Abstract
Structured representation of clinical genetic results is necessary for advancing precision medicine. The Electronic Medical Records and Genomics (eMERGE) Network’s Phase III program initially used a commercially developed XML message format for standardized and structured representation of genetic results for electronic health record (EHR) integration. In a desire to move towards a standard representation, the network created a new standardized format based upon Health Level Seven Fast Healthcare Interoperability Resources (HL7® FHIR®), to represent clinical genomics results. These new standards improve the utility of HL7® FHIR® as an international healthcare interoperability standard for management of genetic data from patients. This work advances the establishment of standards that are being designed for broad adoption in the current health information technology landscape.
Keywords: HL7® FHIR® standard, Clinical genomics, Genetic test results, Interoperability, Electronic health record (EHR), Clinical decision support
1. Introduction
The use of genomic testing in healthcare has grown in recent years, attributed to our increased understanding of the human genome and broader accessibility of tests[1–4]. Typically, the results from genetic testing laboratories are represented in a narrative, unstructured form, delivered as a Portable Document Format (PDF) document, which is simply scanned or uploaded to the electronic health record (EHR)[5]. This approach, though widely accepted and ubiquitous in its use, is primarily targeted to human readers. PDF and other custom structured reporting, though serving the immediate need of delivering genomic test results, hinders widespread reanalysis, data sharing, interoperability, automation, and downstream research efforts. Furthermore, without computational standards we are unable to reliably exchange genomic data and use those data to scale discovery and improve clinical care through automated clinical decision support (CDS)[6]. Because access to relevant genomic data in a standardized computable format has been a major hurdle to widespread implementation of genomic medicine, standards organizations, research networks and healthcare institutions have embarked on multiple projects in the past decade to improve the accessibility of genomic test results as computable data[7–9].
The Electronic Medical Records and Genomics (eMERGE) Network, funded by the National Human Genome Research Institute (NHGRI), is a consortium of U.S. medical research institutions that “develops, disseminates, and applies approaches to research that combine biorepositories with electronic medical record systems for genomic discovery and genomic medicine implementation research”[10]. Phase III of this program, as part of the Network’s overarching goal to integrate genomic test results to the EHR for clinical care, sought to create a proof of concept, standards-based representation of clinical genomic test results using the rapidly evolving Health Level Seven Fast Healthcare Interoperability Resources (HL7® FHIR®2)[11] standard.
1.1. eMERGE background
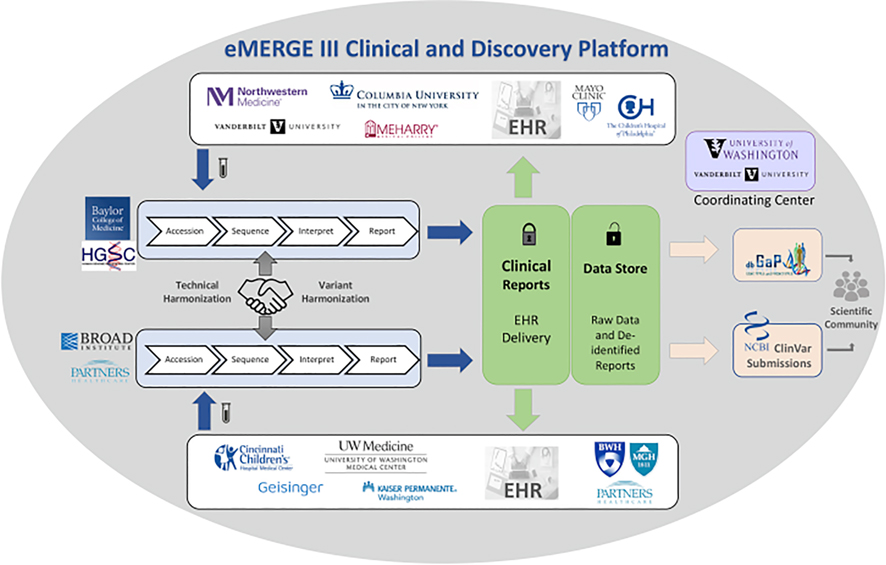
Currently in its fourth phase, the eMERGE Network’s first phase began in September 2007. eMERGE Phase III (September 2015 to March 2020) joined together multiple laboratories and clinics, including two central sequencing and genotyping facilities (CSGs): Baylor College of Medicine Human Genome Sequencing Center (BCM-HGSC) and the Broad Institute (BI) and Partners Laboratory for Molecular Medicine (LMM), as well as eleven study sites. The CSGs performed sequencing, variant interpretation, and clinical report generation. The reports were then returned to study sites and integrated into the respective EHRs [12,13]. As illustrated in Fig. 1[14], the complexity and heterogeneity of Phase III of the Network provided considerable challenges for technical and data harmonization across the CSGs and study sites[15]. The network identified workflow and logistic components to harmonize both data and delivery of results, as well as the need for both a narrative PDF form to satisfy clinicians and regulatory requirements and a custom XML format[15] to represent clinical genetic test results directly in the EHR. This custom format, though opportune for the eMERGE Network at delivering the initial requirements, is not consistent with other open standards and thus not a viable long-term solution to support broader growth and adoption. With the emergence of FHIR® as an interoperable healthcare standard and the work of the HL7® Clinical Genomics Workgroup (CG WG) towards the creation of a FHIR® Genomics Reporting Implementation Guide (GR IG)[16], the Network decided on a specification based on the GR IG in an effort to contribute to and validate the nascent GR IG.
Fig. 1. The eMERGE III Network.
The Network comprised 11 study sites, 2 central sequencing and genotyping facilities (CSGs) and a coordinating center (CC) with samples sent from the study sites to the CSGs. Clinical genetic results were returned to the study sites after sequencing, variant classification and harmonization at the CSGs with raw data and de-identified results being returned to the CC for research. Both the clinical and research data flow is illustrated in this figure[14].
1.2. FHIR® background
FHIR®[17], a standard for exchanging healthcare information electronically, builds on the widely used HL7® Version 2[18] messaging standard and leverages common, modern web technologies that facilitate the development and integration of applications into clinical environments. FHIR® has gained traction since its inception in 2012 as an open API standard for interoperability, continuing to grow with programs such as the FHIR® Accelerator Program[19], the healthcare pledge by technology companies to adopt emerging standards such as FHIR® for interoperability[20], along with NIH notices encouraging the use of FHIR® for clinical and research interoperability[21,22]. Clinical software vendors are also increasingly implementing support for FHIR®-based APIs into EHRs and laboratory information management systems (LIMS), enabling clinical lab test results to be rendered as discrete data elements that can be used to drive CDS.
Building upon the standard[23], the FHIR® community has worked to support its implementation for clinical genetics and genomics. In November 2019, the HL7® CG WG published the first release of the GR IG, which is intended to support “all aspects[16]” of reporting clinical genomics data. The Implementation Guides are supported by the ability of FHIR® to customize resources (basic building blocks which represent key entities) into profiles (focused customization or extension and use of a resource), which provide use in specific contexts, like genetics and genomics. Additionally, FHIR® inherently supports extensions of resources (the ability to add additional attributes)[24].The current release of the GR IG[16], based on FHIR® R4[23], has been balloted as a Standard for Trial Use (STU), which is intended to be vetted through implementation and pilot testing by early adopters. This process is critically important to the advancement of the standard as feedback drives improvements in the specification before it becomes more widely adopted, which limits the ability to make breaking changes as gaps are filled.
1.3. Project objectives
In 2019 the eMERGE Network aimed to improve structured data standards and take advantage of newly emerging FHIR® capabilities. Three milestones were established: 1. Develop a computable and standardized clinical reporting specification for eMERGE with FHIR®; 2. Create a proof-of-concept implementation pilot, generating eMERGE clinical genetic reports with the eMERGE FHIR® Specification; and 3. Establish EHR interoperability with FHIR®-enabled secure ingestion of genetic results and implement clinical decision support use cases.
Recognizing the emerging nature of this landscape, the network limited the scope of its effort to building a baseline upon which future work can be continued. Here we describe these efforts, focusing on identifying lessons learned and future considerations on how organizations can begin to adopt the FHIR® genomics standard, illustrating the level of expertise and effort required to do so, and demonstrating the positive impact that such efforts can have on the development of international standards.
2. Methods
Fulfillment of the objectives and the milestones set forth for this project was managed as a cross-network collaboration with the two CSGs first developing the eMERGE FHIR® Specification. The BCM-HGSC acted in its role of a diagnostic laboratory to generate a sample set of clinical genetic reports utilizing the specification, and study site Northwestern University (NU) and non-clinical affiliate Johns Hopkins University (JHU) acted as provider facilities that implemented a proof-of-concept EHR integration and clinical care pilot to demonstrate the viability of the eMERGE FHIR® Specification. These steps are illustrated in Fig. 2 and detailed further in this section.
Fig. 2. Methods for specification and pilot projects development.
The two columns represent complementary work streams - the leftmost dedicated to the specification development, and the right on use cases for pilot projects development. The solid arrows represent the stepwise or iterative direction of the respective processes.
2.1. eMERGE FHIR® Specification development
The two CSGs established the following guiding principles for this project, taking advantage of their respective experiences:
Structured content - All content from the narrative PDF eMERGE reports and all eMERGE standard reporting use cases should be captured in structured format and as meaningful data elements without losing content and context;
Alignment with HL7® FHIR® and GR IG - All eMERGE concepts and associated elements shall be aligned with GR IG and FHIR® Standards and extended as required;
Computationally reliable representation of results - An optimal computational form for each data element shall be determined, prioritizing eMERGE pilot objectives for EHR integration and CDS. All specimen types and genetic data elements related to the resulting observations must be based on reference sequences, coordinates and structures that consistently and accurately reflect the lab methods used to align the raw data, determine coverage and call the variants; and
Codify concepts when reasonable - Concepts should be codified using FHIR® and GR IG guidance. eMERGE concepts that extend beyond the FHIR® and GR IG guidance should be codified if possible and within reason.
The development of the eMERGE FHIR® Specification consisted of the following steps -
Identify data elements using existing results from a comprehensive set of eMERGE reporting use cases;
Map eMERGE report elements and structures both semantically and structurally to GR IG resources and profiles; perform an analysis, identify issues that require further resolution, and propose resolutions; and
-
Harmonize and document finalized decisions informed by
Harmonizing changes with GR IG
Documenting resolutions requiring custom profiles & extensions
Incorporating feedback from BCM-HGSC lab pilot development
The steps defined above were accomplished by first analyzing eMERGE narrative reports representing a range of use cases. Next, working with domain experts (geneticists from both the CSGs), the semantic meaning and structure of these concepts were verified and all data elements appearing on the reports were associated with the proper concepts. This step was critical to assuring that at a minimum both CSGs agreed with the conceptual meaning of all granular elements on their reports, minimizing subsequent misrepresentation of the elements.
The next step was to determine how best to computationally represent these concepts. A gap analysis was performed between each eMERGE concept and its corresponding GR IG resources and profiles. The aim of this effort was to synchronize GR IG resources and profiles to related eMERGE reporting concepts. Gaps, questions and issues arising during this process were publicly documented and harmonized between the eMERGE CSGs and the HL7® FHIR® community. This reconciliation and harmonization process resulted in the eMERGE FHIR® Specification, documented on the specification’s website[26]. This process also contributed to enhancements and changes of the GR IG itself.
2.2. Use case pilot projects development
We conducted a proof-of-concept implementation pilot that represented real clinical genetic testing reports using the eMERGE FHIR® Specification in EHRs. The pilot project included three use cases: 1. BCM-HGSC - demonstrate the ability to generate sample reports that conform to the eMERGE FHIR® Specification; 2. NU - use the reports from use case 1 to simulate pharmacogenomics clinical decision support; 3. JHU - use the reports from use case 1 to document variant associated phenotypes. Appendix A contains additional details about the pilot and steps conducted at each use case site. The pilot further allowed ratification of the eMERGE FHIR® Specification via iterative feedback from the test sites.
3. Results
3.1. eMERGE FHIR® Specification development
3.1.1. Identification of eMERGE report concepts and elements
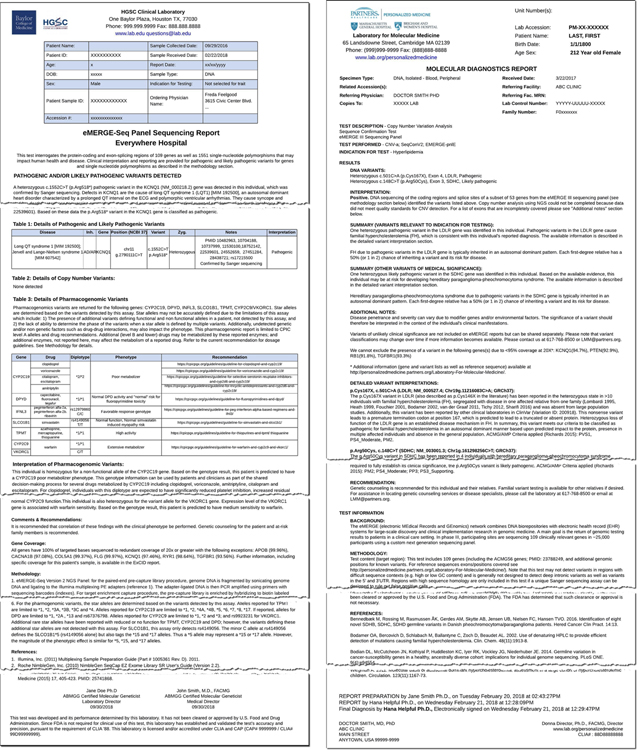
The first step towards the creation of the eMERGE FHIR® Specification was an “As Is” analysis of the existing genetic reports to inventory all eMERGE reporting concepts and elements. To this end, we compiled a set of all-inclusive representative reports from both the CSGs (see Fig. 3 for an example report from each CSG) to ensure use cases requiring unique report concepts and elements were included. Table 1 contains the list of principal eMERGE reporting scenarios.
Fig. 3. BCM-HGSC & LMM eMERGE Narrative Report Examples.
Reports illustrate the content and structure of a typical positive genetic report generated by each of the CSGs.
Table 1. eMERGE Reporting Use Cases.
This table lists all of the eMERGE reporting use cases. The first column is a breakdown of reporting result types and the second column lists more specific outcomes for the corresponding reporting result type. The specialized reporting use cases are specified for the associated eMERGE study sites.
| Standard Reporting Use Cases | |
| Diagnostic Gene Panel | |
| Reported Variants | Single-nucleotide variants (SNVs), Insertions or deletions (InDels), copy number variants (CNVs) |
| Medically Significant Findings for Indication | Pathogenic, Likely Pathogenic, Uncertain Significance |
| Secondary Findings | Medically significant findings in a gene unrelated to the indication for testing |
| Overall Interpretations | Positive, Negative, Inconclusive |
| Pharmacogenomic Gene Panel | |
| PGx Phenotype Findings | Efficacy, Metabolism, Transporter Function |
| Reported Diplotypes | Simple, Complex, Wildtype, Non-Wild Type |
| Specialized Reporting Use Cases | |
| LDLR Scores-Polygenic Risk Scores | Mayo Clinic, an eMERGE Study Site |
| Research Only Reports | Columbia University, an eMERGE Study Site |
Using selected reports for these associated use cases, the structure and composition of the reports was analyzed, and a set of data elements was assembled (Fig. 4), resulting in 18 core concepts and around 100 fundamental data elements. This analysis and documentation of the existing eMERGE report content served as the foundation for the design of the eMERGE FHIR® Specification.
Fig. 4. BCM-HGSC general report layout and detailed mapping.
Illustrates the composition and layout of the data concepts within the eMERGE reports with each of the concepts coded by color and more precisely by number. Note: though Fig. 4 is the BCM-HGSC format, BI/LMM also included the same concepts in a slightly modified layout. The numbering also correlates to the concepts in Fig. 5.
In an effort to manage the pilot within allotted resources and time, the scope of this effort was confined to the Standard Reporting Use Cases and result delivery, while including provisions for future expansion.
3.1.2. eMERGE report to FHIR® GR IG - mapping and analysis
The next step in the development of the eMERGE FHIR® Specification was the mapping of eMERGE report concepts and elements to the GR IG. Adopting the GR IG’s guidance[16], all major eMERGE report concepts were aligned to the GR IG resources and profiles, followed by a granular mapping of every eMERGE report element to a corresponding FHIR® resource element.
3.1.2.1. Mapping eMERGE report concepts to FHIR® resources and profiles.
The GR IG provided the guidance for driving the mapping of the eMERGE report concepts to its resources, profiles and extensions. Our first attempt at mapping resulted in several key structural and organizational questions; as these questions and issues were fundamental to the design of the eMERGE FHIR® Specification, a significant amount of time was spent to identify long and short-term resolutions.
The end result was a set of 21 noteworthy issues and associated resolutions, documented as part of the eMERGE Specification[25]. These issues and their resolutions (Table 2) vary in scope and complexity with proposed solutions being driven by balancing the timeline for delivering the eMERGE pilot against the ideal solution as noted.
Table 2. Issues & Resolutions.
This table lists all of the noteworthy issues that required investigation and collaboration with the HL7® CG WG to resolve. The three columns show the issue label, category (Major, Minor) and resolution status (Clarification - GR IG or FHIR® solution, GR IG Change - CG WG modified GR IG to support, Workaround - extension or non-standard approach, Deferred - deferred workaround but submitted for future consideration). Detailed description for each of these issues is available on the Issues & Resolutions[25] page of the eMERGE FHIR® Specification’s website[26].
| Issue | Cat. | Resolution |
|---|---|---|
| #1 Composite Reporting | Major | GR IG Change |
| #2 Nested Results | Major | Clarification |
| #3 Test Information | Major | Clarification |
| #4 Interpretation Summary | Major | Workaround |
| #5 Gene/Region Coverage | Major | Workaround |
| #6 Recommendations | Major | Clarification |
| #7 Secondary Findings | Major | Clarification |
| #8 Variant Data Types | Major | Deferred |
| #9 Report Comments | Minor | Clarification |
| #10 Confirmation Testing | Major | Workaround |
| #11 Pathogenicity Phenotypes | Minor | GR IG Change |
| #12 Pathogenicity Values | Minor | GR IG Change |
| #13 Report Category | Minor | GR IG Change |
| #14 Assessed Med Citations | Major | Workaround |
| #15 Sign-Out v Sent Date | Minor | Workaround |
| #16 Recommended Followup Reasons | Minor | Workaround |
| #17 Recommended Followup Codes | Minor | GR IG Change |
| #18 Disclaimers | Major | Workaround |
| #19 Patient Internal ID | Minor | Clarification |
| #20 Patient Age | Major | Workaround |
| #21 Research Flag | Minor | Deferred |
Addressing and resolving these issues resulted in the mapping and structural design of the specification, illustrated in Fig. 5. As illustrated, the root profile of the specification is the GenomicsReport; this is the key resource that encapsulates the ServiceRequest for the test, the Observations that constitute the results (i.e. findings or implications of the test), the Tasks that include clinical care recommendations, and the Grouper Profile to organize and manage composite resulting (i.e. GenePanel and PGx results). Other major resources attached to the GenomicsReport include the Patient for whom the test is being ordered, the associated Specimen, the Practitioner ordering the test, the Organization (i.e. Diagnostic Laboratory performing the test) and the Practitioner interpreting the results of the test.
Fig. 5. Structural Design of the eMERGE FHIR® Specification.
A color-coded organizational mapping of the FHIR® GR IG resources and profiles for eMERGE, including custom eMERGE extensions in blue (X6, X7). The color and numbering maps back to the report layout and example in Fig. 4 for easier reference. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
3.1.2.2. Mapping eMERGE report attributes to FHIR® resource elements.
Step 2 involved a granular mapping of every eMERGE report attribute to an equivalent field in the FHIR® resources identified in the previous step. This was a laborious process which in addition to requiring precise and careful mapping of the fields themselves, also required determining naming systems and assignment of coding systems, codes and values. Online documentation[27] includes the complete set of eMERGE FHIR® resources and its associated elements, with a summary table in Appendix B. Furthermore, gap analysis at this step revealed the need for additional fields such as summary interpretation text, test disclaimer etc. that were not available in the GR IG. Though we documented these as feature requests in HL7®’s Jira[28], to satisfy the immediate needs of the project, we created these fields as FHIR® Extensions. The full list of Extensions[29] is available on the eMERGE FHIR® Specification’s website[26].
3.1.3. Harmonization and documentation
The eMERGE FHIR® Specification project was initiated while the HL7® CG WG was still working on its first draft of GR IG. Though the draft was a good start for the eMERGE Specification, it did not encompass all the eMERGE use cases. Documentation for the eMERGE FHIR® Specification is available on the eMERGE FHIR® Specification’s website [26]; the sample reports generated via an implementation of the specification are available on GitHub[30].
3.2. Use case pilot projects development
The principal goals of the pilot were a) for the CSG BCM-HGSC to demonstrate the successful generation of sample reports adhering to the eMERGE FHIR® Specification, b) to successfully ingest these reports to FHIR® servers at the recipient sites, and c) to demonstrate use of the FHIR® formatted data for NU and JHU’s respective use cases. In total, 194 FHIR® reports were created by BCM-HGSC and ingested by FHIR® servers at both NU and JHU. Both institutions were able to utilize the FHIR® reports to demonstrate their respective use cases. Fig. 6 illustrates the software components and data flow with the full details of the results of the pilot described in Appendix A.
Fig. 6. eMERGE III FHIR® Pilot.
FHIR® reports for the eMERGE pilot samples were generated by BCM-HGSC and pushed to NU and JHU’s FHIR® servers. Data was extracted from these FHIR® reports for the NU PGx and JHU Variant Associated Phenotype use cases respectively.
4. Discussion
Through this eMERGE pilot effort we developed a standards-based specification for representing clinical genetic and genomic test results and demonstrated its viability with EHR integration and clinical use cases. This project supported the development of the FHIR® Specification for genomics by basing the eMERGE FHIR® Specification on the GR IG, and subsequently demonstrating use of the FHIR® standard for genomic medicine. Through a data quality assessment framework (Appendix A), we confirmed that the data structures of the FHIR®-formatted reports were acceptable for the two CDS use cases. Additionally, the deployment provided several lessons that guide future evolution of the FHIR® Specification.
The CG WG released the first standard trial use (STU1) of the GR IG in November 2019, nearly a year after the work described in this manuscript began. As an early adopter, we worked collaboratively with the CG WG to further inform the design of the STU1 specification as it matured. As our experience demonstrated, feedback from early adopters contributes significantly to the development of a draft specification, but adopters must be flexible as the standard evolves in parallel.
The level of detail in the GR IG, which uses a series of FHIR® profiles designed to reflect the complexity of clinical genomic test results, requires adopters to have an in-depth understanding of the domain as well as time and resources to implement the complex specification itself. This could be a deterrent to adoption for vendors, laboratories, or consumers that have relatively simple data.
The CG WG, recognizing that the complexity of the GR IG could be a barrier to adoption, is actively working with stakeholders to refine the specification. The experience of this project demonstrates the potential value of working directly with adopters to identify minimum viable products that are based on local use cases. Reciprocally, the CG WG would benefit significantly from concerted efforts by potential adopters (i.e. vendors, laboratories, healthcare systems, health IT engineers) through participation and collaboration to help inform the roadmap and development of the GR IG. Examples of such initiatives include ONC Sync for Genes[31], PGx CDS with FHIR® and CDS Hooks[32], HLA Reporting with FHIR®[33], Variant Interpretation for the Cancer Consortium[34], and mCODE[35].
4.1. Future considerations
The issues we addressed during this project varied in scale in both importance as well as complexity. From these we have selected four key possibilities for further development: variant data types, interpretation summary text, gene/region coverage, and PGx results representation. A comprehensive description of all the issues can be found in the eMERGE FHIR® Specification[25].
4.1.1. Variant data types
Community standards are required to define variants in a manner that will allow science and medicine to adopt and develop tools and procedures to track and attach information to variants at the scale demanded by genetic testing. Efforts to describe and identify variants include: Human Genome Variation Society (HGVS) nomenclature[36], International System for Human Cytogenetic Nomenclature (ISCN) nomenclature[37], and Variant Call File (VCF) formats[38]. Public repositories and registries established by various authorities have evolved over the past decade or more to provide identifiers for variants in these systems (e.g. dbSNP[39], ClinVar[40], ClinGen Allele Registry[41], COSMIC[42]).
The notion of a definitional or reference variant does not exist in FHIR® or the GR IG, with FHIR® and the CG WG lacking the data types and resources needed to build these referenceable variants. The CG WG’s proposed resolution is to develop FHIR® profiles based on the Observation resource. This begins to address the concern of separating the variant observation from the other assertion profiles, like disease causality or therapeutic implications of metabolism or efficacy. However, it does not provide a pragmatic computational standard for representing the variants referenced within those observed variants and associated assertions.
The Genomic Knowledge Standards (GKS) Workstream of the Global Alliance for Genomic Health (GA4GH) is developing and expanding the Variation Representation Specification[43,44] (VRS) to address the need for standards for computationally sharing variation. Instituting such a model in FHIR® will significantly reduce the adoption risks caused by the complexity and unguided extensibility of the current GR IG and FHIR® Specifications. As such, the growing collaboration between the CG WG and the GA4GH GKS Workstream represents a promising step forward at introducing the concepts, resources and data types needed in the FHIR® Specification to improve the viability of implementing use cases related to variation in FHIR® systems.
4.1.2. Interpretation summary text
While structured and coded results are of great importance to the computational utility of results, unstructured text will always play a significant role in conveying information between humans. Though there are a number of text attributes available throughout the GR IG resources, the genetics community requires the ability to associate an interpretation summary with the overall results of the report and with one or more classified variants. It is our recommendation that the CG WG consider all of the important kinds of text fields needed to support clinical genetic test results and assure that there is a mechanism to do so, starting with an interpretation summary text field.
4.1.3. Gene / region coverage
Because testing methods vary, it is important to provide a quantitative representation of the precise molecular sequenced regions covered and the quality of coverage for each region. Perhaps more importantly, this information clearly identifies which regions of the genome were not covered but may be relevant for the indication for testing. Receiving systems would then be able to accurately determine whether a patient may need follow-up testing to interrogate additional regions, and results across cohorts and studies would be more comparable.
4.1.4. PGx results representation
Key challenges to computationally representing PGx results are 1) to include the assayed variants that underlie the derived haplotype/diplotype assertions and 2) to maintain a clear distinction between the case-independent PGx knowledge statements about the haplotype/diplotype states and the patient-specific PGx implications expressed in the interpretation, which are based on the assay findings. The GR IG should provide a mechanism for defining variants at various levels that can be referenced either in the context of a patient observation or independently in a generalizable knowledge base. Also, it is currently difficult to convey the underlying assayed variants using the existing specification; if this could be accomplished then clinical systems could identify previously reported PGx results that may be impacted by future knowledge updates. This design approach supports the need for variant data types as discussed in section 4.1.1.
5. Conclusion
With this project, we have developed and demonstrated the use of a structured, computable and interoperable HL7® FHIR® standard for eMERGE clinical reporting and have demonstrated the feasibility of implementing a standardized representation of clinical genetic and genomic results in the EHR. We have helped move the needle forward for the HL7® Clinical Genomics Workgroup in its development of the Genomics Reporting Implementation Guide by identifying and contributing to several growth areas[25] in order to effectively fulfill the return of clinical genomics results.
The two pilot use cases helped us understand the landscape to use FHIR®-formatted reports for genomic medicine and its challenges. Establishing a standard, though fundamental, is but one step towards the effective use of genomic medicine in the clinic. In order to achieve this vision, additional work is needed not only to mature and stabilize a standard for structured representation of clinical genomic data but also with establishing engagement and collaboration between domain experts. Myriad perspectives are needed, including that of clinicians and geneticists, standards bodies such as HL7®, EHR and other clinical system vendors, diagnostic laboratories, and health IT. These perspectives will further identify genomic medicine use cases and build standards and systems to support these use cases as well as target integration and interoperability amongst users, generators and consumers of clinical genomic data. Tenacious continuation of such undertakings and joint complementary efforts is the path towards supporting standardization and effective use of genomic data in the clinic.
Supplementary Material
Acknowledgements
1. Members of the HL7® Clinical Genomics Workgroup;
2. Members of the eMERGE EHRI Workgroup;
3. Steve Ordhal contributed through Microsoft’s Partner Network that provides enhanced services to Johns Hopkins University.
4. Members of the BCM-HGSC Clinical Laboratory.
Funding
This work was conducted under Phase III of the eMERGE Network, which was initiated and funded by the NHGRI through the following grants: U01HG008657 (Kaiser Permanente Washington/University of Washington); U01HG008685 (Brigham and Women’s Hospital); U01HG008672 (Vanderbilt University Medical Center); U01HG008666 (Cincinnati Children’s Hospital Medical Center); U01HG006379 (Mayo Clinic); U01HG008679 (Geisinger Clinic); U01HG008680 (Columbia University Health Sciences); U01HG008684 (Children’s Hospital of Philadelphia); U01HG008673 (Northwestern University); U01HG008701 (Vanderbilt University Medical Center serving as the Coordinating Center); U01HG008676 (Partners Healthcare/Broad Institute); U01HG008664 (Baylor College of Medicine); and U54MD007593 (Meharry Medical College).
Footnotes
Declaration of Competing Interest
The authors declare the following financial interests/personal relationships which may be considered as potential competing interests: [Luke V. Rasmussen has a patent GENERATING DATA IN STANDARDIZED FORMATS AND PROVIDING RECOMMENDATIONS that is no longer being pursued. Samuel J. Aronson, Hana Zouk and Heidi L. Rehm are employed by Mass General Brigham which receives royalties on sales of GeneInsight software. David R. Crosslin is a consultant for UnitedHealth Group. Richard A. Gibbs declares that Baylor College of Medicine receives payments from Baylor Genetics Laboratories, which provides services for genetic testing; Baylor College of Medicine is part owner of Codified Genomics. Eric Venner is a cofounder of Codified Genomics, which provides variant interpretation services. All other authors declare no competing interests.]
Appendix A. Supplementary data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jbi.2021.103795.
References
- [1].Manolio TA, Chisholm RL, Ozenberger B, Roden DM, Williams MS, Wilson R, Bick D, Bottinger EP, Brilliant MH, Eng C, Frazer KA, Korf B, Ledbetter DH, Lupski JR, Marsh C, Mrazek D, Murray MF, O’Donnell PH, Rader DJ, Relling MV, Shuldiner AR, Valle D, Weinshilboum R, Green ED, Ginsburg GS, Implementing genomic medicine in the clinic: the future is here, Genet. Med 15 (2013) 258–267, 10.1038/gim.2012.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Manolio TA, Green ED, Leading the way to genomic medicine, Am. J. Med. Genet. C Semin, Med. Genet 166 (2014) 1–7, 10.1002/ajmg.c.31384. [DOI] [PubMed] [Google Scholar]
- [3].Green ED, Gunter C, Biesecker LG, Di Francesco V, Easter CL, Feingold EA, Felsenfeld AL, Kaufman DJ, Ostrander EA, Pavan WJ, Phillippy AM, Wise AL, Dayal JG, Kish BJ, Mandich A, Wellington CR, Wetterstrand KA, Bates SA, Leja D, Vasquez S, Gahl WA, Graham BJ, Kastner DL, Liu P, Rodriguez LL, Solomon BD, Bonham VL, Brody LC, Hutter CM, Manolio TA, Strategic vision for improving human health at The Forefront of Genomics, Nature 586 (2020) 683–692, 10.1038/s41586-020-2817-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Yang Y, Muzny DM, Xia F, Niu Z, Person R, Ding Y, Ward P, Braxton A, Wang M, Buhay C, Veeraraghavan N, Hawes A, Chiang T, Leduc M, Beuten J, Zhang J, He W, Scull J, Willis A, Landsverk M, Craigen WJ, Bekheirnia MR, Stray-Pedersen A, Liu P, Wen S, Alcaraz W, Cui H, Walkiewicz M, Reid J, Bainbridge M, Patel A, Boerwinkle E, Beaudet AL, Lupski JR, Plon SE, Gibbs RA, Eng CM, Molecular findings among patients referred for clinical whole-exome sequencing, JAMA 312 (2014) 1870–1879, 10.1001/jama.2014.14601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Shirts BH, Salama JS, Aronson SJ, Chung WK, Gray SW, Hindorff LA, Jarvik GP, Plon SE, Stoffel EM, Tarczy-Hornoch PZ, Van Allen EM, Weck KE, Chute CG, Freimuth RR, Grundmeier RW, Hartzler AL, Li R, Peissig PL, Peterson JF, Rasmussen LV, Starren JB, Williams MS, Overby CL, CSER and eMERGE: current and potential state of the display of genetic information in the electronic health record, J. Am. Med. Inform. Assoc. JAMIA 22 (2015) 1231–1242, 10.1093/jamia/ocv065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Williams MS, Taylor CO, Walton NA, Goehringer SR, Aronson S, Freimuth RR, Rasmussen LV, Hall ES, Prows CA, Chung WK, Fedotov A, Nestor J, Weng C, Rowley RK, Wiesner GL, Jarvik GP, Del Fiol G, Genomic Information for Clinicians in the Electronic Health Record: Lessons Learned From the Clinical Genome Resource Project and the Electronic Medical Records and Genomics Network, Front. Genet 10 (2019) 1059, 10.3389/fgene.2019.01059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Watkins M, Eilbeck K, FHIR Lab Reports: using SMART on FHIR and CDS Hooks to increase the clinical utility of pharmacogenomic laboratory test results, AMIA Summits Transl. Sci. Proc 2020 (2020) 683–692 (accessed October 22, 2020), https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7233102/. [PMC free article] [PubMed] [Google Scholar]
- [8].Alterovitz G, Warner J, Zhang P, Chen Y, Ullman-Cullere M, Kreda D, Kohane IS, SMART on FHIR Genomics: facilitating standardized clinico-genomic apps, J. Am. Med. Inform. Assoc. JAMIA 22 (2015) 1173–1178, 10.1093/jamia/ocv045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Ryu B, Shin S-Y, Baek R-M, Kim J-W, Heo E, Kang I, Yang JS, Yoo S, Clinical Genomic Sequencing Reports in Electronic Health Record Systems Based on International Standards: Implementation Study, J. Med. Internet Res 22 (2020), 10.2196/15040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Electronic Medical Records and Genomics (eMERGE) Network, Genome.Gov. (2020). https://www.genome.gov/Funded-Programs-Projects/Electronic-Medical-Records-and-Genomics-Network-eMERGE (accessed August 26, 2020).
- [11].Summary - FHIR v4.0.1, (2020). https://www.hl7.org/fhir/summary.html (accessed October 22, 2020).
- [12].Wiesner GL, Kulchak Rahm A, Appelbaum P, Aufox S, Bland ST, Blout CL, Christensen KD, Chung WK, Clayton EW, Green RC, Harr MH, Henrikson N, Hoell C, Holm IA, Jarvik GP, Kullo IJ, Lammers PE, Larson EB, Lindor NM, Marasa M, Myers MF, Peterson JF, Prows CA, Ralston JD, Milo Rasouly H, Sharp RR, Smith ME, Van Driest SL, Williams JL, Williams MS, Wynn J, Leppig KA, Returning Results in the Genomic Era: Initial Experiences of the eMERGE Network, J. Pers. Med 10 (2020), 10.3390/jpm10020030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Lessons learned from the eMERGE Network: balancing genomics in discovery and practice, Hum. Genet. Genomics Adv 2 (2021) 100018. 10.1016/j.xhgg.2020.100018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].eMERGE Consortium. Electronic address: agibbs@bcm.edu, eMERGE Consortium, Harmonizing Clinical Sequencing and Interpretation for the eMERGE III Network, Am. J. Hum. Genet 105 (2019) 588–605. 10.1016/j.ajhg.2019.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Aronson S, Babb L, Ames D, Gibbs RA, Venner E, Connelly JJ, Marsolo K, Weng C, Williams MS, Hartzler AL, Liang WH, Ralston JD, Devine EB, Murphy S, Chute CG, Caraballo PJ, Kullo IJ, Freimuth RR, Rasmussen LV, Wehbe FH, Peterson JF, Robinson JR, Wiley K, Overby Taylor C, eMERGE Network EHRI Working Group, Empowering genomic medicine by establishing critical sequencing result data flows: the eMERGE example, J. Am. Med. Inform. Assoc. JAMIA 25 (2018) 1375–1381, 10.1093/jamia/ocy051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].http://hl7.org/fhir/uv/genomics-reporting/index.html, (n.d.).
- [17].Overview - FHIR v4.0.1, (n.d.). https://www.hl7.org/fhir/overview.html (accessed April 3, 2021).
- [18].HL7 Standards Product Brief - HL7 Version 2 Product Suite | HL7 International, (2020). https://www.hl7.org/implement/standards/product_brief.cfm?product_id=185 (accessed October 30, 2020). [Google Scholar]
- [19].HL7® FHIR® Accelerator Program | HL7 International, (2020). http://www.hl7.org/about/fhir-accelerator/index.cfm (accessed October 22, 2020). [Google Scholar]
- [20].HL7, Cloud Providers Unite for Healthcare Interoperability, (2020). http://blog.hl7.org/cloud-providers-unite-for-healthcare-interoperability-fhir (accessed October 22, 2020).
- [21].https://grants.nih.gov/grants/guide/notice-files/NOT-OD-19-122.html, (n.d.).
- [22].https://grants.nih.gov/grants/guide/notice-files/NOT-OD-20-146.html, (n.d.).
- [23].Index - FHIR v4.0.1, (n.d.). http://hl7.org/fhir/ (accessed March 19, 2021).
- [24].Extensibility - FHIR v4.0.1, (n.d.). https://www.hl7.org/fhir/extensibility.html (accessed April 3, 2021).
- [25].Issues & Resolutions — emerge-fhir-spec 6e81023 documentation, (2020). https://emerge-fhir-spec.readthedocs.io/en/1.0/issues_and_resolutions.html# (accessed October 4, 2020).
- [26].eMERGE Results FHIR Specification — emerge-fhir-spec 6e81023 documentation, (2020). https://emerge-fhir-spec.readthedocs.io/en/1.0/index.html (accessed September 25, 2020).
- [27].Catalogue — emerge-fhir-spec 6e81023 documentation, (2020). https://emerge-fhir-spec.readthedocs.io/en/1.0/artifacts/index.html# (accessed September 17, 2020).
- [28].https://jira.hl7.org/projects/FHIR/issues/FHIR-29410?filter=allissues, (n.d.).
- [29].Extensions — emerge-fhir-spec 6e81023 documentation, (2020). https://emerge-fhir-spec.readthedocs.io/en/1.0/artifacts/extensions.html (accessed September 18, 2020).
- [30].emerge-ehri/fhir-specification, GitHub. (2020). https://github.com/emerge-ehri/fhir-specification (accessed September 26, 2020). [Google Scholar]
- [31].Sync for Genes | HealthIT.gov, (2020). https://www.healthit.gov/topic/sync-genes (accessed October 4, 2020). [Google Scholar]
- [32].Dolin RH, Boxwala A, Shalaby J, A Pharmacogenomics Clinical Decision Support Service Based on FHIR and CDS Hooks, Methods Inf. Med 57 (2018) e115–e123, 10.1055/s-0038-1676466. [DOI] [PubMed] [Google Scholar]
- [33].HLA-REPORTING\Home - FHIR v4.0.1, (2020). http://fhir.b12x.org/ig/hla-reporting/ (accessed October 16, 2020).
- [34].Alterovitz G, Heale B, Jones J, Kreda D, Lin F, Liu L, Liu X, Mandl KD, Poloway DW, Ramoni R, Wagner A, Warner JL, Genomics FHIR, enabling standardization for precision medicine use cases, npj Genomic Med. 5 (2020) 1–4, 10.1038/s41525-020-0115-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].HL7.FHIR.US.MCODE\Home - FHIR v4.0.1, (2020). http://build.fhir.org/ig/HL7/fhir-mCODE-ig/branches/master/index.html (accessed October 16, 2020).
- [36].https://varnomen.hgvs.org/, HGVS. (n.d.).
- [37].Simons A, Shaffer LG, Hastings RJ, Cytogenetic Nomenclature: Changes in the ISCN 2013 Compared to the 2009 Edition, Cytogenet. Genome Res. 141 (2013) 1–6. 10.1159/000353118. [DOI] [PubMed] [Google Scholar]
- [38].Danecek P, Auton A, Abecasis G, Albers CA, Banks E, DePristo MA, Handsaker RE, Lunter G, Marth GT, Sherry ST, McVean G, Durbin R, The variant call format and VCFtools, Bioinformatics 27 (2011) 2156–2158, 10.1093/bioinformatics/btr330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Home - SNP - NCBI, (2020). https://www.ncbi.nlm.nih.gov/snp/ (accessed December 4, 2020).
- [40].ClinVar, (2020). https://www.ncbi.nlm.nih.gov/clinvar/ (accessed December 4, 2020).
- [41].Pawliczek P, Patel RY, Ashmore LR, Jackson AR, Bizon C, Nelson T, Powell B, Freimuth RR, Strande N, Shah N, Paithankar S, Wright MW, Dwight S, Zhen J, Landrum M, McGarvey P, Babb L, Plon SE, Milosavljevic A, on behalf of the Clinical Genome (ClinGen) Resource, ClinGen Allele Registry links information about genetic variants, Hum. Mutat 39 (2018) 1690–1701, 10.1002/humu.23637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Cosmic, COSMIC - Catalogue of Somatic Mutations in Cancer, (2020). https://cancer.sanger.ac.uk/cosmic (accessed December 4, 2020).
- [43].GA4GH Variation Representation Specification — GA4GH Variation Representation Specification HEAD documentation, (2020). https://vr-spec.readthedocs.io/en/1.1/ (accessed November 18, 2020).
- [44].The GA4GH Variation Representation Specification (VRS): a Computational Framework for the Precise Representation and Federated Identification of Molecular Variation | bioRxiv, (2021). 10.1101/2021.01.15.426843v1 (accessed January 26, 2021). [DOI]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.