Abstract
Clonal spread and horizontal transfer in the spread of vancomycin resistance genes were investigated. Multiplex PCR, pulsed-field gel electrophoresis (PFGE), hybridization of enterococcal plasmids with the vanA and vanB probes, and sequencing of a fragment of vanB were used in the analysis. Before May 1996, 12 vancomycin-resistant Enterococcus faecium (VRE) isolates were found in Finland. Between May 1996 and October 1997, 156 VRE isolates were found in the Helsinki area. Between December 1997 and April 1998, fecal samples from 359 patients were cultured for VRE. One new case of colonization with VRE was found. During the outbreak period, 88% (137 of 155) of the VRE isolates belonged to two strains (VRE types I and II), as determined by PFGE. Each VRE type I isolate possessed vanB, and five isolates also had vanA. Of the 34 VRE type II isolates, 27 possessed vanA and 7 possessed vanB. Fifteen of 21 (71%) ampicillin-resistant, vancomycin-sensitive E. faecium (VSE) isolates found during and after the outbreak period in one ward were also of type II. Two VSE type II isolates were found in the hospital before the outbreak in 1995. By PFGE, the three groups (vanA, vanB, or no van gene) of type II shared the same band differences with the main type of VRE type II with vanA. None of the differences was specific to or determinative for any of the groups. Our material suggests that vanA and vanB incorporate into an endemic ampicillin-resistant VSE strain.
Until 1990 few Enterococcus faecium isolates were resistant to ampicillin, and vancomycin resistance was unknown in Finland. The rate of ampicillin resistance in E. faecium increased from 17% in 1992 to 74% in 1995 in Meilahti Hospital, which is part of the Helsinki University Central Hospital (HUCH) in Helsinki, Finland. The first vancomycin-resistant E. faecium (VRE) strain was detected in 1992. Until May 1996 only 12 VRE isolates had been detected in Finland.
Between May 1996 and October 1997 VRE was isolated from 156 patients in six hospitals in the Helsinki area. The majority (88%) of the isolates belonged to two outbreak strains. By April 1998 the outbreak had ceased, as only one new VRE isolate had been found. The outbreak is one of the largest successfully controlled outbreaks of VRE.
The appearance of VRE prompted us to perform a prospective citywide survey of its emergence in the Helsinki area. The large, but still controlled outbreak in hospitals with few previous VRE isolates enabled us to study the epidemiology of VRE in detail. Our aim was to assess the contribution of clonal spread as well as horizontal transfer in the dissemination of van resistance genes. In previous studies, numerous pulsed-field gel electrophoresis (PFGE) types carrying the van gene were isolated from each patient (21), transfer of resistance plasmids to different strains had occurred (26), and the VRE strain had altered its van genotype (27). As opposed to earlier studies, we compared the VRE outbreak strains to the VSE isolates collected before, during, and after the VRE outbreak period in one of the outbreak wards.
MATERIALS AND METHODS
Setting.
VRE isolates were collected from the Meilahti Hospital and two other hospitals belonging to HUCH, two primary care hospitals, and one of the two city hospitals. Most of the isolates were found in Meilahti Hospital. Vancomycin-sensitive E. faecium (VSE) isolates were collected from the hematological ward. The hematological ward of HUCH is situated in Meilahti Hospital and has 24 adult in-patient beds.
VRE isolates.
A total of 157 VRE isolates recovered from separate patients in the Helsinki area between May 1996 and April 1998 were studied. The first isolate from each patient was included in the study. Motility-positive species of enterococci were excluded from the epidemiologic analysis. Twelve VRE isolates collected between 1992 and 1995 were also analyzed. Control strains were Enterococcus casseliflavus ATCC 25788 and Enterococcus faecalis ATCC 51299.
VSE isolates.
Altogether, 56 VSE isolates from separate patients were studied. Fifteen ampicillin-resistant VSE isolates were collected from clinical specimens during patients' stays in Meilahti Hospital from 1988 to 1995. Forty-one VSE isolates were collected from patients during or soon after their stay in the hematological ward (most of the patients had also stayed in other wards). Of the 41 VSE isolates, 33 were resistant to ampicillin. Of the 33 ampicillin-resistant VSE isolates, 12 were collected before the VRE outbreak between September 1994 and November 1995, 11 were isolated during the outbreak between September 1996 and September 1997, and 10 were collected after the outbreak in February and March 1998. These 10 isolates were collected from patients who had their first admission to Meilahti Hospital after new VRE cases could not be found in Meilahti Hospital. None of these patients had been in any of the hospitals where VRE isolates were found during the study period. Of the 41 VSE isolates, 8 were sensitive to ampicillin. These isolates were found in the hematological ward between March 1997 and March 1998.
Screening for VRE.
In November 1996 we initiated a survey for VRE using selective media. Fecal cultures were inoculated into Enterococcosel broth (BBL, Cockeysville, Md.) supplemented with 8 μg of vancomycin per ml, and the broth was incubated at 37°C for 24 h. Fecal cultures and broths with visible growth were plated onto selective agar containing 75 μg of neomycin per ml, 7.5 μg of vancomycin per ml, and 50 IU of mycostatin per ml. Conventional biochemical reactions as outlined by Facklam and Collins (7) were used to identify the organisms. Testing for susceptibility to vancomycin, teicoplanin, and ampicillin was performed by disc diffusion methods as described by the National Committee for Clinical Laboratory Standards (13). Enterococcal isolates with vancomycin inhibition zone diameters of ≤16 mm were presumptively considered to be VRE (23). The susceptibilities of all VRE isolates to vancomycin and teicoplanin were tested by the Etest (AB Biodisk, Solna, Sweden).
Multiplex PCR.
Microbiologically identified VRE isolates were further characterized by multiplex PCR (6, 15) with six primer pairs to identify vanA (5), vanB (20), vanC1 (10), vanC2-vanC3 (14), ddl E. faecalis, and ddl E. faecium encoding d-alanyl-d-alanyl ligase (6). Approximately 200 to 1,000 ng (1 μl) of DNA was added to a PCR mixture (99 μl) containing 10 mM Tris-HCl, 50 mM KCl, 2.5 mM MgCl2, 0.01% gelatin, 0.1% Triton X-100, 0.25 mM (each) the four deoxyribonucleotide triphosphates, 100 pmol of each primer, and 2 U of Taq DNA polymerase. Amplification of DNA was performed in a PTC-100 programmable thermal controller (Mj Research Inc.) by using predenaturation at 94°C for 2 min, followed by 30 cycles of 1 min at 94°C, 1 min at 54°C, and 1 min at 72°C and 72°C for 10 min for the last cycle. Amplicons were analyzed by electrophoresis on a 1.7% agarose gel (SeaKem ME Agarose; FMC BioProducts, Rockland, Maine), and the gels were stained with ethidium bromide (0.5 μg/ml).
PFGE.
For PFGE, organisms were lysed as described previously (12), and the DNA was digested with the restriction endonuclease SmaI (Boehringer Mannheim, Mannheim, Germany). Electrophoresis was performed with a CHEF-DR III System (Bio-Rad Laboratories, Hercules, Calif.) by using 1.0% SeaKem agarose in 0.5× Tris-borate-EDTA. Running conditions consisted of two ramps used in sequence. Ramp A was 1 to 11s with a run time of 15 h. Ramp B was 11 to 30s with a run time of 15 h. E. faecium type II isolates were also run with another program. Ramp A was 1 to 9 s with a run time of 24 h. Ramp B was 9 to 30 s with a run time of 9 h. The voltage gradient was 6 V/cm. Previously published guidelines for interpreting chromosomal DNA restriction patterns produced by PFGE were used for the interpretation of PFGE findings (24).
Extraction and digestion of genomic and plasmid DNAs.
The plasmids were extracted by a modified alkaline lysis technique (25) and were digested with ClaI restriction enzyme (Boehringer Mannheim, Mannheim, Germany). The genomic DNA was extracted from enterococci by the GES (guanidium thiocyanate) (19) method and was digested with ClaI. The extracted DNA was separated by agarose gel electrophoresis (0.8% SeaKem agarose in 0.5× Tris-borate-EDTA) at 2.1 V/cm for 20 h (genomic DNA) or 6.4 V/cm for 2.5 h (plasmid DNA). The molecular sizes of the digested enterococcal plasmids were determined by comparing them with the sizes of fragments of phage λ DNA digested with EcoRI and HindIII (Boehringer Mannheim, Mannheim, Germany).
Hybridization with vanA and vanB probes.
Southern blots of ClaI-digested plasmid DNA, ClaI-digested genomic DNA, and undigested DNA separated by PFGE were prepared with a VacuGene blotter (Pharmacia LKB, Milton Keynes, United Kingdom). The vanA and vanB probes consisted of 732- and 635-bp intragenic fragments of the vanA and vanB genes, respectively (6); both of them were labeled with digoxigenin (Boehringer, Mannheim, Germany). All DNA hybridizations were performed at 68°C as recommended by the manufacturer. Blots were then given two low-stringency washes in 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)–0.1% sodium dodecyl sulfate (SDS) at room temperature followed by two high-stringency washes in 0.1× SSC–0.1% SDS at 68°C (15 min each) before hybrids were detected. Hybridization studies were used for the analysis of different subtypes of VRE type I and II isolates in order to determine the locations of van resistance genes.
PCR product sequencing.
We sequenced a 550-bp internal fragment from vanB (positions 223 to 772) from nine vanB isolates. Approximately 2 to 3 μg of the PCR product was purified with a QIAquick PCR purification kit (QiaGen GmbH, Hilden, Germany) and was eluted with 50 μl of the elution buffer provided with the kit. A total of 0.2 to 0.3 μg of the purified PCR product and 0.4 μg of the sequencing primer (6) were mixed. The DNA sequence was determined in both the 5′ to 3′ and the 3′ to 5′ directions with an ABI PRISM kit or a Big Dye Terminator kit (Perkin-Elmer Applied Biosystems Division, Foster City, Calif.) according to the manufacturer's instructions, and reactions were run on an ABI 373 A or 377 sequencer (Perkin-Elmer Applied Biosystems Division).
RESULTS
Epidemiologic investigation.
VRE was isolated from a total of 157 hospitalized patients during the period from May 1996 through April 1998. All isolates were identified as E. faecium. Two patients in the infectious disease ward carried VRE in June and July 1996. The outbreak began in October 1996 in Meilahti Hospital, where several patients in two wards were found to have VRE colonization. From January to March 1997, the VRE isolates were encountered in three other hospitals in the Helsinki area. Ninety-one percent of the new VRE isolates were encountered in surveillance fecal samples, while 9% were found in clinical samples. All E. faecium isolates with disc diffusion zone diameters of <17 mm for vancomycin were confirmed to be VRE by the Etest and multiplex PCR.
PFGE of VRE isolates.
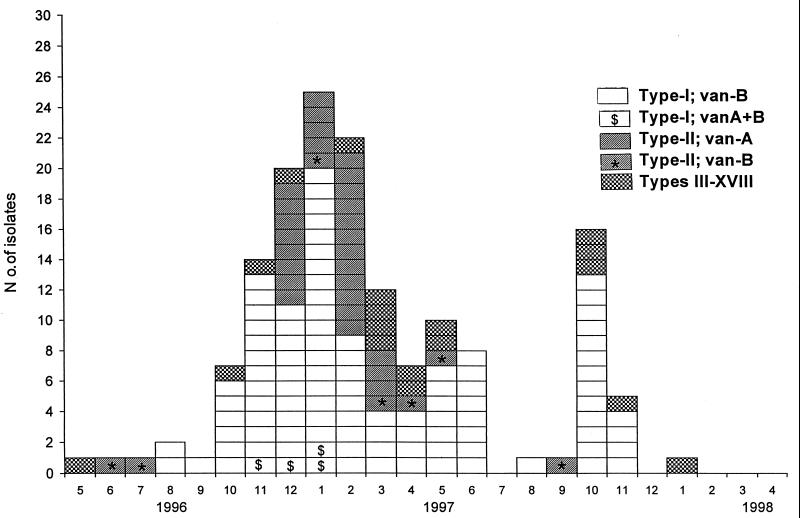
Of the 157 VRE isolates collected from separate patients, 155 were stored for studies of molecular epidemiology. In Fig. 1, their karyotypes and resistance genotypes are presented by the date of the first positive culture for the isolates. Sixty-six percent of the isolates were of karyotype I, 22% were of karotype II, and 18 isolates were of 16 other karyotypes (types III to XVIII). Six patients colonized with VRE were subsequently found to be colonized with both type I and type II isolates. None of the 12 VRE isolates collected from 1992 to 1995 belonged to type I or II.
FIG. 1.
PFGE patterns and resistance genotypes of 155 VRE isolates in the Helsinki area.
van genotypes.
All 103 VRE type I isolates possessed the vanB gene, but in addition, 5 isolates had the vanA gene (vanA+B isolates) (Fig. 1). One of the vanA+B isolates was not collected from the patient's first VRE-positive fecal sample.
Of the 34 VRE type II isolates, 27 contained the vanA gene and 7 contained the vanB gene. All the isolates with only vanB had the VanB phenotype (i.e., were teicoplanin susceptible), and correspondingly, all the vanA isolates had the VanA phenotype.
Furthermore, 18 isolates that belonged to neither karyotype I nor karyotype II were found. They displayed 16 different karyotypes; 9 had vanA and 9 had vanB.
Location of vanA resistance gene.
The undigested DNAs (containing both chromosomal and plasmid DNAs) of seven subtypes of VRE type II strains possessing vanA were separated by PFGE and were transferred to nylon membranes for hybridization with the vanA probe. Five isolates showed a band of 120 kb which hybridized with the vanA probe. The undigested DNAs of two VRE type II isolates repeatedly did not show any bands by PFGE; the chromosomal DNA presumably stayed in the well. ClaI digests of plasmids derived from these two isolates did not hybridize with the vanA probe, but ClaI digests of genomic DNA hybridized with the probe, indicating the chromosomal location of vanA in two isolates.
All five VRE type I isolates with vanA+B hybridized with the 120-kb band. The occurrence of the 120-kb band was associated with an extra band when SmaI-digested DNA was separated by PFGE, suggesting that SmaI-digested plasmid fragment of 90 kb was present. Instead of this 90-kb fragment, a <40-kb band hybridized with the vanA probe. Furthermore, two of nine unique VRE types with vanA were found to have the 120-kb band which hybridized with the vanA probe.
Location of vanB resistance gene.
The undigested DNAs of 5 subtypes of VRE type II and 10 subtypes of VRE type I (all with vanB) did not show any bands by PFGE. Similarly, ClaI digests of plasmids derived from these isolates did not hybridize with the vanB probe. The absence of plasmid fragments that hybridized with the vanB probe suggests the chromosomal locations of the vanB genes.
Occurrence of type I and II isolates in the hematological ward.
As isolates with closely related PFGE types (less than four band differences) contained different van resistance determinants, the hematological ward was investigated for the presence of both VRE and VSE isolates of these PFGE types.
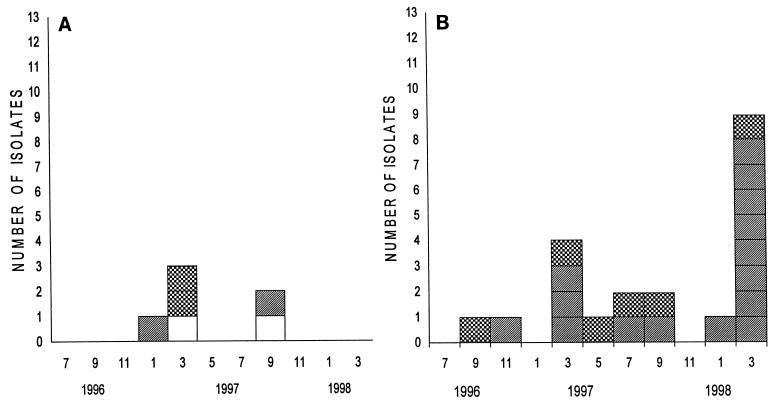
Two VRE type I isolates, two VRE type II isolates (one with vanA and one with vanB), and two sporadic VRE isolates were found in the ward during 1997 (Fig. 2A).
FIG. 2.
Karyotypes of VRE (A) and ampicillin-resistant VSE (B) isolates from patients in the hematological ward. □, type I; ▨, type II (vanA and vanB in panel A; no van gene in panel B); , other types.
None of the 12 ampicillin-resistant VSE isolates found in the hematological ward during 1994 and 1995 resembled the outbreak strains encountered from 1996 to 1998 (data not shown). Between September 1996 and October 1997 6 of 11 (55%) ampicillin-resistant VSE isolates were of type II (Fig. 2B). By November 1997, the outbreak of VRE had ceased in Meilahti Hospital. In February and March 1998, 9 of 10 (90%) ampicillin-resistant VSE isolates were still of type II. None of the ampicillin-resistant VSE isolates showed type I PFGE patterns. Furthermore, eight ampicillin-sensitive VSE isolates differed from the outbreak strains and from one another.
Occurrence of type I and II isolates in Meilahti Hospital before outbreak of VRE.
Two of 15 ampicillin-resistant VSE isolates showed banding patterns similar to those of the type II isolates. They were isolated in April and in November 1995, respectively.
Comparison of PFGE banding patterns of type I and type II isolates.
A total of 96% (99 of 103) and 97% (33 of 34) of VRE type I and II isolates, respectively, showed banding patterns that differed by less than four bands from the pattern for the most frequently isolated subtype (main type containing vanA) when bands within the 80- to 300-kb range (10 bands) were analyzed (Table 1). We could not separate more bands in the 30-h PFGE run. When the bands in the 45- to 300-kb range (16 bands) were included in analysis, 65% (22 of 34) of VRE type II isolates were closely related (33-h PFGE program).
TABLE 1.
PFGE band differences among the outbreak strains
| Karyotype | No. of isolates with the following
band differencesa:
|
|||
|---|---|---|---|---|
| 0–1 bands | 2–3 bands | 4–5 bands | 6–7 bands | |
| VRE type I, vanB (+vanA)b | 86 | 13 | 4 | |
| VRE type II, vanAa | 21 | 6 | ||
| VRE type II, vanBb | 3 | 3 | 1 | |
| VSE type II, van negativeb | 11 | 6 | ||
| VRE type II, vanAc | 13 | 5 | 8 | 1 |
| VRE type II, vanBc | 1 | 3 | 3 | |
| VSE type II, van negativecd | 6 | 7 | 3 | |
Band differences were compared by comparison with the bands for the most commonly isolated subtype within VRE type I or II.
Ten bands (80 to 300 kb) were analyzed.
Sixteen bands (45 to 300 kb) were analyzed.
One isolate was lost; all VSE isolates were resistant to ampicillin.
The most frequently isolated subtype of VSE type II differed from the main type by only one 90-kb band. The subtypes of VSE type II showed almost as many band differences from the main type as the subtypes of VRE type II. Six similar band differences from the main type of VRE type II were encountered in both VRE and VSE type II isolates.
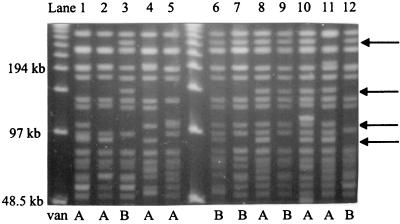
The PFGE banding patterns of VRE type II isolates containing either vanA or vanB are compared in Fig. 3. In addition to the 90-kb band often present in type II isolates with vanA, none of the band differences was specific to determinative for type II isolates with vanA or type II isolates with vanB.
FIG. 3.
PFGE profiles of type II isolates with vanA or vanB. The main type of VRE type II is in lane 1. The vanA and vanB groups share the same band differences compared to the bands for the main type (arrows). By comparing lane 1 to lane 4, it could be seen that patterns differ by one band if only the 10 largest bands are included in the analysis but by 4 bands if 16 bands were analyzed.
Sequencing.
We sequenced 550-bp readable DNA sequences internal to the vanB gene for three VRE type I isolates, three VRE type II isolates, one epidemiologically unrelated Finnish isolate, and two Swedish isolates. All six VRE type I and II isolates had identical vanB amplicon sequences. The epidemiologically unrelated Finnish isolate differed from these by 1 bp. Two Swedish isolates differed from the VRE type I and II isolates by 2 bp, leading to changes in two amino acids. The VRE type I and II isolates exhibited 23-bp changes (4.2%), leading to seven amino acid changes, compared to the vanB sequence of reference strain V583. However, the six VRE type I and II isolates had sequences identical to the vanB sequence of isolate 55 (GenBank accession no. U94528) described by Patel et al. (16).
DISCUSSION
Conditions used to study the epidemiology of VRE isolates in the Helsinki area.
Before May 1996, only 12 cases of colonization with VRE had been verified in Meilahti Hospital (and in Finland). None of the isolates belonged to karyotype I or II. A hematological ward was screened for VRE between June 1994 and October 1996, and only two VRE isolates were found (22) (data not shown). Close cooperation with two infection disease specialists enabled us to monitor the simultaneous spread of two outbreak strains. Between December 1997 and March 1998, fecal samples from 359 patients were screened for VRE. Only one new case of VRE infection was found, indicating that the outbreak was controlled.
Clonal spread or horizontal transfer in the spread of van resistance genes?
As 88% (137 of 155) of VRE isolates were one of two outbreak strains, it seems clear that clonal spread is the main mechanism in the spread of van resistance genes. Both the intrahospital and the interhospital transmission of VanB (2, 11, 18) or VanA (4, 8, 17) E. faecium has previously been documented. As identically sized plasmids containing the vanA gene had incorporated into five E. faecium type I isolates with vanB and as seven type II isolates contained vanB instead of vanA, it is evident that not all of our VRE type I or II isolates carry the same van resistance determinant. Whether the van resistance determinants in type I and II isolates have a common origin remains partly unsolved. At least two VRE type II isolates with vanA did not contain the 120-kb plasmid. On the contrary, two isolates with unique VRE types had the 120-kb plasmid with vanA. At the beginning of the outbreak, some patients were found to carry both of the outbreak strains simultaneously, giving rise to the possibility that the van resistance genes could have mixed between the two outbreak strains.
How could the vanA and vanB genes be found in separate isolates of same PFGE type?
Woodford et al. (27) have reported that VRE strains can alter their van genotypes during an outbreak. Our seven VRE type II isolates with vanB differed from the strain reported by Woodford et al. (27) in that our isolates were closely or possibly related (one to five band differences), vanB was chromosomally located (it was not located on the plasmid), and no intermediate (vanA+B) isolate was found. Only one of the seven patients was shown to be colonized with an isolate with vanA later during the hospitalization. Each type II isolate with vanB could be classified as a unique subtype. The VRE type II isolates with vanB were more scattered in terms of both the date of the first positive culture (Fig. 1) and the isolation location (four different wards in two hospitals) compared to the type II isolates with vanA. The infection control practitioners could not find any common link between patients who were colonized with VRE type II isolates with vanB. We could not distinguish type II isolates with vanB either from epidemiologically unrelated strain types or from type I isolates with vanB by sequencing the vanB gene. The DNA sequence data prove that all our isolates belong to the same vanB2 subtype of the vanB ligase gene (3). There is probably too little variability within the vanB2 subtype for use in outbreak investigations.
Do endemic ampicillin-resistant VSE isolates explain the epidemiology of VRE type II?
None of the 12 VRE isolates collected from separate patients from 1992 to 1995 belonged to type I or II. However, one of these patients was colonized with an ampicillin-resistant VSE isolate of type II in 1995. During the outbreak period VSE type II was isolated from a clinical specimen from one patient after an isolation of VRE type II with vanA. In the hematological ward, the number of VSE type II isolates increased at the same time that the VRE type II isolates spread in Meilahti Hospital. Five months after the last isolation of VRE type II in the hematological ward (Fig. 2), the proportion of VSE type II isolates among all ampicillin-resistant VSE isolates had increased to 90%. Thus, the hospital infection control measures had succeeded in eliminating VRE type II isolates, but VSE type II remained the predominant strain. In the hematological ward, all eight ampicillin-sensitive VSE isolates were unrelated to outbreak strains, proving that the occurrence of the type II strain was more common among both ampicillin- and vancomycin-resistant E. faecium isolates. Kapala et al. (9) have recently shown that an outbreak due to conjugative transfer of vanA genes into a hospital's endemic VSE strain had occurred. Perlada et al. (18) have also stated that some of the VSE isolates were similar to the outbreak VRE strain.
What does subtyping of type I and type II isolates teach us about the clonality of E. faecium?
The isolates from three groups (vanA, vanB, and no van gene) of E. faecium type II shared the same band differences when their bands were compared to the bands for the main type of VRE type II. None of the differences provided to be specific to or determinative for isolates belonging to any of the groups (Fig. 3; see arrows). The distribution of subtypes in the three groups is centered upon a main type to which the subtypes are very similar (VSE isolates lack a 90-kb band), supporting the fact that the isolates have a common origin. The existence of E. faecium type II isolates proves that the great number of subtypes among VRE isolates is caused not only by van genes but also by VSE isolates with similar variations. Bonten et al. (1) recently reported that VRE isolates are genetically closely related (three or fewer band differences by PFGE) or very different (eight or more band differences), providing empirical evidence that PFGE can be used to study the epidemiology of VRE endemicity. In our study, 12 (35%) VRE type II isolates were classified as possibly related (four to seven band differences) within the 45- to 300-kb range and contained either vanA or vanB (Fig. 3; compare lane 1 to lanes 3 and 4). For the clonal isolates in our collection, there could be up to six to seven band differences compared to the bands for the main type. The greatest number of band differences was seen for a group of type II isolates with vanB, probably due to the suggested chromosomal location of vanB and the lack of the 90-kb band associated with vanA. In addition, groups of vanB isolates and isolates with no van gene shared the same band differences when their bands were compared to those for VRE type II isolates.
Conclusions.
We are doubtful whether an E. faecium strain could be named as the carrier of the van resistance determinant. When one E. faecium strain predominates in a hospital environment, transferable van resistance genes can be incorporated into the clone that has been endemic there for many years. In our study one E. faecium clone had persisted in Meilahti Hospital from 1995 to 1998. Some of the isolates of this clone contained the vanA gene and some contained the vanB gene, while the others remained vancomycin sensitive. PFGE analysis and determination of the van genes are sufficient when strains from smaller outbreaks are analyzed. The transfer of van resistance genes in a conjugative transposon to endemic antibiotic-resistant E. faecium clones necessitates transposon structure mapping if full epidemiologic knowledge is needed.
ACKNOWLEDGMENTS
This work was supported in part by the HUCH Institute.
Raija Lahdenperä, Eila Ketolainen, Riitta-Liisa Skogberg, Elina Siren, and Nina Klinger are acknowledged for excellent technical assistance.
REFERENCES
- 1.Bonten M J, Hayden M K, Nathan C, Rice T W, Weinstein R A. Stability of vancomycin-resistant enterococcal genotypes isolated from long-term-colonized patients. J Infect Dis. 1998;177:378–382. doi: 10.1086/514196. [DOI] [PubMed] [Google Scholar]
- 2.Boyce J M, Opal S M, Chow J W, Zervos M J, Patter-Bynoe G, Sherman C B, Romulo R L C, Fortna S, Medeiros A A. Outbreak of multidrug-resistant Enterococcus faecium with transferable vanBclass vancomycin resistance. J Clin Microbiol. 1994;32:1148–1153. doi: 10.1128/jcm.32.5.1148-1153.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dahl K H, Simonsen G S, Olsvik O, Sundsfjord A. Heterogeneity in the vanBgene cluster of genomically diverse clinical strains of vancomycin-resistant enterococci. Antimicrob Agents Chemother. 1999;43:1105–1110. doi: 10.1128/aac.43.5.1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dunne W M, Jr, Wang W. Clonal dissemination and colony morphotype variation of vancomycin resistant Enterococcus faeciumisolates in metropolitan Detroit, Michigan. J Clin Microbiol. 1997;35:388–392. doi: 10.1128/jcm.35.2.388-392.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dutka-Malen S, Leclercq R, Coutant V, Duval J, Courvalin P. Phenotypic and genotypic heterogeneity of glycopeptide resistance determinants in gram-positive bacteria. Antimicrob Agents Chemother. 1990;34:1875–1879. doi: 10.1128/aac.34.10.1875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dutka-Malen S, Evers S, Courvalin P. Detection of glycopeptide resistance genotypes and identification to the species level of clinically relevant enterococci by PCR. J Clin Microbiol. 1995;33:24–27. doi: 10.1128/jcm.33.1.24-27.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Facklam R R, Collins M D. Identification of Enterococcusspecies isolated from human infection by a conventional test scheme. J Clin Microbiol. 1989;27:731–734. doi: 10.1128/jcm.27.4.731-734.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Handwerger S, Raucher B, Altarac D, Monka J, Marchione S, Singh K V, Murray B E, Wolff J, Walters B. Nosocomial outbreak due to Enterococcus faeciumhighly resistant to vancomycin, penicillin, and gentamycin. Clin Infect Dis. 1993;16:750–755. doi: 10.1093/clind/16.6.750. [DOI] [PubMed] [Google Scholar]
- 9.Kapala M M, Willey B M, Arce F, Large G, McGeer A, Low D E. Program and abstracts of the 38th Interscience Conference on Antimicrobial Agents and Chemotherapy. Washington, D.C: American Society for Microbiology; 1998. In vitro evidence of an outbreak due to conjugative transfer of vanA genes into a hospital's endemic vancomycin-susceptible enterococci (VSE), abstr. H-141; p. 355. [Google Scholar]
- 10.Leclercq R, Dutka-Malen S, Duval J, Courvalin P. Vancomycin resistance gene vanC is specific to Enterococcus gallinarum. Antimicrob Agents Chemother. 1992;36:2005–2008. doi: 10.1128/aac.36.9.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Moreno F, Grota P, Crisp C, Magnon K, Melcher G P, Jorgensen J H, Patterson J E. Clinical and molecular epidemiology of vancomycin resistant Enterococcus faeciumduring its emergence in a city in southern Texas. Clin Infect Dis. 1995;21:1234–1237. doi: 10.1093/clinids/21.5.1234. [DOI] [PubMed] [Google Scholar]
- 12.Murray B E, Singh K V, Heath J D, Sharma B R, Weinstock G M. Comparison of genomic DNAs of different enterococcal isolates using restriction endonucleases with infrequent recognition sites. J Clin Microbiol. 1990;28:2059–2063. doi: 10.1128/jcm.28.9.2059-2063.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.National Committee for Clinical Laboratory Standards. National Committee for Clinical Laboratory Standards. Performance standards for antimicrobial susceptibility testing. NCCLS document M100-S9. Wayne, Pa: National Committee for Clinical Laboratory Standards; 1999. [Google Scholar]
- 14.Navarro F, Courvalin P. Analysis of genes encoding d-alanine:d-alanine ligase-related enzymes in Enterococcus casseliflavus and Enterococcus flavescens. Antimicrob Agents Chemother. 1994;38:1788–1793. doi: 10.1128/aac.38.8.1788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Patel R, Uhl J R, Kohner P, Hopkins M K, Cockerill F R., III Multiplex PCR detection of vanA, vanB, vanC-1, and van C2/3genes in enterococci. J Clin Microbiol. 1997;35:703–707. doi: 10.1128/jcm.35.3.703-707.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Patel R, Uhl J R, Kohner P. DNA sequence variation within vanA, vanB, vanC-1, and vanC-2/3 genes of clinical Enterococcusisolates. Antimicrob Agents Chemother. 1998;42:202–205. doi: 10.1128/aac.42.1.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pegues D A, Pegues C F, Hibberd P L, Ford D S, Hooper D C. Emergence and dissemination of a highly vancomycin-resistant vanA strain of Enterococcus faeciumat a large teaching hospital. J Clin Microbiol. 1997;35:1565–1570. doi: 10.1128/jcm.35.6.1565-1570.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Perlada D E, Smulian G, Cushion M T. Epidemiology and antibiotic susceptibility of enterococci in Cincinnati, Ohio: a prospective citywide survey. J Clin Microbiol. 1997;35:2342–2347. doi: 10.1128/jcm.35.9.2342-2347.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pitcher D G, Saunders N A, Owen R J. Rapid extraction of bacterial genomic DNA with guanidium thiocyanate. Lett Appl Microbiol. 1989;8:151–156. [Google Scholar]
- 20.Quintiliani R, Jr, Evers S, Courvalin P. The vanBgene confers various levels of self-transferable resistance to vancomycin in enterococci. J Infect Dis. 1993;167:1220–1223. doi: 10.1093/infdis/167.5.1220. [DOI] [PubMed] [Google Scholar]
- 21.Schoonmaker D J, Bopp L H, Baltch A L, Smith R P, Rafferty M E, George M. Genetic analysis of multiple vancomycin-resistant Enterococcusisolates obtained serially from two long-term patients. J Clin Microbiol. 1998;36:2105–2108. doi: 10.1128/jcm.36.7.2105-2108.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Suppola J P, Volin L, Valtonen V V, Vaara M. Overgrowth of Enterococcus faeciumin the feces of patients with hematologic malignancies. Clin Infect Dis. 1996;23:694–697. doi: 10.1093/clinids/23.4.694. [DOI] [PubMed] [Google Scholar]
- 23.Swenson J M, Ferraro M J, Sahm D F, Charache P, Tenover F C the National Committee for Clinical Laboratory Standards Working Group on Enterococci. New vancomycin disk diffusion breakpoints for enterococci. J Clin Microbiol. 1992;30:2525–2528. doi: 10.1128/jcm.30.10.2525-2528.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tenover F C, Arbeit R D, Goering R V, Michelsen P A, Murray B E, Persing D H. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J Clin Microbiol. 1995;33:2233–2239. doi: 10.1128/jcm.33.9.2233-2239.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Woodford N, Morrison D, Cookson B, Georges R S. Comparison of high-level gentamicin-resistant Enterococcus faeciumisolates from different continents. Antimicrob Agents Chemother. 1993;37:681–684. doi: 10.1128/aac.37.4.681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Woodford N, Morrison D, Johnson A P, Bateman A C, Hastings J G M, Elliott S J, Cookson B. Plasmid-mediated vanBglycopeptide resistance in enterococci. Microb Drug Resist. 1995;1:235–239. doi: 10.1089/mdr.1995.1.235. [DOI] [PubMed] [Google Scholar]
- 27.Woodford N, Chadwick P R, Morrison D, Cookson B D. Strains of glycopeptide-resistant Enterococcus faecium can alter their vangenotypes during an outbreak. J Clin Microbiol. 1997;35:2966–2968. doi: 10.1128/jcm.35.11.2966-2968.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]