Abstract
DNA interstrand cross-links (ICLs) represent lethal DNA damage, because they block transcription, replication, and segregation of DNA. Because of their genotoxicity, agents inducing ICLs are often used in antitumor therapy. The repair of ICLs is complex and involves proteins belonging to nucleotide excision, recombination, and translesion DNA repair pathways in Escherichia coli, Saccharomyces cerevisiae, and mammals. We cloned and analyzed mammalian homologs of the S. cerevisiae gene SNM1 (PSO2), which is specifically involved in ICL repair. Human Snm1, a nuclear protein, was ubiquitously expressed at a very low level. We generated mouse SNM1−/− embryonic stem cells and showed that these cells were sensitive to mitomycin C. In contrast to S. cerevisiae snm1 mutants, they were not significantly sensitive to other ICL agents, probably due to redundancy in mammalian ICL repair and the existence of other SNM1 homologs. The sensitivity to mitomycin C was complemented by transfection of the human SNM1 cDNA and by targeting of a genomic cDNA-murine SNM1 fusion construct to the disrupted locus. We also generated mice deficient for murine SNM1. They were viable and fertile and showed no major abnormalities. However, they were sensitive to mitomycin C. The ICL sensitivity of the mammalian SNM1 mutant suggests that SNM1 function and, by implication, ICL repair are at least partially conserved between S. cerevisiae and mammals.
DNA interstrand cross-links (ICLs) prevent strand separation, thereby physically blocking transcription, replication, and segregation of DNA. In bacterial and yeast cells, the presence of one unrepaired ICL can be lethal (36, 40). Humans are exposed to environmental ICL agents, such as furocoumarins, from plants and cosmetics (50). Due to their extreme genotoxicity, agents that induce cross-links in DNA are widely used in antitumor therapy. Examples include cisplatin, mitomycin C (MMC), and derivatives of nitrogen mustard, such as melphalan and cyclophosphamide. Most of these agents cause a number of different lesions in DNA, including monoadducts, DNA-protein cross-links, and DNA intrastrand cross-links and ICLs. The latter are the main cause of cell death, although they constitute only a small percentage of the total number of adducts (6, 36). Resistance of tumors to cross-linking agents can be caused by a variety of mechanisms, including increased repair of ICLs (1, 5, 31). Therefore, an understanding of how ICLs are repaired is important.
To elucidate the mechanism of ICL repair in yeast, genetic screens have been performed to find genes specifically involved in ICL repair. A number of snm and pso mutants have been isolated after screening for strains with increased sensitivity to nitrogen mustard and 8-methoxypsoralen plus UVA light, respectively (25, 48). Some of the mutants isolated are sensitive to several classes of DNA-damaging agents, while others are almost exclusively sensitive to ICL agents. The gene mutated in one of these strains is SNM1, which is allelic with PSO2 and encodes a nuclear 76-kDa protein (9, 23, 47). snm1 mutants are sensitive to a number of agents that cause ICLs, but they are only mildly sensitive to monofunctional alkylating agents and 254-nm UV light (UV254 nm) and are not hypersensitive to gamma rays (6, 25, 49). In exponentially growing cells, the expression of SNM1 can be induced by ICL agents or UV254 nm (61). Overexpression of SNM1 results in an increased resistance to nitrogen mustard and cisplatin (22).
Many genes involved in DNA damage repair are conserved between yeast and higher eukaryotes. One way to elucidate the mechanisms of ICL repair in mammalian cells is by studying mammalian homologs of Saccharomyces cerevisiae genes specifically involved in ICL repair. A human homolog of SNM1 has been isolated, and a comparison of its predicted protein product (hSnm1) to S. cerevisiae Snm1 (ScSnm1) has revealed 39% similarity, excluding gaps (43). The gene is located on chromosome 10q25, a region often found to be rearranged in tumors (4, 52, 58). Here, we report on the characterization of mammalian SNM1 homologs.
MATERIALS AND METHODS
hSNM1 cDNA constructs.
The cDNA of human SNM1 (hSNM1) (kindly provided by N. Nomura) was used to make several constructs containing tags at the 5′ and 3′ termini of hSNM1 (Table 1). For this purpose, the 5′ terminus of hSNM1 was modified to generate an XhoI restriction enzyme site just in front of the expected initiation codon, thereby deleting the 900-bp 5′ untranslated region that contains 15 additional ATG sequences. The following tags were added to hSnm1: an N-terminal histidine tag (His10; single-letter amino acid code: MGHHHHHHHHHHGGSR), a C-terminal hemagglutinin tag (HA; PGGYPYDVPDYAS), and a C-terminal histidine-hemagglutinin tag (PHHHHHHGGSAYPYDVPDYAS). Parts of the hSNM1 cDNA were subcloned into pEGFPC vectors (expressing green fluorescent protein [GFP]; Clontech) (Table 1).
TABLE 1.
Characteristics of the mammalian SNM1 constructs
| Construct | Vector | Gene | Tag(s)a |
|---|---|---|---|
| hSNM1-HA | pPGK-p(A) | hSNM1 cDNA | HA |
| His-hSNM1-HA | pFastBac1 | hSNM1 cDNA | His10, HA |
| GFP-hSNM1 | pEGFPC2 | hSNM1 cDNA | GFP, His6HA |
| GFP-570hSNM1 | pEGFPC3 | hSNM1 cDNA bp 1–570 | GFP |
| GFP-1515hSNM1 | pEGFPC2 | hSNM1 cDNA bp 1–1515 | GFP |
| GFP-ChSNM1 | pEGFPC3 | hSNM1 cDNA bp 1652–3123 | GFP, His6HA |
| TrcHis-hSNM1 | pTrcHisC | hSNM1 cDNA bp 1930–2285 | His6 |
| mSNM1neo | pBluescript II KS | mSNM1 genomic disruption | NA |
| mSNM1hyg | pBluescript II KS | mSNM1 genomic disruption | NA |
| mSNM1C-GFP | pBluescript II KS | mSNM1-genomic cDNA fusion | GFP |
NA, not applicable.
Subcellular localization of hSNM1.
The constructs GFP-hSNM1, GFP-570hSNM1, GFP-1515hSNM1, and GFP-ChSNM1 were transfected into CHO9 cells. To obtain stable transfectants, cells were split after 1 day and subjected to G418 selection (1 mg/ml). Single colonies expressing GFP-hSnm1 proteins were expanded. The GFP-hSNM1 construct was also microinjected into multinucleated fibroblasts according to previously described procedures (27).
Generation of anti-hSnm1 antibodies.
A 354-bp PstI-HindIII fragment from the hSNM1 cDNA was subcloned into pTrcHisC (Table 1). The fusion protein derived from this plasmid contained amino acids 644 to 763 of hSnm1 fused to a His6 tag and was produced in Escherichia coli strain DH5α. The protein was present in the insoluble fraction and was dissolved in 6 M urea. It was purified on a Ni-nitrilotriacetic acid column, eluted with 200 mM imidazole, and used to immunize two rabbits. The detection limit of the polyclonal antibodies was determined by immunoblotting using a range of 1 to 1,000 ng of antigen and was shown to be below 1 ng. The antibodies were affinity purified using fusion protein immobilized on a nitrocellulose filter.
Expression of recombinant hSnm1.
The cDNA encoding His10-hSnm1-HA was subcloned into pFastBac1 (GibcoBRL) (Table 1). The resulting plasmid was used to create recombinant viruses and to produce the protein in Sf21 cells as described by the manufacturer. Protein extracts of these cells were made as described previously (54).
Construction of mSNM1 targeting vectors.
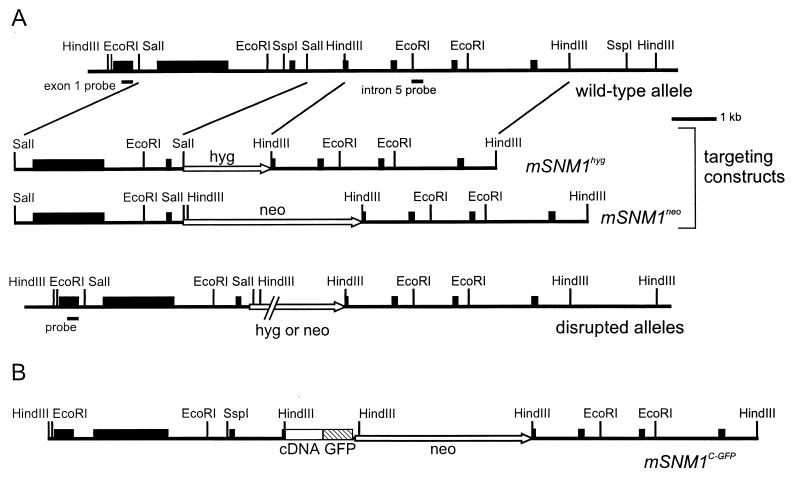
A mouse testis cDNA library was hybridized with a 2.7-kb EcoRI hSNM1 cDNA fragment. The resulting murine SNM1 (mSNM1) cDNA clone was sequenced and used to screen a lambda phage genomic library made from mouse strain 129/Sv. Genomic fragments hybridizing to the mSNM1 cDNA were subcloned in pBluescript II KS (Stratagene). The locations and intron and exon borders of the first five exons were determined by restriction analysis and DNA sequencing. Targeting constructs were made by cloning a 4-kb SalI fragment encompassing exons 2 and 3 and a 5-kb HindIII fragment encompassing part of exon 4 and exons 5 to 7 in pBluescript II KS. Between these fragments, a cassette containing either a neomycin (neo) resistance gene or a hygromycin (hyg) resistance gene was inserted. This step resulted in the partial deletion of exon 4. These targeting constructs are referred to as mSNM1neo and mSNM1hyg, respectively (Table 1) (see Fig. 3A).
FIG. 3.
Characterization of the mSNM1 genomic locus and generation of mSNM1 knockout and knockin constructs. The first seven exons are indicated by boxes. The lines below exon 1 and intron 5 indicate the positions of the probes used to screen for the disrupted alleles and the presence of the knockin allele. The locations of selected restriction sites are shown. (A) Schematic representation of part of the genomic mSNM1 locus and the gene targeting constructs. The top line represents part of the mSNM1 locus. The middle lines represent the targeting constructs and show the positions of the selectable marker genes for neomycin (neo) and hygromycin (hyg) in exon 4, indicated by the arrows. The bottom line represents the disrupted genomic locus. (B) Genomic construct for the rescue of mSNM1−/− cells. The genomic-cDNA mSNM1 fusion construct containing a GFP tag (hatched box) is shown. mSNM1 cDNA is represented by a white box, and the neo selectable marker gene is represented by an arrow.
ES cell culture and electroporation.
Embryonic stem (ES) cells were electroporated with the mSNM1hyg targeting construct and cultured on gelatinized dishes as described previously (17). The cells were split 24 h after electroporation, and hygromycin B was added to a final concentration of 200 μg/ml. After 7 to 10 days, colonies were isolated and expanded. Genomic DNA from individual clones was digested with SpeI or HindIII and analyzed by DNA blotting using the flanking exon 1 probe (see Fig. 3A). DNA from targeted clones with the expected hybridization pattern was subsequently digested with SspI and hybridized with the internal intron 5 probe to confirm proper homologous integration. To obtain ES cell lines carrying a disruption in both mSNM1 alleles, an mSNM1hyg-targeted ES cell line was electroporated with the mSNM1neo targeting construct. After selection with G418 (200 μg/ml) for 7 to 11 days, colonies were isolated and expanded. The isolated DNA was digested with HindIII and hybridized with exon 1 DNA to identify cell lines containing two targeted mSNM1 alleles.
Rescue of mSNM1−/− cells by hSNM1 cDNA constructs.
The hSNM1-HA cDNA was subcloned into pPGK-p(A) to express the gene under the control of the phosphoglycerate kinase (PGK) promoter (Table 1). This cDNA expression construct was coelectroporated with a puromycin-expressing plasmid into mSNM1neo/hyg cells. Clones were selected with puromycin (1 μg/ml) for 10 days. Integration of the cDNA construct was confirmed by DNA blotting.
Rescue of mSNM1−/− cells by mSNM1 genomic constructs.
A GFP-mSNM1 targeting fusion construct was made under the control of the mSNM1 promoter (see Fig. 3B). To obtain the expression of mSNM1 with 3′-terminal GFP, the 3′ terminus of mSNM1 cDNA was modified by PCR to create an EcoRV site in front of the stop codon. In this site, the GFP cDNA was cloned, and the borders were sequenced. Then, the 5.6-kb genomic mSNM1 HindIII fragment encompassing exons 1 to 4 was cloned into the cDNA, thereby fusing exon 4 from the genomic fragment with cDNA exons 4 to 9. Behind the endogenous polyadenylation signal, a neo cassette and the 5-kb mSNM1 HindIII fragment were cloned (see Fig. 3B). The construct, named mSNM1C-GFP, was transfected into mSNM1hyg-targeted ES cells, and clones were selected with G418 (200 μg/ml) and expanded. The isolated DNA was digested with SspI and hybridized with the intron 5 probe to screen for cell lines containing the homologously integrated target DNA.
Cell survival assays.
The sensitivity of ES cells to increasing doses of DNA-damaging agents was determined by measuring their colony-forming ability as described before (17). The cloning efficiency of untreated cells varied between 10 and 30%. Cells were incubated in drug-containing media for 1 h. Sensitivity to 8-methoxypsoralen plus UVA light was determined as follows. Dishes were incubated for 30 min in medium with 8-methoxypsoralen in the dark. Then, the cells were irradiated with 12 kJ of 320- to 380-nm UVA light at 2 mW/cm2 in phosphate-buffered saline containing 8-methoxypsoralen. All measurements were performed in triplicate.
Miscellaneous methods.
Reverse transcription (RT)-PCR, immunofluorescence microscopy, generation of mSNM1 mutant mice, and in vivo MMC survival assays were performed according to standard procedures and as previously described (17, 18, 55).
RESULTS
Comparison of Snm1 from different species.
A database search using either ScSnm1 or hSnm1 as a query sequence revealed the existence of homologs of Snm1 in many different species, including Schizosaccharomyces pombe, Aspergillus niger, Arabidopsis thaliana, Caenorhabditis elegans, Drosophila melanogaster, Danio rerio, and Rattus norvegicus (Fig. 1). We isolated and sequenced the mouse homolog (mSnm1) as described below. In addition, we identified part of a second human and mouse Snm1 homolog that we refer to as hSnm1B or mSnm1B, respectively, and a third human Snm1 homolog, called hSnm1C. Comparison of these different mammalian homologs showed that hSnm1 and mSnm1 displayed the highest degree of similarity to ScSnm1. hSnm1B and mSnm1B showed a high degree of similarity to each other but less similarity to ScSnm1. In A. thaliana, three Snm1 homologs were also found, but these were not clear homologs of hSnm1, hSnm1B, or hSnm1C. The length of the protein varied between different homologs and species. Some homologs showed similarity in the N-terminal region of Snm1, including the putative Zn finger domain present in ScSnm1. Other homologs did not contain this Zn finger domain and showed no significant similarity in the N-terminal region. Deletion of the Zn finger in S. cerevisiae did not render cells more sensitive to nitrogen mustard (23). The middle part of the protein was least conserved among all species, while conservation was highest in the C-terminal region. Eight regions stood out in particular, and we refer to these as motifs (Fig. 1). However, because these motifs appeared unique for Snm1, no clues regarding their function could be obtained through database searches.
FIG. 1.
Comparison of the deduced amino acid sequences of SNM1 homologs. The amino acid sequences of the C termini of S. cerevisiae (Sc), A. thaliana (At), mouse (m), and human (h) Snm1 are aligned and shown in the single-letter code. The second human and mouse Snm1 homologs, called Snm1B, and the third human homolog, called Snm1C, are incomplete. A more extensive version of this figure, including Snm1 homologs from additional species, can be found at http://www.eur.nl/fgg/ch1. Identical and similar amino acids are shown in black and gray boxes, respectively. Dots denote gaps; question marks denote unknown sequences. Amino acid positions are shown on the left. The eight motifs with high conservation are underlined. GenBank accession numbers are X64004 (S. cerevisiae), X98130 (A. thaliana), D42045 (human), AL137856 (hSnm1B), AA315885 and AA306797 (hSnm1C), AF241240 (mouse), and AI530740 and AI503687 (mSnm1B).
Subcellular localization of hSnm1.
The subcellular localization of hSnm1 was determined by microinjecting human fibroblasts and by transfecting CHO9 cells with a GFP-hSNM1 fusion construct (Table 1 and Fig. 2). As a control, the pEGFPC2 vector was transfected. In this case, GFP was diffusely present throughout the cell. In contrast, the location of GFP-hSnm1 was clearly restricted to the nucleus after both microinjection and transfection, as revealed by inspection of the cells under a fluorescence microscope after one to several days (data not shown). GFP-hSnm1 was excluded from the nucleolus. In cells expressing large amounts of protein, there was also weak staining of the cytoplasm. We conclude that hSnm1 is a nuclear protein. A few days after transfection or microinjection, many of the cells expressing large amounts of GFP-hSnm1 underwent morphological changes that were consistent with apoptosis. This finding indicates that Snm1 could be toxic when overexpressed in cells; this notion would also explain why extensive attempts to generate stable protein-expressing clones after G418 selection failed. Transfection of a series of GFP-hSNM1 fusion constructs revealed that the hSnm1 nuclear localization signal is located within the N-terminal 190 amino acids, which corresponds to the potential sequence for the nuclear localization signal at about amino acid 20 (Fig. 2).
FIG. 2.
Subcellular localization of hSnm1. Schematic representation of the different GFP-hSNM1 fusion constructs transfected into CHO9 cells. The upper line shows the hSNM1 cDNA with the putative nuclear localization signal (NLS), the Zn finger, and the conserved motifs. Below are the different hSNM1 constructs tagged with 5′-terminal GFP and the localization of their fluorescence signal in the cells (−, absent; + through +++, present in increasing amounts).
Generation of mSNM1-disrupted mouse cells.
Screening of a mouse testis cDNA library with the hSNM1 cDNA yielded a mouse cDNA clone spanning the 3′-terminal 2 kb of mSNM1. This clone was sequenced and used as a probe to obtain genomic mouse DNA. Two clones that spanned the first seven exons of the mSNM1 genomic locus were obtained. The locus was characterized by restriction site mapping, PCR, and sequencing of the first two exons (Fig. 3A). A targeting construct was made by subcloning into pBluescript II KS a 4-kb SalI fragment encompassing exons 2 and 3 and a 5-kb HindIII fragment encompassing part of exon 4 and exons 5 to 7 with a selectable marker gene in between. In this way, part of intron 3 and 25 bp of exon 4 were deleted. This procedure resulted in elimination of the C-terminal 287 amino acids of mSnm1, which contain most of the conserved motifs (Fig. 1). Moreover, aberrant RNA splicing that skips the targeted exon would result in a frameshift mutation. Disruption constructs were made containing the neo or the hyg selectable marker gene (mSNM1neo and mSNM1hyg, respectively).
The mSNM1hyg construct was electroporated into ES cells. Four independent cell lines containing one disrupted mSNM1 allele were identified by DNA blotting. The second wild-type allele was disrupted by targeting with the mSNM1neo construct. Three double-knockout cell lines in which the remaining wild-type allele was replaced by mSNM1neo were obtained (data not shown).
RT-PCRs with RNAs isolated from mSNM1+/− and mSNM1−/− cells were performed using primers for exons 2 and 7. RNA from mSNM1+/− cells showed a clear signal from the wild-type mSNM1 allele, and both cell lines showed three weak signals from the knockout allele (data not shown). These three bands were cloned and sequenced. One product would result in a frameshift and a premature stop codon. The other two products, one containing an insert from the neo cassette and the other splicing around exon 4, used an alternative splice site in exon 5 which restored the reading frame. Both would result in disruption of conserved motifs 2 and 3 of mSnm1 (Fig. 1). No significant difference in growth rate between mSNM1+/+ ES cells and all mSNM1+/− and mSNM1−/− cell lines was detected. We conclude that, similar to the situation for S. cerevisiae, disruption of mSNM1 in mouse cells results in viable cells (47).
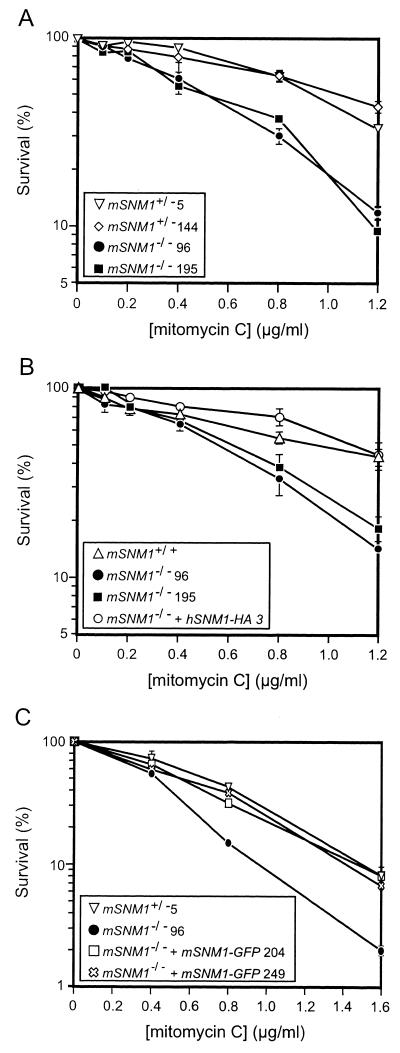
mSNM1−/− ES cells are sensitive to MMC.
S. cerevisiae snm1 mutants are sensitive to DNA ICL agents but not to gamma rays and only slightly to UV light (25, 49). We therefore investigated the effects of these DNA-damaging agents on the survival of mSNM1−/− ES cells. These cells were found to be approximately twofold more sensitive to MMC than mSNM1-proficient cells, which were otherwise isogenic (Fig. 4A). There was no difference between mSNM1+/+ and mSNM1+/− cells. In contrast, we did not find any significant sensitivity to 8-methoxypsoralen plus UVA, cisplatin, melphalan, UV254 nm, methyl methanesulfonate, and gamma rays (Table 2). As a control, ERCC1−/− ES cells were also treated with these agents and found to be sensitive to UV254 nm and all ICL agents tested (Table 2), as expected from the behavior of ERCC1−/− mouse embryonic fibroblasts and ERCC1 mutant CHO cells (14). ERCC1−/− cells were not sensitive to methyl methanesulfonate and gamma rays (Table 2).
FIG. 4.
Effect of MMC on wild-type and mutant mSNM1 ES cells. ES cells were treated with MMC for 1 h (see Materials and Methods). All measurements were performed in triplicate. (A) Clonogenic survival of mSNM1+/− and mSNM1−/− ES cells after treatment with increasing concentrations of MMC. (B) Rescue of the MMC sensitivity of mSNM1−/− cells by hSNM1-HA. An mSNM1−/− cell line containing randomly integrated hSNM1-HA cDNA expression constructs was obtained as described in Materials and Methods. The survival of this cell line was compared to the survival of mSNM1+/+ and mSNM1−/− ES cells. (C) Rescue of the MMC sensitivity of mSNM1−/− cells by mSNM1C-GFP. Two mSNM1C-GFP/− cell lines were obtained as described in Materials and Methods. The survival of these cell lines was compared to the survival of mSNM1+/− and mSNM1−/− ES cells. Error bars represent standard errors of the mean. Numbers following symbol definitions (figure insets), cell line isolation number.
TABLE 2.
Sensitivity of mSNM1−/− and ERCC1−/− ES cells to various genotoxic agents
| Genotoxic agent | Dose | Sensitivitya of the following cells:
|
|
|---|---|---|---|
| mSNM1−/− | ERCC1−/− | ||
| MMC | 0.1–1.6 μg/ml | Sens | Sens |
| Melphalan | 0.5–12 μM | Wt | ND |
| 8-Methoxypsoralen plus UVA light | 0.1–100 ng/ml | Wt | Sens |
| Cisplatin | 0.25–4 μg/ml | Wt | Sens |
| Gamma rays | 1–8 Gy | Wt | Wt |
| Methyl methanesulfonate | 1–2.5 mM | Wt | Wt |
| UV254 nm | 2–12 J/m2 | Wt | Sens |
Wt, sensitivity equal to that of wild-type cells; Sens, sensitivity higher than that of wild-type cells; ND, sensitivity not determined.
To prove that the phenotype of the mSNM1−/− ES cells was caused exclusively by the disruption of mSNM1, cDNA rescue experiments were performed. mSNM1neo/hyg cells were electroporated with a cDNA construct expressing HA-tagged hSnm1 together with a plasmid expressing the dominant selectable marker for puromycin. Puromycin-resistant cell lines that had integrated hSNM1-HA were fully corrected for the MMC sensitivity of mSNM1−/− ES cells (Fig. 4B).
mSNM1 is expressed at low levels in ES cells.
To investigate the expression of mSNM1 in ES cells, we generated a GFP-tagged version of mSNM1 under the control of the endogenous mSNM1 promoter (Fig. 3B and Table 1). For this purpose, a targeting construct, mSNM1C-GFP, was made by cloning mSNM1 cDNA exons 4 to 9 with 3′-terminal GFP behind genomic exon 4. As a consequence, the mRNA consisted of a fusion of four genomic exons and the cDNA with 3′-terminal GFP and was transcribed under the control of the endogenous mSNM1 promoter. mSNM1C-GFP was transfected into mSNM1+/− ES cells, and clones containing tagged and knockout mSNM1 alleles were obtained.
mSNM1C-GFP was expressed and functional, because the MMC sensitivity of mSNM1−/− ES cells was complemented by the presence of mSNM1C-GFP (Fig. 4C). Amplification by RT-PCR using an mSNM1 primer and a GFP primer spanning at least one intron revealed the expression of mSNM1C-GFP at the RNA level (data not shown). However, fluorescence microscopy failed to detect the GFP signal. Given the detection limit, this result implies that there are less than 10,000 molecules of mSnm1 per cell (45). We were also unable to detect either endogenous or transfected mSnm1 by immunoblot analysis with anti-GFP (Clontech) and anti-hSnm1 antibodies, even though anti-hSnm1 antibodies were highly sensitive, with a detection limit of about 5,000 Snm1 molecules per cell (data not shown). We cannot exclude the possibility, however, that our anti-hSnm1 antibodies were unable to recognize mSnm1.
Similarly, we tried to detect the presence of hSnm1-HA expressed under the control of the PGK promoter in mSNM1−/− ES cells; this construct can rescue the MMC sensitivity of these cells (Fig. 4B). Although both anti-hSnm1 and anti-HA antibodies recognized the protein when it was overproduced in Sf21 cells, we could not detect the protein in ES cells by either immunoblot or immunofluorescence microscopy (data not shown). Taken together, these results indicate that Snm1 protein levels in ES cells are very low, even when the protein is expressed under the control of the relatively strong PGK promoter.
Generation of mSNM1−/− mice.
The experiments described above show that disruption of mSNM1 is compatible with normal ES cell growth. Subsequently, we investigated whether mSnm1 is required for normal mouse development. Cells from four targeted ES clones, carrying the mSNM1hyg allele, were injected into C57BL/6 blastocysts, and 11 chimeric mice were obtained. The disrupted mSNM1 allele was transmitted to the mouse germ line. F1 heterozygous offspring were intercrossed, and F2 offspring were genotyped by DNA blotting, PCR analysis, or both (data not shown). Among 78 genotyped animals, 20 mSNM1+/+, 30 mSNM1+/−, and 28 mSNM1−/− animals were identified. This outcome is compatible with normal Mendelian segregation of the disrupted mSNM1 allele. Thus, disruption of mSNM1 does not result in embryonic or neonatal lethality. No statistically significant difference in weight was observed among mSNM1+/+, mSNM1+/−, and mSNM1−/− littermates (data not shown). Importantly, the mSNM1−/− mice exhibited no macroscopic abnormalities up to at least 12 months of age. Both mSNM1−/− males and females were fertile.
mSNM1−/− mice are sensitive to MMC.
We tested whether the sensitivity of mSNM1−/− ES cells to the DNA ICL agent MMC was also found in mSNM1−/− mice. mSNM1+/− and mSNM1−/− mice were treated with various doses of MMC, ranging from 7.5 to 15 mg/kg of body weight. Injection of 10 and 15 mg/kg in female mice resulted in enhanced sensitivity in mSNM1−/− mice (Fig. 5). In addition, with 15 mg/kg, a shorter latency period in mSNM1−/− mice was observed. In male mice, 7.5 mg/kg was lethal for 30% of the knockout mice, while all heterozygous mice survived. At 10 mg/kg, only 10% of the mSNM1−/− mice survived treatment, while 70% of the mSNM1+/− mice survived (Fig. 5). These results show that the hypersensitivity to MMC found in mSNM1−/− ES cells is also present in mSNM1−/− mice.
FIG. 5.
MMC sensitivity of mSNM1−/− mice. Shown are survival curves for mSNM1+/− and mSNM1−/− mice after a single intraperitoneal injection of the indicated amounts of MMC. (A) Survival of 12 mSNM1+/− and 8 mSNM1−/− female mice after injection with a dose of 10 mg of MMC per kg. (B) Survival of 11 mSNM1+/− and 5 mSNM1−/− female mice after injection with 15 mg of MMC per kg. (C) Survival of 6 mSNM1+/− and 7 mSNM1−/− male mice after injection with 7.5 mg of MMC per kg. (D) Survival of 16 mSNM1+/− and 9 mSNM1−/− male mice after injection with 10 mg of MMC per kg.
DISCUSSION
Characterization of mammalian SNM1.
We have analyzed mammalian homologs of the S. cerevisiae SNM1 gene, which specifically provides resistance to ICL agents. Like yeast snm1 knockout cells, mSNM1−/− ES cells and mice are viable, showing that SNM1 is not essential for viability (21). The mice do not show any major abnormalities and are fertile. mSNM1−/− ES cells and mice are sensitive to MMC, but the cells are not sensitive to UV254 nm, methyl methanesulfonate, or gamma rays, in agreement with the results obtained with S. cerevisiae snm1 cells (25, 49). These results argue for a specific role of mammalian SNM1 in the cellular response to ICL agents, similar to the role of yeast SNM1.
hSnm1 resides in the nucleus (Fig. 2), similar to its yeast homolog, a location expected for a DNA repair protein (23). It has a putative nuclear localization signal at the N terminus, and there may be another signal in the C terminus of the protein. The gene is expressed in all tissues tested (43). hSNM1 contains an exceptionally long, 911-bp 5′-terminal untranslated region, containing several ATGs in all reading frames, which might cause inefficient translation. We could not detect any hSnm1-HA or mSnm1-GFP after transfection into mSNM1−/− cells, although the detection limit of our antibodies was below 1 ng and the fluorescence microscopy detection limit of GFP is about 10,000 molecules per cell (45). This result suggests that the level of expression of Snm1 is very low, in agreement with the codon usage and the lack of a TATA box. S. cerevisiae SNM1 is also expressed at a very low level (47). Nevertheless, we could complement the MMC sensitivity of mSNM1−/− cells with hSnm1-HA and mSnm1-GFP; thus, the protein levels must have been sufficient, and the HA and GFP tags at the C terminus of Snm1 must not have interfered with its function.
Cross-link repair pathways.
The formation and repair of ICLs in cells are complex processes, with major differences between ICL agents. The sensitivity of a cell to a certain ICL agent is dependent on all steps in the processing of ICLs, ranging from uptake and metabolization to damage recognition and repair (6). A number of major DNA damage repair pathways are involved in the repair of ICLs. In S. cerevisiae, mutants in the nucleotide excision repair (NER), recombination repair, and translesion repair pathways are sensitive to ICL agents (24). Moreover, a number of ICL-sensitive mutants, such as pso2 to pso4 mutants, do not quite fit into one of these pathways (23). The major repair pathway of ICLs in S. cerevisiae is supposed to start with incision by the NER system, resulting in a DNA double-strand break (DSB), in contrast to the incision in E. coli, which does not result in a DSB (13, 28, 40, 59). The DSB can be repaired by the recombination repair pathway (2, 28). Repair may also occur via the translesion repair pathway, involving RAD6 and RAD18 (51). Yeast translesion repair pathway mutants are not defective in the repair of an ICL on a plasmid, in contrast to recombination repair pathway mutants, suggesting that chromatin structure can have an important influence (39). On the other hand, rad52 mutants, which are impaired in recombination repair, are hardly sensitive to nitrogen mustard, suggesting that the repair pathway used may depend on the ICL agent (51).
In mammals, a large number of mutant cell lines are known to be sensitive to ICL agents (11). Many of the known mutated genes in those cell lines belong to the NER pathway, the recombination repair pathway, and/or the error-prone postreplication repair pathway (11, 17, 32). A number of genes cannot be attributed to one of the known repair pathways (7, 11). The molecular mechanisms of mammalian ICL repair are still unknown, but they are probably similar to those of S. cerevisiae and E. coli ICL repair. Incision of the ICLs is supposed to be performed by some of the NER proteins, as mutations in ERCC1 result in a decrease in the incision of ICLs and reduced repair-associated replication (35). In contrast to the situation in S. cerevisiae, where no significant differences among NER mutants are found, most mammalian NER-deficient cells are only moderately sensitive to ICL agents (11). However, ERCC1/XPF mutants are among the most MMC-sensitive cell lines. Therefore, in addition to their role in NER, ERCC1 and XPF are likely to be involved in the incision of ICLs or subsequent recombination, independent of the other NER genes. The sensitivity of the other NER mutants might be caused solely by a defect in the repair of monoadducts. Hamster cell lines with an MMC sensitivity similar to that of ERCC1 mutants are irs1 and irs1SF (11). The genes mutated in these cell lines are XRCC2 and XRCC3, which are paralogs of the recombination repair gene RAD51 and are important for chromosomal stability and DSB repair (12, 30, 37, 46, 56, 57). The mechanism by which these genes work is still unknown, but they could have an important role in recombination repair of ICLs.
Another group of genes involved in the response to ICL agents consists of the Fanconi anemia (FANC) genes. FANC is characterized by developmental abnormalities, pancytopenia of blood cells, and a predisposition to cancer (16). FANC cells are sensitive to ICL agents and to oxidative DNA damage and show cell cycle abnormalities (7). To date, eight complementation groups have been found, and three genes have been cloned, FANCA, FANCC, and FANCG (15, 19, 29, 38, 53). FANCC knockout mice are very sensitive to MMC, although otherwise, their phenotype is very mild (8, 10, 60). The possibility that SNM1 is one of the remaining FANC genes cannot be excluded.
Role of Snm1 in ICL repair.
The results for S. cerevisiae SNM1 show that Snm1 is not required for all ICL repair. S. cerevisiae snm1 cells show synergism with mutants from both the translesion and the recombination repair pathways with regard to sensitivity to ICL agents, suggesting that Snm1 functions in an alternative pathway (24, 51). Moreover, snm1 cells are capable of repairing an ICL on a plasmid, suggesting that the activity of Snm1 may be related to the modulation of chromosomal structure (39). The sensitivity of snm1 mutants to all ICL agents tested suggests that Snm1 is involved in an ICL agent-independent step and therefore probably after the formation of ICLs (6, 25, 49). This notion is consistent with the fact that snm1 mutants are capable of creating DSBs after treatment with 8-methoxypsoralen plus UVA light, in contrast to NER mutants, which are not able to incise the DNA near the ICL (40, 42). snm1 mutants are, however, epistatic with NER mutants, suggesting that they play a role in this repair pathway (24, 51). Snm1 may be involved in restoring the continuity of the DNA, because snm1 cells are not able to repair the DSBs formed (40). Alternatively, Snm1 could also play a more indirect role, in the regulation of ICL repair.
mSNM1 knockout ES cells are specifically sensitive to MMC but not to cisplatin, melphalan, or 8-methoxypsoralen plus UVA light, in contrast to S. cerevisiae snm1 cells. Therefore, mammalian SNM1 is probably not essential for a general ICL repair pathway. Mammalian cells could have different proteins for the recognition and repair of different ICLs, while yeast cells could depend on a single protein. Alternatively, some translesion repair pathways in mammals could be more efficient than those in yeast, taking over most ICL repair in mSNM1 knockout ES cells but being insufficient to repair MMC-induced ICLs. We cannot exclude the possibility of the presence in our knockout ES cells of some mutated mSnm1 that could fulfill some of the functions of the protein and thereby mitigate the phenotype. However, this notion is not a likely explanation of the mild phenotype, because two highly conserved motifs were deleted. As the sequence of the protein does not give clues to its function, we can only speculate about its role. mSnm1 could be involved in the activation of MMC, decreasing the number of ICLs or other damage caused by this agent. Alternatively, it could be responsible for the recognition of MMC-induced ICLs and other structurally related ICLs. These explanations would be inconsistent with the yeast snm1 phenotype, although a direct comparison is not possible because of the existence of multiple mammalian SNM1 homologs and because MMC has not been tested in S. cerevisiae. It is also possible that Snm1 is involved in a regulatory pathway, influencing the response of the cell to the damage caused by MMC. The most attractive assumption for the function of mSnm1 is a direct role in the repair of ICLs, as suggested by the results for its homolog in S. cerevisiae.
Induction of mouse Rad51 foci is normal in mSNM1−/− cells.
In S. cerevisiae, snm1 and rad52 mutants are synergistic with respect to treatment with ICL agents (24, 51). The RAD52 group of recombination repair genes, of which the main members are RAD51, RAD52, and RAD54, is also involved in ICL repair in mammals (17, 33, 44). On treatment with MMC, mouse Rad51 forms nuclear foci (20). Immunofluorescence on mSNM1 knockout ES cells after MMC treatment showed normal focus formation of Rad51 (data not shown). In contrast, XRCC3 and mouse RAD54 (mRAD54) mutants do not show Rad51 foci after treatment with cisplatin and MMC, respectively (3, 55). These results suggest either that Snm1 is required for the ICL recombination repair pathway after the involvement of Rad51 or that it plays a role independent of the recombination repair pathway.
Redundancy of mammalian Snm1 function.
The relatively mild phenotype of mSNM1−/− ES cells and mice is reminiscent of the phenotype found for mRAD54−/− cells and mice, which are also about twofold more sensitive to MMC than wild-type cells and mice (17, 18). This mild phenotype can probably be attributed in part to the existence of parallel pathways for the repair of ICLs. In addition, known homologs for both mSNM1 and mRAD54 could take over part of the function (Fig. 1) (26). In fact, redundancy is a phenomenon that is quite common in mammalian DNA damage repair pathways. The NER gene RAD23, the translesion repair gene RAD6, and the recombination repair gene RAD51 have several mammalian homologs (34, 41, 57). These duplications provide cells with the possibility of functional differentiation in the repair of different types of damage, in different phases of the cell cycle, or in different cell types. Moreover, the cell can fall back on the alternative repair pathway in case of dysfunction of one of the proteins involved, a strategy which may prevent inappropriate repair and cell transformation. It will be important to analyze the other SNM1 homologs and to look at the effects of ICL agents on double knockout cells. Furthermore, study of cells with mutations in SNM1 and other genes functioning in ICL repair, such as mRAD54, will yield information on the relative importance of the different ICL repair pathways and the level of redundancy between different pathways. Nevertheless, the sensitivity of mSNM1 knockout cells to MMC shows that there is no complete redundancy in mammalian cells. Both the sequence conservation and the functional conservation with yeast Snm1 underline the significance of SNM1 in ICL repair.
ACKNOWLEDGMENTS
We thank W. Vermeulen for performing the microinjection experiment.
This work was supported by grants from The Netherlands Organization for Scientific Research (NWO) and the Dutch Cancer Society. R.K. is a fellow of The Royal Netherlands Academy of Arts and Sciences.
REFERENCES
- 1.Alaoui-Jamali M, Loubaba B B, Robyn S, Tapiero H, Batist G. Effect of DNA-repair-enzyme modulators on cytotoxicity of l-phenylalanine mustard and cis-diamminedichloroplatinum (II) in mammary carcinoma cells resistant to alkylating drugs. Cancer Chemother Pharmacol. 1994;34:153–158. doi: 10.1007/BF00685933. [DOI] [PubMed] [Google Scholar]
- 2.Averbeck D, Dardalhon M, Magana-Schwencke N, Meira L B, Meniel V, Boiteux S, Sage E. New aspects of the repair and genotoxicity of psoralen photoinduced lesions in DNA. J Photochem Photobiol B. 1992;14:47–63. doi: 10.1016/1011-1344(92)85082-6. [DOI] [PubMed] [Google Scholar]
- 3.Bishop D K, Ear U, Bhattacharyya A, Calderone C, Beckett M, Weichselbaum R R, Shinohara A. Xrcc3 is required for assembly of Rad51 complexes in vivo. J Biol Chem. 1998;273:21482–21488. doi: 10.1074/jbc.273.34.21482. [DOI] [PubMed] [Google Scholar]
- 4.Bockmuhl U, Petersen S, Schmidt S, Wolf G, Jahnke V, Dietel M, Petersen I. Patterns of chromosomal alterations in metastasizing and nonmetastasizing primary head and neck carcinomas. Cancer Res. 1997;57:5213–5216. [PubMed] [Google Scholar]
- 5.Bramson J, McQuillan A, Aubin R, Alaoui-Jamali M, Batist G, Christodoulopoulos G, Panasci L C. Nitrogen mustard drug resistant B-cell chronic lymphocytic leukemia as an in vivo model for crosslinking agent resistance. Mutat Res. 1995;336:269–278. doi: 10.1016/0921-8777(94)00063-c. [DOI] [PubMed] [Google Scholar]
- 6.Brendel M, Ruhland A. Relationships between functionality and genetic toxicology of selected DNA-damaging agents. Mutat Res. 1984;133:51–85. doi: 10.1016/0165-1110(84)90003-4. [DOI] [PubMed] [Google Scholar]
- 7.Buchwald M, Moustacchi E. Is Fanconi anemia caused by a defect in the processing of DNA damage? Mutat Res. 1998;408:75–90. doi: 10.1016/s0921-8777(98)00024-x. [DOI] [PubMed] [Google Scholar]
- 8.Carreau M, Gan O I, Liu L, Doedens M, McKerlie C, Dick J E, Buchwald M. Bone marrow failure in the Fanconi anemia group C mouse model after DNA damage. Blood. 1998;91:2737–2744. [PubMed] [Google Scholar]
- 9.Cassier-Chauvat C, Moustacchi E. Allelism between pso1-1 and rev3-1 mutants and between pso2-1 and snm1 mutants in Saccharomyces cerevisiae. Curr Genet. 1988;13:37–40. doi: 10.1007/BF00365754. [DOI] [PubMed] [Google Scholar]
- 10.Chen M, Tomkins D J, Auerbach W, McKerlie C, Youssoufian H, Liu L, Gan O, Carreau M, Auerbach A, Groves T, Guidos C J, Freedman M H, Cross J, Percy D H, Dick J E, Joyner A L, Buchwald M. Inactivation of Fac in mice produces inducible chromosomal instability and reduced fertility reminiscent of Fanconi anaemia. Nat Genet. 1996;12:448–451. doi: 10.1038/ng0496-448. [DOI] [PubMed] [Google Scholar]
- 11.Collins A R. Mutant rodent cell lines sensitive to ultraviolet light, ionizing radiation and cross-linking agents: a comprehensive survey of genetic and biochemical characteristics. Mutat Res. 1993;293:99–118. doi: 10.1016/0921-8777(93)90062-l. [DOI] [PubMed] [Google Scholar]
- 12.Cui X, Brenneman M, Meyne J, Oshimura M, Goodwin E H, Chen D J. The XRCC2 and XRCC3 repair genes are required for chromosome stability in mammalian cells. Mutat Res. 1999;434:75–88. doi: 10.1016/s0921-8777(99)00010-5. [DOI] [PubMed] [Google Scholar]
- 13.Dardalhon M, Averbeck D. Pulsed-field gel electrophoresis analysis of the repair of psoralen plus UVA induced DNA photoadducts in Saccharomyces cerevisiae. Mutat Res. 1995;336:49–60. doi: 10.1016/0921-8777(94)00037-7. [DOI] [PubMed] [Google Scholar]
- 14.de Boer J, Hoeijmakers J H. Cancer from the outside, aging from the inside: mouse models to study the consequences of defective nucleotide excision repair. Biochimie. 1999;81:127–137. doi: 10.1016/s0300-9084(99)80045-5. [DOI] [PubMed] [Google Scholar]
- 15.de Winter J P, Waisfisz Q, Rooimans M A, van Berkel C G, Bosnoyan-Collins L, Alon N, Carreau M, Bender O, Demuth I, Schindler D, Pronk J C, Arwert F, Hoehn H, Digweed M, Buchwald M, Joenje H. The Fanconi anaemia group G gene FANCG is identical with XRCC9. Nat Genet. 1998;20:281–283. doi: 10.1038/3093. [DOI] [PubMed] [Google Scholar]
- 16.Digweed M, Sperling K. Molecular analysis of Fanconi anaemia. Bioessays. 1996;18:579–585. doi: 10.1002/bies.950180709. [DOI] [PubMed] [Google Scholar]
- 17.Essers J, Hendriks R W, Swagemakers S M A, Troelstra C, de Wit J, Bootsma D, Hoeijmakers J H J, Kanaar R. Disruption of mouse RAD54 reduces ionizing radiation resistance and homologous recombination. Cell. 1997;89:195–204. doi: 10.1016/s0092-8674(00)80199-3. [DOI] [PubMed] [Google Scholar]
- 18.Essers J, van Steeg H, de Wit J, Swagemakers S M A, Vermeij M, Hoeijmakers J H J, Kanaar R. Homologous and non-homologous recombination differentially affect DNA damage repair in mice. EMBO J. 2000;19:1703–1710. doi: 10.1093/emboj/19.7.1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fanconi Anaemia/Breast Cancer Consortium. Positional cloning of the Fanconi anaemia group A gene. Nat Genet. 1996;14:324–328. doi: 10.1038/ng1196-324. [DOI] [PubMed] [Google Scholar]
- 20.Haaf T, Golub E I, Reddy G, Radding C M, Ward D C. Nuclear foci of mammalian Rad51 recombination protein in somatic cells after DNA damage and its localization in synaptonemal complexes. Proc Natl Acad Sci USA. 1995;92:2298–2302. doi: 10.1073/pnas.92.6.2298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Haase E, Riehl D, Mack M, Brendel M. Molecular cloning of SNM1, a yeast gene responsible for a specific step in the repair of cross-linked DNA. Mol Gen Genet. 1989;218:64–71. doi: 10.1007/BF00330566. [DOI] [PubMed] [Google Scholar]
- 22.Henriques J A, Brendel M. The role of PSO and SNM genes in DNA repair of the yeast Saccharomyces cerevisiae. Curr Genet. 1990;18:387–393. doi: 10.1007/BF00309906. [DOI] [PubMed] [Google Scholar]
- 23.Henriques J A, Brozmanova J, Brendel M. Role of PSO genes in the repair of photoinduced interstrand cross-links and photooxidative damage in the DNA of the yeast Saccharomyces cerevisiae. J Photochem Photobiol B. 1997;39:185–196. doi: 10.1016/s1011-1344(97)00020-1. [DOI] [PubMed] [Google Scholar]
- 24.Henriques J A, Moustacchi E. Interactions between mutations for sensitivity to psoralen photoaddition (pso) and to radiation (rad) in Saccharomyces cerevisiae. J Bacteriol. 1981;148:248–256. doi: 10.1128/jb.148.1.248-256.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Henriques J A, Moustacchi E. Isolation and characterization of pso mutants sensitive to photo-addition of psoralen derivatives in Saccharomyces cerevisiae. Genetics. 1980;95:273–288. doi: 10.1093/genetics/95.2.273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hiramoto T, Nakanishi T, Sumiyoshi T, Fukuda T, Matsuura S, Tauchi H, Komatsu K, Shibasaki Y, Inui H, Watatani M, Yasutomi M, Sumii K, Kajiyama G, Kamada N, Miyagawa K, Kamiya K. Mutations of a novel human RAD54 homologue, RAD54B, in primary cancer. Oncogene. 1999;18:3422–3426. doi: 10.1038/sj.onc.1202691. [DOI] [PubMed] [Google Scholar]
- 27.Hoeijmakers J H J. Use of microneedle injection to study DNA repair in mammalian cells. In: Friedberg E C, Hanawalt P C, editors. A laboratory manual of research procedures. Vol. 3. New York, N.Y: Marcel Dekker, Inc.; 1988. pp. 133–150. [Google Scholar]
- 28.Jachymczyk W J, von Borstel R C, Mowat M R, Hastings P J. Repair of interstrand cross-links in DNA of Saccharomyces cerevisiae requires two systems for DNA repair: the RAD3 system and the RAD51 system. Mol Gen Genet. 1981;182:196–205. doi: 10.1007/BF00269658. [DOI] [PubMed] [Google Scholar]
- 29.Joenje H, Oostra A B, Wijker M, di Summa F M, van Berkel C G, Rooimans M A, Ebell W, van Weel M, Pronk J C, Buchwald M, Arwert F. Evidence for at least eight Fanconi anemia genes. Am J Hum Genet. 1997;61:940–944. doi: 10.1086/514881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Johnson R D, Liu N, Jasin M. Mammalian XRCC2 promotes the repair of DNA double-strand breaks by homologous recombination. Nature. 1999;401:397–399. doi: 10.1038/43932. [DOI] [PubMed] [Google Scholar]
- 31.Johnson S W, Perez R P, Godwin A K, Yeung A T, Handel L M, Ozols R F, Hamilton T C. Role of platinum-DNA adduct formation and removal in cisplatin resistance in human ovarian cancer cell lines. Biochem Pharmacol. 1994;47:689–697. doi: 10.1016/0006-2952(94)90132-5. [DOI] [PubMed] [Google Scholar]
- 32.Kaiser P, Mansour H A, Greeten T, Auer B, Schweiger M, Schneider R. The human ubiquitin-conjugating enzyme UbcH1 is involved in the repair of UV-damaged, alkylated and cross-linked DNA. FEBS Lett. 1994;350:1–4. doi: 10.1016/0014-5793(94)00656-3. [DOI] [PubMed] [Google Scholar]
- 33.Kanaar R, Hoeijmakers J H J, van Gent D C. Molecular mechanisms of DNA double-strand break repair. Trends Cell Biol. 1998;8:483–489. doi: 10.1016/s0962-8924(98)01383-x. [DOI] [PubMed] [Google Scholar]
- 34.Koken M H, Reynolds P, Jaspers-Dekker I, Prakash L, Prakash S, Bootsma D, Hoeijmakers J H. Structural and functional conservation of two human homologs of the yeast DNA repair gene RAD6. Proc Natl Acad Sci USA. 1991;88:8865–8869. doi: 10.1073/pnas.88.20.8865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Larminat F, Bohr V A. Role of the human ERCC-1 gene in gene-specific repair of cisplatin-induced DNA damage. Nucleic Acids Res. 1994;22:3005–3010. doi: 10.1093/nar/22.15.3005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lawley P D, Phillips D H. DNA adducts from chemotherapeutic agents. Mutat Res. 1996;355:13–40. doi: 10.1016/0027-5107(96)00020-6. [DOI] [PubMed] [Google Scholar]
- 37.Liu N, Lamerdin J E, Tebbs R S, Schild D, Tucker J D, Shen M R, Brookman K W, Siciliano M J, Walter C A, Fan W, Narayana L S, Zhou Z Q, Adamson A W, Sorensen K J, Chen D J, Jones N J, Thompson L H. XRCC2 and XRCC3, new human Rad51-family members, promote chromosome stability and protect against DNA cross-links and other damages. Mol Cell. 1998;1:783–793. doi: 10.1016/s1097-2765(00)80078-7. [DOI] [PubMed] [Google Scholar]
- 38.Lo Ten Foe J R, Rooimans M A, Bosnoyan-Collins L, Alon N, Wijker M, Parker L, Lightfoot J, Carreau M, Callen D F, Savoia A, Cheng N C, van Berkel C G, Strunk M H, Gille J J, Pals G, Kruyt F A, Pronk J C, Arwert F, Buchwald M, Joenje H. Expression cloning of a cDNA for the major Fanconi anaemia gene, FAA. Nat Genet. 1996;14:320–323. doi: 10.1038/ng1196-320. [DOI] [PubMed] [Google Scholar]
- 39.Magana-Schwencke N, Averbeck D. Repair of exogenous (plasmid) DNA damaged by photoaddition of 8-methoxypsoralen in the yeast Saccharomyces cerevisiae. Mutat Res. 1991;251:123–131. doi: 10.1016/0027-5107(91)90222-a. [DOI] [PubMed] [Google Scholar]
- 40.Magana-Schwencke N, Henriques J A, Chanet R, Moustacchi E. The fate of 8-methoxypsoralen photoinduced crosslinks in nuclear and mitochondrial yeast DNA: comparison of wild-type and repair-deficient strains. Proc Natl Acad Sci USA. 1982;79:1722–1726. doi: 10.1073/pnas.79.6.1722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Masutani C, Sugasawa K, Yanagisawa J, Sonoyama T, Ui M, Enomoto T, Takio K, Tanaka K, van der Spek P J, Bootsma D, et al. Purification and cloning of a nucleotide excision repair complex involving the xeroderma pigmentosum group C protein and a human homologue of yeast RAD23. EMBO J. 1994;13:1831–1843. doi: 10.1002/j.1460-2075.1994.tb06452.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Meniel V, Magana-Schwencke N, Averbeck D. Preferential repair in Saccharomyces cerevisiae rad mutants after induction of interstrand cross-links by 8-methoxypsoralen plus UVA. Mutagenesis. 1995;10:543–548. doi: 10.1093/mutage/10.6.543. [DOI] [PubMed] [Google Scholar]
- 43.Nagase T, Miyajima N, Tanaka A, Sazuka T, Seki N, Sato S, Tabata S, Ishikawa K, Kawarabayasi Y, Kotani H, et al. Prediction of the coding sequences of unidentified human genes. III. The coding sequences of 40 new genes (KIAA0081-KIAA0120) deduced by analysis of cDNA clones from human cell line KG-1 (supplement) DNA Res. 1995;2:51–59. doi: 10.1093/dnares/2.1.51. [DOI] [PubMed] [Google Scholar]
- 44.Paques F, Haber J E. Multiple pathways of recombination induced by double-strand breaks in Saccharomyces cerevisiae. Microbiol Mol Biol Rev. 1999;63:349–404. doi: 10.1128/mmbr.63.2.349-404.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Patterson G H, Knobel S M, Sharif W D, Kain S R, Piston D W. Use of the green fluorescent protein and its mutants in quantitative fluorescence microscopy. Biophys J. 1997;73:2782–2790. doi: 10.1016/S0006-3495(97)78307-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pierce A J, Johnson R D, Thompson L H, Jasin M. XRCC3 promotes homology-directed repair of DNA damage in mammalian cells. Genes Dev. 1999;13:2633–2638. doi: 10.1101/gad.13.20.2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Richter D, Niegemann E, Brendel M. Molecular structure of the DNA cross-link repair gene SNM1 (PSO2) of the yeast Saccharomyces cerevisiae. Mol Gen Genet. 1992;231:194–200. doi: 10.1007/BF00279791. [DOI] [PubMed] [Google Scholar]
- 48.Ruhland A, Haase E, Siede W, Brendel M. Isolation of yeast mutants sensitive to the bifunctional alkylating agent nitrogen mustard. Mol Gen Genet. 1981;181:346–351. doi: 10.1007/BF00425609. [DOI] [PubMed] [Google Scholar]
- 49.Ruhland A, Kircher M, Wilborn F, Brendel M. A yeast mutant specifically sensitive to bifunctional alkylation. Mutat Res. 1981;91:457–462. doi: 10.1016/0165-7992(81)90052-x. [DOI] [PubMed] [Google Scholar]
- 50.Scott B R, Pathak M A, Mohn G R. Molecular and genetic basis of furocoumarin reactions. Mutat Res. 1976;39:29–74. doi: 10.1016/0165-1110(76)90012-9. [DOI] [PubMed] [Google Scholar]
- 51.Siede W, Brendel M. Interactions among genes controlling sensitivity to radiation (RAD) and to alkylation by nitrogen mustard (SNM) in yeast. Curr Genet. 1982;5:33–38. doi: 10.1007/BF00445738. [DOI] [PubMed] [Google Scholar]
- 52.Simon R, Burger H, Brinkschmidt C, Bocker W, Hertle L, Terpe H J. Chromosomal aberrations associated with invasion in papillary superficial bladder cancer. J Pathol. 1998;185:345–351. doi: 10.1002/(SICI)1096-9896(199808)185:4<345::AID-PATH109>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- 53.Strathdee C A, Gavish H, Shannon W R, Buchwald M. Cloning of cDNAs for Fanconi's anaemia by functional complementation. Nature. 1992;356:763–767. doi: 10.1038/356763a0. [DOI] [PubMed] [Google Scholar]
- 54.Swagemakers S M A, Essers J, de Wit J, Hoeijmakers J H J, Kanaar R. The human Rad54 recombinational DNA repair protein is a double-stranded DNA-dependent ATPase. J Biol Chem. 1998;273:28292–28297. doi: 10.1074/jbc.273.43.28292. [DOI] [PubMed] [Google Scholar]
- 55.Tan T L, Essers J, Citterio E, Swagemakers S M, de Wit J, Benson F E, Hoeijmakers J H, Kanaar R. Mouse Rad54 affects DNA conformation and DNA-damage-induced Rad51 foci formation. Curr Biol. 1999;9:325–328. doi: 10.1016/s0960-9822(99)80142-0. [DOI] [PubMed] [Google Scholar]
- 56.Tebbs R S, Zhao Y, Tucker J D, Scheerer J B, Siciliano M J, Hwang M, Liu N, Legerski R J, Thompson L H. Correction of chromosomal instability and sensitivity to diverse mutagens by a cloned cDNA of the XRCC3 DNA repair gene. Proc Natl Acad Sci USA. 1995;92:6354–6358. doi: 10.1073/pnas.92.14.6354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Thacker J. A surfeit of RAD51-like genes? Trends Genet. 1999;15:166–168. doi: 10.1016/s0168-9525(99)01733-3. [DOI] [PubMed] [Google Scholar]
- 58.Tong C Y, Ng H K, Pang J C, Hui A B, Ko H C, Lee J C. Molecular genetic analysis of non-astrocytic gliomas. Histopathology. 1999;34:331–341. doi: 10.1046/j.1365-2559.1999.00603.x. [DOI] [PubMed] [Google Scholar]
- 59.Van Houten B. Nucleotide excision repair in Escherichia coli. Microbiol Rev. 1990;54:18–51. doi: 10.1128/mr.54.1.18-51.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Whitney M A, Royle G, Low M J, Kelly M A, Axthelm M K, Reifsteck C, Olson S, Braun R E, Heinrich M C, Rathbun R K, Bagby G C, Grompe M. Germ cell defects and hematopoietic hypersensitivity to gamma-interferon in mice with a targeted disruption of the Fanconi anemia C gene. Blood. 1996;88:49–58. [PubMed] [Google Scholar]
- 61.Wolter R, Siede W, Brendel M. Regulation of SNM1, an inducible Saccharomyces cerevisiae gene required for repair of DNA cross-links. Mol Gen Genet. 1996;250:162–168. doi: 10.1007/BF02174175. [DOI] [PubMed] [Google Scholar]