Abstract
Heteroduplex analysis has been used extensively to identify allelic variation among mammalian genes. It provides a rapid and reliable method for determining and cataloging minor differences between two closely related DNA sequences. We have adapted this technique to distinguish among strains or clonal types of Porphyromonas gingivalis. The ribosomal intergenic spacer region (ISR) was amplified directly from a subgingival plaque sample by PCR with species-specific primers, avoiding the need for culturing the bacteria. The PCR products were then directly compared by heteroduplex analysis with known strains of P. gingivalis for identification. We identified 22 distinct but closely related heteroduplex types of P. gingivalis in 1,183 clinical samples. Multiple strains were found in 34% of the samples in which P. gingivalis was detected. Heteroduplex types were identified from these multistrain samples without separating them by culturing or molecular cloning. PCR with species-specific primers and heteroduplex analysis makes it possible to reliably and sensitively detect and identify strains of P. gingivalis in large numbers of samples.
Periodontitis is a widespread chronic infectious disease that occurs in as much as 40% of the population (2). Although several bacteria have been implicated in the disease process, Porphyromonas gingivalis is the organism most consistently associated with disease (10, 12, 25, 29, 34). In order to understand the bacterial etiology of periodontal disease, assays to readily identify subjects who harbor the bacteria and to distinguish among strains within a bacterial species are essential. Rapid methods that are reproducible, are relatively easy, and allow the differentiation of a large number of strains are needed. These assays will also be important in order to track bacteria for transmission studies and for understanding population structures of P. gingivalis. A number of approaches for identifying strains of P. gingivalis have been reported, including serotyping (5, 23), ribotyping (5, 13, 23, 31), arbitrarily primed PCR (AP-PCR) (20, 31), and whole genomic digest (7, 31). These methods vary in the number of strains or clonal types that they are capable of resolving, from a few (serotyping) to a large number (whole genomic digest). However, all of these methods require isolating the bacteria away from other oral microorganisms and culturing in vitro. This is a costly and labor-intensive process that may introduce a bias for predominant or easily cultivated strains.
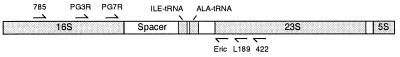
We have previously reported an assay for identification of the periodontal pathogens Actinobacillus actinomycetemcomitans and P. gingivalis based on PCR amplification of the ribosomal DNA (rDNA) intergenic spacer region (ISR) (Fig. 1) (16). Species-specific primers allow the amplification of the ISR directly from a plaque sample without culturing. We now describe a method that uses heteroduplex analysis (8, 26) of the PCR products from the ISR to identify the strains or clonal types of P. gingivalis present in a subgingival plaque sample. With heteroduplex analysis, similar but nonidentical DNA fragments can be distinguished by polyacrylamide gel electrophoresis. DNA fragments are mixed, heated to melt the strands, and then cooled. Both the original homoduplexes and heteroduplexes form. The small areas of mismatch in the heteroduplexes cause them to migrate more slowly so that they are seen as additional bands by polyacrylamide gel electrophoresis. Characteristic heteroduplex patterns can be used to identify genetic variants. Using heteroduplex analysis we have identified 22 unique but closely related P. gingivalis types, many from samples containing multiple strains. This entire assay is rapid and accurate and can be done without culturing the bacteria. It can also be used to estimate the relative abundances of multiple clonal types present in a sample. Data can be stored for future comparisons with other samples.
FIG. 1.
The P. gingivalis ribosomal operon. The ISR used for PCR is shown as the open bar containing two tRNA genes.
MATERIALS AND METHODS
Bacterial strains.
P. gingivalis ATCC 33277, ATCC 49417, and ATCC 53978 were obtained from the American Type Culture Collection (Manassas, Va.). W50, 381, and A7A1 were provided by Joseph Zambon (Buffalo, N.Y.). HG 1691 was provided by Robert Schifferle (Buffalo, N.Y.). 23A4 was provided by Denis Mayrand (Quebec, Quebec, Canada). W83 was provided by Margaret Duncan (Boston, Mass.).
Plaque samples.
Human volunteers for this institutionally approved study were recruited from the dental clinics of the Ohio State University College of Dentistry, the University of Texas Health Science Center at San Antonio, and the University of California, San Francisco, as well as from the Student Health Center of the Ohio State University.
Pooled subgingival dental plaque samples were collected from each subject. Excess saliva was removed with a cotton roll or gauze pad to minimize collection of transient contaminating bacteria. A sterile, medium endodontic paper point (Caulk-Dentsply) was placed in the mesial sulcus of each tooth for 10 s. All teeth present were sampled, and points were pooled in a sterile 2-ml microcentrifuge tube and frozen.
PCR.
DNA was isolated directly from the frozen sample as previously described in a final volume of 20 μl (16). Samples were analyzed for the presence of P. gingivalis by using a nested, two-step PCR procedure with a P. gingivalis-specific primer for the second amplification (16). This assay can detect 10 cells or fewer (16). Figure 1 is a representation of the bacterial rDNA operon with the relative locations and orientations of the primers used. The primers have been described previously (10, 16, 19). PCR was performed with 2.5 U of Taq polymerase in a total volume of 100 μl in a buffer containing 50 mM KCl, 10 mM Tris-HCl (pH 8.8), 3 mM MgCl2, 0.1% Triton X-100, and 0.2 mM each deoxynucleoside triphosphate. The first PCR was carried out with 4 μl of DNA and primers 785 and 422 for 27 cycles of 92°C for 1 min, 42°C for 2 min, and 72°C for 3 min. The second PCR amplification was carried out with 2 μl of the product from the first amplification and primers PG3R and L189 for 27 cycles of 92°C for 1 min, 52°C for 2 min, and 72°C for 3 min.
Positive samples were prepared for heteroduplex analysis by reamplifying the original PCR products with the primers PG7R and eric. PCR conditions were the same as those for the second PCR except that 21 cycles were carried out.
Heteroduplex analysis.
Heteroduplexes were formed by mixing amplified DNA from two samples in a final volume of 8 to 12 μl in a 0.5-ml microcentrifuge tube. A small amount of mineral oil was overlaid. The mixture was incubated at 95°C for 5 min to melt double strands and then cooled to 25°C at the rate of 1°C per min in a thermal cycler to reanneal. The tubes were then placed on ice, and gel loading buffer was added.
Heteroduplexes were detected by polyacrylamide gel electrophoresis (10% acrylamide, 39:1 N,N′-methylenebisacrylamide) in 1× Tris-borate-EDTA. All gels (9 by 6.5 cm) were run in a Bio-Rad apparatus at constant voltage for 3.5 h at 120 V. Gels were stained with ethidium bromide and visualized with UV light. Analysis was performed as previously described (16) on a Macintosh computer with the NIH Image program (22).
To identify the heteroduplex types present in a sample, amplified DNA fragments from samples were duplexed to a panel of DNA fragments generated from laboratory strains, and characteristic migration patterns were noted. To determine if multiple strains were present within a single sample, the sample was prepared as described above without being mixed with other DNA. Heteroduplex types found in samples containing multiple strains of P. gingivalis could be determined by the same method that was used for identifying samples that contained only one strain.
RESULTS
PCR and heteroduplex analysis allowed us to rapidly identify strains of P. gingivalis from a large number of plaque samples. DNA was isolated directly from the sample and used as a template for PCRs with species-specific primers. The target of the PCR was the internal transcribed ISR between the 16S and 23S genes in the rDNA operon (Fig. 1). The PCR products were then directly compared by heteroduplex analysis. With this approach, it was not necessary to culture the bacteria in order to rapidly determine the presence or absence of P. gingivalis in oral samples and to identify the strain or strains of P. gingivalis present.
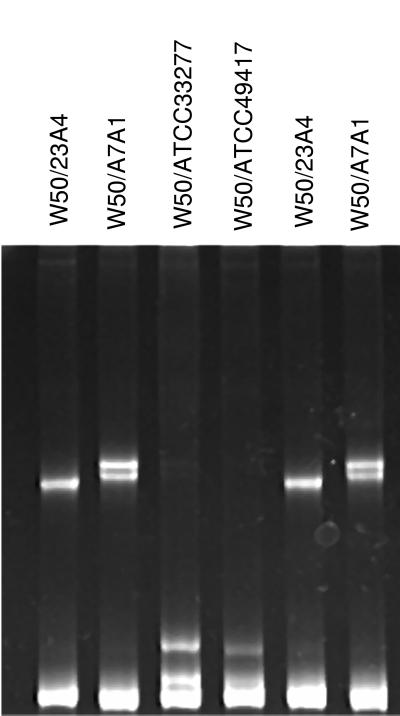
Figure 2 shows P. gingivalis W50 (ATCC 53978) compared with four other laboratory strains of P. gingivalis by heteroduplex analysis. PCR products from strain W50 were combined with the PCR products from each of the other four strains, heated to denature the strands, cooled to re-form double-stranded DNA, and analyzed by polyacrylamide gel electrophoresis. When strains W50 and 23A4 were duplexed, a single heteroduplex band was observed in addition to the homoduplex band at the bottom of the gel (first lane). Formation of heteroduplexes with A7A1, ATCC 33277, or ATCC 49417 (second to fourth lanes) resulted in two heteroduplex bands. The two bands represent reciprocal heteroduplexes that are resolved by polyacrylamide gel electrophoresis. The migration patterns of the heteroduplex bands are characteristic of the two duplexed strains and provide strain identification. The last two lanes contain DNA from the same strains shown in the first two lanes, demonstrating that the migration patterns are reproducible.
FIG. 2.
P. gingivalis W50 (ATCC 53978) compared with four other laboratory strains of P. gingivalis by heteroduplex analysis. The first four lanes contain W50 DNA duplexed with strains 23A4, A7A1, ATCC 33277, and ATCC 49417, respectively. The last two lanes are duplicates of the first two lanes.
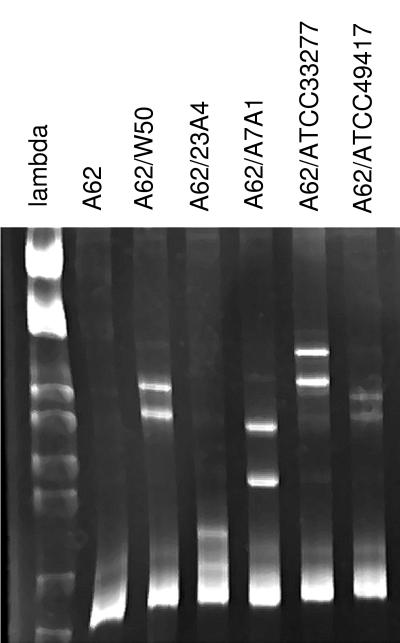
In order to identify strains from clinical samples, PCR products were generated with P. gingivalis-specific primers as outlined above. The resultant DNA fragments were then duplexed with PCR products from a panel of known laboratory strains, and migration patterns were compared to those obtained from known strains. Figure 3 is an example of the panel of comparisons generated to identify the strain present in a plaque sample. The PCR product was duplexed in separate reactions with DNA from five known strains of P. gingivalis and analyzed by polyacrylamide gel electrophoresis. The second lane contains DNA from the unknown strain by itself. It contains only a single homoduplex band. The characteristic heteroduplex pattern generated by combination with known strains is shown in subsequent lanes. The sample was identified as heteroduplex group hA62 based on the migration patterns formed with these five strains as well as other known strains. Type hA62 represents a heteroduplex type that we have detected in clinical samples but have not matched to a characterized laboratory strain. We have identified this heteroduplex type in approximately 8% of samples from several studies.
FIG. 3.
P. gingivalis rDNA spacer region PCR product amplified from a plaque sample duplexed with DNA from five different known strains of P. gingivalis and analyzed by polyacrylamide gel electrophoresis. The first lane contains DNA size markers: phage lambda digested with the restriction enzymes EcoRI and HindIII. The second lane contains ISR DNA from the clinical sample, A62, by itself. The remaining five lanes contain the A62 PCR product duplexed with DNA from P. gingivalis ATCC 53978 (W50), 23A4, A7A1, ATCC 33277, and ATCC 49417, respectively.
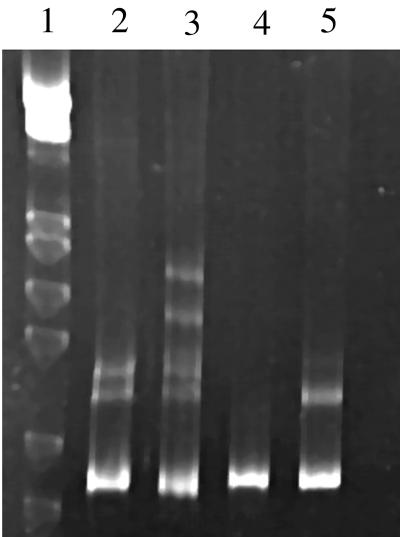
Many of the plaque samples examined in our studies were found to contain multiple strains of P. gingivalis (Table 1). To determine the number of strains present in each sample, PCR products generated from plaque samples with P. gingivalis-specific primers were heated and cooled to form heteroduplexes in the absence of added reference strain DNA. After polyacrylamide gel electrophoresis, the number of bands observed in addition to the homoduplex band indicated the number of strains present in the sample. Because the formation of each heteroduplex can result in either one or two new bands, the presence of one or two bands in addition to the homoduplex band indicates that there are two strains present in the sample. The presence of three to six bands indicates that there are three strains present in the sample. The gel in Fig. 4 shows examples of samples containing one, two, and three heteroduplex types of P. gingivalis.
TABLE 1.
Detection of multiple strains of P. gingivalis in clinical samples
| Study population | No. positive/ no. of subjects (%) | No. (%) of positive samples with ≥2 strains | No. (%) of positive samples with ≥3 strains |
|---|---|---|---|
| Subjects recruited from clinics of colleges of dentistry | |||
| The Ohio State University | 92/148 (62) | 28 (30) | 6 (7) |
| University of California, San Francisco | 67/74 (91) | 45 (67) | 2 (3) |
| University of Texas Health Science Center at San Antonio | 55/104 (53) | 15 (27) | 0 (0) |
| Ohio State University students newly immigrated to the United States | 211/292 (72) | 92 (44) | 20 (9) |
| Randomly selected subjects from extended families in Columbus, Ohio | 236/565 (42) | 48 (20) | 4 (2) |
| Total | 661/1,183 (56) | 228 (34) | 32 (5) |
FIG. 4.
Clinical samples examined for the presence of multiple strains of P. gingivalis by heteroduplex analysis. DNA amplified from each sample was examined by itself. The first lane contains DNA size markers: phage lambda digested with the restriction enzymes EcoRI and HindIII. The sample in lane 4 formed only the homoduplex band, indicating the presence of a single strain. The samples in lanes 2 and 5 each show a heteroduplex doublet, indicating the presence of two strains of P. gingivalis. The sample in lane 3, with two heteroduplex doublets, contains three strains of P. gingivalis.
We have examined five diverse study populations for the presence of multiple heteroduplex types of P. gingivalis (Table 1), for a total sample size of 661 colonized individuals. The number of samples in each cohort that contained multiple strains varied between 20 and 67% of colonized individuals, with 34% of all the positive samples containing multiple strains. This demonstrates that the presence of multiple strains of P. gingivalis is very common. In the majority of samples with multiple strains, we detected only two strains; only 5% of the positive samples showed three strains.
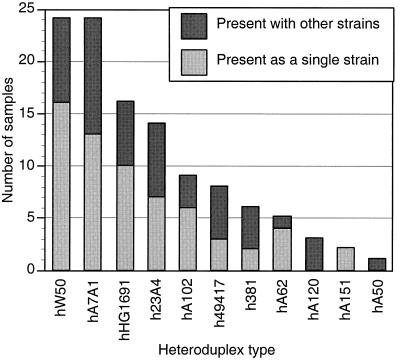
We identified the individual heteroduplex types present in the cohort of 148 subjects recruited from the Ohio State University College of Dentistry clinics listed in Table 1. Most of these subjects were adults with a history of mild to moderate periodontitis. Ninety-two (62%) of the samples were positive for P. gingivalis, and 28 (30%) of those had multiple strains. Figure 5 shows the frequency distribution of heteroduplex types found in this cohort. Because many subjects had multiple strains, the number of strains shown in the figure is greater than the number of samples analyzed.
FIG. 5.
Frequency distribution of P. gingivalis heteroduplex types found in subgingival plaque samples collected from subjects randomly selected from the dental clinics of The Ohio State University. Ninety-two heteroduplex type identifications were made in 64 samples.
From all samples analyzed a total of 22 distinct heteroduplex types of P. gingivalis were identified. Many of the commonly occurring types matched previously characterized laboratory strains that were available to us and were named accordingly. However, several novel patterns were observed. Sequence analysis confirmed that these were within the phylogenetic cluster of P. gingivalis (27). A listing of heteroduplex types, characteristic heteroduplex patterns, and ISR sequence data (27) is available online (15).
DISCUSSION
The development of a rapid, inexpensive, and reliable assay to differentiate among strains of P. gingivalis is important for epidemiologic studies necessary for our understanding of the transmission and pathogenicity of P. gingivalis. Previous studies have relied on techniques that are dependent on culturing, lowering the sensitivity and limiting the number of samples that could be analyzed. We have developed an assay with species-specific primers for PCR to identify subjects who are positive for P. gingivalis, followed by heteroduplex analysis to identify the strain or strains of P. gingivalis present in the sample. This system makes it possible to carry out large-scale studies with sensitive detection thresholds and offers the advantage of the ability to distinguish a large number of strains.
Sequence analysis of the 16S gene in the rDNA operon is a powerful method for differentiating among bacterial species and determining the phylogenetic relationships among these species (4, 6). However, the 16S gene does not provide the sequence diversity necessary to differentiate among strains within a bacterial species. The ISR between the 16S and 23S genes in the rDNA operon is a noncoding region, and lack of selective pressure has resulted in considerable sequence diversity. The ISR has been demonstrated to provide the diversity necessary to distinguish among strains of several species (11, 14, 24, 33). Because it is flanked by the relatively conserved and well-characterized 16S and 23S genes, PCR primers for amplification of the ISR DNA for heteroduplex or other sequence analysis can be identified. In addition, species-specific primers to the 16S gene can be used to amplify the P. gingivalis ISR from a sample containing a mixture of bacteria and other cells.
Heteroduplex analysis provides a rapid method for identifying differences between two closely related sequences. It has provided an effective tool for identifying polymorphic differences in human genes where the target DNA fragment is well characterized, as in cystic fibrosis (8, 26). We have adapted this technology for the identification of strains of P. gingivalis directly from DNA isolated from subgingival plaque samples. Heteroduplex analysis provided a reliable method for strain identification using PCR products and made it possible to identify the multiple strains often found in clinical samples.
Unknown samples that were positive for P. gingivalis were first examined by heteroduplex analysis of the amplified DNA by itself to determine the number of strains. Next, the sample was mixed with DNA from a series of known strains, and the resulting patterns seen by polyacrylamide gel electrophoresis were compared to patterns generated from mixtures of known strains. By carefully choosing comparison strains that yielded the most distinctive patterns, we were able to minimize the number of trials needed to determine the identity of unknown clonal types. For heteroduplex analysis of plaque samples, the best results were obtained with PCR products that were amplified for a minimum number of cycles. Excessive amplification caused the bands to spread out and appear fuzzy after polyacrylamide gel electrophoresis. DNA isolated from pure cultures gave clearer bands than PCR products from complex oral samples. The clearest bands were obtained from ISR fragments cloned into a plasmid and transformed into Escherichia coli. This cloned DNA was routinely used for duplexing of clinical samples. Other technical considerations were important as well. It was critical to have PCR products with little or no background, because light background bands were difficult to distinguish from heteroduplex bands. In addition, if the entire DNA sample was not completely hybridized before electrophoresis, two single-stranded bands appeared at the top of the gel. We routinely used 10% polyacrylamide, although other gel matrices designed especially for heteroduplex analysis are available. The alternative matrices can sometimes give slightly different migration patterns that may aid in identification. On the other hand, we found them more difficult to use.
No cross-species formation of heteroduplexes with the P. gingivalis ISR has been observed. For example, the ISRs from the closely related species Porphyromonas salivosa and Bacteroides macacae do not form heteroduplexes with the ISR from P. gingivalis (data not shown). This is due to the lack of homology between these and other closely related species except in the tRNAs. Even if cross-species formation of heteroduplexes were possible, it would not be a problem with PCR amplification of the ISR using species-specific primers. If products from other species were amplified with the P. gingivalis-specific primers, they would be identified as different by the size variation in the ISR seen by agarose gel electrophoresis. All heteroduplex types found in this study have been confirmed as P. gingivalis by sequencing.
A number of approaches for identifying strains of P. gingivalis have been reported, including serotyping (5, 23), ribotyping (5, 13, 23, 31), whole genomic digest (7, 31), AP-PCR (20, 31), and sequence analysis of the ISR (27). The investigators using these approaches have reported a wide range in the number of clonal types detected, from a very few (based on serotyping) to an almost infinite number of types (based on whole genomic digest). It is likely that other methods could resolve closely related clonal types that are not resolved by heteroduplex analysis. However, resolving P. gingivalis isolates into 22 groups offers sufficient discrimination for clinical studies and avoids the confusion arising from too many categories. Heteroduplex analysis offered good resolution of the laboratory strains tested. Strains W50 and W83 were indistinguishable from one another by heteroduplex analysis. Strains W50 and W83 are also unresolved by sequence analysis of the ISR (27), AP-PCR (3, 21), fimbrial restriction fragment length polymorphism analysis (17), genomic DNA fingerprinting (18), and serotyping (5, 32). It appears that they may be the same strain or very closely related. Strains 381 and ATCC 33277 were not resolved by heteroduplex analysis of the ISR, Southern blotting (1), serotyping (5), genomic DNA fingerprinting (18), or AP-PCR (3) but have been resolved based on infectivity and metabolic requirements (9) and on ISR sequencing (27). All other laboratory strains tested were resolved by heteroduplex analysis.
The number of strains present in a sample was ascertained by the number of bands seen by polyacrylamide gel electrophoresis. Annealing of the products generated by melting two similar but nonidentical DNA fragments results in the reformation of the two original homoduplexes as well as two types of reciprocal heteroduplexes. Because many, but not all, reciprocal heteroduplexes are resolved from each other by polyacrylamide gel electrophoresis, the presence of two strains in a sample can result in one or two distinct heteroduplex bands in addition to the homoduplex band. Based on the number of possible heteroduplex configurations, three strains will result in the presence of three to six bands. Determining the number of strains in samples where more than three strains are present is more difficult, because there is overlap between the numbers of bands possible when four or five strains are present. In practice, however, we did not encounter samples with a large number of visible heteroduplex bands, suggesting that it is very uncommon to have several strains present at high levels. In our study populations, most of the samples contained no more than two strains, and it was very rare that we were able to detect more than three strains in a single sample. No other large epidemiologic studies have been performed, but in small studies with eight or nine subjects, similar results have been reported (28, 30, 35).
The migration patterns characteristic of the heteroduplexes were very reproducible, and data could be stored for subsequent comparison with new samples as they were analyzed. Within a single laboratory, heteroduplex analysis offers an efficient means for identifying and cataloging strains. Indexing of heteroduplex groups to reference strain DNA would allow easy comparison of data between laboratories. For positive identification of clonal types between laboratories, particularly novel clonal types, and for phylogenetic studies, sequence analysis of the ISR provides additional resolving power (27). However, samples that contain multiple strains of P. gingivalis are more easily examined by using heteroduplex analysis.
Based on data from laboratory strains, heteroduplex types that made up at least 10% of the P. gingivalis population in the samples could be easily detected. The heteroduplex method offers a distinct advantage for detection of minor strains over previously reported methods that relied on culturing. Based on the binomial probability distribution, in order to have a 95% certainty of encountering a strain that occurs as 10% of the total, 29 colonies would have to be tested. Many more colonies would have to be screened to estimate the relative proportions of strains. With heteroduplex analysis the relative amount of each strain can be determined by comparing the relative intensities of heteroduplex bands with NIH Image. However, heteroduplex bands seen by polyacrylamide gel electrophoresis rarely have the expected relative intensities compared to those of homoduplex bands. Therefore, in order to obtain an accurate determination of the relative amount of each clonal type present in a sample, it is necessary to compare results to standard curves.
PCR amplification with species-specific primers and heteroduplex analysis provides a rapid and powerful method for identifying clonal types or strains of P. gingivalis, even in the presence of multiple strains. Twenty-two heteroduplex types were identified in 661 colonized subjects, and more than one type of P. gingivalis was detected in 34% of these subjects.
ACKNOWLEDGMENTS
We thank the individuals listed in Materials and Methods for providing strains.
This work was supported by NIH grant DE10467.
REFERENCES
- 1.Barkocy-Gallagher G A, Han N, Patti J M, Whitlock J, Progulske-Fox A, Lantz M S. Analysis of the prtP gene encoding porphypain, a cysteine proteinase of Porphyromonas gingivalis. J Bacteriol. 1996;178:2734–2741. doi: 10.1128/jb.178.10.2734-2741.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brown L J, Oliver R C, Loe H. Periodontal diseases in the U.S. in 1981: prevalence, severity, extent, and role in tooth mortality. J Periodontol. 1989;60:363–370. doi: 10.1902/jop.1989.60.7.363. [DOI] [PubMed] [Google Scholar]
- 3.Chen C, Slots J. Clonal analysis of Porphyromonas gingivalis by the arbitrarily primed polymerase chain reaction. Oral Microbiol Immunol. 1994;9:99–103. doi: 10.1111/j.1399-302x.1994.tb00042.x. [DOI] [PubMed] [Google Scholar]
- 4.Field K G, Olsen G J, Lane D J, Giovannoni S J, Ghiselin M T, Raff E C, Pace N R, Raff R A. Molecular phylogeny of the animal kingdom. Science. 1988;239:748–753. doi: 10.1126/science.3277277. [DOI] [PubMed] [Google Scholar]
- 5.Fisher J G, Zambon J J, Genco R J. Identification of serogroup-specific antigens among Bacteroides gingivalis. J Dent Res. 1987;66:222. [Google Scholar]
- 6.Fox G E, Stackebrandt E, Hespell R B, Gibson J, Maniloff J, Dyer T A, Wolfe R S, Balch W E, Tanner R S, Magrum L J, Zablen L B, Blakemore R, Gupta R, Bonen L, Lewis B J, Stahl D A, Luehrsen K R, Chen K N, Woese C R. The phylogeny of prokaryotes. Science. 1980;209:457–463. doi: 10.1126/science.6771870. [DOI] [PubMed] [Google Scholar]
- 7.Genco R J, Loos B G. The use of genomic DNA fingerprinting in studies of the epidemiology of bacteria in periodontitis. J Clin Periodontol. 1991;18:396–405. doi: 10.1111/j.1600-051x.1991.tb02307.x. [DOI] [PubMed] [Google Scholar]
- 8.Glavac D, Dean M. Applications of heteroduplex analysis for mutation detection in disease genes. Hum Mutat. 1995;6:281–287. doi: 10.1002/humu.1380060402. [DOI] [PubMed] [Google Scholar]
- 9.Grenier D, Mayrand D. Selected characteristics of pathogenic and nonpathogenic strains of Bacteroides gingivalis. J Clin Microbiol. 1987;25:738–740. doi: 10.1128/jcm.25.4.738-740.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Griffen A L, Becker M R, Lyons S R, Moeschberger M L, Leys E J. Prevalence of Porphyromonas gingivalis and periodontal health status. J Clin Microbiol. 1998;36:3239–3242. doi: 10.1128/jcm.36.11.3239-3242.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Griffen A L, Leys E J, Fuerst P A. Strain identification of Actinobacillus actinomycetemcomitans using the polymerase chain reaction. Oral Microbiol Immunol. 1992;7:240–243. doi: 10.1111/j.1399-302x.1992.tb00032.x. [DOI] [PubMed] [Google Scholar]
- 12.Haffajee A D, Cugini M A, Tanner A, Pollack R P, Smith C, Kent R L, Jr, Socransky S S. Subgingival microbiota in healthy, well-maintained elder and periodontitis subjects. J Clin Periodontol. 1998;25:346–353. doi: 10.1111/j.1600-051x.1998.tb02454.x. [DOI] [PubMed] [Google Scholar]
- 13.Hillman J D, Maiden M F, Pfaller S P, Martin L, Duncan M J, Socransky S S. Characterization of hemolytic bacteria in subgingival plaque. J Periodontal Res. 1993;28:173–179. doi: 10.1111/j.1600-0765.1993.tb01066.x. [DOI] [PubMed] [Google Scholar]
- 14.Kambhampati S, Rai K S. Temporal variation in the ribosomal DNA nontranscribed spacer of Aedes albopictus (Diptera: Culicidae) Genome. 1991;34:293–297. doi: 10.1139/g91-047. [DOI] [PubMed] [Google Scholar]
- 15.Leys, E. J., and A. L. Griffen. 9 June 1999, revision date. Data. [Online.] http://www.dent.ohio-state.edu/griffen_leys. [21 June 1999, last date accessed.]
- 16.Leys E J, Griffen A L, Strong S J, Fuerst P A. Detection and strain identification of Actinobacillus actinomycetemcomitans by nested PCR. J Clin Microbiol. 1994;32:1288–1294. doi: 10.1128/jcm.32.5.1288-1294.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Loos B G, Dyer D W. Restriction fragment length polymorphism analysis of the fimbrillin locus, fimA, of Porphyromonas gingivalis. J Dent Res. 1992;71:1173–1181. doi: 10.1177/00220345920710050901. [DOI] [PubMed] [Google Scholar]
- 18.Loos B G, Mayrand D, Genco R J, Dickinson D P. Genetic heterogeneity of Porphyromonas (Bacteroides) gingivalis by genomic DNA fingerprinting. J Dent Res. 1990;69:1488–1493. doi: 10.1177/00220345900690080801. . (Erratum, 69:1623.) [DOI] [PubMed] [Google Scholar]
- 19.McClellan D L, Griffen A L, Leys E J. Age and prevalence of Actinobacillus actinomycetemcomitans in children. J Dent Res. 1995;74:587. [Google Scholar]
- 20.Menard C, Brousseau R, Mouton C. Application of polymerase chain reaction with arbitrary primer (AP-PCR) to strain identification of Porphyromonas (Bacteroides) gingivalis. FEMS Microbiol Lett. 1992;74:163–168. doi: 10.1111/j.1574-6968.1992.tb05360.x. [DOI] [PubMed] [Google Scholar]
- 21.Menard C, Mouton C. Clonal diversity of the taxon Porphyromonas gingivalis assessed by random amplified polymorphic DNA fingerprinting. Infect Immun. 1995;63:2522–2531. doi: 10.1128/iai.63.7.2522-2531.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.NIH Image, version 1.62. 13 April 1999, posting date. [Online.] National Institutes of Health. http://rsb.info.nih.gov/nih-image/. [21 June 1999, last date accessed.]
- 23.Parent R, Mouton C, Lamonde L, Bouchard D. Human and animal serotypes of Bacteroides gingivalis defined by crossed immunoelectrophoresis. Infect Immun. 1986;51:909–918. doi: 10.1128/iai.51.3.909-918.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Peng W, Anderson T J, Zhou X, Kennedy M W. Genetic variation in sympatric Ascaris populations from humans and pigs in China. Parasitology. 1998;117:355–361. doi: 10.1017/s0031182098003102. [DOI] [PubMed] [Google Scholar]
- 25.Preus H R, Anerud A, Boysen H, Dunford R G, Zambon J J, Loe H. The natural history of periodontal disease. The correlation of selected microbiological parameters with disease severity in Sri Lankan tea workers. J Clin Periodontol. 1995;22:674–678. doi: 10.1111/j.1600-051x.1995.tb00825.x. [DOI] [PubMed] [Google Scholar]
- 26.Ravnik-Glavac M, Glavac D, Dean M. Sensitivity of single-strand conformation polymorphism and heteroduplex method for mutation detection in the cystic fibrosis gene. Hum Mol Genet. 1994;3:801–807. doi: 10.1093/hmg/3.5.801. [DOI] [PubMed] [Google Scholar]
- 27.Rumpf R W, Griffen A L, Wen B G, Leys E J. Sequencing of the ribosomal intergenic spacer region for strain identification of Porphyromonas gingivalis. J Clin Microbiol. 1999;37:2723–2725. doi: 10.1128/jcm.37.8.2723-2725.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Saarela M, Stucki A M, von Troil-Linden B, Alaluusua S, Jousimies-Somer H, Asikainen S. Intra- and inter-individual comparison of Porphyromonas gingivalis genotypes. FEMS Immunol Med Microbiol. 1993;6:99–102. doi: 10.1111/j.1574-695X.1993.tb00310.x. [DOI] [PubMed] [Google Scholar]
- 29.Tanner A C, Haffer C, Bratthall G T, Visconti R A, Socransky S S. A study of the bacteria associated with advancing periodontitis in man. J Clin Periodontol. 1979;6:278–307. doi: 10.1111/j.1600-051x.1979.tb01931.x. [DOI] [PubMed] [Google Scholar]
- 30.Teanpaisan R, Douglas C W, Eley A R, Walsh T F. Clonality of Porphyromonas gingivalis, Prevotella intermedia and Prevotella nigrescens isolated from periodontally diseased and healthy sites. J Periodontal Res. 1996;31:423–432. doi: 10.1111/j.1600-0765.1996.tb00511.x. [DOI] [PubMed] [Google Scholar]
- 31.Van Steenbergen T J, Menard C, Tijhof C J, Mouton C, De Graaff J. Comparison of three molecular typing methods in studies of transmission of Porphyromonas gingivalis. J Med Microbiol. 1993;39:416–421. doi: 10.1099/00222615-39-6-416. [DOI] [PubMed] [Google Scholar]
- 32.van Winkelhoff A J, Appelmelk B J, Kippuw N, de Graaff J. K-antigens in Porphyromonas gingivalis are associated with virulence. Oral Microbiol Immunol. 1993;8:259–265. doi: 10.1111/j.1399-302x.1993.tb00571.x. [DOI] [PubMed] [Google Scholar]
- 33.Wakefield A E. Genetic heterogeneity in human-derived Pneumocystis carinii. FEMS Immunol Med Microbiol. 1998;22:59–65. doi: 10.1111/j.1574-695X.1998.tb01187.x. [DOI] [PubMed] [Google Scholar]
- 34.Wolff L F, Aeppli D M, Pihlstrom B, Anderson L, Stoltenberg J, Osborn J, Hardie N, Shelburne C, Fischer G. Natural distribution of 5 bacteria associated with periodontal disease. J Clin Periodontol. 1993;20:699–706. doi: 10.1111/j.1600-051x.1993.tb00694.x. [DOI] [PubMed] [Google Scholar]
- 35.Zhang Y J, Yasui S, Yoshimura F, Ishikawa I. Multiple restriction fragment length polymorphism genotypes of Porphyromonas gingivalis in single periodontal pockets. Oral Microbiol Immunol. 1995;10:125–128. doi: 10.1111/j.1399-302x.1995.tb00132.x. [DOI] [PubMed] [Google Scholar]