Abstract
Non-alcoholic fatty liver disease (NAFLD) is the most common hepatic pathology in industrialized countries, affecting about 25% of the general population. NAFLD is a benign condition, however, it could evolve toward more serious diseases, including non-alcoholic steatohepatitis (NASH), fibrosis, cirrhosis, and finally, hepatocellular carcinoma (HCC). Liver biopsy is still the gold standard for NAFLD diagnosis. Due to the risks associated with liver biopsy and the impossibility to apply it on a large scale, it is now necessary to identify non-invasive biomarkers, which may reliably identify patients at higher risk of progression. Therefore, several lines of research have tried to address this issue by identifying novel biomarkers using omics approaches, including lipidomics, metabolomics and RNA molecules’ profiling. Thus, in this review, we firstly report the conventional biomarkers used in clinical practice for NAFL and NASH diagnosis as well as fibrosis staging, and secondly, we pay attention to novel biomarkers discovered through omics approaches with a particular focus on RNA biomarkers (microRNAs, long-noncoding RNAs), showing promising diagnostic performance for NAFL/NASH diagnosis and fibrosis staging.
Keywords: biomarkers, ncRNAs, NAFLD
1. Introduction
Non-alcoholic fatty liver disease (NAFLD) is the most common hepatic pathology in industrialized countries, and it is associated with an increase in morbidity and mortality. NAFLD represents the hepatic manifestation of metabolic syndrome, which is considered a systemic disease since it affects different organs, including pancreas [1], kidney, adipose tissue and gut [2,3,4]. It is estimated that NAFLD affects about 25% of the general population [5], although the real prevalence is unknown because of the lack of specific and sensitive diagnostic tests. NAFLD includes several pathological conditions, ranging from simple steatosis to non-alcoholic steatohepatitis (NASH), the latter characterized by hepatic necroinflammation and rapid progression toward fibrosis, cirrhosis, and finally, hepatocellular carcinoma (HCC) [5,6,7,8]. NAFLD diagnosis is an exclusion diagnosis in which hepatic biopsy is still the gold standard to discriminate simple steatosis from NASH and to stage fibrosis. Over the last few years, the identification of non-invasive biomarkers to differentiate simple steatosis from NASH and stage fibrosis has become one of the most important aims of research in the hepatological field. This issue is very relevant because of the high prevalence of NAFLD in the general population, the potential progression toward more serious diseases and the impossibility to apply hepatic biopsy on a large scale.
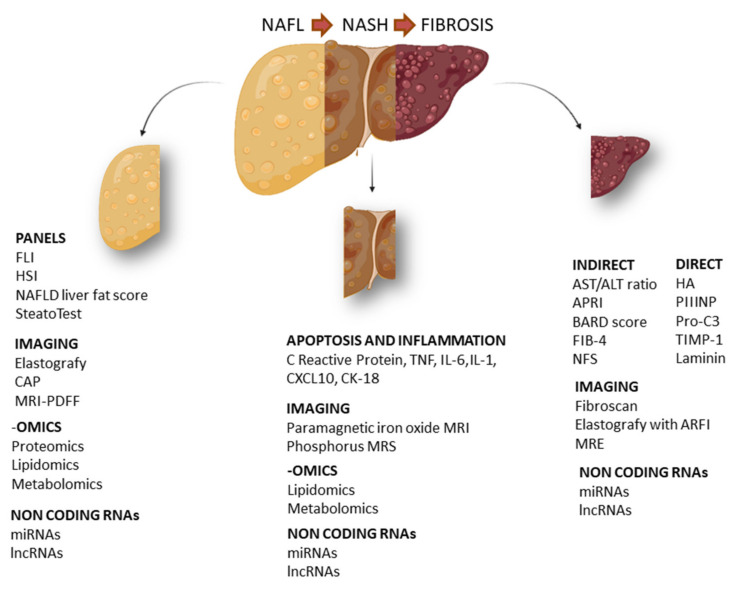
In this article, we aim to analyze both routine biomarkers currently used in clinical practice and novel biomarkers for the NAFLD spectrum, including non-alcoholic fatty liver (NAFL), NASH and fibrosis. In particular, we focus our dissertation on clinical biomarkers, their combination, imaging biomarkers and biomarkers identified through high-throughput approaches (Figure 1).
Figure 1.
Schematic representation of the main circulating biomarkers in NAFLD. Specific biomarkers for NAFL (non-alcoholic fatty liver), NASH (non-alcoholic steatohepatitis) and hepatic fibrosis. FLI (fatty liver index), HSI (hepatic steatosis index), CAP (controlled attenuation parameter), MRI-PDFF (magnetic resonance imaging proton density fat fraction), TNF (tumor necrosis factor), IL-6 (interleukin 6), IL-1 (interleukin 1), CXCL10 (C-X-C motif chemokine ligand 10), CK18 (cytokeratin 18), APRI (AST to platelet ratio index), BARD score (BMI, AST:ALT ratio, presence of diabetes), FIB-4 (fibrosis 4), NFS (NAFLD fibrosis score), HA (hyaluronic acid), PIIINP (N-terminal type III collagen pro-peptide), Pro-C3 (C-terminal cleavage site of N-terminal type II collagen pro-peptide), TIMP-1 (tissue inhibitor of metalloproteinases 1), MRE (magnetic resonance elastography).
2. NAFL Biomarkers
NAFL diagnosis involves the determination of hepatic steatosis (defined as the histological presence of triglyceride accumulation in 5% of hepatocytes) and exclusion of other causes of hepatic damage in people not assuming or assuming limited amounts of alcohol < 20/30 g per day respectively, for women and men [9,10]. Several non-invasive methods attempting to replace hepatic biopsies for NAFL diagnosis have been investigated [11,12].
2.1. Serum Biomarkers and Relative Panels
During the last few years, several indexes and scores have been elaborated to diagnose hepatic steatosis [13,14]. FLI (fatty liver index) includes BMI, waist circumference, serum triglycerides and GGT (gamma-glutamyl transferase). FLI has a moderate diagnostic performance (AUC = 0.84) in discriminating the presence of lipids in the liver as well as echographic analysis [15]; however, it has low accuracy in identifying several grades of steatosis [16]. HSI (hepatic steatosis index) is a panel of biomarkers that includes ALT/AST, BMI, sex and presence of type 2 diabetes, and this index has a moderate diagnostic power for hepatic steatosis identification [17] that considerably decreases in obese children (AUC = 0.67) [18]. Furthermore, similarly to FLI, it has a low accuracy in identifying several grades of hepatic steatosis [16]. A more efficient and sensitive method is the NAFLD liver fat score; through magnetic resonance, this method determines liver fat and combines different parameters, including the presence of metabolic syndrome and type 2 diabetes, insulin level, AST and ALT/AST. This score has a good diagnostic performance to diagnose hepatic steatosis with an AUC of 0.87 [19], however, the inclusion of insulin level, which is not a routinely used test, limits its use in clinical practice. SteatoTest is a panel including more specific parameters to diagnose steatosis. It combines six FibroTest elements (α2-macroglobulin, haptoglobin, apolipoprotein A1, GGT, total bilirubin and ALT) in addition to BMI, cholesterol, triglycerides and glycemia. SteatoTest is adjusted for sex and age, but it has a moderate accuracy to predict hepatic steatosis confirmed by hepatic biopsy (AUC = 0.80) [20]. This test is not widely used because it is expensive and is not able to discriminate different levels of steatosis.
2.2. Imaging Biomarkers
Imaging methods are often used in clinical practice and are used for NAFL diagnosis. The most used imaging method is echography, which is not expensive and is easy to perform. A meta-analysis demonstrated that elastography has high accuracy in discriminating moderate from severe steatosis (AUC = 0.93) [21]. However, this method has some limits: it is not able to detect steatosis when it is less than 20%, it is influenced by the presence of fibrosis and it is operator-dependent [11]. CAP (controlled attenuation parameter) detects the grade of steatosis, measuring the level of ultrasound attenuation due to the presence of fat. It has been reported that CAP has an AUC of 0.82 in detecting every grade of steatosis versus the absence of steatosis [22]. CAP has some limits, including a low sensibility for the low grade of steatosis, and it is operator-dependent. Magnetic resonance imaging proton density fat fraction (MRI-PDFF) is a non-invasive method that maps liver fat. MRI-PDFF is more precise than CAP in detecting several steatosis grades in NAFLD patients (AUC = 0.99) [23]; however, it is time-consuming, very expensive and needs qualified personnel.
2.3. Omics-Based Biomarkers
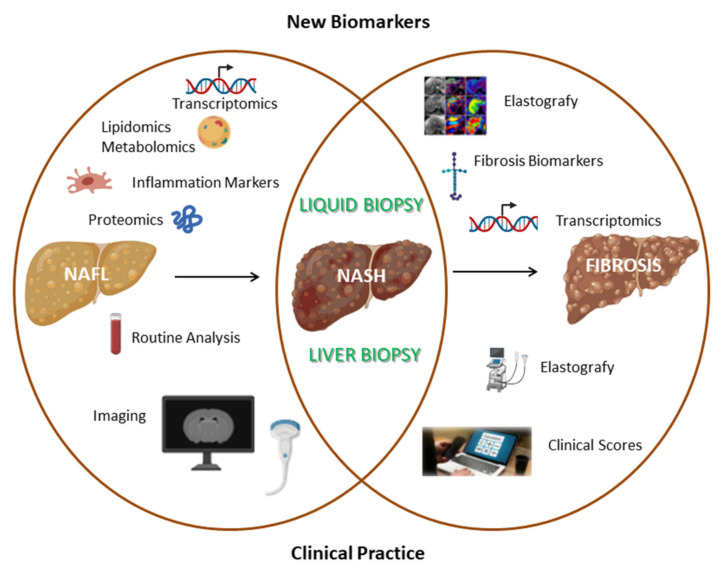
“Omics technologies”, through the detection of thousands of different molecules, can identify novel biomarkers that could be useful for NAFL diagnosis. Proteomics simultaneously analyzes a wide range of proteins by using a small volume of biological fluids. A study conducted on 70 patients (35 controls and 35 NAFL) identified 20 protein peaks in NAFL versus control (sensitivity 89% and specificity 83%); furthermore, through this approach, it has been reported that NAFL patients had a higher basal hemoglobin level [24]. In another study, through a more sensitive proteomic technique (LFQP), 605 differentially expressed proteins were observed in NAFL patients, however this method failed in discriminating simple steatosis patients with respect to NASH [25]. Metabolomics has been applied to identify the metabolic profile specific for NAFL, such as bile acids and glutathione, whose levels are altered during NAFLD onset [26]. Lipidomic-based studies identify the alteration of lipid species levels, for instance, short-chain fatty acids and eicosanoids [13]. A study performed on 679 patients reported that three specific lipids (defined as “lipid triplet”) can identify NAFL, however the diagnostic performance is limited (AUC = 0.71–0.74) [27]. Omics-based biomarkers could also be applied to evaluate disease severity; for instance, a study conducted on 467 NAFLD patients identified a signature of triglycerides that was able to differentiate NAFL from NASH (AUROC = 0.79) [28]. However, these methods have some drawbacks: a validation cohort is often lacking, the patient sample size is limited, and they often fail in discriminating several grades of steatosis and disease progression (Figure 2).
Figure 2.
Comparison between commonly used clinical methods and innovative research fields aimed at identifying novel non-invasive biomarkers for simple steatosis, NASH and fibrosis biomarkers.
3. NASH Biomarkers
NASH patients have a higher probability of developing cirrhosis and of dying from cardiovascular or liver-related causes [29,30]. Therefore, it is extremely important to identify NASH and fibrosis patients early among NAFLD patients in order to reduce the mortality. Thus, several lines of research attempted to develop non-invasive tests to discriminate NAFL from NASH. To achieve this aim, they mainly focused on biochemical and molecular biomarkers involved in specific pathways of progression from NAFL to NASH. Signaling pathways related to NASH development include apoptosis, oxidative stress, inflammation networks and adiponectin-mediated signals.
3.1. Protein Serum and Plasma Biomarkers Associated with Apoptosis and Inflammation
Apoptosis plays a fundamental role in the hepatic damage observed in NASH, and cytokeratin-18 (CK-18) is a biomarker of this process. CK-18 is indeed the main protein constituting hepatocyte intermediate filaments. When these cells undergo to apoptosis, CK-18 is cleaved by caspase 3 and CK-18 fragments are released at the extracellular level [31]. Serum CK-18 fragment levels are easily measurable through the ELISA assay. A meta-analysis of 11 studies highlighted that CK-18 has an overall sensitivity of 66% and an overall specificity of 82%, and therefore the limited accuracy of this biomarker prevents its clinical use for screening analysis and NASH definition [32]. CK-18 has been combined with other indicators in a biomarker panel aiming to increase the diagnostic performance for NASH patient identification. M30 serum levels deriving from CK-18 have been added to the quantification of the serum apoptosis-mediating surface antigen sFAS, involved in the activation of the extrinsic pathway of apoptosis in hepatocytes. The combination of these two markers determined a considerable increase in diagnostic performance, reaching an AUC value of 0.79–0.93. However, this study was conducted on a small sample, and thus this result needs further validation [33]. Younossi et al., combining CK-18 and relative fragments with adiponectin and resistin dosage in a cohort of 101 patients, obtained AUC values between 0.73 and 0.91. This study was conducted on a small sample of obese patients, and therefore other validations are needed [34]. Another study demonstrated that the combination of fibroblast growth factor 12 (FGF12) and CK-18 values increases sensitivity and specificity to values higher than 90% [35].
Since steatohepatitis is characterized by a chronic inflammation state, several studies evaluated markers of inflammation, such as C-reactive protein (CPR), TNF, IL-6, IL-1, IL-1RA (receptor antagonist protein) and CXCL10 (C-X-C motif chemokine ligand 10). However, these studies obtained inconsistent and weak results and a lack of specificity for the NASH pathological condition [36]. Ajmera et al. analyzed the diagnostic performance of 32 plasmatic biomarkers, most of them associated with inflammation, in a cohort of 648 patients. Among the analyzed biomarkers, after clinical and metabolic parameters’ adjustment, only the activated plasminogen activator inhibitor (APAI-1) maintained its predictive power for NASH [37].
3.2. Omics-Based Biomarkers
Oxidative stress is one of the key mechanisms of hepatic damage, and leads to lipid oxidation products that can be detected in serum samples.
Lipidomics studies performed using mass spectrometry led to the identification of specific products deriving from fatty acids associated with NASH. In particular, arachidonic acid oxidation products, including 11-hydroxyeicosatetraenoic acid (11-HETE), and linoleic acid oxidation products such as hydroxyoctadecadienoic acid (HODE) and 9 and 13 oxo-octadecadienoic acids (oxo-ODE), have been identified as biomarkers of NASH. Linoleic acid and 13-HODE ratios have been added to other clinical parameters (age, BMI and AST) in order to construct the oxNASH panel. This panel has a sensitivity of 81% and specificity of 97% in NASH versus not NASH discrimination [38].
In another study performed through the metabolomics approach, an increase in serum taurine- and glycine-conjugated primary and secondary bile acids in NASH patients after feeding was observed [39].
3.3. Imaging Biomarkers
Routine imaging methods such as abdominal ultrasonography, CT or MRI are able to discriminate NAFL from NASH. Several techniques based on MRI have been tested in animals and in pilot studies conducted in humans for NASH diagnosis. The superparamagnetic iron oxide MRI can detect defective Kupffer cell uptake function in in vivo models and in humans affected by NASH. However, the current protocol involves repeated scanning over the course of 72 h, which represents a logistic limit of this method [40]. Hepatocyte membrane turnover and intracellular ATP that are altered in NASH can be detected through phosphorus MRS. In more detail, alpha-nucleotide triphosphate/total phosphate ratio has an AUC value of 0.71 for NASH patient discrimination [41]. Finally, the diagnostic performance of hepatic stiffness, measured through magnetic resonance elastography, needs to be evaluated in a larger cohort and adjusted for fibrosis stages [42] (Figure 2).
4. Fibrosis Biomarkers
NASH-related fibrosis includes different stages, ranging from absent fibrosis (F0) to cirrhosis (F4). From a clinical point of view, fibrosis is defined as clinically significant (F2–F4) or advanced/severe fibrosis (F3–F4). It is widely known that there are several risk factors that are able to predict fibrosis onset, including age, severe obesity, type 2 diabetes mellitus, high AST/ALT ratio, hypertension, dyslipidemia and the presence of the metabolic syndrome [43,44,45].
According to a study published in hepatology, the hepatic fibrosis stage could be the most important factor in determining NAFLD prognosis and preventing the risk of progression toward cirrhosis and related complications [46].
To stage fibrosis in NAFLD patients, it is important to identify novel non-invasive biomarkers. The most used biomarkers in clinical practice are classified into (a) indirect biomarkers of hepatic fibrosis, (b) direct biomarkers of hepatic fibrosis and (c) imaging biomarkers for fibrosis.
4.1. Indirect Biomarkers for Hepatic Fibrosis
Indirect biomarkers do not directly measure fibrogenesis or fibrinolysis, but are often associated with risk factors for fibrosis; furthermore, in order to increase their limited accuracy, two or more of them are combined in panels.
AAR (AST:ALT ratio) and APRI (AST:platelet ratio index) are the simplest indexes to calculate, but they have low accuracy in discriminating advanced stage 3 fibrosis (AUC AST:ALT ratio: between 0.66 and 0.74; AUC APRI: 0.74) [47,48]. The BARD score is based on the sum of three parameters (BMI, AST:ALT ratio, presence of diabetes), and it has a moderate accuracy in identifying F3 fibrosis subjects (AUC between 0.69 and 0.81) [49,50]. The FIB-4 index (fibrosis index based on four factors) involves a more complicated calculation, and thus online calculators are often used: it includes age, AST, ALT and platelet count, and it has a good diagnostic performance (AUC: 0.80 in identifying F3 fibrosis stage subjects) [51]. The NAFLD fibrosis score (NFS) [52] is obtained by combing age, presence of diabetes, body mass index, platelets, albumin and AAR. This score has been validated in several studies and it has an estimated AUC of between 0.75 and 0.83 in F3 fibrosis detection [53].
Although these scores have less accuracy than direct specific fibrosis markers, they are the most commonly used indexes for screening thanks to their wide applicability.
4.2. Fibrosis Biomarkers
Fibrosis biomarkers include molecules directly involved in fibrogenesis and/or fibrinolysis. Hepatic fibrosis is determined by the accumulation of extracellular matrix components, where hyaluronic acid (HA) is the most represented [54]. Serum levels of HA have an AUC of 0.87 for stage F2 fibrosis and 0.92 for cirrhosis [55]. Other components of the extracellular matrix include collagen, proteoglycans, elastin, fibronectin and laminin. Serum procollagen III amino-terminal peptide (PIIINP) originates from novel collagen III biosynthesis or existing collagen III fibril degradation. PIIINP levels alone represent a good diagnostic biomarker for fibrosis [56], but it has been demonstrated that Pro-C3 (the PIIINP neo-epitope) reflects the effective production of collagen III biosynthesis [57]; furthermore, the increase of serum Pro-C3 levels is correlated with NASH and fibrosis [58]. The circulating level of metalloprotease-1 inhibitor (TIMP1) influences extracellular matrix composition, wound healing and reflects the alteration of tissue matrix remodeling during hepatic fibrogenesis and fibrinolysis [59]. TIMP1 has excellent diagnostic performance (AUC: 0.97) in the discrimination of obese patients with NASH-related fibrosis with respect to matched controls [60]. Laminin is the most abundant glycoprotein in the basal membrane and its serum levels are able to identify the presence of fibrosis in NAFLD patients with excellent diagnostic performance (AUC: 0.87) [61].
The enhanced liver fibrosis test (ELF) is a panel analyzing direct biomarkers of fibrosis; specifically, it detects extracellular matrix components. ELF is based on the detection of three fibrosis biomarkers (HA, TIMP1 and PIIINP) and it has excellent diagnostic power in advanced fibrosis identification in adult and pediatric NAFLD patients (AUC 0.93 and 0.99, respectively) [62,63]. Another instrument for fibrosis detection is the FibroTest, that is obtained through the combination of five biomarkers (haptoglobin, α2-macroglobulin, Apolipoprotein A1, total bilirubin and GGT). The diagnostic value of the FibroTest has been evaluated in a wide cohort of NAFLD patients and reached an AUC of 0.88 [63].
The NAFLD FibroMeter is an index including weight, prothrombin index and serum level of ALT, AST, ferritin and fasting glucose. In two different studies conducted in Asia and Europe, the NAFLD FibroMeter surpassed the other serum test for F2 patient discrimination (AUC: 0.76) and F3 patient discrimination (AUC: 0.77) [64,65]. Finally, the Hepascore, composed of age, sex, gender, bilirubin serum level, GGT, HA and α2m, has a good diagnostic performance (AUC: 0.82) to diagnose advanced fibrosis (stages F3–F4) in NAFLD patients [66].
Although these indexes have a diagnostic performance superior to indirect biomarkers of fibrosis, their large-scale use is limited because of the reduced number of known direct fibrosis biomarkers, their cost and the use of a patented formula.
4.3. Imaging Biomarkers of Fibrosis
Over the last ten years, the progress in imaging methods improved the possibility to quantify, in a non-invasive manner, hepatic fibrosis, revolutionizing hepatic disease clinical management. The FibroScan or VTCE (vibration controlled transient elastography) is based on the use of an echographic probe that transmits a vibratory wave propagating through the liver, evaluating liver elasticity, and the velocity of wave propagation is directly proportional to tissue rigidity. Results are expressed in kilopascal (kPa). Usually, the mean value of ten measurements in healthy subjects is between 1.5 and 7.5 kPa, while values > 10.5 kPa are an index of the presence of fibrosis and advanced fibrosis [67]. Unfortunately, VCTE or M probes are less reliable in severe obese NAFLD patients, because adipose tissue attenuates both elastic waves and ultrasound [68]. Therefore, to overcome this issue, a novel probe “XL” has been developed for patients with BMI > 30 kg/m2. The diagnostic performances of FibroScan probes M and XL for advanced fibrosis are 0.88 and 0.85, respectively [69].
Elastography conducted through the ARFI method (acoustic radiation force impulse) is an alternative method to estimate hepatic tissue rigidity that can be integrated with a conventional ultrasound probe, detecting stiffness during routine echography. A region of interest is excited mechanically using acoustic impulses. Hepatic rigidity is expressed in m/s, and this method has a limited range (0.5–4.4 m/s) that limits the definition of a cut-off value to discriminate specific stages of fibrosis. It has been reported that the diagnostic accuracy of elastography with the ARFI technique is higher in discriminating severe fibrosis and cirrhosis (AUC: 0.74–0.97 for F3; AUC: 0.78–0.89 for F4) rather than less severe stages (AUC: 0.70–0.83 for F2) [70].
Magnetic resonance electrography (MRE) is another non-invasive technique to detect fibrosis in NAFLD patients. It combines magnetic resonance with a sound wave to create a visible map (elastogram) that shows hepatic tissue rigidity [71,72]. In more detail, a meta-analysis of 9 studies including 232 NAFLD patients highlighted that MRE can detect fibrosis with high accuracy, independently from hepatic inflammation, and BMI (differently from FibroScan) with an AUC of 0.86–0.91 for each stage of fibrosis [73]. This method has a higher accuracy than FibroScan in detecting F2 fibrosis (AUC: 0.86–0.89 versus AUC: 0.84) and F4 fibrosis (AUC: 0.88–0.97 versus AUC: 0.95) [74]. However, its large-scale application as a screening test is limited by cost and the small number of identified molecules (Figure 2).
5. RNA Biomarkers
In biological fluids, there are RNA molecules belonging to different classes, including messenger RNAs (mRNAs), long-noncoding RNAs (lncRNAs), circular RNAs (circRNAs) and microRNAs (miRNAs). The presence of cell-free RNAs in body fluids stimulated the interest in studying these molecules as novel biomarkers of diagnosis, prognosis, monitoring and therapeutic response of several kinds of pathologies [75,76,77,78].
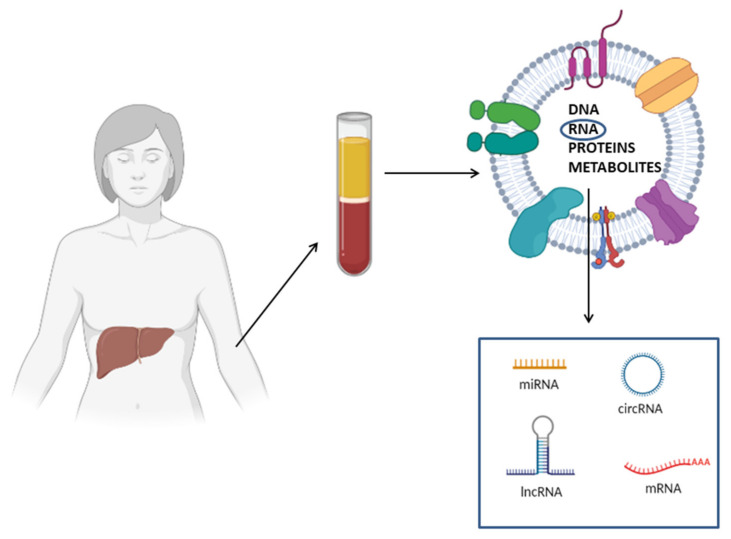
A key feature making cf-RNAs excellent biomarkers is their high stability: these molecules are protected by endogenous RNases and are resistant to different external insults, such as multiple thaw–draw cycles, boiling, extreme PH conditions, prolonged storage and exogenous RNase treatment [79,80]. Circulating RNAs are extremely stable because they are not present in a free form in the circulation; instead, they are encapsulated in membranous vesicles (micro-vesicles, exosomes, apoptotic bodies), or alternatively complexed to RNA binding proteins (i.e., NPM1, Argonauta 2) or associated with lipoproteins. All these mechanisms protect RNAs from degradation [81]. Another aspect making RNA molecules optimal biomarkers is that the techniques used for their detection are extremely sensitive. Differently from proteins, nucleic acids are detected and quantified through PCR-based methods that determine an exponential increase of the initial template. Consequently, protein detection techniques are much less sensitive with respect to methods for nucleic acid detection. In theory, even a single RNA molecule could be detected through quantitative PCR [82]. Finally, it is important to highlight that several studies reported that the differential expression profiles of circulating RNAs are correlated to different physiological and pathological states [83]. These studies support the use of RNA molecules as non-invasive biomarkers; furthermore, their whole expression profile can be analyzed through high-throughput methods, including next-generation sequencing (NGS) and microarray, and thus, analysis of these molecules has a key role in biomarker discovery [82] (Figure 3).
Figure 3.
Schematic representation of circulating RNA biomarkers for NAFLD diagnosis. Circulating RNAs (e.g., mRNAs, miRNAs, lncRNAs and circRNAs) can be embedded in membranous vesicles, associated with RNA binding protein or in a free form. Membranous vesicles, besides RNA, contain other macromolecules, including DNA, proteins and metabolites.
5.1. Circulating RNAs as NAFLD Biomarkers in the General Population
Most of the studies concerning circulating RNAs as NAFL biomarkers are limited to miRNAs. Akuta et al. analyzed miR-122 expression levels in NAFL patients who underwent two serial biopsies. They observed that, in patients showing an improvement of their histopathological scores, serum miR-122 expression levels decreased at the second biopsy with respect to the first biopsy. Furthermore, there was a significant correlation between miR-122 expression levels and histopathological score variations. The authors also observed an association between miR-122 expression levels and routine clinical parameters such as AST and ALT [84]. A study reported that miR-21 levels were also decreased in NAFL patients with respect to controls [85]. Yamada et al. analyzed the expression of 5 miRNAs involved in lipid homeostasis (miR-21, -34a, -122, -145, -451) in a total of 403 subjects. Serum levels of four of these miRNAs were higher in NAFL subjects with respect to controls (92/403) (miR-21, -34a, -122, -451); furthermore, miR-122 expression levels correlated with steatosis grades [86].
In another study, more advanced techniques were used; through deep sequencing, miRNA expression profiles were evaluated in 20 NAFL patients and 20 controls. Subsequently, differentially expressed miRNA were validated in both a wider cohort (90 controls versus 152 NAFL) and an external cohort (80 controls versus 103 NAFL). The analyzed miRNA panel (miR-122, -1290, -27b, -192) showed a high NAFL diagnostic accuracy (AUC: 0.86, sensitivity = 85.55%, specificity = 73.3%). Furthermore, their sensitivity and specificity was superior to ALT and FIB-4 [87]. Raitoharju et al. demonstrated that blood miR-122 and -885 were upregulated in a cohort of 871 subjects with ultra-sonographically diagnosed fatty liver. MiR-122 and miR-885 were upregulated in NAFLD subjects and miR-122 diagnostic performance was similar to transaminase diagnostic performance. Moreover, through the metabolomic approach, they demonstrated that miR-122 expression was positively correlated with some lipoprotein classes (VLDL, IDL, LDL), while miR-885 expression was inversely associated with HDL cholesterol [88]. Another study reported that miR-181d, -99a, -197 and -146b expression levels were lower in NAFL subjects with respect to controls; furthermore, miR-197 and -146b levels negatively correlated with inflammation grade, while miR-181d and -99a levels were inversely associated with GGT in NASH patients [89]. Finally, recent studies reported that miR-29, -1296, -132 and -135 were deregulated in NAFL patients and correlated with the risk of disease onset [90,91,92,93,94]. Only one study, instead, evaluated lncRNAs as an NAFL biomarker. Long-noncoding RNAs are transcripts longer than 200 nucleotides lacking a long protein-coding open reading frame (ORFs) that are involved in a myriad of cellular processes through the regulation of gene expression at epigenetic, transcriptional, post-transcriptional, translational and post-translational levels [95]. The ARSR lncRNA has been reported to be upregulated in serum of NAFL patients versus NASH patients; furthermore, in NASH in vivo models, it has been demonstrated that this lncRNA is involved in hepatic lipogenesis [96].
Circulating extracellular vesicles (exosomes and ectosomes) containing miRNAs, mRNAs, proteins and DNA molecules could be used as NAFLD biomarkers [12]. Ectosomes are vesicles of various size (0.1–1 mm in diameter) that bud directly from the plasma membrane and are shed to the extracellular space [97]. Once released, ectosomes bind to recipient cells and deliver their informative cargo. In NAFL subjects, an increase in ectosomes in monocyte and natural-killer cell surfaces and a decrease in neutrophyl and endothelian cell surfaces have been observed. Production of exosomes and other extracellular vesicles are increased in patients with NASH [98], and it has recently been suggested that a specific protein signature in serum extracellular vesicles may be used to diagnose NASH non-invasively [99] (Figure 4).
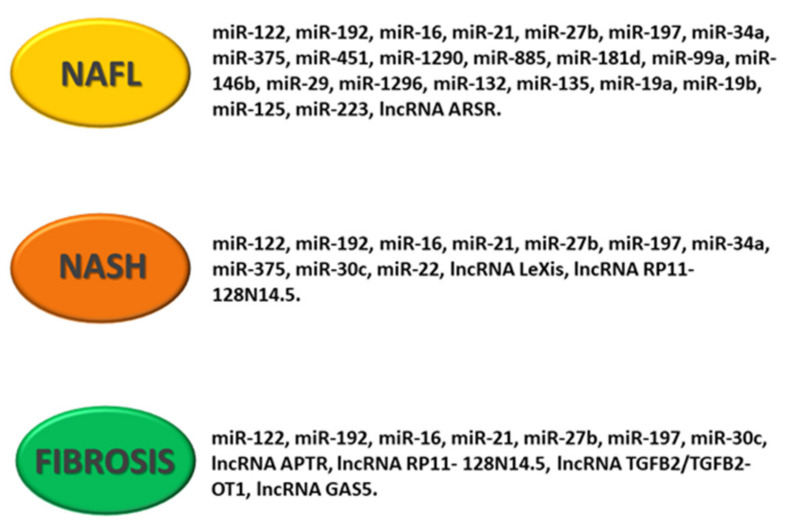
Figure 4.
Schematic representation of deregulated noncoding RNA pattern in NAFLD. The figure shows specific miRNAs and lncRNAs associated with NAFL (non-alcoholic fatty liver), NASH (non-alcoholic steatohepatitis) and hepatic fibrosis.
5.2. Circulating RNAs for Differential Diagnosis between NAFL and NASH
As highlighted for NAFL, and also as far as NASH is concerned, most data concerning circulating RNAs are related to miRNAs. The first attempt to identify circulating miRNAs, such as NAFL/NASH differential diagnosis biomarkers, was conducted by Cermelli et al. in 2011. In more detail, they reported an increase of miRNA-122, -16 and -34a expression levels in 34 NAFLD patients with respect to 19 controls. On average, miR-122 presented a 7.2-fold change upregulation, miR-34a presented a 5.3-fold change upregulation and miR-16 was undetectable in control samples, while pathological samples had 1000 copies per mL [100]. The results obtained sub-stratifying the NAFLD population are more interesting; through this analysis, the authors found that miRNA-122 and -34a further increased in NASH patients with respect to simple steatosis patients, with a fold change of about two and three, respectively. Moreover, through ROC curve analysis, it has been demonstrated that miRNA-122 and -34a have a high diagnostic performance (miRNA-122 AUC: 0.93; miRNA-34a AUC: 0.96) in distinguishing NAFLD patients versus controls, but this decreased in the NASH versus NAFL comparison (respectively, 0.70 and 0.76) [100].
Another important study conducted by Pirola et al. in 2015 aimed to identify NAFL versus NASH differential diagnosis biomarkers. They profiled the expression of 84 miRNAs in patients with NAFL, NASH and controls (n = 48), and successively, they confirmed the upregulation (FC > 2) of a six-miRNA panel in an independent cohort (n = 96): miR-122, -192, -19a, -19b, -125 and -375. Only three of them, miR-122, -192 and -375, correlated with histological severity and were significantly overexpressed in NASH patients with respect to NAFL patients (AUC: 0.69, 0.68 and 0.72 respectively, in NAS > 5 versus NAS < 5 discrimination) [101].
miR-122 and -192 were further validated as indexes of histological severity by Becker et al. [102].
In more detail, they analyzed the expression of four miRNAs, i.e., miR-122, -192, -21 and -223, in 137 NAFLD patients with moderate or severe obesity (87 NASH and 50 simple steatosis (SS)) compared to 61 controls. Furthermore, they performed correlation analysis between miRNA expression level and routine biomarkers such as AST, ALT and CK-18-Asp396 fragments (released by hepatocytes during apoptosis).
Among the analyzed miRNAs, miR-122 and miR-192 were upregulated in NASH patients with severe obesity with respect to SS patients with severe obesity. However, this deregulation was not confirmed in the NASH/SS comparison in patients with moderate obesity. miR-21 was upregulated in NASH patients with respect to SS patients with moderate or severe obesity. miR-223 was upregulated in NASH patients with respect to controls, but it was not able to discriminate between the two pathological conditions [102].
Moreover, the expression levels of miR-122 and -192 had a positive correlation with ALT, CK-18-Asp396 and the NAS score, and miR-21 positively correlated with ALT and the NAS score. Subsequently, serum concentrations of these microRNAs have been used to elaborate a scoring system ranging from 0 to 3 in order to determine the diagnostic performance for NAFL versus NASH discrimination. Through these analyses, Becker et al. found that the combination between the miRNA-based scoring system and CK-18-Asp396 reached an AUC of 0.83, with a sensitivity of 93% and a specificity of 83% [102].
In a subsequent study [103], Liu et al. observed an increase of miR-122, -192, -34a and -16 expression in 48 biopsy-proven NAFLD patients with respect to 37 controls; among these miRNAs, miR-122, -16 and -34a were able to discriminate NASH (n = 31) versus NAFL (n = 17) patients. It is important to highlight that miR-34a had a higher diagnostic power and sensitivity for NASH (AUC: 0.81, sensitivity: 0.70, specificity: 0.86) than other already known biomarkers, such as ALT (AUC: 0.68, sensitivity: 0.84, specificity: 0.94), CK-18-M30 and CK-18-M60 (M30 AUC: 0.70, sensitivity: 0.41, specificity: 0.94; M60 AUC: 0.72, sensitivity: 0.52, specificity: 0.94).
Finally, a more recent study concerning miRNAs as progression biomarkers of hepatic steatosis was conducted in 2018 by Lopez-Riera et al. Analyzing miRNAs already reported to be deregulated in NAFLD, they demonstrated that miRNA-27b, -34a, -22, -122 and -192 and miRNA-30c, -16 and -197 are reduced in the serum of patients with severe disease with respect to simple steatosis patients. Furthermore, miR-34a/mir-197 and miR-27b/miR-30c ratios showed the best diagnostic performance for the identification of NASH, defined as SAF ≥ 2 (AUC: 0.81) or NAS score ≥ 5 (AUC: 0.79) [104]. Therefore, to date, based on the above-reported data, we can affirm that the upregulation of miRNA-122, -192 and -34a is consistent in several studies. Further validation in larger cohorts could lead to the introduction of these RNA-based biomarkers in clinical practice for the identification of patients with a high risk for NAFL to NASH progression.
Few studies evaluated lncRNAs as NASH biomarkers. Park et al. analyzed the expression level of Lexis lncRNA in plasma samples of 35 biopsy-proven NASH patients versus 9 NAFL patients. They found that Lexis was increased in NASH patients versus NAFL patients, with AUC: 0.74, sensitivity: 54.3% and specificity: 100% [105].
In a study published in 2019 by our research group [106], after microarray analysis, the upregulation of RP11-128N14.5 lncRNA was validated in patients with severe NAFLD (NAS score ≥ 5 versus NAS score ≤ 4), with a diagnostic power of 0.71 (sensitivity = 73.7%, specificity = 70.4%) superior to AST (AUC = 0.66) and ALT (AUC = 0.37) (Figure 4).
5.3. Circulating RNAs for Fibrosis Stage Definition
It is widely known that miRNAs are involved in NAFLD pathogenesis and their deregulation has been reported in NASH-related fibrosis [107]. Although there are many studies reporting miRNA roles in pathogenesis, very few studies evaluated circulating miRNAs as biomarkers of fibrosis in NAFLD populations.
The expression of miR-122 has been analyzed in 67 NAFLD patients with several degrees of fibrosis. miR-122 levels were significantly lower in patients with severe fibrosis versus mild fibrosis. The diagnostic performance of miR-122 was also evaluated and compared with other fibrosis biomarkers, such as hyaluronic acid and type IV collagen. miR-122 had a stronger diagnostic accuracy (AUC: 0.82) versus hyaluronic acid and type IV collagen [108].
On the contrary, Pirola et al. demonstrated that miR-122 was upregulated in patients with advanced fibrosis F2–F3 versus patients with fibrosis stages F0–F1, with an AUC of 0.61. Although this value is not high enough for a biomarker, it is important to highlight that miR-122 efficiency was higher than CK-18 (AUC: 0.49), ALT (AUC: 0.59) and AST (AUC: 0.64) in fibrosis prediction [101].
In 2018, López-Riera et al. [104] analyzed the expression of a set of miRNAs previously reported to be deregulated in NAFLD. miRNA expression was analyzed stratifying the patient cohort according to fibrosis stages in a two-group comparison (F ≤ 2 or F > 2) or in a five-group comparison (F0, F1, F2, F3, F4, F5). Through this latter comparison, they observed that miR-122 and miR-192 had an increased expression trend in mild fibrosis (F1 and F2) and a decreasing expression trend in severe fibrosis (F3 and F4). Thus, in this study, miR-122 had an opposite expression trend than in Miyaaki’s study [108], and the lack of consistency of mir-122 data limits its use as a biomarker for severe fibrosis prediction among NAFLD patients [104]. In the same study, another miRNA that had an opposite trend with respect to literature data was miR-16. López-Riera et al. [104] reported that miR-16 was downregulated in F3–F4 fibrosis; however, Liu et al. [103] reported that miR-16 was upregulated and had a good diagnostic performance in fibrosis prediction (AUC: 0.71) [103]. In addition, López-Riera et al. reported that circulating miRNA-21 and -27b are upregulated, while miRNA-30c is downregulated in F ≤ 2 versus F > 2 fibrosis patients. However, diagnostic performances of the analyzed miRNAs for severe fibrosis identification did not overcome conventional fibrosis algorithms. The best-performing miRNA was miR-30c, showing an AUC value of 0.72; moreover, the miR-27b plus miR-30c combination and the miR-27b plus miR-197 combination slightly increased the diagnostic performance, reaching AUC values of 0.770 and 0.75, respectively [104].
To date, few studies have taken into account the dosage of lncRNAs as NASH-related liver fibrosis biomarkers. Yu et al. demonstrated that APTR (Alu-mediated p21 transcriptional regulator) serum levels were upregulated in 34 biopsy-proven cirrhosis patients with respect to 24 controls [109]; thus, APTR could represent a potential biomarker of NASH-related hepatic cirrhosis.
In our previously published study in 2019 [106], after microarray analysis, we validated the upregulation of serum TGFB2/TGFB2-OT1 lncRNA in F3–F4 stages with respect to F0–F2 patients. TGFB2/TGFB2-OT1 lncRNA showed an AUC value of 0.80 in F3–F4 versus F0–F2 discrimination (sensitivity = 65%, specificity = 81.3%). In addition, when TGFB2-OT1 expression levels were combined with FIB-4 or FibroScan, there was an improvement in diagnostic performance.
Finally, Han et al. [110] analyzed the expression of GAS5 lncRNA in plasma samples of patients with several fibrosis stages. GAS5 plasma levels were increased in patients with advanced fibrosis with respect to patients with absent or moderate fibrosis (F < 2), and the expression of GAS5 positively correlated with several fibrosis stages (from F0 to F3). However, GAS5 was upregulated in F3 versus F < 2, and its expression was downregulated in NAFLD patients with cirrhosis (F4) versus patients with advanced fibrosis (F3). This is the first study that demonstrated that GAS5 levels are altered during fibrosis progression and cirrhosis development (Figure 4).
Our study provides novel high-throughput data concerning the expression of non-coding RNA in serum of NAFLD patients and lays the foundation for the identification of novel biomarkers that could represent a future alternative to hepatic biopsy.
6. Conclusions
To date, NAFLD is a risk for global health because of its morbidity, mortality and high prevalence in the general population [111].
The possibilities to correctly identify NASH patients among NAFLD subjects and to stage fibrosis are important clinical challenges. Currently, hepatic biopsy is still the gold standard to diagnose NASH and stage fibrosis. However, because of risks related to biopsy, clinicians often use biochemical and imaging tests, even though they are characterized by limited diagnostic performance [13,14,21]. The identification of novel non-invasive methods is fundamental because it could make the monitoring of treatment response and disease progression easier. Over the last few years, several classes of molecules such as specific proteins or metabolites have been proposed to diagnose NAFL, NASH and stage fibrosis. Their limited diagnostic performance restricted the clinical application of these markers for NAFLD screening and fibrosis staging. Nevertheless, it is important to highlight that several studies demonstrated that the combination of several serum biomarkers in panels increased their diagnostic performance. Therefore, this combined approach could allow, in the future, clinical application for steatosis diagnosis and staging.
The introduction of novel “omics” technologies could allow the identification of novel non-invasive biomarkers for NAFLD patient identification, NASH versus NAFL patient discrimination and fibrosis staging. Nevertheless, these “omics studies” need to be validated in larger and more heterogeneous cohorts before being used in clinical practice.
Acknowledgments
We wish to thank the Scientific Bureau of the University of Catania for language support. This study was in keeping with the objectives of the project “DEGENER-action”, Department of Clinical and Experimental Medicine University of Catania.
Author Contributions
Conceptualization, S.D.M., A.F. and A.S.; data curation, S.D.M., A.F. and A.S.; writing—original draft preparation, S.D.M., A.F., A.S., R.M. and S.P.; writing—review and editing, S.D.M., A.F., A.S., A.D.P., R.S., R.M. and S.P.; project administration, S.P. and F.P. All authors have read and agreed to the published version of the manuscript.
Funding
This review received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Filippello A., Scamporrino A., Di Mauro S., Malaguarnera R., Di Pino A., Scicali R., Purrello F., Piro S. Direct Effects of D-Chiro-Inositol on Insulin Signaling and Glucagon Secretion of Pancreatic Alpha Cells. Biomolecules. 2020;10:1404. doi: 10.3390/biom10101404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Armutcu F., Akyol S., Vura H. Metabolic Syndrome is an Important Cornerstone in the Health-disease Line and Pathological Organ Interaction. J. Cell. Signal. 2020;1:70–75. [Google Scholar]
- 3.Filippello A., Di Mauro S., Scamporrino A., Malaguarnera R., Torrisi S.A., Leggio G.M., Di Pino A., Scicali R., Purrello F., Piro S. High Glucose Exposure Impairs L-Cell Differentiation in Intestinal Organoids: Molecular Mechanisms and Clinical Implications. Int. J. Mol. Sci. 2021;22:6660. doi: 10.3390/ijms22136660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Filippello A., Urbano F., Di Mauro S., Scamporrino A., Di Pino A., Scicali R., Rabuazzo A.M., Purrello F., Piro S. Chronic Exposure to Palmitate Impairs Insulin Signaling in an Intestinal L-cell Line: A Possible Shift from GLP-1 to Glucagon Production. Int. J. Mol. Sci. 2018;19:3791. doi: 10.3390/ijms19123791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Younossi Z.M., Koenig A.B., Abdelatif D., Fazel Y., Henry L., Wymer M. Global epidemiology of nonalcoholic fatty liver disease-Meta-analytic assessment of prevalence, incidence, and outcomes. Hepatology. 2016;64:73–84. doi: 10.1002/hep.28431. [DOI] [PubMed] [Google Scholar]
- 6.Di Mauro S., Ragusa M., Urbano F., Filippello A., Di Pino A., Scamporrino A., Pulvirenti A., Ferro A., Rabuazzo A.M., Purrello M., et al. Intracellular and extracellular miRNome deregulation in cellular models of NAFLD or NASH: Clinical implications. Nutr. Metab. Cardiovasc. Dis. 2016;26:1129–1139. doi: 10.1016/j.numecd.2016.08.004. [DOI] [PubMed] [Google Scholar]
- 7.Scicali R., Di Pino A., Urbano F., Ferrara V., Marchisello S., Di Mauro S., Scamporrino A., Filippello A., Rabuazzo A.M., Purrello F., et al. Analysis of steatosis biomarkers and inflammatory profile after adding on PCSK9 inhibitor treatment in familial hypercholesterolemia subjects with nonalcoholic fatty liver disease: A single lipid center real-world experience. Nutr. Metab. Cardiovasc. Dis. 2021;31:869–879. doi: 10.1016/j.numecd.2020.11.009. [DOI] [PubMed] [Google Scholar]
- 8.Masarone M., Rosato V., Aglitti A., Bucci T., Caruso R., Salvatore T., Sasso F.C., Tripodi M.F., Persico M. Liver biopsy in type 2 diabetes mellitus: Steatohepatitis represents the sole feature of liver damage. PLoS ONE. 2017;12:e0178473. doi: 10.1371/journal.pone.0178473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chalasani N., Younossi Z., Lavine J.E., Charlton M., Cusi K., Rinella M., Harrison S.A., Brunt E.M., Sanyal A.J. The diagnosis and management of nonalcoholic fatty liver disease: Practice guidance from the American Association for the Study of Liver Diseases. Hepatology. 2018;67:328–357. doi: 10.1002/hep.29367. [DOI] [PubMed] [Google Scholar]
- 10.Rinaldi L., Pafundi P.C., Galiero R., Caturano A., Morone M.V., Silvestri C., Giordano M., Salvatore T., Sasso F.C. Mechanisms of Non-Alcoholic Fatty Liver Disease in the Metabolic Syndrome. A Narrative Review. Antioxidants. 2021;10:270. doi: 10.3390/antiox10020270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhou J.H., Cai J.J., She Z.G., Li H.L. Noninvasive evaluation of nonalcoholic fatty liver disease: Current evidence and practice. World J. Gastroenterol. 2019;25:1307–1326. doi: 10.3748/wjg.v25.i11.1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Piazzolla V.A., Mangia A. Noninvasive Diagnosis of NAFLD and NASH. Cells. 2020;9:1005. doi: 10.3390/cells9041005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wong V.W., Adams L.A., de Ledinghen V., Wong G.L., Sookoian S. Noninvasive biomarkers in NAFLD and NASH—current progress and future promise. Nat. Rev. Gastroenterol. Hepatol. 2018;15:461–478. doi: 10.1038/s41575-018-0014-9. [DOI] [PubMed] [Google Scholar]
- 14.Kogachi S., Noureddin M. Noninvasive Evaluation for Nonalcoholic Fatty Liver Disease and Nonalcoholic Steatohepatitis. Clin. Ther. 2021;43:455–472. doi: 10.1016/j.clinthera.2021.01.012. [DOI] [PubMed] [Google Scholar]
- 15.Bedogni G., Bellentani S., Miglioli L., Masutti F., Passalacqua M., Castiglione A., Tiribelli C. The Fatty Liver Index: A simple and accurate predictor of hepatic steatosis in the general population. BMC Gastroenterol. 2006;6:33. doi: 10.1186/1471-230X-6-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fedchuk L., Nascimbeni F., Pais R., Charlotte F., Housset C., Ratziu V., Group L.S. Performance and limitations of steatosis biomarkers in patients with nonalcoholic fatty liver disease. Aliment. Pharmacol. Ther. 2014;40:1209–1222. doi: 10.1111/apt.12963. [DOI] [PubMed] [Google Scholar]
- 17.Lee J.H., Kim D., Kim H.J., Lee C.H., Yang J.I., Kim W., Kim Y.J., Yoon J.H., Cho S.H., Sung M.W., et al. Hepatic steatosis index: A simple screening tool reflecting nonalcoholic fatty liver disease. Dig. Liver Dis. 2010;42:503–508. doi: 10.1016/j.dld.2009.08.002. [DOI] [PubMed] [Google Scholar]
- 18.Koot B.G., van der Baan-Slootweg O.H., Bohte A.E., Nederveen A.J., van Werven J.R., Tamminga-Smeulders C.L., Merkus M.P., Schaap F.G., Jansen P.L., Stoker J., et al. Accuracy of prediction scores and novel biomarkers for predicting nonalcoholic fatty liver disease in obese children. Obesity. 2013;21:583–590. doi: 10.1002/oby.20173. [DOI] [PubMed] [Google Scholar]
- 19.Kotronen A., Peltonen M., Hakkarainen A., Sevastianova K., Bergholm R., Johansson L.M., Lundbom N., Rissanen A., Ridderstrale M., Groop L., et al. Prediction of non-alcoholic fatty liver disease and liver fat using metabolic and genetic factors. Gastroenterology. 2009;137:865–872. doi: 10.1053/j.gastro.2009.06.005. [DOI] [PubMed] [Google Scholar]
- 20.Poynard T., Ratziu V., Naveau S., Thabut D., Charlotte F., Messous D., Capron D., Abella A., Massard J., Ngo Y., et al. The diagnostic value of biomarkers (SteatoTest) for the prediction of liver steatosis. Comp. Hepatol. 2005;4:10. doi: 10.1186/1476-5926-4-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hernaez R., Lazo M., Bonekamp S., Kamel I., Brancati F.L., Guallar E., Clark J.M. Diagnostic accuracy and reliability of ultrasonography for the detection of fatty liver: A meta-analysis. Hepatology. 2011;54:1082–1090. doi: 10.1002/hep.24452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Karlas T., Petroff D., Sasso M., Fan J.G., Mi Y.Q., de Ledinghen V., Kumar M., Lupsor-Platon M., Han K.H., Cardoso A.C., et al. Individual patient data meta-analysis of controlled attenuation parameter (CAP) technology for assessing steatosis. J. Hepatol. 2017;66:1022–1030. doi: 10.1016/j.jhep.2016.12.022. [DOI] [PubMed] [Google Scholar]
- 23.Park C.C., Nguyen P., Hernandez C., Bettencourt R., Ramirez K., Fortney L., Hooker J., Sy E., Savides M.T., Alquiraish M.H., et al. Magnetic Resonance Elastography vs Transient Elastography in Detection of Fibrosis and Noninvasive Measurement of Steatosis in Patients With Biopsy-Proven Nonalcoholic Fatty Liver Disease. Gastroenterology. 2017;152:598–607. doi: 10.1053/j.gastro.2016.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yu C., Xu C., Xu L., Yu J., Miao M., Li Y. Serum proteomic analysis revealed diagnostic value of hemoglobin for nonalcoholic fatty liver disease. J. Hepatol. 2012;56:241–247. doi: 10.1016/j.jhep.2011.05.027. [DOI] [PubMed] [Google Scholar]
- 25.Bell L.N., Theodorakis J.L., Vuppalanchi R., Saxena R., Bemis K.G., Wang M., Chalasani N. Serum proteomics and biomarker discovery across the spectrum of nonalcoholic fatty liver disease. Hepatology. 2010;51:111–120. doi: 10.1002/hep.23271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kalhan S.C., Guo L., Edmison J., Dasarathy S., McCullough A.J., Hanson R.W., Milburn M. Plasma metabolomic profile in nonalcoholic fatty liver disease. Metab. Clin. Exp. 2011;60:404–413. doi: 10.1016/j.metabol.2010.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Oresic M., Hyotylainen T., Kotronen A., Gopalacharyulu P., Nygren H., Arola J., Castillo S., Mattila I., Hakkarainen A., Borra R.J., et al. Prediction of non-alcoholic fatty-liver disease and liver fat content by serum molecular lipids. Diabetologia. 2013;56:2266–2274. doi: 10.1007/s00125-013-2981-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mayo R., Crespo J., Martinez-Arranz I., Banales J.M., Arias M., Minchole I., Aller de la Fuente R., Jimenez-Aguero R., Alonso C., de Luis D.A., et al. Metabolomic-based noninvasive serum test to diagnose nonalcoholic steatohepatitis: Results from discovery and validation cohorts. Hepatol. Commun. 2018;2:807–820. doi: 10.1002/hep4.1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ekstedt M., Hagstrom H., Nasr P., Fredrikson M., Stal P., Kechagias S., Hultcrantz R. Fibrosis stage is the strongest predictor for disease-specific mortality in NAFLD after up to 33 years of follow-up. Hepatology. 2015;61:1547–1554. doi: 10.1002/hep.27368. [DOI] [PubMed] [Google Scholar]
- 30.Galiero R., Caturano A., Vetrano E., Cesaro A., Rinaldi L., Salvatore T., Marfella R., Sardu C., Moscarella E., Gragnano F., et al. Pathophysiological mechanisms and clinical evidence of relationship between Nonalcoholic fatty liver disease (NAFLD) and cardiovascular disease. Rev. Cardiovasc. Med. 2021;22:755–768. doi: 10.31083/j.rcm2203082. [DOI] [PubMed] [Google Scholar]
- 31.Eguchi A., Wree A., Feldstein A.E. Biomarkers of liver cell death. J. Hepatol. 2014;60:1063–1074. doi: 10.1016/j.jhep.2013.12.026. [DOI] [PubMed] [Google Scholar]
- 32.Kwok R., Tse Y.K., Wong G.L., Ha Y., Lee A.U., Ngu M.C., Chan H.L., Wong V.W. Systematic review with meta-analysis: Non-invasive assessment of non-alcoholic fatty liver disease--the role of transient elastography and plasma cytokeratin-18 fragments. Aliment. Pharmacol. Ther. 2014;39:254–269. doi: 10.1111/apt.12569. [DOI] [PubMed] [Google Scholar]
- 33.Tamimi T.I., Elgouhari H.M., Alkhouri N., Yerian L.M., Berk M.P., Lopez R., Schauer P.R., Zein N.N., Feldstein A.E. An apoptosis panel for nonalcoholic steatohepatitis diagnosis. J. Hepatol. 2011;54:1224–1229. doi: 10.1016/j.jhep.2010.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Younossi Z.M., Jarrar M., Nugent C., Randhawa M., Afendy M., Stepanova M., Rafiq N., Goodman Z., Chandhoke V., Baranova A. A novel diagnostic biomarker panel for obesity-related nonalcoholic steatohepatitis (NASH) Obes. Surg. 2008;18:1430–1437. doi: 10.1007/s11695-008-9506-y. [DOI] [PubMed] [Google Scholar]
- 35.Shen J., Chan H.L., Wong G.L., Choi P.C., Chan A.W., Chan H.Y., Chim A.M., Yeung D.K., Chan F.K., Woo J., et al. Non-invasive diagnosis of non-alcoholic steatohepatitis by combined serum biomarkers. J. Hepatol. 2012;56:1363–1370. doi: 10.1016/j.jhep.2011.12.025. [DOI] [PubMed] [Google Scholar]
- 36.Fitzpatrick E., Dhawan A. Noninvasive biomarkers in non-alcoholic fatty liver disease: Current status and a glimpse of the future. World J. Gastroenterol. 2014;20:10851–10863. doi: 10.3748/wjg.v20.i31.10851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ajmera V., Perito E.R., Bass N.M., Terrault N.A., Yates K.P., Gill R., Loomba R., Diehl A.M., Aouizerat B.E., Network N.C.R. Novel plasma biomarkers associated with liver disease severity in adults with nonalcoholic fatty liver disease. Hepatology. 2017;65:65–77. doi: 10.1002/hep.28776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Feldstein A.E., Lopez R., Tamimi T.A., Yerian L., Chung Y.M., Berk M., Zhang R., McIntyre T.M., Hazen S.L. Mass spectrometric profiling of oxidized lipid products in human nonalcoholic fatty liver disease and nonalcoholic steatohepatitis. J. Lipid Res. 2010;51:3046–3054. doi: 10.1194/jlr.M007096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ferslew B.C., Xie G., Johnston C.K., Su M., Stewart P.W., Jia W., Brouwer K.L., Barritt A.S.T. Altered Bile Acid Metabolome in Patients with Nonalcoholic Steatohepatitis. Dig. Dis. Sci. 2015;60:3318–3328. doi: 10.1007/s10620-015-3776-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Smits L.P., Coolen B.F., Panno M.D., Runge J.H., Nijhof W.H., Verheij J., Nieuwdorp M., Stoker J., Beuers U.H., Nederveen A.J., et al. Noninvasive Differentiation between Hepatic Steatosis and Steatohepatitis with MR Imaging Enhanced with USPIOs in Patients with Nonalcoholic Fatty Liver Disease: A Proof-of-Concept Study. Radiology. 2016;278:782–791. doi: 10.1148/radiol.2015150952. [DOI] [PubMed] [Google Scholar]
- 41.Abrigo J.M., Shen J., Wong V.W., Yeung D.K., Wong G.L., Chim A.M., Chan A.W., Choi P.C., Chan F.K., Chan H.L., et al. Non-alcoholic fatty liver disease: Spectral patterns observed from an in vivo phosphorus magnetic resonance spectroscopy study. J. Hepatol. 2014;60:809–815. doi: 10.1016/j.jhep.2013.11.018. [DOI] [PubMed] [Google Scholar]
- 42.Chen J., Talwalkar J.A., Yin M., Glaser K.J., Sanderson S.O., Ehman R.L. Early detection of nonalcoholic steatohepatitis in patients with nonalcoholic fatty liver disease by using MR elastography. Radiology. 2011;259:749–756. doi: 10.1148/radiol.11101942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dixon J.B., Bhathal P.S., O’Brien P.E. Nonalcoholic fatty liver disease: Predictors of nonalcoholic steatohepatitis and liver fibrosis in the severely obese. Gastroenterology. 2001;121:91–100. doi: 10.1053/gast.2001.25540. [DOI] [PubMed] [Google Scholar]
- 44.Angulo P., Keach J.C., Batts K.P., Lindor K.D. Independent predictors of liver fibrosis in patients with nonalcoholic steatohepatitis. Hepatology. 1999;30:1356–1362. doi: 10.1002/hep.510300604. [DOI] [PubMed] [Google Scholar]
- 45.Ratziu V., Giral P., Charlotte F., Bruckert E., Thibault V., Theodorou I., Khalil L., Turpin G., Opolon P., Poynard T. Liver fibrosis in overweight patients. Gastroenterology. 2000;118:1117–1123. doi: 10.1016/s0016-5085(00)70364-7. [DOI] [PubMed] [Google Scholar]
- 46.Angulo P. Long-term mortality in nonalcoholic fatty liver disease: Is liver histology of any prognostic significance? Hepatology. 2010;51:373–375. doi: 10.1002/hep.23521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sheth S.G., Flamm S.L., Gordon F.D., Chopra S. AST/ALT ratio predicts cirrhosis in patients with chronic hepatitis C virus infection. Am. J. Gastroenterol. 1998;93:44–48. doi: 10.1111/j.1572-0241.1998.044_c.x. [DOI] [PubMed] [Google Scholar]
- 48.Shaheen A.A., Myers R.P. Diagnostic accuracy of the aspartate aminotransferase-to-platelet ratio index for the prediction of hepatitis C-related fibrosis: A systematic review. Hepatology. 2007;46:912–921. doi: 10.1002/hep.21835. [DOI] [PubMed] [Google Scholar]
- 49.Shah A.G., Lydecker A., Murray K., Tetri B.N., Contos M.J., Sanyal A.J., Nash Clinical Research Network Comparison of noninvasive markers of fibrosis in patients with nonalcoholic fatty liver disease. Clin. Gastroenterol. Hepatol. 2009;7:1104–1112. doi: 10.1016/j.cgh.2009.05.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.McPherson S., Stewart S.F., Henderson E., Burt A.D., Day C.P. Simple non-invasive fibrosis scoring systems can reliably exclude advanced fibrosis in patients with non-alcoholic fatty liver disease. Gut. 2010;59:1265–1269. doi: 10.1136/gut.2010.216077. [DOI] [PubMed] [Google Scholar]
- 51.Sterling R.K., Lissen E., Clumeck N., Sola R., Correa M.C., Montaner J., Sulkowski M.S., Torriani F.J., Dieterich D.T., Thomas D.L., et al. Development of a simple noninvasive index to predict significant fibrosis in patients with HIV/HCV coinfection. Hepatology. 2006;43:1317–1325. doi: 10.1002/hep.21178. [DOI] [PubMed] [Google Scholar]
- 52.Angulo P., Hui J.M., Marchesini G., Bugianesi E., George J., Farrell G.C., Enders F., Saksena S., Burt A.D., Bida J.P., et al. The NAFLD fibrosis score: A noninvasive system that identifies liver fibrosis in patients with NAFLD. Hepatology. 2007;45:846–854. doi: 10.1002/hep.21496. [DOI] [PubMed] [Google Scholar]
- 53.Musso G., Gambino R., Cassader M., Pagano G. Meta-analysis: Natural history of non-alcoholic fatty liver disease (NAFLD) and diagnostic accuracy of non-invasive tests for liver disease severity. Ann. Med. 2011;43:617–649. doi: 10.3109/07853890.2010.518623. [DOI] [PubMed] [Google Scholar]
- 54.Gudowska M., Gruszewska E., Panasiuk A., Cylwik B., Flisiak R., Swiderska M., Szmitkowski M., Chrostek L. Hyaluronic acid concentration in liver diseases. Clin. Exp. Med. 2016;16:523–528. doi: 10.1007/s10238-015-0388-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Suzuki A., Angulo P., Lymp J., Li D., Satomura S., Lindor K. Hyaluronic acid, an accurate serum marker for severe hepatic fibrosis in patients with non-alcoholic fatty liver disease. Liver Int. 2005;25:779–786. doi: 10.1111/j.1478-3231.2005.01064.x. [DOI] [PubMed] [Google Scholar]
- 56.Monarca A., Petrini C., Perolini S., Pozzi F., Adelasco L., Natangelo R., Croce G. Procollagen-type III peptide serum concentrations in alcoholic and non-alcoholic liver disease. Ric. Clin. Lab. 1985;15:167–171. doi: 10.1007/BF03029835. [DOI] [PubMed] [Google Scholar]
- 57.Nielsen M.J., Nedergaard A.F., Sun S., Veidal S.S., Larsen L., Zheng Q., Suetta C., Henriksen K., Christiansen C., Karsdal M.A., et al. The neo-epitope specific PRO-C3 ELISA measures true formation of type III collagen associated with liver and muscle parameters. Am. J. Transl. Res. 2013;5:303–315. [PMC free article] [PubMed] [Google Scholar]
- 58.Leeming D.J., Grove J.I., Kaye P., Hoad C., Francis S., Nielsen M.J., Karsdal M.A., Guha I.N., Aithal G.P. Estimation of serum “true collagen type III formation” (Pro-C3) levels as a marker of non-alcoholic steatohepatitis in a prospective cohort. J. Hepatol. 2017;1((Suppl. S1)):S154. [Google Scholar]
- 59.Hemmann S., Graf J., Roderfeld M., Roeb E. Expression of MMPs and TIMPs in liver fibrosis—A systematic review with special emphasis on anti-fibrotic strategies. J. Hepatol. 2007;46:955–975. doi: 10.1016/j.jhep.2007.02.003. [DOI] [PubMed] [Google Scholar]
- 60.Abdelaziz R., Elbasel M., Esmat S., Essam K., Abdelaaty S. Tissue Inhibitors of Metalloproteinase-1 and 2 and Obesity Related Non-Alcoholic Fatty Liver Disease: Is There a Relationship. Digestion. 2015;92:130–137. doi: 10.1159/000439083. [DOI] [PubMed] [Google Scholar]
- 61.Santos V.N., Leite-Mor M.M., Kondo M., Martins J.R., Nader H., Lanzoni V.P., Parise E.R. Serum laminin, type IV collagen and hyaluronan as fibrosis markers in non-alcoholic fatty liver disease. Braz. J. Med. Biol. Res. 2005;38:747–753. doi: 10.1590/s0100-879x2005000500012. [DOI] [PubMed] [Google Scholar]
- 62.Guha I.N., Parkes J., Roderick P., Chattopadhyay D., Cross R., Harris S., Kaye P., Burt A.D., Ryder S.D., Aithal G.P., et al. Noninvasive markers of fibrosis in nonalcoholic fatty liver disease: Validating the European Liver Fibrosis Panel and exploring simple markers. Hepatology. 2008;47:455–460. doi: 10.1002/hep.21984. [DOI] [PubMed] [Google Scholar]
- 63.Nobili V., Parkes J., Bottazzo G., Marcellini M., Cross R., Newman D., Vizzutti F., Pinzani M., Rosenberg W.M. Performance of ELF serum markers in predicting fibrosis stage in pediatric non-alcoholic fatty liver disease. Gastroenterology. 2009;136:160–167. doi: 10.1053/j.gastro.2008.09.013. [DOI] [PubMed] [Google Scholar]
- 64.Boursier J., de Ledinghen V., Leroy V., Anty R., Francque S., Salmon D., Lannes A., Bertrais S., Oberti F., Fouchard-Hubert I., et al. A stepwise algorithm using an at-a-glance first-line test for the non-invasive diagnosis of advanced liver fibrosis and cirrhosis. J. Hepatol. 2017;66:1158–1165. doi: 10.1016/j.jhep.2017.01.003. [DOI] [PubMed] [Google Scholar]
- 65.Loong T.C., Wei J.L., Leung J.C., Wong G.L., Shu S.S., Chim A.M., Chan A.W., Choi P.C., Tse Y.K., Chan H.L., et al. Application of the combined FibroMeter vibration-controlled transient elastography algorithm in Chinese patients with non-alcoholic fatty liver disease. J. Gastroenterol. Hepatol. 2017;32:1363–1369. doi: 10.1111/jgh.13671. [DOI] [PubMed] [Google Scholar]
- 66.Boursier J., Vergniol J., Guillet A., Hiriart J.B., Lannes A., Le Bail B., Michalak S., Chermak F., Bertrais S., Foucher J., et al. Diagnostic accuracy and prognostic significance of blood fibrosis tests and liver stiffness measurement by FibroScan in non-alcoholic fatty liver disease. J. Hepatol. 2016;65:570–578. doi: 10.1016/j.jhep.2016.04.023. [DOI] [PubMed] [Google Scholar]
- 67.Wilder J., Patel K. The clinical utility of FibroScan((R)) as a noninvasive diagnostic test for liver disease. Med. Devices. 2014;7:107–114. doi: 10.2147/MDER.S46943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bonder A., Tapper E.B., Afdhal N.H. Contemporary assessment of hepatic fibrosis. Clin. Liver Dis. 2015;19:123–134. doi: 10.1016/j.cld.2014.09.007. [DOI] [PubMed] [Google Scholar]
- 69.Xiao G., Zhu S., Xiao X., Yan L., Yang J., Wu G. Comparison of laboratory tests, ultrasound, or magnetic resonance elastography to detect fibrosis in patients with nonalcoholic fatty liver disease: A meta-analysis. Hepatology. 2017;66:1486–1501. doi: 10.1002/hep.29302. [DOI] [PubMed] [Google Scholar]
- 70.Cassinotto C., Boursier J., de Ledinghen V., Lebigot J., Lapuyade B., Cales P., Hiriart J.B., Michalak S., Bail B.L., Cartier V., et al. Liver stiffness in nonalcoholic fatty liver disease: A comparison of supersonic shear imaging, FibroScan, and ARFI with liver biopsy. Hepatology. 2016;63:1817–1827. doi: 10.1002/hep.28394. [DOI] [PubMed] [Google Scholar]
- 71.Venkatesh S.K., Wang G., Teo L.L., Ang B.W. Magnetic resonance elastography of liver in healthy Asians: Normal liver stiffness quantification and reproducibility assessment. J. Magn. Reson. Imaging. 2014;39:1–8. doi: 10.1002/jmri.24084. [DOI] [PubMed] [Google Scholar]
- 72.Lee Y., Lee J.M., Lee J.E., Lee K.B., Lee E.S., Yoon J.H., Yu M.H., Baek J.H., Shin C.I., Han J.K., et al. MR elastography for noninvasive assessment of hepatic fibrosis: Reproducibility of the examination and reproducibility and repeatability of the liver stiffness value measurement. J. Magn. Reson. Imaging. 2014;39:326–331. doi: 10.1002/jmri.24147. [DOI] [PubMed] [Google Scholar]
- 73.Singh S., Venkatesh S.K., Wang Z., Miller F.H., Motosugi U., Low R.N., Hassanein T., Asbach P., Godfrey E.M., Yin M., et al. Diagnostic performance of magnetic resonance elastography in staging liver fibrosis: A systematic review and meta-analysis of individual participant data. Clin. Gastroenterol. Hepatol. 2015;13:440–451.e6. doi: 10.1016/j.cgh.2014.09.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Imajo K., Kessoku T., Honda Y., Tomeno W., Ogawa Y., Mawatari H., Fujita K., Yoneda M., Taguri M., Hyogo H., et al. Magnetic Resonance Imaging More Accurately Classifies Steatosis and Fibrosis in Patients With Nonalcoholic Fatty Liver Disease Than Transient Elastography. Gastroenterology. 2016;150:626–637.e7. doi: 10.1053/j.gastro.2015.11.048. [DOI] [PubMed] [Google Scholar]
- 75.Drag M.H., Kilpelainen T.O. Cell-free DNA and RNA-measurement and applications in clinical diagnostics with focus on metabolic disorders. Physiol. Genomics. 2021;53:33–46. doi: 10.1152/physiolgenomics.00086.2020. [DOI] [PubMed] [Google Scholar]
- 76.Di Mauro S., Scamporrino A., Fruciano M., Filippello A., Fagone E., Gili E., Scionti F., Purrazzo G., Di Pino A., Scicali R., et al. Circulating Coding and Long Non-Coding RNAs as Potential Biomarkers of Idiopathic Pulmonary Fibrosis. Int. J. Mol. Sci. 2020;21:8812. doi: 10.3390/ijms21228812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Scicali R., Di Pino A., Pavanello C., Ossoli A., Strazzella A., Alberti A., Di Mauro S., Scamporrino A., Urbano F., Filippello A., et al. Analysis of HDL-microRNA panel in heterozygous familial hypercholesterolemia subjects with LDL receptor null or defective mutation. Sci. Rep. 2019;9:20354. doi: 10.1038/s41598-019-56857-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Barbagallo C., Di Martino M.T., Grasso M., Salluzzo M.G., Scionti F., Cosentino F.I.I., Caruso G., Barbagallo D., Di Pietro C., Ferri R., et al. Uncharacterized RNAs in Plasma of Alzheimer’s Patients Are Associated with Cognitive Impairment and Show a Potential Diagnostic Power. Int. J. Mol. Sci. 2020;21:7644. doi: 10.3390/ijms21207644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Happel C., Ganguly A., Tagle D.A. Extracellular RNAs as potential biomarkers for cancer. J. Cancer Metastasis Treat. 2020;6:32. doi: 10.20517/2394-4722.2020.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zhu H., Fan G.C. Extracellular/circulating microRNAs and their potential role in cardiovascular disease. Am. J. Cardiovasc Dis. 2011;1:138–149. [PMC free article] [PubMed] [Google Scholar]
- 81.Pos O., Biro O., Szemes T., Nagy B. Circulating cell-free nucleic acids: Characteristics and applications. Eur. J. Hum. Genet. 2018;26:937–945. doi: 10.1038/s41431-018-0132-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Turchinovich A., Baranova A., Drapkina O., Tonevitsky A. Cell-Free Circulating Nucleic Acids as Early Biomarkers for NAFLD and NAFLD-Associated Disorders. Front. Physiol. 2018;9:1256. doi: 10.3389/fphys.2018.01256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Velu V.K., Ramesh R., Srinivasan A.R. Circulating MicroRNAs as Biomarkers in Health and Disease. J. Clin. Diagn. Res. 2012;6:1791–1795. doi: 10.7860/JCDR/2012/4901.2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Akuta N., Kawamura Y., Suzuki F., Saitoh S., Arase Y., Fujiyama S., Sezaki H., Hosaka T., Kobayashi M., Suzuki Y., et al. Analysis of association between circulating miR-122 and histopathological features of nonalcoholic fatty liver disease in patients free of hepatocellular carcinoma. BMC Gastroenterol. 2016;16:141. doi: 10.1186/s12876-016-0557-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Sun C., Huang F., Liu X., Xiao X., Yang M., Hu G., Liu H., Liao L. miR-21 regulates triglyceride and cholesterol metabolism in non-alcoholic fatty liver disease by targeting HMGCR. Int. J. Mol. Med. 2015;35:847–853. doi: 10.3892/ijmm.2015.2076. [DOI] [PubMed] [Google Scholar]
- 86.Yamada H., Suzuki K., Ichino N., Ando Y., Sawada A., Osakabe K., Sugimoto K., Ohashi K., Teradaira R., Inoue T., et al. Associations between circulating microRNAs (miR-21, miR-34a, miR-122 and miR-451) and non-alcoholic fatty liver. Clin. Chim. Acta. 2013;424:99–103. doi: 10.1016/j.cca.2013.05.021. [DOI] [PubMed] [Google Scholar]
- 87.Tan Y., Ge G., Pan T., Wen D., Gan J. A pilot study of serum microRNAs panel as potential biomarkers for diagnosis of nonalcoholic fatty liver disease. PLoS ONE. 2014;9:e105192. doi: 10.1371/journal.pone.0105192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Raitoharju E., Seppala I., Lyytikainen L.P., Viikari J., Ala-Korpela M., Soininen P., Kangas A.J., Waldenberger M., Klopp N., Illig T., et al. Blood hsa-miR-122-5p and hsa-miR-885-5p levels associate with fatty liver and related lipoprotein metabolism-The Young Finns Study. Sci. Rep. 2016;6:38262. doi: 10.1038/srep38262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Celikbilek M., Baskol M., Taheri S., Deniz K., Dogan S., Zararsiz G., Gursoy S., Guven K., Ozbakir O., Dundar M., et al. Circulating microRNAs in patients with non-alcoholic fatty liver disease. World J. Hepatol. 2014;6:613–620. doi: 10.4254/wjh.v6.i8.613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Jampoka K., Muangpaisarn P., Khongnomnan K., Treeprasertsuk S., Tangkijvanich P., Payungporn S. Serum miR-29a and miR-122 as Potential Biomarkers for Non-Alcoholic Fatty Liver Disease (NAFLD) Microrna. 2018;7:215–222. doi: 10.2174/2211536607666180531093302. [DOI] [PubMed] [Google Scholar]
- 91.Yu F., Wang X., Zhao H., Hao Y., Wang W. Decreased Serum miR-1296 may Serve as an Early Biomarker for the Diagnosis of Non-Alcoholic Fatty Liver Disease. Clin. Lab. 2019;65 doi: 10.7754/Clin.Lab.2019.190335. [DOI] [PubMed] [Google Scholar]
- 92.He Z., Yang J.J., Zhang R., Li H.T., Wu L., Jiang F., Jia W.P., Hu C. Circulating miR-29b positively correlates with non-alcoholic fatty liver disease in a Chinese population. J. Dig. Dis. 2019;20:189–195. doi: 10.1111/1751-2980.12716. [DOI] [PubMed] [Google Scholar]
- 93.Zong Y., Yan J., Jin L., Xu B., He Z., Zhang R., Hu C., Jia W. Relationship between circulating miR-132 and non-alcoholic fatty liver disease in a Chinese population. Hereditas. 2020;157:22. doi: 10.1186/s41065-020-00136-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Jiang H., Qian Y., Shen Z., Liu Y., He Y., Gao R., Shen M., Chen S., Fu Q., Yang T. Circulating microRNA135a3p in serum extracellular vesicles as a potential biological marker of nonalcoholic fatty liver disease. Mol. Med. Rep. 2021;24:1–12. doi: 10.3892/mmr.2021.12137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Di Mauro S., Salomone F., Scamporrino A., Filippello A., Morisco F., Guido M., Lembo V., Cossiga V., Pipitone R.M., Piro S., et al. Coffee Restores Expression of lncRNAs Involved in Steatosis and Fibrosis in a Mouse Model of NAFLD. Nutrients. 2021;13:2952. doi: 10.3390/nu13092952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Zhang M., Chi X., Qu N., Wang C. Long noncoding RNA lncARSR promotes hepatic lipogenesis via Akt/SREBP-1c pathway and contributes to the pathogenesis of nonalcoholic steatohepatitis. Biochem. Biophys. Res. Commun. 2018;499:66–70. doi: 10.1016/j.bbrc.2018.03.127. [DOI] [PubMed] [Google Scholar]
- 97.Cocucci E., Meldolesi J. Ectosomes and exosomes: Shedding the confusion between extracellular vesicles. Trends Cell Biol. 2015;25:364–372. doi: 10.1016/j.tcb.2015.01.004. [DOI] [PubMed] [Google Scholar]
- 98.Kornek M., Lynch M., Mehta S.H., Lai M., Exley M., Afdhal N.H., Schuppan D. Circulating microparticles as disease-specific biomarkers of severity of inflammation in patients with hepatitis C or nonalcoholic steatohepatitis. Gastroenterology. 2012;143:448–458. doi: 10.1053/j.gastro.2012.04.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Povero D., Yamashita H., Ren W., Subramanian M.G., Myers R.P., Eguchi A., Simonetto D.A., Goodman Z.D., Harrison S.A., Sanyal A.J., et al. Characterization and Proteome of Circulating Extracellular Vesicles as Potential Biomarkers for NASH. Hepatol. Commun. 2020;4:1263–1278. doi: 10.1002/hep4.1556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Cermelli S., Ruggieri A., Marrero J.A., Ioannou G.N., Beretta L. Circulating microRNAs in patients with chronic hepatitis C and non-alcoholic fatty liver disease. PLoS ONE. 2011;6:e23937. doi: 10.1371/journal.pone.0023937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Pirola C.J., Fernandez Gianotti T., Castano G.O., Mallardi P., San Martino J., Mora Gonzalez Lopez Ledesma M., Flichman D., Mirshahi F., Sanyal A.J., Sookoian S. Circulating microRNA signature in non-alcoholic fatty liver disease: From serum non-coding RNAs to liver histology and disease pathogenesis. Gut. 2015;64:800–812. doi: 10.1136/gutjnl-2014-306996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Becker P.P., Rau M., Schmitt J., Malsch C., Hammer C., Bantel H., Mullhaupt B., Geier A. Performance of Serum microRNAs -122, -192 and -21 as Biomarkers in Patients with Non-Alcoholic Steatohepatitis. PLoS ONE. 2015;10:e0142661. doi: 10.1371/journal.pone.0142661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Liu X.L., Pan Q., Zhang R.N., Shen F., Yan S.Y., Sun C., Xu Z.J., Chen Y.W., Fan J.G. Disease-specific miR-34a as diagnostic marker of non-alcoholic steatohepatitis in a Chinese population. World J. Gastroenterol. 2016;22:9844–9852. doi: 10.3748/wjg.v22.i44.9844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Lopez-Riera M., Conde I., Quintas G., Pedrola L., Zaragoza A., Perez-Rojas J., Salcedo M., Benlloch S., Castell J.V., Jover R. Non-invasive prediction of NAFLD severity: A comprehensive, independent validation of previously postulated serum microRNA biomarkers. Sci. Rep. 2018;8:10606. doi: 10.1038/s41598-018-28854-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Park J.G., Kim G., Jang S.Y., Lee Y.R., Lee E., Lee H.W., Han M.H., Chun J.M., Han Y.S., Yoon J.S., et al. Plasma Long Noncoding RNA LeXis is a Potential Diagnostic Marker for Non-Alcoholic Steatohepatitis. Life. 2020;10:230. doi: 10.3390/life10100230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Di Mauro S., Scamporrino A., Petta S., Urbano F., Filippello A., Ragusa M., Di Martino M.T., Scionti F., Grimaudo S., Pipitone R.M., et al. Serum coding and non-coding RNAs as biomarkers of NAFLD and fibrosis severity. Liver Int. 2019;39:1742–1754. doi: 10.1111/liv.14167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Sun C., Fan J.G., Qiao L. Potential epigenetic mechanism in non-alcoholic Fatty liver disease. Int. J. Mol. Sci. 2015;16:5161–5179. doi: 10.3390/ijms16035161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Miyaaki H., Ichikawa T., Kamo Y., Taura N., Honda T., Shibata H., Milazzo M., Fornari F., Gramantieri L., Bolondi L., et al. Significance of serum and hepatic microRNA-122 levels in patients with non-alcoholic fatty liver disease. Liver Int. 2014;34:e302–e307. doi: 10.1111/liv.12429. [DOI] [PubMed] [Google Scholar]
- 109.Yu F., Zheng J., Mao Y., Dong P., Li G., Lu Z., Guo C., Liu Z., Fan X. Long non-coding RNA APTR promotes the activation of hepatic stellate cells and the progression of liver fibrosis. Biochem. Biophys. Res. Commun. 2015;463:679–685. doi: 10.1016/j.bbrc.2015.05.124. [DOI] [PubMed] [Google Scholar]
- 110.Han M.H., Lee J.H., Kim G., Lee E., Lee Y.R., Jang S.Y., Lee H.W., Chun J.M., Han Y.S., Yoon J.S., et al. Expression of the Long Noncoding RNA GAS5 Correlates with Liver Fibrosis in Patients with Nonalcoholic Fatty Liver Disease. Genes. 2020;11:545. doi: 10.3390/genes11050545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Estes C., Razavi H., Loomba R., Younossi Z., Sanyal A.J. Modeling the epidemic of nonalcoholic fatty liver disease demonstrates an exponential increase in burden of disease. Hepatology. 2018;67:123–133. doi: 10.1002/hep.29466. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.