Abstract
Cathepsin B (CTSB), a lysosomal cysteine protease’s high expression and activity, has been reported to cause poor-quality embryos in porcine and bovine. Nevertheless, CTSB functions in mice granulosa cells remain to explore. To discuss the CTSB functional role in follicular dynamics, we studied apoptosis, proliferation, cell cycle progression, and related signaling pathways in primary mouse granulosa cells transfected with small interference RNA specific to CTSB (siCTSB) for 48 h. Further, mRNA and protein expression of cell proliferation regulators (Myc and cyclin D2), apoptosis regulators (caspase 3, caspase 8, TNF-α, and Bcl2), steroidogenesis-related genes (FSHR and CYP11A1), and autophagy markers (LC3-I and ATG5) were investigated. In addition, the effect of CTSB on steroidogenesis and autophagy was also examined. Flow cytometry analysis assay displayed that silencing of CTSB decreased the early and total apoptosis rate by downregulating TNF-α, caspase 8, and caspase 3, and upregulating Bcl2. By regulating Myc and cyclin D2 expression and activating the p-Akt and p-ERK pathways, CTSB knockdown increased GC proliferation and number. A significant decline in estradiol and progesterone concentrations was observed parallel to a significant decrease in autophagy-related markers LC3-I and ATG5 compared to the control group. Herein, we demonstrated that CTSB serves as a proapoptotic agent and plays a critical role in folliculogenesis in female mice by mediating apoptosis, autophagy, proliferation, and steroidogenesis. Hence, CTSB could be a potential prognostic agent for female infertility.
Keywords: CTSB, mice, proliferation, apoptosis, granulosa cells
1. Introduction
Female reproduction pathways are the driving forces in evolution, including folliculogenesis, ovulation, fertilization, embryo development, parturition, and lactation [1]. The central functional units of the mammalian ovary, including granulosa cells (GCs) and theca cells (TCs), experience functional, morphological, and physiological changes during folliculogenesis; a spontaneous, most complex, and intricate reproductive phenomenon [2].
During the development of ovarian follicles, all stages of follicular atresia are associated with the apoptosis or death of granulosa cells (GCs). Based on this observation, GC apoptosis or death is considered the primary mechanism underlying follicular atresia [3,4]. In mammalian ovarian follicular development, only limited follicles reach ovulation, while the rest suffer from atresia at various stages of development. [5]. In parallel to apoptosis, nonapoptotic forms of programmed cell death, such as autophagy and necrotic-like cell death, have also been observed in the antral follicles of geese and quails [6]. Autophagy is an internal bulk degradation system in which autophagosomes, a part of the cytoplasm enclosed in double membrane-bound structures, mature and unite with lysosomes for degradation [7,8]. Autophagy promotes cell death by excessive self-digestion and degradation of essential cellular constituents [6], and various stimuli that induce apoptosis could trigger autophagy [9,10]. In humans, the exposure of an oxidized low-density lipoprotein has caused granulosa cells’ death by autophagy via inducing endothelial cell apoptosis [11]. Taken together, autophagy may be involved in folliculogenesis, as granulosa cells are the primary site of apoptosis during follicle atresia [12]. Previously, a study has demonstrated induction of autophagy in granulosa cells during folliculogenesis and a strong correlation with apoptosis in rat granulosa cells [6].
Despite overwhelming evidence for ovarian follicular atresia, the cellular and molecular mechanisms underlying this condition remain unknown. Therefore, it is imperative to comprehend dynamically regulated ovarian follicular growth, proliferation, and apoptosis. In addition, identifying novel regulatory molecules with an underlying GC function mechanism is critical to understand folliculogenesis thoroughly. A thorough understanding of folliculogenesis could aid in the development of novel molecular diagnostic and therapeutic approaches to combat the ever-increasing infertility problem in mammals.
Intracellular proteins are degraded in lysosomes by a lysosomal cysteine protease called CTSB [13], which controls various biological processes such as cell death, proliferation, migration, and cancer [14]. Before incorporating into the acidic lysosome environment, a CTSB (enzyme precursor) inactive form is converted into an active form through post-translational modifications [15,16]. CTSB active form has heavy and light-chain subunits linked by disulfide with a molecular weight of 30 kDa [17], which critically regulate various physiological and pathological processes, including initiating apoptosis and extracellular matrix remodeling. CTSB, either directly or indirectly, plays a critical role in activating the apoptotic pathway via initiator caspases rather than executioner caspases [18]. Indirect regulation of caspases by CTSB is mediated through mitochondrial membrane degradation, which translocates apoptosis-inducing components from the mitochondria to the cytoplasm [19].
Different cells have been reported to expressing the CTSB, such as cumulus–oocyte complex (COCs) and embryos in bovine and porcine [20,21,22]. CTSB activity and high protein levels were determined in poor-quality bovine and porcine embryos. In particular, inhibiting CTSB activity reversed these effects and improved preimplantation embryos in bovine and porcine models [20,23]. In addition, poor quality and heat-shocked bovine oocytes have higher CTSB activity than controls. Inhibiting CTSB activity increases the rate of development and improves embryo quality after in vitro fertilization (IVF) [23,24]. Taken together, the regulation of CTSB can serve as a promising tool to produce high-quality embryos in-vitro. These tidbits of evidence support the role of CTSB signaling by regulating folliculogenesis and embryogenesis.
Despite CTSB, expression is high in GC, and its functional role in folliculogenesis has not been elucidated. Herein, for the first time, we investigated the biological role of CTSB by employing small interference specific to CTSB in mice primary GC in vitro. Oocyte development and steroidogenesis were hypothesized to be controlled by CTSB in the GC. These processes were also examined in terms of the mechanisms by which the CTSB regulates them.
In this present study, we evaluated the silencing effects of CTSB on mouse primary GCs apoptosis, proliferation, cell cycle progression, and steroidogenesis with the underlying mechanism. Our results indicated that CTSB-KD suppressed apoptosis, autophagy, and steroidogenesis; increased proliferation; and enhanced the cell cycle progression through the AKT/ERK pathway by modulating FSHR and CYP11A1.
2. Results
2.1. siRNA Successfully Represses CTSB Expression
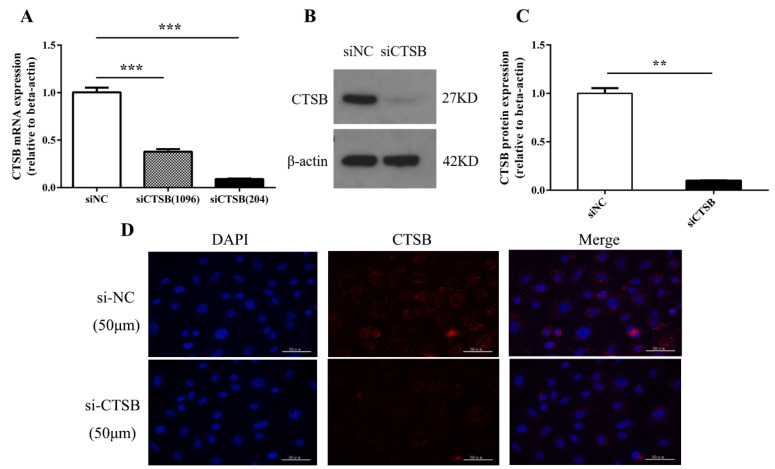
To uncover the biological function of CTSB in murine GCs, we designed and synthesized two different siRNAs: siCTSB (1096) and siCTSB (204). In brief, RT-qPCR, Western Blot, and immunofluorescence staining were used to determine siCTSB transcription and translation. The results shown in Figure 1 demonstrated that expression of CTSB is successfully inhibited in transfected murine GCs compared to control. The knockdown efficiency of CTSB mRNA level by siCTSB (204) transfection reached 91.16% (Figure 1A), compared to control siRNA-transfected cells. We selected siCTSB (204) for subsequent experiments because it was the most effective siRNA. Compared with the control group (1.00 ± 0.03), the CTSB protein relative expression of the siCTSB group (0.10 ± 0.01) was significantly lower (p < 0.001; Figure 1B,C).
Figure 1.
CTSB-KD inhibits CTSB expression in mouse GCs in vitro, both at the protein and mRNA levels. (A) CTSB mRNA expression was successfully decreased in mouse GCs transfected with siCTSB (100 nM). (B–D) GCs transfected with siCTSB (100 nM) exhibited a significant decrease in expression of CTSB at both the protein and mRNA levels in mouse GCs. Protein was determined by Western blotting and immunofluorescence, and mRNA levels by RT-qPCR relative to endogenous control β-actin. The results are indicated as means ± SEM of three independent experiments. ** p < 0.01, *** p < 0.001. siNC, negative control siRNA; CTSB, cathepsin B; GC, granulosa cells; siCTSB; and cathepsin B siRNA.
2.2. CTSB Depletion Suppresses Apoptosis in Murine GCs In Vitro
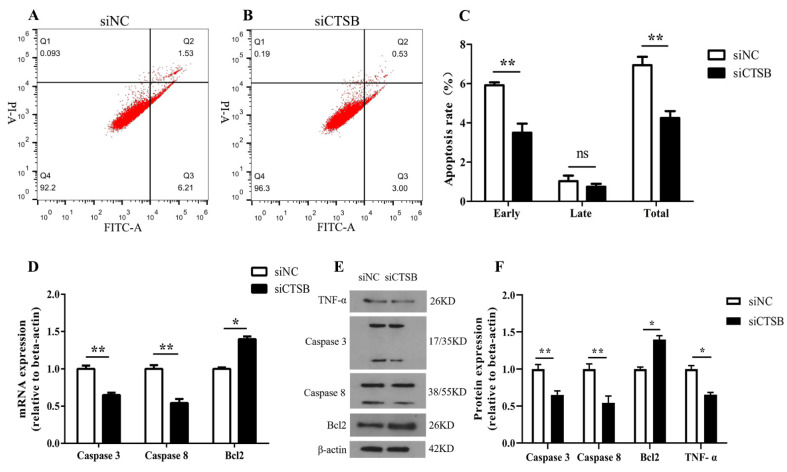
The apoptosis rate was determined by transfecting murine GCs for 48 h with siCTSB or NC. Resulted showed that siCTSB-treated GCs had a significantly lower early apoptosis rate (3.50 ± 0.46) and total apoptosis rate (4.24 ± 0.36) compared to the control (early apoptosis rate, 5.91 ± 0.15; total apoptosis rate, 6.94 ± 0.42), respectively (p < 0.01; (Figure 2A–C).
Figure 2.
In vitro, CTSB KD inhibits apoptosis and suppresses the expression of apoptosis regulators in mouse GC. (A) (siNC) and (B) (siCTSB). Flow cytometry analysis was performed to detect dead cells in control and transfected cells (5 × 105 cells per 6-well plate) using the annexin V-APC stain. (C) The rates of early, late, and total apoptosis were assessed. (D) In GCs transfected with siCTSB, the expression levels of apoptosis marker genes caspase 3, caspase 8, and Bcl-2 were measured using RT-qPCR. (E,F). The expression levels of apoptosis marker genes (caspase 3, caspase 8, Bcl-2, and TNF) in siCTSB and NC-transfected GCs were determined using Western blotting against β-actin as an endogenous control. All results, at least from three independent experiments, are presented as means ± SEMs. * p < 0.05, ** p < 0.01. Early apoptosis = lower right (LR); late apoptosis = upper right (UR); viable cells = lower left (LL); necrotic cells = upper left (UL); FITC, v-fluorescein isothiocyanate; PI, propidium iodine; siNC, negative control.
Next, beneath molecular mechanism of murine GCs apoptosis mediated by CTSB, the expression level of apoptotic regulators (downstream) was examined, both at the mRNA and protein level. Caspase 8 and caspase 3 were inhibited in siCTSB-treated GC compared to control (p < 0.01), while Bcl2 expression was promoted (p < 0.05). A significant reduction in TNF protein was also observed in the siCTSB group compared to the control groups (p < 0.05; Figure 2D–F). These results suggested that CTSB is involved in intrinsic apoptotic pathways in mouse GCs.
2.3. CTSB Downregulation Promotes Cell Proliferation and Affects Cell Cycle Progression in Mice GCs
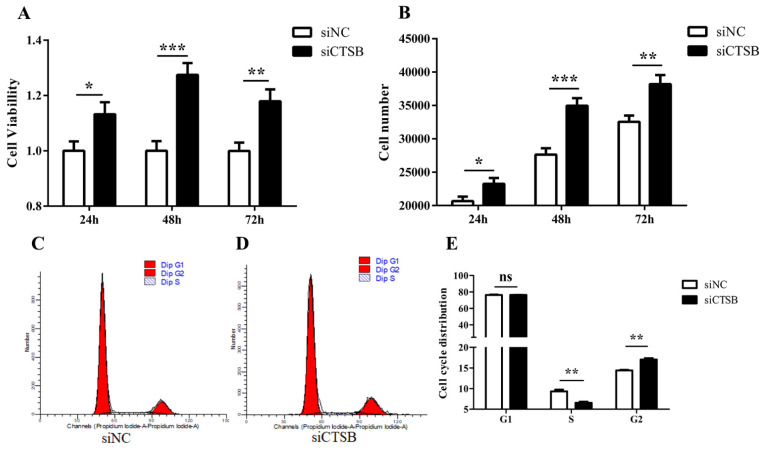
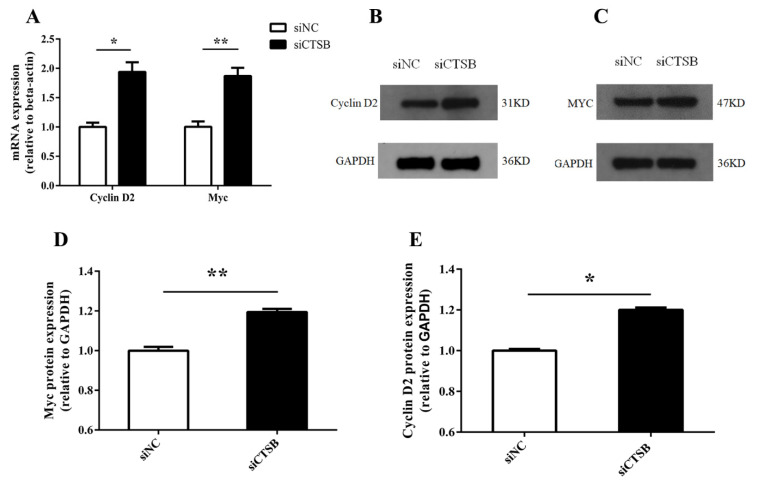
After small interference-mediated downregulation of CTSB in murine GCS in vitro, GCs were evaluated for cell proliferation, cell number, and progression in the cell cycle. The CCk8 and cell-counting results of different time points (24, 48, and 72 h) showed that increase in GCs cell proliferation and cell number (0.11 ± 0.03 and 8200 ± 3166, respectively) was highly significantly (p < 0.01) at 48 h of transfection in si-CTSB-treated cells compared to that of the control (Figure 3A,B). Hence, we used the 48-h time point in the subsequent experiments. Flow cytometry analysis for cell cycle indicated the cell cycle was arrested in si-CTSB-treated cells compared to the control, as there was a significant decrease in S (2.78%) and increase in the G2 (2.63%) phase GCs (p < 0.01) (Figure 3C–E). We also examined the expression of downstream proliferation markers (Myc and cyclin D2) in mice to determine how CTSB controls GC proliferation. RT-qPCR and Western blot results shown in Figure 4 described a highly significant increase in Myc compared to its target gene, cyclin D2, at mRNA and protein expression levels (p < 0.01, and p < 0.05, respectively).
Figure 3.
In vitro, CTSB depletion increased proliferation and modulated cell cycle progression in mouse GC. (A,B) The CCK-8 kit was used to assess the proliferation rate of GCs transfected with siCTSB and NC at 24, 48, and 72 h. The viability of the cells was determined by the concentration of formazan dye, a cell viability indicator. A multimode plate reader with absorbance at 450 nm was used to read the plates, and cell numbers were counted using an automated cell counter. (C–E) Mouse GC was transfected with siCTSB and incubated for 48 h before being saturated in PI, and the FACS and GC cycle were determined. All results are presented as means ± SEM. Significance difference, * p < 0.05, ** p < 0.01, *** p < 0.001; siCTSB, cathepsin B siRNA; GC, granulosa cells; siNC, negative control siRNA; ns, non-significant.
Figure 4.
In vitro effect of CTSB depletion on expression levels (mRNA and protein) of proliferation and cell cycle marker genes in mouse GC. (A) Expression levels of Myc and cyclin D2 genes in GC transfected with siRNA and NC were determined by RT-qPCR against endogenous control β-actin. (B–E) siCTSB promoted the protein expression level of Myc and cyclin D2 in mouse GCs against an endogenous control GAPDH. The data were presented as means ± SEM of three independent experiments. Significant differences, * p < 0.05, ** p < 0.01; siCTSB, Cathepsin B siRNA; GC, granulosa cells; siNC, negative control siRNA.
2.4. Downregulation of CTSB Mediates Mouse GC Proliferation by Activation of the ERK and Akt Phosphorylation Pathways
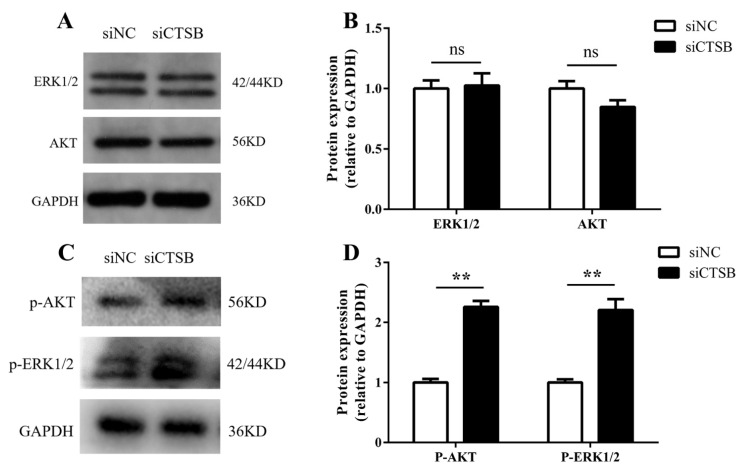
Following the CTSB footprints in GC proliferation, the pathways involved in CTSB’s functions in GC proliferation were investigated further. Western blotting was used to determine total and the phosphorylation levels of ERK1/2 and Akt serine–threonine kinase (Akt), downstream signaling pathways leading to cell proliferation. Total ERK1/2 expression between NC and siCTSB groups confirmed that expression of ERK1/2 was stable between two groups. In contrast to cells treated with siNC, CTSB-depleted cells had higher phosphorylation levels of Akt and ERK1/2 (p < 0.01; Figure 5). These results show that CTSB could be involved in regulating GC’s survival through ERK1/2 expression.
Figure 5.
In vitro, CTSB downregulation activates ERK1/2 phosphorylation in mouse GCs. GCs transfected with siCTSB and siNC were harvested 48-h post-transfection for protein extracts. The level of total AKT/ERK 1/2 (A,B) and phosphorylated AKT/ERK 1/2 (C,D) proteins was quantified by Western blotting against endogenous control GAPDH. The data from three independent experiments were presented as means ± SEM, ** p < 0.01. siCTSB, cathepsin B siRNA; GCs, granulosa cells. siNC, negative control siRNA.
2.5. Steroidogenesis and Autophagy-Related Gene Expression Were Altered by CTSB Depletion in Mouse GCs In Vitro
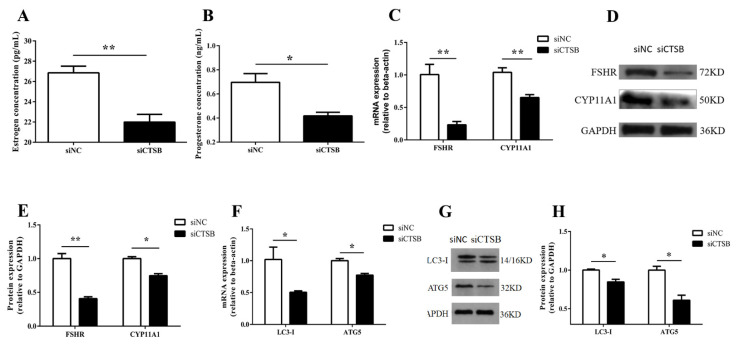
Whether knocking down of CTSB expression influenced the secretion of steroid hormones from GCs. To reveal the facts, at 48 h of transfection with siCTSB or NC, basal estradiol and progesterone production were detected in GCs culture media for steroidogenesis. As shown in Figure 6A,B, si-CTSB-treated GCs had a highly significant lower concentration of estradiol (26.86 ± 0.66 pg/mL), whereas the decrease in the progesterone (0.70 ± 0.07 ng/mL) concentration was significant lower compared to control (estradiol: 22.00 ± 0.755 pg/mL; progesterone: 0.42 ± 0.03 ng/mL) (p < 0.01, and p < 0.05), respectively. Mechanistically, a follicle-stimulating hormone receptor (FSHR), the cytochrome p450 family 11 subfamily A member 1 (CYP11A1), was downregulated both at mRNA, and protein expression levels in CTSB-depleted GCs (p < 0.01, and p < 0.05, respectively, Figure 6C–E). These results indicate that CTSB signaling regulates steroidogenesis via FSHR and CYP11A1 and plays an important in oocyte development and intra-ovarian function. The higher progesterone estrogen ratio in culture media of transfected granulosa cells indicated that CTSB knockdown could promote GC luteinization. Further, we examined the mRNA expression of autophagy-related markers (LC-3-I and ATG5) to infer CTSB knockdown effects on autophagy. As shown in Figure 6F, the mRNA and protein expression level of LC3-I (0.50 ± 0.02) and ATG5 (0.77 ± 0.026) were significantly decreased in the treatment group compared to the control (1.01 ± 0.19 vs. 1.00 ± 0.03, respectively). These results suggested that CTSB knockdown regulates steroidogenesis and autophagy through FSHR, CYP11A1, and LC3-I, ATG5, respectively.
Figure 6.
In vitro, CTSB deficiency altered the expression (mRNA and protein) of folliculogenesis and steroidogenesis marker genes in mouse GC. (A,B) At 48 h of GC transfection with siCTSB and NC, culture media was harvested, and estradiol and progesterone concentration was measured with an ELISA kit. (C–E) FSHR and CYP11A1, as well as (F–H) LC3-I and ATG5, were quantified at the mRNA and protein level through real-time PCR and Western blotting following siCTSB transfection. Results from three independent experiments were shown as means ± SEM. * p < 0.05, ** p < 0.01. siNC, negative control siRNA; siCTSB, cathepsin B siRNA; GCs, granulosa cells.
3. Discussion
Despite CTSB potentially being involved in both normal and pathological functions, there are well-established roles in carcinogenesis (hepatocellular carcinomas [25], colon cancer [26], esophageal adenocarcinoma [27], pancreatic adenocarcinoma [28], cellular functions apoptosis [29], oxidative stress [30], and autophagy [31]). In different cells, CTSB was detected, including the bovine and porcine cumulus–oocyte complex (COCs) and embryos [20,21,22]. Despite mammalian ovary exhibited significant interplays and expression of CTSB, the knowledge about its biological functions and signaling related to female reproduction is still in its infancy. Interestingly, CTSB expression was found negatively correlated to bovine embryos quality. However, inhibition of CTSB activity decreased the developmental competency of preimplantation embryos both in bovine and porcine [23]. Consequently, CTSB was supposed to play a key role in GC functions, such as apoptosis, proliferation, cell cycle progression, and intra-ovarian functions. siRNA is widely used to knock down the target gene in any cell type with ease in delivery and convenience to use [32]. Using siRNA, the transcriptional and post-transcriptional abundance of CTSB was disrupted in mouse GCs to investigate its physiological function. In this experiment, siCTSB effectively suppressed CTSB expression at both the mRNA and protein levels (Figure 1).
Proliferation and apoptosis of GCs are essential physiological processes for cells [33,34]. A follicle’s fate is ultimately determined by the crosstalk between death and survival signals from GCs [2,35]. The endo/lysosomal compartment has been reported to cause cell death, particularly by cathepsin (Cts), which can regulate apoptosis [36]. Nevertheless, cellular context and Cts type determine Cts’ positive or negative influence on cell death [36,37,38,39]. It has been reported that Cts, B, H, L, and S can cleave classic caspase substrates, such as procaspase-1, -3, and -8 [40,41], and release proapoptotic mitochondrial enzymes cytochrome c [42,43], thus activating caspases and apoptosis. CTSB was reported to cause apoptosis through initiator caspases rather than executor caspases either directly or indirectly [18]. Previously, Guicciardi et al. found that CTSB might be involved in TNF-alpha-triggered apoptosis by releasing mitochondrial cytochrome c [43]. These studies concluded that cathepsin B has a vital role in TNF- α-induced apoptosis; however, exact mechanisms remain to be explored.
In this regard, the potency of CTSB in regulating mice GCs is not investigated. Findings of the current study described that CTSB downregulation suppressed apoptosis in mice GCs in vitro. In CTSB-depleted GCs, mRNA expression and protein level of apoptosis-related marker genes; caspase 8, an initiator caspase of extrinsic apoptotic pathway; and caspase 3, a key apoptosis executor among its family expression levels were reduced (Figure 2D–F). Furthermore, CTSB KD also decreased the expression of TNF-α in CTSB-depleted GCs (Figure 2E,F). CTSB mediated a decline in GCs apoptosis and marker genes, which is consistent with previous studies that suggest that CTSB signaling in mice GCs can activate apoptotic pathway mediated by apoptosis initiator caspase 8 and concomitantly its interaction with apoptosis executioner caspase 3, thereby promoting apoptosis through extrinsic or caspase-8-mediated apoptosis. Previously, inhibition of CTSB was reported to reduce the apoptotic nuclei in the cumulus cell layer of IVM oocytes, probably due to the release of CTSB from lysosome into cumulus cells or oocytes in response to some stress, and thereby promoting apoptosis. Moreover, Xin Shan et al. reported that upregulation of CTSB expression had an association with upregulated apoptosis signaling pathway in atretic follicle granulosa cells (AFGCs) compared with healthy follicle granulosa cells (HFGCs) in porcine [44]. In this study, CTSB-depleted GCs from mice were shown to have reduced apoptosis for the first time, indicating that CTSB is a proapoptotic factor in immature cells.
CTSB-depleted granulosa cells were examined for proliferation in vitro. In this experimental study, different time points 24, 48, and 72, 48 h post-transfection, were slightly modified after being established in our lab to explore the GC proliferation [45], suggesting that the cells’ inherent biochemical and physiological properties depict cell proliferation. CTSB-depleted granulosa cells at 48 h had a significantly greater proliferation capacity and cell numbers than the NC group, suggesting that CTSB has antiproliferative behavior in mouse GC (Figure 3A,B). CTSB knockdown in mice GC increased G2/M cell numbers and decreased S-phase cell numbers, according to cell cycle assays (Figure 3C–E).
However, pieces of evidence have shown the diverse results of this study in some cancer cells. Li et al. found that knocking out CTSB inhibits cell proliferation in the human cholangiocarcinoma cell line QBC939 [46], and mice lacking CTSB had reduced cell proliferation in mammary carcinomas and their lung metastases. In cancerous tissues and cells, CTSB silencing inhibited proliferation. Despite not being precisely similar, these findings support our results due to survival effects of CTSB depletion.
Presently, the molecular mechanism to influence mouse GC proliferation mediated by CTSB signaling is not understood. Thereby, to further examine how CTSB regulates GC proliferation, our study showed that mRNA and protein expression levels of the proliferation-related genes, Myc and cyclin D2, were increased in response to CTSB depletion (Figure 4), reinforcing our proliferation assay results.
In gastric cancer cells, Myc upregulation was reported to increase the cells in the G2/M stage and decrease S-phase cells [47], agreeing with our findings. Opposite to the cyclin D1 conflicting data, the cyclin D2 (CCND2) gene is a recognized target gene of Myc. Myc interacts with CCND2 promoter in humans through a single highly conserved E-box element in vivo [48,49]. It induces histone acetylation in a TRRAP-dependent manner and induces CCND2 mRNA and protein expression [49]. Cell proliferation was retrained with inhibition of CCND2 [50], and overexpression of CCND2 induces cell proliferation [51]. Herein, depletion of CTSB increased Myc and CCDN2 expressions; thereby, Myc promoted S/G2-phase cell cycle progression by upregulating the transcription of the CCND2 gene.
C-myc is known as a positive regulator of cyclin D2 transcription [48,49] and can be phosphorylated at Thr58 and Ser 62 by ERK1/2 [52,53]. ERK and AKT are essential in cell proliferation, death, and differentiation in almost all cell types [54]. AKT, also known as protein kinase B (PKB), mediates the phosphatidylinositol 3-kinase (PI3K) signaling pathway. The PI3K/AKT pathway regulates cell growth, proliferation, survival, motility, and invasion [55]. In addition to suppressing the functions of proapoptotic proteins, ERK1/2 can promote cell survival by enhancing the activity of anti-apoptotic molecules. Mcl-1, an anti-apoptotic member of the Bcl-2 family, is phosphorylated at Thr163 by ERK1/2, thus increasing its stability and enhancing its anti-apoptotic activity [56]. Hence, we examined the AKT/ERK1/2 phosphorylation level in CTSB silencing GCs. The findings of the current study, which show the increase in the phosphorylation level of AKT/ERK1/2 (Figure 5C) and the increased expression of BCl2 (Figure 2D,E) along with c-myc and cyclin D2 (Figure 4A–D), infer that CTSB expression negatively regulates the proliferation of GC by AKT/ERK1/2 pathway.
Nevertheless, it remains to know if CTSB regulates steroidogenesis in GCs. In this study, we have demonstrated that CTSB downregulation induces a more significant decrease in E2 than P4 in GC culture media in vitro (Figure 6A,B). This finding suggests that CTSB could regulate the differential concentration of P4 and estradiol. For the first time, this study demonstrated that CTSB could regulate the secretion of E2 and P4 in GCs, and FSHR and CYP11A1 may mediate its effects. As shown in Figure 6C–E, transcription and translation levels of FSHR and CYP11A1 were significantly decreased in mice GCs treated with siCTSB. FSHR overexpression in granulosa cells was linked to the upregulation of proapoptotic genes and increased cell death than cells expressing relatively low FSHR levels [57]. Hence, low FSHR expression levels predominantly exhibited proliferative signals due to preferential activation of ERK signaling through B-arrestin. Altogether, these findings are consistent with the CTSB-KD-mediated low expression of FSHR and proliferative signals through ERK1/2 in the current study. However, further research is warranted to explore this detailed mechanism for CTSB.
The P450scc enzyme encoded by CYP11A1 catalyzes the first and rate-limiting step in steroidogenesis by converting cholesterol to pregnenolone, a precursor to all other steroid hormones [58]. In other words, P4 biosynthesis is primarily dependent on CYP11A1; therefore, an aberrant expression of CYP11A1 could fluctuate progesterone hormones levels [59]. FSHR expresses in ovarian granulosa cells to stimulate follicular maturation throughout the menstrual cycle and promote estradiol synthesis by aromatization [60]. The current study’s findings suggested that downregulation of CTSB could decrease the concentration of estradiol and P4 by reducing the expression of FSHR and CYP11A1, respectively.
The induction of autophagy in human granulosa cells was first described by Duerrschmidt et al. [11]. Some other studies in the PMSG rats model of follicular atresia have reported that autophagy in follicular atresia strongly correlates with apoptosis, as LC3 and cleaved caspase-3 expression was positively correlated with apoptosis [6]. Further, lysosomes are also involved in the degradation of regulators of steroidogenesis in the ovary, such as LH–LHR and FSH–FSHR complexes [61]. Taken together, considering the possibility of autophagy’s role in steroidogenesis in GCS, we examined the autophagy-related markers in CTSB silencing granulosa cells. The results revealed a significant decrease in mRNA expression of autophagy marker genes LC3-1 and ATG5. A similar trend of decrease in caspase 3 and LC3-1 in CTSB-depleted GCs is consistent with previous studies, suggesting that CTSB might be regulating GCs through autophagy and apoptosis. CTSB-KD mediating a decrease in steroidogenesis, CYP11A1, and ATG5 in GCs is also consistent with a previous study in mice which described a link between the deletion of the core autophagy gene ATG5 as well as the reduced steroid levels and steroid deficiency phenotype mediated by CYP11A1 [61,62]. However, further research in GCs from different stages follicles is warranted to explore the possible mechanism of CTSB in this context.
4. Materials and Methods
4.1. Management of Experimental Animals
This study was approved by the research approved by the Research Animal Ethics Committee of Huazhong Agricultural University (HZAUMO-2021–0016), and each experiment in this study was carried out in accordance with animal welfare standards. The experimental mice were purchased from Hubei Provincial Center for Disease Control. The mice were housed at Huazhong Agricultural University’s experimental animal center, where they had unlimited access to water and food and were subjected to a 12-h light/dark cycle.
4.2. Mouse GCs Isolation and Culture
The ovaries were collected from female KM (21 days old). Following an F12 (DMEM/F12) wash of pooled ovaries using needle puncture methods, GCs were isolated and purified three times using centrifugation (1000 rpm; 5 min) and DMEM/F12 wash. Pellets were saved after the supernatants were discarded. GCs were cultured in 6-well plates in culture media (DMEM/F12) with 10% FBS and 1% penicillin/streptomycin (Hyclone) and incubated for 48 h (37 °C; 5% CO2). During the incubation, culture media were replaced once at 24 h.
4.3. Cathepsin B (CTSB) siRNA Transfection
Both siRNA and NC-siRNA (without any target sequence) were purchased from Shanghai GenePharma Co., Ltd. (Shanghai, China). The cells were transfected with 100 nM of siRNA compared to NC using Lipofectamine RNAiMAX reagent in Opti-MEM medium (Life Technology, Inc., Carlsbad, CA, USA) instructions. Cells were harvested 48 h after transfection for mRNA and protein expression. Small RNA interference sequences specific to CTSB (siCTSB) used in this study are given in Table 1.
Table 1.
Primer sequences for quantitative real-time PCR and si-CTSB sequences.
| Gene Name | AC/NO | Primer Sequences | Production Length (bp) | Annealing Temp. (°C) |
|---|---|---|---|---|
| β-actin | NM_007393 | F: GTGACGTTGACATCCGTAAAGA | 287 | 60 |
| R: GTAACAGTCCGCCTAGAAGCAC | ||||
| Caspase 3 | NM_009810 | F: GTCTGACTGGAAAGCCGAAAC | 205 | 59.5 |
| R: GACTGGATGAACCACGACCC | ||||
| Caspase 8 | NM_001080126 | F: TCTCGGAATCGGTAGCAAACC | 173 | 60 |
| R: AGAAGAGCTGTAACCTGTGGC | ||||
| Cyclin D2 | NM_009829.3 | F: TACCTCCCGCAGTGTTCCTA | 158 | 60 |
| R: GCCAAGAAACGGTCCAGGTA | ||||
| MYC | NM_001177352.1 | F: GTTGGAAACCCCGCAGACAG | 264 | 60.5 |
| R: GTAGCGACCGCAACATAGGA | ||||
| CYP11A1 | NM_001346787.1 | F: TACTAACCTAGCCCGCCTCG | 163 | 60.5 |
| R: CTCCTGCGCATAGAGAGAGC | ||||
| FSHR | NM_013523.3 | F: AACACTTGCCAGCCTTTCAC | 184 | 60 |
| R: TGGGTTCCGTTGAATGCACA | ||||
| CTSB | NM_007798.3 | F: CAATGGCCGAGTCAACGTG | 176 | 59 |
| R: TGGTGTATGGTAAGCAGCCT | ||||
| BCL2 | NM_009741.5 | F: GAACTGGGGGAGGATTGTGG | 194 | 60 |
| R: GCATGCTGGGGCCATATAGT | ||||
| LC3-I | NM_025735.3 | F:AGGAGAAGGATGAAGACGGA | 160 | 57 |
| R:CCACTGGGGACTGAAATAGC | ||||
| ATG5 | NM_001358596.1 | F:CAGTGTGATCCCGGCAGA | 199 | 58 |
| R:GAGTAAAGCAAGTTGGAATTCG | ||||
| si-CTSB (204) | F: GCUGUCGGAUGACCUGAUUTT | |||
| R: AAUCAGGUCAUCCGACAGCTT | ||||
| si-CTSB (1096) | F: UCAGAAAUUGUGGCUGGAATT | |||
| R: UUCCAGCCACAAUUUCUGATT |
4.4. RNA Extraction and Reverse-Transcription Polymerase Chain Reaction (RT-PCR)
Cells were lysed 48 h after transfection to extract total RNA using a kit method (Total RNA Kit I (200); R6834-02; Omega Bio-Tek, Norcross, GA, USA) following the manufacturer’s instructions for subsequent cDNA synthesis. Spectrophotometry was used to determine the quality and quantity of RNA (Nanodrop 2000 Analyzer; Thermo Scientific, Wilmington, DE, USA). Finally, the kit method (FastKing RT Kit; TianGen Biotech, Co., Ltd., Beijing, China) was used for reverse transcription of 1 μg of qualified total RNA (260/280; 1.8–2.1), according to manufacturer instructions.
4.5. Quantitative Real-Time PCR (qRT-PCR)
Primers listed (Table 1) were designed using Primer 5.0 for this study. Gene expressions levels were determined by QRT-PCR (Light Cycler 480 Multiwell Plate 96; Roche, Indianapolis, IN, USA) using Bio-Rad (Bio-Rad, Inc., Hercules, CA, USA) against β-actin or GAPDH as an endogenous control. Each 10 μL of the reaction mixture was comprised cDNA (1 μL), RNase-free water (3 μL), sense and antisense primers (1 μL), and SYBR-Green Master Mix (QIAGEN, Hilden, Germany) (5 μL).
4.6. Protein Extraction and Western Blotting
GCs transfected with siCTSB and NC in a 6-well plate were placed on ice, washed with PBS, followed by cell lysis using a lysis buffer RIPA (Servicebio, Wuhan, China) augmented with phosphorylase inhibitor (1 mM) and phenylmethanesulphonyl fluoride (PMSF). Proteins were isolated by SDS-PAGE (12%) and subsequently transferred to polyvinylidene difluoride membranes (PVDF; Immobilon-P, Millipore) via electrophoresis. Preliminary, the membranes were incubated with 5% skimmed milk diluted in TBS for 2 h, and subsequently incubated (4 °C, overnight) with these primary antibodies: Bcl2 (4223S), CTSB (31718), cyclin D2 (3741P), p-ERK1/2 (4370S), LC3-I (2475S) (1:1000, CST, Danvers, MA, USA), caspase 3 (A19654), CYP11A1 (A1713), FSHR (A1480) (1:1000, Abclonal-Tech, Wuhan, China), TNF-α (17590-1-AP), AKT (10176-2-AP) (1:1000, Proteintech Group, lnc., Wuhan, China), ERK (1:1000, AF0155; Affinity Biologicals Inc. USA), p-Akt (Ab81283), ATG5 (Ab109490) (1:2000; Abcam, Cambridge, UK); Abcam, Cambridge, UK), MYC (1:1000, NBP2-25147SS, Novus, CO, USA), caspase 8 (1:1000, BS1387, Bioworld, Nanjing, China), GAPDH (1:1000, 60004-1-Ig; Proteintech Group, lnc. Wuhan, China), and β-actin (1:500, BM0627; BosterBio, Wuhan, China) as a control. Target proteins in each sample were determined using enhanced chemiluminescence (NCI5079; Bio-Rad, USA). WB-images were captured by Gel-Pro analyzer version 4 (Media Cybernetics, Rockville, MD, USA), and Image J software was used to determine bands, respectively. The data were normalized toβ-actin or GAPDH.
4.7. Cell Apoptosis Detection
Flow cytometry was used to determine the apoptosis rate in mouse GCs cultured in 6-well plates 48 h after transfection with si-CTSB and NC using the FITC/ PI Apoptosis Detection Kit (KeyGEN Biotech, Nanjing, China) as directed by the manufacturer. The trypsin-lysed cells were washed twice with PBS before being resuspended in a binding buffer solution. The cells were stained (FITC and PI; 15 min, room temperature). Finally, flow cytometry was used to determine the apoptosis rate using the FACSVerse Calibur (BD Biosciences, San Jose, CA, USA) according to the manufacturer’s protocol. The apoptosis assay results were validated by examining the levels of mRNA and protein expression of apoptosis-related molecules.
4.8. Cell Counts and Proliferation Assay
Si-CTSB and NC-transfected GC were seeded in a 96-well plate and incubated for proliferation studies in a culture medium for 48 h. The cells that were trypsin-lysed were then counted using an automated cell counter (BIO-RAD Laboratories, Inc., TC20TM, Hercules, CA, USA). The kit method determined the capacity of GC proliferation (CCK-8; Dojindo Molecular Technologies, Inc., Rockville, MD, USA). As directed by the manufacturer, the viability of the cells was determined by the concentration of formazan dye, a cell viability indicator. In brief, each well-received 10% culture media with a 100-L CCK-8 solution was incubated for 3 h at 37 °C with 5% CO2 and saturated humidity. The GCs were then loaded onto a multimode plate reader (PerkinElmer, EnSpire, Waltham, MA, USA) to determine each experimental group’s absorbance at 450 nm.
4.9. Cellular Immunofluorescence
Using 4% paraformaldehyde in PHEM (0.5% Triton X-100), GCS were fixed for 45 min after PBS wash for immunofluorescence. Afterward, blocking was performed using PBS (2% BSA; 0.05% Tween-20, 1 h, Rm. Temp.). After that, GCs were incubated with a CTSB antibody (overnight at 4 °C) (1:800, CST, Danvers, MA, USA). Following three 10-min washes in PBS (0.05 percent Tween-20), GCs were incubated for one hour at 37 °C with Cy3-labeled goat anti-rabbit (Boster; 1:100) or TRITC-conjugated goat anti-human (Proteintech;1:50). PBS (1 g/mL DAPI; 10 min at room temperature) labeled the DNA. Finally, using DABCO, GC’s were mounted on glass slides for confocal microscopy (Zeiss LSM 510 META, Carl Zeiss Imaging, and Germany) equipped with a plan apochromat 63×/1.4 oil DIC objective. Zeiss LSM Image Browser and Adobe Photoshop were used to process the confocal images (Adobe Systems Inc., San Jose, CA, USA).
4.10. Cell Cycle Assay Middling
Following 48 h of transfection with si-CTSB and NC, mouse primary GCs were exposed to 0.25 percent trypsin (37 °C, 3 min) and centrifuged (4000 rpm, 5 min). Following a 70 percent ethanol wash and an overnight incubation at 4 °C, harvested cells were stained with RNase A and PI solution (100 μL and 400 μL, respectively; 30 min). Cell cycle progression was assessed by the FACSVerse Calibur (BD Biosciences, San Jose, CA, USA). ModFit was used to analyze the proportion of the cell cycle in different phases across three independent experiments.
4.11. Hormones Assay
Culture media were collected from si-CTSB and NC, transfected GCs at 48 h, and subjected to centrifugation (1000 g, 20 min) to ascertain hormone concentration. ELISA Kit (MLBIO Biotechnology Co., Ltd.; Shanghai, China) specific to mice was used to measure estradiol-17β (E2) and progesterone (P4) concentrations.
4.12. Statistical Analysis
Data indicated as mean ± SEM were obtained from three independent replicates at least. The statistically significant difference between groups was determined using paired-samples t-test in Graph-Pad. The cut-off value adjusted to a statistically significant difference was p < 0.05.
5. Conclusions
Our results established that siRNA-mediated downregulation of CTSB decreased the TNFa-mediated apoptosis and autophagy. Small interference-mediated downregulation of CTSB suppressed GC apoptosis; therefore, it can be concluded that CTSB serves as proapoptotic in mice ovarian cells. Furthermore, inhibiting CTSB promotes mouse GC proliferation via activation of the p-Akt and p-ERK1/2 pathways and significant changes in cell cycle progression. In this study, CTSB silencing induces a decrease in steroids hormones (progesterone and estradiol) secretion by suppressing the critical genes for steroid synthesis (FSHR and CYP11A1). Despite a novel role of CTSB, it is demonstrated herein that, other than recognized biological functions, additional experiments are warranted to confirm these findings in vivo.
Altogether, by illustrating the physiological role of the CTSB gene in an ovarian cell, this study suggests that future research should consider CTSB’s potential to improve fertility in female mammals and its use to develop new therapeutic approaches.
Author Contributions
Conceptualization and methodology, L.Y., C.C. and M.J.A.; software and validation, T.Y.; formal analysis, C.D.; investigation, C.C. and M.J.A.; resources, A.L.; data curation, X.Z.; writing—original draft preparation, C.C.; writing—review and editing, A.L. and M.J.A.; visualization, X.Z.; supervision, L.Y.; project administration, L.Y.; funding acquisition, L.Y. All authors have read and agreed to the published version of the manuscript.
Funding
This research was supported by China Agriculture Research System of MOF and MARA.
Institutional Review Board Statement
This study was approved by the Research Animal Ethics Committee of Huazhong Agricultural University (no, HZAUMO-2018–060, January 2018).
Informed Consent Statement
Not applicable.
Data Availability Statement
The authors confirm that the data supporting the findings of this study are available within the article.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Heatwole H. The Heat Dissipation Limit Theory and Evolution of Life Histories in Endotherms—Time to Dispose of the Disposable Soma Theory?—Supplementary Data. Integr. Comp. Biol. 2012;50:793–807. doi: 10.1093/icb/icq049. [DOI] [PubMed] [Google Scholar]
- 2.Worku T., Wang K., Ayers D., Wu D., Ur Rehman Z., Zhou H., Yang L. Regulatory roles of ephrinA5 and its novel signaling pathway in mouse primary granulosa cell apoptosis and proliferation. Cell Cycle. 2018;17:892–902. doi: 10.1080/15384101.2018.1456297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hongshusui Y.S., Weili Z.B., Jinghetan M.J. Apoptosis in Granulosa cells during follicular atresia: Relationship with steroids and insulin-like growth factors. Cell Res. 2004;14:341–346. doi: 10.1038/sj.cr.7290234. [DOI] [PubMed] [Google Scholar]
- 4.Hughes F.M., Gorospe W.C. Biochemical identification of apoptosis (programmed cell death) in granulosa cells: Evidence for a potential mechanism underlying follicular atresia. Endocrinology. 1991;129:2415–2422. doi: 10.1210/endo-129-5-2415. [DOI] [PubMed] [Google Scholar]
- 5.Byskov A.G. Follicular atresia. In: Jones R.E., editor. The Vertebrate Ovary: Comparative Biology and Evolution. Plenum Press; New York, NY, USA: 1978. pp. 533–562. [Google Scholar]
- 6.Choi J.Y., Jo M.W., Lee E.Y., Yoon B.-K., Choi D.S. The role of autophagy in follicular development and atresia in rat granulosa cells. Fertil. Steril. 2010;93:2532–2537. doi: 10.1016/j.fertnstert.2009.11.021. [DOI] [PubMed] [Google Scholar]
- 7.Klionsky D.J., Emr S.D. Autophagy as a regulated pathway of cellular degradation. Science. 2000;290:1717–1721. doi: 10.1126/science.290.5497.1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Levine B., Klionsky D.J. Development by self-digestion: Molecular mechanisms and biological functions of autophagy. Dev. Cell. 2004;6:463–477. doi: 10.1016/S1534-5807(04)00099-1. [DOI] [PubMed] [Google Scholar]
- 9.Hwang S.O., Lee G.M. Nutrient deprivation induces autophagy as well as apoptosis in Chinese hamster ovary cell culture. Biotechnol. Bioeng. 2008;99:678–685. doi: 10.1002/bit.21589. [DOI] [PubMed] [Google Scholar]
- 10.Kessel D., Reiners J.J., Jr. Initiation of apoptosis and autophagy by the Bcl-2 antagonist HA14-1. Cancer Lett. 2007;249:294–299. doi: 10.1016/j.canlet.2006.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Duerrschmidt N., Zabirnyk O., Nowicki M., Ricken A., Hmeidan F.A., Blumenauer V., Borlak J., Spanel-Borowski K. Lectin-like oxidized low-density lipoprotein receptor-1-mediated autophagy in human granulosa cells as an alternative of programmed cell death. Endocrinology. 2006;147:3851–3860. doi: 10.1210/en.2006-0088. [DOI] [PubMed] [Google Scholar]
- 12.Billig H., Furuta I., Hsueh A. Estrogens inhibit and androgens enhance ovarian granulosa cell apoptosis. Endocrinology. 1993;133:2204–2212. doi: 10.1210/endo.133.5.8404672. [DOI] [PubMed] [Google Scholar]
- 13.AJ B. Kirschke H: Cathepsin B, cathepsin H and cathepsin L. Methods Enzym. 1981;80 Pt C:535–561. doi: 10.1016/s0076-6879(81)80043-2. [DOI] [PubMed] [Google Scholar]
- 14.Mohamed M.M., Sloane B.F. Multifunctional enzymes in cancer. Nat. Rev. Cancer. 2006;6:764–775. doi: 10.1038/nrc1949. [DOI] [PubMed] [Google Scholar]
- 15.Buhling F., Fengler A., Brandt W., Welte T., Ansorge S., Nagler D.K. Part II: Cellular Endopeptidases: New Cathepsins; Results from Knock-out-mice; Regulatory Aspects-26. Review: Novel Cysteine Proteases of the Papain Family. Adv. Exp. Med. Biol. 2000;477:241–254. doi: 10.1007/0-306-46826-3_26. [DOI] [PubMed] [Google Scholar]
- 16.Turk B., Turk D., Turk V. Lysosomal cysteine proteases: More than scavengers. Biochim. Biophys. Acta (BBA) Protein Struct. Mol. Enzymol. 2000;1477:98–111. doi: 10.1016/S0167-4838(99)00263-0. [DOI] [PubMed] [Google Scholar]
- 17.Klose A., Zigrino P., Dennhöfer R., Mauch C., Hunzelmann N. Identification and discrimination of extracellularly active cathepsins B and L in high-invasive melanoma cells. Anal. Biochem. 2006;353:57–62. doi: 10.1016/j.ab.2006.01.037. [DOI] [PubMed] [Google Scholar]
- 18.Vancompernolle K., Van Herreweghe F., Pynaert G., Van De Craen M., De Vos K., Totty N., Sterling A., Fiers W., Vandenabeele P., Grooten J. Atractyloside-induced release of cathepsin B, a protease with caspase-processing activity. FEBS Lett. 1998;438:150–158. doi: 10.1016/S0014-5793(98)01275-7. [DOI] [PubMed] [Google Scholar]
- 19.Bossy-Wetzel E., Newmeyer D.D., Green D.R. Mitochondrial cytochrome c release in apoptosis occurs upstream of DEVD-specific caspase activation and independently of mitochondrial transmembrane depolarization. EMBO J. 1998;17:37–49. doi: 10.1093/emboj/17.1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim S.-H., Zhao M.-H., Liang S., Cui X.-S., Kim N.-H. Inhibition of cathepsin B activity reduces apoptosis by preventing cytochrome c release from mitochondria in porcine parthenotes. J. Reprod. Dev. 2015;61:261–268. doi: 10.1262/jrd.2015-019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bettegowda A., Patel O.V., Lee K.B., Park K.E., Salem M., Yao J., Ireland J.J., Smith G.W. Identification of novel bovine cumulus cell molecular markers predictive of oocyte competence: Functional and diagnostic implications. Biol. Reprod. 2008;79:301–309. doi: 10.1095/biolreprod.107.067223. [DOI] [PubMed] [Google Scholar]
- 22.Balboula A., Yamanaka K., Sakatani M., Hegab A., Zaabel S., Takahashi M. Intracellular cathepsin B activity is inversely correlated with the quality and developmental competence of bovine preimplantation embryos. Mol. Reprod. Dev. 2011;77:1031–1039. doi: 10.1002/mrd.21250. [DOI] [PubMed] [Google Scholar]
- 23.Balboula A., Yamanaka K., Sakatani M., Hegab A., Zaabel S., Takahashi M. Cathepsin B Activity is Related to the Quality of Bovine Cumulus Oocyte Complexes and Its Inhibition Can Improve Their Developmental Competence. Mol. Reprod. Dev. 2010;77:439–448. doi: 10.1002/mrd.21164. [DOI] [PubMed] [Google Scholar]
- 24.Balboula A.Z., Yamanaka K., Sakatani M., Kawahara M., Hegab A.O., Zaabel S.M., Takahashi M. Cathepsin B activity has a crucial role in the developmental competence of bovine cumulus-oocyte complexes exposed to heat shock during in vitro maturation. Reproduction. 2013;146:407–417. doi: 10.1530/REP-13-0179. [DOI] [PubMed] [Google Scholar]
- 25.Ruan J., Zheng H., Rong X., Rong X., Zhang J., Fang W., Zhao P., Luo R. Over-expression of cathepsin B in hepatocellular carcinomas predicts poor prognosis of HCC patients. Mol. Cancer. 2016;15:17. doi: 10.1186/s12943-016-0503-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chan A.T., Baba Y., Shima K., Nosho K., Chung D.C., Hung K.E., Mahmood U., Madden K., Poss K., Ranieri A., et al. Cathepsin B Expression and Survival in Colon Cancer: Implications for Molecular Detection of Neoplasia. Cancer Epidemiol. Biomark. Prev. 2010;19:2777–2785. doi: 10.1158/1055-9965.EPI-10-0529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hughes S.J., Glover T.W., Zhu X.X., Kuick R., Thoraval D., Orringer M.B., Beer D.G., Hanash S. A novel amplicon at 8p22-23 results in overexpression of cathepsin B in esophagel adenocarcinoma. Proc. Natl. Acad. Sci. USA. 1998;95:12410–12415. doi: 10.1073/pnas.95.21.12410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Niedergethmann M., Wostbrock B., Sturm J.W., Willeke F., Post S., Hildenbrand R. Prognostic Impact of Cysteine Proteases Cathepsin B and Cathepsin L in Pancreatic Adenocarcinoma. Pancreas. 2004;29:204. doi: 10.1097/00006676-200410000-00005. [DOI] [PubMed] [Google Scholar]
- 29.Sendler M., Maertin S., John D., Persike M., Weiss F.U., Krueger B., Wartmann T., Wagh P., Halangk W., Schaschke N., et al. Cathepsin B Activity Initiates Apoptosis via Digestive Protease Activation in Pancreatic Acinar Cells and Experimental Pancreatitis. J. Biol. Chem. 2016;291:14717–14731. doi: 10.1074/jbc.M116.718999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bai H., Yang B., Yu W., Xiao Y., Yu D., Zhang Q. Cathepsin B links oxidative stress to the activation of NLRP3 inflammasome. Exp. Cell Res. 2017;362:180–187. doi: 10.1016/j.yexcr.2017.11.015. [DOI] [PubMed] [Google Scholar]
- 31.Iwama H., Mehanna S., Imasaka M., Hashidume S., Nishiura H., Yamamura K.-I., Suzuki C., Uchiyama Y., Hatano E., Ohmuraya M. Cathepsin B and D deficiency in the mouse pancreas induces impaired autophagy and chronic pancreatitis. Sci. Rep. 2021;11:6596. doi: 10.1038/s41598-021-85898-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tai W. Chemical modulation of siRNA lipophilicity for efficient delivery. J. Control. Release. 2019;307:98–107. doi: 10.1016/j.jconrel.2019.06.022. [DOI] [PubMed] [Google Scholar]
- 33.Sargent K.M., Lu N., Clopton D.T., Pohlmeier W.E., Brauer V.M., Ferrara N., Silversides D.W., Cupp A.S. Loss of Vascular Endothelial Growth Factor A (VEGFA) Isoforms in Granulosa Cells Using pDmrt-1-Cre or Amhr2-Cre Reduces Fertility by Arresting Follicular Development and by Reducing Litter Size in Female Mice. PLoS ONE. 2015;10:e0116332. doi: 10.1371/journal.pone.0116332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Quirk S.M., Cowan R.G., Harman R.M., Hu C.-L., Porter D.A. Ovarian follicular growth and atresia: The relationship between cell proliferation and survival. J. Anim. Sci. 2004;82((Suppl. S13)):E40. doi: 10.2527/2004.8213_supplE40x. [DOI] [PubMed] [Google Scholar]
- 35.Kaipia A., Hsueh A.J. Regulation of ovarian follicle atresia. Annu. Rev. Physiol. 1997;59:349–363. doi: 10.1146/annurev.physiol.59.1.349. [DOI] [PubMed] [Google Scholar]
- 36.Chwieralski C., Welte T., Bühling F. Cathepsin-regulated apoptosis. Apoptosis. 2006;11:143–149. doi: 10.1007/s10495-006-3486-y. [DOI] [PubMed] [Google Scholar]
- 37.Boya P., Andreau K., Poncet D., Zamzami N., Perfettini J.-L., Metivier D., Ojcius D., Jaattela M., Kroemer G. Lysosomal membrane permeabilization induces cell death in a mitochondrion-dependent fashion. J. Exp. Med. 2003;197:1323–1334. doi: 10.1084/jem.20021952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Johansson A.-C., Appelqvist H., Nilsson C., Kågedal K., Roberg K., Öllinger K. Regulation of apoptosis-associated lysosomal membrane permeabilization. Apoptosis. 2010;15:527–540. doi: 10.1007/s10495-009-0452-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Droga-Mazovec G., Bojic L., Petelin A., Ivanova S., Repnik U., Salvesen G.S., Stoka V., Turk V., Turk B. Cysteine cathepsins trigger caspase-dependent cell death through cleavage of bid and antiapoptotic Bcl-2 homologues. J. Biol. Chem. 2008;283:19140–19150. doi: 10.1074/jbc.M802513200. [DOI] [PubMed] [Google Scholar]
- 40.Johnson D. Noncaspase proteases in apoptosis. Leukemia. 2000;14:1695–1703. doi: 10.1038/sj.leu.2401879. [DOI] [PubMed] [Google Scholar]
- 41.Leist M., Jäättelä M. Triggering of apoptosis by cathepsins. Cell Death Differ. 2001;8:324–326. doi: 10.1038/sj.cdd.4400859. [DOI] [PubMed] [Google Scholar]
- 42.Zhao M., Antunes F., Eaton J.W., Brunk U.T. Lysosomal enzymes promote mitochondrial oxidant production, cytochrome c release and apoptosis. Eur. J. Biochem. 2003;270:3778–3786. doi: 10.1046/j.1432-1033.2003.03765.x. [DOI] [PubMed] [Google Scholar]
- 43.Guicciardi M.E., Deussing J., Miyoshi H., Bronk S.F., Svingen P.A., Peters C., Kaufmann S.H., Gores G.J. Cathepsin B contributes to TNF-α–mediated hepatocyte apoptosis by promoting mitochondrial release of cytochrome c. J. Clin. Investig. 2000;106:1127–1137. doi: 10.1172/JCI9914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shan X., Yu T., Yan X., Wu J., Fan Y., Guan X., Fang F., Lin Y., Zhang Y., Li Y., et al. Proteomic analysis of healthy and atretic porcine follicular granulosa cells. J. Proteom. 2020;232:104027. doi: 10.1016/j.jprot.2020.104027. [DOI] [PubMed] [Google Scholar]
- 45.Talpur H.S., Worku T., Rehman Z.U., Dad R., Bhattarai D., Bano I., Liang A., He C., Yang L. Knockdown of melatonin receptor 1 and induction of follicle-stimulating hormone on the regulation of mouse granulosa cell function. Reprod. Biol. 2017;17:380–388. doi: 10.1016/j.repbio.2017.10.005. [DOI] [PubMed] [Google Scholar]
- 46.Li J.-X., Ding X.-M., Han S., Wang K., Jiao C.-Y., Li X.-C. mir-637 inhibits the proliferation of cholangiocarcinoma cell QBC939 through interfering CTSB expression. Eur. Rev. Med Pharmacol. Sci. 2018;22:1265–1276. doi: 10.26355/eurrev_201803_14467. [DOI] [PubMed] [Google Scholar]
- 47.Zhang L., Hou Y., Ashktorab H., Gao L., Xu Y., Wu K., Zhai J., Zhang L. The impact of C-MYC gene expression on gastric cancer cell. Mol. Cell. Biochem. 2010;344:125–135. doi: 10.1007/s11010-010-0536-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bouchard C., Thieke K., Maier A., Saffrich R., Hanley-Hyde J., Ansorge W., Reed S., Sicinski P., Bartek J., Eilers M. Direct induction of cyclin D2 by Myc contributes to cell cycle progression and sequestration of p27. EMBO J. 1999;18:5321–5333. doi: 10.1093/emboj/18.19.5321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bouchard C., Dittrich O., Kiermaier A., Dohmann K., Menkel A., Eilers M., Lüscher B. Regulation of cyclin D2 gene expression by the Myc/Max/Mad network: Myc-dependent TRRAP recruitment and histone acetylation at the cyclin D2 promoter. Genes Dev. 2001;15:2042–2047. doi: 10.1101/gad.907901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang L., Liu X., Jin H., Guo X., Xia L., Chen Z., Bai M., Liu J., Shang X., Wu K., et al. MiR-206 inhibits gastric cancer proliferation in part by repressing CyclinD2. Cancer Lett. 2013;332:94–101. doi: 10.1016/j.canlet.2013.01.023. [DOI] [PubMed] [Google Scholar]
- 51.Chen S., Shimoda M., Chen J., Matsumoto S., Grayburn P.A. Transient overexpression of cyclin D2/CDK4/GLP1 genes induces proliferation and differentiation of adult pancreatic progenitors and mediates islet regeneration. Cell Cycle. 2012;11:695–705. doi: 10.4161/cc.11.4.19120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Alvarez E., Northwood I.C., Gonzalez F.A., Latour D.A., Seth A., Abate C., Curran T., Davis R.J. Pro-Leu-Ser/Thr-Pro is a consensus primary sequence for substrate protein phosphorylation. Characterization of the phosphorylation of c-myc and c-jun proteins by an epidermal growth factor receptor threonine 669 protein kinase. J. Biol. Chem. 1991;266:15277–15285. doi: 10.1016/S0021-9258(18)98613-8. [DOI] [PubMed] [Google Scholar]
- 53.Sears R., Nuckolls F., Haura E., Taya Y., Tamai K., Nevins J.R. Multiple Ras-dependent phosphorylation pathways regulate Myc protein stability. Genes Dev. 2000;14:2501–2514. doi: 10.1101/gad.836800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lefloch R., Pouyssegur J., Lenormand P. Single and combined silencing of ERK1 and ERK2 reveals their positive contribution to growth signaling depending on their expression levels. Mol. Cell. Biol. 2008;28:511–527. doi: 10.1128/MCB.00800-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cao Z., Liao Q., Su M., Huang K., Jin J., Cao D. AKT and ERK dual inhibitors: The way forward? Cancer Lett. 2019;459:30–40. doi: 10.1016/j.canlet.2019.05.025. [DOI] [PubMed] [Google Scholar]
- 56.Lu Z., Xu S. ERK1/2 MAP kinases in cell survival and apoptosis. IUBMB Life. 2006;58:621–631. doi: 10.1080/15216540600957438. [DOI] [PubMed] [Google Scholar]
- 57.Casarini L., Crépieux P. Molecular mechanisms of action of FSH. Front. Endocrinol. 2019;10:305. doi: 10.3389/fendo.2019.00305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hu M.-C., Hsu H.-J., Guo I.-C., Chung B.-C. Function of Cyp11a1 in animal models. Mol. Cell. Endocrinol. 2004;215:95–100. doi: 10.1016/j.mce.2003.11.024. [DOI] [PubMed] [Google Scholar]
- 59.Chien Y., Cheng W.-C., Wu M.-R., Jiang S.-T., Shen C.-K.J., Chung B.-C. Misregulated progesterone secretion and impaired pregnancy in Cyp11a1 transgenic mice. Biol. Reprod. 2013;89:91. doi: 10.1095/biolreprod.113.110833. [DOI] [PubMed] [Google Scholar]
- 60.Macklon N.S., Fauser B. Follicle development during the normal menstrual cycle. Maturitas. 1998;30:181–188. doi: 10.1016/S0378-5122(98)00072-3. [DOI] [PubMed] [Google Scholar]
- 61.Ye X., Li Y., Wang Z., Andersen C.L. Functions of lysosomes in mammalian female reproductive system. Reprod. Dev. Med. 2020;4:109. doi: 10.4103/2096-2924.288025. [DOI] [Google Scholar]
- 62.Yoshii S.R., Kuma A., Akashi T., Hara T., Yamamoto A., Kurikawa Y., Itakura E., Tsukamoto S., Shitara H., Eishi Y., et al. Systemic analysis of Atg5-null mice rescued from neonatal lethality by transgenic ATG5 expression in neurons. Dev. Cell. 2016;39:116–130. doi: 10.1016/j.devcel.2016.09.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The authors confirm that the data supporting the findings of this study are available within the article.