Abstract
The relatively high frequency of marine mammal stranding events in the Philippines provide many research opportunities. A select set of stranders (n = 21) from 2017 to 2018 were sampled for bacteriology and histopathology. Pertinent tissues and bacteria were collected from individuals representing eight cetacean species (i.e. Feresa attenuata, Kogia breviceps, Globicephala macrorhynchus, Grampus griseus, Lagenodelphis hosei, Peponocephala electra, Stenella attenuata and Stenella longirostris) and were subjected to histopathological examination and antibiotic resistance screening, respectively. The antibiotic resistance profiles of 24 bacteria (belonging to genera Escherichia, Enterobacter, Klebsiella, Proteus, and Shigella) that were isolated from four cetaceans were determined using 18 antibiotics. All 24 isolates were resistant to at least one antibiotic class, and 79.17% were classified as multiple antibiotic resistant (MAR). The MAR index values of isolates ranged from 0.06 to 0.39 with all the isolates resistant to erythromycin (100%; n = 24) and susceptible to imipenem, doripenem, ciprofloxacin, chloramphenicol, and gentamicin (100%; n = 24). The resistance profiles of these bacteria show the extent of antimicrobial resistance in the marine environment, and may inform medical management decisions during rehabilitation of stranded cetaceans. Due to inadequate gross descriptions and limited data gathered by the responders during the stranding events, the significance of histopathological lesions in association with disease diagnosis in each cetacean stranding or mortality remained inconclusive; however, these histopathological findings may be indicative or contributory to the resulting debility and stress during their strandings. The findings of the study demonstrate the challenges faced by cetacean species in the wild, such as but not limited to, biological pollution through land-sea movement of effluents, fisheries interactions, and anthropogenic activities.
Introduction
The surveillance of wildlife health is part of an early warning system for detecting the emergence or resurgence of disease threats. In the case of cetacean populations in the Philippines, perhaps the most practical way of investigating their health is through their stranding events. A marine mammal is considered stranded when it runs aground, or in a helpless position such as when it is ill, weak, or simply lost [1]. While the event itself deserves attention, as it is not normal for any marine mammal to strand for no apparent reason, each stranded individual can give information on the abundance, distribution, health, and other ecological characteristics of its free-living counterparts [2], as well as threats faced by its population [3]. It is important that stranding events be responded to as quickly as possible, since some stranded animals may quickly die depending on the size of the animal and extent of human intervention [4].
Biases exist in investigating the factors involved in cetacean strandings; easy-to-detect circumstances such as obvious injuries (especially those intentionally inflicted by humans) are likely to be more reported, whereas the role of diseases or parasites may be underestimated. The capacity to detect the presence of pathogens or parasites of stranded cetaceans depends on resources, such as the presence of a stranding network with the capability to respond to stranding events as well as availability of expertise for conducting necropsy and other protocols for case investigation. Nonetheless, whether or not a pathological condition is the underlying cause of a stranding, stranded animals are good representatives for monitoring wildlife health. Also, while live strandings provide good biological samples for laboratory analyses, a dead or decomposing carcass on the beach is just as useful in providing specimens and other information as demonstrated in previous studies.
The available literature on bacteria that were isolated from marine mammals worldwide support the significance of investigating Gram-negative species and their antibiotic resistance or susceptibility. Antibiotic susceptibility patterns have been described for populations and individuals of Atlantic bottlenose dolphins, Pacific bottlenose dolphins, Risso’s dolphins, California sea lions, beluga whale, sea otters and pinnipeds [5–8]. Strains of zoonotic bacteria resistant to multiple antibiotics used for human and animal treatments were isolated from these animals, and some of those bacteria were recognized by the American Biological Safety Association (ABSA) as human pathogens. Associations between increased prevalence of antibiotic resistant bacteria in marine mammals and proximity to human activities were strongly suggested [5, 7, 9–11]. The antibiotic susceptibility profiles of bacteria isolated from cetaceans found in the Philippines where previously reported, wherein more than half of the bacteria (n = 14) had single or multiple resistances to a selection of antibiotics [12].
On the other hand, histopathological assessments proved to be useful in determining probable causes of death or debility of stranded cetaceans worldwide [13–17]. Tissue lesions help confirm parasitic and bacterial infections, co-morbidities, physical injuries (e.g., brought about by fisheries or human interactions) and bioaccumulation of chemical compounds (e.g., persistent organic pollutants) in cetaceans [16, 18–22]. Histopathological assessment is a practical and informative tool that provides pathological evidence and reinforces the necropsy conducted in dead cetaceans as part of the stranding response.
In this study, swab and tissue samples collected from cetaceans that stranded locally from February 2017-April 2018 were subjected to bacterial isolation (with subsequent antibiotic resistance screening) and histopathological assessment. Data on antibiotic resistant bacteria, parasites, and tissue lesions in cetaceans are valuable in evaluating the factors that may be associated with their local stranding events, observed to have increased in recent years [23, 24]. Of the 29 confirmed species in the country, 28 were reported to have stranded from 2005–2018 [24]. A yearly average of 105 cetacean strandings occurred in the country from 2014 to 2018 [24]; 229 events were recorded by the Philippine Marine Mammal Stranding Network (PMMSN) in collaboration with the Bureau of Fisheries and Aquatic Resources (BFAR) from 2017 (n = 121) to 2018 (n = 108) involving 118 dead and 108 live (n = 3 unknown) stranders.
Materials and methods
All biological samples were collected in coordination with PMMSN and the Marine Mammal Research and Stranding Laboratory (MMRSL) of the Institute of Environmental Science and Meteorology (IESM), University of the Philippines, Diliman (UPD). The marine mammal stranding response and tissue collection is a nationwide effort which is part of the Memorandum of Agreement (MOA) between PMMSN and BFAR. Laboratory work was done at Microbial Ecology of Terrestrial and Aquatic Systems Laboratory (METAS), Institute of Biology, UPD.
Sample collection
Cetaceans that stranded in the Philippines from February 2017 to April 2018 were opportunistically sampled for tissues and swabs by veterinarians, prosectors, or biologists who were trained by PMMSN in collaboration with BFAR. Swabs were collected from routine and non-routine sites depending on animal disposition and physical preservation, i.e., based on the expanded version of the Code system established by the Smithsonian Institution’s Marine Mammal Events Program [1]. For routine sites, swab samples were collected from the blowhole and anus of live cetaceans. For blowhole area, swabs were inserted into the hole during a breath, gently moved along the wall, and removed during the next breath in live stranders. Whenever possible, exhaled breath condensate (blow) was collected by lowering a sterile petri dish directly over the blowhole and the dish was swabbed afterwards. Anal swabs were collected by inserting rayon swabs into the anal orifice, and gently swabbing the area. Swab samples were also taken from blowhole and anal areas of freshly dead individuals. Swab samples from non-routine sites (e.g., lesions, organs, and abdominal or thoracic fluid) were also obtained from both live and dead animals especially in relation to suspected infection. Tissues were obtained during necropsy following the procedures of Pugliares et al., 2007 [25]. Stranded cetaceans were characterized in terms of species, sex, age class, stranding type, stranding site, and stranding season. Data gathered from the stranding and necropsy reports were include in the analysis.
Histopathological assessment
Tissue samples (< 1 cm3 each) were preserved in 10% neutral buffered formalin, processed by paraffin-embedded technique, sectioned at 5 μm, and subjected to hematoxylin and eosin (H&E) staining. Tissue sectioning and H&E staining technique were performed at Providence Hospital, Quezon City where tissue sections were stained with hematoxylin in water, dehydrated using a series of increasing concentrations of alcohol, and applied with eosin as a counterstain. Stained specimens were passed through xylol and toluol before mounting [26]. Using light microscopy, stained tissue samples were observed for the following: inflammation; fibrosis; granuloma lesions; edema; presence of cysts; endothelial damage (including endothelial deposits); presence of macrophages; granules; microthrombi formation; and hemorrhage.
Bacterial isolation and antibiotic resistance screening
Swab samples in transport media (e.g., Amies) were stored at 4°C and were sent to the laboratory within 18–24 h. Swabs were then enriched in Tryptic Soy Broth (TSB) for 18–24 h at 37°C. From the enriched media, inocula were streaked on MacConkey Agar (MCA) plates. Morphologically distinct Gram-negative colonies were sub-cultured and purified. Bacterial smears of pure cultures were Gram-stained according to Brown and Smith (2015) [27]. Gram-negative bacterial isolates were subjected to 16S rRNA gene sequencing-based identification and antibiotic resistance screening.
Pure bacterial isolates were identified using 16S rRNA gene amplification. Bacterial DNA was extracted from the purified isolates using either the GF-1 Bacterial DNA Extraction Kit (Vivantis Technologies) following manufacturer’s instructions, or the Boil Lysis Method following Ahmed and Dablool (2017) [28]. The universal 16S rRNA bacterial gene was amplified from the DNA of isolates through polymerase chain reaction (PCR). The primers used for targeting the 16S rRNA gene were 27F (5’-AGAGTTTGATCCTGGCTCAG-3’) and 1541R (5’-AAGGAGGTGATCCANCCRCA-3’) [29, 30]. The PCR reaction mix consisted of: dNTPs, MgCl2, Taq DNA polymerase, DNA template, forward and reverse primers, and nuclease-free water. The thermal cycler conditions were as follows: initial denaturation for 2 min at 95°C, 30 cycles of denaturation for 30 s at 94°C, annealing for 30 s at 55–60°C, extension for 30 s at 72°C, and final extension for 7 min at 72°C. Positive controls (E. coli ATCC® 25922) and blanks (DNA-free templates) were included. PCR products were subjected to agarose gel electrophoresis (AGE) to detect target DNA band. PCR products were then sent to Macrogen (South Korea) for DNA purification and sequencing. PreGap4 and Gap4 (Staden Package 2.0) were used to obtain the consensus sequences [31]. Sequence homologies were determined using NCBI BLASTn search and further analyses were done using BioEdit [32, 33].
Kirby-Bauer Disk Diffusion Assay [34] was performed to determine the sensitivity of the bacterial isolates to antibiotics (Table 1). These antibiotics were chosen based on (1) inclusion in the priority list of WHO for antibiotic resistance research; (2) known use in agriculture and aquaculture; (3) reported susceptibility profiles of bacteria isolated from marine animals worldwide; (4) use during rehabilitation of stranded marine mammals; and (5) known spectrum activity [5, 35–38]. To ensure that only acquired resistances will be observed, antibiotics to which the bacterial isolates have intrinsic resistances were excluded in the assay. The reactions of the isolates to the antibiotics were described as Susceptible (S), Intermediate (I), or Resistant (R) based on Clinical and Laboratory Standards Institute (CLSI) M31-A2 (2002), M100-S24 (2014), and European Committee on Antimicrobial Susceptibility Testing (EUCAST) v 8.0 (2018). E. coli ATCC® 25922 was used as the control [39–41]. Multiple Antibiotic Resistance (MAR) Index values were computed using the formula: (# of resistant antibiotics / total # of antibiotics tested) [40]. MAR indices greater than 0.2 were interpreted to come from sources where antibiotics are often used [35, 42, 43]. Also, MAR isolates were interpreted as those that are resistant to three or more antibiotic classes [44].
Table 1. Antibiotics used in the Kirby-Bauer Disk Diffusion Assay.
| Antibiotic class | Antibiotics |
|---|---|
| Carbapenems | Imipenem |
| Meropenem | |
| Ertapenem | |
| Doripenem | |
| Penicillins | Ampicillin |
| Cephems | Cephalothin |
| Ceftriaxone | |
| Cefoxitin | |
| Fluoroquinolones | Moxifloxacin |
| Ciprofloxacin | |
| Ofloxacin | |
| Aminoglycosides | Amikacin |
| Gentamicin | |
| Tetracyclines | Tetracyclines |
| Oxytetracyclines | |
| Phenicols | Chloramphenicol |
| Folate pathway inhibitors | Trimethoprim-sulfamethoxazole |
| Macrolides | Erythromycin |
Results
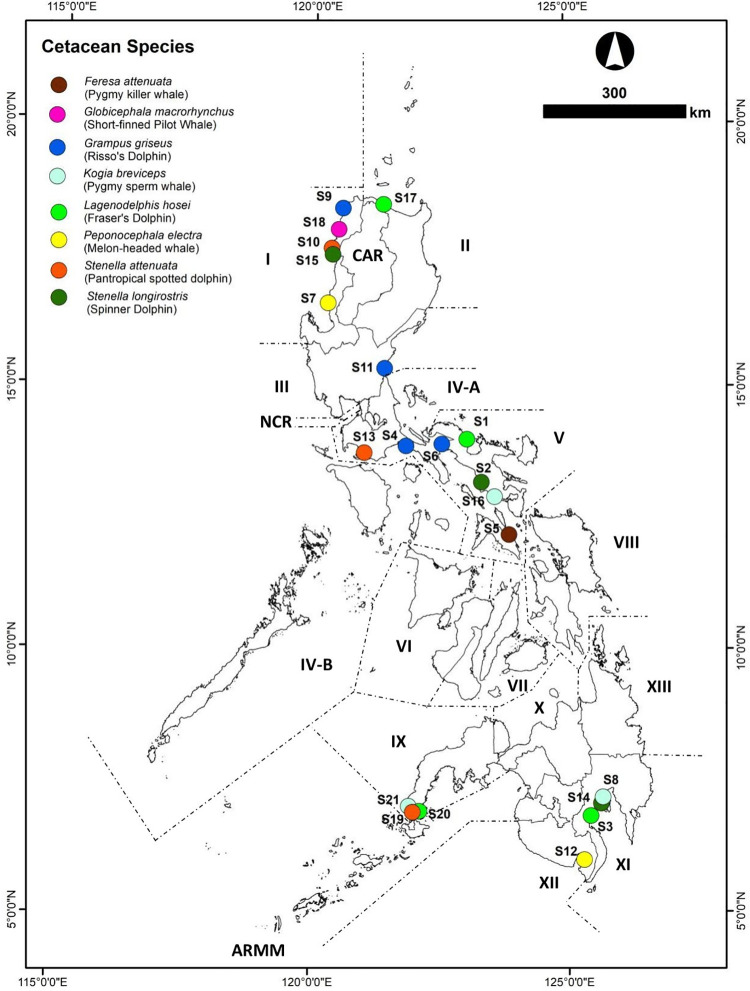
In this study, tissue samples and bacterial isolates were obtained from 21 stranded cetaceans representing eight species (Feresa attenuata, Kogia breviceps, Globicephala macrorhynchus, Grampus griseus, Lagenodelphis hosei, Peponocephala electra, Stenella attenuata, and Stenella longirostris) (Fig 1). Of the 21 select cases sampled, 15 were originally live stranders and six (6) fresh dead. These stranded cetaceans came mainly from Luzon (n = 14) and Mindanao (n = 7). The stranded marine mammals sampled consisted of four (4) Fraser’s, four (4) Risso’s, three (3) spinner, and three (3) pantropical spotted dolphins, and one (1) short-finned pilot, three (3 pygmy sperm, and two (2) melon-headed whales. Samples came from 16 females, four (4) males and two (2) undetermined. By age class, the samples were composed of 15 adults, five (5) subadults, and one (1) neonate (Table 2).
Fig 1. Sites of cetacean stranding events from February 2017-April 2018.
S1 –S21: Cetacean Strander Codes; I-XIII, CAR, NCR and ARMM: Administrative Regions in the Philippines (Reprinted from Philippines—Subnational Administrative Boundaries under a CC BY license, with permission from The Humanitarian Data Exchange, original copyright 2020).
Table 2. Stranded cetaceans sampled for the study (2017–2018).
| Strander Code | PMMSN Code | Species | Region | Date of Stranding | Stranding Type | Age Class | Condition | Sex |
|---|---|---|---|---|---|---|---|---|
| S01 | Lh03R5270217 | Fraser’s dolphin Lagenodelphis hosei | V | 27-Feb-17 | Single | Adult | Alive | Male |
| S02 | Sl21R5040317 | Spinner dolphin Stenella longirostris | V | 04-Mar-17 | Single | Adult | Alive | Female |
| S03 | Lh03R11010317 | Fraser’s dolphin Lagenodelphis hosei | XI | 09-Mar-17 | Single | Adult | Dead | Female |
| S04 | Gg04R4A290317 | Risso’s dolphin Grampus griseus | IV-A | 29-Mar-17 | Single | Subadult | Alive (Died) | Male |
| S05 | Fa02R5020517 | Pygmy killer whale Feresa attenuata | V | 02-May-17 | Mass | Adult | Alive (Died) | Unknown |
| S06 | Gg15R5090517 | Risso’s dolphin Grampus griseus | V | 09-May-17 | Single | Adult | Alive (Died) | Unknown |
| S07 | Pe04R1300417 | melon-headed whale Peponocephala electra | I | 30-April-17 | Single | Adult | Dead | Female |
| S08 | Kb07R11160517 | Pygmy sperm whale Kogia breviceps | XI | 16-May-17 | Single | Adult | Alive | Male |
| S09 | Gg10R1150617 | Risso’s dolphin Grampus griseus | I | 15-Jun-17 | Single | Neonate | Alive | Female |
| S10 | Sa18R1210617 | pantropical spotted dolphin Stenella attenuata | I | 21-Jun-17 | Single | Subadult | Dead | Female |
| S11 | Gg02R3230617 | Risso’s dolphin Grampus griseus | III | 23-Jun-17 | Single | Adult | Dead | Female |
| S12 | Pe06R12030717 | melon-headed whale Peponocephala electra | XII | 03-Jul-17 | Single | Adult | Alive | Male |
| S13 | Sa03R4A280717 | pantropical spotted dolphin Stenella attenuata | IV-A | 28-Jul-17 | Single | Subadult | Alive | Female |
| S14 | Sl06R11310817 | spinner dolphin Stenella longirostris | XI | 31-Aug-17 | Single | Subadult | Alive | Female |
| S15 | Sl23R1300917 | spinner dolphin Stenella longirostris | I | 30-Sep-17 | Single | Subadult | Dead | Female |
| S16 | Kb02R5091117 | pygmy sperm whale Kogia breviceps | V | 09-Nov-17 | Single | Adult | Alive | Female |
| S17 | Lh04R2011217 | Fraser’s dolphin Lagenodelphis hosei | II | 01-Dec-17 | Single | Adult | Alive | Female |
| S18 | Gm11R151217 | short-finned pilot whale Globicephala macrorhynchus | I | 05-Dec-17 | Single | Adult | Alive | Female |
| S19 | Sa03R9160118 | pantropical spotted dolphin Stenella attenuata | IX | 16-Jan-18 | Single | Adult | Alive | Female |
| S20 | Lh01R9170418 | Fraser’s dolphin Lagenodelphis hosei | IX | 17-Apr-18 | Single | Adult | Dead | Female |
| S21 | Kb01R9260418 | pygmy sperm whale Kogia breviceps | IX | 27-Apr-18 | Single | Adult | Alive | Female |
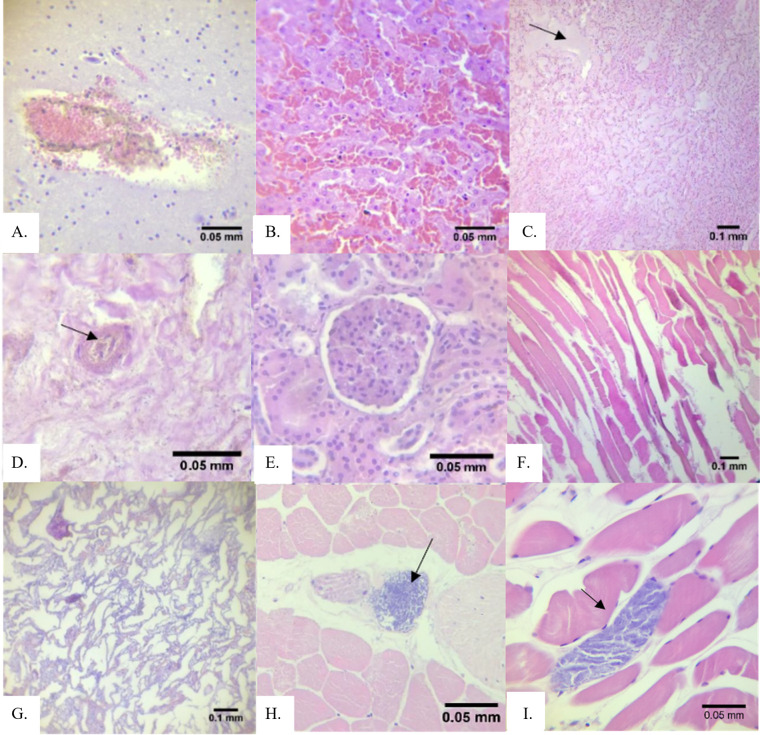
A total of 73 tissue samples representing 6 organs (brain, cardiac muscle, kidney, skeletal muscle, liver, lungs) were obtained from 21 stranded cetaceans: 3 spotted dolphins (S. attenuata), 3 spinner dolphins (S. longirostris), 4 Fraser’s dolphins (L. hosei), 4 Risso’s dolphins (G. griseus), 3 pygmy sperm whales (K. breviceps), 2 melon-headed whales (P. electra), 1 pygmy killer whale (F. attenuata) and 1 short-finned pilot whale (G. macrorhynchus). Of these animals, 19 (90.48%) showed lesions in the organs tissues collected (Table 3 and Fig 2). Most of these cetaceans were adults; there was only one neonate. Unidentified cysts and putative Sarcocystis sp. were observed in some tissues with prevalence rates of 47.62% and 9.52% respectively. Some of the unidentified cysts are hypothesized to be other coccidian cysts based on observed structures (e.g., size, shape, thick or thin membrane, etc.) very similar to any stage of reference species (e.g., Toxoplasma), however in the absence of confirmatory methods such as immunohistochemical staining, these cysts are labeled as “unidentified”, as observed in H & E stained tissues. Also, P. delphini cysts in the muscle-blubber region and nematodes in the stomach were seen during gross necropsy and the reported identification was confirmed by the authors.
Table 3. Histopathological findings/remarks on cetaceans that stranded in the Philippines from February to April 2018.
| Strander No. | Species | Sex | Age Class | Findings |
|---|---|---|---|---|
| S101 | Stenella attenuata | Female | Subadult | moderate congestion, hemorrhage, and membranous glomerulopathy in the kidney; unidentified cysts in the skeletal muscle; moderate to severe congestion in the liver; atelectasis in the lungs; no apparent lesion in brain and cardiac muscle |
| S13 | Stenella attenuata | Female | Subadult | glomerulopathy and edema in the kidney; no apparent lesion in brain and cardiac muscle |
| S19 | Stenella attenuata | Female | Adult | severe congestion in the cardiac muscle; hemorrhage, severe congestion, glomerulopathy in the kidney |
| S02 | Stenella longirostris | Female | Adult | moderate congestion in the brain; no apparent lesion in cardiac muscle, kidney, and skeletal muscle |
| S14 | Stenella longirostris | Female | Subadult | moderate congestion for cardiac muscle; glomerulopathy with lymphocytic aggregation and unidentified cysts in the kidney; unidentified cyst and Sarcocystis cyst in the skeletal muscle |
| S152 | Stenella longirostris | Female | Subadult | no apparent lesions in the brain, cardiac muscle, skeletal muscle, liver, and lungs |
| S013 | Lagenodelphis hosei | Male | Adult | moderate congestion in the brain; unidentified cyst in the skeletal muscle; no apparent lesions in the cardiac muscle and kidney |
| S034 | Lagenodelphis hosei | Female | Adult | moderate congestion in the brain and cardiac muscle; unidentified cysts in the cardiac muscle; severe congestion, hemorrhage, and edema in the kidney; no apparent lesion in skeletal muscle |
| S17 | Lagenodelphis hosei | Female | Adult | glomerulopathy and edema in the kidney; unidentified cyst in the skeletal muscle; |
| S20 | Lagenodelphis hosei | Female | Adult | severe congestion in the brain, cardiac muscle and kidney; glomerulopathy in the kidney |
| S04 | Grampus griseus | Male | Subadult | severe congestion in the cardiac muscle and kidney; no apparent lesion in brain and skeletal muscle |
| S06 | Grampus griseus | Unknown | Adult | atrophy and Zenker’s necrosis in the skeletal muscle; no apparent lesion in brain and cardiac muscle |
| S09 | Grampus griseus | Female | Neonate | severe congestion in the cardiac muscle; unidentified cyst in the skeletal muscle; severe diffused hepatic sinusoidal congestion in the liver; severe congestion and focal pulmonary edema in the lungs; no apparent lesion in kidney |
| S115 | Grampus griseus | Female | Adult | no apparent lesion in cardiac muscle |
| S07 | Peponocephala electra | Female | Adult | swollen glomerulus and hemosiderosis in the kidney; unidentified cysts in the skeletal muscle; no apparent lesion in cardiac muscle |
| S12 | Peponocephala electra | Male | Adult | membranous glomerulopathy in the kidney; hepatic edema in the liver; pulmonary edema in the lungs; no apparent lesion in brain, cardiac muscle, and skeletal muscle |
| S086 | Kogia breviceps | Male | Adult | putative Sarcocystis cyst in the skeletal muscle; no apparent lesion in brain and cardiac muscle |
| S16 | Kogia breviceps | Female | Adult | severe congestion in the brain; unidentified cyst in the cardiac muscle; hemorrhage and severe congestion in the kidney |
| S21 | Kogia breviceps | Female | Adult | moderate congestion in the cardiac muscle; hemorrhage and glomerulopathy in the kidney |
| S05 | Feresa attenuata | Unknown | Adult | moderate to severe congestion; hemorrhage and hemosiderosis in the brain; unidentified cysts and hemosiderosis in the kidney; no apparent lesion in skeletal muscle |
| S18 | Globicephala macrorhynchus | Female | Adult | moderate congestion in the cardiac muscle; hemorrhage and glomerulopathy in the kidney; unidentified cyst in the skeletal muscle; no apparent lesion in the brain |
1 acoustic trauma likely cause of stranding.
2 only subadult animal without any apparent lesions in organs examined.
3 unidentified parasites on eyes and P. delphini cyst in the skeletal muscle seen during necropsy.
4 shark attack likely cause of stranding.
5 only adult animal without apparent lesions.
6 P. delphini cysts in the muscle-blubber and nematodes in the stomach seen during necropsy.
Observed tissues include brain, cardiac muscle, kidney, skeletal muscle, liver, and lungs tissues; tissues not mentioned in the findings are those that were not available for histopathological observation.
Fig 2. Histopathological lesions observed in tissues of 21 cetaceans that stranded in the Philippines (2017–2018).
(A) hemorrhage in S03 kidney; (B) severe congestion in S11 liver; (C) edema in S12 liver; (D) hemosiderosis characterized by the presence of brown granular pigments in S05 kidney; (E) glomerulopathy in S14 kidney; (F) Zenker’s necrosis characterized by hyaline degeneration, loss of striations, and muscle fiber waviness in S06 skeletal muscle; (G) atelectasis (collapsed alveoli) in S10 lungs; (H) unidentified cyst in skeletal muscle of S14; and (I) putative Sarcocystis cyst in skeletal muscle of S08.
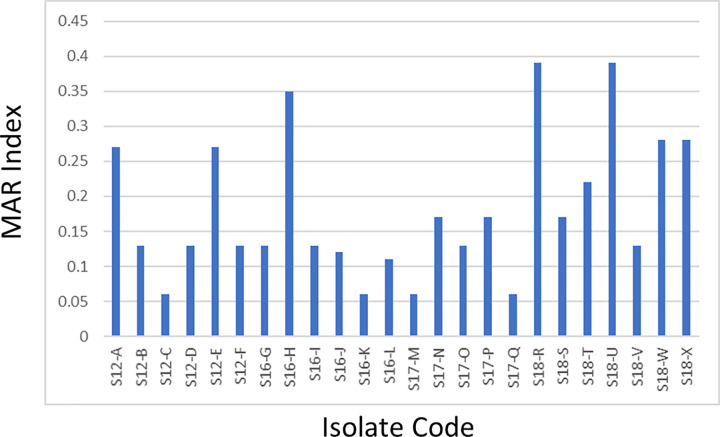
A total of 24 Gram-negative bacteria that belong to the family Enterobacteriaceae were isolated from four cetaceans (S12, S16, S17, S18). Based on 16S rRNA gene, these isolates were confirmed to have 98–100% sequence similarities to species belonging to the following genera: Escherichia (n = 6), Enterobacter (n = 8), Klebsiella (n = 5), Proteus (n = 4), and Shigella (n = 1) (Table 4). These isolates were resistant to at least one antibiotic class tested, and 79.17% were classified as multiple antibiotic resistant (i.e., resistant to at least three antibiotic classes). The MAR index values of the isolates ranged from 0.06 to 0.39. (Fig 3).
Table 4. Genotypic identification of bacteria isolated from stranded cetaceans.
| Source Cetacean (Code) | Swab Site | Isolate Code | Nearest Phylogenetic Affiliation (% Sequence Similarity) | NCBI* Accession Number |
|---|---|---|---|---|
| S12 (Peponocephala electra, adult) | urine | S12-A | Enterobacter cloacae (99%) | MH101512.1 |
| S12-B | Klebsiella aerogenes (99%) | CP024883.1 | ||
| S12-C | Escherichia hermannii (98%) | JN644551.1 | ||
| S12-D | Enterobacter sp. (99%) | KC236445.1 | ||
| S12-E | Enterobacter cloacae (100%) | KY492312.1 | ||
| S12-F | Enterobacter cloacae (99%) | JN644583.1 | ||
| S16 (Kogia breviceps, adult) | blowhole | S16-G | Enterobacter ludwigii (99%) | JQ659806.1 |
| S16-H | Escherichia hermannii (99%) | JN644551.1 | ||
| S16-I | Enterobacter cloacae (99%) | KM538690.1 | ||
| S16-J | Klebsiella pneumoniae (99%) | FO203501.1 | ||
| S16-K | Klebsiella pneumoniae (99%) | KJ803907.1 | ||
| S16-L | Shigella sp. (99%) | KU362661.1 | ||
| S17 (Lagenodelphis hosei adult) | genital Slit | S17-M | Klebsiella pneumoniae (99%) | CP020847.1 |
| S17-N | Escherichia coli (99%) | AP017620.1 | ||
| blowhole | S17-O | Enterobacter cloacae (99%) | CP010512.1 | |
| S17-P | Escherichia coli (99%) | JQ661149.1 | ||
| wound | S17-Q | Klebsiella quasipneumoniae (99%) | CP014696.2 | |
| S18 (Globicephala macrorhynchus, adult) | anus | S17-R | Proteus mirabilis (99%) | CP015347.1 |
| brainstem | S17-S | Proteus mirabilis (99%) | CP015347.1 | |
| cerebellum | S17-T | Proteus mirabilis (99%) | CP004022.1 | |
| lungs | S18-U | Escherichia fergusonii (99%) | KJ803900.1 | |
| blowhole | S18-V | Enterobacter tabaci (99%) | NR_146667.2 | |
| S18-W | Proteus mirabilis (99%) | CP015347.1 | ||
| S18-X | Escherichia coli (99%) | CP027060.1 |
*National Center for Biotechnology Information.
Fig 3. MAR index values of Enterobacteriaceae from sampled cetaceans.
Discussion
Lesions in tissues of stranded cetaceans
Histopathological assessment of tissues is a very useful tool to identify factors causing the death of stranded cetaceans or determine the cause of their stranding events [15, 45–47]. However, our histopathological findings were mainly limited to indicative debility and stress during the stranding events because of the inadequate gross descriptions available from the stranding and necropsy reports, prohibiting us to link these findings to disease processes particularly those contributory to cetacean mortalities. As the stranding network (and the country in general) is still building the expertise in performing necropsy for investigating the death of stranded cetaceans, we tried to gather scientific information by performing histopathological observations of available tissues as ancillary to the stranding report. We recognize that there is inadequate information which will help us ascertain the cause of death of the stranded animal, but at the same time deem our findings useful in providing information about the health of the cetaceans.
In general, lesions in tissues of cetaceans were associated with bycatch, trauma, parasitic and bacterial infections, and presence of persistent organic pollutants in stranded cetaceans [13–15, 17, 20, 48–50]. A previous study in the Philippines involved the histopathological assessment of renal tissues which corroborated the results of molecular and culture methods for a suggested case of leptospirosis in a melon-headed whale (Peponocephala electra) [51].
When a cetacean strands, it is highly likely to have congestion in the liver and other organs due to the pressure from the weight of its body lying on the thorax as well as immobility preventing venous circulation [52]. It may be noted that organ congestion is the most observed type of lesion in this study, i.e., observed in at least one organ tissue of 13 cetaceans (62%). Several factors can also put cetaceans in stressful situations which can induce stress myopathy and possibly cause congestion and hemorrhage [53]. The stranding event itself can induce trauma and stress myopathy on the animal, causing congestion, hemorrhage, and skeletal and cardiac muscle degeneration such as in the case of Zenker’s necrosis [15, 53, 54] in the skeletal muscle of an adult Risso’s dolphin. However, we cannot corroborate these assumptions with other evidence.
Congestion in brain and kidneys of cetaceans has also been associated with acoustic trauma [55]. One of the ways to confirm acoustic trauma is through histological observations of the inner ears [56]. Acoustic trauma was suggested as the cause of some previously reported cetacean stranding events in the Philippines, possibly due to blast fishing activities near the stranding sites [23, 57]. There is a growing concern on marine environment being compromised by human activities (e.g. underwater explosions, seismic exploration, shipping, operation of naval sonar) which affects the physiology, communication, behavior and energetics of several population of marine species [58–60]. Anthropogenic noise is now recognized as a major global pollutant and is acknowledged as an environmental stressor [58]. Thus, future efforts should include histopathological examinations of the inner ear.
Glomerulopathy was observed in 10 out of 17 cetaceans (59%) with kidney tissues available for observation, and is the most observed kidney tissue lesion (10 out of 14 with lesions or 71%). Comparably, membranous glomerulonephritis was a common finding among stranded cetaceans in Brazil [17]. This lesion was suggested in other studies to be associated with microbial infections or chronic exposure of cetaceans to metals such as cadmium, copper, and zinc, but this remains speculative in our case due to the lack of toxicological analyses and conclusive diagnoses of infections or diseases [50, 61].
Parasites in cetaceans may predispose these animals to bacterial infections, cardiovascular complications, septicemia and other conditions, which are also frequently reported as probable causes of death during their stranding events [16, 62, 63]. Here, we are reporting the detection of cysts in the observed tissues of cetaceans. There is no known histopathological report on cysts such as for example, T. gondii and Sarcocystis sp., in tissues sampled from cetaceans that stranded in different sites in the Philippines, although there are earlier reports on T. gondii detection using serological and molecular methods [51, 64]. However, as mentioned, we did not perform confirmatory methods for the identification of the cysts, and so we refer to them as either “unidentified” or “putative”. A better understanding of the biology, epidemiology, and pathogenesis of tissue-encysting coccidian organisms that parasitize marine mammals is needed to properly assess the risks and burden of protozoal disease in aquatic ecosystems [65–67]. The transmission of these parasites is still poorly understood in marine mammals, although it is known that they are found in striated muscles of intermediate hosts [68–71]. The most likely modes of transmission of these parasites to aquatic animals are via ingestion of water-borne oocysts or sporocysts originating from sewage runoff or through infected prey [65, 66, 72–74]. During the past two decades, coccidian infections have been detected in marine mammals that stranded along the coast of the northeastern Pacific Ocean [65, 75]. These infections include encephalitis, myositis, hepatitis and myocarditis [66, 67].
In addition, the presence of P. delphini in the muscles and blubber of stranded Fraser’s dolphin (L. hosei) and pygmy sperm whale (K. breviceps) was reported in the necropsy reports. This parasite has been documented in many cetacean species, commonly in the subcutaneous blubber with typical concentration in the perigenital region [76]. Siquier and Le Bas (2003) suggested that Fraser’s dolphins (Lagenodelphis hosei) could act as intermediate or accidental hosts for P. delphini, and that definitive host infection could occur through predation. There is a need for more evidence to confirm the role of cetaceans in the life cycle of this parasite [77]. The consumption of muscles containing these parasites is one of the major routes of transmission to humans. This route of transmission is unlikely to involve cetaceans in the Philippines, as hunting and killing of marine mammals are prohibited under Section 4 of Republic Act 9147 (Wildlife Resources Conservation and Protection Act of the Philippines). Still, there were local reports of fishermen butchering cetaceans for food consumption (pers comm., BFAR Region V).
Antibiotic resistant bacteria from stranded cetaceans
Overall, the bacterial isolates have resistances to carbapenems and third-generation cephalosporins. Enterobacteriaceae resistant to carbapenems and third-generation cephalosporins are considered a research priority for the discovery of new antibiotic agents [38]. As the “last line of defense” against multiple antibiotic resistant bacteria, the detection of carbapenem-resistant strains is a troubling point of concern as carbapenems are fourth- generation antibiotics recommended for critical Gram-negative infections [78]. To the best knowledge of the authors, only Greig et al. (2007) had so far used imipenem and meropenem for antibiotic susceptibility tests on bacteria isolated from cetaceans. Greig et al. reported imipenem-resistant E. coli in bottlenose dolphins, but all of their isolates were still susceptible to meropenem at the time [5].
All isolates were most resistant to erythromycin. The high frequency of resistance against this antibiotic is said to be due to acquired macrolide–lincosamide–streptogramin B (MLS) resistance genes, which is common among Enterobacteriaceae [79, 80]. More than 50% of the isolates were also resistant to cephalothin, ampicillin, and moxifloxacin. It must be noted that the isolated Klebsiella spp., and Escherichia spp., bacterial species often reported as pathogenic to cetaceans, were resistant to erythromycin [81, 82]. Similarly, high resistance to erythromycin, cephalothin, and ampicillin of E. coli isolated from bottlenose dolphins in Florida and South Carolina was reported [5, 6]. Extra-intestinal pathogenic E. coli isolated from resident killer whales of San Juan Islands, Washington, were found to be resistant to aminoglycosides, sulfonamides, and tetracycline [83]. Resistances against cephalothin and ampicillin were also observed in bacteria isolated from dolphins, whales, and seals in the Northeastern United States Coast [35]. An overall high prevalence (88%) of resistance to at least one antibiotic was found among bacteria isolated from wild bottlenose dolphins in Florida, with highest resistances against erythromycin followed by ampicillin [6]. A previous study on antibiotic susceptibility patterns of bacteria isolated from stranded cetaceans in the Philippines reported the highest resistance (47%) to cefazolin [12]. Susceptibilities to amikacin and gentamicin were also reported among bacteria isolated from marine mammals in Florida, South Carolina, and Northeastern US Coast [5, 35].
Based on these findings, the choice of antibiotics for treating bacterial infections (most commonly pneumonia: pers comm., PMMSN veterinarians) caused by Enterobacteriaceae in locally stranded cetaceans under rehabilitation should consider the susceptibility and/or resistance profiles of bacteria. This may be possibly done through the stranding response being carried out by PMMSN, wherein such profiles can be provided by the collaborating microbiologists (e.g., the authors of this study) to the veterinarians handling the medical management of cetaceans. In the case of the bacteria isolated from cetaceans sampled in the present study, carbapenems are the most effective antibiotic. However, this information must be interpreted with caution, as the bacteria were not significantly associated with any clinical presentation of infection or disease in the cetacean.
The cetacean species sampled in this study generally inhabit deep waters, but their physiology entails a regular need to surface to sequester oxygen from the air for breathing, thus exposing themselves to sewage outflows and other forms of pollution that eventually reach them from the nearby coast [84–86]. The presence of bacteria (and associated antibiotic resistances) in these cetaceans indicate biological pollution and presence of antibiotic resistance in their habitats [87–89]. In this study, 33.33% of the isolates from cetaceans had MAR indices greater than 0.2, suggesting that the isolates may have developed resistance from sources that the cetaceans were exposed to, such as bodies of water highly polluted with antibiotics, including domestic, industrial and hospital sewage outflows, water-treatment facilities, and the like [85, 86]. As the use of antibiotics stems from anthropogenic activities, this implies the need to regulate and monitor the use and improper disposal of antibiotics to water bodies.
Conclusion
Twenty-one cetaceans that stranded in different parts of the Philippines were sampled for bacterial isolation and antibiotic resistance screening as well as histopathological assessment of available tissues. In the absence of conclusive data on the specific causes of the mortality or morbidity of the cetaceans in relation to the stranding event, the histopathological findings just provide clues on possible involvement of factors (e.g., acoustic trauma, stress, etc.) that may have affected the health of cetaceans rendering them to strand or die, or possible effects of the stranding event itself on the animal. Bacteriological findings showed more than 50% of the isolated bacteria are multiple antibiotic resistant and that all of them are resistant to erythromycin and susceptible to imipenem, doripenem, ciprofloxacin, chloramphenicol, and gentamicin. While these information may be helpful in the medical management of stranded cetaceans during rehabilitation, they also indicate the extent of antimicrobial resistance in the marine environment. As sentinels, cetaceans demonstrate the threats faced by their populations in the wild, and monitoring their health through stranded representatives is a practical approach that can help improve conservation efforts. As local stranding network expands and veterinary and research expertise improve, more robust data from bacteriological and histopathological assessments of cetaceans are expected to be available in the coming years.
Acknowledgments
We thank the Philippine Marine Mammal Stranding Network (PMMSN) and the Bureau of Fisheries and Aquatic Resources (BFAR) for the nationwide cetacean stranding response. Likewise, we thank Honey Leen M. Laggui for help in the preparation of Fig 1 and Christopher Torno, DVM for valuable comments.
Data Availability
All relevant data are within the manuscript.
Funding Statement
MCMO and LVA received funding for the study through project grants BIO-19-1-05 (Natural Sciences Research Institute, University of the Philippines Diliman) for the conduct of methodology and 171704SOS (Office of the Vice Chancellor for Research and Development, University of the Philippines Diliman) for sample collection respectively. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Geraci JR, Lounsbury VJ. Marine mammals ashore: a field guide for strandings. Baltimore: National Aquarium in Baltimore, Inc, 2005. [Google Scholar]
- 2.Bossart GD. Marine mammals as sentinels for oceans and human health. Vet. Pathol. 2011;2(4): 3–6. doi: 10.1177/0300985810388525 [DOI] [PubMed] [Google Scholar]
- 3.Leeney RH, Amles R, Broderick AC, Witt MJ, Loveridge J, Doyle J. et al. Spatio-temporal analysis of cetacean strandings and bycatch in a UK fisheries hotspot. Biodivers. Conserv. 2008;17: 2323–2338. [Google Scholar]
- 4.Evans K, Thresher R, Warneke RM, Bradshaw CJA, Pook M, Thiele D. et al., Periodic variability in cetacean strandings: Links to large-scale climatic events. Biol. Lett. 2005;1: 147–150. doi: 10.1098/rsbl.2005.0313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Greig TW, Bemiss JA, Lyon BR, Bossart GD, Fair PA. Prevalence and diversity of antibiotic resistant Escherichia coli in bottlenose dolphins (Tursiops truncatus) from the Indian River Lagoon, Florida, and Charleston Harbor Area, South Carolina, Aquat. Mamm. 2007;33(2): 185–194. [Google Scholar]
- 6.Schaefer AM, Goldstein JD, Reif JS, Fair PA, Bossart GD. Antibiotic-resistant organisms cultured from Atlantic bottlenose dolphins (Tursiops truncatus) inhabiting estuarine waters of Charleston, SC and Indian River Lagoon, FL. Ecohealth, 2009;6: 33–41. doi: 10.1007/s10393-009-0221-5 [DOI] [PubMed] [Google Scholar]
- 7.Brownstein D, Miller MA, Oates SC, Byrne BA, Jang S, murray MJ, et al. Antimicrobial susceptibility of bacterial isolates from sea otters (Enhydra lutris). J Wildl. Dis. 2011;47(2): 278–292. doi: 10.7589/0090-3558-47.2.278 [DOI] [PubMed] [Google Scholar]
- 8.Wallace CC, Yund PO, Ford TE, Matassa KA, Bass AL. Increase in antimicrobial resistance in bacteria isolated from stranded marine mammals of the Northwest Atlantic. Ecohealth. 2013;10: 201–210. doi: 10.1007/s10393-013-0842-6 [DOI] [PubMed] [Google Scholar]
- 9.Bogomolni AL, Gast RJ, Ellis JC, Dennett M, Pugliares KR, Lentell BJ, et al. Victims or vectors: a survey of marine vertebrate zoonoses from coastal waters of the Northwest Atlantic. Dis Aquat Organ. 2008;81: 13–38. doi: 10.3354/dao01936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stoddard RA, Atwill ER, Gulland FMD, Miller MA, Dabritz HA, Paradies DM, et al. Risk factors for infection with pathogenic and antimicrobial-resistant fecal bacteria in northern elephant seals in California. Public Health Rep. 2008;123: 360–370. doi: 10.1177/003335490812300316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Miller MA, Byrne BA, Jang SS, Dodd EM, Dorfmeier E, Harris MD, et al. Enteric bacterial pathogen detection in southern sea otters (Enhydra lutris nereis) is associated with coastal urbanization and freshwater runoff. Vet. Res. 2010;41(1): 1–13. doi: 10.1051/vetres/2009049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Obusan MCM, Aragones LV, Rivera WL, Siringan MAT. Antibiotic susceptibility patterns of bacteria isolated from cetaceans stranded in the Philippines. Aquat. Mamm. 2018;44(5): 568–579. [Google Scholar]
- 13.McFee WE, Lipscomb TP. Major pathologic findings and probable causes of mortality in bottlenose dolphins in South Carolina from 1993 to 2006. J. Wildl. Dis. 2009;45(3): 575–593. doi: 10.7589/0090-3558-45.3.575 [DOI] [PubMed] [Google Scholar]
- 14.Gonzales-Viera O, Chavera A, Yaipén-Llanos C, Perales-Camacho, R. Histopathological aspects and etiology of pneumonias in stranded marine mammals from Lima, Peru. Braz. J. Vet. Pathol, 2011;4(1): 23–29. [Google Scholar]
- 15.Díaz-Delgado J, Fernández A, Sierra E, Sacchini S, Andrada M, Vela AI, et al. Pathologic findings and causes of death of stranded cetaceans in the Canary Islands (2006–2012). PLoS One. 2018; 13(10):e0204444. doi: 10.1371/journal.pone.0204444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guimarães JP, Febronio AMB, Vergara-Parente JE, Werneck MR. Lesions associated with Halocercus brasiliensis Lins de Almeida, 1933 in the lungs of dolphins stranded in the northeast of Brazil. J. Parasitol. 2015;101(2): 248–251. doi: 10.1645/14-513.1 [DOI] [PubMed] [Google Scholar]
- 17.Domiciano IG, Domit C, Broadhurst MK, Koch MS, Bracarense APF. Assessing disease and mortality among small cetaceans stranded at a world heritage site in southern Brazil. PloS one. 2016;11(2). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Arbelo M, Espinosa de los Monteros A, Herráez P, Andrada M, Sierra E Rodríguez F, et al. Pathology and causes of death of stranded cetaceans in the Canary Islands (1999–2005). Dis. Aquat. Organ. 2013; 103(2): 87–99. doi: 10.3354/dao02558 [DOI] [PubMed] [Google Scholar]
- 19.Ko FC, WE NY, Chou LS. Bioaccumulation of persistent organic pollutants in stranded cetaceans from Taiwan coastal waters. J. Hazard. Mater. 2014;277: 127–133. doi: 10.1016/j.jhazmat.2013.12.057 [DOI] [PubMed] [Google Scholar]
- 20.Bondoc JL, Aragones LV, Masangkay JS. Hematological, macroscopic and microscopic findings in two-stranded whales (Mesoplodon densirostris and Kogia sima) and Possible Causes of Deaths. Philipp. J. Vet. Med. 2017;54(1): 63–69. [Google Scholar]
- 21.Giorda F, Ballardini M, Di Guardo G, Pintore MD, Grattarola C, Iulini B, et al. Postmortem findings in cetaceans found stranded in the Pelagos Sanctuary, Italy, 2007–14. J. Wildl. Dis. 2017;53(4): 795–803. doi: 10.7589/2016-07-150 [DOI] [PubMed] [Google Scholar]
- 22.Li WT, Chang HW, Chen MH, Chiou HY, Liou BY, Pang VF, et al. Investigation of silver (Ag) deposition in tissues from stranded cetaceans by autometallography (AMG). Environ. Pollut. 2018;235: 534–545. doi: 10.1016/j.envpol.2018.01.010 [DOI] [PubMed] [Google Scholar]
- 23.Aragones LV, Roque MAA, Flores MB, Encomienda RP, Laule GE, Espinos BG, et al. The Philippine marine mammal strandings from 1998 to 2009: animals in the Philippines in peril?. Aquat. Mamm. 2010;36(3): 219. [Google Scholar]
- 24.Aragones LV, Laggui HLM. Marine Mammal Strandings in the Philippines from 2017 to 2018: Initial Biennial Analysis (Technical). Quezon City: A PMMSN Publication. 2019. Available from www.pmmsn.org. [Google Scholar]
- 25.Pugliares KR, Bogomolni A, Touhey KM, Herzig SM, Harry CT, Moore MJ. Marine mammal necropsy: An Introductory guide for stranding responders and field biologists. Woods Hole, MS. Woods Hole Oceanographic Institution Technical Document. WHOI-2007-06. [Google Scholar]
- 26.Ross MH. Pawlina W. Histology: A Text and Atlas. Wolters Kluwer/Lippincott Williams & Wilkins. Philadelphia. 2011. pp. 996. [Google Scholar]
- 27.Brown A, Smith H. Gram Staining. In Benson’s Microbiological Applications: Laboratory Manual in General Microbiology. New York, NY. McGraw-Hill Education. 2015. pp. 105–110. [Google Scholar]
- 28.Ahmed OB, Dablool AS. Quality improvement of the DNA extracted by boiling method in gram negative bacteria. Int J Bioassays. 2017;6(4): 5347–9. [Google Scholar]
- 29.Lane DJ. 16S/23S rRNA sequencing. In Nucleic Acid Techniques in Bacterial Systematics. Edited by Stackebrandt E. and Goodfellow M. Chichester: Wiley. 1991 [Google Scholar]
- 30.Suzuki MT, Giovannoni SJ. Bias caused by template annealing in the amplification of mixtures of 16s rRNA genes by PCR. Appl. Environ. Microbiol. 1996;62: 625–630. doi: 10.1128/aem.62.2.625-630.1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bonfield JK, Smith KF, Staden R. A new DNA sequence assembly program. Nucleic Acids Res. 1995;23: 4992–4999. doi: 10.1093/nar/23.24.4992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J. Mol. Biol. 1990;215(3): 403–410. doi: 10.1016/S0022-2836(05)80360-2 [DOI] [PubMed] [Google Scholar]
- 33.Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl. Acids. Symp. Ser. 1999;41: 95–98. [Google Scholar]
- 34.Hudzicki J. Kirby-Bauer disk diffusion susceptibility test protocol. 2009. [Cited 2017 Aug] Available from: http://www.asmscience.org/content/education/protocol/protocol.3189 [Google Scholar]
- 35.Rose JM, Gast RJ, Bogomolni A, Ellis JC, Lentell BJ, Touhey K, et al. Occurrence and patterns of antibiotic resistance in vertebrates off the Northeastern United State coast. FEMS Microbiol. Ecol. 2009;67: 421–431. doi: 10.1111/j.1574-6941.2009.00648.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Managaki S, Murata A, Takada H, Tuyen BC, Chiem NH. Distribution of macrolides, sulfonamides, and trimethoprim in tropical waters: Ubiquitous occurrence of veterinary antibiotics in the Mekong Delta. Environ. Sci, Technol. 2007; 41(23): 8004–8010. [DOI] [PubMed] [Google Scholar]
- 37.Baticados MCL, Paclibare JO. The use of chemotherapeutic agents in aquaculture in the Philippines. In Shariff M., Subasinghe R. P., & Arthur J. R. (Eds.), Diseases in Asian Aquaculture, In: proceedings of the first Symposium on Diseases in Asian Aquaculture: 1990 Nov 26–29, Bali, Indonesia. Makati, Metro Manila, Philippines: Fish Health Section, Asian Fisheries Society. 1992. 531–549. [Google Scholar]
- 38.World Health Organization. Global priority list of antibiotic-resistant bacteria to guide research, discovery, and development of new antibiotics. 2017. [Cited 2017 Sep 1] Available from: http://www.who.int/medicines/ publications/WHO-PPL-Short_Summary_25Feb-ET_NM_WHO.pdf?ua = 1. [Google Scholar]
- 39.NCCLS. Performance standards for antimicrobial disk diffusion and dilution susceptibility tests for bacteria isolated from animals: Approved standard-2nd edition. NCCLS document M31-A2. Pennsylvania: National Committee for Clinical Laboratory Standard; 2002. [Google Scholar]
- 40.CLSI. Performance standards for antimicrobial susceptibility testing: Twenty-fourth International Supplement. CLSI document M100-S24. Pennsylvania: Clinical and Laboratory Standards Institute; 2014. [Google Scholar]
- 41.The European Committee on Antimicrobial Susceptibility Testing. Breakpoint tables for interpretation of MICs and zone diameters. 2018. Version 8.0 [Cited 2020 November 18]. Available from http://www.eucast.org. [Google Scholar]
- 42.Krumperman PH. Multiple antibiotic resistance indexing of Escherichia coli to identify high-risk sources of fecal contamination of foods. Appl. Environ. Microbiol. 1983;46(1): 165–170. doi: 10.1128/aem.46.1.165-170.1983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sandhu R, Dahiya S, Sayal P. Evaluation of multiple antibiotic resistance (MAR) index and doxycycline susceptibility of Acinetobacter species among inpatients. Indian J. Microbiol. Res. 2016;3(3): 299. [Google Scholar]
- 44.Magiorakos AP, Srinivasan A, Carey RB, Carmeli Y, Falagas ME, Giske CG, et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 2011;18(3): 268–281. doi: 10.1111/j.1469-0691.2011.03570.x [DOI] [PubMed] [Google Scholar]
- 45.Sierra E, Fernández A, Felipe-Jiménez I, Zucca D, Díaz-Delgado J, Puig-Lozano R, et al. Histopathological differential diagnosis of meningoencephalitis in cetaceans: Morbillivirus, Herpesvirus, Toxoplasma gondii, Brucella sp., and Nasitrema sp. Front Vet Sci. 2020; 7(650): 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Alvarado-Rybak Mario; Toro Frederick; Abarca Paulette; Paredes Enrique; Español-Jiménez Sonia; Seguel Mauricio. Pathological findings in cetaceans sporadically stranded along the Chilean Coast. Front. Mar. Sci. 2020; 7(684): 1–10.32802822 [Google Scholar]
- 47.Seguel M, George RC, Maboni G, Sanchez S, Page-Karjian A, Wirth E, et al. Pathologic findings and causes of death in bottlenose dolphins Tursiops truncatus stranded along the Georgia coast, USA (2007–2013). Dis Aquat Organ. 2020; 141:25–38. [DOI] [PubMed] [Google Scholar]
- 48.Jaber JR, Pérez J, Arbelo M, Andrada M, Hidalgo M, Gómez-Villamandos JC, et al. Hepatic lesions in cetaceans in the Canary Islands. Vet Pathol, 2004;41: 147–153. doi: 10.1354/vp.41-2-147 [DOI] [PubMed] [Google Scholar]
- 49.Cozzi B, Mazzariol S, Podesta M, Zotti A. Diving Adaptations of the Cetacean Skeleton. The Open Zoology Journal. 2009;2: 24–32. [Google Scholar]
- 50.Gonzales-Viera O, Ruoppolo V, Marigo J, Carvalho VL, Groch KR, Bertozzi CP, et al. Renal lesions in cetacenas from Brazil. J. Comp. Path. 2015; 1–10. doi: 10.1016/j.jcpa.2015.02.004 [DOI] [PubMed] [Google Scholar]
- 51.Obusan MCM, Villanueva RMD, Siringan MAT, Rivera WL, Aragones LV. Leptospira spp. and Toxoplasma gondii in stranded representatives of wild cetaceans in the Philippines. BMC Vet Res. 2019;15(1): 372. doi: 10.1186/s12917-019-2112-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cornaglia E, Rebora L, Gili C, Di Guardo G. Histopathological and immunohistochemical studies on cetaceans found stranded on the coast of Italy between 1990 and 1997. J. Vet Med A Physiol. Pathol. Clin. Med. 2000;47(3): 129–142. doi: 10.1046/j.1439-0442.2000.00268.x [DOI] [PubMed] [Google Scholar]
- 53.Câmara N, Sierra E, Fernández-Maldonado C, Espinosa de los Monteros A, Arbelo M, Fernández A, et al. Stress cardiomyopathy in stranded cetaceans: a histological, histochemical and immunohistochemical study. Vet. Rec. 2019; 185: 694. doi: 10.1136/vr.105562 [DOI] [PubMed] [Google Scholar]
- 54.IJsseldijk LL, van Neer A, Deaville R, Begeman L, van de Bildt M, van de Brand JMA, et al. Beached bachelors: An extensive study on the largest recorded sperm whale Physeter macrocephalus mortality event in the north Sea. PLoS One, 2018;13(8): e0201221. doi: 10.1371/journal.pone.0201221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Peng C, Zhao X, Liu G. Noise in the sea and its impacts on marine organisms. Int. J Env. Res. Pub. He. 2015;12(10): 12304–12323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Morell M, Brownlow A, McGovern B, Raverty S.A, Shadwick RE, Andre M. Implementation of a method to visualize noise-induced hearing loss in mass stranded cetaceans. Sci Rep. 2017; 7(41848): 1–8. doi: 10.1038/srep41848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pacini AF, Nachtigall PE, Smith AB, Suarez LJ, Magno C, Laule GE, et al. Evidence of hearing loss due to dynamite fishing in two species of odontocetes. In Proceedings of Meetings on Acoustics 4ENAL. Acoustical Society of America. 2016;27(1): 010043 [Google Scholar]
- 58.Rako-Gospie N, Picciulin M. Underwater Noise: Sources and Effects on Marine Life. In C. Sheppard, World Seas: An Environmental Evaluation. London, UK: Elsevier. 2019, pp. 367–389. [Google Scholar]
- 59.Samuel Y, Morreale SJ, Clark CW, Greene CH, Richmond ME. Underwater, low-frequency noise in a coastal sea turtle habitat. J Acoust Soc Am. 2005. 1465–1472. doi: 10.1121/1.1847993 [DOI] [PubMed] [Google Scholar]
- 60.Weilgart LS. The impacts of anthropogenic ocean noise on cetaceans and implications for management. Canadian Journal of Zoology. 2007. 1091–1116. [Google Scholar]
- 61.Alpers CE, Fogo AB. Kidney and its collecting system. In Kumar V. A., Robbins Basic Pathology. Philadelphia: Elsevier Saunders. 2013. pp. 517–549 [Google Scholar]
- 62.Jepson PD, Baker JR, Kuiken T, Simpson VR, Kennedy S, Bennett PM. Pulmonary pathology of harbour porpoises stranded in England and Wales between 1990 and 1996. Vet Rec. 2000;146: 721–728. doi: 10.1136/vr.146.25.721 [DOI] [PubMed] [Google Scholar]
- 63.Marigo J, Ruoppolo V, Rosas FCW, Valente ALS, Oliveira MR, Dias RA, et al. Helminths of Sotalia guianensis (Cetacea: Delphinidae) from the south and southeastern coasts of Brazil. J. Wildl. Dis. 2010;46(2): 599–602. doi: 10.7589/0090-3558-46.2.599 [DOI] [PubMed] [Google Scholar]
- 64.Obusan MCM, Aragones LV, Salibay CC, Siringan MAT, Rivera WL. Occurrence of Human Pathogenic Bacteria and Toxoplasma gondii in cetaceans stranded in the Philippines. Aquatic Mammals. 2015;41(2): 149–166. [Google Scholar]
- 65.Gibson AK, Raverty S, Lambourn DM, Huggins J, Magargal SL, Grigg ME. Polyparasitism is associated with increases disease severity in Toxoplasma gondii-infected marine mammal sentinel species. PLoS Negl Trop Dis. 2011;5(5): e1142. doi: 10.1371/journal.pntd.0001142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Girard YA, Johnson CK, Fritz HM, Shapiro K, Packham AE, Melli AC, et al. Detection and characterization of diverse coccidian protozoa shed by California sea lions. Int J Parasitol Parasites Wildl. 2016;5(1): 5–16. doi: 10.1016/j.ijppaw.2015.11.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Miller M.A., Miller W.A., Conrad P.A., James E.R., Melli A.C., Leutenegger C.M., et al. Type X Toxoplasma gondii in wild mussel and terrestrial carnivores from coastal California: New linkages between terrestrial animals, runoff and toxoplasmosis of sea otters. Int. J. Parasitol. 2008;38: 1319–1328. doi: 10.1016/j.ijpara.2008.02.005 [DOI] [PubMed] [Google Scholar]
- 68.Rosonke S, Brown SR, Tornquist SJ, Snyder SP, Garner MM, Blythe LL. Encephalomyelitis associated with a Sarcocystis neurona-like organism in a sea otter. J. Am. Vet. Med. Assoc. 1999;215: 1839–1842. [PubMed] [Google Scholar]
- 69.Miller MA, Crosbie PR, Sverlow K, Hanni K, Barr BC, Kock N, et al. Isolation and characterization of Sarcocystis from brain tissue of a free-living southern sea otter (Enhydra lutris nereis) with fatal meningoencephalitis. Parasitol. Res. 2001;87: 252–257. doi: 10.1007/s004360000340 [DOI] [PubMed] [Google Scholar]
- 70.Kirillova V, Prakas P, Calero-Bernal R, Gavarane I, Fernandez-Garcia JL, Martinez-Gonzalez M, et al. Identification and genetic characterization of Sarcocystis arctica and Sarcocytis lutrae in red foxes (Vulpes vulpes) from Baltic States and Spain. Parasites Vectors. 2018; 11(173): 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Costa-Silva S, Sacristan C, Gonzales-Viera O, Diaz-Delgado J, Sanchez-Sarmiento AM, Marigo J, et al. Toxoplasma gondii in cetaceans of Brazill: a histopathological and immunohistochemical survey. Rev. Bras. Parasitol., Vet. 2019;28(3): 395–402. [DOI] [PubMed] [Google Scholar]
- 72.Dubey JP, Hamir AN. Immunohistochemical confirmation of Sarcocystis neurona infections in racoons, mink, cat, skunk, and pony. J. Parasitol. 2000;86: 1150–1152. doi: 10.1645/0022-3395(2000)086[1150:ICOSNI]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- 73.Dubey JP, Chapman JL, Rosenthal BM, Mense M, Schueler RL. Clinical Sarcocystis neurona, Sarcocystis canis, Toxoplasma gondii, and Neospora caninum infections in dogs. Vet Parasitol. 2006;137: 36–49. doi: 10.1016/j.vetpar.2005.12.017 [DOI] [PubMed] [Google Scholar]
- 74.Dubey JP, Zarnke R, Thomas NJ, Wong SK, Van Bonn W, Briggs M, et al. Toxoplasma gondii, Neospora caninum, Sarcocystis neurona, and Sarcocystis canis-like infections in marine mammals. Vet Parasitol. 2003;116: 275–296. doi: 10.1016/s0304-4017(03)00263-2 [DOI] [PubMed] [Google Scholar]
- 75.Barbosa L, Johnson CK, Lamborun DM, Gibson AK, Haman KH, Huggins JL, et al. A novel Sarcocytis neurona genotype XIII is associated with severe encephalitis in an unexpectedly broad range of mairne mammals from the northeastern Pacific Ocean. Int. J. Parasitol. 2015;45: 595–603. doi: 10.1016/j.ijpara.2015.02.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Aznar FJ, Agusti C, Littlewood DTJ, Raga JA, Olson PD. Insight into the role of cetaceans in the life cycle of the tetraphyllideans (Platyhelminthes:Cestoda). Int. J. Parasitol. 2007;37(2): 243–255. doi: 10.1016/j.ijpara.2006.10.010 [DOI] [PubMed] [Google Scholar]
- 77.Siquier GF, Le Bas. Morphometrical categorization of Phylobothrium delphini (Cestoidea, Tetraphyllidea) cysts from Fraser’s dolphin, Lagenodelphis hosei (Cetacea, Delphinidae). Lat. Am. J. Aquat. Mamm. 2003;2(2): 95–100. [Google Scholar]
- 78.Zheng B, Dai Y, Liu Y, Shi W, Dai E, Han Y, et al. Molecular epidemiology and risk factors of carbapenem-resistant Klebsiella pneumoniae infections in Eastern China. Front Microbiol. 2017;8: 1061. doi: 10.3389/fmicb.2017.01061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Andremont A, Gerbaud G, Courvalin P. Plasmid-mediated high-level resistance to erythromycin in Escherichia coli. Antimicrob. Agents Chemother. 1986;29(3): 515–518. doi: 10.1128/AAC.29.3.515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Van Hoek AHAM, Mevius D, Guerra B, Mullany P, Roberts AP, Aarts HJM. Acquired antibiotic resistance genes: an overview. Front Microbiol. 2011;2(203): 1–27. doi: 10.3389/fmicb.2011.00203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Whitaker DM, Reichley S, Griffin M.J, Prager K, Richey CA, Kenelty KV, et al. Hypermucoviscous Klebsiella pneumoniae isolates from stranded and wild caught marine mammals of the US Pacific coast: Determination of prevalence, phenotype and genotype. J. Wildl Dis. 2018;54(4): 659–670. doi: 10.7589/2017-07-178 [DOI] [PubMed] [Google Scholar]
- 82.Seguel M, Gottdenker NL, Colegrove K, Johnson S, Struve C, Howerth EW. Hypervirulent Klebsiella pneumoniae in California sea lions (Zalophus californianus): pathologic findings in natural infections. Vet Pathol. 2017;54(5): 846–850. doi: 10.1177/0300985817705172 [DOI] [PubMed] [Google Scholar]
- 83.Roberts MC. One Health Approach for Identification of Sources/Reservoir of Multidrug Resistant Bacteria in Wild Animals and their Environment. J Integr OMICS, 2019;9(2). [Google Scholar]
- 84.International Union for Conservation of Nature. The IUCN red list of threatened species. 2017 [Cited 2018 May 08]. Available from http://www.iucnredlist.org.
- 85.Kooyman GL. Respiratory adaptations in marine mammals. Integr. Comp. Biol. 1973;13(2): 457–468. [Google Scholar]
- 86.Lourenço NGGS, Takahashi CK, Lopes TF, Lopes CAM. Environmental parameters and antimicrobial susceptibility of Enterobacteriaceae isolated from estuarine waters of São Vicente, São Paulo state, Brazil. J Venom. Anim Toxins Incl. Trop. Dis. 2007;13(2): 472–478. [Google Scholar]
- 87.Hatosy SM, Martiny AC. The ocean as a global reservoir of antibiotic resistance genes. Appl. Environ. Microbiol. 2015;81(21): 7593–7599. doi: 10.1128/AEM.00736-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Barkovskii AL, Babb CM, Hurley D, Shin E. Origins and environmental mobility of antibiotic resistance genes, virulence factors and bacteria in a tidal creek’s watershed. J. Appl Microbiol. 2015;118(3): 764–776. doi: 10.1111/jam.12735 [DOI] [PubMed] [Google Scholar]
- 89.Maravić A, Skočibušić M, Cvjetan S, Šamanić I, Fredotović Ž, Puizina J. Prevalence and diversity of extended-spectrum-β-lactamase-producing Enterobacteriaceae from marine beach waters. Mar. Pollut. Bull. 2015;90(1–2): 60–67. doi: 10.1016/j.marpolbul.2014.11.021 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All relevant data are within the manuscript.