Abstract
Group B Streptococcus (GBS) is an important pathogen of neonatal infections, and the clonal complex (CC)-17/serotype III GBS strain has emerged as the dominant strain. The clinical manifestations of CC17/III GBS sepsis may vary greatly but have not been well-investigated. A total of 103 CC17/III GBS isolates that caused neonatal invasive diseases were studied using a new approach based on clustered regularly interspaced short palindromic repeats (CRISPR) loci and restriction fragment length polymorphism (RFLP) analyses. All spacers of CRISPR loci were sequenced and analyzed with the clinical presentations. After CRISPR-RFLP analyses, a total of 11 different patterns were observed among the 103 CRISPR-positive GBS isolates. GBS isolates with the same RFLP patterns were found to have highly comparable spacer contents. Comparative sequence analysis of the CRISPR1 spacer content revealed that it is highly diverse and consistent with the dynamics of this system. A total of 29 of 43 (67.4%) spacers displayed homology to reported phage and plasmid DNA sequences. In addition, all CC17/III GBS isolates could be categorized into three subgroups based on the CRISPR-RFLP patterns and eBURST analysis. The CC17/III GBS isolates with a specific CRISPR-RFLP pattern were more significantly associated with occurrences of severe sepsis (57.1% vs. 29.3%, p = 0.012) and meningitis (50.0% vs. 20.8%, p = 0.009) than GBS isolates with RFLP lengths between 1000 and 1300 bp. Whole-genome sequencing was also performed to verify the differences between CC17/III GBS isolates with different CRISPR-RFLP patterns. We concluded that the CRISPR-RFLP analysis is potentially applicable to categorizing CC17/III GBS isolates, and a specific CRISPR-RFLP pattern could be used as a new biomarker to predict meningitis and illness severity after further verification.
Keywords: group B Streptococcus, CRISPR, multilocus sequence typing, antimicrobial resistance, phage
1. Introduction
Streptococcus agalactiae (Group B Streptococcus; GBS) is part of the commensal flora in the human genitourinary and gastrointestinal tracts, but GBS is one of the most important pathogens that cause neonatal sepsis and meningitis and sometimes adult invasive diseases [1,2]. The colonization rate of GBS in pregnant women is approximately 18–35%, the infection rate in neonates is approximately 0.4 to 1.1 cases per 1000 live births, and the GBS-sepsis-attributable mortality rate is approximately 10–18% [3,4,5]. The molecular and epidemiological information on GBS isolates in colonized women and their children has been the focus of numerous studies [6,7,8] because these data are especially important for the prevention of neonatal GBS sepsis and the development of effective GBS vaccination [9,10,11].
The differences in capsular polysaccharides (CPS) define GBS isolates into ten different serotypes (Ia, Ib, and II to IX), with serotype III emerging as the most common strain that causes neonatal sepsis in most countries [5,11,12]. In addition, multilocus sequence typing (MLST) has been developed to optimize the discrimination of specific GBS strains to study the epidemiology of GBS infection [11,12,13,14]. The sequence types (STs) are identified by MLST using a combination of alleles for seven housekeeping genes, and genetically related STs could be clustered into clonal complexes (CCs) following phylogenetic analyses. This subtyping system consists of CPS, and STs are now applied by the majority of current GBS studies [11,12,13,14].
Clustered regularly interspaced short palindromic repeats (CRISPR) are composed of short (25–40 base pairs) direct repeats interspaced by nonrepetitive similar-sized sequences called spacers [15,16]. CRISPR-associated protein-9 (Cas9)-mediated genome modification is a novel RNA-domain-containing endonuclease-based genome editing technology for a variety of biological and therapeutic applications [17,18,19]. CRISPR/Cas9-based genomic editing of various bacterial strains, such as Escherichia coli, Clostridia, and Streptomycetes, has been performed to investigate the effects of gene deletion, insertion, replacement, and gene regulation [19,20]. CRISPR loci are characterized by a dynamic, rapidly evolving, and polymorphic composition because the system is able to capture novel spacers derived from mobile genetic elements (MGEs) in the environment [18,19,20]. Therefore, spacer polymorphism and acquisition of novel spacers of CRISPR loci can be epidemiological markers for transmission route identification and bacterial genotyping. Recent studies have also documented that CRISPR analysis is an applicable tool to follow maternal GBS colonization and separate GBS isolates with greater discriminating power than traditional subtyping systems [20,21,22]. The CRISPR loci of different GBS strains are highly diverse and reveal the dynamic nature of this system [20,21,22]. However, the characteristics of the CRISPR loci of the same GBS strains have not yet been investigated. In this study, we aimed to investigate the homogeneity of CC17/III GBS strains from neonatal invasive diseases.
2. Results
From January 2006 to December 2018, 103 nonduplicate CC17/III GBS isolates from 103 neonates with invasive GBS disease in CGMH were collected for analyses. There were a total of 8 patients with early-onset disease and 95 patients with late-onset disease. The median (interquartile range (IQR)) gestational age and birth weight of this cohort were 39.0 (37.0–40.0) weeks and 2940 (2655.0–3360.0) g, respectively. Only 19 (18.4%) were premature neonates (GA < 37 weeks), and only 4 neonates were very low birth weight infants (BBW < 1500 g). The onset of GBS sepsis occurred at 29.0 (15.0–51.0) (median (IQR)) days of life. There were four cases of very late GBS sepsis (disease onset > 90 days old). Among these patients, 29 (28.1%) had meningitis. MLST analysis and CPS serotyping were performed to confirm that all these GBS isolates were ST17/III GBS strains.
2.1. CRISPR-RFLP and CRISPR Direct Repeat Analysis
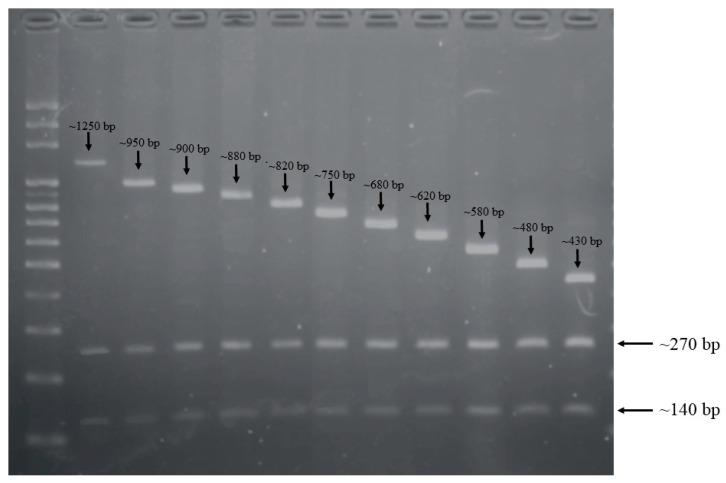
For each isolate, we performed CRISPR-RFLP with the restriction enzyme DdeI. The resulting restriction fragments were separated according to length by gel electrophoresis (Figure 1). After RFLP and electrophoresis, all PCR results of CRISPR1 from these GBS isolates showed three bands, with lengths of CRISPR1 sequences ranging from 140 to 1250 bp. The common 140 and 270 bp fragments were part of the type II-A CRISPR-associated protein and hypothetical protein, respectively. Among all GBS isolates, a total of 11 different gel band patterns were detected by CRISPR-RFLP (Table 1). Because previous studies showed that GBS isolates with the same RFLP pattern may contain similar CRISPR spacer compositions [21], the CRISPR compositions of all isolates were sequenced and analyzed to document this phenomenon. In addition, the PCR products of CRISPR-RFLP were sequenced, and we found that GBS isolates with the same RFLP pattern also had similar spacer compositions (Table 1). Most of the spacers were identified in previous studies, and the same represented numbers were coded [23]. We found that 82 out of 103 (79.6%) GBS isolates contained seven spacers in a conserved order (101, 102, 49, 53, 54, 55, and 56). A total of 18 new spacers were found in this cohort, submitted to GeneBank, and assigned as S1 to S18. We found that all of the newly identified spacers were in GBS strains with larger-molecular-weight CRISPR-RFLP bands. In addition, it is well-known that new spacers are inserted at the 5′ end of the array in most cases, and we suggest that these spacers may have been acquired recently after verification by checking the positions of the spacers.
Figure 1.
Restriction fragment length polymorphism (RFLP) results of CRISPR1 from 103 CC17/III GBS isolates. The length of CRISPR1 sequences after restriction enzyme digestions ranged from 140 to 1250 bp. The 140 and 270 bp fragments existed in all CC17/III GBS isolates, and the third fragment with spacers had lengths ranging from 430 to 1250 bp.
Table 1.
Correlation of the CRISPR-RFLP pattern with spacer compositions.
| Length of CRISPR-RFLP (bp) | No. of Isolates (%) | Fragment Length (bp) | No. of Isolates (%) | CRISPR1 Spacers | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ~1660 | 1 (1) | ~1250 | ~270 | ~140 | 1 | (1) | 476 | 449 | S2 | S3 | 398 | S4 | 119 | 120 | 82 | 82 | 83 | 84 | 85 | 86 | 87 | 7 |
| ~1360 | 2 (1.9) | ~950 | ~270 | ~140 | 2 | (1.9) | S5 | S11 | S12 | S13 | S14 | S15 | 101 | 102 | 49 | 53 | 54 | 55 | 56 | |||
| ~1310 | 2 (1.9) | ~900 | ~270 | ~140 | 2 | (1.9) | S11 | S12 | S13 | S14 | S15 | 101 | 102 | 49 | 53 | 54 | 55 | 56 | ||||
| ~1290 | 2 (1.9) | ~880 | ~270 | ~140 | 1 | (1) | 277 | S6 | 984 | 254 | 101 | 102 | 49 | 53 | 54 | 55 | 56 | |||||
| 1 | (1) | S7 | 724 | S8 | 243 | 101 | 102 | 49 | 53 | 54 | 55 | 56 | ||||||||||
| ~1230 | 23 (22.3) | ~820 | ~270 | ~140 | 22 | (21.4) | 171 | 984 | 254 | 101 | 102 | 49 | 53 | 54 | 55 | 56 | ||||||
| 1 | (1) | S6 | 984 | 254 | 101 | 102 | 49 | 53 | 54 | 55 | 56 | |||||||||||
| ~1160 | 27 (26.2) | ~750 | ~270 | ~140 | 14 | (13.6) | 984 | 254 | 101 | 102 | 49 | 53 | 54 | 55 | 56 | |||||||
| 3 | (2.9) | 171 | 984 | 254 | 101 | 102 | 53 | 54 | 55 | 56 | ||||||||||||
| 3 | (2.9) | S18 | 243 | 101 | 102 | 49 | 53 | 54 | 55 | 56 | ||||||||||||
| 2 | (1.9) | S9 | 243 | 101 | 102 | 49 | 53 | 54 | 55 | 56 | ||||||||||||
| 2 | (1.9) | S17 | 243 | 101 | 102 | 49 | 53 | 54 | 55 | 56 | ||||||||||||
| 1 | (1) | 171 | 984 | 254 | 102 | 49 | 53 | 54 | 55 | 56 | ||||||||||||
| 1 | (1) | S19 | 243 | 101 | 102 | 49 | 53 | 54 | 55 | 56 | ||||||||||||
| 1 | (1) | S11 | S12 | 101 | 102 | 49 | 53 | 54 | 55 | 56 | ||||||||||||
| ~1090 | 21 (20.4) | ~680 | ~270 | ~140 | 20 | (19.4) | 243 | 101 | 102 | 49 | 53 | 54 | 55 | 56 | ||||||||
| 1 | (1) | S10 | S1 | S12 | S13 | S14 | S15 | 101 | 56 | |||||||||||||
| ~1030 | 4 (3.9) | ~620 | ~270 | ~140 | 1 | (1) | 243 | 101 | 102 | 49 | 53 | 54 | 55 | |||||||||
| 1 | (1) | 101 | 102 | 49 | 53 | 54 | 55 | 56 | ||||||||||||||
| 1 | (1) | 243 | 102 | 49 | 53 | 54 | 55 | 56 | ||||||||||||||
| 1 | (1) | 243 | 101 | 49 | 53 | 54 | 55 | 56 | ||||||||||||||
| ~990 | 5 (4.9) | ~580 | ~270 | ~140 | 2 | (1.9) | 724 | 49 | 53 | 54 | 55 | 56 | ||||||||||
| 1 | (1) | 243 | 101 | 102 | 49 | 55 | 56 | |||||||||||||||
| 1 | (1) | 171 | 49 | 53 | 54 | 55 | 56 | |||||||||||||||
| 1 | (1) | 243 | 101 | 102 | 54 | 55 | 56 | |||||||||||||||
| ~890 | 12 (11.7) | ~480 | ~270 | ~140 | 12 | (11.7) | 49 | 53 | 54 | 55 | 56 | |||||||||||
| ~840 | 4 (3.9) | ~430 | ~270 | ~140 | 4 | (3.9) | 171 | 984 | 254 | 56 | ||||||||||||
Highlight in yellow, spacer (viral DNA, phage); orange, spacer (plasmid); green, spacer (chromosomal sequences); blue, spacer (unmatched).
Among the 103 CC17/III GBS isolates, a total of 966 repeats were identified, including a highly conserved typical repeat presenting in all isolates and representing 89.3% of the 966 repeats identified, and 100% of the internal repeats. BLASTn and CRISPRdb were used to analyze the typical repeat sequence and investigate whether it had homology to other bacterial sequences. We found that the typical repeat sequence (5′-GTTTTAGAGCTGTGCTGTTTCGAATGGTTCCAAAAC-3′) and terminal repeat sequence (5′-GTTTTAGAGCTGTGCTGTTATTATGCTAGGACATCA-3′) were identical to those found by Lier et al. [24]. Therefore, this typical repeat showed sequence similarity to other typical repeats found in species of the Streptococcus genus [25].
2.2. CRISPR1-Based Subgroups of CC17/III GBS Isolates and Spacer Analysis
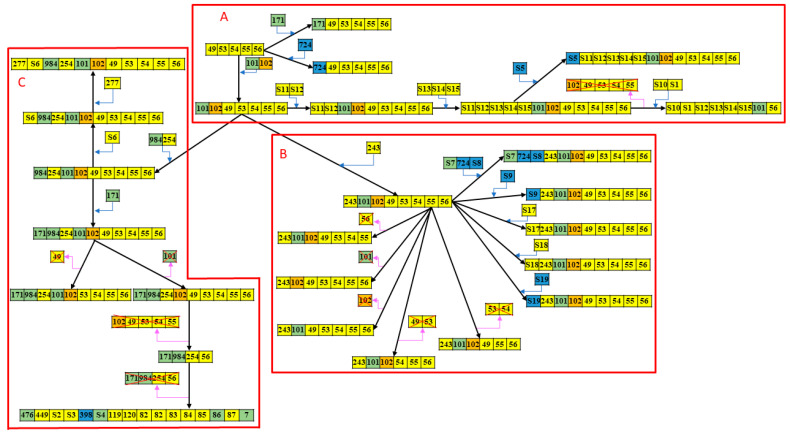
The phylogenetic analysis of 103 GBS isolates was carried out using eBURST software based on the diversity and relatedness of CRISPR type, as shown in Figure 2. All 103 GBS strains were divided into three phylogenetically distinct groups. The patient demographics and clinical presentations of the three subgroups are presented in Table 2. We found that the patient demographics were not significantly different between the three subgroups. However, neonatal sepsis caused by GBS isolates in group A had significantly higher rates of severe sepsis (63.6% vs. 30.9%, p = 0.002) and meningitis (50.0% vs. 22.2%, p = 0.015) than GBS isolates in groups B and C. In addition, the CRISPR-RFLP band lengths of GBS isolates in group A were mostly <1000 bp or >1300 bp, whereas those in group B and C were between 1000 and 1300 bp. Therefore, we found that the CC17/III GBS isolates with RFLP lengths ≥1300 bp and <1000 bp were more significantly associated with the occurrence of severe sepsis (57.1% vs. 29.3%, p = 0.012) and meningitis (50.0% vs. 20.8%, p = 0.009) than GBS isolates with RFLP lengths between 1000 and 1300 bp. The overall mortality of this cohort was 3.9% (a total of four patients died).
Figure 2.
Phylogenetic relationship of 103 CRISPR-positive GBS strains constructed on 43 CRISPR types based on eBURST analysis. All 103 CC17/III GBS isolates can be categorized into three major subgroups (A–C) based on the CRISPR-RFLP array and eBURST analysis. The differences between compositions of CRISPR and gain or loss of spacers are marked and their relationships are presented.
Table 2.
Clinical presentations of 103 cases of GBS invasive diseases categorized based on the length of the CRISPR-RFLP array and eBURST analysis.
| Group | A | B | C |
|---|---|---|---|
| Case number, n (% of total) | 22 (21.4) | 34 (33.0) | 47 (45.6) |
| RFLP patterns | |||
| <1000 bp | 15 (68.2) | 2 (5.9) | 4 (8.5) |
| 1000–1300 bp | 3 (13.6) | 32 (94.1) | 42 (89.4) |
| >1300 bp | 4 (18.2) | 0 (0) | 1 (2.1) |
| Birth body weight (g) | 2914.8 ± 430.2 | 2880.0 ± 644.5 | 3002.6 ± 683.0 |
| Gestational age (weeks) | 38.5 ± 1.8 | 37.6 ± 3.1 | 38.0 ± 3.1 |
| Prematurity | 3 (13.6) | 8 (23.5) | 8 (17.0) |
| Sex (male/female) | 7 (31.8)/15 (68.2) | 24 (70.6)/10 (29.4) | 18 (38.3)/29 (61.7) |
| Any chronic comorbidity | 1 (4.5) | 3 (8.8) | 5 (10.6) |
| Antibiotic susceptibility | |||
| Erythromycin (R) | 17 (77.3) | 2 (82.4) | 47 (100.0) |
| Clindamycin (R) | 15 (68.2) | 2 (70.6) | 47 (100.0) |
| Clinical presentations | |||
| Severe sepsis * | 14 (63.6) ** | 7 (20.6) | 18 (38.3) |
| Meningitis | 11 (50.0) ** | 6 (17.6) | 12 (25.5) |
| Neurological sequelae | 6 (27.3) | 4 (11.8) | 6 (12.8) |
| Final mortality | 1 (4.5) | 1 (0) | 2 (4.3) |
All data are presented as number (%); R: resistance; RFLP: restriction fragment length polymorphism; * Severe sepsis is defined as the presence of septic shock, respiratory failure requiring intubation, and/or multiple organ failure; ** p < 0.05.
All 103 GBS strains were sensitive to penicillin, ampicillin, and vancomycin. None of these strains displayed cefotaxime resistance. On the other hand, only 8.7% (n = 9) and 14.6% (n = 15) of all GBS strains were susceptible to erythromycin and clindamycin, respectively. Most of the erythromycin-resistant GBS isolates (51.1%) were also not susceptible to clindamycin (94.6%, 87 isolates, p < 0.001 by Pearson’s χ2 test). In addition, all the GBS isolates in group C were antibiotic resistant, which was relatively higher than other groups.
BLASTn analyses were performed to analyze the homologous sequences of all the spacers to determine their potential source and function (Table 3). The CRISPR spacers are generally segments of DNA captured from invasive plasmids or phages. We found that more than 86% of these spacers had similar sequences in the NCBI database, with 65% (n = 28) of these spacers showing homology to phage sequences, 18.6% (n = 8) to the GBS chromosome, and only one spacer to a plasmid sequence. Only one spacer (2.3%) showed homology to plasmid, which indicated that horizontal gene transfer by plasmids was uncommon in CC17/III GBS strains. In addition, seven spacers showed homology to the same phage LF2, five spacers showed homology to the same phage Javan52, and three spacers showed homology to the same phage Javan48. Among those with matched MGEs, the majority of the spacers (24/29, 82.8%) displayed 100% homology to the corresponding sequences in phages or plasmids, and only five spacers had between one and seven base mutations. The correspondences between spacers and patients’ clinical conditions are presented in Supplementary Table S1 and no specific conclusions can be documented.
Table 3.
Spacer sequences of all CRISPR loci in the 103 GBS isolates and the potential sources and function using BLASTN analysis.
| Spacer | Sequence | Homology Analysis | Homology Percentage | Function |
|---|---|---|---|---|
| Viral DNA (phages), 65.1% (n = 28) | ||||
| 49 | TGCTAAAAGGTAAATTTAACATTCCAGGTA | Streptococcus phage LF2 | 100% | major tail protein |
| 53 | TATTTGATAGCGGTAACGGGTCATATACAA | Streptococcus phage Javan284 | 76% | putative C5 methylase |
| 54 | TGGTGGTATTTATAATGTACGAGCAAATCG | Streptococcus phage Javan52 | 100% | tail fibers protein |
| 55 | GATAAAAAGTGGGAGCTGAATTAAAAGGCA | Streptococcus phage Javan52 | 100% | hypothetical protein |
| 56 | ATTTGAACGATTTTTATATTCCTGATATGT | Streptococcus phage Javan516 | 100% | lysin, N-acetylmuramoyl-l-alanine amidase |
| 82 | CGTACCATCTATCAATTTACCGCAAGCTGT | Streptococcus phage LF2 | 100% | hypothetical protein |
| 83 | CCGATTATTTCCTACATAATACGCACGTTT | Streptococcus phage LF2 | 100% | hypothetical protein |
| 84 | AACTGTGTGATACTTTTCGTTTTTTTCTTT | Streptococcus phage Javan7 | 100% | hypothetical protein |
| 85 | TTTTATAAGTGATAGAGTGTGCAACACCGT | Streptococcus phage JX01 | 100% | putative minor structural protein |
| 87 | TTCTAAATGCTGGTGACTGCTTTGCATAAA | Streptococcus phage LF2 | 100% | hypothetical protein |
| 119 | GCGATGATGGTAAGTCATCATGGACAGCGT | Streptococcus phage Javan48 | 100% | tail fibers protein |
| 120 | TTTTACACACGATGTCAGATATAATGTCAA | Streptococcus phage Javan10 | 100% | membrane protein |
| 243 | TTGACCGCTCGTCCATTTTTTTAATGTAAA | Streptococcus phage Javan48 | 100% | tail fibers protein |
| 254 | ACCTTGCTCCGATGACACCATCGCGAACCT | Streptococcus phage Javan52 | 100% | tail fibers protein |
| 277 | AATTGATTGCCGTTAAAACCGATAGAGGA | Streptococcus phage LF2 | 100% | structural protein |
| 449 | TAAAATCCTGAAACAGAATGGGATTGATAT | Streptococcus phage Javan90 | 82% | antirepressor protein |
| S1 | AATTGATACATTGCAACGTCTAGCAGGAGC | Streptococcus phage LF2 | 100% | tail length tape-measure protein |
| S2 | GTGTGTTCTTCATTTTTATCAAACCAAAA | Streptococcus phage Javan471 | 100% | hypothetical protein |
| S3 | AAGAAATTCGGTAGAGACCCCAGACTCAT | Streptococcus phage Javan46 | 100% | tail length tape-measure protein |
| S6 | ATTAAATCTTCTTTTGAAGTTACTGTACGT | Staphylococcus phage pSco-10 | 70% | hypothetical protein |
| S10 | TTTATATTGTTCAGAAGAATGCCGCAAAAA | Streptococcus phage Javan44 | 100% | Phage-associated protein |
| S11 | TGTGTACGTTGCCTTTCCGTCAGCACCAGC | Streptococcus phage Javan52 | 100% | tail fibers protein |
| S12 | CCATAAACTTGCCAGTAGATGTGTCACGCT | Streptococcus phage Javan648 | 100% | hypothetical protein |
| S13 | ACCATTCGAAGTAGCTAGTTTGATTTCGTA | Streptococcus phage LF2 | 100% | tail fibers protein |
| S14 | TGTCGATGGTGTTCAAATACAAATGTTTTC | Streptococcus phage Javan52 | 100% | hypothetical protein |
| S15 | CTTTACCATTATTGATTTGTTCTTGCTTTT | Streptococcus phage Javan478 | 100% | hypothetical protein |
| S17 | TCGCAATAATTACTATATGCTTAAGCGGAG | Streptococcus phage Javan7 | 96% | Phage protein |
| S18 | AATCCAACAAAAACAACTTGCTTTAAATAA | Streptococcus phage Javan48 | 100% | membrane protein |
| Plasmid, 2.3% (n = 1) | ||||
| 102 | CTGTTCATAAAGAGCAACTAGTGGCAACAT | Bacillus megaterium strain YC4-R4 plasmid unnamed2 | 70% | hypothetical protein |
| Chromosomal sequences, 18.6% (n = 8) | ||||
| 7, 86, 101, 171, 476, 984, S4, S7 | GBS chromosome | x | x | |
| Unmatched, 14.0% (n = 6) | ||||
| 398, 724, S5, S8, S9, S19 | Unmatch | x | x | |
2.3. WGS Analyses of CC17/III GBS Isolates with Different CRISPR-RFLP Patterns
WGS was conducted in CC17/III GBS isolates with different CRISPR-RFLP patterns in order to verify the results (Supplementary Figure S1). A total of eighteen CC17/III GBS isolates, including six in group A, six in group B, and six in group C, were chosen to investigate the genetic differences that may potentially cause severe sepsis and meningitis. We found that the genes of component systems CovS/R, capsular serotypes, presence of pili island genes, and most virulence genes were not significantly different between all these CC17/III GBS isolates. The only differences were the antibiotic-resistance genes. We found that the significant difference was the presence of one integrative and conjugative element, ICESag37, carrying multiple antibiotic resistant genes and virulence genes, which was found in antimicrobial-resistant CC17/III GBS isolates only and absent in antimicrobial-susceptible CC17/III GBS isolates. The presence of ICESag37 was not significantly different between different CRISPR-RFLP patterns and between the three subgroups. Additionally, the presence of ICESag37 was not associated with the occurrence of severe sepsis and meningitis [6].
3. Discussion
In this study, we investigated the molecular characteristics of invasive GBS strains by comparing the CRISPR1 loci of 103 CC17/III GBS isolates from 103 different neonates over an 11 year period. We aimed to assess the diversity of spacer content of the same GBS serotype over time and evaluate whether the CRISPR1 locus can be an epidemiological marker, since it can reflect the evidence of phage infection in vivo and is potentially associated with biological functions [17,18,19,20]. Based on CRISPR-RFLP analysis and CRISPR1 locus diversity, we can divide the invasive CC17/III GBS strains into several categories because GBS isolates with the same CRISPR-RFLP length have nearly 100% similarity in spacer compositions. Based on different CRISPR subtypes, certain specific subtypes of CC17/III GBS isolates were significantly associated with the occurrence of bacterial meningitis, severe sepsis, and septic shock.
Previous studies have demonstrated that the approach of using CRISPR-RFLP analysis to compare the CRISPR compositions, followed by sequencing and CRISPR array analyses, can be useful in clinical applications of screening GBS isolates and following maternal or neonatal carriage [20,21,22,25]. In these studies, CRISPR analyses were performed in a large number of GBS isolates of different serotypes and sequence types from different subjects, and there was a strong agreement between traditional MLST and CRISPR genotyping [20,21,22,24]. In addition, distinctive features of CRISPR loci were identified between different clonal complexes that are representative of different GBS strains [22,25]. It is well-known that the CC17/III GBS strain has emerged as the major pathogen causing neonatal invasive diseases and meningitis because this hypervirulent strain has specific virulence factors and adhesion molecules that can assist in invasion [26,27,28]. However, the clinical manifestations of all CC17/III GBS strains may vary greatly at presentation, as some of them present with fulminant sepsis and meningitis. These large variations have not been investigated, and this is the first study to use the CRISPR array and spacer compositions to subgroup these invasive CC17/III GBS isolates. In contrast to a previous study that found less spacer variation within the CC17 isolates [22,24], we documented that the diversity within the CC17 isolates was much higher, which can be explained partly by the significantly larger number of CC17 isolates in this study.
Investigation into the likelihood of phage infection can be performed in vivo through alterations in the spacer content and addition at the leader site of CRISPR1 [17,18,19,25]. The spacers of CRISPR are obtained from foreign DNA and can protect bacteria from attack by homologous phages or plasmids in the environment. Previous studies compared interspecies CRISPR loci and documented that the number of spacers is inversely correlated with prophage acquisition [29]. Because GBS is a significant bacterial pathogen of neonates and immunocompromised adults, continual genome evolution may result in the emergence of new virulent strains [29,30,31]. In our cohort, identical CRISPR1 loci among CC17 strains from different subjects were observed, which contrasts with previous studies that suggested the spacer content of the CRISPR1 locus is generally specific for isolates from a given subject [21,22]. In addition, this longitudinal study was performed using GBS strains over a long period of 11 years. Therefore, our study demonstrated the predominance of certain clones to emerge as the major strain causing neonatal invasive diseases in our institute, which is consistent with a recent study that found that a few clones comprised the majority of the invasive GBS isolates [32]. Further studies that enroll more CC17 GBS strains from multiple centers in Taiwan are warranted to document this phenomenon.
It is known that spacer acquisition at the 5′ leader-end plays an important role for CRISPR array diversity, but in vitro spacer acquisition has never been found in CC17/III GBS isolates [22,24]. Most of our CC17/III GBS isolates (95.1%, n = 98) had spacer compositions of 49-53-54-55-56, and we found that 81.6% (n = 80) of them had spacer acquisition, whereas 11.7% (n = 12) had spacer loss. Our data support the recent findings of Pastuszka et al. that spacer acquisition, sometimes accompanied with the loss of old spacers, occurs and may be more than ~10% of the ST17 clones [33]. Currently, only very few studies are available regarding the CC17/III GBS lineage [34], and this phenomenon requires further confirmation after enrolling a larger number of strains.
To the best of our knowledge, this is the first study to find a significant relationship between the CRISPR-RFLP pattern and clinical manifestations of GBS invasive diseases. Interestingly, GBS isolates with RFLP lengths between 1000 and 1300 bp were associated with a lower likelihood of severe sepsis and meningitis. In contrast, GBS isolates with RFLP length ≥1300 bp and ≤1000 bp were significantly more likely to cause severe illness, and GBS isolates within the same subgroup based on the RFLP length were always genetically related to each other. In addition, GBS isolates with the same CRISPR-RFLP pattern were found to have similar CRISPR loci and spacer sequences, as documented in this study and others [21]. Various virulence factors, including HvgA, pilus, and Srr1/Srr2, have been associated with meningitis through enhanced adhesion to epithelial cells and penetration of the blood–brain barrier [27,35,36], but the differences in these gene sequences were not associated with higher illness or meningitis in our cohort. We hypothesize that the CRISPR/Cas system in CC17/III GBS isolates can affect at least some biological functions that may contribute to the occurrence of meningitis. In addition, the CRISPR-RFLP assay may potentially be used as a new biomarker to predict clinical meningitis after further verification.
In conclusion, the diversity of the CRISPR1 sequences among CC17/III GBS isolates was demonstrated in this study, which represents a powerful and applicable subgrouping method for better investigation of specific clone dissemination. The CRISPR diversity is much higher even within a given GBS clonal complex [23], and involves spacer gain, random loss, and occasional duplication. Within the CC17/III GBS strains, a specific sublineage, which is identifiable by the ubiquitous CRISPR1 locus and spacer organization, is potentially associated with a higher severity of illness and a higher likelihood of meningitis. Further studies that enroll more clinical CC17/III GBS isolates from multiple centers are needed to verify these findings, since it is applicable to use the CRISPR-RFLP assay to identify the most dangerous CC17/III GBS strain in clinical practice and prompt more aggressive therapeutic strategies in advance.
4. Materials and Methods
4.1. Bacterial Isolates
From January 2008 to December 2018, all GBS strains isolated from blood and/or cerebrospinal fluid of neonates with invasive GBS diseases were obtained from the bacterial library of Chang Gung Memorial Hospital (CGMH, Taoyuan, Taiwan). All identifications of initial GBS isolates and storage in the bacterial library were based on the standard procedures: MALDI-TOF MS (Bruker corporation, Karlsruhe, Germany) was used for documentation of all GBS isolates (https://www.ncbi.nlm.nih.gov/pmc/articles/PMC8328194/, accessed on 24 February 2021). All GBS strains were genotyped using MLST and CPS serotyping in our previous studies [37,38]. Among them, CC17/III GBS isolates were retrieved to analyze their CRISPR loci. The clinical features of all neonates with CC17/III GBS invasive diseases were collected and followed [37,38]. This study was approved by the ethics committee of CGMH, and a waiver of informed consent for anonymous data collection was approved.
4.2. DNA Extraction
Genomic DNA was extracted following enzymatic lysis with lysozyme (Sigma, North Liberty, IA, USA), and a QIAamp DNA Mini kit (QIAGEN, Dusseldorf, Germany) was used. We prepared a bacterial suspension of 1.5 McFarland standard in 250 µL water containing 20 U of lysozyme, 2 mM EDTA (Protech, Taipei, Taiwan), 1.2% Triton (Sigma, North Liberty, IA, USA), and 20 mM Tris buffer. The DNA-containing supernatants were collected after incubation of the suspension at 56 °C for 30 min and 96 °C for 15 min. Repeated centrifugation of the lysates at 6000× g for 1 min was performed after buffer AW1 and AW2 (QIAGEN, Dusseldorf, Germany) were added.
4.3. CRISPR1 Locus Amplification
CRISPR1 locus amplification was performed in a TProfessionalTRIO thermocycler (Biometra, Gottingen, Germany) using CRISPR1 SEQ-F (GAAGACTCTATGATTTACCGC) and CRISPR1 SEQ-R (CAGCAATCACTAAAAGAACCAAC) primers targeting the CRISPR1-flanking regions, similar to previous studies [21]. PCR amplification was performed in a total volume of 25 μL, which contained 0.5 μM of each primer, 0.2 mM deoxynucleoside triphosphates (dNTPs), 0.02 U/μL HotStarTaq polymerase kit (QIAGEN, Dusseldorf, Germany), 1× PCR buffer, and 1 μL extracted DNA (0.05 μg/μL). The PCR mixtures were heated to 94 °C for 5 min, followed by 40 cycles of a denaturation step at 94 °C for 30 s, an annealing step at 58 °C for 30 s, an elongation step at 72 °C for 3 min, and a final extension step at 72 °C for 10 min. PCR amplification was verified by electrophoretic migration in a 1% agarose gel.
4.4. Restriction Fragment Length Polymorphism (RFLP) Analysis
We used NEBcutter V2.0 (http://nc2.neb.com/NEBcutter2/, accessed on 23 February 2021) to select the restriction enzymes according to previously sequenced CRISPR1 arrays. DdeI and MnlI were selected to test most of the CRISPR1 arrays. The RFLP analyses were performed on PCR products of the above CRISPR1 locus amplification, which contained the repeats and spacers. Restriction digests were performed in a total volume of 10 μL, containing 0.1 U/μL DdeI, 1× NEBuffer, and 4 μL extracted DNA (0.05 μg/μL) in a TProfessionalTRIO thermocycler (Biometra, Gottingen, Germany). Restriction products were analyzed by electrophoretic migration in a 1.5% agarose gel. The CRISPR1 locus was then sequenced in all CC17/III GBS isolates with different sizes and/or different patterns in RFLP.
4.5. CRISPR1 Locus Sequencing and Analysis
The DNA sequences of all CRISPR1 loci were analyzed and assembled using Sequencing analysis software v.5.3.1 (Applied Biosystems, Waltham, MA, USA). All the PCR products were sequenced in both the forward and reverse directions to obtain double-stranded sequences. All sequences of the spacers, repeats, and flanking regions were assembled using SeqMan software (DNASTAR, Madison, WS, USA) and then submitted to the CRISPR-finder database (https://crispr.i2bc.paris-saclay.fr/Server/, accessed 23 February 2021) (the spacer Id in the database: 2493099). Then spacer sequences were compared to CRISPRtionary (https://crispr.i2bc.paris-saclay.fr/CRISPRcompar/Dict/Dict.php, accessed 23 February 2021), and graphic representations of spacers were produced as colored cells in Excel spreadsheets. The CRISPR spacers were aligned in the CRISPR-finder database and named manually. New spacers identified in the GBS strains were assigned a unique number preceded by the letter “S”.
Based on the sequences of CRISPR spacers, BLASTn (default parameters, nr database, and short queries) was used to identify phage genes, plasmids, or GBS chromosome harboring sequences with homology to the spacers. The phage–host interactions and the evolutionary changes between phages and bacteria could be speculated on after examining the arrangement of spacers. The phylogenetic relationship of the GBS strains based on CRISPR diversity was constructed using Bionumeriucs 7.0 software (DNASTAR, Madison, WS, USA).
4.6. MLST and Capsule Genotyping
The MLST and capsule genotyping of the CC17/III GBS isolates were performed previously based on the standard protocol in previous studies [39].
4.7. Antimicrobial Susceptibility Testing
All GBS isolates were rated for susceptibility to seven antibiotics, including erythromycin, penicillin, clindamycin, vancomycin, ampicillin, cefotaxime, and teicoplanin, according to the guidelines of the Clinical and Laboratory Standards Institute for the microdilution minimum inhibitory concentration (MIC) method [40]. The double-disk diffusion test was applied to identify inducible clindamycin resistance.
4.8. Whole-Genome Sequencing (WGS)
WGS was conducted to investigate the genomic differences between CC17/III GBS isolates with different RFLP patterns and CRISPR1 locus. WGS was performed using both PacBioTM SMRT (Pacific Biosciences, Menlo Park, CA, USA) [41] and MiSeqTM (Illumina, San Diego, CA, USA) [42] sequencing technologies. The sequencing library was prepared using a TruSeq DNA LT Sample Prep Kit (Illumina, San Diego, CA, USA) for the Illumina MiSeq system. Genomic libraries were generated using Nextera XT kits (Illumina, San Diego, CA, USA). All the sequencing processes were performed using a DNA-sequencing kit 4.02v2 (QIAGEN, Dusseldorf, Germany) and SMRT cell 8 Pac (QIAGEN, Dusseldorf, Germany). The circlator tool (v1.4.0) (Cambridge, UK) was used to correct and linearize the genome, and QUAST (v4.5) (San Diego, CA, USA) was applied to evaluate the assembled genome quality.
Acknowledgments
All authors thank Chiao-Ching Chiang for keeping the database of our NICU, and all nursing staff working in our NICUs for keeping extremely detailed patient records, which contributed greatly to the completion of this research.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/ijms222111626/s1.
Author Contributions
Conceptualization: J.-F.H., J.-J.L. and M.-H.T.; data collection and verification: J.-F.H., C.L., S.-M.C., H.-R.H., M.-C.C. and M.-Y.L.; formal analysis: J.-F.H., J.-J.L., C.L. and L.-C.L.; funding acquisition: J.-F.H. and J.-J.L.; investigation: J.-F.H., H.-R.H., C.L. and L.-C.L.; methodology: M.-Y.L., S.-M.C., H.-R.H. and M.-C.C.; supervision: J.-F.H. and M.-H.T.; writing—original draft: J.-F.H. and J.-J.L.; writing—review and editing: M.-H.T. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by a grant from Chang Gung Memorial Hospital (CMRPG3L0621, CMRPG3D1241, and CMRPG3E1491).
Institutional Review Board Statement
This study was approved by the Institutional Review Board of Chang Gung Memorial Hospital (certificate no. 201701494A3, approved on 16 November 2017).
Informed Consent Statement
Patient consent was waived due to all patient records and information were anonymized and deidentified prior to analysis.
Data Availability Statement
The datasets used/or analyzed during the current study are available from the corresponding author on reasonable request.
Conflicts of Interest
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Joubrel C., Tazi A., Six A., Dmytruk N., Touak G., Bidet P., Raymond J., Trieu-Cuot P., Fouet A., Kernéis S., et al. Group B streptococcus neonatal invasive infections, France 2007–2012. Clin. Microbiol. Infect. 2015;21:910–916. doi: 10.1016/j.cmi.2015.05.039. [DOI] [PubMed] [Google Scholar]
- 2.Lin C., Chu S.M., Wang H.C., Yang P.H., Huang H.R., Chiang M.C., Fu R.H., Tsai M.H., Hsu J.F. Complicated Streptococcus agalactiae sepsis with/without meningitis in young infants and newborns: The clinical and molecular characteristics and outcomes. Microorganisms. 2021;9:2094. doi: 10.3390/microorganisms9102094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kadambari S., Trotter C.L., Heath P.T., Goldacre M.J., Pollard A.J., Goldacre R. Group B Streptococcal disease in England (1998-2017): A population based observational study. Clin. Infect Dis. 2021;72:e791–e798. doi: 10.1093/cid/ciaa1485. [DOI] [PubMed] [Google Scholar]
- 4.Al Luhidan L., Madani A., Albanyan E.A., Saif S.A., Nasef M., AlJohani S., Madkhali A., al Shaalan M., Alalola S. Neonatal group B Streptococcus infection in a tertiary care hospital in Saudi Arabia: A 13-year experience. Pediatr. Infect. Dis. J. 2019;38:731–734. doi: 10.1097/INF.0000000000002269. [DOI] [PubMed] [Google Scholar]
- 5.Medugu N., Iregbu K., Parker R., Plemmons J., Singh P., Audu L., Efetie E., Davies H., Manning S. Group B streptococcal colonization and transmission dynamics in pregnant women and their newborns in Nigeria: Implications for prevention strategies. Clin. Microbiol. Infect. 2017;23:673.e9–673.e16. doi: 10.1016/j.cmi.2017.02.029. [DOI] [PubMed] [Google Scholar]
- 6.Hsu J.F., Tsai M.H., Lin L.C., Chu S.M., Lai M.Y., Huang H.R., Chiang M.C., Yang P.H., Lu J.J. Genomic characterization of serotype III/ST-17 Group B Streptococcus strains with antimicrobial resistance using whole genome sequencing. Biomedicines. 2021;9:1477. doi: 10.3390/biomedicines9101477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li S., Huang J., Chen Z., Guo D., Yao Z., Ye X. Antibiotic prevention for maternal group B Streptococcal colonization on neonatal GBS-related adverse outcomes: A meta-analysis. Front. Microbiol. 2017;8:374. doi: 10.3389/fmicb.2017.00374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mukhopadhyay S., Dukhovny D., Mao W., Eichenwald E.C., Puopolo K.M. 2010 perinatal GBS prevention guideline and resource utilization. Pediatrics. 2014;133:196–203. doi: 10.1542/peds.2013-1866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Creti R., Imperi M., Berardi A., Pataracchia M., Recchia S., Alfarone G., Baldassarri L. Neonatal group B streptococcus infections: Prevention strategies, clinical and microbiologic characteristics in 7 years of surveillance. Pediatr. Infect. Dis. J. 2017;36:256–262. doi: 10.1097/INF.0000000000001414. [DOI] [PubMed] [Google Scholar]
- 10.Alhhazmi A., Hurteau D., Tyrrell G.J. Epidemiology of invasive group B streptococcal disease in Alberta, Canada, from 2003 to 2013. J. Clin. Microbiol. 2016;54:1774–1781. doi: 10.1128/JCM.00355-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li S., Wen G., Cao X., Guo D., Yao Z., Wu C., Ye X. Molecular characteristics of Streptococcus agalactiae in a mother-baby prospective cohort study: Implication for vaccine development and insights into vertical transmission. Vaccine. 2018;36:1941–1948. doi: 10.1016/j.vaccine.2018.02.109. [DOI] [PubMed] [Google Scholar]
- 12.Guan X., Mu X., Ji W., Yuan C., He P., Zhang L., Huang Y., Li J., Chen J., Zhong H., et al. Epidemiology of invasive group B streptococcal disease in infants from urban area of South China, 2011–2014. BMC Infect. Dis. 2018;18:14. doi: 10.1186/s12879-017-2811-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Morozumi M., Wajima T., Takata M., Iwata S., Ubukata K. Molecular characteristics of Group B Streptococci isolated from adults with invasive infections in Japan. J. Clin. Microbiol. 2016;54:2695–2700. doi: 10.1128/JCM.01183-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li C., Sapugahawatte D., Yang Y., Wong K., Lo N., Ip M. Multidrug-Resistant Streptococcus agalactiae Strains Found in Human and Fish with High Penicillin and Cefotaxime Non-Susceptibilities. Microorganisms. 2020;8:1055. doi: 10.3390/microorganisms8071055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hazafa A., Mumtaz M., Farooq M.F., Bilal S., Chaudhry S.N., Firdous M., Naeem H., Ullah M.O., Yameen M., Mukhtiar M.S., et al. CRISPR/Cas9: A powerful genome editing technique for the treatment of cancer cells with present challenges and future directions. Life Sci. 2020;263:118525. doi: 10.1016/j.lfs.2020.118525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang F., Wen Y., Guo X. CRISPR/Cas9 for genome editing: Progress, implications and challenges. Hum. Mol. Genet. 2014;23:R40–R46. doi: 10.1093/hmg/ddu125. [DOI] [PubMed] [Google Scholar]
- 17.Ma Y., Zhang L., Huang X. Genome modification by CRISPR/Cas9. FEBS J. 2014;281:5186–5193. doi: 10.1111/febs.13110. [DOI] [PubMed] [Google Scholar]
- 18.Zhang C., Quan R., Wang J. Development and application of CRISPR/Cas9 technologies in genomic editing. Hum. Mol. Genet. 2018;27:R79–R88. doi: 10.1093/hmg/ddy120. [DOI] [PubMed] [Google Scholar]
- 19.Cho S., Shin J., Cho B.-K. Applications of CRISPR/Cas System to Bacterial Metabolic Engineering. Int. J. Mol. Sci. 2018;19:1089. doi: 10.3390/ijms19041089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Spencer B.L., Deng L., Patras K.A., Burcham Z.M., Sanches G.F., Nagao P.E., Doran K.S. Cas9 contributes to group B Streptococcal colonization and diseases. Front. Microbiol. 2019;10:1930. doi: 10.3389/fmicb.2019.01930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Beauruelle C., Pastuszka A., Mereghetti L., Lanotte P. Group B Streptococcus vaginal carriage in pregnant women as deciphered by clustered regularly interspaced short palindromic repeats analysis. J. Clin. Microbiol. 2018;56:e01949-17. doi: 10.1128/JCM.01949-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Beauruelle C., Pastuszka A., Horvath P., Perrotin F., Mereghetti L., Lanotte P. CRISPR: A Useful Genetic Feature to Follow Vaginal Carriage of Group B Streptococcus. Front. Microbiol. 2017;8:1981. doi: 10.3389/fmicb.2017.01981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lopez-Sanchez M.J., Sauvage E., Da Cunha V., Clermont D., Ratsima Hariniaina E., Gonzalez-Zorn B., Poyart C., Rosinski-Chupin I., Glaser P. The highly dynamic crispr1 system of streptococcus agalactiae controls the diversity of its mobilome. Mol. Microbiol. 2012;85:1057–1071. doi: 10.1111/j.1365-2958.2012.08172.x. [DOI] [PubMed] [Google Scholar]
- 24.Lier C., Baticle E., Horvath P., Haguenoer E., Valentin A.-S., Glaser P., Mereghetti L., Lanotte P. Analysis of the type II-A CRISPR-Cas system of Streptococcus agalactiae reveals distinctive features according to genetic lineages. Front. Genet. 2015;6 doi: 10.3389/fgene.2015.00214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McGinn J., Marraffini L.A. Molecular mechanisms of CRISPR–Cas spacer acquisition. Nat. Rev. Genet. 2018;17:7–12. doi: 10.1038/s41579-018-0071-7. [DOI] [PubMed] [Google Scholar]
- 26.Renard A., Barbera L., Courtier-Martinez L., Dos Santos S., Valentin A.-S., Mereghetti L., Quentin R., Van Der Mee-Marquet N.L. phiD12-Like Livestock-Associated Prophages Are Associated with Novel Subpopulations of Streptococcus agalactiae Infecting Neonates. Front. Cell. Infect. Microbiol. 2019;9:166. doi: 10.3389/fcimb.2019.00166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bianchi-Jassir F., Paul P., To K.-N., Carreras-Abad C., Seale A.C., Jauneikaite E., Madhi S.A., Russell N.J., Hall J., Madrid L., et al. Systematic review of Group B Streptococcal capsular types, sequence types and surface proteins as potential vaccine candidates. Vaccine. 2020;38:6682–6694. doi: 10.1016/j.vaccine.2020.08.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vuillemin X., Hays C., Plainvert C., Dmytruk N., Louis M., Touak G., Saint-Pierre B., Adoux L., Letourneur F., Frigo A., et al. Invasive group B Streptococcus infections in non-pregnant adults: A retrospective study, France, 2007–2019. Clin. Microbiol. Infect. 2020;27:129.e1–129.e4. doi: 10.1016/j.cmi.2020.09.037. [DOI] [PubMed] [Google Scholar]
- 29.Hargreaves K.R., Flores C.O., Lawley T.D., Clokie M.R.J. Abundant and Diverse Clustered Regularly Interspaced Short Palindromic Repeat Spacers in Clostridium difficile Strains and Prophages Target Multiple Phage Types within This Pathogen. mBio. 2014;5:e01045-13. doi: 10.1128/mBio.01045-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen S.L. Genomic Insights Into the Distribution and Evolution of Group B Streptococcus. Front. Microbiol. 2019;10:1447. doi: 10.3389/fmicb.2019.01447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Martins E., Pedroso-Roussado C., Cristino J.M., Ramirez M., The Portuguese Group for the Study of Streptococcal Infections. Oliveira H., Vaz T., Gião M., Ferreira R., Silva A.C., et al. Streptococcus agalactiae Causing Neonatal Infections in Portugal (2005–2015): Diversification and Emergence of a CC17/PI-2b Multidrug Resistant Sublineage. Front. Microbiol. 2017;8:499. doi: 10.3389/fmicb.2017.00499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gajic I., Plainvert C., Kekic D., Dmytruk N., Mijac V., Tazi A., Glaser P., Ranin L., Poyart C., Opavski N. Molecular epidemiology of invasive and non-invasive group B Streptococcus circulating in Serbia. Int. J. Med Microbiol. 2018;309:19–25. doi: 10.1016/j.ijmm.2018.10.005. [DOI] [PubMed] [Google Scholar]
- 33.Pastuszka A., Beauruelle C., Camiade E., Rousseau G.M., Moineau S., Mereghetti L., Horvath P., Lanotte P. Functional Study of the Type II-A CRISPR-Cas System of Streptococcus agalactiae Hypervirulent Strains. CRISPR J. 2021;4:233–242. doi: 10.1089/crispr.2020.0145. [DOI] [PubMed] [Google Scholar]
- 34.Mosterd C., Rousseau G.M., Moineau S. A short overview of the CRISPR-Cas adaptation stage. Can. J. Microbiol. 2021;67:1–12. doi: 10.1139/cjm-2020-0212. [DOI] [PubMed] [Google Scholar]
- 35.Gori A., Harrison O.B., Mlia E., Nishihara Y., Chan J.M., Msefula J., Mallewa M., Dube Q., Swarthout T.D., Nobbs A.H., et al. Pan-GWAS of Streptococcus agalactiae highlights lineage-specific genes associated with virulence and Niche adaptation. mBio. 2020;11:e00728-20. doi: 10.1128/mBio.00728-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Campisi E., Rosini R., Ji W., Guidotti S., Rojas-López M., Geng G., Deng Q., Zhong H., Wang W., Liu H., et al. Genomic analysis reveals multi-drug resistance clusters in group B Streptococcus CC17 hypervirulent isolates causing neonatal invasive disease in southern mainland China. Front. Microbiol. 2016;7:1265. doi: 10.3389/fmicb.2016.01265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tsai M.-H., Hsu J.-F., Lai M.-Y., Lin L.-C., Chu S.-M., Huang H.-R., Chiang M.-C., Fu R.-H., Lu J.-J. Molecular Characteristics and Antimicrobial Resistance of Group B Streptococcus Strains Causing Invasive Disease in Neonates and Adults. Front. Microbiol. 2019;10:264. doi: 10.3389/fmicb.2019.00264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kao Y., Tsai M.H., Lai M.Y., Chu S.M., Huang H.R., Chiang M.C., Fu R.H., Lu J.J., Hsu J.F. Emerging serotype III sequence type 17 group B streptococcus invasive infection in infants: The clinical characteristics and impacts on outcomes. BMC Infect. Dis. 2019;19:538. doi: 10.1186/s12879-019-4177-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Björnsdóttir E., Martins E., Erlendsdóttir H., Haraldsson G., Cristino J.M., Kristinsson K., Ramirez M. Changing epidemiology of group B streptococcal infections among adults in Iceland: 1975–2014. Clin. Microbiol. Infect. 2015;22:379–e9. doi: 10.1016/j.cmi.2015.11.020. [DOI] [PubMed] [Google Scholar]
- 40.Clinical and Laboratory Standards Institute . Performance Standards for Antimicrobial Susceptibility Testing, Twenty-Fourth Informational Supplement, M100-S24. SLCI; Wayne, MI, UAS: 2014. [Google Scholar]
- 41.Metcalf B.J., Chochua S., Gertz R.E., Jr., Hawkins P.A., Ricaldi J., Li Z., Walker H., Tran T., Rivers J., Mathis S., et al. Short-read whole genome sequencing for determination of antimicrobial resistance mechanisms and capsular serotypes of current invasive Streptococcus agalactiae recovered in the USA. Clin. Microbiol. Infect. 2017;23:574.e7–574.e14. doi: 10.1016/j.cmi.2017.02.021. [DOI] [PubMed] [Google Scholar]
- 42.Shelburne S.A., Sahasrabhojane P., Saldaña M., Yao H., Su X., Horstmann N., Thompson E., Flores A.R. Streptococcus mitisStrains Causing Severe Clinical Disease in Cancer Patients. Emerg. Infect. Dis. 2014;20:762–771. doi: 10.3201/eid2005.130953. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used/or analyzed during the current study are available from the corresponding author on reasonable request.