Abstract
Oral mucosal colonization and infection with Candida are common in patients receiving radiation therapy for head and neck cancer. Infection is marked by oral pain and/or burning and can lead to significant patient morbidity. The purpose of this study was to identify Candida strain diversity in this population by using a chromogenic medium, subculturing, molecular typing, and antifungal susceptibility testing of clinical isolates. These results were then correlated with clinical outcome in patients treated with fluconazole for infection. Specimens from 30 patients receiving radiation therapy for head and neck cancer were cultured weekly for Candida. Patients exhibiting clinical infection were treated with oral fluconazole. All isolates were plated on CHROMagar Candida and RPMI medium, subcultured, and submitted for antifungal susceptibility testing and molecular typing. Infections occurred in 27% of the patients and were predominantly due to Candida albicans (78%). Candida carriage occurred in 73% of patients and at 51% of patient visits. Yeasts other than C. albicans predominated in carriage, as they were isolated from 59% of patients and at 52% of patient visits. All infections responded clinically, and all isolates were susceptible to fluconazole. Molecular typing showed that most patients had similar strains throughout their radiation treatment. One patient, however, did show the acquisition of a new strain. With this high rate of infection (27%), prophylaxis to prevent infection should be evaluated for these patients.
Oropharyngeal candidiasis is a common infection in patients receiving cancer therapies. Oral mucosal colonization (up to 93%) and infection (ranging from 17 to 29%) with Candida are particularly common in patients receiving radiation therapy for head and neck cancer (3, 17, 24). Compromised salivary function secondary to destruction of glandular tissue by radiation is thought to be a major factor leading to Candida infection (3, 5). This infection is marked by oral pain and/or burning and can lead to significant patient morbidity (3).
Determination of the epidemiology of Candida isolates in patients receiving head and neck radiation has traditionally involved taking individual cultures and identifying yeasts to the species level, as well as using techniques to identify specific yeast strains with serotyping and biotyping. In the past Candida albicans has been by far the most predominant organism isolated (24). However, taking a single culture and performing the above-mentioned tests may not be sensitive in distinguishing all C. albicans strains or non-C. albicans Candida present at colonization and/or infection (9, 10). It appears from other populations, primarily human immunodeficiency virus (HIV) patients, that by employing subculturing of multiple colonies from each culture and by using molecular techniques with these subcultures, a more accurate picture of the epidemiology of these organisms can be developed (9, 10, 15, 19). Also, the use of a chromogenic medium will allow the identification of multiple Candida species on the basis of colony color (14).
Molecular typing techniques have been very useful for determining the identities of serial isolates with recurrent candidal infections (9, 10, 15, 19). In many patients, a single unique strain of Candida may persist over many months and be associated with recurrent infection or colonization (15, 18, 19). This suggests that recurrence may be due to failure of therapy to eradicate the initial organism. However, in other studies with cancer patients and with HIV-infected patients, a new strain of C. albicans or a different Candida species with recurrent infection or colonization has been observed (15, 19). As many as 50% of HIV patients may develop infection due to strains distinct from that causing the initial infection (15, 19). Molecular typing techniques have also been helpful in determining Candida strain diversity of individual cultures (10, 15, 19). HIV patients may have as many as three different Candida strains from any one culture of colonization or infection (15, 19).
Fluconazole is the predominant medication utilized to treat oropharyngeal candidiasis. It has been studied extensively for HIV patients (12, 16). Development of resistance to fluconazole in these patients has become a growing concern and usually is correlated with the degree of immunosuppression and the total dose of drug (1, 20, 21). Fluconazole has also been used effectively to treat this infection in patients receiving head and neck radiation, as the predominant organism has been C. albicans (3, 17). However, recently an increase in non-C. albicans Candida has been reported in head and neck cancer patients receiving radiation therapy (17). The issue of resistance to fluconazole has not been evaluated closely for this patient population. If resistance to fluconazole does occur, its occurrence may be due to the presence of yeasts other than C. albicans which are less susceptible to fluconazole, or it may result from the development of resistance in a previously susceptible C. albicans strain. While both of these resistance mechanisms may occur in patients with HIV infection (15), the operative mechanism in patients with head and neck cancer has not been established.
Oral Candida strain diversity, as determined by using a chromogenic medium, subculturing, and molecular techniques, and the potential effectiveness of fluconazole treatment for oropharyngeal candidiasis, as determined by antifungal susceptibility testing, have not been closely studied for patients receiving radiation to treat head and neck cancer. The purpose of this study was to evaluate Candida diversity and to correlate it with fluconazole treatment in this patient population.
MATERIALS AND METHODS
Patients.
Thirty patients receiving a 6-week course of radiation therapy for treatment of head and neck cancer were enrolled in the study. Patients were examined for signs of oropharyngeal candidiasis at baseline and weekly thereafter during their radiation treatment. Oral cultures were obtained at each visit and from any clinical infection. Infection was defined as positive clinical signs of white intraoral plaques confirmed by use of a 10% KOH preparation and/or positive culture. Patients with infection were treated with fluconazole (200 mg [loading dose] and 100 mg/day for 7 days to 14 days). If a clinical cure was not achieved in 14 days, the dosage was increased up to 800 mg/day to achieve a clinical cure (21). Fungal cultures employed an oral swab and a swish sample of 10 ml of normal saline instilled in the mouth for 10 s and then collected in a sterile container. These samples were plated on CHROMagar Candida- and RPMI-based medium (20). Yeasts were identified by standard techniques. For all cultures, three to five yeast colonies from primary plates were picked and stored on Sabouraud dextrose slants for antifungal susceptibility testing and molecular typing.
Antifungal susceptibility testing.
Three to five colonies from each sample were submitted for broth macro- and microdilution MIC determinations, to correlate with appearance on fluconazole-containing chromogenic medium as either susceptible or resistant (14). Testing of fluconazole MICs by the National Committee for Clinical Laboratory Standards (NCCLS) method was performed at the Fungus Testing Laboratory, University of Texas Health Science Center, San Antonio (6, 13).
Genotypic identification tests.
All C. albicans isolates were karyotyped. Selected isolates were also identified by using restriction fragment length polymorphism (RFLP) analysis and the moderately repetitive Ca 3 probe.
(i) Electrophoretic karyotyping.
C. albicans isolates were plated on Sabouraud dextrose agar and grown at 30°C for 48 h. Colonies were suspended in 2 ml of 75 mM NaCl–25 mM EDTA to a turbidity of approximately a 2.0 McFarland standard. The suspension was centrifuged at 230 × g for 10 min, and the pellet was resuspended in 1 ml of 75 mM NaCl–25 mM EDTA. Plugs were made by mixing together at 37°C (i) 1 ml of 1.5% low-melting-point agarose in 125 mM EDTA (pH 7.5), (ii) 75 μl of 2,000-U/ml Zymolyase-20T (ICN Biomedicals, Inc., Aurora, Ohio), and (iii) 1 ml of the cell suspension. This mixture was distributed into plug molds and refrigerated for 1 h at 4°C. Spheroplasts were made by placing plugs in 4 ml of 0.5 M EDTA (pH 9.0)–7.5% β-mercaptoethanol. Plugs were incubated overnight at 37°C and then rinsed with 5 ml of 50 mM EDTA, pH 7.5. Four milliliters of ESP (0.5 M EDTA, 10% sarcosyl, 20 μg of proteinase K per ml) solution was added to each tube and incubated at 50°C overnight. Plugs were refrigerated at 4°C for 1 h. The chromosomes were resolved on a 1.0% low-melting-point agarose gel with a contour-clamped homogenous electric field (CHEF) (CHEF-DR III; Bio-Rad, Hercules, Calif.). The switching times used for CHEF electrophoresis were (i) 120 s at 4.5 V/cm for 21 h; (ii) 300 s at 4.5 V/cm for 18 h; and (iii) 300 s at 3.4 V/cm for 28 h. After electrophoresis, the gels were stained with ethidium bromide, illuminated under UV light, and photographed (2).
(ii) RFLP.
RFLP patterns were generated by digestion of genomic DNA with SfiI or EcoRI (Boehringer Mannheim, Indianapolis, Ind.) and subsequent separation of DNA fragments by pulsed-field gel electrophoresis. Briefly, plugs prepared as described above were incubated in the presence of the corresponding restriction endonuclease. After digestion, the plugs were loaded in wells of a 0.8% agarose gel. This gel was placed in a CHEF gel chamber (DR III; Bio-Rad), and electrophoresis was performed with the following parameters: for SfiI, pulse times were ramped from 5 to 35 s for 24 h at 6 V/cm, and for EcoRI, pulse times were ramped from 5 to 35 s for 18 h at 4.5 V/cm. After the run, gels were stained with ethidium bromide and photographed (26).
(iii) Southern hybridization with the moderately repetitive probe Ca 3.
The materials present in the RFLP gels were transferred to nylon membranes (Nytran; Schleicher and Schuell, Keene, N.H.) overnight by using a Turboblotter apparatus and 20× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) buffer (Schleicher and Schuell). Subsequently, materials present in the nylon membranes were hybridized with a Ca 3 probe radioactively labeled by random priming (Random Primers DNA Labeling System; Gibco-BRL, Gaithersburg, Md.). Prehybridization and hybridization were performed with Rapid-hyb buffer (Amersham Life Science Inc., Arlington Heights, Ill.) according to the manufacturer's instructions. The membranes were then washed and exposed to autoradiography film (Du Pont, Wilmington, Del.) (23).
(a) Documentation.
Pictures of the gels or films were scanned with a Kodak EDAS 120 digital camera and imaging processing system. For preparation of the figures, digital images were processed by using the Photo Shop program (Adobe Systems Inc., Mountain View, Calif.).
(b) Visual analysis of band patterns.
The fingerprints obtained were compared for similarity by visual inspection of band patterns. Sizes of DNA fragments amplified by PCR were determined by direct comparison with the DNA marker (100-bp ladder; Gibco-BRL). Fingerprints were considered highly similar when all visible bands obtained had the same migration distance for each isolate. Variations in the intensity and shape of bands among isolates were not considered differences. The presence or absence of more than two distinct bands was considered a difference (11).
(c) Computer-assisted analysis of fingerprinting patterns.
All fingerprints were analyzed with the Molecular Analyst fingerprinting software (Bio-Rad) by using band-based cluster analysis. Dendrograms were generated by the hierarchic unweighted pair group method using arithmetic averages cluster algorithm. Fingerprint analysis and the methods and algorithms used in this study were performed according to the manufacturer's instructions.
RESULTS
Patient diseases and radiation doses are listed in Table 1. The radiation dose at the last visit in this study ranged from 1,000 to 7,200 cGy, with a mean of 5,595 cGy. Four patients did not complete the study, as they dropped out prior to finishing their radiation treatment. Candida infections occurred in 8 of 30 patients (27%) and at 9 of 185 patient visits (5%). C. albicans alone was isolated in seven of nine epidoses of infection (78%). C. albicans and a yeast other than C. albicans were isolated in one of nine episodes of infection (11%). A yeast other than C. albicans alone was isolated in one of nine episodes of infection (11%). Overall, 22 of 30 patients (73%) and 95 of 185 patient visits (51%) were positive for Candida carriage (infection and colonization). C. albicans alone was isolated from 9 of 22 patients (41%) and at 46 of 95 patient visits (48%). Yeasts other than C. albicans were detected in 13 of 22 patients (59%) with positive cultures and at 49 of 95 patient visits with positive cultures (52%). Ten different yeast species were isolated (Tables 2 and 3). Mixed cultures of C. albicans and other yeasts were isolated from 6 of 22 patients (27%) and at 15 of 95 patients visits (16%). Yeasts other than C. albicans alone were isolated from 7 of 22 patients (32%) and at 34 of 95 patient visits (36%) (Table 2).
TABLE 1.
Patient diagnosis and radiation dose
| Patient no. | Diagnosisa | Radiation (cGy) |
|---|---|---|
| 1 | SCCA, lymph node | 4,800 |
| 2 | SCCA, neck | 6,000 |
| 3 | SCCA, supraglottis | 6,640 |
| 4 | SCCA, tongue | 2,000b |
| 5 | SCCA, tongue and tonsil | 5,000 |
| 6 | SCCA, epiglottis | 5,000 |
| 7 | SCCA, tongue | 1,000b |
| 8 | SCCA, floor of mouth | 5,000 |
| 9 | SCCA, floor of mouth | 6,000 |
| 10 | SCCA, supraglottis | 6,200 |
| 11 | SCCA, tonsil | 6,400 |
| 12 | SCCA, larynx and vallecula | 5,400 |
| 13 | Lymphoma, tonsil | 4,000 |
| 14 | SCCA, alveolar ridge and palate | 6,600 |
| 15 | SCCA, hypopharynx | 6,600 |
| 16 | SCCA, epiglottis | 7,200 |
| 17 | SCCA, tongue | 2,700b |
| 18 | SCCA, parotid | 4,800b |
| 19 | SCCA, floor of mouth | 6,600 |
| 20 | SCCA, pyriform sinus | 7,200 |
| 21 | SCCA, supraglottis | 6,200 |
| 22 | SCCA, hypopharynx | 6,200 |
| 23 | Neuroendocrine of parotid | 6,200 |
| 24 | SCCA, tongue | 6,800 |
| 25 | SCCA, tonsil | 6,120 |
| 26 | SCCA, neck | 6,000 |
| 27 | SCCA, vocal cord | 6,400 |
| 28 | SCCA, pyriform sinus | 5,600 |
| 29 | SCCA, pyriform sinus | 6,600 |
| 30 | SCCA, vocal cord | 6,600 |
SCCA, squamous cell carcinoma.
Patient withdrew before completion of radiation.
TABLE 2.
Oral Candida infection and/or carriage in patients receiving radiation for head and neck cancer
| Status and organism(s) | No. positive/total (% positive)
|
|
|---|---|---|
| Patients | Patient visits | |
| Infection | ||
| Total | 8/30 (27) | 9/185 (5) |
| C. albicans only | 6/8 (75) | 7/9 (78) |
| C. albicans and non-C. albicans | 1/8 (13) | 1/9 (11) |
| Non-C. albicans only | 1/8 (13) | 1/9 (11) |
| Carriage | ||
| Total | 22/30 (73) | 95/185 (51) |
| C. albicans only | 9/22 (41) | 46/95 (48) |
| C. albicans and non-C. albicans | 6/22 (27) | 15/95 (16) |
| Non-C. albicans only | 7/22 (32) | 34/95 (36) |
TABLE 3.
Susceptibilities of yeasts other than C. albicans to fluconazole as determined by NCCLS broth macrodilution testing
| Patient no. | Organism | No. of visits | Fluconazole MIC (μg/ml)
|
|
|---|---|---|---|---|
| 24 h | 48 h | |||
| 1a | C. dubliniensis | 5 | ≤0.125–0.25 | ≤0.125–0.5 |
| 2a | C. rugosa | 3 | 4 | 4–8 |
| 7 | C. glabrata | 2 | 1–16 | 1–16 |
| C. tropicalis | 1 | 2–64 | 4–64 | |
| 10 | C. kefyr | 4 | ≤0.125–0.25 | 0.25–1 |
| 11a | C. krusei | 3 | 16 | 32–64 |
| 13 | C. glabrata | 5 | 2–32 | 8–64 |
| C. krusei | 4 | 32 | >64 | |
| C. famata | 2 | 0.25 | 0.5–1 | |
| 14 | C. glabrata | 7 | 2–32 | 8–64 |
| Saccharomyces cerevisiae | 2 | 0.5–4 | 1–8 | |
| 15 | C. krusei | 3 | 16 | 32–64 |
| 19 | C. krusei | 2 | 64 | >64 |
| Hansenula anomala | 1 | 1–2 | 1–2 | |
| 24a | S. cerevisiae | 6 | 2–4 | 4 |
| 27 | C. glabrata | 3 | 4 | 16–64 |
| C. krusei | 3 | 32 | 64 | |
| 28 | Cryptococcus albidus | 1 | 16–32 | >64 |
| 29 | C. krusei | 4 | 8–32 | 16–64 |
The patient experienced clinical infection.
All infections involving C. albicans alone responded to fluconazole therapy at a dosage of 100 mg/day for 7 to 14 days. All of the C. albicans isolates recovered on symptomatic infection as well as asymptomatic colonizing isolates were susceptible to fluconazole in vitro (Table 4). Patient 7 showed fluconazole MICs of >64 μg/ml but these probably represented trailing endpoints, as the organism was susceptible to fluconazole on chromogenic agar containing fluconazole (22). Patient 20 exhibited one relapse of infection, but the organism remained susceptible and responded clinically to standard fluconazole therapy at 100 mg/day.
TABLE 4.
Susceptibilities of C. albicans isolates to fluconazole as determined by NCCLS broth macrodilution testing
| Patient no. | No. of visits | Fluconazole MIC (μg/ml)
|
|
|---|---|---|---|
| 24 h | 48 h | ||
| 1a | 3 | ≤0.125–0.25 | ≤0.125–1 |
| 3a | 3 | 0.25–0.5 | 0.5–2 |
| 6a | 4 | ≤0.125–0.25 | 0.25–2 |
| 7 | 1 | 64b | 64b |
| 11a | 5 | 0.25–0.5 | 0.25–1 |
| 15 | 3 | ≤0.125–0.25 | 0.25 |
| 16 | 7 | 0.25–0.5 | 0.25–2 |
| 17 | 4 | ≤0.125–0.5 | 0.25–1 |
| 18 | 1 | 0.25 | 0.25–0.5 |
| 19 | 1 | 0.25 | 0.25 |
| 20a | 6 | 0.25 | 0.25–0.5 |
| 23 | 5 | ≤0.125–0.5 | 0.25–1 |
| 24a | 7 | ≤0.125–1 | 0.25–2 |
| 25a | 6 | 0.25 | 0.25–1 |
| 26 | 5 | 0.25 | 0.25–0.5 |
The patient experienced clinical infection.
These were determined to be trailing endpoints, as the organism was susceptible to fluconazole on chromogenic agar.
There was a wide range in the susceptibility of yeasts other than C. albicans, with MICs ranging from <0.125 to >64 μg/ml (Table 3). Patient 2, infected with Candida rugosa, required 200 mg of fluconazole per day to achieve a clinical cure. His fluconazole MICs were elevated (4 to 8) but still within the susceptible range. Patient 1 exhibited a mixed infection and colonization pattern with C. albicans and Candida dubliniensis. Treatment of the infection eradicated the C. albicans after 7 days, but clinical signs were still present and C. dubliniensis was cultured. After an additional 7 days of therapy, the patient was clinically cured and all cultures were negative for yeasts. All isolates of both organisms were susceptible to fluconazole.
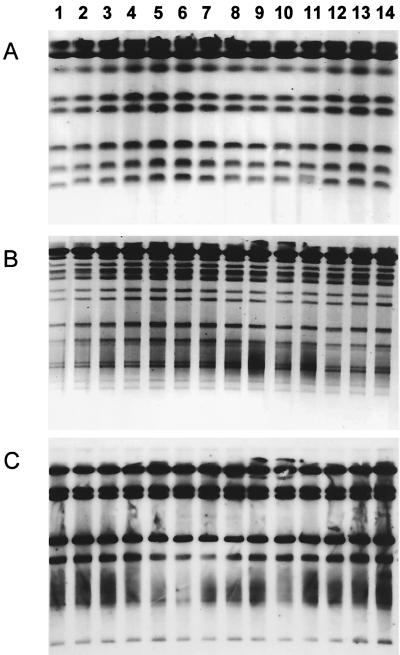
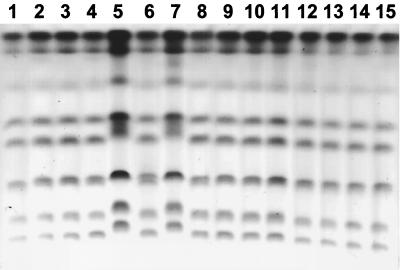
Molecular typing was performed for all 12 patients who showed persistent C. albicans carriage. Karyotyping was performed on isolates from all 12 patients. RFLP analysis and Southern hybridization with the moderately repetitive Ca 3 probe were performed on isolates from selected patients. Serial isolates appeared to be similar for all patients except patient 24, who displayed two distinct strains. In this patient common strains were seen at all visits except visit three, where a new strain emerged with the original strain and clinical infection was present. The patient was successfully treated with fluconazole, and the new strain was eradicated. However, the patient remained colonized with the original strain for four more visits. All of these isolates appeared to be susceptible to fluconazole in vitro. Figure 1 shows karyotype, RFLP, and Southern hybridization results for patient 20. Figure 2 shows karyotype results for patient 24.
FIG. 1.
Isolates from patient 20. Each lane represents a separate subisolate collected over six visits when cultures were positive. (A) Karyotype; (B) RFLP analysis with SfiI digestion of genomic DNA; (C) fingerprinting analysis with the Ca 3 probe. All isolates appear to be similar.
FIG. 2.
Isolates from patient 24. Each lane represents the karyotype analysis of a separate subisolate collected over seven visits when cultures were positive. Lanes 5 and 7 show the emergence of a new strain at visit 3. All other isolates appear to be similar, including that in lane 6, which was also from visit 3.
DISCUSSION
The epidemiology of C. albicans and other yeasts from the oropharynx of patients receiving radiation for head and neck cancer is quite varied. C. albicans was the predominant organism associated with symptomatic infection. C. albicans alone was present in all but two of nine infections. It is of interest that in a mixed infection of C. albicans and the recently described oral pathogen C. dubliniensis, the C. dubliniensis was the presumed causative agent of the infection, as clinical infection persisted with the presence of C. dubliniensis alone. C. dubliniensis has been associated with oropharyngeal candidiasis (25) and recently has been isolated from HIV-infected patients in North America (8). However, to our knowledge this is the first report of an oral infection with C. dubliniensis in a patient receiving head and neck radiation. The other infection due to yeasts other than C. albicans alone was due to C. rugosa and was an isolate which was susceptible in vitro to fluconazole and clinically responded to fluconazole therapy.
Candida colonization was common and was detected in 73% of patients and in 51% of patient visits. However, colonization due to C. albicans alone occurred in only 48% of all positive cultures and 41% of positive patients. Yeasts other than C. albicans alone were detected in 36% of all positive cultures and 32% of patients. Fifteen patients were free of yeast at the initial visit. Eight patients remained free of yeast colonization, and seven patients became colonized after the initial visit. This data strengthens earlier reports of a significant role for yeasts other than C. albicans in oropharyngeal carriage in patients receiving head and neck radiation (17). Infection was due to yeasts other than C. albicans in 25% of the patients, and one of these infections required an increased dose of fluconazole to achieve a clinical response. Thus, it would seem prudent to search for these organisms in patients who do not respond to standard therapy. If culturing is employed, the use of chromogenic agar may be helpful in this identification (14). Antifungal susceptibility testing may also be useful in guiding antifungal therapy (21). Fluconazole appeared to be very effective in treating all infected patients. All infections were treated successfully with the standard dose, except for the one patient with C. rugosa, which responded to 200 mg of fluconazole. Of the C. albicans isolates, all appeared to be susceptible in vitro to fluconazole except the one that apparently displayed trailing endpoints. One of the eight patients with infection relapsed once with the same strain of C. albicans, but both episodes of infection were successfully treated with fluconazole at 100 mg/day. In contrast, yeasts other than C. albicans exhibited significant increases in fluconazole MICs. Ten of 12 patients with non-C. albicans isolates showed increased fluconazole MICs, ranging from 4 to >64 μg/ml.
Molecular characterization of serial isolates from multiple cultures showed that the majority of patients retain similar strains of C. albicans over time. However, emergence of new strains does occur. Interestingly, in patient 24 the emergence of a new strain corresponded to clinical infection although the original strain was present as well. All other cultures before and after the infection were associated with colonization and showed the original strain only. All C. albicans isolates were susceptible to fluconazole and responded to fluconazole therapy.
Other patient groups with an opportunistic infection rate of 27% (e.g., bone marrow transplantation patients and HIV-infected patients) are commonly considered for prophylaxis (4, 7, 27). Oral mucositis associated with radiation therapy is typically very painful and can affect oral intake and even limit the radiation dosage. The relative role of candidiasis in this condition is not well defined. Could elimination or significant reduction of oropharyngeal candidiasis reduce the morbidity associated with radiation-induced mucositis? Prophylaxis of all patients may be problematic, as the potential for selection of resistant organisms would exist. However, preemptive therapy may be more appropriate. All eight patients who developed infection in this study were colonized prior to infection. Also, they all developed their infections after they had received at least 1,000 cGy of radiation. If only such patients are targeted for preemptive therapy, this would appear to cover all who will develop infection but to greatly reduce drug exposure among all patients. Our group is now evaluating preemptive therapy in this population.
ACKNOWLEDGMENTS
This work was supported by a grant from Pfizer Inc. (to S.W.R.); by Public Health Service grants 1 R01 DE11381 (to T.F.P.), 1 R29 AI42401 (to J.L.L.-R.), and M01-RR-01346 for the Frederic C. Bartter General Clinical Research Center; and by a Department of Veterans Affairs Postdoctoral Fellowship in Dental Research (to S.W.R.). Chromogenic medium was provided by Chromagar Co. (Paris, France).
REFERENCES
- 1.Boken D J, Swindels S, Rinaldi M G. Fluconazole-resistant Candida albicans. Clin Infect Dis. 1993;17:1018–1021. doi: 10.1093/clinids/17.6.1018. [DOI] [PubMed] [Google Scholar]
- 2.Doebbling B N, Lehmenn R F, Hollis R J, Wu L C, Widmer A F, Voss A, Pfaller M A. Comparison of pulsed-field gel electrophoresis with isoenzyme profiles as a typing system for Candida tropicalis. Clin Infect Dis. 1993;16:377–383. doi: 10.1093/clind/16.3.377. [DOI] [PubMed] [Google Scholar]
- 3.Fotos P G, Hellstein J W. Candida and candidosis. Dent Clin N Am. 1992;36:857–878. [PubMed] [Google Scholar]
- 4.Goodman J L, Winston D J, Greenfield R A, et al. A controlled trial of fluconazole to prevent fungal infections in patients undergoing bone marrow transplantation. N Engl J Med. 1992;362:845–851. doi: 10.1056/NEJM199203263261301. [DOI] [PubMed] [Google Scholar]
- 5.Haveman C W, Redding S W. Dental management and treatment of xerostomic patients. Texas Dent J. 1998;115:43–56. [PubMed] [Google Scholar]
- 6.Isenberg H D, editor. Clinical microbiology procedures handbook. Washington, D.C: American Society for Microbiology; 1992. [Google Scholar]
- 7.Just-Nübling G, Gentschew G, Mebner K, et al. Fluconazole prophylaxis of recurrent oral candidiasis in HIV-positive patients. Eur J Clin Microbiol Infect Dis. 1991;10:917–921. doi: 10.1007/BF02005444. [DOI] [PubMed] [Google Scholar]
- 8.Kirkpatrick W R, McAtee R K, Lopez-Ribot J L, Fothergill A W, McCarthy D I, Rinaldi M D, Patterson T F. Detection of Candida dubliniensis in oropharyngeal samples from human immunodeficiency virus-infected patients in North America by primary CHROM agar Candida screening and susceptibility testing of isolates. J Clin Microbiol. 1998;36:3007–3012. doi: 10.1128/jcm.36.10.3007-3012.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.López-Ribot J L, AcAtee R K, Lee L N, Kirkpatrick W R, White T C, Sanglard D, Patterson T F. Distinct patterns of gene expression associated with development of fluconazole resistance in serial Candida albicans isolates from human immunodeficiency virus-infected patients with oropharyngeal candidiasis. Antimicrob Agents Chemother. 1998;42:2932–2937. doi: 10.1128/aac.42.11.2932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.López-Ribot, J. L., R. K. McAtee, L. N. Lee, W. R. Kirkpatrick, M. G. Rinaldi, and T. F. Patterson. Molecular mechanisms of resistance to azoles in Candida albicans: multiple resistant phenotypes coexist during the same episode of oropharyngeal candidiasis in human immunodeficiency virus-infected patients.Antimicrob. Agents Chemother., in press. [DOI] [PMC free article] [PubMed]
- 11.Magee P T, Bowdin L, Staudinger J. Comparison of molecular typing methods for Candida albicans. J Clin Microbiol. 1992;30:2674–2679. doi: 10.1128/jcm.30.10.2674-2679.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Meunier F, Aoun M, Gerard M. Therapy for oropharyngeal candidiasis in the immunocompromised host: a randomized double-blind study of fluconazole vs ketoconazole. Rev Infect Dis. 1990;12(Suppl. 3):S364–S368. doi: 10.1093/clinids/12.supplement_3.s364. [DOI] [PubMed] [Google Scholar]
- 13.National Committee for Clinical Laboratory Standards. Reference method for broth dilution antifungal susceptibility testing of yeasts; approved standard. NCCLS document M27-A. Wayne, Pa: National Committee for Clinical Laboratory Standards; 1997. [Google Scholar]
- 14.Patterson T F, Revankar S G, Kirkpatrick W R, Dib O, Fothergill A W, Redding S W, McGough D A, Rinaldi M G. A simple method for detecting fluconazole-resistant yeasts with chromogenic agar. J Clin Microbiol. 1996;34:1794–1797. doi: 10.1128/jcm.34.7.1794-1797.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pfaller M A, Rhine-Chalberg J, Redding S W, Smith J, Farinacci G, Fothergill A W, Rinaldi M G. Variations in fluconazole susceptibility and electrophoretic karyotype among oral isolates of Candida albicans from patients with AIDS and oral candidiasis. J Clin Microbiol. 1994;32:59–64. doi: 10.1128/jcm.32.1.59-64.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pons V, Greenspan D, Debruin M the Multicenter Study Group. Therapy for oropharyngeal candidiasis in HIV-infected patients: a randomized prospective multicenter study of oral fluconazole versus clotrimazole troches. J Acquir Immune Defic Syndr. 1993;6:2322–2326. [PubMed] [Google Scholar]
- 17.Ramirez-Amador V, Silverman S, Mayer P, Tyler M, Quivery J. Candidal colonization and oral candidiasis in patients undergoing oral and pharyngeal radiation therapy. Oral Surg Oral Med Oral Pathol Oral Radiol Endod. 1997;84:149–153. doi: 10.1016/s1079-2104(97)90061-5. [DOI] [PubMed] [Google Scholar]
- 18.Redding S, Smith J, Farinacci G, Rinaldi M, Fothergill A, Rhine-Chalberg J, Pfaller M. Resistance of Candida albicans to fluconazole during treatment of oropharyngeal candidiasis in a patient with AIDS. Clin Infect Dis. 1994;18:240–242. doi: 10.1093/clinids/18.2.240. [DOI] [PubMed] [Google Scholar]
- 19.Redding S W, Pfaller M A, Messer S A, Smith J A, Prows J, Bradley L L, Fothergill A W, Rinaldi M G. Variations in fluconazole susceptibility and DNA subtyping of multiple Candida albicans colonies from patients with AIDS and oral candidiasis suffering one or more episodes of infection. J Clin Microbiol. 1997;37:1761–1765. doi: 10.1128/jcm.35.7.1761-1765.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Revankar S G, Kirkpatrick W R, McAtee R, Dib O P, Fothergill A W, Redding S W, McGough D A, Rinaldi M G, Patterson T F. Detection and significance of fluconazole resistance in oropharyngeal candidiasis in HIV-infected patients. J Infect Dis. 1996;174:821–827. doi: 10.1093/infdis/174.4.821. [DOI] [PubMed] [Google Scholar]
- 21.Revankar S G, Dib O P, Kirkpatrick W R, McAtee R K, Fothergill A W, Rinaldi M G, Redding S W, Patterson T F. Clinical evaluation and microbiology of oropharyngeal infection due to fluconazole resistant Candida in human immunodeficiency virus-infected patients. Clin Infect Dis. 1998;26:960–963. doi: 10.1086/513950. [DOI] [PubMed] [Google Scholar]
- 22.Revankar S G, Kirkpatrick W R, McAtee R F, Fothergill A W, Redding S W, Rinaldi M G, Patterson T F. Interpretation of trailing endpoints in antifungal susceptibility testing by the National Committee for Clinical Laboratory Standards method. Anticrob Agents Chemother. 1998;36:153–156. doi: 10.1128/jcm.36.1.153-156.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schmid J, Voss E, Soll D R. Computer-assisted methods for assessing strain relatedness in Candida albicans by fingerprinting with the moderately repetitive sequence Ca 3. J Clin Microbiol. 1990;28:1236–1243. doi: 10.1128/jcm.28.6.1236-1243.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Silverman S, Luangjarmekorn L, Greenspan D. Occurrence of oral Candida in irradiated head and neck cancer patients. J Oral Med. 1984;39:194–196. [PubMed] [Google Scholar]
- 25.Sullivan D, Haynes K, Billie J, Boerlin P, Rodero L, Lloyd S, Herman M, Coleman D. Widespread geographic distribution of oral Candida dubliniensis strains in human immunodeficiency virus-infected individuals. J Clin Microbiol. 1997;35:960–964. doi: 10.1128/jcm.35.4.960-964.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vasquez J A, Beckley A, Sobel J D, Zervos J J. Comparison of restriction enzyme analysis and pulsed-field gradient gel electrophoresis as typing systems for Candida albicans. J Clin Microbiol. 1991;29:962–967. doi: 10.1128/jcm.29.5.962-967.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Winston D J, Chandrasekar P H, Lazarus H M, Goodman J L, Silber J L, Horowitz H, et al. Fluconazole prophylaxis of fungal infection in patients with acute leukemia. Results of a placebo-controlled, double-blind, multicenter trail. Ann Intern Med. 1993;118:495–503. doi: 10.7326/0003-4819-118-7-199304010-00003. [DOI] [PubMed] [Google Scholar]