Abstract
Multilocus sequence typing and antigen gene sequencing were used to investigate an outbreak of meningococcal disease in a university in the United Kingdom. The data obtained showed that five distinct Neisseria meningitidis strains belonging to the ET-37 complex were present in the student population during the outbreak. Three of these strains were not associated with invasive disease, and two distinct strains caused invasive disease, including several fatalities. The initial case of the disease cluster was caused by a strain distinct from that responsible for at least two subsequent cases and two cases remote from the university, which were epidemiologically linked to the outbreak. These observations were consistent with pulsed-field gel electrophoresis data, but the sequence data alone were sufficient to resolve the strains involved in the disease cluster. Interpretation of the nucleotide sequence data was more straightforward than interpretation of the fingerprint patterns, and the sequence data provided information on the genetic differences among the isolates.
Management of meningococcal disease outbreaks is complicated by their unpredictability, the rapidity and severity of the disease, and the resulting public anxiety (4). In Europe and North America, most pathological infections with Neisseria meningitidis are sporadic, endemic cases among infants. Disease clusters also occur, and these often affect teenagers and young adults attending schools, universities, or other institutions. Timely and accurate isolate characterization is essential to distinguish genuine disease outbreaks, comprising related cases with a common epidemiological source, from clusters of temporally and geographically proximate but unrelated cases. Although identification of the serogroup of the outbreak strain is essential, given the availability of vaccines against some, but not all, capsular polysaccharides, serogrouping alone is not sufficient for outbreak identification. Serotyping and serosubtyping (9), based on subcapsular protein antigens, are also inadequate for this purpose, particularly for serogroup B and C meningococci, which are responsible for the majority of infections: an increasing proportion of isolates are reported as not typeable (NT) or not subtypeable (12, 14, 15).
Disease outbreaks in educational and military institutions are often caused by meningococci belonging to the electrophoretic type 37 (ET-37) complex (25, 26). The ET-37 complex, in common with other hypervirulent lineages of meningococci, was first identified by multilocus enzyme electrophoresis (5, 17, 19), a technique unsuited to rapid outbreak investigation. Multilocus sequence typing (MLST), which is similar in concept to multilocus enzyme electrophoresis but which uses nucleotide sequence determination to identify alleles of housekeeping genes, can rapidly identify hypervirulent meningococci (13). This report describes the use of MLST (10, 13) and antigen gene sequencing (8, 16) in the investigation of a meningococcal disease cluster at a British university (27).
MATERIALS AND METHODS
Nucleotide sequence determination.
Chromosomal DNA was extracted from meningococcal cells grown as described previously (8) by the Isoquick DNA extraction procedure (Orca Research). Amplification and sequencing of the MLST genes were as described previously (13), with the addition of primers for amplification (primer fumC-A1 [5′-CAC CGA ACA CGA CAC GAT GG-3′] and primer fumC-A2 [5′-ACG ACC AGT TCG TCA AAC TC-3′]) and sequencing (primer fumC-S1 [5′-TCG GCA CGG GTT TGA ACA GC-3′] and primer fumC-S2 [5′-CAA CGG CGG TTT CGC GCA AC-3′]) of the fumC gene. Amplification and sequencing of the porA and porB genes were as described previously (21, 24). Sequencing of the tbpB gene was as described previously (18). Sequencing reactions were performed with Big Dye terminators (PE Biosystems), and the products were separated and detected with an Applied Biosystems Prism 377 automated sequencer. The sequences were assembled with the sequence analysis package of Staden (20).
PFGE fingerprinting.
Approximately 10 N. meningitidis colonies, grown overnight on Columbia agar (Oxoid Ltd.) supplemented with 5% horse blood in a atmosphere of 5% CO2, were suspended in 150 μl of cell suspension buffer (20 mM sodium chloride, 50 mM EDTA, 10 mM Tris-Cl [pH 7.2]). The suspension was mixed with 150 μl of 2.0% agarose for pulsed-field gel electrophoresis (PFGE) plugs (Sigma) at 50°C, and the mixture was immediately dispensed in two PFGE plug molds (Bio-Rad). Once set, the plugs were expelled into 1 ml of lysis buffer (1 mg of lysozyme per ml, 50 mM sodium chloride, 0.2% sodium deoxycholate, 0.5% sodium lauryl sarcosine, 10 mM Tris-Cl [pH 7.2]) and were incubated at 37°C overnight. The plugs were washed three times for 30 min each time with 2 ml of TE buffer (50 mM EDTA, 20 mM Tris-Cl [pH 8.0]) at room temperature and were then digested overnight at 55°C in 1 ml of proteinase K reaction buffer (1 mg of proteinase K per ml, 0.2% sodium deoxycholate, 1% sodium lauryl sarcosine, 100 mM EDTA [pH 8.0]). Finally, the plugs were washed three times with TE buffer.
The plugs were cut into pieces that were washed once in 0.1 × TE buffer for 1 h and were equilibrated for ≥3 h in 300 μl of the appropriate enzyme-specific restriction buffer (New England Biolabs). The buffer was replaced, and approximately 10 U of the appropriate restriction endonuclease (SpeI, NheI, or SfiI) (New England Biolabs) plus 0.1% acetylated bovine serum albumin (New England Biolabs) was added. After overnight incubation at the temperature recommended for the enzyme by the manufacturer, restriction digestion was stopped by replacing the reaction buffer with TE buffer. The digestion products were separated with a Genepath System (Bio-Rad) operated in accordance with the manufacturer's instructions. Program 13, for the separation of fragments of 25 to 400 kb, was used for all endonuclease digestion products. The gels were stained with 0.5 mg of ethidium bromide (Sigma) per ml, visualized with UV illumination, and photographed with a digital camera. Experiments with several replicate gels were performed for each restriction endonuclease.
Nucleotide sequence accession numbers.
The sequences of the tbpB alleles from the ET-37 complex isolate, strain 1, and strain 2 and from strains 3, 4, and 5 have been given GenBank accession no. AJ250244 and AJ250243, respectively.
RESULTS
Epidemiology.
During 19 days there were six university-associated cases of meningococcal disease with three deaths. Case 1 occurred 2 weeks before cases 2 to 6, which were diagnosed over 5 days, and cases 1 to 5 involved students from the same accommodation complex. Serogroup C meningococci were isolated from cases 1, 3, and 6; cases 2 and 5 were confirmed as having serogroup C meningococcal infections by PCR (2); and case 4 was diagnosed solely on clinical grounds. Two patients from other towns (remote cases 1 and 2) were epidemiologically suspected as being part of the outbreak. During outbreak investigation and control, 587 students were examined for meningococcal carriage: 147 meningococci were isolated, and 6 of these were serogroup C.
MLST.
All of the isolates from cases and five of the six isolates from carriers were shown by MLST analysis to belong to the ET-37 complex, but the isolates were not identical. Those from case 1, carrier 1, and carrier 2 were sequence type (ST)-11, which had previously been reported to be characteristic of ET-37 complex isolates (13). The other isolates from cases, including those from the remote cases, were ST-50 (differentiated from ST-11 by the aroE locus), two isolates from carriers (carriers 4 and 5) were ST-51 (which differed from ST-11 at the fumC and pdhC loci), and the isolate from carrier 3 was ST-52 (which varied from ST-11 at the abcZ locus) (Table 1 and Table 2).
TABLE 1.
STs found among serogroup C strains associated with the outbreak
TABLE 2.
Properties of ET-37 meningococci isolated during the outbreak
| Isolate description | Serological characterization | ST | porA VR1,VR2a | porB alleleb | tpbB allele |
|---|---|---|---|---|---|
| ET-37 complex isolate from United Kingdom (1993) | C:2a:P1.5 | 11 | 5a,10d | 2-2 | 2 |
| Strain 1 | |||||
| Case 1 | C:2a:NT | 11 | 5,2c | 2-36 | 2 |
| Carrier 1 (case 1 contact) | C:2a:NT | 11 | 5,2c | 2-36 | 2 |
| Strain 2 (outbreak strain) | |||||
| Case 3 | C:NT:P1.5 | 50 | 5a,10d | 2-37 | 2 |
| Case 6 | C:NT:P1.5 | 50 | 5a,10d | 2-37 | 2 |
| Remote case 1 | C:NT:P1.5 | 50 | 5a,10d | 2-37 | 2 |
| Remote case 2 | C:NT:P1.5 | 50 | 5a,10d | 2-37 | 2 |
| Strain 3, carrier 2 | C:NT:P1.5,2 | 11 | 5,2 | 2-39 | 1 |
| Strain 4, carrier 3 | C:2a:NT | 52 | NAd | 2-2 | 1 |
| Strain 5 | |||||
| carrier 4 | C:2a:P1.15 | 51 | 19d,15 | 2-38 | 1 |
| carrier 5 | C:2a:P1.15 | 51 | 19d,15 | 2-38 | 1 |
Antigen gene sequences.
Further discrimination among the isolates was apparent from the nucleotide sequences of the entire porB gene, which encodes the serotyping antigen, and the variable regions (VRs) of the porA gene, which determine meningococcal serosubtype (Table 2). This confirmed that the isolate from case 1 was distinct from the isolates from the other cases and showed that while it was identical to the isolate from carrier 1 (strain 1), it was distinct from the isolate from carrier 2 (strain 3). The strain isolated from the two remaining university-associated cases (cases 3 and 6) and the two remote patients (strain 2), which was ST-50, was also characterized by distinct porA and porB genes. While this strain possessed the same porA VRs as a previously characterized ET-37 complex isolate from the United Kingdom (Table 1), it was distinguishable by its ST and porB allele. The remaining isolates from carriers (strain 4 and strain 5), which were indistinguishable from each other, were also distinct on the basis of their antigen genes (Table 2). The ET-37 complex isolate and the strain 1 and strain 2 isolates possessed identical tbpB alleles (numbered 2 in Table 2; GenBank accession no. AJ250244), which was distinct from the tbpB allele (numbered 1 in Table 2; GenBank accession no. AJ250243) shared by the remaining isolates (strains 3, 4, and 5).
PFGE fingerprinting.
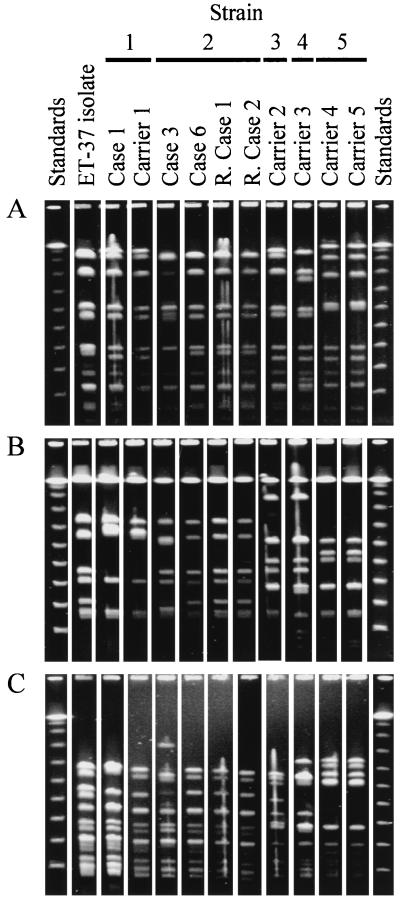
The 11 isolates examined gave related fingerprint patterns with each of the three endonucleases that were used in the present study and that have previously been shown to identify relationships among meningococci (3). The isolates from case 1 and carrier 1 (Strain 1) were indistinguishable from each other. Their NheI fingerprint patterns were also observed for the other ST-11 isolate (strain 3 from carrier 2; Fig. 1A), which was distinguished from them by distinct SpeI and SfiI fingerprints (Fig. 1B and C). They also had SpeI fingerprint patterns identical to those of three of the strain 2 isolates (from case 6, remote case 1, and remote case 2; Fig. 1C) but were distinguished from these isolates by their NheI and SfiI fingerprints. These three strain 2 isolates had identical fingerprint patterns, but the remaining strain 2 isolate (from patient 3) differed from them by two bands for each fingerprint. All three fingerprints for the strain 4 isolate (from carrier 3) were unique. For all endonuclease fingerprints, the isolates from carrier 4 and carrier 5 (strain 5) were identical and unique to these two isolates.
FIG. 1.
PFGE fingerprint analysis of the isolates. Isolates are identified by the same labelling as in Table 1 (except for “R. Case,” which stands for “remote case”). Standards were phage lambda concatamers. (A) Fingerprints generated with restriction endonuclease NheI; (B) fingerprints generated with restriction endonuclease SfiI; (C) fingerprints generated with restriction endonuclease SpeI. The results shown in each panel are from the same gel, and the lanes appear in the same order as on the original gel with the exception of those for remote case 2 (R. Case 2), which were moved from a position adjacent to the right-hand standard track for ease of comparison. To account for higher sample loading, the photographic contrast of the first two sample lanes of panel C (ET-37 isolate and Case 1) was adjusted independently of that for the rest of the gel in the preparation of this figure.
DISCUSSION
The data obtained in the present study indicated that an ET-37 complex variant, strain 1 (Table 2), was responsible for at least four cases of meningococcal disease suspected as being part of an outbreak. Although samples from the three cases for which no meningococci were isolated (cases 2, 4, and 5) were not available for MLST in this particular outbreak investigation, two of these cases were shown to be serogroup C by a PCR assay (2). Given their temporal and geographic proximity to cases 3 and 6, it is likely, although not proven, that strain 2 was the causative organism of these cases. As MLST and antigen gene sequencing are PCR-based techniques, they can potentially be applied directly to culture-negative clinical specimens in future studies. The sequence data further demonstrated that while the disease in case 1 was also caused by an ET-37 complex variant, it was not part of the outbreak and that, with the exception of carrier 1, a close contact of case 1, neither of the disease-causing strains was recovered from carriers. Finally, these sequence data showed that three distinct ET-37 complex strains (strains 3 to 5) were circulating among the student population during the outbreak without causing disease.
The sequence data provided resolution similar to that observed by PFGE fingerprinting, but the data were more readily interpreted and were easily compared with data obtained in other studies. The single anomaly between the sequence data and the PFGE fingerprints was the different patterns observed with one of the strain 2 isolates (from case 3) and the remaining isolates. The two band differences seen in each PFGE fingerprint might have been the result of a chromosome rearrangement, which would have the effect of changing several fingerprint patterns while the strain retained its ST and antigen genes. Chromosome rearrangements are known within the pathogenic Neisseria (6) and have been observed among ET-37 complex variants (11); such rearrangements have a potentially confusing effect on the interpretation of PFGE fingerprint data.
An advantage of sequence-based isolate characterization is the identification of genetic variation among isolates, which is not possible in PFGE fingerprint analyses, from which the genetic reasons for pattern changes cannot be easily deduced without additional data. Three of the housekeeping gene changes among the STs, those in abcZ, aroE, and pdhC, were likely to have resulted from horizontal genetic exchange (Fig. 2). A single nucleotide change in the fumC allele that distinguished ST-51 from the other STs could have been a point mutational change; however, as this polymorphism is present in other fumC alleles, it is possible that this change was also due to a genetic exchange event.
FIG. 2.
Polymorphic sites present in the genes analyzed. For each of the sequence types, the variant bases of the variant MLST loci compared to the sequence of the allele present in ST-11 are shown. The vertical numbers represent the base position within the MLST allele. For the porA genes, the entire sequence for VR1 and VR2 are given and their sequences are compared to the sequences defining serotype P1.5,2. The variant nucleotides of the five porB alleles compared with the sequence of porB2-2 are given, and the vertical numbers indicate the base position in an alignment of all known porB alleles. Allele porB2-39 was distinguished from allele porB2 by a four-codon deletion (data not shown) at base positions 604 to 615 inclusive. In all cases a base identical to that of the first sequence is shown with a period, a polymorphism is shown with the appropriate letter, and an alignment gap is shown with a hyphen.
Two of the porB polymorphisms, those distinguishing porB2-36 and porB2-38 from porB2-2, were unlikely to have changed the immunogenicity of the encoded proteins, and strains 1 and 5 serotyped as 2a. The porB2-39 allele in the NT isolate from carrier 2 (strain 3) was identical to the porB2-2 allele except for a 12-bp deletion in predicted loop V of the protein (7). The other porB variant from an NT isolate, porB2-37, was a distinct allele that had probably been introduced by horizontal genetic exchange. Among the strains that were not subtypeable, the porA gene of strain 1 had been inactivated by an IS1301 insertion (1), and no porA gene could be amplified from strain 4, indicating that porA may have been deleted in this meningococcus. The porA allele conferring the P1.19d,15 subtype on strain 5 was probably the result of a horizontal genetic exchange event, and the differences between the P1.5a,10- and P1.5,2-encoding alleles may have been the result of the accumulation of point mutations within the ET-37 complex over time. The tbpB alleles were sufficiently diverse that it is probable that this also represented a horizontal genetic exchange event.
These results demonstrated that MLST and antigen gene sequencing can provide high-resolution data applicable to epidemiological investigations of meningococcal disease outbreaks. In addition to identifying strains belonging to the hypervirulent ET-37 complex, these approaches distinguished strains identical by serological means or antigen gene sequencing (Table 2), and the data obtained by these approaches were more easily analyzed than PFGE fingerprint patterns (Fig. 1). Furthermore, it is not necessary that sequence analyses be performed simultaneously, and the sequence analyses of the housekeeping and antigen genes can be performed sequentially as clinical specimens become available. A final advantage is that the PCR sequencing-based approaches have potential application when no isolate is available, which is not possible for many approaches including PFGE fingerprint analysis.
REFERENCES
- 1.Arhin F F, Moreau F, Coulton J W, Mills E L. Sequencing of porA from clinical isolates of Neisseria meningitidis defines a subtyping scheme and its genetic regulation. Can J Microbiol. 1998;44:56–63. [PubMed] [Google Scholar]
- 2.Borrow R, Claus H, Guiver M, Smart L, Jones D M, Kaczmarski E B, Frosch M, Fox A J. Non-culture diagnosis and serogroup determination of meningococcal B and C infection by a sialyltransferase (siaD) PCR ELISA. Epidemiol Infect. 1997;118:111–117. doi: 10.1017/s0950268896007261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bygraves J A, Maiden M C J. Analysis of the clonal relationships between strains of Neisseria meningitidis by pulsed field gel electrophoresis. J Gen Microbiol. 1992;138:523–531. doi: 10.1099/00221287-138-3-523. [DOI] [PubMed] [Google Scholar]
- 4.Cartwright K A V, editor. Meningococcal disease. Chichester, United Kingdom: John Wiley & Sons; 1995. [Google Scholar]
- 5.Caugant D A, Mocca L F, Frasch C E, Froholm L O, Zollinger W D, Selander R K. Genetic structure of Neisseria meningitidis populations in relation to serogroup, serotype, and outer membrane protein pattern. J Bacteriol. 1987;169:2781–2792. doi: 10.1128/jb.169.6.2781-2792.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5a.Chan, M.-S. 1998, posting date. MLST Website. [Online.] http://mlst.zoo.ox.ac.uk [6 October 1999, last date accessed.]
- 6.Dempsey J A, Wallace A B, Cannon J G. The physical map of the chromosome of a serogroup A strain of Neisseria meningitidis shows complex rearrangements relative to the chromosome of the two mapped strains of the closely related species N. gonorrhoeae. J Bacteriol. 1995;177:6390–6400. doi: 10.1128/jb.177.22.6390-6400.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Derrick J P, Urwin R, Suker J, Feavers I M, Maiden M C J. Structural and evolutionary inference from molecular variation in Neisseria porins. Infect Immun. 1999;67:2406–2413. doi: 10.1128/iai.67.5.2406-2413.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Feavers I M, Suker J, McKenna A J, Heath A B, Maiden M C J. Molecular analysis of the serotyping antigens of Neisseria meningitidis. Infect Immun. 1992;60:3620–3629. doi: 10.1128/iai.60.9.3620-3629.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Frasch C E, Zollinger W D, Poolman J T. Serotype antigens of Neisseria meningitidis and a proposed scheme for designation of serotypes. Rev Infect Dis. 1985;7:504–510. doi: 10.1093/clinids/7.4.504. [DOI] [PubMed] [Google Scholar]
- 10.Holmes E C, Urwin R, Maiden M C J. The influence of recombination on the population structure and evolution of the human pathogen Neisseria meningitidis. Mol Biol Evol. 1999;16:741–749. doi: 10.1093/oxfordjournals.molbev.a026159. [DOI] [PubMed] [Google Scholar]
- 11.Jelfs J, Munro R, Ashton F, Rawlinson W, Caugant D A. Proceedings of the Eleventh Pathogenic Neisseria Conference. Paris, France: Editions E.D.K.; 1998. Global study of variation in a new variant of the ET-37 complex of Neisseria meningitidis; p. 5. [Google Scholar]
- 12.Maiden M C J. The impact of molecular techniques on the study of meningococcal disease. In: Woodford N, Johnson A P, editors. Molecular bacteriology: protocols and clinical applications. Totowa, N.J: Humana Press; 1998. pp. 265–291. [DOI] [PubMed] [Google Scholar]
- 13.Maiden M C J, Bygraves J A, Feil E, Morelli G, Russell J E, Urwin R, Zhang Q, Zhou J, Zurth K, Caugant D A, Feavers I M, Achtman M, Spratt B G. Multilocus sequence typing: a portable approach to the identification of clones within populations of pathogenic microorganisms. Proc Natl Acad Sci USA. 1998;95:3140–3145. doi: 10.1073/pnas.95.6.3140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Maiden M C J, Feavers I M. Meningococcal typing. J Med Microbiol. 1994;40:157–158. doi: 10.1099/00222615-40-3-157. [DOI] [PubMed] [Google Scholar]
- 15.Maiden M C J, Feavers I M. Population genetics and global epidemiology of the human pathogen Neisseria meningitidis. In: Baumberg S, Young J P W, Wellington E M H, Saunders J R, editors. Population genetics of bacteria. Cambridge, United Kingdom: Cambridge University Press; 1995. pp. 269–293. [Google Scholar]
- 16.Maiden M C J, Suker J, McKenna A J, Bygraves J A, Feavers I M. Comparison of the class 1 outer membrane proteins of eight serological reference strains of Neisseria meningitidis. Mol Microbiol. 1991;5:727–736. doi: 10.1111/j.1365-2958.1991.tb00743.x. [DOI] [PubMed] [Google Scholar]
- 17.Olyhoek T, Crowe B A, Achtman M. Clonal population structure of Neisseria meningitidis serogroup A isolated from epidemics and pandemics between 1915 and 1983. Rev Infect Dis. 1987;9:665–682. doi: 10.1093/clinids/9.4.665. [DOI] [PubMed] [Google Scholar]
- 18.Rokbi B, Mignon M, Caugant D A, Quentin-Millet M J. Heterogeneity of tbpB, the transferrin-binding protein B gene, among serogroup B Neisseria meningitidis strains of the ET-5 complex. Clin Diagn Lab Immunol. 1997;4:522–529. doi: 10.1128/cdli.4.5.522-529.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Selander R K, Caugant D A, Ochman H, Musser J M, Gilmour M N, Whittam T S. Methods of multilocus enzyme electrophoresis for bacterial population genetics and systematics. Appl Environ Microbiol. 1986;51:837–884. doi: 10.1128/aem.51.5.873-884.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Staden R. The Staden sequence analysis package. Mol Biotechnol. 1996;5:233–241. doi: 10.1007/BF02900361. [DOI] [PubMed] [Google Scholar]
- 21.Suker J, Feavers I M, Achtman M, Morelli G, Wang J-F, Maiden M C J. The porA gene in serogroup A meningococci: evolutionary stability and mechanism of genetic variation. Mol Microbiol. 1994;12:253–265. doi: 10.1111/j.1365-2958.1994.tb01014.x. [DOI] [PubMed] [Google Scholar]
- 22.Suker J, Feavers I M, Maiden M C J. Monoclonal antibody recognition of members of the P1.10 variable region family: implications for serological typing and vaccine design. Microbiology. 1996;142:63–69. doi: 10.1099/13500872-142-1-63. [DOI] [PubMed] [Google Scholar]
- 23.Urwin R. Ph.D. thesis. United Kingdom: University of Staffordshire, Stoke-on-Trent; 1998. [Google Scholar]
- 24.Urwin R, Feavers I M, Jones D M, Maiden M C J, Fox A J. Molecular variation of meningococcal serotype 4 antigen genes. Epidemiol Infect. 1998;121:95–101. doi: 10.1017/s0950268898008942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vogel U, Morelli G, Zurth K, Claus H, Kriener E, Achtman M, Frosch M. Necessity of molecular techniques to distinguish between Neisseria meningitidis strains isolated from patients with meningococcal disease and from their healthy contacts. J Clin Microbiol. 1998;36:2465–2470. doi: 10.1128/jcm.36.9.2465-2470.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang J-F, Caugant D A, Morelli G, Koumaré B, Achtman M. Antigenic and epidemiological properties of the ET-37 complex of Neisseria meningitidis. J Infect Dis. 1993;167:1320–1329. doi: 10.1093/infdis/167.6.1320. [DOI] [PubMed] [Google Scholar]
- 27.Williams J N, Jones G R, Christodoulides M, Jolley K, Heckels J E. Proceedings of the Eleventh International Pathogenic Neisseria Conference. Paris, France: Editions E.D.K.; 1998. Investigation into an outbreak of meningococcal infection at Southampton University; p. 254. [Google Scholar]