Abstract
Sets of oligonucleotide primers were designed according to the sequences of the open reading frames (ORFs) ORF1 and ORF2 of the prototype nonpathogenic PK-15 strain of porcine circovirus (PCV) type 1 (PCV-1). By the PCR performed with the various primer sets, genomic DNA or RNA from other bacterial or viral pathogens of the respiratory tracts of pigs could not be amplified. A positive amplification reaction could be visualized with DNA extracted from a viral suspension containing as few as 10 viral particles per ml. No DNA fragment could be amplified from lysates of continuous porcine cell lines (PT, ST, and PFT cells) known to be negative for PCV. When tested with clinical samples from pigs, the results of the single PCR method showed nearly 93% (13 of 14 samples) correlation with histopathological and immunohistochemical findings. Interestingly, subclinical PCV infections could be detected by single PCR with clinical samples that have been submitted from animals with irrelevant cases of respiratory and/or enteric problems. On the basis of the nucleotide sequences of PCV strains (PCV-2) recently associated with outbreaks of postweaning multisystemic wasting syndrome (PWMS) in Quebec, Canada, pig farms, other primers were designed from the PCV-1 genome, and these primers failed to amplify genomic fragments specific to the ORF1 or ORF2 genes of clinical isolates associated with PWMS but amplified DNA from the PCV-1 strain. Two rapid multiplex PCR (mPCR) methods have been developed to distinguish between both genotypes of PCV. By those two mPCR methods, (i) species-specific primer pairs were used to amplify a DNA fragment of 488 bp specific for the ORF2 genes of both genotypes, whereas a 375-bp fragment was amplified from the ORF1 gene of the PCV-1 strain only, or (ii) species-specific primer pairs were used to amplify a DNA fragment of 646 bp specific for the ORF1 genes of both genotypes, whereas a 425-bp fragment was amplified from the ORF2 gene of the PCV-1 strain only. By both mPCR methods, a PCV-2 infection was demonstrated in tissues of 94.2% (33 of 35) of the sick pigs tested, in agreement with previous findings showing the close association of this new genotype of PCV with outbreaks of PMWS in Europe and North America. On the other hand, a PCV-1 infection was confirmed in only 5.7% (2 of 35) of the pigs, and confirmation of a mixed infection with PCV-2 was obtained by a single PCR with PCV-2-specific primers.
Postweaning multisystemic wasting syndrome (PMWS) is an apparently new disease of 5- to 8-week-old pigs being diagnosed with increasing frequency in Canada (4, 11, 15), the United States (2, 5, 17), and Europe (2, 18, 29). Clinically, the disease is characterized by progressive weight loss, respiratory signs (tachypnea, dyspnea), and jaundice (2, 11, 25). Consistent macroscopic and histological lesions include lymphocytic to granulomatous interstitial pneumonia, lymphadenopathy, and, less frequently, lymphocytic to granulomatous hepatitis and nephritis. Distinctively, multinucleated giant cells and basophilic cytoplasmic inclusion bodies are observed mostly in lymph nodes, tonsils, and Peyer's patches of the ileum (2, 11, 17, 25). Previous investigators have reported morbidity rates ranging from 5 to 50% in affected herds, with close to 100% mortality in sick piglets (25). There is growing evidence that a porcine circovirus (PCV) is associated with PMWS (2, 5, 11, 17, 25), and quite recently, a Canadian group has succeeded in reproducing typical signs and lesions of PMWS with the supernatant of PCV-infected cell cultures (12). Interestingly, the PCV strain isolated from animals with PMWS has been shown, by its pathogenesis and genome sequence, to differ from the nonpathogenic PCV strain (14, 22, 24). The latter was first detected as a noncytopathic contaminant of a continuous pig kidney (PK-15) cell line (33) that failed to reproduce clinical disorders or lesions after being inoculated into healthy pigs (1, 30).
The PK-15 PCV strain (a PCV type 1 [PCV-1] strain) was characterized as a small nonenveloped virus that contains a single-stranded circular DNA genome of 1.76 kb and was classified in a newly recognized virus family, the Circoviridae (23), along with chicken anemia virus (34), beak-and-feather disease virus of psittacine birds (27), and several plant viruses (3, 16, 28). No recognized DNA sequence homologies or common antigenic determinants exist between PCV and the currently recognized circoviruses (25). Recently, it was suggested that the family members be reclassified into three groups, with beak-and-feather disease virus and PCV remaining members of the Circoviridae family and chicken anemia virus and plant circoviruses being grouped into new distinct families (26).
Until recently, several methods have been used for the detection of PCV without discriminating among the different strains: virus isolation in cell cultures, electron microscopy, in situ hybridization, and immunohistochemical staining with hyperimmune serum (2, 11, 17, 25). Serological surveys by indirect immunofluorescence, immunoperoxidase, or enzyme-linked immunosorbent assays indicate that antibodies to PCV-1 are very common in North American and European pig herds (2, 10, 14, 18, 23, 32). Type-specific serological tests are needed to study the prevalence of the PMWS-associated PCV strain. Recently, the use of PCR for the detection of the PCV genome in tissues of infected pigs has been described (12, 25) and has enabled comparative sequencing studies of the genomes of both nonpathogenic (PCV-1) and pathogenic or PMWS-associated (PCV-2) strains. Overall, the genome of PCV-1 and those of PMWS-associated PCV-2 strains isolated in the United States, Canada, and France have been found to share only 68% sequence identity, with their first halves (nucleotide [nt] positions 1 to 900) having over 82% sequence homology and their second halves (nt 901 to 1768 or 1759) having only 62% homology (14, 22, 25). The combined use of PCR for amplification of the entire genome and determination of the EcoRI digestion patterns also allowed discrimination of both types of PCV strains (22, 25). In this report, we describe the development and application of a rapid multiplex PCR (mPCR) assay with primer sets capable of detecting and typing PCV in clinical respiratory samples.
MATERIALS AND METHODS
Cells and PCV reference strains.
PK-15 cells (PK-15 is a continuous pig kidney cell line [ATCC CCL33] persistently infected with PCV-1) were kindly provided to us by A. Afshar, Animal Diseases Research Institute, Agriculture Canada, Nepean, Ontario, Canada. Cells were grown as monolayers in 75- or 150-cm2 tissue culture flasks in minimum essential medium (MEM) supplemented with 5% fetal bovine serum, 1 mM glutamine, 1% sodium pyruvate, 100 U of penicillin per ml, and 100 μg of streptomycin per ml. Virus growth was monitored by immunofluorescence (25) with a rabbit hyperimmune serum produced to purified PCV-1 by immunization protocols described elsewhere (6, 7). For the purposes of virus and viral DNA purification, viral yield was improved by treating the cells with 300 mM glucosamine (31). The virus was harvested by freezing and thawing infected cells three times and removing cellular debris by centrifugation at 5,000 × g for 15 min. Following one extraction step with an equal volume of trichlorotrifluoroethane (Freon; Fisher Scientific, Dorval, Quebec, Canada) to remove lipoproteins and cellular membranes, virus was pelleted by differential ultracentrifugation at 100,000 × g for 3 h through a cushion of a 30% (wt/vol) sucrose solution. Concentrated virus was then purified by CsCl isopycnic gradient ultracentrifugation as described previously (7). After an overnight centrifugation at 100,000 × g, virus band was collected and the material was checked by negatively stained electron microscopy (8).
The IAF-614 and IAF-4370 strains of PCV-2 were isolated from the lungs of 5- to 9-week-old pigs in Quebec with clinical signs and lesions compatible with PMWS. Both strains were propagated on PK-15A cells (PK-15 A is a PK-15 cell line shown to be free of PCV) obtained from the National Veterinary Service Laboratory, U.S. Department of Agriculture, Ames, Iowa. Three other cell lines, PT and ST-148 (swine testicle) and PFT (porcine fallopian tube) (7), were used as negative controls.
Viruses and bacteria used in specificity tests.
The porcine mycoplasmas and walled bacteria used in the specificity tests are listed in Table 1. Mycoplasma hyopneumoniae, Mycoplasma hyorhinis, and Mycoplasma fermentens were cultivated in modified Friis medium (13). Gram-positive and gram-negative bacteria commonly associated with respiratory disorders in pigs were grown in nutrient broths and were kindly provided to us by S. Messier, Faculty of Veterinary Medicine, University of Montreal, St-Hyacinthe, Quebec, Canada. Pig viruses used as controls included the IAF-Klop strain of porcine reproductive and respiratory syndrome virus (PRRSV), the Purdue strain of porcine transmissible gastroenteritis virus (ATCC VR763), the IAF-Q890 strain of porcine encephalomyocarditis virus, the NADL-2 strain of porcine parvovirus, and the A/Sw/Quebec/5393/91 (swQC-91) strain of swine influenza virus. The origins, cultivation, and purification procedures for these reference viruses have been described elsewhere (6–8, 20).
TABLE 1.
Bacterial and viral strains used in the PCR specificity tests
| Species | Strain | PCR resultsa |
|---|---|---|
| Mycoplasma hyopneumoniae | ATCCb | − |
| Mycoplasma hyorhinis | ATCC | − |
| Mycoplasma flocculare | ATCC | − |
| Mycoplasma arginini | ATCC | − |
| Acholeplasma laidlawii | ATCC | − |
| Escherichia coli | Field strain | − |
| Haemophilus parasuis | Field strain | − |
| Streptococcus suis | Field strain | − |
| Actinobacillus suis | Field strain | − |
| Actinobacillus pleuropneumoniae | Serotype 1 | − |
| Actinobacillus pleuropneumoniae | Serotype 5 | − |
| Bordetella bronchiseptica | Field strain | − |
| Pasteurella multocida | Field strain | − |
| PRRSV | IAF-Klop | − |
| Transmissible gastroenteritis virus | Purdue | − |
| Encephalomyocarditis virus | IAF-Q890 | − |
| Porcine parvovirus | NADL-2 | − |
| Porcine influenza virus | A/Sw/Quebec/5393/91 | − |
| Porcine circovirus | PK-15 | + |
+, amplification; −, no amplification.
ATCC, American Type Culture Collection.
Clinical specimens or samples.
Fresh, as well as frozen, specimens of lungs, lymph nodes, spleens, and tonsils of 5- to 12-week-old pigs suffering from respiratory problems accompanied by progressive weight loss were obtained from regional Animal Pathology Laboratories of the Ministry of Agriculture in Quebec, as well as from the Veterinary Faculty Diagnostic Laboratory, University of Montreal, St-Hyacinthe, Quebec, Canada. Affected pigs originated from farrow-to-finish operations in southern Quebec. In four of these animals, previously forwarded to the Department of Pathology, Western College of Veterinary Medicine, University of Saskatchewan, Saskatoon, Saskatchewan, Canada, the presence of PCV antigen in the lungs, spleens, and lymph nodes was confirmed by immunoperoxidase staining with rabbit hyperimmune serum (11).
Nucleic acid extraction.
Viral genomic DNA was extracted either from 400 μl of lysates of PK-15 cells persistently infected with PCV-1, CsCl gradient-purified viral preparation, or 100 mg of fresh or frozen clinical specimens from dyspneic piglets with a commercial DNA extraction kit (Tripure; Boehringer Mannheim, Laval, Quebec, Canada) according to the manufacturer's directions. To eliminate any contamination in the harvesting process, lung tissues collected from specific-pathogen-free pigs were processed simultaneously. Following extraction, the pelleted DNA was resuspended in 300 μl of 8 mM NaOH with 0.1 M HEPES and was kept at −20°C until use.
Single PCR.
In preliminary experiments, five oligonucleotide primers suitable for single PCR were selected from a published PCV-1 sequence (GenBank accession no. U49186) (19, 23). Primer pairs were selected in open reading frame (ORF) ORF1 regions for which 86 to 100% nt sequence identities have been identified between PCV-1 and PCV-2 strains. Different combinations were evaluated in order to obtain amplification of DNA fragments corresponding to the ORF1 genes of both genotypes ranging from 375 to 865 bp in length. All primers chosen were 16- to 21-mers and had G+C contents of less than or equal to 55%. The sequences of the oligonucleotide primers used for single PCR and their positions on the virus genome are described in Table 2. A set of oligonucleotide primers (primers ORF2.PCV2.S4 and ORF2.PCV2.AS4) was also designed in order to permit specific amplification of a DNA fragment of 493 bp in length of the ORF-2 of PCV-2 only (Table 2). Both primers were selected in ORF2 regions showing less than 31 and 36% nt sequence identities between PCV-1 and PCV-2 strains. Amplification was carried out in a 100-μl reaction mixture containing 2.5 μl of PK-15 DNA suspension or purified viral DNA, 20 mM Tris-HCl, 50 mM KCl, 1.5 mM MgCl2 (Gibco-BRL), each deoxynucleoside triphosphate (dNTP) at a concentration of 200 μM, 50 pmol of each primer, and 2.5 U of Taq DNA polymerase (Gibco-BRL). Amplification with a DNA Engine thermocycler (model PT-100 with hot bonnet; MJ Research) consisted of 5 cycles at 94°C for 1 min, 42°C for 1 min, and 72°C for 1.50 min, followed by 30 cycles at 94°C for 1 min, 50°C for 1 min, and 72°C for 1.50 min. The PCR was ended with a final elongation step of 10 min at 72°C. Amplicons were detected by electrophoresing 10-μl aliquots through 2% agarose gels (Boehringer Mannheim) in TAE (0.04 M Tris-acetate [pH 8.5], 0.002 M EDTA) in the presence of ethidium bromide for approximately 30 min at 10 V/cm and photographing the gels under UV illumination.
TABLE 2.
Sequences of oligonucleotides used for single PCR and their localization on the virus genomea
| Primer | Sense | Sequence (5′–3′) | Genomic position | Template | % Homology between PCV-1 and PCV-2 |
|---|---|---|---|---|---|
| ORF1.PCV1.S1 | + | GCCAAGCAAGAAAAGC | 47–63 | ORF1 | 93 |
| ORF1.PCV1.S2 | + | GAGGTGGGTGTTCACCCT | 82–97 | ORF1 | 94 |
| ORF1.PCV1.AS1 | − | CGTTACAGGGAACTGCTC | 445–462 | ORF1 | 89 |
| ORF1.PCV1.AS2 | − | GTGTACAGCTGTCTTCCAATC | 527–547 | ORF1 | 86 |
| ORF1.PCV1.AS5 | − | GTGGATTGTTCTCCAGCAGTC | 892–912 | ORF1 | 86 |
| ORF1.PCV1.AS6 | − | CACACAGTCTCAGTAGATCATCC | 706–728 | ORF1 | 100 |
| ORF2.PCV2.S4 | + | CACGGATATTGTAGTCCTGGT | 1093–1114 | ORF2 | 36 |
| ORF2.PCV2.AS4 | + | CCGCACCTTCGGATATACTGTC | 1565–1586 | ORF2 | 31 |
Nucleotide sequences and localization on the PCV-1 or PCV-2 genome were given in reference to the sequences of the prototype ATCC CCL-33 strain of PCV-1 (GenBank accession no. 49186) (23) and the sequences of the PMWS-associated PCV-2 strain isolated in Manitoba, Canada (GenBank accession no. AF027217) (14).
mPCR.
Two mPCR methods were set up in order to permit detection and differentiation of both genotypes of PCV. The primer pairs used in the mPCR methods, their genomic position, the templates amplified (ORF1 or ORF2), and the sizes of the expected amplified products are depicted in Table 3. In each mPCR method, two primer pairs were used in order to permit simultaneous amplification of DNA fragments of the ORF1 and ORF2 genes of the PCV genome. For the first mPCR method, 2.5 μl of DNA sample was added to 97.5 μl of a reaction mixture containing 20 mM Tris-HCl (pH 8.5), 2 mM MgCl2, 50 mM KCl, each dNTP at a concentration of 400 μM, and 2.5 U of Taq polymerase. The amplification reaction consisted of 35 cycles of 94°C for 1 min, 53°C for 1 min, and 65°C for 3 min. The PCR was ended with a final elongation step of 10 min at 72°C. For the second mPCR method, the reaction mixture was similar to that for the first one except that only 300 μM each dNTP and 1.5 U of Taq polymerase were added. The amplification program was, instead, 33 cycles at 94°C for 1 min, 53°C for 1 min, and 65°C for 5 min.
TABLE 3.
Description and genomic positions of four sets of primers designed for mPCR according to PCV-1 sequencea
| mPCR | Primer | Sequence (5′–3′) | Genomic position | Template | Amplicon (bp) | % Homology between PCV-1 and PCV-2 | Specificity for strain:
|
|
|---|---|---|---|---|---|---|---|---|
| PCV-1 | PCV-2 | |||||||
| 1 | ORF1.PCV1.S3 | CGGAGAGGAAGGTTTGGAAG | 172–191 | ORF1 | 375 | 60 | + | − |
| ORF1.PCV1.AS2 | GTGTACAGCTGTCTTCCAATC | 527–547 | ORF1 | 86 | ||||
| ORF2.PCV1.S | ATGACGTGGCCAAGGAGGCGTTAC | 1723–1700 | ORF2 | 488 | 92 | + | + | |
| ORF2.PCV.1AS | GTGAAGTACCTGGAGTGGTAGG | 896–916 | ORF2 | 91 | ||||
| 2 | ORF1.PCV1.S2 | GAGGTGGGTGTTCACCCT | 82–97 | ORF1 | 646 | 94 | + | + |
| ORF1.PCV1.AS6 | GGATGATCTACTGAGACTGTGTG | 706–728 | ORF1 | 100 | ||||
| ORF2.PCV1.S1 | GGGTTACAAAGTTGGCATCCAAG | 1327–1350 | ORF2 | 407 | 70 | + | − | |
| ORF2.PCV1.AS1 | CACTTTTATAGGATGACGTGG | 1714–1734 | ORF2 | 52 | ||||
Nucleotide sequences and their localization on the PCV-1 genome were given in reference to those of the sequence of the prototype ATCC CCL-33 strain of PCV-1 (GenBank accession no. 49186) (23).
Sequence analysis.
Purified DNA amplicons of two randomly chosen Quebec field strains of PCV-2 were cloned into the TA cloning plasmid pCR 2.1 (Invitrogen Corporation, San Diego, Calif.) as described previously (9). Both strands of three full-length clones derived from independent PCRs were sequenced to produce the final sequences. The sequences of the PCV genomes were analyzed by computer programs: McVector (International Biotechnologies, Inc., New Haven, Conn.) and GeneWorks (IntelliGenetics Inc., Mountain View, Calif.).
Virus isolation.
Specimens from a total of 18 animals that were found to be PCV positive by PCR were processed to attempt virus isolation. Samples of 100 mg of pooled tissue (lungs, liver, spleen, lymph nodes) were homogenized in 10 ml of MEM containing 100 U of penicillin per ml, 100 μg of streptomycin per ml, and 100 μg of amphotericin B (Fungizone; Gibco) per ml. Tissue homogenates were clarified by centrifugation at 2,500 × g for 60 min at 4°C and were extracted once with 2/3 volume of trichlorotrifluoroethane. Following another centrifugation step at 3,000 × g for 15 min, the upper phases were collected and filtered through 0.2-μm-pore-size membranes. Aliquots of 0.5 ml of the homogenates were inoculated onto confluent monolayers of PCV-free PK-15A cells in 25-cm2 tissue culture flasks. After an adsorption period of 1 h and 30 min at 37°C, 10 ml of MEM containing 1 mM glutamine and antibiotics was added to the infected cultures. After 2 days of incubation at 37°C, monolayers were rinsed twice with phosphate-buffered saline and were incubated for 30 min in MEM containing 300 mM d-glucosamine. The monolayers were rinsed and were then reincubated in MEM until day 5 postinfection. Monolayers were stained for circoviral antigen after two successive passages by using the rabbit polyclonal serum and indirect immunofluorescence.
Nucleotide sequence accession numbers.
The nucleotide sequence accession numbers (EMBL/GeneBank/DDBJ) are AF118095 for strain IAF-614 and AF118097 for strain IAF-4370.
RESULTS
Pathological findings.
Since January 1998, outbreaks of respiratory signs and wasting affecting 5- to 12-week-old piglets have been reported in farrow-to-finish pig farms in southern Quebec. A mild diarrhea could also be observed in approximately 20% of the affected pigs. At necropsy, macroscopic lesions were mostly confined to the lower respiratory tract. Lungs were usually noncollapsed and were mottled red to pale tan with a rubbery texture. Enlarged bronchial, inguinal, and mesenteric lymph nodes were observed in approximately 40% of the affected pigs. Occasionally, the spleen was also enlarged, and whitish necrotic foci could be observed in the kidney cortex and medulla. Microscopic examination of tissues from the necropsied pigs revealed lesions of lymphohistiocytic interstitial pneumonitis, with moderate to severe depletion of tonsillar and splenic lymphoid tissue. In most of these animals, the presence of cytoplasmic basophilic inclusion bodies was demonstrated in macrophage-like cells in the tonsils, lymph nodes, and lungs. In a few animals, indirect immunofluorescence staining revealed PRRSV antigen in the lung, but in more than 50% of the animals the presence of PRRSV could be demonstrated in the lungs and lymph nodes by PCR and virus isolation in cultures of porcine alveolar macrophages (20, 21). From April 1998 to February 1999, pooled organs (lungs, spleens, lymph nodes, livers, and tonsils) collected from nearly 40 affected pigs from 30 pig operations were forwarded to our laboratory to investigate the possibility of a PCV infection.
Efficacy of the PCR method for detection of PCV.
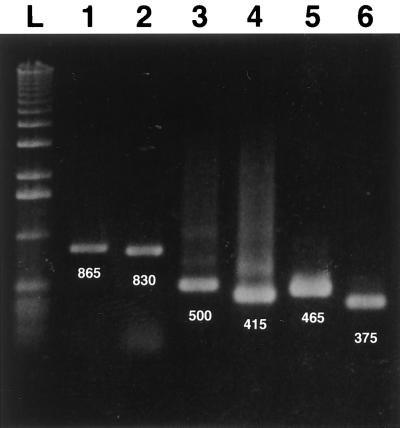
In preliminary experiments, an evaluation of the efficacy of a single PCR method in detecting PCV genomic DNA in clinical specimens was done (23). Sets of oligonucleotide primers were first designed according to the sequence of the ORF1 gene of the PCV-1 strain. As illustrated in Fig. 1, the six primer pairs used in this study, from a combination of five different primers covering the entire ORF1, permitted amplification of genomic fragments of the expected size when tested with genomic DNA extracted from CsCl gradient-purified PCV-1. The primer pairs ORF1.PCV1.S1 and ORF1.PCV1.AS5, ORF1.PCV1.S2 and ORF1.PCV1.AS5, ORF1.PCV1.S1 and ORF1.PCV1.AS2, ORF1.PCV1.S1 and ORF1.PCV1.AS1, ORF1.PCV1.S2 and ORF1.PCV1.AS2, and ORF1.PCV1.S2 and ORF1.PCV1.AS1 yielded DNA amplicons of 865, 830, 500, 415, 465, and 375 bp in length, respectively. The specificities of the PCR products were confirmed by testing the primer pair ORF1.PCV1.S1 and ORF1.PCV1.AS1 and the primer pair ORF1.PCV1.S2 and ORF1.PCV1.AS2 with DNA or RNA templates from other viral and bacterial pathogens commonly associated with respiratory disorders in pigs. As expected, no amplicon product was obtained with heterologous pathogens or with RNA prepared from mock-infected pig cells (PK-15, PT, ST-148, and PFT cells) or distilled water (Table 1).
FIG. 1.
Electrophoretic profiles of DNA amplicons obtained by single PCR with genomic DNA extracted from CsCl gradient-purified PCV-1. The six primer pairs used to amplify ORF1 of the PCV-1 strain yielded DNA amplicons of the expected size. Lane 1, primers ORF1.PCV1.S1 and ORF1.PCV1.AS5 (865 bp); lane 2, primers ORF1.PCV1.S2 and ORF1.PCV1.AS5 (830 bp); lane 3, ORF1.PCV1.S1 and ORF1.PCV1.AS2 (500 bp); lane 4, primers ORF1.PCV1.S1 and ORF1.PCV1.AS1 (415 bp); lane 5, primers ORF1.PCV1.S2 and ORF1.PCV1.AS2 (465 bp); and lane 6, primers ORF1.PCV1.S2 and ORF1.PCV1.AS1 (375 bp); lane L, 1-kb DNA ladder.
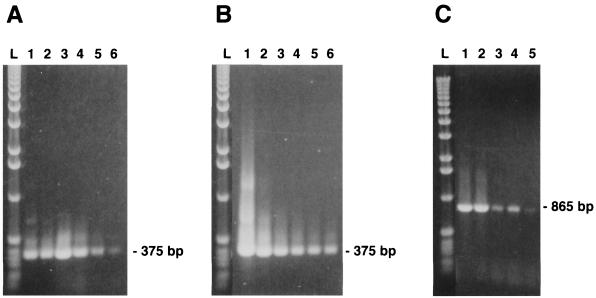
To evaluate the sensitivity of the single PCR method in detecting viral genomic DNA, primer pair ORF1.PCV.S1 and ORF1.PCV1.AS5 (865 bp amplicon) and primer pair ORF1.PCV1.S2 and ORF1.PCV1.AS1 (375 bp amplicon) were tested with 10-fold dilutions of DNA extracted from 100 μl of a purified PCV suspension (preadjusted to 106 viral particles/ml, as determined by negatively stained electron microscopy) and from persistently infected PK-15 cells (adjusted to 106 cells/ml). As illustrated in Fig. 2A and C, independently of the size of amplicon, a positive amplification reaction could be visualized with the 10−5 dilution of the purified viral preparation, corresponding to approximately 10 viral particles per ml. On the other hand, a positive reaction was obtained with the 10−6 dilution of DNA extracted from persistently infected PK-15 cells, suggesting that amplification of the viral genome was not compromised by excessive amounts of cellular nucleic acids (Fig. 2B).
FIG. 2.
Sensitivity of the single PCR method in detecting virus-specific DNA extracted from 10-fold dilutions (10−1 to 10−6; lanes 1 to 6, respectively) of purified viral preparation of PCV-1 (preadjusted to 106 viral particles per ml) (A and C) or from PK-15 cells infected with PCV-1 (total of 106 cells) (B). The primer pairs used were chosen in order to permit amplification of ORF1 fragments of either 375 bp (primers ORF1.PCV1.S2 and ORF1.PCV1.AS1) or 865 bp (primers ORF1.PCV1.S1 and ORF1.PCV1.AS5) in length. Lane l, 1-kb DNA ladder.
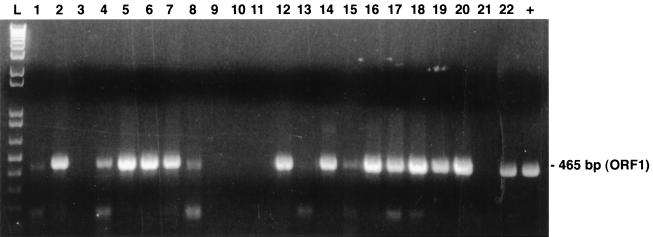
The single PCR method was then tested for its efficacy at detection of PCV from crude clinical specimens. Table 4 summarizes the data obtained from tests with 22 lung specimens that have been forwarded to our laboratory for virological investigations. A preliminary diagnosis of PCV infection (PMWS) has been suggested for 14 of these specimens on the basis of clinical and histological findings (presence of intracytoplasmic basophilic inclusion bodies in macrophage-like cells, detection of PCV antigen by the immunoperoxidase reaction), whereas a diagnosis of PMWS for two other specimens from animals with irrelevant cases of respiratory and/or enteric disorders was doubtful. Specimens from other six animals with irrelevant cases of infection or from clinically healthy pigs were also tested as controls. Specific primer pair ORF1.PCV1.S2 and ORF1.PCV1.AS2 yielded a PCR product of the expected size (465 bp) for 13 of the 14 pigs with histopathological lesions typical of PCV infection and for 1 of the 2 pigs with irrelevant infections and a doubtful result (Fig. 3). Overall, three of the eight pigs with irrelevant clinical infections were found to be positive by PCR. As expected, a negative reaction was obtained with lung tissues from a specific-pathogen-free pig, as well as PCV-free PK-15 cells, which were used as negative controls that were processed simultaneously for DNA extraction. The PCR results were reproducible when other sections of the tested clinical samples were tested, including samples from the three positive pigs with irrelevant clinical infections. The specific primer pair ORF1.PCV1.S2 and ORF1.PCV1.AS2 was more effective at detecting PCV DNA (16 of 22 pigs) than primer pair ORF1.PCV1.S1 and ORF1.PCV1.AS1 (only 8 of 22 pigs).
TABLE 4.
Correlation between histopathological findings and detection of PCV genomic DNA in lungs and lymphoid organs by single PCR with ORF1 primers
| No. of clinical specimensa | Presence of histological lesions
|
No. of specimens positive by single PCR | |
|---|---|---|---|
| Inclusion bodies | Immunoperoxidase reaction | ||
| 14b | + | + | 13 |
| 2c | − | Dd | 1 |
| 6c | − | − | 2 |
The specimens comprised lungs, lymph nodes, and spleen specimens.
Eight of 14 specimens were found to be PRRSV positive.
Submitted because of respiratory and/or enteric problems.
D, doubtful.
FIG. 3.
PCR products resulting from enzymatic amplification of PCV genomic DNA extracted from infected porcine lung tissue. Clinical samples with histopathological lesions suggestive of a PCV infection (presence of basophilic inclusion bodies) (Table 4) were treated as described in the Materials and Methods section, and then extracted DNA was tested by the single PCR method with primers ORF1.PCV1.S2 and ORF1.PCV1.AS2, which yielded DNA amplicons of 465 bp in length. Lanes 1 to 22, samples from 22 pigs, respectively; lane L, 1-kb DNA ladder; +, DNA from a purified PCV-1 strain.
mPCR.
The nt sequences of two Quebec PCV isolates, IAF-614 and IAF-4370, for which the entire genome could be amplified as two overlapping fragments with primers flanking the ORF1 and ORF2 of the np PCV strain (19, 23), were determined. Analysis of sequencing data showed nearly 99% nt sequence identities with previously published sequences of PCV-2 strains reported to be associated with PMWS outbreaks in the United States, France, and Germany (14, 22, 24). Accordingly, both Quebec strains were found to have 1,768 bp of DNA and shared approximately 76% sequence identity with the reference PCV-1 strain (23). As described previously (14, 24), greater variation was found within the ORF2 part of the genome, which had less than 67% homology with the genome of the PCV-1 strain. The homology in ORF1 between both Quebec field isolates and the PCV-1 strain was close to 83% at the nt level.
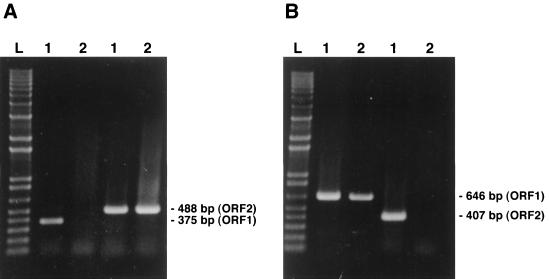
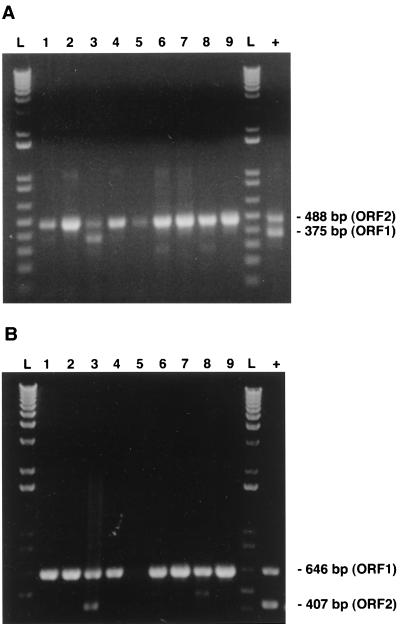
With the aim of distinguishing between both genotypes of PCV that may be present in clinical specimens, two mPCR approaches were set up. The primers were designed to permit amplification of either ORF1 or ORF2 DNA fragments for PCV-2 field isolates (type-specific primers) but to permit amplification of both regions in the case of PCV-1 field isolates. All the primers chosen were 18- to 22-mers and had G+C contents of less than or equal to 55%. Annealing temperatures of 50 to 60°C were selected in preliminary attempts to give maximum product yield and specificity. The sequences were also chosen to avoid the formation of dimers either within or between pairs; no significant theoretical mispriming was identified on any template. Primers that were used for amplication of PCV-1 only were selected in ORF1 or ORF2 regions with less than 70% nt identities between both genotypes, with most variations being located at the 5′ termini of the primers (Table 3). The specific electrophoretic patterns of the amplified products obtained for the two PCV genotypes by both mPCR approaches are depicted in Fig. 4. As expected from the genomic sequences determined previously, primer pair ORF1.PCV1.S3 and ORF1.PCV1.AS2 and primer pair ORF2.PCV1.S and ORF2.PCV1.AS, which were used simultaneously for the first mPCR method, yielded DNA amplicons of 488 bp specific for the ORF2 genes of both genotypes, whereas a 375-bp fragment was amplified from the ORF1 of the PCV-1 strain only (Fig. 4A). On the other hand, primer pair ORF1.PCV1.S2 and ORF1.PCV1.AS6 and primer pair ORF2.PCV1.S1 and ORF2.PCV1.AS1 used for the second mPCR method yielded DNA amplicons of 646 bp specific for the ORF1 genes of both genotypes, whereas a 407-bp fragment was amplified from the ORF2 of the PCV-1 strain only (Fig. 4B). Electrophoretic profiles of the mPCR products obtained with lung homogenates from 9 of the 16 PCV-infected pigs previously identified by the single PCR method are illustrated in Fig. 5. Only one of the specimens showed an electrophoretic profile similar to that of the PCV-1 strain when products from both ORFs could be amplified. For routine diagnosis purpose, the second mPCR method was selected since the ORF1 has been found by several investigators to be more conserved than ORF2 among both types of PCV (14, 22, 25), which was also the case for both Quebec strains sequenced. When tested with field clinical samples forwarded to our laboratory from April 1998 to February 1999, 33 of 35 PCR-positive samples had mPCR profiles typical of those of PCV-2 strains when only the ORF1 amplicons could be visualized. In two of these clinical samples, genomic fragments of both ORFs could be amplified, and thus, they were considered to be infected with PCV-1.
FIG. 4.
Strain specificity of the oligonucleotide primers used in both mPCR methods. As expected from the genomic sequences of PCV-1 and PCV-2, primer pair ORF1.PCV1.S3 and ORF1.PCV1.AS2 and primer pair ORF2.PCV1.S and ORF2.PCV1.AS which were used in the first mPCR method (A) yielded DNA amplicons of 488 bp specific for the ORF2 genes of both genotypes, whereas a 375-bp fragment was amplified for the ORF1 gene of the PCV-1 strain only. In the second mPCR-2 method (B), primer pair ORF1.PCV1.S2 and ORF1.PCV1.AS6 and primer pair ORF2.PCV1.S1 and ORF2.PCV1.AS1 yielded DNA amplicons of 646 bp specific for the ORF1 genes of both genotypes, whereas a 407-bp fragment was amplified for the ORF2 gene of the PCV-1 strain only. Lane L, 1-kb DNA ladder; lanes 1 and 2, genotypes 1 and 2 of PCV, respectively.
FIG. 5.
Detection and typing of PCV in a panel of positive clinical specimens (lung tissue; lanes 1 to 9) by both mPCR methods. As described in the Materials and Methods section, in both mPCR methods, two sets of primers were used simultaneously with DNA extracted from lungs of the affected pigs to amplify fragments of the ORF1 and ORF2 genes of PCV-1 but only the ORF1 or ORF2 fragment of PCV-2. (A and B) For only one of the clinical samples tested, an mPCR profile of a PCV-1 strain was observed. As expected, by the mPCR1 method (A), a fragment of 488 bp specific for the ORF2 genes of strains of both genotypes could be amplified for all positive samples, whereas for only one sample (lane 3), the ORF1 fragment (375 bp) could be amplified. By the mPCR2 method (B), a fragment of 646 bp specific for the ORF1 genes of both genotypes could be amplified for all positive samples, whereas for only the same sample described in panel A, the 407-bp fragment specific for the ORF1 of PCV-1 could be amplified (lane 3). Lane L, 1-kb DNA ladder; lane +, DNA extracted from purified PCV-1.
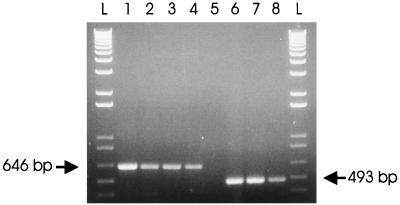
To address the question of whether the two animals positive for the PCV-1 pattern may also be infected with PCV-2, samples from both animals were further tested by a single PCR with primer pair ORF2.PCV2.S4 and ORF2.PCV2.AS4, the sequences of which have been deduced from specific sequences of the ORF2 of PCV-2 (Table 2). As expected, a DNA fragment of 493 bp in length could be amplified from Quebec PCV-2 isolates, isolates IAF-614 and IAF-4370, as well as other clinical specimens already found to be positive for PCV-2 (data not shown), but not from DNA extracted from PK-15 cells persistently infected with PCV-1 (Fig. 6). By this single PCR approach, amplicons were also obtained from the two animals previously shown to be positive for PCV-1, suggesting a mixed infection with both genotypes of PCV.
FIG. 6.
Differentiation of PCV-1 and PCV-2 strains by single PCR with universal and type 2-specific primer pairs. Primer pair ORF1.PCV1.S2 and ORF1.PCV1.AS6 (lanes 1 to 4) yielded a DNA amplicon of 646 bp specific for the ORF1 genes of both genotypes, whereas primer pair ORF2.PCV2.S4 and ORF2.PCV2.AS4 (lanes 5 to 8) yielded a DNA amplicon of 493 bp specific for the ORF2 gene of PCV-2 only. Lanes 1 and 5, DNA extracted from PK-15 cells persistently infected with the PCV-1 strain; lanes 2 and 6, clinical specimen positive for PCV-1 by the mPCR; lanes 3 and 7, Quebec PCV-2 strain IAF-614; lanes 4 and 8, Quebec PCV-2 strain IAF-4370; lanes L, 1-kb DNA ladder; lane +, DNA extracted from purified PCV-1.
Virus isolation.
Attempts were made to isolate in cell cultures PCV from 18 clinical samples previously found to be positive for PCV-2 by mPCR. After two successive passages and an incubation period of 5 days, no cytopathic effect was observed in PK-15A cell cultures inoculated with pooled tissue homogenates from the mPCR-positive animals. However, for seven of these samples (38.8%), detection of viral antigen in the cytoplasm of infected cells could be detected by indirect immunofluorescence with specific rabbit hyperimmune serum. For these samples, the presence of PCV-2 in culture supernatants was detected by mPCR after the second passage.
DISCUSSION
Several reports dealing with the occurrence of a new genotype of PCV associated with PMWS outbreaks in Europe and North America have recently been published (14, 22, 24). Although PCR was reported to be a useful tool for detecting PCV in clinical specimens from naturally and experimentally infected pigs (12, 14, 25), an investigation such as the one described herein was needed to establish the sensitivity and specificity of the technique in comparison with those of other routine diagnostic procedures that have been used for the detection of PCV, mainly virus isolation in cell cultures, in situ hybridization, and immunohistochemical staining with hyperimmune serum (2, 11, 17, 25). The data obtained in the present study confirmed the specificity of the PCR method since no DNA amplicons could be obtained from DNA or RNA prepared from bacterial and viral pathogens commonly associated with respiratory disorders in pigs by using primer sets corresponding to different regions of the ORF1 gene of the PCV-1 strain. These primers were in fact chosen from regions of the polymerase gene, which is highly conserved among both genotypes of PCV (14, 23). The sensitivity was comparable to those of PCR methods that have previously been reported for the detection of other small DNA viruses such as porcine parvoviruses (24). By negatively stained electron microscopy, it was estimated that the highest dilution from which PCV DNA could be detected by the single PCR assay contained approximately 10 viral particles per ml, which would correspond to as little as 5 to 50 pg of viral genomic DNA. A comparable sensitivity was obtained with primer pairs covering one-third (375 bp) or the entire (approximately 900 bp) polymerase gene. When tested with clinical samples from pigs, the results of the single PCR method showed a nearly 93% (13 of 14 samples) correlation with histopathological and immunohistochemical findings. Interestingly, PCV infection could also be detected by single PCR with three clinical samples that have been submitted from pigs with irrelevant respiratory and/or enteric problems. Since the positive PCR results were reproducible, even when other sections from the same specimens were tested, the data obtained were suggestive of subclinical PCV infections. In Canada, a seroprevalence of nearly 70% was reported in the early 1990s in the absence of clinical symptoms (10). However, due to the lack of a type-specific serological test, the genotype involved in asymptomatic or subclinical infections could not be identified.
By the single PCR method, the lungs appeared to be the most suitable organs for the recovery of PCV from PMWS-affected pigs, as similar data were obtained with pooled organ homogenates (data not shown). Amplification products of the expected sizes were obtained for all Quebec field isolates by PCR amplification of the ORF1 gene with different sets of primer pairs, which suggests that this first half of the PCV genome is highly conserved among field isolates, in agreement with previous findings by other investigators (14, 22, 25). Data also demonstrated that amplification of the viral genome was not compromised by excessive amounts of cellular nucleic acids (Fig. 2B).
Although our preliminary findings further demonstrated the suitability of the single PCR method for the identification of PCV-positive clinical samples, this technique was unable to differentiate which genotype was in fact involved. Sequencing analyses demonstrated that two representative Quebec isolates of PCV that were associated with outbreaks of PMWS, detected by single PCR, and selected at random differed significantly from the nonpathogenic PCV-1 strain. Both Quebec field isolates instead showed nearly 99% nt sequence identities with previously published sequences of PCV-2 strains reported to be associated with PMWS outbreaks in the United States, France, and Germany (14, 22, 25). These two representative Quebec strains displayed only 76% nt sequence identities with the reference PCV-1 strain (23).
A technique for differentiating the nonpathogenic and pathogenic strains is needed since serological surveys have demonstrated that infection with PCV is very common in North American and European pig herds (2, 10, 23, 30, 32). The possibility that both genotypes of PCV may coexist in the same pig should also be considered. By comparing the nucleotide sequences of the two Quebec PMWS-associated PCV strains with that of the reference PCV-1 strain (23), oligonucleotide primers were designed from within regions of ORF1 or ORF2 of PCV-1 to permit PCR amplification of strains of both genotypes or only PCV-1. By the two mPCR methods investigated, amplification of both ORF1 and ORF2 genomic fragments from clinical samples was suggestive of a PCV-1 infection, whereas in the case of a PCV-2 infection, only one of the two genomic regions was amplified. However, a major drawback of these two mPCR methods is that the primers used probably cannot detect PCV-2 DNA in the presence of a PCV-1 infection, so that a mixed infection with both genotypes could be misdiagnosed. On the basis of the data obtained for 35 animals that were found to be PCV positive by the mPCRs, PCV-2 infection was demonstrated in tissues of 94.2% (33 of 35) of sick animals tested, in agreement with previous findings showing the close association of this new genotype of PCV with outbreaks of PMWS in Europe and North America (12, 14, 25). On the other hand, a PCV-1 infection was demonstrated for only 5.7% (2 of 35) of the animals; thus, for a minority of the specimens tested by mPCR, further analyses were needed to eliminate the possibility of a mixed infection with both PCV genotypes. For this purpose, a single PCR was conduced with PCV-2-specific primers designed from ORF2 regions showing less than 35% nt identities with the sequence of PCV-1. Specific amplification of PCV-2 was obtained with all positive clinical specimens tested, including both PCV-1-positive specimens, thus confirming a mixed infection with both PCV genotypes. Consequently, in the absence of type-specific serological tests, this additional single PCR method should routinely be done when a PCV-1 profile is obtained by mPCR.
Interestingly, data from the mPCR method could be obtained within 1 or 2 days, whereas the more classical methods, such as virus isolation in cell cultures, followed by serotyping, require at least 2 weeks. In the present study, PCV could be isolated in cell cultures from only one-third of the PCR-positive samples tested. For all samples, a distinct cytopathic effect was not observed, so that serological identification by indirect immunofluorescence was required for final diagnosis. The mPCR method thus appeared to be more convenient and reliable for routine diagnosis of PCV infection.
ACKNOWLEDGMENTS
We thank Judith Caron, Hélène Drolet, and Nicole Sawyer for technical assistance. We also thank the following veterinarians and pathologists from the Ministère de l'Agriculture, des Pêcheries et de l'Alimentation du Québec for assistance with obtaining the clinical samples and involvement in histopathological studies: René Sauvageau, André Bourgault, and René Larochelle. The collaboration of W. L. Mengeling from the National Veterinary Service Laboratory, U.S. Department of Agriculture, Ames, Iowa, and A. Afshar from the Animal Diseases Research Institute, Agriculture Canada, Nepean, Ontario, Canada, in providing the PCV-free and PCV-infected PK-15 cell lines was greatly appreciated.
This research was partly funded by the Ministère de l'Agriculture, des Pêcheries et de l'Alimentation du Québec and the Fondation Armand-Frappier, Laval, Québec, Canada.
REFERENCES
- 1.Allan G M, McNeilly F, Cassidy J P, Reilly G A C, Adair B, Ellis W A, McNulty M S. Pathogenesis of porcine circovirus: experimental infections of colostrum deprived piglets and examination of pig foetal material. Vet Microbiol. 1995;44:49–64. doi: 10.1016/0378-1135(94)00136-k. [DOI] [PubMed] [Google Scholar]
- 2.Allan G M, McNeilly F, Kennedy S, Daft B, Clarke E G, Ellis J A, Haines D M, Meeham B M, Adair B M. Isolation of porcine circovirus-like viruses from pigs with a wasting disease in the USA and Europe. J Vet Diagn Invest. 1998;10:3–10. doi: 10.1177/104063879801000102. [DOI] [PubMed] [Google Scholar]
- 3.Boevink P, Chu P W G, Keese P. Sequence of subterranean clover stunt virus DNA: affinities with the geminiviruses. Virology. 1995;207:354–361. doi: 10.1006/viro.1995.1094. [DOI] [PubMed] [Google Scholar]
- 4.Clark E G. Post-weaning multisystemic wasting syndrome. Proc Am Assoc Swine Pract. 1997;28:499–501. [Google Scholar]
- 5.Daft B, Nordhausen R W, Latimer K S, Niagro F D. Interstitial pneumonia and lymphadenopathy associated with circoviral infection in a six-week-old piglet. Proc Am Assoc Vet Lab Diagn. 1996;39:32. [Google Scholar]
- 6.Dea S, Garzon S. Identification of coronaviruses by the use of indirect protein-A immunogold electron microscopy. J Vet Diagn Invest. 1991;3:297–305. doi: 10.1177/104063879100300405. [DOI] [PubMed] [Google Scholar]
- 7.Dea S, Bilodeau R, Sauvageau R A, Martineau G P. Outbreaks of respiratory and reproductive problems associated with encephalomyocarditis virus in Quebec pig farms. J Vet Diagn Invest. 1991;3:275–282. doi: 10.1177/104063879100300401. [DOI] [PubMed] [Google Scholar]
- 8.Dea S, Bilodeau R, Monpetit C, Sauvageau R A, Martineau G P. Antigenic variant of swine influenza virus causing proliferative and necrotizing pneumonia in pigs. J Vet Diagn Invest. 1992;4:380–392. doi: 10.1177/104063879200400403. [DOI] [PubMed] [Google Scholar]
- 9.Dea S, Michaud L, Milane G. Comparison of bovine coronavirus isolates associated with neonatal calf diarrhea and winter dysentery in adult dairy cattle in Quebec. J Gen Virol. 1995;76:1263–1270. doi: 10.1099/0022-1317-76-5-1263. [DOI] [PubMed] [Google Scholar]
- 10.Dulac G C, Ahmad A. Porcine circovirus antigens in PK-15 cell line (ATCC CCL-33) and evidence of antibodies to circovirus in Canadian pigs. Can J Vet Res. 1989;53:431–433. [PMC free article] [PubMed] [Google Scholar]
- 11.Ellis J, Hassard L, Clark E, Harding J, Allan G, Wilson P, Strokappe J, Martin K, McNeilly F, Meehan B, Todd D, Haines D. Isolation of circovirus from lesions of pigs with postweaning multisystemic wasting syndrome. Can Vet J. 1998;39:44–51. [PMC free article] [PubMed] [Google Scholar]
- 12.Ellis J, Krakowka S, Lairmore M, Haines D, Bratanich A, Clark E, Allan G, Konoby C, Hassard L, Meehan R, Martin K, Harding J, Kennedy S, McNeilly F. Reproduction of lesions of postweaning multisystemic wasting syndrome in gnotobiotic piglets. J Vet Diagn Invest. 1999;11:3–14. doi: 10.1177/104063879901100101. [DOI] [PubMed] [Google Scholar]
- 13.Friis N F. Some recommendations concerning primary isolation of Mycoplasma suipneumoniae and Mycoplasma flocculare. Nord Veterinaer-med. 1975;27:337–339. [PubMed] [Google Scholar]
- 14.Hamel A L, Lin L L, Nayar G P S. Nucleotide sequence of porcine circovirus associated with postweaning multisystemic wasting syndrome in pigs. J Virol. 1998;72:5262–5267. doi: 10.1128/jvi.72.6.5262-5267.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Harding J. Post-weaning multisystemic wasting syndrome (PMWS): preliminary epidemiology and clinical presentation. Proc Am Assoc Swine Pract. 1997;28:503. [Google Scholar]
- 16.Harding R M, Burns T M, Hafner G, Dietzgen R G, Dale J L. Nucleotide sequence of one component of the banana bunchy top virus genome contains a putative replicase gene. J Gen Virol. 1993;74:323–328. doi: 10.1099/0022-1317-74-3-323. [DOI] [PubMed] [Google Scholar]
- 17.Kiupel M, Stevenson G W, Mittal S K, Clark E G, Haines D M. Circovirus-like viral associated disease in weaned pigs in Indiana. Vet Pathol. 1998;35:303–307. doi: 10.1177/030098589803500411. [DOI] [PubMed] [Google Scholar]
- 18.LeCann P, Albian E, Madec F, Carilet R, Jestin A. Piglet wasting disease. Vet Rec. 1997;141:660. [PubMed] [Google Scholar]
- 19.Mankertz J, Buhk H-J, Blaess G, Mankertz A. Transcription analysis of porcine circovirus (PCV) Virus Genes. 1998;16:267–276. doi: 10.1023/a:1008022521329. [DOI] [PubMed] [Google Scholar]
- 20.Mardassi H, Athanassious R, Mounir S, Dea S. Porcine reproductive and respiratory syndrome virus: morphological, biochemical and serological characteristics of Quebec isolates associated to acute and chronic outbreaks of PRRS. Can J Vet Res. 1994;58:55–64. [PMC free article] [PubMed] [Google Scholar]
- 21.Mardassi H, Wilson L, Mounir S, Dea S. Detection of porcine reproductive and respiratory syndrome virus and efficient differentiation between Canadian and European strains by reverse transcription and PCR amplification. J Clin Microbiol. 1994;32:2197–2203. doi: 10.1128/jcm.32.9.2197-2203.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Meehan B H, McNeilly F, Todd D, Kennedy S, Jewhurst V A, Ellis J A, Hassard L, Clark E G, Haines D M, Allan G M. Characterization of novel circovirus DNAs associated with wasting syndromes in pigs. J Gen Virol. 1998;79:2171–2179. doi: 10.1099/0022-1317-79-9-2171. [DOI] [PubMed] [Google Scholar]
- 23.Meehan B H, Creelan J L, McNulty M S, Todd D. Sequence of porcine circovirus DNA: affinities with plant circoviruses. J Gen Virol. 1997;78:221–227. doi: 10.1099/0022-1317-78-1-221. [DOI] [PubMed] [Google Scholar]
- 24.Molitor T W, Oraveerakul K, Zhang Q Q, Choi C S, Ludeman L R. Polymerase chain reaction (PCR) amplification for the detection of porcine parvovirus. J Virol Methods. 1991;32:201–211. doi: 10.1016/0166-0934(91)90051-z. [DOI] [PubMed] [Google Scholar]
- 25.Morozov I, Sirinarumitr T, Sorden S D, Halbur P G, Morgan M K, Yoon K-J, Paul P S. Detection of a novel strain of porcine circovirus in pigs with postweaning multisystemic wasting syndrome. J Clin Microbiol. 1998;36:2535–2541. doi: 10.1128/jcm.36.9.2535-2541.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Niagro F D, Forsthoefel A N, Lawther R P, Kamalanathan L, Ritchie B W, Latimer K S, Lukert P D. Beak and feather disease virus porcine circovirus genomes: intermediates between the geminiviruses and plant circovirus. Arch Virol. 1998;143:1723–1744. doi: 10.1007/s007050050412. [DOI] [PubMed] [Google Scholar]
- 27.Ritchie B W, Niagro F D, Luckert P D, Steffens W L, Latimer K S. Characterization of a new virus from cockatoos with psittacine beak and feather disease. Virology. 1989;171:83–88. doi: 10.1016/0042-6822(89)90513-8. [DOI] [PubMed] [Google Scholar]
- 28.Rohde W, Randles J W, Langridge P, Harold D. Nucleotide sequence of a circular single stranded DNA associated with coconut foliar decay virus. Virology. 1990;176:648–651. doi: 10.1016/0042-6822(90)90038-s. [DOI] [PubMed] [Google Scholar]
- 29.Segales J, Sitjar M, Domingo M, Dee S, DelPozo M, Noval R, Sacristan C, DeLasHeras A, Ferro A, Latimer K S. First report of post-weaning multisystemic wasting syndrome in pigs in Spain. Vet Rec. 1997;141:600–601. [PubMed] [Google Scholar]
- 30.Tischer I, Mields W, Wolff D, Vagt M, Greim W. Studies on epidemiology and pathogenicity of porcine circovirus. Arch Virol. 1986;91:271–276. doi: 10.1007/BF01314286. [DOI] [PubMed] [Google Scholar]
- 31.Tischer I, Peters D, Rasch R, Pociuli S. Replication of porcine circovirus-induction by glucosamine and cell-cycle dependence. Arch Virol. 1987;96:39–57. doi: 10.1007/BF01310989. [DOI] [PubMed] [Google Scholar]
- 32.Tischer I, Bode L, Peters D, Pociuli S, Germann B. Distribution of antibodies to porcine circovirus in swine populations of different breeding farms. Arch Virol. 1995;140:737–743. doi: 10.1007/BF01309961. [DOI] [PubMed] [Google Scholar]
- 33.Tischer I, Rasch R, Tochtermann G. Characterization of papovavirus- and picornavirus-like particles in permanent pig kidney cell lines. Zentralbl Bakteriol Parasitenkd Infektionkr Hyg Abt 1 Orig. 1974;26:153–167. [PubMed] [Google Scholar]
- 34.Todd D, Creelan J L, Mackie D P, Rixon F, McNulty M S. Purification and biochemical characterization of chicken anaemia agent. J Gen Virol. 1990;71:819–823. doi: 10.1099/0022-1317-71-4-819. [DOI] [PubMed] [Google Scholar]