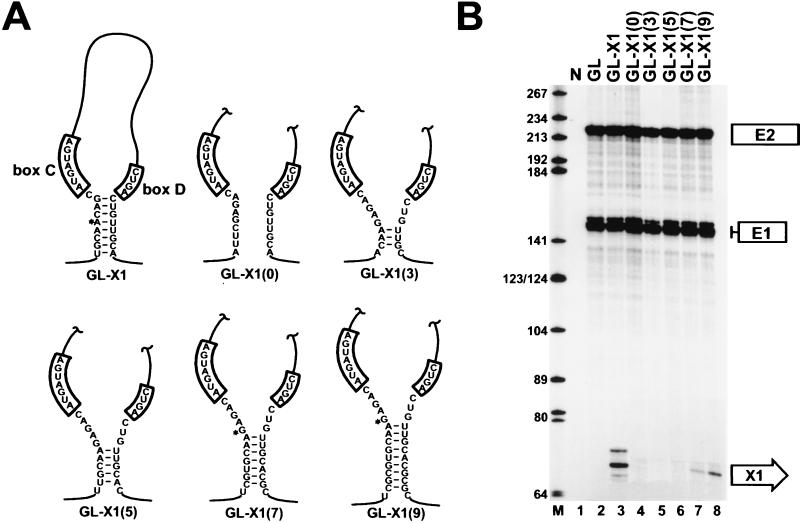
To exclude the formal possibility that internal sequence and/or structural elements located in the U46 snoRNA might contribute to the processing of this snoRNA, we constructed an artificial box C/D snoRNA, called X1 RNA. Apart from the essential box C and D motifs and the 3′-terminal nucleotides that were designed to form an 8-bp helix with sequences preceding the C box, the sequence of the X1 RNA was randomly generated. The artificial X1 locus was inserted into the ClaI and MluI sites of the pGLCXM expression vector (Fig. 2A) and transfected into COS7 cells. A predicted two-dimensional structure of the 5′,3′-terminal stem-box C/D domain of the putative pre-X1 RNA is shown in Fig. 4A. In the GL-X1(0) construct, base pairing of the terminal stem of X1 RNA was disrupted (Fig. 4A). In the GL-X1(3), GL-X1(5), GL-X1(7), and GL-X1(9) constructs, novel helices of increasing length were constructed outside of the predicted mature X1 RNA sequences.
FIG. 4.
Processing of an artificial box C/D snoRNA from the second intron of the human β-globin pre-mRNA. (A) Schematic representation of artificial box C/D snoRNAs. The X1, X1(0), X1(3), X1(5), X1(7), and X1(9) artificial snoRNAs were expressed in COS7 cells within the second intron of the human globin pre-mRNA. Asterisks indicate the 5′-terminal nucleotides of the processed X1, X1(7), and X1(9) RNAs. (B) RNase mapping of the accumulation of X1 artificial snoRNAs. RNAs extracted from COS7 cells transfected with the GL, GL-X1, GL-X1(0), GL-X1(3), GL-X1(5), GL-X1(7), or GL-X1(9) expression construct were mapped with sequence-specific RNA probes as indicated above the lanes. Lane N, control mapping performed with RNAs obtained from nontransfected cells; lane M, size markers in nucleotides. See the legend to Fig. 2 for designations.
RNase A/T1 mapping of RNAs extracted from COS7 cells transfected with the GL-X1 construct detected a 70- to 72-nucleotide-long RNA (Fig. 4B, lane 3) that was absent from cells transfected with the pGLCXM expression vector (lane 2). Since the size of the accumulating RNA corresponded to the predicted length of the mature X1 RNA, we concluded that the artificial snoRNA was processed from the globin pre-mRNA. This conclusion was further corroborated by primer extension analysis using an X1-specific oligo primer (data not shown). The accumulation of X1 RNA further supported the idea that the processing of box C/D snoRNAs can rely exclusively on the canonical box C/D-stem core motif (6, 44, 57). Again, predictably enough (6, 9, 20, 57), disruption of the terminal stem of X1 RNA completely abolished RNA accumulation (Fig. 4B, lane 4). More importantly, introduction of an external helix of 7 bp (Fig. 4B, lane 7) but not 3 nor 5 bp (lanes 5 and 6) restored the accumulation of X1 RNA. Extension of the length of the external helix up to 9 bp [GL-X1(9) construct] further improved the accumulation of the mature X1 RNA (Fig. 4B, lane 8), demonstrating that an intronic helix can substitute for the canonical terminal stem during the processing of mammalian intron-encoded box C/D snoRNAs. The 5′ termini of X1 RNAs excised from the GL-X1, GL-X1(7), and GL-X1(9) transcripts were determined by primer extension analysis (data not shown). In each case, the accumulating artificial snoRNA featured five 5′-terminal nucleotides preceding the C box motif, indicating that selection of the snoRNA 5′ terminus is independent of the position of the stem structure.
Compilation of mammalian intronic pre-snoRNA sequences lacking a canonical 5′,3′-terminal stem.
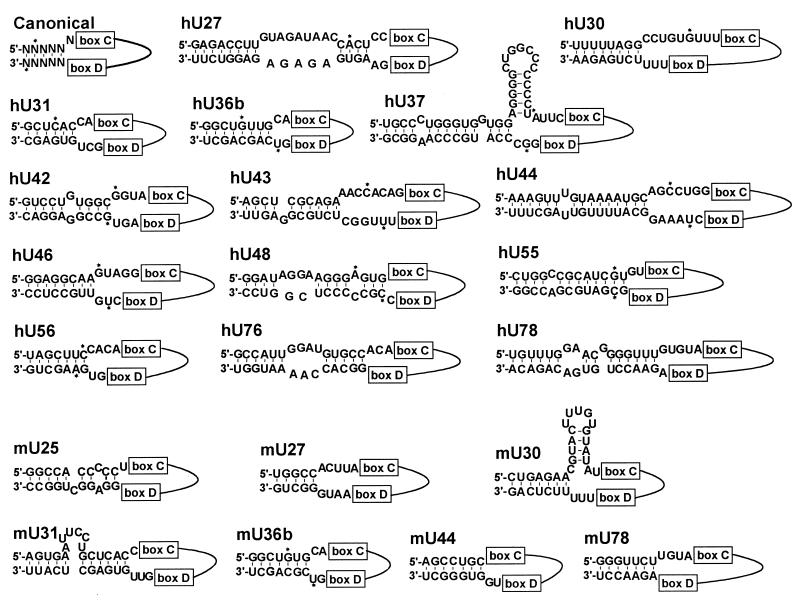
A closer inspection of the available human and mouse precursor sequences of box C/D intronic snoRNAs revealed that each snoRNA that lacks a 5′,3′-terminal stem possesses upstream and downstream intronic sequences which are able to form short stem structures (Fig. 5). The length of these putative double helices varies between 5 bp (mouse U27) and 16 bp (human U44). Unfortunately, the 5′- and 3′-terminal nucleotides have been experimentally determined for only a few snoRNAs (Fig. 5). Nevertheless, the available data show that in some instances, the putative external stem may include nucleotides that are preserved in the 5′- and/or 3′-terminal sequences of mature snoRNAs (e.g., hU31, hU36b, hU42, hU48, hU55, and mU36b). In other instances, the proposed external stem is composed of intronic sequences that are fully degraded during snoRNA formation (e.g., hU30, hU37, hU43, hU44, and hU46). While snoRNAs with a canonical box C/D-stem core motif possess five or six 3′-terminal nucleotides after the D box motif, snoRNAs lacking a terminal stem usually feature only two unpaired 3′-terminal nucleotides. In both groups of snoRNAs, the C box is preceded by four or, more frequently, five nucleotides (Fig. 5). In summary, the presence of potential external helices in the pre-RNAs of mammalian intron-encoded box C/D snoRNAs lacking a canonical 5′,3′-terminal stem further underlines the functional importance of intronic stem structures in snoRNA processing and accumulation.
FIG. 5.
Proposed structures of the intron-snoRNA junction regions of human and mouse pre-mRNAs encoding box C/D snoRNAs without a canonical 5′,3′-terminal stem. Sequences of the human (h) U27, U30, and U31 (54), U36b (18), U37 (38), U42, U43, U44, U46, U48, U55, and U56 (26), and U76 and U78 (47) pre-snoRNAs and the mouse (m) U25, U27, U30, and U31 (54), U36b (18), and U44 and U78 (47) pre-snoRNAs were taken from the literature. Solid lines represent snoRNA sequences located between the C and D box motifs. Asterisks indicate the experimentally determined 5′,3′-terminal nucleotides of mature snoRNAs.
An internal helix structure supports the accumulation of X1 artificial snoRNA.
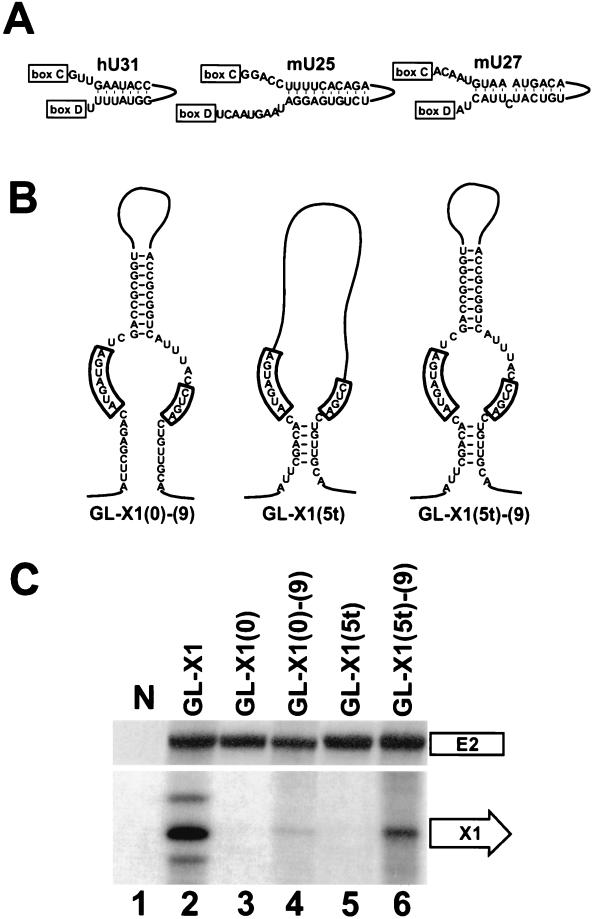
Although the data presented thus far are consistent with the idea that external helices play an important role in the biogenesis of box C/D snoRNAs, it remains questionable whether external helices alone could support the accumulation of some naturally occurring snoRNAs. The minimal length of the external stem capable of supporting production of the X1 artificial snoRNA was established as 7 bp (Fig. 4). Therefore, it seems unlikely that, for example, processing of the human U31 or the mouse U25 and U27 snoRNAs, which possess only 5- or 6-bp-long external helices, could rely exclusively on such short interactions (Fig. 5). Computer-aided modeling of the human U31 and the mouse U25 and U27 snoRNAs revealed that they contain 7- to 10-bp-long internal helices that could be formed by snoRNA sequences located between the C and D boxes (Fig. 6A). Moreover, extension of the structural investigation to other snoRNAs lacking a canonical 5′,3′-terminal stem showed that these snoRNAs frequently possess potential internal stem structures (data not shown).
FIG. 6.
Processing of artificial box C/D snoRNAs carrying internal stem structures. (A) Proposed internal stems of the human (h) U31 and the mouse (m) U25 and U27 snoRNAs. For other details, see the legend to Fig. 5. (B) Schematic representation of artificial X1 snoRNAs. (C) Accumulation of X1 RNAs processed from the human β-globin pre-mRNA. RNA samples obtained from COS7 cells transfected with the GL-X1, GL-X1(0), GL-X1(0)-(9), GL-X1(5t), or GL-X1(5t)-(9) expression construct were analyzed by RNase A/T1 mapping. For other details, see the legends to Fig. 2 and 4.
We assayed whether internal helices could support the accumulation of intron-encoded box C/D snoRNAs. To this end, processing from chimeric globin pre-mRNAs carrying the X1 artificial box C/D snoRNA either possessing or lacking an internal helix structure was investigated with COS7 cells (Fig. 6B and C). Consistent with our previous observation (Fig. 4), the X1(0) artificial snoRNA, which carries neither a terminal nor an external stem, failed to accumulate in COS7 cells (Fig. 6C, lane 3). However, when an internal stem of 9 bp was introduced into the X1(0) snoRNA the resulting X1(0)-(9) snoRNA was correctly, although inefficiently, processed from the globin pre-mRNA (Fig. 6B, lane 4).
Finally, we tested whether internal and external stem structures could function in a cooperative manner. The X1(5t) artificial snoRNA possesses the potential to form a terminal stem of 5 bp in a noncanonical configuration. Namely, the D box and the putative terminal stem of X1(5t) are separated by an unpaired nucleotide (Fig. 6B). Upon expression in COS7 cells, the noncanonical terminal stem of X1(5t) failed to support snoRNA processing from the globin pre-mRNA (Fig. 6C, lane 5). In contrast, when the noncanonical terminal stem of the X1(5t) snoRNA was supplemented with a 9-bp-long internal stem (Fig. 6B), the resulting X1(5t)-(9) snoRNA was faithfully processed (Fig. 6C, lane 6). Significantly, the level of accumulation of the X1(5t)-(9) snoRNA was comparable to that of the X1 snoRNA, carrying a canonical 5′,3′-terminal stem (Fig. 6C, lane 2). These results demonstrate that an internal stem structure can compensate for the lack of a functional terminal stem structure and that internal helices may function in concert with external elements during snoRNA processing.
DISCUSSION
Most box C/D snoRNAs are processed from intronic or polycistronic pre-snoRNAs by exonucleolytic activities (see above). The box C/D terminal core motif, including the phylogenetically conserved nucleotides of boxes C and D and the base-paired 5′,3′-terminal stem, plays a pivotal role in the biogenesis and function of these snoRNAs. The box C/D-stem motif directs the correct processing of the snoRNA (6, 9, 57, 60), provides metabolic stability for the mature snoRNA (6, 20, 31), directs the nucleolar localization of the snoRNA (29, 36, 44), and functions in the rRNA 2′-O-methylation reaction (10, 26, 27). The multiple functions of the box C/D-stem motif are accomplished by snoRNP proteins which directly or indirectly bind to this structural motif (8, 14, 28, 58). In vitro reconstitution experiments suggested that, in addition to the conserved C and D boxes, the adjacent 5′,3′-terminal stem also plays a fundamental role in snoRNP assembly (8, 58). Consistent with this conclusion, the terminal stem is essential for the processing (57, 60) and accumulation (9, 20, 31) of box C/D snoRNAs.
Contrary to the observed essentiality of the 5′,3′-terminal stem, many box C/D snoRNAs lack a terminal helix (26, 32, 54). Elements underlying the accumulation of these snoRNAs remain conjectural. In this study, systematic modeling of pre-mRNA introns hosting a box C/D snoRNA suggested that the presence of an external intronic stem is a common feature of mammalian snoRNAs lacking a terminal stem (Fig. 5). Indeed, in vivo processing studies demonstrated that an intronic stem is required for the accumulation of the human U46 snoRNA, which features no terminal stem (Fig. 2 and 3). Apparently, the nucleotide composition of the external stem of U46 snoRNA has no functional importance, since replacement of the cognate intronic stem of U46 snoRNA with an artificial helix influenced neither the efficiency nor the fidelity of snoRNA excision (Fig. 3). The above conclusion was further corroborated by the fact that, in the presence of an external stem, the X1 artificial snoRNA was efficiently processed from the β-globin pre-mRNA (Fig. 4). Together, these findings suggest that the processing of mammalian box C/D snoRNAs lacking a canonical 5′,3′-terminal stem is supported by external stem structures that are fully or partially formed by intronic sequences flanking the snoRNA.
The notion that external stem structures can facilitate the processing of intronic box C/D snoRNAs reinforces the previous idea that bringing boxes C and D into close proximity is an important requirement for snoRNA accumulation (52, 57). Most likely, appropriate spatial arrangement of the C and D boxes promotes binding of the snoRNP core proteins that protect the snoRNA sequences from exonucleolytic degradation. Since the intronic sequences involved in external helix formation are degraded during snoRNA excision, we propose that folding of the C and D boxes together is essential only for snoRNA processing and that snoRNP proteins bound to the C and D boxes provide metabolic stability for the mature snoRNA. This idea also implies that, contrary to the previous belief, the canonical 5′,3′-terminal stem functions mainly, if not exclusively, in the processing of box C/D snoRNAs. Production of mature intronic snoRNAs can be considered a compromise of two competitive processes, namely, exonucleolytic intron degradation and snoRNP assembly on the intronic precursor RNA. In vitro snoRNP reconstitution experiments suggested that the canonical box C/D core motif, in which the terminal helix is immediately followed by the D box and is separated from the C box by one unpaired nucleotide (Fig. 5), represents the most favorable structural configuration for snoRNP assembly (8, 58). We assume that the accumulation of intronic snoRNAs with a canonical 5′,3′-terminal stem depends mainly on the level of synthesis of the host pre-mRNA, since these snoRNAs are processed with high efficacy. In contrast, the expression of snoRNAs lacking a canonical terminal stem can depend greatly on the structural conformation of the pre-snoRNA (Fig. 4 and 6). In other words, intronic pre-snoRNAs with weak snoRNP binding capacity are more likely to be degraded by processing exonucleases; therefore, these snoRNAs may accumulate less efficiently (Fig. 4).
Previously, it has been hypothesized that the processing of box C/D snoRNAs lacking a 5′,3′-terminal stem is facilitated by internal stem structures that are formed by snoRNA sequences located between the C and D boxes (57). Although an internal helix introduced into the X1 artificial snoRNA restored a detectable level of snoRNA accumulation, it failed to support the efficient production of the X1 RNA (Fig. 6). This result indicates that folding of the C and D boxes together by an internal stem could not entirely fulfill the structural requirements of efficient snoRNA processing. However, in concert with a short noncanonical terminal stem, the same internal helix could support the efficient processing of X1 RNA, even though the noncanonical stem was unable to sustain snoRNA processing alone (Fig. 6). This result suggests that internal stem structures, in conjunction with other structural elements, such as noncanonical terminal or external stem structures, may significantly influence the accumulation of box C/D snoRNAs.
Very recently, processing of the yeast U18 and snR38 intron-encoded snoRNAs has been reported to be dependent on external intronic stem structures (56), suggesting that parallels can be drawn between the processing mechanisms of yeast and mammalian intronic snoRNAs lacking a terminal stem. Indeed, a closer examination of yeast polycistronic and intronic box C/D snoRNAs devoid of a canonical terminal stem revealed that these snoRNAs are flanked by inverted repeat sequences that, in principle, are able to form helix structures (56; data not shown). In contrast to the mammalian introns, large parts of the yeast snoRNA host introns can be folded into long rod-shaped structures that are built of 27 (U18) to 59 (snR39 and snR59) bp forming small internal loops and bulges (data not shown). Remarkably, the snoRNAs are located invariantly on the top of the long intronic stem structure. The functional importance of this unique organization of the yeast snoRNA host introns, if any, remains to be established.
In conclusion, in vivo processing experiments performed with the human U46 snoRNA and an artificial box C/D snoRNA revealed that the biogenesis of mammalian intron-encoded box C/D snoRNAs is a more complex process than has been anticipated. Besides the previously characterized box C/D-stem core motif, additional cis-acting elements located within the snoRNA and/or in the flanking intronic sequences may contribute to the processing and accumulation of this class of snoRNAs. Most likely, the external and internal stem structures, like the canonical 5′,3′-terminal stem, promote the folding of C and D boxes into close proximity with each other and thereby facilitate the assembly of the snoRNP core complex on the pre-snoRNA.
ACKNOWLEDGMENTS
We thank Y. de Preval for the synthesis of oligonucleotides.
X.D. was supported by Ministère de l'Education Nationale, de la Recherche, et de la Technologie. This work was supported by the Centre Nationale de la Recherche Scientifique, Université Paul Sabatier, Toulouse, France, and by grants from la Ligue Nationale Contre le Cancer and l'Association pour la Recherche sur le Cancer.
REFERENCES
- 1.Allmang C, Petfalski E, Podtelejnikov A, Mann M, Tollervey D, Mitchell P. The yeast exosome and human PM-Scl are related complexes of 3′→5′ exonucleases. Genes Dev. 1999;13:2148–2158. doi: 10.1101/gad.13.16.2148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Allmang C, Kufel J, Chanfreau G, Mitchell P, Petfalski E, Tollervey D. Functions of the exosome in rRNA, snoRNA and snRNA synthesis. EMBO J. 1999;18:5399–5410. doi: 10.1093/emboj/18.19.5399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bachellerie J-P, Michot B, Nicoloso M, Balakin A, Ni J, Fournier M J. Antisense snoRNAs: a family of nucleolar RNAs with long complementarities to rRNA. Trends Biochem Sci. 1995;20:261–264. doi: 10.1016/s0968-0004(00)89039-8. [DOI] [PubMed] [Google Scholar]
- 4.Balakin A G, Smith L, Fournier M J. The RNA world of the nucleolus: two major families of small RNAs defined by different box elements with related functions. Cell. 1996;86:823–834. doi: 10.1016/s0092-8674(00)80156-7. [DOI] [PubMed] [Google Scholar]
- 5.Bortolin M-L, Ganot P, Kiss T. Elements essential for accumulation and function of small nucleolar RNAs directing site-specific pseudouridylation of ribosomal RNAs. EMBO J. 1999;18:457–469. doi: 10.1093/emboj/18.2.457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Caffarelli E, Fatica A, Prislei S, De Gregorio E, Fragapane P, Bozzoni I. Processing of the intron-encoded U16 and U18 snoRNAs: the conserved C and D boxes control both the processing reaction and the stability of the mature snoRNA. EMBO J. 1996;15:1121–1131. [PMC free article] [PubMed] [Google Scholar]
- 7.Caffarelli E, Arese M, Santoro B, Fragapane P, Bozzoni I. In vitro study of processing of the intron-encoded U16 snoRNA in Xenopus laevis. Mol Cell Biol. 1994;14:2966–2974. doi: 10.1128/mcb.14.5.2966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Caffarelli E, Losito M, Giorgi C, Fatica A, Bozzoni I. In vitro identification of nuclear factors interacting with the conserved elements of box C/D small nucleolar RNAs. Mol Cell Biol. 1998;18:1023–1028. doi: 10.1128/mcb.18.2.1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cavaillé J, Bachellerie J-P. Processing of fibrillarin-associated snoRNAs from pre-mRNA introns: an exonucleolytic process exclusively directed by the common stem-box terminal structure. Biochimie. 1996;78:443–456. doi: 10.1016/0300-9084(96)84751-1. [DOI] [PubMed] [Google Scholar]
- 10.Cavaillé J, Nicoloso M, Bachellerie J-P. Targeted ribose methylation of RNA in vivo directed by tailored antisense RNA guides. Nature. 1996;383:732–735. doi: 10.1038/383732a0. [DOI] [PubMed] [Google Scholar]
- 11.Cecconi F, Mariottini P, Amaldi F. The Xenopus intron-encoded U17 snoRNA is produced by exonucleolytic processing of its precursor in oocytes. Nucleic Acids Res. 1995;23:4670–4676. doi: 10.1093/nar/23.22.4670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chanfreau G, Rotondo G, Legrain P, Jacquier A. Processing of a dicistronic small nucleolar RNA precursor by the RNA endonuclease Rnt1. EMBO J. 1998;17:3726–3737. doi: 10.1093/emboj/17.13.3726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chanfreau G, Legrain P, Jacquier A. Yeast RNase III as a key processing enzyme in small nucleolar RNA metabolism. J Mol Biol. 1998;284:975–988. doi: 10.1006/jmbi.1998.2237. [DOI] [PubMed] [Google Scholar]
- 14.Fatica A, Galardi S, Altieri F, Bozzoni I. Fibrilarin binds directly and specifically to U16 box C/D snoRNA. RNA. 2000;6:88–95. doi: 10.1017/s1355838200991623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fragapane P, Prislei S, Michienzi A, Caffarelli E, Bozzoni I. A novel small nucleolar RNA (U16) is encoded inside a ribosomal protein intron and originates by processing of the pre-mRNA. EMBO J. 1993;12:2921–2928. doi: 10.1002/j.1460-2075.1993.tb05954.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ganot P, Caizergues-Ferrer M, Kiss T. The family of box ACA small nucleolar RNAs is defined by an evolutionarily conserved secondary structure and ubiquitous sequence elements essential for RNA accumulation. Genes Dev. 1997;11:941–956. doi: 10.1101/gad.11.7.941. [DOI] [PubMed] [Google Scholar]
- 17.Ganot P, Bortolin M-L, Kiss T. Site-specific pseudouridine formation in preribosomal RNA is guided by small nucleolar RNAs. Cell. 1997;89:799–809. doi: 10.1016/s0092-8674(00)80263-9. [DOI] [PubMed] [Google Scholar]
- 18.Gilley J, Fried M. Evolution of U24 and U36 snoRNAs encoded within introns of vertebrate rpL7a gene homologs: unique features of mammalian U36 variants. DNA Cell Biol. 1998;17:591–602. doi: 10.1089/dna.1998.17.591. [DOI] [PubMed] [Google Scholar]
- 19.Goodall G J, Wiebauer K, Filipowicz W. Analysis of pre-mRNA processing in transfected plant protoplasts. Methods Enzymol. 1990;181:148–161. doi: 10.1016/0076-6879(90)81117-d. [DOI] [PubMed] [Google Scholar]
- 20.Huang G M, Jarmolowski A, Struck J C R, Fournier M J. Accumulation of U14 small nuclear RNA in Saccharomyces cerevisiae requires box C, box D, and a 5′,3′-terminal stem. Mol Cell Biol. 1992;12:4456–4463. doi: 10.1128/mcb.12.10.4456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hughes J M X, Ares M., Jr Depletion of U3 small nucleolar RNA inhibits cleavage in the 5′ external transcribed spacer of yeast pre-ribosomal RNA and impairs formation of 18S ribosomal RNA. EMBO J. 1991;10:4231–4239. doi: 10.1002/j.1460-2075.1991.tb05001.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kass S, Tyc K, Steitz J A, Sollner-Webb B. The U3 small nucleolar ribonucleoprotein functions in the first step of preribosomal RNA processing. Cell. 1990;60:897–908. doi: 10.1016/0092-8674(90)90338-f. [DOI] [PubMed] [Google Scholar]
- 23.Kiss T, Bortolin M L, Filipowicz W. Characterization of the intron-encoded U19 RNA, a new mammalian small nucleolar RNA that is not associated with fibrillarin. Mol Cell Biol. 1996;16:1391–1400. doi: 10.1128/mcb.16.4.1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kiss T, Filipowicz W. Small nucleolar RNAs encoded by introns of the human cell cycle regulatory gene RCC1. EMBO J. 1993;12:2913–2920. doi: 10.1002/j.1460-2075.1993.tb05953.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kiss T, Filipowicz W. Exonucleolytic processing of small nucleolar RNAs from pre-mRNA introns. Genes Dev. 1995;9:1411–1424. doi: 10.1101/gad.9.11.1411. [DOI] [PubMed] [Google Scholar]
- 26.Kiss-László Z, Henry Y, Bachellerie J-P, Caizergues-Ferrer M, Kiss T. Site-specific ribose methylation of preribosomal RNA: a novel function for small nucleolar RNAs. Cell. 1996;85:1077–1088. doi: 10.1016/s0092-8674(00)81308-2. [DOI] [PubMed] [Google Scholar]
- 27.Kiss-László Z, Henry Y, Kiss T. Sequence and structural elements of methylation guide snoRNAs essential for site-specific ribose methylation of pre-rRNA. EMBO J. 1998;17:797–807. doi: 10.1093/emboj/17.3.797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lafontaine D L J, Tollervey D. Nop58 is a common component of the box C+D snoRNPs that is required for snoRNP stability. RNA. 1999;5:455–467. doi: 10.1017/s135583829998192x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lange T S, Borovjagin A, Maxwell E S, Gerbi S A. Conserved boxes C and D are essential nucleolar localization elements of U14 and U8 snoRNAs. EMBO J. 1998;17:3176–3187. doi: 10.1093/emboj/17.11.3176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li H V, Zagorski J, Fournier M J. Depletion of U14 small nuclear RNA (snR128) disrupts production of 18S rRNA in Saccharomyces cerevisiae. Mol Cell Biol. 1990;10:1145–1152. doi: 10.1128/mcb.10.3.1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li H V, Fournier M J. U14 function in Saccharomyces cerevisiae can be provided by large deletion variants of yeast U14 and hybrid mouse-yeast U14 RNAs. EMBO J. 1992;11:683–689. doi: 10.1002/j.1460-2075.1992.tb05100.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lowe T M, Eddy S R. A computational screen for methylation guide snoRNAs in yeast. Science. 1999;283:1168–1171. doi: 10.1126/science.283.5405.1168. [DOI] [PubMed] [Google Scholar]
- 33.Lyman S K, Gerace L, Baserga S L. Human Nop5/Nop58 is a component common to the box C/D small nucleolar ribonucleoproteins. RNA. 1999;5:1597–1604. doi: 10.1017/s1355838299991288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Maxwell E S, Fournier M J. The small nucleolar RNAs. Annu Rev Biochem. 1995;35:897–934. doi: 10.1146/annurev.bi.64.070195.004341. [DOI] [PubMed] [Google Scholar]
- 35.Morrissey J P, Tollervey D. Yeast snR30 is a small nucleolar RNA required for 18S rRNA synthesis. Mol Cell Biol. 1993;13:2469–2477. doi: 10.1128/mcb.13.4.2469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Narayanan A, Speckmann W, Terns R, Terns M P. Role of the box C/D motif in localization of small nucleolar RNAs to coiled bodies and nucleoli. Mol Biol Cell. 1999;10:2131–2147. doi: 10.1091/mbc.10.7.2131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ni J, Tien A L, Fournier M J. Small nucleolar RNAs direct site-specific synthesis of pseudouridine in ribosomal RNA. Cell. 1997;89:565–573. doi: 10.1016/s0092-8674(00)80238-x. [DOI] [PubMed] [Google Scholar]
- 38.Nicoloso M, Qu L-H, Michot B, Bachellerie J-P. Intron-encoded antisense small nucleolar RNAs: the characterization of nine novel species points to their role as guides for the 2′-O-ribose methylation of rRNAs. J Mol Biol. 1996;260:178–195. doi: 10.1006/jmbi.1996.0391. [DOI] [PubMed] [Google Scholar]
- 39.Ooi S L, Samarsky D A, Fournier M J, Boeke J D. Intronic snoRNA biosynthesis in Saccharomyces cerevisiae depends on the lariat-debranching enzyme: intron length effects and activity of precursor snoRNA. RNA. 1998;4:1096–1110. doi: 10.1017/s1355838298980785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Peculis B A, Steitz J A. Disruption of U8 nucleolar snRNA inhibits 5.8S and 28S rRNA processing in the Xenopus oocyte. Cell. 1993;73:1233–1245. doi: 10.1016/0092-8674(93)90651-6. [DOI] [PubMed] [Google Scholar]
- 41.Petfalski E, Dandekar T, Henry Y, Tollervey D. Processing of the precursors to small nucleolar RNAs and rRNAs requires common components. Mol Cell Biol. 1998;18:1181–1189. doi: 10.1128/mcb.18.3.1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Prislei S, Michienzi A, Presutti C, Fragapane P, Bozzoni I. Two different snoRNAs are encoded in introns of amphibian and human L1 ribosomal protein genes. Nucleic Acids Res. 1993;21:5824–5830. doi: 10.1093/nar/21.25.5824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Qu L-H, Henras A, Lu Y-J, Zhou H, Zhou W X, Zhu Y-Q, Zhao J, Henry Y, Caizergues-Ferrer M, Bachellerie J-P. Seven novel methylation guide small nucleolar RNAs are processed from a common polycistronic transcript by Rat1p and RNase III in yeast. Mol Cell Biol. 1999;19:1144–1158. doi: 10.1128/mcb.19.2.1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Samarsky D A, Fournier M J, Singer R H, Bertrand E. The snoRNA box C/D motif directs nucleolar targeting and also couples snoRNA synthesis and localization. EMBO J. 1998;17:3747–3757. doi: 10.1093/emboj/17.13.3747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 46.Savino R, Gerbi S A. In vivo disruption of Xenopus U3 snRNA affects ribosomal RNA processing. EMBO J. 1990;9:2299–2308. doi: 10.1002/j.1460-2075.1990.tb07401.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Smith M S, Steitz J A. Classification of gas5 as a multi-small-nucleolar-RNA (snoRNA) host gene and a member of the 5′-terminal oligopyrimidine gene family reveals common features of snoRNA host genes. Mol Cell Biol. 1998;18:6897–6909. doi: 10.1128/mcb.18.12.6897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sollner-Webb B, Tycowski K T, Steitz J A. Ribosomal RNA processing in eukaryotes. In: Zimmermann R A, Dahlberg A E, editors. Ribosomal RNA. Structure, evolution, processing, and function in protein biosynthesis. Boca Raton, Fla: CRC Press, Inc.; 1996. pp. 469–490. [Google Scholar]
- 49.Tollervey D, Kiss T. Function and synthesis of small nucleolar RNAs. Curr Opin Cell Biol. 1997;9:337–342. doi: 10.1016/s0955-0674(97)80005-1. [DOI] [PubMed] [Google Scholar]
- 50.Tyc K, Steitz J A. U3, U8 and U13 comprise a new class of mammalian snRNPs localized in the nucleolus. EMBO J. 1989;8:3113–3119. doi: 10.1002/j.1460-2075.1989.tb08463.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tycowski K T, Smith C M, Shu M-D, Steitz J A. A small nucleolar RNA required for site-specific ribose methylation of rRNA in Xenopus. Proc Natl Acad Sci USA. 1996;93:14480–14485. doi: 10.1073/pnas.93.25.14480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tycowski K T, Shu M-D, Steitz J A. A small nucleolar RNA is processed from an intron of the human gene encoding ribosomal protein S3. Genes Dev. 1993;6:1120–1130. doi: 10.1101/gad.7.7a.1176. [DOI] [PubMed] [Google Scholar]
- 53.Tycowski K T, Shu M-D, Steitz J A. Requirement for intron-encoded U22 small nucleolar RNA in 18S ribosomal RNA maturation. Science. 1994;266:1558–1561. doi: 10.1126/science.7985025. [DOI] [PubMed] [Google Scholar]
- 54.Tycowski K T, Shu M-D, Steitz J A. A mammalian gene with introns instead of exons generating stable RNA products. Nature. 1996;379:464–466. doi: 10.1038/379464a0. [DOI] [PubMed] [Google Scholar]
- 55.Villa T, Ceradini F, Presutti C, Bozzoni I. Processing of the intron-encoded U18 small nucleolar RNA in the yeast Saccharomyces cerevisiae relies on both exo- and endonucleolytic activities. Mol Cell Biol. 1998;18:3376–3383. doi: 10.1128/mcb.18.6.3376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Villa T, Ceradini F, Bozzoni I. Identification of a novel element required for processing of intron-encoded box C/D small nucleolar RNAs in Saccharomyces cerevisiae. Mol Cell Biol. 2000;20:1311–1320. doi: 10.1128/mcb.20.4.1311-1320.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Watkins N J, Leverette R D, Xia L, Andrew M T, Maxwell E S. Elements essential for processing intronic U14 snoRNA are located at the termini of the mature snoRNA sequence and include conserved nucleotide boxes C and D. RNA. 1996;2:118–133. [PMC free article] [PubMed] [Google Scholar]
- 58.Watkins N J, Newman D R, Kuhn J F, Maxwell E S. In vitro assembly of the mouse U14 snoRNP core complex and identification of a 65-kDa box C/D-binding protein. RNA. 1998;5:582–593. doi: 10.1017/s1355838298980128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Weinstein L B, Steitz J A. Guided tours: from precursor snoRNA to functional snoRNP. Curr Opin Cell Biol. 1999;11:378–384. doi: 10.1016/S0955-0674(99)80053-2. [DOI] [PubMed] [Google Scholar]
- 60.Xia L, Watkins N J, Maxwell E S. Identification of specific nucleotide sequences and structural elements required for intronic U14 snoRNA processing. RNA. 1997;3:17–26. [PMC free article] [PubMed] [Google Scholar]