Abstract
Chronic obstructive pulmonary disease (COPD) is an incurable and prevalent respiratory disorder that is characterized by chronic inflammation and emphysema. COPD is primarily caused by cigarette smoke (CS). CS alters numerous cellular processes, including the post-transcriptional regulation of mRNAs. The identification of RNA-binding proteins (RBPs), microRNAs (miRNAs), and long non-coding RNAs (lncRNAs) as main factors engaged in the regulation of RNA biology opens the door to understanding their role in coordinating physiological cellular processes. Dysregulation of post-transcriptional regulation by foreign particles in CS may lead to the development of diseases such as COPD. Here we review current knowledge about post-transcriptional events that may be involved in the pathogenesis of COPD.
Keywords: post-transcriptional regulation, RNA binding proteins, miRNAs, COPD
1. Introduction
Human organ systems rely on the dynamics of gene expression to regulate homeostasis, cell survival, fate and differentiation, as well as responses to stress and environmental signals [1]. Eukaryotic cells have developed sophisticated mechanisms to produce and use the transcripts with optimum efficacy through their life cycle. When RNA is synthesized in the nucleus, its biogenesis, translocation to the cytosol, and interaction with proteins and other components are necessary to achieve their encoding function. All these steps undergo post-transcriptional regulation of that initial messenger RNA (mRNA), and these steps comprise an important part of overall gene and protein expression. Post-transcriptional regulation is a coordinated process that takes place when the RNA is transcribed, but before it is translated into protein. Factors that associate with and regulate target mRNAs at the post-transcriptional level are RNA-binding proteins (RBPs), microRNA (miRNA), and long non-coding RNA (lncRNA) [2,3]. In mammalian cells, the fate of mRNA is controlled by almost 2000 RBPs [4] and ~2300 miRNAs [5]. In humans, there are approximately 172,216 lncRNAs [6], and 27,919 lncRNAs have been identified in a variety of human primary cells [7]. These factors dynamically modulate mRNAs during biological processes, and their dysregulation is likely to be involved in pathological processes. Cigarette smoke (CS) causes a variety of chronic lung disorders, including chronic obstructive pulmonary disease (COPD) and lung cancer. CS is responsible for approximately 70% of COPD cases [8] and 90% of lung cancer cases [9], and remains a major cause of morbidity and mortality worldwide. The pathogenesis of diseases associated with smoking involves the dysregulation of numerous cellular and physiological pathways, such as proliferation, apoptosis, and inflammation [9,10,11,12,13,14]. These processes are controlled at the post-transcriptional level via the regulation of mRNAs. In this review, we will discuss RBPs, miRNAs, and lncRNAs that regulate the post-transcriptional modifications of mRNAs, and their involvement in normal physiology. We will then highlight post-transcriptional regulatory mechanisms that are dysregulated in response to smoke, and, thus, may be implicated in the pathogenesis of COPD.
2. Post-Transcriptional Regulation of mRNAs
2.1. RNA Binding Proteins (RBPs)
RBPs are a group of over 2000 proteins, each possessing multiple RNA binding domains, and which are known to be involved in RNA decay [4]. RBPs associate with RNA transcripts and form ribonucleoprotein (RNP) complexes after transcription. Some RBPs bind early during RNA synthesis to precursor mRNA (pre-mRNA) and remain bound to the pre-mRNA until its degradation or translation, whereas other RBPs recognize and bind to pre-mRNA for specific processes, such as splicing, stability, transport, and cellular localization [15]. The diversity of RBP functions suggest that several RNA-binding domains (RBD) are responsible for RNA recognition and for recruitment to specific RNA targets [16,17]. RBPs contain one or multiple RNA-binding domains, such as the RNA-recognition motif (RRM), K-homology domain (KH), double-stranded RBD (dsRBD), zinc fingers (Znf), DEAD box helicase domain, among others. There is diversity in the specificity and affinity of RBD interaction with RNA [4,18,19]. Some RBPs with dsRBD interact with the phosphate-sugar backbone of their RNA targets [4,18]. Other RBPs, such as those with RRMs, interact in a sequence-specific manner with the nucleotide base and shape complementarity of the RNA [4,18,19,20]. In this section, we will explore the role of RBPs in different aspects of RNA biology.
2.1.1. Biological Functions of RBPs
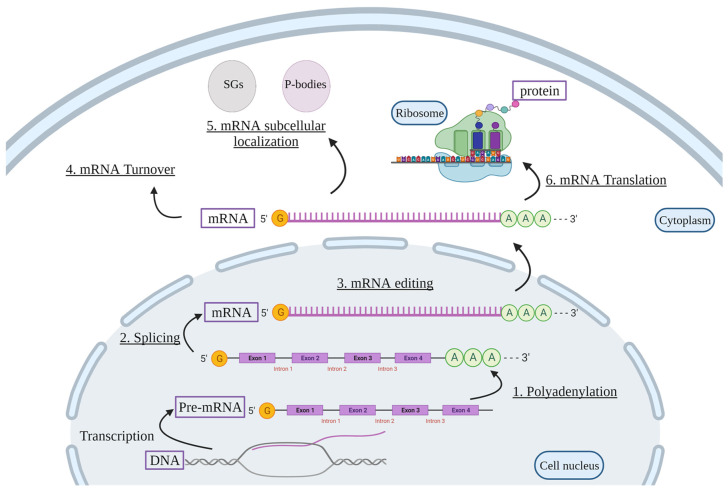
RBP-mediated post-transcriptional regulation is essential for proper cellular function, and its perturbation can lead to the development of disease. For example, the fragile X syndrome of mental retardation is caused by a defect in the RBP fragile X mental retardation protein, which is important for normal brain development [21]. The fate of RNA, from transcription to translation, is highly dependent on RBP-mediated polyadenylation, pre-mRNA splicing, as well as mRNA editing, turnover, subcellular localization, and translation (Figure 1).
Figure 1.
Cellular functions of RBPs. RBPs are involved in post-transcriptional regulation of target mRNAs. Pre-mRNA is first transcribed from the DNA. Then, RBPs regulate the production of mature mRNA via polyadenylation (1), splicing (2), and mRNA editing (3). RBPs can also regulate mRNA stability (4) and mRNA subcellular localization (5) within the cell in SGs or P-bodies, as well as mRNA translation into proteins (6). Pre-mRNA, precursor mRNA; SGs, stress granules; P-bodies, processing-bodies. Created with BioRender.com.
Polyadenylation
Polyadenylation of pre-mRNA is an essential processing event for RNA nuclear export, stability, and translation. Polyadenylation is a maturation step in which all pre-mRNAs in eukaryotic cells, except mRNA encoding histones, receive poly(A) tails of around 200 adenine (A) nucleotides to their 3’ end by a multiprotein machinery complex [22,23]. This occurs in a coupled cleavage reaction whereby the pre-mRNA is first cleaved between AAUAAA sequences upstream and U/GU rich sequences downstream of the cleavage site, followed by the addition of a polyadenosine tail. The cleavage and polyadenylation machinery consists of four multi-subunit protein complexes: the cleavage and polyadenylation specificity factor (CPSF); the cleavage stimulation factor (CstF); and mammalian cleavage factors I and II (CFIm and CFIIm) [24]. The CPSF protein complex consists of six protein subunits that are vital for cleavage of pre-mRNA and interaction with AAUAAA sequences [23,24]. CstF consists of three subunits that interact with the downstream element and upstream site of the pre-mRNA [22,24,25]. CFIm and CFIIm are required for the cleavage step [25,26]. Then, poly (A) polymerase, stimulated by CPSF and the RBP nuclear poly(A) binding protein (PABPN1), adds the poly(A) tail to the cleavage product of the synthesized pre-mRNA molecule to produce mature mRNA [27,28]. The best-characterized mRNA-stabilizing factor, hu antigen R (HuR)/embryonic lethal abnormal vision Drosophila-like (ELAVL1), is also involved in the polyadenylation step. HuR is a ubiquitously-expressed RBP which is predominantly nuclear, but shuttles between the nucleus and cytoplasm. HuR blocks polyadenylation of the simian virus 40 late (SVL) poly(A) site that has U-rich sequences both upstream and downstream of the cleavage site. This leads to a decrease of SVL poly(A) site-containing mRNA, and an increase of pre-mRNA levels [29].
Pre-mRNA Splicing
Splicing of pre-mRNA is a step of gene expression in which introns (noncoding sequences) are removed, and exons (coding sequences) are assembled by the spliceosome. The spliceosome is a ribonucleoprotein complex composed of five small nuclear RNAs (snRNAs), U1, U2, U4, U5, and U6, and more than 50 protein factors, such as U2 auxiliary factor and SR (serine-arginine rich) proteins [30,31]. Some exons are constitutively spliced [32]. However, many exons are alternatively spliced, in which more than one mRNA can be generated from a single pre-mRNA. At least 74% of human multi-exon genes express several mRNAs through alternative splicing (AS) [33]. Studies using high-throughput sequencing showed that ~95% of multi-exon genes undergo AS [34,35]. RBPs also regulate this process, including SR proteins and heterogeneous nuclear ribonucleoproteins (hnRNPs) [36]. In human cells, hnRNPs are the most abundant RBPs that regulate AS of pre-mRNAs. Genome-wide analysis showed that more than half of all AS events are regulated by six major hnRNP proteins: A1; A2/B1; H1; F; M; and U [37]. One of the RBPs involved in this process is HuR, which can promote exon 6 skipping of the apoptosis receptor Fas pre-mRNA through the inhibition of the U2 auxiliary factor 65 kDa association with the upstream 3’ splice site—this leads to the production of the soluble isoform of Fas that prevents apoptosis [38].
mRNA Editing
RNA editing is a type of RNA modification characterized by the alteration of site-specific RNA sequences from that encoded in DNA [39]. The RNA codon and protein sequence are changed if the editing occurs in the coding region [40]. When editing occurs in the noncoding regions, it may affect splicing, stability, or translation of the mRNA [41,42]. RNA editing, mediated by adenosine deaminases acting on RNA (ADAR) proteins, involves adenosine (A) deamination to inosine (I) that is then recognized as guanosine by the translational apparatus [39,41,43,44]. ADARs contain two or three RNA binding domains (RBDs), and a highly conserved deaminase domain [45]. Three ADAR proteins, ADAR1, ADAR2, and ADAR3, are present in humans [44,45,46,47]. A-to-I editing can occur in noncoding regions of the RNA, in Alu repeats, which are good substrates for ADAR proteins [42,48,49].
mRNA Turnover
The translation of mRNA is coupled with its stability and decay. RBPs that regulate mRNA stability are either mRNA decay activators or mRNA stabilizers. Activators of mRNA decay recognize the constitutive decay AU- and GU-rich elements of their target mRNAs, and affect its cellular levels by several mechanisms [50]. For example, tristetraprolin (TTP), also known as zinc finger protein 36, is an RBP that promotes mRNA decay [51]. TTP promotes deadenylation of tumor necrosis factor alpha (TNF-α) mRNA and its degradation upon exposure to lipopolysaccharide (LPS) [52,53,54]. TTP also downregulates numerous inflammatory mRNAs, such as interleukin (IL)-6, IL-2, and cyclooxygenase-2 (COX-2/ PTGS2) [55,56,57]. Other RNA-binding proteins implicated in mRNA decay include KH-type splicing regulatory protein (KSRP) [58], Roquin [59], and ARE/poly(U)-binding/degradation factor 1 (AUF1) [60,61,62]. Conversely, other RBPs act as mRNA stabilizers, and impede mRNA degradation. One of the best known RBPs with a regulatory influence on mRNA stability is HuR. Besides HuR regulation of pre-mRNA polyadenylation and splicing, HuR also targets mRNAs which have U- or AU-rich elements (AREs) in the 3’-untranslated region (UTR)—these mRNA typically encode proteins involved in cell proliferation, differentiation, migration, apoptosis, inflammation, and fibrosis [63,64,65,66,67,68,69,70].
mRNA Subcellular Localization
Localization of mRNA is critical for protein synthesis. Stress granules (SGs) and processing (P-) bodies are cytoplasmic RNA granules consisting of aggregates of ribonucleoprotein complexes. SGs and P-bodies are assembled in stressed and in unstressed cells, respectively [71]. SGs sequester mRNAs for storage and translational silencing [72]. In this context, HuR also regulates the subcellular localization of mRNAs. In human osteoarthritis chondrocytes, for example, in response to IL-1β, PTGS2 mRNA is sequestered in SGs by HuR, thereby decreasing protein levels due to a delay in translation [73]. SGs also contain other RPBs involved in RNA metabolism, including poly(A)-binding protein (PABP), T-cell intracellular antigen 1 (TIA-1), and TTP [74]. In contrast, P-bodies contain mRNAs targeted for degradation, and the RBPs are involved in this process [75]. For instance, Roquin suppresses inducible co-stimulator (ICOS) expression, which prevents autoimmunity through its association with P-bodies in T-helper cells [76].
mRNA Translation
The regulation of mRNA translation controls gene expression in the cytoplasm. Numerous proteins, including RBPs, regulate mRNA location and assembly into ribosomes for protein synthesis. For example, the RBP PABP that binds to stable mRNA also interacts with eukaryotic initiation factors 4E (eIF4E), whereby the 48S and 80S ribosome initiation complex are assembled and translation is initiated [77]. Another example is the RBP TIA-1, which represses translation of various mRNAs, including PTGS2 mRNA [78]. Some RBPs can inhibit or promote translation depending on the context. HuR promotes the translation of prothymosin α, an enhancer of cell survival, in response to irradiation [79], but can inhibit the translation of p27, a cyclin-dependent kinase inhibitor, and Wnt5a, a non-transforming Wnt protein, during proliferation and oncogenesis, respectively [80,81].
2.2. miRNAs
miRNA are small non-coding RNAs (~22 nucleotides) that affect gene expression [82,83]. In the nucleus, miRNAs are synthesized from primary miRNA which are processed to precursor miRNA (pre-miRNA) by ribonuclease III (RNase III), Drosha, and the RBP DiGeorge syndrome critical region 8 (DGCR8). The resulting pre-miRNA is exported to the cytoplasm by the nuclear transport factor exportin-5. The pre-miRNA is then further processed to ~22-nt miRNA by Dicer, another RNase III enzyme [83,84]. miRNAs pair to the 3′ UTR of mRNA by partial sequence matching after being incorporated into the RNA-induced silencing complex (RISC). This leads to direct post-transcriptional repression by inhibiting translation and/or inducing mRNA decay [85,86]. In 1993, lin-4, a developmental regulator, was the first miRNA identified in Caenorhabditis elegans [87,88]. At the present time, about 2300 miRNAs have been discovered in humans, around half of which are annotated in miRbase [5]. The importance of the RNA interference (RNAi) machinery in mammals is highlighted in studies where Dicer or DGCR8 were knocked out, leading to embryonic lethality [89,90].
Biological Function of miRNAs
The function of miRNAs has been explored in murine knockout models, which have revealed important roles for miRNA in various biological processes, including development and immunity. For example, targeted ablation of miRNA-1-2, a muscle-specific miRNA, leads to cardiac morphogenetic and electrophysiologic defects [91]. Furthermore, the knockout of miRNA-155 causes defects in adaptive immunity [92]. miR-223 is a myeloid-specific miRNA that targets Mef2c, a transcription factor which promotes myeloid progenitor proliferation. miR-223 null mice have marked neutrophilia, and, consequently, develop pulmonary inflammation and exaggerated tissue destruction in response to LPS [93]. Some miRNAs have multiple essential functions. An example is miR-17~92: these knockout mice die postnatally with heart defects and lung hypoplasia, but also exhibit defects in B cell development [94].
2.3. LncRNA
LncRNA are a class of ncRNA that are more than 200 nucleotides in length and do not translate into protein [95,96,97]. lncRNAs can be classified based on their genomic proximity to protein-coding genes [97,98]. lncRNA are sub-grouped into five main classes, including: (1) antisense lncRNAs or natural antisense transcripts (NATs); (2) sense lncRNAs; (3) intronic lncRNAs; (4) long intergenic RNAs (lincRNAs); and (5) bidirectional lncRNAs [97,98]. The most common classes of lncRNAs in humans are antisense and intergenic lncRNAs [98].
Biological Function of lncRNA
Based on the molecular mechanisms of action, lncRNA can also be categorized as decoy, scaffold, and guide [97,99]. Decoy lncRNA bind and capture different molecules, including proteins, transcription factors, and other regulatory RNA, which results in inhibition of functions [97,99]. Decoy lncRNA can positively or negatively affect transcription [97,99]. Decoy lncRNA can titrate and prevent transcription factors or repressors from binding their target gene promoters [97,99]. Scaffold lncRNA have binding sites that interact with distinct effector molecules, and, thus, can serve as a platform for connecting various protein complexes [97,99]. lncRNA can also guide chromatin to specific genomic regions to regulate gene expression [100,101,102], increase DNA methylation by binding with DNA methyltransferases [103], participate in AS by directly interacting with splicing factors or proteins [104], or directly interact with either RNA polymerase II (Pol II) or transcription factors [100,101,105]. lncRNA can also regulate post-transcriptional events [100,101,105] to control mRNA function by changing its stability, splicing patterns, and translation [105]. lncRNA can alter mRNA stability by interacting with a specific sequence motif of an RBP, resulting in the formation of lncRNA-protein complexes [106]. As such, lncRNA have numerous cellular functions including regulating proliferation, differentiation, and survival.
3. Post-Transcriptional Regulation in COPD
3.1. COPD Pathogenesis
COPD is a leading cause of chronic morbidity and mortality worldwide [107]. The World Health Organization (WHO) lists COPD as the third leading cause of death [108], with its prevalence expected to increase by more than 30% in the coming decade. The Global Initiative for Chronic Obstructive Lung Disease (GOLD) defines COPD as a lung disease characterized by progressive and irreversible airflow limitation, which is usually associated with an abnormal inflammatory response in the airways and lungs to noxious particles or gases. The clinical presentation in COPD patients includes cough, sputum production, and/or dyspnea [109]. Chronic airflow limitation is due to both emphysema, which is the irreversible destruction of the gas-exchanging alveoli, and chronic bronchitis, a disease entity characterized by the presence of a productive cough for at least three consecutive months during the last two consecutive years [110]. The risk factors for the development of COPD include a combination of genetic susceptibility and exposure to environmental toxicants [111].
The main cause of COPD is CS [111]. Globally, there are around 1.3 billion tobacco smokers [112]. CS is a complex combination of thousands of chemicals (approximately 7000 individual components) of which at least 158 have known toxicological properties [113,114]. The components with the strongest correlations to disease development are polycyclic aromatic hydrocarbons (PAHs) and N-nitrosamines. Other components that are associated with pulmonary toxicity include free radicals, catechols, and aldehydes [9]. Beyond CS, additional risk factors for COPD include childhood asthma and respiratory infections, exposure to ambient and biomass air pollution, exposure to second-hand smoke, and occupational exposure to dust and fumes [8,110,111,115]. Genetic factors are associated with the development of COPD. The most notable of these is deficiency of alpha-1 antitrypsin (α1AT), which accounts for approximately 1–2% of COPD cases [111,115]. Clinical symptoms can develop in patients many years after starting smoking, with COPD commonly diagnosed in people over the age of 50 years, with the highest frequency at approximately 70 years [116].
Mechanistically, the development of COPD is initiated by inflammation caused by repeated exposure to CS, which induces a pulmonary inflammatory response in several cell types, including epithelial cells, fibroblasts, and macrophages [10,11,12]. Repeated exposure to CS leads to the additional recruitment of innate and adaptive immune cells, including neutrophils, macrophages, and lymphocytes. This, in turn, amplifies the expression of inflammatory mediators, such as TNF-α, IL-6, C-C motif ligand 2 (CCL2), CCL7, C-X-C motif ligand 1 (CXCL1), CXCL5, CXCL8 (IL-8), leukotriene (LT) B4, and COX-2 [11,117,118,119,120,121,122]. In addition to inflammation, other pathogenic mechanisms involved in COPD include an imbalance between proteases and antiproteases, as well as heightened oxidative stress in the lungs [123]. Repeated exposure to CS causes the release of proteases. The increased production of lung proteases, such as neutrophil elastase (NE), and the resulting apoptosis of alveolar septal cells lead to the destruction of alveolar walls, causing emphysema [124]. Moreover, proteases, such as NE, cathepsin G, and proteinase-3 promote mucus secretion by increasing the number of goblet cells, stimulating degranulation in these cells, and causing the enlargement of submucosal glands. The combination of mucus hypersecretion, inflammation in the airway walls and lumen, fibrosis formation around the small airways, and loss of lung elastic recoil leads to the narrowing of these airways, leading to airflow obstruction [118,124,125] (Figure 2). Finally, the inflammation in COPD further increases during acute exacerbations, which are defined as a worsening of day-to-day symptoms, and are predominantly caused by bacterial or viral infection [117]. Exacerbations in COPD are strongly correlated with an increase in hospitalization and mortality, and a decrease in lung function [116].
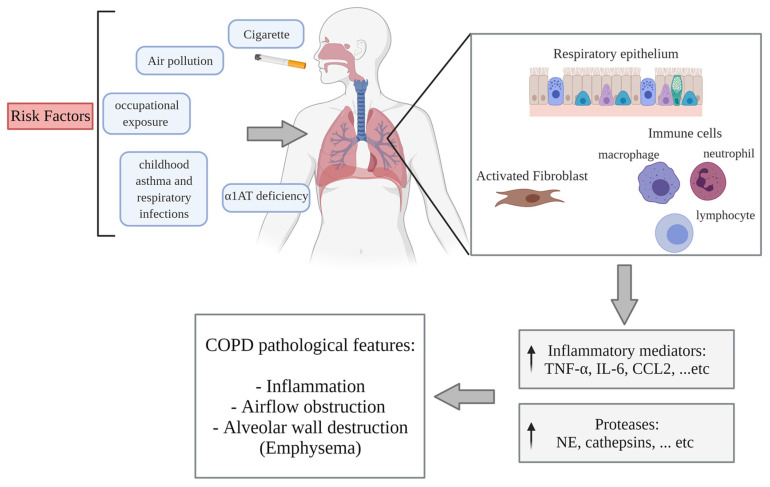
Figure 2.
Overview of the etiology and pathogenesis of COPD. Risk factors for the development of COPD include: cigarette smoke; air pollution; occupational exposures; childhood asthma; respiratory infections; and alpha 1-anti-trypsin (α1AT) deficiency. Upon exposure to inhaled toxicants, lung structural cells, including epithelial cells and fibroblasts, as well as alveolar macrophages, are activated. These cells produce inflammatory mediators to recruit additional inflammatory cells, such as neutrophils, macrophages, and lymphocytes, to the site of exposure. This augments the expression of inflammatory mediators, such as TNF-α, IL-6, CCL2, CCL7, CXCL1, CXCL5, CXCL8 (IL-8), LTB4, and COX-2, and releases proteases, such as NE, cathepsins, and MMPs. This cascade of events can lead to chronic pulmonary inflammation, airflow obstruction, and alveolar wall destruction (emphysema) in a susceptible individual. Created with BioRender.com.
No disease-modifying therapies exist for COPD. After smoke exposure and in COPD, transcriptional regulation alters the expression of inflammatory mediators, such as IL-6, COX-2, TNF-α, IL-1β, and IL-8 [126]. However, post-transcriptional regulation of mRNA has emerged as an important factor in the overall regulation of gene and protein expression in response to environmental exposures. A better understanding of the mechanistic underpinnings of post-transcriptional regulation of mRNA could lead to the development of new targeted therapies for COPD. Here, we summarize the current state-of-knowledge of post-transcriptional regulation that is applicable to pathogenic mechanisms implicated in the response to CS and in the development of COPD.
3.2. RBPs
3.2.1. The Response of RBPs to CS
RBPs appear to play a role in the cellular response to CS. We have previously shown that lung fibroblasts produce COX-2 in response to CS [11,121], and that the aryl hydrocarbon receptor (AhR) destabilizes PTGS2 mRNA by preventing HuR translocation into the cytoplasm [127]. It is now well-described that COX-2, and other inflammatory mediators that are increased in COPD [11,117,118,119,120,121,122], are regulated by HuR [128]. Furthermore, HuR controls IL-8 secretion from human bronchial epithelial cells exposed to cigarette smoke extract (CSE) in combination with human rhinovirus (HRV) [129]. HRV infection is a common trigger of virus-associated COPD exacerbations that are correlated with persistent lung inflammation [130,131]. Thus, HuR may be involved in the early pathogenic events, such as inflammation, that are associated with the development of COPD and/or exacerbations.
Another RBP that has been studied in the context of smoking is RNA-binding motif protein 5 (RBM5). The gene of RBM5, also known as H37 or Luca15, located in chromosomal region 3p21.3, is frequently deleted in heavy smokers and lung cancer patients [132,133]. Rbm5 loss-of-function (heterozygous) mice exposed to the tobacco carcinogen 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone (NNK) develop more aggressive lung cancer [134]. In cells exposed to CSE, RBM5 mRNA and protein levels are decreased, and β-catenin is increased. β-catenin is a key player in canonical Wnt signaling, whose activation induces genes involved in cell differentiation [135]. β-catenin is increased in proximal airway epithelium in COPD, with activation of Wnt/β-catenin signaling increasing epithelial-to-mesenchymal transition (EMT) [136]. EMT is a process where epithelial cells gradually lose cellular polarity and adhesiveness, and acquire migratory capacity and invasiveness, like that in a mesenchymal phenotype. EMT is increased in bronchial epithelial cells from COPD patients, which contributes to fibrosis formation around the small airways, leading to airflow obstruction [137]. Although the evidence is indirect, these studies raise the prospect that RBM5 could regulate EMT in COPD through the β-catenin pathway. At this writing, the expression and function of RBM5 in COPD is unknown.
3.2.2. The Regulation of RBPs in COPD
Elucidation of changes in the expression and the function of RBPs may suggest putative pathogenetic roles for them. Targeting RBPs could also be a novel therapeutic strategy. However, only a handful of studies directly investigate RBPs in COPD. One of these was a genome-wide association study which identified iron-responsive element binding protein 2 (IRP2 or IREB2), an RNA-binding protein that regulates cellular iron homeostasis, as a COPD susceptibility gene. IRP2 mRNA and protein levels are elevated in lungs from COPD subjects [138,139,140,141], and IRP2 expression is increased in the lungs of mice chronically exposed to CS. Furthermore, the knockout of IRP2 protected mice from CS-induced pulmonary inflammation and impairment of airway mucociliary clearance. Mechanistically, IRP2 in the lungs induces mitochondrial dysfunction by promoting mitochondrial iron loading and cytochrome c oxidase [142]. Previous observations have shown that iron deposition is increased in lungs from severe COPD patients, as well as in response to CS [143,144], which may be regulated by the elevation of IRP2.
Another RBP studied in the context of COPD is HuR, where its expression is increased in the airway epithelium from smokers with or without COPD [145]. This suggests that this increase is likely due to smoking, a notion further supported by a separate study showing that HuR expression is similar in the bronchial epithelium from both COPD subjects and smokers without COPD [146]. Mechanistically, recent studies support a role for HuR in the pathogenesis of COPD. For example, HuR stabilizes zinc finger E-box binding homeobox 1 (ZEB-1), a transcription factor involved in EMT. The expression of ZEB-1 is increased in the airway epithelium from COPD [145]. This suggests the possibility that HuR may be involved in the pathogenesis of COPD by regulating EMT.
Finally, the RBP AUF1, which participates in mRNA decay, is decreased in the bronchial epithelium from COPD subjects compared to smokers without COPD. Analysis of a microarray from a primary airway epithelium of COPD revealed that AUF1 target genes are upregulated, including those associated with inflammation [146]. Although this suggests that AUF1 may regulate the expression of inflammatory genes involved in COPD, direct regulation by AUF1 of these downstream mRNA, and its implications for the pathogenesis of COPD, remain to be investigated. Recently, a mapping profile of RBPs indicated that most RBP genes are downregulated in the small airway epithelium of those with COPD, comparing to non-smokers and smokers [147]. Overall, these studies raise the possibility that RBPs may be involved in the development of COPD. As little is known about the direct role of RBPs in COPD per se, below we summarize studies which have examined related mechanisms associated with the development of this disease.
RBPs in Inflammation
CS causes direct damage to airway and alveolar epithelial cells, which leads to the recruitment of inflammatory cells, and the release of numerous inflammatory mediators, including TNF-α [9,11,117,118,119,120,121,148], the overexpression of which induces pulmonary inflammation and airspace enlargement [149]. An RBP known to regulate TNF-α expression is TTP. TTP is generally an anti-inflammatory RBP, as TTP knockout mice have a proinflammatory phenotype [150]. TTP promotes mRNA decay of TNF-α by binding to AREs present within the 3’ UTR [52]. TTP also destabilizes other mRNAs associated with inflammation, including PTGS2, IL-6, CXCL8, and CCL2 [151,152]. Glucocorticoids, which are used clinically in COPD, elevate mRNA and protein levels of TTP that are crucial for glucocorticoid-mediated inhibition of TNF-α mRNA [153]. Glucocorticoid inhibition of other inflammatory genes, including CCL2, CCL7, CXCL1, and CXCL5, is also abrogated in TTP-knockout cells [154]. Overall, these studies suggest that TTP target mRNAs encode proteins responsible for the inflammatory response associated with COPD.
AUF1 is another RBP that induces the decay of target mRNAs, including TNF-α. AUF1 knockout mice are susceptible to endotoxin challenge due to TNF-α and IL-1β overproduction [155]—these mice also exhibit chronic dermatitis with age, concomitant with TNF-α and IL-1β overexpression [156]. Given that AUF1 expression is decreased in COPD [146], and that many inflammatory mediators regulated by AUF1 are also upregulated in COPD, it is possible that dysregulation of AUF1 may contribute to the inflammatory response associated with this disease.
While AUF1 and TTP may be important in controlling inflammation by promoting mRNA decay, other RBPs, such as HuR, stabilize mRNA associated with inflammation, such as CCL2 and TNF-α, in pulmonary epithelial cells [69], as well as macrophages [157]. HuR itself is also activated by TNF-α, as evidenced by its translocation to the cytoplasm following treatment with TNF-α and IL-4 to regulate CCL-11 (eotaxin) mRNA levels [158]. This may be relevant in the context of COPD, as eotaxin is an inflammatory chemokine whose expression is elevated in blood from COPD patients [159]. Thus, these studies highlight HuR as a key player in post-transcriptional gene regulation of inflammatory mRNAs.
RBPs in Apoptosis and Protease Expression
Emphysema is characterized by the loss of lung structural cells, including alveolar epithelial cells and fibroblasts [109,160]. Mechanistically, emphysema is thought to develop because of CS-induced apoptotic cell death [13,14]. Evidence for this comes from studies where intra-tracheal administration of active caspase-3 induced epithelial cell apoptosis, elastolytic activity in bronchoalveolar lavage (BAL), and airspace enlargement in mouse lungs [161]. Other proteins, such as vascular endothelial growth factor (VEGF), help alveolar cells withstand damage by CS. Experimentally, the blocking of VEGF receptors stimulates apoptosis of alveolar cells, and induces an emphysema-like pathology [162,163]. In COPD, the level of VEGF is decreased, which may be a contributing factor to the development of emphysema in people [164]. Many of the RPBs mentioned above that regulate inflammation also have roles in apoptosis. For example, TTP destabilizes VEGF mRNA [165]. In contrast, hnRNP L stabilizes VEGF expression [166]. hnRNP L is a multifunctional splicing factor that is involved in the regulation of alternate splicing and mRNA stability [166,167]. Interestingly, the knockout of hnRNP L in hematopoietic stem cells causes cell death through caspase-dependent pathways [168], raising the possibility that the downregulation of VEGF and upregulation of cell death in COPD could be regulated by hnRNP L.
In addition to aberrant cell death, lung tissue destruction in COPD is mediated by proteases. Activated neutrophils are a potent source of proteases, such as NE, cathepsin G, proteinase-3, MMP-8, and MMP-9, all of which can contribute to the destruction of alveolar walls [169,170]. Macrophages also secrete MMP-9 and MMP-12, as well as cathepsins L and S [171]. RBPs implicated in the regulation of proteases include TTP, which destabilizes MMP-9 and MMP-2 mRNAs [172], and HuR, which binds to MMP-9 mRNA to stabilize its expression [173]. Changes in the expression of RBPs can greatly impact their function and, thus, regulation of protease expression. For example, nitric oxide and IL-10 can both reduce HuR expression and its subsequent binding to MMP-9 mRNA [174,175].
3.2.3. Interplay of RBPs
Dynamic interactions between RBPs may fine-tune post-transcriptional modifications of common mRNAs. RBPs can either cooperate or compete to bind target mRNAs. For example, ADAR1 cooperates with HuR, and forms an RNA-dependent complex, which regulates the stability of ADAR1 targets in human B cells [176]. ADAR1 mediates A-to-I editing of cathepsin S (CTSS) [177], a cysteine protease associated with the remodeling/degradation of connective tissue and basement membrane [178]. HuR also binds to the 3′ UTR of CTSS mRNA, and controls its stability and expression [177]. Interestingly, CTSS is elevated in smokers and COPD patients [179]. HuR and TIA-1 can also interact to impact mRNA encoding programmed cell death 4 (PDCD4), a tumor suppressor that induces apoptosis. Here, increasing TIA-1 prevents HuR from binding to the PDCD4 mRNA, whereas decreasing TIA-1 induces HuR binding to the PDCD4 mRNA [180]. Furthermore, TTP interacts with PABP1 in activated primary mouse bone-marrow-derived-macrophages, and represses the translation of TTP target mRNAs involved in the inflammatory response [181]. Together, these studies illustrate the interplay of RBPs in the regulation of various post-transcriptional processes involved in physiological and pathological mechanisms.
3.3. miRNA
Another aspect of post-transcriptional regulatory mechanisms of relevance for smoke-induced lung disease are miRNAs. Thus, miRNAs are being pursued as therapeutic targets or, through their utility, as a diagnostic tool. For example, let-7, miR-206, and miR-16 are downregulated in lung cancer. The observation of miR-16 is of interest, as overexpression of miR-16 prevents cell proliferation and invasion [182,183,184,185]. Interestingly, a miR-16 mimic (the TargomiR drug, MesomiR-1) is in a phase 1 trial for patients with malignant pleural mesothelioma and lung cancer. Thus far, this trial has shown safety, as well as initial signs of response [186,187]. These studies suggest that miRNAs play a key role in the progression of diseases, and that targeting miRNAs as a diagnostic or as a therapeutic target for disease is attractive. Numerous studies have now comprehensively interrogated changes in miRNA expression caused by smoking and/or in COPD [188,189,190,191,192,193,194,195,196,197,198,199,200,201,202,203,204,205,206]. Smoking alters miRNA expression, as shown by observations in human smokers, as well as in lungs of mice and rats exposed to CS [190,191]. One such study showed that 34 miRNAs are differentially expressed between never-smokers, smokers, and COPD subjects, including 8 miRNAs that were downregulated when compared with never-smokers [197]. Some miRNAs are altered in the context of COPD itself, and the nature of this dysregulation may be cell-type specific, as observed for miR-199a-5p. Though miR-199a-5p expression is reduced in T-cells from COPD patients [198], miR-199a-5p expression is elevated in lung tissue [199].
As the severity of COPD increases, miRNA expression changes. For example, miR-206, miR-133a-5p, and miR-133a-3p are upregulated in extracellular vesicles of plasma from moderate COPD patients compared to mild and severe patients—these miRNAs are involved in hundreds of biological processes that are associated with COPD features [196]. miRNAs are differentially expressed based on emphysema severity, and these changes correlate with the changes in the expression of their predicted mRNA targets [205]. Some of the miRNAs altered by emphysema severity include miR-34c, miR-34b, miR-133b, and miR-149, which are reduced in lung tissue from COPD patients with moderate emphysema compared to mild disease [206]. Thus, there is differential expression of miRNAs in COPD and/or in response to CS [188,189,190,191,192,193,194,195,196,197,198,199,200,201,202,203,204,205,206], which may, in turn, perturb downstream pathways controlling pathological processes, such as inflammation and cell survival.
3.3.1. miRNA and Pathogenic Mechanisms of COPD
Experimental studies profiling miRNA expression have shed light on the possible biological significance of dysregulated miRNA to COPD pathogenesis, including their ability to regulate smoke-induced inflammation. In one such study of miRNA expression in current and never-smokers’ bronchial airway epithelium, it was found that 28 miRNAs are differentially expressed. The most downregulated miRNA in smokers was miR-218—its expression is also decreased in primary bronchial epithelial cells exposed to cigarette smoke condensate [188]. The mature form of miR-218 is generated from two separate loci, miR-218-1 and miR-218-2 [207]. miR-218 targets MAFG, a transcription factor that is elevated in smokers and in response to CS [188], and induces pro-inflammatory gene transcription [208]. miR-218-5p (miR-218-2) is also significantly downregulated in lung tissue from smokers and COPD patients, as well as in response to experimental smoke exposure. Overexpression of miR-218-5p in normal human bronchial epithelial cells exposed to CSE reduces the mRNA and the protein levels of CCL20 and CXCL8 [200], chemokines that are involved in the pathogenesis of COPD [118,209]. Furthermore, inhibiting miR-218-5p in mice exposed to smoke worsens smoke-induced inflammation [200]. These findings indicate an important role of miR-218-5p in the CS-induced inflammatory response.
Other miRNA implicated in regulating inflammation are miR-181c, miR-145, miR146a, and miR-16, all of which are downregulated in response to CS and/or in COPD [190,201,202,210]. Overexpression of miR-181c in mice exposed to CS reduces neutrophil infiltration, in conjunction with IL-6 and CXCL8 expression in the lungs. Overexpression of miR-181c in primary human bronchial epithelial cells treated with CSE also decreases IL-6 and CXCL8 expression [201]. miR-145 is also downregulated in the lungs of mice exposed to CS [190]. Outside the lungs, miR-145 overexpression represses the release of IL-6 and CXCL8 [211], as well as VEGF and MMP-9 [212]. Finally, miR-146a is downregulated in the serum of COPD patients with acute exacerbations when compared with stable COPD patients and healthy controls, which, in turn, is negatively correlated with inflammatory cytokines [202]. Mechanistically, we found that miR-146a suppresses cigarette smoke-induced COX-2 protein expression in murine lung fibroblasts [203]. miR-146a also suppresses COX-2 in lung fibroblasts from COPD subjects upon IL-1β/TNF-α stimulation, and, therefore, reduces prostaglandin (PG)E2 production [204]. miR-16 is another miRNA whose expression is decreased in lung fibroblasts from heavy smokers and COPD patients compared to those from non-smokers [210]—miR-16 also silences COX-2 [213]. These findings suggest that the loss of this miRNA can eventually enhance COX-2 expression in response to smoke.
Other miRNAs are increased by smoking, include miR-101, miR-144 [192], miR-135b [193,194], and miR-223. miR-223 leads to a decrease in histone deacetylase 2 (HDAC2) expression, which alters the expression of pro-inflammatory chemokines [195]. miR-101 and miR-144 are higher in human bronchial epithelial cells exposed to CS, and suppresses expression of the cystic fibrosis transmembrane regulator (CFTR), a chloride channel which maintains airway surface fluid homeostasis [192]. miR-101 is also upregulated in lungs of mice exposed to CS [192].
Alveolar macrophages are among the first cell types to respond to smoke inhalation, as these innate immune cells patrol the luminal surface of alveoli [171]. Alveolar macrophages are implicated in the development of COPD, and show impaired phagocytosis of pathogens and efferocytosis of apoptotic cells, a feature that might contribute to worsening inflammation [171,214]. Smoking also changes miRNA expression in alveolar macrophages [189]. In alveolar macrophages from current and never smokers, 43 miRNAs are downregulated in smokers. One of these miRNAs is miR-452. Mechanistically, inhibition of miR-452 induces the expression of MMP-12 [189], a protease that is upregulated in alveolar macrophages from smokers [189,215].
In addition to miRNA playing fundamental roles in inflammation, many miRNAs are also implicated in regulating cell survival. Both miR-23a and miR-421, for example, repress caspase-9 and caspase-3, respectively [216,217]. miR-421 is downregulated in response to smoke [191], whereas the activity of caspase-3 is increased by smoke exposure [218]. This raises the possibility that downregulation of miR-421 in response to smoke may upregulate apoptosis.
3.3.2. Interplay of RBPs with miRNAs
miRNAs and RBPs interact to further fine-tune post-transcriptional regulatory mechanisms. For example, hnRNP K increases PTGS2 mRNA stability by reducing miR-16 binding to the 3′UTR of PTGS2 mRNA [219], and overexpression of HuR suppresses the ability of miR-16 to promote PTGS2 mRNA decay [220]. HuR also promotes the translation of STAT3 mRNA in myotubes exposed to IFN-γ and TNF-α by binding to its 3′UTR—this binding interferes with miR-330–mediated translation inhibition [221]. HuR could also work cooperatively with miRNA to downregulate gene expression. An example of this is the ability of HuR to promote the interaction of let-7- loaded RISC with the 3′UTR of the proto-oncogene MYC mRNA to repress its expression [222]. Other examples include the ability of the RBP Pumilio-1 to bind to 3′UTR of p27 (CDKN1B; cyclin dependent kinase inhibitor 1b) mRNA, and facilitate its association with miR-221/222 to destabilize p27 expression [223]. While these examples are outside the context of COPD, the discovery of crosstalk between RBPs and miRNAs provides support for their dynamic regulation of gene expression associated with cellular mechanisms whose dysregulation contributes to the pathogenesis of chronic lung disease development.
3.4. LncRNA in COPD
Altered expression of lncRNA is now thought to be involved in the pathogenesis of COPD. For example, a previous study found that 109 lncRNA are differentially expressed in the lungs of mice exposed to CS, of which 51 lncRNAs were significantly upregulated, and 58 were significantly downregulated. Gene ontology analysis of potential lncRNA target protein-coding genes showed enrichment in pathways involved in the cellular response to interferon-beta [224]. Furthermore, genome-wide expression analysis of lncRNAs in lung tissue from non-smokers and smokers with/without COPD showed differential expression of hundreds of lncRNAs in COPD, independent of smoking [225]. Another study found that 8376 lncRNAs are differentially expressed in COPD lung, of which 3939 are upregulated, and 4437 are downregulated [226]. However, the mechanism of how lncRNA affect the pathogenesis of COPD is not fully understood.
3.4.1. LncRNA and Pathogenic Mechanisms of COPD
Dysregulation of lncRNA expression suggests they have roles in the pathogenesis of CS-related diseases. For example, lncRNA are upregulated by smoking: this includes smoke- and cancer–associated lncRNA–1 (SCAL1) [227], and lung cancer progression–association transcript 1 (LCPAT1) [228]. LCPAT1 is involved in smoke-induced DNA damage [228], and SCAL1 protects against CS–induced toxicity [227], suggesting that SCAL1 serves as a protective mechanism against smoke. Metastasis-associated in lung adenocarcinoma transcript 1 (MALAT1) [229] and HOX transcript antisense RNA (HOTAIR) [230] are also upregulated in response to CS, and involved in CS-induced EMT.
LncRNAs that are involved in EMT are also increased in COPD, including MALAT1 [231] and taurine-upregulated gene 1 (TUG1) [226]. The knockdown of MALAT1 in human lung fibroblasts reduces the expression of fibronectin and α-smooth muscle actin, proteins that are involved in fibrogenesis, in response to transforming growth factor β (TGF-β) [231]. The knockdown of TUG1 also decreases the expression of fibronectin and α-smooth muscle actin in human lung epithelial cells and lung fibroblasts exposed to TGF-β [226]. These data suggest that lncRNAs may regulate fibrosis formation around the small airways in COPD, in part, via the regulation of EMT.
LncRNAs are also involved in inflammation, such as maternally expressed gene 3 (MEG3) [232], MALAT1 [233], and HOTAIR [234]. Silencing of MEG3 inhibits apoptosis, and reduces inflammatory mediators in human bronchial epithelial cells exposed to CSE [232]. Interestingly, MEG3 is elevated in blood samples from COPD patients and smokers [232], suggesting that MEG3 may regulate CS-induced inflammation and apoptosis. Additionally, in macrophages exposed to LPS, the expression of MALAT1 is upregulated, which, in turn, interacts with nuclear factor kappa B (NF-κB) in the nucleus, and reduces the expression of inflammatory cytokines [233]. The expression of HOTAIR is also upregulated in in cardiomyocytes exposed to LPS. Conversely, HOTAIR induces inflammatory response by activating NF-κB [234], and regulates oxidative stress and apoptosis [235]. It is currently not known whether the increase in MALAT1 and HOTAIR expression from CS [229,230] regulates inflammation and/or apoptosis in COPD pathogenesis.
3.4.2. Interplay of RBPs with lncRNA
LncRNA are known to interact with RBPs, a feature which might be important in understanding the functional role of RBPs and lncRNAs in the development of lung diseases. For example, the lncRNA c-Myc-upregulated (MYU) associates with hnRNP K to stabilize the mRNA of cyclin-dependent kinase 6 (CDK6), which promotes G1/S transition of the cell cycle. HnRNP K also inhibits miR-16 binding to the 3′UTR of CDK6 [236]. Although hnRNP K stabilizes PTGS2 mRNA by reducing the association of miR-16 to the PTGS2 3′UTR [219], hnRNP K may collaborate with MYU to regulate the stability of PTGS2 mRNA. Another example of such an association is the lncRNA functional intergenic repeating RNA element (FIRRE), which interacts with hnRNP U to stabilize vascular cell adhesion molecule 1 (VCAM1), a cell adhesion molecule that is involved in inflammation [237]. Finally, the lncRNA lincRNA regulator of reprogramming (Linc-RoR) stabilizes the proto-oncogene MYC mRNA by interacting with two RBPs. Linc-RoR interacts with hnRNP I to stabilize MYC mRNA, and interacts with the destabilizing RBP AUF1 to inhibit its binding to MYC mRNA [238]. This suggests that Linc-RoR may control the competition of the two RBPs for MYC mRNA. The discovery of the interplay between RBPs and lncRNAs highlights their regulation of post-transcriptional mechanisms of target mRNAs. This would be another step in understanding the landscape of genes that are involved in the pathogenesis of chronic lung disease.
4. Conclusions
RBPs, miRNA, and lncRNA are examples of post-transcriptional regulons that may be involved in COPD pathogenesis. To date, many studies have largely focused on transcriptional regulatory pathways implicated in the development of COPD. However, it is increasingly apparent that post-transcriptional regulation of gene expression adds a dynamic layer of complexity to chronic diseases, as RBPs regulate polyadenylation, pre-mRNA splicing, RNA modification, nuclear export, localization, and turnover of target mRNAs. Similarly, miRNA and lncRNA typically regulate post-transcriptional repression of target mRNAs. RBPs, miRNA, and lncRNA function independently, or may operate cooperatively or competitively, adding complexity to the system. Altered function of post-transcriptional regulation may contribute to the development of chronic diseases, particularly those caused by environmental exposures, such as COPD, and future work should address these mechanisms. In this regard, both RBPs and miRNAs represent potential therapeutic targets. For instance, MS-444 is a small molecule inhibitor that interferes with the RNA binding activity of HuR [239], and has been shown to exhibit antitumor activity in vitro and in vivo [240]. miRNAs are also targeted as disease treatment, such as let-7, whose exogenous delivery reduces tumor development in a mouse model of lung cancer [241]. Therefore, post-transcriptional regulation of protein expression deserves serious consideration in therapeutic strategies for smoke-related diseases, such as COPD.
Abbreviations
| α1AT | Alpha 1-anti-trypsin |
| ADAR | Adenosine deaminases acting on RNA |
| AhR | Aryl hydrocarbon receptor |
| AUF1 | AU-binding factor 1 |
| ARE | AU-rich elements |
| AS | Alternative splicing |
| BAL | Bronchoalveolar lavage |
| CCL | C-C motif ligand |
| COPD | Chronic obstructive pulmonary disease |
| COX-2/PTGS2 | Cyclooxygenase-2 |
| CS | Cigarette smoke |
| CSE | Cigarette smoke extract |
| CXCL | C-X-C motif ligand |
| EMT | Epithelial-to-mesenchymal transition |
| GOLD | Global initiative for chronic obstructive lung disease |
| GR | Glucocorticoid receptors |
| hnRNP | Heterogeneous nuclear ribonucleoprotein |
| HOTAIR | HOX transcript antisense RNA |
| HuR | Hu antigen R |
| IL | Interleukin |
| IRP2 | Iron-responsive element binding protein 2 |
| LCPAT1 | Lung cancer progression–association transcript 1 |
| Linc-RoR | LincRNA regulator of reprogramming |
| LncRNA | Long non-coding RNAs |
| LPS | Lipopolysaccharide |
| MALAT1 | Metastasis-associated in lung adenocarcinoma transcript 1 |
| MEG3 | Maternally expressed gene 3 |
| MiRNA/MiR | MicroRNA |
| MMP | Matrix metalloproteinase |
| NE | Neutrophil Elastase |
| NF-κB | Nuclear Factor Kappa B |
| PABP | Poly(A)-binding protein |
| RBD | RNA-binding domain |
| RBM5 | RNA-binding motif protein 5 |
| RBP | RNA-binding protein |
| RISC | RNA-induced silencing complex |
| RNP | Ribonucleoprotein |
| SCAL1 | Smoke and cancer–associated lncRNA–1 |
| SGs | Stress granules |
| TIA-1 | T-cell Intracellular Antigen 1 |
| TNF-α | Tumor necrosis factor α |
| TTP | Tristetraprolin |
| UTR | Untranslated region |
| VEGF | Vascular endothelial growth factor |
Author Contributions
Manuscript writing, review and editing: N.A., D.H.E. and C.J.B.; writing: A.A. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the Canada Foundation for Innovation (CFI), the Canadian Institutes for Health Research (CIHR; Project Grants PJT-168836 and PJT-162273), and the Natural Sciences and Engineering Research Council of Canada (NSERC). C.J.B. was supported by a salary award from the Fonds de recherche du Quebec-Sante (FRQ-S). N.A. was supported by a scholarship from Taibah University, Saudi Arabia. A.A. was supported by a scholarship from the Ministry of Education, Saudi Arabia.
Conflicts of Interest
The authors declare that they have no conflict of interests.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Orphanides G., Reinberg D. A unified theory of gene expression. Cell. 2002;108:439–451. doi: 10.1016/S0092-8674(02)00655-4. [DOI] [PubMed] [Google Scholar]
- 2.Keene J.D. RNA regulons: Coordination of post-transcriptional events. Nat. Rev. Genet. 2007;8:533–543. doi: 10.1038/nrg2111. [DOI] [PubMed] [Google Scholar]
- 3.He R.Z., Luo D.X., Mo Y.Y. Emerging roles of lncRNAs in the post-transcriptional regulation in cancer. Genes Dis. 2019;6:6–15. doi: 10.1016/j.gendis.2019.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Corley M., Burns M.C., Yeo G.W. How RNA-Binding Proteins Interact with RNA: Molecules and Mechanisms. Mol. Cell. 2020;78:9–29. doi: 10.1016/j.molcel.2020.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Alles J., Fehlmann T., Fischer U., Backes C., Galata V., Minet M., Hart M., Abu-Halima M., Grasser F.A., Lenhof H.P., et al. An estimate of the total number of true human miRNAs. Nucleic Acids Res. 2019;47:3353–3364. doi: 10.1093/nar/gkz097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fang S., Zhang L., Guo J., Niu Y., Wu Y., Li H., Zhao L., Li X., Teng X., Sun X., et al. NONCODEV5: A comprehensive annotation database for long non-coding RNAs. Nucleic Acids Res. 2018;46:D308–D314. doi: 10.1093/nar/gkx1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hon C.C., Ramilowski J.A., Harshbarger J., Bertin N., Rackham O.J., Gough J., Denisenko E., Schmeier S., Poulsen T.M., Severin J., et al. An atlas of human long non-coding RNAs with accurate 5’ ends. Nature. 2017;543:199–204. doi: 10.1038/nature21374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tan W.C., Sin D.D., Bourbeau J., Hernandez P., Chapman K.R., Cowie R., FitzGerald J.M., Marciniuk D.D., Maltais F., Buist A.S., et al. Characteristics of COPD in never-smokers and ever-smokers in the general population: Results from the CanCOLD study. Thorax. 2015;70:822–829. doi: 10.1136/thoraxjnl-2015-206938. [DOI] [PubMed] [Google Scholar]
- 9.Bhalla D.K., Hirata F., Rishi A.K., Gairola C.G. Cigarette smoke, inflammation, and lung injury: A mechanistic perspective. J. Toxicol. Environ. Health B Crit. Rev. 2009;12:45–64. doi: 10.1080/10937400802545094. [DOI] [PubMed] [Google Scholar]
- 10.Stampfli M.R., Anderson G.P. How cigarette smoke skews immune responses to promote infection, lung disease and cancer. Nat. Rev. Immunol. 2009;9:377–384. doi: 10.1038/nri2530. [DOI] [PubMed] [Google Scholar]
- 11.Martey C.A., Pollock S.J., Turner C.K., O’Reilly K.M., Baglole C.J., Phipps R.P., Sime P.J. Cigarette smoke induces cyclooxygenase-2 and microsomal prostaglandin E2 synthase in human lung fibroblasts: Implications for lung inflammation and cancer. Am. J. Physiol. Lung Cell Mol. Physiol. 2004;287:L981–L991. doi: 10.1152/ajplung.00239.2003. [DOI] [PubMed] [Google Scholar]
- 12.Li C.J., Ning W., Matthay M.A., Feghali-Bostwick C.A., Choi A.M. MAPK pathway mediates EGR-1-HSP70-dependent cigarette smoke-induced chemokine production. Am. J. Physiol. Lung Cell Mol. Physiol. 2007;292:L1297–L1303. doi: 10.1152/ajplung.00194.2006. [DOI] [PubMed] [Google Scholar]
- 13.Kosmider B., Messier E.M., Chu H.W., Mason R.J. Human alveolar epithelial cell injury induced by cigarette smoke. PLoS ONE. 2011;6:e26059. doi: 10.1371/journal.pone.0026059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Baglole C.J., Bushinsky S.M., Garcia T.M., Kode A., Rahman I., Sime P.J., Phipps R.P. Differential induction of apoptosis by cigarette smoke extract in primary human lung fibroblast strains: Implications for emphysema. Am. J. Physiol. Lung Cell Mol. Physiol. 2006;291:L19–L29. doi: 10.1152/ajplung.00306.2005. [DOI] [PubMed] [Google Scholar]
- 15.Dreyfuss G., Kim V.N., Kataoka N. Messenger-RNA-binding proteins and the messages they carry. Nat. Rev. Mol. Cell Biol. 2002;3:195–205. doi: 10.1038/nrm760. [DOI] [PubMed] [Google Scholar]
- 16.Burd C.G., Dreyfuss G. Conserved structures and diversity of functions of RNA-binding proteins. Science. 1994;265:615–621. doi: 10.1126/science.8036511. [DOI] [PubMed] [Google Scholar]
- 17.Glisovic T., Bachorik J.L., Yong J., Dreyfuss G. RNA-binding proteins and post-transcriptional gene regulation. FEBS Lett. 2008;582:1977–1986. doi: 10.1016/j.febslet.2008.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lunde B.M., Moore C., Varani G. RNA-binding proteins: Modular design for efficient function. Nat. Rev. Mol. Cell Biol. 2007;8:479–490. doi: 10.1038/nrm2178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lorković Z.J. RNA Binding Proteins. Landes Bioscience; Austin, TX, USA: 2012. p. 162. [Google Scholar]
- 20.Auweter S.D., Oberstrass F.C., Allain F.H. Sequence-specific binding of single-stranded RNA: Is there a code for recognition? Nucleic Acids Res. 2006;34:4943–4959. doi: 10.1093/nar/gkl620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Richter J.D., Zhao X. The molecular biology of FMRP: New insights into fragile X syndrome. Nat. Rev. Neurosci. 2021;22:209–222. doi: 10.1038/s41583-021-00432-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Takagaki Y., MacDonald C.C., Shenk T., Manley J.L. The human 64-kDa polyadenylylation factor contains a ribonucleoprotein-type RNA binding domain and unusual auxiliary motifs. Proc. Natl. Acad. Sci. USA. 1992;89:1403–1407. doi: 10.1073/pnas.89.4.1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shimberg G.D., Michalek J.L., Oluyadi A.A., Rodrigues A.V., Zucconi B.E., Neu H.M., Ghosh S., Sureschandra K., Wilson G.M., Stemmler T.L., et al. Cleavage and polyadenylation specificity factor 30: An RNA-binding zinc-finger protein with an unexpected 2Fe-2S cluster. Proc. Natl. Acad. Sci. USA. 2016;113:4700–4705. doi: 10.1073/pnas.1517620113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Neve J., Patel R., Wang Z., Louey A., Furger A.M. Cleavage and polyadenylation: Ending the message expands gene regulation. RNA Biol. 2017;14:865–890. doi: 10.1080/15476286.2017.1306171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Minvielle-Sebastia L., Keller W. mRNA polyadenylation and its coupling to other RNA processing reactions and to transcription. Curr. Opin. Cell Biol. 1999;11:352–357. doi: 10.1016/S0955-0674(99)80049-0. [DOI] [PubMed] [Google Scholar]
- 26.Dettwiler S., Aringhieri C., Cardinale S., Keller W., Barabino S.M. Distinct sequence motifs within the 68-kDa subunit of cleavage factor Im mediate RNA binding, protein-protein interactions, and subcellular localization. J. Biol. Chem. 2004;279:35788–35797. doi: 10.1074/jbc.M403927200. [DOI] [PubMed] [Google Scholar]
- 27.Wahle E. A novel poly(A)-binding protein acts as a specificity factor in the second phase of messenger RNA polyadenylation. Cell. 1991;66:759–768. doi: 10.1016/0092-8674(91)90119-J. [DOI] [PubMed] [Google Scholar]
- 28.Bienroth S., Keller W., Wahle E. Assembly of a processive messenger RNA polyadenylation complex. EMBO J. 1993;12:585–594. doi: 10.1002/j.1460-2075.1993.tb05690.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhu H., Zhou H.L., Hasman R.A., Lou H. Hu proteins regulate polyadenylation by blocking sites containing U-rich sequences. J. Biol. Chem. 2007;282:2203–2210. doi: 10.1074/jbc.M609349200. [DOI] [PubMed] [Google Scholar]
- 30.Staley J.P., Guthrie C. Mechanical devices of the spliceosome: Motors, clocks, springs, and things. Cell. 1998;92:315–326. doi: 10.1016/S0092-8674(00)80925-3. [DOI] [PubMed] [Google Scholar]
- 31.Zahler A.M., Lane W.S., Stolk J.A., Roth M.B. SR proteins: A conserved family of pre-mRNA splicing factors. Genes Dev. 1992;6:837–847. doi: 10.1101/gad.6.5.837. [DOI] [PubMed] [Google Scholar]
- 32.Will C.L., Luhrmann R. Spliceosome structure and function. Cold Spring Harb. Perspect. Biol. 2011;3:a003707. doi: 10.1101/cshperspect.a003707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Johnson J.M., Castle J., Garrett-Engele P., Kan Z., Loerch P.M., Armour C.D., Santos R., Schadt E.E., Stoughton R., Shoemaker D.D. Genome-wide survey of human alternative pre-mRNA splicing with exon junction microarrays. Science. 2003;302:2141–2144. doi: 10.1126/science.1090100. [DOI] [PubMed] [Google Scholar]
- 34.Pan Q., Shai O., Lee L.J., Frey B.J., Blencowe B.J. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat. Genet. 2008;40:1413–1415. doi: 10.1038/ng.259. [DOI] [PubMed] [Google Scholar]
- 35.Wang E.T., Sandberg R., Luo S., Khrebtukova I., Zhang L., Mayr C., Kingsmore S.F., Schroth G.P., Burge C.B. Alternative isoform regulation in human tissue transcriptomes. Nature. 2008;456:470–476. doi: 10.1038/nature07509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang Z., Burge C.B. Splicing regulation: From a parts list of regulatory elements to an integrated splicing code. RNA. 2008;14:802–813. doi: 10.1261/rna.876308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Huelga S.C., Vu A.Q., Arnold J.D., Liang T.Y., Liu P.P., Yan B.Y., Donohue J.P., Shiue L., Hoon S., Brenner S., et al. Integrative genome-wide analysis reveals cooperative regulation of alternative splicing by hnRNP proteins. Cell Rep. 2012;1:167–178. doi: 10.1016/j.celrep.2012.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Izquierdo J.M. Hu antigen R (HuR) functions as an alternative pre-mRNA splicing regulator of Fas apoptosis-promoting receptor on exon definition. J. Biol. Chem. 2008;283:19077–19084. doi: 10.1074/jbc.M800017200. [DOI] [PubMed] [Google Scholar]
- 39.Bass B.L. RNA editing and hypermutation by adenosine deamination. Trends Biochem. Sci. 1997;22:157–162. doi: 10.1016/S0968-0004(97)01035-9. [DOI] [PubMed] [Google Scholar]
- 40.Schaub M., Keller W. RNA editing by adenosine deaminases generates RNA and protein diversity. Biochimie. 2002;84:791–803. doi: 10.1016/S0300-9084(02)01446-3. [DOI] [PubMed] [Google Scholar]
- 41.Valente L., Nishikura K. ADAR gene family and A-to-I RNA editing: Diverse roles in posttranscriptional gene regulation. Prog. Nucleic Acid Res. Mol. Biol. 2005;79:299–338. doi: 10.1016/S0079-6603(04)79006-6. [DOI] [PubMed] [Google Scholar]
- 42.Athanasiadis A., Rich A., Maas S. Widespread A-to-I RNA editing of Alu-containing mRNAs in the human transcriptome. PLoS Biol. 2004;2:e391. doi: 10.1371/journal.pbio.0020391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Keller W., Wolf J., Gerber A. Editing of messenger RNA precursors and of tRNAs by adenosine to inosine conversion. FEBS Lett. 1999;452:71–76. doi: 10.1016/S0014-5793(99)00590-6. [DOI] [PubMed] [Google Scholar]
- 44.Gerber A.P., Keller W. RNA editing by base deamination: More enzymes, more targets, new mysteries. Trends Biochem. Sci. 2001;26:376–384. doi: 10.1016/S0968-0004(01)01827-8. [DOI] [PubMed] [Google Scholar]
- 45.Bass B.L. RNA editing by adenosine deaminases that act on RNA. Annu. Rev. Biochem. 2002;71:817–846. doi: 10.1146/annurev.biochem.71.110601.135501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bass B.L., Nishikura K., Keller W., Seeburg P.H., Emeson R.B., O’Connell M.A., Samuel C.E., Herbert A. A standardized nomenclature for adenosine deaminases that act on RNA. RNA. 1997;3:947–949. [PMC free article] [PubMed] [Google Scholar]
- 47.Chen C.X., Cho D.S., Wang Q., Lai F., Carter K.C., Nishikura K. A third member of the RNA-specific adenosine deaminase gene family, ADAR3, contains both single- and double-stranded RNA binding domains. RNA. 2000;6:755–767. doi: 10.1017/S1355838200000170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Levanon E.Y., Eisenberg E., Yelin R., Nemzer S., Hallegger M., Shemesh R., Fligelman Z.Y., Shoshan A., Pollock S.R., Sztybel D., et al. Systematic identification of abundant A-to-I editing sites in the human transcriptome. Nat. Biotechnol. 2004;22:1001–1005. doi: 10.1038/nbt996. [DOI] [PubMed] [Google Scholar]
- 49.Kim D.D., Kim T.T., Walsh T., Kobayashi Y., Matise T.C., Buyske S., Gabriel A. Widespread RNA editing of embedded alu elements in the human transcriptome. Genome Res. 2004;14:1719–1725. doi: 10.1101/gr.2855504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schoenberg D.R., Maquat L.E. Regulation of cytoplasmic mRNA decay. Nat. Rev. Genet. 2012;13:246–259. doi: 10.1038/nrg3160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Blackshear P.J. Tristetraprolin and other CCCH tandem zinc-finger proteins in the regulation of mRNA turnover. Pt 6Biochem. Soc. Trans. 2002;30:945–952. doi: 10.1042/bst0300945. [DOI] [PubMed] [Google Scholar]
- 52.Carballo E., Lai W.S., Blackshear P.J. Feedback inhibition of macrophage tumor necrosis factor-alpha production by tristetraprolin. Science. 1998;281:1001–1005. doi: 10.1126/science.281.5379.1001. [DOI] [PubMed] [Google Scholar]
- 53.Lai W.S., Carballo E., Strum J.R., Kennington E.A., Phillips R.S., Blackshear P.J. Evidence that tristetraprolin binds to AU-rich elements and promotes the deadenylation and destabilization of tumor necrosis factor alpha mRNA. Mol. Cell Biol. 1999;19:4311–4323. doi: 10.1128/MCB.19.6.4311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kratochvill F., Machacek C., Vogl C., Ebner F., Sedlyarov V., Gruber A.R., Hartweger H., Vielnascher R., Karaghiosoff M., Rulicke T., et al. Tristetraprolin-driven regulatory circuit controls quality and timing of mRNA decay in inflammation. Mol. Syst. Biol. 2011;7:560. doi: 10.1038/msb.2011.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sauer I., Schaljo B., Vogl C., Gattermeier I., Kolbe T., Muller M., Blackshear P.J., Kovarik P. Interferons limit inflammatory responses by induction of tristetraprolin. Blood. 2006;107:4790–4797. doi: 10.1182/blood-2005-07-3058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ogilvie R.L., Abelson M., Hau H.H., Vlasova I., Blackshear P.J., Bohjanen P.R. Tristetraprolin down-regulates IL-2 gene expression through AU-rich element-mediated mRNA decay. J. Immunol. 2005;174:953–961. doi: 10.4049/jimmunol.174.2.953. [DOI] [PubMed] [Google Scholar]
- 57.Phillips K., Kedersha N., Shen L., Blackshear P.J., Anderson P. Arthritis suppressor genes TIA-1 and TTP dampen the expression of tumor necrosis factor alpha, cyclooxygenase 2, and inflammatory arthritis. Proc. Natl. Acad. Sci. USA. 2004;101:2011–2016. doi: 10.1073/pnas.0400148101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Winzen R., Thakur B.K., Dittrich-Breiholz O., Shah M., Redich N., Dhamija S., Kracht M., Holtmann H. Functional analysis of KSRP interaction with the AU-rich element of interleukin-8 and identification of inflammatory mRNA targets. Mol. Cell Biol. 2007;27:8388–8400. doi: 10.1128/MCB.01493-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Leppek K., Schott J., Reitter S., Poetz F., Hammond M.C., Stoecklin G. Roquin promotes constitutive mRNA decay via a conserved class of stem-loop recognition motifs. Cell. 2013;153:869–881. doi: 10.1016/j.cell.2013.04.016. [DOI] [PubMed] [Google Scholar]
- 60.Paschoud S., Dogar A.M., Kuntz C., Grisoni-Neupert B., Richman L., Kuhn L.C. Destabilization of interleukin-6 mRNA requires a putative RNA stem-loop structure, an AU-rich element, and the RNA-binding protein AUF1. Mol. Cell Biol. 2006;26:8228–8241. doi: 10.1128/MCB.01155-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gratacos F.M., Brewer G. The role of AUF1 in regulated mRNA decay. Wiley Interdiscip. Rev. RNA. 2010;1:457–473. doi: 10.1002/wrna.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.White E.J., Brewer G., Wilson G.M. Post-transcriptional control of gene expression by AUF1: Mechanisms, physiological targets, and regulation. Biochim. Biophys. Acta. 2013;1829:680–688. doi: 10.1016/j.bbagrm.2012.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wang W., Caldwell M.C., Lin S., Furneaux H., Gorospe M. HuR regulates cyclin A and cyclin B1 mRNA stability during cell proliferation. EMBO J. 2000;19:2340–2350. doi: 10.1093/emboj/19.10.2340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Figueroa A., Cuadrado A., Fan J., Atasoy U., Muscat G.E., Munoz-Canoves P., Gorospe M., Munoz A. Role of HuR in skeletal myogenesis through coordinate regulation of muscle differentiation genes. Mol. Cell Biol. 2003;23:4991–5004. doi: 10.1128/MCB.23.14.4991-5004.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.van der Giessen K., Di-Marco S., Clair E., Gallouzi I.E. RNAi-mediated HuR depletion leads to the inhibition of muscle cell differentiation. J. Biol. Chem. 2003;278:47119–47128. doi: 10.1074/jbc.M308889200. [DOI] [PubMed] [Google Scholar]
- 66.Dormoy-Raclet V., Menard I., Clair E., Kurban G., Mazroui R., Di Marco S., von Roretz C., Pause A., Gallouzi I.E. The RNA-binding protein HuR promotes cell migration and cell invasion by stabilizing the beta-actin mRNA in a U-rich-element-dependent manner. Mol. Cell Biol. 2007;27:5365–5380. doi: 10.1128/MCB.00113-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ishimaru D., Ramalingam S., Sengupta T.K., Bandyopadhyay S., Dellis S., Tholanikunnel B.G., Fernandes D.J., Spicer E.K. Regulation of Bcl-2 expression by HuR in HL60 leukemia cells and A431 carcinoma cells. Mol. Cancer Res. 2009;7:1354–1366. doi: 10.1158/1541-7786.MCR-08-0476. [DOI] [PubMed] [Google Scholar]
- 68.Doller A., Huwiler A., Muller R., Radeke H.H., Pfeilschifter J., Eberhardt W. Protein kinase C alpha-dependent phosphorylation of the mRNA-stabilizing factor HuR: Implications for posttranscriptional regulation of cyclooxygenase-2. Mol. Biol. Cell. 2007;18:2137–2148. doi: 10.1091/mbc.e06-09-0850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Fan J., Ishmael F.T., Fang X., Myers A., Cheadle C., Huang S.K., Atasoy U., Gorospe M., Stellato C. Chemokine transcripts as targets of the RNA-binding protein HuR in human airway epithelium. J. Immunol. 2011;186:2482–2494. doi: 10.4049/jimmunol.0903634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bai D., Gao Q., Li C., Ge L., Gao Y., Wang H. A conserved TGFbeta1/HuR feedback circuit regulates the fibrogenic response in fibroblasts. Cell Signal. 2012;24:1426–1432. doi: 10.1016/j.cellsig.2012.03.003. [DOI] [PubMed] [Google Scholar]
- 71.Adjibade P., Mazroui R. Control of mRNA turnover: Implication of cytoplasmic RNA granules. Semin. Cell Dev. Biol. 2014;34:15–23. doi: 10.1016/j.semcdb.2014.05.013. [DOI] [PubMed] [Google Scholar]
- 72.Mahboubi H., Stochaj U. Cytoplasmic stress granules: Dynamic modulators of cell signaling and disease. Biochim. Biophys. Acta Mol. Basis Dis. 2017;1863:884–895. doi: 10.1016/j.bbadis.2016.12.022. [DOI] [PubMed] [Google Scholar]
- 73.Ansari M.Y., Haqqi T.M. Interleukin-1beta induced Stress Granules Sequester COX-2 mRNA and Regulates its Stability and Translation in Human OA Chondrocytes. Sci. Rep. 2016;6:27611. doi: 10.1038/srep27611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kedersha N., Ivanov P., Anderson P. Stress granules and cell signaling: More than just a passing phase? Trends Biochem. Sci. 2013;38:494–506. doi: 10.1016/j.tibs.2013.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Jain S., Parker R. The discovery and analysis of P Bodies. Adv. Exp. Med. Biol. 2013;768:23–43. doi: 10.1007/978-1-4614-5107-5_3. [DOI] [PubMed] [Google Scholar]
- 76.Glasmacher E., Hoefig K.P., Vogel K.U., Rath N., Du L., Wolf C., Kremmer E., Wang X., Heissmeyer V. Roquin binds inducible costimulator mRNA and effectors of mRNA decay to induce microRNA-independent post-transcriptional repression. Nat. Immunol. 2010;11:725–733. doi: 10.1038/ni.1902. [DOI] [PubMed] [Google Scholar]
- 77.Kahvejian A., Svitkin Y.V., Sukarieh R., M’Boutchou M.N., Sonenberg N. Mammalian poly(A)-binding protein is a eukaryotic translation initiation factor, which acts via multiple mechanisms. Genes Dev. 2005;19:104–113. doi: 10.1101/gad.1262905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Dixon D.A., Balch G.C., Kedersha N., Anderson P., Zimmerman G.A., Beauchamp R.D., Prescott S.M. Regulation of cyclooxygenase-2 expression by the translational silencer TIA-1. J. Exp. Med. 2003;198:475–481. doi: 10.1084/jem.20030616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lal A., Kawai T., Yang X., Mazan-Mamczarz K., Gorospe M. Antiapoptotic function of RNA-binding protein HuR effected through prothymosin alpha. EMBO J. 2005;24:1852–1862. doi: 10.1038/sj.emboj.7600661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kullmann M., Gopfert U., Siewe B., Hengst L. ELAV/Hu proteins inhibit p27 translation via an IRES element in the p27 5’UTR. Genes Dev. 2002;16:3087–3099. doi: 10.1101/gad.248902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Leandersson K., Riesbeck K., Andersson T. Wnt-5a mRNA translation is suppressed by the Elav-like protein HuR in human breast epithelial cells. Nucleic Acids Res. 2006;34:3988–3999. doi: 10.1093/nar/gkl571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lee Y., Jeon K., Lee J.T., Kim S., Kim V.N. MicroRNA maturation: Stepwise processing and subcellular localization. EMBO J. 2002;21:4663–4670. doi: 10.1093/emboj/cdf476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Tomari Y., Zamore P.D. MicroRNA biogenesis: Drosha can’t cut it without a partner. Curr. Biol. 2005;15:R61-4. doi: 10.1016/j.cub.2004.12.057. [DOI] [PubMed] [Google Scholar]
- 84.Han J., Lee Y., Yeom K.H., Nam J.W., Heo I., Rhee J.K., Sohn S.Y., Cho Y., Zhang B.T., Kim V.N. Molecular basis for the recognition of primary microRNAs by the Drosha-DGCR8 complex. Cell. 2006;125:887–901. doi: 10.1016/j.cell.2006.03.043. [DOI] [PubMed] [Google Scholar]
- 85.Kawamata T., Tomari Y. Making RISC. Trends Biochem. Sci. 2010;35:368–376. doi: 10.1016/j.tibs.2010.03.009. [DOI] [PubMed] [Google Scholar]
- 86.Fabian M.R., Sonenberg N., Filipowicz W. Regulation of mRNA translation and stability by microRNAs. Annu. Rev. Biochem. 2010;79:351–379. doi: 10.1146/annurev-biochem-060308-103103. [DOI] [PubMed] [Google Scholar]
- 87.Lee R.C., Feinbaum R.L., Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-Y. [DOI] [PubMed] [Google Scholar]
- 88.Wightman B., Ha I., Ruvkun G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell. 1993;75:855–862. doi: 10.1016/0092-8674(93)90530-4. [DOI] [PubMed] [Google Scholar]
- 89.Bernstein E., Kim S.Y., Carmell M.A., Murchison E.P., Alcorn H., Li M.Z., Mills A.A., Elledge S.J., Anderson K.V., Hannon G.J. Dicer is essential for mouse development. Nat. Genet. 2003;35:215–217. doi: 10.1038/ng1253. [DOI] [PubMed] [Google Scholar]
- 90.Wang Y., Medvid R., Melton C., Jaenisch R., Blelloch R. DGCR8 is essential for microRNA biogenesis and silencing of embryonic stem cell self-renewal. Nat. Genet. 2007;39:380–385. doi: 10.1038/ng1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Zhao Y., Ransom J.F., Li A., Vedantham V., von Drehle M., Muth A.N., Tsuchihashi T., McManus M.T., Schwartz R.J., Srivastava D. Dysregulation of cardiogenesis, cardiac conduction, and cell cycle in mice lacking miRNA-1-2. Cell. 2007;129:303–317. doi: 10.1016/j.cell.2007.03.030. [DOI] [PubMed] [Google Scholar]
- 92.Rodriguez A., Vigorito E., Clare S., Warren M.V., Couttet P., Soond D.R., van Dongen S., Grocock R.J., Das P.P., Miska E.A., et al. Requirement of bic/microRNA-155 for normal immune function. Science. 2007;316:608–611. doi: 10.1126/science.1139253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Johnnidis J.B., Harris M.H., Wheeler R.T., Stehling-Sun S., Lam M.H., Kirak O., Brummelkamp T.R., Fleming M.D., Camargo F.D. Regulation of progenitor cell proliferation and granulocyte function by microRNA-223. Nature. 2008;451:1125–1129. doi: 10.1038/nature06607. [DOI] [PubMed] [Google Scholar]
- 94.Ventura A., Young A.G., Winslow M.M., Lintault L., Meissner A., Erkeland S.J., Newman J., Bronson R.T., Crowley D., Stone J.R., et al. Targeted deletion reveals essential and overlapping functions of the miR-17 through 92 family of miRNA clusters. Cell. 2008;132:875–886. doi: 10.1016/j.cell.2008.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Mercer T.R., Dinger M.E., Mattick J.S. Long non-coding RNAs: Insights into functions. Nat. Rev. Genet. 2009;10:155–159. doi: 10.1038/nrg2521. [DOI] [PubMed] [Google Scholar]
- 96.Jarroux J., Morillon A., Pinskaya M. History, Discovery, and Classification of lncRNAs. Adv. Exp. Med. Biol. 2017;1008:1–46. doi: 10.1007/978-981-10-5203-3_1. [DOI] [PubMed] [Google Scholar]
- 97.Dhanoa J.K., Sethi R.S., Verma R., Arora J.S., Mukhopadhyay C.S. Long non-coding RNA: Its evolutionary relics and biological implications in mammals: A review. J. Anim. Sci. Technol. 2018;60:25. doi: 10.1186/s40781-018-0183-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Tsagakis I., Douka K., Birds I., Aspden J.L. Long non-coding RNAs in development and disease: Conservation to mechanisms. J. Pathol. 2020;250:480–495. doi: 10.1002/path.5405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Ma H., Hao Y., Dong X., Gong Q., Chen J., Zhang J., Tian W. Molecular mechanisms and function prediction of long noncoding RNA. Sci. World J. 2012;2012:541786. doi: 10.1100/2012/541786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Marchese F.P., Raimondi I., Huarte M. The multidimensional mechanisms of long noncoding RNA function. Genome Biol. 2017;18:206. doi: 10.1186/s13059-017-1348-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Dykes I.M., Emanueli C. Transcriptional and Post-transcriptional Gene Regulation by Long Non-coding RNA. Genom. Proteom. Bioinform. 2017;15:177–186. doi: 10.1016/j.gpb.2016.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Gil N., Ulitsky I. Regulation of gene expression by cis-acting long non-coding RNAs. Nat. Rev. Genet. 2020;21:102–117. doi: 10.1038/s41576-019-0184-5. [DOI] [PubMed] [Google Scholar]
- 103.Song C., Xiong Y., Liao W., Meng L., Yang S. Long noncoding RNA ATB participates in the development of renal cell carcinoma by downregulating p53 via binding to DNMT1. J. Cell Physiol. 2019;234:12910–12917. doi: 10.1002/jcp.27957. [DOI] [PubMed] [Google Scholar]
- 104.Keren H., Lev-Maor G., Ast G. Alternative splicing and evolution: Diversification, exon definition and function. Nat. Rev. Genet. 2010;11:345–355. doi: 10.1038/nrg2776. [DOI] [PubMed] [Google Scholar]
- 105.Zhang X., Wang W., Zhu W., Dong J., Cheng Y., Yin Z., Shen F. Mechanisms and Functions of Long Non-Coding RNAs at Multiple Regulatory Levels. Int. J. Mol. Sci. 2019;20:5573. doi: 10.3390/ijms20225573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Lee S., Kopp F., Chang T.C., Sataluri A., Chen B., Sivakumar S., Yu H., Xie Y., Mendell J.T. Noncoding RNA NORAD Regulates Genomic Stability by Sequestering PUMILIO Proteins. Cell. 2016;164:69–80. doi: 10.1016/j.cell.2015.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Vestbo J., Hurd S.S., Agusti A.G., Jones P.W., Vogelmeier C., Anzueto A., Barnes P.J., Fabbri L.M., Martinez F.J., Nishimura M., et al. Global strategy for the diagnosis, management, and prevention of chronic obstructive pulmonary disease: GOLD executive summary. Am. J. Respir. Crit. Care Med. 2013;187:347–365. doi: 10.1164/rccm.201204-0596PP. [DOI] [PubMed] [Google Scholar]
- 108.World Health Organization The Top 10 Causes of Death. [(accessed on 28 October 2021)]. Available online: https://www.who.int/en/news-room/fact-sheets/detail/the-top-10-causes-of-death.
- 109.Global Strategy for the Diagnosis. Management and Prevention of COPD Global Initiative for Chronic Obstructive Lung Disease (GOLD) 2020. [(accessed on 28 October 2021)]. Available online: https://www.google.com/url?sa=t&rct=j&q=&esrc=s&source=web&cd=&ved=2ahUKEwiFyoOAqOrzAhVeQvEDHVvzCuYQjBB6BAgPEAE&url=https%3A%2F%2Fgoldcopd.org%2Fwp-content%2Fuploads%2F2020%2F03%2FGOLD-2020-POCKET-GUIDE-ver1.0_FINAL-WMV.pdf&usg=AOvVaw3jGwvv-lRohtDCg1_pHTST.
- 110.Pauwels R.A., Rabe K.F. Burden and clinical features of chronic obstructive pulmonary disease (COPD) Lancet. 2004;364:613–620. doi: 10.1016/S0140-6736(04)16855-4. [DOI] [PubMed] [Google Scholar]
- 111.Decramer M., Janssens W., Miravitlles M. Chronic obstructive pulmonary disease. Lancet. 2012;379:1341–1351. doi: 10.1016/S0140-6736(11)60968-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.World Health Organization Tobacco. [(accessed on 28 October 2021)]. Available online: https://www.who.int/news-room/fact-sheets/detail/tobacco.
- 113.FDA Chemicals in Cigarettes: From Plant to Product to Puff. [(accessed on 28 October 2021)]; Available online: https://www.fda.gov/tobacco-products/products-ingredients-components/chemicals-cigarettes-plant-product-puff.
- 114.Fowles J., Dybing E. Application of toxicological risk assessment principles to the chemical constituents of cigarette smoke. Tob. Control. 2003;12:424–430. doi: 10.1136/tc.12.4.424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Athanazio R. Airway disease: Similarities and differences between asthma, COPD and bronchiectasis. Clinics. 2012;67:1335–1343. doi: 10.6061/clinics/2012(11)19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.King P.T. Inflammation in chronic obstructive pulmonary disease and its role in cardiovascular disease and lung cancer. Clin. Transl. Med. 2015;4:68. doi: 10.1186/s40169-015-0068-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Barnes P.J. Inflammatory mechanisms in patients with chronic obstructive pulmonary disease. J. Allergy Clin. Immunol. 2016;138:16–27. doi: 10.1016/j.jaci.2016.05.011. [DOI] [PubMed] [Google Scholar]
- 118.Sethi S., Mahler D.A., Marcus P., Owen C.A., Yawn B., Rennard S. Inflammation in COPD: Implications for management. Am. J. Med. 2012;125:1162–1170. doi: 10.1016/j.amjmed.2012.06.024. [DOI] [PubMed] [Google Scholar]
- 119.Donnelly L.E., Barnes P.J. Chemokine receptors as therapeutic targets in chronic obstructive pulmonary disease. Trends Pharm. Sci. 2006;27:546–553. doi: 10.1016/j.tips.2006.08.001. [DOI] [PubMed] [Google Scholar]
- 120.Frankenberger M., Eder C., Hofer T.P., Heimbeck I., Skokann K., Kassner G., Weber N., Moller W., Ziegler-Heitbrock L. Chemokine expression by small sputum macrophages in COPD. Mol. Med. 2011;17:762–770. doi: 10.2119/molmed.2010.00202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Sheridan J.A., Zago M., Nair P., Li P.Z., Bourbeau J., Tan W.C., Hamid Q., Eidelman D.H., Benedetti A.L., Baglole C.J. Decreased expression of the NF-kappaB family member RelB in lung fibroblasts from Smokers with and without COPD potentiates cigarette smoke-induced COX-2 expression. Respir. Res. 2015;16:54. doi: 10.1186/s12931-015-0214-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Chen Y., Chen P., Hanaoka M., Droma Y., Kubo K. Enhanced levels of prostaglandin E2 and matrix metalloproteinase-2 correlate with the severity of airflow limitation in stable COPD. Respirology. 2008;13:1014–1021. doi: 10.1111/j.1440-1843.2008.01365.x. [DOI] [PubMed] [Google Scholar]
- 123.Bagdonas E., Raudoniute J., Bruzauskaite I., Aldonyte R. Novel aspects of pathogenesis and regeneration mechanisms in COPD. Int. J. Chron. Obs. Pulm. Dis. 2015;10:995–1013. doi: 10.2147/COPD.S82518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Owen C.A. Roles for proteinases in the pathogenesis of chronic obstructive pulmonary disease. Int. J. Chron. Obs. Pulm. Dis. 2008;3:253–268. doi: 10.2147/COPD.S2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Barnes P.J., Shapiro S.D., Pauwels R.A. Chronic obstructive pulmonary disease: Molecular and cellular mechanisms. Eur. Respir. J. 2003;22:672–688. doi: 10.1183/09031936.03.00040703. [DOI] [PubMed] [Google Scholar]
- 126.Ahn K.S., Aggarwal B.B. Transcription factor NF-kappaB: A sensor for smoke and stress signals. Ann. N. Y. Acad. Sci. 2005;1056:218–233. doi: 10.1196/annals.1352.026. [DOI] [PubMed] [Google Scholar]
- 127.Zago M., Sheridan J.A., Nair P., Rico de Souza A., Gallouzi I.E., Rousseau S., Di Marco S., Hamid Q., Eidelman D.H., Baglole C.J. Aryl Hydrocarbon Receptor-Dependent Retention of Nuclear HuR Suppresses Cigarette Smoke-Induced Cyclooxygenase-2 Expression Independent of DNA-Binding. PLoS ONE. 2013;8:e74953. doi: 10.1371/journal.pone.0074953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Srikantan S., Gorospe M. HuR function in disease. Front. Biosci. 2012;17:189–205. doi: 10.2741/3921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Hudy M.H., Proud D. Cigarette smoke enhances human rhinovirus-induced CXCL8 production via HuR-mediated mRNA stabilization in human airway epithelial cells. Respir. Res. 2013;14:88. doi: 10.1186/1465-9921-14-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.George S.N., Garcha D.S., Mackay A.J., Patel A.R., Singh R., Sapsford R.J., Donaldson G.C., Wedzicha J.A. Human rhinovirus infection during naturally occurring COPD exacerbations. Eur. Respir. J. 2014;44:87–96. doi: 10.1183/09031936.00223113. [DOI] [PubMed] [Google Scholar]
- 131.Owuor N., Nalamala N., Gimenes J.A., Jr., Sajjan U.S. Rhinovirus and COPD airway epithelium. Pulm. Crit. Care Med. 2017:2. doi: 10.15761/PCCM.1000139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Angeloni D. Molecular analysis of deletions in human chromosome 3p21 and the role of resident cancer genes in disease. Brief. Funct. Genom. Proteomic. 2007;6:19–39. doi: 10.1093/bfgp/elm007. [DOI] [PubMed] [Google Scholar]
- 133.Timmer T., Terpstra P., van den Berg A., Veldhuis P.M., Ter Elst A., Voutsinas G., Hulsbeek M.M., Draaijers T.G., Looman M.W., Kok K., et al. A comparison of genomic structures and expression patterns of two closely related flanking genes in a critical lung cancer region at 3p21.3. Eur. J. Hum. Genet. 1999;7:478–486. doi: 10.1038/sj.ejhg.5200334. [DOI] [PubMed] [Google Scholar]
- 134.Jamsai D., Watkins D.N., O’Connor A.E., Merriner D.J., Gursoy S., Bird A.D., Kumar B., Miller A., Cole T.J., Jenkins B.J., et al. In vivo evidence that RBM5 is a tumour suppressor in the lung. Sci. Rep. 2017;7:16323. doi: 10.1038/s41598-017-15874-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Hao Y.Q., Su Z.Z., Lv X.J., Li P., Gao P., Wang C., Bai Y., Zhang J. RNA-binding motif protein 5 negatively regulates the activity of Wnt/beta-catenin signaling in cigarette smoke-induced alveolar epithelial injury. Oncol. Rep. 2015;33:2438–2444. doi: 10.3892/or.2015.3828. [DOI] [PubMed] [Google Scholar]
- 136.Carlier F.M., Dupasquier S., Ambroise J., Detry B., Lecocq M., Bietry-Claudet C., Boukala Y., Gala J.L., Bouzin C., Verleden S.E., et al. Canonical WNT pathway is activated in the airway epithelium in chronic obstructive pulmonary disease. EBioMedicine. 2020;61:103034. doi: 10.1016/j.ebiom.2020.103034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Milara J., Peiro T., Serrano A., Cortijo J. Epithelial to mesenchymal transition is increased in patients with COPD and induced by cigarette smoke. Thorax. 2013;68:410–420. doi: 10.1136/thoraxjnl-2012-201761. [DOI] [PubMed] [Google Scholar]
- 138.Bhattacharya S., Srisuma S., Demeo D.L., Shapiro S.D., Bueno R., Silverman E.K., Reilly J.J., Mariani T.J. Molecular biomarkers for quantitative and discrete COPD phenotypes. Am. J. Respir. Cell Mol. Biol. 2009;40:359–367. doi: 10.1165/rcmb.2008-0114OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.DeMeo D.L., Mariani T., Bhattacharya S., Srisuma S., Lange C., Litonjua A., Bueno R., Pillai S.G., Lomas D.A., Sparrow D., et al. Integration of genomic and genetic approaches implicates IREB2 as a COPD susceptibility gene. Am. J. Hum. Genet. 2009;85:493–502. doi: 10.1016/j.ajhg.2009.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Qiu W., Cho M.H., Riley J.H., Anderson W.H., Singh D., Bakke P., Gulsvik A., Litonjua A.A., Lomas D.A., Crapo J.D., et al. Genetics of sputum gene expression in chronic obstructive pulmonary disease. PLoS ONE. 2011;6:e24395. doi: 10.1371/journal.pone.0024395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Hardin M., Zielinski J., Wan E.S., Hersh C.P., Castaldi P.J., Schwinder E., Hawrylkiewicz I., Sliwinski P., Cho M.H., Silverman E.K. CHRNA3/5, IREB2, and ADCY2 are associated with severe chronic obstructive pulmonary disease in Poland. Am. J. Respir. Cell Mol. Biol. 2012;47:203–208. doi: 10.1165/rcmb.2012-0011OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Cloonan S.M., Glass K., Laucho-Contreras M.E., Bhashyam A.R., Cervo M., Pabon M.A., Konrad C., Polverino F., Siempos I.I., Perez E., et al. Mitochondrial iron chelation ameliorates cigarette smoke-induced bronchitis and emphysema in mice. Nat. Med. 2016;22:163–174. doi: 10.1038/nm.4021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Ghio A.J., Hilborn E.D., Stonehuerner J.G., Dailey L.A., Carter J.D., Richards J.H., Crissman K.M., Foronjy R.F., Uyeminami D.L., Pinkerton K.E. Particulate matter in cigarette smoke alters iron homeostasis to produce a biological effect. Am. J. Respir. Crit. Care Med. 2008;178:1130–1138. doi: 10.1164/rccm.200802-334OC. [DOI] [PubMed] [Google Scholar]
- 144.Philippot Q., Deslee G., Adair-Kirk T.L., Woods J.C., Byers D., Conradi S., Dury S., Perotin J.M., Lebargy F., Cassan C., et al. Increased iron sequestration in alveolar macrophages in chronic obstructive pulmonary disease. PLoS ONE. 2014;9:e96285. doi: 10.1371/journal.pone.0096285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Sun J., Gu X., Wu N., Zhang P., Liu Y., Jiang S. Human antigen R enhances the epithelial-mesenchymal transition via regulation of ZEB-1 in the human airway epithelium. Respir. Res. 2018;19:109. doi: 10.1186/s12931-018-0805-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Ricciardi L., Col J.D., Casolari P., Memoli D., Conti V., Vatrella A., Vonakis B.M., Papi A., Caramori G., Stellato C. Differential expression of RNA-binding proteins in bronchial epithelium of stable COPD patients. Int. J. Chron. Obs. Pulm. Dis. 2018;13:3173–3190. doi: 10.2147/COPD.S166284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Ricciardi L., Giurato G., Memoli D., Pietrafesa M., Dal Col J., Salvato I., Nigro A., Vatrella A., Caramori G., Casolaro V., et al. Posttranscriptional Gene Regulatory Networks in Chronic Airway Inflammatory Diseases: In silico Mapping of RNA-Binding Protein Expression in Airway Epithelium. Front. Immunol. 2020;11:579889. doi: 10.3389/fimmu.2020.579889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Hogg J.C., Timens W. The pathology of chronic obstructive pulmonary disease. Annu. Rev. Pathol. 2009;4:435–459. doi: 10.1146/annurev.pathol.4.110807.092145. [DOI] [PubMed] [Google Scholar]
- 149.Thomson E.M., Williams A., Yauk C.L., Vincent R. Overexpression of tumor necrosis factor-alpha in the lungs alters immune response, matrix remodeling, and repair and maintenance pathways. Am. J. Pathol. 2012;180:1413–1430. doi: 10.1016/j.ajpath.2011.12.020. [DOI] [PubMed] [Google Scholar]
- 150.Taylor G.A., Carballo E., Lee D.M., Lai W.S., Thompson M.J., Patel D.D., Schenkman D.I., Gilkeson G.S., Broxmeyer H.E., Haynes B.F., et al. A pathogenetic role for TNF alpha in the syndrome of cachexia, arthritis, and autoimmunity resulting from tristetraprolin (TTP) deficiency. Immunity. 1996;4:445–454. doi: 10.1016/S1074-7613(00)80411-2. [DOI] [PubMed] [Google Scholar]
- 151.Prabhala P., Ammit A.J. Tristetraprolin and its role in regulation of airway inflammation. Mol. Pharm. 2015;87:629–638. doi: 10.1124/mol.114.095984. [DOI] [PubMed] [Google Scholar]
- 152.Mino T., Takeuchi O. Post-transcriptional regulation of cytokine mRNA controls the initiation and resolution of inflammation. Biotechnol. Genet. Eng. Rev. 2013;29:49–60. doi: 10.1080/02648725.2013.801236. [DOI] [PubMed] [Google Scholar]
- 153.Smoak K., Cidlowski J.A. Glucocorticoids regulate tristetraprolin synthesis and posttranscriptionally regulate tumor necrosis factor alpha inflammatory signaling. Mol. Cell Biol. 2006;26:9126–9135. doi: 10.1128/MCB.00679-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Ishmael F.T., Fang X., Galdiero M.R., Atasoy U., Rigby W.F., Gorospe M., Cheadle C., Stellato C. Role of the RNA-binding protein tristetraprolin in glucocorticoid-mediated gene regulation. J. Immunol. 2008;180:8342–8353. doi: 10.4049/jimmunol.180.12.8342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Lu J.Y., Sadri N., Schneider R.J. Endotoxic shock in AUF1 knockout mice mediated by failure to degrade proinflammatory cytokine mRNAs. Genes Dev. 2006;20:3174–3184. doi: 10.1101/gad.1467606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Sadri N., Schneider R.J. Auf1/Hnrnpd-deficient mice develop pruritic inflammatory skin disease. J. Investig. Derm. 2009;129:657–670. doi: 10.1038/jid.2008.298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Dean J.L., Wait R., Mahtani K.R., Sully G., Clark A.R., Saklatvala J. The 3’ untranslated region of tumor necrosis factor alpha mRNA is a target of the mRNA-stabilizing factor HuR. Mol. Cell Biol. 2001;21:721–730. doi: 10.1128/MCB.21.3.721-730.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Atasoy U., Curry S.L., Lopez de Silanes I., Shyu A.B., Casolaro V., Gorospe M., Stellato C. Regulation of eotaxin gene expression by TNF-alpha and IL-4 through mRNA stabilization: Involvement of the RNA-binding protein HuR. J. Immunol. 2003;171:4369–4378. doi: 10.4049/jimmunol.171.8.4369. [DOI] [PubMed] [Google Scholar]
- 159.Bradford E., Jacobson S., Varasteh J., Comellas A.P., Woodruff P., O’Neal W., DeMeo D.L., Li X., Kim V., Cho M., et al. The value of blood cytokines and chemokines in assessing COPD. Respir. Res. 2017;18:180. doi: 10.1186/s12931-017-0662-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Morissette M.C., Parent J., Milot J. Alveolar epithelial and endothelial cell apoptosis in emphysema: What we know and what we need to know. Int. J. Chron. Obs. Pulm. Dis. 2009;4:19–31. [PMC free article] [PubMed] [Google Scholar]
- 161.Aoshiba K., Yokohori N., Nagai A. Alveolar wall apoptosis causes lung destruction and emphysematous changes. Am. J. Respir. Cell Mol. Biol. 2003;28:555–562. doi: 10.1165/rcmb.2002-0090OC. [DOI] [PubMed] [Google Scholar]
- 162.Kasahara Y., Tuder R.M., Taraseviciene-Stewart L., Le Cras T.D., Abman S., Hirth P.K., Waltenberger J., Voelkel N.F. Inhibition of VEGF receptors causes lung cell apoptosis and emphysema. J. Clin. Investig. 2000;106:1311–1319. doi: 10.1172/JCI10259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Kasahara Y., Tuder R.M., Cool C.D., Lynch D.A., Flores S.C., Voelkel N.F. Endothelial cell death and decreased expression of vascular endothelial growth factor and vascular endothelial growth factor receptor 2 in emphysema. Pt 1Am. J. Respir. Crit. Care Med. 2001;163:737–744. doi: 10.1164/ajrccm.163.3.2002117. [DOI] [PubMed] [Google Scholar]
- 164.Kanazawa H., Tochino Y., Asai K., Hirata K. Simultaneous assessment of hepatocyte growth factor and vascular endothelial growth factor in epithelial lining fluid from patients with COPD. Chest. 2014;146:1159–1165. doi: 10.1378/chest.14-0373. [DOI] [PubMed] [Google Scholar]
- 165.Essafi-Benkhadir K., Onesto C., Stebe E., Moroni C., Pages G. Tristetraprolin inhibits Ras-dependent tumor vascularization by inducing vascular endothelial growth factor mRNA degradation. Mol. Biol. Cell. 2007;18:4648–4658. doi: 10.1091/mbc.e07-06-0570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Shih S.C., Claffey K.P. Regulation of human vascular endothelial growth factor mRNA stability in hypoxia by heterogeneous nuclear ribonucleoprotein L. J. Biol. Chem. 1999;274:1359–1365. doi: 10.1074/jbc.274.3.1359. [DOI] [PubMed] [Google Scholar]
- 167.Heiner M., Hui J., Schreiner S., Hung L.H., Bindereif A. HnRNP L-mediated regulation of mammalian alternative splicing by interference with splice site recognition. RNA Biol. 2010;7:56–64. doi: 10.4161/rna.7.1.10402. [DOI] [PubMed] [Google Scholar]
- 168.Gaudreau M.C., Grapton D., Helness A., Vadnais C., Fraszczak J., Shooshtarizadeh P., Wilhelm B., Robert F., Heyd F., Moroy T. Heterogeneous Nuclear Ribonucleoprotein L is required for the survival and functional integrity of murine hematopoietic stem cells. Sci. Rep. 2016;6:27379. doi: 10.1038/srep27379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Hoenderdos K., Condliffe A. The neutrophil in chronic obstructive pulmonary disease. Am. J. Respir. Cell Mol. Biol. 2013;48:531–539. doi: 10.1165/rcmb.2012-0492TR. [DOI] [PubMed] [Google Scholar]
- 170.Jasper A.E., McIver W.J., Sapey E., Walton G.M. Understanding the role of neutrophils in chronic inflammatory airway disease. F1000Research. 2019;8 doi: 10.12688/f1000research.18411.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Kapellos T.S., Bassler K., Aschenbrenner A.C., Fujii W., Schultze J.L. Dysregulated Functions of Lung Macrophage Populations in COPD. J. Immunol Res. 2018;2018:2349045. doi: 10.1155/2018/2349045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Van Tubergen E.A., Banerjee R., Liu M., Vander Broek R., Light E., Kuo S., Feinberg S.E., Willis A.L., Wolf G., Carey T., et al. Inactivation or loss of TTP promotes invasion in head and neck cancer via transcript stabilization and secretion of MMP9, MMP2, and IL-6. Clin. Cancer Res. 2013;19:1169–1179. doi: 10.1158/1078-0432.CCR-12-2927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Huwiler A., Akool E.S., Aschrafi A., Hamada F.M., Pfeilschifter J., Eberhardt W. ATP potentiates interleukin-1 beta-induced MMP-9 expression in mesangial cells via recruitment of the ELAV protein HuR. J. Biol. Chem. 2003;278:51758–51769. doi: 10.1074/jbc.M305722200. [DOI] [PubMed] [Google Scholar]
- 174.Akool E.S., Kleinert H., Hamada F.M., Abdelwahab M.H., Forstermann U., Pfeilschifter J., Eberhardt W. Nitric oxide increases the decay of matrix metalloproteinase 9 mRNA by inhibiting the expression of mRNA-stabilizing factor HuR. Mol. Cell Biol. 2003;23:4901–4916. doi: 10.1128/MCB.23.14.4901-4916.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Krishnamurthy P., Rajasingh J., Lambers E., Qin G., Losordo D.W., Kishore R. IL-10 inhibits inflammation and attenuates left ventricular remodeling after myocardial infarction via activation of STAT3 and suppression of HuR. Circ. Res. 2009;104:e9–e18. doi: 10.1161/CIRCRESAHA.108.188243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Wang I.X., So E., Devlin J.L., Zhao Y., Wu M., Cheung V.G. ADAR regulates RNA editing, transcript stability, and gene expression. Cell Rep. 2013;5:849–860. doi: 10.1016/j.celrep.2013.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Stellos K., Gatsiou A., Stamatelopoulos K., Perisic Matic L., John D., Lunella F.F., Jae N., Rossbach O., Amrhein C., Sigala F., et al. Adenosine-to-inosine RNA editing controls cathepsin S expression in atherosclerosis by enabling HuR-mediated post-transcriptional regulation. Nat. Med. 2016;22:1140–1150. doi: 10.1038/nm.4172. [DOI] [PubMed] [Google Scholar]
- 178.Brown R., Nath S., Lora A., Samaha G., Elgamal Z., Kaiser R., Taggart C., Weldon S., Geraghty P. Cathepsin S: Investigating an old player in lung disease pathogenesis, comorbidities, and potential therapeutics. Respir. Res. 2020;21:111. doi: 10.1186/s12931-020-01381-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Andrault P.M., Schamberger A.C., Chazeirat T., Sizaret D., Renault J., Staab-Weijnitz C.A., Hennen E., Petit-Courty A., Wartenberg M., Saidi A., et al. Cigarette smoke induces overexpression of active human cathepsin S in lungs from current smokers with or without COPD. Am. J. Physiol. Lung Cell Mol. Physiol. 2019;317:L625–L638. doi: 10.1152/ajplung.00061.2019. [DOI] [PubMed] [Google Scholar]
- 180.Wigington C.P., Jung J., Rye E.A., Belauret S.L., Philpot A.M., Feng Y., Santangelo P.J., Corbett A.H. Post-transcriptional regulation of programmed cell death 4 (PDCD4) mRNA by the RNA-binding proteins human antigen R (HuR) and T-cell intracellular antigen 1 (TIA1) J. Biol. Chem. 2015;290:3468–3487. doi: 10.1074/jbc.M114.631937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Zhang X., Chen X., Liu Q., Zhang S., Hu W. Translation repression via modulation of the cytoplasmic poly(A)-binding protein in the inflammatory response. Elife. 2017;6:e27786. doi: 10.7554/eLife.27786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Takamizawa J., Konishi H., Yanagisawa K., Tomida S., Osada H., Endoh H., Harano T., Yatabe Y., Nagino M., Nimura Y., et al. Reduced expression of the let-7 microRNAs in human lung cancers in association with shortened postoperative survival. Cancer Res. 2004;64:3753–3756. doi: 10.1158/0008-5472.CAN-04-0637. [DOI] [PubMed] [Google Scholar]
- 183.Sun C., Liu Z., Li S., Yang C., Xue R., Xi Y., Wang L., Wang S., He Q., Huang J., et al. Down-regulation of c-Met and Bcl2 by microRNA-206, activates apoptosis, and inhibits tumor cell proliferation, migration and colony formation. Oncotarget. 2015;6:25533–25574. doi: 10.18632/oncotarget.4575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Ke Y., Zhao W., Xiong J., Cao R. Downregulation of miR-16 promotes growth and motility by targeting HDGF in non-small cell lung cancer cells. FEBS Lett. 2013;587:3153–3157. doi: 10.1016/j.febslet.2013.08.010. [DOI] [PubMed] [Google Scholar]
- 185.Chen T., Xiao Q., Wang X., Wang Z., Hu J., Zhang Z., Gong Z., Chen S. miR-16 regulates proliferation and invasion of lung cancer cells via the ERK/MAPK signaling pathway by targeted inhibition of MAPK kinase 1 (MEK1) J. Int. Med. Res. 2019;47:5194–5204. doi: 10.1177/0300060519856505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Kao S.C., Fulham M., Wong K., Cooper W., Brahmbhatt H., MacDiarmid J., Pattison S., Sagong J.O., Huynh Y., Leslie F., et al. A Significant Metabolic and Radiological Response after a Novel Targeted MicroRNA-based Treatment Approach in Malignant Pleural Mesothelioma. Am. J. Respir. Crit. Care Med. 2015;191:1467–1469. doi: 10.1164/rccm.201503-0461LE. [DOI] [PubMed] [Google Scholar]
- 187.van Zandwijk N., Pavlakis N., Kao S.C., Linton A., Boyer M.J., Clarke S., Huynh Y., Chrzanowska A., Fulham M.J., Bailey D.L., et al. Safety and activity of microRNA-loaded minicells in patients with recurrent malignant pleural mesothelioma: A first-in-man, phase 1, open-label, dose-escalation study. Lancet Oncol. 2017;18:1386–1396. doi: 10.1016/S1470-2045(17)30621-6. [DOI] [PubMed] [Google Scholar]
- 188.Schembri F., Sridhar S., Perdomo C., Gustafson A.M., Zhang X., Ergun A., Lu J., Liu G., Zhang X., Bowers J., et al. MicroRNAs as modulators of smoking-induced gene expression changes in human airway epithelium. Proc. Natl. Acad. Sci. USA. 2009;106:2319–2324. doi: 10.1073/pnas.0806383106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Graff J.W., Powers L.S., Dickson A.M., Kim J., Reisetter A.C., Hassan I.H., Kremens K., Gross T.J., Wilson M.E., Monick M.M. Cigarette smoking decreases global microRNA expression in human alveolar macrophages. PLoS ONE. 2012;7:e44066. doi: 10.1371/journal.pone.0044066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Izzotti A., Calin G.A., Arrigo P., Steele V.E., Croce C.M., De Flora S. Downregulation of microRNA expression in the lungs of rats exposed to cigarette smoke. FASEB J. 2009;23:806–812. doi: 10.1096/fj.08-121384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Izzotti A., Larghero P., Longobardi M., Cartiglia C., Camoirano A., Steele V.E., De Flora S. Dose-responsiveness and persistence of microRNA expression alterations induced by cigarette smoke in mouse lung. Mutat. Res. Fundam. Mol. Mech. Mutagenes. 2011;717:9–16. doi: 10.1016/j.mrfmmm.2010.12.008. [DOI] [PubMed] [Google Scholar]
- 192.Hassan F., Nuovo G.J., Crawford M., Boyaka P.N., Kirkby S., Nana-Sinkam S.P., Cormet-Boyaka E. MiR-101 and miR-144 regulate the expression of the CFTR chloride channel in the lung. PLoS ONE. 2012;7:e50837. doi: 10.1371/journal.pone.0050837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Rogers S., de Souza A.R., Zago M., Iu M., Guerrina N., Gomez A., Matthews J., Baglole C.J. Aryl hydrocarbon receptor (AhR)-dependent regulation of pulmonary miRNA by chronic cigarette smoke exposure. Sci. Rep. 2017;7:40539. doi: 10.1038/srep40539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Halappanavar S., Nikota J., Wu D., Williams A., Yauk C.L., Stampfli M. IL-1 receptor regulates microRNA-135b expression in a negative feedback mechanism during cigarette smoke-induced inflammation. J. Immunol. 2013;190:3679–3686. doi: 10.4049/jimmunol.1202456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Leuenberger C., Schuoler C., Bye H., Mignan C., Rechsteiner T., Hillinger S., Opitz I., Marsland B., Faiz A., Hiemstra P.S., et al. MicroRNA-223 controls the expression of histone deacetylase 2: A novel axis in COPD. J. Mol. Med. 2016;94:725–734. doi: 10.1007/s00109-016-1388-1. [DOI] [PubMed] [Google Scholar]
- 196.Carpi S., Polini B., Nieri D., Dubbini N., Celi A., Nieri P., Neri T. Expression Analysis of Muscle-Specific miRNAs in Plasma-Derived Extracellular Vesicles from Patients with Chronic Obstructive Pulmonary Disease. Diagnostics. 2020;10:502. doi: 10.3390/diagnostics10070502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Van Pottelberge G.R., Mestdagh P., Bracke K.R., Thas O., van Durme Y.M., Joos G.F., Vandesompele J., Brusselle G.G. MicroRNA expression in induced sputum of smokers and patients with chronic obstructive pulmonary disease. Am. J. Respir. Crit. Care Med. 2011;183:898–906. doi: 10.1164/rccm.201002-0304OC. [DOI] [PubMed] [Google Scholar]
- 198.Chatila W.M., Criner G.J., Hancock W.W., Akimova T., Moldover B., Chang J.K., Cornwell W., Santerre M., Rogers T.J. Blunted expression of miR-199a-5p in regulatory T cells of patients with chronic obstructive pulmonary disease compared to unaffected smokers. Clin. Exp. Immunol. 2014;177:341–352. doi: 10.1111/cei.12325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Mizuno S., Bogaard H.J., Gomez-Arroyo J., Alhussaini A., Kraskauskas D., Cool C.D., Voelkel N.F. MicroRNA-199a-5p is associated with hypoxia-inducible factor-1alpha expression in lungs from patients with COPD. Chest. 2012;142:663–672. doi: 10.1378/chest.11-2746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Conickx G., Mestdagh P., Avila Cobos F., Verhamme F.M., Maes T., Vanaudenaerde B.M., Seys L.J., Lahousse L., Kim R.Y., Hsu A.C., et al. MicroRNA Profiling Reveals a Role for MicroRNA-218-5p in the Pathogenesis of Chronic Obstructive Pulmonary Disease. Am. J. Respir. Crit. Care Med. 2017;195:43–56. doi: 10.1164/rccm.201506-1182OC. [DOI] [PubMed] [Google Scholar]
- 201.Du Y., Ding Y., Chen X., Mei Z., Ding H., Wu Y., Jie Z. MicroRNA-181c inhibits cigarette smoke-induced chronic obstructive pulmonary disease by regulating CCN1 expression. Respir. Res. 2017;18:155. doi: 10.1186/s12931-017-0639-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Chen B.B., Li Z.H., Gao S. Circulating miR-146a/b correlates with inflammatory cytokines in COPD and could predict the risk of acute exacerbation COPD. Medicine. 2018;97:e9820. doi: 10.1097/MD.0000000000009820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Zago M., Rico de Souza A., Hecht E., Rousseau S., Hamid Q., Eidelman D.H., Baglole C.J. The NF-kappaB family member RelB regulates microRNA miR-146a to suppress cigarette smoke-induced COX-2 protein expression in lung fibroblasts. Toxicol. Lett. 2014;226:107–116. doi: 10.1016/j.toxlet.2014.01.020. [DOI] [PubMed] [Google Scholar]
- 204.Sato T., Liu X., Nelson A., Nakanishi M., Kanaji N., Wang X., Kim M., Li Y., Sun J., Michalski J., et al. Reduced miR-146a increases prostaglandin E(2)in chronic obstructive pulmonary disease fibroblasts. Am. J. Respir. Crit. Care Med. 2010;182:1020–1029. doi: 10.1164/rccm.201001-0055OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Christenson S.A., Brandsma C.A., Campbell J.D., Knight D.A., Pechkovsky D.V., Hogg J.C., Timens W., Postma D.S., Lenburg M., Spira A. miR-638 regulates gene expression networks associated with emphysematous lung destruction. Genome Med. 2013;5:114. doi: 10.1186/gm519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Savarimuthu Francis S.M., Davidson M.R., Tan M.E., Wright C.M., Clarke B.E., Duhig E.E., Bowman R.V., Hayward N.K., Fong K.M., Yang I.A. MicroRNA-34c is associated with emphysema severity and modulates SERPINE1 expression. BMC Genom. 2014;15:88. doi: 10.1186/1471-2164-15-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Yang Y., Ding L., Hu Q., Xia J., Sun J., Wang X., Xiong H., Gurbani D., Li L., Liu Y., et al. MicroRNA-218 functions as a tumor suppressor in lung cancer by targeting IL-6/STAT3 and negatively correlates with poor prognosis. Mol. Cancer. 2017;16:141. doi: 10.1186/s12943-017-0710-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Wheeler M.A., Clark I.C., Tjon E.C., Li Z., Zandee S.E.J., Couturier C.P., Watson B.R., Scalisi G., Alkwai S., Rothhammer V., et al. MAFG-driven astrocytes promote CNS inflammation. Nature. 2020;578:593–599. doi: 10.1038/s41586-020-1999-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Demedts I.K., Bracke K.R., Van Pottelberge G., Testelmans D., Verleden G.M., Vermassen F.E., Joos G.F., Brusselle G.G. Accumulation of dendritic cells and increased CCL20 levels in the airways of patients with chronic obstructive pulmonary disease. Am. J. Respir. Crit. Care Med. 2007;175:998–1005. doi: 10.1164/rccm.200608-1113OC. [DOI] [PubMed] [Google Scholar]
- 210.Andriani F., Majorini M.T., Mano M., Landoni E., Miceli R., Facchinetti F., Mensah M., Fontanella E., Dugo M., Giacca M., et al. MiR-16 regulates the pro-tumorigenic potential of lung fibroblasts through the inhibition of HGF production in an FGFR-1- and MEK1-dependent manner. J. Hematol. Oncol. 2018;11:45. doi: 10.1186/s13045-018-0594-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.O’Leary L., Sevinc K., Papazoglou I.M., Tildy B., Detillieux K., Halayko A.J., Chung K.F., Perry M.M. Airway smooth muscle inflammation is regulated by microRNA-145 in COPD. FEBS Lett. 2016;590:1324–1334. doi: 10.1002/1873-3468.12168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Chen Y., Sun Y., Rao Q., Xu H., Li L., Chang C. Androgen receptor (AR) suppresses miRNA-145 to promote renal cell carcinoma (RCC) progression independent of VHL status. Oncotarget. 2015;6:31203–31215. doi: 10.18632/oncotarget.4522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Agra Andrieu N., Motino O., Mayoral R., Llorente Izquierdo C., Fernandez-Alvarez A., Bosca L., Casado M., Martin-Sanz P. Cyclooxygenase-2 is a target of microRNA-16 in human hepatoma cells. PLoS ONE. 2012;7:e50935. doi: 10.1371/journal.pone.0050935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Vlahos R., Bozinovski S. Role of alveolar macrophages in chronic obstructive pulmonary disease. Front. Immunol. 2014;5:435. doi: 10.3389/fimmu.2014.00435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Woodruff P.G., Koth L.L., Yang Y.H., Rodriguez M.W., Favoreto S., Dolganov G.M., Paquet A.C., Erle D.J. A distinctive alveolar macrophage activation state induced by cigarette smoking. Am. J. Respir. Crit. Care Med. 2005;172:1383–1392. doi: 10.1164/rccm.200505-686OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Shang J., Yang F., Wang Y., Wang Y., Xue G., Mei Q., Wang F., Sun S. MicroRNA-23a antisense enhances 5-fluorouracil chemosensitivity through APAF-1/caspase-9 apoptotic pathway in colorectal cancer cells. J. Cell Biochem. 2014;115:772–784. doi: 10.1002/jcb.24721. [DOI] [PubMed] [Google Scholar]
- 217.Zhou Y., Cheng X., Wan Y., Chen T., Zhou Q., Wang Z., Zhu H. MicroRNA-421 Inhibits Apoptosis by Downregulating Caspase-3 in Human Colorectal Cancer. Cancer Manag. Res. 2020;12:7579–7587. doi: 10.2147/CMAR.S255787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Damico R., Simms T., Kim B.S., Tekeste Z., Amankwan H., Damarla M., Hassoun P.M. p53 mediates cigarette smoke-induced apoptosis of pulmonary endothelial cells: Inhibitory effects of macrophage migration inhibitor factor. Am. J. Respir. Cell Mol. Biol. 2011;44:323–332. doi: 10.1165/rcmb.2009-0379OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Shanmugam N., Reddy M.A., Natarajan R. Distinct roles of heterogeneous nuclear ribonuclear protein K and microRNA-16 in cyclooxygenase-2 RNA stability induced by S100b, a ligand of the receptor for advanced glycation end products. J. Biol. Chem. 2008;283:36221–36233. doi: 10.1074/jbc.M806322200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Young L.E., Moore A.E., Sokol L., Meisner-Kober N., Dixon D.A. The mRNA stability factor HuR inhibits microRNA-16 targeting of COX-2. Mol. Cancer Res. 2012;10:167–180. doi: 10.1158/1541-7786.MCR-11-0337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Mubaid S., Ma J.F., Omer A., Ashour K., Lian X.J., Sanchez B.J., Robinson S., Cammas A., Dormoy-Raclet V., Di Marco S., et al. HuR counteracts miR-330 to promote STAT3 translation during inflammation-induced muscle wasting. Proc. Natl. Acad. Sci. USA. 2019;116:17261–17270. doi: 10.1073/pnas.1905172116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Kim H.H., Kuwano Y., Srikantan S., Lee E.K., Martindale J.L., Gorospe M. HuR recruits let-7/RISC to repress c-Myc expression. Genes Dev. 2009;23:1743–1748. doi: 10.1101/gad.1812509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Kedde M., van Kouwenhove M., Zwart W., Oude Vrielink J.A., Elkon R., Agami R. A Pumilio-induced RNA structure switch in p27-3’ UTR controls miR-221 and miR-222 accessibility. Nat. Cell Biol. 2010;12:1014–1020. doi: 10.1038/ncb2105. [DOI] [PubMed] [Google Scholar]
- 224.Zhang H., Sun D., Li D., Zheng Z., Xu J., Liang X., Zhang C., Wang S., Wang J., Lu W. Long non-coding RNA expression patterns in lung tissues of chronic cigarette smoke induced COPD mouse model. Sci Rep. 2018;8:7609. doi: 10.1038/s41598-018-25702-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Bi H., Zhou J., Wu D., Gao W., Li L., Yu L., Liu F., Huang M., Adcock I.M., Barnes P.J., et al. Microarray analysis of long non-coding RNAs in COPD lung tissue. Inflamm. Res. 2015;64:119–126. doi: 10.1007/s00011-014-0790-9. [DOI] [PubMed] [Google Scholar]
- 226.Tang W., Shen Z., Guo J., Sun S. Screening of long non-coding RNA and TUG1 inhibits proliferation with TGF-beta induction in patients with COPD. Int. J. Chron. Obs. Pulm. Dis. 2016;11:2951–2964. doi: 10.2147/COPD.S109570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Thai P., Statt S., Chen C.H., Liang E., Campbell C., Wu R. Characterization of a novel long noncoding RNA, SCAL1, induced by cigarette smoke and elevated in lung cancer cell lines. Am. J. Respir. Cell Mol. Biol. 2013;49:204–211. doi: 10.1165/rcmb.2013-0159RC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Gao S., Lin H., Yu W., Zhang F., Wang R., Yu H., Qian B. LncRNA LCPAT1 is involved in DNA damage induced by CSE. Biochem. Biophys. Res. Commun. 2019;508:512–515. doi: 10.1016/j.bbrc.2018.11.171. [DOI] [PubMed] [Google Scholar]
- 229.Lu L., Luo F., Liu Y., Liu X., Shi L., Lu X., Liu Q. Posttranscriptional silencing of the lncRNA MALAT1 by miR-217 inhibits the epithelial-mesenchymal transition via enhancer of zeste homolog 2 in the malignant transformation of HBE cells induced by cigarette smoke extract. Toxicol. Appl. Pharm. 2015;289:276–285. doi: 10.1016/j.taap.2015.09.016. [DOI] [PubMed] [Google Scholar]
- 230.Liu Y., Luo F., Xu Y., Wang B., Zhao Y., Xu W., Shi L., Lu X., Liu Q. Epithelial-mesenchymal transition and cancer stem cells, mediated by a long non-coding RNA, HOTAIR, are involved in cell malignant transformation induced by cigarette smoke extract. Toxicol. Appl. Pharm. 2015;282:9–19. doi: 10.1016/j.taap.2014.10.022. [DOI] [PubMed] [Google Scholar]
- 231.Hu T.J., Huang H.B., Shen H.B., Chen W., Yang Z.H. Role of long non-coding RNA MALAT1 in chronic obstructive pulmonary disease. Exp. Med. 2020;20:2691–2697. doi: 10.3892/etm.2020.8996. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 232.Lei Z., Guo H., Zou S., Jiang J., Kui Y., Song J. Long non-coding RNA maternally expressed gene regulates cigarette smoke extract induced lung inflammation and human bronchial epithelial apoptosis via miR-149-3p. Exp. Med. 2021;21:60. doi: 10.3892/etm.2020.9492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Zhao G., Su Z., Song D., Mao Y., Mao X. The long noncoding RNA MALAT1 regulates the lipopolysaccharide-induced inflammatory response through its interaction with NF-kappaB. FEBS Lett. 2016;590:2884–2895. doi: 10.1002/1873-3468.12315. [DOI] [PubMed] [Google Scholar]
- 234.Wu H., Liu J., Li W., Liu G., Li Z. LncRNA-HOTAIR promotes TNF-alpha production in cardiomyocytes of LPS-induced sepsis mice by activating NF-kappaB pathway. Biochem. Biophys. Res. Commun. 2016;471:240–246. doi: 10.1016/j.bbrc.2016.01.117. [DOI] [PubMed] [Google Scholar]
- 235.Liu J., Huang G.Q., Ke Z.P. Silence of long intergenic noncoding RNA HOTAIR ameliorates oxidative stress and inflammation response in ox-LDL-treated human macrophages by upregulating miR-330-5p. J. Cell Physiol. 2019;234:5134–5142. doi: 10.1002/jcp.27317. [DOI] [PubMed] [Google Scholar]
- 236.Kawasaki Y., Komiya M., Matsumura K., Negishi L., Suda S., Okuno M., Yokota N., Osada T., Nagashima T., Hiyoshi M., et al. MYU, a Target lncRNA for Wnt/c-Myc Signaling, Mediates Induction of CDK6 to Promote Cell Cycle Progression. Cell Rep. 2016;16:2554–2564. doi: 10.1016/j.celrep.2016.08.015. [DOI] [PubMed] [Google Scholar]
- 237.Lu Y., Liu X., Xie M., Liu M., Ye M., Li M., Chen X.M., Li X., Zhou R. The NF-kappaB-Responsive Long Noncoding RNA FIRRE Regulates Posttranscriptional Regulation of Inflammatory Gene Expression through Interacting with hnRNPU. J. Immunol. 2017;199:3571–3582. doi: 10.4049/jimmunol.1700091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Huang J., Zhang A., Ho T.T., Zhang Z., Zhou N., Ding X., Zhang X., Xu M., Mo Y.Y. Linc-RoR promotes c-Myc expression through hnRNP I and AUF1. Nucleic Acids Res. 2016;44:3059–3069. doi: 10.1093/nar/gkv1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Meisner N.C., Hintersteiner M., Mueller K., Bauer R., Seifert J.M., Naegeli H.U., Ottl J., Oberer L., Guenat C., Moss S., et al. Identification and mechanistic characterization of low-molecular-weight inhibitors for HuR. Nat. Chem. Biol. 2007;3:508–515. doi: 10.1038/nchembio.2007.14. [DOI] [PubMed] [Google Scholar]
- 240.Blanco F.F., Preet R., Aguado A., Vishwakarma V., Stevens L.E., Vyas A., Padhye S., Xu L., Weir S.J., Anant S., et al. Impact of HuR inhibition by the small molecule MS-444 on colorectal cancer cell tumorigenesis. Oncotarget. 2016;7:74043–74058. doi: 10.18632/oncotarget.12189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Trang P., Medina P.P., Wiggins J.F., Ruffino L., Kelnar K., Omotola M., Homer R., Brown D., Bader A.G., Weidhaas J.B., et al. Regression of murine lung tumors by the let-7 microRNA. Oncogene. 2010;29:1580–1587. doi: 10.1038/onc.2009.445. [DOI] [PMC free article] [PubMed] [Google Scholar]